Abstract
A species-specific quantitative PCR (qPCR) assay was combined with two novel water-sampling methods and compared with the mouse bioassay for the quantitative detection of S. japonicum in surface waters. The novel methods were capable of capturing cercariae and, with subsequent analysis through qPCR, detecting the presence of a minimum of 1 cercaria.
Schistosoma japonicum infects human and animal hosts by means of a free-living larval stage, cercariae, which are released by amphibious intermediate host snails (Oncomelania hupensis) present in fecally contaminated surface water (17, 18). Currently an estimated 800,000 humans are infected with S. japonicum, and 30 million people remain at risk in the People's Republic of China (11, 22), contributing to substantial ill health (8), developmental effects (6, 13), and socioeconomic impacts (10, 21). With the success of China's mass chemotherapeutic control campaigns, difficulties detecting increasingly rare and low-concentration transmission sites are posing challenges to achieving elimination (16). Traditional environmental detection methods, including microscopy-based techniques and animal bioassays, are of limited utility for surveillance. A sentinel mouse bioassay, the current method for detecting cercariae in environmental water samples, requires extensive resources and time (up to 6 weeks), prohibiting its use in large-scale screening programs. Environmental detection methods using molecular assays have previously been described for Schistosoma cercariae (3, 4) and we recently described a highly sensitive S. japonicum-specific quantitative PCR (qPCR) assay for the detection of cercariae in a laboratory setting (2, 5). This study reports a field application and comparison of the qPCR assay with the mouse bioassay to determine effectiveness under field conditions. It aims to provide a complete detection technique capable of quantifying cercariae in natural water samples.
During routine epidemiology research activities conducted by the Institute of Parasitic Disease (IPD), two novel water-sampling apparatuses, a direct sampler and a siphon sampler (Fig. 1 and 2, respectively), were used to collect environmental samples in potential transmission foci within eight villages with reemerging schistosomiasis located in China's Sichuan Province. These samples were analyzed using an S. japonicum-specific qPCR assay, and the results were compared with those of the mouse bioassay. At each sampling site (n = 49), a mouse bioassay (19) was carried out in parallel with the direct sampling device. In four of these sites, where a sufficient hydrostatic head existed, the siphon method was also deployed. For every mouse bioassay two samples were collected using our newly developed samplers, one for each day of the 2-day mouse bioassay exposure. In total, 49 mouse bioassays and 108 environmental water samples were collected. The mice were maintained for 6 weeks to allow for the maturation of parasites into adult worms and then euthanized, and adult schistosomes were recovered and counted, if found. Mouse bioassays were linked to environmental samples through a unique site identification code for subsequent comparison. The animal use protocols were approved by the Institutional Animal Care and Use Committee of the Sichuan Institute of Parasitic Disease, which maintains an Animal Welfare Assurance with the U.S. National Institutes of Health, and by the Animal Care and Use Committee of the University of California at Berkeley.
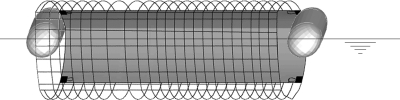
FIG. 1.
Front view of direct sampling apparatus, where the gray section is the sampling filter (30 cm by 10 cm) supported by a cage that filters macroscopic material while providing housing for the sampling material. The cylindrical floats position the sampler so that the water surface transects the filter material.
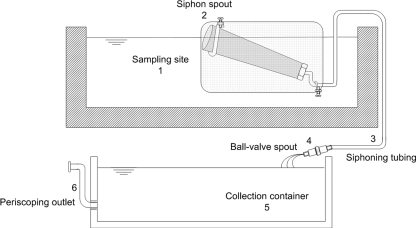
FIG. 2.
Cross-sectional view of siphon sampling. The siphon sampler is deployed in a location where the sampling site of interest (1) is elevated, making it is possible to establish a siphon gradient. The siphon spout (2) is initiated and then positioned so that it skims the top centimeter of water through the use of floats (not pictured). The water is directed into the collection container (5) (45 cm by 30 cm by 15 cm) by a length of plastic siphoning tubing (3) connected by a ball-valve spout (4). Excess water is drained from the center of the water column by the periscoping attachment (6).
