Abstract
Average lifespan has increased over the last centuries, as a consequence of medical and environmental factors, but maximal life span remains unchanged. Better understanding of the underlying mechanisms of aging and determinants of life span will help to reduce age-related morbidity and facilitate healthy aging. Extension of maximal life span is currently possible in animal models with measures such as genetic manipulations and caloric restriction (CR). CR appears to prolong life by reducing oxidative damage. Reactive oxygen species (ROS) have been proposed to cause deleterious effects on DNA, proteins, and lipids, and generation of these highly reactive molecules takes place in the mitochondria. But ROS is also positively implicated in cellular stress defense mechanisms, and formation of ROS a highly regulated process controlled by a complex network of intracellular signaling pathways. There are endogenous anti-oxidant defense systems that have the potential to partially counteract ROS impact. In this review, we will describe pathways contributing to the regulation of the age related decline in mitochondrial function and their impact on longevity.
Keywords: Mitochondria, longevity, lifespan, ROS, oxidative damage, caloric restriction
INTRODUCTION
Within the last few hundred years, the average human lifespan has increased substantially in the world and transformed the demography of most industrialized countries thanks to substantial improvements in medical and environmental factors. In contrast to our achievements in average lifespan, the maximal lifespan of humans remains unchanged. The average lifespan continues to increase, largely due to advances in preventive and therapeutic medicine, as well as improvements in nutrition and technology. It appears that the maximal life span may be genetically programmed or determined by inevitable deleterious cellular changes that that occur with age. These deleterious cellular changes typify the senescent phenotype, leading eventually to death.
Many types of interventions, including genetic manipulations and caloric restriction (CR), have been shown to increase the maximal life span in animal models. However, it remains to be determined whether the results from these elegant animal experiments will eventually help to extend the human life span. Hypotheses have been proposed to explain the biological processes regarding senescence, and one important theory proposes that the formation of reactive oxygen species (ROS) is the major generator of cellular damage and senescence [1, 2]. These deleterious effects from ROS could slowly accumulate over years and lead to dysfunction of several cellular functions. The mitochondrion is the main organelle to produce ROS, but as important as the energy generation in the mitochondrion is for survival, the mitochondrial ROS production has some negative consequences for age related intracellular changes. However, ROS is not only a waste of oxidative phosphorylation in the respiratory chain, but is also highly regulated signal molecules involved in cellular stress response.
MITOCHONDRIAL FUNCTION AND ROS FORMATION
The main role of Mitochondria is to generate energy rich phosphate bonds in the form of adenosine triphosphate (ATP). This ATP critical for several energy demanding processes in the cell, and the mitochondria's crucial role in the cells homeostasis is obvious. When carbon substrates enter the tricarboxylic acid cycle (TCA cycle), nicotinamide adenine dinucleotide (NAD+) and flavin adenine dinucleotide (FAD) are reduced to NADH and FADH2 within the matrix or through glycolysis in the cytosol, and promote electron flow though the respiratory chain. During the electron transfer process, protons are released from complexes I, III, and IV to the intermembrane space, and a proton gradient is formed across the inner-membrane, and complex V (ATP synthase) harvests the energy from this gradient to form ATP from ADP and P.
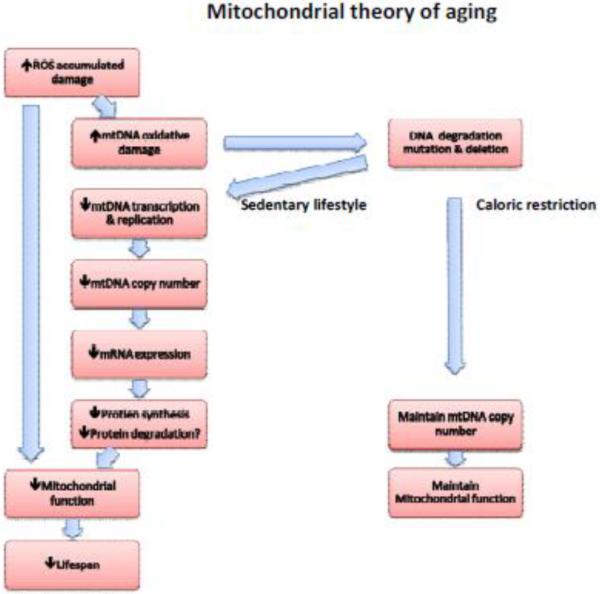
During the necessary oxygen dependent ATP production in the respiratory chain, there also is generation of reactive oxygen species (ROS) as a natural byproduct. ROS are highly reactive molecules containing valence shell electrons, as a product of electron leak in complex I and complex III where oxygen is reduced to form the superoxide radical[3]. Superoxide will form hydrogen peroxide, which is the major contributor to oxidative damage. ROS are implicated in several important cell signaling pathways during oxidative stress, and can induce damage to DNA, amino acids in proteins, and fatty acids. The close regional relationship between mitochondrial ROS production and mitochondrial-DNA (mtDNA) has given inspiration to the mitochondrial free radical theory of aging[1]. (Fig. 1)
Fig. 1.
A proton gradient is formed over the inner mitochondrial membrane by the respiratory chain. This proton gradient, achieved by oxidizing NADH and FADH2 from the TCA cycle, is crucial for complex IV to generate ATP from ADP + P. But during this process, single electrons generate superoxide by interacting with oxygen at complex I and III. The free radicals are then converted to oxygen and water by SOD1-2 and catalase. Uncoupling proteins in the mitochondrial innermembrane can decrease the proton gradient, and thereby decrease the ROS formation.
The mitochondrial free radical theory of aging, where ROS is believed to be a detrimental byproduct from the respiratory chain without any regulatory mechanism and that prompted, substantial research by many investigators to identify the potential beneficial effects of antioxidant treatment. Some studies reveal no effect of antioxidant treatment on health-beneficial outcomes as cancer[4, 5], diabetes[6] and cardiovascular disease[7], while other studies find a negative effect on outcome parameters as cancer[8–10] and life span in humans[11, 12]. One study has shown that glucose restriction increases life span in C. Elegans and that the underlying mechanism behind this extension can be altered by antioxidant treatment (13). It seems that ROS may have some advantageous effects on longevity by inducing increased stress defense systems [13]. Furthermore, the health beneficial effect of exercise on insulin sensitivity and increased endogenous antioxidant defense capacity is also impaired with antioxidant treatment [14]. The increased defense against cellular damage induced by low ROS levels is believed to reduce the overall stress level and thereby be implicated in life span extension. A favorable effect of a low dose of poison is called hormesis, and because of the similarity to the beneficial effect of ROS is this mechanism named mitohormesis [15]. There are contradictory reports on the impact of exogenous antioxidants such as Vitamin C on mitochondrial biogenesis and endogenous antioxidant defense stimulated by exercise [16, 17] and more definitive studies are needed to address this issue.
MITOCHONDRIAL THEORY OF AGING
Mitochondria are unique organelles in the cell because they posses their own DNA. Multiple copies of mitochondrial DNA (mtDNA) exist within every mitochondrion, and encode 2 ribosomal RNAs, 22 transfer RNAs and 13 proteins in the respiratory chain [18, 19]. But most mitochondrial proteins are encoded by the nuclear genome and are actively transported to its location in the organelle. Unlike nuclear DNA, mtDNA does not have histones, but form protein-DNA complexes (nucleoids) in the mitochondrial matrix[20]. This nucleoid formation protects against oxidative damage, but not in the same way as histones, and therefore mtDNA is more susceptible to ROS mediated damage than nuclear DNA. The damage to mtDNA consists of modifications as 8-oxo-2'-deoxyguanosine[21], FapyG, FapyA[22, 23], and thymine glycol[24, 25], which can lead to point mutations[26, 27], deletions[28], strand breaks[29, 30], and block DNA transcription and replication[24, 25, 31]. Any of these modifications to mtDNA could be critical in the aging process, and it is evident that oxidative damage to DNA increases with age, and perhaps leads to increased mutations and deletions seen in senescence [32–34].
To investigate whether these changes are a consequence of aging or induce the aging process, investigators have employed knock-in mice expressing a defective mitochondrial DNA polymerase. This knock-in of the defective polymerase resulted in a fivefold increase in levels of point mutations and increased amounts of deleted mtDNA. The mutations in mtDNA are associated with reduced lifespan and early onset of age related alterations without increasing ROS levels [35, 36]. These data support the notion that aging is a consequence of mitochondrial modifications over time. Although these mutations in the mitochondrial genome increase with age, the mutation rate may not be high enough to cause any physiological effect [37]. Moreover, the mRNA abundance of not only mitochondrial encoded genes, but also those mitochondrial proteins encoded by the nuclear genes in skeletal muscle are also reported to be reduced with age [32, 38]
An accumulation of alterations in the displacement loop (D-loop) [39–45], which controls mtDNA transcription and replication, could cause the progressive decrease of mtDNA copy numbers observed with age. In vitro studies have shown that oxidative damage also causes mtDNA strand breaks which result in mtDNA degradation [29, 30, 46, 47]. Since mtDNA replication is regulated by transcription factors encoded by nuclear DNA, it is critical that nuclear mediated replication in mitochondrial DNA occurs to maintain mtDNA abundance.
