Abstract
The capacity of bacteria to survive and grow at alkaline pH values is of widespread importance in the epidemiology of pathogenic bacteria, in remediation and industrial settings, as well as in marine, plant-associated and extremely alkaline ecological niches. Alkali-tolerance and alkaliphily, in turn, strongly depend upon mechanisms for alkaline pH homeostasis, as shown in pH shift experiments and growth experiments in chemostats at different external pH values. Transcriptome and proteome analyses have recently complemented physiological and genetic studies, revealing numerous adaptations that contribute to alkaline pH homeostasis. These include elevated levels of transporters and enzymes that promote proton capture and retention (e.g. the ATP synthase and monovalent cation/proton antiporters), metabolic changes that lead to increased acid production, and changes in the cell surface layers that contribute to cytoplasmic proton retention. Targeted studies over the past decade have followed up the long-recognized importance of monovalent cations in active pH homeostasis. These studies show the centrality of monovalent cation/proton antiporters in this process while microbial genomics provides information about the constellation of such antiporters in individual strains. A comprehensive phylogenetic analysis of both eukaryotic and prokaryotic genome databases has identified orthologes from bacteria to humans that allow better understanding of the specific functions and physiological roles of the antiporters. Detailed information about the properties of multiple antiporters in individual strains is starting to explain how specific monovalent cation/proton antiporters play dominant roles in alkaline pH homeostasis in cells that have several additional antiporters catalyzing ostensibly similar reactions. New insights into the pH-dependent Na+/H+ antiporter NhaA that plays an important role in Escherichia coli have recently emerged from the determination of the structure of NhaA. This review highlights the approaches, major findings and unresolved problems in alkaline pH homeostasis, focusing on the small number of well-characterized alkali-tolerant and extremely alkaliphlic bacteria.
Keywords: pH homeostasis, Na+(K+)/H+ antiporters, alkaline pH homeostasis, NhaA, Mrp, MdfA, Tet(L)
1. Introduction
Bacteria must maintain a cytoplasmic pH that is compatible with optimal functional and structural integrity of the cytoplasmic proteins that support growth. Most non-extremophilic bacteria grow over a broad range of external pH values, from 5.5–9.0, and maintain a cytoplasmic pH that lies within the narrow range of pH 7.4–7.8 [1–4]. Hence, they are able to acidify or alkalinize the cytoplasm relative to the external milieu. The consequences of being unable to do so are striking. Caloramator fervidus (previously named Clostridium fervidus [5]) possesses bioenergetic cycles that are entirely Na+-coupled and thus it lacks active H+ extrusion or uptake systems that can support pH homeostasis. As a result, it grows only within the narrow pH range of 6.3–7.7 that corresponds to an optimal cytoplasmic pH [6]. Enterococcus (formerly Streptococcus) faecalis can grow in the presence of ionophores that inhibit the H+ circulation required for pH homeostasis if the cells are grown in a rich medium that is K+-replete, low in Na+ and has a pH near pH 7.6 [7]. Acid pH homeostasis has been extensively studied and recently reviewed [8–12]. Here we review bacterial alkaline pH homeostasis, which also features prominently in life cycles of pathogens [13, 14], as well as ecologically and industrially important bacteria [15–17].
A shift to an alkaline environment, like a shift to an acid environment, is stressful for bacteria as shown by how Escherichia coli responds to alkali with SOS and heat shock-like responses [18–20]. It is important to distinguish between survival and growth in response to alkali. Survival involves no net increase in viable cells at the high test-pH and is usually accompanied by some loss of viability. Survival is monitored by counts of viable cells as colony-forming units on a neutral medium. Since survival is often monitored during stationary phase, it should be noted that cultures that are not well-buffered exhibit a marked rise in pH in many conventional growth media, e.g. LB [21]. Growth is accompanied by a logarithmic increase in viable cell count under the alkaline condition although the growth rate is reduced relative to the rate at the optimal neutral pH [20, 22]. Turbidity (A600) is frequently used to monitor growth, but viable counts are preferable here too because changes in shape as well as a non-viable cell component can introduce artifacts into the absorbance. The distinction between survival and growth at high pH raises the question of whether the underlying mechanisms are identical, overlapping or completely different.
Many natural habitats for microbes have high pH. Marine bacteria grow in alkaline marine environments at ~ pH 8.2 [23] and other bacteria spend part of their life cycle in marine environments, where they must survive or grow [24, 25]. Within the human body, high pH (≥10) occurs at the pancreatic duct just below the pylorus [20, 26]; thus enteric flora must exhibit substantial alkali-resistance. Steady-state high pH is found consistently in regions of many insect guts [20, 26–28]. A haloalkaliphilic archaeon Natronococcus is part of the termite gut flora [29], where a pH of 12 is reached [27]. Additional, extremely alkaline environments include saline alkaline soda lakes, non-saline alkaline lakes and man-made enrichments that are by-products of industrial activities such as the indigo dye production facilities from which the first alkaliphiles were isolated [30–34].
A large number of adaptive strategies are deployed for alkaline pH homeostasis and will be discussed in this review both in the context of the methods used to identify them and the context of the organism in which they are found. The strategies include: (i) increased metabolic acid production through amino acid deaminases and sugar fermentation; (ii) increased ATP synthase that couples H+ entry to ATP generation; (iii) changes in cell surface properties and (iv) increased expression and activity of monovalent cation/proton antiporters. Among these strategies, monovalent cation/proton antiporters play an essential and dominant role in alkaline pH homeostasis in many bacteria in addition to roles in Na+ and volume homeostasis [3, 35–41] Similarly, Na+/H+ antiporters play essential roles in homeostasis of pH, Na+ and volume in eukaryotic cells and organelles [37, 42–45].
Bacteria have multiple monovalent cation/proton antiporters (e.g. at least four in E. coli [2, 37], five in B. subtilis [46–49] and four in alkaliphilic Bacillus strains [40, 50–52] see Fig. 1). Detailed information about individual Na+/H+ and Na+(K+)/H+ antiporters is beginning to provide insights into the basis for the dominant role of specific antiporters in pH homeostasis of individual bacterial strains. This review will focus on several Gram-negative and Gram-positive bacteria in which studies of individual antiporters as well as genomic, global proteome and/or transcriptome information is available in relation to alkaline pH homeostasis. The methodological approaches used, the major findings and the major unresolved questions will be discussed.
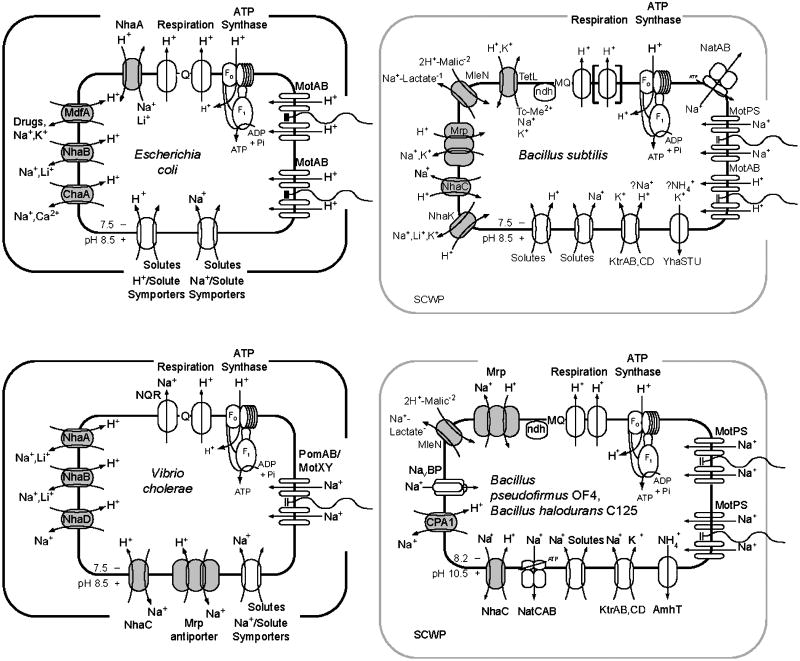
Figure 1.
Na+/H+ antiporters, the Na+ cycle and selected H+- and K+-translocating proteins in membranes of four physiologically distinct bacteria. Transporters of the inner membrane of two Gram-negative neutralophilic examples, E. coli and V. cholerae, are schematically depicted within the inner membrane surrounded by the outer membrane. E. coli: a respiratory chain with Q representing ubiquinone or meanquinone, ATP synthase (F0F1) and flagellar motor that are all H+-coupled [236, 237]; both H+- and Na+-coupled solute symporters [238, 239]; and four monovalent cation/proton antiporters, one of which is also a multidrug/proton antiporter [37, 122, 144, 238]. V. cholerae: a respiratory chain that extrudes both Na+ and H+ and H+-coupled ATP synthase; Na+-coupled solute symporters and polar flagellar motor [13, 173] (whereas marine Vibrio species also have H+-coupled lateral flagella [240]); and at least five monovalent cation/proton antiporters; an annotated NhaP and MleN are not shown ([14, 37, 241], www.membranetransport.org). Transporters in Gram-positive neutralophilic B. subtilis and the alkaliphilic species B. pseudofirmus OF4 and Bacillus halodurans C-125 are shown in the cytoplasmic membrane surrounded by a peptidoglycan layer that has associated SCWP (secondary cell wall polymers). The Bacillus species have H+-extruding respiratory chains (MQ=menaquinone), H+-coupled ATP synthases [88, 242, 243], an ABC-type Na+ efflux system [244, 245], and a CPA:2-type K+-extruding system that has been shown to also extrude NH4+ in B. pseudofirmus OF4 [80, 81]. B. subtilis: additionally has both H+- and Na+-coupled solute symporters and flagellar motors [214, 246], five characterized monovalent cation/proton antiporters [47–49, 193, 247] and two Ktr-type K+- uptake systems whose ion-coupling properties are not established [136]. B. pseudofirmus OF4 has exclusively Na+-coupled solute symporters and flagellar rotors [88, 214], two monovalent cation/proton antiporters thus far partially characterized [40] and a voltage-gated Na+ channel that plays a role in supporting pH homeostasis as well as chemotaxis and motility[215]. The B. halodurans C-125 genome indicates the presence of MleN and a CPA1 type antiporter [52].
2. Assays of pH homeostasis
The cytoplasmic pH of bacteria has usually been calculated from a pre-set external pH and the ΔpH across the cytoplasmic membrane of the cell. Most assays of the ΔpH, acid inside relative to outside, use labeled probes that are membrane-permeant weak bases. These weak bases distribute across the membrane according to the ΔpH and therefore accumulate within the more acidic cytoplasm that bacteria maintain when the external pH is high [53, 54]. The weak base is either radiolabeled or is fluorescent, with fluorescence quenching occurring as the probe is internalized. Weak acids are similarly used to measure a ΔpH, alkaline inside relative to outside; it is best to do both a weak acid and weak base distribution assay when the ΔpH is small. 31P-NMR has been used over a limited range of pH values to assess cytoplasmic pH in E. coli, but the conditions for such assays do not always replicate normal growth conditions because of the high cell density needed [3]. The cytoplasmic pH of bacteria has also been assayed using pH-sensitive fluorescent reporter proteins or dyes, such as green fluorescent proteins [12] or BCECF [55–57].
Measurements of the cytoplasmic pH of cells growing or surviving at different external pH values provide information about the limits of alkaline pH homeostasis when correlated with the viable count (at a permissive pH) of the bacterium. A rigorous assessment of the pH homeostatic capacity is further obtained from pH shift experiments, which are optimally conducted under carefully controlled pre- and post-shift pH values [58]. An alkaline shift in the external medium can be imposed on growing cells in pH-controlled batch cultures ([58] and Fig. 2A) or in pH stat (a chemostat in which pH is strictly controlled) [54, 59, 60]. Alternatively, cells are harvested at particular growth stages and equilibrated in pre-shift buffer until the cytoplasmic and external pH values are approximately the same; the shift is then initiated under specific conditions of energization and cation content ([51, 61] and Fig. 2B). Survival data should be collected by pre- and post-shift viable counts (at a permissive pH) to be sure that survival is sufficient for meaningful cytoplasmic pH measurements. There are several parameters that can be varied in the protocols: (i) the size and range of the shift; (ii) the time over which the shift is imposed, i.e. sudden vs. gradual; (iii) the rate and duration of the measurements; (iv) the ion composition of the medium. Since, in general, the mechanism of alkaline pH homeostasis involves cation/H+ antiporters, the ion specificity of pH homeostasis can identify which antiporter might be involved and how.
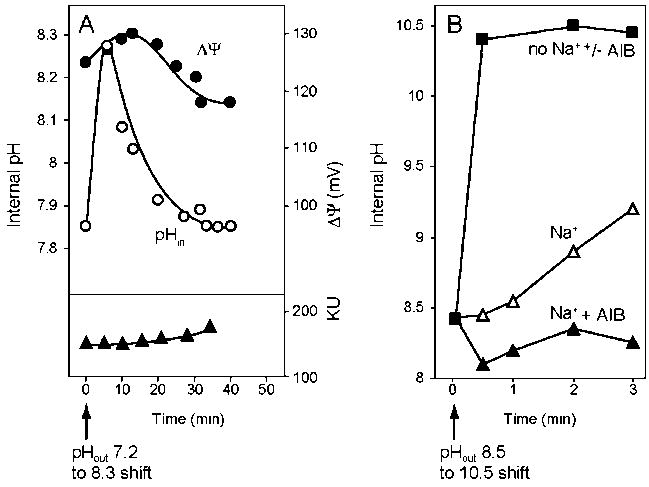
Figure 2.
Alkaline pH shift experiments with E. coli and alkaliphilic B. pseudofirmus RAB. A. E. coli cells were grown logarithmically at pH 7.2 in minimal glycerol medium batch cultures before the pH of the medium was adjusted to 8.3 over 30 seconds. A rapid method of cytoplasmic pH and ΔΨ determination in intact cells has made it possible to follow the time course and sequence of changes occuring following the pH shift [58]. Growth was also assed (Klett units). The data are replotted from Figure 3 in reference [58] with permission from the American Society for Microbiology. B. Washed pH 10.5-grown cells of B. pseudofirmus RAB were equilibrated in potassium carbonate buffer at pH 8.5 and then rapidly shifted to pH 10.5 buffer with either 50 mM potassium carbonate (“no Na+”, with or without added AIB), 50 mM sodium carbonate (“Na+”) or 50 mM sodium carbonate + 500 μM AIB. The cytoplasmic pH was monitored after the shift. The figure is a modified version of Figure 3 from reference [61], with permission from the American Society for Biochemistry and Molecular Biology.
3. Methods for identifying the proteins responsible for pH homeostasis
3.1. Global approaches
3.1.1. Proteomics
Two dimensional gel electrophoresis of the proteome of bacteria grown at various pH values identifies changes in the level of individual proteins in response to a change in external pH. The proteins are identified by mass spectrometry. Findings from such screens are well-represented by the study of Stancik et al. [62] in E. coli. Under aerobic conditions (in LB medium, which contains tryptone, yeast extract and NaCl), the abundance of a variety of proteins is increased by high pH, including the periplasmic protein YceI, MalE and OmpA porins and the virulence-associated OmpX porin [62]. These changes presumably lead to metabolic patterns that are adaptive. For example, MalE would increase availability of an acid-generating carbohydrate. Several enzymes of amino acid catabolism are also increased in abundance at alkaline pH, including TnaA (tryptophan deaminase) and TnaB (the tryptophan transporter) as well as serine deaminase (SdaA). Deaminases provide an acid-generating mechanism that is adaptive to alkaline challenge just as decarboxylases promote alkalinization that is adaptive for an acid challenge [12, 63, 64], as predicted by early work by Gale and colleagues [65].
Increased production of TnaA has also been implicated in a specific and remarkable defense against alkali when anaerobic E. coli uses TMAO as its terminal electron acceptor. Use of TMAO generates an alkaline reduction product and in apparent “anticipation” of that alkaline challenge, the Tor phospho-relay system that detects TMAO increases expression of tnaA. This results in a functional TnaA- and tryptophan-dependent defense against alkali [66]. It has not yet been shown whether deaminase activities lower cytoplasmic pH, external pH or both. Overall the proteomic data are consistent with a model in which E. coli modulates multiple metabolic pathways of amino acid consumption so as to minimize the effect of an increase in the external pH [63]. A mechanism based on a change of external pH itself can only be realistic when the culture volume is small and/or the fluid layer surrounding the cells is unstirred so that its composition is not in equilibrium with that of the bulk external fluid. In general, the internal volume of cells is much smaller than the external volume. Hence, modest changes in intracellular acid or base production and/or in the protons pumped out of or taken up by the cytoplasm can have a significant impact on maintenance of a constant intracellular pH.
The proteome of a B. subtilis mutant disrupted in Bs-mrpA (a Na+(K+)/H+ antiporter, see 5.3.4.) and grown at high pH shows elevated levels of salt stress proteins as well as of the ATP synthase [67]. The latter is consistent with the alkali-adaptive value of ATP synthase inferred in E. coli ([20] and see 3.1.2.1.). By contrast, ATP synthase genes are down-regulated during acid-adaptation of H. pylori [68] and in fermentative bacteria, the H+-pumping, hydrolytic mode of the ATPase is a critical piece of acid-adaptation [69–71].
3.1.2. Genomics - microarrays
In this approach cDNA is synthesized labeled and hybridized to antisense genomic DNA arrays. Positive hybridization signals quantitatively reflect the level of gene expression and can be compared as a function of growth pH or before and after a shift in external pH.