The direct sampler used passive water flow to filter and capture cercariae (Fig. 1). For each sample, one 300-count nylon filter (Yaohua Screen & Filter Cloth Co. Ltd., Zhejiang, China) was fastened to the inner wall of the sampler. External floats ensured consistent sampling depth and positioned the sampler so that the surface of the water column transected the filter (see the supplemental material). Upon withdrawal of the sample, the filter screen was removed from the housing, placed into a BD Falcon tube containing 50 ml of 98% ethanol, transported to the laboratory, and maintained at 4°C until being processed. The siphon sampler (Fig. 2), made from a gravel cleaner (Warmtone, WT-228) and a 16-liter collection container, used a siphoning action to collect and concentrate a water sample. In order to filter macroscopic debris and provide structure to the siphon head, a supporting wire mesh cage was built (see the supplemental material) to allow the collection of a surface sample of the water column while maintaining the siphoning action (Fig. 2). After the container was full, the collection concentrated the sample by draining water from the center of the water column via a periscoping outlet. This was required to achieve efficient concentration of cercariae, which are surface-seeking when alive and sink rapidly when dead.
The siphon was initiated using a hand-cranked pump and then rotated into the sampling position and secured. At the conclusion of siphon sampling, the siphon was processed as follows: (i) an 8-cm by 10-cm strip of 300-count filter material was used to skim the surface of the water collected in the collection container, and (ii) the remaining water was poured through a funnel lined with a strip of 300-count nylon filter material. The filters were placed in individual 50-ml BD Falcon tubes as mentioned above.
To further concentrate and isolate cercariae, the filters were removed from the ethanol and placed into a clean 50-ml BD Falcon tube. The tube containing ethanol was centrifuged at 3,000 rpm for 10 min to pellet any particles. The supernatant was transferred to the tube containing the filter and vortexed for 1 min. The contents of the filter paper were scraped into the ethanol using a sterile spatula, after which the filter was discarded and the two pellets were combined in one 50-ml BD Falcon tube for further DNA extraction.
To detect S. japonicum by qPCR, DNA standards were prepared as follows. Live, S. japonicum-infected Oncomelania hupensis (subspecies hupensis) (obtained from the Biomedical Research Institute [BRI]-NIAID) were maintained according to BRI-recommended housing and environmental procedures (1, 9, 14). Prior to DNA extraction, snails were washed and crushed into a sterile petri dish using a glass microscope slide. Cercariae were liberated from the snail tissue by teasing apart the secondary sporocyst using fine forceps and a needle (Y. Liang and F. Lewis, unpublished material). Five hundred cercariae were counted using an inverted light microscope and washed into a 50-ml BD Falcon tube using 98% ethanol. Samples were stored at −20°C prior to DNA extraction.
DNA from these cercariae, and from environmental samples, was extracted using the QIAamp stool kit (Qiagen, CA). Samples were centrifuged at 3,000 rpm for 10 min, after which the supernatant was discarded and the pellets were air dried. A modified QIAamp stool kit protocol was employed with the following modifications: (i) samples were lysed with 3.5 ml of buffer ASL and 40 μl proteinase K, and (ii) the lysate was incubated at 95°C for 2 h with vortexing at 15-min intervals. The manufacturer's protocol for purification of the sample through the spin column was followed. However, purified DNA was eluted once with 100 μl of buffer AE in the final step. DNA content was quantified using a spectrophotometer (Nanodrop, ND-1000). All purified DNA samples were stored at −80°C until downstream analysis by qPCR.
To ensure stable transportation of those DNA samples at ambient temperature to Atlanta, GA, the DNA was stabilized using a Whatman card protocol. A 100-μl volume was applied to a Whatman FTA minicard (Whatman Inc., NJ), subsequently dried on the benchtop, and then sealed in a multibarrier pouch containing a desiccant. Positive and negative controls were included in each preparation using known positive S. japonicum miricidial DNA or distilled water, respectively.
