Abstract
We determined the substrate specificities of the protein tyrosine phosphatases (PTPs) PTP1B, RPTPα, SHP-1, and SHP-2 by on-bead screening of combinatorial peptide libraries and solution-phase kinetic analysis of individually synthesized phosphotyrosyl (pY) peptides. These PTPs exhibit different levels of sequence specificity and catalytic efficiency. The catalytic domain of RPTPα has very weak sequence specificity and is approximately two orders of magnitude less active than the other three PTPs. The PTP1B catalytic domain has modest preference for acidic residues on both sides of pY, is highly active towards multiply phosphorylated peptides, but disfavors basic residues at any position, a Gly at the pY−1 position, or a Pro at the pY+1 position. By contrast, SHP-1 and SHP-2 share similar but much narrower substrate specificities, with a strong preference for acidic and aromatic hydrophobic amino acids on both sides of the pY residue. An efficient SHP-1/2 substrate generally contains two or more acidic residues on the N-terminal side and one or more acidic residues on the C-terminal side of pY, but no basic residues. Subtle differences exist between SHP-1 and SHP-2 in that SHP-1 has a stronger preference for acidic residues at the pY−1 and pY+1 positions and the two SHPs prefer acidic residues at different positions N-terminal to pY. A survey of the known protein substrates of PTP1B, SHP-1, and SHP-2 shows an excellent agreement between the in vivo dephosphorylation pattern and the in vitro specificity profiles derived from library screening. These results suggest that different PTPs have distinct sequence specificity profiles and the intrinsic activity/specificity of the PTP domain is an important determinant of the enzyme’s in vivo substrate specificity.
Keywords: Combinatorial library, catalytic activity, kinetics, phosphotyrosine, PTP, substrate specificity
Protein-tyrosine phosphatases (PTPs) are a large family of enzymes (the human genome encodes 107 PTPs) that catalyze the hydrolysis of phosphotyrosine (pY) in proteins to tyrosine and inorganic phosphate (1). Once thought to be promiscuous “housekeeping” enzymes, PTPs are now known to be actively involved in cell signaling and have been described as having “exquisite substrate specificity” in vivo. However, in contrast to their well-established catalytic mechanism, their physiological substrates and functions remain poorly defined. Substantial evidence suggests that the substrate specificity of PTPs is controlled by both the intrinsic sequence specificity of the catalytic domain and the presence of targeting domains, which direct the PTP activity to their physiological targets and/or proper cellular locations (2).
PTP1B was the first PTP discovered and is one of the most studied PTPs. It is ubiquitously expressed and localized on the cytoplasmic face of the endoplasmic reticulum (3). PTP1B acts as an important regulator of metabolic signaling. Mice lacking Ptp1b exhibit enhanced insulin, leptin, and growth hormone sensitivity and resistance to diet-induced diabetes and obesity (4). Ptp1b is also required for Neu-induced breast cancer (5, 6). Accordingly, PTP1B is a promising target for the treatment of multiple diseases. A large number of phosphoproteins, including receptors, non-receptor protein tyrosine kinases, and other cytoplasmic proteins have been identified as PTP1B substrates in vivo (7). Examination of the peptide sequences flanking the pY sites dephosphorylated by PTP1B in these proteins, however, does not reveal any simple consensus sequence. This raises the question of whether the catalytic domain of PTP1B has any sequence specificity and how its in vivo substrate specificity is achieved.
SHP-1 and SHP-2 are non-transmembrane, Src homology 2 (SH2) domain-containing PTPs, which play critical roles in many signaling pathways and disease processes (8, 9). The two enzymes share >55% sequence identity, a similar 3D structure, and a common regulatory mechanism (10, 11). However, they often perform opposite cellular functions, with SHP-1 typically acting as a negative regulator of signaling, whereas SHP-2 is a positive regulator. Some of the functional differences may be attributed to the SH2 domains, which display overlapping but non-identical binding specificities and may therefore direct their respective PTP domains to different pY proteins (12). However, chimeras in which the PTP domains of SHP-1 and SHP-2 were swapped failed to exhibit the same biological activity as the wild-type enzymes (13, 14). These and other observations have led to the hypothesis that the catalytic domains of SHP-1 and SHP-2 may have distinct sequence specificities.
RPTPα is a representative receptor-like PTP that contains two tandem PTP domains. Both PTP domains are catalytically active, but the membrane-proximal domain (D1) is four to five orders of magnitude more active against pY peptides than the membrane-distal domain (D2) (15–17). The absence of two critical catalytic residues in the D2 domain – the tyrosine from the pY recognition loop (also known as the KNRY motif) and the aspartic acid from the WPD loop (which acts as a general acid/base during catalysis) – was shown to be responsible for the decreased activity. The existing data suggest that RPTPα-D1 is responsible for the enzyme’s in vivo phosphatase activity, whereas the D2 domain regulates the activity/specificity of the D1 domain through protein-protein interactions. RPTPα forms catalytically inactive homodimers under physiological conditions and the D2 domain has been shown to be critical for stable dimer formation (18). RPTPα-D2 has also been shown to directly bind to Src kinase and is required for the dephosphorylation and activation of Src by RPTPα (19). The catalytic activity and substrate specificity of RPTPα has been examined using a small panel of pY peptides (17, 20). These studies revealed that RPTPα is substantially less active than most of the other PTPs and exhibits only modest sequence specificity.
Because information on the specificity profile of a PTP would be useful for identifying its physiological substrates and designing selective PTP inhibitors, there have been numerous attempts to define the intrinsic sequence specificities of PTPs. Early studies involved individual kinetic assays of synthetic pY peptides derived from known pY proteins in solution (17, 21–25). More recently, microarrays containing these pY peptides have been employed to profile PTP specificities (26, 27). These methods are inherently limited because a PTP can potentially interact with 3–5 residues on either side of pY (28–31). Complete characterization of the PTP would require the synthesis and assay of a prohibitively large number of peptides (206–2010). Therefore, great efforts have been devoted to the development of various combinatorial approaches. However, due to the lack of stable association between an active PTP and a pY peptide, library screening usually has been based on binding instead of catalysis, using either an inactive PTP mutant or a non-hydrolyzable pY analogue (32–35). Other approaches employed include inverse alanine scanning (36), ECLPISE (37), tagging PTP products via cleavage with α-chymotrypsin (38), or using phosphocumarin as a hydrolyzable pY analogue (39). Although the above approaches have provided valuable information on the specificity of a few PTPs [e.g., PTP1B (27, 32, 36)], specificity data are generally lacking for most of the other PTPs. To date, there is only very limited information on the substrate specificity of RPTPα (17, 20). Two earlier studies on SHP-1 and SHP-2 generated conflicting conclusions (25, 40). By screening a 361-member peptide library (XXpY) by mass spectrometry (ECLIPSE), we showed that SHP-1 prefers an acidic residue (Asp or Glu) at the pY−2 position and, to a lesser extent, at the pY−1 position as well (relative to pY, which is designated as position 0) (37). Screening a kinase-treated phage-display peptide library for binding to a catalytically inactive SHP-1 mutant also revealed a preference for Glu at the pY−2 position (35).
We have recently developed a general methodology to systematically profile the sequence specificity of PTPs (41, 42). The validity of the method has recently been demonstrated by profiling the substrate specificity of the TULA family of PTPs (which putatively utilize a histidine as the catalytic nucleophile) (43). In this work, we report an improved version of the library screening method and its application to determine the sequence specificities of PTP1B, RPTPα, SHP-1, and SHP-2. Our results show that RPTPA and PTP1B are broad-specificity enzymes with no simple consensus sequence, whereas SHP-1 and SHP-2 share similar specificities for peptides containing acidic and hydrophobic residues on both the N- and C-terminal sides of pY.
Materials and Methods
Materials
Fmoc-protected L-amino acids were purchased from Advanced ChemTech (Louisville, KY), Peptides International (Louisville, KY), or NovaBiochem (La Jolla, CA). O-Benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU) and 1-hydroxybenzotriazole hydrate (HOBt) were from Peptides International. All solvents and other chemical reagents were obtained from Aldrich, Fisher Scientific (Pittsburgh, PA), or VWR (West Chester, PA) and were used without further purification unless noted otherwise. N-(9-Fluorenylmethoxycarbonyloxy) succinimide (Fmoc-OSu) was from Advanced ChemTech. Phenyl isothiocyanate (PITC) was purchased in 1-mL sealed ampoules from Sigma-Aldrich, and a freshly opened ampoule was used in each experiment. Nα-Fmoc-O-t-butyl-3,5-difluorotyrosine [Fmoc-F2Y(tBu)-OH] was synthesized as described previously (42). Polyethylene glycol acrylamide (PEGA) resin (0.4 mmol/g, 150–300 μm in water) was from Peptide International. Clear amide resin (90 μm, 0.23 mmol/g) was from Advanced ChemTech. α-Cyano-4-hydroxycinnamic acid was purchased from Sigma and recrystallized prior to use.
Purification of Tyrosinase and PTPs
Tyrosinase was expressed in Escherichia coli transformed with the prokaryotic expression vector pET-22b(+) containing the gene for Streptomyces antibioticus tyrosinase and a C-terminal six-histidine tag (44). E. coli BL21(DE3)-RP CodonPlus cells were grown at 37 °C in LB medium containing ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) until the OD600 reached 0.6, when protein expression was induced by the addition of isopropyl-thio-β-D-galactoside (1 mM) and CuSO4 (1 mM). After 6 h of incubation at 30 °C, the cells were harvested and the tyrosinase was purified from the crude lysate by metal-affinity chromatography on a Co2+ (Talon) column, following the manufacturer’s instructions. The purified tyrosinase was dialyzed in 20 mM Tris, pH 8.3, 100 mM NaCl, and 0.1 mM CuSO4, concentrated, mixed with an equal volume of glycerol, and stored at −80 °C. The catalytic domains of SHP-1 [SHP-1(ΔSH2)] and SHP-2 [SHP-2(ΔSH2)] were expressed in E. coli and purified as previously described (45). PTP1B (amino acids 1–321) and RPTPα (amino acids 224–802) were expressed in E. coli and purified as previously described (46, 47). Protein concentration was determined by the Bradford method, using bovine serum albumin (BSA) as the standard.
Synthesis of Peptide Libraries
Library I (SAX5pYAABBRM-resin) was synthesized on 5 g of amino PEGA resin (0.4 mmol/g, 150–300 μm in water). All manipulations were performed at room temperature unless otherwise noted. The invariant positions (pYAABBRM and SA) were synthesized with 4 equivalents of Fmoc-amino acids using HBTU/HOBt/N-methylmorpholine (NMM) as the coupling reagents, and the coupling reaction was terminated after negative ninhydrin tests. For the synthesis of random residues, the resin was split into 19 equal portions and each portion was coupled twice, each with 5 equivalents of a different Fmoc-amino acid/HBTU/HOBt/NMM for 1 h (except for F2Y, for which 2.5 equivalents were used and coupled only once). To facilitate sequence determination by mass spectrometry, 5% (mol/mol) CD3CO2D was added to the coupling reactions of leucine and lysine, whereas 5% CH3CD2CO2D was added to the coupling reaction of Nle (48). The resin-bound library was washed with dichloromethane and deprotected using a modified reagent K [7.5% phenol, 5% water, 5% thioanisole, 2.5% ethanedithiol and 1% anisole in trifluoroacetic acid (TFA)] at room temperature for 2 h. The library was washed exhaustively with TFA, DCM and DMF and stored in DMF at −20 °C until use. Libraries II–V were synthesized in a similar manner. Libraries VI and VII were similarly prepared but with the following modifications. Prior to coupling of the first random position (X1) of library VI (ANNX6X5X4X3X2X1AABBRRM), the resin was split into two equal aliquots. The first half of resin was coupled with 2.5 equivalents of Fmoc-pY-OH, whereas the second half was further split into 19 equal portions, each of which was coupled to a different amino acid. For positions X2–X6, the resin was equally split into 20 portions and each was coupled to Fmoc-pY-OH or the 19 amino acids used for library I. To construct the random sequence region of library VII (which contains reduced Arg and Lys contents), the resin was split into 19 aliquots; two aliquots each contained 0.58% of the total resin mass and were coupled with Fmoc-Arg or Fmoc-Lys, while the remaining 17 aliquots each contained 5.8% of the resin and were coupled to the other 17 Fmoc-amino acids.
Library Screening
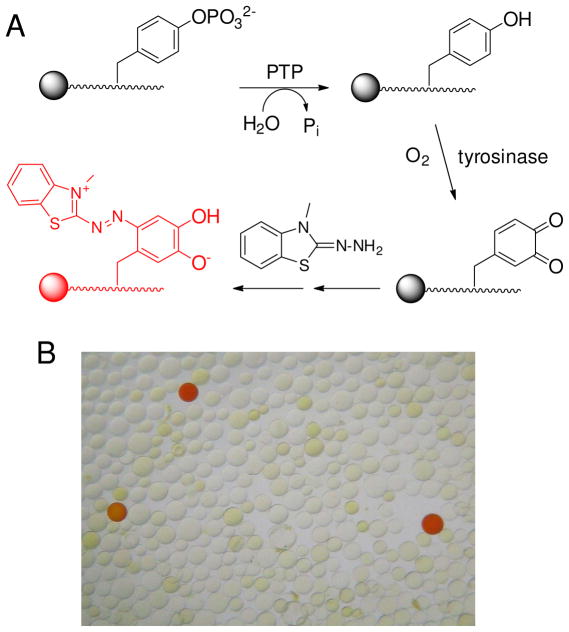
In a typical screening experiment, 15 mg of the proper peptide library (dry weight, ~40,000 beads) was placed in a plastic micro-BioSpin column (2 mL, Bio-Rad) and extensively washed with DMF and ddH2O. The resin was blocked for 1 h with blocking buffer (30 mM Hepes, pH 7.4, 150 mM NaCl, 0.01% Tween 20, and 0.1% gelatin). The library was then incubated with the proper PTP (final concentration 0.5–50 nM) in blocking buffer containing 5 mM tris(carboxyethyl)phosphine (total volume 0.8 mL) at room temperature for 5–20 min with gentle mixing. The resin was drained, washed with 0.1 M KH2PO4 (pH 6.8), and resuspended in 1.6 mL of 0.1 M KH2PO4 (pH 6.8) containing 1.2 μM tyrosinase and 6 mM 3-methyl-2-benzothiazolinonehydrazone (MBTH). The resulting mixture was incubated at room temperature with gentle mixing and exposure to air. Intense pink/red color typically developed on positive beads after 20–60 min. The positive beads were retrieved from the library using a micropipette under a dissecting microscope and sequenced by partial Edman degradation-mass spectrometry (PED-MS) (48). Control experiments without PTPs produced no red beads under otherwise identical conditions.
Synthesis of Selected Peptides
Individual peptides were synthesized on 100 mg of CLEAR-amide resin using standard Fmoc/HBTU/HOBt chemistry. For the coupling reaction of pY, 2.0 equivalents of Fmoc-Tyr(PO3H2)-OH were employed, whereas 4.0 equivalents were used for all other amino acids. The resin-bound peptides were washed with dichloromethane, cleaved from the resin, and deprotected using modified reagent K at room temperature for 2 h. After evaporation of solvents, the mixture was triturated three times with 20 volumes of cold Et2O. The precipitate was collected and dried under vacuum. The crude peptides were purified by reversed-phase HPLC on a semi-preparative C18 column. The identity of each peptide was confirmed by MALDI-TOF mass spectrometric analysis.
