Abstract
The ATPase activity of chloroplast and bacterial F1-ATPase is strongly inhibited by both the endogenous inhibitor ϵ and tightly bound ADP. Although the physiological significance of these inhibitory mechanisms is not very well known for the membrane-bound F0F1, these are very likely to be important in avoiding the futile ATP hydrolysis reaction and ensuring efficient ATP synthesis in vivo. In a previous study using the α3β3γ complex of F1 obtained from the thermophilic cyanobacteria, Thermosynechococcus elongatus BP-1, we succeeded in determining the discrete stop position, ∼80° forward from the pause position for ATP binding, caused by ϵ-induced inhibition (ϵ-inhibition) during γ rotation (Konno, H., Murakami-Fuse, T., Fujii, F., Koyama, F., Ueoka-Nakanishi, H., Pack, C. G., Kinjo, M., and Hisabori, T. (2006) EMBO J. 25, 4596–4604). Because γ in ADP-inhibited F1 also pauses at the same position, ADP-induced inhibition (ADP-inhibition) was assumed to be linked to ϵ-inhibition. However, ADP-inhibition and ϵ-inhibition should be independent phenomena from each other because the ATPase core complex, α3β3γ, also lapses into the ADP-inhibition state. By way of thorough biophysical and biochemical analyses, we determined that the ϵ subunit inhibition mechanism does not directly correlate with ADP-inhibition. We suggest here that the cyanobacterial ATP synthase ϵ subunit carries out an important regulatory role in acting as an independent “braking system” for the physiologically unfavorable ATP hydrolysis reaction.
Keywords: ATP Synthase, ATPases, Energetics, Enzyme Inactivation, F1F0-ATPase, ϵ-Inhibition, ADP-Inhibition
Introduction
F0F1-ATP synthase synthesizes ATP from ADP and inorganic phosphate using the proton motive force (pmf),2 which is generated across the cytoplasmic membranes of bacteria, thylakoid membranes of chloroplasts, and inner membranes of mitochondria by respiratory or photosynthetic electron transport systems (1–3). The complex consists of the membrane-embedded portion F0 and the water-soluble portion F1. F0, the proton translocation device, is composed of three different subunits, a, b, and c with a stoichiometry of a1b2c10–15 (1, 4–6). F1 is composed of five different subunits designated α to ϵ with a stoichiometry of α3β3γ1δ1ϵ1 (7). The minimum catalytic core as F1-ATPase is α3β3γ (8–10). As a prevailing enzyme catalysis mechanism, the rotary catalysis mechanism was first proposed by Boyer and co-workers (11). Following the x-ray crystal structure analysis of mitochondrial F1 (12), the rotation of the γ subunit during ATP hydrolysis was studied by the biochemical cross-linking experiment (13), the polarized spectroscopic technique (14). Finally, rotation of the γ subunit was visualized at a single-molecule level using a microprobe attached to the rotating γ subunit (15). Thorough analysis of rotation of the γ subunit revealed that the γ subunit rotates with a discrete 120° step per single-molecule ATP consumption and that this 120° step consists of further 80° and 40° substeps (16, 17). The 80° substep rotation of γ occurs by ATP binding, whereas ATP cleavage and the release of one of its products, phosphate, result in a subsequent 40° substep (17, 18).
Activity of the ATP synthase is highly regulated in vivo and in vitro. Among them, ADP-inhibition of the ATPase activity is a common regulatory mechanism that has been found to occur in all ATP synthase examined so far. The ATP hydrolysis reaction of F1-ATPase is strongly inhibited by tight binding of ADP-Mg to the catalytic site(s) (9, 19–21). Single-molecule analysis showed that F1 often lapses into an inactive ADP-induced inhibition (ADP-inhibition) state during rotation, and the inhibition has been assigned to the pause of rotation at 80° (22).
The intrinsic inhibitory function of ϵ, which is well characterized in bacteria and chloroplasts, is known as further regulation of the ATP hydrolysis reaction (23–25). The C-terminal α-helical domain of the ϵ subunit has been suggested to be important in conferring its inhibitory function (26, 27) and can adopt two distinct conformations: the extended conformation and the retracted conformation (28). In the extended conformation, the ϵ subunit inhibits the ATPase activity of F0F1 but has no significant effect on ATP synthesis (28, 29). Transition from the retracted conformation to the extended conformation is thought to act as an intrinsic regulation system (29, 30).
The significance of the C-terminal domain of the ϵ subunit on the inhibition is especially remarkable in the case of CF1 obtained from spinach thylakoids. When the effect of the structural mutant ϵ subunit containing a deleted C-terminal domain (ϵΔC) was examined, the ATPase activity of CF0CF1 was found to be 6-fold higher than that of the enzyme inhibited by the wild-type ϵ (31), whereas the ATPase activity of EF0EF1 in the presence of ϵΔC was only 1∼2-fold higher compared with EF0EF1 with the wild-type ϵ subunit (32).
In a previous study, we showed that the pausing angular position of γ in ϵ-inhibition is identical to that observed for ADP-inhibition, but distinctly different from the waiting position for ATP binding (33). Based on this result, we suggested that ϵ-inhibition is closely related to ADP-inhibition. Feniouk et al. also reported that the ADP-inhibition of TF0TF1 is enhanced by the ϵ subunit (34) and vice versa, inhibitory ADP-induced ϵ-inhibition in α3β3γ from Bacillus PS3 was confirmed by the single-molecule experiment (35). However, at their origin, ADP-inhibition and ϵ-inhibition must be independent phenomena from each other because even the α3β3γ subcomplex lapses into the ADP-inhibition state without the ϵ subunit. Hence, the relationship between these two inhibition systems is still unclear. In this study, we aimed to determine whether ϵ-inhibition can act independently of ADP-inhibition or whether ϵ-inhibition requires ADP-inhibition. The kinetic difference of the inhibited enzymes derived by these inhibition mechanisms is critical in understanding the regulation of the rotary motor enzyme when physiological conditions fluctuate.
