Abstract
Circulating microRNAs (miRNA) are relatively stable in plasma and are a new class of disease biomarkers. Here we present evidence that high-density lipoprotein (HDL) transports endogenous miRNAs and delivers them to recipient cells with functional targeting capabilities. Cellular export of miRNAs to HDL was demonstrated to be regulated by neutral sphingomyelinase. Reconstituted HDL injected into mice retrieved distinct miRNA profiles from normal and atherogenic models. HDL delivery of both exogenous and endogenous miRNAs resulted in the direct targeting of mRNA reporters. Furthermore, HDL-mediated delivery of miRNAs to recipient cells was demonstrated to be scavenger receptor BI-dependent. The human HDL-miRNA profile from normal subjects is significantly different than familial hypercholesterolemia subjects. Notably, HDL-miRNA from atherosclerotic subjects induced differential gene expression, with significant loss of conserved mRNA targets in cultured hepatocytes. Collectively, these observations suggest that HDL participates in a mechanism of intercellular communication involving the transport and delivery of miRNAs.
Until recently, the predominant view of intercellular communication was that it was limited to cell-to-cell adhesion conduits (gap junctions) or secreted signals, such as hormones and neurotransmitters. New studies have revealed that plasma membrane-derived vesicles, namely exosomes and microvesicles, can transfer proteins, mRNAs, and microRNAs (miRNAs) from donor cells to recipient cells1–4. miRNAs are short non-coding regulatory RNAs that modulate biological homeostasis, by controlling gene expression though mRNA targeting and translational repression5–7. Circulating miRNAs as a whole are a new class of biomarkers and have recently been used for a diverse set of diseases8.
Specific lipids found on lipoproteins, such as phosphatidylcholine (PC), have been shown to form stable ternary complexes with nucleic acids9–11. Infrared spectrometry, small-angle x-ray scattering, and other physical techniques have demonstrated that the interaction between RNA-DNA with zwitterionic lipids is mediated by divalent cation bridging9–15. A wide variety of nucleic acid-lipid complexes have been used for the delivery of genetic material to cells both in vitro and in vivo. In fact, liposomes containing apolipoprotein A-I (apoA-I), the main protein component of high-density lipoprotein (HDL), have been used for the systemic and specific delivery of small interfering RNA (siRNA) in animal models13.
The resemblance of these artificial gene delivery vehicles to native HDL and the biophysical interaction between nucleic acids with HDL phospholipids combined with the existence of circulating miRNAs prompted us to investigate whether endogenous miRNAs are associated with HDL in plasma. In this study, we present evidence that HDL not only transports endogenous miRNAs, which can differ with disease states, but can also deliver miRNAs to recipient cells with functional gene regulatory consequences. This study, therefore, reveals a potential new role for HDL in gene regulation and intercellular communication.
RESULTS
Human HDL contains small RNAs
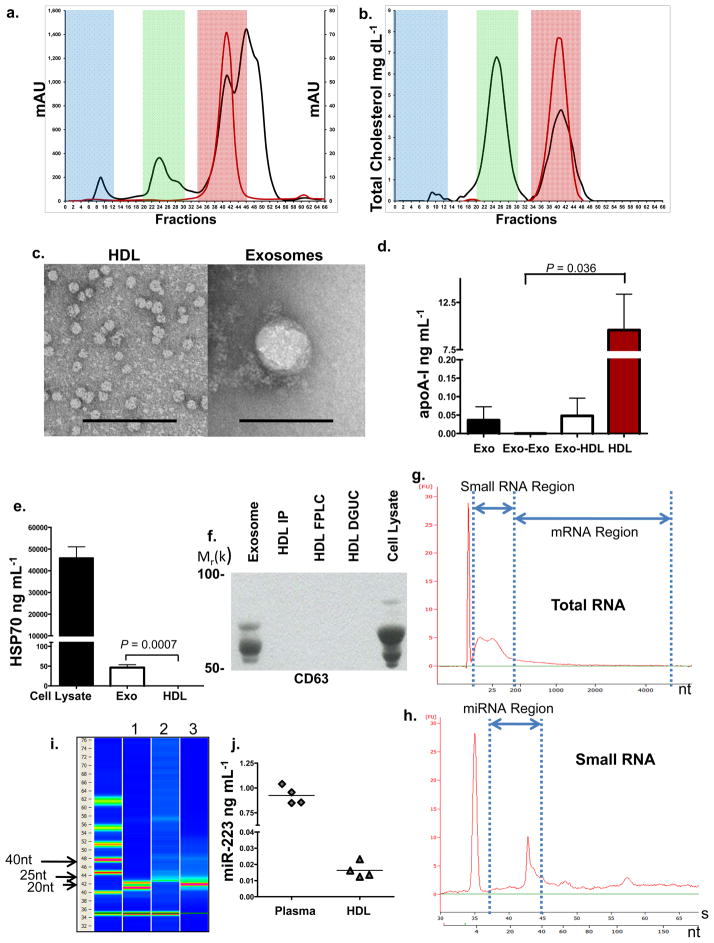
Highly-purified fractions of HDL were prepared from human plasma in a 3-step protocol, consisting of density gradient ultracentrifugation (DGUC) (d=1.063–1.21 g mL−1), followed by fast-protein liquid chromatography (FPLC), and anti-apoA-I immunoprecipitation (HDL-IP). Exosomes, which have a density (d=1.13–1.19 g mL−1) similar to HDL, are significantly larger in size (50–100 nm) than HDL (8–12 nm) and thus should be effectively removed by FPLC gel filtration, as well as by HDL-IP16, 17. The FPLC profile of human plasma (Fig. 1a) was used to define relative size zones associated with HDL (red), low-density lipoprotein (LDL) (green), and very-low density lipoprotein (VLDL) or exosomes (blue). The FPLC profile of purified HDL (red line) illustrates the absence of any particles larger than HDL (Fig. 1a), and the distribution of total cholesterol in the FPLC fractions further validates the purity of isolated HDL particles (Fig. 1b). HDL isolated by the three-step sequential procedure was observed by transmission electron microscopy (TEM) to be of uniform size (10–12 nm) and shape (Fig. 1c). Purified HDL particles were enriched for apoA-I (Fig. 1d), and were negative for classic exosomal protein markers HSP70 (Fig. 1e), CD63 (Fig. 1f), and ICAM-1 (Fig. S1a)18–20. Total RNA, isolated from highly-purified HDL, contained small RNAs (Fig. 1g–i) but was devoid of long mRNAs. Fluorescent quantification (RiboQuant) of gel-extracted HDL RNA, 15 to 30nts in length, confirmed small RNA in the purified HDL pool (Fig. S1b). The small RNA fraction is likely miRNA, as evidenced by its approximate length (22–26nt) observed in RNA gels, Bioanalyzer Small RNA characterization (Fig. 1g–i), and the presence of the widely-expressed miRNA, hsa-miR-223, in purified HDL (Fig. 1j)21, 22.
Figure 1. HDL RNA Analysis.
a.) Human plasma and HDL FPLC protein distribution (mAU). Red shading, HDL; green shading, LDL; and blue shading, VLDL-exosome. Black line, human plasma; red line, purified HDL. b.) Human plasma and HDL FPLC total cholesterol distribution (mg dL−1). Black line, plasma; red line, purified HDL. c.) TEM of HDL and exosomes, Scale bar = 100 nm. d.) Quantification of ApoA-I (ng mL−1) protein. Exo, exosomes (n=3); Exo-Exo, FPLC exosome zone from total exosome preparations (n=3); Exo-HDL, FPLC HDL zone from total exosome preparations (n=4); HDL, purified from plasma (n=5). Data are means +/− s.e.m. e.) Quantification of HSP70 protein (ng mL−1), Huh7 cell lysates (n=4); Exo, exosomes (n=4); and HDL, purified from plasma (n=4). Data are means +/− s.e.m. f.) Western blot, Anti-CD63. Purified exosomes; HDL IP, HDL immunoprecipitation; HDL FPLC; HDL DGUC, HDL density-gradient ultracentrifugation; Huh7 cell lysate. Uncropped image of blot (Fig. S6) g.) Bioanalyzer Pico analysis of HDL total RNA. h.) Bioanalyzer Small RNA analysis of HDL total RNA. i.) Digital gel electrophoresis of HDL small RNAs. Lane 1, miRNA standards 22 & 25 nts; Lane 2, HDL total RNA; Lane 3, miR-223 positive control. j.) Quantification of hsa-miR-223 levels (ng mL−1). n=4.
Distinct HDL-miRNA signature in disease
Total RNA was isolated from highly-purified HDL from normal and familial hypercholesterolemia (FH) subjects (Table S1). HDL-miRNA profiles were analyzed using an amplification-based microarray platform (Applied Biosystems) and normalized (to individual array mean Ct) abundances of HDL-miRNAs were ranked (Table 1, Table S2). The most abundant miRNAs associated with HDL from normal subjects were hsa-miR-135a* (relative quantitative value, RQV=121.9), hsa-miR-188-5p (RQV=17.1), and hsa-miR-877 (RQV=11.1); and all three were in the top ten rankings for all normal subjects (Table 1). Most importantly, the normal HDL-miRNA profile was distinctly different (R=0.4598, P<0.0001) than the purified exosome-miRNA profile (Table S3) from matched subjects (Fig. S1c), including differences in content, abundances, and top-ranked miRNAs.
Table 1.