PTP Assay
PTP activity assays were performed with synthetic pY peptides as substrates in a quartz microcuvette. A typical reaction (total volume 120 μL) contained 100 mM Hepes, pH 7.4, 50 mM NaCl, 2 mM EDTA, 1 mM TCEP, 0.1 mg/mL BSA, and 0–1.4 mM pY peptide. The reaction was initiated by the addition of PTP (final concentration 5–50 nM) and monitored continuously at 282 nm (Δε = 1102 M−1 cm−1) on a UV-VIS spectrophotometer. The initial rates were calculated from the early regions of the reaction progress curves (typically <60 s). Data fitting against the Michaelis-Menten equation V = Vmax · [S]/(KM + [S]) or the simplified equation V = kcat[E][S]/KM (when KM ≫ [S]) gave the kinetic constants kcat, KM, and/or kcat/KM. For a few peptides (the alanine-scan series) against PTP1B, the kinetic constants were obtained by directly fitting the reaction progress curves against equation
where t is time, p is the product concentration at time t, E is the total enzyme concentration, and p∞ is the product concentration at infinity (49).
Results
Development of an Improved Library Screening Method
In our original method (41), pY peptide libraries were synthesized on TentaGel resin. Library screening involved oxidization of the exposed tyrosine by mushroom tyrosinase and derivatization of the orthoquinone product with biotin hydrazide. Subsequent addition of streptavidin-alkaline phosphatase conjugate and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) produced turquoise color on positive beads. Unfortunately, TentaGel resin is poorly permeable to macromolecules such as proteins (50), preventing their ready access to the immobilized substrates. In this work, we replaced the TentaGel resin with 150–300 μm (wet bead size) PEGA resin (cross-linked acrylamide and polyethylene glycol), which is permeable to proteins up to 35 kDa (50–52). However, PEGA resin is incompatible with the coloration method involving streptavidin-alkaline phosphatase and BCIP, because the hydrophobic dye formed does not deposit well on the hydrophilic resin. We thus developed another coloration method, by trapping the orthoquinone in situ with MBTH to form a red pigment on positive beads (42) (Figure 1). The color intensity of a bead is directly proportional to the amount of PTP reaction. To facilitate the oxidation reaction, we replaced mushroom tyrosinase, which exists as a tetramer of 120 kDa (53), with the smaller (32 kDa) tyrosinase from bacterium S. antibioticus (44). To determine whether the bacterial enzyme has any sequence specificity, a peptide library previously used for another study (54), MX5LNBBRM-PEGA resin where B is β-alanine and X is any of the 18 proteinogenic amino acids (except for Lys and Arg), was treated with excess tyrosinase and MBTH (6 mM). Approximately 20% of the library beads became red colored (theoretically, 23% of the library beads/peptides contain at least one tyrosine). Sequencing analysis of 90 randomly selected positive beads showed that each positive sequence contained at least one tyrosine (Table S1 in Supporting Information) and the tyrosinase has no obvious sequence selectivity. Trp and Phe were somewhat underrepresented among the most colored beads (Figure S1), likely because the hydrophobic peptides rendered the beads swell poorly in the aqueous buffer and were poorly accessible to the enzyme.
Figure 1.
(A) Reactions involved in library screening. (B) A portion of the peptide library after treatment with SHP-1 and tyrosinase/MBTH (viewed under a dissecting microscope).
N-Terminal Specificity of PTP1B
We chose to test our method on PTP1B, whose sequence specificity has been relatively well characterized (32, 36). Kinetic (22) and structural studies (28–31) have shown that the active site of PTPs makes contacts with 3–5 residues on each side of pY and the N-terminal sequence is generally more dominant in defining the substrate specificity. We decided to first determine the N-terminal specificity by screening the library, SAX5pYAABBRM-resin [library I, where B is β-alanine and X is 3,5-difluorotyrosine (F2Y or Y), norleucine (Nle or M), or any of the 17 proteinogenic amino acids excluding Met, Cys and Tyr] (Table 1). The C-terminal specificity was next defined by screening a second library that features random sequences on the C-terminal side of pY. The linker sequence BBRM was added at the C-terminus to facilitate mass spectrometric analysis (by introducing the positively charged Arg residue) and cleavage from the resin (after Met with CNBr). The dipeptides SA (at the N-terminus) and AA (C-terminal to pY) were intended to minimize any potential bias caused by the positive charges associated with the free N-terminus and the C-terminal Arg, respectively. Substitution of Nle for Met in the random region prevents internal cleavage during peptide release with CNBr, whereas replacement of Tyr with F2Y is necessary to avoid false positives during library screening. Substitution of F2Y for Tyr does not significantly alter the catalytic activity of PTPs (42). The library has a theoretical diversity of 2.48 × 106 and was synthesized on 5 g of PEGA resin (~1.5 × 107 beads).
Table 1.
Composition of Peptide Libraries Used in this Work
| Library No. | Library Design | Amino Acid Composition at Random Positions (X) |
|---|---|---|
| I | SAXXXXXpYAABBRM | Ala, Arg, Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| II | W[S/T][D/E][D/E]VpYXXXXLNBBRM | Ala, Arg, Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| III | WAAAApYXXXXXNNBBRM | Ala, Arg, Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| IV | AXXXXXpYXXXXXNNBBRM | Ala, Arg, Asn, Asp, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| V | AXXXXXpYXXXXXEEBBRM | Ala, Arg, Asn, Asp, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val |
| VI | ANNX6X5X4X3X2X1AABBRRM | Ala, Arg, Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys, Nle, Phe, Pro, Ser, Thr, Trp, Val, and pY (50% at X1 position) |
| VII | Alloc-ASXXXXXpYAABBRM | Ala, Arg (10-fold reduced content), Asn, Asp, F2Y, Gly, Gln, Glu, His, Ile, Leu, Lys (10-fold reduced content), Nle, Phe, Pro, Ser, Thr, Trp, Val |
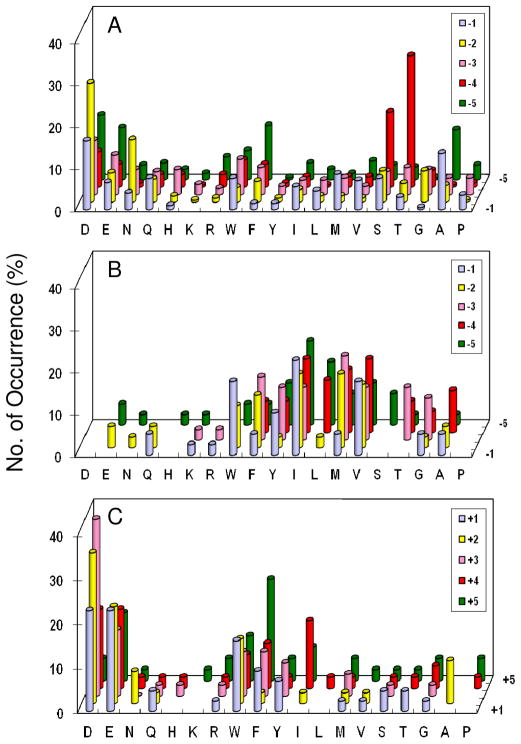
A total of 225 mg of library I (~570,000 beads) was screened against PTP1B (1–50 nM) in 15 separate experiments. The most intensely colored beads were removed manually from the library and individually sequenced by PED-MS to give 322 sequences (Table S2). These sequences can be readily separated into two different classes. The class I peptides resemble the preferred PTP1B substrates previously reported and are generally rich in acidic amino acids, whereas the class II peptides are rich in basic residues (Arg and Lys). Since the class II peptides have significantly poorer activities toward PTP1B than the class I peptides (vide infra), we consider them as “false” positives; they were selected from the library likely because the beads displaying these positively charged sequences swelled better in the screening buffer and/or recruited more PTP1B to their surfaces (vide infra). Inspection of the class I sequences reveals that PTP1B has very broad specificity, accepting most amino acids at pY−1 to pY−5 positions (Figure 2a). However, PTP1B does show a modest preference for acidic residues at all random positions (especially an Asp at position pY−2) and Thr/Ser at position pY−4. At the pY−1 position, while PTP1B does not have strong preference for a particular amino acid, it strongly disfavors basic amino acids; out of the 202 preferred class I substrates, none contained an Arg or Lys at this position. Gly and His are also disfavored at this position. These results are in agreement with the previous reports (32, 36).
Figure 2.
Histograms showing the sequence specificity of PTP1B. (A) Plot of the most preferred substrates selected from library I (N-terminal to pY); (B) plot of PTP1B-resistant sequences (colorless beads) from library I; and (C) plot of the most preferred substrates from library III (C-terminal to pY). The y axis represents the percentage of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (pY−5 to pY+5, on the z axis). M, Nle; Y, F2Y.
Additional information on the poor substrates of an enzyme provides a more complete picture of the specificity profile. The poor substrates of a PTP can be identified by exhaustively treating a peptide library with a high concentration of PTP for an extended period of time and analyzing the colorless beads. Treatment of library I (15 mg) with 1.0 μM PTP1B for 2 h resulted in the coloration of ~99% of the library beads. Fifty randomly selected colorless beads were sequenced by PED-MS. In general, the poor substrates were rich in hydrophobic residues and had few hydrophilic residues (Table S3 and Figure 2b). Again, we believe that the lack of reactivity of the hydrophobic sequences was because the beads containing these peptides swelled poorly in the aqueous buffer, rendering the peptides inaccessible to the PTP and/or tyrosinase. Fortunately, false negatives of this kind do not affect the validity of our library screening results, as these hydrophobic sequences are unlikely to be found on protein surfaces to act as PTP substrates in vivo. This result also suggests that PTP1B has broad substrate specificity, capable of dephosphorylating almost any pY peptide, albeit with different catalytic efficiencies.
C-Terminal Specificity of PTP1B
Library II (15 mg), W[S/T][D/E][D/E]VpYX4LNBBRM-resin (where X is the same set of 19 amino acids as in library I) (Table 1), which features the preferred sequences of PTP1B N-terminal to pY, was screened against PTP1B to give 17 most preferred sequences (Table S4). PTP1B primarily selected acidic and hydrophobic aromatic residues especially Trp. To determine whether the acidic sequences N-terminal to pY biased the C-terminal profile, we synthesized and screened a third library, WA4pYX5NNBBRM-resin (library III; X is the same set of 19 amino acids described above) (Table 1). The selected sequences also fell into two different classes (Table S4 and Figure 2c). The class I peptides were similar to those selected from library II and contained a combination of acidic and hydrophobic aromatic residues (Table S4). The most frequently selected motifs had a hydrophobic residue followed by two acidic residues at positions pY+1 to pY+3 (e.g., WDD, FDD, and YDE) or its inverted sequence (e.g., DDF, DEF, and EEW). The class II peptides were extremely rich in basic amino acids. The strong influence of the N-terminal sequence on the C-terminal sequences selected suggests potential sequence covariance between the N- and C-terminal sequences. This possibility was further investigated by screening PTP1B against library IV, AX5pYX5NNBBRM-resin (where X is the same set of amino acids but no F2Y), which is randomized at positions pY−5 to pY+5 (Table 1). Note that library IV has a theoretical diversity of 6.1 × 1012; the number of sequences screened (~2.1 × 105 total) represented only a tiny fraction of the sequence space. Nevertheless, screening of library IV confirmed the results obtained from libraries I–III and provided important new insights. When screening was carried out under normal salt conditions (150 mM NaCl), the sequences selected by PTP1B were overwhelmingly rich in basic residues (class II in Table 2). Screening at 500 mM NaCl greatly decreased the number of class II sequences and increased the number of acidic (class I) peptides (Table 2). The class I peptides were similar to those from library I, showing an overrepresentation of Asp, Glu, and other small hydrophilic residues at the pY−2 position and Ser, Thr, Glu, and Asp at the pY−4 position. The dramatic effect of salt concentration on the nature of sequences suggests that the Arg- and Lys-containing sequences were selected because of better accessibility of PTP1B to the beads carrying the Arg- and Lys-rich peptides. Presumably, the positive charges on the peptides repel each other and increase the distances between adjacent peptides. This in turn improves the swelling of these beads and makes the peptides more accessible to the enzyme. The positively charged beads may also recruit more PTP1B (pI 6.8) to their surfaces than beads carrying neutral or acidic peptides (55). To test this hypothesis, we screened PTP1B against library V, AX5pYX5EEBBRM-resin (where X is the same set of amino acids but no F2Y) (Table 1). This library is identical library IV, except that the Asn-Asn motif C-terminal to the random region was replaced by Glu-Glu. Screening of library V (30 mg) against PTP1B resulted in exclusively class I sequences (Table 2). It is likely that the two Glu residues increased the accessibility of the beads carrying acidic peptides through charge-charge repulsion and decreased the accessibility of beads with Arg- and Lys-rich peptides (by neutralizing the positive charges). Taken together, these results suggest that PTP1B has broad specificity C-terminal to pY as well, with a modest preference for acidic and hydrophobic residues. Interestingly, PTP1B strongly disfavors a proline at the pY+1 position. Out of the 112 class I and 86 class II peptides selected from libraries II-V (Table 2 and S4), only one sequence (PTLGDpRPRVAQ in Table 2) contained a proline at position pY+1. Vetter et al. reported a similar finding (36). The overall agreement of our results with the reported PTP1B data validates our library screening method.
Table 2.
