MATERIALS AND METHODS
Animals
Transgenic mice with floxed-Ar (Arflox, official symbol Artm1Verh), on a 129/Sv genetic background, were obtained from the Catholic University of Leuven, Belgium [1]. Both males and females were crossed with mice carrying the jsd mutation on a C57BL/6 background, and the offspring were intercrossed for several generations to generate Arflox and jsd homozygous double mutant females (Arflox/flox, Utp14bjsd/jsd). Mice expressing cre recombinase under the control of the anti-Mullerian hormone (Amh) promoter (Amh-cre mice,cre+/−, official symbol Tg(Amh-cre)8815Reb, on a 129/Sv background were obtained from the University of Washington, Seattle [3]. Both males and females (cre+/−) were crossed with mice carrying the jsd mutation on a C3H-B6–129 (HB129) mixed background [21] for one or more generations to generate double-mutant males that were heterozygous for both Amh-cre and jsd (cre+/−, Utp14b+/jsd). The Arflox/flox, Utp14bjsd/jsd females were then mated with cre+/−, Utp14bjsd/+ males to produce the triple-mutant test model males (Arflox/Y,cre+/−, Utp14bjsd/jsd, SCARKO-jsd). The jsd mice used as controls carried Arflox. Genotyping of SCARKO-jsd and control jsd mice was described earlier [20]. Note that the same breeding protocol was used as in the earlier report [20], although some of the background strains were misidentified in that report. All animal studies were approved by the M.D. Anderson Cancer Center Institutional Animal Care and Use Committee.
Testicular Histology
After the mice were euthanized, testes were removed and fixed in Bouin's solution and embedded in methacrylate. The blocks were sectioned and stained with hematoxylin and periodic acid–Schiff reagent.
Stereology Analysis
Stereological analysis using the optical dissector approach [22] was performed on 25-μm thick sections with Stereo Investigator version 8.0 software (MicroBrightField, Inc., Williston, VT) under a stage-controlled microscope (Leica DMLB 100S, Leica Microsystems, Germany) using a 100× oil immersion lens. A 60 μm × 60 μm counting frame was positioned at multiple sampling sites across the section by the systematic random sampling method. The total sampling volume (Vs) for a given section was calculated as the product of counting frame area, number of sampling sites, and dissector height (15 μm). The total numbers of germ, Sertoli, interstitial, vascular smooth muscle, and peritubular myoid cells per testis (Nt) were then calculated as follows: Nt = Ns × (Vt/Vs), where Ns is the number of nuclei of the specific cells counted in the total sampling volume and Vt is the testis volume, which was determined by dividing the testis weight by the testicular tissue density of 1.05 g/cm3 [23].
RNA Preparation and Microarray Hybridization
Testis tissue samples were removed from three 12-wk-old control jsd and three 12-wk-old SCARKO-jsd mice and immediately placed in RNAlater (Qiagen, Valencia, CA) and stored at −20°C until total RNA was extracted using the RNeasy Mini kit (Qiagen). To normalize both microarray and qRT-PCR data on a per testis basis, we added 4 μl of a 1:10 000 dilution of B. subtilis RNA containing the dap gene with an artificial poly A tail (GeneChip Poly-A RNA Control Kit, Affymetrix, Santa Clara, CA) to each testis sample. We then extracted total RNA using the RNeasy Mini kit. RNA quality was determined by both electrophoresis and by 230-, 260-, and 280-nm absorption readings (NanoDrop, Wilmington, DE).
Ten micrograms of total RNA from each sample was labeled using a GeneChip One-Cycle Target Labeling Kit (Affymetrix) to produce labeled cRNA, which was then hybridized to the Affymextrix GeneChip Mouse Genome M430 2.0 array (Affymetrix). Production of cDNA and cRNA, hybridization to arrays, and evaluation of hybridization quality were completed by the Laboratory for Biotechnology and Bioanalysis I at Washington State University, Pullman, WA.