EXPERIMENTAL PROCEDURES
Materials
Biotin-PEAC5-maleimide was purchased from Dojin (Kumamoto, Japan). ATP, phosphoenolpyruvate, and BSA were obtained from Sigma. Pyruvate kinase, lactate dehydrogenase, and NADH were purchased from Roche Diagnostics. Other chemicals were of the highest grade commercially available.
Preparation of Proteins
In the previous single-molecule study of the cyanobacterial F1, we used a mutant, α3β3γrot, for the rotation study, which consisted of αC144S/C194S, His10-βC53S, and γC90S/G112C/A125C (33). In this study, we prepared a new mutant complex, α3β3γArot using His10-αC144A/C194A, His10-βC53A, and γC90A/G112C/A125C. In addition, a His10 tag was introduced onto α to strengthen the binding of the complex to the glass plate during the single-molecule experiments, and the cysteines on the original complex were substituted into alanine, conferring higher structural stability to the complex compared with α3β3γrot. The stability of the complex was confirmed by gel filtration as described elsewhere. The higher structural stability of α3β3γArot compared with α3β3γrot is mainly due to His10 onto α (supplemental Fig. S1). All cysteines on the complex were substituted with alanine by the Mega-primer method (36) using the following mutation primers: 5′-CCCCTGCACCTGGTATTGTCCAGCGCAAATCTGTGGCGGAGCCATTGCAAACGGG-3′ for αC144A; 5′-CAATCTTGAACCAAAAGGGCCAAGACGTGATTGCGGTGTATGTGGCCATTGGTCAAAAAGCC-3′ for αC194A; 5′-CGCGGCCGGCTTAGACGTGGCTGTAACCGCGGAAGTGCAACAACTCCTTGGCG-3′ for βC53A; and 5′-GGCGCTGCTGGTGGTAACAGGCGATCGCGGGCTGGCGGGCGGTTACAACACTAATGTCATTCGCC-3′ for γC90A. His10 was introduced to the N terminus of α using the following primer: 5′-CGCGCCCATGGAATCTAAGAAGGAGATATACATATGCATCACCATCATCACCATCACCATCACCATATGGTAAGTATCCGACCCG-3′. The subcomplexes α3β3γArot and the ϵ subunit were expressed and purified as described (33) and stored at −80 °C. The yield of the α3β3γArot complex was 3.0 mg/g cells, which was much higher than that of the former complex α3β3γrot (0.3 mg/g cells). This higher yield should be accomplished by alanine mutation and the additional histidine tag on the α subunit. The ATPase activity and the sensitivity for ϵ-inhibition of the α3β3γArot were almost the same as that of α3β3γrot, but demonstrated slightly higher sensitivity to lauryl dimethylamine oxide (LDAO) (supplemental Table S1).
Restoration of the Rotation of Inhibited Complex by Magnetic Tweezers
Magnetic tweezers were constructed and manipulated as described (37). The magnetic field was measured with a gaussmeter (421 Gaussmeter; Lake Shore), and the measuring magnetic field 10 mm above the sample was about 200 G. The rotation assay was carried out as described (33) with slight modifications. Rotation of the attached 250-nm diameter duplex magnetic beads (nanomag-D streptavidin; Micromod) on the γ subunit was monitored with a conventional optical microscope type IX71 (Olympus, Tokyo, Japan) with a 100× objective lens, and the images were recorded by digital video recorder. Recorded images were analyzed by custom software, Trans viewer, prepared by Yusung Kim.
Measurement of ATPase Activity
ATPase activity was measured in the presence of an ATP-regenerating system (38) in 50 mm HEPES-KOH, pH 8.0, 100 mm KCl, 2.5 mm MgCl2, 100 μm ATP, 50 μg/ml pyruvate kinase, 50 μg/ml lactate dehydrogenase, 2 mm phosphoenolpyruvate, and 0.2 mm NADH. The assay was carried out at 25 °C. The rate of ATP hydrolysis after addition of the enzyme was determined by monitoring the decrease in NADH absorption at 340 nm using a spectrophotometer.
Apparent Binding Affinity of the ϵ Subunit to the Complex
The proportion of the complex-bound ϵ subunit was estimated from the extent of the inhibition of the ATPase activity of the α3β3γ complex. ATP hydrolysis was initiated by addition of the complex to the reaction mixture, monitored for 4.5 min, and various concentrations of the ϵ subunit added. The final concentration of the α3β3γ complex in the reaction mixture was fixed at 1 nm. The rate of ATP hydrolysis in the steady state was determined from 350 to 400 s after addition of the ϵ subunit. The observed Kd value of the ϵ subunit for the α3β3γ complex was calculated from the inhibition curve as described (33).
Estimation of the Stiffness of the α3β3γ Complex
Fluctuation of the γ subunit was monitored as fluctuation of 340-nm diameter duplex streptavidin-coated beads attached to the γ subunit using a dark-field microscope (IX 70; Olympus, Tokyo, Japan) equipped with a mercury lamp and a high speed camera (Hi-DcamII; NAC Image Technology, Tokyo) at 500 frames/s. Recorded images were analyzed using our original software as described above. Torsional stiffness of the γ subunit was calculated from the rotary fluctuations as described (39).
Removal of the Bound Nucleotides on the α3β3γArot and α3β3γArotϵ Complexes
Removal of the bound nucleotides on the α3β3γArot and α3β3γArotϵ complexes was carried out as described (40), and the amount of bound nucleotides was determined by the method reported (41). The stability of the complexes was then analyzed by gel filtration chromatography. To investigate structural recovery of α3β3γArot, 0.3 mg of the nucleotide-depleted α3β3γArot complex was incubated with 2 mm ADP or with the wild-type ϵ subunit (10 times molar excess of the α3β3γArot complex) or with the C-terminal α-helices truncated ϵ subunit at room temperature for 2 h. The incubated complex was then loaded onto the Superdex 200 HPLC column (GE Healthcare) equilibrated with 50 mm HEPES-KOH, pH 8.0, and 100 mm KCl. The elution was monitored by absorbance at 280 nm.