Preferred PTP1B Substrates Selected from Libraries IV (AX5pYX5NN) and library V (AX5pYX5EE)a
| Library IV (150 mM NaCl) | |
|---|---|
| Class I | NRSRMpYQRGLF |
| GTDHApYHDNLL | NHQQApYRVRRI |
| ADVFQpYKRNRW | PARXXpYSRIRR |
| IINSMpYRRGHH | QRGQNpYRRAVR |
| FTQNTpYTRRKH | RSNLRpYSRRIM |
| IISNQpYRFARR | RRRMTpYGSFGT |
| RRFSKpYRVDGR | |
| Class II | RRNXXpYFLKRA |
| RRRXXpYNTKMM | RKIGLpYRHLRM |
| ANRRFpYHRRRR | RKFGNpYRRHVS |
| AIAGIpYNRRKA | RHAGFpYINRRS |
| DGNXXpYRGTRR | RVGLGpYQKFAR |
| FQRFFpYRQSQR | STNGRpYFEIAQ |
| FKNRIpYGARRA | TAKNRpYSRNRR |
| GFAGGpYRRRHG | TRQQGpYRVKRR |
| HFPASpYTIRRK | VRRAFpYRTIGH |
| INRNIpYSRRXX | VNASRpYRRHKL |
| IERRLpYKRRVH | XXRNVpYRRRGF |
| IGHXXpYGRRKK | XXKGLpYSRAVR |
| MPRSApYMRRIK | XNSAFpYTRQKW |
| NSSRSpYIQRKR | |
| Library IV (500 mM NaCl) | |
| Class I | SSVIDpYFRKNT |
| WNGMDpYDNFVG | NGFDSpYNRFTR |
| IQSNDpYGIIHG | PTLGDpYPRVAQ |
| MEIEFpYGDAIP | EMQSMpYNRSFG |
| AEWDPpYGLDFI | |
| ESLELpYTEMSE | Class II |
| STWDMpYFGTAP | EENHIpYRRSRQ |
| HSSDTpYNEDSM | KERQTpYVRHHS |
| QDDWIpYFDTFQ | LRDKLpYMQRKR |
| FFEXXpYEVGWG | XESKPpYDFKRR |
| FEVWApYQSDXX | DSREGpYESHNR |
| MQSWIpYDDAWV | XIFGApYSRRRV |
| ASFSPpYNXXEL | VTLRApYNRRNI |
| AQWTXpYEFWEP | KSWRFpYRTKPQ |
| KVDTDpYDTRDG | XKHRGpYKKRWH |
| MWETDpYFDKGF | RTNSGpYMTRKK |
| PTHDDpYLQHRR | RRAVGpYSNRRH |
| WFSSEpYSMNDR | KASAIpYRRFAX |
| NTGDHpYITTFK | PKRAMpYSMIRR |
| KDANTpYIHSGR | VRRRMpYKNTWM |
| FETHVpYGNDSK | IRFGNpYGARKR |
| KLDNApYIDGFG | AFSWPpYKSRRR |
| KKDSApYNHLNM | RMRGQpYRRKVH |
| ASWSNpYMKHRH | IKRRRpYGSNRK |
| NSFTSpYTRAGW | GQIASpYRIRWR |
| VFENMpYRRHKR | MRAPVpYRRART |
| APQTVpYRKDWD | RSFXVpYRVRXX |
| Library V (150 mM NaCl) | |
| Class I | NFISQpYSQWDT |
| QTEPMpYSDKWD | FFGEWpYGNDRS |
| GSLTVpYEPMWE | FMGFEpYTIPEQ |
| TQLGEpYGGWHN | FNAPMpYEEWVG |
| SGWAFpYGSPER | EQTFSpYQDMWF |
| PESDApYSDWFQ | LIILSpYGWNDN |
| WEMDVpYQAHAH | NMWFWpYATPNE |
| PDGTPpYQWNED | KVEDWpYGESFP |
| EFEAFpYGQFEH | MGQDGpYGWNFE |
| AWEAVpYGEPED | NDMXXpYEWKVD |
| VAWMNpYGNQLD | XVNMVpYSMDFN |
| FFDGEpYTWDHT | REENLpYTQHWE |
M, Nle; X, unidentified amino acid.
PTP1B Prefers Multiply Phosphorylated Peptides
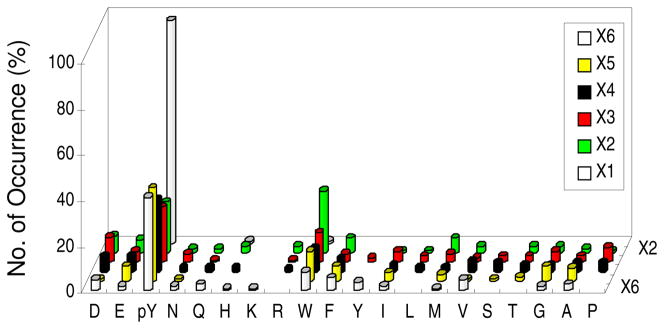
PTP1B was shown to be highly active toward peptides containing tandem pY motifs such as the DpY1162pYRK motif of insulin receptor (30, 56). To determine whether the pYpY motif is a specific recognition motif of PTP1B, we designed library VI, ANNX6X5X4X3X2X1AABBRRM, in which X1–X6 were Nle, F2Y, or any of the 17 amino acids excluding Cys, Met, and Tyr and X1 was enriched in pY content (50% total) (Table 1). A total of 43 mg of library VI was screened against PTP1B to give 129 most preferred substrates, which fall into two different classes (Table S5). The class I peptides (107 total) were surprisingly rich in pY at all six random positions (Figure 3). Among the class I peptides, 98% contained a pY at the X1 position, while 22, 24, 31, 42, and 41% of the sequences contained a pY at the X2 to X6 positions, respectively. One hundred and two peptides (96%) contained two or more pY residues, with most of them having a pY at the X1 position and another pY at the X5 or X4 position. Other than pY, PTP1B showed little selectivity for other amino acids, except for an overrepresentation of Trp at the X2 (pY−1) position. Forty-six peptides (43%) contained the pYpY motif; however, most of them had three or more pY residues. Among the 38 peptides that contained two pY residues, only two of them had a pYpY motif (Table S5). These results indicate that PTP1B generally prefers multiply phosphorylated peptides as substrates but has no special preference for the pYpY motif. Furthermore, none of the pYpY motifs had Arg or Lys after the second pY residue, as is present in the DpYpYRK motif of insulin receptor. The class II peptides (22 total) each contained a single pY residue, predominantly at the X1 position (Table S5). Their rich Arg and Lys contents suggest that they were selected due to the bead swelling and/or enzyme recruiting effects.
Figure 3.
Histograms showing PTP1B’s specificity toward multiply phosphorylated peptides. The y axis represents the number of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (X1 to X6, on the z axis). M, Nle; Y, F2Y.
N-Terminal Specificity of SHP-1 and SHP-2
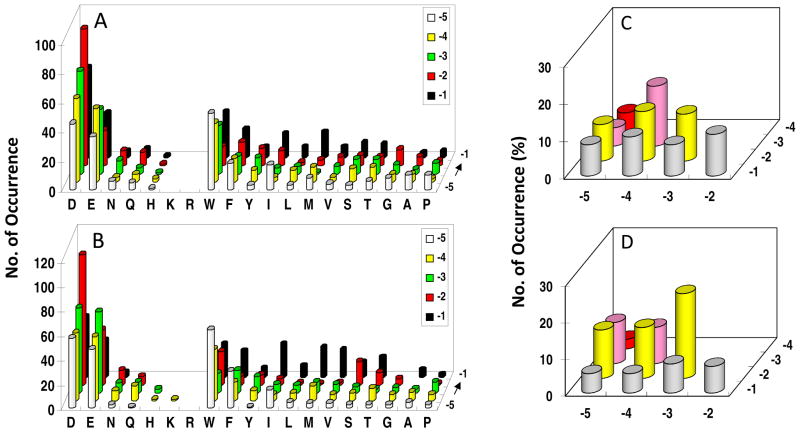
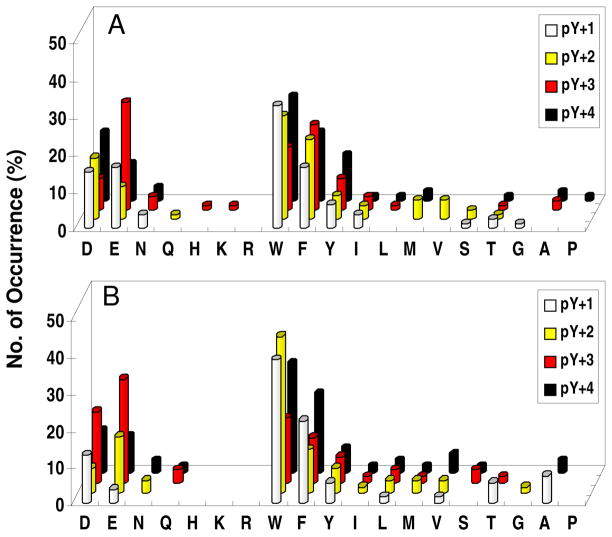
Library I was screened against the PTP domains of SHP-1 and SHP-2 (lacking SH2 domains). A total of 90 mg of library I (~240,000 beads) was screened against SHP-1 in six separate experiments, yielding 230 unambiguous sequences (Table S6). Similarly, 120 mg of the library was screened against SHP-2 to give 255 most preferred sequences (Table S7). SHP-1 and SHP-2 have similar specificity profiles, and the most striking feature is their overwhelming preference for acidic residues (especially Asp) (Figure 4a, b). More than 90% of the selected sequences for each PTP (208/230 for SHP-1 and 233/255 for SHP-2) contained at least two acidic residues, with some of them having three or four acidic residues. Among the ~6% peptides that had only one Asp or Glu, most also contained one or more Asn, Gln, Ser, or Thr. By contrast, positively charged residues were almost completely absent from the preferred substrates; out of the 485 selected sequences, only one SHP-2 substrate (FKDDI) had a Lys and only a few contained His (Table S6 and S7). Interestingly, the acidic residues were selected at all of the random positions (pY−1 to pY−5), although both PTPs most prefer an Asp at the pY−2 position. At the pY−1 position, SHP-1 and SHP-2 also exhibit significant preference for hydrophobic residues (Trp, Phe, Ile, Nle, and Val), but disfavor Gly and Pro. At positions pY−3 to pY−5, aromatic hydrophobic residues (Trp and Phe) are second most preferred by both PTPs (after acidic residues).
Figure 4.
Histograms showing a comparison of the sequence specificity of SHP-1 (A and C) and SHP-2 (B and D) on the N-terminal side of pY. In (A) and (B), the y axis represents the number of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (pY−5 to pY−1, on the z axis). M, Nle; Y, F2Y. In (C) and (D), the y axis represents the percentage of selected peptides that contained acidic residues at a particular combination of positions within the peptides (pY−1 to pY−4 on the z axis and pY−2 to pY−5 on the x axis). Only peptides containing two acidic residues (143 sequences for SHP-1 and 151 sequences for SHP-2) were used for the analysis in (C) and (D).
Closer examination of the selected sequences revealed some subtle differences between SHP-1 and SHP-2. First, SHP-1 has a stronger preference for acidic residues at the pY−1 position than SHP-2, which selected a larger number of aliphatic hydrophobic residues at this position (Figure 4a, b). At position pY−2, the trend is opposite, with SHP-2 showing stronger selectivity for Asp and Glu. Second, although the two enzymes selected a similar percentage of peptides containing multiple acidic residues (90.4 and 91.3% for SHP-1 and SHP-2, respectively), the distribution of the acidic residues among the five random positions (pY−1 to pY−5) was significantly different. To illustrate this point, we selected the sequences that contained two acidic residues from Table S6 and S7 and examined the location of their acidic residues (Figure 4c, d). SHP-1 shows a relatively even distribution of acidic residues among the ten possible combinations, with a slight preference for the −3/−4 combination (acidic residues at pY−3 and pY−4 positions). SHP-2, on the other hand, strongly favors the −2/−3 combination, followed by the −2/−4, −2/−5, −3/−5, and −3/−4 combinations, but disfavors the −4/−5 combination.
C-Terminal Specificity of SHP-1 and SHP-2
The C-terminal specificity was first evaluated by screening library II, which contained the preferred sequences of SHP-1/2 on the N-terminal side of pY. Screening 45 mg of library II against SHP-1 and SHP-2 produced 71 and 51 most preferred (and complete) sequences, respectively (Table S8). The two PTPs also showed similar specificity profiles on the C-terminal side of pY, selecting mostly hydrophobic aromatic and acidic residues, as they did on the N-terminal side (Figure 5). However, unlike the N-terminal side where acidic residues were more frequently selected than hydrophobic residues, both PTPs prefer hydrophobic over acidic residues C-terminal to pY. The vast majority of the selected sequences contained one (66% for SHP-1 and 80% for SHP-2) or two acidic residues (28% for SHP-1 and 16% for SHP-2), while only a few sequences (4/71 for SHP-1 and 2/51 for SHP-2) had zero or more than two acidic residues at the pY+1 to pY+4 positions. Next, SHP-1 and SHP-2 were screened against library III (WA4pYX5). Both PTPs selected acidic and hydrophobic aromatic residues at positions pY+1 to pY+5 from this library as well (Figure S2 and Table S9). However, when a hydrophobic sequence was on the N-terminal side, they exhibited a greater preference for acidic amino acids on the C-terminal side. For SHP-1, 33% and 54% of the selected sequences had two and three acidic residues, respectively, whereas for SHP-2, the corresponding values were 32% and 59%. These results indicate that peptide sequences on both sides of pY contribute to overall catalytic efficiency and there is a significant degree of covariance between the N- and C-terminal sequences. We therefore screened SHP-1 and SHP-2 against library IV (X5pYX5NN). Examination of the selected sequences revealed that most of the preferred substrates contained acidic residues on both sides of pY, confirming the results obtained from screening libraries I–III (Table S10). A few peptides contained no or only one acidic residue (though they often contain one or more Asn, Gln, Ser, and/or Thr) on the N-terminal side of pY, but then their C-terminal sequences had two or more acidic residues (e.g., SWIDFpYEDQVA and VWNNLpYDPFED in Table S10). Thus, acidic sequences on the C-terminal side can also render a peptide a good SHP-1/2 substrate. Some of the selected substrates had an occasional Arg and/or Lys on either side of pY (e.g., AHDRVpYDWDDF and DDSMLpYEFDRV), suggesting that an Arg- and/or Lys-containing peptide can still be a good substrate of SHP-1/2 as long as it contains acidic residues (four or more) to compensate for the positive charge associated with the Arg and/or Lys. Screening of SHP-1 against library VI resulted in similar sequences to those of PTP1B, except that SHP-1 did not select any Arg- and Lys-rich sequences (Table S11). Most of the selected substrates contained two or more pY residues, but relatively few of them had tandem pY motifs (especially among peptides that contained two pY residues). A notable difference from PTP1B was that screening libraries I and IV against SHP-1 and SHP-2 under high salt conditions (300 and 500 mM NaCl) did not cause any obvious change in their specificity profiles (data not shown). On the basis of the results from all four libraries, we conclude that SHP-1 and SHP-2 are more specific than PTP1B, and an efficient substrate of SHP-1 or SHP-2 should have a total of three or four acidic residues, preferably with two or three of them N-terminal to pY and one or two on the C-terminal side. Finally, the results from libraries II–IV revealed a subtle and yet recurring difference between SHP-1 and SHP-2, i.e., SHP-1 always selected a higher percentage of acidic residues than SHP-2 at the pY+1 position (Figures 5 and S3; Table S10).
Figure 5.
Histograms showing a comparison of the sequence specificity of SHP-1 (A) and SHP-2 (B) on the C-terminal side of pY. The y axis represents the percentage of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (pY+1 to pY+4, on the z axis). M, Nle; Y, F2Y.
Poor Substrates of SHP-1 and SHP-2
Screening of library II (15 mg each) against SHP-1 and SHP-2 (1.0 μM) resulted in ~1% colorless beads in each experiment. Thirty to 50 of the colorless beads were randomly selected and sequenced by PED-MS. In general, the poor substrates had very hydrophobic sequences and few acidic residues (Table S12 and Figure S3). Approximately 50% of the sequences contained one or more Arg, Lys, and/or His. Similar results were obtained from screening library I against SHP-1 and SHP-2 (data not shown). This is notably different from the poor substrates of PTP1B, which were almost exclusively hydrophobic sequences and had few Arg or Lys residues (Figure 2b). These observations reinforce the earlier conclusion that SHP-1 and SHP-2 prefer acidic and exclude basic sequences as substrates. They also demonstrate that under “forced” conditions, SHP-1 and SHP-2 are capable of dephosphorylating most pY peptides, albeit at drastically different catalytic efficiencies.