3.1.2.1. E. coli
When grown in a buffered LBK medium (highly buffered LB in which KCl replaces the NaCl) at pH 7.0, E. coli cells exhibit a generation time of 18 min vs. pH 8.7, where the cells exhibit a generation time of 25 min [20]. Microarray studies show that among the genes repressed at the alkaline pH are genes encoding two respiratory chain complexes that pump protons outward during electron transport, cyo and nuo, as well as flagellar and chemotaxis genes [20]. Induced genes include those encoding the F1F0-ATP synthase that imports protons during ATP synthesis, and the alternate cyd-encoded terminal oxidase, that generates a PMF without outward proton pumping. These observations indicate that alkali challenge, under growth conditions, changes the gene expression profile: robust oxidative phosphorylation together with the ATP synthase maximize proton retention and proton capture by the cell. This elegant study also shows that pH differentially regulates a large number of periplasmic and envelope proteins. Although the experimental conditions are not directly comparable, it is interesting to note an apparent difference in expression patterns in a microarray study conducted with E. coli in sea water (where the organism survives but does not grow) [25] vs. a glucose-containing laboratory medium [72]. It had earlier been found that the sea water survival condition induces RpoS-controlled genes [73], a type of regulation usually involved in stationary phase ([74, 75] and see 3.2.3.). The microarray data further show that genes involved in cell division and nucleotide biosynthesis are repressed, consistent with cessation of growth. On the other hand, motility and chemotaxis genes as well as many metabolic genes are induced in sea water [72]; under these conditions induction rather than repression of motility and chemotaxis may support the search by the surviving E. coli cells for a niche that can support growth.
3.1.2.2. B. subtilis
An alkaline shift protocol has been applied to B. subtilis [60]. In LB medium at pH of 6.2, B. subtilis initially grows rapidly and acidifies the medium. This is followed by a slower growth rate during which the medium is alkalinized to about pH 7.5. B. subtilis does not survive a sudden shift to pH ≥ 9.3 imposed just before the growth slow-down phase. A more modest shift, to pH 9.0, results in a growth arrest for five hours during which is a significant loss of viable cells, while the medium acidifies gradually. Growth resumes at a slower rate when the outside pH is about 8.5, reaching the same final cell density as the nonshifted control.
Microarray studies have been conducted using the same shift protocol, comparing the pre-shift gene expression profile and the profile after a shift to pH 8.9 [60]. A group of the up-regulated genes is under the control of σw, one of the extracytoplasmic function family of σ factors (ECF) that commonly regulate groups of genes in response to extracytoplasmic stimuli [76]. It has been proposed that the induction of the σw regulon reflects an intersection of alkali-stress with cell wall stress and Na+-stress ([77] see also 7.). Additional genes that are up-regulated after the alkaline shock include Bs-mrp [60] genes that encode the Mrp Na+(K+)/H+ antiporter ([41, 49, 78, 79] and see 5.3.4). This is important since, as noted below (3.1.4.), the micro-array technique often does not detect significant up-regulation of genes encoding membrane transporters including antiporters of known involvement in alkali-stress. Another transporter gene locus that is up-regulated in the alkaline shock experiment is the YhaSTU locus that catalyzes K+ efflux in B. subtilis and is regulated in a complex way by alkaline pH and Na+ stress [80]. YhaSTU may extrude NH4+ (in addition to K+), as does its alkaliphile homologue AmhT [81]. This could contribute to acidification of the medium (Fig. 1).
3.1.3. lacZ fusions
In this approach in-frame promoter or genes encoding protein fusions are randomly introduced to the genome by transposition with the lacZ reporter gene [82]. The strains bearing the fusion are identified genetically and by the activity of the reporter gene lacZ. Depending on the fusion type, LacZ activity reflects transcriptional or translational regulation or both. In E. coli, a novel alkali-induced gene locus, alx, was identified by this approach [83]; its product was shown to be a membrane-associated redox modulator [62]. In a comparable gene fusion approach, plasmid-based libraries of promoter-reporter fusions (to a luxCDABE reporter) were introduced into E. coli and used to detect promoters that were activated in sea water, including the rpoS-dependent promoters [73]. The promoter fusion approach has thus highlighted interplay of alkali-stress with both stationary phase stress and oxidative stress (see 7.).
3.1.4. Caveats concerning use of the global approaches
The global approaches provide a valuable integrated picture of responses of genes and proteins to various growth conditions and stresses. However, there are several drawbacks that especially pertain to integral membrane proteins. The proteomic approach cannot appropriately resolve many of these proteins, which constitute around 20% of the bacterial proteome. The major reason is that integral membrane proteins are very hydrophobic proteins and often cross the membrane several times in a zig-zag fashion; their hydrophobicity commonly introduces artifacts in 2D as well as in ordinary gel electrophoresis [84, 85] and some preparatory steps used for 2D samples (e.g. boiling) can induce their aggregation [84]. This is compounded by the fact that the proteins of greatest interest are generally present at very low levels in the membrane (e.g. the Ec-NhaA antiporter constitutes < 0.2% of the E. coli membrane proteins [84]). Another problem in applying microarray approaches to membrane proteins such as transporters is that small changes in expression (e.g. 2–3 fold [46]) can have a significant physiological effect, but many microarray studies fail to capture such changes.
Another caveat is that neither proteome nor transcriptome studies can identify proteins that are regulated at the level of activity, a feature that is important in antiporters that participate in alkaline pH homeostasis (see 4.3.). As yet, this property can be discerned only by combining classical genetics (including gene fusion approach) with biochemical approaches. It is not surprising therefore that very few antiporter genes or proteins have been identified in alkali-adaptation screens by any of the global approaches. Yet, their importance has unequivocally been shown by targeted studies of specific antiporter genes and proteins.
3.2. Focus on single genes and their products of potential interest for alkaline pH homeostasis
3.2.1. Identification of genes/proteins involved in alkaline pH homeostasis
For targeted identification of specific genes to be practical, a clue is needed as to which gene/protein might be involved in alkaline pH homeostasis. The global proteomic and genomic assays can provide such clues, but critical insights are often obtained from physiological studies. When Na+/H+ antiporters were described by West and Mitchell in intact E. coli cells [86], their potential for a role in adaptation to alkali pH was noted, initiating research on the Na+/H+ antiporters. The absolute requirement for Na+ for pH homeostasis in alkaliphilic bacteria and diminished Na+/H+ antiporter activity in non-alkaliphilic mutants then provided strong support for an antiporter based pH homeostasis mechanism [39, 87, 88]. Study of a Li+ resistant mutant [89] led to the first cloning of an antiporter gene, Ec-nhaA of E. coli [90, 91] and its regulator Ec-nhaR [92]. Ec-NhaA is the main antiporter of E. coli and many enterobacteria [37].
The genome project provided a flood of eukaryotic and prokaryotic putative antiporter genes of which products were grouped by Saier and colleagues in the monovalent cation proton antiporter superfamily (CPA) http://tcdb.ucsd.edu/tcdb/ [93]. A recent phylogenetic analysis of database information shows that NhaA belongs to CPA2, a sub family of the monovalent cation/H+ antiporters, and its orthologs exist in very many prokaryotic genomes as well as eukaryotic genomes including that of humans [45]. The CPA1 family includes the main eukaryotic NHE Na+/H+ antiporters and has many prokaryotic orthologs such as Mj-NhaP1 ([45] and see 4.3.2.).
The Mrp-type antiporters, classified as the CPA3 faimly, are the main antiporters of alkaliphilic bacilli [94]. They were discovered when a partial mrp/sha operon was found to complement a pH homeostasis-negative and Na+-sensitive phenotypes of an alkaliphilic Bacillus halodurans C-125 mutant [50]. Transpositional insertion libraries of B. subtilis for Na+- and alkali-sensitive mutants led to the identification of Bs-Tet(L) as a multifunctional (tetracycline-metal+)(Na+)(K+)/H+ antiporter with a role in pH homeostasis [47]. Once antiporter-deficient strains of E. coli were constructed, functional complementation screens have become a common route to clone new antiporters genes and study the encoded antiporters ([95–98] and see 3.2.2.).
3.2.2. Changing the dosage of the candidate gene to probe its physiological role
The physiological role of a putative antiporter in alkaline pH homeostasis is best inferred from the effect of changes in the dosage of the gene on survival and growth at alkaline pH. For example, the Ec-nhaA carried on multicopy plasmid confers upon E. coli a markedly increased growth capacity in the presence of high Li+ [90] or Na+ at neutral pH or growth at alkaline pH in the presence of Na+ [95, 98]. Deletion of Ec-nhaA produces a strain (NM81) that is sensitive to alkaline pH in the presence of Na+ [95]. This alkaline-sensitive strain was used to clone Ec-NhaB, a house-keeping antiporter in E. coli [99], leading to the generation of the double nhaA and nhaB deletion strain (EP432, [100]). E. coli EP432 was used to clone a third antiporter of E. coli, Ec-chaA, that has Na+(Ca2+)/H+ activity ([101, 102], Figure 1). E. coli strains NM81, EP432 and two triple mutants in all three antiporters strains, KNabc and TO114 [103, 104], have been extensively used to identify Na+(K+)/H+ antiporter genes from diverse bacteria and to conduct studies of the properties of the antiporters encoded [37, 105]. However, it should be stressed that for non-E. coli genes this is a heterologous system that cannot provide actual evidence for their roles in the native host. The effects of changing gene dosage in alkali-stressed pH homeostasis and the properties of the antiporters must be examined in the native host, in the correct context.
3.2.3. Studies of gene regulation provide further physiological insights: Ec-NhaR, E. coli nhaA regulator, as a case study
The pattern of transcriptional regulation of Ec-nhaA in E. coli offers a particularly interesting demonstration that the mechanisms regulating a single antiporter in response to alkalinization can be totally different when supporting logarithmic growth than when supporting survival during stationary phase. The expression pattern of the reporter gene lacZ of a nhaA-lacZ fusion and Northern analyses of exponentially growing cells show that transcription of Ec-nhaA is specifically dependent on Na+ and increases at alkaline pH [106]. This regulatory system involves the positive regulator Ec-NhaR [92, 107–109] of the LysR family of regulatory proteins [110–113], which is Na+- and Li+-specific and not modulated by ionic strength or osmolarity. Intracellular rather than extracellular Na+ is the signal for induction [109]. The binding site for Ec-NhaR on the Ec-nhaA was identified using purified Ec-NhaR in both gel retardation and foot printing assays [108]. Ec-NhaR binds the DNA under both inducing and non-inducing conditions. Na+ changes Ec-NhaR conformation so that its footprint on Ec-nhaA is altered in a Na+-specific and alkali-dependent fashion [108]. Two Ec- nhaA promoters, P1 and P2, were identified by primer extension analysis; only P1 is involved in the Ec-NhaR- and Na+-dependent regulation of Ec-nhaA [114].
In stationary phase, Ec-nhaA becomes part of the rpoS regulon and its expression is from the P2 promoter rather than from the P1 promoter used during logarithmic growth [114]. RpoS is a sigma factor that is recruited to the RNA polymerase to transcribe stationary phase specific genes, the rpoS regulon [74, 75, 115, 116]. Accordingly, the reporter activity of P2-lacZ fusion is induced in stationary phase. Primer extension studies show that this induction is dependent neither on Na+ nor on NhaR, but on RpoS [114]. Most importantly, the rpoS dependent P2 is responsible for the survival of the cells in the stationary phase in the presence of Na+, at alkaline pH and at combined high Na+ and alkaline pH [114], the most stressful condition [117].
4. What properties underpin the ability of certain antiporters to support alkaline pH homeostasis?
Almost all bacteria have multiple monovalent cation/proton antiporters that form part of complex H+ and Na+ cycles in these cells (Figure 1). As best studied in E. coli and B. subtilis, these antiporters do not contribute equally to alkaline pH homeostasis. This raises the question of what unique properties are needed for an antiporter to play a major role in this process. Properties of the antiporter that are considered include: the stoichiometry and kinetics of the exchange; the KD of substrate binding; positive and negative effectors of the activity; structure-function relationships and the reaction mechanism. Many of the properties of antiporters have been studied in right-side-out [118] and everted membrane vesicle systems [119], often using expression of the antiporter from a plasmid in appropriate antiporter-deficient strains of E. coli [96]. The use of these membrane preparations facilitates rapid screening of many conditions and properties.
However, there are two general caveats with respect to the properties determined in everted membrane vesicles. First, it is impossible to exclude the possibility that proteins, other than the test antiporter, are affecting the assay results. Second, if the membranes are from E. coli and the antiporter is from a different bacterial source, the properties may be altered by the phospholipid and heterologous protein complement of the membranes. A striking example of this is the change in cation coupling of the Bacillus stearothermophilus and Bacillus caldotenax L-glutamate transporters from 1Na+ + 1H+ in the native membranes to ≥2H+ in E. coli membranes [120]. Therefore, it is of great importance to corroborate major findings using proteoliposomes, preferably made from the phospholipids of the natural bacterial host, into which purified antiporter is reconstituted. Among the antiporters involved in alkaline pH homeostasis, this was first accomplished with Ec-NhaA [84] and has since been achieved with several others [121, 122].
4.1. Electrogenicity
4.1.1. Importance of electrogenicity for Na+/H+ antiport-dependent pH homeostasis
For pH homeostasis (maintenance of intracellular pH around 7.6) at external alkaline pH (>7.6), net cytoplasmic accumulation of H+ must occur. When Na+/H+ antiporters are used for pH homeostasis, the antiport has to be electrogenic rather than electroneutral [39, 98, 123]. An electroneutral exchange of cytoplasmic Na+ for external H+ is a 1:1 exchange in which no net flux of charge is involved, so the ΔΨ component of the PMF cannot contribute to energization of the exchange. Bacteria, like other living cells, exclude Na+ from the cytoplasm by diverse extrusion mechanisms because it is cytotoxic [117]. Therefore there is no outwardly directed Na+ gradient to support electroneutral Na+/H+ antiport. Hence, an electroneutral antiport can equilibrate the ΔpH component of the PMF, but cannot catalyze a net H+ accumulation needed to acidify the cytoplasm at alkaline pH. Electrogenic Na+/H+ antiporters exchange a larger number of entering H+ than the number of exiting Na+ during a turnover, so that net positive charge moves inward. Therefore, the ΔΨ (negative inside relative to the outside) drives the inward H+ movement and cytoplasmic acidification relative to the outside can be achieved.
4.1.2. Determination of coupling stoichiometries
As exemplified by Ec-NhaA and Ec-NhaB, the coupling stoichiometries of antiport must be determined in proteoliposomes containing the purified antiporter proteins [124, 125]. Two approaches have been used: in a kinetic approach, the efflux of Na+ and influx of H+ are measured in parallel and the stoichiometry is calculated from the ratio of the fluxes under conditions in which the ΔΨ and the ΔpH (or ΔpNa+) created by the antiporter do not limit its rate [124]; in a steady-state thermodynamic approach, various Na+ chemical gradients are imposed across the proteoliposomal membrane under conditions in which the ΔpH is collapsed. The antiporter-dependent membrane potential that develops is measured, allowing calculation of stoichiometry assuming thermodynamic equilibrium and complete coupling [124]. Based on both methods, the coupling stoichiometry of Ec-NhaA is 2H+/1Na+ [124]. The stoichiometry of Ec-NhaB is 1.5H+/1Na+ [125]. These findings suggest that this small difference in electrogenicity underpins the ability of Ec-NhaA but not Ec-NhaB to support growth of E. coli at alkaline pH in the presence of Na+ [100].
4.1.3. Assessing electrogenicity vs. electroneutrality without defining coupling stoichiometry
For other Na+/H+ and Na+(K+)/H+ antiporters that function in alkaline pH homeostasis, precise coupling stoichiometries have not yet been determined. However, electrogenicity vs. electroneutrality of antiport activity is inferred from the effect of selectively abolishing the ΔΨ or the ΔpH component of the PMF. ΔΨ is abolished by permeant anions or cations. Commonly, the potassium ionophore valinomycin is used together with K+ to abolish ΔΨ. ΔpH is usually dissipated by using low concentrations of nigericin; K+ must be present and high concentrations of nigericin must be avoided because electrogenic exchange can occur under those conditions and abolish both components of the PMF [126, 127]. Use of both nigericin and valinomycin together, in the presence of K+, or use of a protonophore (CCCP) abolishes both the ΔΨ and ΔpH and serves as a negative control. Abolition of the ΔΨ is accompanied by an increase in ΔpH and vice versa [1, 128]. Therefore, abolition of the ΔΨ leads to stimulation of electroneutral antiporters while abolition of the ΔpH inhibits these antiporters. Electrogenic antiporters are inhibited by abolition of the ΔΨ even though the ΔpH increases; antiport inhibition occurs because when ΔpH (directed outward in everted membrane vesicles) is the sole driving force, the charge that is transported by an electrogenic antiporter produces a ΔΨ, positive out, that limits the rate of the antiport in the everted system. Two more approaches are used to demonstrate an electrogenic antiport. One is to impose a gradient of the cationic substrate and use ΔΨ probes to show that an antiporter- and cation-dependent electrical potential is generated [124, 129, 130]; it should be noted that chloride ion is permeant in E. coli membranes and should not be included in buffers where ΔΨ is being measured [131]. The second approach is to drive the antiport with an imposed H+ or a Na+ chemical gradient and show that permeant ions increase the rate of the antiporter [132].