A previously described qPCR assay was modified and used to amplify an 85-bp S. japonicum-specific putative deoxyribodipyrimidine photolyase (PL) gene segment (5). qPCR was performed in 25-μl volumes containing 1× TaqMan Universal PCR master mix (Applied Biosystems Inc., CA), 500 nM final concentrations of both the forward (PL-F) and reverse (PL-R) primers (Sigma-Aldrich, MO), a 240 nM final concentration of the FAM (6-carboxyfluorescein)-TAMRA (6-carboxytetra-methylrhodamine)-labeled probe (Sigma-Aldrich, MO), and 5 μl of extracted DNA as a template. The DNA content of the 500-cercaria standard was 2.7 ng/μl as measured by the spectrophotometer. The quantitative PCR parameters were set as follows: 2 min at 50°C and 10 min at 95°C, followed by 45 cycles alternating between 15 s at 95°C and 1 min at 60°C. qPCR standards, created using 5-fold serial dilutions of DNA extracted from 500 cercariae, were run with each plate. To confirm the expected size of the amplicon (85 bp), some qPCR products were separated by gel electrophoresis on a 2% ethidium bromide-stained agarose gel.
The results of the 49 mouse bioassays and a total of 108 water samples (94 direct sampler and 14 siphon samples) were analyzed and compared. Descriptive statistics for these water samples are included in Tables S2 and S3 in the supplemental material. For the mouse bioassay, 6 weeks after exposure, a total of 239 mice were euthanized and examined for the presence of S. japonicum infection. All mice (n = 239) were negative for the presence of adult worms in the mesenteric vessels, suggesting that those water sources were not contaminated. However, when evaluated by qPCR, 7 of the 108 field samples were positive for S. japonicum, with threshold cycle (CT) values corresponding to a cercarial count ranging between ∼1 cercaria and 2.5 cercariae. Of the 94 samples obtained by the direct sampling apparatus, 5 were positive, while among the 14 samples obtained by the siphoning mechanism, 2 showed positive results (Table 1). Positive qPCR products run in 2% agarose gels confirmed the expected size of bands (85 bp) (see Fig. S1 in the supplemental material). No PCR product was detected from negative nontemplate controls. The DNA standards showed consistent correlation between CT values and concentration of cercarial DNA, while replicates of no-template controls (NTCs) did not generate any CT value.
TABLE 1.
Infection status of villages among sentinel mice and qPCR analysis of environmental samples in June 2009
| Village (no. of sites) | Resulta |
||||
|---|---|---|---|---|---|
| Mouse bioassay | Direct sampling |
Siphon sampling |
|||
| Day 1 | Day 2 | Day 1 | Day 2 | ||
| Village A (n = 8) | 0/8 | 0/8 | 2/8 | 1/2 | 0/4 |
| Village B (n = 6) | 0/6 | 0/6 | 1/6 | ||
| Village C (n = 6) | 0/6 | 1/6 | 0/6 | 0/2 | 0/2 |
| Village D (n = 6) | 0/6 | 0/6 | 0/6 | ||
| Village E (n = 4) | 0/4 | 0/4 | 1/4 | ||
| Village F (n = 6) | 0/5 | 0/5 | 0/5 | ||
| Village G (n = 6) | 0/6 | 0/6 | 0/6 | ||
| Village H (n = 6) | 0/6 | 0/6 | 0/6 | 1/2 | 0/2 |
| Total | 0/47 | 1/47 | 4/47 | 2/6 | 0/8 |
No. of positive samples/total no. of samples.
This report introduces two novel water sampling protocols that, when coupled with a species-specific qPCR assay, are designed to provide a complete protocol for the rapid, cost-effective, and high-throughput detection of S. japonicum cercariae in water. The sampling methods evaluated here were designed to exploit biologic characteristics of S. japonicum cercariae, such as phototaxic and surface-seeking behavior, to maximize the capture of any cercariae present in surface water. As well, during the design process additional logistic considerations were taken into account in developing prospective techniques for field application in resource-limited settings (see Table S1 in the supplemental material). Results of the sample analysis demonstrated the effectiveness of water sampling techniques to capture cercariae and the sensitivity of the qPCR assay to amplify an S. japonicum target sequence. Comparison of these results with those of mouse bioassays replicated the in vitro findings of the previous protocol, which could detect DNA equivalent to less than one cercaria. Due to the absence of mouse infection, it is not possible to evaluate the true sensitivity of this assay; these findings warrant further evaluation in areas in which the infection is highly endemic to confirm this relationship.