Specificity of RPTPα
Initial screening of RPTPα against library I resulted in almost exclusively positively charged sequences (Table S13). To determine whether this selectivity is an intrinsic property of the enzyme or was the result of preferential enzyme recruitment/resin swelling, we synthesized library VII (Alloc-ASX5pYAABBRM) (Table 1), which is structurally similar to library I but features a 10-fold reduction in the Arg and Lys content. This reduces the probability of multiple Arg and/or Lys residues in a sequence and therefore the effect of preferential enzyme recruitment/bead swelling on library screening. Library VII (total 60 mg) was screened against RPTPα (5 nM) and the most intensely colored beads were sequenced to give 104 sequences (Table S14). RPTPα showed only very weak N-terminal sequence selectivity, with some preference for Thr and Ser at the pY−2 and pY−5 positions and a slight preference for acidic residues at pY−2 to −4 positions. Other amino acids were selected with essentially equal frequencies at all of the random positions (Figure 6). Arg and Lys were underrepresented at all five positions. However, the number of Arg/Lys selected (4 total) was similar to the expected value of the unselected library [104 × 5 × 2/(19 × 10) = 5.5 Arg and/or Lys expected]. These observations suggest that RPTPα readily tolerates, but has no special preference for, the positively charged residues and that the Arg- and Lys-rich sequences selected from library I was the result of preferential enzyme recruitment/bead swelling.
Figure 6.
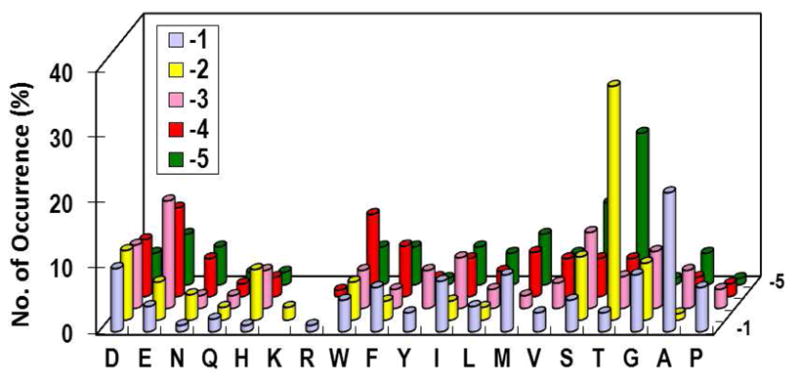
Substrate specificity of RPTPα on the N-terminal side of pY. The y axis represents the percentage of selected peptides that contained a particular amino acid (x axis) at a given position within the peptide (pY−1 to pY−4, on the z axis). M, Nle; Y, F2Y.
To assess whether RPTPα is selective on the C-terminal side of pY, we screened the enzyme against library IV (X5pYX5). Under normal salt conditions (150 mM NaCl), the selected sequences were overwhelmingly positively charged on both sides of pY (Table S15). Increasing the salt concentration to 300 mM had no significant effect, whereas at 500 mM NaCl, the selected sequences showed a decrease in the Arg and Lys content. The selected peptides were highly diverse in sequence (though there was an overrepresentation of hydrophobic residues at the pY−1 position) and no obvious consensus sequence could be derived. On the basis of these observations and solution-phase kinetic studies (vide infra), we conclude that RPTPα also lacks significant selectivity on the C-terminal side of pY.
Kinetic Properties of Selected Peptides toward PTP1B
To further evaluate the library screening results, we resynthesized four representative sequences selected against PTP1B (Ac-ASSDDpYAA-NH2 and Ac-DTADVpYAA-NH2 in class I and Ac-ARKRIpYAA-NH2 and Ac-RRISTpYAA-NH2 in class II) and determined their kinetic constants in the solution phase (Table 3, entries 1–4). The two class I peptides are indeed excellent substrates of PTP1B, having kcat/KM values of 1.8–3.5 × 106 M−1 s−1. The class II peptides also are good substrates, although their kcat/KM values were nearly an order of magnitude lower than those of the class I substrates. To assess the contribution of each position to the overall catalytic activity, an alanine-scanning study was carried out with peptide Ac-TDADVpYAA-NH2 (Table 3, entries 2 and 5–8). In keeping with PTP1B’s broad specificity, further substitutions caused only modest decreases in the catalytic activity. Mutation of the pY−1 Val had the largest effect (3-fold), followed by Asp→Ala mutation at position pY−2 (~2-fold). Substitution of Ala at the more distal positions including the Thr at position pY−4 had smaller effects (<2-fold). The importance of C-terminal residues was next assessed by kinetic analysis of a series of artificially designed substrates (Table 3, entries 9–16). Starting with peptide Ac-AAAApYAAAA-NH2 [which has a kcat/KM value of 1.3 × 106 M−1 s−1 (36)], introduction of acidic (Ac-AAAAApYEEVH-NH2) vs basic sequences C-terminal to pY (Ac-AAAAApYRHRR-NH2) resulted in a 7.9-fold difference in activity. The same trend was observed with peptides Ac-RTLAApYEEVH-NH2, Ac-RTLAApYAAAA-NH2, and Ac-RTLAApYRFQK-NH2 (which have kcat/KM values of 8.4 × 105, 5.1 × 105, 2.2 × 105 M−1 s−1, respectively). Similarly, replacement of the C-terminal AA of peptide 2 with acidic sequences DWEF and GEFTI increased the kcat/KM value by 1.7- and 1.4-fold, respectively (Table 3, compare entries 2, 15, and 16). Finally, nine peptides selected from library VI were tested to confirm PTP1B’s preference for multiply phosphorylated peptides (Table 3, entries 17–25). They contained one (e.g., Ac-AKFEDTpYAA-NH2), two (e.g., Ac-AVWEFpYpYAA-NH2 and Ac-AWSpYADpYAA-NH2), or three pY residues (e.g., Ac-ApYpYWTApYAA-NH2). Note that treatment of the peptides containing more than one pY with PTP1B is expected to produce a mixture of products and therefore the reported kcat and KM values are apparent ones. The mutli-pY peptides are generally more efficient substrates than the mono-pY peptides described above. For example, peptide Ac-AWSpYADpYAA-NH2 has a kcat/KM value of 2.35 × 107 M−1 s−1. To our knowledge, this is the most active PTP1B substrate known to date. Peptide Ac-ApYpYWTApYAA-NH2 is also an excellent substrate (kcat/KM = 1.51 × 107 M−1 s−1). None of the pYpY-containing peptides tested (Ac-AVWEFpYpYAA-NH2, Ac-ApYpYEVSpYAA-NH2, and Ac-ApYpYWTApYAA-NH2) had unusually high activities compared to the other peptides in this series, supporting our conclusion that while PTP1B clearly has preference for multiple pY peptide substrates, there is no special preference for tandem pY sequences.
Table 3.
Kinetic Properties of PTP1B against pY Peptides
| Entry No. | Peptide Sequencea | kcat (s−1) | KM (μM) | kcat/KM (μM−1 s−1) |
|---|---|---|---|---|
| 1 | ASSDDpYAA | 33 ± 1 | 18 ± 1 | 1.8 |
| 2 | DTADVpYAA | 31 ± 3 | 8.9 ± 1 | 3.5 |
| 3 | ARKRIpYAA | - | >500 | 0.14 |
| 4 | RRISTpYAA | 78 ± 2 | 90 ± 7 | 0.87 |
| 5 | DTADApYAA | 38 ± 2 | 35 ± 2 | 1.1 |
| 6 | DTAAVpYAA | 32 ± 1 | 18 ± 0.14 | 1.8 |
| 7 | DAADVpYAA | 32 ± 4 | 10 ± 1 | 3.1 |
| 8 | ATADVpYAA | 31 ± 1 | 12 ± 2 | 2.5 |
| 9 | AAAApYAAAA | - | - | 1.3b |
| 10 | AAAAApYEEVH | 34 ± 2 | 32 ± 7 | 1.1 |
| 11 | AAAAApYRHRR | 33 ± 3 | 234 ± 37 | 0.14 |
| 12 | RTLAApYAAAA | 33 ± 3 | 66 ± 17 | 0.51 |
| 13 | RTLAApYEEVH | 36 ± 0 | 43 ± 1 | 0.84 |
| 14 | RTLAApYRFQK | 37 ± 3 | 165 ± 18 | 0.22 |
| 15 | DTADVpYDWEF | 29 ± 5 | 5.2 ± 1 | 5.8 |
| 16 | DTADVpYGEFTI | 33 ± 3 | 7.0 ± 1 | 4.8 |
| 17 | AKFEDTpYAA | 17 ± 1 | 3.5 ± 1 | 4.9 |
| 18 | AVWEFpYpYAA | 26 ± 1 | 2.9 ± 0.6 | 8.9 |
| 19 | AYTEpYTpYAA | 17 ± 1 | 2.9 ± 0.5 | 6.0 |
| 20 | AWSpYADpYAA | 21 ± 1 | 0.91 ± 0.08 | 23.5 |
| 21 | AVpYADHpYAA | 24 ± 1 | 3.1 ± 0.6 | 7.8 |
| 22 | ApYFPIWpYAA | 20 ± 1 | 3.5 ± 0.4 | 5.9 |
| 23 | ApYYAESpYAA | 16 ± 1 | 3.1 ± 0.4 | 5.1 |
| 24 | ApYpYEVSpYAA | 19 ± 1 | 2.0 ± 0.4 | 9.5 |
| 25 | ApYpYWTApYAA | 26 ± 1 | 1.7 ± 0.2 | 15.1 |
All peptides contained an N-terminal acetyl group and a C-terminal amide.
Data from ref. 36. Data reported represented the mean ± SD from three or more independent sets of experiments.
Kinetic Properties of Selected Peptides toward SHP-1 and SHP-2
Four of the preferred SHP-1 substrates from Table S7 (Ac-WAGDDpYAA-NH2, Ac-FDIDIpYAA-NH2, Ac-EIFDFpYAA-NH2, and Ac-FYDIDpYAA-NH2) were individually synthesized and assayed against SHP-1 and SHP-2. All four peptides are excellent substrates of SHP-1, with kcat/KM values of 1.2–2.5 × 106 M−1 s−1 (Table 4, entries 1–4). They are also efficient substrates of SHP-2, although the kcat/KM values are 2–3-fold lower than those of SHP-1. Next, we attempted to design an optimal substrate for SHP-1/2 by using the specificity information gained from the library screening. Starting with peptide Ac-SASASpYSASA-NH2 (Ser was used to improve aqueous solubility), replacement of the neutral N-terminal sequence with an acidic motif, WDEDF, increased the kcat/KM values by 54- and 61-fold for SHP-1 and SHP-2, respectively (Table 4, entries 5 and 6). Note that peptide 6 (Ac-WDEDFpYSASA-NH2) is a better substrate for SHP-2 (by 1.6-fold), confirming our earlier observation that SHP-2 prefers acidic residues at the pY−2 and pY−3 positions (Figure 4d). Substitution of an acidic sequence C-terminal to pY (Ac-SASASpYDWEF-NH2) also increased the activity by 19-fold for SHP-1 and 10-fold for SHP-2. Introduction of acidic sequences on both sides of pY produced an exceptionally active substrate, Ac-WDEDFpYDWEF-NH2, with kcat/KM values of 2.3 × 107 and 7.0 × 106 M−1 s−1 for SHP-1 and SHP-2, respectively. Thus, replacement of the C-terminal sequence of peptide 6 by an acidic motif further increased its activity to SHP-1 and SHP-2 by 33- and 6.2-fold, respectively. This peptide represents the most active substrate of SHP-1 and SHP-2 known to date. As expected, changing the N-terminal sequence of the peptide to a positively charged sequence (WRKRF) decreased the activity by three to four orders of magnitude (Table 4, entries 8 and 9). A positively charged sequence on the C-terminal side of pY also reduced activity, though by a smaller magnitude (20–100-fold) (Table 4, entries 8 and 10). The significance of the selected aromatic residues (Trp and Phe) was assessed by comparing the activities of peptides Ac-WDEDFpYAA-NH2, Ac-DWEDFpYAA-NH2, and their variants in which the Trp was replaced by Phe or Ala (Table 4, entries 11–16). While substitution of Phe for Trp had minimal effect on their activities toward SHP-2, substitution of Ala resulted in 1.3–1.6-fold reduction in activity. Therefore, hydrophobic residues such as Trp and Phe do contribute to PTP binding and catalysis, likely via hydrophobic and/or cation-π interactions. Kinetic analysis against multiply phosphorylated peptides showed that SHP-1 has no special selectivity for the tandem pY motif (Table 4, entries 17–20). These solution-phase kinetic results are in excellent agreement with the on-bead library screening data and confirm our earlier conclusions that: 1) SHP-1 and SHP-2 strongly prefer acidic (and to a less extent hydrophobic) residues on both sides of pY for efficient catalysis and 2) SHP-1 has a stronger preference for acidic residues at the pY+1 position than SHP-2.
Table 4.
Kinetic Properties of SHP-1 and SHP-2 against pY Peptides
| Entry No. | Peptide Sequencea | SHP-1 | SHP-2 | ||||
|---|---|---|---|---|---|---|---|
| kcat (s−1) | KM (μM) | kcat/KM (μM−1 s−1) | kcat (s−1) | KM (μM) | kcat/KM (μM−1 s−1) | ||
| 1 | WAGDDpYAA | 57 ± 2 | 45 ± 5 | 1.3 | 36 ± 3 | 68 ± 15 | 0.53 |
| 2 | FDIDIpYAA | 33 ± 1 | 13 ± 1 | 2.5 | 30 ± 2 | 39 ± 5 | 0.77 |
| 3 | EIFDFpYAA | 32 ± 2 | 27 ± 4 | 1.2 | 26 ± 1 | 54 ± 6 | 0.48 |
| 4 | FYDIDpYAA | 66 ± 2 | 28 ± 2 | 2.4 | 50 ± 4 | 66 ± 13 | 0.75 |
| 5 | SASASpYSASA | 26 ± 4 | 1990 ± 420 | 0.013 | 42 ± 5 | 2300 ± 400 | 0.018 |
| 6 | WDEDFpYSASA | 37 ± 2 | 53 ± 10 | 0.70 | 50 ± 3 | 46 ± 10 | 1.1 |
| 7 | SASASpYDWEF | 12 ± 1 | 48 ± 9 | 0.25 | 35 ± 1 | 180 ± 7 | 0.19 |
| 8 | WDEDFpYDWEF | 75 ± 2 | 3.2 ± 0.3 | 23 | 43 ± 1 | 6.3 ± 0.4 | 6.8 |
| 9 | WRKRFpYDWEF | 1.7 ± 1.0 | 780 ± 640 | 0.0022 | 1.3 ± 0.2 | 560 ± 140 | 0.0023 |
| 10 | WDEDFpYRWKF | 42 ± 2 | 150 ± 15 | 0.28 | 73 ± 7 | 210 ± 35 | 0.34 |
| 11 | WDEDFpYAA | 48 ± 2 | 43 ± 2 | 1.1 | |||
| 12 | FDEDFpYAA | 30 ± 2 | 31 ± 6 | 1.0 | |||
| 13 | ADEDFpYAA | 28 ± 1 | 34 ± 2 | 0.82 | |||
| 14 | DWEDFpYAA | 30 ± 1 | 17 ± 2 | 1.7 | |||
| 15 | DFEDFpYAA | 37 ± 1 | 23 ± 2 | 1.6 | |||
| 16 | DAEDFpYAA | 30 ± 1 | 28 ± 3 | 1.1 | |||
| 17 | AKFEDTpYAA | 19 ± 2 | 58 ± 13 | 0.34 | |||
| 18 | AVWEFpYpYAA | 24 ± 2 | 7.5 ± 2 | 3.3 | |||
| 19 | AYTEpYTpYAA | 17 ± 1 | 9.1 ± 1 | 1.9 | |||
| 20 | AWSpYADpYAA | 13 ± 1 | 22 ± 5 | 0.58 | |||
All peptides contain an N-terminal acetyl group and a C-terminal amide. Data reported represented the mean ± SD from three or more independent sets of experiments.
