Abstract
Enterotoxigenic Escherichia coli (ETEC) is a Gram-negative enteric pathogen that causes profuse watery diarrhea through the elaboration of heat-labile and/or heat-stable toxins. Virulence is also dependent upon the expression of adhesive pili and afimbrial adhesins that allow the pathogen to adhere to the intestinal epithelium or mucosa. Both types of enterotoxins are regulated at the level of transcription by cyclic AMP (cAMP) receptor protein (CRP). To further our understanding of virulence gene regulation, an in silico approach was used to identify putative CRP binding sites in the genome of H10407 (O78:H11), an ETEC strain that was originally isolated from the stool of a Bangledeshi patient with cholera-like symptoms circa 1971. One of the predicted binding sites was located within an intergenic region upstream of tibDBCA. TibA is an autotransporter and afimbrial adhesin that is glycosylated by TibC. Expression of the TibA glycoprotein was abolished in an H10407 crp mutant and restored when crp was provided in trans. TibA-dependent aggregation was also abolished in a cyaA::kan strain and restored by addition of exogenous cAMP to the growth medium. DNase I footprinting confirmed that the predicted site upstream of tibDBCA is bound by CRP. Point mutations within the CRP binding site were found to abolish or significantly impair CRP-dependent activation of the tibDB promoter. Thus, these studies demonstrate that CRP positively regulates the expression of the glycosylated afimbrial adhesin TibA through occupancy of a binding site within tibDBp.
More than 100 genes in Escherichia coli and many other bacterial species are regulated by cyclic AMP (cAMP) receptor protein (CRP) through direct and indirect pathways (12, 33, 36, 42, 75, 76). For CRP, DNA binding is cAMP dependent and the position of a CRP binding site relative to RNA polymerase largely determines whether it functions as an activator or a repressor of a particular gene or operon. With respect to the transcription start site, CRP binding sites centered at or near −62, −72, −83, or −93 are typical for class I CRP-dependent promoters. Class II promoters have a binding site centered at or near −41.5 (16, 24, 50). Class III promoters are more complex in that they usually require an additional transcription factor or multiple CRP binding sites (24). In comparison to class I and class II promoters, class III and CRP-repressed promoters display a greater range of binding site positions (33).
In addition to regulating the expression of metabolic and housekeeping genes, CRP has been shown to regulate the expression of many virulence factors. For example, CRP positively regulates the expression of Pla, a surface-exposed protease of Yersinia pestis that is necessary for the establishment of primary pneumonic plague and facilitates dissemination of the bacterium during bubonic plague (46, 49). In Vibrio vulnificus, a marine bacterium that causes gastroenteritis when ingested with contaminated seafood, CRP positively regulates the expression of a cytolytic hemolysin and a heme receptor (18, 61). The Vibrio cholerae toxin is negatively regulated by CRP through an indirect pathway involving the second regulator, TcpP (47, 70). In contrast, expression of the heat-labile toxin of enterotoxigenic E. coli (ETEC) is repressed by CRP when it binds to an operator centered directly over the toxin promoter's −35 hexamer (10). The Salmonella enterica serovar Typhimurium Stn enterotoxin is negatively regulated by CRP (51). This is not surprising, because Stn is similar to both cholera and heat-labile toxin.
Adherence to biotic and abiotic surfaces is another important trait of many pathogenic bacteria, and several studies have shown that many types of adhesive pili are CRP regulated. For example, the toxin-coregulated pilus of V. cholerae is negatively regulated by CRP (70). The bundle-forming pilus of enteropathogenic E. coli may also be repressed by CRP (26, 63). In Serratia marcescens and uropathogenic E. coli, CRP negatively regulates the expression of type I fimbriae (41, 59). CRP positively regulates the expression of ETEC K99 and 987P pili through direct and indirect pathways, respectively (25, 54). CRP apparently also regulates the expression of other ETEC pili such as CFA/I, CS1, CS2, and CS3 because their expression is subject to catabolite repression (29, 43). In this study, we show that CRP also regulates the expression of the ETEC tibDBCA locus, which encodes a glycosylated autotransporter (TibA) that facilitates adherence to mammalian cells, autoaggregation, and biofilm formation (52, 53, 69).
MATERIALS AND METHODS
Strains and plasmids.
The relevant features of the E. coli strains used in this study are shown in Table 1. The tibDB promoter region from −594 through +77, with the numbering relative to the tibD ATG start codon, was amplified from the chromosome of ETEC strain H10407 (20, 30) with primers SN879 and SN882. The sequences of oligonucleotide primers are listed in Table 2. The 0.7-kb PCR product was digested with XbaI and EcoRI and then ligated into the same sites of the pHKLac1z polylinker to construct pTibDBLac1 [tibDBp(−594 to +77)::lacZ]. Plasmid pHKLac1z is a promoterless Lac reporter plasmid carrying the pir-dependent R6Kγ origin of replication, aadA conferring resistance to spectinomycin and streptomycin, and attPHK022. Read-through from flanking sequences is prevented by transcriptional terminators located immediately upstream of the polylinker and downstream of lacZ. Plasmid pTibDBLac1 served as the PCR template for oligonucleotide-directed mutagenesis of the CRP binding site tibDBo. In brief, the plasmid was subjected to inverse PCR with primer pair SN892 and SN893 or SN890 and SN891. The PCR products were digested with XhoI and then circularized with T4 DNA ligase. The resulting binding sites, tibDBo1 and tibDBo2, carry XhoI sites in the promoter-distal and promoter-proximal regions of the CRP binding site, respectively. DNA sequencing confirmed that no other mutations were present in the tibDB promoter fragments. To avoid the possibility of PCR errors outside the sequenced regions, the mutagenized promoter fragments were cloned into pHKLac1z as described above. The resulting reporter plasmids, pTibDBLac2 and pTibDBLac3, carry tibDBo1 and tibDBo2, respectively. Lac reporter strains were constructed by integration of reporter plasmids into the chromosomal attBHK022 site of BW25113 (22) and its isogenic crp::kan progeny JW5702 (5) by site-specific recombination with IntHK022 (37). Single integrants were verified by colony PCR as previously described (37).