Human and mouse HDL-miRNA signatures in health and atherosclerosis.
| Human HDL miRNAs | ||||||
|---|---|---|---|---|---|---|
| Normal | Familial Hypercholesterolemia | |||||
| Normalized Abundance | Top Ten | Top Ten | ||||
| miRNA | (+/−SD) | P/A | miRNA | (+/−SD) | P/A | |
| 1 | miR-135a* | 121.9+/−41.3 | 6 / 6 | miR-223 | 3014553.4+/−5939444 | 5 / 5 |
| 2 | miR-188-5p | 17.1+/−5.7 | 6 / 6 | miR-105 | 13671.6+/−26536 | 4 / 5 |
| 3 | miR-877 | 11.1+/−5.2 | 6 / 6 | miR-106a | 437.3+/−812.3 | 4 / 5 |
| 4 | miR-223 | 8.4+/−6.9 | 6 / 6 | miR-17 | 436.8+/−815.4 | 3 / 5 |
| 5 | miR-760 | 5.9+/−2.1 | 5 / 6 | miR-135a* | 223.7+/−347.6 | 5 / 5 |
| 6 | miR-375 | 5.4+/−1.6 | 6 / 6 | miR-372 | 170.3+/−203.3 | 2 / 5 |
| 7 | miR-106a | 5.1+/−3.1 | 4 / 6 | miR-489 | 170.3+/−204 | 2 / 5 |
| 8 | miR-138-1* | 4.7+/−1.1 | 6 / 6 | miR-24 | 123.8+/−192.4 | 3 / 5 |
| 9 | miR-625* | 4.7+/−3.2 | 3 / 6 | miR-193b | 120.3+/−194.4 | 2 / 5 |
| 10 | miR-17 | 3.5+/−1.1 | 4 / 6 | let-7g | 106.9+/−201.7 | 1 / 5 |
| Significant Differential Abundance on Familial Hypercholesterolemia HDL | ||||||
| miRNA | Increased Abundance Fold Change | p-value | miRNA | Decreased Abundance Fold Change | p-value | |
| 1 | miR-223 | 3780.64 | 0.020 | miR-625* | −10.95 | 0.005 |
| 2 | miR-24 | 65.27 | 0.002 | miR-520c-3p | −5.43 | 0.013 |
| 3 | miR-342-3p | 30.35 | 0.037 | miR-509-3p | −3.74 | 0.012 |
| 4 | let-7b | 12.27 | 0.043 | miR-573 | −3.58 | 0.007 |
| 5 | miR-412 | 9.51 | 0.006 | miR-632 | −3.30 | 0.004 |
| 6 | miR-222 | 8.20 | 0.016 | miR-188-5p | −2.71 | 0.005 |
| 7 | miR-106a | 7.85 | 0.045 | miR-572 | −2.48 | 0.008 |
| 8 | miR-218 | 7.80 | 0.010 | miR-323-3p | −2.47 | 0.012 |
| 9 | miR-191 | 4.66 | 0.009 | miR-135a* | −2.41 | 0.049 |
| 10 | miR-20a | 19.56 | 0.066 | miR-877 | −2.26 | 0.021 |
| 11 | miR-186 | 10.94 | 0.161 | miR-138-1* | −2.00 | 0.037 |
| 12 | miR-17 | 7.64 | 0.103 | miR-526b* | −5.85 | 0.051 |
| miRNA | OFF (+/−SD) | P/A | miRNA | ON (+/−SD) | P/A | |
| 1 | miR-639 | 1.83+/−1.6 | 2 / 6 | miR-105 | 13671.6+/−26536 | 4 / 5 |
| 2 | miR-584 | 1.76+/−0.9 | 3 / 6 | miR-489 | 170.3+/−204.0 | 4 / 5 |
| 3 | miR-643 | 1.2 | 1 / 6 | let-7g | 106.9+/−201.7 | 4 / 5 |
| 4 | miR-766 | 0.62 | 1 / 6 | miR-146b-3p | 73.7+/−112.7 | 3 / 5 |
| 5 | miR-597 | 0.59+/−.2 | 3 / 6 | miR-339-5p | 50.7+/−0.0 | 1 / 5 |
| 6 | miR-211 | 37+/−56.3 | 3 / 5 | |||
| 7 | let-7e | 26.3+/−51.6 | 4 / 5 | |||
| 8 | miR-223* | 26+/−35.5 | 2 / 5 | |||
| 9 | miR-661 | 13.1+/−0.0 | 1 / 5 | |||
| 10 | miR-99a | 13+/−17.7 | 2 / 5 | |||
| 11 | miR-302b | 8.8+/−14.8 | 3 / 5 | |||
| Mouse HDL miRNAs | ||||||
| WT Chow | Apoe −/− HFD | |||||
| Normalized Abundance | ||||||
| miRNA | (+/−SD) | P/A | miRNA | (+/−SD) | P/A | |
| 1 | miR-375 | 48916.24+/−49386.5 | 3 / 3 | miR-383 | 942017.3+/−1332214 | 1 / 4 |
| 2 | miR-223 | 3905.9+/−5671.8 | 3 / 3 | miR-188-5p | 928408.4+/−1856815 | 1 / 4 |
| 3 | miR-17 | 973.8 | 1 / 3 | miR-293 | 222741.1 | 1 / 4 |
| 4 | miR-135a* | 257.42+/−196.9 | 3 / 3 | miR-223 | 12356.26+/−15251.4 | 4 / 4 |
| 5 | miR-709 | 122.5 | 1 / 3 | miR-375 | 312.78+/−2129 | 4 / 4 |
| 6 | miR-466d-3p | 119.4+/−167.34 | 1 / 3 | miR-135a* | 138.1546+/−114.1 | 4 / 4 |
| 7 | miR-141* | 47.16+/−31.6 | 3 / 3 | miR-720 | 136.1308+/−144.1 | 2 / 4 |
| 8 | miR-764-5p | 30.5 | 1 / 3 | miR-466d-3p | 125.6375+/−145.8 | 2 / 4 |
| 9 | miR-720 | 15.6 | 1 / 3 | miR-709 | 58.083+/−2.8 | 2 / 4 |
| 10 | miR-760 | 9.04+/−4.5 | 2 / 3 | miR-764-5p | 31.63+/−5.4 | 2 / 4 |
| Significant Differential Abundance on Apoe −/− HFD HDL | ||||||
| miRNA | Increased Abundance Fold Change | p-value | miRNA | Decreased Abundance Fold Change | p-value | |
| 1 | miR-188-5p | 747272.10 | 0.43 | miR-17 | −107.69 | na |
| 2 | miR-203 | 21.89 | na | miR-375 | −15.71 | 0.05 |
| 3 | miR-685 | 13.78 | 0.09 | miR-99b* | −14.03 | na |
| 4 | miR-24 | 9.13 | 0.30 | miR-801 | −3.23 | na |
| 5 | miR-720 | 8.75 | na | miR-875-5p | −2.55 | 0.19 |
| 6 | miR-877 | 5.77 | 0.48 | miR-30e* | −2.42 | 0.33 |
| 7 | miR-34c | 3.99 | 0.46 | miR-141* | −2.30 | 0.23 |
| 8 | miR-146a | 3.91 | 0.51 | miR-709 | −2.10 | na |
| 9 | miR-378 | 3.33 | 0.05 | miR-30e | −1.95 | 0.03 |
| 10 | miR-223 | 3.16 | 0.41 | miR-135a* | −1.88 | 0.35 |
| OFF | miRNA | ON | P/A | |||
| 1 | miR-383 | 942017.3+/−1332214 | 2 / 4 | |||
| 2 | miR-293 | 222741.1 | 1 / 4 | |||
| 3 | miR-704 | 1.7 | 1 / 4 | |||
| 4 | miR-211 | 1.34+/−0.47 | 2 / 4 | |||
Italicized n.s.; P/A, number of individual profiles specific miRNA is present in top ten ranking / total arrays; SD, standard deviation.
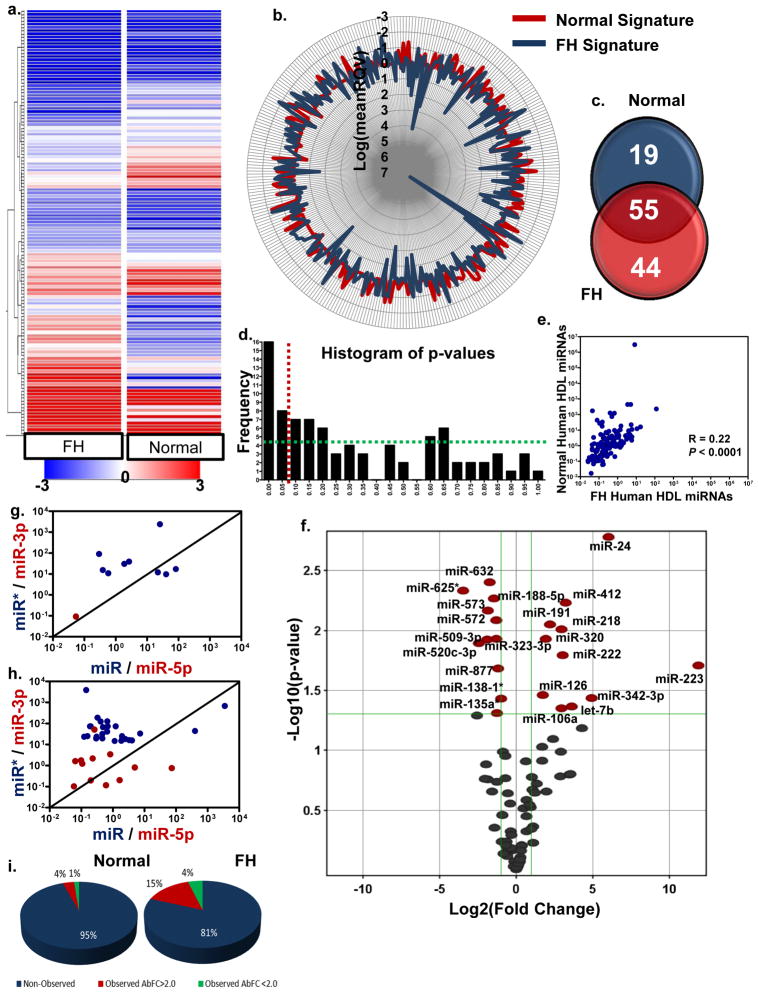
The most abundant FH HDL-miRNAs were hsa-miR-223 (RQV=3,014,553), hsa-miR-105 (RQV=13671.6), and hsa-miR-106a (RQV=437.8) (Table 1). HDL from FH subjects had a greater concentration of the most prominent miRNAs (Fig. 2a,b) and contained more individual miRNAs than normal subjects (99 to 74, ≥3 call per group) (Fig. 2c). For example, the most abundant FH HDL-miRNA, hsa-miR-223, was the fourth-most abundant miRNA associated with normal HDL (Table 1); however, hsa-miR-223 levels increased 3780.6-fold with FH disorder (P=0.019). The frequency of significant and low p-values was greater than random distribution, which suggests differential HDL-miRNA signatures in health and disease (Fig. 2d). Furthermore, HDL-miRNA signatures from normal and diseased (FH) subjects were determined to be distinct (R=0.2253, P<0.0001) (Fig. 2e). A volcano plot shows that 22 HDL-miRNAs were significant (P<0.05) differentially (2-fold) abundant with FH, not including miRNAs that were completely unique (ON-OFF) for each condition (Fig. 2f).