Kinetic Properties of Selected Peptides toward RPTPα
Eight of the pY peptides in Tables 2 and 3 were chosen for assay against RPTPα to assess the library screening results (Table 5, entries 1–8). These peptides contained acidic, neutral, or basic residues on either side of pY. Three of the peptides (EADTApY, ASSDDpY, and WDEDFpY) are similar to some of the sequences selected against RPTPα (EIDTApY, TSFDDpY, and WDFDFpY in Table S14, respectively). Remarkably, despite their very different sequences, all eight peptides have similar activities toward RPTPα, with kcat/KM values in the range of 1.2–8.0 × 104 M−1 s−1. These activities are approximately two orders of magnitude lower than those of PTP1B, SHP-1, and SHP-2, indicating that RPTPα is intrinsically less active. The lower catalytic activity is caused by high KM values, which were ~1 mM for the peptide substrates tested, while the kcat values (~30 s−1) were similar to those of PTP1B, SHP-1, and SHP-2.
Table 5.
Kinetic Properties of RPTPA against pY Peptides
| Entry No. | Peptide Sequence | kcat (s−1) | KM (mM) | kcat/KM (mM−1 s−1) |
|---|---|---|---|---|
| 1 | EADTApYAAa | 26 ± 1 | 1.4 ± 0.1 | 19 |
| 2 | ASSDDpYAAa | ND | ND | 13 |
| 3 | WDEDFpYSASAa | ND | ND | 69 |
| 4 | ARKRIpYAAa | 39 ± 1 | 0.49 ± 0.03 | 80 |
| 5 | RRISTpYAAa | 49 ± 3 | 1.2 ± 0.1 | 41 |
| 6 | SASASpYSASAa | 23 ± 4 | 0.53 ± 0.15 | 43 |
| 7 | AAAAApYEEVHa | ND | ND | 39 |
| 8 | AAAAApYRHRRa | 30 ± 2 | 2.6 ± 0.2 | 12 |
| 9 | TATEPQpYQPGENLb | 9.7 ± 0.2 | 1.07 ± 0.03 | 9.1 |
| 10 | TSTEPQpYQPGENLb | 8.9 ± 0.5 | 1.16 ± 0.14 | 7.7 |
| 11 | LIEDNEpYTARQGAb | 8.3 ± 0.2 | 0.77 ± 0.02 | 11 |
| 12 | Phenyl phosphate | ND | ND | 0.020 |
| 13 | pNPP | 0.54 ± 0.28 | 2.7 ± 0.3 | 0.19 |
These peptides contained an N-terminal acetyl group and a C-terminal amide.
Data from ref. 20. Data reported represented the mean ± SD from three or more independent sets of experiments.
Comparison with In Vivo Dephosphorylation Pattern
To determine whether the sequence specificity defined by library screening is biologically relevant, we examined the pY sites in previously reported PTP1B, SHP-1, and SHP-2 substrate proteins. RPTPα was not subjected to this analysis, because few protein substrates of RPTPα are currently known and still less is known about the specific sites of dephosphorylation. Out of the numerous known or putative PTP1B substrates reported (7, 57–65), the actual site(s) of dephosphorylation by PTP1B has been determined or inferred for 18 proteins, resulting in 25 pY sites (Table 6). Inspection of these in vivo PTP1B sites reveals that PTP1B does not have any recognizable consensus sequence, although there are some obvious trends. On the N-terminal side of pY, there is an overrepresentation of acidic (Asp, Glu, and pSer) and small residues (e.g., Ala, Gly, Pro, Ser, and Thr). The great majority of the substrates (21/25) contain at least one acidic residue at the pY−1 to pY−5 position, whereas basic residues are rare. On the C-terminal side, 18 sites have acidic or neutral sequences at positions pY+1 to pY+4, while none of the 25 substrate sites have a basic residue at the pY+1 position. These features are in agreement with the library screening data.
Table 6.
Reported PTP1B, SHP-1, and SHP-2 Substrates and Their Dephosphorylation Sites
| Substrate | Reported or predicted SHP-1 or SHP-2 site(s) | Ref. |
|---|---|---|
| PTP1B | ||
| Caveolin-1* | DSEGHLpY14TVPIR | 57 |
| Cortactin* | TEPEPVpY446SMEAA, PPpSpSPIpY421EDAAP | 58, 59 |
| CSF-1R* | ICTIDVpY944MIMVK | 60 |
| EGFR | TAENAEpY1197LRVAP | 59 |
| IR* | DIYETDpY1162pYRKGGK | 4 |
| JAK2* | LPQDKEpY1007pYKVKE | 61 |
| LPP | PSSGQIpY245GPGPR, GRYYEPpY301pYAAGPS, SEGDTApY318GQQVQ | 59 |
| PDGFR | KDESIDpY751VPMLD | 59 |
| PLCγ1 | GTAEPDpY771GALYE | 59 |
| p120ctn | PSRQDVpY257GPQPQ, RFHPEPpY280GLEDD | 59 |
| p62Dok | GpSPPALpY295AEPLD | 59 |
| p120RasGAP | VDGKEIpY451NTIRR | 59 |
| c-Src* | TSTEPQpY527QPGEN | 62, 63 |
| SHIP2 | TLSEVDpY1136APGPG | 59 |
| STAM2 | SEPEPVpY291IDEDK, VNEAPVpY371SVYSK, APVYSVpY374SKLHP, TVSNPTpY461MNQNS | 64 |
| STAT5a | AKAVDGpY694VKPQI | 65 |
| Tyk2* | VPEGHEpY1054pYRVRE | 61 |
| ZO-1 | EQPAPApY1164EVHNR | 59 |
| SHP-2 | ||
| ASK1† | ? | 79 |
| EGFR* | VVDADEpY992LIPQQ | 66 |
| FAK* | VSETDDpY397AEIID, YMEDSTpY576pY577KASKG | 67 |
| GAB1 | EADGELpY285VFNTP, PTPGNTpY317QIPRT, EELDENpY447VPMNP, HDSEENpY589VPMNP | 72 |
| HER2* | LVDAEEpY1023LVPQQ | 68 |
| MVP† | ? | 80 |
| PAG | MVEDCLpY181ETVKE, KAEFAEpY227ASVDR, EEPEPDpY387EAIQT | 73 |
| Paxillin | LSEETPpY31SYPTG, GEEEHVpY118SFPNK | 73, 74 |
| PDGFRβ* | KDESVDpY751VPMLD, DIESSNpY771MAPYD | 69 |
| PZR | KSESVVpY263ADIRK | 75 |
| p190A* | NEEENIpY1105SVPHD | 70 |
| p190B | HPDYEEpY306INLEG, MDPSDNpY1091AEPID, GYSDEIpY1109VVPDD | 76 |
| SIRPα | DTNDITpY428ADLNL, PNNHTEpY452ASIQT, SEDTLTpY469ADLDM | 77, 78 |
| Sprouty 1* | IRGSNEpY53TEGPS, INVNNNpY89EHRHT | 71 |
| STAT1† | ? | 81 |
| STAT5a† | ? | 82 |
| SHP-1 | ||
| Actin† | ? | 94 |
| BLNK | DDFDSDpY72ENPDE, HSDSEMpY84VMPAE, ENADDSpY96EPPPV, lEDEADpY178VVPVE, EDNDENpY189IHPTE | 85 |
| p120ctn | GQIVETpY112TEEDP, GYDDLDpY296GMMSD | 86 |
| CD22 | MEDGISpY762TTLRF, CDDTVTpY796SALHK, EDEGIHpY822SELIQ, AQENVDpY842VILKH | 87 |
| CD72 | DDGEITpY39ENVQV | 88 |
| p62Dok | MLENSLpY146SPTWE, PKEDPIpY362DEPEG, NPATDDpY409AVPPP | 89 |
| Lck* | LIEDNEpY394TAREG | 83 |
| PIR-B† | ? | 90 |
| SHP-1* | KGQESEpY536GNITY | 84 |
| SIRPα | DTNDITpY428ADLNL, PNNHTEpY452ASIQT, SEDTLTpY469ADLDM | 90 |
| SLP76 | SFEEDDpY113ESPND, GEDDGDpY128ESPNE, VEDDADpY145EPPPS | 91 |
| Syk | PMDTEVpY348ESPYA, EVYESPpY352ADPEE | 92 |
| Vav1 | VGDEDIpY142SGLSD, EEDEDLpY160DCVEN, AEGDEIpY174EDLMR, EDYSEpY844C | 93 |
The listed pY motifs in these proteins have been established as PTP1B, SHP-1, or SHP-2 substrates.
No known pY sites match the specificity of SHP-1 or SHP-2.
Out of the 16 reported SHP-2 substrates, the in vivo dephosphorylation sites have been determined for epidermal growth factor receptor (EGFR, pY992) (66), focal adhesion kinase (FAK, pY397 and pY576/577) (67), HER2 (pY1023 and pY1127) (68), platelet-derived growth factor receptor (PDGFRβ, pY771 and pY751) (69), p190A RhoGAP (pY1105) (70), and Sprouty 1 (pY53 and pY89) (71). With the exception of Sprouty 1, whose pY motifs contain a mixture of Glu, Asn, and Ser [Note that similar motifs were selected by SHP-2 from libraries I and IV (Table S8 and S11)], all other SHP-2 reactive sites contain two or more acidic residues N-terminal to pY. Although the sites of action by SHP-2 in GAB1 (72), PAG (73), paxillin (73, 74), PZR (75), p190B RhoGAP (76), and SIRPα (77, 78) are yet unknown, they each contain one or more experimentally established pY motifs that closely match the specificity profile of SHP-2 (Table 5). We propose that these pY motifs are the likely sites dephosphorylated by SHP-2. Four of the reported SHP-2 substrates, ASK1 (79), MVP (80), STAT1 (81), and STAT5 (82), do not contain established pY sites that match SHP-2 specificity. Further studies are needed to determine the SHP-2 sites in these proteins or whether they are genuine SHP-2 substrates.
A similar survey of the literature revealed 13 putative SHP-1 substrates (Table 6). Among them, pY394 of Lck (83) and pY536 of SHP-1 (84) have been reported to be dephosphorylated by SHP-1 in vivo. These pY motifs contain three and two acidic residues, respectively, N-terminal to pY. Nine of the proteins, including BLNK (85), p120CTN (86), CD22 (87), CD72 (88), p62Dok (89), SIRPα (90), SLP76 (91), Syk (92), and Vav1 (93), each contain one or more established pY motifs that match the specificity profile of SHP-1. We suggest these motifs as the most probable sites of action by SHP-1 in vivo. For the two remaining proteins, actin (94) and PIR-B (92), there are no reported pY sites that match the specificity profile of SHP-1. We noticed that a significantly greater fraction of the SHP-1 substrates contain an acidic residue at the pY+1 position (9/29) as compared to SHP-2 (3/25). Overall, there is an excellent correlation between the sequence specificity of SHP-1/2 and their in vivo dephosphorylation pattern. These results suggest that at least for SHP-1/2, the sequence specificity of the PTP domain is an important determinant of their in vivo specificity against protein substrates.
Discussion
Comparison to Other Library Screening Methods
Screening of combinatorial libraries for catalytic activity is often technically challenging; in the case of PTPs, the difficulty lies in differentiating the PTP product (i.e., tyrosine) from a complex mixture of pY peptides. Most previous methods simplified this problem by screening for binding instead of catalysis and using catalytically impaired PTP mutants or non-hydrolyzable pY analogues (26, 32–35). However, a high-affinity peptide need not be a good substrate, as it may bind to the PTP active site in a non-productive manner (and not undergo catalysis) or an area other than the active site. Conversely, a good substrate may not bind tightly. Moreover, many of the earlier methods provided only a relative preference for certain amino acids at a given position, but not individual sequences. These methods are problematic with enzymes that recognize multiple classes of sequences. Our approach offers several advantages over the earlier methods. First, our library screening is genuinely based on PTP catalysis (instead of binding) and capable of identifying both the optimal (most colored beads) and poor (colorless beads) substrates. Second, our method gives individual sequences and is therefore able to reveal sequence contextual effects and the presence of multiple consensus sequences. This feature is critical for determining the specificity profiles of the PTPs in this study, none of which exhibit a simple consensus sequence. Third, unlike some of the earlier methods that were limited to relatively small libraries (<103 in diversity) (27, 32, 36, 37), the current method has no theoretical limit for library size.
The disadvantage of our method (which is common to all on-bead screening methods) is that it can potentially be biased by non-specific events, such as differential bead swelling or enzyme recruitment. Highly hydrophobic sequences make beads swell poorly in the aqueous screening buffer and may result in false negatives (e.g., apparently “poor” substrates of PTP1B). Fortunately, false negatives of this type are not a major concern for PTPs, as they are unlikely to be present on protein surfaces to act as PTP substrates in vivo. Arg- and Lys-rich peptides, on the other hand, represent a more serious problem. These peptides are highly positively charged under our screening conditions (pH 7.4), and their mutual repulsion presumably facilitates bead swelling, making the pY peptides on the beads more accessible to the PTPs. These beads may also recruit more PTP protein onto their surfaces, if the PTP is negatively charged or contains a negatively charged surface patch (55). Obviously, the impact of these biases on library screening depends on the specificity of the PTP in question. For a highly specific PTP, these factors should have a minimal effect on the screening result because the positively charged peptides, even with the advantage of better swelling and/or more recruited PTP, would not be dephosphorylated fast enough to compete with the most preferred substrates. This is indeed the case with SHP-1 and SHP-2, which did not select Arg/Lys-rich sequences under any of the conditions tested. For less specific PTPs, the biases caused by differential bead swelling and/or enzyme recruitment may dominate the activity differences. For instance, when library I was screened against RPTPα, which has only weak sequence selectivity (Table 5), Arg- and Lys-rich sequences were selected exclusively (Table S13). PTP1B has a modest level of sequence selectivity (in between those of RPTPα and SHP-1/2) and it selected a mixture of genuinely preferred substrates (class I) and biased sequences (class II) (Table S2). This scenario provides a possible explanation for the selection of Arg/Lys-containing sequences in our previous study (41), which employed a peptide library on TentaGel resin. TentaGel is impermeable to large proteins such as PTP1B (50), and gaining access to the immobilized pY peptides may be the rate-limiting step of the screening reaction cascade. Consistent with this notion, the previous study required treatment with 1 μM PTP1B for 6 h to generate sufficient color intensity on beads, whereas in the present work, a typical screening reaction involved treatment with 1–5 nM PTP1B for 20 min. The Arg and Lys residues likely increased bead swelling of and/or PTP recruiting to the positive beads.
