Abstract
Mass spectrometry imaging (MSI), a rapidly growing subfield of chemical imaging, employs mass spectrometry (MS) technologies to create single- and multi-dimensional localization maps for a variety of atoms and molecules. Complimentary to other imaging approaches, MSI provides high chemical specificity and broad analyte coverage. This powerful analytical toolset is capable of measuring the distribution of many classes of inorganics, metabolites, proteins and pharmaceuticals in chemically and structurally complex biological specimens in vivo, in vitro, and in situ. The MSI approaches highlighted in this Methods in Molecular Biology volume provide flexibility of detection, characterization, and identification of multiple known and unknown analytes. The goal of this chapter is to introduce investigators who may be unfamiliar with MS to the basic principles of the mass spectrometric approaches as used in MSI. In addition to guidelines for choosing the most suitable MSI method for specific investigations, cross-references are provided to the chapters in this volume that describe the appropriate experimental protocols.
1. Introduction
Understanding the functioning of a cell, organ, or organism, whether normal or pathological, often relies on the ability to follow the spatial localization of molecules and atoms with sufficient spatial and temporal resolution. Success can be facilitated by mass spectrometry imaging (MSI), a robust analytical approach that combines the chemical information content of MS detection with chemical imaging technology. MSI allows the simultaneous localization of hundreds of analytes in a variety of biological samples, from defined morphological structures to entire organisms, without analyte preselection using antibody labeling. The metabolome, peptidome, and proteome can be investigated with MSI, providing spatial and chemical information for literally hundreds to thousands of analytes, ranging from small molecules to large proteins. Although implementation requires specialized hardware, software, and experimental protocols, including sample preparation methods, a range of features, along with recent advances in instrumentation, have contributed to an increased interest in using MSI for chemical imaging.
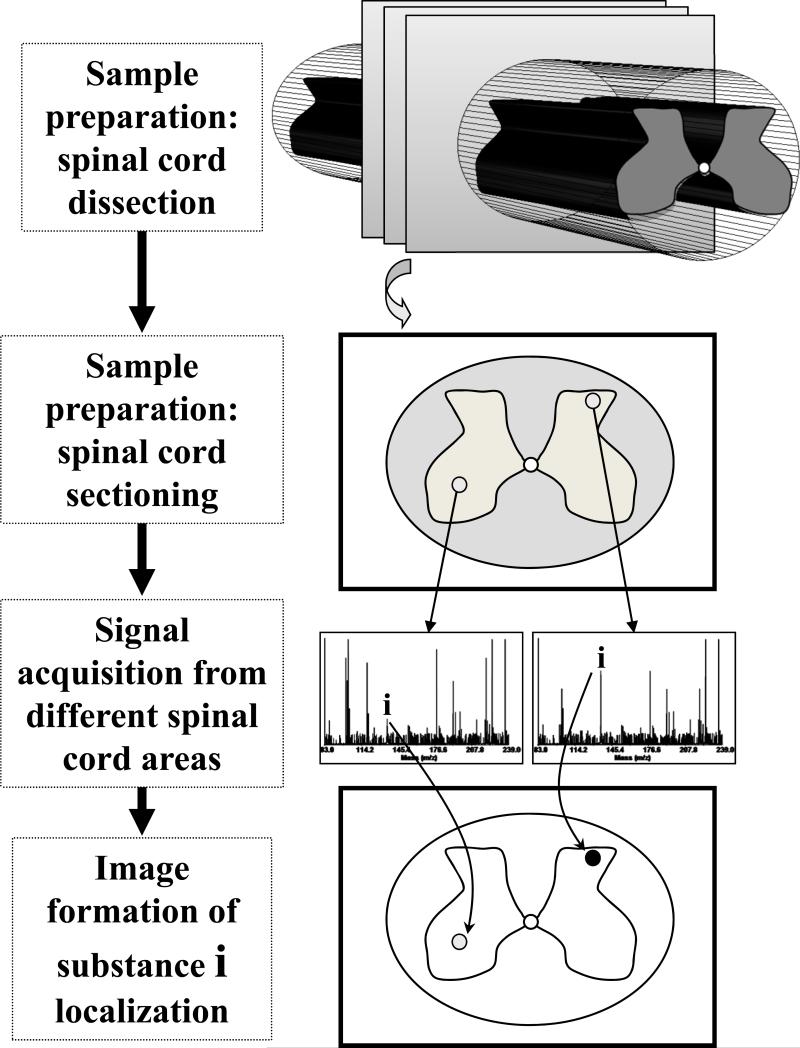
In MSI, a specimen is typically interrogated by a beam of light or ions called a microprobe. As a result, multiple analytes are moved from solid or liquid to gas phase, ionized, and characterized according to their mass-to-charge ratio. The microprobe is then moved to another location and the process repeated. If the microprobe is scanned across the surface at a sufficient number of regularly spaced points, chemical images for each analyte are created (Fig. 1). A chemical image presents not only information about analyte localization, but also provides signal intensity information, often encoded by color or grayscale. Depending on the analytical method used and sample properties, signal intensity often correlates with the absolute or relative concentration of each analyte. Although MSI approaches can differ in the software and sample preparation protocols used, they share many features of common MS measurements and thus, are often employed in MS or proteomics facilities. However, MSI can easily be implemented in many biological and neuroscientific laboratories.
Fig. 1.
Schematic of the generalized mass spectrometry imaging process utilizing the microprobe mode of operation. The major steps involved include sample preparation and interrogation with a probe, signal acquisition, processing, and final image formation. Distribution of a single or multiple analytes is determined in an array of spots covering the sample surface. Only two mass spectra containing signals from different analytes are shown in the signal acquisition step. Detected signal intensity is often encoded by color or grayscale. Here, the more intense signal is presented in the color black, which provides the most contrast from the background color.
In addition to the discussion of the basics of mass spectrometric technologies presented herein, the reader will be directed, as appropriate, to cross-referenced chapters in this volume that provide more specifics on the MSI approaches. Even greater detail on both MS and MSI can be found in several excellent books and reviews (1-10).
2. Mass Spectrometry (MS)
Mass, considered a fundamental property of matter (11), is defined as a measure of a body's inertia, or resistance to a change in body motion upon application of an external force. Having knowledge of a particle's exact mass and some details about its structure, is sufficient to identify an element or polyatomic substance. MS is one of most precise, fast, and reliable methods available to determine the molecular and atomic masses of analytes with high accuracy in a single measurement.
2.1 Principles of Mass Spectrometry
A key element of MS is based on the ability of magnetic and/or electric fields to influence the motion of charged atoms and molecules, typically in a vacuum, in relation to their mass and charge. Newton's second law is a fundamental basis for MS, where the resistance of a particle to a change in its motion (inertia) exerted by a force, and the proportionality of this change to the intensity of the applied force, are used to determine a particle's mass. Indeed, the trajectories of two identically moving particles of the same charge will change differently under the influence of identical electric and/or magnetic field(s) if the particles have different masses. The motion of the heavier particle will be affected less than the lighter one. As a result, the final relative spatial positions of the moving particles will depend on their inertial mass (m) and, for differently charged particles, on their charge (z). A particle's charge is an important factor as the amount of force developed by a field relates directly to the charge. The motion of the higher-charged particle will be affected more by the fields than the lesser-charged particle's motion, when all parameters, except charge, are the same. Therefore, by controlling the forces, predicting particle trajectories, and selectively detecting particles, one can deconvolve the motions into each distinct particle's mass-to-charge ratio, or m/z. Results of mass spectrometric investigation are typically presented in the form of mass spectra, where m/z values are plotted along the x-axis and the relative intensity of the charged particle signals against the y-axis.
In what follows, the major instrument components used in MS measurements are described. Why is knowledge about the mass spectrometric process important to a biologist interested in MSI? Although it may not seem obvious, understanding of the technology ensures selection of the optimum measurement approach and the most appropriate protocols for a specific project. As explained below, many of the measurement limitations experienced in an MSI investigation are directly related to the characteristics of the mass measurement process selected.