4.2. Substrate and cation specificity
Alkaline pH homeostasis can be achieved by antiporters other than the Na+(Li+)/H+ specific antiporters. An example is the electroneutral Bs-MleN, a paralogue of the Na+/H+ antiporter Bs-NhaC [133]. Bs-MleN catalyzes 2H+-Malate−2/Na+-Lactate−1 exchange (Fig. 1). When driven by inwardly directed malate and outwardly directed lactate gradients, the external malate enters with H+ and the cytoplasmic lactate exits with Na+. Therefore the exchange supports net cytoplasmic H+ accumulation although a role in pH homeostasis of B. subtilis has not yet been tested [133]. In theory, an electroneutral K+/H+ antiport could also support net H+ uptake since bacteria usually maintain an outwardly directed K+ gradient that could serve as the driving force [134–136]. However, since bacterial growth is inhibited when cytoplasmic K+ is depleted, even a carefully regulated electroneutral K+/H+ antiporter might be a liability. On the other hand, when an electrogenic Na+(K+)/H+ antiporter such as multifunctional Bs-Tet(L) and Bs-Mrp antiporter [47, 49, 121] use K+, the outwardly directed K+ chemical gradient is an additional component to the driving force provided by the PMF. Involvement of K+/H+ antiport in alkaline pH homeostasis has been reported in several bacteria but the responsible genes and proteins have not been identified and characterized [137–139]. NhaK, a B. subtilis transporter belonging to the widely distributed CPA1 antiporter family [140], catalyzes K+/H+ antiport at alkaline pH [48]. This recent finding suggests that the K+/H+ antiport activity observed in other bacteria could depend upon other members of this transporter group, i.e. NhaK, NhaP and NhaG homologs [48].
4.3. pH regulation at the level of antiporter activity
The antiporters that are critically involved in alkaline pH homeostasis should be equipped with sensors of the external and/or cytoplasmic pH. They then must be able to transduce this environmental signal to produce a change in antiporter activity, so as to maintain pH homeostasis [98]. Failure to activate the antiporter will result in cytoplasmic alkalinization at alkaline extracellular pH [141]. Failure to shut off the antiporter could lead to over-acidification at lower pH values and energy depletion at higher pH values [132]. Indeed, one of the most characteristic properties of both eukaryotic and prokaryotic Na+/H+ antiporters is pH-dependent regulation of their activity at the protein level [37, 43, 142–147].
4.3.1. E. coli NhaA as a paradigm
Ec-NhaA is a paradigm for a prokaryotic, pH-regulated antiporter [144]. Thus, in proteoliposomes containing pure Ec-NhaA as the only protein, Ec-NhaA exhibits a pH response [84] that is identical to that in the native membrane [98]. It shuts off below pH 6.5. As the pH is shifted up to pH 8.5, the NhaA Vmax increases over three orders of magnitude but does not increase further as pH is raised above pH 9. The pK of the pH response of Ec-NhaA is 7.6 that equals the “set point” for the intracellular pH [1]. At intracellular pH values above the “set point”, Ec-NhaA will be activated so as to restore pH homeostasis. At intracellular pH below the “set point” it will be inactivated and it shuts off at pH 6.5, a mechanism that avoids over-acidification of the cytoplasm. When the set point of Ec-NhaA is altered by mutation the result is a change in the pH profile of growth. For example, the G338S mutation of Ec-NhaA results in an antiporter that has lost pH control [132]. The protein is active throughout the range of pH 6.5–9. The G338S mutant is highly pH sensitive at alkaline pH, presumably because the excessive activity at high pH depletes the cells of energy [132]. The pH-dependence of the Ec-NhaA protein with an H225R mutation is shifted towards acidic pH [141, 148] while that of an E252C mutant is shifted towards alkaline pH [149]. These changes are accompanied by changes in the pH profiles of growth in the same direction [141, 149].
Insights have been gained into how the Ec-NhaA “pH sensor” transduces the pH signal into a change in antiporter activity. There is only partial overlap between the amino acid residues that affect pH regulation and those that affect transport activity [144, 150, 151]. This implies that, as in eukaryotic Na+/H+ antiporters, there is a pH regulatory site that is distinct from the active site [43, 144, 152, 153]. In Ec-NhaA many of the amino acids that are involved in pH regulation cluster at the N-terminus [154] and the loop between TMS VIII-IX [85, 144, 149] that, as revealed by cross-linking, are in close proximity [144, 155]. Furthermore alkali-dependent activation of Ec-NhaA is accompanied by a conformational change involving the N-terminus [154] and the loop between TMS VIII-IX [85, 149].
4.3.2. Antiporter pH profiles: variations on the theme
Correlations between growth and the pH-dependence of antiporters have been noted in other bacteria. In B. subtilis, the Na+(K+)/H+ antiport activity of Bs-Tet(L) exhibits a lower apparent Km for Na+ at pH 8.5 than 7.5 while the apparent Km for the tetracycline-metal complex is higher at pH 8.5 than 7.5 [143]. These pH-dependent differences are adaptive, since Tc is most toxic in the lower pH range (because it enters cells by diffusion of the protonated form). On the other hand, Na+ is more cytotoxic at the higher pH [117]. The NhaP Na+/H+ antiporter from Methanococcus jannaschii, a hyperthermophilic archaeon, exhibits no activity at pH ≥8.0 when expressed in E. coli and cannot complement antiporter-deficient E. coli strains at elevated pH in the presence of Na+. However, Mj-NhaP shows increasing activity as the pH is reduced to 6.0, correlating with the growth profile of its natural host [156]. NhaA from Helicobacter pylori, which is highly homologous to Ec-NhaA, lacks pH regulation and exhibits high activity from pH 6.5–8.5 when assayed in E. coli vesicles [157]. It will be interesting to study the still controversial pH homeostasis mechanism of this bacterium [158, 159]. Notably, there are exceptions to these striking correlations between the pH profile of Ec-NhaA and other antiporters and the growth pH profiles of their natural hosts; this is consistent with existence of other participants in the adaptation to pH.
4.4. Kinetics
A fast antiporter such as Ec-NhaA (turn over of 89000 min−1 [84]) is advantageous for pH homeostasis because it can rapidly meet the challenge of alkali up to pH 9 once it is activated by an elevated cytoplasmic pH. E. coli, cells have been subjected to alkaline shock, during growth at pH 7.2 in a minimal medium with glycerol as carbon source, by imposition of a pH shift in the external pH to 8.3 over 30 seconds ([58, 59], Figure 2A). In the first sample taken after completion of the shift, the imposed pH gradient is observed, with the cytoplasmic pH at the pre-shift value of 7.8. Rapid cytoplasmic alkalinization and an increase in ΔΨ then occur. Within 4 minutes after the shift, the ΔpH is near zero and the cytoplasmic pH is almost as alkaline as the new external pH. During this period, growth is very slow but recovers when the ΔpH is reestablished. More rapid and/or larger shifts in external pH (to pH ≤ 9) result in longer and more complete growth arrest and a much slower growth rate upon recovery [58, 59]. Hence, E. coli experiences and survives substantial cytoplasmic alkalinization after an alkaline shift. Little or no growth occurs while the cytoplasmic pH is above 8.0. Most likely, this cytoplasmic alkalinization is the pH signal that leads to tremendous activation of Ec-NhaA, which in turn establishes a cytoplasmic pH that supports substantial growth. Failure of one or more elements of the cell division mechanism may normally lead to growth arrest in E. coli at pHin ≥ 8.3 ([58] and see 3.1.2.1.).
The extreme alkaliphile, Bacillus pseudofirmus OF4 (formerly Bacillus firmus OF4 [160]) can successfully respond to an even larger and more sudden alkaline shift (e.g. from pH 8.5 to 10.5) without a detectable alkalinization of the cytoplasm as long as Na+ is available to the Bs-Mrp antiporter [61]. This is achieved by including a solute that is co-transported with Na+ in the reaction mixture as shown for B. pseudofirmus RAB in Figure 2B (and see 6.3.). B. pseudofirmus RAB, an obligately alkaliphilic strain, is closely related to the facultatively alkaliphilic B. pseudofirmus OF4 that has been used more extensively in recent studies since its non-alkaliphilic mutants are viable [161, 162]. In addition to the robust pH homeostasis upon a sudden alkaline shift, B. pseudofirmus OF4 differs from E. coli by growing when the cytoplasmic pH exceeds 8.5, i.e. when the external pH > 10.5 ([163] and see 6.1).
4.5. Meeting the challenge of the low external H+ concentration
While a high H+/Na+ stoichiometry makes it possible for an antiporter to use the ΔΨ to acidify the cytoplasm at alkaline pH, it does not solve the problem of capture of the H+ by the antiporter when the external concentration of H+ is very low. No features have yet been explicitly described could trap protons on the surface, comparable to those suggested for some membrane-embedded electron transport- and light-driven proton pumps [164–166]. Structural data such as those described below may reveal testable candidates.
4.6. New insights from the structure of Ec-NhaA
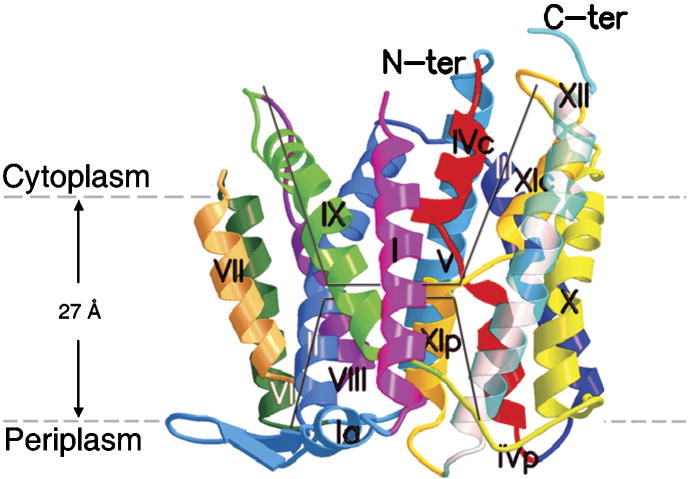
The crystal structure of NhaA, in the down-regulated conformation found at low pH, has recently been determined at 3.45 Å resolution [167] and (Fig. 3). The NhaA architecture is comprised of 12 TMS (Trans Membrane Segments). A negatively charged ion funnel formed of TMSs II, IX, IVc and V, opens to the cytoplasm and ends in the middle of the membrane at the putative ion binding site. There, a unique assembly of short helices of TMS IV and TMS XI are connected and crossed by extended chains. This TMS IV/TMS XI assembly creates a delicately balanced electrostatic environment in the middle of the membrane. A shallow negatively-charged funnel formed by TMSs II, VIII and XIp opens to the periplasm [167] and (Fig. 3). The two funnels point to each other but are separated by a group of densely packed hydrophobic residues forming a periplasmic barrier. On the basis of the structure, it has been proposed that binding of the charged substrates causes electric imbalance allowing for a rapid alternating access mechanism. This ion exchange machinery is regulated by a conformational change elicited by a pH signal perceived at the pH sensor located at the cytoplasmic funnel entry and transduced to the TMSs IV/XI assembly by helix IX [167] and (Fig. 3). This working model is strongly supported by genetic and biochemical data. At the N-terminus of helix IX and in its vicinity pH induced conformational changes occur [85, 149, 154] and many mutations affect the pH response of NhaA [144]. Many residues in TMS IV of the TMS IV/XI assembly are conserved in NhaA protein family and their mutations dramatically affect the apparent Km for Na+ and Li+ [150, 151]. Mutations in conserved residues in TMS XI of the assembly affect the pH response [144]. Furthermore the need for reorientation of helix IVc with respect to XIp is strongly supported by the double mutant F136C/S342C [151]. This mutant forms a disulfide bond that inhibits transport, while reducing the bond restores activity in a reversible manner [151]. Extensive cross-linking data are in accordance with the close contacts between the TMSs of the TMSs IV and XI assembly and their crucial role for activity and pH regulation [150, 151]. The most informative example are the mutants G338S [132] and G338C [151] of the extended chain XI that completely remove the pH control and produce NhaA variants fully active in a pH-independent manner. In the inactive double mutant (T132C/G338C) both pH control and activity can be partially restored by re-establishing the physical interaction between the extended chains by chemical cross-linking [151].
Figure 3.
Overall architecture of NhaA. A ribbon presentation of the NhaA molecule viewed parallel to the membrane (grey broken line) is shown. The 12 TMSs are labeled with Roman numerals. N and C indicate the N and C termini. Note the unique and novel fold formed by unwinding of helices VI and XI in the middle of the membrane. The remaining short helices at the periplasmic or cytoplasmic sides of the membrane are denoted by p or c respectively. The cytoplasmic and periplasmic funnels are depicted by black line. For further details see [167].
The architecture of the Na+/H+ antiporter NhaA reveals the structural basis for the mechanism of Na+/H+ exchange and its unique regulation by pH. The physical separation between the “pH sensor” and the exchange machinery revealed by the structure entails long-range, pH induced conformational changes for pH activation. This observation is in agreement with biochemical data obtained from both prokaryotic [144] and eukaryotic [142, 153] Na+/H+ antiporters. Furthermore since the structure of NhaA represents a novel fold it contributes to the understanding of the architecture of membrane proteins in general.
5. Alkaline pH homeostasis in neutralophilic bacteria
5.1 pH homeostasis in E. coli, a combined effort of NhaA and MdfA antiporters
Even though Ec-NhaA is a paradigm of a pH-regulated antiporter that is suitable for pH homeostasis, Ec-nhaA deletion mutants can grow at alkaline pH as long as no Na+ is added to the medium [98]. These observations suggest that either pH homeostasis is not required under low Na+ conditions or that another system exists in E. coli that controls intracellular pH in the absence of Ec-NhaA. Very recently this puzzle was solved. A deletion of mdfA, a multidrug transporter gene, results in modest alkali-sensitivity [122]. Furthermore, mdfA expression from a multi-copy plasmid enables E. coli to grow, albeit slowly and to a low yield, at pH values as high as 10 in a complex medium. The enhancing effect of Ec-mdfA over-expression on pH homeostasis is illustrated in an experiment which measures the response to a gradual alkaline shift from pH 7.5 to 8.3 or 9.3 of an Ec-mdfA deletion mutant, in comparison with a transformant of the same E. coli strain over-expressing Ec-mdfA from a plasmid (Table 1). Deletion of Ec-mdfA has a marginal effect on the response to the smaller shift but results in significantly reduced pH homeostasis in response to the larger pH shift. No significant capacity for cytoplasmic pH homeostasis is observed by over-expression of MdfA when tested without added Na+ or K+, but upon a shift to pH 9.3 in the presence of either ion, multi-copy mdfA supports maintenance of a cytoplasmic pH of 7.8–7.9 ([122] and Table 1).
Table 1.
E. coli MdfA confers a capacity for Na+- and K+-dependent pH homeostasis during an alkaline shift in the external pHa
| Cytoplasmic pH after a gradual shift in pHout | ||||||
|---|---|---|---|---|---|---|
| pH 7.0→8.3 | pH 7.0→9.3 | |||||
| CholineCl | NaCl | KCl | CholineCl | NaCl | KCl | |
| E. coli ΔmdfA mutant, control | 7.9 | 7.8 | 7.8 | 9.3 | 9.3 | 9.3 |
| E. coli ΔmdfA mutant, mdfA expressed from plasmid | 7.6 | 7.6 | 7.6 | 9.3 | 7.9 | 7.8 |
Cytoplasmic pH was measured by the methylamine accumulation assay after cells were shifted from a growth medium at pH 7.0 to growth media at the final pH values indicated and had established logarithmic growth [122].
In both everted membrane vesicles and MdfA proteoliposomes, MdfA catalyzes Na+/H+ antiport and K+/H+ antiport activity in addition to the earlier described efflux of multiple drug substrates in exchange for H+ [122]. The findings suggest that both Ec-NhaA and Ec-MdfA participate in pH homeostasis of E. coli. Ec-NhaA, with its very high and pH regulated activity (pH 7.6 “set point”), has a major function in the pH range up to pH 9 ([144] and 4.3.1.). Ec-MdfA extends the pH tolerance up to pH 10 and also takes over when Ec-nhaA is deleted. These observations indicate that the presence of the right antiporter is sufficient to enable a non-alkaliphilic bacterium to grow, albeit suboptimally, at an extremely high pH on a rich medium.
Why is Ec-NhaA unable to support E. coli growth at pH >9 while Ec-MdfA can? Possibly the contribution of Ec-NhaA is limited by its ability to capture external H+ at the most alkaline edge of the E. coli pH range. Since Ec-MdfA can utilize K+ as well as Na+ in contrast to Na+(Li+)-specific Ec-NhaA, Ec-MdfA might also have an advantage in the extremely alkaline range in being able to utilize the outwardly directed gradient of K+. We predict that an E. coli mutant deleted of both Ec-nhaA and Ec-mdfA will be a pH-conditional mutant, but such a strain is yet to be constructed. The existence of additional antiporters in E. coli cannot be excluded: two CPA1 type antiporters were deduced from the E. coli genome for which no associated Na+/H+ antiporter activity was demonstrated [168]. Many CPA1 type Na+/H+ antiporters are found in diverse bacteria [48, 169–171] and, as noted, additional ones such as those in E. coli may function as K+/H+ antiporters [48].
5.2. The ecological importance of the antiporters in E. coli and V. cholerae
Enterobacteria such as E. coli and Vibrio cholerae are released into the alkaline sea water and estuaries during their life cycles. In these environments they often survive rather than grow [24, 25]. Measurements of survival of E. coli mutants show that expression of nhaA from the P2 promoter, via the rpoS regulon, is essential for the survival of E. coli in salinic environments at alkaline pH ([114] and 3.2.3.).