Several modifications to the previously published protocol were required for effective field application. For example, the QIAamp stool kit, which is used for the extraction of S. japonicum DNA from stool samples (12), was chosen on account of the high turbidity of water in endemic sites, expected sample volume and composition, and evidence of qPCR inhibitors in previous environmental samples (5). Some modifications to the qPCR protocol were also required to take into account sampling and extraction protocol modifications. The template volume and numbers of cycles were modified from the previous qPCR protocol, allowing for improved detection of standards, as well as unknown samples, while maintaining the consistent absence of amplification in the no-template control. Similar cycling parameters have been used and published elsewhere (7, 15, 20).
In addition to demonstrating improved sensitivity over the bioassay, the qPCR technique provides an opportunity to overcome the cost and time limitations of the mouse method. According to research records, each mouse bioassay costs approximately $100, while the total cost of all qPCR assay consumables amounts to less than $15 per sample. Due to the expense of the mouse bioassay, it cannot be deployed in sufficient quantity to elucidate the spatial and temporal variability of the parasite. Even at the village level, high variability exists between individual sites (19); even where multiple assays are deployed annually, only a crude estimate of risk can be calculated. Moreover, the qPCR technique, performed in as little as 14 active hours, has a significantly shorter turnaround time than the mouse bioassay. These qualities improve the ability of public health workers to initiate prompt intervention and control measures for contaminated sites, reducing the risk of infection for populations living in areas of S. japonicum endemicity.
Despite the benefits of a qPCR technique, there are several important limitations. First, standard qPCR techniques are unable to assess if captured cercariae are live and/or infective, whereas the mouse bioassay detects only live and infective cercariae. However, even the presence of inactive cercariae can indicate a risk of infection, as cercarial shedding is occurring in the locale. Additionally, the use of qPCR applications requires a capital investment for specialized equipment and reliable environmental controls, limiting its application in severely resource-constrained settings. However, the sampling and analysis techniques presented here provide a foundation for eventual replacement of the mouse bioassay as the gold standard technique for cercarial detection in natural water.
Supplementary Material
Acknowledgments
This work was supported in part by the International Foundation for Ethical Research and by the Ecology of Infectious Disease program of the National Science Foundation under grant no. 0622743. Mouse bioassays were supported by the National Institute for Allergy and Infectious Diseases grant no. R01AI068854. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful for the assistance of David Blair of James Cook University and Glen C. McGugan, Jr., and Fred Lewis with the Biomedical Research Institute, Rockville, MD, who supplied schistosomes and snail hosts under a U.S. National Institute of Allergy and Infectious Diseases supply contract (N01-A1-30026). We appreciate the assistance of Yiliu Chen and Gregory Ian Spain for their work with sample collection and analysis. We also thank Dongchuan Qiu of the Institute of Parasitic Diseases, Sichuan Center for Disease Control and Prevention, Chengdu, People's Republic of China, for his support and assistance.