2.2 Mass Spectrometers
Mass spectrometers consist of several functionally distinct components (Fig. 2)—the ion source, mass analyzer, and detector—with resulting signal processing/output. As mentioned above, the characteristics of these components are what combine to give the overall figures of merit of the measurement and determine the most appropriate system for a given analytical challenge. More than one ion source, mass analyzer, and detector are often integrated into a single mass spectrometer, thereby providing greater flexibility in instrumental operating modes and enabling different types of analytes to be characterized. Importantly, most mass spectrometers can be used for MSI if the system allows ion signal acquisition from a number of discretely selected points and if the spatial positions of the specimen can be recorded.
Fig. 2.
Principal schematic of a mass spectrometer with atmospheric pressure ionization (API). In many mass spectrometers, the ion source operates under a vacuum, while in API, the ionization occurs under ambient/atmospheric pressure.
2.2.1 Ion Sources
Perhaps no other part of the measurement process affects the resulting data more significantly than the ionization step. Unless originally in the gas phase, analytes need to be vaporized from a solid or liquid phase, ionized, and transferred into the vacuum system of the mass analyzer. This vaporization process, often called desorption (sputtering), can be fairly energetic, and can degrade/transform the analytes before they are characterized. Vaporization can be achieved by a variety of techniques, including heating the samples, exposing them to a high electric field or laser irradiation, and/or via bombardment with fast atoms, or atomic or molecular ions. Ionization of analytes may occur before, during, and after desorption. Therefore, the term desorption/ionization is commonly used to describe the entire event. Obviously, only desorption is required to produce gaseous neutrals, which can be ionized later using several post-ionization processes. Depending on the characteristics of the sample and desorption/ionization processes, different sets of neutral elements, smaller molecular fragments or intact molecules, as well as their ions, are generated.
The resulting ions have either a positive or negative charge, and so the polarity of the electric field applied at the ion source can determine which ions are injected into the mass analyzer. Importantly, some analytes are more likely to form positive ions and so are easier to characterize in the positive ion mode; others that form negative ions are more easily characterized in the negative ion mode. Moreover, the same sample analysis using different desorption/ionization approaches, even when coupled to the same mass analyzer, may yield different results.
Selecting the appropriate desorption/ionization approach is critical. That decision can result in analyte classes of a specific m/z range either being efficiently or poorly ionized, and it can often govern the spatial resolution of the MSI method, needed sample preparation steps, throughput and sensitivity of the analysis. Thus, many of the protocol chapters in this volume describe specific ionization approaches and their corresponding sample preparation steps in great detail.
Four of the most commonly used desorption/ionization methods for MSI are secondary ion mass spectrometry (SIMS), desorption electrospray ionization (DESI), matrix-assisted laser desorption/ionization (MALDI) and laser ablation (LA) with post-ionization. Other desorption/ionization approaches such as laser desorption/ionization (LDI) (see Chapter 9, (12)), desorption/ionization on silicon (DIOS) (13), electrospray ionization (ESI) (14), and nanostructure-initiator mass spectrometry (NIMS) (15, 16) also have great potential in MSI. Importantly, many mass spectrometers equipped with a MALDI ion source can be used with related ionization processes such as LDI, DIOS, LA, and laser-NIMS. Frequently, a specific ion source arrangement is optimized for a specific mass analyzer; for example, MALDI is often interfaced to a time-of-flight (TOF) mass analyzer (described below) although it can also be used with ICR-based instruments.
Although many instruments are designed for a specific ion source, several can integrate a range of sources. For instance, greater flexibility can be achieved with instruments that allow analysis of ions generated at atmospheric (ambient) pressure (AP), where the ion source does not require the vacuum conditions typically needed for the operation of most mass analyzers. AP sources enable investigation of samples containing volatile compounds that are incompatible with ion sources operating at vacuum. Some AP desorption/ionization systems provide interchangeable ion sources, including MALDI, ESI, and DESI, making these systems more flexible; however, this flexibility may be associated with tradeoffs in performance specifications such as sensitivity. These modular ion sources are available from a variety of vendors, as well as manufactured according to researcher specifications (see Chapter 9 for an example of a laboratory-built ion source). In the following, the principles of ion generation using different ion sources are briefly described. Only the major ionization modes used for MSI are highlighted here; other sources frequently applied to mass spectrometry are not common in imaging at this time.
2.2.1.1 SIMS
In SIMS, a beam of fast-moving ions is aimed at the sample (Fig. 3). On impact, these ions eject (or sputter) a variety of particles from the sample surface, including secondary ions. The bombardment process results in formation of gas phase neutrals, ions of both polarities, electrons, and photons. A number of primary ions are used in SIMS, including lightweight monoatomic ions such as Au+, Ar+, Xe+, O-, O2+, Ga+, and Cs+. Larger polyatomic cluster ion beams, such as Aun+, Bin+, C60+, and SF5+5, are also employed; several of these result in an improved mass range of detection and sensitivity for heavier analytes. This increase in detection mass range is related to the characteristics of larger primary ions, which create higher ion currents and higher energy ion beams. Laser post-ionization has become an important tool for increased ion production with semisoft desorption by the cluster C60+ ion beam (17). Depending on the type of ion gun, the energy of the projectiles is in the keV - MeV range, which is sufficient to form atomic and molecular ions with m/z up to 2000 in experiments with biological specimens. When working with biological tissues, molecular fragments such as histidine-related m/z 110, phospholipid-related m/z 184, among others, are often detected. Primary atomic ion beams can be focused to the nanometer diameter. As an example, 40 nm lateral resolution has been achieved with a Ga ion gun (18). Often, the larger cluster ion beams cannot be focused as well as their smaller ion counterparts and therefore, the probed sample spot can exceed 1 μm2. A defocused ion beam is useful in MSI, with SIMS instruments capable of operating in the microscope mode. In this mode, the entire investigated area is probed with primary ions; secondary ions are transferred to the detector with their relative position preserved, resulting in the creation of a stigmatic 2D image. Much more common is the microprobe mode in which ion signals are collected from individual spots on an area of interest and the corresponding image recreated pixel-by-pixel.
Fig. 3.
Schematic of SIMS ion source operation.
Which primary ion flux should be considered for a particular MSI experiment? Investigation of biological specimens by SIMS is typically carried out in the static regime, which is when less than 1% of the sample surface, and about a nanometer of sample depth, is interrogated/modified by ion bombardment. It corresponds to a primary ion dose that does not exceed 1013 ions/cm2. Higher-dose bombardment, or the dynamic mode, leads to greater sample alteration due to analyte layer mixing and more molecule fragmentation. The dynamic mode is useful for investigating elemental composition and depth profiling. Good results in sample depth profiling were recently achieved with large cluster primary ions (19). SIMS sputtering/ionization performance can be improved by coating the sample surface with metals and organic substances (see Chapter 11).
Mass spectrometers equipped with SIMS ion sources are broadly used in MSI of element, metabolite and pharmaceutical localization (see Chapters 4, 6, 11, 15, and 16), as well as quantitative measurements of drug localization in normal and pathology-affected tissues (see Chapter 6). SIMS imaging is very effective in imaging metabolite distribution in biological systems. Such investigation requires significant attention to preserving analyte localization. Therefore, sample preparation of hydrated specimens requires special consideration and is described in the above-mentioned chapters.
2.2.1.2 Laser Desorption Approaches
Several desorption/ionization approaches use light to vaporize and ionize analytes from liquid and solid samples. Ultraviolet (UV) and infrared (IR) lasers (e.g., from the 193 nm excimer to the 10.6 μm TEA-CO2 laser) have been employed in laser-based ion sources. The appropriate approach is selected based on sample and/or analyte characteristics, such as mass and ionization properties; options include LA, LDI, laser-NIMS, DIOS, or MALDI.
The LA approach is popular in elemental analyses of specimens where the laser is set to produce light with a flux sufficient enough to completely vaporize a sample region (see Chapter 3 and (10)). However, output of analyte ions can be low and post-ionization methods such as inductively coupled plasma (ICP) are needed.