The Na+ cycle of V. cholerae plays a role (by a mechanism that is not yet understood) in the virulence of V. cholerae [13, 172] and is expected to play a role in survival of this pathogen in salinic-alkaline environments throughout its life cycle. The genes that participate in Na+ efflux of the V. cholerae Na+ cycle include the nqr operon that encodes an electron transport-linked Na+ pump, the NADH-quinone oxidoreductase complex (NQR) that is also found in marine Vibrio species such as Vibrio alginolyticus ([173, 174], Figure 1). In addition, V. cholerae possesses genes encoding three putative Na+/H+ antiporters that have been studied, Vc-NhaA, Vc-NhaB [14], Vc-NhaD [175]; four nhaC paralogues, a mrp operon, nhaP and mleN are also predicted from genomics (www.membranetransport.org) but have not yet been explored. Vc-nhaA and Vc-nhaB are homologous to the respective E. coli genes and are also found in V. alginolyticus [176, 177] and pathogenic marine Vibrio parahaemolyticus [103, 178]. Vc-nhaD is homologous to that of V. parahaemolyticus [175, 179, 180].
In contrast to E. coli, where a mutant inactivated in the two antiporter genes, nhaA and nhaB, is more susceptible to NaCl than each of the respective single mutants [100], the inactivation of three putative antiporters (Vc-nhaABD) does not alter the exponential growth of V. cholerae in the presence of high Na+ concentrations and has only a slight effect on the bacterial survival in the stationary phase. In contrast, all mutants lacking Vc-nhaA show a Li+ sensitive phenotype during the exponential phase of growth and a marked decrease in survival of the triple mutant is observed in the stationary phase in the presence of Li+ [14]. The capacity of the NQR pump of V. cholerae to specifically extrude Na+ but not Li+ [13, 174] may explain the higher contribution of the V. cholerae Na+/H+ antiporters to Li+ resistance as compared to Na+ resistance. In the absence of three of the Na+/H+ antiporters, the NQR pump can compensate for the Na+/H+ but not the Li+/H+ antiport activity, resulting in a Li+ sensitive but not a Na+ sensitive phenotype. Experiments with a quinone analogue that inhibits NQR support this idea [14]. Although the inhibitor has no effect on the growth of wild type V. cholerae, it greatly inhibits growth of both the Vc-nhaA and Vc-nhaABD mutants in the presence of elevated Na+ and pH [14]. These results suggest that NhaA is involved in the Na+ and H+ homeostasis of V. cholerae at alkaline conditions but its contribution is revealed only when the Na+ pump activity of NQR is inhibited.
5.3. B. subtilis has interlocking H+, Na+ and K+ cycles that support moderate alkali-resistance
5.3.1. Multi-functional antibiotic-resistance proteins Tet(L) and Tet(K) catalyzing (Tetracycline)(Na+)(K+)/H+ antiport
Tet(L) and Tet(K) genes are close homologues. Bs-Tet(L) is encoded in the B. subtilis chromosome and in plasmids of Gram-positive bacteria [181–183]. When expressed from a single chromosomal copy, the Bs-Tet(L) gene confers resistance only to very low levels of tetracycline (Tc), but when the gene is amplified under selective pressure or expressed from a plasmid, clinically relevant levels of antibiotic-resistance are observed [46, 47, 184, 185]. Sa-Tet(K) is encoded in plasmids that can be incorporated in the chromosome of the human pathogen Staphylococcus aureus; there it supports a high level of antibiotic-resistance [186]. Bs-Tet(L) and Sa-Tet(K) form a branch of the 14-TMS member of the drug/H+ efflux antiporters of the Major Facilitator Superfamily of membrane transporters [187]. They both catalyze Tc-Me/H+ antiport. The Tc is complexed with a divalent metal ion (magnesium, manganese, cobalt are optimal) the Tc-Me complex bears a single net positive charge [143, 188–190]. Bs-Tet(L) catalyzes Na+/H+ and K+/H+ antiport as well as Tc-Me/H+ antiport in both membrane vesicles from antiporter-deficient E. coli and in proteoliposomes reconstituted with purified Bs-Tet(L) [143, 191]. Sa-Tet(K) also catalyzes these multiple antiport activities, as demonstrated in membrane vesicles [130, 191]. Just like their monovalent cation substrates; neither Tc nor the divalent metal ion can be effluxed alone by Bs-Tet(L) or Sa-Tet(K). Competitive patterns of inhibition are evident between the three efflux substrates [191]. Data from site-directed mutagenesis experiments support the hypothesis that antiport occurs by a ping-pong mechanism using a single translocation pathway. However, the Tc-Me complex probably requires more contacts than Na+ or K+ for binding and positioning through the translocation pathway [191].
The Bs-Tet(L)-mediated antiport activities are electrogenic, with entry of a greater number of H+ than cytoplasmic Na+, K+ or Tc-Me exiting per turnover [121, 130, 143]; the precise stoichiometry has not yet been determined. An additional catalytic capacity of Bs-Tet(L) is observed when the external pH is elevated and the combined ΔΨ and external K+ concentration (relative to cytoplasmic or intra-vesicular [K+]) produces an inwardly directed electrochemical K+ gradient. Under these conditions, K+ and H+ may serve as coupling ions that are taken up in exchange for Na+ or Tc-Me [191].
5.3.2. Bs-Tet(L) has a major role in Na+(K+)/H+ antiport-dependent pH homeostasis in B. subtilis
Deletion of Bs-tet(L) in B. subtilis results in a tetracycline- and alkali-sensitive phenotype and reduced ability to grow on media with suboptimal [K+] at elevated pH ([46, 192, 193] and 3.2.1.). When pH 7.5-equilibrated wild type cells are subjected to a shift in external pH to 8.5, they maintain a cytoplasmic pH of 7.5–7.6 in a Na+- or K+-dependent manner; Bs-tet(L) deletion strains completely lose this capacity (Table 2), but it is restored in transformants with either Bs-tet(L) or Sa-tet(K)-bearing plasmids [46]. The physiological functions of Bs-Tet(L) that are unrelated to the antibiotic are highly integrated into the physiology of B. subtilis, a conclusion supported by a transcription-level regulatory component that is mediated by Na+ and elevated pH [46] that is distinct from the post-translational regulation by Tc [194].
Table 2.
Capacities of Bacillus subtilis and alkaliphilic Bacillus pseudofirmus OF4 for Na+- or K+-dependent pH homeostasis during a moderate or extreme alkaline shift in external pHa
| Cytoplasmic pH, 10 min after a sudden shift | ||||||
|---|---|---|---|---|---|---|
| pH 7.5→8.5 | pH 8.5→10.5 | |||||
| CholineCl | NaCl | KCl | Na2CO3 | K2CO3 | ||
| B. subtilis | Wild type | 8.5 | 7.5 | 7.6 | 10.5 | 10.5 |
| Δtet(L) | 8.5 | 8.5 | 8.5 | --- | --- | |
| ΔmrpA | 8.5 | 7.6 | 7.8 | --- | --- | |
| B. pseudofirmus OF4 | Wild type | 8.5 | 7.5 | 8.4 | 8.2 | 10.5 |
For the pH 7.5→8.5 shift, washed logarithmic phase cells, grown on and energized with malate, were equilibrated at pH 7.5 in the presence of 100 mM choline Cl, KCl or NaCl before the external pH was rapidly raised to 8.5. For the pH 8.5→10.5 shift, comparable cells were equilibrated in 50 mM Na2CO3 or K2CO3 buffer at pH 8.5 before the external pH was rapidly raised to 10.5. In both sets of shift experiments, the cytoplasmic pH was determined after 10 min. from a methylamine accumulation assay as described earlier [46, 49].
5.3.3. Convergence of antibiotic/multi-drug resistance and alkali-resistance
The similar roles of the multi-drug efflux protein Ec-MdfA (see 5.1.) and Tc efflux protein Bs-Tet(L) in Na+- and K+-dependent pH homeostasis and alkali-tolerance in their native hosts raises the questions of how and why this convergence of functions arises. Genomic evidence suggests that the 14-TMS Bs-Tet(L) and Sa-Tet(K) efflux antiporters evolved independently from the Gram-negative, 12-TMS Tn10-type Tet proteins [195, 196]. They may have evolved in a pre-existing 12-TMS transporter by insertion of the two central TMS, VII and VIII [197]. This idea is supported by the loss of Tc-Me efflux capacity and retention of all the Na+(K+)/H+ and K+ uptake functions of Bs-Tet(L) by a synthetic “Bs-Tet(L)-12” which lacks those central TMS [198]. Interestingly, the conjoining of Tc-Me2+ efflux activity with Na+/(K+)/H+ antiport activity may be an even more widespread phenomenon since a gene encoding a new member of the Tet(L)/Tet(K) group tet38, has recently been found in the S. aureus chromosome [199] and the deduced product of another recently described Tc-resistance gene, tet35, from Vibrio harveyi exhibits 28% identity and even greater similarity to NhaC from alkaliphilic B. pseudofirmus RAB [200].
The particular combination, in one transporter, of drug-multidrug/H+ and Na+(K+)/H+ antiport has arisen multiple times, i.e. in Ec-MdfA and Bs-Tet(L) and perhaps Tet35. Apparently this combination of antiport activities is readily achievable by protein evolution and bacteria are likely to be advantaged by having such a multifunctional antiporter. The Na+(K+)/H+ antiport could enhance the fitness of the host under conditions of alkaline pH, leading to persistence of the antiporter gene even when its drug substrate is absent [17]. Further study of the similar physiological roles of an Mdr, Ec-MdfA and of the single drug efflux protein, Tet(L), in the alkali-tolerance of E. coli and B. subtilis, respectively, will afford insights into general design principles of these multifunctional drug transport proteins and into the physiological networks in which they function.
5.3.4. Contributions of the Bs-Mrp antiporter and additional B. subtilis antiporters to pH homeostasis
The Bs-Mrp (also called Sha) antiporter is part of a large incompletely understood and unique antiporter family that is encoded by operons containing 6–7 mrp genes and has been designated CPA3 [41, 140]. In contrast to the central, Na+-specific role of Mrp in pH homeostasis in alkaliphilic Bacillus species (see 6.2.1.), in B. subtilis, Bs-Mrp hardly plays any role in Na+- and K+-dependent pH homeostasis (Table 2) while it plays a major role in Na+-resistance [49]. This critical role in Na+-resistance probably accounts for the appearance of Bs-mrp genes on a list of essential genes of B. subtilis compiles from assessments in LB medium, which contains added Na+ [201]; a Bs-mrp null mutant is viable on LBK medium, in which Na+ is replaced by K+ ([78] and 3.1.2.2.). The basis for a more dominant role for Bs-Tet(L) in pH homeostasis and for Bs-Mrp in Na+-resistance is not yet clear. Nor is the basis for the apparent complexity of Mrp understood [41], but perhaps Mrp proteins have more than a single Na+(K+)/H+ antiporter activity [202] and/or additional catalytic activities as suggested for MrpE [203] and supported by the involvement of Bs-MrpF in cholate efflux [49, 78].
B. subtilis has at least three additional Na+/H+ antiporters. The Bs-MleN antiporter couples this exchange to a malate uptake and lactate efflux and contributes to growth at low PMF, i.e. in the presence of inhibitory concentrations of uncouplers ([133] and 4.2.). Bs-NhaC (YheL), another Na+/H+ antiporter, does not have a prominent role in wild-type pH homeostasis; its expression is increased in Bs-tet(L) deletion mutants, suggesting that it has a back-up role [193]. The newly characterized Bs-NhaK is a Na+(K+)(Li+)/H+ antiporter [48].. The precise contributions of these diverse antiporters to the physiology of B. subtilis are a puzzle to be solved. In the epiphytic niches of B. subtilis, there are variable pH-salt conditions, diverse plant metabolites that serve as growth substrates and numerous potential growth inhibitors [204]. They create different combinations of alkali, salinity and toxins that could account for the complexity of the Na+ cycle and its interactions with H+ and K+ cycles (Figure 1).
6. Alkaliphilic bacilli: the Na+-cycle and other adaptations to extremely alkaline conditions
6.1. The centrality of active pH homeostasis in alkaliphily
Extremely alkaliphilic Bacillus species grow optimally at pH values of 10 and above [88, 205]. Obligate alkaliphiles (Bacillus alcalophilus, B. pseudofirmus RAB) are better adapted to pH values above 11 than facultative strains [206] but facultative alkaliphiles are more useful for studies of the relationship of specific gene products to alkaliphily because non-alkaliphilic mutant strains can grow at pH 7–8.5. Among the most intensively studied facultative alkaliphiles (B. pseudofirmus OF4, B. halodurans C-125, and Bacillus cohnii YN2000), B. pseudofirmus OF4 is the most alkaliphilic, grows in semi-defined and defined media at extremely alkaline pH and is genetically accessible to targeted gene deletions [51, 52, 206, 207]. B. pseudofirmus OF4 grows with a doubling time of 54 minutes at pH 7.5 vs. a doubling time of 38 minutes at external pH values from 8.5–10.6 in chemostat cultures containing semi-defined malate medium ([163], Figure 4). At external pH values of 7.5, 8.5 and 9.5, the cytoplasmic pH remains close to 7.5, i.e. the alkaliphile produces a ΔpH, acid in, that rises from zero to 2.0 pH units (shown in parentheses in Figure 4). Although the ΔpH at pH 10.6 is 2.3 units, larger than at pH 9.5, it does not keep pace with the external pH increase and the cytoplasmic pH rises to 8.3 at external pH of 10.6. The cytoplasmic pH and the doubling time increase almost in parallel above pH 10.6. Neither the ΔΨ nor the total PMF correlates similarly with the increasing doubling time in the high pH range. At the highest external pH examined, pH 11.4, the doubling time is more than 10 hours and many of the cells form chains with multiple septa.
Figure 4.
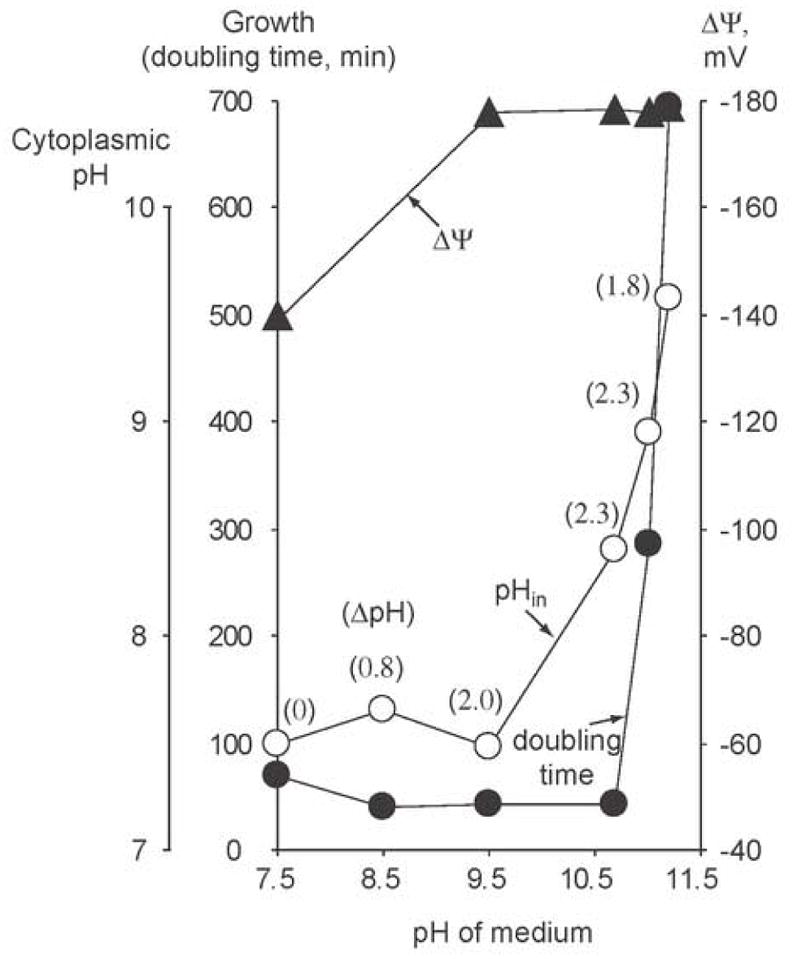
Generation time and PMF parameters of B. pseudofirmus OF4 as a function of external pH in continuous cultures of malate-containing medium. Cells were grown in semi-defined malate containing medium maintained at the indicated pH values. tg, doubling time was calculated from the dilution rates in the chemostat and PMF parameters were determined as described in reference [88]; the data are replotted from Figure 1 of this reference with permission from Blackwell Publishing Ltd.
Major inferences can be drawn from the chemostat study about alkaliphily and pH homeostasis in extremophile Bacillus growing under non-fermentative growth conditions. First, generation of a ΔpH, acid in, of 2–2.3 units is consistent with a remarkable pH homeostasis capacity that was also evident in pH shift experiments (Figure 2B); it is not known what prevents further increases in the ΔpH at external pH of ≥ 10.6. Second, the cells grow optimally even after the cytoplasmic pH increases significantly beyond 7.5. At external pH of 10.6 a doubling time of 38 minutes is still observed when the cells have a cytoplasmic pH of 8.3. We hypothesize that alkaliphiles have greater tolerance than non-alkaliphiles for elevated cytoplasmic pH (see the E. coli shift experiment in Figure 2A and 4.4.). This is consistent with an intruiging structural study showing that a cytoplasmic enzyme of alkaliphilic B. alcalophilus exhibits features not found in neutralophile homologues but are associated with extracellular enzymes that function at very high pH values [208]. Third, above a cytoplasmic pH of about 8.5, increasing cytoplasmic pH correlates well with a decreasing growth rate. As in E. coli cells ([58], 4.4. and 3.1.2.1.) elements of cell division may be more sensitive to alkalinization of the cytoplasm than other cellular processes. Cell wall stress could also be an important contributor to setting the upper limit for growth in the alkaliphiles.