TABLE 1.
Bacterial strains
| Strain | Relevant features | Reference |
|---|---|---|
| H10407 | ETEC O78:H11:K80 TibA+ CFA/I+ LT+ STh+ STp+ | 30 |
| H10407 Δcrp | H10407 Δcrp | This study |
| HB101 | recA13 lacY1 | 12a |
| BW25113 | ΔlacZ4787(::rrnB-4) | 22 |
| JW5702 | BW25113 crp::kan | 5 |
| JW3778 | BW25113 cyaA::kan | 5 |
| GPM1710 | BW25113 attBHK022::pTibDBLac1 [tibDBo tibDBp::lacZ] | This study |
| GPM1713 | JW5702 attBHK022::pTibDBLac1 [tibDBo tibDBp::lacZ] | This study |
| GPM1605 | BW25113 attBHK022::pTibDBLac2 [tibDBo1 tibDBp::lacZ] | This study |
| GPM1607 | JW5702 attBHK022::pTibDBLac2 [tibDBo1 tibDBp::lacZ] | This study |
| GPM1604 | BW25113 attBHK022::pTibDBLac3 [tibDBo2 tibDBp::lacZ] | This study |
| GPM1606 | JW5702 attBHK022::pTibDBLac3 [tibDBo2 tibDBp::lacZ] | This study |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| SN879 | GCATCTAGAAATGCCGGGCTGGCTG |
| SN882 | GACGAATTCGTCTGCGTCTCTCAG |
| SN884 | GACGAATCAGTGCAGAGTAACGCG |
| SN885 | GACTCTAGATTTTAGCTTTTGTCTGG |
| SN890 | GACTCGAGAATTTTTAATTAATTGTTGTTTGC |
| SN891 | GTCTCGAGTATAAATCAATAAAACTCTCG |
| SN892 | GTCTCGAGTTTATATCAAACAATTTTTAATTAATTG |
| SN893 | GACTCGAGAAACTCTCGTATCAAATCCCTTC |
Underlined nucleotides indicate primer-template mismatches that add sites for restriction endonucleases.
Plasmid pDCRP is an ampicillin-resistant derivative of pBR322 that expresses crp from its own promoter (7). Plasmid pDU9 is a Δcrp subclone of pDCRP (7). Plasmid pSE186 (39) was constructed by cloning crp into the vector pHG165, which confers resistance to ampicillin (71). The complete tib locus, tibDBCA, was cloned into pHG165 to construct pET109 (28). Plasmid pET142 carries the same 3.6-Kb tibDBCA141 fragment as pET146 (28) but in pHG165 rather than pACYC184. To construct pGPM1121 (tibB60CA); the tibDB promoter region, tibD, and the first 59 codons of tibB were excised from pET109 as a 3.5-Kb BglII fragment.
β-Galactosidase assays.
Lac reporter strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) with 30 μg/ml streptomycin, 30 μg/ml spectinomycin, and, as required, 100 μg/ml ampicillin. Cells were harvested from stationary-phase cultures, lysed, and assayed for β-galactosidase activity as previously described (58).
Aggregation assays.
Strains BW25113 and JW3778 harboring pET109, pET142, or pGPM1121 were cultured aerobically with agitation to stationary phase at 37°C in LB10 (10 g/liter tryptone, 5 g/liter yeast extract, 0.58 g/liter NaCl) with 100 μg/ml ampicillin. This low-salt medium was used for aggregation assays because we have observed that TibA-dependent aggregation, but not necessarily TibA expression, is progressively inhibited by increasing concentrations of NaCl. High salt concentrations have also been reported to disrupt self-interactions of AIDA-I, another autoaggregating adhesin of diarrheagenic E. coli (34). For qualitative comparisons, representative cultures of four or more independent replicates were photographed immediately after vigorous mixing and 3 h after standing at room temperature without agitation. A spectrophotometer was used to measure the absorbance at 600 nm of aliquots from the upper portion of selected cultures. The aggregation factor was calculated as the difference between the mixed and static absorbance readings.
DNase I footprinting.
CRP was purified to 89% purity, as determined by lab-on-chip analysis (Agilent Technologies), on an immobilized metal ion affinity column as previously described (10, 74). For DNase I footprinting of the coding strand, a DNA template from −334 through +57 was produced by PCR with primer SN885 and 32P-end-labeled primer SN882 (Table 2). For the noncoding strand, a PCR product from −576 through −70 was generated with primer SN879 and 32P-end-labeled primer SN884. The PCR products, whose numbering is relative to the tibD initiating codon, were purified on nondenaturing acrylamide gels as previously described (60). Following their purification, the DNA templates were equilibrated with or without CRP for 30 min at 37°C in 10 mM Tris-Cl (pH 7.6), 50 mM KCl, 1 mM dithiothreitol, 2 ng/μl poly(dI-dC), 0.4 mM MgCl2, 0.2 mM CaCl2, 100 μg/ml bovine serum albumin, 0.5 mM cAMP. Once equilibrated, DNA templates were digested with DNase I at a final concentration of 100 ng/μl for 1 min at 37°C. Cleavage reactions were terminated by the addition of approximately 10 volumes of 570 mM NH4 acetate, 50 μg/ml tRNA, 80% (vol/vol) ethanol. DNA was then precipitated on dry ice, washed with 70% (vol/vol) ethanol, and suspended in 4 μl of 80% (vol/vol) formamide, 50 mM Tris-borate (pH 8.3), 1 mM EDTA, 0.1% (wt/vol) xylene cyanol, 0.1% (wt/vol) bromophenol blue. After denaturation at 85°C for 5 min, aliquots were separated on sequencing gels alongside TC and GA sequence ladders (57). Gels of primer extension and footprinting reactions were exposed to phosphor screens (Bio-Rad Laboratories), which were then scanned with a phosphorimager (GE Healthcare). Digital densitometry was done using ImageJ (1) running under Mac OS X (Apple Inc.).
