SUMMARY
High-content screening for gene profiling has generally been limited to single cells. Here, we explore an alternative approach—profiling gene function by analyzing effects of gene knockdowns on the architecture of a complex tissue in a multicellular organism. We profile 554 essential C. elegans genes by imaging gonad architecture and scoring 94 phenotypic features. To generate a reference for evaluating methods for network construction, genes were manually partitioned into 102 phenotypic classes, predicting functions for uncharacterized genes across diverse cellular processes. Using this classification as a benchmark, we developed a robust computational method for constructing gene networks from high-content profiles based on a network context-dependent measure that ranks the significance of links between genes. Our analysis reveals that multi-parametric profiling in a complex tissue yields functional maps with a resolution similar to genetic interaction-based profiling in unicellular eukaryotes—pinpointing subunits of macromolecular complexes and components functioning in common cellular processes.
INTRODUCTION
A major challenge of the post-genomic era is to translate the parts lists generated by genome sequencing into maps of the pathways that execute cellular processes. Approaches to do this combine systematic gene inhibition with functional tests that span a continuum—from single readout assays to complex assays that interrogate a broad spectrum of cellular processes. Whereas single readout assays identify pathways that impact a specific process (Mathey-Prevot and Perrimon, 2006), complex assays can be used to construct functional networks from collections of genes with diverse cellular roles. Two approaches have emerged for distilling complex phenotypes for phenotypic profiling: genetic interaction profiling and high-content screening. Although the methodologies are distinct, both strategies translate the consequences of inhibiting gene activity into phenotypic profiles that can be compared to generate a map of the functional relationships between genes (Boone et al., 2007; Collins et al., 2009; Conrad and Gerlich, 2010; Piano et al., 2002; Sönnichsen et al., 2005).
Genetic interaction profiling was pioneered in budding yeast, using a comprehensive deletion library of non-essential genes and collections of hypomorphic alleles of essential genes (Boone et al., 2007; Collins et al., 2009). Genetic interaction profiling captures the consequences of inhibiting a gene by measuring the effect on growth rate of pairwise inhibitions with each of the other genes in the collection. This analysis generates quantitative interaction profiles for each gene that can be clustered to reveal functionally significant relationships. A genome-scale genetic interaction map was recently constructed for S. cerevisiae (Costanzo et al., 2010), and maps have also been generated for subsets of gene implicated in specific processes—such as RNA processing, chromosome biology, proteasome function, and the secretory pathway (Breslow et al., 2008; Collins et al., 2007; Schuldiner et al., 2005; Wilmes et al., 2008).
In metazoans, genetic interaction profiling is difficult to implement because comprehensive libraries of deletion/hypomorphic strains do not exist and developing reproducible high-throughput methods to quantify fitness is a formidable barrier (Gunsalus, 2008). Consequently, high-content screening is the primary method for mapping functional gene networks in animal cells. In a high-content screen, light microscopy is used to assess phenotypes arising from gene inhibition by RNA-mediated interference, and the phenotype is captured by scoring a large parameter set (Conrad and Gerlich, 2010). The depth of the phenotypic profile is based on the biological complexity of the assay and the nature and accuracy of parameter scoring. To date, the requirement for high-resolution imaging has generally limited high-content screening to single cells or early embryos.
C. elegans is a prototype metazoan system for the functional mapping of essential genes (Piano et al., 2006). C. elegans has ~20,000 genes, of which ~2,500 are essential for embryo production or viability (WormBase release WS210; Harris et al., 2010). In a set of pioneering high-content screens, time-lapse Differential Interference Contrast (DIC) microscopy was used to film the early divisions of embryos following individual inhibitions of specific subsets of C. elegans genes (Gönczy et al., 2000; Piano et al., 2000; Zipperlen et al., 2001). This was extended to a full-genome screen that generated high-content phenotypic profiles for ~500 essential genes (Sönnichsen et al., 2005). These profiles were combined with protein-protein interaction and expression profiling data to create a first-generation integrative map that linked 305 essential C. elegans genes in a “multiple support” network that grouped genes into modules involved in specific processes including spindle assembly, chromosome segregation, nuclear envelope dynamics, cortical dynamics, and centrosome function (Gunsalus et al., 2005). Despite the success of these studies, a large collection of essential genes could not be profiled because their inhibition results in sterility of the treated worm. Thus, the 554 genes in the “sterile” collection, which control fundamental cellular processes such as membrane trafficking, translation, proteasome function, and cortical remodeling, were largely absent from this analysis.
To fill this gap in the analysis of the C. elegans essential gene set, we profiled the 554 sterile genes by imaging syncytial gonad architecture at high-resolution following gene knockdown and scoring 94 phenotypic parameters. To generate a reference for evaluating computational methods for network construction, genes were manually partitioned into 102 phenotypic classes, predicting functions for 106 of the 116 uncharacterized genes in the collection. Using the manual classification as a benchmark, we developed a robust computational method for constructing gene networks from high-content profiles based on a network context-dependent measure that ranks the significance of functional links between genes. This method allowed us to integrate our data with that from the prior high-content embryo-filming dataset to generate a network representation of 818 essential C. elegans genes that can be viewed at multiple levels of functional resolution.
RESULTS
Phenotypic Profiling Based on The Morphology of a Complex Tissue: the C. elegans Gonad
Around 900 C. elegans genes are required for embryo production and/or for the early embryonic cell divisions (Sönnichsen et al., 2005); this collection includes the majority of genes essential for basic processes common to all cells. Because their inhibition leads to sterility, 554 of these genes could not be profiled by embryo filming. Of the 554 sterile genes, 166 were unnamed, indicating no prior characterization (Fig. 1A). For each unnamed gene, we determined whether the predicted product is a member of a KOG (eukaryotic orthologous group; Tatusov et al., 2003) and used the Ensembl database to determine if it has orthologs across species. Of the 166 unnamed genes, 50 had characterized orthologs that predicted a function for the C. elegans protein (Unnamed-Group I in Table S2). The remaining 116 were either members of KOGs of unknown function, had no predicted orthologs, or had multiple C. elegans paralogs (Unnamed-Group II in Table S2); we refer to these 116 genes as “uncharacterized” (Fig 1A).