Figure 2. Human HDL carries distinct miRNA signatures in health and disease.
a.) Hierarchal clustering heatmap of human HDL miRNAs from normal (n=6) and FH (familial hypercholesterolemia) (n=5) subjects. Blue to red, color range gradient of mean abundance (−3 to 3). b.) StarGlyph distribution of each miRNA observed on HDL. Log values of the mean RQV (relative quantitative value). Red, normal; blue, FH HDL. c.) Venn diagram of normal and FH HDL miRNA totals. miRNA must be observed on ≥ 3 arrays within class. d.) Histogram of frequency distribution of p-values. Red line, P=0.05. Green line illustrates uniform distribution of p-values. e.) Spearman non-parametric correlation between normal and FH HDL miRNA profiles. f.) Volcano plot of significant differentially abundant miRNAs on FH HDL compared to normal HDL. Red marks, >2-fold change (Log2); P<0.05 (−Log10). g.) Expression (mean) ratios of miRNA pairs (miR/miR*, blue; miR-5p/miR-3p, red). Black line represents expression ratio of 1 (x=y) and similar abundance of both strands of pairs. Log10 scale. g.) Human HDL-miRNA pairs from normal subjects h.) Human HDL-miRNA pairs from FH subjects. i.) Pie charts illustrating strand observations (percentages) for normal (left) and FH (right) HDL. Blue, ≤1 strand of miRNA pair was observed; red, both strands of miRNA pair was observed but absolute fold change (AbFC) >2.0; green, both strands of pair were observed and had relatively equal abundance (AbFC<2.0).
Each strand of a duplex miRNA, processed from the pre-miRNA, has the potential to be a distinct functional miRNA. Expression ratios of the two strands are variable and the active strand can originate from both the 5p and 3p end of the precursor hairpin. To investigate the HDL-miRNA expression (abundance) ratios, we analyzed the ratio of mature miRNAs (miR) and their less abundant duplex strands (miR*), including analysis of 5p to 3p miRNA ratios (Fig. 2g–i). Our array platform allowed for the analysis of 197 miRNA pairs; however, only 10 pairs were observed in HDL from normal subjects, with a mean ratio (absolute fold change) of 17. Similar abundance levels (absolute fold change <2.0) of both strands were only observed in 3 of the 10 pairs (Fig. 2g,i). Collectively, the abundance levels of both strands were relatively equal in less than 2% of all pairs analyzed (Fig. 2g,i). Similar results were observed in FH subjects (Fig. 2h,i); therefore, the vast majority of miRNA pairs were only represented by one strand on HDL.
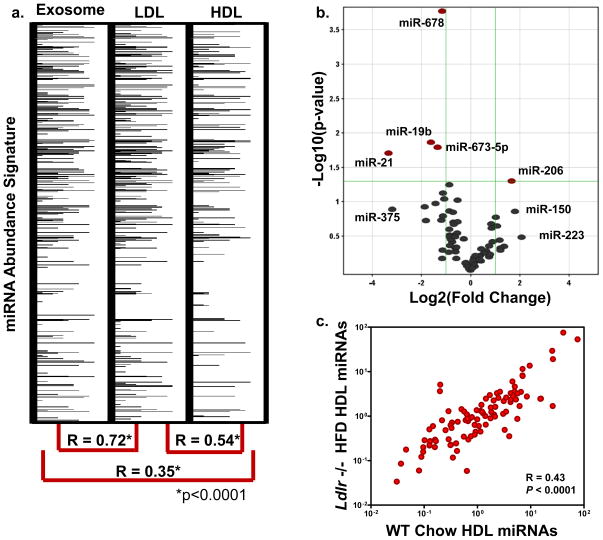
LDL has similar physico-chemical properties as HDL; however, LDL is larger in size (22 nm), differential in protein composition, and its generation and maturation are distinct from HDL. Highly pure fractions of LDL were also found to contain miRNAs (Fig. 3a). Nonetheless, the LDL-miRNA signature is more closely aligned with the exosome-miRNA signature (R=0.72, P<0.0001) than HDL (R=0.54, P<0.0001) when quantified in the same subject (Fig. 3a, Table S4). FH, a genetic disorder of the LDL receptor (LDLR), results in extreme atherosclerosis in homozygous cases23. As described above, FH subjects exhibit a significant differential HDL-miRNA profile (Fig. 2, Table 1,S2) compared to normal subjects. To further investigate this observation, LDL receptor (Ldlr) null mice were placed on a high fat diet (HFD) and the HDL-miRNA profile was compared to wild-type (WT) controls (Table S5,S6). Similar to humans, loss of Ldlr resulted in a significant change to the mouse HDL-miRNA profile; however, the total number of miRNAs significantly altered was less in Ldlr−/− mice (5) than human FH disorder (22) (Fig. 3b;Table S5,S6). Overall, the Ldlr−/− HFD HDL-miRNA profile was less distinct from WT controls (R=0.43, P<0.0001) than human FH HDL profile was from normal subjects (R=0.22, P<0.0001) (Fig. 2e,3c), which may be a function of the degree of atherosclerosis.
Figure 3. The role of LDL and the LDL receptor in HDL-miRNA signatures.
a.) miRNA abundance signatures of human exosomes, LDL, and HDL from same subject. Spearman non-parametric correlation between each profile. *p<0.0001. miRNAs are numerically ranked top-down and abundances are represented by horizontal band intensity. b.) Volcano plot of significant (P<0.05) differential (>2.0-fold) HDL-miRNA abundances in Ldlr −/− HFD mice (n=3) compared to WT controls (n=3). Red marks, >2-fold change (Log2); P<0.05 (−Log10). c.) Spearman non-parametric correlation between Ldlr −/− HFD mice and WT controls. R=0.43, P<0.0001.
Native HDL incorporates small RNAs in vitro
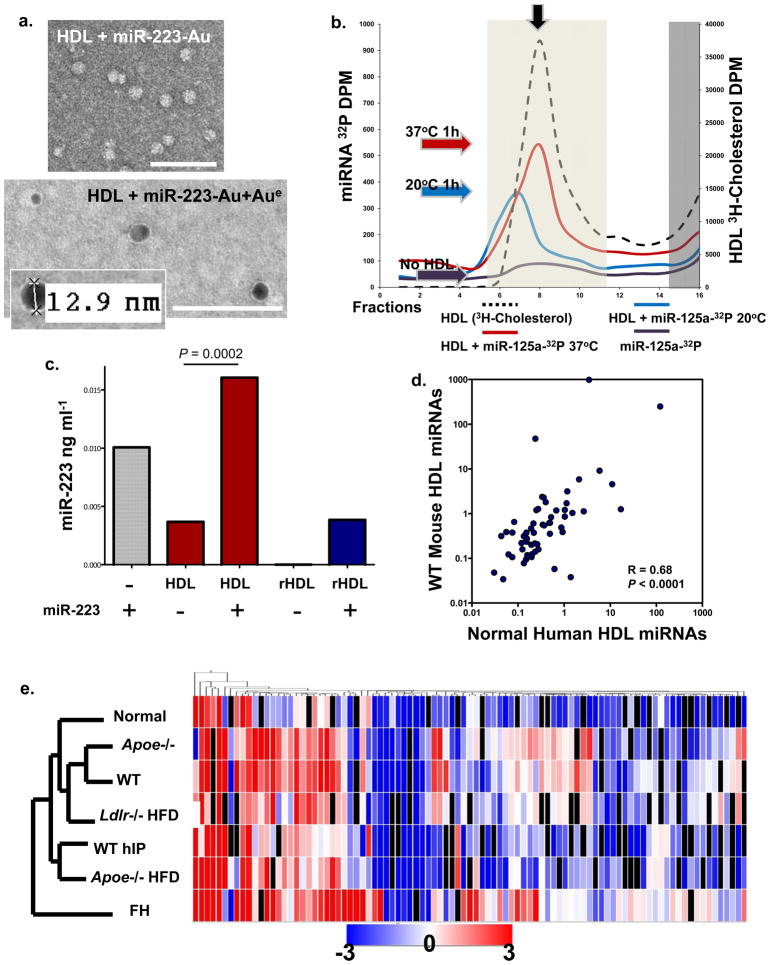
To observe miRNA bound to native HDL, miR-223 was labeled with 1.4 nm monoamino Nanogold, incubated with human HDL (1h, 37°C), and complexes were purified by HDL-IP. Gold-enhanced Nanogold-labeled miRNA-HDL complexes were electron dense and measured to be 9–13 nm in diameter (Fig. 4a). Gold enhancement of Nanogold-labeled antibodies on a grid under the same conditions yielded large dense grains, some greater than 100 nm in diameter (Fig. S2a). This suggests that the Nanogold-labeled miRNA may be buried within the HDL to which it is bound, resulting in reduced exposure to the gold enhancement reagents. To further determine if miRNAs associate with or bind to native HDL in vitro, radiolabeled (32P) miRNA mimetics were used in FPLC gel-filtration assays. Radiolabeled miRNAs were incubated with HDL at 37°C, and the complexes were separated by FPLC (S-200 column). The association of miRNA (32P-miR-125a) with native HDL (3H-cholesterol, black arrow) was temperature-influenced as shown by binding at 37°C versus 20°C (Fig. 4b). To quantify HDL-miRNA incorporation, native HDL and reconstituted HDL (rHDL) (Fig. S2b) were incubated with miR-223 and HDL-miRNA complexes were isolated by HDL-IP. Both native (P=0.0002) and rHDL contained significantly more miR-223 after complexing in vitro, as determined by quantitative PCR (Fig. 4c). Native human HDL has a natural abundance of endogenous hsa-miR-223; however, rHDL was free of RNA prior to incorporation assays (Fig. 4c).
