Abstract
We reported previously that Go-deficient mice develop severe neurological defects that include hyperalgesia, a generalized tremor, lack of coordination, and a turning syndrome somewhat reminiscent of unilateral lesions of the dopaminergic nigro-striatal pathway. By using frozen coronal sections of serially sectioned brains of normal and Go-deficient mice, we studied the ability of several G protein coupled receptors to promote binding of GTPγS to G proteins and the ability of GTP to promote a shift in the affinity of D2 dopamine receptor for its physiologic agonist dopamine. We found a generalized, but not abolished reduction in agonist-stimulated binding of GTPγS to frozen brain sections, with no significant left–right differences. Unexpectedly, the ability of GTP to regulate the binding affinity of dopamine to D2 receptors (as seen in in situ [35S]sulpiride displacement curves) that was robust in control mice, was absent in Go-deficient mice. The data suggest that most of the effects of the Gi/Go-coupled D2 receptors in the central nervous system are mediated by Go instead of Gi1, Gi2, or Gi3. In agreement with this, the effect of GTP on dopamine binding to D2 receptors in double Gi1 plus Gi2- and Gi1 plus Gi3-deficient mice was essentially unaffected.
Keywords: G protein, knockout mice, homologous recombination, GTP shift, sulpiride
Most G protein coupled receptors (GPCRs) display the so-called GTP-shift phenomenon whereby the affinity of the receptor for its activating ligand—hormone, neurotransmitter, autacoid, cytokine—is low when the receptor is dissociated from the G protein and increases on interacting with the nucleotide-free form of the specific G protein(s) it activates. Although it was originally observed as a decrease in affinity for glucagon in liver membranes (1), it is most commonly tested for indirectly by measuring the displacement of a labeled antagonist—for which the affinity is independent of the association of the receptor with its G protein—by an activating agonist in the absence and presence of a guanine nucleotide (2, 3). The typical result is that a competitive displacement curve obtained in the absence of the guanine nucleotide is to the left of that obtained in its presence. This led to the use of the term “GTP-shift” to describe the conversion of high affinity agonist binding sites into forms of lower affinity (e.g., refs. 4 and 5). The nucleotides commonly used to functionally separate the receptor from the G protein are nonhydrolyzable GTP analogs, such as guanylyl 5′-imidodiphosphate [GMP-P(NH)P] or GTPγS. However, the affinity transition can be elicited equally well with GTP, GDP, or the thio-analogue GDPβS (1, 6), indicating that the GTP-shift is not a measure or consequence of receptor-induced G protein activation. Indeed, a heterotrimeric Gs, whose α subunit carries the Gly210 to Ala mutation and cannot be activated by GTP [or GMP-P(NH)P], is competent for interaction with receptors onto which it confers high agonist affinity that is lost on addition of guanine nucleotides (7, 8).
When assessed for their GTP-shifts in agonist binding, receptors display equilibrium displacement kinetics that vary, requiring either two-state or three-state models to fit the data. Based on biochemical reconstitution and genetic ablation data, the two receptor states are thought to arise as a consequence of an equilibrium between free receptors with low agonist affinity and G protein-associated receptors with high agonist affinity (cf. refs. 9–11). The need for three-state models is less well understood and may be due to interaction of the receptor with more than one G protein, each inducing a distinct high affinity state, to the presence of more than one receptor subtype, to both, or to some other as yet to be determined factor(s). Experimentally, three-state models appear in studies of receptors that are coupled to effectors by one or more of the pertussis toxin (PTX)-sensitive G proteins (i.e., of the Gi/Go family).
In addition to exhibiting GTP-shifts in their binding to agonists, functional GPCRs can of course also be identified by their effect(s) on specific effector systems and, more generally, by their universal function to catalyze the exchange of resident GDP for incoming GTP or GTP analog. Thus, incubation of membranes or tissue slices with agonists in the presence of [35S]GTPγS and Mg2+—required for activation of G proteins—is a sensitive test for the presence of functional receptors and the G protein(s) to which they couple. This has been used to test for central nervous system (CNS) distribution of serotonin receptors in brain slices (12).
Go belongs to the PTX-sensitive G proteins to which the three Gi (Gi1, Gi2, and Gi3) and the sensory G proteins rod and cone transducins and gustducin belong. The o of Go stands for other, because it was the second G protein discovered to be a substrate for the ADP-ribosylating activity of PTX (13). In the CNS, Go is by far the most abundant of all of the G proteins, reaching up to 1% of membrane protein (13). Lower levels are expressed also in endocrine cells and in the heart (14). There are two flavors of Go because of the use of an alternate pair of exons 7 and 8. These exons encode the last third of the protein and include key portions of the guanine nucleotide binding pocket and of the receptor-interacting residues of the C terminus (refs. 15–18; reviewed in ref. 1).