Microarray Analysis
Expression analysis was performed using the Robust Multi-array Averaging function [24] from the Affy package (v1.5.8) through the BioConductor software (http://www.bioconductor.org/ [25]). Then, genes differentially expressed between jsd and SCARKO-jsd mice were identified with Significance Analysis of Microarrays software [26]. The data were deposited into the National Center for Biotechnology Information Gene Expression Omnibus, with accession number GSE20918. Analysis of the levels of dap RNA in the normalized microarray data showed that there were equivalent levels of dap RNA in the jsd and the SCARKO-jsd samples (Supplemental Table S1, all Supplemental Data are available online at www.biolreprod.org). This result demonstrated that the relative values of gene expression obtained in the normalized microarray data were indeed equivalent to those that would be calculated on a per testis basis [27].
Real-time RT-PCR
Testis samples were collected from 12-wk-old jsd and SCARKO-jsd mice (n = 5 for jsd and n = 4 for SCARKO-jsd, which includes the samples used in the microarray), and total RNA was extracted using RNeasy Mini kit (Qiagen), with DNase I treatment to digest genomic DNA. Total RNA (3 μg) was used to generate the cDNA with a Transcriptor First Strand cDNA synthesis kit (Roche Applied Sciences, Indianapolis, IN), which was then used as the template for real-time RT-PCR. Quantitative real-time PCR was performed using the Rotor-Gene 3000 thermocycler (Corbett Research, Sydney, Australia) and SYBR Green (JumpStart Taq ready mix, Sigma, St. Louis, MO). The housekeeping gene, beta-actin, was used to normalize concentration values for each sample. The ratios of beta-actin to dap RNA were identical in the samples from the jsd and the SCARKO-jsd testes (Supplemental Table S1), demonstrating that values presented relative to beta-actin were equivalent to those on a per testis basis. The PCR reaction volume of 10 μl contained 5 μl of SYBR Green JumpStart Taq Mix, 4.8 μl of first-strand cDNA product (1:100 dilution), and 0.1 μl each of specific forward and reverse primers, with a final concentration of 0.1 μM (Sigma Genosys, Houston, TX). Cycling conditions were as follows: 2 min 95°C hold, followed by 40 cycles of 15 sec at 95°C, 1 min at 60°C, and 1 min at 72°C. Fluorescence was measured and acquired at 72°C. The primer sequences are listed in Supplemental Table S2. All samples were run in triplicate. Relative levels of gene expression were calculated using Rotor-Gene 6.0 software.
Gene Ontology Analysis
Genes that were differentially regulated in SCARKO-jsd mouse testis were analyzed by DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov).
DISCUSSION
In this study, we used the selective elimination of AR from Sertoli cells in adult Utp14b mutant (Utp14bjsd/jsd) mice as a unique model system for identifying the androgen-regulated genes in adult Sertoli cells that may be involved in the testosterone-dependent functions of these cells, such as the progression of germ cells through meiosis.
Whereas no system is perfect, our current system fulfills many criteria for an ideal system to uncover important hormonally regulated Sertoli cell genes (Table 2). First, it detects gene expression changes due to loss of androgen action, specifically on the Sertoli cells, whereas other models, such as hpg [8] and Tfm [7], involve reduced androgen action on multiple testicular cells. In the study using the Tfm model, this problem was partially overcome by employing methods to eliminate genes primarily expressed in germ or interstitial cells. Second, it involves the use of adult animals, which is preferable since Sertoli cells from immature mice may not express the genes necessary for the maturation of germ cells, as exemplified by the elimination of Sertoli-cell AR having no effect on germ cell populations in 10-day-old mice [6]. Third, it involves prolonged differential levels of androgen action between the test model and the control, since the SCARKO mice are deficient in AR in Sertoli cells from birth. Fourth, our model showed minimal differences in composition of germ cells from the control, thus eliminating the secondary effects of changing germ cell populations on the expression of genes by the Sertoli cells. Furthermore, the almost total absence of germ cells, which make up the bulk of the cells in a normal testis, increases the efficiency of detection of Sertoli cell genes in the jsd mice. However, the absence of germ cells may be a shortcoming of the model that could result in differences from the normal testis, since germ cells affect gene expression in Sertoli cells and induce stage-specific cyclical changes. Therefore, the Sertoli cells in jsd or SCARKO-jsd mice do experience some different physiological conditions from those in mice with a full complement of germ cells. Also, the current model and others previously reported [1–6] eliminate AR from Sertoli cells in the embryonic testis, which may alter Sertoli cell development during neonatal development and cause some gene expression changes in adult testes, which differ from those that would be observed by elimination of AR specifically in the adult testis. This can be improved in the future by using a conditional knockout model that will knockout AR in Sertoli cells when the animals have entered adulthood.