Figure 4. HDL readily incorporates with miRNAs in vitro and in vivo.
a) TEM image of HDL + Nanogold-labeled miRNA (miR-223-Au) unenhanced (Top); HDL + Nanogold-labeled miRNA (miR-223-Au) gold enhanced (Aue) (2 min) (Bottom). Scale bars = 100 nm. b.) FPLC separation of radiolabeled HDL (3H-cholesterol) -miRNA (32P-miR-125a) complexes. Red line, HDL + 32P-miR-125a (37°C reaction); blue line, HDL + 32P-miR-125a (20°C reaction); purple line, 32P-miR-125a alone; black dash line, 3H-HDL + cold miR-125a. Light shading indicates HDL complex zone, dark shading indicates uncomplexed free radiolabeled 32P-miR-125a and 3H-HDL zone. Colored arrows indicate peak associations, red (37°C), blue (20°C), purple (no HDL). S200 Column. c.) Quantification of HDL-miR-223 incorporation. hsa-miR-223 (ng ml−1) levels post-HDL-IP, miR-223, positive control; native human HDL; native human HDL + miR-223; reconstituted HDL (rHDL); rHDL + miR-223. n=2 d.) Spearman non-parametric correlation between WT mouse and normal human HDL profiles. R=0.68, P<0.0001. e.) Hierarchal clustering heatmap of HDL-miRNA profiles (mean) in wellness, hyperlipidemia, and atherosclerosis (mouse & human). n=7 conditions: FH, familial hypercholesterolemia (n=5); Apoe−/− HFD (n=4); WT hIP (n=3), rHDL retrieved from wild-type (WT) mice (n=3); Ldlr −/− HFD (n=3); WT chow diet (n=3); Apoe−/− chow diet (n=3); and Normal human HDL (n=6).
Reconstituted HDL retrieves miRNAs in vivo
To determine if rHDL (free of RNA) has the capacity to be loaded with miRNAs in vivo, human rHDL (150 μg) was injected intravenously into WT or apolipoprotein E (Apoe−/−) null mice on normal chow or HFD. After 6h post-injection, rHDL was recovered from mouse plasma by anti-human apoA-I immunoprecipitation and mouse miRNAs were profiled by amplification-based microarrays. Injected rHDL retrieved 110, 162, and 130 unique miRNAs from WT, Apoe−/− chow, and Apoe−/− HFD plasma, respectively (Table 1,S7; Fig. S3). Although WT and Apoe−/− HFD mice shared 92 HDL-miRNAs, the profiles were distinct and the levels of the most abundant miRNAs were greatly altered in the atherosclerosis mouse model (Apoe−/− HFD) (Fig. S3b, Table 1,S7). HDL-miRNA profiling of rHDL isolated from Apoe−/− mice on chow diet was used to determine if HDL-miRNA signature changes were associated with vascular arterial disease or simply hyperlipidemia, although hypercholesterolemia ultimately results in atherogenesis. The HDL-miRNA profile from Apoe−/− HFD mice was determined to be significantly different than WT and Apoe−/− chow mice (Fig. S3c,d). Taken together these results suggest that HDL binds to miRNAs in vivo and that atherosclerosis in mice results in altered HDL-miRNA signatures.
Homology between human and mouse HDL profiles
Although miR-223 is one of the top most abundant HDL-miRNAs in mice and humans, its concentration was increased 3.2-fold in Apoe−/− HFD mice, 4.2-fold in Ldlr−/− HFD mice, and 3780-fold with FH compared to controls (Table 1,S1–S7, Fig. S3). miR-24 was also increased in both Apoe−/− HFD (9.13-fold) and FH (65.3-fold) compared to controls. Furthermore, miR-211 was not detectable in HDL fractions from either normal subjects or WT mice; however, miR-211 was abundant in Apoe−/− HFD and FH HDL (Table 1). Conversely, miR-135a*, one of the most abundant miRNAs observed on HDL, was reduced in humans (−2.41-fold) and mice (Apoe−/− HFD −1.9-fold) with atherosclerosis (Table 1). To assess homology between the representative groups of health and disease (atherosclerosis), the complete mean miRNA profiles were filtered to only miRNAs conserved between humans and mice. The mean abundances of the conserved miRNAs were significantly correlated (R=0.682, p<0.0001) between healthy groups (normal humans and WT mice) (Fig. 4d). Hierarchal clustering of the filtered mean HDL profiles from all assayed conditions (n=7) resulted in the separation of the FH profile; however, the human FH profile was most tightly clustered with the Apoe−/− HFD mouse profile (Fig. 4e). Similarly, the normal human, Apoe−/− chow, and WT mouse profiles were clustered furthest from the FH signature. Furthermore, 11 miRNAs, conserved between humans and mice, were found on HDL of every healthy subject or mouse analyzed (Table S8). These data suggest that both humans and mice transport a consistent group of conserved miRNAs on HDL in wellness and disease.
HDL transfers miRNAs to recipient cells
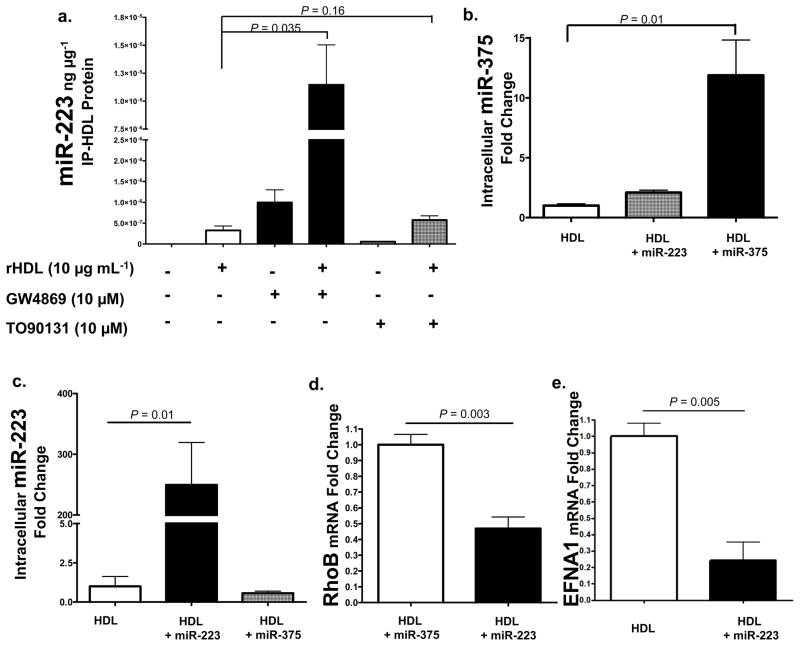
To investigate the mechanisms of miRNA export to HDL, we examined the role of ATP-binding cassette transporter A1 (ABCA1) and neutral sphingomyelinase 2 (nSMase2) pathways. Biogenesis of HDL is mediated through ABCA1 cellular efflux of cholesterol and phospholipids to nascent HDL24. To test if ABCA1 mediates the efflux of small RNAs during the maturation of HDL, we induced overexpression of ABCA1 with the Liver-X-Receptor alpha (LXRα) agonist TO90131. J774 mouse macrophages were treated with 10 μM TO90131 and rHDL (10 μg mL−1) in lipoprotein-depleted serum (LPDS). After 48h, rHDL was isolated from culture media by anti-human apoA-I immunoprecipitation and total RNA was purified. Quantitative PCR was used to measure the levels of exported miR-223 as normalized to rHDL-IP total protein. The addition of rHDL alone facilitated the export of miR-223 and the induction of ABCA1 by TO90131 increased the abundance of miR-223 on rHDL 1.8-fold (P=0.16) (Fig. 5a). Recently, a study has demonstrated that inhibition of the ceramide signaling pathway through GW4869 chemical inhibition of nSMase2 resulted in the attenuation of cellular miRNA export by exosomes and a significant reduction of specific extracellular miRNAs25. To determine if nSMase2 regulates the export of miRNAs to HDL, J774 macrophages were treated with 10 μM GW4869 for 48h. Contrary to previous results obtained in HEK293 cells measuring exosomal miRNAs, inhibition of nSMase2 significantly increased the amount of miR-223 exported to HDL (35.3-fold, P=0.035) from macrophages. Tandem mass spectrometry and Western blotting failed to identify previously described miRNA binding proteins on HDL, including nucleophosmin and Argonaute family members (Fig. S4b, Table S9).