The decrease in mtDNA abundance occurs in sedentary people and may be responsible for reduced mRNA abundance of mitochondrial encoded proteins, possibly in a tissue specific manner [32, 38, 48, 49]. The mRNA abundance does not always correlate to protein synthesis, but it is evident that the mitochondrial protein synthesis rate is reduced with aging[38, 50–53]. It is not known how the protein degradation rate is regulated, but the mitochondrial Lon protease is reduced with aging[54], and indirect evidence from whole body measurements support the possibility of reduced degradation of proteins with aging [55]. This could slow the clearance rate of non-functional mitochondrial proteins, and thereby be the cause of impaired function of several mitochondrial proteins and overall mitochondrial function [48, 49, 53]. Aging related decreased protein turnover could further lead to increased ROS exposure time, causing the elevated levels of damaged proteins through carbonylation and nitrotyrosylation[56–58].
Not all studies find this decline in muscle mitochondrial function with aging [59–64]. ATP production has been extensively used to asses overall mitochondrial function with aging, and several investigators have observed an inverse relationship between age and ATP production[32, 65–67], but this observation is not consistent[63, 64, 68–70]. The reason for these discrepancies is not entirely clear, but differences in activity levels and relatively small numbers of participants studied may explain the differences. (Fig. 1)
CALORIC RESTRICTION, EXERCISE AND EXTENSION OF LIFE SPAN
Caloric restriction (CR) has been known for 75 years to extend lifespan in rodents [71], and it is currently the only known non-pharmacological intervention with this effect. The prolongation of lifespan has been examined in a variety of species, and the effect of CR is conserved across a broad range of species, from yeast to rats[72]. There are still no data in humans, but there are ongoing studies of CR in monkeys. The primary outcome on longevity of these monkeys is yet to be revealed, but the preliminary data indicate a beneficial effect on age related changes such as insulin resistance[73, 74], atherosclerosis[75], oxidative damage[76], cancer, and age-related mortality[77]. Although no human studies comparable to animals have been performed, Holloszy's group has reported that people who deliberately follow a CR diet for a long period and maintain their weight at a low level have several beneficial effects. They have low levels of inflammation markers, including C-reactive protein and Tumor-Necrosis-Factor-α, low LDL and high HDL in serum, and 40% reduction of carotid artery intima-media thickness as a marker of reduced atherosclerosis[78]. Left ventricular diastolic stiffness is reduced, suggesting a lower level of fibrosis in the heart[79].
Furthermore, insulin sensitivity indexes are significantly higher in the CR group compared to controls on western diet[80]. However, it remains to be determined whether these people live longer or may succumb to diseases such as pulmonary infection and die of respiratory failure as has been observed in other frail people. (Fig. 2)
Fig. 2.
The free radical theory of aging: ROS formation leads to damaged mitochondrial DNA and proteins. The ROS induced damage reduces mitochondrial function and lifespan. This reduction in mitochondrial function with aging can be prevented by CR, and can prolong lifespan in several species.
Exercise increased mitochondrial content and capacity[81] through increased expression of mitochondrial proteins encoded by nuclear and mitochondrial genomes[82]. Furthermore, mtDNA copy number is also significantly increased by exercise training in mice[82] and humans[38]. Endurance exercise give rise to beneficial adaptations in human muscle on factors impaired with aging. These changes are also evident in trained elderly people, there like young people increase their maximal oxygen consumption, mitochondrial content, and enzyme activity[83–85]. The age dependent decline of mtDNA copy number and mitochondrial proteins persist despite a high aerobic exercise level, but at a higher expression level in trained subjects[38]. The exercise induced improvement in mitochondrial function is similar to the effect seen by CR, and it seems likely that exercise can help delay the onset of many age related mitochondrial changes, but it remains to be determined if it has any impact on lifespan.
Because of the reduced oxidative damage reported in CR animals, we conducted studies to examine the effect on mitochondria in rats. CR increased transcripts of genes involved in ROS scavenging, tissue development, and energy metabolism, while decreased expression of genes involved in signal transduction, stress response and structural and contractile proteins[86]. High fat diet had the opposite effect of CR, by reducing expression of genes involved in ROS scavenging[87]. The diet induced alterations in transcripts involved in ROS scavenging underline that mitochondria could contribute to regulation of longevity, and to understand the regulation of these mechanisms, a greater detail of mitochondrial function and ROS regulation is needed.
UNCOUPLING PROTEINS
ROS production is dependent on the mitochondrial membrane potential, Δψ. High mitochondrial membrane potential will induce a greater potential for backflow of electrons in the respiratory chain, and this increases the ROS production because of the electron leak in complex I and complex III[3, 88, 89]. The mitochondrial membrane potential is partly regulated by uncoupling proteins, UCP1-3 (UCP1 in brown adipose tissue[90] UCP2 is broadly expressed[91–93] and UCP-3 is highly expressed in skeletal muscle tissue[94, 95]).
Uncoupling proteins are located in the inner membrane and shuttle protons from the intermembrane space to the matrix, before they pass complex IV to form ATP. This shuttle function decreases ROS production by reducing the membrane potential[96, 97]. But ROS are also shown to increase proton conductance over the mitochondrial membrane by activation of mammalian UCPs[98–100], and it is a possibility that this is a regulatory mechanism to decrease the abundance of free radicals and their effects in the cell. It seems that UCPs can play a role in the regulation of mitochondrial membrane potential and thereby be a key factor of ROS generation, and this is the basics of the “uncoupling–to-survive” hypothesis[101]. According to the hypothesis, uncoupling ATP synthesis from oxygen consumption has an important role in the regulation of aging, and has been supported by several studies over the last decade. UCP3 abundance decreases with aging[102], and knockout increases markers of oxidative stress in mice[103, 104], while overexpression causes a blunted age increased ROS production[105]. Experimental evidence of a positive correlation between longevity and uncoupling of proton flux was found in a comparison of mice from the upper and lower quartile of metabolic intensity. Metabolic intensity was calculated as the ratio daily food intake per body weight, and showed that higher metabolic intensity is associated with a significantly longer lifespan, higher oxygen consumption in rest, and a higher uncoupling in the mitochondria of the upper quartile[106]. However, this approach has important limitations because the body weight includes both metabolically active and inactive (Eg; triglyceride stores) tissues. Even when adjusted for lean tissue, there are limitations related to the variability of metabolic activity in different lean tissues of the body (Eg; muscle VS liver).
The above data are based on metabolic intensity, and the correlation to uncoupling and lifespan supports a hypothesis that individual mice with higher metabolism have greater mitochondrial uncoupling and longer life. This hypothesis opposes the notion that reducing oxidative metabolism reduces the flux of electrons across the electron transport chain, with a consequent decrease in ROS production that may occur in CR. However, it supports the notion that increasing oxidative metabolism causes a greater flux of electrons through the electron transport chain, with a consequent decrease in ROS production[107]. CR is well known for its ability to prolong lifespan in several species, and is proven to increase mRNA expression and protein content of UCP2-3 in mice[108, 109]. It is possible to mimic the increased longevity effect of CR by treating mice with the uncoupling agent 3,4-dinitrophenol, which decreased ROS production and oxidative damage[110]. By overexpression of human UCP2 in fly neurons, it has been demonstrated that ROS production is decreased and oxidative damage is reduced, together with increased state 4 respiration as a marker of increased proton leak. These transgenic flies also have significant prolongation of lifespan[111]. These observations were extended in mouse overexpression and knockout models. One study reported that UCP2 knockout is correlated with a significantly lower lifespan than control WT mice[112], while another study reported no effect of UCP2 and UCP3 knockout on altering longevity[113]. The mean survival age in these transgenic mice was not increased by overexpression of human UCP2 alone, but the combination of overexpression both UCP2 and UCP3 cause a slight increase in mean survival. This increase in mean survival was not accompanied by increase in maximal lifespan as seen in CR[113]. The fact that CR is increasing UCP2 and UCP3, but transgenic overexpression lack the increase in maximal lifespan talks against a direct role of UCPs in this process.