The biases described above, although somewhat of a nuisance, do not prevent one from obtaining the genuine substrate specificity of an enzyme. Because our method gives individual sequences, one can readily classify the selected sequences into different families. If an enzyme selects Arg/Lys-rich sequences, it signifies that the swelling/recruiting biases may be in play and the enzyme likely has limited sequence selectivity. To obtain genuine optimal substrates, one can carry out the screening reaction at higher salt concentrations to suppress the biases. Alternatively, one can screen the enzyme against a library with reduced Arg and Lys content. As demonstrated successfully with RPTPα, a 10-fold decrease in the Arg and Lys content greatly reduced the number of peptides containing multiple Arg and Lys residues and largely eliminated the swelling/recruiting biases. Of course, it is possible that an enzyme may genuinely prefer positively charged sequences; in such a case, screening of the enzyme against the library of reduced Arg/Lys content should still enrich the Arg and/or Lys contents of the selected peptides to levels above the statistical averages.
PTPs Have Different Intrinsic Catalytic Efficiencies
The intrinsic catalytic efficiency of a PTP [defined here as the kcat/KM value(s) of the PTP active site toward its optimal peptide substrate(s)] is likely a key determinant of its in vivo function. Previously, comparison of different PTPs with regard to their catalytic efficiencies was not possible, because the optimal substrates of PTPs were not known. The availability of the substrate specificity profiles of PTP1B, RPTPα, SHP-1, and SHP-2 from this study allows us to compare their catalytic efficiencies. The catalytic domains of PTP1B, SHP-1, and SHP-2 all have kcat/KM values in the range of 106–107 M−1 s−1 toward their respective optimal substrates and thus have similar intrinsic catalytic efficiencies (Tables 3 and 4). On the other hand, the intrinsic catalytic efficiency of RPTPα is two to three orders of magnitude lower than those of the other three PTPs (kcat/KM = 104–105 M−1 s−1). The lower catalytic efficiency is mostly due to higher KM values (Table 5). According to the mechanism of the PTP-catalyzed reaction,
the KM value is related to the dissociation constant (KD) of the enzyme-substrate complex by the equation: KM = KD · k3/(k2 + k3). Since RPTPα has KM values of ~1 mM toward all of the peptide substrates tested, the active site of RPTPα must bind the substrates only weakly (KD ≥ 1 mM). This observation, coupled with its minimal sequence selectivity, suggests that RPTPα recognizes peptide substrates primarily through interactions with the pY residue and the peptide backbone of flanking residues. We suspect that other PTPs may have varying levels of intrinsic catalytic efficiencies.
PTPs Have Distinct Substrate Specificity Profiles
Each of the four PTPs investigated in this study has a unique specificity profile, defined as both the range of substrates accepted and the actual sequences preferred by the enzyme. Among the four PTPs, RPTPα has the broadest substrate specificity (least sequence selectivity) and dephosphorylates all pY peptides with similar efficiencies. This conclusion is based on the observation that all 19 amino acids were selected from peptide libraries with similar frequencies at each random position and that there was only a 6.7-fold difference in the kcat/KM value between the most and least reactive substrates tested (Table 5). It has slight preference for hydrophobic residues at the pY−1 position, hydrophilic residues at the pY−2 position, and non-basic residues at the C-terminal side of pY. A unique feature of RPTPα is that it readily accepts acidic, neutral, and basic residues at pY−2 to pY−5 positions.
The catalytic domain of PTP1B also has broad specificity, but is more restrictive than that of RPTPα. Library screening showed that PTP1B accepts essentially all 20 amino acids at each position, except for basic residues (Arg and Lys) at the pY−1 position (and to a lesser extent at the pY−2 position). It has a modest preference for a hydrophilic residue (e.g. Asp, Asn, Ser, Glu, Gln, and Gly) at position pY−2. PTP1B also has a weak selectivity for Thr and Ser at the pY−4 position, although the frequency of Ser/Thr occurrence at this position may have been enhanced by background reaction. We found that prolonged incubation of libraries I, IV, and V under the library screening condition (without PTP) produced a small number of lightly colored beads. Sequencing of these beads showed that they all contained a Ser or Thr at the pY−4 position (data not shown). On the C-terminal side of pY, PTP1B favors acidic over basic residues. Consistent with the screening results, all of the peptides tested including those containing disfavored amino acids (e.g., Ac-ARKRIpYAA-NH2, Ac-RRISTpYAA-NH2, and Ac-AAAAApYRHRR-NH2) were efficient substrates of PTP1B, with kcat/KM values >105 M−1 s−1 (Table 3). Substitution of Ala for the preferred residues (e.g., Asp at pY−2 or Thr at pY−4 position) had only modest effects on the catalytic activity (<2-fold). Further, peptide Ac-AAAApYAAAA-NH2 is only ~3-fold less active than peptide Ac-DTADVpYAA-NH2, one of the most reactive substrates selected from library I. Previous studies by others have also led to the suggestion that PTP1B has only limited sequence selectivity (32, 36).
A previous study showed that the bisphosphorylated insulin receptor peptide (DpY1162pYRK) had ~70-fold higher activity toward PTP1B than the monophosphorylated peptide DpYYRK (36). This and other observations led to the notion that DpYpYR/K may be a specific recognition motif of PTP1B (61). Screening of library VI against PTP1B selected predominantly peptides with two or more pY residues (Table S5), which generally have higher activities than mono-phosphorylated peptides (Table 3). However, PTP1B does not have any special preference for the tandem pY motif. In fact, peptides containing the second pY at position pY−3 to pY−5 had the highest kcat/KM values (Table 3). We believe that the second pY residue simply serves as an acidic residue that interacts with the positively charged residues on the PTP1B surface. The phosphate group is more effective for electrostatic interactions than the Asp/Glu side chain because it is dianionic at physiological pH and forms multi-dentate interactions.
Compared to RPTPα and PTP1B, SHP-1 and SHP-2 have much narrower specificities including much less tolerance for positively charged residues. As an indicator of this difference, the largest activity difference (in kcat/KM value) between any of the peptides tested in this work was 6.7-fold for RPTPα and 168-fold for PTP1B (Table 3, compare peptides 3 and 20), whereas the corresponding differences were 10,500- and 3,000-fold for SHP-1 and SHP-2, respectively (Table 4, compare peptides 8 and 9). SHP-1 and SHP-2 strongly prefer acidic residues and to a less degree hydrophobic aromatic residues on both sides of pY. Surprisingly, the preferred substrates do not exhibit a simple consensus sequence(s); the acidic and hydrophobic residues may be present at any position (from pY−5 to pY+5), though some positions are more preferred than others. This is very different from other enzymes that utilize peptide substrates (e.g., proteases) and illustrates the importance of obtaining individual sequences during library screening against PTPs. SHP-1 and SHP-2 have similar but not identical substrate specificity profiles. While both enzymes prefer acidic sequences, they prefer them at different positions (Figure 4c, d). SHP-1 also has a stronger preference for acidic residues at the pY−1 and pY+1 positions, whereas SHP-2 favors hydrophobic residues at these two positions. A quantitative assessment of the specificity difference between the two enzymes can be made by comparing their activities towards representative pY peptides. For example, the peptide WAGDDpYAA was 2.5-fold more reactive to SHP-1, whereas SHP-2 had 1.6-fold higher activity against the peptide WDEDFpYSASA (Table 4, entries 1 and 6). Thus, differential positioning of the acidic residues at the N-terminal side of pY causes a 4-fold change in the relative activity for SHP-1 and SHP-2. On the C-terminal side, replacement of the neutral SASA motif with acidic motif DWEF increased the activity toward SHP-1 and SHP-2 by 32- and 6.6-fold, respectively, resulting in a 5-fold difference in favor of SHP-1 (Table 4, entries 6 and 8). It is conceivable that a peptide featuring the optimal sequence of SHP-1 on both sides of pY may have at least an order of magnitude higher activity for SHP-1 than SHP-2, and vice versa.
Structural Basis of Differential PTP Substrate Specificity
The crystal structures of many PTPs including RPTPα, PTP1B, SHP-1, and SHP-2 have been determined with and without bound pY peptide substrates (10, 11, 28–31, 47, 56). The PTP active site typically features a small, deep pocket for the pY side chain and relatively flat surfaces around the pocket. Interactions between the pY side chain and the active site pocket provide the majority of the binding energy and dictate the enzyme’s specificity for pY (instead of pS or pT). The additional binding energy, as well as sequence specificity, is provided by interactions between substrate residues flanking the pY and the highly diverse PTP surfaces near the pY pocket. The flat surfaces permit different peptides to bind to the same PTP active site, perhaps by engaging a slightly different surface area in each case. It has been shown that bound pY peptides can assume different conformations to allow a hydrophobic residue at either pY−2 or pY−3 position to occupy the same shallow hydrophobic pocket on the SHP-1 surface (29). PTP1B has also shown this type of plasticity during substrate binding (31). The broad specificity of PTP1B can be explained by the fact that most of the interactions in the E·S complex are mediated by the backbone amides of the peptide substrate (28–31). Only one or two acidic residues N-terminal to pY (in addition to pY) make specific side-chain interactions with the PTP1B surface. The differential preference for acidic residues (and tolerance for basic residues) by the four PTPs can be readily explained by their surface electrostatic potentials. The surfaces surrounding the SHP-1 and SHP-2 active sites are overwhelmingly positively charged (47), allowing them to bind to acidic (negatively charged) peptides through electrostatic interactions. The long-range, non-directional nature of charge-charge interactions makes it possible for acidic residues at different positions to engage in productive interactions with the basic residues on the PTP surface. The surfaces near the PTP1B active site are also overall positively charged but much less so compared to SHP-1 and SHP-2 (47). Hence, PTP1B has modest preference for acidic peptides. Finally, the substrate-binding surface of RPTPα is interspersed with both positively and negatively charged residues and overall neutral (47), consistent with its nearly equivalent preference (tolerance) for acidic and basic peptides as substrates.
Biological Implications
The lack of stringent sequence specificity by the PTP catalytic domains suggests that other factors such as intracellular localization, protein-protein interactions, and/or post-translational regulation must contribute to the in vivo substrate specificity. For SHP-1 and SHP-2, their SH2 domains can bind specific pY proteins and target the PTP domain to the pY proteins and other protein(s) associated with them. The in vivo specificity of SHP-1 and SHP-2 is further enhanced by their auto-inhibitory mechanism, in which the N-terminal SH2 domain binds to and inactivates the PTP active site in an intramolecular manner (10, 11); the PTP domain remains inactive until the N-terminal SH2 domain is engaged with a specific target protein. We have previously shown that the four SH2 domains of SHP-1 and SHP-2 have overlapping but not identical specificities (12). The SH2 domains of SHP-1 and SHP-2 may bind to different pY proteins, different pY motifs on the same protein, or the same pY protein with different affinities. Once bound to the target protein(s), the intrinsic sequence specificity of the PTP domain likely dictates which pY residue to dephosphorylate and how fast the reaction takes place. This combinatorial control of substrate specificity is consistent with the previous observation that replacement of the SHP-1 PTP domain with that of SHP-2 or vice versa did not produce the same phenotype as the respective wild-type enzymes (13, 14).
Despite its lack of significant sequence specificity, RPTPα may act as a highly specific PTP in vivo. Because of its low intrinsic catalytic efficiency, the PTP domain alone may not be able to compete effectively with other PTPs for dephosphorylation of pY proteins. However, if a substrate protein is specifically recruited to the vicinity of RPTPα through protein-protein interaction, the high local concentration would permit the PTP active site to catalyze the reaction at or near the turnover rate (~30 s−1). A number of protein substrates have been reported for RPTPα, including Src family kinases Src, Lck, Lyn, Fyn, and Yes (95–97), channel proteins kv1.1 and kv1.2 (98), and p130cas (99). The kinases and channel proteins are either integral membrane proteins or recruited to the cytoplasmic membrane during cell signaling, where RPTPα is located. p130cas was shown to be co-localized with RPTPα on the cell membrane (99), though it is not yet clear whether membrane localization of p130cas is a condition or a consequence of interaction with the RPTPα PTP domain.
PTP1B is an intriguing case. It has very high intrinsic catalytic efficiency and yet broad specificity, suggesting that it is capable of dephosphorylating a wide variety of proteins. PTP1B is localized to the ER membrane (3, 100); this localization undoubtedly leads to some preference for a subset of pY proteins such as those localized on the ER membrane. Previous studies have shown that calpain-mediated cleavage of PTP1B from the ER membrane in platelets allows it to access different substrates (101, 102). However, this “compartmentalization” may not be sufficient to endow PTP1B with “exquisite” substrate specificity in vivo. A recent crystal structure has shown that PTP1B can form a complex with the insulin receptor at a site different from its site of dephosphorylation (103). Although the physiological relevance of these observations remain to be established, PTP1B might employ additional substrate recruiting strategies via either its catalytic or non-catalytic C-terminal domains. Notably, the C-terminal domain of PTP1B has proline-rich sequences capable of binding to SH3 domain-containing proteins and also has several phosphorylation sites within this region. Moreover, PTP1B activity is regulated spatially within cells (104), possibly as a consequence of oxidation by locally produced reactive oxygen species (105). Nevertheless, our results suggest that PTP1B may have a relatively wide variety of substrates in vivo. This notion is supported by several previous experimental observations. For example, overexpression of a substrate-trapping (D181A) mutant of PTP1B in COS cells increased the pY levels of a large number of proteins (106). The mutant protein also co-immunoprecipitated with a large number of pY proteins (106, 107), many of which have now been identified (Table 6). The reported PTP1B substrates contain highly diverse sequences surrounding the pY residue. A tantalizing hypothesis is that PTP1B acts on many pY proteins in vivo, most of which are shared with other PTPs. The “specific” effects associated with PTP1B ablation may represent the effects on its unique substrates (those that are not shared with other PTPs). In this regard, it is worth noting that PTP1B is exceptionally active toward multiply phosphorylated substrates, a feature that does not appear to be shared by other PTPs. Clearly, additional work is necessary to test this notion and fully understand the in vivo substrate specificity of PTP1B.
Conclusion
By employing an improved combinatorial library method, we have determined the substrate specificity profiles for RPTPα, PTP1B, SHP-1, and SHP-2. Our results show that each PTP has a different degree of sequence specificity and a unique substrate specificity profile. The specificity data should be useful for identifying the physiological substrates of PTPs, especially those with more stringent sequence specificities (e.g., SHP-1 and SHP-2), as well as the specific sites of dephosphorylation by PTPs in their substrate proteins. Our library method should be readily applicable to other PTPs.
Supplementary Material
Acknowledgments
We thank Ms. Jing Zhang for assistance with some of the PTP assays, Dr. Stefan Knapp for the RPTPα DNA construct, and Dr. S. Li for the bacterial tyrosinase expression vector.