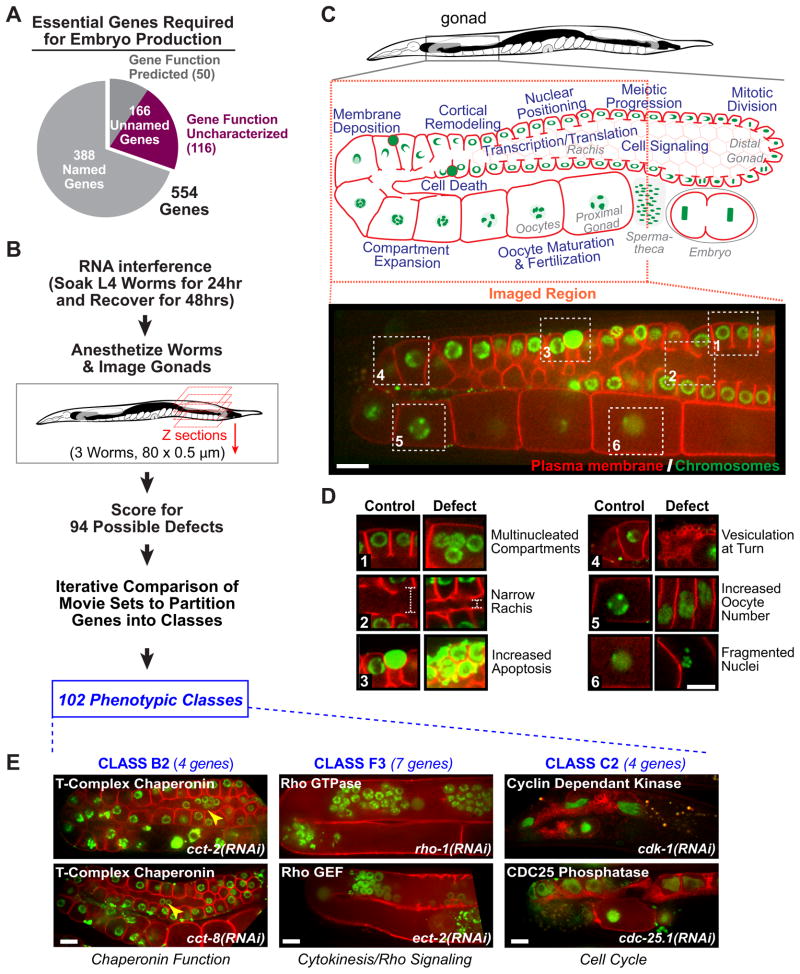
Figure 1. Phenotypic profiling using a high-content assay based on the architecture of a complex tissue.
A. Breakdown of the 554 genes in the sterile collection. B. Screen flow chart. C. The C. elegans gonad is a complex dynamic tissue whose architecture depends on a broad spectrum of interacting cellular processes (blue text on the schematic). Spinning disc confocal microscopy was used to collect an 80 plane two-color z-series of each gonad. The imaged region is indicated (red dashed box) and a sample central z-section from a control gonad is shown. D. Six sample defects are illustrated by pairing the numbered boxed regions from the control gonad in C with the corresponding regions from gonads with the indicated defects. E. Central plane images from two gene knockdowns in 3 phenotypic classes. Knockdowns in Class B2 (left column), which contains chaperonin complex subunits, led to rounded compartments and nuclei that fell out of their compartments (yellow arrowheads). Knockdowns in Class F3 (middle column), which contains genes implicated in Rho GTPase signaling, led to “tubulated” gonads with clustered nuclei. Knockdowns in Class C2 (right column), which contains cell cycle regulators, led to gonads with few compartments/oocytes. Bars, 10 μm. See also Figure S1, Table S1 and Table S2.
To profile the 554 sterile genes, we developed a high-content assay based on 3D two-color fluorescence confocal imaging of the gonad, a complex tissue in the adult C. elegans hermaphrodite (Fig. 1B). The syncytial gonad contains ~1000 meiotic nuclei in cup-shaped compartments open to a common cytoplasmic core. Compartments mature into oocytes as they progress from the distal tip to the proximal region of the gonad adjacent to the spermatheca. Gonad maturation and maintenance involves a broad spectrum of basic cellular processes (Fig. 1C), making this tissue an attractive substrate for high-content profiling. Gonad architecture was analyzed in a strain co-expressing fluorescent markers that target to the plasma membrane (GFP fusion that binds PI4,5P2) and chromosomes (mCherry-histone H2B). Hermaphrodites were soaked in dsRNA against a target gene for 24 hours beginning at the late L4 larval stage, when the gonad has almost achieved its full complement of nuclei (Kimble and Crittenden, 2005). After 48 hours recovery, gonad architecture was assessed in triplicate by anesthetizing worms and imaging one gonad per worm.
Binary phenotypic profiles were generated by scoring the set of 3 image stacks per target gene for 94 possible defects. The movie set for each gene was inspected for each of the 94 defects (Fig. 1D; for a complete list with examples see Table S1), assigning a “0” when the defect was absent and a “1” when the defect was present in at least 2 of the 3 movies. All image stacks were analyzed by the same pair of individuals, who viewed and scored them together; image stacks were indexed by RNA number, making their analysis blind to gene identity. In the 24 cases where the three movies were not consistent, the experiment was repeated (see Fig. S1, Suppl. Experimental Procedures, and Table S6 for details on screen design and scoring methods).
Generation of a Benchmark for Constructing Gene Networks Based on Gonad Architecture Phenotypes
Initial attempts using the raw parameter dataset for automated clustering broadly grouped genes, but failed to partition them at a resolution similar to what could be achieved through blinded manual classification by an experienced investigator. To develop a better computational method, we began by manually partitioning the genes into classes to generate a reference that we could use to evaluate computational methods for network construction. Manual partitioning placed the 554 sterile genes into 102 phenotypic classes (Table S2 contains a description, sample image, and gene list for each class). For organizational purposes, the 102 classes were grouped into 29 broad categories (labeled A–Z, AA, AB and AC) that each contain classes sharing one or more prominent defects. Movies and class designations can be accessed via the Phenobank website (http://worm.mpi-cbg.de/phenobank_gonad).
The manually defined phenotypic classes contained characterized genes with common annotated molecular functions (Fig. 1E), indicating that similar gonad architecture phenotypes reflect similar molecular functions. Manual partitioning placed 106 of the 116 uncharacterized genes into phenotypic classes containing characterized genes, leading to predictions for their functions. Prior to using the manually defined classes as a benchmark to optimize computational methods for network analysis, we validated the functional predictions arising from our classification by following up on 8 uncharacterized genes in 5 classes.