ROS SCAVENGING
When the mitochondrial inner membrane potential is high enough to induce ROS production, the antioxidant superoxide dismutase (SOD) is responsible for the enzymatic process of converting superoxide to hydrogen peroxide, 202− + 2H+ → H2O2 + O2. This dismutation reaction makes it possible for catalase, glutathione peroxidase, and peroxiredoxins to catalyze the degradation of hydrogen peroxide to water and oxygen, H2O2 → H2O + O2. SOD is found in two compartment specific isoforms; copper-zinc superoxide dismutase (ZnSOD, SOD1) and manganese superoxide dismutase (MnSOD, SOD2). SOD1 is responsible for superoxide consumption in the cytosol and extracellular matrix while SOD2's catalytic activity takes place in the mitochondrial matrix [89]. These dismutation enzymes make it possible for the cell to harvest the energy yield from the proton gradient without the potential damaging effect of ROS production. A link between cellular ROS abundance and longevity has been investigated in several studies with manipulation of the oxidant scavenging system. Slowing of telomere shortening has been reported in cultured fibroblasts when superoxide dismutase is elevated[114], and in this is also evident in relatively shortlived[115], but not long lived strains of drosophila melanogaster when superoxide dismutase and catalase are overexpressed[116]. Moreover, elevated expression of superoxide dismutase and catalase are present in the long-lived AGE-1 mutants of caenorhabditis elegans (C. Elegans) compared to normal[117, 118]. It seems that enhancing antioxidant capacity is capable of significantly prolonging the lifespan in several species, but decreasing the capacity has given conflicting results. Knock-down of copper-zinc superoxide dismutase induces senescence in human fibroblasts[119]. In mice knockout models there are, however, conflicting results: some studies with knockout of glutathione peroxidase or superoxide dismutase 1 or 2 show no alteration on lifespan[120–122], while others find significant reduction with knock out of superoxide dismutase 2[123]. The reduced lifespan by knock out of SOD2 can be counteracted by overexpression of human UCP2, probably because of reduced need for the enzymatic dismutation process when the UCP2 induced proton leak reduces membrane potential[112].
P66Shc AND ROS GENERATION
There will, however, always be some degree of oxidative damage particularly in mitochondrial proteins, lipids, and DNA, independent of antioxidant buffering capacity. The oxidative stress is also evident from elevated oxidation levels in proteins (carbonylation), lipids (peroxidation), and DNA in older humans[124]. The superoxide production rate is generally regarded as having a constant positive relationship with oxygen consumption, but since the ROS production is dependent on membrane potential, this relationship is presumably more complex. As mentioned earlier, the membrane potential is regulated by several mechanisms, and furthermore ROS is also produced by other sources than the backward flow of electrons in the respiratory chain. It is now evident that it is not only the direct damaging effect of ROS on DNA, proteins, and lipids, but also a highly regulated interaction of ROS on distinctive signaling pathways related to aging.
The majority of ROS production is created as a consequence of backward electron flow in the respiratory chain, but the 66 kda isoform of Shc (p66Shc) does function as a oxidoreductase and shuttles electrons to molecular oxygen to form superoxide[125], and cultivated p66Shc knock out cells have reduced ROS formation and damage[125–127]. P66Shc is shown to be a critical factor in programmed cell death, and targeted mutation of the gene in mice is correlated to increased stress resistance and prolonged lifespan. Furthermore, mouse embryo fibroblasts from p66Shc knock out mice are resistant to apoptosis under oxidative stress[128]. In mammals, the proto-oncogene Shc is expressed in three isoforms; 46, 52 and 66 kda[129]. All isoforms interact with tyrosine-phosphorylated proteins on the cytosolic domain of growth factor receptors, where they, upon activation, are phosphorylated and form complexes with Grb2[130–132]. The 66 kda isoform does not activate the signal in the Ras, Raf-1, MAPK pathway as the other isoforms, but is rather a competitive antagonist[133–136].
About 1/5 of p66Shc is in close contact with mitochondria in fibroblasts, but because of translocation, this ratio increases during oxidative stress[137]. The serine residue on position 36 (ser36) in the N-terminal domain of p66Shc is phosphorylated as a critical post-translational activation site upon oxidative stress[138]. After phosphorylation and translocation the protein is bound to a complex with Heat Shock Protein 70 (HSP70), and this binding destabilizes by pro-apoptopic stimuli, and the monomeric active form of p66Shc is released[137, 139]. In the mitochondria, reduction of oxygen can then be mediated by p66Shc binding to cytochrome c. This interaction shuttles electrons to oxygen and forms free radicals[125]. These modifications are independent of membrane potential and create ROS in a regulated pattern.
Moreover p66Shc is an essential downstream target of p53 stress induced elevation of oxidants, cytochrome release, and apoptosis, and p53 increases p66Shc protein levels by enhancing stability[127]. Consequently, it seem like p53-p66Shc is a sensor of oxidative damage and high intensity oxidative signals and drives the cell against apoptosis.
The activation of these pathways induces an increased probability of outer mitochondrial membrane permeabiliazation as a consequence of the increased ROS levels[125]. Normally, cytochrome c forms a complex with the anionic phospholipid cardiolipin in the mitochondrial innermembrane, but during oxidative stress cardiolipin will be peroxidized and the affinity for cytochrome c decreased. The lower affinity will release cytochrome c from its membrane localization and impair the respiratory chain, mediating electron backflow and an increase in the ROS production rate. This increased ROS production will further increase the oxidation of cardiolipin and amplify the oxidative damage to the mitochondria[140–142]. As a part of this process, the inner membrane becomes permeable to cytochrome c, as a consequence of mitochondrial transition pore opening and concomitant swelling of the organelle[125]. The abundance of cytochrome c in cytoplasm is one of the factors that triggers apoptosis. The formation of a complex called apoptosome comprised of cytochrome c, Apaf-1, and procaspase 9 in the cytosol, will activate procaspase 9, and this will induce further downstream activation of caspases executing the apoptotic degradation of DNA and proteins in the cell[142–144].
FOXO MEDIATED CELL SURVIVAL
P66Shc is not only interacting in the redox activity of the cell by its function as oxidoreductase, but is also interacting in this system by inhibiting forkhead/winged helix box gene group O (FOXO) induced transcription of ROS scavenging enzymes[126]. FOXO is a highly conserved group of transcription factors involved in a wide range of adaptations to oxidative and metabolic stress. The FOXO family consist of four genes in mammals (FOXO1, FOXO3, FOXO4 and FOXO6)[145, 146]. This family of transcription factors is expressed in a wide range of tissues and binds to TTGTTTAC recognition motif[147]. In invertebrates, the FOXO transcription factor induces prolongation of lifespan[148–151], and in mammals is the protein family involved in detoxification of reactive oxygen species[126], tumor suppression[152], energy metabolism[153, 154], erythroid differentiation[155], and myogenesis[156]. Furthermore, FOXO3 is genetically associated with longevity in humans[157, 158]. They induce the effect via promotion of ROS scavenging enzymes[126, 159], cell cycle arrest[152], DNA repair[160], and apoptosis[148, 161]. Overexpression of FOXO1 in muscle tissue is, however, not capable of altering longevity in transgenic mice[162].
The activity of these transcription factors is primarily dependent on subcellular location, and this is regulated by a broad range of post-translational modifications; phosphorylations, acetylations and ubiquitinations. These regulation mechanisms are highly complex and sometimes directly antagonistic. (Fig. 3)
Fig. 3.
FOXO transcription factors need DNA contact to induce transcription of target genes, and the activity is primarily dependent on subcellular location. AKT dependent phosphorylation mediates export from the nucleus, while AMPK phosphorylation on other amino acid residues activates transcription. Acetylation inactivates FOXO transcription, while deacetylation by sirt1 and sirt3 has the opposite effect.
The insulin/IGF-1, PI3K, Akt pathway is, to a large extent, involved in lifespan regulation. Suppression of the pathway increases lifespan, and upregulation shortens lifespan in a broad range of species[163]. Insulin and IGF-I receptors vary significantly amongst mammals and indicate a more complicated regulation of lifespan than in many other species. It has been shown that IGF-1 and insulin have different impacts on lifespan modulation[164].
Upregulating glucose signaling shortens lifespan in C. Elegans [165], while downregulation of the signal transduction by mutation of daf-2 (insulin/IGF-1 receptor gene) in nematodes doubles their lifespan[166], and a mutation in the same receptor can increase the lifespan in drosophila melanogaster up to 80%[167]. A similar lifespan extension is seen in mice with decreased IGF-1 signaling[168, 169], and increased SOD and catalase are observed as downstream effects of the reduced IGF-I signal[128]. Downstream from the insulin/IGF-1 receptor, the active phosphorylated form of AKT is crucial in the negative regulation of FOXO3, via exclusion of the transcription factors from the nucleus[148, 149, 170]. Insulin and insulin like growth factors induce phosphorylation on FOXO3 (T32 and S253)[170, 171] via Akt, and this generates a binding site for the chaperone 14-3-3[172]. The binding of chaperones further promotes active transport of the complex out of the nucleus, probably via a conformational change that helps expose the FOXO nuclear export sequence[173]. The phosphorylation is not only responsible for export of the transcription factors to the cytoplasm, but also prevents re-entry to the nucleus via introduction of a negative charge in the residues that accounts for the FOXO nuclear localization signal, and by alterations in the flexibility of the same region[174]. The regulation of FOXO activity is not only regulated through subcellular compartmentation, but also the Akt dependent phosphorylation of FOXO1 (S256; equivalent to FOXO3ser253) which reduces the positive charge in the DNA-binding domain. The DNA-binding is compromised in in vitro binding assays through this change in charge[175, 176]. Additionally, EAK-7 is also shown to inhibit FOXO transcription without influencing subcellular localization[177].