Abbreviations
- Abu (U)
2-aminobutyrate
- F2Y
3,5-difluorotyrosine
- Fmoc-OSu
N-(9-fluorenylmethoxycarbonyloxy) succinimide
- HBTU
O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- HOBt
1-hydroxybenzotriazole hydrate
- MBTH
3-methyl-2-benzothiazolinonehydrazone
- Nle (M)
norleucine
- OBOC
one-bead-one-compound
- PED-MS
partial Edman degradation-mass spectrometry
- PEGA
polyethyleneglycol acrylamide
- PITC
phenyl isothiocyanate
- PTP
protein tyrosine phosphatase
- pY
phosphotyrosine
- TFA
trifluoroacetic acid
Footnotes
This work was supported by grants from the National Institutes of Health (CA132855, GM062820, CA49132, and CA49152). L.R. was supported by a predoctoral fellowship from China Scholarship Council affiliated with the Ministry of Education of P. R. China. T.M.M. was supported by a predoctoral fellowship from the NIH Chemistry-Biology Interface Training Program (T32GM08512).
Supporting Information Available: Tables containing the peptides sequences selected against the three PTPs and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Tonks NK, Neel BG. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr Opin Cell Biol. 2001;13:182–195. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 3.Frangioni JV, Beahm PH, Shifrin V, Jost CA, Neel BG. The nontransmembrane tyrosine phosphatase PTP1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- 4.Elchelby M, Payette P, Michaliszyn E, Cromlish W, Collins S, Lee Loy A, Normandin D, Cheng A, Himms-Hagen J, Chan CC, Ramanchandran C, Gresser MJ, Tremblay ML, Kennedy BP. Increased insulin sensitivity and obesity resistance in mice lacking the protein phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 5.Julien SG, Dube N, Read M, Penney J, Paquet M, Han YX, Kennedy BP, Muller WJ, Tremblay ML. Protein tyrosine phosphatase 1B deficiency or inhibition delays Erb2-induced mammary tumorigenesis and protects from lung metastasis. Nat Genet. 2007;39:339–346. doi: 10.1038/ng1963. [DOI] [PubMed] [Google Scholar]
- 6.Bentires-Alj M, Neel BG. Protein-tyrosine phosphatase 1B is required for HER2/Neu-indeuced breast cancer. Cancer Res. 2007;67:2420–2424. doi: 10.1158/0008-5472.CAN-06-4610. [DOI] [PubMed] [Google Scholar]
- 7.Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochem J. 2007;402:1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neel BG, Gu H, Pao LI. SH2-domain-containing protein tyrosine phosphatases. Handbook of Cell Signaling. 2003;1:707–728. [Google Scholar]
- 9.Mohi MG, Neel BG. The role of Shp2 (PTPN11) in cancer. Curr Opin Genet Dev. 2007;17:23–30. doi: 10.1016/j.gde.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Liu L, He D, Song X, Liang X, Zhao ZJ, Zhou GW. Crystal structure of human protein tyrosine phosphatase SHP-1. J Biol Chem. 2003;278:6516–6520. doi: 10.1074/jbc.M210430200. [DOI] [PubMed] [Google Scholar]
- 12.Sweeney MC, Wavreille AS, Park J, Butchar JP, Tridandapani S, Pei D. Decoding protein-protein interactions through combinatorial chemistry: sequence specificity of SHP-1, SHP-2, and SHIP SH2 domains. Biochemistry. 2005;44:14932–14947. doi: 10.1021/bi051408h. [DOI] [PubMed] [Google Scholar]
- 13.O’Reilly AM, Neel BG. Structural determinants of SHP-2 function and specificity in Xenopus mesoderm induction. Mol Cell Biol. 1998;18:161–177. doi: 10.1128/mcb.18.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenev T, Keilhack H, Tomic S, Stoyanov B, Stein-Gerlach M, Lammers R, Krivtsov AV, Ullrich A, Böhmer FD. Both SH2 domains are involved in interactions of SHO-1 with the epidermal growth factor receptor but cannot confer receptor-directed activity to SHP-1/SHP-2 chimera. J Biol Chem. 1997;272:5966–5973. doi: 10.1074/jbc.272.9.5966. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Pallen CJ. The receptor-like protein tyrosine phosphatase HPTP alpha has two active catalytic domains with distinct substrate specificities. EMBO J. 1991;10:3231–3237. doi: 10.1002/j.1460-2075.1991.tb04886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim KL, Ng CH, Pallen CJ. Catalytic activation of the membrane distal domain of protein tyrosine phosphatase epsilon, but not CD45, by two point mutations. Biochim Biophys Acta. 1999;1434:275–283. doi: 10.1016/s0167-4838(99)00189-2. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Buis A, den Hertog J, Zhang ZY. Comparative kinetic analysis and substrate specificity of the tandem catalytic domains of the receptor-like protein tyrosine phosphatase α. J Biol Chem. 1997;272:6994–7002. doi: 10.1074/jbc.272.11.6994. [DOI] [PubMed] [Google Scholar]
- 18.Jiang GQ, den Hertog J, Hunter T. Receptor-like protein tyrosine phosphatase alpha homodimerizes on the cell surface. Mol Cell Biol. 2000;20:5917–5929. doi: 10.1128/mcb.20.16.5917-5929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vacaru AM, den Hertog J. Catalytically active membrane-distal phosphatase domain of receptor protein-tyrosine phosphatase α is required for Src activation. FEBS J. 2010;277:1562–1570. doi: 10.1111/j.1742-4658.2010.07584.x. [DOI] [PubMed] [Google Scholar]
- 20.Ng DHW, Jabali MD, Maiti A, Borodchak P, Harder KW, Brocker T, Malissen B, Jirik FR, Johnson P. CD45 and RPTPα display different protein tyrosine phosphatase activities in T lymphocytes. Biochem J. 1997;327:867–876. doi: 10.1042/bj3270867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho HJ, Ramer SE, Itoh M, Winkler DG, Kitas E, Bannwarth W, Burn P, Saito H, Walsh CT. Purification and characterization of a soluble catalytic fragment of the human transmembrane leukocyte antigen related (LAR) protein tyrosine phosphatase from an Escherichia coli expression system. Biochemistry. 1991;30:6210–6216. doi: 10.1021/bi00239a019. [DOI] [PubMed] [Google Scholar]
- 22.Zhang ZY, Maclean D, McNamara DJ, Sawyer TK, Dixon JE. Protein tyrosine phosphatase substrate specificity: size and phosphotyrosine positioning requirements in peptide substrates. Biochemistry. 1994;33:2285–2290. doi: 10.1021/bi00174a040. [DOI] [PubMed] [Google Scholar]
- 23.Harder KW, Owen P, Wong LKH, Aebersold R, Clark-Lewis I, Jirik FR. Characterization and Kinetic Analysis of the intracellular domain of human protein tyrosine phosphatase β (HPTPβ) using synthetic phosphopeptides. Biochem J. 1993;298:395–401. doi: 10.1042/bj2980395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bobko M, Wolfe HR, Saha A, Dolle RE, Fisher DK, Higgins TJ. CD45 Protein Tyrosine Phosphatase: Determination of minimal peptide length for substrate recognition and synthesis of some tyrosine-based electrophiles as potential active-site directed irreversible inhibitors. Bioorg Med Chem Lett. 1995;5:353–356. [Google Scholar]
- 25.Dechert U, Affolter M, Harder KW, Matthews J, Owen P, Clark-Lewis I, Thomas ML, Aebersold R, Jirik FR. Comparison of the specificity of bacterially expressed cytoplasmic protein-tyrosine phosphatases SHP and SH-PTP2 towards synthetic phosphopeptide substrates. Eur J Biochem. 1995;231:673–681. doi: 10.1111/j.1432-1033.1995.0673d.x. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Tan LP, Gao L, Yao SQ. High-throughput screening of catalytically inactive mutants of protein tyrosine phosphatases (PTPs) in a phosphopeptide microarray. Chem Commun. 2009;6:677–679. doi: 10.1039/b817853d. [DOI] [PubMed] [Google Scholar]
- 27.Kohn M, Gutierrez-Rodriguez M, Jonkheijm P, Wetzel S, Wacker R, Schroeder H, Prinz H, Niemeyer CM, Breinbauer R, Szedlacsek SE, Waldmann H. A Microarray Strategy for Mapping the Substrate Specificity of Protein Tyrosine Phosphatase. Angew Chem Int Ed. 2007;46:7700 –7703. doi: 10.1002/anie.200701601. [DOI] [PubMed] [Google Scholar]
- 28.Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Cheng Z, Niu T, Liang X, Zhao ZJ, Zhou GW. Structural basis for substrate specificity of protien-tyrosine phosphatase SHP-1. J Biol Chem. 2000;275:4066–4071. doi: 10.1074/jbc.275.6.4066. [DOI] [PubMed] [Google Scholar]
- 30.Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell. 2000;6:1401–1412. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 31.Sarmiento M, Puius YA, Vetter SW, Keng YF, Wu L, Zhao Y, Lawrence DS, Almo SC, Zhang ZY. Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition. Biochemistry. 2000;39:8171–8179. doi: 10.1021/bi000319w. [DOI] [PubMed] [Google Scholar]
- 32.Pellegrini MC, Liang H, Mandiyan S, Wang K, Yuyev A, Vlattas I, Sytwu T, Li YC, Wennogle LP. Mapping the subsite preferences of protein tyrosine phosphatase PTP1B using combinatorial chemistry approaches. Biochemistry. 1998;37:15598–15606. doi: 10.1021/bi981427+. [DOI] [PubMed] [Google Scholar]
- 33.Huyer G, Kelly J, Moffat J, Zamboni R, Jia Z, Gresser MJ, Ramachandran C. Affinity selection from peptide libraries to determine substrate specificity of protein tyrosine phosphatases. Anal Biochem. 1998;258:19–30. doi: 10.1006/abio.1997.2541. [DOI] [PubMed] [Google Scholar]
- 34.Espanel X, van Huijsduijnen RH. Applying the SPOT peptide synthesis procedure to the study of protein tyrosine phosphatase substrate specificity: probing for the heavenly match in vitro. Methods. 2005;35:64–72. doi: 10.1016/j.ymeth.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Walchli S, Espanel X, Harrenga A, Rossi M, Cesareni G, van Huijsduijnen HR. Probing protein tyrosine phosphatase substrate specificity using a phosphotyrosine-containing phage library. J Biol Chem. 2004;279:311–318. doi: 10.1074/jbc.M307617200. [DOI] [PubMed] [Google Scholar]
- 36.Vetter SW, Keng YF, Lawrence DS, Zhang ZY. Assessment of protein tyrosine phosphatase 1B substrate specificity using “inverse alanine scanning”. J Biol Chem. 2000;275:2265–2268. doi: 10.1074/jbc.275.4.2265. [DOI] [PubMed] [Google Scholar]
- 37.Wang P, Fu H, Snavley DF, Freitas MA, Pei D. Screening combinatorial libraries by mass spectrometry. 2 Identification of optimal substrates of protein tyrosine phosphatase SHP-1. Biochemistry. 2002;41:6202–6210. doi: 10.1021/bi025591f. [DOI] [PubMed] [Google Scholar]
- 38.Cheung YW, Abell C, Balasubramanian S. A combinatorial approach to identifying protein tyrosine phosphatase substrates from a phosphotyrosine peptide library. J Am Chem Soc. 1997;119:9568–9569. [Google Scholar]
- 39.Mitra S, Barrios AM. Identifying selective protein tyrosine phosphatase substrates and inhibitors from a fluorogenic, combinatorial peptide library. ChemBioChem. 2008;9:1216–1219. doi: 10.1002/cbic.200800046. [DOI] [PubMed] [Google Scholar]
- 40.Mishra AK, Zhang A, Niu T, Yang J, Liang X, Zhao ZJ, Zhou GW. Substrate specificity of protein tyrosine phosphatase: Differential behavior of SHP-1 and SHP-2 towards signal regulation protein SIRPα1. J Cell Biochem. 2002;84:840–846. doi: 10.1002/jcb.10090. [DOI] [PubMed] [Google Scholar]
- 41.Garaud M, Pei D. Substrate profiling of protein tyrosine phosphatase PTP1B by screening a combinatorial peptide library. J Am Chem Soc. 2007;129:5366–5367. doi: 10.1021/ja071275i. [DOI] [PubMed] [Google Scholar]
- 42.Gopishetty B, Ren L, Waller TM, Wavreille AS, Lopez M, Thakkar A, Zhu J, Pei D. Synthesis of 3,5-difluorotyrosine-containing peptides: Application in substrate profiling of protein tyrosine phosphatases. Org Lett. 2008;10:4605–4608. doi: 10.1021/ol801868a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Ren L, Kim S, Carpino N, Daniel JL, Kunapuli SP, Tsygankov AY, Pei D. Determination of the Substrate Specificity of Protein Tyrosine Phosphatase TULA-2 and Identification of Syk as a TULA-2 Substrate. J Biol Chem. 2010;285:31268–31276. doi: 10.1074/jbc.M110.114181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S, Zeng D. Chemoenzymatic enrichment of phosphotyrosine-containing peptides. Angew Chem Int Ed. 2007;46:4751–4753. doi: 10.1002/anie.200700633. [DOI] [PubMed] [Google Scholar]
- 45.Pei D, Neel BG, Walsh CT. Overexpression, purification, and characterization of SHPTP1, a Src homology 2-containing protein-tyrosine-phosphatase. Proc Natl Acad Sci U S A. 1993;90:1092–1096. doi: 10.1073/pnas.90.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J, Pei D. trans-β-Nitrostyrene derivatives as slow-binding inhibitors of protein tyrosine phosphatases. Biochemistry. 2004;43:15014–15021. doi: 10.1021/bi0486233. [DOI] [PubMed] [Google Scholar]
- 47.Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Müller S, Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thakkar A, Wavreille AS, Pei D. Traceless capping agent for peptide sequencing by partial edman degradation and mass spectrometry. Anal Chem. 2006;78:5935–5938. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- 49.Zhang ZY, Maclean D, Thieme-Sefler AM, Roeske RW, Dixon JE. A continuous spectrophotometric and fluorimetric assay for protein tyrosine phosphatase using phosphotyrosine-containing peptides. Anal Biochem. 1993;211:7–15. doi: 10.1006/abio.1993.1224. [DOI] [PubMed] [Google Scholar]
- 50.Kress J, Zanaletti R, Amour A, Ladlow M, Frey JG, Bradley M. Enzyme accessibility and solid supports: Which molecular weight enzymes can be used on solid support? An investigation using confocal Raman microscopy. Chem Eur J. 2002;8:3769–3772. doi: 10.1002/1521-3765(20020816)8:16<3769::AID-CHEM3769>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 51.Ulijn RV, Baragaña B, Halling PJ, Flitsch SL. Protease-catalyzed peptide synthesis on solid support. J Am Chem Soc. 2002;124:10988–10989. doi: 10.1021/ja026912d. [DOI] [PubMed] [Google Scholar]
- 52.Ulijn RV, Bisek N, Halling P, Flitsch SL. Understanding protease catalyzed solid phase peptide synthesis. Org Biomol Chem. 2003;1:1277–1281. doi: 10.1039/b211890d. [DOI] [PubMed] [Google Scholar]
- 53.Jolley RL, Jr, Nelson RM, Robb DA. The multiple forms of tyrosinase. J Biol Chem. 1969;244:3251–3257. [PubMed] [Google Scholar]