RESULTS
Observation of the Restoration of Rotation of F1-ATPase in ADP-inhibition and ϵ-inhibition with Magnetic Tweezers
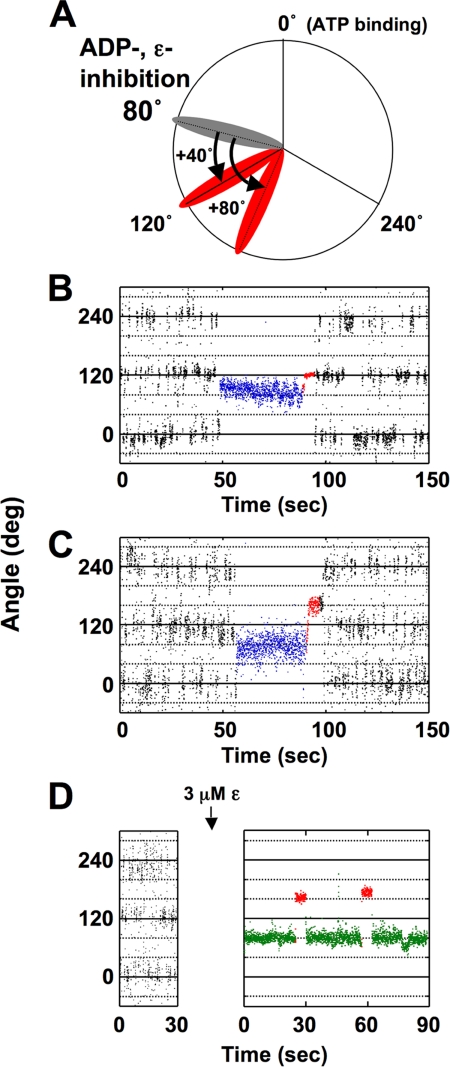
The restoration of rotation of the ADP-inhibited TF1 by pushing the γ subunit to the direction of ATP hydrolysis with magnetic tweezers has already been reported (37), and this method appeared useful for studying the relevance between ADP-inhibition and ϵ-inhibition. By using the newly prepared α3β3γArot complex, we first investigated the frequency of restoration of rotation of ADP-inhibited F1 (Fig. 1, A–C). The γ subunit of ADP-inhibited α3β3γArot was forced at arbitrary angles from the position of the ADP-inhibition (80°) for 5 s (Fig. 1A) and then released from the tweezers. When rotation of F1-ATPase was not restored by forcing, the beads attached to the γ subunit returned to the original position of ADP-inhibition. When rotation of α3β3γArot was observed at 250 nm ATP, discrete 120° steps were detected as an oscillating trace (Fig. 1B). In addition, the α3β3γArot complex spontaneously lapsed into the ADP-inhibited form at 80° forward from that for ATP binding. We then investigated the frequency of restoration of rotation of the ADP-inhibited α3β3γArot by 40° forcing (Fig. 1B) and 80° forcing (Fig. 1C), respectively. The frequencies of restoration of γ rotation were 64% for 40° forcing, and 86% for 80° forcing (Table 1). Frequencies of the restoration of rotation at these forcing angles were almost same as those observed for TF1 (37). We then examined the restoration of rotation of the ϵ-inhibited F1 by 80° forcing. Because the frequency of the restoration of rotation of the ADP-inhibited F1 by 80° forcing was high (86%), we expected that the restoration of rotation of the ϵ-inhibited F1 must be observed by the same 80° forcing, if ϵ-inhibition requires ADP-inhibition. Rotation of α3β3γArot was observed for 30 s at 250 nm ATP, and the buffer containing 3 μm ϵ subunit was infused into the objective chamber. As observed in our previous experiment, rotation of the γ subunit completely stopped at 80° forward from the angular position for ATP binding (Fig. 1D) by infusion of the ϵ subunit. However, no restoration of rotation was observed by 80° forcing in ϵ-inhibition, which was very different from that of ADP-inhibition. The results obtained for the restoration of γ rotation with magnetic tweezers are summarized in Table 1. These results strongly suggest that ϵ-inhibition does not require ADP-inhibition.
FIGURE 1.
Manipulation of rotation of the γ subunit in the ADP-inhibited and the ϵ-inhibited α3β3γArot complex with magnetic tweezers. A, scheme of mechanical manipulation of the γ subunit with magnetic tweezers. During the observation of rotation, α3β3γArot spontaneously fell into ADP-inhibition. When the ATP binding position prior to ADP-inhibition was settled as 0°, the position of ADP-inhibition was 80° (gray). The bead attached to the γ subunit of this α3β3γArot complex was then forced to 40° or 80° from the angle for the ADP-inhibition with magnetic tweezers, and the position of the beads reached to 120° or 160° (red; we designated this manipulation as 40° or 80° forcing). B, time course of the restoration of rotation of the ADP-inhibited α3β3γArot complex by 40° forcing with magnetic tweezers. The γ subunit in the α3β3γArot complex rotated with 120° steps at 250 nm ATP, which can be seen as an oscillating trace. During the observation of rotation, α3β3γArot spontaneously fell into ADP-inhibition (blue colored traces). Red traces show the manipulation time (5 s) using the magnetic tweezers. C, time course of the restoration of the rotation of the ADP-inhibited α3β3γArot complex by 80° forcing with magnetic tweezers. ADP-inhibition (blue traces) was forced by using the magnetic tweezers (red traces). D, time course of the 80° forcing of the ϵ-inhibited α3β3γArot with magnetic tweezers. The ϵ-inhibition (green traces) was forced by using the magnetic tweezers (red traces).