As mentioned above, p66shc interacts in FOXO transcriptional regulation of ROS scavenging enzymes. In p66shc−/− fibroblasts, the phosphorylation of FOXO3 (T32 and S253) is inhibited after oxidative stress, and therefore FOXO inhibition is dependent on active p66shc after oxidative stress in wild-type fibroblasts[126]. Furthermore, the binding of FOXO to the promoter region of the catalase gene is regulated via this pathway, which is consistent with studies demonstrating that the FOXO3 induces MnSOD expression[126, 159], and the FOXO homolog DAF-16 is a transcription factor for several antioxidant scavenger genes (SOD, catalase and OLD-1) in C. Elegans[178, 179].
FOXO transcription factors are not only negatively regulated through phosphorylation, but are also activated by phosphorylation on other amino acid residues. Under periods with low energy availability, AMP activated protein kinase (AMPK) is activated. This is due to formation of AMP as a consequence of the of the reduced ATP generation from the respiratory chain. The allosteric activator AMP binds to the γ-subunit of AMPK and induces a conformational change that activates the catalytic α-subunit[180]. AMP activated AMPK induces phosphorylation on all four isoforms of FOXO, with a preference for FOXO3[181]. The phosphorylation of FOXO3 leads upregulated transcriptional activity. This AMPK dependent upregulation is due to phosphorylation on several sites in mammalian cells; in vitro FOXO3 T179, S399, S413, S555, S588, and S626, and in vivo S413 and S588[181]. The activation of FOXO by AMPK increases the expression of specific target genes involved in stress resistance and energy balance. This AMPK dependent activation does not upregulate transcription in all FOXO3 target genes, but is more selective to energy adaptation genes, and it seems not to regulate FOXO1 in the same way[181, 182]. During caloric restriction, AMPK extends the lifespan in C.Elegans via activation of the forkhead transcription factor DAF-16[183], and this could be an important pathway in response to caloric restriction in several species.
The post-translational regulation of FOXO transcription factors is, as mentioned above, not only through phosphorylation, but acetylation also plays a significant role in the activity regulation. The opposing action of acetylases and deacetylases is a continuous process. The acetylase involved in the regulation of FOXO is cyclic response element (CREB)-binding protein (CBP)[184], while the deacetylation is mediated via the NAD-dependent class III deacetylase silent information regulator T1 (sirt1)[185, 186]. The DNA-binding ability of FOXO3 is reduced by the CBP acetylation of lysin residue 245, and the AKT induced phosphorylation of S253 on FOXO1 is enhanced when the protein is acetylated[187, 188]. Overall, acetylation is enhancing and interacting with AKT phosphorylation induced inhibition of FOXO, while sirt1 deacetylation enhances FOXO activity.
SIRTUINS: SIRT1 AND SIRT3 IMPORTANT FACTORS IN LONGEVITY
Sirtuins (named from silent mating-type information regulating two (Sir2) in yeast) are a class of proteins that act as deacetylases or mono-ribosyltransferases, and have a highly conserved catalytic groove from E. Coli to humans[189]. While sirt1 inhibits FOXO and p53 induced apotosis[186], FOXO3 and p53 promote increased sirt1 expression during nutrient deprivation. This increase in sirt1 transcription is achieved by two p53 binding sites in the sirt1 promoter, after physical interaction between FOXO3 and p53[190].
The family of sirtuins is expressed in a variety of mammalian tissues, and consists of seven members, sirt1–7, where sirt1 is the most extensively studied[191]. Sirt1–3 and -5 catalyze lysine deacetylation on target proteins, via cleavage of NAD+ into nicotinamide (NAM) and 1-O-acetyl-ADP-ribose[192, 193]. NAD+ hydrolysis is essential for the reaction, and this dependency of sirtuins on NAD+ links the catalytic activity directly to energy balance. This dependency of NAD+ abundance on enzymatic activity makes sirt1 and -3 act as a fuel-sensing regulators of metabolic adaptations to nutrient deprivation[194]. NAM is a NAD+ antagonist, by binding to the catalytic core domain of sirtuins[195]. CR increases activity of the NAM salvage pathway, which will reduce the abundance of NAM and increases the abundance of NAD+. The decrease in NAM is induced by the nicotinamidase PNC1 in yeast, which has been shown to highly regulate lifespan[195–197], and consistent with this is PNC1 homologs in flies and worms also correlated to longevity[198, 199]. In higher eukaryotes, the rate limiting enzyme in the NAM salvage pathway, NAMPT, is also shown to regulate NAD+ levels and thereby sirtuin activity in human cell lines[200, 201]. Sirtuin activity is not only regulated by NAD+ and NAM levels, but sirt1 activity is also inhibited by direct binding of DBC1 in the catalytic grove in in vitro[202] and in in vivo[203]. It is evident that reduced binding of DBC1 to sirt1, significantly increased deacetylase activity after starvation in mice, but not much is known about the regulation of this interaction[203]. Sirt1 deacetylation of p53 is upregulated by complex formation with AROS[204] and Necdin[205].
Furthermore, sirt1 activity is also regulated by post-translational modifications, such as sumoylations and phosphorylations. Mutation of the sirt1 sumoylation site lysine 734 or desumoylation by SENP1 reduces the deacetylase activity, and cells depleted of SENP1 have improved resistance to stress induced apoptosis. This indicates that sumoylation is an important post-translational modification that increases sirt1 activity in cells, and thereby prevents acetylation and activation of apoptotic proteins[206]. Interaction of cJUN N-terminal kinase (JNK1) and sirt1 induces phosphorylation on three sites of sirt1; Ser27, Ser 47 and Thr530. These phosphorylation sites were shown to increase enzymatic activity via deacetylation of histone H3, but not p53, which further illustrates the complex regulation of this pathway[207].
Deacetylation by sirt1 is evident in FOXO1, -3 and -4[185, 186, 208]. Sirt1 induced increase in FOXO transcription activity does not include all FOXO target genes, like AMPK activation.
Sirt1 induces cell cycle arrest by transcriptional silencing though physical modification of DNA due to histone deacetylation[194]. Sirt1 increase expression of genes involved in stress resistance, including the FOXO target gene MnSOD[209], and inhibits the pro-apoptopic FOXO target genes BIM and FAS[186]. Sirt1 is not only inhibiting FOXO induced apoptosis, but is also downregulating the p53 induced apoptosis via deacetylation[210–212]. Furthermore, sirt1 deacetylation of two critical lysine residues on the DNA repair factor Ku70 improves the binding of Ku70 to Bax, which is involved in the initiation of apoptosis. The formation of the Ku70/Bax complex keeps Bax away from the mitochondria, and thereby increases cell survival[213]. This parallel inhibition of FOXO, p53, and Bax induced apoptosis, and enhanced expression of cell cycle arrest and stress resistance genes is vital for sirt1's ability to enhance lifespan in a wide range of organisms.
Activation of Sirt1 does not only affect mitochondria via increased resistance to oxidative stress and inhibition of apoptosis, but also directly interacts with mitochondrial biogenesis. As mentioned above, sirt1 and AMPK exhibit cross-talk on FOXO transcription factors, which is not surprising since these two proteins posses energy sensing properties. This cross-talk is also present in mitochondrial biogenesis, where the interaction between proteins can potentiate reciprocal activity. The upstream activator of AMPK Liver Kinase B1 (LKB1) is deacetylated on lysine 48 by sirt1, thereby mediating migration from the nucleus to the cytosol[214]. As already described, AMPK is activated by AMP-binding on the γ-subunit during metabolic stress, but this activation is dependent on LKB1[215, 216] or calcium/calmodulin kinase kinase-β (CaMKKβ)[217] phosphorylation of AMPK threonine residue 172 on the α-catalytic subunit[218]. LKB1 is the primary upstream AMPK kinase, which increases activity in response to energy deprived states. Alternatively, AMPK is capable of activating sirt1 by upregulating NAMPT, and thereby increasing NAD+ and decrease NAM[219]. An important factor in mitochondrial biogenesis is PGC1α, and it has been shown that sirt1 deacetylation of PGC1α increase target gene expression in a NAD+ dependent manner[220]. Furthermore deacetylation of PGC1α by sirt1 was shown to improve mitochondrial function in mice, with significant increase in aerobic capacity[221], and this regulation is dependent on AMPK[222]. The increased mitochondrial function by sirt1-PGC1α is achieved by upregulation of genes for oxidative phosphorylation and mitochondrial biogenesis[221]. (Fig. 4) Indeed, sirt1 induces transcription of mitochondrial fatty acid oxidative genes via PGC1α deacetylation[223]. There has recently been identified an interaction of sirt1, PGC1α, and mitochondrial transcription factor A (TFAM) in close proximity to the D-loop of mtDNA recognized by TFAM[224]. This suggests that the sirt1-PGC1α interaction is highly involved in both nuclear and mitochondrial transcription of DNA, and may represent a similar regulation and cross-talk of the nuclear and mitochondrial genome.
Fig. 4.
AMPK increases transcription of PNC1/NAMPT during metabolic stress, and thereby increases NAD+ and further increases sir2/sirt1 activity. Activated sir2/sirt1 deacetylates the upstream AMPK kinase LKB1, which further increases AMPK activity. This interdependency is crucial in mitochondrial biogenesis through increased PGC1α expression and activity.