Figure 5. HDL transfers miRNAs to recipient cells with functional targeting.
a.) nSMase2 regulates miRNA export to HDL. Quantification of HDL-miR-223 levels (qPCR) normalized to IP-rHDL protein (ng μg−1). rHDL, reconstituted HDL. GW4869, chemical inhibitor of nSMase2; TO90131, LXRα agonist induces ABCA1 expression. n=3. Data are means +/− s.e.m. b.) Quantification of intracellular miR-375 levels (qPCR) in hepatocytes (Huh7) treated with HDL (80 μg mL−1) alone (n=4), HDL+miR-223 (80 μg mL−1) (n=4), or HDL+miR-375 (80 μg mL−1) (n=4). Data are means +/− s.e.m. c.) Quantification of intracellular miR-223 levels (qPCR) in hepatocytes (Huh7) treated with HDL alone (80 μg mL−1) (n=4), HDL+miR-223 (80 μg mL−1) (n=4), or HDL+miR-375 (80 μg mL−1) (n=4). Data are means +/− s.e.m. d.) Quantification of RhoB mRNA levels (fold change) in hepatocytes (Huh7) treated with HDL+miR-375 (80 μg mL−1) (n=4) or HDL+miR-223 (80 μg mL−1) (n=4). Data are means +/− s.e.m. e.) Quantification of Ephrin A1 mRNA levels (fold change) in hepatocytes (Huh7) treated with HDL alone (80 μg mL−1) (n=3) or HDL+miR-223 (80 μg mL−1) (n=3). Data are means +/− s.e.m.
To test whether HDL has the capacity to deliver small RNAs to recipient cells, native HDL was incorporated with exogenous miRNAs and introduced to cultured hepatocytes (Huh7). In cells treated with HDL-miR-375 complexes, intracellular levels of miR-375 increased 11.8-fold (P=0.01) (Fig. 5b). Cellular delivery of HDL incorporated with miR-223 also significantly (P=0.012) increased intracellular miR-223 levels (250-fold) (Fig. 5c), which resulted in the significant loss of miR-223 mRNA targets (Fig. 5d,e). miR-223 has recently been experimentally validated to target the Ras homolog gene family, member B (RhoB) with two 3′UTR seed target sites26. In our study, overexpression of miR-223 in hepatocytes by transient transfection (100 nM) confirmed the direct targeting of RhoB by miR-223 (60% loss, p<0.05) (Fig. S4a). More importantly, delivery of miR-223 by HDL also reduced RhoB mRNA levels by 53% (P=0.003) (Fig. 5d). Ephrin A1 (EFNA1), a GPI-anchored receptor tyrosine kinase ligand expressed in the liver and up-regulated in hepatocarcinoma, also harbors a conserved miR-223 seed target site within its 3′UTR27. In our study, HDL-mediated miR-223 delivery resulted in a significant (P=0.005) 76% loss of EFNA1 mRNA (Fig. 5e). Taken together, these results suggest that native HDL can readily associate with exogenous small RNAs and deliver genetic material to recipient cells with functional targeting capabilities, leading to altered gene expression.
HDL-miRNA delivery is SR-BI-dependent
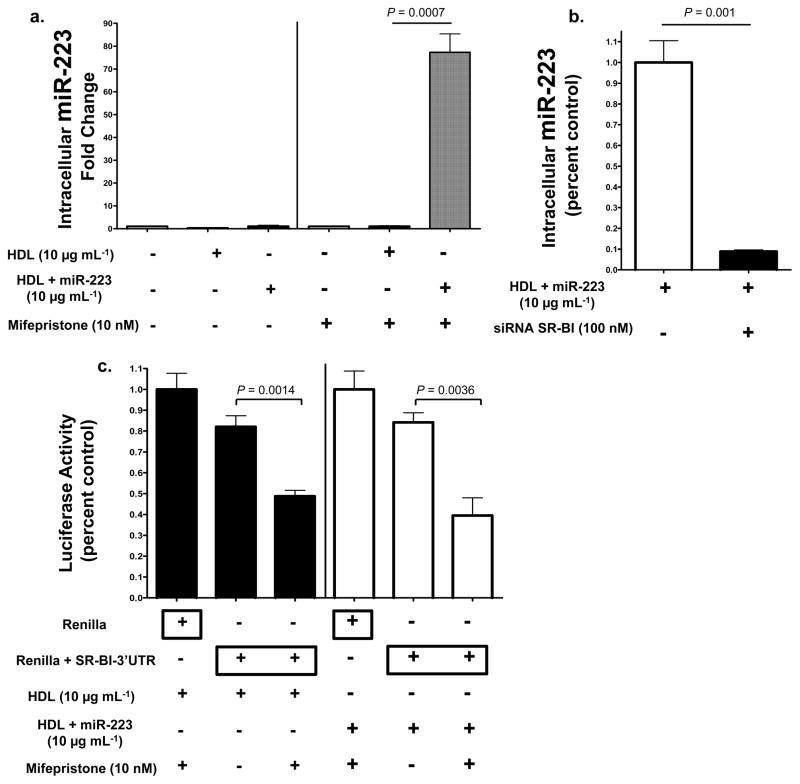
To test whether HDL miRNAs are selectively transferred by SR-BI, loss-of-function and gain-of-function studies were utilized. Baby hamster kidney (BHK) cells, which do not express significant quantities of SR-BI, were stably transfected with an inducible human SR-BI vector. Both non-induced and induced BHK cells were incubated with native human HDL (10 μg mL−1) or HDL-miR-223 complexes for 48h in LPDS. Intracellular levels of hsa-miR-223 were 69-fold higher (P=0.0007) in SR-BI expressing cells treated with HDL-miR-223 complexes compared to HDL alone (Fig. 6a, Fig. S5a). In siRNA knock-down studies, loss of SR-BI resulted in a significant reduction (89% loss, P=0.014) of HDL-miR-223 delivery, as quantified by intracellular miR-223 levels (Fig. 6b). To determine if SR-BI-dependent HDL-miRNA delivery results in the direct targeting of specific mRNAs, luciferase reporter assays were used in conjunction with the inducible SR-BI system in BHK cells. HDL complexed with exogenous miR-223 directly targeted Renilla-SR-BI-3′UTR luciferase after induced SR-BI-dependent HDL delivery (P=0.0036) (Fig. 6c). SR-BI harbors a highly conserved miR-223 target site within its 3′UTR (Fig. 4c). Interestingly, native human HDL delivered sufficient concentrations of endogenous miRNAs, most likely miR-223, to significantly (P=0.0014) repress the Renilla-SR-BI-3′UTR luciferase reporters in SR-BI expressing cells (Fig. 6c). These results suggest that HDL delivery of both endogenous and exogenous miRNAs through the SR-BI receptor alters gene expression. SID1 transmembrane family member 1 (SIDT1) has previously been demonstrated to mediate the uptake of extracellular dsRNA and cholesterol-conjugated siRNAs28, 29. SIDT1 (siRNA) knockdown failed, however, to result in a significant loss of HDL-miR-223 delivery as quantified by intracellular miR-223 levels (Fig. S5).
Figure 6. HDL-miRNA delivery is SR-BI-dependent.
a.) Quantification of intracellular hsa-miR-223 levels (fold change). Transfected BHK cells, pSwitch human SR-BI inducible (mifepristone 10 nM) expression system, treated with HDL alone (10 μg mL−1) (n=3) or HDL+miR-223 (10 μg mL−1) (n=3). Data are means +/− s.e.m. b.) Quantification of HDL-miR-223 delivery, as determined by intracellular miR-223 levels (qPCR, miR-223 standard curve). Data reported as percent control. Human hepatocytes (Huh7) transfected with SR-BI siRNA (100 nM, On-target Plus pool) (n=3) or mock reagent (n=3), prior to HDL+miR-223 (10 μg mL−1) treatment. Data are means +/− s.e.m. c.) Renilla luciferase activity normalized to Firefly (transfection control) luciferase activity. BHK cells transfected with pSwitch human SR-BI inducible (mifepristone 10 nM) expression system treated with HDL (80 μg mL−1) (n=4) or HDL+miR-223 (80 μg mL−1) (n=4). Renilla-SR-BI-3′UTR luciferase reporter activities reported as fold changes to Renilla controls. Data are means +/− s.e.m.
Intercellular communication by HDL
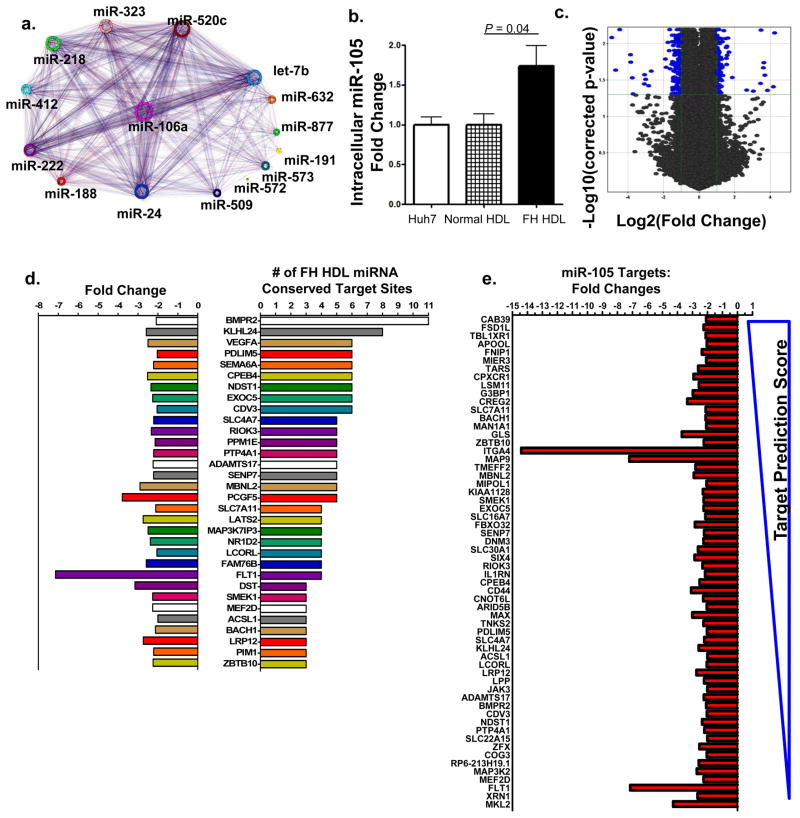
In silico target prediction (TargetScan5.1) identified mRNAs that are conserved putative targets of the 22 differentially abundant miRNAs on FH HDL30, 31. The predicted impact upon recipient cell gene expression and phenotype attributed to FH HDL-miRNA delivery was conceptualized by the FH HDL-miRNA target network interactome (Fig. 7a). The most significant pathways predicted to be affected consist of multiple inflammation and signaling pathways (Table S10). To further examine whether endogenous levels of miRNAs transported on HDL are sufficient to directly alter gene expression in recipient cells, HDL from FH and normal subjects were used to treat hepatocytes (Huh7). Huh7 cells endogenously express hsa-miR-105 and intracellular levels failed to increase with treatment of normal (healthy) HDL that lacks hsa-miR-105 (Fig. 7b). In contrast, treatment with FH HDL, which has abundant levels of hsa-miR-105 (Table 1), resulted in a significant (P=0.042) increase in intracellular hsa-miR-105 levels (Fig. 7b).