TABLE 1.
Frequency of mechanical restoration of the ADP-inhibited and ϵ-inhibited α3β3γ complex
The α3β3γ complex in ADP-inhibition or ϵ-inhibition was stalled at the angle indicated from its inhibition with magnetic tweezers. The number of the particles that showed rotation at each of the stall angles was counted and is shown as “Activated.” Those that did not show rotation were defined as “Not activated.” ND, not detected.
| Stall angle | Activated | Not activated | Frequency of activation |
|---|---|---|---|
| % | |||
| 40°(ADP-inhibition) | 9 | 5 | 64 (n = 14) |
| 80°(ADP-inhibition) | 12 | 2 | 86 (n = 14) |
| 80°(ϵ-inhibition) | ND | 10 | ND (n = 10) |
Effect of LDAO on ϵ-Inhibition
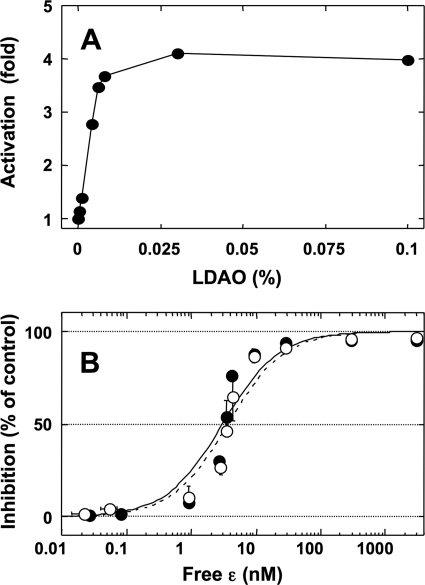
LDAO has been reported to stimulate the ATP hydrolysis activity of F1 or F0F1 (42), and the effect is thought to promote the release from ADP-inhibition (43). In addition, several biochemical studies on EF1 or EF0EF1 showed that the restoration from ADP-inhibition by LDAO obviously decrease the extent of ϵ-inhibition (42, 44, 45) without dissociation of the ϵ subunit (46), and therefore these two inhibitions were regarded as closely linked systems. To confirm the relationship between ϵ-inhibition and ADP-inhibition on cyanobacterial F1, we investigated the effect of LDAO on ϵ-inhibition of α3β3γArot. At first, the extent of stimulation of ATPase activity of α3β3γArot by LDAO was measured using the ATP-regenerating system at 100 μm ATP. LDAO stimulated the ATPase activity of α3β3γArot up to 4-fold, and the stimulation effect was saturated at 0.1% LDAO (Fig. 2A). Next, the effect of LDAO on ϵ-inhibition was investigated by measuring the change of the apparent Kd values for the interaction between ϵ and α3β3γArot. The apparent Kd values were determined from the extent of inhibition of ATPase activity of the complex at various concentrations of the ϵ subunit in the presence or the absence of LDAO (33). If ϵ-inhibition requires ADP-inhibition, release from ADP-inhibition by LDAO must increase the apparent Kd of the ϵ subunit to the α3β3γArot. The apparent Kd values of the ϵ subunit obtained for α3β3γArot in the presence and absence of LDAO were, however, 3.0 ± 0.3 nm and 3.7 ± 0.4 nm, respectively (Fig. 2B). In contrast to the results from EF1 α3β3γ, LDAO did not affect ϵ-inhibition in cyanobacterial α3β3γ. However, as the theoretical curve obtained from these Kd values did not fit to the experimental data very well (Fig. 2B), there may be a further undefined relationship between the ATPase activity and ϵ binding. Because the data obtained in the presence and in the absence of LDAO showed a similar tendency, these apparent discrepancies must be not due to ADP-inhibition or LDAO effects. Again, the result in this study indicates that ϵ-inhibition does not require ADP-inhibition.
FIGURE 2.
Effect of LDAO on the inhibition of ATPase activity of the α3β3γArot complex by the ϵ subunit. A, ATPase activity of α3β3γArot was measured in the presence of various concentrations of LDAO at 25 °C. The measurement was initiated by the addition of 1 nm α3β3γArot to the reaction mixture containing 100 μm ATP. Steady-state activities were then determined from the slope from 350 to 380 s after addition of the enzyme, and the results obtained from two independent experiments were averaged. The ATP hydrolysis activity in the absence of LDAO was 3.6 units/mg for α3β3γArot. B, extent of inhibition of ATP hydrolysis activities of α3β3γArot by the ϵ subunit in the presence (filled circles) and the absence (open circles) of LDAO measured at various concentrations of the ϵ subunit. The concentration of LDAO used was 0.1%. ATP hydrolysis was initiated by the addition of the complex to the reaction mixture and monitored for 4.5 min, and various concentrations of the ϵ subunit were then added. Steady-state activities were determined from the slope from 400 to 450 s following ϵ addition. The percentages of inhibition of ATPase activity of α3β3γArot by the ϵ subunit were plotted against the concentration of free ϵ subunit. The results of three independent experiments were averaged. The vertical and horizontal error bars in the figure indicate the S.D. for the extent of ϵ-inhibition and the concentration of free ϵ, respectively. The titration curves in the presence (solid line) and the absence (dotted line) of LDAO were fitted with the hyperbolic equation, y = ((A × ([ϵ]free)/(Kd + [ϵ]free)), where y represents the percentage of inhibition, A is the maximum inhibition (%), and Kd is the equilibrium dissociation constant for the ϵ subunit.