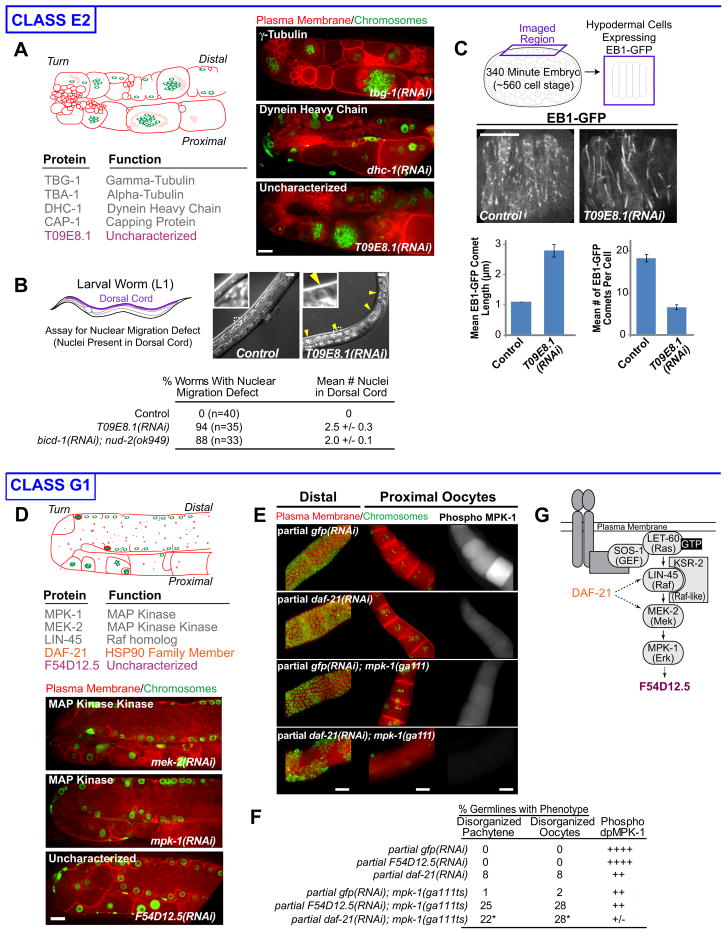
The first class (E2) predicted a role for T09E8.1 in the microtubule cytoskeleton (Fig. 2A). Unlike other class E2 genes, partial T09E8.1 inhibition did not lead to defects in the early embryonic cell divisions (not shown). In dividing cells, the microtubule cytoskeleton is organized by centrosomes, whereas the gonad is dominated by non-centrosomal microtubule arrays (Zhou et al., 2009). Thus, we hypothesized that T09E8.1 is specifically required for non-centrosomal microtubule array formation. Hypodermal cells, born late in embryogenesis, also have non-centrosomal microtubule arrays that function in nuclear positioning (Fridolfsson and Starr, 2010). Partial T09E8.1 inhibition in the hypodermis led to defects in nuclear positioning (Fig. 2B), as well as a ~3 fold reduction in the number and an increase in the length of the GFP-EBP-1 “comets” that mark growing microtubule ends (Fig. 2C). We conclude that T09E8.1, which we name noca-1 (for non-centrosomal array), plays an important role in the organization of the non-centrosomal microtubule arrays in the gonad and hypodermis.
Figure 2. Validation of manual classification I.
Class E2: A. List of the genes in Class E2. Schematic and central plane images illustrate the class phenotype. B. Nuclei are in the dorsal cords of T09E8.1(RNAi) (yellow arrowheads), but not control, L1 worms (insets 2.4X), reflecting a hypodermal cell nuclear migration defect. Table shows quantification of the nuclear migration defect. The effects of simultaneous inhibition of the dynein-regulatory proteins BICD-1 and NUD-2 quantified from the dataset in Fridolfsson et al. (2010) are shown for comparison. C. The effect of T09E8.1 knockdown on microtubule arrays in the hypodermis (schematic) was monitored by timelapse imaging of an EB1-GFP fusion (Fridolfsson and Starr, 2010). Bar graphs show the mean length (left) and number (right) of EB1-GFP comets. Error bars are the SE.
Class G1: D. Class G1 contains 3 characterized genes implicated in MAPK signaling (gray), 1 characterized gene not previously implicated in MAPK signaling (orange), and 1 uncharacterized gene (purple). Schematic and central plane images illustrate the class phenotype. E. Partial RNAi of daf-21, or GFP as a control, was performed in the presence or absence of a weak mpk-1 loss-of-function mutation (ga111). Gonads were fixed and stained for chromosomes (DAPI, green), the plasma membrane (SYN-4 and PTC-1, red), and activated MPK-1 (Phospho-MPK-1, right panels). All analysis was performed in the rrf-1(pk1417) background in which RNAi is effective in the gonad, but not in the surrounding somatic cells. F. Quantification of phenotypes resulting from partial RNAi of daf-21 or F54D12.5 in the mpk-1(ga111) background (see Table S3). * Percentages do not include the 21% of germlines that showed a severe MPK-1 “null” phenotype. G. Schematic places DAF-21 and F54D12.5 in the MAPK signaling pathway. Error bars are the SE. Bars, 10 μm. See also Figure S2, Table S2 and Table S3
The second class (G1) predicted a role for F54D12.5 and DAF-21, an hsp90-family chaperone, in the MAPK signaling pathway (Fig. 2D). We tested these predictions using a genetic test. Worms homozygous for the reduction-of-function allele mpk-1(ga111) exhibit normal gonad morphology, even though phosphorylated MPK-1 levels, a readout for pathway activity, are reduced compared to controls (Fig. 2E,F; Lee et al., 2007). Partial knockdown of daf-21 or F54D12.5, under conditions that did not result in a morphological phenotype in control worms, led to a strong MAPK knockdown phenotype in the mpk-1(ga111) background (Fig. 2E,F; Fig. S2, Table S3). RNAi of daf-21 reduced phosphorylated MPK-1 levels in control and let-60 gain-of-function worms (Fig. 2E,F; Table S3), indicating that DAF-21 acts at the level of or downstream of LET-60/Ras in the MAPK pathway (Fig. 2G). F54D12.5 RNAi did not reduce phosphorylated MPK-1 levels, and F54D12.5 contains 2 potential MPK-1 docking sites suggesting that it is an MPK-1 substrate (Fig. 2G). We conclude that DAF-21 and F54D12.5 function at different points in the MAPK signaling pathway and name F54D12.5, eom-1, for enhancer of mpk-1(ga111).