Primer extension.
Total RNA was harvested from Lac reporter strains GPM1710, GPM1713, GPM1604, GPM1605, GPM1606, and GPM1607 grown to mid-log phase in LB medium at 37°C as described elsewhere (11). 32P-end-labeled primer SN884 was combined with 15 to 40 μg of total RNA, and the solution was heated for 5 min at 65°C and then cooled on ice for 2 min. The annealed primer was then extended with SuperScript III reverse transcriptase according to the manufacturer's protocol (Invitrogen). Aliquots of each reaction mixture were separated on sequencing gels alongside dideoxy chain-terminated sequencing ladders (66).
In silico analyses.
Mauve (21) was used to align the chromosomes of ETEC strain H10407 (GenBank accession no. FN649414) and K-12 strain MG1655 (accession no. U00096) (9, 20). A data set and nucleotide frequency table of 139 aligned CRP binding sites were downloaded from the RegTransBase database (44) (http://regtransbase.lbl.gov/). The frequency table was then used to calculate a position-weighted matrix as described previously (62). The position-weighted matrix was then used by our own software (DNAEntropy) to search for and rank putative CRP binding sites within select regions of the H10407 chromosome.
Membrane fractionation.
Outer membrane fractions were isolated as previously described (28). Briefly, bacteria were cultured aerobically at 37°C in LB medium to late log phase, harvested by centrifugation, and then lysed by two passages through a French press. Inner and outer membranes were separated by sucrose density ultracentrifugation (67). The protein concentration of membrane fractions was determined by the Bradford method (13) with reagents purchased from Bio-Rad Laboratories.
Protein electrophoresis.
Electrophoresis of membrane fractions was performed under denaturing conditions by the method of Laemmli (48). Gels were stained for proteins with Coomassie blue or for glycoprotein as described below. For immunoblotting, proteins separated by electrophoresis were transferred to nitrocellulose and then blocked with casein filler solution (10 mM Tris-buffered saline [pH 7.4], 2% [wt/vol] casein, 0.1% [wt/vol] sodium azide). Blocked filters were incubated for 1 h with a 1:200 dilution (in casein filler) of rabbit polyclonal anti-TibA antibody (52), washed in Tris-buffered saline containing 0.05% (vol/vol) Triton X-100, incubated for 1 h with alkaline phosphatase-conjugated protein A (Sigma) (1:2,000 dilution in casein filler), rewashed, and then detected with nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate toluidine salt (NBT-BCIP; Roche).
Detection of glycoproteins.
Glycoproteins were detected on nitrocellulose membranes using method B of the digoxigenin (DIG) glycan detection kit according to the manufacturer's recommendations (Roche). Briefly, proteins were separated by SDS-PAGE and then transferred to nitrocellulose. Membranes were washed in phosphate-buffered saline (PBS; 50 mM potassium phosphate [pH 6.5], 150 mM NaCl), and carbohydrates were oxidized with sodium metaperiodate. Oxidized carbohydrates were labeled with DIG-conjugated hydrazide. DIG-labeled proteins were visualized using alkaline phosphatase-conjugated anti-DIG antibodies and then detected with NBT-BCIP.
Digital images.
Digital images were cropped and scaled using Canvas X (ACD Systems) running under Mac OS X (Apple Inc.). The same software was also used to adjust brightness and contrast uniformly across digital images as necessary for visual clarity.
RESULTS
Identification of a CRP binding site upstream of tibDB.
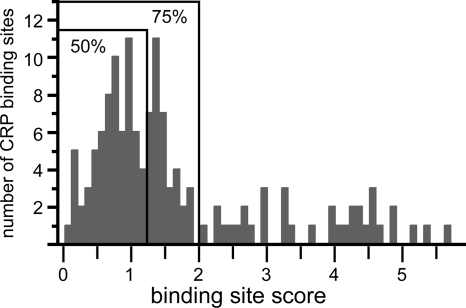
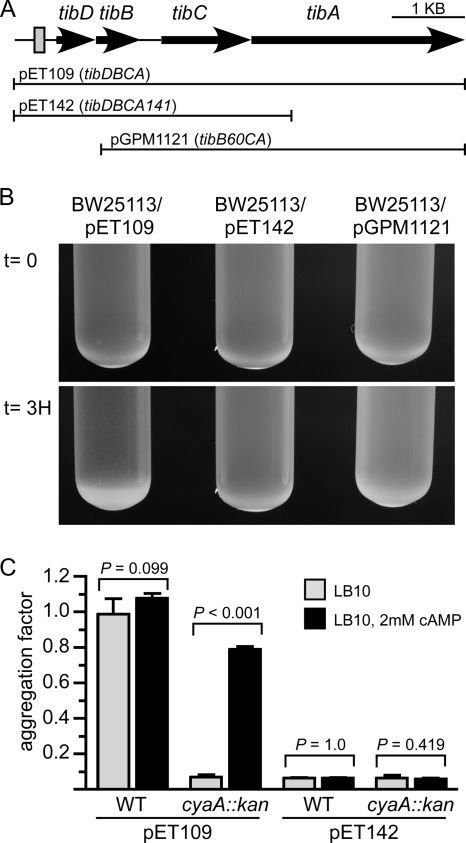
CRP regulates the expression of several ETEC virulence factors, including enterotoxins and certain types of adhesive pili (10, 25, 29, 43, 54). Thus, we reasoned that the further elucidation of the CRP regulon in ETEC could advance our understanding of this pathogen. Since microarrays containing an ETEC genome were not available for this study, we sought to identify CRP-regulated genes using a bioinformatics approach. To do this, we used a frequency table compiled from 139 known CRP binding sites to calculate a position-weighted matrix. The matrix was then used to search the genome of ETEC strain H10407 for potential CRP binding sites. Putative binding sites were ranked by the product of the Shannon redundancy index and Berg-von Hippel function (62). To limit the identification of false positives, we also restricted our analysis to putative sites ranking below a Shannon redundancy index of 2.0 because low values indicate a better fit to the data set of known CRP binding sites than high values. Although this cutoff value is arbitrary, 75% of the 139 characterized CRP binding sites in the RegTransBase database score below this threshold (Fig. 1). Since CRP binding sites have already been extensively characterized in K-12 strains, we further restricted our analysis to pathogen-specific regions of the chromosome. These unique regions were identified by aligning the H10407 and K-12 chromosomes (35, 36, 45). Among the putative CRP binding sites that we identified, one is located within a 16-Kb insertion at min 44.8 relative to the MG1655 chromosome. Features flanking this insertion include the CP4-44 cryptic prophage and sbmC (http://www.ecogene.org/). The putative CRP binding site, tibDBo, is located within a 372-bp intergenic region upstream of tibDB (Fig. 2 A). Relative to the 139 binding sites that define the position-weighted matrix, tibDBo ranks within the 50th percentile with a score of 1.1 (Fig. 1).
