Abstract
A thiophene-based tripodal receptor has been synthesized and its complexes with nitrate and iodide have determined by single-crystal X-ray analysis. In the nitrate complex, one nitrate is encapsulated in a selective orientation forming a C3 symmetric complex, which is bonded to three protonated secondary amines with six NH···O bonds. The anion is coordinated in a plane perpendicular to the principal rotation axis passing through the tertiary nitrogen of the receptor and the nitrogen of the encapsulated nitrate. High-level DFT calculations support the crystallographic results demonstrating that an adduct with trigonal binding of three oxygen atoms is more stable than that of one oxygen atom of the encapsulate nitrate. On the other hand, in the structure of the iodide complex, all three iodides lie outside the cavity. 1H NMR titration studies indicate that the receptor forms a 1:1 complex with nitrate with a binding constant of K = 315 M−1 in chloroform, showing a moderate selectivity over halides and perchlorate.
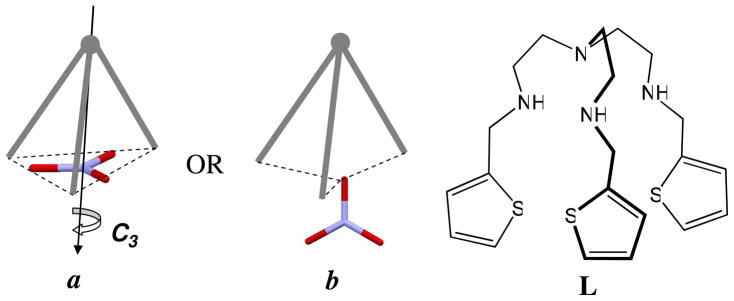
Because of the ubiquitous presence of nitrate in the environment and its impact in ground water contamination,1 there has been considerable interest in selective recognition of nitrate with synthetic hosts.2 However, the inherent trigonal geometry of the nitrate anion often makes it difficult to design strong and selective hosts which are complementary to its three-fold symmetry.3 Previous crystallographic studies have suggested that macromonocycles bind two nitrates from both sides of the macrocyclic plane4 or fold about a single nitrate ion.5 Ansyln and coworkers reported an elegant amide-based macrocycle where three converging hydrogen bonds were used for selective binding of nitrate anions.6 Several years later, the same group designed tren-based receptors featuring a C3v symmetric cavity with a single Cu(II) along the C3 axis, which provided excellent shape and size complementarity to an anion with a tetrahedral geometry.7 This compound showed strong selectivity for phosphate at neutral pH, while for nitrate, the observed binding constant was less than 100 M−1. Since the nitrate ion is planar, with a negative charge delocalized about the trigonally-arranged oxygen atoms, simple tripodal frameworks should be ideal candidates for a nitrate. In the case of trigonal binding (Scheme 1), a nitrate can interact with a host either through all three oxygens in binding mode a (C3 symmetry) or through a single oxygen in binding mode b (no C3 symmetry). Because of the topological matching of nitrate with a tripodal framework as well as the participation of the increased number of NH···O interactions, mode a is expected to be more stable. This hypothesis was supported by earlier work of Bowman-James8 and co-workers in a study on bicyclic receptor. More recently, Bianchi and coworkers reported that the simple tris(2-aminoethyl)amine (tren) formed a nitrate complex showing binding mode b.9 Theoretical calculations suggest binding mode a is more stable than the other binding mode in the gas phase, while mode b is the preferred conformation in solution.9 Although, a number of simple acyclic tren-derived receptors10 or metal containing receptors11 have been reported as nitrate-binding hosts, to the best of our knowledge, none of them is known to topologically host a nitrate showing a C3 symmetric complex. Herein, we report structural evidence of a C3 symmetric nitrate complex with a new tripodal receptor L complemented with the results of theoretical and solution studies.
Scheme 1.
Trigonal binding of nitrate (binding modes a and b) and the tripodal host (L). A C3 nitrate complex is expected from binding mode a.