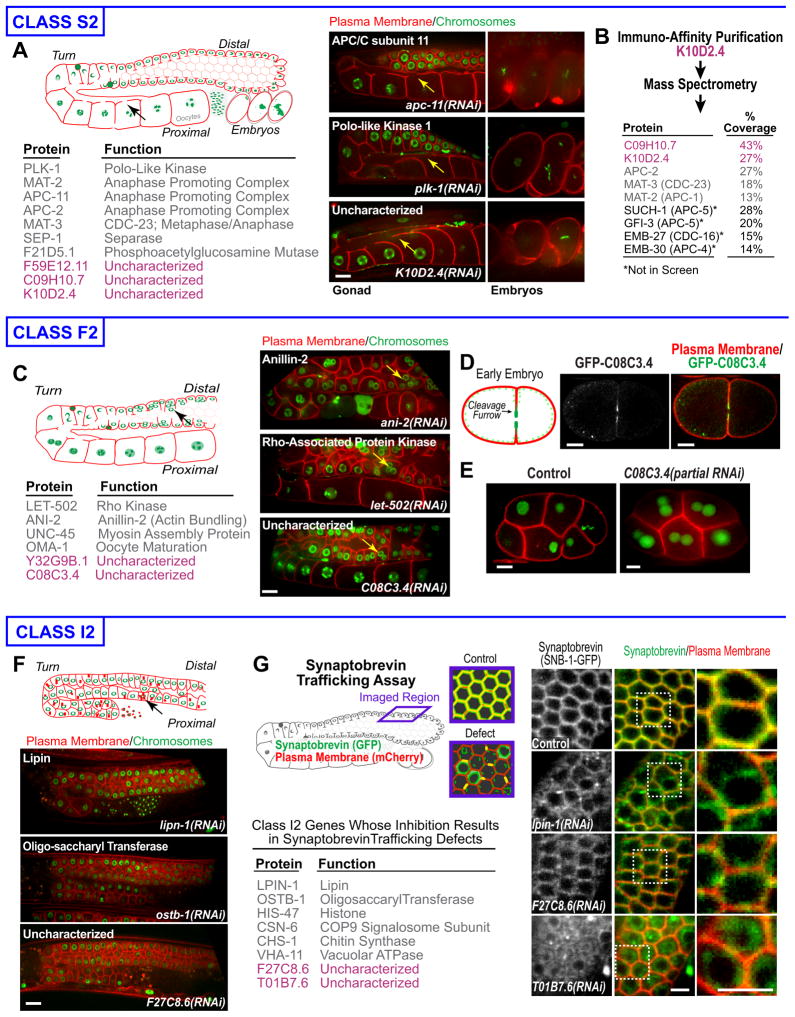
The third class (S2) predicted roles for three uncharacterized proteins in the anaphase-promoting complex/cyclosome (APC/C; Fig. 3A). Immuno-affinity purification of one uncharacterized gene product, K10D2.4, from C. elegans extracts followed by mass spectrometry recovered seven APC components (Fig. 3B). The product of another uncharacterized Class S2 gene, C09H10.7, was also recovered—indicating that both K10D2.4 and C09H10.7 are APC/C subunits. K10D2.4 was recently identified as a metazoan-specific component of the APC/C (Hubner et al., 2010; Hutchins et al., 2010; Kops et al., 2010) and the gene was named emb-1/apc-16. We name C09H10.7, apc-17.
Figure 3. Validation of manual classification II.
Class S2: A. List of the genes in Class S2. Schematic and central plane images of gonads (left) and adjacent embryos (right) illustrate the class phenotype. B. List of relevant proteins identified by mass spectrometry in an immuno-affinity purification of K10D2.4 from worm extracts, along with percent coverage. Proteins encoded by the uncharacterized Class S2 genes (purple) are listed, along with APC components in Class S2 (gray) and additional APC components not in the screen (black).
Class F2: C. List of the genes in Class F2. Schematic and central plane images illustrate the class phenotype. D. Embryo co-expressing GFP-C08C3.4 (green) and an mCherry tagged plasma membrane probe (red) during the first cell division. E. Central plane images of control and C08C3.4(RNAi) embryos expressing mCherry-histone H2B (green) and a GFP plasma membrane probe (red). Multiple nuclei in each cell in the C08C3.4(RNAi) embryo are due to cytokinesis failure.
Class I2: F. Schematic and central plane images illustrate the Class I2 phenotype, which includes punctate debris containing the plasma membrane probe (arrow in schematic). G. Schematic of the trafficking assay. List of Class I2 genes with defects in the SNB-1-GFP trafficking assay. Images of the assay for 1 characterized and 2 uncharacterized Class I2 genes. Bars are 10μm in A-F and 5 μm in G. See also Table S2.
The fourth class (F2) predicted a role for the BTB-domain containing protein C08C3.4 in cortical remodeling/cytokinesis (Fig. 3C). A GFP fusion with C08C3.4 localized to the contractile ring at the tip of the cleavage furrow in dividing embryos (Fig. 3D), and embryos partially depleted of C08C3.4 exhibited cytokinesis defects (Fig. 3E). We conclude that C08C3.4 is required for cortical remodeling in the gonad and cytokinesis in embryos and name the gene cyk-7.
The fifth class (I2) phenotype included debris labeled with the plasma membrane probe, suggesting a role in membrane trafficking (Fig. 3F). We tested for a trafficking function by imaging compartment boundaries in a strain co-expressing a mCherry labeled plasma membrane probe and a GFP fusion with SNB-1, a SNARE trafficked through the endomembrane system and delivered to the plasma membrane (Fig. 3G). Compartment boundaries in control worms have SNB-1-GFP and the plasma membrane probe and are yellow. Trafficking defects prevent SNB-1-GFP from reaching the plasma membrane, leading to red compartment boundaries. 8 of the 15 Class I2 genes, including F27C8.6 and T01B7.6, exhibited defects in the SNB-1 assay (Fig. 3G). We name F27C8.6 and T01B7.6 trcs-1 and trcs-2, respectively, for (transport to the cell surface).