Active mechanisms of pH homeostasis have a less exclusive role during alkaliphile growth on fermentable carbon sources. When wild type B. pseudofirmus OF4 cells are grown on and energized with glucose instead of malate during a pH 8.5 –> 10.5 shift (the same shift protocol as in Table 2), the cytoplasmic pH in the absence of Na+ is 9.2 instead of 10.5 as observed in the presence of malate [209]. This suggests that an antiporter-independent capacity such as intracellular acid production can partially replace the active transport-based mechanism.
6.2. The antiporters of the alkaliphile Na+ cycle
In support of the essential role of Na+/H+ antiporters in alkiliphily, alkaliphile pH homeostasis is strictly dependent on the presence of Na+ (Figure 2B and Table 2, [40, 87, 210]). We hypothesize that the alkaliphiles use Na+-specific antiporters in order to avoid the risk of lowering their cytoplasmic K+ concentration adversely during the robust, ongoing antiport that is required at high pH. Assays of the total, PMF-dependent 22Na+ efflux capacity of whole cells of B. subtilis and B. pseudofirmus OF4 have been conducted under conditions that were at the upper pH range for B. subtilis and the lower end for the alkaliphile; the alkaliphile exhibits about 10-fold higher capacity for PMF-dependent Na+ efflux (that is presumed to represent antiport activity) than the neutralophile [211].
6.2.1. Alkaliphile Mrp antiporter
Full mrp operons are widely distributed in bacterial genomes and usually contain 6–7 genes whose products (Mrp A, B, C, D, E, F, G) are all predicted to be hydrophobic membrane proteins [41]. The full Bp-mrp operon has been cloned and expressed in antiporter-deficient E. coli strains in which fluorescence-based assays demonstrate Na+-specific monovalent cation proton antiport [212]. Active Mrp antiporters have not yet been purified and studied in sufficient biochemical detail to indicate any adaptations that may help to meet the major challenge of adequate proton capture at pH ≥ 10. Nevertheless, genetic data assign roles in alkiliphily and Na+ resistance to mrp. A non-alkaliphilic mutant of B. halodurans C-125 no longer grows above pH 9.5, has a cytoplasmic pH of 10.4 when shifted to that external pH and exhibits no ΔΨ-dependent Na+/H+ antiport in native vesicles although significant ΔpH-dependent antiport remains ([50, 213] and 3.2.1.). This mutant has a single point mutation in Bh-mrpA that could be corrected by cross-over from a partial clone of the mrp operon, leading to the restoration of normal pH homeostasis and alkaliphily. Deletions of Bp-mrp in B. pseudofirmus OF4 have thus far been lethal (Ito, M., unpublished data) but a Bs-mrp null mutant and in-frame deletion mutants of each Bs-mrp gene of B. subtilis are viable [78]. More robust Na+ extrusion is probably necessary throughout the pH range of the alkaliphile than in B. subtilis, since all the ion-coupled solute symporters and flagellar motors of the alkaliphile are Na+ coupled (Fig. 1) [88, 214].
6.2.2. Bp-NhaC and additional antiporters of alkaliphlic Bacillus
Bp-NhaC plays a role in pH homeostasis at pH 10.5 under Na+-limiting conditions although its major role is in Na+ extrusion at lower pH values [51]. Based on genomic data, B. halodurans C-125 [52] is predicted to have, in addition to Bh-nhaC and Bh-mrp, a homologue of the Bs-mleN gene that encodes the 2H-malate/Na-lactate antiporter ([133] and see 4.2.) and a member of the CPA-1 family of antiporters whose closest bacterial homologue is the NhaP Na+/H+ antiporters [169, 171].
6.3. Re-entry routes for Na+ are required for optimal pH homeostasis
The essential role of Na+/H+ antiporters in alkaliphily also makes routes that recycle Na+ essential to provide substrate for ongoing antiporter activity. One such route is Na+ coupled transporters that increase Na+. During a pH 8.5 –-> 10.5 shift in external pH in the presence of 100 mM Na+, cells of B. pseudofirmus RAB exhibit a cytoplasmic pH that increases from just below 8.5, at 30 seconds after the shift, to pH 9.1 at 3 minutes (Fig. 2B). When the non-metabolizable amino acid AIB that is co-transported with Na+ is included in the shift buffer, pH homeostasis is greatly enhanced, with an initial reduction in the cytoplasmic pH to about 8.2, followed by a return to the initial pre-shift pH ([61], Fig. 2B).
The observed role of Na+/solute symporter in providing cytoplasmic Na+ for the antiporters raises the question of how alkaliphiles achieve significant (although not optimal) Na+/H+-dependent pH homeostasis when solutes that enter with Na+ are limiting or omitted. A Na+-channel that is activated at alkaline cytoplasmic pH was postulated [4, 88]. Recently, this hypothesis has been supported by studies of two Na+ channels of B. pseudofirmus OF4, Bp-MotPS and Bp-NavBP [4, 88, 214, 215]. MotPS is the Na+-translocating motility channel that was earlier suggested as a strong candidate for a role in Na+-entry in support of pH homeostasis [216]. NavBP, encoded by ncbA, is a member of a newly characterized family of novel voltage-gated, ion-selective Na+ channels in bacteria [215, 217, 218]. The effect of single and double deletions of Bp-motPS and the Bp-ncbA show that Bp-NavBP plays a significant role in supporting pH homeostasis while Bp-MotPS plays an ancillary role [215]. Bp-NavBP also has roles in motility and chemotaxis, which are observed only at high pH in the alkaliphile [163, 214, 215, 219]. Consistently, the amplitude of channel-mediated current is increased and its activation is potentiated by high pH [215]. The detailed unraveling of interactions between Bp-NavBP and the chemotaxis apparatus and of the interplay between NavBP signaling and pH homeostasis will be an interesting new chapter of alkaliphile physiology that will have applications to the other salt- and alkaline-tolerant bacteria in which this family of channels has been identified [218].
6.4. Alkali-adpative strategies of alkaliphiles apart from the Na+ cycle
6.4.1. A role for cell surface properties
Secondary cell wall polymers (SCWP) that are associated with the peptidoglycan layer of Gram-positive bacteria have been implicated in an ancillary role in Bacillus alkaliphily ([55, 220, 221] and Fig. 1). B. halodurans C-125 has teichuronic acid and teichuronopeptide as the major SCWP [222]. A mutant strain that does not produce teichuronic acids shows no defect in growth at pH 10 or 10.5, whereas a mutant lacking both teichuronic acids and teichuronopeptide grows poorly at pH 10.5. At pH 10, growth is impaired but still substantial, correlating with a slightly higher cytoplasmic pH than wild type [55]. The major SCWP in B. pseudofirmus OF4 is an S-layer layer, encoded by the highly expressed slpA gene. At pH 7.5, deletion of slpA leads to significantly better growth in the mutant than wild type. A modest growth defect is observed at pH 10.5 that is exacerbated at pH 11. These observations suggest that the alkaliphile produces a polymer whose costly production reduces its growth rate at pH 7.5 but provides a small advantage in the event of a sudden alkaline shift [221]. In both B. halodurans C-125 and B. pseudofirmus OF4, SCWP are neither necessary nor sufficient substitutes for active antiporter-based pH homeostasis. The positive effect that these acidic SCWP do provide upon alkaline challenge is presumed to result from their ability to bind cations, perhaps even trapping them near the membrane surface. This may enhance the availability of H+ and Na+ for pH homeostasis and bioenergetic work [221, 223, 224].
Another cell surface feature that should be noted is the presence of high levels of cardiolipin as well as squalene in membranes of alkaliphilic Bacillus, with high pH-dependent increases in the cardiolipin [225]. Models have been advanced for a role of cardiolipin in trapping protons at the membrane surface [226] and for squalene in lowering H+ permeability of the lipid bilayer [227], roles that are yet to be probed experimentally.
6.4.2. Involvement of the oxidative phosphorylation machinery
The bioenergetic capacities of a panel of Bp-ATP synthase mutants in comparison with wild type B. pseudofirmus OF4 show that ATP synthesis capacity is a significant factor in overall pH homeostasis [228]. This is also inferred from microarray studies of E. coli and B. subtilis ([20, 67] and see 3.1.2.). The alkaliphile ATP synthase is further adapted for alkaliphily in having features that minimize loss of H+ to the bulk through the synthase when cells experience a sudden alkaline shift [228, 229].
In contrast to the repression of H+-extruding terminal oxidase Cyo in E. coli growing at elevated pH ([20] and see 3.1.2.), the H+-extruding Cta terminal oxidase of B. pseudofirmus OF4 is up-regulated at the alkaline edge of the pH range [230, 231]. In fact, general properties of alkaliphilic Bacillus species include: alkali-dependent increases in level of H+-extruding respiratory chain components; and low redox potentials of alkaliphile respiratory chain components relative to neutralophile homologues [206, 232–234]. Both these properties presumably relate to the importance of generating of a ΔΨ that is higher in alkaliphiles than in most bacteria (compare the B. pseudofirmus OF4 ΔΨ in Figure 3 with that of E. coli in Figure 2). This capacity is especially important at high pH, where the burden of ongoing electrogenic antiport and the higher energetic cost of ATP synthesis increase the demands upon ΔΨ generation [88].
7. The interaction between alkaline stress and other stresses
Alkaline stress has significant interplay with many other stresses. These overlaps complicate a linear association of a protein with defense against alkali per se on the basis of its activation or induction upon alkaline challenge. Several examples follow. First, there is an overlap between alkaline stress and salt stress. It is due to the significant increase in the Na+ cytotoxicity as the pH rises [117]. Moreover, the toxicity of Na+ is highly dependent upon the status of the cytoplasmic K+ concentration [22]. Second, alkaline pH has interplay with cell wall stress [77], because of the alkali-vulnerability of the subset of cell wall biosynthetic enzymes that are exposed on the outside of the membrane. Third and fourth, interplay of an alkaline challenge with that of high temperature arises because high temperature alters the H+ permeability more than Na+ permeability in many bacteria [235] and interplay with changes in osmolarity arises because some monovalent cation/proton antiporters have roles in osmoadaptation [98]. Fifth, interplay between alkaline stress and the stress of entry into stationary phase is strikingly illustrated by a switch in the regulatory mechanism of transcription of Ec-nhaA in E. coli ([114] and 3.2.3.). Finally, the induction by alkaline conditions of an E. coli protein, Alx, that is involved in redox stress, adds another stress intersection to the list [62, 83].
9. Closing remarks
The ability of bacteria to survive or to grow under alkaline conditions is of widespread ecological, industrial and epidemiological importance. The total repertoire and the overlap of proteins required for survival vs. growth are still being elucidated. They encompass proteins that contribute to structural, metabolic as well as bioenergetic adaptations. Active alkaline pH homeostasis that is centrally important in both survival and growth depends upon monovalent cation/proton antiporters, whose identification is generally achieved by combining genetic and biochemical approaches. Genomic and proteomic information is providing a wealth of information about the complexity of interlocking H+ and Na+ cycles that function in individual bacterial strains. An intricate interplay between alkaline and other stresses regulates the level of expression and activity of these antiporters. Studies of novel antiporters and channels involved in alkaliphily both of neutralophiles and extremely alkaliphilic species continue to reveal strategies that turn out to have counterparts across a broad range of important bacteria. Further biochemical studies are required to define the specific mechanisms that underpin the efficacy of the physiologically important antiporters and, in some cases, their capacity to catalyze multiple activities such as drug or multi-drug/H+ antiport as well as Na+(K+)/H+ antiport. Given the insights provided by the novel NhaA structure, determination of additional antiporter structures will be needed to clarify structural principles related to the mechanism of Na+/H+, its regulation by pH and capacity for cation capture.
Acknowledgments
We are grateful to Talia Swartz for help with the figures and comments on the text. Studies that form the basis for this review were supported by research grants from the Israel Science Foundation and the German Israeli Foundation for Scientific Research and Development (to E.P.), grants from the Y. Leon Benoziyo Institute for Molecular Medicine at the Weizmann Institute of Science and the Israel Cancer Research Foundation (to E.B.), a Grant-in-Aid for Scientific Research on Priority Areas ‘Genome Biology’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.I.), and research grants GM28454 and GM52837 from the National Institutes of Health (to T.A.K.). E.P. is the Adelina and Massimo DellaPergola Professor of Life Sciences.
Abbreviations
- ΔpH
transmembrane pH gradient
- ΔΨ
transmembrane electrical gradient
- AIB
α-aminoisobutyric acid
- BCECF
2′,7′-bis-(2-carboxyethyl)-5(and-6)-carboxyfluorescein
- LB
Luria-Bertani medium
- PMF
proton-motive force, a transmembrane electrochemical gradient of protons composed of the ΔpH (alkaline inside) and ΔΨ (negative inside)
- SCWP
secondary cell wall polymers
- SMF
sodium-motive force
- Tc
tetracycline
- TMAO
trimethylamine N-oxide
- TMS
trans membrane segment
- MDR
multi-drug transporter
- CPA
cation proton antiporters
- Ec
Escherichia coli
- Vc
Vibrio cholerae
- Mj
Methanococcus jannaschii
- Bs
Bacillus subtilis
- Bh
Bacillus halodurans C-125
- Bp
Bacillus pseudofirmus OF4
- Sa
Staphylococcus aureus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Padan E, Zilberstein D, Rottenberg H. The proton electrochemical gradient in Escherichia coli cells. Eur J Biochem. 1976;63:533–541. doi: 10.1111/j.1432-1033.1976.tb10257.x. [DOI] [PubMed] [Google Scholar]
- 2.Padan E, Zilberstein D, Schuldiner S. pH homeostasis in bacteria. Biochim Biophys Acta. 1981;650:151–166. doi: 10.1016/0304-4157(81)90004-6. [DOI] [PubMed] [Google Scholar]
- 3.Slonczewski JL, Rosen BP, Alger JR, Macnab RM. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci USA. 1981;78:6271–6275. doi: 10.1073/pnas.78.10.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth IR. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985;49:359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 6.Speelmans G, Poolman B, Abee T, Konings WN. Energy transduction in the thermophilic anaerobic bacterium Clostridium fervidus is exclusively coupled to sodium ions. Proc Natl Acad Sci USA. 1993;90:7975–7979. doi: 10.1073/pnas.90.17.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harold FM, Van Brunt J. Circulation of H+ and K+ across the plasma membrane is not obligatory for bacterial growth. Science. 1977;197:372–373. doi: 10.1126/science.69317. [DOI] [PubMed] [Google Scholar]
- 8.Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri,and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matin A. pH homeostasis in acidophiles. Novartis Found Symp. 1999;221:152–163. doi: 10.1002/9780470515631.ch10. [DOI] [PubMed] [Google Scholar]
- 10.Stingl K, Uhlemann EM, Schmid R, Altendorf K, Bakker EP. Energetics of Helicobacter pylori and its implications for the mechanism of urease-dependent acid tolerance at pH 1. J Bacteriol. 2002;184:3053–3060. doi: 10.1128/JB.184.11.3053-3060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachs G, Weeks DL, Melchers K, Scott DR. The gastric biology of Helicobacter pylori. Annu Rev Physiol. 2003;65:349–369. doi: 10.1146/annurev.physiol.65.092101.142156. [DOI] [PubMed] [Google Scholar]
- 12.Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 13.Hase CC, Fedorova ND, Galperin MY, Dibrov PA. Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol Mol Biol Rev. 2001;65:353–370. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herz K, Vimont S, Padan E, Berche P. Roles of NhaA, NhaB, and NhaD Na+/H+ antiporters in survival of Vibrio cholerae in a saline environment. J Bacteriol. 2003;185:1236–1244. doi: 10.1128/JB.185.4.1236-1244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones BE, Grant WD, Duckworth AW, Owenson GG. Microbial diversity of soda lakes. Extremophiles. 1998;2:191–200. doi: 10.1007/s007920050060. [DOI] [PubMed] [Google Scholar]