The exact roles of Go as a signal transducer are rather poorly understood. In contrast to other G proteins, it appears that most of the effects of Go activation may be due to the generation of Gβγ subunits, which proceed to regulate positively or negatively a plethora of cellular functions that includes activation of phospholipase Cβ (19), inwardly rectifying K channels (20), type 2 and 4 adenylyl cyclase (cf. with ref. 21) and types β and γ of phosphatidylinositol 3-kinases (22, 23), as well as the inhibition of type 1 adenylyl cyclase (cf. with ref. 21) and several subtypes of voltage-activated Ca2+ channels (24, 25). In contrast, the role of Goα is rather controversial, having been reported to inhibit some adenylyl cyclases, activate certain types of hippocampal K+ channels, and more recently to interact with a GTPase activating protein (GAP) acting on rap1A (26). Constitutively active Goα transforms NIH 3T3 cells acting via the Stat3 pathway (27). Taken together, Goα seems to play roles in cellular proliferation, transformation, and/or differentiation (see also ref. 28).
GPCR's are generally spoken of as Gs-, Gq-, or Gi/Go-coupled, indicating their preference/specificity for one or the other type of G proteins. This type of classification satisfactorily predicts major consequences of GPCR activation for those receptors coupled by the Gs and Gq family of G proteins (e.g., stimulation of adenylyl cyclase vs. that of phospholipase Cβ). In the case of Gi/Go, however, the effects of receptors on cells are more likely to differ if Gi1 or Gi3 are activated than if Go is activated or, when Go is absent, if Gi2 is activated. Like Go, Gi2 is expressed at higher levels than Gi1 and Gi3 (14, 29). Gi1 and Gi3, because of their much lower expression levels are likely to be acting for the most part via their α subunits, whereas receptors activating Go or Gi2 are likely to elicit immediate effects primarily via Gβγ (Go) or via both Gβγ and Gα (Gi2). Indeed, receptors recognize this difference and appear to exhibit very high degrees of specificity toward the α subunit of the G protein(s) they activate, including distinguishing between Gαo1 and Gαo2 splice variants (30, 31). This high degree of specificity extends also to the recognition of the G12 and G13 class of G proteins that regulate cytoskeletal aspects of cell function. Receptors regulate the cytoskeleton independently of their effects on Gs, Gq, and/or Gi/Go, giving rise to finely tuned cellular responses to GPCR stimulation. It is not known whether G12 and G13 induce GTP-shifts.
The effects of dopamine (DA) are mediated by the D1-like D1 and D5 receptors and the D2-like D2, D3, and D4 receptors. Functional variants affecting the third intracellular loop—believed to play a key role in G protein specificity—have been described for the D2 and D4 receptors, and in the mouse also for the D3 receptor. D2 receptor variants arise from alternative splicing and are designated as D2S (short) and D2L (long) to denote the presence of an extra stretch of 29 amino acids. D2L and D2S are coexpressed, with D2L being the predominant form. D4 receptor variants arise from the expansion of a 16-aa (48-bp) repeat. In humans, the predominant form is the four-repeat D4.4, followed by the D4.2 and the D4.7 forms. Up to 11 repeats have been found. In the mouse, the use of alternative splice acceptor sites generates two forms differing by 21 amino acids in their third intracellular loop. None of the D2-like receptor variants has so far been associated with differences in expression or pharmacological properties (reviewed in refs. 32 and 33). D1- and D2-like receptors are coupled to their effectors by Gs and the Gi/Go proteins, respectively (34). The cellular effects of D2-like receptors vary with the cell type in which they are expressed and include inhibition of adenylyl cyclase, activation of certain types of K+ channel, inhibition of Ca2+ currents, activation of the MAP kinase pathway, stimulation of cell proliferation, and inhibition of gene transcription. As expected for Gi/Go-coupled receptors, the effects of D2-like receptors are uncoupled by PTX (reviewed in refs. 33 and 35). Although the potential of the various D2-like receptors to differentially activate one or the other of the Gi/Go proteins has been studied rather extensively, no clear picture has emerged.