LDI utilizes lower intensity (I) laser pulses, I ≤ 10 MW/cm2; metabolites and relatively large intact molecules, like peptides and some proteins, can be desorbed and ionized. An increase in energy leads to more molecules and atoms being desorbed into the gas phase, but results in a higher level of molecule fragmentation and loss of chemical information. IR LDI with electrospray post-ionization has proven to be a powerful approach to determine 2D and 3D localization of metabolites in live plant leaves (see Chapter 9). Switching to a shorter wavelength UV laser in the LDI ion source helps improve imaging lateral resolution to 10 μm and has been used to investigate distribution of UV-absorbing metabolites in plant tissues (12). Interestingly, covering the sample with colloid silver and interrogating with LDI results in production of intact ions of metabolites that have bound silver atoms.
Since its introduction in the early 20th century by Sir Joseph John Thomson, mass spectrometry has been mostly applied to analyses of smaller analytes, with fewer successes in peptide and protein research. The limiting factor had been that it is difficult to vaporize and ionize intact proteins. However, this completely changed about 20 years ago with the invention of soft laser desorption by Koichi Tanaka (20), matrix-assisted laser desorption/ionization by Franz Hillenkamp and Michael Karas (21), as well as a critical demonstration of the applicability of electrospray ionization to analyses of large molecules by Bennett Fenn (22) (as described below); these ionization approaches opened the door to widespread use of MS to measure molecules ranging from low molecular weight metabolites to megadalton proteins, and even to small structures such as viruses. Recognizing the impact of these achievements, Tanaka and Fenn were awarded the Nobel Prize in chemistry in 2002.
MALDI is widely employed in MSI. LDI and LA experiments have demonstrated that the surrounding analyte environment is critical for the desorption process. Therefore, mixing of analytes with a light absorbing matrix was shown to be beneficial for the process (Fig. 4) (21). Upon laser irradiation, the matrix “explodes,” carrying intact analytes, some of which are ionized. Importantly, for successful desorption/ionization, analyte particles should be in close proximity to the matrix/buffer molecules and be spatially separated from each other, thus making the relative concentration of analyte-to-matrix ratio a critical factor to consider. More matrix is needed for analysis of the same number of larger molecules as compared to smaller ones. Application of matrix to the sample surface is a critical step in MSI because it may contribute to unwanted analyte spatial redistribution.
Fig. 4.
Schematic of ion generation in the MALDI ion source.
Which matrices should be used? For MSI, one often uses solid or liquid low molecular mass organic matrices such as 2,5-dihydroxybenzoic acid (DHB) and alpha-cyano cinnamic acid (CHCA), or their mixtures (23, 24). A variety of MALDI matrices have been introduced over the years. It is interesting that different matrices have some specificity in promoting desorption/ionization of particular types of analytes. For example, 9-aminoacridine (9-AA) is preferable for working with metabolites (25-27). DHB is often used for desorption/ionization of peptides and proteins, and 3-hydroxypicolinic acid (3-HPA) makes possible oligonucleotide analysis (28). For each MALDI matrix, analyte type and sample type, the polarity (positive or negative) of the MALDI ion source must be determined experimentally in order to achieve the best results. Consider for instance that metabolite analysis with 9-AA achieves better results in negative ion mode, while protein analysis with DHB performs better in positive ion mode. Mixing of the matrix solution with a solution containing analytes, or applying matrix solution onto a sample and allowing the analytes to mix with the matrix, precedes matrix analyte cocrystallization and interrogation with the laser. Solid matrix crystals can be also applied to dry or semidry samples with successful outcomes. However, relative spatial localization of analyte and matrix molecules is not always clear. Interestingly, DHB matrix solution not only works well for analyte extraction, it can also serve as a preservation media. The antioxidant properties of DHB and the acidic environment of the media likely combine to achieve biologically active peptide preservation during several years of storage in the solution (29).
The MALDI process produces predominantly singly-charged ions, thereby facilitating the processing and understanding of resulting data. Neutrals, singly-charged and multiply-charged ions, ions with elemental adducts, and those with water loss are also formed, and may represent the dominant signal for a particular analyte. The ramification of producing singly-charged ions is important in the discussion of mass analyzers that follows. One of the exemplary facets of MALDI is that it produces some of the best detection limits of any ionization method. Moreover, it performs well across a broad mass range in a wide number of analyte and sample types, including dried droplets on a metal target and tissue samples on a glass slide.
The diameter of the laser beam used to probe the sample surface typically determines the effective spatial resolution of a measurement performed in microprobe mode. Obviously, the laser beam diameter can be reduced by focusing the beam to smaller dimensions. However, as the laser beam diameter is reduced, it illuminates a smaller area, fewer molecules of each analyte are present within the probe beam, and so fewer molecules are ionized at each location. Therefore, smallest diameter beams are rarely practical because the amount of analyte that can be desorbed and ionized from a smaller sample area is not sufficient for detection and high-accuracy mass measurement. Consequently, the laser probe diameter for the analyses of proteins and peptides usually is larger than ten microns.
2.2.1.3 Desorption Electrospray Ionization Mass Spectrometry
DESI, a relatively new ionization method that allows surfaces to be characterized under ambient conditions (30), offers a number of interesting characteristics for imaging. A dynamic liquid buffer interface is created on top of the sample by bombarding the sample surface with charged microdroplets, gas phase molecules and charged clusters of solvents (e.g., a methanol/water/acetic acid mixture) (Fig. 5). This desorbs and ionizes analytes via several means, including electric field-related mechanisms. The analytes enter the electrostatic field formed between the spray emitter and mass spectrometer inlet. More gaseous ions are generated at this stage by desorption/ionization, similar to what takes place during ESI; however, the precise mechanisms are still being determined (31, 32). DESI ion sources operate in either negative or positive ion modes. DESI produces singly-charged ions as well as series of multiply-charged ions for a single analyte. The larger the analyte, the more complex the multiply-charged ion series will be. Even though the presence of an analyte in multiple charge states complicates qualitative and quantitative data analysis, it often assists with fragmentation of analyte molecules, as well as permits the detection of analytes over an extended mass range. Perhaps most importantly, production of gaseous ions with DESI often does not require sample preparation and can be performed on almost any surface.
Fig. 5.
Schematic of the desorption electrospray ionization (DESI) process (see text for details).
2.2.2 Mass Analyzers
2.2.2.1 General Principles
After the ions are created, the next step is to transfer them to the mass analyzer. Depending on ionization mechanisms, sampling requirements, and detection sensitivity and selectivity requisites, the injection of analyte ions into the mass analyzer can be done immediately after their desorption/ionization/post-ionization, or after a delay. A delay is implemented for different purposes, including delivery of ions from the ion source to a distantly located mass analyzer (DESI), focusing of ions, or equalization of the energies of identical ions (MALDI). The reasons for some of these arrangements are discussed under the specific mass analyzers described below. After selecting the appropriate ionization source for an experiment, the second most important decision is choosing the right mass analyzer. Playing a key role in the separation of generated ion source ions according to their mass, charge and, in some cases, shape, the mass analyzer also guides ions towards the detector, or away from it. To be individually recognized, packets of ions of the same mass and charge are separated in space/time from ions of different mass and charge.
Two ions with the same mass-to-charge ratio cannot be distinguished by the mass analyzer, as would be the case with stereoisomers. However, these molecules can often be distinguished by breaking the ions apart and investigating the ion fragments with another mass analyzer. To accomplish this, mass spectrometers can be configured to store, filter and fragment ions. Because each of these functions may require additional hardware, contemporary multiplexed mass spectrometer systems are often hybrids and include more than one mass analyzer, capable of multistage operation. This multiplex approach is generally referred to as tandem mass spectrometry, or MS/MS, to describe the two stages of analysis. Fragmentation of ions is important for analyte identification. Due to the existence of a variety of isobaric molecules, having knowledge of the exact mass is not enough to identify an analyte. Tandem MS improves identification via fragmentation because the tandem mass spectrum may provide a unique profile of the product ions, representing a molecular fingerprint. In fact, tandem MS of resulting fragments is also possible; in this case, the term MS3 is used. This concept can be carried out even further, to create “MS to the n” (MSn), applied when n stages of fragmentation/separation are involved in the analysis of a single substance. Although MS12 has been performed (33), MS3 is sufficient for identification of some structurally similar lipid species (34). Historically, the names of many mass analyzers reflect the mechanisms of ion manipulation, details of hardware engineering, or principles of analyte mass determination. Multiplexed mass analyzers have led to the need to abbreviate the lengthy names of these MS systems. As a result, the lexicon of mass spectrometrists has become filled with QTOF, QqQ, oTOF, BEBE, QQQHQCQ, and others -- acronyms which will be explained later in the text.