TABLE 2.
Comparisons of models used for microarray analysis of androgen regulated genes in Sertoli cells.
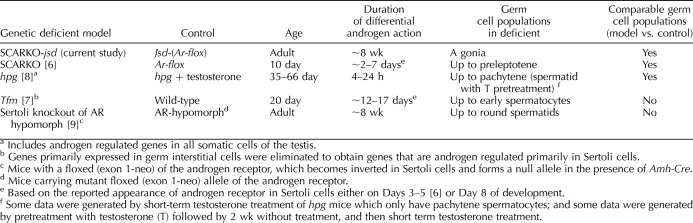
As reviewed above (Table 2), the various model systems used to identify androgen-regulated genes in Sertoli cells employ different conditions. We propose that the genes that are AR-regulated under multiple conditions are more likely to be directly regulated by AR and be the ones that are regulated by AR in the normal adult testis. Conversely, the ones that are regulated by AR only in one specific model system may be false positives, indirect effects, or in populations other than Sertoli cells, and they may not be regulated by AR in the Sertoli cells of the normal adult testis. In Table 3, we have compared the changes in expression of the highly regulated genes in SCARKO-jsd (Table 1) with the regulation of these genes in two other studies of androgen-regulated Sertoli-cell gene expression [6, 7]. Of the 21 genes, only three downregulated genes, Rhox5, Corin, and Drd4, and one upregulated gene, Fhod3, are qualitatively regulated in the same direction in all three systems. The androgen regulation of Rhox5 has been well studied, occurs in rat as well as mouse [28], and is a direct target of AR [29]. Using the PATSER tool (http://rsat.ulb.ac.be/rsat/) [30] with a cutoff of Weight Score 7 (P <10−4), we have identified at least one potential ARE binding site for all of the genes in Table 1 (from −5000 bp upstream of the transcription start site to the end of transcription) except for Akr1c12 (data not shown). An attempt to determine whether these genes were regulated similarly in the rat using the irradiated rat testis model [31] was not informative, as these genes either could not be found on the rat 230 2.0 expression array or their expression was too low to be detected on that array.
TABLE 3.
Comparison of regulation of genes strongly regulated by Sertoli-cell AR in adult jsd mice with the AR regulation of these genes in immature SCARKO and Tfm mice.
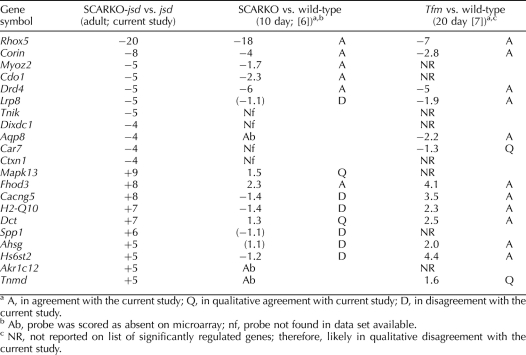
The present results can also be used to test the robustness of reports of effects of AR elimination on changes in expression of functional groups of genes, such as junctional complex proteins, proteases and protease inhibitors, proteins involved in vitamin A metabolism, and solute carriers [6, 7, 9, 32] in Sertoli cells (Supplemental Table S4). Some major proteins involved in the tight junctions between Sertoli cells [33], such as occludin (Ocln), gelsolin (Gsn), and claudin 11 (Cldn11), are not AR-regulated in the jsd model. Only alpha actinin 3 (Actn3) and thrombospondin 1 (Thsb1) showed consistent downregulation in several Sertoli AR knockout models. In addition, none of the proteases or protease inhibitors or genes regulating the vitamin A levels showed the same regulation by elimination of AR in the jsd model as in other models; only eppin (Spinlw1), which was downregulated 8-fold in 10-day-old SCARKO mice, and alcohol dehydrogenase (Adh1), which was upregulated 9-fold in 20-day-old Tfm mice, showed the same direction of regulation, but these changes were only 1.4- and 1.7-fold, respectively. Finally, none of the solute transport proteins, except the cationic amino acid transporter Slc7a4, were regulated by elimination of Sertoli-cell AR in the jsd model. These results suggest that caution should be exercised in making broad generalizations as to groups of genes regulated by AR in Sertoli cells, when only one model of a specific age and germ cell composition is used.