Figure 7. Atherosclerotic HDL induces differential gene expression through miRNA transfer.
a.) Conceptualization of FH HDL-miRNA mRNA target interactome. Putative targets of differentially abundant FH HDL-miRNAs (unique colors) and their association to other miRNAs and targets b.) Quantification of intracellular (Huh7) hsa-miR-105 (fold change) after human FH (n=4) or normal (n=4) HDL (80 μg mL−1) delivery. Data are means +/− s.e.m. c.) Volcano plot illustrating significant differential gene expression (mRNA) changes (blue marks) attributed to FH HDL-miRNA delivery, compared to normal HDL. −Log10 (Benjamini-Hochberg corrected p-value) p<0.05; Log2 (fold change) >2-fold. n=3. d.) Down-regulated genes (32) due to FH HDL-miRNA delivery with ≥ 3 predicted target sites of differential FH HDL-miRNAs. Left, negative fold changes; right, number of FH HDL-miRNA conserved target sites within mRNA 3′UTRs. e.) Down-regulated genes (60) due to FH HDL-miRNA delivery that are putative targets of FH HDL specific hsa-miR-105. Predicted targeting score (TargetScan) range (−0.89 top to 0.04 bottom). n=3.
FH HDL-miRNA delivery induced significant gene expression changes in human hepatocytes compared to normal HDL (Fig. 7c). High-level analysis of the whole-genome microarray data resolved 217 significant (correctedP<0.05) differentially (>2-fold) expressed genes altered by FH HDL treatment (Fig. 7c, Table S11). Most (87%) of the down-regulated genes (79/91) are putative targets of the 22 differentially abundant miRNAs present of FH HDL. Many (32) of the significantly (correctedP<0.05) down-regulated (>2-fold) mRNAs harbored 3 or more conserved target sites within 3′UTRs for differential FH HDL-miRNAs (Fig. 7d). Furthermore, 60% of the significantly down-regulated mRNAs were putative targets of the FH-specific hsa-miR-105 (Fig. 7e).
DISCUSSION
A key major finding of this study is that HDL transports endogenous miRNAs. Although the classic view of HDL is that it is a delivery vehicle for the return of excess cellular cholesterol to the liver for excretion, a wide variety of biological functions, outside of reverse cholesterol transport, have recently been ascribed to HDL32, 33. Furthermore, it is now recognized that HDL is much more complex in terms of differential lipids and proteins that it transports34. This study, therefore, has revealed yet another constituent transported by HDL, namely miRNAs, and a possible new mechanism whereby some of the biological effects of HDL could be mediated.
The exact process of how HDL is loaded with miRNAs and what proteins if any facilitate this association are not known; however, small RNAs (25nt) have previously been shown to complex with zwitterionic liposomes, specifically PC35. Previous biophysical studies suggest that HDL could simply bind to extracellular plasma miRNAs through divalent cation bridging9, 12, 14, 15, 35. In these studies, interactions between DNA molecules and zwitterionic PCs resulted in conformational shifts in PC head groups and subsequent altered orientations of aliphatic chains, thus facilitating the incorporation of the DNA molecules into the protected space12. In the case of HDL this could lead to a tighter association with miRNAs, and possibly shield bound miRNAs from external RNases8. Furthermore, Langmuir monolayer model systems have shown that nucleic acids easily penetrate lipid monolayers at low surface pressures12. Due to its high radius of curvature, HDL has a much lower surface tension than other lipoproteins, which is known to facilitate its binding to several of its other constituents36. Results presented here suggest that nSMase2 and most likely the ceramide pathway repress miRNA export to HDL. Overexpression of nSMase2 and activation of the ceramide pathway have previously been shown to induce exosome release from cells and trigger cellular export of miRNAs25, 37. These results suggest that the export of specific miRNAs through the exosomal pathway and the HDL pathway may be distinct mechanisms, possibly opposing, although both pathways are likely regulated by nSMase2 activity and ceramide synthesis.
One implication of this study is that HDL-miRNAs could potentially serve as novel diagnostic markers in much the same way that exosome miRNAs have been used38–42. HDL could simply be a depot or carrier for circulating miRNAs and their presence on HDL may not necessarily relate to the function of HDL in atherosclerosis or lipid metabolism. If so, miRNAs on HDL could perhaps be used diagnostically for a wide variety of diseases besides atherosclerosis. Nonetheless, many of the genes significantly altered in response to atherosclerotic HDL-miRNA delivery have a role lipid metabolism, inflammation, and atherosclerosis; including NDST143, NR1D244, 45, BMPR245, VEGFA, and FLT146.
Recently, apoA-I containing liposomes have been used to deliver siRNAs to the liver, with functional targeting and gene expression changes13, 47; however, the exact mechanism of siRNA transfer from apoA-I liposomes to hepatocytes was not determined. Furthermore, HDL loaded with cholesterol-conjugated siRNAs was shown to be 8-15X more effective in delivering siRNA-mediated silencing than cholesterol-conjugated siRNAs alone29. Similar to our observations in this study for HDL-miRNAs, the delivery of HDL-associated lipophilic-conjugated siRNAs was also determined to be SR-BI-dependent29. SR-BI-mediated transfer of HDL-miRNAs would likely prevent the delivery of miRNAs into the lysosomal pathway and instead divert it into the cytoplasm where it would be expected to be more stable with increased functional integrity and potential to alter gene expression. Overall these results suggest that artificial HDL delivery strategies that incorporate lipophilic-siRNA conjugates may likely depend on a naturally occurring lipoprotein-RNA delivery pathway.
One of the most abundant miRNAs that was found in both human and mouse HDL, and increased with atherosclerosis, was the highly conserved miR-22348. Expression of miR-223 in various diseases and its effect on biological processes has been extensively investigated6, 22, 40, 49–51. For example, miR-223 has been found to be involved in the regulation of glucose metabolism in cardiomyocytes, cell cycle auto-regulatory loops, ischemia-reperfusion injury, granulopoiesis, and osteoclast differentiation50, 52–55. Multiple targets of miR-223 have been experimentally validated, including RhoB26 and EFNA154. EFNA1 has been shown to promote hepatocellular carcinoma and is a marker for liver cancer27, 56. In our study, the delivery of miR-223 by HDL significantly reduced EFNA1 and RhoB mRNA levels in hepatocytes. Taken together, increases in specific intracellular miRNA levels, reduction of specific mRNA targets, and the direct repression of luciferase 3′UTR reporters establish that HDL can deliver miRNAs to cells and directly alter gene expression. Furthermore, our results suggest that the delivery of endogenous miRNAs are sufficient to directly target mRNAs and alter gene expression, although some of the observed gene changes from FH HDL treatment could also be related to protein or lipid compositional differences between normal and FH HDL preparations.
In summary, a possible new intercellular communication pathway, involving the transport and cellular delivery of miRNAs by HDL, has been described. This novel role for HDL potentially elevates lipoproteins to mediators of systemic gene expression apart from their role in cholesterol dynamics. The discovery of the HDL-mediated miRNA delivery pathway raises many intriguing possibilities for better understanding the pathogenesis of atherosclerosis and possible new treatment strategies, involving modifications to the miRNA content of HDL for altering gene expression.
METHODS
HDL Purification
Blood was collected from normal (n=20) and homozygous FH (n=7) patients at the NIH Clinical Center as approved by NIH NHLBI IRB. FH subjects were used as a human atherosclerosis model of vascular disease. HDL was isolated from human plasma using sequential density (KBr) gradient ultracentrifugation (DGUC), 330,000 × g, followed by extensive dialysis at 4°C in 50 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 2 mM CaCl2, pH 7.0. FPLC was used to purify HDL from DGUC (d=1.063–1.21 g ml−1) fractions. Superose 6 10–30 (Fraction 41 = 31.69 mLs) or Sephacryl 200 30–100 (GE Healthcare) columns were used with EDTA-free FPLC buffer (10 mM Tris, 150 mM NaCl, 0.02% sodium azide). Immuno-affinity purification of HDL was achieved as previously described57, using goat anti-human apoA-I antibodies (11A-G1, Academy BioMedical Company) or anti-mouse apoA-I (FL-267, Santa Cruz Biotechnology) conjugated to Sepharose-4B beads. Exosomes were isolated from human plasma by high speed ultracentrifugation, as previously described16, 58. Exosome preparations were depleted of lipoproteins by FPLC fractionation (>LDL). For FPLC detection of radiolabeled miRNA, 3′-phohsphates were removed (calf intestine phosphatase) from miR-125a-5p followed by 3′-radiolabeling with 32P-γ-ATP (2 mCi mL−1) by T4 polynucleotide kinase, 1h 37°C. Unincorporated radiolabel was removed by NAP-5 columns.