As mentioned above, MS uses electric and/or magnetic fields to determine the accurate mass of ionized atoms and molecules. The force developed by the fields on charged particles is described by Lorentz force law: F = q(E + v × B), where F is the force applied to the ion, q is the charge of the ion, E is the electric field, and v × B is the vector cross-product of the ion velocity and applied magnetic field. Change in motion of the charged particle under influence of Lorentz force follows Newton's second law: F = ma where m is the mass of the charged particle and a is the acceleration. Combining these two equations gives an expression for the motion of charged particles under the influence of the fields: (m/q)a = (E + v × B).
It is evident from this equation that either mass-to-charge is measured, or knowledge of the charge is required, in order to determine the mass using magnetic and/or electric fields. The mass spectra represent the distribution of signals of different mass-to-charge ratios, or m/z. The m/z is a dimensionless parameter where m is the mass of the particle and z represents the number of charges; z is proportional to the charge of a single proton (e; z = q/e). Therefore, separation and detection of ions in the mass spectrometer, as well as their representation in the resulting mass spectra, are done in order of their mass/charge ratio. Signals of heavier molecules with a larger charge may occur on the x-coordinate (m/z coordinate) of a mass spectrum earlier than the signal of a lighter but less-charged one. Obviously, if all analyzed ions were singly charged, this would not be important. For example, MALDI produces predominantly singly-charged ions. However, multiply-charged ions are formed by DESI (and at times with MALDI). Additional steps are needed to take into account the charge on a particle; this deconvolution process is typically done automatically for most instruments. However, for lower quality measurements, manual manipulation may be necessary.
One way that mass analyzers can be categorized is by the way they manipulate ions -- continuous or pulsed. Continuous mass analyzers maintain a constant flow of ions into the analyzer and are well suited for interface to continuous ion sources such as DESI. MALDI uses a pulsed laser for desorption/ionization, making it compatible with pulsed mass analyzers. Several pulsed mass analyzers, such as ion cyclotron resonance, time-of-flight and ion traps, can accommodate continuous sources and accumulate the ions and introduce them in pulses. The accumulation/storage time depends on system capabilities and can range from microseconds to seconds. Magnetic sector (B), electric sector (E), and quadrupole (Q) mass filters are continuous mass analyzers, while time-of-flight (TOF), quadrupole ion trap (QIT), linear ion trap, Orbitrap, and ion cyclotron resonance (ICR) are discontinuous, or pulsed analyzers. While several of the standard configurations have been mentioned, other combinations are possible; with effort, most ion sources can be linked to most mass analyzers.
Obviously, an ion needs to last as long as its analysis takes in the mass analyzer. Ion lifetime is related to the quality of the vacuum system, the requirements for which vary with each mass analyzer. The length of ion flight path and strength of the ion guiding fields, as well as the measurement accuracy and resolution needed, combine to govern the quality of vacuum required. A longer flight path may need a higher vacuum so as to reduce the number of unwanted collisions between analyte ions and gaseous environmental particles. These collisions result in a change in ion motion trajectory and/or velocity and consequently, distortion of signal or even ion recombination and loss of signal. Although such effects can be negative, they can also be beneficial, depending on whether the ions need to be kinetically cooled, or to achieve controlled fragmentation. Typically, fragmentation is performed at selected locations in the instrument by adding a collision gas into the ion flight path collision cell or directly into the mass analyzer.
Resolution is one of the primary figures of merit provided by a mass analyzer and is the parameter that often governs the purchase price of a system (Fig. 6). The higher the resolution, the better the instrument will perform when assigning chemical structures to specific signals. Another important characteristic of MS measurement is mass accuracy, which represents the closeness of the measured mass to the true mass. Mass resolution and mass accuracy for the same ions are predominantly determined by characteristics of the mass analyzers and ion detectors.
Fig. 6.
Figures of merit for defining mass analyzers: accuracy, resolution (R), and resolving power (RP). These parameters are used to compare the performance of different MS approaches. An idealized mass spectrum containing two peaks with m/z values M and M1 shown in the center of the figure. The resolution represents how close the m/z of two peaks can be before they overlap and cannot be distinguished. Resolving power is the inverse, and is a measure of the ability of a mass spectrometer to separate different ions. Signal processing, such as filtering and baseline correction, influence final data sets and hence, the resolution value calculated from raw data will be different from that from processed data. ΔM can be measured at other signal intensity levels, e.g., 10%. ΔMFWHM represents the full width of the peak at half its maximum (FWHM) height; ppm = parts per million. Importantly, there are a variety of resolution and resolving power definitions which may even contradict each other (http://goldbook.iupac.org, (7, 9)).
High mass resolution is important for detection of isotopic distributions (Fig. 7A). A majority of polyatomic compounds in biological specimens are composed of different isotopes of the same combination of atoms. For a singly-charged molecule, a set of signals (peaks) differing by 1 m/z unit will appear in the mass spectra. The first peak in the distribution corresponds to a compound consisting of the lightest atomic isotopes, such as 12C, 14N, 16O, and 1H. For example, the isotope 12C has a 98.9% abundance while the abundance of 13C is only 1.1%. The mass of the molecule having the lightest atomic isotopes is called the monoisotopic mass. Of course, for multiply-charged analytes, the spacing of these isotopic signals depends on the charge (e.g., for a doubly-charged analyte, the spacing will be 0.5 Da instead of 1 Da). Knowledge of the isotopic distribution aids in analyte characterization. Fig. 7B shows a mass spectrum with two unambiguously detected ions of m/z 1864.3 and m/z 1882.3, where the first ion is 18 units lighter, in good accordance with a loss of water molecule. Analyses of the peak pattern distribution in the signal series starting with m/z 1864.3 indicates that a compound with putative m/z 1866.5 exists in the sample. Unusual patterns of isotopic distribution also may occur due to poor instrument performance and/or incorrect instrument settings.
Fig. 7.
High resolution mass spectrometry enables determination of isotopic distributions. A. Overlaid mass spectra of a compound with a monoisotopic mass of 1802 Da (monoprotonated [M+H] = m/z 1803) and an average mass of 1802.6. Differing resolutions of detection are indicated. B. Mass spectrum containing two different m/z ions and one putative ion at m/z 1866.5. C. Mass spectrum of the isotopic distribution of a singly-charged molecular ion. D. Mass spectrum of the isotopic distribution of a doubly-charged molecular ion.
Determination of mass for multiply-charged ions is possible by analysis of the isotopic pattern distribution, where spacing between peaks is related to ion charge. Fig. 7C shows a mass spectrum with 1 m/z spacing, indicative of a singly-charged ion, while Fig. 7D presents a pattern with 0.5 m/z spacing, corresponding to a doubly-charged ion. The number of charges on the ion depends on many parameters, including the ionization approach used and the size of the particle. Larger proteins ionized with ESI may routinely have a charge of +30. Obviously, the mass analyzer requires exceptional resolving power to determine the isotopic distribution in this case, as the peaks are separated by 0.033 Da. Large ions with more charges are easier to manipulate by electric and/or magnetic fields, as well as fragment for sequence determination. However, because different molecules of the same compound may have different charge states, complex charge-state distributions create more complex mass spectra. This problem can be solved by forcing production of maximally charged ions with addition of glycerol or m-nitrobenzyl alcohol to the electrospray solution (35). Currently, the phenomenon of multiply-charged ions exists mostly for DESI-MSI. MALDI, SIMS, and LA desorption/ionization approaches produce predominantly singly-charged ions. Different principles are used to achieve high resolution, high throughput and measurement accuracy among available mass analyzers. The following sections contain brief descriptions of the mass analyzers used in MSI.