FIG. 1.
In silico identification of a CRP binding site upstream of tibDBCA. Histogram of characterized CRP binding sites. A position-weighted matrix was calculated from a frequency table derived from 139 aligned CRP binding sites. The matrix was then used to score each of the 139 binding sites as the product of the Shannon redundancy index and the Berg-von Hippel function. Low scores indicate a better fit to the collection of known binding sites than high scores. Bound regions enclose 50% and 75% of the 139 binding sites.
FIG. 2.
TibA-dependent aggregation. (A) Graphic depicting the chromosomal tib locus of ETEC strain H10407. The predicted CRP binding site upstream of tibDB is represented by a shaded rectangle. Solid lines represent regions of the tib locus carried by plasmids pET109 (tibDBCA), pET142 (tibDBCA141), and pGPM1121 (tibB60CA). (B) Representative stationary-phase cultures photographed immediately after mixing (t = 0) and 3 h after standing without agitation (t = 3H). (C) The absorbance (600 nm) of the upper fraction from stationary-phase cultures was read immediately after mixing and after the cultures were allowed to stand for 3 h without agitation. The difference between the two absorbance readings was used as the aggregation factor. Error bars represent the standard deviations of the means (n = 4). P values were calculated using the Student t test. WT, wild-type K-12 strain BW25113; cyaA::kan, K-12 strain JW3778.
TibA expression is dependent upon CRP.
Since bacteria expressing TibA autoaggregate and settle out of solution (69), we evaluated TibA expression by monitoring the aggregation of K-12 strains that were transformed with various plasmids. We observed that BW25113 aggregated and settled out of solution more rapidly when it harbored pET109 (tibDBCA) than when it harbored pET142 (tibDBCA141) (Fig. 2B). This result was expected and confirms that this assay reflects TibA-dependent aggregation, because tibA141 is a deletion of the tibA 3′ coding region (Fig. 2A). In addition to tibA, aggregation was found to be dependent upon tibDB and/or the tibDB promoter region because BW25113/pGPM1121 (tibB60CA) failed to aggregate (Fig. 2B). This result also demonstrates that a tib-encoded promoter is required for TibA expression, as opposed to a vector-encoded promoter, because pGPM1121 (tibB60CA) was constructed by deletion of an internal tib fragment from pET109 (tibDBCA) without disruption of flanking vector sequences. In addition, this tib-encoded promoter must lie upstream of tibB.
Having established that aggregation is an indicator of TibA expression from a tib-encoded promoter, we next compared aggregation of a cyaA::kan strain to that of an isogenic wild-type strain. Unlike the wild-type strain harboring pET109 (tibDBCA), which readily aggregated, aggregation was abolished in the cyaA::kan strain unless cAMP was provided exogenously (Fig. 2C). Since CRP requires cAMP for its activity, these results suggest that CRP is a positive regulator of TibA expression. As expected, strains harboring pET142 (tibDBCA141) failed to aggregate regardless of genetic background or exogenous cAMP.
In addition to aggregation assays with K-12 strains, we also evaluated the expression of TibA in ETEC strain H10407 and its isogenic H10407 Δcrp derivative. In agreement with our aggregation assays, we found by Western blotting that TibA expression was abolished in H10407 Δcrp (Fig. 3 A). Transformation of H10407 Δcrp with the CRP expression plasmid pSE186 restored TibA expression. In contrast, TibA was not expressed when H10407 Δcrp was transformed with the vector control pHG165. Since TibA is glycosylated by TibC, Western blot results were mirrored by glycoprotein staining (Fig. 3B). Taken together, the results of our aggregation assays, Western blotting, and glycoprotein staining demonstrate that CRP is required for the expression of TibA.
FIG. 3.
TibA expression in H10407 and H10407 Δcrp. Outer membranes were purified from the following strains (by lane): 1, HB101/pET109 (tibDBCA); 2, H10407; 3, H10407 Δcrp/pSE186 (crp+); 4, H10407 Δcrp/pHG165 (vector); and 5, H10407 Δcrp. Plasmid pHG165 is the vector control for both pET109 and pSE186. Outer membrane fractions were then separated by SDS-PAGE and subjected to immunoblotting with anti-TibA polyclonal antibody (A), glycoprotein staining (B), or Coomassie blue staining (C). As indicated, lane 6 contains molecular mass standards of 117 and 97 kDa. TibA has an expected molecular mass of 98 kDa, which is increase to ∼104 kDa by glycosylation (69).