The ligand L was synthesized from the reaction of tris(2-aminoethyl)-amine with 2-thiophene aldehyde in ethanol followed by the reduction with NaBH4. The nitrate complex, [H3L(NO3)](NO3)2, was obtained from the reaction of L with nitric acid in methanol. Recrystallization of the salt from an aqueous solution resulted in rod-like crystals suitable for X-ray analysis.12 Crystallographic results reveal that the ligand is triprotonated to adopt a conical shape in which three secondary nitrogens (N2, N3 and N4) form an almost perfect equilateral triangle (N2···N3 = 4.946(4), N3···N4 = 4.960(4) and N4···N2 = 5.000(4) Å), forming an ideal environment for hydrogen bonding interactions in a trigonal binding mode. As shown in Figure 1, the protonated ligand is found to encapsulate one nitrate anion in its cavity with six NH···O bonds, which is consistent with electrostatic potential calculations reported earlier3 providing six distinct binding sites of nitrate for D-H interactions.
Figure 1.
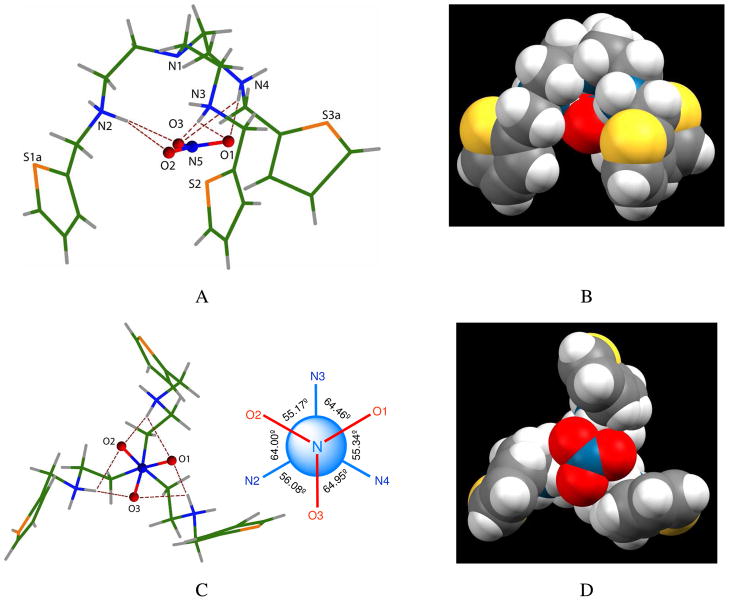
Crystal structure of [H3L(NO3)]2+ motif showing one encapsulated nitrate with six NH···O bonds in binding mode a. Side views (A and B) and top-down views (C and D) are shown. B and D are the space filling models of the perspective views of A and C, respectively. Relative dihedral angles of [H3L]3+ with the encapsulated nitrate are shown to the right of C.
The nitrate with trigonal planar geometry lies essentially parallel to a plane formed by the three secondary nitrogens, showing a dihedral angle of 1.6(2)° and an interplanar distance of 1.607(3) Å (Figure 2). Further inspection of this structure shows that the overall conformation of the complex has C3 symmetry with the principal axis passing through the nitrogen of nitrate and the tertiary nitrogen of the host. Each oxygen has two hydrogen bonds from secondary amines in a staggered conformation with respect to the three nitrogens (Figure 1C), in which the dihedral angles (60 ± 5°) are about 60°. The synergistic bonding between the three nitrate oxygen atoms with the protonated amines allows the anion to be strongly pulled inside the charged cavity. The three NH···O bonds tend to be linear (D—H···O angles are 171.4–175.9°), suggesting the formation of strong, orientationally-directed hydrogen bonds (Table 1). Two other spectator nitrates remain outside the cavity (not shown), and are involved in hydrogen bonding interactions with amines. All three thiophene units exhibit disorder over two positions related by approximate two-fold rotations about C-C bonds linking them to aliphatic groups. The major component, shown in Figure 1, has populations in the range 0.515(4) – 0.587(5) (see Supporting Information).