Torsional Stiffness of α3β3γ in ϵ-Inhibition Compared with That of ADP-Inhibition
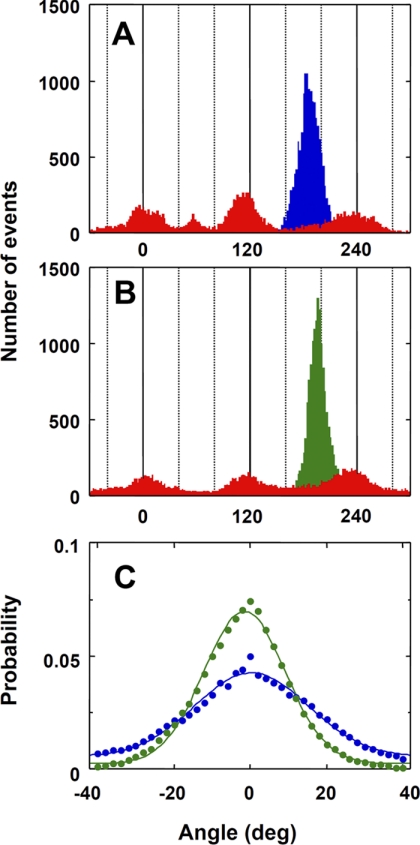
Because the apparent Kd value of the ϵ subunit to α3β3γArot was not affected very much by LDAO, it seems likely that ADP-inhibition and ϵ-inhibition are independent phenomena of each other. To explore further the relationship between them, we compared the torsional stiffness of α3β3γArot in ϵ-inhibition with that of ADP-inhibition by the single-molecule analysis. Torsional stiffness of α3β3γArot in ϵ-inhibition and ADP-inhibition was estimated by measuring thermal fluctuations of the beads attached to the α3β3γArot complex (Fig. 3, A and B) as described (39). The fluctuations of the beads were observed for 30 s with 2-ms time resolution. The probability distributions of fluctuation of the beads in ϵ-inhibition and ADP-inhibition were then fitted using a single Gaussian function (Fig. 3C). We found that the resulting distribution for ADP-inhibition was broader than that for ϵ-inhibition, and the standard deviations, σ, were 14.8 for ADP-inhibition and 10.6 for ϵ-inhibition, respectively. The torsional stiffness of α3β3γArot was calculated using variance, σ2, from the Gaussian distribution, as described in the legend to Fig. 3, and the determined stiffnesses, κ, were 119.6 pNnm for ϵ-inhibition and 61.4 pNnm for ADP-inhibition, respectively. The results clearly indicate that the ϵ subunit physically stiffens α3β3γ complex more strictly than ADP-inhibition.
FIGURE 3.
Stiffness of the F1 in ADP-inhibition and ϵ-inhibition. A and B, histograms of angular position of the 340-nm duplex beads at 250 nm ATP (red), in ADP-inhibition (blue in A) or in ϵ-inhibition (green in B). Both the stop angular positions of the γ subunit inhibited at ADP-inhibited form and by the ϵ subunit were 80° forward from that for ATP binding. The fluctuations of the beads in ADP-inhibition or ϵ-inhibition were observed for 30 s with a 2-ms time resolution. C, probability distribution of the fluctuation of the beads in ADP-inhibition (blue) and ϵ-inhibition (green) fitted using a single Gaussian function. The stiffness of the γ subunit, κ, was calculated from the equation: σ2 = kBTκ−1, where σ2 is a variance from Gaussian distribution as described (39).
Effect of ADP and the ϵ Subunit on the Structural Stability of Bound Nucleotide-depleted α3β3γ Complex
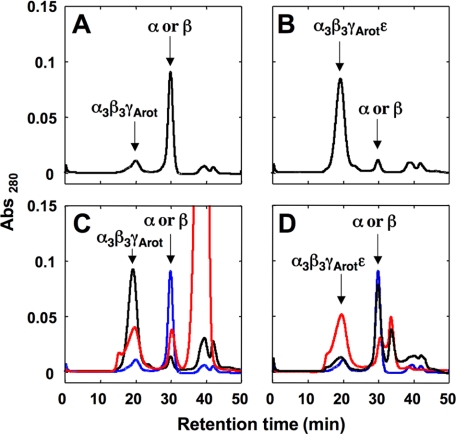
To examine how the ϵ subunit affects the nucleotide binding at the catalytic or noncatalytic sites, we depleted the bound nucleotides from α3β3γArot and α3β3γArotϵ as described (40). As shown in supplemental Fig. S2, A and B, most of the bound nucleotides on α3β3γArot and α3β3γArotϵ were removed from the complex by this procedure. Stabilities of these nucleotide-depleted α3β3γArot and α3β3γArotϵ complexes were then analyzed as described in the legend of Fig. 4. Although depletion of bound nucleotides from CF1 without the ϵ subunit did not result in structural instability of the complex (40), the cyanobacterial α3β3γArot complex was highly unstable when the bound nucleotides were depleted (Fig. 4A). In contrast, the stability of α3β3γArotϵ was in no way affected by the amount of bound nucleotides (Fig. 4B). These unexpected phenomena allowed us to investigate the effect of the ϵ subunit on the structural stability of the complex, which might be useful in differentiating between the impact of ϵ binding and nucleotide binding on stability of the complex structure. The nucleotide-depleted α3β3γArot complex was incubated in the presence and absence of 2 mm ADP for 2 h at room temperature, and the stability of the complex was analyzed as described. The structural stability of the nucleotide-depleted α3β3γArot was restored by incubation with ADP (Fig. 4C). Next, the nucleotide-depleted α3β3γArot complex was incubated with the ϵ subunit or the mutant ϵ subunit lacking the C-terminal α-helical part, rendering it unable to inhibit ATPase activity (26, 27, 31). Recovery of the structural stability of the nucleotide-depleted α3β3γArot complex by addition of the ϵ subunit was only observed when the full-length ϵ subunit was used (Fig. 4D), indicating that the C-terminal α-helical part of the ϵ subunit is important for both the inhibition of ATPase activity of α3β3γArot and the structural stability of the complex. The structural stability of α3β3γwt was also measured (supplemental Fig. S3). The result for α3β3γwt was not much different from that for α3β3γArot, suggesting that the effect of the C-terminal α-helical part of the ϵ subunit on the stability of α3β3γArot is not an experimental artifact of the mutation.