Another protein in the sirtuin family is the mitochondrial sirt3[225]. It is not as well investigated as sirt1, but is also associated with longevity. While sirt1 has been shown to regulate lifespan in a number of species, sirt3 is the only sirtuin genetically linked to longevity in humans[226], and its abundance decreases with age[38]. This decrease can partly be counter acted by exercise[38].
Sirt3−/− mice have a higher risk of developing mammary tumors[227], however, they have no signs of other disorders compared to their wild-type controls under normal conditions[228]. But during oxidative stress, ROS production increases and ATP levels decrease in sirt3−/− cardiomyocytes[229], and sirt3 has also been shown to decrease hypertrophic signalling in the cardiomyocytes, via inhibition of AKT in a sirt3-LKB1-AMPK dependent manner during NAD+ treatment in mice[230]. During fasting sirt3−/− transgenic mice have hyperacetylated long-chain acyl coenzyme A dehydrogenase (LCAD) on lysine 42, and thereby reduced levels of fatty-acid oxidation, and accumulation of fatty-acid intermediates in the liver. This sirt3 dependent regulation of lipid metabolism could be responsible for the decrease in ATP production in sirt3−/− transgenic mice[231]. Sirt3 could be an important factor in regulating ROS abundance, since it promotes FOXO3 transport to the nucleus where the transcription factor as already mentioned, increases the production of MnSOD and catalase[232]. Increased ROS levels correlate to a gradual decrease in the integrity of mtDNA in livers from sirt3−/− mice at 20, 36 and 58 weeks, and it has been shown that sirt3 acts as a tumor suppressor[227].
Sirt3's suppression of tumor formation goes well together with the observation that sirt3−/− mice develop mammary tumors.
SUMMARY AND CONCLUSIONS
The biology of aging remains to be fully understood. Here, we review the mitochondrial interaction with pathways and potential mechanisms that cause damage to DNA, proteins, and tissues with aging. The effect of CR appears to be related to reduction of ROS induced cellular damage. There is increasing evidence that chronic exercise can counteract the age-related decline in mitochondrial DNA abundance and functions besides preserving the expression of SIRT3, which is a longevity gene. Reducing caloric intake and preventing excessive accumulation of adipose tissue and regular exercise programs have many beneficial effects that may extend the average life span and contribute to healthy aging. However, like CR, exercise also remains to be demonstrated as a measure to extend life span. Further studies in human and animal models of the various longevity pathways are needed to determine whether human life span can be extended while maintaining relatively normal functions of organs.
Fig. 5.

Acknowledgements
Support was provided by NIH grants RO1-AG09531, UL1-RR024150, and the David Murdock-Dole Professorship (K.S.N). MHV is supported by a grant from the Aarhus University and Aarhus University Hospital, Denmark.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20(4):145–7. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 2.Linnane AW, et al. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1(8639):642–5. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg ER, et al. A clinical trial of antioxidant vitamins to prevent colorectal adenoma. Polyp Prevention Study Group. N Engl J Med. 1994;331(3):141–7. doi: 10.1056/NEJM199407213310301. [DOI] [PubMed] [Google Scholar]
- 5.Gaziano JM, et al. Vitamins E and C in the prevention of prostate and total cancer in men: the Physicians' Health Study II randomized controlled trial. JAMA. 2009;301(1):52–62. doi: 10.1001/jama.2008.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, et al. Long-term beta-carotene supplementation and risk of type 2 diabetes mellitus: a randomized controlled trial. JAMA. 1999;282(11):1073–5. doi: 10.1001/jama.282.11.1073. [DOI] [PubMed] [Google Scholar]
- 7.Sesso HD, et al. Vitamins E and C in the prevention of cardiovascular disease in men: the Physicians' Health Study II randomized controlled trial. JAMA. 2008;300(18):2123–33. doi: 10.1001/jama.2008.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardia A, et al. Efficacy of antioxidant supplementation in reducing primary cancer incidence and mortality: systematic review and meta-analysis. Mayo Clin Proc. 2008;83(1):23–34. doi: 10.4065/83.1.23. [DOI] [PubMed] [Google Scholar]
- 9.Bairati I, et al. A randomized trial of antioxidant vitamins to prevent second primary cancers in head and neck cancer patients. J Natl Cancer Inst. 2005;97(7):481–8. doi: 10.1093/jnci/dji095. [DOI] [PubMed] [Google Scholar]
- 10.Hercberg S, et al. Antioxidant supplementation increases the risk of skin cancers in women but not in men. J Nutr. 2007;137(9):2098–105. doi: 10.1093/jn/137.9.2098. [DOI] [PubMed] [Google Scholar]
- 11.Vivekananthan DP, et al. Use of antioxidant vitamins for the prevention of cardiovascular disease: meta-analysis of randomised trials. Lancet. 2003;361(9374):2017–23. doi: 10.1016/S0140-6736(03)13637-9. [DOI] [PubMed] [Google Scholar]
- 12.Bjelakovic G, et al. Antioxidant supplements for prevention of mortality in healthy participants and patients with various diseases. Cochrane Database Syst Rev. 2008;(2):CD007176. doi: 10.1002/14651858.CD007176. [DOI] [PubMed] [Google Scholar]
- 13.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6(4):280–93. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Ristow M, et al. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc Natl Acad Sci U S A. 2009;106(21):8665–70. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45(6):410–8. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Gomez-Cabrera MC, et al. Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am J Clin Nutr. 2008;87(1):142–9. doi: 10.1093/ajcn/87.1.142. [DOI] [PubMed] [Google Scholar]
- 17.Wadley GD, McConell GK. High-dose antioxidant vitamin C supplementation does not prevent acute exercise-induced increases in markers of skeletal muscle mitochondrial biogenesis in rats. J Appl Physiol. 2010;108(6):1719–26. doi: 10.1152/japplphysiol.00127.2010. [DOI] [PubMed] [Google Scholar]
- 18.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 19.Bibb MJ, et al. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–80. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 20.Gilkerson RW. Mitochondrial DNA nucleoids determine mitochondrial genetics and dysfunction. Int J Biochem Cell Biol. 2009;41(10):1899–906. doi: 10.1016/j.biocel.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 21.de Souza-Pinto NC, et al. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61(14):5378–81. [PubMed] [Google Scholar]
- 22.Jaruga P, et al. Endogenous oxidative DNA base modifications analysed with repair enzymes and GC/MS technique. Nucleic Acids Res. 2000;28(6):E16. doi: 10.1093/nar/28.6.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, et al. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280(49):40544–51. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 24.Ide H, Kow YW, Wallace SS. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13(22):8035–52. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark JM, Beardsley GP. Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry. 1987;26(17):5398–403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 26.Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325(Pt 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33(1):1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet. 2005;14(7):893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oka S, et al. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. Embo J. 2008;27(2):421–32. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shokolenko I, et al. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37(8):2539–48. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatahet Z, Purmal AA, Wallace SS. Oxidative DNA lesions as blocks to in vitro transcription by phage T7 RNA polymerase. Ann N Y Acad Sci. 1994;726:346–8. doi: 10.1111/j.1749-6632.1994.tb52847.x. [DOI] [PubMed] [Google Scholar]
- 32.Short KR, et al. Decline in skeletal muscle mitochondrial function with aging in humans. Proc Natl Acad Sci U S A. 2005;102(15):5618–23. doi: 10.1073/pnas.0501559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton ML, et al. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98(18):10469–74. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Remmen H, Hamilton ML, Richardson A. Oxidative damage to DNA and aging. Exerc Sport Sci Rev. 2003;31(3):149–53. doi: 10.1097/00003677-200307000-00009. [DOI] [PubMed] [Google Scholar]
- 35.Trifunovic A, et al. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci U S A. 2005;102(50):17993–8. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 37.Sciacco M, et al. Distribution of wild-type and common deletion forms of mtDNA in normal and respiration-deficient muscle fibers from patients with mitochondrial myopathy. Hum Mol Genet. 1994;3(1):13–9. doi: 10.1093/hmg/3.1.13. [DOI] [PubMed] [Google Scholar]
- 38.Lanza IR, et al. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57(11):2933–42. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michikawa Y, et al. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286(5440):774–9. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 40.Calloway CD, et al. The frequency of heteroplasmy in the HVII region of mtDNA differs across tissue types and increases with age. Am J Hum Genet. 2000;66(4):1384–97. doi: 10.1086/302844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, et al. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc Natl Acad Sci U S A. 2001;98(7):4022–7. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Bo R, et al. Evidence and age-related distribution of mtDNA D-loop point mutations in skeletal muscle from healthy subjects and mitochondrial patients. J Neurol Sci. 2002;202(1–2):85–91. doi: 10.1016/s0022-510x(02)00247-2. [DOI] [PubMed] [Google Scholar]
- 43.Del Bo R, et al. High mutational burden in the mtDNA control region from aged muscles: a single-fiber study. Neurobiol Aging. 2003;24(6):829–38. doi: 10.1016/s0197-4580(02)00233-6. [DOI] [PubMed] [Google Scholar]
- 44.Theves C, et al. Detection and quantification of the age-related point mutation A189G in the human mitochondrial DNA. J Forensic Sci. 2006;51(4):865–73. doi: 10.1111/j.1556-4029.2006.00163.x. [DOI] [PubMed] [Google Scholar]