In the present study, we examined coronal sections of brains of mice in which we had disrupted the Go gene for the consequence of this disruption on three parameters: (i) in situ binding of GPTγS in response to several CNS agonists, (ii) bulk expression of D2-like receptors by analyzing in situ binding of the receptor antagonist iodosulpiride, and (iii) the D2 receptor GTP-shift in dopamine binding. In these mice, the Go protein has been inactivated by disruption of its sixth exon (36). Mice defective in Go were found to have multiple neurological deficiencies and tend to live less than 3 weeks, dying within the first and second day post partum and around weaning. The few that survive are hyperactive and develop a turning syndrome, the cause of which is unknown (36). In addition to generally poor health and ill-defined neurologic problems, Go-deficient mice have so far revealed two specific roles for Go: (i) It is required for muscarinic inhibition of cardiac voltage-gated calcium channel(s) (14); and (ii) it is obligatory for transduction of the photoreceptor signal in bipolar-ON cells (37).
Materials and Methods
Reagents.
[35S]GTPγS and [125I]sulpiride were obtained from New England Nuclear and Amersham Pharmacia, respectively. GDP and GTP were obtained from Roche Molecular Biochemicals. All other reagent grade chemicals were obtained from Sigma or Fisher.
Gα-Deficient Mice.
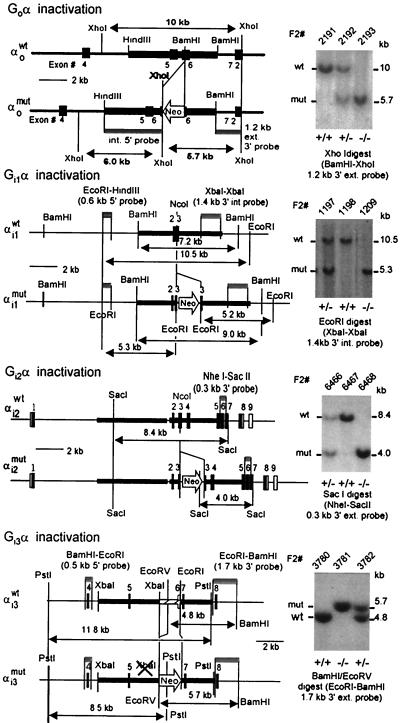
Mice deficient in Go (36), Gi1 (G.B. and L.B., unpublished data), Gi2 (38), and Gi3 (K.S. and L.B., unpublished data) were generated by introducing a neomycin resistance gene into the respective genes following the strategies described in refs. 39 and 40. Predicted restriction enzyme digest maps and confirmatory Southern analysis of tail biopsies of the wild-type mice and mice heterozygous and homozygous for the disruption of the four genes is shown in Fig. 1. Mice with combined deficiencies in Gi1 plus Gi3 and Gi1 plus Gi2 were generated by interbreeding.
Figure 1.
Restriction maps of the wild-type and disrupted genes encoding Gαi1, Gαi2, Gαi3, and Gαo and representative Southern analysis of tail biopsies of wild-type and targeted mice. Shown are the probes and sizes of fragments used to diagnose the structures of the genes.
Serial Coronal Sectioning of Mouse Brains.
Mutant and wild-type mice were anesthetized, killed by cervical dislocation, and decapitated. The brains were dissected and frozen in isopentane cooled to −4°C with dry ice. Frozen mouse brains (≈1.0 cm long in the anterior-posterior axis) were serially cut at −2°C into 10-μm-thick coronal sections in a cryostat-microtome (HM500 M Microm, Zeiss) and thaw-mounted onto 20 sets of Fisherbrand SuperFrost Plus slides, each set receiving every 20th section and thus representing the whole brain at 200-μm intervals. Brain sizes varied somewhat so that each set contained between 35 and 50 cuts. Slides were stored at −8°C in a tightly closed box until used.
Agonist-Stimulated [35S]GTPγS Binding to Coronal Brain Sections on Glass Slides.
Previously reported guidelines (12, 41) for the study of serotonin receptor activity in guinea pig brain slices were followed. Briefly, for assessment of agonist-stimulated binding of GTPγS to brain sections, the slides were brought to room temperature and overlaid with 1.0 ml of binding buffer (50 mM Hepes (pH 7.5), 3 mM MgCl2, 100 mM NaCl, 0.2 mM EGTA, 0.2 mM DTT, 2 mM GDP) for 5 min at room temperature. The buffer was then exchanged for 1.0 ml binding buffer containing in addition 0.1 nM [35S]GTPγS (1000–1500 Ci/mM; 1 Ci = 37 GBq) and the appropriate agonist. After an incubation of 60 min at room temperature, the slides were washed twice for 3 min with ice-cold washing buffer (50 mM Hepes (pH 7.5), 1 mM MgCl2), dipped briefly in ice-cold double-distilled water, air dried, and exposed to Kodak BIOMAX MR film.