FIGURE 4.
Effect of ADP and the ϵ subunit on the stability of the bound nucleotide-depleted α3β3γArot complex. A and B, effects of depletion of the bound nucleotides on the stability of the α3β3γArot (A) and the α3β3γArotϵ (B) complexes were investigated as described under “Experimental Procedures.” C, 0.3 mg of the nucleotide-depleted α3β3γArot complex was incubated without (blue) or with (red) 2 mm ADP at room temperature for 2 h, and the incubated complexes were loaded onto the Superdex 200 HPLC column. Black shows the case of α3β3γArot without nucleotide depletion. D, effect of the ϵ subunit on the stability of the nucleotide-depleted α3β3γArot complex was also investigated. 0.3 mg of the nucleotide-depleted α3β3γArot complex was incubated without ϵ (blue), with wild-type ϵ (10 × mol of the α3β3γArot complex, red) or with C-terminal α-helices truncated ϵ (10 × mol of the α3β3γArot complex, black) at room temperature for 2 h, and the stability of α3β3γArot was investigated described above.
DISCUSSION
No restoration of rotation was observed by 80° forcing in ϵ-inhibition, which was very different from ADP-inhibition. Therefore, ϵ-inhibition may not require ADP-inhibition to inhibit the ATPase activity of F1. The frequency of restoration of rotation of the ADP-inhibited TF1 increases with the stall angle from the position of ADP-inhibition, which was interpreted as a decrease in the binding affinity to the inhibitory ADP caused by the forced movement of the γ subunit (37). If the forced conformational change and the decrease in the binding affinity for ADP occur also in our cyanobacterial F1 complex, there is a possibility that the ϵ subunit increases binding affinity of ADP to F1. In this case, ADP may not be released by 80° stalling or immediately rebind to the F1 after release caused by the forced rotation, and no restoration of the rotation must be observed in the presence of the ϵ subunit. Consistent with this assumption, an increase of the binding affinity of Mg-ATP and Mg-ADP to the high affinity catalytic site(s) of α3β3γ of EF1 caused by the addition of the ϵ subunit has already been reported (47). In contrast, the ϵ subunit weakened the binding affinity of Mg-ADP to each catalytic site, especially to the high affinity site(s) in the case of the α3β3γ complex of TF1 (48). Hence, there must be a variety of ϵ effects on the nucleotide binding affinity based on the difference of the origin of the ϵ subunits (47, 48).
In this study, the difference in nucleotide binding affinity both on α3β3γ and α3β3γϵ could not be precisely determined because the nucleotide-depleted α3β3γArot complex of cyanobacteria was obviously unstable (Fig. 4A). When mechanical manipulation of the γ subunit in the ϵ-inhibited α3β3γ complex was investigated in the presence of the ATP-regenerating system in our experiments shown in Fig. 1D, inhibitory ADP released from the complex was likely immediately converted to ATP. Rebinding of ADP to α3β3γϵ is therefore highly unlikely. This suggests that binding of the ϵ subunit to the complex only affects the off-rate of the inhibitory ADP, if the ϵ subunit increases binding affinity of ADP to the α3β3γ complex of the cyanobacteria. As mentioned, the restoration from ADP-inhibition by LDAO an obvious decrease in the extent of ϵ-inhibition (42, 44, 45) without dissociation of the ϵ subunit (46) was reported, and these findings might be good evidence that these two inhibitions are closely linked to each other. In contrast, LDAO-assisted restoration from ADP-inhibition had no effect on ϵ-inhibition of the ATPase activity of cyanobacterial F1 (Fig. 2B), suggesting that these two inhibition mechanisms are not directly related. The γ subunit of the cyanobacterial F1, chloroplast one too, contains the inserted region (25∼40 amino acids) that is absent from bacteria and mitochondria, and the region plays an important role in the sensitivity of ϵ-inhibition (33, 49). The cause of the different result of the LDAO effect on ϵ-inhibition between EF1 and cyanobacterial F1 in this study might be attributed to the specific inserted region on the γ subunit of cyanobacteria.
The other possible event inhibited at 80° by the ϵ subunit is thought to be the phosphate-releasing step (50). Indeed, it has been previously found that binding of the ϵ subunit to EF1 inhibits ATPase activity by decreasing the off-rate of phosphate (51). Furthermore, a conformational change of the ϵ subunit dependent on the addition of phosphate has been reported (52). However, the recovery of the structural stability of the nucleotide-depleted complex by the ϵ subunit required the C-terminal α-helical part of the ϵ subunit but not nucleotide or phosphate (Fig. 4D). Although the above phenomena do not explain the effect of inhibition of ATPase activity by the ϵ subunit directly, there is a possibility that the ϵ-inhibition can act independently of ADP-inhibition or inhibition of phosphate release.
The C terminus of the ϵ subunit reaches to the central cavity of F1 in the extended conformation (53). Our result in this study therefore indicates that ϵ-inhibition of ATPase activity must be simply caused by steric hindrance of rotation of the γ subunit, and this inhibition alone may be sufficient to stop rotation without other events, ADP-inhibited form and inhibition of phosphate release, which also occur at 80° from ATP binding in cyanobacterial enzyme. Indeed, the torsional stiffness of α3β3γ in ϵ-inhibition was stronger than that in ADP-inhibition (Fig. 3). However, the stiffness of α3β3γ containing the ϵ subunit (119.6 pNnm) was too high to force the rotation of ATP synthase by the reported pmf value 200 mV for ATP synthesis, which is equivalent to 56∼72 pNnm on 10∼14 c-ring. Recently, an increase of the stiffness of α3β3γ by the ϵ subunit was also shown for TF1-ATPase (54). However, the conformational change of the ϵ subunit in chloroplast CF1, and probably that in cyanobacteria too, is induced when the pmf is formed across the thylakoid membranes (55), and the C-terminal α-helical part of the ϵ subunit would ultimately be exposed (56). This implies that the ϵ subunit in CF0CF1 may not induce higher stiffness of the enzyme during the ATP synthesis reaction in chloroplasts. This assumption is supported by the finding that ATP synthesis activity of CF0CF1 did not change significantly when the C-terminal domain of the ϵ subunit was deleted (53), although ATP synthesis activities of TF0TF1 (ϵΔC) and EF0EF1 (ϵΔC) are higher than that of the wild-type enzyme complex (57, 58). The cause of the conformational change of chloroplast-type ϵ required to overcome a high stiffness remains unclear.