- 45.McInerny SC, Brown AL, Smith DW. Region-specific changes in mitochondrial D-loop in aged rat CNS. Mech Ageing Dev. 2009;130(5):343–9. doi: 10.1016/j.mad.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Wilson VL, et al. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262(21):9948–51. [PubMed] [Google Scholar]
- 47.Crawford DR, Abramova NE, Davies KJ. Oxidative stress causes a general, calcium-dependent degradation of mitochondrial polynucleotides. Free Radic Biol Med. 1998;25(9):1106–11. doi: 10.1016/s0891-5849(98)00143-9. [DOI] [PubMed] [Google Scholar]
- 48.Barazzoni R, Short KR, Nair KS. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J Biol Chem. 2000;275(5):3343–7. doi: 10.1074/jbc.275.5.3343. [DOI] [PubMed] [Google Scholar]
- 49.Welle S, et al. Reduced amount of mitochondrial DNA in aged human muscle. J Appl Physiol. 2003;94(4):1479–84. doi: 10.1152/japplphysiol.01061.2002. [DOI] [PubMed] [Google Scholar]
- 50.Marcus DL, Ibrahim NG, Freedman ML. Age-related decline in the biosynthesis of mitochondrial inner membrane proteins. Exp Gerontol. 1982;17(5):333–41. doi: 10.1016/0531-5565(82)90033-x. [DOI] [PubMed] [Google Scholar]
- 51.Marcus DL, et al. Effect of inhibitors and stimulators on isolated liver cell mitochondrial protein synthesis from young and old rats. Exp Gerontol. 1982;17(6):429–35. doi: 10.1016/s0531-5565(82)80004-1. [DOI] [PubMed] [Google Scholar]
- 52.Bailey PJ, Webster GC. Lowered rates of protein synthesis by mitochondria isolated from organisms of increasing age. Mech Ageing Dev. 1984;24(2):233–41. doi: 10.1016/0047-6374(84)90074-5. [DOI] [PubMed] [Google Scholar]
- 53.Rooyackers OE, et al. Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proc Natl Acad Sci U S A. 1996;93(26):15364–9. doi: 10.1073/pnas.93.26.15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bota DA, Davies KJ. Protein degradation in mitochondria: implications for oxidative stress, aging and disease: a novel etiological classification of mitochondrial proteolytic disorders. Mitochondrion. 2001;1(1):33–49. doi: 10.1016/s1567-7249(01)00005-8. [DOI] [PubMed] [Google Scholar]
- 55.Henderson GC, et al. Higher muscle protein synthesis in women than men across the lifespan, and failure of androgen administration to amend age-related decrements. Faseb J. 2009;23(2):631–41. doi: 10.1096/fj.08-117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feng J, et al. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J Gerontol A Biol Sci Med Sci. 2008;63(11):1137–52. doi: 10.1093/gerona/63.11.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fugere NA, Ferrington DA, Thompson LV. Protein nitration with aging in the rat semimembranosus and soleus muscles. J Gerontol A Biol Sci Med Sci. 2006;61(8):806–12. doi: 10.1093/gerona/61.8.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaleel A, et al. Identification of de novo synthesized and relatively older proteins -- Accelerated oxidative damage to de novo synthesized ApoA-1 in type 1 diabetes. Diabetes. doi: 10.2337/db10-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsson L, Sjodin B, Karlsson J. Histochemical and biochemical changes in human skeletal muscle with age in sedentary males, age 22--65 years. Acta Physiol Scand. 1978;103(1):31–9. doi: 10.1111/j.1748-1716.1978.tb06187.x. [DOI] [PubMed] [Google Scholar]
- 60.Grimby G, et al. Morphology and enzymatic capacity in arm and leg muscles in 78–81 year old men and women. Acta Physiol Scand. 1982;115(1):125–34. doi: 10.1111/j.1748-1716.1982.tb07054.x. [DOI] [PubMed] [Google Scholar]
- 61.Aniansson A, et al. Muscle morphology, enzymatic activity, and muscle strength in elderly men: a follow-up study. Muscle Nerve. 1986;9(7):585–91. doi: 10.1002/mus.880090702. [DOI] [PubMed] [Google Scholar]
- 62.Trappe SW, et al. Skeletal muscle characteristics among distance runners: a 20-yr follow-up study. J Appl Physiol. 1995;78(3):823–9. doi: 10.1152/jappl.1995.78.3.823. [DOI] [PubMed] [Google Scholar]
- 63.Rasmussen UF, et al. Experimental evidence against the mitochondrial theory of aging. A study of isolated human skeletal muscle mitochondria. Exp Gerontol. 2003;38(8):877–86. doi: 10.1016/s0531-5565(03)00092-5. [DOI] [PubMed] [Google Scholar]
- 64.Rasmussen UF, et al. Human skeletal muscle mitochondrial metabolism in youth and senescence: no signs of functional changes in ATP formation and mitochondrial oxidative capacity. Pflugers Arch. 2003;446(2):270–8. doi: 10.1007/s00424-003-1022-2. [DOI] [PubMed] [Google Scholar]
- 65.Karakelides H, et al. Age, obesity, and sex effects on insulin sensitivity and skeletal muscle mitochondrial function. Diabetes. 59(1):89–97. doi: 10.2337/db09-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conley KE, Jubrias SA, Esselman PC. Oxidative capacity and ageing in human muscle. J Physiol. 2000;526(Pt 1):203–10. doi: 10.1111/j.1469-7793.2000.t01-1-00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petersen KF, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300(5622):1140–2. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trounce I, Byrne E, Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. Lancet. 1989;1(8639):637–9. doi: 10.1016/s0140-6736(89)92143-0. [DOI] [PubMed] [Google Scholar]
- 69.Barrientos A, et al. Absence of relationship between the level of electron transport chain activities and aging in human skeletal muscle. Biochem Biophys Res Commun. 1996;229(2):536–9. doi: 10.1006/bbrc.1996.1839. [DOI] [PubMed] [Google Scholar]
- 70.Brierly EJ, et al. Mitochondrial function in muscle from elderly athletes. Ann Neurol. 1997;41(1):114–6. doi: 10.1002/ana.410410120. [DOI] [PubMed] [Google Scholar]
- 71.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition. 1989;5(3):155–71. discussion 172. [PubMed] [Google Scholar]
- 72.Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17(3):313–21. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- 73.Lane MA, Ingram DK, Roth GS. Calorie restriction in nonhuman primates: effects on diabetes and cardiovascular disease risk. Toxicol Sci. 1999;52(2 Suppl):41–8. doi: 10.1093/toxsci/52.2.41. [DOI] [PubMed] [Google Scholar]
- 74.Kemnitz JW, et al. Dietary restriction increases insulin sensitivity and lowers blood glucose in rhesus monkeys. Am J Physiol. 1994;266(4 Pt 1):E540–7. doi: 10.1152/ajpendo.1994.266.4.E540. [DOI] [PubMed] [Google Scholar]
- 75.Verdery RB, et al. Caloric restriction increases HDL2 levels in rhesus monkeys (Macaca mulatta) Am J Physiol. 1997;273(4 Pt 1):E714–9. doi: 10.1152/ajpendo.1997.273.4.E714. [DOI] [PubMed] [Google Scholar]
- 76.Zainal TA, et al. Caloric restriction of rhesus monkeys lowers oxidative damage in skeletal muscle. Faseb J. 2000;14(12):1825–36. doi: 10.1096/fj.99-0881com. [DOI] [PubMed] [Google Scholar]
- 77.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325(5937):201–4. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fontana L, et al. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci U S A. 2004;101(17):6659–63. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meyer TE, et al. Long-term caloric restriction ameliorates the decline in diastolic function in humans. J Am Coll Cardiol. 2006;47(2):398–402. doi: 10.1016/j.jacc.2005.08.069. [DOI] [PubMed] [Google Scholar]
- 80.Fontana L, Klein S, Holloszy JO. Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age (Dordr) 32(1):97–108. doi: 10.1007/s11357-009-9118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holloszy JO. Biochemical adaptations in muscle. Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J Biol Chem. 1967;242(9):2278–82. [PubMed] [Google Scholar]
- 82.Chow LS, et al. Impact of endurance training on murine spontaneous activity, muscle mitochondrial DNA abundance, gene transcripts, and function. J Appl Physiol. 2007;102(3):1078–89. doi: 10.1152/japplphysiol.00791.2006. [DOI] [PubMed] [Google Scholar]
- 83.Coggan AR, et al. Skeletal muscle adaptations to endurance training in 60- to 70-yr-old men and women. J Appl Physiol. 1992;72(5):1780–6. doi: 10.1152/jappl.1992.72.5.1780. [DOI] [PubMed] [Google Scholar]
- 84.Short KR, et al. Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes. 2003;52(8):1888–96. doi: 10.2337/diabetes.52.8.1888. [DOI] [PubMed] [Google Scholar]
- 85.Menshikova EV, et al. Effects of exercise on mitochondrial content and function in aging human skeletal muscle. J Gerontol A Biol Sci Med Sci. 2006;61(6):534–40. doi: 10.1093/gerona/61.6.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sreekumar R, et al. Effects of caloric restriction on mitochondrial function and gene transcripts in rat muscle. Am J Physiol Endocrinol Metab. 2002;283(1):E38–43. doi: 10.1152/ajpendo.00387.2001. [DOI] [PubMed] [Google Scholar]
- 87.Sreekumar R, et al. Impact of high-fat diet and antioxidant supplement on mitochondrial functions and gene transcripts in rat muscle. Am J Physiol Endocrinol Metab. 2002;282(5):E1055–61. doi: 10.1152/ajpendo.00554.2001. [DOI] [PubMed] [Google Scholar]
- 88.Staniek K, Nohl H. Are mitochondria a permanent source of reactive oxygen species? Biochim Biophys Acta. 2000;1460(2–3):268–75. doi: 10.1016/s0005-2728(00)00152-3. [DOI] [PubMed] [Google Scholar]
- 89.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120(4):483–95. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Nicholls DG, Locke RM. Thermogenic mechanisms in brown fat. Physiol Rev. 1984;64(1):1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 91.Pecqueur C, et al. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress, and evidence for translational regulation. J Biol Chem. 2001;276(12):8705–12. doi: 10.1074/jbc.M006938200. [DOI] [PubMed] [Google Scholar]
- 92.Richard D, et al. Distribution of the uncoupling protein 2 mRNA in the mouse brain. J Comp Neurol. 1998;397(4):549–60. [PubMed] [Google Scholar]
- 93.Richard D, et al. Uncoupling protein 2 in the brain: distribution and function. Biochem Soc Trans. 2001;29(Pt 6):812–7. doi: 10.1042/0300-5127:0290812. [DOI] [PubMed] [Google Scholar]
- 94.Vidal-Puig A, et al. UCP3: an uncoupling protein homologue expressed preferentially and abundantly in skeletal muscle and brown adipose tissue. Biochem Biophys Res Commun. 1997;235(1):79–82. doi: 10.1006/bbrc.1997.6740. [DOI] [PubMed] [Google Scholar]
- 95.Boss O, et al. Uncoupling protein-3: a new member of the mitochondrial carrier family with tissue-specific expression. FEBS Lett. 1997;408(1):39–42. doi: 10.1016/s0014-5793(97)00384-0. [DOI] [PubMed] [Google Scholar]
- 96.Negre-Salvayre A, et al. A role for uncoupling protein-2 as a regulator of mitochondrial hydrogen peroxide generation. Faseb J. 1997;11(10):809–15. [PubMed] [Google Scholar]