Figure 2.
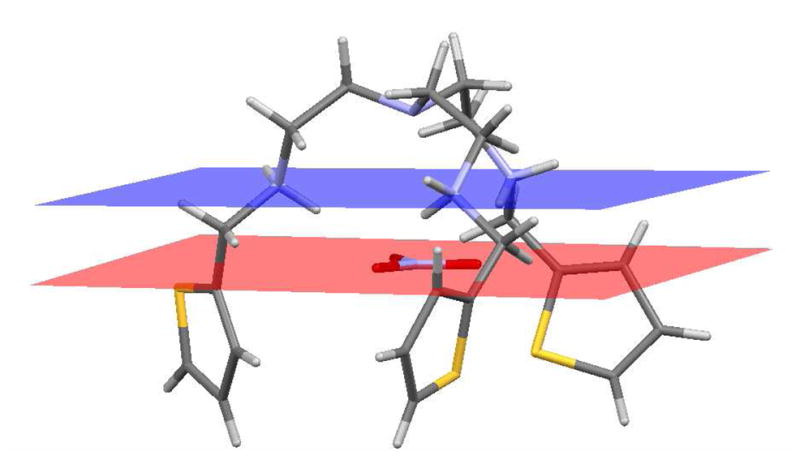
Crystal structure of the nitrate complex showing the trigonal plane formed by NO3− lying parallel to the plane formed by the three secondary nitrogens with an interplanar distance of 1.607(3) Å.
Table 1.
Hydrogen bonding parameters (A, °)
| D—H···O | D···O Exp. |
D···O Calc. |
∠DHO Exp. |
∠DHO Calc. |
|---|---|---|---|---|
| N3—H32N··· O1 | 3.044(4) | 2.893 | 129.0 | 115.14 |
| N4—H41N···O1 | 2.877(3) | 2.806 | 171.7 | 172.65 |
| N2—H21N···O2 | 3.010(4) | 2.893 | 121.9 | 115.14 |
| N3—H32N···O2 | 2.846(4) | 2.806 | 171.4 | 172.65 |
| N4—H41N···O3 | 3.058(4) | 2.893 | 117.7 | 115.14 |
| N2—H21N···O3 | 2.895(4) | 2.806 | 175.9 | 172.65 |
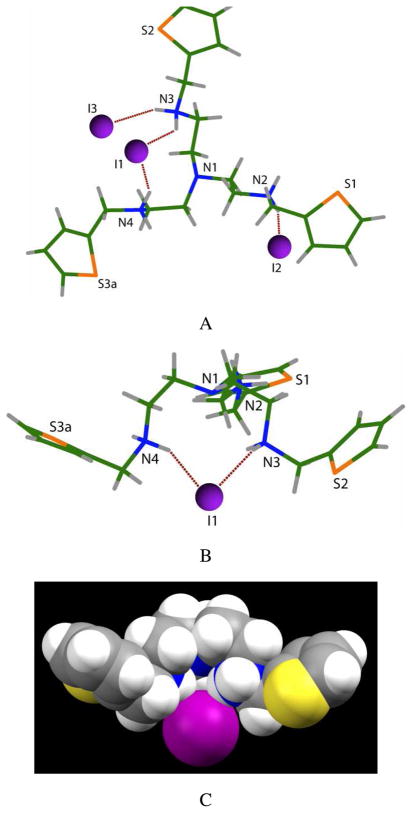
The iodide complex of [H3L](I)3 as obtained from the reaction of L with hydroiodic acid gave X-ray crystals from slow evaporation of a solution of the salt dissolved in water and ethanol (1:2 v/v, 1 mL) at room temperature.12 Crystallographic analysis suggests that the ligand is again triprotonated, adopting a non-crystallographic C3 symmetric conformation (Figure 3).13 However, in this structure the iodide is not fully encapsulated. The host molecule is relatively flat, and one iodide is hydrogen bonded with two protonated amines N3 and N4 (Figure 3A and B). The protons on the third nitrogen (N2) are directed outward from the cavity formed by the three secondary nitrogens. As shown in Figure 3B and C, the iodide (I1) lies above the cavity at the distance of 4.233 Å from the apical nitrogen (N1). The other two anions are singly bonded to N2 and N3. Such bonding patterns were previously observed in the bromide complex of (N,N′,N″-tris(2-benzylaminoethyl)amine).10c The NH···I interactions range from 3.460(3) to 3.553(4) Å, which are comparable to those observed in the iodide complex of tiny octaazacryptand (3.476(4) – 3.632(4) Å).14
Figure 3.