CRP binds to tibDBo in vitro.
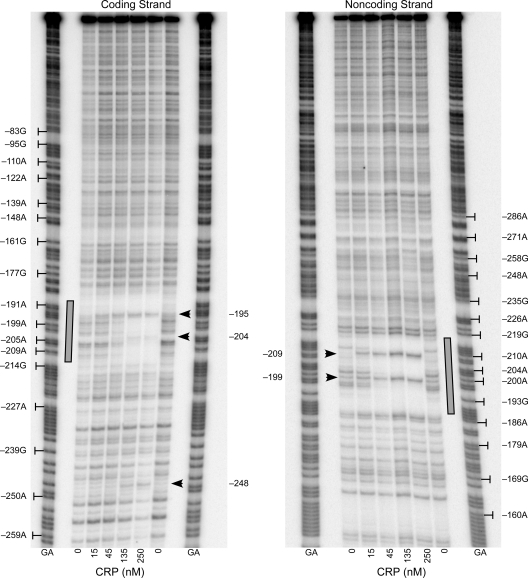
As expected from its relatively low score, in vitro DNase I footprinting confirmed that CRP binds to the site predicted by in silico analysis (Fig. 4). With respect to the tibD start codon, CRP protected a region from −212 through −188 on the coding strand and from −218 through −187 on the noncoding strand. CRP-dependent DNase I hypersensitive sites were also observed at −204 and −195 on the coding strand and −209 and −199 on the noncoding strand. These hypersensitive sites may indicate a distortion of the DNA helix, which is expected because CRP has been shown to bend DNA by as much as 90 degrees (68, 72). An additional hypersensitive site was observed at −248 on the coding strand. Curiously, this site lies approximately three helical turns upstream of the DNase I footprint of bound CRP. Since hypersensitivity is often visually apparent before protection, the hypersensitive site at −248 may indicate the presence of an unsaturated low-affinity CRP binding site. This phenomenon is evident on the noncoding strand at position −209, which displays hypersensitivity at 15 nM CRP in the absence of substantial protection of other bases within the site (Fig. 4).
FIG. 4.
CRP binds to a site upstream of tibDB. DNase I footprints of CRP bound to coding and noncoding strands of tibDBp are shown. The CRP binding site, tibDBo, is highlighted by shaded rectangles. Arrowheads indicate CRP-dependent DNase I hypersensitive sites. Numbering is relative to the tibD start codon. GA, Maxam-Gilbert sequence ladders.
CRP activates the tibDB promoter.
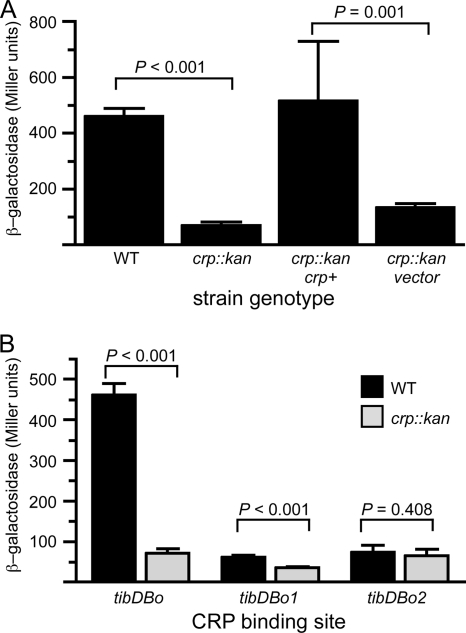
To evaluate the biological relevance of the CRP binding site, we integrated a tibDBp::lacZ reporter plasmid into the chromosomal attBHK022 site of wild-type and crp::kan K-12 strains. Our analysis revealed that expression of β-galactosidase was 7-fold lower in the crp::kan reporter strain than in the wild-type reporter strain (Fig. 5 A). Expression of β-galactosidase was restored to wild-type levels when CRP was provided in trans to the crp::kan mutant. As expected, the vector control plasmid pDU9 had no effect in the same genetic background. We also evaluated the effects of point mutations within binding site tibDBo (Fig. 5B). Mutagenized binding site tibDBo1 carries six mutations in the promoter-distal portion of the CRP binding site, while tibDBo2 carries five mutations in the promoter-proximal portion (Fig. 6 ). We found that tibDBo1 and tibDBo2 abolish CRP's ability to activate tibDBp (Fig. 5B). Thus, these results demonstrate that occupancy of tibDBo, which was predicted in silico and subsequently confirmed by DNase I footprinting, results in CRP-dependent activation of the tibDB promoter.
FIG. 5.
The tibDB promoter is activated by CRP. β-Galactosidase activities of tibDBp::lacZ reporters integrated into the chromosomal attBHK022 site of wild-type strain BW25113 or crp::kan strain JW5702 are shown. Assays were conducted in triplicate on two successive days (n = 6), and error bars show the standard deviations of the means. P values were calculated using the Student t test. (A) Reporter strains GPM1710 (WT) and GPM1713 (crp::kan) carrying attBHK022::pTibDBLac1 (tibDBo). For complementation assays, GPM1713 was transformed with pDCRP (crp+) or pDU9 (vector). (B) tibDBp activity in wild-type (WT) and crp::kan reporter strains carrying the natural CRP binding site tibDBo or the mutagenized binding sites tibDBo1 and tibDBo2.
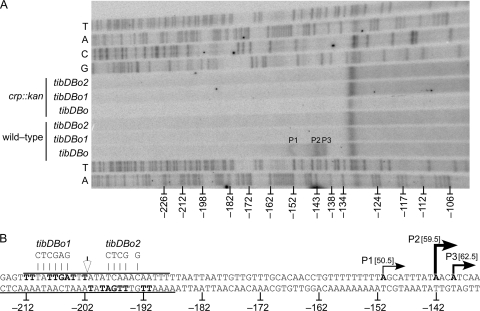
FIG. 6.