Since Rhox5 was strongly downregulated in the absence of AR in SCARKO-jsd testes, we investigated whether genes reported to be regulated by Rhox5 in other models would also be regulated by AR in adult jsd testes. However, of the 13 genes reported to be regulated by Rhox5 [34, 35], only Tmem176a showed a change in SCARKO-jsd testis in the same direction as predicted, but the change was very small (only −1.3-fold downregulation), and one gene, Ins2, which had been reported to be upregulated by Rhox5 [34], was actually upregulated 2.5-fold in SCARKO-jsd Sertoli cells, cells whose Rhox5 was dramatically reduced (Supplemental Table S3). Thus, these results suggest that the regulation of these genes by Rhox5 is highly dependent on the developmental stage and environment of the Sertoli cells and/or they are also regulated by AR-dependent mechanisms independently of Rhox5, which overcome the effects of the Rhox5 regulation. In either case, it does not appear that changes in expression of Rhox5-regulated genes are responsible for the dislocation of Sertoli cell nuclei in SCARKO-jsd mice.
To investigate whether the Sertoli cell AR-regulated genes identified in multiple model systems are important for spermatogenesis, we reviewed the effects of targeted disruption. Only homozygous mice with targeted disruption of Lrp8 [36] and Rhox5 [37], but not those with null mutation for Corin, Myoz2, Drd4, and Thbs1, have been reported to exhibit male infertility. However, in the case of Lrp8 null mice, spermatogenesis proceeds normally and the sperm defects develop in the epididymis; in Rhox5 null mice, germ cells pass through meiosis and the mice still retain some fertility. Since there is no evidence that any specific androgen-regulated gene is critical for the completion of spermatogenesis, it is likely that when AR is eliminated from Sertoli cells, spermatogenesis is disrupted as a result of multiple gene alterations.
The biological significance of the dramatic overrepresentation of a group of genes involved in bone mineralization among the Sertoli cell AR–regulated genes in jsd testes is not clear. It is tempting to speculate that since the Sertoli cells and bone cells are both involved in tissue remodeling processes, there is some overlap in gene expression. Three of the genes identified (Spp1, Ank, and Cd276) are preferentially expressed in the Sertoli cells, and hence alterations of their expression in testis could be directly affected by changes in the Sertoli cells. Also, since bone mineralization is stimulated by androgens, it would be expected that bone mineralization genes would be preferentially found among an androgen-regulated set. However, only one of the genes identified in the group (Spp1, also known as osteopontin) is known to be downregulated by androgen [38]. Alternatively, it is quite possible that, although these genes all share the “bone mineralization” functional category, it is their other functions, including cell adhesion, signal transduction, and cell differentiation and/or proliferation, that make them stand out in this analysis, and these functions explain the disrupted Sertoli cell nuclear localization and detachment from the basement membrane in the SCARKO-jsd testes [20].
The neuron differentiation genes may have been identified by ontology analysis due to the same principle. Their functions include cell adhesion (Alcam, Nrcam, Ret), signal transduction (Alcam, Ank3, Mcf2, Tiam1), cell differentiation and/or proliferation (Bex1, Gas7, Id4), and cytoskeleton formation (Mtap1b, Nrcam), which may have been responsible for the alteration of Sertoli cell nuclear localization in SCARKO-jsd mouse testis. It is also possible that the cell elongation and induction of asymmetry, which occur during both neuron differentiation and Sertoli cell maturation (Fig. 1C), involve the same cellular processes, which appear to be lost in the SCARKO-jsd testes.