The hypothesis, which shows that ϵ-inhibition can act independently of ADP-inhibition and inhibition of phosphate release in cyanobacterial enzyme, is further supported by the result that the recovery of the structural stability of the complex by the ϵ subunit does not require nucleotide or phosphate binding (Fig. 4). However, the steric hindrance caused by the ϵ subunit may not be sufficient to stop rotation of the γ subunit in other bacterial F1 probably because of the lack of the inserted region on the γ subunit of chloroplast-type F1. Therefore, in bacterial F1, except for the cyanobacterial one, a greater opportunity to lapse into ADP-inhibition or inhibition of phosphate release caused by the binding of the ϵ subunit to γ must be originally required to stop rotation of the γ subunit to the ATP hydrolysis direction, which must confer a strong advantage to avoiding futile ATP consumption in the cells.
Acknowledgments
We thank C. S. Harwood (University Iowa), M. Ikeuchi (University of Tokyo), and T. Suzuki and M. Yoshida (ATP Synthesis Regulation Project, International Cooperative Research Project, Japan Science and Technology Agency) for providing suitable experimental materials. We also thank B. Feniouk, K. Kinosita, Jr., E. Muneyuki, K. Yokoyama, K. Motohashi, F. Motojima, and M. Kobayashi-Imashimizu for fruitful discussion.
This work was supported by Grant-in-aid for Scientific Research 18074002 (to T. H.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan and by Management Expenses Grants for National Universities Corporations from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- pmf
- proton motive force
- LDAO
- lauryl dimethylamine oxide
- TF1 and TF0TF1
- F1 and F0F1 from Bacillus PS3
- ADP-inhibition
- ADP-induced inhibition
- ϵ-inhibition
- ϵ-induced inhibition.
REFERENCES
- 1. Senior A. E. (1990) Annu. Rev. Biophys. Biophys. Chem. 19, 7–41 [DOI] [PubMed] [Google Scholar]
- 2. Boyer P. D. (1997) Annu. Rev. Biochem. 66, 717–749 [DOI] [PubMed] [Google Scholar]
- 3. Yoshida M., Muneyuki E., Hisabori T. (2001) Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 4. Stock D., Leslie A. G., Walker J. E. (1999) Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 5. Seelert H., Poetsch A., Dencher N. A., Engel A., Stahlberg H., Müller D. J. (2000) Nature 405, 418–419 [DOI] [PubMed] [Google Scholar]
- 6. Meier T., Polzer P., Diederichs K., Welte W., Dimroth P. (2005) Science 308, 659–662 [DOI] [PubMed] [Google Scholar]
- 7. Yoshida M., Sone N., Hirata H., Kagawa Y., Ui N. (1979) J. Biol. Chem. 254, 9525–9533 [PubMed] [Google Scholar]
- 8. Kaibara C., Matsui T., Hisabori T., Yoshida M. (1996) J. Biol. Chem. 271, 2433–2438 [DOI] [PubMed] [Google Scholar]
- 9. Matsui T., Muneyuki E., Honda M., Allison W. S., Dou C., Yoshida M. (1997) J. Biol. Chem. 272, 8215–8221 [DOI] [PubMed] [Google Scholar]
- 10. Hisabori T., Kato Y., Motohashi K., Kroth-Pancic P., Strotmann H., Amano T. (1997) Eur. J. Biochem. 247, 1158–1165 [DOI] [PubMed] [Google Scholar]
- 11. Gresser M. J., Myers J. A., Boyer P. D. (1982) J. Biol. Chem. 257, 12030–12038 [PubMed] [Google Scholar]
- 12. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 13. Duncan T. M., Bulygin V. V., Zhou Y., Hutcheon M. L., Cross R. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10964–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sabbert D., Engelbrecht S., Junge W. (1996) Nature 381, 623–625 [DOI] [PubMed] [Google Scholar]
- 15. Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 16. Yasuda R., Noji H., Kinosita K., Jr., Yoshida M. (1998) Cell 93, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 17. Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. (2001) Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 18. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Minkov I. B., Fitin A. F., Vasilyeva E. A., Vinogradov A. D. (1979) Biochem. Biophys. Res. Commun. 89, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 20. Bar-Zvi D., Shavit N. (1982) J. Bioenerg. Biomembr. 14, 467–478 [DOI] [PubMed] [Google Scholar]
- 21. Feldman R. I., Boyer P. D. (1985) J. Biol. Chem. 260, 13088–13094 [PubMed] [Google Scholar]
- 22. Hirono-Hara Y., Noji H., Nishiura M., Muneyuki E., Hara K. Y., Yasuda R., Kinosita K., Jr., Yoshida M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13649–13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nelson N., Nelson H., Racker E. (1972) J. Biol. Chem. 247, 7657–7662 [PubMed] [Google Scholar]
- 24. Richter M. L., Patrie W. J., McCarty R. E. (1984) J. Biol. Chem. 259, 7371–7373 [PubMed] [Google Scholar]