2.2.2.2 Time-of-Flight (TOF) Instruments
The mechanism of operation of a TOF mass analyzer is fairly straightforward (Fig. 8). In what is known as the linear mode, ions are extracted from the ion source and unidirectionally accelerated by short pulses of electrostatic field, entering and moving in a drift space containing no field; all ions are accelerated with the same kinetic energy. Thus, lighter ions of the same charge will move faster than heavier ones, and therefore, come to the detector earlier; obviously, separation takes place in space and detection in time. One of drawbacks of the linear mode is that the ions initially do not all have the exact same position and velocity, so that there is a spread in ion arrival times at the detector. This spatial distribution leads to formation of broad, lower amplitude signals at the detector, resulting in reduced resolution and at times, lower sensitivity of detection.
Fig. 8.
Schematics of co-axial geometry time-of-flight (TOF) mass analyzer operation in linear and reflector modes. A simplified equation describing the TOF process is presented in the insert. m = mass of the ion, V = acceleration voltage, d = length of flight path, t = time from the moment of ion acceleration to the detection event.
There are two approaches used in TOF mass analyzers to improve resolution. One is delayed extraction (delayed injection into the mass analyzer, also called pulsed ion extraction) of the ions formed in the MALDI ion source. This delay reduces the energy spread of the same m/z ions. Delayed extraction parameters can be adjusted by the operator, with a longer delay time needed for improved detection of larger molecules. Another approach utilizes reflecting ion optics (reflector or reflectron mode) such as ion mirrors -- a set of evenly spaced electrodes encompassing space on the ion path (Fig. 8). A single linear electric field with higher potential energy than the source potential is formed around each electrode. Ions are flying in these fields and are reflected (repulsed). As a result, resolution improves due to two factors. First, there is an increase in ion path length and thus, a greater distance between packets of ions; second, there is a reduction in the spread of kinetic energies of different particles of the same m/z. More energetic ions will travel longer paths in the field space than lower kinetic energy ions; therefore, they will be focused as they leave the ion mirror area, arriving at the detector in a more temporally compact packet. Both approaches are typically used when metabolites, peptides and small- and medium-sized proteins are analyzed. However, the linear mode of operation with delayed extraction remains the preferred option for analysis of large molecules and molecular aggregates.
TOF analyzers are common in MSI applications because of their speed of operation and wide m/z range. They allow analysis of large singly-charged molecular ions produced by MALDI (see Chapters 7 - 11, 14 - 20, 22 - 24, 26, and 27).
2.2.2.3 Sector Instruments
Magnetic sector and electrostatic sector mass analyzers are well suited for operation with continuous ion sources; the trajectories of moving ions are curved by forces developed by the electric or magnetic fields (Fig. 9). The extent of this curvature depends on an ion's m/z. Sector analyzers can be used to monitor a single ion with high resolution. A narrow slit is installed between the detector and ion analyzer; the position of the slit determines which ion is detected. A narrower slit improves mass resolution but decreases sensitivity. Mass resolution is also dependent on the cross-section of the incoming ion beam, the m/z ion kinetic energy spread, and the radius of ion trajectory. Different m/z ions can be recorded simultaneously by using multiple detectors (or a detector array). Due to fast ion transmission and low level of interaction between ions in the beam (e.g., minimal space-charge effects leading to ion-ion repulsions), sector analyzers are capable of quantitative measurements. Ions approach the sector analyzer as focused or defocused beams. The latter can be refocused with a direction-focusing approach using a magnetic sector mass analyzer (Fig. 9, inset). Electrostatic mass analyzers are efficient ion kinetic energy filters whereas magnetic sector analyzers are capable of filtering ions with differing momentum (Fig. 9). Therefore, hybrid instruments combining these two mass analyzers enable double-focusing and may achieve 100,000 resolution. Double-focusing instruments have been employed to image the distribution of elements by LA-ICP MSI (see Chapter 3). Magnetic sector SIMS instruments (see Chapter 6), like the CAMECA IMS 7f, are capable of distinguishing such ions as 56Fe and 28Si2, and allow direct ion microscopy (stigmatic mode imaging) and scanning microprobe mode imaging.
Fig. 9.
General schematic of a sector mass analyzer. Ions extracted from the ion source are accelerated by an electrostatic field (accelerating potential, V) and enter the sector analyzer with velocity, v. Electric (electric flux density, E) or magnetic (magnetic flux density, B) fields bend the trajectory of the ions into curved paths with radius, r. Trajectories of ions with larger m/z are affected more than smaller ones. An illustration of the direction-focusing ion beam approach in a magnetic sector mass analyzer is shown in the insert. Due to the dependence of the radius of an ion's trajectory on its kinetic energy (E) in the electrostatic sector mass analyzer, and on its momentum (mv) in the magnetic sector mass analyzer, the systems are also referred to as ion energy and ion momentum filters.
2.2.2.4 Quadrupole (Q) Instruments
The quadrupole analyzer is compatible with continuous ion sources such as DESI. Although the upper m/z range of the quadrupole is not high, neither is DESI well suited for desorbing high molecular weight analytes, and so the two approaches work well together. The quadrupole, as its name suggests, consists of four precisely aligned metal hyperbolic or cylindrical rods (Fig. 10). Superimposed direct current (DC) and oscillating radiofrequency (RF) electric fields are used to create conditions where ions of only a certain m/z (typically a 1 m/z mass transmission window) will have a stable trajectory inside the device and therefore pass through it. Other ions of lower and higher m/z will leave the analyzer prematurely or collide with the rods and skeleton. Therefore, the quadrupole mass analyzer is a form of mass filter. The ion path in the quadrupole starts as circular transforms to complex spiral-like propagation inside the field. Depending on the analyzer's design, stable trajectories for ions of a particular m/z can be achieved in a oscillating electric field by setting the appropriate RF frequency, and RF and DC voltage amplitudes. Importantly, simultaneous change of DC and RF voltage amplitudes allows transmission of ions of differing m/z. Keeping the RF/DC amplitude ratio constant, while gradually and simultaneously changing the amplitudes, helps to scan/transmit ions in a broad m/z range with a high level of selectivity. Simultaneous increase in RF and DC amplitudes is necessary to transmit ions with larger m/z. Altering the optimal ratio will increase or decrease the m/z window, thus impacting the selectivity of detection and mass resolution.
Fig. 10.
The quadrupole mass analyzer is an ion filtering device that creates an oscillating electric field between four rods. A. Schematic of a quadrupole mass analyzer. -(U + Vcos ω t) and +(U + Vcosωt) are cumulative potential created by superimposed direct current potential (U) and radiofrequency current potential (Vcosωt). B. The encircled equation describes an area of stability for a particular m/z ion trajectory (ω = circular frequency; r0 = field radius) depicted on the graph.
The quadrupole mass analyzer is relatively small, efficient and affordable. They are widely used in gas chromatography MS (GC-MS) and liquid chromatography MS (LC-MS). A variety of hybrid instruments exist with multiple quadrupole mass filters installed in the ion path, including systems as complex as QQQHQCQ (Q - Quadrupole lens; H - Homogeneous magnetic sector; C - Cylindrical electric sector). However, triple quads are most common. In this case, the first Q selects a narrow m/z range, the second Q fragments it with RF field energy and/or collision gas, and the third Q scans and passes the resulting fragments towards the detector. The application of quadrupole analyzers to MSI typically has been in the role of ion filter or collision cell in hybrid instruments (see Chapters 9 and 23).
2.2.2.5 Ion Traps
It was recognized that the approach of creating a stable ion path using DC and RF electric fields could also be used for ion storage. Ion traps can accumulate and spatially contain ions, as well as release them for detection. Extended storage times allow opportunities for ion fragmentation, detection and sorting. How does one prevent the ion cloud from dissipating and colliding with mass analyzer surfaces? Additional fields guide the ions along complex trajectories in a relatively small space. Different approaches for forming these forces distinguish the types of ion traps. Quadrupole ion traps (linear and 3D) utilize electrostatic fields and RF potentials (a Paul ion trap), and the Orbitrap (discussed in detail below) operates with electrostatic fields (a Kingdon trap).