The follow up work on these five classes demonstrates that gonad architecture has sufficient resolution to functionally classify genes across a broad spectrum of essential cellular functions. It also validates the manual classification, establishing it as a benchmark for evaluating computational methods for network construction.
Development of a Network Context-Dependent Method to Evaluate the Significance of Functional Links Between Genes Based on High-Content Phenotypic Profiles
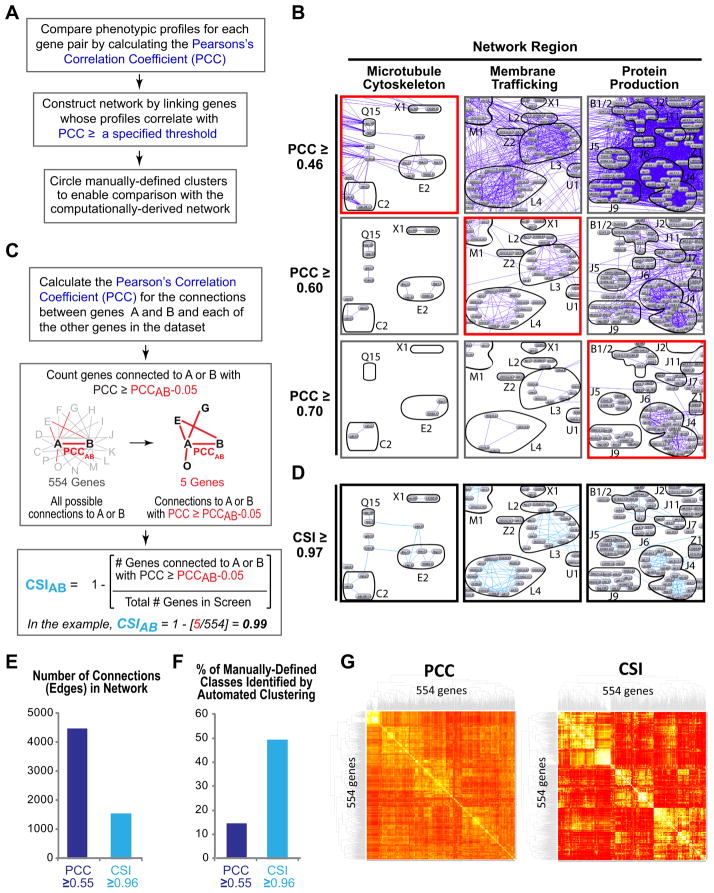
Our initial efforts using automated clustering were unable to partition genes at a resolution comparable to what could be achieved by an experienced investigator. To circumvent this limitation, we used the manually-defined classes as a tool to develop a robust computational method for constructing gene networks based on high-content parameter profiles. We first compared the phenotypic profiles by calculating the Pearson’s Correlation Coefficient (PCC) for each pair of genes. The resulting network was visualized using N-Browse, an interactive Java-based tool (Kao and Gunsalus, 2008), to display connections between genes whose profiles were correlated with a PCC greater than or equal to a specified threshold (Fig. 4A,B; dark blue lines connecting grey gene nodes). To assess the effectiveness of this approach, we circled gene clusters that corresponded to our manually-defined classes. This approach revealed that the optimal PCC threshold for viewing functionally relevant connections (red outlined boxes in Fig. 4B) varied substantially between different network neighborhoods. This variability was due to the varying nature of the profiled phenotypes and the extent to which they are captured by the parameter set, the extent to which scored parameters are related versus independent, and the fact that profiles with more features often exhibit more variance. Viewing the entire network at a single PCC threshold is not possible because for some regions the threshold is too low and the view is cluttered with non-specific connections (Fig 4B, images to the right of the red boxed images), and for other regions the threshold is too high, yielding an empty network in which many meaningful connections are absent (Fig. 4B, images to the left of red boxed images).
Figure 4. Constructing gene networks using Connection Specificity Index (CSI) instead of the PCC reduces connection noise and allows connections of similar functional significance to be viewed across the entire network at a uniform threshold.
A. Flowchart of the steps used to construct the gene networks in B. B. Gene networks were constructed by displaying connections (dark blue lines) between genes whose knockdown profiles were correlated with a PCC ≥ three specified thresholds (0.46, top; 0.60, middle; 0.70, bottom). Each column shows a network region, labeled based on the primary function of the genes in that region. To compare the computational network to the manually-defined phenotypic classes, gene groups from manually-assigned classes were circled and labeled. The optimal PCC threshold at which significant connections were displayed and non-specific connections were filtered out (red boxes) was different for different network regions. C. Method used to calculate the CSI. D. Gene network showing the same regions in B constructed by displaying connections (light blue lines) with a CSI ≥ 0.97. E. Bar graph showing the number of connections in gene networks constructed using PCC or CSI thresholds of 0.55 or 0.96, respectively. These thresholds were chosen because they are the highest thresholds that retain most of the genes in the network; genes drop out of the network when they no longer make any connections with a PCC/CSI that exceeds the specified threshold. F. Bar graph showing the percent of the 49 manually-defined phenotypic classes containing 4 or more genes identified by an automated clustering algorithm (MINE) in networks constructed using PCC or CSI thresholds of 0.55 or 0.96, respectively. G. Heatmap dendograms of the sterile gene set constructed based on the PCC or the CSI. See also Figure S3 and Table S4.
To circumvent the limitations of PCC-based analysis, we developed a measure that ranks the significance of functional links between genes based on network context. For each pair of genes A and B, we assigned a Connection Specificity Index (CSI) by: 1) calculating the PCC for the connections between A and B and each of the other genes in the dataset, 2) counting the number of genes connected to A or B with PCC ≥ PCCAB μ 0.05 (i.e. at a level comparable to or better than the correlation between A and B, correlations with a PCC up to 0.05 less than PCCAB were considered similar—this offset was determined empirically); 3) dividing this number by the total number of genes in the screen (554); and 4) subtracting the result from 1.0 (Fig. 4C). The CSI is equivalent to the fraction of genes in the dataset whose profiles are less similar to those of A and B than the profile of A is to the profile of B. For example, a CSI of 0.97 means that the similarity between A and B is highly specific: only ~3% of gene knockdowns have profiles with comparable or higher similarity to either A or B. Since the CSI scales uniformly with functional significance across the entire network, connections of a similar level of significance can simultaneously be displayed at a single CSI threshold (Fig. 4D).