- 97.Kowaltowski AJ, Costa AD, Vercesi AE. Activation of the potato plant uncoupling mitochondrial protein inhibits reactive oxygen species generation by the respiratory chain. FEBS Lett. 1998;425(2):213–6. doi: 10.1016/s0014-5793(98)00231-2. [DOI] [PubMed] [Google Scholar]
- 98.Echtay KS, et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415(6867):96–9. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 99.Echtay KS, et al. Superoxide activates mitochondrial uncoupling protein 2 from the matrix side. Studies using targeted antioxidants. J Biol Chem. 2002;277(49):47129–35. doi: 10.1074/jbc.M208262200. [DOI] [PubMed] [Google Scholar]
- 100.Murphy MP, et al. Superoxide activates uncoupling proteins by generating carbon-centered radicals and initiating lipid peroxidation: studies using a mitochondria-targeted spin trap derived from alpha-phenyl-N-tert-butylnitrone. J Biol Chem. 2003;278(49):48534–45. doi: 10.1074/jbc.M308529200. [DOI] [PubMed] [Google Scholar]
- 101.Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35(6–7):811–20. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 102.Kerner J, et al. Aging skeletal muscle mitochondria in the rat: decreased uncoupling protein-3 content. Am J Physiol Endocrinol Metab. 2001;281(5):E1054–62. doi: 10.1152/ajpendo.2001.281.5.E1054. [DOI] [PubMed] [Google Scholar]
- 103.Brand MD, et al. Oxidative damage and phospholipid fatty acyl composition in skeletal muscle mitochondria from mice underexpressing or overexpressing uncoupling protein 3. Biochem J. 2002;368(Pt 2):597–603. doi: 10.1042/BJ20021077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vidal-Puig AJ, et al. Energy metabolism in uncoupling protein 3 gene knockout mice. J Biol Chem. 2000;275(21):16258–66. doi: 10.1074/jbc.M910179199. [DOI] [PubMed] [Google Scholar]
- 105.Nabben M, et al. The effect of UCP3 overexpression on mitochondrial ROS production in skeletal muscle of young versus aged mice. FEBS Lett. 2008;582(30):4147–52. doi: 10.1016/j.febslet.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 106.Speakman JR, et al. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell. 2004;3(3):87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- 107.Nicholls DG, Ferguson SJ. Bioenergetics 3. Academic Press; London: 2002. [Google Scholar]
- 108.Bevilacqua L, et al. Effects of short- and medium-term calorie restriction on muscle mitochondrial proton leak and reactive oxygen species production. Am J Physiol Endocrinol Metab. 2004;286(5):E852–61. doi: 10.1152/ajpendo.00367.2003. [DOI] [PubMed] [Google Scholar]
- 109.Bevilacqua L, et al. Long-term caloric restriction increases UCP3 content but decreases proton leak and reactive oxygen species production in rat skeletal muscle mitochondria. Am J Physiol Endocrinol Metab. 2005;289(3):E429–38. doi: 10.1152/ajpendo.00435.2004. [DOI] [PubMed] [Google Scholar]
- 110.Caldeira da Silva CC, et al. Mild mitochondrial uncoupling in mice affects energy metabolism, redox balance and longevity. Aging Cell. 2008;7(4):552–60. doi: 10.1111/j.1474-9726.2008.00407.x. [DOI] [PubMed] [Google Scholar]
- 111.Fridell YW, et al. Targeted expression of the human uncoupling protein 2 (hUCP2) to adult neurons extends life span in the fly. Cell Metab. 2005;1(2):145–52. doi: 10.1016/j.cmet.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 112.Andrews ZB, Horvath TL. Uncoupling protein-2 regulates lifespan in mice. Am J Physiol Endocrinol Metab. 2009;296(4):E621–7. doi: 10.1152/ajpendo.90903.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McDonald RB, et al. Characterization of survival and phenotype throughout the life span in UCP2/UCP3 genetically altered mice. Exp Gerontol. 2008;43(12):1061–8. doi: 10.1016/j.exger.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 114.Serra V, et al. Extracellular superoxide dismutase is a major antioxidant in human fibroblasts and slows telomere shortening. J Biol Chem. 2003;278(9):6824–30. doi: 10.1074/jbc.M207939200. [DOI] [PubMed] [Google Scholar]
- 115.Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263(5150):1128–30. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 116.Orr WC, et al. Effects of overexpression of copper-zinc and manganese superoxide dismutases, catalase, and thioredoxin reductase genes on longevity in Drosophila melanogaster. J Biol Chem. 2003;278(29):26418–22. doi: 10.1074/jbc.M303095200. [DOI] [PubMed] [Google Scholar]
- 117.Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90(19):8905–9. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vanfleteren JR, De Vreese A. Rate of aerobic metabolism and superoxide production rate potential in the nematode Caenorhabditis elegans. J Exp Zool. 1996;274(2):93–100. doi: 10.1002/(SICI)1097-010X(19960201)274:2<93::AID-JEZ2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 119.Blander G, et al. Superoxide dismutase 1 knock-down induces senescence in human fibroblasts. J Biol Chem. 2003;278(40):38966–9. doi: 10.1074/jbc.M307146200. [DOI] [PubMed] [Google Scholar]
- 120.Ho YS, et al. Mice deficient in cellular glutathione peroxidase develop normally and show no increased sensitivity to hyperoxia. J Biol Chem. 1997;272(26):16644–51. doi: 10.1074/jbc.272.26.16644. [DOI] [PubMed] [Google Scholar]
- 121.Melov S, et al. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat Genet. 1998;18(2):159–63. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 122.Reaume AG, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13(1):43–7. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 123.Lebovitz RM, et al. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93(18):9782–7. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mecocci P, et al. Age-dependent increases in oxidative damage to DNA, lipids, and proteins in human skeletal muscle. Free Radic Biol Med. 1999;26(3–4):303–8. doi: 10.1016/s0891-5849(98)00208-1. [DOI] [PubMed] [Google Scholar]
- 125.Giorgio M, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122(2):221–33. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 126.Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295(5564):2450–2. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- 127.Trinei M, et al. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21(24):3872–8. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 128.Migliaccio E, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402(6759):309–13. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 129.Luzi L, et al. Evolution of Shc functions from nematode to human. Curr Opin Genet Dev. 2000;10(6):668–74. doi: 10.1016/s0959-437x(00)00146-5. [DOI] [PubMed] [Google Scholar]
- 130.Ravichandran KS. Signaling via Shc family adapter proteins. Oncogene. 2001;20(44):6322–30. doi: 10.1038/sj.onc.1204776. [DOI] [PubMed] [Google Scholar]
- 131.Yokote K, et al. Direct interaction between Shc and the platelet-derived growth factor beta-receptor. J Biol Chem. 1994;269(21):15337–43. [PubMed] [Google Scholar]
- 132.Skolnik EY, et al. The SH2/SH3 domain-containing protein GRB2 interacts with tyrosine-phosphorylated IRS1 and Shc: implications for insulin control of ras signalling. Embo J. 1993;12(5):1929–36. doi: 10.1002/j.1460-2075.1993.tb05842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Migliaccio E, et al. Opposite effects of the p52shc/p46shc and p66shc splicing isoforms on the EGF receptor-MAP kinase-fos signalling pathway. Embo J. 1997;16(4):706–16. doi: 10.1093/emboj/16.4.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pacini S, et al. p66SHC promotes apoptosis and antagonizes mitogenic signaling in T cells. Mol Cell Biol. 2004;24(4):1747–57. doi: 10.1128/MCB.24.4.1747-1757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xi G, Shen X, Clemmons DR. p66shc negatively regulates insulin-like growth factor I signal transduction via inhibition of p52shc binding to Src homology 2 domain-containing protein tyrosine phosphatase substrate-1 leading to impaired growth factor receptor-bound protein-2 membrane recruitment. Mol Endocrinol. 2008;22(9):2162–75. doi: 10.1210/me.2008-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xi G, Shen X, Clemmons DR. p66shc inhibits insulin-like growth factor-I signaling via direct binding to Src through its polyproline and Src homology 2 domains, resulting in impairment of Src kinase activation. J Biol Chem. 285(10):6937–51. doi: 10.1074/jbc.M109.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Orsini F, et al. The life span determinant p66Shc localizes to mitochondria where it associates with mitochondrial heat shock protein 70 and regulates trans-membrane potential. J Biol Chem. 2004;279(24):25689–95. doi: 10.1074/jbc.M401844200. [DOI] [PubMed] [Google Scholar]
- 138.Pinton P, et al. Protein kinase C beta and prolyl isomerase 1 regulate mitochondrial effects of the life-span determinant p66Shc. Science. 2007;315(5812):659–63. doi: 10.1126/science.1135380. [DOI] [PubMed] [Google Scholar]
- 139.Nemoto S, et al. The mammalian longevity-associated gene product p66shc regulates mitochondrial metabolism. J Biol Chem. 2006;281(15):10555–60. doi: 10.1074/jbc.M511626200. [DOI] [PubMed] [Google Scholar]
- 140.Mootha VK, et al. A reversible component of mitochondrial respiratory dysfunction in apoptosis can be rescued by exogenous cytochrome c. Embo J. 2001;20(4):661–71. doi: 10.1093/emboj/20.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao Y, Wang ZB, Xu JX. Effect of cytochrome c on the generation and elimination of O2*- and H2O2 in mitochondria. J Biol Chem. 2003;278(4):2356–60. doi: 10.1074/jbc.M209681200. [DOI] [PubMed] [Google Scholar]
- 142.Ow YP, et al. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9(7):532–42. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 143.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 144.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8(5):405–13. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 145.Biggs WH, 3rd, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome. 2001;12(6):416–25. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- 146.Jacobs FM, et al. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278(38):35959–67. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 147.Furuyama T, et al. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349(Pt 2):629–34. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lin K, et al. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–22. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 149.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389(6654):994–9. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 150.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11(24):1975–80. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 151.Hwangbo DS, et al. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429(6991):562–6. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 152.Medema RH, et al. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404(6779):782–7. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 153.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423(6939):550–5. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 154.Kim MS, et al. Role of hypothalamic Foxo1 in the regulation of food intake and energy homeostasis. Nat Neurosci. 2006;9(7):901–6. doi: 10.1038/nn1731. [DOI] [PubMed] [Google Scholar]