Identification of CRP-dependent transcription start sites. Numbering is relative to the tibD initiating codon. (A) Primer extension was used to map the transcription start sites of tibDBp in wild-type and crp::kan reporter strains carrying the natural CRP binding site, tibDBo, or the mutagenized binding sites, tibDBo1 and tibDBo2. Lanes T, A, C, and G contain dideoxy chain-terminated sequencing ladders produced with the same primer as the primer extension reactions. The band running across all lanes near −131 is an artifact. (B) Sequences of the tibDB promoter region and the mutagenized binding sites tibDBo1 and tibDBo2. Overlines and underlines indicate the extent of CRP DNase I footprints. The inverted repeat within the binding site is shown in bold. Filled arrows and bold type denote the positions of the three transcription start sites. The weight of each line/arrow reflects the relative proportions of each primer extension product. The distance between the center of the CRP binding site (unfilled arrow) and each transcription start site is shown in brackets.
Identification of CRP-dependent transcription start sites.
After determining that CRP activates tibDBp, we used primer extension to map the promoter's CRP-dependent transcription start sites (Fig. 6A). P2, the most abundant primer extension product by digital densitometry, mapped to an adenosine at −142 relative to the tibD start codon (Fig. 6B). A less pronounced primer extension product, P3, mapped to an adenosine at −139, followed by a minor product, P1, that corresponds to an adenosine at −151. Although only one representative reaction is shown, the relative intensities of the three bands (P2 > P3 > P1) was reproducible. In addition, all three bands are CRP dependent because they were not observed with RNA harvested from crp::kan strains (Fig. 6A). The CRP binding site mutations tibDBo1 and tibDBo2 also abolished the three primer extension products, even in the crp+ background (Fig. 6A). However, β-galactosidase assays demonstrate that the tibDB promoter is not completely silenced by these mutations or in the crp::kan background. Thus, the absence of primer extension products does not indicate a complete absence of transcription. It does, however, indicate that the transcripts are below the limits of detection by primer extension. In addition to the three bona fide primer extension products, we also observed a band near position −131 that was present in all primer extension reactions, including the dideoxy sequencing reactions, which utilized the same primer (Fig. 6A). This observation was confirmed by densitometric analyses and indicates that the band near −131 is a primer or gel running artifact.
DISCUSSION
In this study, we have shown that CRP activates the tibDB promoter by occupying a binding site upstream of tibD. We have also identified three CRP-dependent primer extension products in the tibDB promoter region. The CRP binding site is centered 59.5 bp upstream of P2, the most abundant primer extension product, and 62.5 bp upstream of P3, the second most abundant product (Fig. 6B). The distance of these transcription start sites to the CRP binding site suggests that tibDBp is primarily a class I promoter (14). With regard to P1, the minor primer extension product, the 50.5 bp between the CRP binding site center and the transcription start site is ambiguous and will require additional experimentation before P1 can be classified with confidence. Additional experimentation will also be required to identify the −10 and −35 hexamers of each promoter. Nevertheless, it is clear from these studies that CRP binds to tibDBo, activates the tibDB promoter, and is required for TibA expression. Furthermore there is no evidence of an additional CRP-dependent promoter within or downstream of tibB, because a strain carrying pGPM1121 (tibB60CA) did not display TibA-dependent autoaggregation.
Transposon mutagenesis and subcloning of the tib locus has shown that TibB is required for the expression of TibA (28). TibA is a 104-kDa protein that utilizes a carboxy-terminal autotransporter domain to reach the outer membrane, where it functions as an adhesin to mammalian cells (38, 53). TibA also mediates bacterial aggregation and biofilm formation (69). Adherence to mammalian cells, but not aggregation and biofilm formation, is dependent upon glycosylation of TibA by TibC (28, 69). Although tibDB are naturally located in cis with tibCA (Fig. 2A), TibA is also expressed when tibDB are provided in trans (28). Moreover, TibB and/or TibD apparently function at the level of transcription, because they are not required for the expression or membrane localization of TibA when it is expressed from a heterologous promoter such as lacp. TibB is probably a DNA binding protein because it contains a LuxR-like DNA binding helix-turn-helix motif near its carboxy terminus. We have also identified an 18-bp spaced inverted repeat (CAACGACTAAAGTCGTTG) centered 196 bp upstream of tibC within the intergenic region between tibDB and tibCA (Fig. 2A). This spaced inverted repeat may function as a TibB binding site, because other proteins with LuxR DNA binding motifs have been shown to bind spaced inverted repeats (23, 55, 73).
The translation of tibB is likely coupled to that of tibD because the two genes overlap by 4 bp. The tibD gene encodes a 20.4-kDa protein that is predicted to be cytoplasmic because it lacks a signal peptide and transmembrane helices. Residues 59 to 80 are predicted to form a helix-turn-helix motif (19), and this motif is contained within a larger region with similarity to the PF07180 (DUF1401) superfamily, whose members include GrlA and CaiF (32). GrlA is encoded within, and positively regulates other genes within, the locus of enterocyte attachment and effacement of enterohemorrhagic E. coli, enteropathogenic E. coli, and Citrobacter rodentium (6, 27, 40, 64). Additionally, GrlA has been shown to positively regulate the expression of enterohemolysin in enterohemorrhagic E. coli (65). CaiF is an activator of the E. coli caiTABCDE and fixABCX operons, which are required for carnitine metabolism under anaerobic conditions (15). Although this suggests that TibD is a transcription factor, it is not yet known if TibD is required for tibCA expression, because Tn5 insertions within tibD undoubtedly also disrupted TibB expression. Nevertheless, our results, when combined with previous studies that have shown that TibB is required for TibA expression (28), indicate that CRP positively regulates the expression of tibCA through an indirect pathway involving TibB and possibly TibD. The expression of the TibA-like AIDA-I adhesin has also been reported to be subject to catabolite repression (8). However, it is not yet known if this is a direct or indirect effect.