- 16.Horikoshi K. Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev. 1999;63:735–750. doi: 10.1128/mmbr.63.4.735-750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krulwich TA, Lewinson O, Padan E, Bibi E. Do physiological roles foster persistence of drug/multidrug-efflux pumps? A case study. Nat Microbiol Rev. 2005;3:566–572. doi: 10.1038/nrmicro1181. [DOI] [PubMed] [Google Scholar]
- 18.Schuldiner S, Agmon V, Brandsma J, Cohen A, Friedman E, Padan E. Induction of SOS functions by alkaline intracellular pH in Escherichia coli. J Bacteriol. 1986;168:936–939. doi: 10.1128/jb.168.2.936-939.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taglicht D, Padan E, Oppenheim AB, Schuldiner S. An alkaline shift induces the heat shock response in Escherichia coli. J Bacteriol. 1987;169:885–887. doi: 10.1128/jb.169.2.885-887.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell MJ, Finkel SE. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J Bacteriol. 2003;185:7044–7452. doi: 10.1128/JB.185.24.7044-7052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harel-Bronstein M, Dibrov P, Olami Y, Pinner E, Schuldiner S, Padan E. MH1, a second-site revertant of an Escherichia coli mutant lacking Na+/H+ antiporters (ΔnhaAΔnhaB)regains Na+ resistance and a capacity to excrete Na+ in a ΔμH+-independent fashion. J Biol Chem. 1995;270:3816–3822. doi: 10.1074/jbc.270.8.3816. [DOI] [PubMed] [Google Scholar]
- 23.Harvey HW. The chemistry and fertility of sea weters. Cambridge University Press; London: 1955. [Google Scholar]
- 24.Colwell RR, Huq A. In: Vibrio cholerae and cholera: molecular to global perspectives. Wachsmuth IK, Blake PA, Olsvik O, editors. ASM Press; Washington, D.C.: 1994. pp. 117–133. [Google Scholar]
- 25.Rozen Y, Belkin S. Survival of enteric bacteria in seawater. FEMS Microbiol Rev. 2001;25:513–529. doi: 10.1111/j.1574-6976.2001.tb00589.x. [DOI] [PubMed] [Google Scholar]
- 26.Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ, Hardcastle JD. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brune A, Kuhl M. pH profiles of the extremely alkaline hindguts of soil-feeding termites (Isoptera: Termitidae) determined with microelectrodes. J Insect Physiol. 1996;42:1121–1127. [Google Scholar]
- 28.Lemke T, Stingl U, Egert M, Friedrich MW, Brune A. Physicochemical conditions and microbial activities in the highly alkaline gut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae) Appl Environ Microbiol. 2003;69:6650–6658. doi: 10.1128/AEM.69.11.6650-6658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donovan SE, Purdy KJ, Kane MD, Eggleton P. Comparison of Euryarchaea strains in the guts and food-soil of the soil-feeding termite Cubitermes fungifaber across different soil types. Appl Environ Microbiol. 2004;70:3884–3892. doi: 10.1128/AEM.70.7.3884-3892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horikoshi K. Microorganisms in alkaline environments. VCH Publishers Inc; New York: 1991. [Google Scholar]
- 31.Jones PC, Fillingame RH. Genetic fusions of subunit c in the F0 sector of H+-transporting ATP synthase. Functional dimers and trimers and determination of stoichiometry by cross-linking analysis. J Biol Chem. 1998;273:29701–29705. doi: 10.1074/jbc.273.45.29701. [DOI] [PubMed] [Google Scholar]
- 32.Ma Y, Zhang W, Xue Y, Zhou P, Ventosa A, Grant WD. Bacterial diversity of the Inner Mongolian Baer Soda Lake as revealed by 16S rRNA gene sequence analyses. Extremophiles. 2004;8:45–51. doi: 10.1007/s00792-003-0358-z. [DOI] [PubMed] [Google Scholar]
- 33.Rees HC, Grant WD, Jones BE, Heaphy S. Diversity of Kenyan soda lake alkaliphiles assessed by molecular methods. Extremophiles. 2004;8:63–71. doi: 10.1007/s00792-003-0361-4. [DOI] [PubMed] [Google Scholar]
- 34.Tiago I, Chung AP, Verissimo A. Bacterial diversity in a nonsaline alkaline environment: heterotrophic aerobic populations. Appl Environ Microbiol. 2004;70:7378–7387. doi: 10.1128/AEM.70.12.7378-7387.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Padan E, Schuldiner S. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim Biophys Acta. 1994;1185:129–151. doi: 10.1016/0005-2728(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 36.Padan E, Gerchman Y, Rimon A, Rothman A, Dover N, Carmel-Harel O. Novartis Found Symp. Vol. 221. Wiley; Chichester: 1999. pp. 183–196. [DOI] [PubMed] [Google Scholar]
- 37.Padan E, Venturi M, Gerchman Y, Dover N. Na+/H+ antiporters. Biochim Biophys Acta. 2001;1505:144–157. doi: 10.1016/s0005-2728(00)00284-x. [DOI] [PubMed] [Google Scholar]
- 38.Kitada M, Kosono S, Kudo T. The Na+/H+ antiporter of alkaliphilic Bacillus sp. Extremophiles. 2000;4:253–258. doi: 10.1007/s007920070010. [DOI] [PubMed] [Google Scholar]
- 39.Krulwich TA, Ito M, Gilmour R, Hicks DB, Guffanti AA. Energetics of alkaliphilic Bacillus species: physiology and molecules. Adv Microb Physiol. 1998;40:401–438. doi: 10.1016/s0065-2911(08)60136-8. [DOI] [PubMed] [Google Scholar]
- 40.Krulwich TA, Guffanti AA, Ito M. Novartis Found Symp. Vol. 221. Wiley; Chichester: 1999. Mechanisms by which bacterial cells respond to pH; pp. 167–182. [DOI] [PubMed] [Google Scholar]
- 41.Swartz H, Ikewada S, Ishikawa O, Ito M, Krulwich TA. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles. 2005 doi: 10.1007/s00792-005-0451-6. in press. [DOI] [PubMed] [Google Scholar]
- 42.Counillon L, Pouyssegur J. The expanding family of eucaryotic Na+/H+ exchangers. J Biol Chem. 2000;275:1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Orlowski J, Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflugers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 44.Fliegel L. The Na+/H+ exchanger isoform 1. The international Journal of biochemistry and cell biology. 2005;37:33–37. doi: 10.1016/j.biocel.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am J Physiol Cell Physiol. 2005;288:C223–39. doi: 10.1152/ajpcell.00360.2004. [DOI] [PubMed] [Google Scholar]
- 46.Cheng J, Guffanti AA, Wang W, Krulwich TA, Bechhofer DH. Chromosomal tetA(L) gene of Bacillus subtilis: regulation of expression and physiology of a tetA(L) deletion strain. J Bacteriol. 1996;178:2853–2860. doi: 10.1128/jb.178.10.2853-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng J, Guffanti AA, Krulwich TA. The chromosomal tetracycline resistance locus of Bacillus subtilis encodes a Na+/H+ antiporter that is physiologically important at elevated pH. J Biol Chem. 1994;269:27365–27371. [PubMed] [Google Scholar]
- 48.Fujisawa M, Kusomoto A, Wada Y, Tsuchiya T, Ito M. NhaK, a novel monovalent cation/H+ antiporter of Bacillus subtilis. Arch Microbiol. 2005 doi: 10.1007/s00203-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 49.Ito M, Guffanti AA, Oudega B, Krulwich TA. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J Bacteriol. 1999;181:2394–2402. doi: 10.1128/jb.181.8.2394-2402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamamoto T, Hashimoto M, Hino M, Kitada M, Seto Y, Kudo T, Horikoshi K. Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol Microbiol. 1994;14:939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 51.Ito M, Guffanti AA, Zemsky J, Ivey DM, Krulwich TA. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J Bacteriol. 1997;179:3851–3857. doi: 10.1128/jb.179.12.3851-3857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rottenberg H. The measurement of membrane potential and pH in cells, organelles, and vesicles. Methods Enzymol. 1979;55:547–569. doi: 10.1016/0076-6879(79)55066-6. [DOI] [PubMed] [Google Scholar]
- 54.Padan E, Schuldiner S. Intracellular pH regulation in bacterial cells. Methods Enzymol. 1986;125:337–352. doi: 10.1016/s0076-6879(86)25029-6. [DOI] [PubMed] [Google Scholar]
- 55.Aono R, Ito M, Machida T. Contribution of the cell wall component teichuronopeptide to pH homeostasis and alkaliphily in the alkaliphile Bacillus lentus C-125. J Bacteriol. 1999;181:6600–6606. doi: 10.1128/jb.181.21.6600-6606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsujimoto K, Semadeni M, Huflejt M, Packer L. Intracellular pH of halobacteria can be determined by the fluorescent dye 2′, 7′-bis(carboxyethyl)-5(6)-carboxyfluorescein. Biochem Biophys Res Commun. 1988;155:123–129. doi: 10.1016/s0006-291x(88)81058-1. [DOI] [PubMed] [Google Scholar]
- 57.Molenaar D, Abee T, Konings WN. Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim Biophys Acta. 1991;1115:75–83. doi: 10.1016/0304-4165(91)90014-8. [DOI] [PubMed] [Google Scholar]
- 58.Zilberstein D, Agmon V, Schuldiner S, Padan E. Escherichia coli intracellular pH, membrane potential, and cell growth. J Bacteriol. 1984;158:246–252. doi: 10.1128/jb.158.1.246-252.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zilberstein D, Agmon V, Schuldiner S, Padan E. The sodium/proton antiporter is part of the pH homeostasis mechanism in Escherichia coli. J Biol Chem. 1982;257:3687–3691. [PubMed] [Google Scholar]
- 60.Wiegert T, Homuth G, Versteeg S, Schumann W. Alkaline shock induces the Bacillus subtilis sigma(W) regulon. Mol Microbiol. 2001;41:59–71. doi: 10.1046/j.1365-2958.2001.02489.x. [DOI] [PubMed] [Google Scholar]
- 61.Krulwich TA, Federbush JG, Guffanti AA. Presence of a nonmetabolizable solute that is translocated with Na+ enhances Na+-dependent pH homeostasis in an alkalophilic Bacillus. J Biol Chem. 1985;260:4055–4058. [PubMed] [Google Scholar]
- 62.Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blankenhorn D, Phillips J, Slonczewski JL. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J Bacteriol. 1999;181:2209–2216. doi: 10.1128/jb.181.7.2209-2216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186:6032–6041. doi: 10.1128/JB.186.18.6032-6041.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gale EF, Epps HMR. The effect of the pH of the medium during growth on the enzymatic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem J. 1942;36:600–619. doi: 10.1042/bj0360600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bordi C, Theraulaz L, Mejean V, Jourlin-Castelli C. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol Microbiol. 2003;48:211–223. doi: 10.1046/j.1365-2958.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- 67.Kosono S, Asai K, Sadaie Y, Kudo T. Altered gene expression in the transition phase by disruption of a Na+/H+ antiporter gene (shaA) in Bacillus subtilis. FEMS Microbiol Lett. 2004;232:93–99. doi: 10.1016/S0378-1097(04)00037-0. [DOI] [PubMed] [Google Scholar]
- 68.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Acid-adaptive genes of Helicobacter pylori. Infect Immun. 2003;71:5921–5939. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belli WA, Marquis RE. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl Environ Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi H, Suzuki T, Unemoto T. Streptococcal cytoplasmic pH is regulated by changes in amount and activity of a proton-translocating ATPase. J Biol Chem. 1986;261:627–630. [PubMed] [Google Scholar]
- 71.Kuhnert WL, Zheng G, Faustoferri RC, Quivey RG., Jr The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J Bacteriol. 2004;186:8524–8528. doi: 10.1128/JB.186.24.8524-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rozen Y, Larossa RA, Templeton LJ, Smulski DR, Belkin S. Gene expression analysis of the response by Escherichia coli to seawater. Antonie Van Leeuwenhoek. 2002;81:15–25. doi: 10.1023/a:1020500821856. [DOI] [PubMed] [Google Scholar]
- 73.Rozen Y, Dyk TK, LaRossa RA, Belkin S. Seawater Activation of Escherichia coli Gene Promoter Elements: Dominance of rpoS Control. Microb Ecol. 2001;42:635–643. doi: 10.1007/s00248-001-1012-x. [DOI] [PubMed] [Google Scholar]
- 74.Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- 75.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 77.Cao M, Wang T, Ye R, Helmann JD. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol Microbiol. 2002;45:1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x. [DOI] [PubMed] [Google Scholar]
- 78.Ito M, Guffanti AA, Wang W, Krulwich TA. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J Bacteriol. 2000;182:5663–5670. doi: 10.1128/jb.182.20.5663-5670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kosono S, Morotomi S, Kitada M, Kudo T. Analyses of a Bacillus subtilis homologue of the Na+/H+ antiporter gene which is important for pH homeostasis of alkaliphilic Bacillus sp. C-125. Biochim Biophys Acta. 1999;1409:171–175. doi: 10.1016/s0005-2728(98)00157-1. [DOI] [PubMed] [Google Scholar]
- 80.Fujisawa M, Wada Y, Ito M. Modulation of the K+ efflux activity of Bacillus subtilis YhaU by YhaT and the C-terminal region of YhaS. FEMS Microbiol Lett. 2004;231:211–217. doi: 10.1016/S0378-1097(03)00959-5. [DOI] [PubMed] [Google Scholar]
- 81.Wei Y, Southworth TW, Kloster H, Ito M, Guffanti AA, Moir A, Krulwich TA. Mutational loss of a K+ and NH4+ transporter affects the growth and endospore formation of alkaliphilic Bacillus pseudofirmus OF4. J Bacteriol. 2003;185:5133–5147. doi: 10.1128/JB.185.17.5133-5147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slonczewski JL, Gonzalez TN, Bartholomew FM, Holt NJ. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J Bacteriol. 1987;169:3001–3006. doi: 10.1128/jb.169.7.3001-3006.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bingham RJ, Hall KS, Slonczewski JL. Alkaline induction of a novel gene locus, alx, in Escherichia coli. J Bacteriol. 1990;172:2184–2186. doi: 10.1128/jb.172.4.2184-2186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 85.Gerchman Y, Rimon A, Padan E. A pH-dependent conformational change of NhaA Na+/H+ antiporter of Escherichia coli involves loop VIII-IX, plays a role in the pH response of the protein, and is maintained by the pure protein in dodecyl maltoside. J Biol Chem. 1999;274:24617–24624. doi: 10.1074/jbc.274.35.24617. [DOI] [PubMed] [Google Scholar]
- 86.West IC, Mitchell P. Proton/sodium ion antiport in Escherichia coli. Biochem J. 1974;144:87–90. doi: 10.1042/bj1440087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McLaggan D, Selwyn MH, Sawson AP. Dependence of Na+ of control of cytoplasmic pH in a facultative alkalophile. FEBS Lett. 1984:254–258. [Google Scholar]
- 88.Krulwich TA. Alkaliphiles: ‘basic’ molecular problems of pH tolerance and bioenergetics. Mol Microbiol. 1995;15:403–410. doi: 10.1111/j.1365-2958.1995.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 89.Niiya S, Yamasaki K, Wilson TH, Tsuchiya T. Altered cation coupling to melibiose transport in mutants of Escherichia coli. J Biol Chem. 1982;257:8902–8906. [PubMed] [Google Scholar]
- 90.Goldberg EB, Arbel T, Chen J, Karpel R, Mackie GA, Schuldiner S, Padan E. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:2615–2619. doi: 10.1073/pnas.84.9.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karpel R, Olami Y, Taglicht D, Schuldiner S, Padan E. Sequencing of the gene ant which affects the Na+/H+ antiporter activity in Escherichia coli. J Biol Chem. 1988;263:10408–10414. [PubMed] [Google Scholar]
- 92.Carmel O, Dover N, Rahav-Manor O, Dibrov P, Kirsch D, Karpel R, Schuldiner S, Padan E. A single amino acid substitution (Glu134-->Ala) in NhaR1 increases the inducibility by Na+ of the product of nhaA, a Na+/H+ antiporter gene in Escherichia coli. EMBO J. 1994;13:1981–1989. doi: 10.1002/j.1460-2075.1994.tb06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang AB, Lin R, Studley WK, Tran CV, Saier MH. Phylogeny as a guide to structure and function of membrane transport proteins. Mol Membr Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- 94.Swartz TH, Ito M, Hicks DB, Nuqui M, Guffanti AA, Krulwich TA. The Mrp Na+/H+ antiporter increases the activity of the malate:quinone oxidoreductase of an Escherichia coli respiratory mutant. J Bacteriol. 2005;187:388–391. doi: 10.1128/JB.187.1.388-391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s) J Biol Chem. 1989;264:20297–20302. [PubMed] [Google Scholar]
- 96.Padan E, Schuldiner S. Molecular physiology of the Na+/H+ antiporter in Escherichia coli. J Exp Biol. 1994;196:443–456. doi: 10.1242/jeb.196.1.443. [DOI] [PubMed] [Google Scholar]
- 97.Padan E, Schuldiner S. Na+/H+ antiporters, molecular devices that couple the Na+ and H+ circulation in cells. J Bioenerg Biomembr. 1993;25:647–669. doi: 10.1007/BF00770252. [DOI] [PubMed] [Google Scholar]
- 98.Padan E, Schuldiner S. In: The Handbook of Biological Physics, Vol. II. Transport processes in membranes. Konings WN, Kaback HR, Lolkema J, editors. Elsevier Science; Amsterdam: 1996. pp. 501–531. [Google Scholar]