Footnotes
Published ahead of print on 28 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bruce, J. I., and Y.-S. Liang. 1992. Cultivation of schistosomes and snails for researchers in the United States of America and other countries. J. Med. Appl. Malacol. 4:13-30. [Google Scholar]
- 2.Driscoll, A. J., J. L. Kyle, and J. Remais. 2005. Development of a novel PCR assay capable of detecting a single Schistosoma japonicum cercaria recovered from Oncomelania hupensis. Parasitology 131:497-500. [DOI] [PubMed] [Google Scholar]
- 3.Hamburger, J., Y. X. Xu, R. M. Ramzy, J. Jourdane, and A. Ruppel. 1998. Development and laboratory evaluation of a polymerase chain reaction for monitoring Schistosoma mansoni infestation of water. Am. J. Trop. Med. Hyg. 59:468-473. [DOI] [PubMed] [Google Scholar]
- 4.Hertel, J., J. Hamburger, B. Haberl, and W. Haas. 2002. Detection of bird schistosomes in lakes by PCR and filter-hybridization. Exp. Parasitol. 101:57-63. [DOI] [PubMed] [Google Scholar]
- 5.Hung, Y. W., and J. Remais. 2008. Quantitative detection of Schistosoma japonicum cercariae in water by real-time PCR. PLoS Negl. Trop. Dis. 2:e337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jukes, M. C., et al. 2002. Heavy schistosomiasis associated with poor short-term memory and slower reaction times in Tanzanian schoolchildren. Trop. Med. Int. Health 7:104-117. [DOI] [PubMed] [Google Scholar]
- 7.Kimura, B., S. Kawasaki, H. Nakano, T. Fujii. 2001. Rapid, quantitative PCR monitoring of growth of Clostridium botulinum type E in modified-atmosphere-packaged fish. Appl. Environ. Microbiol. 67:206-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leenstra, T., et al. 2006. Schistosoma japonicum reinfection after praziquantel treatment causes anemia associated with inflammation. Infect. Immun. 74:6398-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis, F. 2001. Schistosomiasis. Curr. Protoc. Immunol. 19:19.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, Y. S., Z. Y. Zhao, M. Ellis, and D. P. McManus. 2005. Applications and outcomes of periodic epidemiological surveys for schistosomiasis and related economic evaluation in the People's Republic of China. Acta Trop. 96:266-275. [DOI] [PubMed] [Google Scholar]
- 11.Liang, S., C. Yang, B. Zhong, and D. Qiu. 2006. Re-emerging schistosomiasis in hilly and mountainous areas of Sichuan, China. Bull. World Health Organ. 84:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lier, T., et al. 2006. Novel real-time PCr for detection of Schistosoma japonicum in stool. Southeast Asian J. Trop. Med. Public Health 37:257-264. [PubMed] [Google Scholar]
- 13.McGarvey, S. T., et al. 1993. Child growth, nutritional status, and schistosomiasis japonica in Jiangxi, People's Republic of China. Am. J. Trop. Med. Hyg. 48:547-553. [DOI] [PubMed] [Google Scholar]
- 14.McGugan, G. C., Y. Liang, and F. Lewis. 1 August 2010. NIAID Schistosomiasis Resource Center. Standard Operating Procedures. http://www.schisto-resource.org/sop.htm.
- 15.Noble, R. T., et al. 2006. Multititered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California. Appl. Environ. Microbiol. 72:1604-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remais, J., A. Akullian, L. Ding, and E. Seto. 2010. Analytical methods for quantifying environmental connectivity for the control and surveillance of infectious disease spread. J. R. Soc. Interface 7:1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross, A. G., et al. 2002. Schistosomiasis. N. Engl. J. Med. 346:1212-1220. [DOI] [PubMed] [Google Scholar]
- 18.Ross, A. G., et al. 2001. Schistosomiasis in the People's Republic of China: prospects and challenges for the 21st century. Clin. Microbiol. Rev. 14:270-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spear, R. C., et al. 2004. Spatial and temporal variability in schistosome cercarial density detected by mouse bioassays in village irrigation ditches in Sichuan, China. Am. J. Trop. Med. Hyg. 71:554-557. [PubMed] [Google Scholar]
- 20.Wichman, D., et al. 2009. Diagnosing schistosomiasis by detection of cell-free parasite DNA in human plasma. PLoS Negl. Trop. Dis. 3:e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, X. H., et al. 2002. Studies of impact on physical fitness and working capacity of patients with advanced Schistosomiasis japonica in Susong County, Anhui Province. Acta Trop. 82:247-252. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, X. N., et al. 2007. Epidemiology of schistosomiasis in the People's Republic of China, 2004. Emerg. Infect. Dis. 13:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.