Transmission Electron Microscopy
HDL was dialyzed in 10 mM NH4HCO3 and adsorbed onto 300 mesh copper or nickel grids with formvar and carbon support films made hydrophilic by glow discharge. Adsorbed samples were negatively stained with 1% uranyl acetate and imaged with a JEM 1200EX electron microscope (JEOL USA) and AMT XR-60 digital camera (Advanced Microscopy Techniques). After absorption, Nanogold-conjugated miRNA (HDL complexes) were gold enhanced (2 min), as per manufacturer’s protocol. Prior to TEM, miRNA was labeled with 1.4 nm monoamino Nanogold (Nanoprobes) according to the manufacturer’s protocol. Briefly, miRNA was oxidized with NaIO4 (200 nmol) in solution (20 mM PIPES, pH 7 buffer) and quenched (glycerol, 5 min) oxidized products were collected (NAP-5 chromatography) and precipitated overnight, −20°C in 2.5 volumes of 100% ethanol, 1/15 volumes of 3M sodium acetate, pH 5.2 and 1 μl of 20 mg ml−1 glycogen. Precipitated miRNA was washed twice with cold 70% ethanol, dried, resuspended in water, and incubated with 300 μM monoamino Nanogold dissolved in DMSO. Nanogold-miRNA products were reduced with freshly prepared 20 mg ml−1 borane tert-butylamine, quenched with acetone, and separated from unconjugated-miRNA by FPLC.
Preparation of Reconstituted HDL
rHDL was prepared using cholate dialysis method with slight modifications59. Briefly, L-α-phosphatidylcholine (131401C, Avanti Polar Lipids) was evaporated under nitrogen, dried under vacuum, and resuspended with 1X PBS. Freshly prepared sodium cholate (30 mg ml−1) was added at 0.8 molar ratio (phospholipid to sodium cholate), vortexed, and incubated at 37°C for 15 min. Purified human apoA-I (>95% pure) was added to resuspension at 1:60 mol ratio (apoA-I to PC), incubated for 1h 37°C, and sodium cholate was removed by dialysis. Human rHDL particles were determined to be 13.8 −/+2.3 nm in diameter as measured by non-invasive dynamic back scattering (173°) (ZetasizerNanoZS, Malvern Instruments).
Animal Studies
Human rHDL (150 μg) was injected intravenously, retro-orbital site, into age-matched c57BL/6 (WT) or Apoe−/− mice at 16 weeks. Plasma and tissues were collected 6h post injection. Native HDL was isolated from plasma of WT and Ldlr−/− mice (16 weeks of age), as described. Mice were fed either a normal chow (NIH-31) or Western (42% calories from fat, Harlan Teklad) diet (28 days) prior to injection or sampling.
Gene Expression Studies
Total RNA was isolated from cells, HDL, and exosomes by Qiazol and miRNAeasy kits (Qiagen) and quantified by spectrophotometry (Nanodrop) and fluorometry (Qubit, Invitrogen). Small RNA was quantified by size specific (15–30nt) gel extraction and RiboQuant fluorescent RNA detection methods (Quant-It, Invitrogen). Total RNA and small RNA were assessed by Agilent 2100 Bioanalyzer using Total RNA Pico Kits and Small RNA Assay Kits, respectively (Agilent Technologies). Gene expression profiling was completed using Affymetrix GeneChip microarray assays according to protocols (Affymetrix). Briefly, 1 μg total RNA was reverse transcribed (T7-dT primers), amplified, and biotinylated (Affymetrix IVT Labeling Kit). Samples were hybridized to HG-U133 Plus 2.0 human whole genome microarrays (Affymetrix), washed, stained (Affymetix Fluidics Station) and scanned (Affymetrix GeneChip Scanner), as per instructions. Microarray results were validated by individual TaqMan gene assays. For real-time PCR, total RNA was reverse transcribed using random primers (mRNA) or specific miRNA primers and TaqMan Reverse Transcription or MicroRNA Reverse Transcription Kits (Applied Biosystems), as per instructions. Real-time PCR (7300 Real-Time PCR System, Applied Biosystems) and TaqMan probes (Applied Biosystems) were used to quantify mRNA and miRNA levels. Absolute quantification (miR-223 standard curve) or relative quantitative values (RQV) were used to determine mRNA and miRNA levels. RQV = 2−dCt. Peptidylprolyl isomerase A (PPIA) (mRNA) and RNU6 (miRNA) were used as housekeeping controls. RQVs are presented as fold changes to experimental baselines.
microRNA microarray
miRNA profiling was completed using TaqMan Array microRNA Assays (Applied Biosystems). Total RNA (≥20 ng) isolated from cell lysates, HDL, LDL, or exosomes was reverse transcribed using either Rodent or Human MegaPlex RT Primer Pools, and cDNA (5 μl) was amplified with Rodent or Human MegaPlex PreAmp Primer Pools and PreAmp Master Mix. Amplified samples were injected into TaqMan Array Rodent (v2.0) or Human (v2.0) MicroRNA Cards and analyzed using a 7900 Real Time PCR System (Applied Biosystems). Taqman MicroRNA Arrays enabled quantification of 667 (human) and 518 (mouse) unique miRNAs (Sanger miRBase v13.0)60. Cts were calculated by SDS 1.2 software (Applied Biosystems) and normalized to individual array mean Ct.
Proteomics
Total protein was isolated from Huh7 cells, HDL, and exosomes using extraction buffer (1X RIPA buffer, 1 mM Na3VO4, and cocktail protease inhibitors). Human apoA-I (MABTECH), HSP70 and ICAM-1 (MesoScale Diagnostics) were quantified using ELISA, as per instructions, and normalized to total protein (μg μL−1). Total protein was quantified by BCA Total Protein Assay Kit (BioRad). Western blotting consisted of 1-D SDS PAGE (4%–12% Bis-Tris gels) separation and semi-dry transfer to nitrocellulose (iBlot, Invitrogen). Primary antibodies (1:200, Santa Cruz) used included anti-human CD63 (MEM-259, Novus Biologicals), anti-human AGO2 (eLF2C H-300), anti-human GAPDH (D-6), and anti-human NPM1 (B23 H-106), followed by mouse or rabbit HRP-linked IgG1 secondary antibodies (1:2000, Santa Cruz) and 1:1 Western Lightning Plus–ECL Chemiluminescence detection. Tandem mass spectrometry (MS:MS) was performed with native HDL isolated by DGUC or DGUC followed by FPLC. HDL (40 μg) was separated by 1-D SDS-PAGE (4%–12% Bis-Tris gel) and underwent in-gel reduction and alkylation. Bands were trypsin digested and extracted peptides were cleaned with C18 monolithic tips (Protea Biosciences). Samples were analyzed on an Orbitrap Velos Mass Spectrometer. Cyclic top 10 precursor ions were selected for tandem mass spectrometry. Peptide IDs were identified with SwissProt (Swiss Bioinformatics Institute) database using MASCOT Search Engine (Matrix Sciences).
Cell Culture
Huh7, HEK293, J774, and BHK cells were cultured in F-12 DMEM, supplemented with streptomycin (100 μg mL−1), penicillin (100 U mL−1), and either 10% FBS or 10% LPDS (DGUC 1.21 bottom fraction), and maintained at 37°C with 5% CO2. J774 macrophages were treated with GW4869 (10 μM, Sigma), TO90131 (10 μM, Sigma), rHDL (10 μg mL−1) for 48h. For transfection, cells were plated at 1 × 105 cells mL−1 for 24h prior to transfection (48h) with DharmaFECT4 (Dharmacon). Transient transfections (100 nM) were conducted with siRNA (targeting pool) against SCARB1 (ON-TARGETplus SMARTpool L-010592-00), siRNA (targeting pool) against SIDT1 (ON-TARGETplus SMARTpool L-020674-01), miR-223 miRIDIAN mimic (C-300580-07), miR-375 miRIDIAN mimic (C-300682-05), and miRIDIAN miRNA mimic negative controls (C-001000-02).
Luciferase Reporters
The complete SR-BI 3′UTR (SCARB1 839bp, GeneTax ID 9606) was cloned into psiCHECK2-dual luciferase reporter construct (Promega) and confirmed by sequencing. BHK+SR-BI-pSwitch vector cells were transiently transfected with 100 ng of psiCHECK2-dual vector alone or psiCHECK2-SR-BI-3′UTR for 24h prior to HDL delivery. Renilla luciferase and Firefly luciferase activities (transfection control), were determined by Dual-Glo luciferase reagents (Promega), as per instructions. BHK cells were stably transfected with SR-BI pSwitch vector (GeneSwitch Technology, Invitrogen), and human SR-BI (SCARB1) gene was induced by mifepristone (10 nM), which served as GeneSwitch System activation element (Invitrogen). BHK-SR-BI inducible cells were also transfected with psiCHECK2-dual vectors and mifepristone (24h) prior (removed) to HDL or HDL-miR-223 (complex from HDL-IP) treatments (80 μg mL−1) in 10% LPDS media (48h).