Side view (A) of the crystal structure of [H3L](I)3 showing hydrogen-bonded iodide. Top-down views (B and C) of [H3L](I)]2+ motif showing one iodide with two NH···O bonds (two iodide anions are omitted for clarity) are shown. View C represents the space filling model of the perspective view of B.
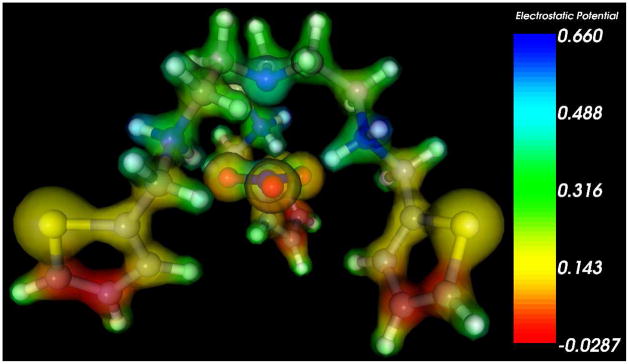
In order to quantitatively understand the unique bonding within the tripodal ligand, density functional theory (DFT) calculations were performed on two different molecular conformations: (1) simultaneous interaction of the protonated amine groups with all three oxygen atoms in binding mode a, and (2) interaction of the three protonated ligand amine groups with a single oxygen atom of NO3− in binding mode b. All quantum chemical calculations were carried out with the recent M06-2X meta-GGA hybrid functional which has been shown to accurately predict the binding energies of ions and other noncovalent bonding interactions in large molecular systems.15 Molecular geometries (including the empty ligand) were completely optimized without constraints at the M06-2X/6-31G(d,p) level of theory, and single-point energies with a very large 6-311+G(d,p) basis set were carried out in the presence of a polarizable continuum model (PCM) solvent model16 to approximate a chloroform environment (chloroform dielectric = 4.71). Figure 4 shows the optimized geometries of binding modes a and b of the NO3− anion electrostatically bound to the protonated tripodal ligand. From the DFT-optimized geometries, we found that the two conformations incorporate different binding conformations and energies. Binding mode a is stabilized by three short hydrogen bonds (O···H =1.8 Å) and three longer O···H interactions (2.3 Å), yielding a C3-symmetry structure while binding mode b is characterized via three hydrogen bonds (1.6 Å average O···H bond distance) to a single oxygen atom. The stabilization energy for each structure was calculated as Es = E (anion-ligand) – E (ligand) – E (anion), and binding energies of −84.8 kcal/mol and −78.1 kcal/mol were obtained for conformations a and b, respectively, in the PCM solvent. While both complexes are stable and display strong electrostatic interactions with the tripodal ligands, conformation a has a stronger binding energy due to a simultaneous electrostatic interaction between all three negatively-charged oxygen atoms (see Figure 5 for the electrostatic potential) compared to only one oxygen atom involved in conformation b.