[125I]Sulpiride Binding to Coronal Brain Sections and Displacement by Agonist in the Presence and Absence of GTP.
Slides prepared as above were brought to room temperature and overlaid with 1.0 ml of ligand binding buffer (50 mM Tris⋅HCl (pH 7.5), 120 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5.7 mM ascorbic acid) containing 0.2 nM [125I]sulpiride (2000 Ci/mmol) and the indicated concentrations of unlabeled dopamine. After an incubation of 30 min at room temperature, the slides were washed twice for 3 min with ice-cold ligand binding buffer, dipped into ice-cold double distilled water, air dried, and autoradiographed by using Kodak BIOMAX MR films for visual inspection. Exposures were quantified by densitometry using a Bio-Rad GS-700 Densitometer.
Results and Discussion
Our previous studies showed that Go-deficient mice have multiple neurologic deficiencies apparent both at the cellular level, where the response of voltage-gated Ca2+ channels in primary dorsal root ganglion cells is altered, and at various levels of integrated physiological activities—including a turning syndrome and loss of the bipolar ON response in the retina (36, 37). To gain further insight into the reasons for the turning syndrome, we studied two G protein-dependent effects in frozen brain slices: agonist mediated GTPγS binding and the GTP-shift of CNS D2-like dopaminergic receptors.
We performed these studies in coronal sections with the hope that left–right differences might show up that could give a clue as to the reason for the turning syndrome. Moreover, by thaw-mounting serially the sequential 10-μm sections onto 20 slides, this technique allows for the splitting of one brain into 20 equivalent “aliquots” (here referred to as sets), that can then be tested for binding in replicates under several conditions (increasing ligand concentrations, presence and absence of unlabeled ligand to control for nonspecific bindings, and test of several agonists). As illustrated in Fig. 2, the approach allows for reliable detection of brain regions in which one or the other agonist activates GPCRs and highlights the fact that different receptors may act on G proteins with different efficacy, leading to differing levels or rates of nucleotide exchange. The rates of nucleotide exchange observed depend of course on the combination of abundance and efficacy. Proper interpretation of the GTPγS binding data awaits, therefore, an analysis of the respective receptor densities.
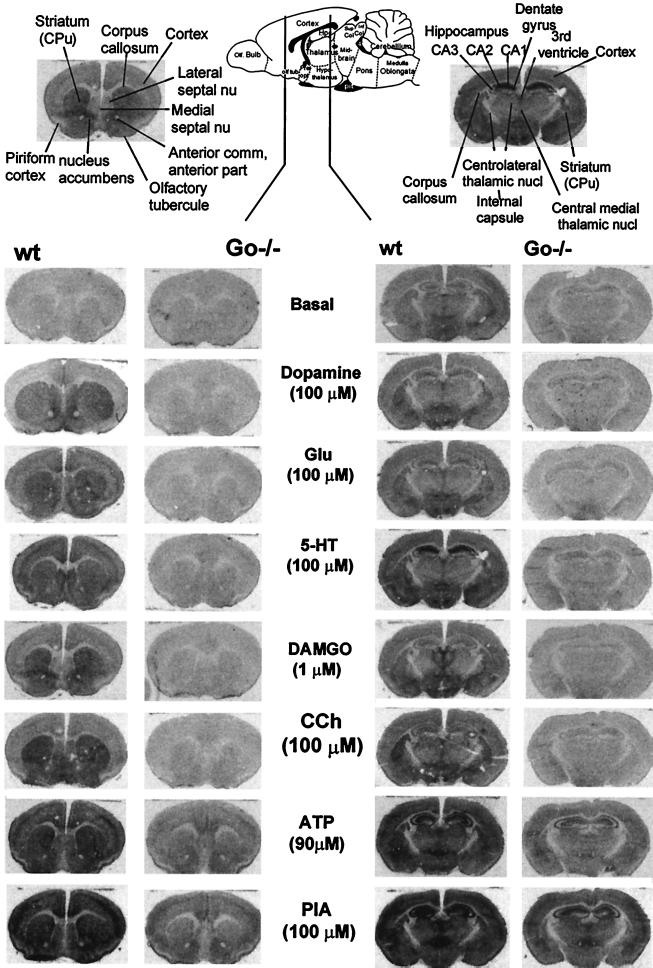
Figure 2.