Informatics
Putative miRNA targets were determined by TargetScan (targetscan.org) prediction software. Low-level and high-level post-analyses of mRNA and miRNA microarray data were performed using GeneSpring GX11.0 (Agilent Technologies). Graphical conceptualization of putative miRNA targets were illustrated by Cytoscape software (cytoscape.org)61. Affymetrix GeneChip signal values and percent calls (present/absent) were calculated by Affymetrix GCOS v1.4. Raw mRNA data were imported into GeneSpringGX11.0 as .cel files and summarizations were determined by LiWong method62. Data were quantile normalized with no baseline transformation, and filtered by expression (20–100)th percentile in raw data and by error (CV<50%). Statistical analyses were performed by unpaired t-tests, p-value computation was asymptotic, and Benjamini-Hochberg FDR was used for multiple-testing corrections. miRNA data files (RQV) were imported as generic single color experiments with custom technology and data were log2 transformed and analyzed using unpaired t-tests. List of significant (P<0.05) differentially (>2.0-fold) abundant miRNAs were used to cross-reference TargetScan database (50th context percentile) for conserved-only predicted mRNA targets (GeneSpring-TargetScanScript). All mRNA and miRNA microarray data have been submitted to NCBI Gene Expression Omnibus database (GSE25425).
Statistics
All results are expressed as means and n= derives from independent experiments. When comparing 2 groups either unpaired t-tests or Mann-Whitney non-parametric tests (two-tailed) were used as determined by normality of data. To determine if data sets come from normal distribution, Shapiro-Wilk W-tests were used (JMP9.0 Software). All correlations between profiles were determined by Spearman non-parametric analyses. For all tests a p-value ≤ 0.05 was considered significant, exact values are listed.
Supplementary Material
Acknowledgments
This study was helped by the NHLBI Electron Microscopy Core, Genomics Core, and Proteomics Core Facilities. Special thanks are extended to Praveen Sethupathy, Ph.D., for target prediction and discussion, Maureen Sampson for her work on patient lipid profiles, and Andrew Loncarich for statistical analysis. This research was supported in total by the Intramural Research Program of the NIH, NHLBI DIR.
Footnotes
AUTHOR CONTRIBUTIONS
K.C.V., B.T.P., and A.T.R designed the research plan and study. K.C.V., B.T.P., and B.M.S. performed all experiments. R.D.S. provided human samples. K.C.V and A.T.R drafted and edited manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
ACCESSION NUMBERS
SCARB1 NM_005505; EFNA1 NM_004428; RHOB NM_004040; SIDT1 NM_017699; PPIA NP_066953; Apoe NC_000073.5; Ldlr NC_000075.5; APOA1 NP_000030; ICAM1 NP_000192; HSP70 HSPA1A NP_005336; CD63 NP_001035123; AGO2 EIF2C2 NP_036286; NPM1 NP_002511; RNU6 NR_004394; hsa-miR-223 (MIR223) NR_029637 MIMAT0000280; hsa-miR-105 (MIR105-1) NR_029521 MIMAT0000102; hsa-miR-375 (MIR375) NR_029867 MIMAT0000728; hsa-miR-135a* (MIR135A1) NR_029677 MIMAT0004595; hsa-miR-188-5p (MIR188) NR_029708 MIMAT0000457; hsa-miR-877 (MIR877) NR_030615 MIMAT0004949; hsa-miR-106a (MIR106A) NR_029523 MIMAT0000103; hsa-miR-125a (MIR125A) NR_029523 MIMAT0000443; hsa-miR-24 (MIR24-1) NR_029496 MIMAT0000080; hsa-miR-211 (MIR211) NR_029624 MIMAT0000268; mmu-miR-223 (Mir223) NR_029801 MIMAT0000665
References
- 1.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 3.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006;20:1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- 5.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 6.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janas T, Janas T, Yarus M. Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res. 2006;34:2128–2136. doi: 10.1093/nar/gkl220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manavbasi Y, Suleymanoglu E. Nucleic acid-phospholipid recognition: Fourier transform infrared spectrometric characterization of ternary phospholipid-inorganic cation-DNA complex and its relevance to chemicopharmaceutical design of nanometric liposome based gene delivery formulations. Arch Pharm Res. 2007;30:1027–1040. doi: 10.1007/BF02993973. [DOI] [PubMed] [Google Scholar]
- 11.Suleymanoglu E. Phospholipid-nucleic acid recognition: developing an immobilized liposome chromatography for DNA separation and analysis. PDA J Pharm Sci Technol. 2006;60:232–239. [PubMed] [Google Scholar]
- 12.Gromelski S, Brezesinski G. DNA condensation and interaction with zwitterionic phospholipids mediated by divalent cations. Langmuir. 2006;22:6293–6301. doi: 10.1021/la0531796. [DOI] [PubMed] [Google Scholar]
- 13.Kim SI, et al. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Mol Ther. 2007;15:1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- 14.McManus JJ, Radler JO, Dawson KA. Does Calcium Turn a Zwitterionic Lipid Cationic? The Journal of Physical Chemistry B. 2003;107:9869–9875. [Google Scholar]
- 15.Mengistu DH, Bohinc K, May S. Binding of DNA to zwitterionic lipid layers mediated by divalent cations. J Phys Chem B. 2009;113:12277–12282. doi: 10.1021/jp904986j. [DOI] [PubMed] [Google Scholar]
- 16.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 17.Lima ES, Maranhao RC. Rapid, Simple Laser-Light-Scattering Method for HDL Particle Sizing in Whole Plasma. Clin Chem. 2004;50:1086–1088. doi: 10.1373/clinchem.2004.032383. [DOI] [PubMed] [Google Scholar]
- 18.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 19.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 20.Conde-Vancells J, et al. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res. 2008;7:5157–5166. doi: 10.1021/pr8004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 22.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 23.Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund-Katz S, Phillips MC. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell Biochem. 2010;51:183–227. doi: 10.1007/978-90-481-8622-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun G, Li H, Rossi JJ. Sequence context outside the target region influences the effectiveness of miR-223 target sites in the RhoB 3′UTR. Nucleic Acids Res. 2010;38:239–252. doi: 10.1093/nar/gkp870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui XD, et al. EFNA1 ligand and its receptor EphA2: potential biomarkers for hepatocellular carcinoma. Int J Cancer. 2010;126:940–949. doi: 10.1002/ijc.24798. [DOI] [PubMed] [Google Scholar]
- 28.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 29.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podrez EA. Antioxidant Properties of High Density Lipoprotein and Atherosclerosis. Clin Exp Pharmacol Physiol. 2010;37:719–725. doi: 10.1111/j.1440-1681.2010.05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinecke JW. The HDL proteome: a marker--and perhaps mediator--of coronary artery disease. J Lipid Res. 2009;50 (Suppl):S167–171. doi: 10.1194/jlr.R800097-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothblat GH, Phillips MC. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr Opin Lipidol. 2010;21:229–238. doi: 10.1097/mol.0b013e328338472d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu D, Rhodes DG. Binding of phosphorothioate oligonucleotides to zwitterionic liposomes. Biochim Biophys Acta. 2002;1563:45–52. doi: 10.1016/s0005-2736(02)00384-x. [DOI] [PubMed] [Google Scholar]
- 36.Qiu X, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 37.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 38.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 39.Wang GK, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 40.Wang JF, et al. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 41.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Kerin MJ. MicroRNAs as Novel Biomarkers for Breast Cancer. J Oncol. 2009;2009:950201. doi: 10.1155/2010/950201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilad S, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacArthur JM, et al. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. J Clin Invest. 2007;117:153–164. doi: 10.1172/JCI29154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishnan SN, Lau P, Burke LJ, Muscat GE. Rev-erbbeta regulates the expression of genes involved in lipid absorption in skeletal muscle cells: evidence for cross-talk between orphan nuclear receptors and myokines. J Biol Chem. 2005;280:8651–8659. doi: 10.1074/jbc.M413949200. [DOI] [PubMed] [Google Scholar]
- 45.Yao Y, et al. High-density lipoproteins affect endothelial BMP-signaling by modulating expression of the activin-like kinase receptor 1 and 2. Arterioscler Thromb Vasc Biol. 2008;28:2266–2274. doi: 10.1161/ATVBAHA.108.176958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreno PR, Purushothaman KR, Sirol M, Levy AP, Fuster V. Neovascularization in human atherosclerosis. Circulation. 2006;113:2245–2252. doi: 10.1161/CIRCULATIONAHA.105.578955. [DOI] [PubMed] [Google Scholar]
- 47.Lee H, et al. Hepatic siRNA delivery using recombinant human apolipoprotein AI in mice. Biochem Biophys Res Commun. 2009;378:192–196. doi: 10.1016/j.bbrc.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 48.Fukao T, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 49.Gentner B, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 50.Eyholzer M, et al. Complexity of miR-223 regulation by CEBPA in human AML. Leuk Res. 2010;34:672–676. doi: 10.1016/j.leukres.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Fazi F, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 52.Pulikkan JA, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res. 2010;86:410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 54.Yu CH, Xu CF, Li YM. Association of MicroRNA-223 expression with hepatic ischemia/reperfusion injury in mice. Dig Dis Sci. 2009;54:2362–2366. doi: 10.1007/s10620-008-0629-8. [DOI] [PubMed] [Google Scholar]
- 55.Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. 2007;101:996–999. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 56.Iida H, et al. Ephrin-A1 expression contributes to the malignant characteristics of {alpha}-fetoprotein producing hepatocellular carcinoma. Gut. 2005;54:843–851. doi: 10.1136/gut.2004.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang L, et al. Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5:3314–3328. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 58.Nieuwland R, et al. Cellular origin and procoagulant properties of microparticles in meningococcal sepsis. Blood. 2000;95:930–935. [PubMed] [Google Scholar]
- 59.Matz CE, Jonas A. Micellar complexes of human apolipoprotein A-I with phosphatidylcholines and cholesterol prepared from cholate-lipid dispersions. J Biol Chem. 1982;257:4535–4540. [PubMed] [Google Scholar]
- 60.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.