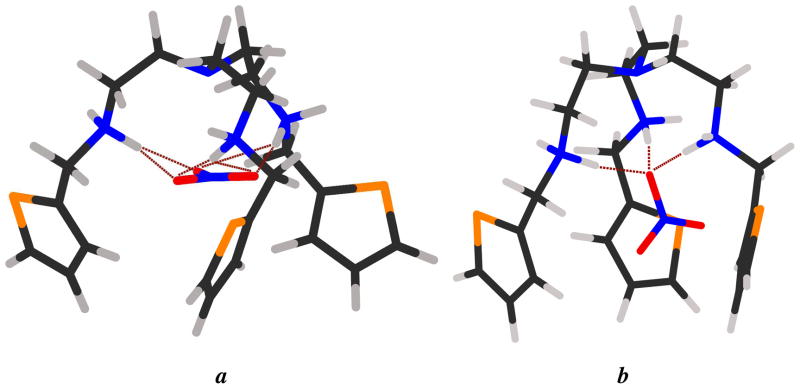
Figure 4.
Optimized DFT geometries of [H3L(NO3)]2+ showing two binding modes (a and b) obtained at the M06-2X/6-31G(d,p) level of theory. (a) Each of the three oxygen atoms in NO3− is hydrogen-bonded to the ligand (bond distance = 2.85 Å), yielding a C3-symmetric structure; (b) a single oxygen atom in NO3− is hydrogen-bonded via three hydrogen bonds (average bond distance = 2.70 Å).
Figure 5.
Electrostatic DFT potential of the NO3− anion electrostatically bound to the triprotonated ligand in PCM chloroform solvent. The complex is stabilized by strong electrostatic interactions between the negatively-charged oxygen atoms in NO3− and the positively-charged protonated amine groups in the tripodal ligand.
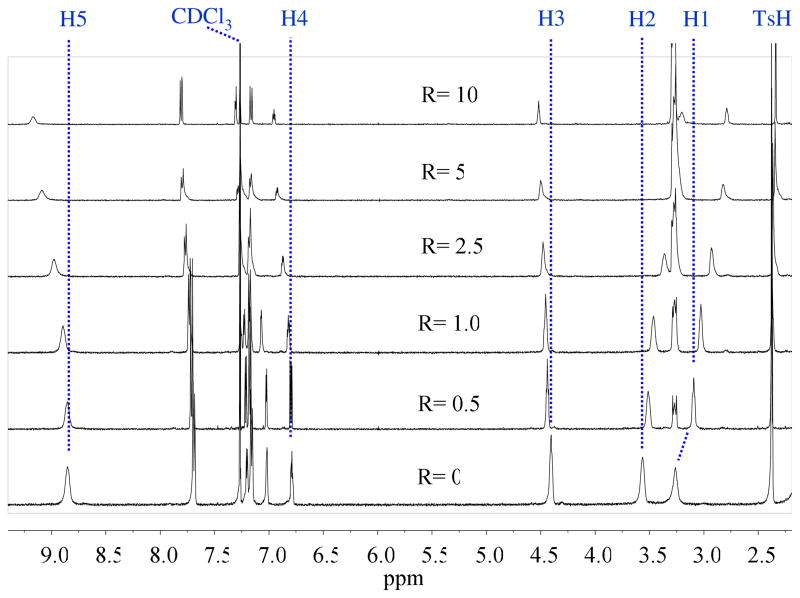
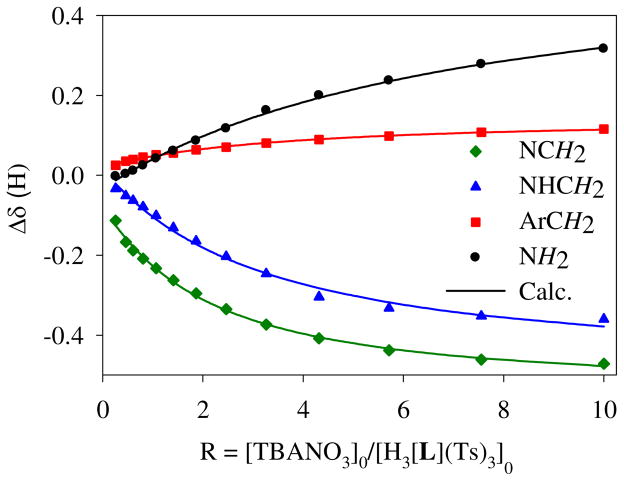
1H NMR titrations were carried out to quantify the interaction of the host for nitrate in solution. The addition of tetrabutylammonium nitrate to the tosylate salt of H3L3+ in CDCl3 led to significant shifts in the resonances of the ligand, with the highest downfield shift for NH protons, suggesting the direct participation of NH protons in nitrate binding (Figure 6). The variation of NMR signals with an increasing amount of nitrate solution provided the best fit to a 1:1 binding model (Figure 7), yielding an association constant of K = 315 M−1 (14.3 kJ/mol).17 The binding constant is higher than that observed for the nitrate binding with the phenyl analogue (K = 35 M−1) under similar conditions.10c The higher affinity in the present case could be due to a directional effect of secondary-amine protons towards the cavity, as indicated by our crystallographic analysis. This value is, however, lower than that observed with the bicyclic cyclophane (K =1000 M−1), where the anion was encapsulated in the confined cavity.6a Titrations were also performed for the receptor to examine its binding affinity towards other halides (F−, Cl−, Br− > and I−) and oxoanions (HSO4−, H2PO4− and ClO4−) using their tetrabutyl ammonium salts in CDCl3. The binding results for the anions, as obtained from non-linear regression analysis, are summarized in Table 2. The results show that the host forms a 1:1 complex with the halides, giving binding constants in the range of K = 50 