Representative autoradiograms depicting basal and agonist-stimulated binding of [35S]GTPγS to 10-μm-thick coronal sections from 2 of 39 CNS regions of wild-type and Go−/− mice. (Upper Center) Schematic drawing of a sagittal section of a rodent brain depicting location of the sections tested for GTPγS binding. (Upper, Left and Right) annotation of anatomical landmarks present in the tested sections as described by Paxinos and Watson (42). Note: (i) There is a generalized decrease in agonist-stimulated GTPγS binding consistent with a generalized decrease in G protein density in Go−/− mice; and (ii) extensive examination of these and several repeat experiments failed to reveal a left–right difference in GTPγS binding in Go−/− mice. The following agonists were tested at the indicated concentrations: DA, dopamine; CCh, carbachol (nonselective muscarinic receptor agonist); Glu, glutamate; 5-HT, 5-hydroxytryptamine or serotonin; DAMGO, D-Ala2, N-Met-Phe4, Gly-ol5-enkephlin (μ selective opioid receptor agonist); PIA, phenylisopropyladenosine (A1 selective adenosine receptor agonist); ATP, adenosine triphosphate.
The most striking effect of the loss of Go on GTPγS binding was the generalized decrease in binding that any of the tested agonists was able to promote. This indicated that all these Gi/Go-coupled receptors are indeed coupled to effectors by Go in their natural CNS environments. Close examination of long exposures revealed in addition that, except for dopamine, all other agonists tested were still able to promote the binding of GTPγS in selected areas over that seen in the absence of agonists (data not shown). This indicates that, by this test, Gi/Go-coupled receptors are indeed coupled to effectors by both types of proteins.
In contrast, we failed to uncover any significant left–right difference in GTPγS binding. This is not to say that there may not be one in a specific area or region. The region may be small and require a higher resolution approach than used here. Further sensory organs, such as the vestibule and the cochlea, were not included in these studies.
Given that dopamine is involved in the control of locomotion and cognition, we specifically tested for consequences of Go-deficiency on the D2-like dopaminergic receptors in the CNS by assessing binding of the D2-like receptor antagonist [125I]sulpiride and its displacement by dopamine. By using iodosulpiride (Kd = 0.9 nM) at 0.2 nM we noted no significant difference in receptor abundance between wild-type and Go−/− mice (Fig. 3). We also noted no significant left–right difference in either wild-type or Go−/− brains, and have therefore not uncovered a reason that would allow for development of a rational hypothesis as to why Go−/− mice turn.
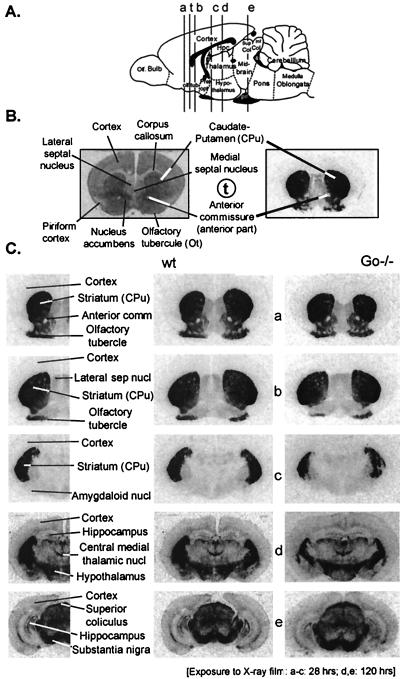
Figure 3.
Binding of the D2 receptor antagonist [125I]iodosulpiride to coronal sections of wild-type and Go−/− mouse brains. (A) Diagram showing location of coronal sections tested. (C Left) Annotation of anatomical landmarks present in the respective sections (42). (C Center and Right) Binding of 0.2 nM 125I-sulpiride (30 min at room temperature) to D2 receptors in the respective sections as revealed by autoradiography. Note that there appears to be no significant difference between wild-type and Go−/− brains in the binding of a subsaturating concentration of the antagonist (0.2 nM).