CRP has also been shown to regulate the expression of other ETEC virulence factors. For example, it has recently been shown that CRP represses the expression of the heat-labile toxin (10). Unlike TibA, which is regulated through an indirect pathway, CRP regulates the expression of the heat-labile toxin by binding to a site centered over the −35 hexamer of the toxin's promoter. CRP also regulates the expression of heat-stable toxins (2, 3, 10, 17, 56). Adhesive pili, such as CFA/I, CS1, CS2, and CS3, have also been reported to be subject to catabolite repression (29, 43). For CS1, CS2, and CS3 pili, the effect is produced indirectly through CRP-dependent expression of the regulator Rns (M. D. Bodero and G. P. Munson, unpublished). Thus, it is now clear that nearly all of the characterized ETEC virulence factors are regulated by CRP through direct or indirect pathways. Since the activity of CRP is inhibited when sugars such as glucose suppress the synthesis of cAMP, the pathogen may use the lumenal concentration of glucose as a cue to differentially express its virulence factors as it moves through the small intestine, as has been suggested previously (10, 25). CRP-repressed virulence factors, such as heat-labile toxin, may be expressed predominately in the duodenum, where the concentration of glucose is relatively high compared to that in the lower sections of the small intestine (31). As the pathogen travels toward the ileum, catabolite repression would be relieved as glucose and other monosaccharides are progressively absorbed by the small intestine. As a result, CRP-dependent adherence factors such as TibA and certain types of pili may not be fully expressed until the pathogen reaches the lower sections of the small intestine. This model for the spatial distribution of ETEC virulence factors is consistent with a murine model of colonization that has shown preferential colonization of the ileum by H10407 (4).
Acknowledgments
We thank Stephen Busby, Susan Egan, and Alan Wolfe for providing strains and plasmids.
This work was supported by NIAID grant number 057648.
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print on 7 January 2011.
REFERENCES
- 1.Abramoff, M. D., P. J. Magalhaes, and S. J. Ram. 2004. Image processing with image. J. Biophotonics Int. 11:36-41. [Google Scholar]
- 2.Alderete, J. F., and D. C. Robertson. 1977. Nutrition and enterotoxin synthesis by enterotoxigenic strains of Escherichia coli: defined medium for production of heat-stable enterotoxin. Infect. Immun. 15:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alderete, J. F., and D. C. Robertson. 1977. Repression of heat-stable enterotoxin synthesis in enterotoxigenic Escherichia coli. Infect. Immun. 17:629-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, K., M. Randolph, and J. Fleckenstein. 2006. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect. Immun. 74:869-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba, T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barba, J., et al. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187:7918-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, A., et al. 1990. Mutations that alter the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res. 18:7243-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berthiaume, F., M. F. Leblond, J. Harel, and M. Mourez. 2010. Growth-phase-dependent expression of the operon coding for the glycosylated autotransporter adhesin AIDA-I of pathogenic Escherichia coli. FEMS Microbiol. Lett. 311:176-184. [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 10.Bodero, M., and G. Munson. 2009. Cyclic AMP receptor protein-dependent repression of heat-labile enterotoxin. Infect. Immun. 77:791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodero, M., M. Pilonieta, and G. Munson. 2007. Repression of the inner membrane lipoprotein NlpA by Rns in enterotoxigenic Escherichia coli. J. Bacteriol. 189:1627-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botsford, J., and J. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Mol. Biol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 13.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 14.Browning, D., and S. Busby. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57-65. [DOI] [PubMed] [Google Scholar]
- 15.Buchet, A., W. Nasser, K. Eichler, and M. Mandrand-Berthelot. 1999. Positive co-regulation of the Escherichia coli carnitine pathway cai and fix operons by CRP and the CaiF activator. Mol. Microbiol. 34:562-575. [DOI] [PubMed] [Google Scholar]
- 16.Busby, S., and R. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 17.Busque, P., A. Letellier, J. Harel, and J. D. Dubreuil. 1995. Production of Escherichia coli STb enterotoxin is subject to catabolite repression. Microbiology 141:1621-1627. [DOI] [PubMed] [Google Scholar]
- 18.Choi, H. K., et al. 2002. Promoter analysis and regulatory characteristics of vvhBA encoding cytolytic hemolysin of Vibrio vulnificus. J. Biol. Chem. 277:47292-47299. [DOI] [PubMed] [Google Scholar]
- 19.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 20.Crossman, L. C., et al. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192:5822-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darling, A. C. E., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datsenko, K., and B. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devine, J. H., G. S. Shadel, and T. O. Baldwin. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC 7744. Proc. Natl. Acad. Sci. U. S. A. 86:5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebright, R. 1993. Transcription activation at class I CAP-dependent promoters. Mol. Microbiol. 8:797-802. [DOI] [PubMed] [Google Scholar]
- 25.Edwards, R., and D. Schifferli. 1997. Differential regulation of fasA and fasH expression of Escherichia coli 987 P fimbriae by enviromental cues. Mol. Microbiol. 25:797-809. [DOI] [PubMed] [Google Scholar]
- 26.Edwards, R. A., and J. L. Puente. 1998. Fimbrial expression in enteric bacteria: a critical step in intestinal pathogenesis. Trends Microbiol. 6:282-287. [DOI] [PubMed] [Google Scholar]
- 27.Elliott, S. J., et al. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1189. [DOI] [PubMed] [Google Scholar]
- 28.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans, D., D. Evans, T. Karjalainen, and C. Lee. 1991. Production of colonization factor antigen II of enterotoxigenic Escherichia coli is subject to catabolite repression. Curr. Microbiol. 23:71-74. [Google Scholar]