to 115 M−1. The magnitude of the binding constants follows as Br− > Cl− > F− > I−. However, the addition of the salt of HSO4− or H2PO4− to the host solution (2 mM) resulted in an immediate precipitation in the NMR tube during the titration; therefore, we failed to measure the binding constants for these two anions. The host was found to interact weakly with ClO4− with a binding constant of K= 55 M−1 as compared to K = 315 M−1 for nitrate. Therefore, the host shows a moderate selectivity for nitrate over perchlorate or other halides.
Figure 6.
1H NMR spectra of [H3L](Ts)3 (2 mM) with an increasing amount of tetrabutyl ammonium nitrate (20 mM) in CDCl3 (H1 = NCH2, H2 = NCH2CH2, H3=ArCH2, H4 = ArH, H5 = NH).
Figure 7.
1H NMR titration curves of [H3L](Ts)3 (2mM) with NO3− in CDCl3. Net changes in the chemical shifts of different protons are shown against an increasing amount of TBANO3 (20 mM).
Table 2.
Binding constants of H3L3+ for anions in CDCl3 at 298 K.a
Estimated deviations are less than 10%.
Binding constant could not be measured due to precipitation during the titration.
In summary, we have shown that a simple tripodal host with a three-fold rotation axis is complementary to topological fitting of a nitrate anion in its cavity to form a C3 symmetric anion complex. The nitrate is held strongly with a total of six hydrogen bonds and oriented in a plane perpendicular to the principal rotation axis passing through both the tertiary nitrogen of the ligand and the nitrogen atom of the nitrate. Our crystallographic results were further supported by theoretical calculations predicting that an adduct with trigonal binding mode (mode a) is energetically more stable than a single binding mode (mode b). Clearly, the hydrogen bonding interactions between the three oxygens of the internal nitrate and the trigonally-oriented NH’s of the host play a role in the stabilization of the structure. Such C3 symmetry is not observed in the iodide structure, where the anionic species fail to bind to the three arms symmetrically. Although, the ligand only showed a moderate selectivity for nitrate over halides and perchlorate, the solid state structure described here represents a unique bonding feature of nitrate anion that has not been structurally identified before with a tren base-ligand.