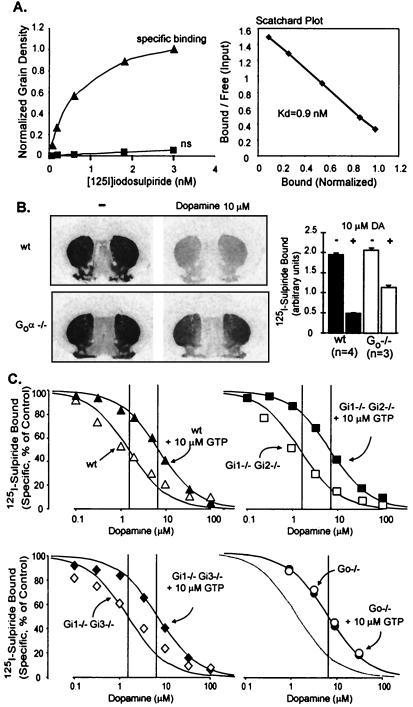
Unexpectedly, however, we noted an essentially total loss of high agonist affinity forms of the D2 receptor system in Go−/− mice (Fig. 4). Reports in the literature on the specificity of D2-like receptors for Gi and Go proteins are unclear and varied with the method used and the test cells, but they have consistently pointed to interaction with both Gi and Go (43–51). Our data add to the literature by clearly pointing to Go as the major target of the D2-like class of dopamine receptors. Moreover, it would appear that Gi proteins either do not substitute for Go, or do so poorly. It should be noted, however, that the data in no way indicate that D2 receptors in their native environment are incapable of signaling through other G proteins. As stated earlier the paper, most (and hence not all) GPCRs exhibit a GTP-shift (e.g., see ref. 51). Furthermore, GTP-shifts vary with the intrinsic efficacy of the agonist (53, 54) and may also vary with the type of G protein they affect. It is thus possible that D2 receptors have evolved to have low affinity for G proteins and act mostly through the highly abundant Go. In the absence of Go, D2 receptor effects are thus likely to be of low efficiency (rather than absent) and mediated by one of the Gi proteins that causes small shifts in their affinity for dopamine.
Figure 4.
GTP-shift of D2 dopamine receptor of the brains of wild-type mice; loss in Go−/− mice, but not Gi1 plus Gi2, or Gi1 plus Gi3−/− mice. Sets of slides with brain sections containing the D2 receptor-rich caudate–putamen regions were analyzed. (A) Scatchard analysis of iodosulpiride binding to D2 receptors in wild-type mice. Coronal sections were overlaid with the indicated concentrations of iodosulpiride without (triangles) or with 100 μM sulpiride (squares), processed as described in Materials and Methods, and autoradiographed. [125I]sulpiride retained was quantified from autoradiograms by densitometry. The data were corrected for nonspecific binding and the binding (grain density per unit area) at 3 nM iodosulpiride was arbitrarily set at 1.0 for normalization purposes. (B) Competitive inhibition by dopamine of 0.2 nM iodosulpiride binding to D2 receptors in wild-type and Go−/− mice. Bars represent means ± SEM of four experiments. Note the higher effectiveness of 10 μM dopamine in inhibiting iodosulpiride binding in wild-type than Go−/− mice due to loss of high agonist affinity form(s) of the D2 receptor in Go−/− mice. (C) Dose–response curves for dopamine inhibition of iodosulpiride binding to D2 receptors in brains of wild-type mice and mice deficient in either Gi1 plus Gi2, Gi1 plus Gi3, or Go. Note that there is a loss of high affinity dopamine binding in Go−/− mice, but not in mice deficient in any of the Gi-type proteins.
Acknowledgments
This work was supported in part by National Institutes of Health Grant DK-19318.
Abbreviations
- GPCR
G protien coupled receptor
- GTPγS
guanylyl 5′-[γ-thio]-triphosphate
- PTX
pertusis toxin
- CNS
central nervous system
References
- 1.Rodbell M, Krans H M J, Pohl S L, Birnbaumer L. J Biol Chem. 1971;246:1872–1876. [PubMed] [Google Scholar]
- 2.Maguire M E, Van Arsdale P M, Gilman A G. Mol Pharmacol. 1976;12:335–339. [PubMed] [Google Scholar]
- 3.Lefkowitz R J, Mullikan D, Caron M G. J Biol Chem. 1976;251:4686–4692. [PubMed] [Google Scholar]
- 4.DeLean A, Stadel J M, Lefkowitz R J. J Biol Chem. 1980;255:7108–7117. [PubMed] [Google Scholar]
- 5.Kent R S, DeLean A, Lefkowitz R J. Mol Pharmacol. 1980;17:14–23. [PubMed] [Google Scholar]