- 99.Pinner E, Padan E, Schuldiner S. Cloning, sequencing, and expression of the nhaB gene, encoding a Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1992;267:11064–11068. [PubMed] [Google Scholar]
- 100.Pinner E, Kotler Y, Padan E, Schuldiner S. Physiological role of nhaBa specific Na+/H+ antiporter in Escherichia coli. J Biol Chem. 1993;268:1729–1734. [PubMed] [Google Scholar]
- 101.Ivey DM, Guffanti AA, Zemsky J, Pinner E, Karpel R, Padan E, Schuldiner S, Krulwich TA. Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J Biol Chem. 1993;268:11296–11303. [PubMed] [Google Scholar]
- 102.Sakuma T, Yamada N, Saito H, Kakegawa T, Kobayashi H. pH dependence of the function of sodium ion extrusion systems in Escherichia coli. Biochim Biophys Acta. 1998;1363:231–237. doi: 10.1016/s0005-2728(97)00102-3. [DOI] [PubMed] [Google Scholar]
- 103.Nozaki K, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem Biophys Res Commun. 1996;222:774–779. doi: 10.1006/bbrc.1996.0820. [DOI] [PubMed] [Google Scholar]
- 104.Ohyama T, Igarashi K, Kobayashi H. Physiological role of the chaA gene in sodium and calcium circulations at a high pH in Escherichia coli. J Bacteriol. 1994;176:4311–4315. doi: 10.1128/jb.176.14.4311-4315.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Padan E, Schuldiner S. Molecular biology of Na+/H+ antiporters: molecular devices that couple the Na+ and H+ circulation in cells. Biochim Biophys Acta. 1994;1187:206–210. doi: 10.1016/0005-2728(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 106.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 107.Rahav-Manor O, Carmel O, Karpel R, Taglicht D, Glaser G, Schuldiner S, Padan E. NhaR, a protein homologous to a family of bacterial regulatory proteins (LysR), regulates nhaAthe sodium proton antiporter gene in Escherichia coli. J Biol Chem. 1992;267:10433–10438. [PubMed] [Google Scholar]
- 108.Carmel O, Rahav-Manor O, Dover N, Shaanan B, Padan E. The Na+-specific interaction between the LysR-type regulator, NhaR, and the nhaA gene encoding the Na+/H+ antiporter of Escherichia coli. EMBO J. 1997;16:5922–5929. doi: 10.1093/emboj/16.19.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dover N, Higgins CF, Carmel O, Rimon A, Pinner E, Padan E. Na+-induced transcription of nhaA, which encodes an Na+/H+ antiporter in Escherichia coli, is positively regulated by nhaR and affected by hns. J Bacteriol. 1996;178:6508–6517. doi: 10.1128/jb.178.22.6508-6517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Storz G, Tartaglia LA, Ames BN. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 111.Schell MA. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 112.Henikoff S, Haughn GW, Calvo JM, Wallace JC. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988;85:6602–6. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Christman MF, Storz G, Ames BN. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989;86:3484–8. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dover N, Padan E. Transcription of nhaA, the main Na+/H+ antiporter of Escherichia coli, is regulated by Na+ and growth phase. J Bacteriol. 2001;183:644–653. doi: 10.1128/JB.183.2.644-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hengge-Aronis R. In: Bacterial Stress Responses. Storz G, Hengge-Aronis R, editors. ASM Press; Washington, DC: 2000. pp. 161–178. [Google Scholar]
- 116.Hengge-Aronis R. Recent insights into the general stress response regulatory network in Escherichia coli. J Mol Microbiol Biotechnol. 2002;4:341–346. [PubMed] [Google Scholar]
- 117.Padan E, Krulwich TA. In: Bacterial Stress Response. Storz G, Hengge-Aronis R, editors. ASM Press; Washington, D.C.: 2000. pp. 117–130. [Google Scholar]
- 118.Kaback HR. Transport in isolated bacterial membrane vesicles. Methods Enzymol. 1974;31:698–709. doi: 10.1016/0076-6879(74)31075-0. [DOI] [PubMed] [Google Scholar]
- 119.Rosen BP. Ion extrusion systems in E. coli. Methods Enzymol. 1986;125:328–386. doi: 10.1016/s0076-6879(86)25028-4. [DOI] [PubMed] [Google Scholar]
- 120.Tolner B, Ubbink-Kok T, Poolman B, Konings WN. Cation-selectivity of the L-glutamate transporters of Escherichia coli, Bacillus stearothermophilus and Bacillus caldotenax: dependence on the environment in which the proteins are expressed. Mol Microbiol. 1995;18:123–133. doi: 10.1111/j.1365-2958.1995.mmi_18010123.x. [DOI] [PubMed] [Google Scholar]
- 121.Cheng J, Hicks DB, Krulwich TA. The purified Bacillus subtilis tetracycline efflux protein TetA(L) reconstitutes both tetracycline-cobalt/H+ and Na+(K+)/H+ exchange. Proc Natl Acad Sci USA. 1996;93:14446–14451. doi: 10.1073/pnas.93.25.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lewinson O, Padan E, Bibi E. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc Natl Acad Sci USA. 2004;101:14073–14078. doi: 10.1073/pnas.0405375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Macnab RM, Castle AM. A variable stoichiometry model for pH homeostasis in bacteria. Biophys J. 1987;52:637–647. doi: 10.1016/S0006-3495(87)83255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Taglicht D, Padan E, Schuldiner S. Proton-sodium stoichiometry of NhaA, an electrogenic antiporter from Escherichia coli. J Biol Chem. 1993;268:5382–5387. [PubMed] [Google Scholar]
- 125.Pinner E, Padan E, Schuldiner S. Kinetic properties of NhaB, a Na+/H+ antiporter from Escherichia coli. J Biol Chem. 1994;269:26274–26279. [PubMed] [Google Scholar]
- 126.Harrington CR, Baddiley J. Synthesis of peptidoglycan and teichoic acid in Bacillus subtilis: role of the electrochemical proton gradient. J Bacteriol. 1984;159:925–933. doi: 10.1128/jb.159.3.925-933.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jolliffe LK, Doyle RJ, Streips UN. The energized membrane and cellular autolysis in Bacillus subtilis. Cell. 1981;25:753–763. doi: 10.1016/0092-8674(81)90183-5. [DOI] [PubMed] [Google Scholar]
- 128.Reenstra WW, Patel L, Rottenberg H, Kaback HR. Electrochemical proton gradient in inverted membrane vesicles from Escherichia coli. Biochemistry. 1980;19:1–9. doi: 10.1021/bi00542a001. [DOI] [PubMed] [Google Scholar]
- 129.Waggoner AS. The use of cyanine dyes for the determination of membrane potentials in cells, organelles, and vesicles. Methods Enzymol. 1979;55:689–695. doi: 10.1016/0076-6879(79)55077-0. [DOI] [PubMed] [Google Scholar]
- 130.Guffanti AA, Cheng J, Krulwich TA. Electrogenic antiport activities of the Gram-positive Tet proteins include a Na+(K+)/K+ mode that mediates net K+ uptake. J Biol Chem. 1998;273:26447–26454. doi: 10.1074/jbc.273.41.26447. [DOI] [PubMed] [Google Scholar]
- 131.Nakamura T, Hsu C, Rosen BP. Cation/proton antiport systems in Escherichia coli: solubilization and reconstitution of ΔpH-driven sodium/proton and calcium/proton antiporters. J Biol Chem. 1986;261:678–683. [PubMed] [Google Scholar]
- 132.Rimon A, Gerchman Y, Kariv Z, Padan E. A point mutation (G338S) and its suppressor mutations affect both the pH response of the NhaA-Na+/H+ antiporter as well as the growth phenotype of Escherichia coli. J Biol Chem. 1998;273:26470–26476. doi: 10.1074/jbc.273.41.26470. [DOI] [PubMed] [Google Scholar]
- 133.Wei Y, Guffanti AA, Ito M, Krulwich TA. Bacillus subtilis YqkI is a novel malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J Biol Chem. 2000;275:30287–30292. doi: 10.1074/jbc.M001112200. [DOI] [PubMed] [Google Scholar]
- 134.Epstein W. The roles and regulation of potassium in bacteria. Prog Nucleic Acid Res Mol Biol. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 135.Bakker EP, Booth IR, Dinnbier U, Epstein W, Gajewska A. Evidence for multiple K+ export systems in Escherichia coli. J Bacteriol. 1987;169:3743–3749. doi: 10.1128/jb.169.8.3743-3749.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Holtmann G, Bakker EP, Uozumi N, Bremer E. KtrAB and KtrCD: Two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol. 2003;185:1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Plack RH, Jr, Rosen BP. Cation/proton antiport systems in Escherichia coli. Absence of potassium/proton antiporter activity in a pH-sensitive mutant. J Biol Chem. 1980;255:3824–3825. [PubMed] [Google Scholar]
- 138.Kakinuma Y, Igarashi K. Potassium/proton antiport system of growing Enterococcus hirae at high pH. J Bacteriol. 1995;177:2227–2229. doi: 10.1128/jb.177.8.2227-2229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nakamura T, Kawasaki S, Unemoto T. Roles of K+ and Na+ in pH homeostasis and growth of the marine bacterium Vibrio alginolyticus. J Gen Microbiol. 1992;138:1271–1276. doi: 10.1099/00221287-138-6-1271. [DOI] [PubMed] [Google Scholar]
- 140.Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 141.Gerchman Y, Olami Y, Rimon A, Taglicht D, Schuldiner S, Padan E. Histidine-226 is part of the pH sensor of NhaA, a Na+/H+ antiporter in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1212–1216. doi: 10.1073/pnas.90.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aronson PS. Kinetic properties of the plasma membrane Na+-H+ exchanger. Annu Rev Physiol. 1985;47:545–560. doi: 10.1146/annurev.ph.47.030185.002553. [DOI] [PubMed] [Google Scholar]
- 143.Guffanti AA, Krulwich TA. Tetracycline/H+ antiport and Na+/H+ antiport catalyzed by the Bacillus subtilis TetA(L) transporter expressed in Escherichia coli. J Bacteriol. 1995;177:4557–4561. doi: 10.1128/jb.177.15.4557-4561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Padan E, Tzubery T, Herz K, Kozachkov L, Rimon A, Galili L. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim Biophys Acta. 2004;1658:2–13. doi: 10.1016/j.bbabio.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 145.Putney LK, Denker SP, Barber DL. The changing face of the Na+/H+ exchanger, NHE1: structure, regulation, and cellular actions. Annu Rev Pharmacol Toxicol. 2002;42:527–52. doi: 10.1146/annurev.pharmtox.42.092001.143801. [DOI] [PubMed] [Google Scholar]
- 146.Wakabayashi S, Shigekawa M, Pouyssegur J. Molecular physiology of vertebrate Na+/H+ exchangers. Physiol Rev. 1997;77:51–74. doi: 10.1152/physrev.1997.77.1.51. [DOI] [PubMed] [Google Scholar]
- 147.Wakabayashi S, Fafournoux P, Sardet C, Pouyssegur J. The Na+/H+ antiporter cytoplasmic domain mediates growth factor signals and controls “H+-sensing”. Proc Natl Acad Sci USA. 1992;89:2424–2428. doi: 10.1073/pnas.89.6.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rimon A, Gerchman Y, Olami Y, Schuldiner S, Padan E. Replacements of histidine 226 of NhaA-Na+/H+ antiporter of Escherichia coli. Cysteine (H226C) or serine (H226S) retain both normal activity and pH sensitivity, aspartate (H226D) shifts the pH profile toward basic pH, and alanine (H226A) inactivates the carrier at all pH values. J Biol Chem. 1995;270:26813–26817. doi: 10.1074/jbc.270.45.26813. [DOI] [PubMed] [Google Scholar]
- 149.Tzubery T, Rimon A, Padan E. Mutation E252C increases drastically the Km value for Na+ and causes an alkaline shift of the pH dependence of NhaA Na+/H+ antiporter of Escherichia coli. J Biol Chem. 2004;279:3265–3272. doi: 10.1074/jbc.M309021200. [DOI] [PubMed] [Google Scholar]
- 150.Galili L, Rothman A, Kozachkov L, Rimon A, Padan E. Transmembrane domain IV is involved in ion transport activity and pH regulation of the NhaA-Na+/H+ antiporter of Escherichia coli. Biochemistry. 2002;41:609–617. doi: 10.1021/bi011655v. [DOI] [PubMed] [Google Scholar]
- 151.Galili L, Herz K, Dym O, Padan E. Unraveling functional and structural interactions between transmembrane domains IV and XI of NhaA Na+/H+ antiporter of Escherichia coli. Journal of Biological Chemistry. 2004;279:23104–23113. doi: 10.1074/jbc.M400288200. [DOI] [PubMed] [Google Scholar]
- 152.Aronson PS, Nee J, Suhm MA. Modifier role of internal H+ in activating the Na+-H+ exchanger in renal microvillus membrane vesicles. Nature. 1982;299:161–163. doi: 10.1038/299161a0. [DOI] [PubMed] [Google Scholar]
- 153.Wakabayashi S, Hisamitsu T, Pang T, Shigekawa M. Kinetic dissection of two distinct proton binding sites in Na+/H+ exchangers by measurement of reverse mode reaction. J Biol Chem. 2003;278:43580–43585. doi: 10.1074/jbc.M306690200. [DOI] [PubMed] [Google Scholar]
- 154.Venturi M, Rimon A, Gerchman Y, Hunte C, Padan E, Michel H. The monoclonal antibody 1F6 identifies a pH-dependent conformational change in the hydrophilic NH2 terminus of NhaA Na+/H+ antiporter of Escherichia coli. J Biol Chem. 2000;275:4734–4742. doi: 10.1074/jbc.275.7.4734. [DOI] [PubMed] [Google Scholar]
- 155.Rimon A, Tzubery T, Galili L, Padan E. Proximity of cytoplasmic and periplasmic loops in NhaA Na+/H+ antiporter of Escherichia coli as determined by site-directed thiol cross-linking. Biochemistry. 2002;41:14897–14905. doi: 10.1021/bi0261342. [DOI] [PubMed] [Google Scholar]
- 156.Hellmer J, Teubner A, Zeilinger C. Conserved arginine and aspartate residues are critical for function of MjNhaP1, a Na+/H+ antiporter of M. jannaschii. FEBS Lett. 2003;542:32–36. doi: 10.1016/s0014-5793(03)00332-6. [DOI] [PubMed] [Google Scholar]
- 157.Inoue H, Sakurai T, Ujike S, Tsuchiya T, Murakami H, Kanazawa H. Expression of functional Na+/H+ antiporters of Helicobacter pylori in antiporter-deficient Escherichia coli mutants. FEBS Lett. 1999;443:11–16. doi: 10.1016/s0014-5793(98)01652-4. [DOI] [PubMed] [Google Scholar]
- 158.Stingl K, Altendorf K, Bakker EP. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 2002;10:70–74. doi: 10.1016/s0966-842x(01)02287-9. [DOI] [PubMed] [Google Scholar]
- 159.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Acid-adaptive genes of Helicobacter pylori. Infection and Immunity. 2003;71:5921–5939. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Takami H, Krulwich TA. Reidentification of facultatively alkaliphilic Bacillus firmus OF4 as Bacillus pseudofirmus OF4. Extremophiles. 2000;4:19–22. doi: 10.1007/s007920050003. [DOI] [PubMed] [Google Scholar]
- 161.Guffanti AA, Finkelthal O, Hicks DB, Falk L, Sidhu A, Garro A, Krulwich TA. Isolation and characterization of new facultatively alkalophilic strains of Bacillus species. J Bacteriol. 1986;167:766–773. doi: 10.1128/jb.167.3.766-773.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Krulwich TA, Ito M, Gilmour R, Sturr MG, Guffanti AA, Hicks DB. Energetic problems of extremely alkaliphilic aerobes. Biochim Biophys Acta. 1996;1275:21–26. doi: 10.1016/0005-2728(96)00044-8. [DOI] [PubMed] [Google Scholar]
- 163.Sturr MG, Guffanti AA, Krulwich TA. Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J Bacteriol. 1994;176:3111–3116. doi: 10.1128/jb.176.11.3111-3116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hosler JP. The influence of subunit III of cytochrome c oxidase on the D pathway, the proton exit pathway and mechanism-based inactivation in subunit I. Biochim Biophys Acta. 2004;1655:332–339. doi: 10.1016/j.bbabio.2003.06.009. [DOI] [PubMed] [Google Scholar]
- 165.Marantz Y, Nachliel E, Aagaard A, Brzezinski P, Gutman M. The proton collecting function of the inner surface of cytochrome c oxidase from Rhodobacter sphaeroides. Proc Natl Acad Sci USA. 1998;95:8590–8595. doi: 10.1073/pnas.95.15.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Riesle J, Oesterhelt D, Dencher NA, Heberle J. D38 is an essential part of the proton translocation pathway in bacteriorhodopsin. Biochemistry. 1996;35:6635–6643. doi: 10.1021/bi9600456. [DOI] [PubMed] [Google Scholar]
- 167.Hunte C, Screpanti M, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;534:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 168.Verkhovskaya ML, Barquera B, Wikstrom M. Deletion of one of two Escherichia coli genes encoding putative Na+/H+ exchangers (ycgO) perturbs cytoplasmic alkali cation balance at low osmolarity. Microbiology. 2001;147:3005–3013. doi: 10.1099/00221287-147-11-3005. [DOI] [PubMed] [Google Scholar]
- 169.Kuroda T, Fujita N, Utsugi J, Kuroda M, Mizushima T, Tsuchiya T. A major Li+ extrusion system NhaB of Pseudomonas aeruginosa: comparison with the major Na+ extrusion system NhaP. Microbiol Immunol. 2004;48:243–250. doi: 10.1111/j.1348-0421.2004.tb03520.x. [DOI] [PubMed] [Google Scholar]
- 170.Gouda T, Kuroda M, Hiramatsu T, Nozaki K, Kuroda T, Mizushima T, Tsuchiya T. nhaG Na+/H+ antiporter gene of Bacillus subtilis ATCC9372, which is missing in the complete genome sequence of strain 168, and properties of the antiporter. J Biochem (Tokyo) 2001;130:711–717. doi: 10.1093/oxfordjournals.jbchem.a003038. [DOI] [PubMed] [Google Scholar]
- 171.Utsugi J, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Cloning and sequencing of a novel Na+/H+ antiporter gene from Pseudomonas aeruginosa. Biochim Biophys Acta. 1998;1398:330–334. doi: 10.1016/s0167-4781(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 172.Hase CC. Virulence and sodium bioenergetics. Trends Microbiol. 2000;8:490–491. doi: 10.1016/s0966-842x(00)01870-9. [DOI] [PubMed] [Google Scholar]