- 155.Bakker WJ, et al. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J Cell Biol. 2004;164(2):175–84. doi: 10.1083/jcb.200307056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hribal ML, et al. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J Cell Biol. 2003;162(4):535–41. doi: 10.1083/jcb.200212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Willcox BJ, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105(37):13987–92. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Flachsbart F, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106(8):2700–5. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kops GJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419(6904):316–21. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 160.Tran H, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296(5567):530–4. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 161.Dijkers PF, et al. FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. J Cell Biol. 2002;156(3):531–42. doi: 10.1083/jcb.200108084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chiba T, et al. Overexpression of FOXO1 in skeletal muscle does not alter longevity in mice. Mech Ageing Dev. 2009;130(7):420–8. doi: 10.1016/j.mad.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 163.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299(5611):1346–51. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 164.Rincon M, et al. The paradox of the insulin/IGF-1 signaling pathway in longevity. Mech Ageing Dev. 2004;125(6):397–403. doi: 10.1016/j.mad.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 165.Lee SJ, Murphy CT, Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 2009;10(5):379–91. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kenyon C, et al. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366(6454):461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 167.Tatar M, et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292(5514):107–10. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 168.Holzenberger M, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–7. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 169.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299(5606):572–4. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 170.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 171.Biggs WH, 3rd, et al. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96(13):7421–6. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Obsilova V, et al. 14-3-3 Protein interacts with nuclear localization sequence of forkhead transcription factor FoxO4. Biochemistry. 2005;44(34):11608–17. doi: 10.1021/bi050618r. [DOI] [PubMed] [Google Scholar]
- 173.Brunet A, et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156(5):817–28. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rena G, et al. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. Embo J. 2002;21(9):2263–71. doi: 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang X, et al. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277(47):45276–84. doi: 10.1074/jbc.M208063200. [DOI] [PubMed] [Google Scholar]
- 176.Tsai WC, et al. Insulin inhibition of transcription stimulated by the forkhead protein Foxo1 is not solely due to nuclear exclusion. Endocrinology. 2003;144(12):5615–22. doi: 10.1210/en.2003-0481. [DOI] [PubMed] [Google Scholar]
- 177.Alam H, et al. EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab. 12(1):30–41. doi: 10.1016/j.cmet.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13(11):1385–93. [PubMed] [Google Scholar]
- 179.Murakami S, Johnson TE. The OLD-1 positive regulator of longevity and stress resistance is under DAF-16 regulation in Caenorhabditis elegans. Curr Biol. 2001;11(19):1517–23. doi: 10.1016/s0960-9822(01)00453-5. [DOI] [PubMed] [Google Scholar]
- 180.Kahn BB, et al. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1(1):15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 181.Greer EL, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282(41):30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 182.Barthel A, et al. Regulation of the forkhead transcription factor FKHR (FOXO1a) by glucose starvation and AICAR, an activator of AMP-activated protein kinase. Endocrinology. 2002;143(8):3183–6. doi: 10.1210/endo.143.8.8792. [DOI] [PubMed] [Google Scholar]
- 183.Greer EL, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17(19):1646–56. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fukuoka M, et al. Negative regulation of forkhead transcription factor AFX (Foxo4) by CBP-induced acetylation. Int J Mol Med. 2003;12(4):503–8. [PubMed] [Google Scholar]
- 185.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101(27):10042–7. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 187.Tsai KL, et al. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007;35(20):6984–94. doi: 10.1093/nar/gkm703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Matsuzaki H, et al. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102(32):11278–83. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5(5):224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306(5704):2105–8. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 191.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273(2):793–8. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 192.Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 193.Tanner KG, et al. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97(26):14178–82. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14(9):1021–6. [PubMed] [Google Scholar]
- 195.Bitterman KJ, et al. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277(47):45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 196.Gallo CM, Smith DL, Jr., Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24(3):1301–12. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Anderson RM, et al. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–5. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Balan V, et al. Life span extension and neuronal cell protection by Drosophila nicotinamidase. J Biol Chem. 2008;283(41):27810–9. doi: 10.1074/jbc.M804681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.van der Horst A, et al. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev. 2007;128(4):346–9. doi: 10.1016/j.mad.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 200.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14(5):661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279(49):50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 202.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451(7178):583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 203.Escande C, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 120(2):545–58. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kim EJ, et al. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28(2):277–90. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 205.Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008;28(35):8772–84. doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9(11):1253–62. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nasrin N, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One. 2009;4(12):e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.van der Horst A, et al. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279(28):28873–9. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 209.Pfluger PT, et al. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105(28):9793–8. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 211.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 212.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. Embo J. 2002;21(10):2383–96. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 214.Lan F, et al. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283(41):27628–35. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hawley SA, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2(4):28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Woods A, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13(22):2004–8. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 217.Tamas P, et al. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J Exp Med. 2006;203(7):1665–70. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hawley SA, et al. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem. 1996;271(44):27879–87. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 219.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20(2):98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 221.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 222.Canto C, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 11(3):213–9. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26(7):1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Aquilano K, et al. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 285(28):21590–9. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Onyango P, et al. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99(21):13653–8. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Rose G, et al. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38(10):1065–70. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 227.Kim HS, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sundaresan NR, et al. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28(20):6384–401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pillai VB, et al. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 285(5):3133–44. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 464(7285):121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sundaresan NR, et al. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119(9):2758–71. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]