- 6.Iyengar R, Abramowitz J, Bordelon-Riser M E, Blume A J, Birnbaumer L. J Biol Chem. 1980;255:10312–10321. [PubMed] [Google Scholar]
- 7.Bourne H R, Beiderman B, Steinberg F, Brothers V M. Mol Pharmacol. 1982;22:204–210. [PubMed] [Google Scholar]
- 8.Lee E, Taussig R, Gilman A G. J Biol Chem. 1992;267:1212–1218. [PubMed] [Google Scholar]
- 9.Cerione R A, Codina J, Benovic J L, Lefkowitz R J, Birnbaumer L, Caron M G. Biochemistry. 1984;23:4519–4525. doi: 10.1021/bi00315a003. [DOI] [PubMed] [Google Scholar]
- 10.Bird S J, Maguire M E. J Biol Chem. 1978;253:8826–8834. [PubMed] [Google Scholar]
- 11.Birnbaumer L, Birnbaumer M. In: Handbook of Biomembranes. Shiniztky M, editor. Vol. 3. Rehovoth, Israel: Balaban Publishers; 1994. pp. 153–252. [Google Scholar]
- 12.Sim L J, Selley D E, Childers S R. Proc Natl Acad Sci USA. 1995;92:7242–7246. doi: 10.1073/pnas.92.16.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sternweis P C, Robishaw J D. J Biol Chem. 1984;259:13806–13813. [PubMed] [Google Scholar]
- 14.Valenzuela D, Han X, Mende U, Fankhauser C, Mashimo H, Huang H, Pfeffer J, Neer E J, Fishman M C. Proc Natl Acad Sci USA. 1997;94:1727–1732. doi: 10.1073/pnas.94.5.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu W H, Rudolph U, Sanford J, Bertrand P, Olate J, Nelson C, Moss L G, Boyd A E, III, Codina J, Birnbaumer L. J Biol Chem. 1990;265:11220–11226. [PubMed] [Google Scholar]
- 16.Strathmann M, Wilkie T, Simon M I. Proc Natl Acad Sci USA. 1990;87:6477–6481. doi: 10.1073/pnas.87.17.6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertrand P, Sanford J, Rudolph U, Codina J, Birnbaumer L. J Biol Chem. 1990;265:18576–18580. [PubMed] [Google Scholar]
- 18.Tsukamoto T, Toyama R, Itoh H, Kozasa T, Matsuoka M, Kaziro Y. Proc Natl Acad Sci USA. 1991;88:2974–2978. doi: 10.1073/pnas.88.8.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camps M, Carozzi A, Schnabel P, Scheer A, Parker P J, Gierschik P. Nature (London) 1992;360:684–686. doi: 10.1038/360684a0. [DOI] [PubMed] [Google Scholar]
- 20.Logothetis D E, Kurachi Y, Galper J, Neer E J, Clapham D E. Nature (London) 1987;327:21–22. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 21.Taussig R, Gilman A G. J Biol Chem. 1996;270:1–4. doi: 10.1074/jbc.270.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Tang X, Downes A G J. J Biol Chem. 1997;272:14193–14199. doi: 10.1074/jbc.272.22.14193. [DOI] [PubMed] [Google Scholar]
- 23.Vanhaessebroeck B, Leevers S J, Panayotu G, Waterfiled M D. Trends Biochem Sci. 1997;22:267–272. doi: 10.1016/s0968-0004(97)01061-x. [DOI] [PubMed] [Google Scholar]
- 24.Herlitze S, Garcia D E, Mackie K, Hille B, Scheuer T, Catterall W A. Nature (London) 1996;380:258–262. doi: 10.1038/380258a0. [DOI] [PubMed] [Google Scholar]
- 25.Qin N, Olcese R, Zhou J, Cabello O, Birnbaumer L, Stefani E. Am J Physiol. 1996;271:C1539–C1545. doi: 10.1152/ajpcell.1996.271.5.C1539. [DOI] [PubMed] [Google Scholar]
- 26.Jordan J D, Carey K D, Stork P J, Iyengar R. J Biol Chem. 1999;274:21507–21510. doi: 10.1074/jbc.274.31.21507. [DOI] [PubMed] [Google Scholar]
- 27.Ram P T, Horvath C M, Iyengar R. Science. 2000;287:142–144. doi: 10.1126/science.287.5450.142. [DOI] [PubMed] [Google Scholar]
- 28.Ram, P.T. & Iyengar, R. (2001) Oncogene, in press. [DOI] [PubMed]