- 30.Evans, D., R. Silver, D. Evans, Jr., D. Chase, and S. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Esherichia coli enterotoxigenic for humans. Infect. Immun. 12:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferraris, R., S. Yasharpour, K. Lloyd, R. Mirzayan, and J. Diamond. 1990. Luminal glucose concentrations in the gut under normal conditions. Am. J. Physiol. 259:G822-837. [DOI] [PubMed] [Google Scholar]
- 32.Finn, R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gama-Castro, S., et al. 2008. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 36:D120-D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girard, V., et al. 2010. Conformation change in a self-recognizing autotransporter modulates bacterial cell-cell interaction. J. Biol. Chem. 285:10616-10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gosset, G., Z. Zhang, S. Nayyar, W. A. Cuevas, and M. H. Saier. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186:3516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grainger, D. C., D. Hurd, M. Harrison, J. Holdstock, and S. J. W. Busby. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 102:17693-17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haldimann, A., and B. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henderson, I., and J. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holcroft, C. C., and S. M. Egan. 2000. Roles of cyclic AMP receptor protein and the carboxyl-terminal domain of the alpha subunit in transcription activation of the Escherichia coli rhaBAD operon. J. Bacteriol. 182:3529-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyoda, S., et al. 2006. The GrlR-GrlA regulatory system coordinately controls the expression of flagellar and LEE-encoded type III protein secretion systems in enterohemorrhagic Escherichia coli. J. Bacteriol. 188:5682-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalivoda, E. J., N. A. Stella, D. M. O'Dee, G. J. Nau, and R. M. Q. Shanks. 2008. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl. Environ. Microbiol. 74:3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanack, K. J., L. J. Runyen-Janecky, E. P. Ferrell, S.-J. Suh, and S. E. H. West. 2006. Characterization of DNA-binding specificity and analysis of binding sites of the Pseudomonas aeruginosa global regulator, Vfr, a homologue of the Escherichia coli cAMP receptor protein. Microbiology 152:3485-3496. [DOI] [PubMed] [Google Scholar]
- 43.Karjalainen, T., D. Evans, and D. Evans. 1991. Catabolite repression of the colonization factor antigen I (CFA/I) operon of Escherichia coli. Curr. Microbiol. 23:307-313. [Google Scholar]
- 44.Kazakov, A. E., et al. 2007. RegTransBase—a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 35:D407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khankal, R., J. W. Chin, D. Ghosh, and P. C. Cirino. 2009. Transcriptional effects of CRP* expression in Escherichia coli. J. Biol. Eng. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim, T.-J., et al. 2007. Direct transcriptional control of the plasminogen activator gene of Yersinia pestis by the cyclic AMP receptor protein. J. Bacteriol. 189:8890-8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 48.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 49.Lathem, W. W., P. A. Price, V. L. Miller, and W. E. Goldman. 2007. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315:509-513. [DOI] [PubMed] [Google Scholar]
- 50.Lawson, C. L., et al. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14:10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim, S., et al. 2003. Molecular analysis of Salmonella enterotoxin gene expression. J. Microbiol. Biotechnol. 13:598-606. [Google Scholar]
- 52.Lindenthal, C., and E. A. Elsinghorst. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lo-Tseng, T., J. Lee, and R. E. Isaacson. 1997. Regulators of Escherichia coli K99 region 1 genes. Adv. Exp. Med. Biol. 412:303-310. [DOI] [PubMed] [Google Scholar]
- 55.Maris, A. E., et al. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 9:771-778. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Cadena, M., L. Guzman-Verduzco, H. Stieglitz, and Y. Kupersztoch-Portnoy. 1981. Catabolite repression of Escherichia coli heat-stable enterotoxin activity. J. Bacteriol. 145:722-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 59.Müller, C., A. Åberg, and J. Straseviçiene. 2009. Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP-cAMP. PLoS Pathog. 5:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munson, G., and J. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 181:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh, M. H., S. M. Lee, D. H. Lee, and S. H. Choi. 2009. Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect. Immun. 77:1208-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Neill, M. C. 1998. A general procedure for locating and analyzing protein-binding sequence motifs in nucleic acids. Proc. Natl. Acad. Sci. U. S. A. 95:10710-10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 64.Russell, R., F. Sharp, and D. Rasko. 2007. QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J. Bacteriol. 189:5387-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saitoh, T., et al. 2008. Transcription of the ehx enterohemolysin gene is positively regulated by GrlA, a global regulator encoded within the locus of enterocyte effacement in enterohemorrhagic Escherichia coli. J. Bacteriol. 190:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schnaitman, C. A. 1970. Protein composition of the cell wall and cytoplasmic membrane of Escherichia coli. J. Bacteriol. 104:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253:1001-1007. [DOI] [PubMed] [Google Scholar]
- 69.Sherlock, O., R. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skorupski, K., and R. K. Taylor. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 94:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stewart, G. S., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 72.Suck, D. 1997. DNA recognition by structure-selective nucleases. Biopolymers 44:405-421. [DOI] [PubMed] [Google Scholar]
- 73.White, C. E., and S. C. Winans. 2007. The quorum-sensing transcription factor TraR decodes its DNA binding site by direct contacts with DNA bases and by detection of DNA flexibility. Mol. Microbiol. 64:245-256. [DOI] [PubMed] [Google Scholar]
- 74.Wickstrum, J., and S. Egan. 2002. Ni+-affinity purification of untagged cAMP receptor protein. Biotechniques 33:728-730. [DOI] [PubMed] [Google Scholar]
- 75.Zhan, L., et al. 2008. The cyclic AMP receptor protein, CRP, is required for both virulence and expression of the minimal CRP regulon in Yersinia pestis biovar microtus. Infect. Immun. 76:5028-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng, D., C. Constantinidou, and J. Hobman. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]