- 25. Aggeler R., Capaldi R. A. (1996) J. Biol. Chem. 271, 13888–13891 [DOI] [PubMed] [Google Scholar]
- 26. Kuki M., Noumi T., Maeda M., Amemura A., Futai M. (1988) J. Biol. Chem. 263, 17437–17442 [PubMed] [Google Scholar]
- 27. Kato-Yamada Y., Bald D., Koike M., Motohashi K., Hisabori T., Yoshida M. (1999) J. Biol. Chem. 274, 33991–33994 [DOI] [PubMed] [Google Scholar]
- 28. Tsunoda S. P., Rodgers A. J., Aggeler R., Wilce M. C., Yoshida M., Capaldi R. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6560–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suzuki T., Murakami T., Iino R., Suzuki J., Ono S., Shirakihara Y., Yoshida M. (2003) J. Biol. Chem. 278, 46840–46846 [DOI] [PubMed] [Google Scholar]
- 30. Iino R., Murakami T., Iizuka S., Kato-Yamada Y., Suzuki T., Yoshida M. (2005) J. Biol. Chem. 280, 40130–40134 [DOI] [PubMed] [Google Scholar]
- 31. Nowak K. F., McCarty R. E. (2004) Biochemistry 43, 3273–3279 [DOI] [PubMed] [Google Scholar]
- 32. Cipriano D. J., Dunn S. D. (2006) J. Biol. Chem. 281, 501–507 [DOI] [PubMed] [Google Scholar]
- 33. Konno H., Murakami-Fuse T., Fujii F., Koyama F., Ueoka-Nakanishi H., Pack C. G., Kinjo M., Hisabori T. (2006) EMBO J. 25, 4596–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feniouk B. A., Suzuki T., Yoshida M. (2007) J. Biol. Chem. 282, 764–772 [DOI] [PubMed] [Google Scholar]
- 35. Tsumuraya M., Furuike S., Adachi K., Kinosita K., Jr., Yoshida M. (2009) FEBS Lett. 583, 1121–1126 [DOI] [PubMed] [Google Scholar]
- 36. Landt O., Grunert H. P., Hahn U. (1990) Gene 96, 125–128 [DOI] [PubMed] [Google Scholar]
- 37. Hirono-Hara Y., Ishizuka K., Kinosita K., Jr., Yoshida M., Noji H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4288–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stiggall D. L., Galante Y. M., Hatefi Y. (1979) Methods Enzymol. 55, 308–315 [DOI] [PubMed] [Google Scholar]
- 39. Sielaff H., Rennekamp H., Wächter A., Xie H., Hilbers F., Feldbauer K., Dunn S. D., Engelbrecht S., Junge W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17760–17765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Digel J. G., Kishinevsky A., Ong A. M., McCarty R. E. (1996) J. Biol. Chem. 271, 19976–19982 [DOI] [PubMed] [Google Scholar]
- 41. Hisabori T., Muneyuki E., Odaka M., Yokoyama K., Mochizuki K., Yoshida M. (1992) J. Biol. Chem. 267, 4551–4556 [PubMed] [Google Scholar]
- 42. Lötscher H. R., deJong C., Capaldi R. A. (1984) Biochemistry 23, 4140–4143 [DOI] [PubMed] [Google Scholar]
- 43. Jault J. M., Dou C., Grodsky N. B., Matsui T., Yoshida M., Allison W. S. (1996) J. Biol. Chem. 271, 28818–28824 [DOI] [PubMed] [Google Scholar]
- 44. Dunn S. D., Tozer R. G., Zadorozny V. D. (1990) Biochemistry 29, 4335–4340 [DOI] [PubMed] [Google Scholar]
- 45. Peskova Y. B., Nakamoto R. K. (2000) Biochemistry 39, 11830–11836 [DOI] [PubMed] [Google Scholar]
- 46. Bragg P. D., Hou C. (1986) Biochim. Biophys. Acta 851, 385–394 [DOI] [PubMed] [Google Scholar]
- 47. Weber J., Dunn S. D., Senior A. E. (1999) J. Biol. Chem. 274, 19124–19128 [DOI] [PubMed] [Google Scholar]
- 48. Yasuno T., Muneyuki E., Yoshida M., Kato-Yamada Y. (2009) Biochem. Biophys. Res. Commun. 390, 230–234 [DOI] [PubMed] [Google Scholar]
- 49. Samra H. S., Gao F., He F., Hoang E., Chen Z., Gegenheimer P. A., Berrie C. L., Richter M. L. (2006) J. Biol. Chem. 281, 31041–31049 [DOI] [PubMed] [Google Scholar]
- 50. Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. (2007) Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 51. Dunn S. D., Zadorozny V. D., Tozer R. G., Orr L. E. (1987) Biochemistry 26, 4488–4493 [DOI] [PubMed] [Google Scholar]
- 52. Mendel-Hartvig J., Capaldi R. A. (1991) Biochemistry 30, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 53. Nowak K. F., Tabidze V., McCarty R. E. (2002) Biochemistry 41, 15130–15134 [DOI] [PubMed] [Google Scholar]
- 54. Saita E., Iino R., Suzuki T., Feniouk B. A., Kinosita K., Jr., Yoshida M. (2010) J. Biol. Chem. 285, 11411–11417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Komatsu-Takaki M. (1989) J. Biol. Chem. 264, 17750–17753 [PubMed] [Google Scholar]
- 56. Johnson E. A., McCarty R. E. (2002) Biochemistry 41, 2446–2451 [DOI] [PubMed] [Google Scholar]
- 57. Masaike T., Suzuki T., Tsunoda S. P., Konno H., Yoshida M. (2006) Biochem. Biophys. Res. Commun. 342, 800–807 [DOI] [PubMed] [Google Scholar]
- 58. Iino R., Hasegawa R., Tabata K. V., Noji H. (2009) J. Biol. Chem. 284, 17457–17464 [DOI] [PMC free article] [PubMed] [Google Scholar]