- 173.Hase CC, Barquera B. Role of sodium bioenergetics in Vibrio cholerae. Biochim Biophys Acta. 2001;1505:169–178. doi: 10.1016/s0005-2728(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 174.Tokuda H, Unemoto T. Characterization of the respiration-dependent Na+ pump in the marine bacterium Vibrio alginolyticus. J Biol Chem. 1982;257:10007–10014. [PubMed] [Google Scholar]
- 175.Dzioba J, Ostroumov E, Winogrodzki A, Dibrov P. Cloning, functional expression in Escherichia coli and primary characterization of a new Na+/H+ antiporter, NhaD, of Vibrio cholerae. Mol Cell Biochem. 2002;229:119–124. doi: 10.1023/a:1017932829927. [DOI] [PubMed] [Google Scholar]
- 176.Nakamura T, Komano Y, Itaya E, Tsukamoto K, Tsuchiya T, Unemoto T. Cloning and sequencing of an Na+/H+ antiporter gene from the marine bacterium Vibrio alginolyticus. Biochim Biophys Acta. 1994;1190:465–468. doi: 10.1016/0005-2736(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 177.Nakamura T, Enomoto H, Unemoto T. Cloning and sequencing of nhaB gene encoding an Na+/H+ antiporter from Vibrio alginolyticus. Biochim Biophys Acta. 1996;1275:157–160. doi: 10.1016/0005-2728(96)00034-5. [DOI] [PubMed] [Google Scholar]
- 178.Kuroda T, Shimamoto T, Inaba K, Tsuda M, Tsuchiya T. Properties and sequence of the NhaA Na+/H+ antiporter of Vibrio parahaemolyticus. J Biochem. 1994;116:1030–1038. doi: 10.1093/oxfordjournals.jbchem.a124624. [DOI] [PubMed] [Google Scholar]
- 179.Nozaki K, Kuroda T, Mizushima T, Tsuchiya T. A new Na+/H+ antiporter, NhaD, of Vibrio parahaemolyticus. Biochim Biophys Acta. 1998;1369:213–220. doi: 10.1016/s0005-2736(97)00223-x. [DOI] [PubMed] [Google Scholar]
- 180.Ostroumov E, Dzioba J, Loewen PC, Dibrov P. Asp(344) and Thr(345) are critical for cation exchange mediated by NhaD, Na+/H+ antiporter of Vibrio cholerae. Biochim Biophys Acta. 2002;1564:99–106. doi: 10.1016/s0005-2736(02)00407-8. [DOI] [PubMed] [Google Scholar]
- 181.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Guillaume G, Ledent V, Moens W, Collard JM. Phylogeny of efflux-mediated tetracycline resistance genes and related proteins revisited. Microb Drug Resist. 2004;10:11–26. doi: 10.1089/107662904323047754. [DOI] [PubMed] [Google Scholar]
- 183.Levy SB. Active efflux, a common mechanism for biocide and antibiotic resistance. J Appl Microbiol. 2002;92:65S–71S. [PubMed] [Google Scholar]
- 184.Ives CL, Bott KF. Cloned Bacillus subtilis chromosomal DNA mediates tetracycline resistance when present in multiple copies. J Bacteriol. 1989;171:1801–1810. doi: 10.1128/jb.171.4.1801-1810.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ives CL, Bott KF. Characterization of chromosomal DNA amplifications with associated tetracycline resistance in Bacillus subtilis. J Bacteriol. 1990;172:4936–4944. doi: 10.1128/jb.172.9.4936-4944.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gillespie MT, May JW, Skurray RA. Detection of an integrated tetracycline resistance plasmid in the chromosome of methicillin-resistant Staphylococcus aureus. J Gen Microbiol. 1986;132:1723–1728. doi: 10.1099/00221287-132-6-1723. [DOI] [PubMed] [Google Scholar]
- 187.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.McMurry L, Petrucci RE, Jr, Levy SB. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yamaguchi A, Udagawa T, Sawai T. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J Biol Chem. 1990;265:4809–4813. [PubMed] [Google Scholar]
- 190.Yamaguchi A, Shiina Y, Fujihira E, Sawai T, Noguchi N, Sasatsu M. The tetracycline efflux protein encoded by the tet(K) gene from Staphylococcus aureus is a metal-tetracycline/H+ antiporter. FEBS Lett. 1995;365:193–197. doi: 10.1016/0014-5793(95)00455-i. [DOI] [PubMed] [Google Scholar]
- 191.Jin J, Guffanti AA, Bechhofer DH, Krulwich TA. Tet(L) and Tet(K) tetracycline-divalent metal/H+ antiporters: characterization of multiple catalytic modes and a mutagenesis approach to differences in their efflux substrate and coupling ion preferences. J Bacteriol. 2002;184:4722–4732. doi: 10.1128/JB.184.17.4722-4732.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Krulwich TA, Jin J, Guffanti AA, Bechofer DH. In: Microbial multidrug efflux. Paulsen IT, Lewis K, editors. Horizon Press; Norfolk, England: 2002. pp. 233–254. [Google Scholar]
- 193.Wang W, Guffanti AA, Wei Y, Ito M, Krulwich TA. Two types of Bacillus subtilis tetA(L) deletion strains reveal the physiological importance of TetA(L) in K+ acquisition as well as in Na+, alkali, and tetracycline resistance. J Bacteriol. 2000;182:2088–2095. doi: 10.1128/jb.182.8.2088-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stasinopoulos SJ, Farr GA, Bechhofer DH. Bacillus subtilis tetA(L) gene expression: evidence for regulation by translational reinitiation. Mol Microbiol. 1998;30:923–932. doi: 10.1046/j.1365-2958.1998.01119.x. [DOI] [PubMed] [Google Scholar]
- 195.Saier MH, Jr, Paulsen IT, Sliwinski MK, Pao SS, Skurray RA, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–274. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 196.Levy SB. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Griffith JK, Baker ME, Rouch DA, Page MG, Skurray RA, Paulsen IT, Chater KF, Baldwin SA, Henderson PJ. Membrane transport proteins: implications of sequence comparisons. Curr Opin Cell Biol. 1992;4:684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- 198.Jin J, Guffanti AA, Beck C, Krulwich TA. Twelve-transmembrane-segment (TMS) version (ΔTMS VII–VIII) of the 14-TMS Tet(L) antibiotic resistance protein retains monovalent cation transport modes but lacks tetracycline efflux capacity. J Bacteriol. 2001;183:2667–2671. doi: 10.1128/JB.183.8.2667-2671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol. 2005;187:2395–23405. doi: 10.1128/JB.187.7.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Teo JW, Tan TM, Poh CL. Genetic determinants of tetracycline resistance in Vibrio harveyi. Antimicrob Agents Chemother. 2002;46:1038–1045. doi: 10.1128/AAC.46.4.1038-1045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O’Reilly M, O’Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mathiesen C, Hagerhall C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim Biophys Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 203.Yoshinaka T, Takasu H, Tomizawa R, Kosona S, Kudo T. A shaE deletion mutant showed lower Na+ sensitivity compared to other deletion mutants in the Bacillus subtilis sodium/hydrogen antiporter (Sha) system. J Biosci Bioeng. 2003;95:306–309. doi: 10.1016/s1389-1723(03)80035-x. [DOI] [PubMed] [Google Scholar]
- 204.Andrews JH, Harris RF. The ecology and biogeography of microorganisms on plant surfaces. Annu Rev Phytopathol. 2000;38:145–180. doi: 10.1146/annurev.phyto.38.1.145. [DOI] [PubMed] [Google Scholar]
- 205.Yumoto I. Bioenergetics of alkaliphilic Bacillus spp. J Biosci Bioeng. 2002;93:342–353. doi: 10.1016/s1389-1723(02)80066-4. [DOI] [PubMed] [Google Scholar]
- 206.Yumoto I. Electron transport system in alkaliphilic Bacillus spp. Recent Res Devel Bacteriol. 2003;1:131–149. [Google Scholar]
- 207.Ito M. In: Encyclopedia of Environmental Microbiology. Bitton G, editor. Wiley; New York: 2002. pp. 133–140. [Google Scholar]
- 208.Dubnovitsky AP, Kapetaniou EG, Papageorgiou AC. Enzyme adaptation to alkaline pH: atomic resolution (1.08 A) structure of phosphoserine aminotransferase from Bacillus alcalophilus. Protein Sci. 2005;14:97–110. doi: 10.1110/ps.041029805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gilmour R, Krulwich TA. Construction and characterization of a mutant of alkaliphilic Bacillus firmus OF4 with a disrupted cta operon and purification of a novel cytochrome bd. J Bacteriol. 1997;179:863–870. doi: 10.1128/jb.179.3.863-870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Krulwich TA, Guffanti AA, Bornstein RF, Hoffstein J. A sodium requirement for growth, solute transport, and pH homeostasis in Bacillus firmus RAB. J Biol Chem. 1982;257:1885–1889. [PubMed] [Google Scholar]
- 211.Krulwich TA, Cheng J, Guffanti AA. The role of monovalent cation/proton antiporters in Na+-resistance and pH homeostasis in Bacillus: an alkaliphile versus a neutralophile. J Exp Biol. 1994;196:457–470. doi: 10.1242/jeb.196.1.457. [DOI] [PubMed] [Google Scholar]
- 212.Ito M, Guffanti AA, Krulwich TA. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett. 2001;496:117–120. doi: 10.1016/s0014-5793(01)02417-6. [DOI] [PubMed] [Google Scholar]
- 213.Kudo T, Hino M, Kitada M, Horikoshi K. DNA sequences required for the alkalophily of Bacillus sp. strain C-125 are located close together on its chromosomal DNA. J Bacteriol. 1990;172:7282–7283. doi: 10.1128/jb.172.12.7282-7283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ito M, Hicks DB, Henkin TM, Guffanti AA, Powers B, Zvi L, Uematsu K, Krulwich TA. MotPS is the stator-force generator for motility of alkaliphilic Bacillus and its homologue is a second functional Mot in Bacillus subtilis. Mol Microbiol. 2004;53:1035–1049. doi: 10.1111/j.1365-2958.2004.04173.x. [DOI] [PubMed] [Google Scholar]
- 215.Ito M, Xu H, Guffanti AA, Wei Y, Zvi L, Clapham DE, Krulwich TA. The voltage-gated Na+ channel NavBP has a role in motility, chemotaxis, and pH homeostasis of an alkaliphilic Bacillus. Proc Natl Acad Sci USA. 2004;101:10566–10571. doi: 10.1073/pnas.0402692101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sugiyama S. Na+-driven flagellar motors as a likely Na+ re-entry pathway in alkaliphilic bacteria. Mol Microbiol. 1995;15:592. doi: 10.1111/j.1365-2958.1995.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 217.Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 218.Koishi R, Xu H, Ren D, Navarro B, Spiller BW, Shi Q, Clapham DE. A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 219.Aono R, Ogino H, Horikoshi K. pH-dependent flagella formation by facultative alkaliphilic Bacillus sp. C-125. Biosci Biotechnol Biochem. 1992;56:48–53. doi: 10.1271/bbb.56.48. [DOI] [PubMed] [Google Scholar]
- 220.Aono R, Ohtani M. Loss of alkalophily in cell-wall-component-defective mutants derived from alkalophilic Bacillus C-125. Isolation and partial characterization of the mutants. Biochem J. 1990;266:933–936. [PMC free article] [PubMed] [Google Scholar]
- 221.Gilmour R, Messner P, Guffanti AA, Kent R, Scheberl A, Kendrick N, Krulwich TA. Two-dimensional gel electrophoresis analyses of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization of an S-layer protein with a role in alkaliphily. J Bacteriol. 2000;182:5969–5981. doi: 10.1128/jb.182.21.5969-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Aono R, Ito M, Horikoshi K. Instability of the protoplast membrane of facultative alkaliphilic Bacillus sp. C-125 at alkaline pH values below the pH optimum for growth. Biochem J. 1992;285:99–103. doi: 10.1042/bj2850099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Calamita HG, Ehringer WD, Koch AL, Doyle RJ. Evidence that the cell wall of Bacillus subtilis is protonated during respiration. Proc Natl Acad Sci USA. 2001;98:15260–15263. doi: 10.1073/pnas.261483798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Koch AL. The pH in the neighborhood of membranes generating a protonmotive force. J Theor Biol. 1986;120:73–84. doi: 10.1016/s0022-5193(86)80018-2. [DOI] [PubMed] [Google Scholar]
- 225.Clejan S, Krulwich TA, Mondrus KR, Seto-Young D. Membrane lipid composition of obligately and facultatively alkalophilic strains of Bacillus spp. J Bacteriol. 1986;168:334–340. doi: 10.1128/jb.168.1.334-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Haines TH, Dencher NA. Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 2002;528:35–39. doi: 10.1016/s0014-5793(02)03292-1. [DOI] [PubMed] [Google Scholar]
- 227.Hauss T, Dante S, Dencher NA, Haines TH. Squalane is in the midplane of the lipid bilayer: implications for its function as a proton permeability barrier. Biochim Biophys Acta. 2002;1556:149–154. doi: 10.1016/s0005-2728(02)00346-8. [DOI] [PubMed] [Google Scholar]
- 228.Wang Z, Hicks DB, Guffanti AA, Baldwin K, Krulwich TA. Replacement of amino acid sequence features of a- and c-subunits of ATP synthases of alkaliphilic Bacillus with the Bacillus consensus sequence results in defective oxidative phosphorylation and non-fermentative growth at pH 10.5. J Biol Chem. 2004;279:26546–26554. doi: 10.1074/jbc.M401206200. [DOI] [PubMed] [Google Scholar]
- 229.Rivera-Torres IO, Krueger-Koplin RD, Hicks DB, Cahill SM, Krulwich TA, Girvin ME. pKa of the essential Glu54 and backbone conformation for subunit c from the H+-coupled F1F0 ATP synthase from an alkaliphilic Bacillus. FEBS Lett. 2004;575:131–135. doi: 10.1016/j.febslet.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 230.Hicks DB, Plass RJ, Quirk PG. Evidence for multiple terminal oxidases, including cytochrome d, in facultatively alkaliphilic Bacillus firmus OF4. J Bacteriol. 1991;173:5010–5016. doi: 10.1128/jb.173.16.5010-5016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Quirk PG, Hicks DB, Krulwich TA. Cloning of the cta operon from alkaliphilic Bacillus firmus OF4 and characterization of the pH-regulated cytochrome caa3 oxidase it encodes. J Biol Chem. 1993;268:678–685. [PubMed] [Google Scholar]
- 232.Lewis RJ, Prince RC, Dutton PL, Knaff DB, Krulwich TA. The respiratory chain of Bacillus alcalophilus and its nonalkalophilic mutant derivative. J Biol Chem. 1981;256:10543–10549. [PubMed] [Google Scholar]
- 233.Hicks DB, Krulwich TA. The respiratory chain of alkaliphilic bacteria. Biochim Biophys Acta. 1995;1229:303–314. doi: 10.1016/0005-2728(95)00024-d. [DOI] [PubMed] [Google Scholar]
- 234.Yumoto I, Takahashi S, Kitagawa T, Fukumori Y, Yamanaka T. The molecular features and catalytic activity of CuA-containing aco3-type cytochrome c oxidase from a facultative alkalophilic Bacillus. J Biochem. 1993;114:88–95. doi: 10.1093/oxfordjournals.jbchem.a124145. [DOI] [PubMed] [Google Scholar]
- 235.Tolner B, Poolman B, Konings WN. Adaptation of microorganisms and their transport systems to high temperatures. Comp Biochem Physiol A Physiol. 1997;118:423–428. doi: 10.1016/s0300-9629(97)00003-0. [DOI] [PubMed] [Google Scholar]
- 236.Blair DF, Berg HC. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 237.Fillingame RH. The proton-translocating pumps of oxidative phosphorylation. Annu Rev Biochem. 1980;49:1079–1113. doi: 10.1146/annurev.bi.49.070180.005243. [DOI] [PubMed] [Google Scholar]
- 238.Wilson TH, Ding PZ. Sodium-substrate cotransport in bacteria. Biochim Biophys Acta. 2001;1505:121–130. doi: 10.1016/s0005-2728(00)00282-6. [DOI] [PubMed] [Google Scholar]
- 239.Abramson J, Iwata S, Kaback HR. Lactose permease as a paradigm for membrane transport proteins. Mol Membr Biol. 2004;21:227–236. doi: 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- 240.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 241.Padan E, Rimon A, Tzubery T, Muller M, Herz K, Galili L. In: The sodium-hydrogen exchanger, from molecule to its role in disease. Karmazyn M, Avkiran M, Fliegel L, editors. Kluver Academic Publishers; Boston: 2003. pp. 91–108. [Google Scholar]
- 242.Lauraeus M, Haltia T, Saraste M, Wikstrom M. Bacillus subtilis expresses two kinds of haem-A-containing terminal oxidases. Eur J Biochem. 1991;197:699–705. doi: 10.1111/j.1432-1033.1991.tb15961.x. [DOI] [PubMed] [Google Scholar]
- 243.Hicks DB, Cohen DM, Krulwich TA. Reconstitution of energy-linked activities of the solubilized F1F0 ATP synthase from Bacillus subtilis. J Bacteriol. 1994;176:4192–4195. doi: 10.1128/jb.176.13.4192-4195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Cheng J, Guffanti AA, Krulwich TA. A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol. 1997;23:1107–1120. doi: 10.1046/j.1365-2958.1997.2951656.x. [DOI] [PubMed] [Google Scholar]
- 245.Wei Y, Guffanti AA, Krulwich TA. Sequence analysis and functional studies of a chromosomal region of alkaliphilic Bacillus firmus OF4 encoding an ABC-type transporter with similarity of sequence and Na+ exclusion capacity to the Bacillus subtilis NatAB transporter. Extremophiles. 1999;3:113–120. doi: 10.1007/s007920050106. [DOI] [PubMed] [Google Scholar]
- 246.Krom BP, Warner JB, Konings WN, Lolkema JS. Transporters involved in uptake of di- and tricarboxylates in Bacillus subtilis. Antonie Van Leeuwenhoek. 2003;84:69–80. doi: 10.1023/a:1024445131925. [DOI] [PubMed] [Google Scholar]
- 247.Pragai Z, Eschevins C, Bron S, Harwood CR. Bacillus subtilis NhaC, an Na+/H+ antiporter, influences expression of the phoPR operon and production of alkaline phosphatases. J Bacteriol. 2001;183:2505–2515. doi: 10.1128/JB.183.8.2505-2515.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]