We evaluated the performance of CSI and PCC threshold networks by comparing the ability of an automated clustering algorithm (MINE, Module Identification in NEtworks; K. Rhrissorrakrai and K.C. Gunsalus, in press) to identify gene clusters corresponding to our manually-defined classes. This analysis revealed that over the useful range of the two parameters, networks generated using CSI thresholds have ~3-fold fewer connections than comparable PCC networks; this noise reduction translates into a substantial improvement in the ability of an automated algorithm to identify functionally relevant gene clusters (Fig. 4E,F; Fig. S3). Approximately half of the manually-defined classes containing 4 or more genes could be identified by automated clustering in a network constructed using the single CSI threshold of 0.96, whereas only 14% could be identified in a comparable PCC network (Fig. 4F). When MINE was allowed to search networks spanning a range of CSI thresholds (0.90 to 0.99), clusters corresponding to ~90% of the manually-defined classes could be identified, compared to 65% for networks spanning a range of PCC thresholds (0.5–1). Heat map dendograms also revealed more sharply defined clusters when using the CSI, indicating that the CSI increases network clarity (Fig. 4G; Table S4).
We conclude that constructing networks using CSI thresholds circumvents the variability in the significance of a strict correlation measure in different network regions that arises due to the complexity of high-content screening parameters. Using the CSI instead of the PCC reduces connection noise and allows connections of a similar functional significance to be viewed across the entire network at a single threshold.
Varying the CSI Threshold Reveals Functional Modularity in the Gene Network at Different Levels of Resolution
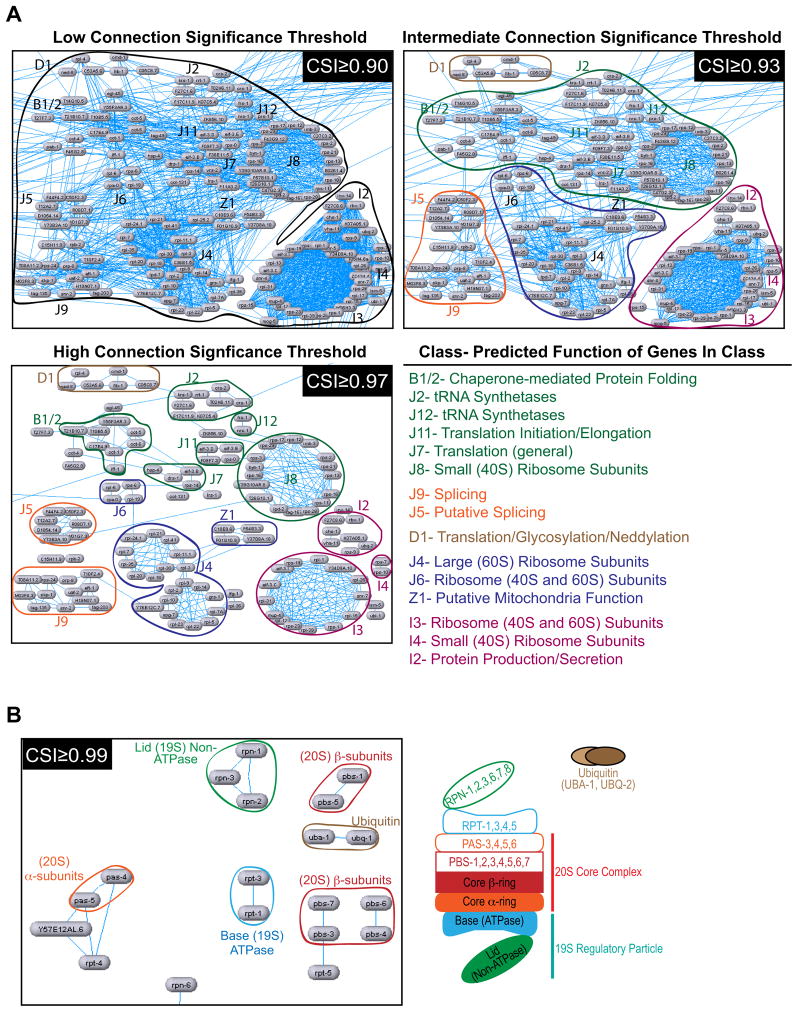
The CSI-based network representation allows exploration of functional modularity at different levels of resolution. This point is illustrated by the region of the gene network involved in protein production (Fig. 5A): a relatively low threshold CSI of 0.90 connects the entire set of genes involved in protein translation, mRNA splicing, and protein folding in a dense meshwork. An intermediate CSI threshold of 0.93 results in a sparser network of more specific connections that defines smaller gene groups. At a high CSI threshold of 0.97, the chaperonins, tRNA synthetases, translation initiation/elongation factors, small ribosome subunits, large ribosome subunits, and splicing factors are resolved into separate clusters. Thus, dialing the CSI up or down reveals functional relationships at different levels of resolution. The automated clusters identified by MINE at three different CSI thresholds are provided in the supplement (Table S5; note that genes can be in multiple clusters).
Figure 5. Varying the CSI threshold reveals functional modularity in the gene network at different levels of resolution.
A. The region of the gene network involved in protein production is shown using three different CSI thresholds to filter displayed connections. The gene clusters apparent at each threshold are circled and gene groups from manually-defined classes are labeled. B. The region of the network involved in protein degradation is shown using the very high CSI threshold of 0.99. The connections that remain link components within specific proteasome subcomplexes (illustrated schematically on the right). See also Figure S4 and Table S5.
We assessed the resolution limit of the gonad architecture assay using the region of the network representing genes involved in protein degradation (Fig. 5B, Fig. S4). Above the very high CSI threshold of 0.99, the connections that remained linked genes within specific proteasome subcomplexes (the Lid, the core β-ring, the core α-ring or the 19S ATPase base; Fig. 5B). We conclude that phenotypic profiling based on complex tissue architecture, coupled with automated construction of CSI-based networks from parameter profiles, is capable of correctly assigning very fine distinctions in protein function.