The 3D quadrupole ion trap (QIT) consists of three hyperbolic-shaped electrodes encompassing the region where the electric field is formed by RF potential applied to the central ring electrode, and DC, supplementary RF or ground potential is established on two end-cap electrodes (Fig. 11A). Ions of an m/z range, determined by amplitude and frequency of the RF fields, are trapped in this region. The trapped ions have stable oscillating trajectories in the QIT until a controlled destabilizing change in the potentials is introduced. Such destabilization can be done for cleanup of QIT space from multiple unwanted ions using broadband waveforms or for sending predetermined m/z ions towards the detector by an RF scan. Similar to the Q analyzer, superimposing RF and DC potentials on the ring electrode allows storage of single m/z ions. This feature is important in MSn experiments where individual analytes can be selected and fragmented, the fragments selected for detection, and/or a specific fragment selected for another round of fragmentation. Typically, MS5 can be achieved . Ion capacity is important for MSn experiments because each subsequent MS stage has less material for analysis than the previous stage. Increased ion storage volume also allows for reduction of space-charge effects that may occur in situations where the concentration of ions is high enough that they start to repel each other. The space-charge effect often determines low mass resolution due to broadening of analyte signals and also for formation of artifacts in mass spectra related to interactions between ions. QIT use for MSI is described in Chapter 13.
Fig. 11.
Quadrupole ion traps provide a versatile approach for storage, fragmentation, and selection of ions. A. Schematic of 3-D quadrupole ion trap consisting of a ring electrode and two end-cap electrodes. Only one ion ejection pathway is shown. A combination of DC and RF potentials is applied to the electrodes. 1 mtorr of helium is typically added to the mass analyzer. B. Schematic of a linear quadrupole ion trap. During ion trapping, DC and RF ion-guiding potentials are maintained on the central electrodes while the end section electrodes maintain stop potential, SP.
A variant of the ion trap is the linear version that creates a cylindrical space for the ion cloud, linear quadruple ion traps (Fig. 11B) provide greater ion storage volume compared to QITs. As a result, more ions can be stored with greater efficiency, allowing detection with high resolution and increased sensitivity for a particular sample. Linear ion traps are called 2D because they utilize a two-dimensional RF field to confine ions radially. This field is developed between four centrally located hyperbolic or cylindrical rods surrounded on both sides by two sets of quadrupole end electrodes, which maintain DC stop potential, thereby preventing ions from leaving the trap axially. Both 2D and 3D quadrupole ion traps can be part of a hybrid set up, or serve as the sole mass analyzer. Pulsed ion sources are common for ion traps in MS analyses that require longer operating sequences. However, different instrumental configurations, as well as high speed operation, allow ions traps to be linked with continuous ion sources without loss of sample (for an example in MSI, see Chapter 25).
2.2.2.6 Ion Cyclotron Resonance (ICR)
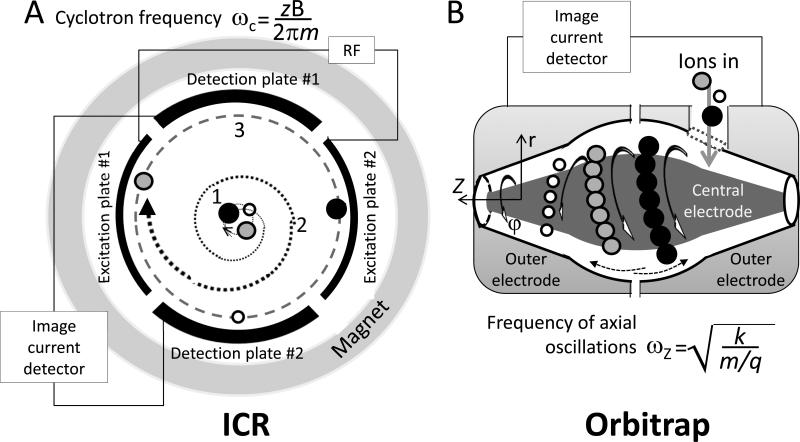
Ion cyclotron resonance uses the principle of ions orbiting in an ICR cell, with each ion having a characteristic frequency that depends on its m/z. A combination of magnetic and electric fields allows separation of ions according to their m/z. The magnetic field, created by a magnet which surrounds the ICR cell, is static and may have different geometries (Fig. 12A). The cell has six electrodes, four of which are used for ion containment and ion path manipulation, and two as trapping electrodes. The ions, whether injected or generated inside the cell, are constrained by the magnetic field as well as the electric fields established on the trapping electrodes. Ions continuously orbit while static electric potential restricts their axial movement. At this stage, the ions form a diffuse ion cloud, comprised of a mix of ions of differing m/z. By applying a sequence of RF pulses to the excitation plates, specific ions are resonantly activated and start to move in a well-defined packet while maintaining the cyclotron trajectory. Using sequential RF pulses, the ion packets can be synchronized. These packets of ions are measured as they repeatedly pass near the detection plates. The detection plates are used to measure image current from the ions, where each m/z has a unique cyclotron frequency that depends on the mass and charge of the ions. Higher m/z ions will appear less frequently. The timing of the image currents represents a frequency recording of the ions in the detection cell; the more transits of each ion, the more accurately the frequencies can be determined. The frequencies are converted into a more conventional mass spectrum using a Fourier transform (FT).
Fig. 12.
High resolution mass spectrometric analyses are achieved with ion cyclotron resonance (ICR) and Orbitrap mass analyzers. A. Schematic of an ICR mass analyzer consisting of a magnet surrounding a cylindrical ICR cell formed by three pairs of electrodes (two trapping electrodes are not shown), electronic circuitries for detection of the ion image current, and generation of RF and DC potentials. Three stages of ion separation and detection are marked with numbers: 1 - initial electrostatic trapping (no RF is applied), 2 - cyclotron motion when RF is used to resonantly excite ions of a particular m/z and move them to higher orbit, and 3 -detection phase when RF is turned off and detection plates are engaged to sense ion packets. The equation shows the dependence of cyclotron frequency on mass (m) and charge (z) of an analyte ion moving in the magnetic field of strength, B. B. Schematic representation of an Orbitrap mass analyzer. A central electrode is surrounded by one outer electrode, which is divided into two halves by nonconductive space. Although different approaches are implemented for ion detection with the Orbitrap, only one is shown here. A simplified equation describing the frequency of axial oscillations in the mass analyzer demonstrates that this parameter - ωZ - can be used for determination of mass (m) to charge (q) ratio. k = field curvature; φ = rotation; Z and r = coordinate axes. Both analyzers are operating at vacuum conditions.
ICR is known for achieving the highest mass resolution (m/Δmx, quoted as resolving power in some publications), as one example, reaching 8 million for 8.6 kDa bovine ubiquitin (36). ICR resolution increases linearly with the increasing strength of the magnetic field. Therefore, large magnets, up to 25 Tesla, are employed in FT-ICR MS (37). A resolution (m/Δm50%) of 200,000 for a 14.5 Tesla instrument at m/z 400 at a 1 Hz measurement rate has been reported (38). ICR mass analyzers became powerful tools in proteomics investigations with the advent of top-down (intact proteins are analyzed) and bottom-up (peptides of enzymatically digested proteins are analyzed) approaches. A large variety of ion fragmentation approaches are available for ICR, although these are less common in MSI. This is due in part to its limited m/z range (typically not over ~10 KDa); however, ICR works well with ESI when multiply charged ions are generated so that a much higher mass range can be interrogated. This limitation is much more severe in MSI when using MALDI because it generates mostly singly charged ions.
2.2.2.7 The Orbitrap
Recently, a compact but powerful mass analyzer -- the Orbitrap -- was introduced (Fig. 12B) (39). Using a balance between the centripetal influence of an electrostatic field developed on a central electrode and the opposite centrifugal force of ions rapidly injected and moving in the mass analyzer, the Orbitrap produces packets of ions according to their m/z. The trajectory of ion motion inside the Orbitrap resembles a complex spiral orbiting alongside the central electrode. The axial component of this motion is dependent on the mass and charge of a particle. Therefore, recording the image current generated by the motion of different m/z ion packets produces a complex record of change of current amplitude over time. Again, a Fourier transform allows the conversion into relative signal intensity versus m/z. The Orbitrap employs relatively new technology and at its current stage of development is capable of producing a resolution of >100,000 at m/z 400 for a 1.5 s acquisition time, or 60,000 at a 1 Hz acquisition rate. This capability allows determination of the localization of analytes with similar m/z values, in particular using the protocol described in Chapter 25.