Supplementary Material
Acknowledgments
National Science Foundation is acknowledged for a CAREER award (CHE-1056927) to MAH. The work was supported by the National Institute of health (G12RR013459). The NMR instrument used for this work was funded by the National Science Foundation (CHE-0821357). Purchase of the diffractometer was made possible by grant No. LEQSF (1999–2000)-ENH-TR-13, administered by the Louisiana Board of Regents.
Footnotes
Supporting Information available: Crystallographic data in CIF format, synthetic procedures, NMR spectra, ORTEP diagrams, experimental procedure for binding studies and binding isotherms. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kavallieratos K, Bryan JC, Sachleben RA, Van Berkel GJ, Espetia OE, Kelly MA, Danby AM, Bowman-James K, Moyer BA. In: Fundamentals and Applications of Anion Separations. Moyer BA, Singh R, editors. Kluwer/Plenum; New York: 2004. pp. 125–150. [Google Scholar]
- 2.(a) Gale PA, Sessler JL, Kral V, Lynch V. J Am Chem Soc. 1996;118:5140–5141. [Google Scholar]; (b) Gale PA. Coord Chem Rev. 2000;199:181–233. [Google Scholar]; (c) Qian Q, Wilson GS, Bowman-James K, Girault H. Anal Chem. 2001;73:497–503. doi: 10.1021/ac000806h. [DOI] [PubMed] [Google Scholar]; (d) Turner DR, Spencer EC, Howard JAK, Tocher DA, Steed JW. Chem Commun. 2004:1352–1353. doi: 10.1039/b402882a. [DOI] [PubMed] [Google Scholar]; (e) McKee V, Nelson J, Town RM. Chem Soc Rev. 2003;32:309–325. doi: 10.1039/b200672n. [DOI] [PubMed] [Google Scholar]; (f) Sessler JL, An D, Cho WS, Lynch V, Marquez M. Chem Eur J. 2005;11:2001–2011. doi: 10.1002/chem.200400894. [DOI] [PubMed] [Google Scholar]; (g) Filby MH, Steed JW. Coord Chem Rev. 2006;250:3200–3218. [Google Scholar]; (h) Portis B, Dey KR, Saeed MA, Powell DR, Hossain MA. Acta Cryst. 2009;E65:o260. doi: 10.1107/S1600536809039166. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Saeed MA, Fronczek FR, Huang MJ, Hossain MA. Chem Commun. 2010;46:404–406. doi: 10.1039/b923013k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Das MC, Ghosh SK, Bharadwaj PK. Cryst Eng Comm. 2010;12:413–419. [Google Scholar]
- 3.Hay BP, Gutowski M, Dixon DA, Garza J, Vargas R, Moyer BA. J Am Chem Soc. 2004;126:7925–7934. doi: 10.1021/ja0487980. [DOI] [PubMed] [Google Scholar]
- 4.(a) Cramer RE, Fermin V, Kuwabara E, Kirkup R, Selman M, Akoi K, Adeyemo A, Yamazaki HJ. Am Chem Soc. 1991;113:7033–7034. [Google Scholar]; (b) Hossain MA, Llinares JM, Powell D, Bowman-James K. Inorg Chem. 2001;40:4710–4720. doi: 10.1021/ic010135l. [DOI] [PubMed] [Google Scholar]
- 5.(a) Papoyan G, Gu K, Wiorkiewicz-Kuczera J, Kuczera K, Bowman-James K. J Am Chem Soc. 1996;118:1354–1364. [Google Scholar]; (b) Valencia L, Bastida R, Garcia-Espana E, Julian-Ortiz JVD, Llinares JM, Macias A, Lourido P. Cryst Growth Des. 2010;10:3418–3423. [Google Scholar]
- 6.(a) Bisson AP, Lynch VM, Monahan MKC, Anslyn EV. Angew Chem Int Ed. 1997;36:2340–2342. [Google Scholar]; (b) Best MD, Tobey SL, Anslyn EV. Coord Chem Rev. 2003;240:3–15. [Google Scholar]