CSI-Enabled Integration of High-Content Data Sets Generates a Global View of the C. elegans Essential Gene Network
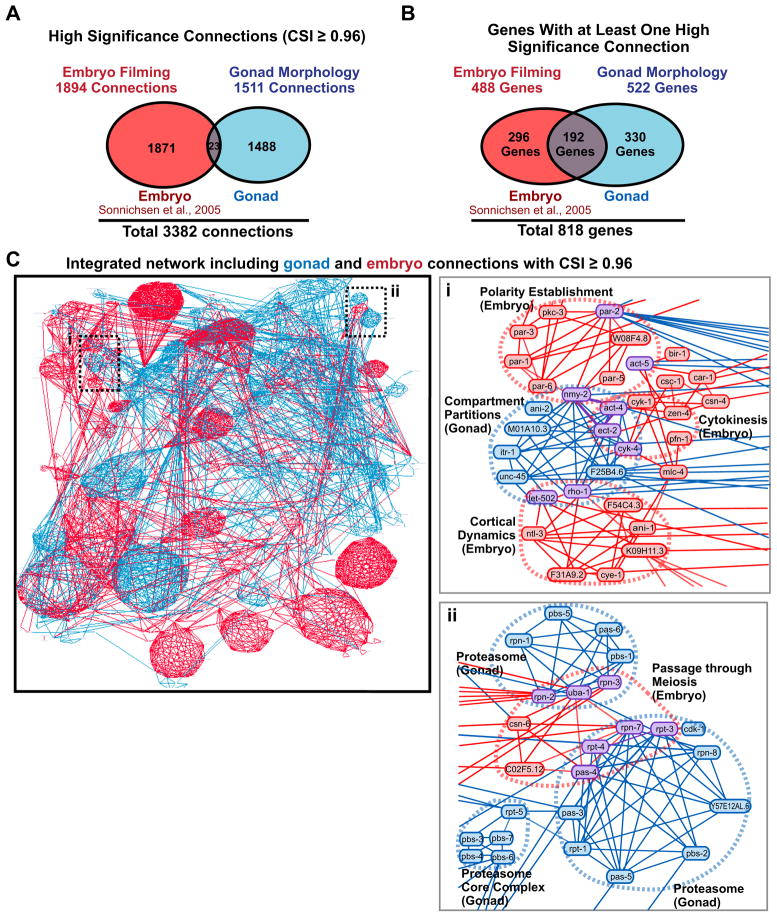
Together, the gonad architecture (554 genes) and time-lapse embryo filming (661 genes) screens provide high-content profiles for 885 essential genes (330 were profiled in both screens). The CSI is ideally suited for integrating these datasets because it filters out low specificity phenotypic links, leading to a network that combines only the significant relationships identified by the two screens. We binarized the phenotypic signatures in the embryo timelapse screen, which were based on scoring for 45 possible defects (Sönnichsen et al., 2005), calculated a CSI for each gene pair, and simultaneously displayed both data sets in N-Browse to create an integrated network. The merged network, or network regions centered on genes of interest, can be viewed at any CSI threshold in N-Browse (instructions and a demo video describing how to use N-Browse can be accessed at http://worm.mpi-cbg.de/phenobank_gonad/nbrowse). At a CSI threshold of 0.96, the integrated network has 3382 high significance connections linking 818 genes (Fig. 6A–C). The combined view provides functional information for ~90% of the genes required for embryo production or the first two embryonic divisions. Despite the presence of 330 genes in common, the relationships identified by the two datasets were highly orthogonal only—23 of the total 3382 phenotypic links are shared (Fig. 6A). This lack of overlap is explained by the majority of the 330 common genes failing to make high-significance connections in the embryo time-lapse screen due to sterility onset (Fig. S5). Overall, the CSI-based network integrates information acquired in the biologically distinct contexts of the gonad and embryo to provide a multi-layered view of the function of 818 genes required for essential cellular processes in a multicellular organism.
Figure 6. The CSI enables integration of high-content data sets to generate a global view of the essential C. elegans gene network.
A. Venn diagram showing the high significance connections identified by the embryo-filming (Sönnichsen et al., 2005) and gonad architecture data. B. Venn diagram showing the genes that make at least one high significance connection identified by each dataset. C. Bird’s eye view of the integrated network combining high-significance connections based on the gonad (blue) and embryo (red) data. Insets (i–ii) highlight regions where the gonad and embryo data intersect (connections identified by both datasets are purple). See also Figure S5.
DISCUSSION
High-content screening is the primary method for mapping functional gene networks in animal cells (Collins et al., 2009; Conrad and Gerlich, 2010; Gunsalus, 2008). A number of high-content screens have classified genes based on cell morphology following gene knockdown (examples include Bakal et al., 2007; Echard et al., 2004; Eggert et al., 2004; Goshima et al., 2007; Liu et al., 2009; Neumann et al., 2010; Piano et al., 2002; Sönnichsen et al., 2005). While successful in identifying genes that contribute to cell division and morphology, the ability of these screens to assess protein function across a breadth of cellular processes falls far short of what has been attained using genetic interaction profiling in fungi. Screen resolution—the ability to discriminate between different biological functions—depends on the information content of the phenotypic assay; therefore, one approach to enhance resolution has been time-lapse imaging (Conrad and Gerlich, 2010). Here, we explored an alternative approach—analyzing the effects of gene knockdowns on complex tissue architecture at a single timepoint in a multicellular organism. We monitored C. elegans gonad architecture following gene knockdown by scoring 94 phenotypic features. Our results demonstrate that the biological complexity of the gonad translates into phenotypes that enable profiling across a wide spectrum of essential cellular processes at a resolution approaching that of genetic interaction profiling in yeasts.
The Sterile Gene Collection: A Prominent Gap in the Functional Genomic Analysis of the C. elegans Essential Gene Set
Sequencing of the C. elegans genome and the discovery of RNAi catalyzed efforts to systematically catalog the functions of its predicted genes (Piano et al., 2006). C. elegans has ~900 essential genes required for embryo production and/or events during the first two embryonic divisions. A subset of these genes was previously characterized by time-lapse imaging of early embryos (Fraser et al., 2000; Gönczy et al., 2000; Piano et al., 2002; Sönnichsen et al., 2005). However, sterility onset following RNAi was a major complication that precluded in-depth characterization of a large number of genes. Our efforts focused on these 554 sterile genes, which we profiled by imaging gonad architecture following RNAi knockdowns and generating a parameterized description of the resulting phenotypes.