2.2.3 Detectors
After the ions are separated or processed, they need to be detected. For several mass analyzers, ion detection is integral to mass analyzer operation as is true for the ICR instrument. Here we briefly describe one of the most common ion detectors.
Moving ions produce signals in detectors on impact or by creating electric currents. A number of types of electron multipliers are used in mass spectrometers. As an example, a microchannel electron multiplier plate has a large array of 5 - 10 micrometer diameter channels which may occupy an area >10 cm2. Ions strike the detector's surface inside an individual channel, thereby inducing secondary electron and photon emission. The efficiency of this process depends on the kinetic energy of incoming ions, incident angle of impact, and detector surface properties. Secondary electrons continue to move toward the detector and strike tunnel walls, again inducing more electrons. This effect can multiply the number of ions by more than a million fold. The process continues along the channel until electrons reach a conductor, which transmits this electric current to amplifiers and further signal processing. Detectors operate at vacuum conditions usually below 10-5 torr. However, new technologies for generation of curved channels in channel electron multipliers has resulted in an ion detector that is operational at 10-2 torr (http://www.detechinc.com/em/quad.htm). Importantly, the ion detectors of some mass spectrometers can bereplaced/updated, translating into significant improvements in detection capabilities.
Why is it important to understand how the detector works? Many mass spectrometers have the option of adjusting detector sensitivity. Increasing the sensitivity may help to observe more compounds and aid in the detection of low-abundance analytes. Unfortunately, detector operation at a higher sensitivity setting can increase the noise level and reduce the lifetime of the detector. In addition, if the detector is set at a higher sensitivity and intense signals are present, this can degrade detector performance. High spatial resolution MS imaging requires significant dynamic range and often the detector is optimized for low analyte levels at each probed spot. Thus, detector performance can degrade during the hundreds of thousands of acquisitions that take place when creating a number of larger ion images.
What is next? The signals produced by detectors are digitized, processed and converted to mass spectra. MS imaging experiments may generate thousands of mass spectra containing information on hundreds of signals acquired from specimens. Obviously, it is not possible to check individual mass spectrum quality. Therefore, data analysis in MSI experiments is mostly done using final ion images. Such analysis may generate false-positive signal detection due to a variety of factors, including issues as shown in Fig. 13. Different baseline correction, denoising, and intelligent peak picking algorithms are implemented for batch mass spectra processing in MSI (40, 41) (see Chapters 1, 22 and 27). Nevertheless, a random check of several of the original mass spectra is an important step during data analysis.
Fig. 13.
Factors complicating analysis of MSI experiment results. A. Formation of alkali metal adducts. Sodium and potassium adducts are marked on the mass spectrum. These can occur in a region-specific manner in an MSI image. B. Detector saturation. Mass spectra acquired with different laser fluencies (the total energy per unit area). Note lost resolution on the major peak and improved detection of lower intensity peaks as a result of fluency increase (lower trace). C. Curved baseline shape, chemical and/or digitization noise may produce false-positive peak detection during the automatic peak picking process. Mass spectrum shows an elevated baseline and high level of chemical noise (left inset) in the lower m/z range, mixed chemical and digitization noises in the middle range (central inset), and only digitization noise in the higher m/z region.
2.2.4 Calibration
Mass spectrometers are complex systems and many parameters can be optimized according to experimental requirements. These adjustments, as well as possible drift in the calibration of different components, may lead to measurement of analyte m/z with a systematic error. Therefore, calibration of the mass scale is often performed using standards, often more than three; this compensates for these deleterious effects and results in a more accurate m/z scale. During automatic or manual calibration, standards with known masses and predictable charge are measured, differences between determined mass and true mass are found, and a set of calibration constants created. These constants become part of the m/z calibration during subsequent measurements performed with the same instrumental settings with unknown analytes.
Internal, external or mass defect-based calibration can be performed (42). External calibration is done with mixtures of standards measured separately from analyzed unknown samples. In contrast, internal calibration requires the presence of calibrants in the sample. Importantly, if the m/z of endogenous analytes are known, they can be used to calibrate the mass spectrometer. This type of calibration is preferable because no increase in complexity of sample occurs and influences from different parameters, such as sample thickness and analyte environment, are accounted for. As an example, trypsin autocleavage molecular ions are used for calibration in proteomics research (43), as is mass defect-based calibration, where peptides of enzymatically cleaved proteins are used for calibration and protein identifications (42, 44). The latter approach is also useful for determination and subsequent elimination of nonpeptide species from peak lists. Calibrant selection is governed by several requirements, including the nature of targeted analytes, investigated m/z range, type of mass spectrometer, and calibration approach. Unique, exogenous calibrants are preferable for internal calibration when endogenous calibrants are not available. Using analytes that can be endogenously present in an investigated sample for calibration is discouraged due to the possibility of ambiguous experimental outcomes, as well as accidental contamination of other samples. High-quality calibration is achieved with calibrants that bracket the mass of the analyte of interest. After their acquisition, individual mass spectra can be recalibrated employing different calibration profiles. However, recalibration of complex MSn spectra, or an entire set of mass spectra obtained during an MS imaging experiment, can be difficult. Therefore, it is important to verify calibration before performing an imaging experiment using the appropriate standards.
3. Conclusions
Mass spectrometry provides a set of tools with versatile and powerful figures of merit. When used under the control of appropriate operating protocols, many MS-platforms can be used for MSI. The large number of data-points in an image, and the complex matrix of working directly with tissue samples, can place severe performance constraints on an MSI experiment. Nonetheless, the rapid increase in the performance of ion sources, mass analyzers, and detectors has led to a new generation of sensitive, high resolution, and relatively easy to use mass spectrometers. These are now providing unmatched capabilities for chemical imaging. The mature mass spectrometry field provides biologists with a variety of novel imaging modalities that are appropriate for a number of applications. While there is no single MS-based tool that is ideal for all analytes, the assortment of instruments now available allows a broad range of samples to be probed. In the following chapters, specific protocols are described in much greater detail.
Acknowledgements
The authors would like to thank Kevin Tucker for helpful discussion of the chapter and Stephanie Baker for her help with the manuscript preparation. This material is based upon work supported by Award No. P30DA018310 from the National Institute on Drug Abuse (NIDA). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIDA or the National Institutes of Health.