- 7.Tobey SL, Jones BD, Anslyn EV. J Am Chem Soc. 2003;125:4026–4027. doi: 10.1021/ja021390n. [DOI] [PubMed] [Google Scholar]
- 8.Mason S, Clifford T, Seib L, Kuczera K, Bowman-James K. J Am Chem Soc. 1998;120:8899–8900. [Google Scholar]
- 9.Bazzicalupi C, Bencini A, Bianchi A, Danesi A, Giorgi C, Valtancoli B. Inorg Chem. 2009;48:2391–2398. doi: 10.1021/ic8013128. [DOI] [PubMed] [Google Scholar]
- 10.(a) Gale P. Acc Chem Res. 2006;39:465–475. doi: 10.1021/ar040237q. [DOI] [PubMed] [Google Scholar]; (b) Danby A, Seib L, Alcock NW, Bowman-James K. Chem Commun. 2000:973–974. [Google Scholar]; (c) Hossain MA, Liljegren JA, Powell D, Bowman-James K. Inorg Chem. 2004;43:3751–3755. doi: 10.1021/ic049762b. [DOI] [PubMed] [Google Scholar]; (d) Arunachalam M, Ghosh P. Inorg Chem. 2010;49:943–951. doi: 10.1021/ic901644h. [DOI] [PubMed] [Google Scholar]
- 11.(a) Sang RL, Xu L. Polyhedron. 2006;25:2167–2174. [Google Scholar]; (b) Deng QL, Lai LH, Han YZ, Miao ZW, Ji AX, Xu XJ. Acta Physico-Chimica Sinica. 1994;10:444–448. [Google Scholar]; (c) Gallego ML, Cano M, Campo JA. Helv Chim Acta. 2005;88:2433–2440. [Google Scholar]
- 12.Crystal data for [H3L(NO3)](NO3)2: C21H33N4S3·3(NO3), M = 623.72, monoclinic, a = 9.906(3) Å, b = 16.193(4) Å, c = 17.823(6) Å, β = 95.252(10)°, V = 2846.9(15) Å3, T = 100.0(5) K, space group P21/n, Z = 4, 20961 reflections measured, 5797 independent reflections (Rint = 0.040). The final R1 value was 0.0618 (I > 2σ(I)). The final R1 value was 0.0997 (all data). Crystal data for [H3L][I]3: C21H33N4S3·3(I), M = 818.39, orthorhombic, a = 10.5433(5) Å, b = 11.4203(6) Å, c = 24.5107(15) Å, V = 2951.3(3) Å3, T = 90.0(5) K, space group P212121, Z = 4, 90447 reflections measured, 10653 independent reflections (Rint = 0.069). The final R1 values were 0.038 (I > 2σ(I)). The final R1 values were 0.048 (all data).
- 13.Isiklan M, Fronczek FR, Hossain MA. Acta Cryst. 2010;E66:o2755. doi: 10.1107/S1600536810039462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hossain MA, Llinares JM, Powell D, Bowman-James K. Supramol Chem. 2002;2:143–151. [Google Scholar]
- 15.(a) Zhao Y, Truhlar DG. Theor Chem Acc. 2008;120:215–241. [Google Scholar]; (b) Wong BM. J Comput Chem. 2009;30:51–56. doi: 10.1002/jcc.21022. [DOI] [PubMed] [Google Scholar]; (c) Hossain MA, Saeed MA, Fronczek FR, Wong BM, Dey KR, Mendy JS, Gibson D. Cryst Growth Des. 2010;10:1478–1481. doi: 10.1021/cg100110f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Saeed MA, Wong BM, Fronczek FR, Venkatraman R, Hossain MA. Cryst Growth Des. 2010;10:1486–1488. doi: 10.1021/cg100161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasi J, Mennucci B, Cammi R. Chem Rev. 2005;105:2999–3094. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 17.Schneider HJ, Kramer R, Simova S, Schneider U. J Am Chem Soc. 1988;110:6442–6448. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.