- 29.Rudolph U, Spicher K, Birnbaumer L. Proc Natl Acad Sci USA. 1996;93:3209–3214. doi: 10.1073/pnas.93.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleuss C, Hescheler J, Ewel C, Rosenthal W, Schultz G, Wittig B. Nature (London) 1991;353:43–48. doi: 10.1038/353043a0. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Clarke I J. J Physiol. 1996;491:21–29. doi: 10.1113/jphysiol.1996.sp021193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seeman P, Van Tol H H M. Trends Pharmacol Sci. 1994;15:264–270. doi: 10.1016/0165-6147(94)90323-9. [DOI] [PubMed] [Google Scholar]
- 33.Huff R M. Cell Signalling. 1996;8:453–459. doi: 10.1016/s0898-6568(96)00074-5. [DOI] [PubMed] [Google Scholar]
- 34.Kebabian J W, Calne D B. Nature (London) 1979;277:93–96. doi: 10.1038/277093a0. [DOI] [PubMed] [Google Scholar]
- 35.Missale C, Nash S R, Robinson S W, Jabar M, Caron M G. Pharmacol Rev. 1998;78:189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 36.Jiang M, Gold M S, Boulay G, Peyton M, Spicher K, Brabet P, Srinivasan Y, Rudolph U, Ellison G, Birnbaumer L. Proc Natl Acad Sci USA. 1998;95:3269–3274. doi: 10.1073/pnas.95.6.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhingra A, Lyubarsky A, Jiang M, Pugh E N, Birnbaumer L, Sterling P, Vardi N. J Neurosci. 2001;20:9053–9058. doi: 10.1523/JNEUROSCI.20-24-09053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudolph U, Brabet P, Hasty P, Bradley A, Birnbaumer L. Transgenic Res. 1993;2:345–355. doi: 10.1007/BF01976176. [DOI] [PubMed] [Google Scholar]
- 39.Rudolph U, Bradley A, Birnbaumer L. Methods Enzymol. 1994;237:366–386. doi: 10.1016/s0076-6879(94)37076-1. [DOI] [PubMed] [Google Scholar]
- 40.Jiang, M., Spicher, K., Boulay, G., Martín-Requero, A., Dye, C., Rudolph, U. & Birnbaumer, L. (2001) Methods Enzymol., in press. [DOI] [PubMed]
- 41.Florio V A, Sternweis P C. J Biol Chem. 1985;260:3477–3483. [PubMed] [Google Scholar]
- 42.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4th Ed. San Diego: Academic; 1998. [Google Scholar]
- 43.Liu Y F, Jakobs K H, Rasenick M M, Albert P R. J Biol Chem. 1994;269:13880–13886. [PubMed] [Google Scholar]
- 44.Senogles S E. J Biol Chem. 1994;269:23120–23127. [PubMed] [Google Scholar]
- 45.Izenwasser S, Cote T E. J Neurochem. 1995;64:1514–1621. doi: 10.1046/j.1471-4159.1995.64041614.x. [DOI] [PubMed] [Google Scholar]
- 46.O'Hara C M, Tang L, Taussig R, Todd R D, O'Malley K L. J Pharmacol Exp Ther. 1996;278:354–360. [PubMed] [Google Scholar]
- 47.Watts V J, Wiens B L, Cumbay M G, Vu M N, Neve R L, Neve K A. J Neurosci. 1998;18:8692–8699. doi: 10.1523/JNEUROSCI.18-21-08692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L X, Burgess L H, Gonzalez A M, Sibley D R, Chiodo L A. Synapse. 1999;31:108–118. doi: 10.1002/(SICI)1098-2396(199902)31:2<108::AID-SYN3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 49.Vanhauwe J F, Fraeyman N, Francken BJ, Luyten WH, Leysen J E. J Pharmacol Exp Ther. 1999;290:908–916. [PubMed] [Google Scholar]
- 50.Obadiah J, Avidor-Reiss T, Fishburn C S, Carmon S, Bayewitch M, Vogel Z, Fuchs S, Levavi-Sivan B. Cell Mol Neurobiol. 1999;19:653–664. doi: 10.1023/A:1006988603199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghahremani M H, Forget C, Albert P R. Mol Cell Biol. 2000;20:1497–1506. doi: 10.1128/mcb.20.5.1497-1506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abramowitz J, Iyengar R, Birnbaumer L. Endocrinology. 1982;110:336–346. doi: 10.1210/endo-110-2-336. [DOI] [PubMed] [Google Scholar]
- 53.Green S A, Holt B D, Ligget S B. Mol Pharmacol. 1992;41:889–893. [PubMed] [Google Scholar]
- 54.Levy F O, Zhu X, Kaumann A J, Birnbaumer L. Proc Natl Acad Sci USA. 1993;90:10798–10802. doi: 10.1073/pnas.90.22.10798. [DOI] [PMC free article] [PubMed] [Google Scholar]