- 54.Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D. Protein N-terminal processing: Substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry. 2010;49:5588–5599. doi: 10.1021/bi1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Tan PH, Zhang Y, Pei D. On-bead screening of combinatorial libraries: Reduction of nonspecific binding by decreasing surface ligand density. J Comb Chem. 2009;11:604–611. doi: 10.1021/cc9000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puius YA, Zhao Y, Sullivan M, Lawrence DS, Almo SC, Zhang ZY. Identification of second aryl phosphate-binding site in protein-tyrosine phosphates 1B: A paradigm for inhibitor design. Proc Natl Acad Sci U S A. 1997;94:13420–13425. doi: 10.1073/pnas.94.25.13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee H, Xie LP, Luo Y, Lee SY, Lawrence DS, Wang XB, Sotgia F, Lisanti MP, Zhang ZY. Identification of phosphocaveolin-1 as a novel protein tyrosine phosphatase 1B substrate. Biochemistry. 2006;45:234–240. doi: 10.1021/bi051560j. [DOI] [PubMed] [Google Scholar]
- 58.Stuible M, Dube N, Tremblay ML. PTP1B regulates cortactin tyrosine phosphorylation by targeting Tyr446. J Biol Chem. 2008;283:15740–15746. doi: 10.1074/jbc.M710534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mertins P, Eberl HC, Renkawitz J, Olsen JV, Tremblay ML, Mann M, Ullrich A, Daub H. Investigation of protein-tyrosine phosphatase 1B function by quantitative proteomics. Mol Cell Proteomics. 2008;7.9:1763–1777. doi: 10.1074/mcp.M800196-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinonen KM, Dube N, Bourdeau A, Lapp WS, Tremblay ML. Protein tyrosine phosphatase 1B negatively regulates macrophage development in intact cells. Proc Natl Acad Sci U S A. 2006;103:2776–2781. doi: 10.1073/pnas.0508563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, Salmeen A, Barford D, Tonks NK. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 62.Liang F, Lee SY, Liang J, Lawrence DS, Zhang ZY. The role of protein-tyrosine phosphatase 1B in integrin signaling. J Biol Chem. 2005;280:24857–24863. doi: 10.1074/jbc.M502780200. [DOI] [PubMed] [Google Scholar]
- 63.Bjorge JD, Pang A, Fujita DJ. Identification of protein-tyrosine phosphatase 1B as the major tyrosine phosphatase activity capable of dephosphorylating and activating c-Src in several human cancer cell lines. J Biol Chem. 2000;275:41439–41446. doi: 10.1074/jbc.M004852200. [DOI] [PubMed] [Google Scholar]
- 64.Stuible M, Abella JV, Feldhammer M, Nossov M, Sangwan V, Blagoev B, Park M, Tremblay ML. PTP1B targets the endosomal sorting machinery. J Biol Chem. 2010;285:23899–23907. doi: 10.1074/jbc.M110.115295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aoki N, Matsuda T. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J Biol Chem. 2000;275:39718–39726. doi: 10.1074/jbc.M005615200. [DOI] [PubMed] [Google Scholar]
- 66.Agazie YM, Hayman MJ. Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol. 2003;23:7875–7886. doi: 10.1128/MCB.23.21.7875-7886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsutsumi R, Takahashi A, Azuma T, Higashi H, Hatakeyama M. Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol Cell Biol. 2006;26:261–276. doi: 10.1128/MCB.26.1.261-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou XD, Agazie YM. Molecular mechanism for SHP2 in promoting HER2-induced signaling and transformation. J Biol Chem. 2009;284:12226–12234. doi: 10.1074/jbc.M900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klinghoffer RA, Kazlauskas A. Identification of a putative Syp substrate, the PDGFβ receptor. J Biol Chem. 1995;270:22208–22217. doi: 10.1074/jbc.270.38.22208. [DOI] [PubMed] [Google Scholar]
- 70.Bregeon J, Loirand G, Pacaud P, Rolli-Derkinderen M. Angiotensin II induces RhoA activation through SHP2-dependent dephosphorylation of the RhoGAP p190A in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2009;297:C1062–C1070. doi: 10.1152/ajpcell.00174.2009. [DOI] [PubMed] [Google Scholar]
- 71.Lesley AJ, Stephanie JT, Michael AS, Mark AK, Rachel KS. Sprouty proteins are in vivo targets of Corkscrew/SHP-2 tyrosine phosphatases. Development. 2006;133:1133–1142. doi: 10.1242/dev.02255. [DOI] [PubMed] [Google Scholar]
- 72.Bard-Chapeau EA, Yuan J, Droin N, Long S, Zhang EE, Nguyen TV, Feng GS. Concerted functions of Gab1 and Shp2 in liver regeneration and hepatoprotection. Mol Cell Biol. 2006;26:4664–4674. doi: 10.1128/MCB.02253-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang SQ, Yang W, Kontaridis MI, Bivona TG, Wen G, Araki T, Luo J, Thompson JA, Schraven BL, Philips MR, Neel BG. Shp2 regulates Src family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13:341–355. doi: 10.1016/s1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 74.Ren Y, Meng S, Mei L, Zhao ZJ, Jove R, Wu J. Roles of Gab1 and SHP2 in Paxillin tyrosine dephosphorylation and Src activation in response to epidermal growth factor. J Biol Chem. 2004;279:8497–8505. doi: 10.1074/jbc.M312575200. [DOI] [PubMed] [Google Scholar]
- 75.Zhao ZJ, Zhao R. Purification and cloning of PZR, a binding protein and putative physiological substrate of tyrosine phosphatase SHP-2. J Biol Chem. 1998;273:29367–29372. doi: 10.1074/jbc.273.45.29367. [DOI] [PubMed] [Google Scholar]
- 76.Kontaridis MI, Eminaga S, Fornaro M, Zito CI, Sordella R, Settleman J, Bennett AM. SHP-2 positively regulates myogenesis by coupling to the rho GTPase signaling pathway. Mol Cell Biol. 2004;24:5340–5352. doi: 10.1128/MCB.24.12.5340-5352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. A family of proteins that inhibit signaling through tyrosine kinase receptors. Nature. 1997;386:181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 78.Noguchi T, Matozaki T, Fujioka Y, Yamao T, Tsuda M, Takada T, Kasuga M. Characterization of a 115-kDa protein that binds to SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in Chinese hamster ovary cells. J Biol Chem. 1996;271:27652–27658. doi: 10.1074/jbc.271.44.27652. [DOI] [PubMed] [Google Scholar]
- 79.Yu L, Min W, He Y, Qin L, Zhang H, Bennett AM, Chen H. JAK2 and SHP2 reciprocally regulate tyrosine phosphorylation and stability of proapoptotic protein ASK1. J Biol Chem. 2009;284:13481–13488. doi: 10.1074/jbc.M809740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koli S, Zito CI, Mossink MH, Wiemer EAC, Bennett AM. The major vault protein is a novel substrate for the tyrosine phosphatase SHP-2 and scaffold protein in epidermal growth factor signaling. J Biol Chem. 2004;279:29374–29385. doi: 10.1074/jbc.M313955200. [DOI] [PubMed] [Google Scholar]
- 81.Wu TR, Hong YK, Wang XD, Ling MY, Dragoi AM, Chung AS, Campbell AG, Han ZY, Feng GS, Chin YE. SHP-2 is a Dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem. 2002;277:47572–47580. doi: 10.1074/jbc.M207536200. [DOI] [PubMed] [Google Scholar]
- 82.Chen Y, Wen R, Yang S, Schuman J, Zhang EE, Yi T, Feng GS, Wang D. Identification of shp-2 as a stat5A phosphatase. J Biol Chem. 2003;278:16520–16527. doi: 10.1074/jbc.M210572200. [DOI] [PubMed] [Google Scholar]
- 83.Chiang GG, Sefton BM. Specific dephosphorylation of tyrosine protein kinase at Tyr-394 by the SHP-1 protein-tyrosine phosphatase. J Biol Chem. 2001;276:23173–23178. doi: 10.1074/jbc.M101219200. [DOI] [PubMed] [Google Scholar]
- 84.Ozawa T, Nakata K, Mizuno K, Yakura H. Negative autoregulation of Src homology region 2-containing phosphatase 1 in rat basophilic leukemia-2H3 cells. International Immunol. 2007;19:1049–1061. doi: 10.1093/intimm/dxm070. [DOI] [PubMed] [Google Scholar]
- 85.Mizuno K, Tagawa Y, Mitomo K, Arimura Y, Hatano N, Katagiri T, Ogimoto M, Yakura H. Src homology region 2-containing phosphatase 1 dephosphorylates B cell linker protein/SH2 domain leukocyte protein of 65 kDa and selectively regulates c-JUN NH2-terminal kinase activation in B cells. J Immunol. 2000;165:1344–1351. doi: 10.4049/jimmunol.165.3.1344. [DOI] [PubMed] [Google Scholar]
- 86.Keilhack H, Hellman U, van Hengel J, van Roy F, Godovac-Zimmermann J, Böhmer FD. The protein-tyrosine phosphatase SHP-1 binds to and dephosphorylates p120 catenin. J Biol Chem. 2000;275:26376–26384. doi: 10.1074/jbc.M001315200. [DOI] [PubMed] [Google Scholar]
- 87.Law CL, Sidorenko SP, Chandran KA, Zhao Z, Shen SH, Fischer EH, Clark EA. CD22 associates with protein tyrosine phosphatase 1C, Syk, and phospholipase C-γ1 upon B cell activation. J Exp Med. 1996;183:547–560. doi: 10.1084/jem.183.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu Y, Nadler MJ, Brennan LA, Gish GD, Timms JF, Fusaki N, Jongstra-Bilen J, Tada N, Pawson T, Wither J, Neel BG, Hozumi N. The B-cell transmembrane protein CD72 binds to and is an in vivo substrate of the protein tyrosine phosphatase SHP-1. Curr Biol. 1998;8:1009–1017. doi: 10.1016/s0960-9822(07)00421-6. [DOI] [PubMed] [Google Scholar]
- 89.Berg KL, Siminovitch KA, Stanley ER. SHP-1 regulation of p62DOK tyrosine phosphorylation in macrophages. J Biol Chem. 1999;274:35855–35865. doi: 10.1074/jbc.274.50.35855. [DOI] [PubMed] [Google Scholar]
- 90.Timms JF, Carlberg K, Gu H, Chen H, Kamatkar S, Nadler MJ, Rohrschneider LR, Neel BG. Identification of major binding protein and substrates for the SH2-containing protein tyrosine phosphatase SHP-1 in macrophages. Mol Cell Biol. 1998;18:3838–3850. doi: 10.1128/mcb.18.7.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Binstadt BA, Billadeau DD, Jevremović D, Williams BL, Fang N, Yi T, Koretzky GA, Abraham RT, Leibson PJ. SLP-76 is a direct substrate of SHP-1 recruited to killer cell inhibitory receptors. J Biol Chem. 1998;273:27518–27523. doi: 10.1074/jbc.273.42.27518. [DOI] [PubMed] [Google Scholar]
- 92.Dustin LB, Plas DR, Wong J, Hu YT, Soto C, Chan AC, Thomas ML. Expression of dominant negative src-homology domain 2-containing protein tyrosine phosphatase-1 results in increased Syk tyrosine kinase activity and B cell activation. J Immunol. 1999;162:2717–2724. [PubMed] [Google Scholar]
- 93.Stebbins CC, Watzl C, Billadeau DD, Leibson PJ, Burshtyn DN, Long EO. Vav1 dephosphorylation by the tyrosine phosphatase SHP-1 as a mechanism for inhibition of cellular cytotoxicity. Mol Cell Biol. 2003;23:6291–6299. doi: 10.1128/MCB.23.17.6291-6299.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baba T, Fusaki N, Shinya N, Iwamatsu A, Hozumi N. Actin tyrosine dephosphorylation by the Src homology 1-containing protein tyrosine phosphatase is essential for actin depolymerization after membrane IgM cross-linking. J Immunol. 2003;170:3762–3768. doi: 10.4049/jimmunol.170.7.3762. [DOI] [PubMed] [Google Scholar]
- 95.Zheng XM, Wang Y, Pallen CJ. Cell transformation and activation of pp60c-src by overexpression of protein tyrosine phosphatase. Nature. 1992;359:336–339. doi: 10.1038/359336a0. [DOI] [PubMed] [Google Scholar]
- 96.Samayawardhena LA, Pallen CJ. PTPα activates Lyn and Fyn and suppresses Hck to negatively regulate FcεRI-dependent mast cell activation and allergic responses. J Immunol. 2010;185:5993–6002. doi: 10.4049/jimmunol.1001261. [DOI] [PubMed] [Google Scholar]
- 97.Le HT, Maksumova L, Wang J, Pallen CJ. Reduced NMDA receptor tyrosine phosphorylation in PTPα-deficient mouse synaptosomes is accompanied by inhibition of four src family kinases and Pyk2: an upstream role for PTPα in NMDA receptor regulation. J Neurochem. 2006;98:1798–1809. doi: 10.1111/j.1471-4159.2006.04075.x. [DOI] [PubMed] [Google Scholar]
- 98.Imbrici P, Tucker SJ, D’Adamo MC, Pessia M. Role of receptor tyrosine phosphatase α (RPTPα) and tyrosine phosphorylation in the serotonergic inhibition of voltage-dependent potassium channels. Eur J Physiol. 2000;441:257–262. doi: 10.1007/s004240000406. [DOI] [PubMed] [Google Scholar]
- 99.Buist A, Blanchetot C, Teroolen LGJ, den Hertog J. Identification of p130cas as an in vivo substrate of receptor protein-tyrosine phosphatase α. J Biol Chem. 2000;275:20754–20761. doi: 10.1074/jbc.M001626200. [DOI] [PubMed] [Google Scholar]
- 100.Woodford-Thomas TA, Rhodes JD, Dixon JE. Expression of a protein tyrosine phosphatase in normal and v-src-transformed mouse 3T3 fibroblasts. J Cell Biol. 1992;117:401–414. doi: 10.1083/jcb.117.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frangioni JV, Oda A, Smith M, Salzman EW, Neel BG. Calpain-catalyzed cleavage and subcellular relocation of protein phosphotyrosine phosphatase 1B (PTP-1B) in human platelets. EMBO J. 1993;12:4843–4856. doi: 10.1002/j.1460-2075.1993.tb06174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kucahy SM, Kim N, Grunz EA, Fay WP, Chishti AH. Double knockouts reveal that protein tyrosine phosphatase 1B is a physiological target of calpain-1 in platelets. Mol Cell Biol. 2007;27:6038–6052. doi: 10.1128/MCB.00522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li S, Depetris RS, Barford D, Chernoff J, Hubbard SR. Crystal structure of a complex between protein tyrosine phosphatase 1B and the insulin receptor tyrosine kinase. Structure. 2005;13:1643–1651. doi: 10.1016/j.str.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 104.Yudushkin IA, Schleifenbaum A, Kinkhabwala A, Neel BG, Schultz C, Bastiaens PIH. Live-cell Imaging of enzyme-substrate interaction reveals spatial regulation of PTP1B. Science. 2007;315:115–119. doi: 10.1126/science.1134966. [DOI] [PubMed] [Google Scholar]
- 105.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxidants and Redox Signaling. 2005;7:560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 106.Flint AJ, Tiganis T, Barford D, Neel BG. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garton AJ, Flint AJ, Tonks NK. Identification of p130cas as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol Cell Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.