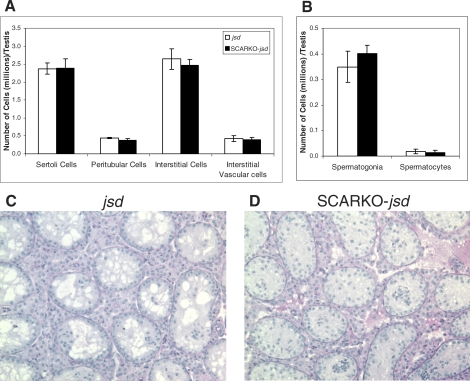
Previous studies have shown that the selective elimination of AR from Sertoli cells disrupted the localization of Sertoli cell nuclei both in adult SCARKO mouse testes [32] and in adult SCARKO-jsd testes [20]. With only a few type A spermatogonia left in the seminiferous tubules, Sertoli cell nuclear localization was more drastically altered in SCARKO-jsd testis [20] than in the SCARKO testis. When we examined genes that were identified as significantly altered in expression in postnatal day 10.5 SCARKO mouse testis and were considered to play roles in Sertoli cell nucleus dislocation [32], including Vim, Lama5, Cldn11, Ocln, Gsn, Cacna1a, and Plat, we found them in SCARKO-jsd testis to be unchanged compared to jsd testis or not expressed at all (Supplemental Table S3). The lack of changes in expression of these genes in SCARKO-jsd mice indicates that they are not the main causes of Sertoli cell nuclear dislocation and their changes in the day-10.5 SCARKO mice could result from a difference between prepubertal and adult animals. Therefore, changes in expression of the bone mineralization (Ank, Spp1, P2rx7, Ahsg, and Cd276) and neuron differentiation (Ank3, Gas7, Tiam1, Olig1, Mtap1b, Ptprz1, Id4, Nrcam, Alcam, Bex1, Ret, Pigt, and Mcf2) genes identified in our array data provide reasonable explanations for the alteration in Sertoli cell nucleus localization. These genes make good candidates with which to further explore the detailed molecular mechanisms of AR regulation of Sertoli cell structure.
The upregulation of a group of genes that are expressed in differentiated germ cells in the SCARKO-jsd mice was surprising, since no morphological changes in germ cell populations were detected (Fig. 1) [20]. Microarray and real-time RT-PCR results demonstrated that the genes known to be expressed in differentiated A spermatogonia and B spermatogonia were increased 2–3 fold when the AR was eliminated from Sertoli cells, whereas an undifferentiated spermatogonial marker remained unchanged and spermatocyte makers were not significantly detectable. These results demonstrate that the presence of AR in Sertoli cells did contribute, to a small extent, to the inhibition of spermatogonial differentiation in jsd mice. One of the histological endpoints that showed no significant difference between SCARKO-jsd and jsd testes [20], the tubule differentiation index, which measures the percentage of tubules with cells at the B spermatogonial stage or later [39], had a large variance in that study so that a 2-fold change may not have been detected. Although there is no 2-fold increase in spermatogonial numbers in SCARKO-jsd testes (Fig. 1), we cannot rule out the possibility of a relative increase in the more differentiated spermatogonial stages in the SCARKO-jsd mice since all spermatogonia were counted together. In any case, the degree of spermatogonial differentiation induced by elimination of AR from the Sertoli cells of jsd mice is minor compared to that caused by total elimination of AR [19] or addition of AR antagonists [18]. This further demonstrates that testosterone acts on the AR in other somatic cell types to produce the factors that inhibit jsd spermatogonial differentiation.
In summary, this study, using a unique model of specific elimination of AR in adult Sertoli cells in a testis essentially lacking germ cells, has identified a group of genes that are consistently AR-regulated in Sertoli cells of different ages and environments and has demonstrated that other previously reported genes are AR-regulated in Sertoli cells only under specific conditions. We have also identified groups of genes common to other biological processes that are regulated by AR in adult Sertoli cells, which may provide insight into the cause of the nuclear dislocation of the Sertoli cells in SCARKO-jsd testis. Finally, the results demonstrate that AR action in Sertoli cells contributes to a small but significant degree to the inhibition of spermatogonial differentiation in jsd mutant mice.