References
- 1.Burlingame AL. Methods in Enzymolology. Elsevier Academic Press; Amsterdam; Boston: 2005. Mass spectrometry: modified proteins and glycoconjugates. [Google Scholar]
- 2.Chance M, editor. Mass Spectrometry Analysis for Protein-Protein Interactions and Dynamics. John Wiley & Sons; Hoboken, N.J.: 2008. [Google Scholar]
- 3.Downard K, editor. Mass Spectrometry of Protein Interactions. Wiley Interscience; Hoboken, N.J.: 2007. [Google Scholar]
- 4.Lipton MS, Páya-Tolic L. Mass Spectrometry of Proteins and Peptides: Methods and Protocols. Humana Press; Springer, distributor; New York, N.Y.; London: 2009. [DOI] [PubMed] [Google Scholar]
- 5.Matthiesen R, editor. Mass Spectrometry Data Analysis in Proteomics. Humana Press; Totowa, N.J.: 2007. [Google Scholar]
- 6.Murphy RC. Mass Spectrometry of Lipids. Plenum Press; New York, NY: 1993. [Google Scholar]
- 7.Sparkman OD. Mass Spec Desk Reference. 2nd ed. Global View Publishing; Pittsburgh, PA: 2006. [Google Scholar]
- 8.Wanner KT, Höfner G, editors. Mass Spectrometry in Medicinal Chemistry: Applications in Drug Discovery. Wiley-VCH; John Wiley (distributor); Weinheim; Chichester: 2007. [Google Scholar]
- 9.McLafferty FW, Turecek F. Interpretation of Mass Spectra. University Science Books; Mill Valley, CA: 1993. [Google Scholar]
- 10.Becker JS. Inorganic Mass Spectrometry: Principles and Applications. John Wiley & Sons, Chichester; England; Hoboken, NJ: 2007. [Google Scholar]
- 11.De Podesta M. Understanding the Properties of Matter. Taylor & Francis; London; New York: 2001. [Google Scholar]
- 12.Holscher D, Shroff R, Knop K, Gottschaldt M, Crecelius A, Schneider B, Heckel DG, Schubert US, Svatos A. Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging on the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J. 2009;60:907–918. doi: 10.1111/j.1365-313X.2009.04012.x. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Guo Z, He L. Mass spectrometry imaging of small molecules using desorption/ionization on silicon. Anal Chem. 2007;79:3535–3541. doi: 10.1021/ac0611465. [DOI] [PubMed] [Google Scholar]
- 14.Van Berkel GJ, Kertesz V, Koeplinger KA, Vavrek M, Kong AN. Liquid microjunction surface sampling probe electrospray mass spectrometry for detection of drugs and metabolites in thin tissue sections. J Mass Spectrom. 2008;43:500–508. doi: 10.1002/jms.1340. [DOI] [PubMed] [Google Scholar]
- 15.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 16.Yanes O, Woo HK, Northen TR, Oppenheimer SR, Shriver L, Apon J, Estrada MN, Potchoiba MJ, Steenwyk R, Manchester M, Siuzdak G. Nanostructure initiator mass spectrometry: tissue imaging and direct biofluid analysis. Anal Chem. 2009;81:2969–2975. doi: 10.1021/ac802576q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willingham D, Kucher A, Winograd N. Molecular depth profiling and imaging using cluster ion beams with femtosecond laser postionization. Appl Surf Sci. 2008;255:831–833. [Google Scholar]
- 18.Sakamoto T, Koizumi M, Kawasaki J, Yamaguchi J. Development of a high lateral resolution TOF-SIMS apparatus for single particle analysis. Appl Surf Sci. 2008;255:1617–1620. [Google Scholar]
- 19.Wucher A, Cheng J, Zheng L, Winograd N. Three-dimensional depth profiling of molecular structures. Anal Bioanal Chem. 2009;393:1835–1842. doi: 10.1007/s00216-008-2596-5. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka K. The origin of macromolecule ionization by laser irradiation (Nobel lecture). Angew Chem Int Ed Engl. 2003;42:3860–3870. doi: 10.1002/anie.200300585. [DOI] [PubMed] [Google Scholar]
- 21.Hillenkamp F, Karas M. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 1990;193:280–295. doi: 10.1016/0076-6879(90)93420-p. [DOI] [PubMed] [Google Scholar]
- 22.Fenn J. Electrospray ionization mass spectrometry: how it all began. J Biomol Tech. 2002;13:101–118. [PMC free article] [PubMed] [Google Scholar]
- 23.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 24.Kaletas BK, van der Wiel IM, Stauber J, Guzel C, Kros JM, Luider TM, Heeren RM. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9:2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 25.Seetharaman V, Royston G. Quantitative detection of metabolites using matrix-assisted laser desorption/ionization mass spectrometry with 9-aminoacridine as the matrix. Rapid Commun Mass Spectrom. 2007;21:2072–2078. doi: 10.1002/rcm.3063. [DOI] [PubMed] [Google Scholar]
- 26.Rachal LV-S, David MH. 9-Aminoacridine as a matrix for negative mode matrix-assisted laser desorption/ionization. Rapid Commun Mass Spectrom. 2002;16:1575–1581. [Google Scholar]
- 27.Edwards JL, Kennedy RT. Metabolomic analysis of eukaryotic tissue and prokaryotes using negative mode MALDI time-of-flight mass spectrometry. Anal Chem. 2005;77:2201–2209. doi: 10.1021/ac048323r. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Sun X, Guo B. Matrix-assisted laser desorption/ionization time-of-flight analysis of low-concentration oligonucleotides and mini-sequencing products. Rapid Commun Mass Spectrom. 2003;17:2354–2360. doi: 10.1002/rcm.1200. [DOI] [PubMed] [Google Scholar]
- 29.Romanova EV, Rubakhin SS, Sweedler JV. One-step sampling, extraction, and storage protocol for peptidomics using dihydroxybenzoic acid. Anal Chem. 2008;80:3379–3386. doi: 10.1021/ac7026047. [DOI] [PubMed] [Google Scholar]
- 30.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 31.Manicke NE, Wiseman JM, Ifa DR, Cooks RG. Desorption electrospray ionization (DESI) mass spectrometry and tandem mass spectrometry (MS/MS) of phospholipids and sphingolipids: ionization, adduct formation, and fragmentation. J Am Soc Mass Spectrom. 2008;19:531–543. doi: 10.1016/j.jasms.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Wiseman JM, Ifa DR, Venter A, Cooks RG. Ambient molecular imaging by desorption electrospray ionization mass spectrometry. Nat Protoc. 2008;3:517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- 33.Lundstrom SL, D'Alexandri FL, Nithipatikom K, Haeggstrom JZ, Wheelock AM, Wheelock CE. HPLC/MS/MS-based approaches for detection and quantification of eicosanoids. Methods Mol Biol. 2009;579:161–187. doi: 10.1007/978-1-60761-322-0_8. [DOI] [PubMed] [Google Scholar]
- 34.Houjou T, Yamatani K, Nakanishi H, Imagawa M, Shimizu T, Taguchi R. Rapid and selective identification of molecular species in phosphatidylcholine and sphingomyelin by conditional neutral loss scanning and MS3. Rapid Commun Mass Spectrom. 2004;18:3123–3130. doi: 10.1002/rcm.1737. [DOI] [PubMed] [Google Scholar]
- 35.Iavarone AT, Jurchen JC, Williams ER. Supercharged protein and peptide ions formed by electrospray ionization. Anal Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi SD, Hendrickson CL, Marshall AG. Counting individual sulfur atoms in a protein by ultrahigh-resolution Fourier transform ion cyclotron resonance mass spectrometry: experimental resolution of isotopic fine structure in proteins. Proc Natl Acad Sci U.S.A. 1998;95:11532–11537. doi: 10.1073/pnas.95.20.11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi SDH, Drader JJ, Hendrickson CL, Marshall AG. Fourier transform ion cyclotron resonance mass spectrometry in a high homogeneity 25 tesla resistive magnet. J Am Soc Mass Spectrom. 1999;10:265–268. [Google Scholar]
- 38.Schaub TM, Hendrickson CL, Horning S, Quinn JP, Senko MW, Marshall AG. High-performance mass spectrometry: Fourier transform ion cyclotron resonance at 14.5 tesla. Anal Chem. 2008;80:3985–3990. doi: 10.1021/ac800386h. [DOI] [PubMed] [Google Scholar]
- 39.Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 40.Jardin-Mathe O, Bonnel D, Franck J, Wisztorski M, Macagno E, Fournier I, Salzet M. MITICS (MALDI Imaging Team Imaging Computing System): a new open source mass spectrometry imaging software. J Proteomics. 2008;71:332–345. doi: 10.1016/j.jprot.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Broersen A, van Liere R, Altelaar AF, Heeren RM, McDonnell LA. Automated, feature-based image alignment for high-resolution imaging mass spectrometry of large biological samples. J Am Soc Mass Spectrom. 2008;19:823–832. doi: 10.1016/j.jasms.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 42.Hjerno K, Hojrup P. Calibration of matrix-assisted laser desorption/ionization time-of-flight peptide mass fingerprinting spectra. Methods Mol Biol. 2007;367:49–60. doi: 10.1385/1-59745-275-0:49. [DOI] [PubMed] [Google Scholar]
- 43.Luo Q, Nieves E, Kzhyshkowska J, Angeletti RH. Endogenous transforming growth factor-beta receptor-mediated Smad signaling complexes analyzed by mass spectrometry. Mol Cell Proteomics. 2006;5:1245–1260. doi: 10.1074/mcp.M600065-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Wolski WE, Farrow M, Emde AK, Lehrach H, Lalowski M, Reinert K. Analytical model of peptide mass cluster centres with applications. Proteome Sci. 2006;4:18. doi: 10.1186/1477-5956-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]