Our analysis generated functional predictions for 106 of the 116 uncharacterized sterile genes. These predictions span a variety of cellular processes including membrane trafficking, glycosylation, fatty acid synthesis, cortical dynamics, chromosome structure/segregation, RNA splicing, mitochondrial function, microtubule cytoskeleton, proteasome function, translation, MAPK signaling, transcription, RNA binding, and the metaphase-anaphase transition. We validated these predictions for 8 genes that play important roles in the microtubule cytoskeleton, MAP kinase signaling, the metaphase-anaphase transition, cortical remodeling/cytokinesis, and membrane trafficking. Although the gonad was used as a “test tube” to functionally profile these genes, subsequent work in the early and later stage embryos indicates that the majority of the essential genes in this collection are broadly important. Of the 554 sterile genes, ~60% have predicted homologs in higher organisms, and more than 50% of the 106 uncharacterized genes have a predicted homolog or conserved domain. Thus, in addition to filling a large gap in the analysis of the C. elegans essential gene set, our findings provide a starting point to address the functions of related genes in humans.
High-Resolution Phenotypic Profiling Based on the Morphology of a Complex Tissue
While the original motivation for our screen was to profile the sterile gene set, a fortuitous byproduct was the realization that a complex tissue, whose structure depends on the dynamic coordination of a broad spectrum of cellular processes, is an ideal substrate for phenotypic profiling. A major finding of our work is that profiles based on complex tissue architecture acquired at a single time point following gene knockdown have greater information depth than profiles derived from timelapse imaging of the first two divisions of the early embryo (a single cell). For example, as shown in Figure 5, the gonad data partitions the set of genes involved in protein production into 15 different classes (chaperones, ribosome components, tRNA synthetases, proteins involved in protein folding, splicing components etc.) whereas none of these distinctions could be made based on embryo filming data. This high level of resolution generally holds throughout the dataset.
We note that the means of inferring function in a single point morphology assay is distinct from that used in the embryo-filming screen. In the embryo screen, gene function was inferred directly from phenotype. For example, defects in spindle assembly, chromosome segregation, or polarity establishment led to assignment to classes implicated in the corresponding processes (Sönnichsen et al., 2005). By contrast, the link between cataloged phenotypic features in the gonad architecture assay and gene function is not direct—we do not know why knockdown of genes with specific functions lead to a specific spectrum of phenotypic features. Instead, protein function is inferred by comparing the profile of phenotypic features to those of other genes in the dataset, a process conceptually analogous to how function is inferred from genetic interaction profiles in budding yeast.
The presence of proteasome components in our dataset offered an opportunity to compare the resolution of our approach to genetic interaction profiling in yeast. The proteasome is a 2.5-MDa macro-molecular machine that contains over 30 different subunits organized into a 20S core complex, composed of α and β rings, and a 19S regulatory particle, containing base and lid sub-complexes (Bochtler et al., 1999). In yeast, distinct genetic interaction profiles were obtained for the subunits of the four proteasome sub-complexes using a sensitive competitive growth assay (Breslow et al., 2008). At a CSI ≤ 0.99 (a high-stringency filter), the connections that remained were between subunits of specific proteasome subcomplexes (α ring, β ring, lid and base; Fig. 5B). This observation indicates phenotypic profiles based on gonad morphology provide resolution sufficient to group subunits of specific proteasome subcomplexes.
At the core of our dataset are profiles composed of parameters visually scored by experienced investigators rather than acquired through automated image analysis. Given the complexity of the substrate—a 3 dimensional tissue in a living organism that can be variably positioned in the worm—and the large spectrum of knockdown phenotypes that can entirely change the properties of the structure (gonad size, shape, position, compartment and nuclei number and morphology) automated parameter scoring would have been exceedingly difficult. However, the exact properties that make automated analysis difficult—the varied and dramatic effects that gene knockdowns can have on gonad architecture—are also the properties that give the assay its profiling power. Although manual parameter scoring could introduce some bias, this was minimized by performing the analysis blinded to gene identity and the by fact that the individual parameters were scored by investigators who were largely oblivious to the larger patterns that would ultimately emerge.
A Robust Computational Method for Constructing Gene Networks From High-Content Screening Data
By evaluating computational methods for network construction using a validated manual classification of a rich phenotypic dataset, we were able to devise a robust computational method for constructing gene networks from high-content phenotypic profiles. This method overcomes two challenges encountered in network analysis of high-content datasets. The first challenge is that the level of profile correlation that is significant varies in different network regions due to the varying nature of the profiled phenotypes and the extent to which they are captured by the parameter set. The second challenge is that commonly encountered (and thus less informative) phenotypes can generate a “hairball” of connections that obscures meaningful functional links. At the center of the method we developed to overcome these challenges is a simple metric—the CSI, which is a network context-dependent measure that ranks the significance of functional links between genes. Compared to the Pearson’s Correlation Coefficient, constructing networks based on the CSI reduces non-specific connection noise, improves network clarity, and allows connections of a similar functional significance to be simultaneously viewed across the entire network at a single threshold.
Ranking connection significance allows exploration of the gene network at different levels of functional resolution and integration of high-content screening data from different sources. We demonstrate the usefulness of the CSI by using it to integrate the high-content data from our gonad architecture screen with that from the prior embryo-filming screen, to generate an integrated network that provides a multi-layered view of 818 genes in the C. elegans essential gene set. The phenotypic profiles in these datasets are composed of parameters that were visually scored rather than measured through automated image analysis. However, constructing networks based on phenotypic profiles faces the same challenges, regardless of whether parameters are scored through manual or automated means. Consequently, we anticipate that the CSI-based method described here will be of equal utility in analyzing and integrating datasets composed of parameters acquired through automated analysis.
EXPERIMENTAL PROCEDURES
C. elegans Strains
Strains are listed in Table S7. The strains OD95, UD299, DP38, OD70, MSN142, NL2098, and BS3623 were previously described (Arur et al., 2009; Essex et al., 2009, Fridolfsson and Star, 2010; Kachur et al., 2008; Maduro and Pilgrim, 1995; Shi et al., 2010; Sijen et al., 2001). OD447 was generated by using a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad Laboratories; Praitis et al., 2001) to bombard a construct containing the C08C3.4 genomic locus cloned into the SpeI site of pIC26 (Cheeseman et al., 2004) into DP38. OD449 was generated by mating OD447 with OD70.
RNA Production
Templates for dsRNA production were generated by using primers with tails containing the T3 and T7 promoters to amplify to amplify a 500–1000 bp region of the corresponding gene from genomic DNA. When possible, the oligo pairs used by Sönnichsen et al. (2005) were chosen. New oligos were designed for genes not in the Sönnichsen screen and when the Sönnichsen oligo pairs amplified introns or regions smaller or larger than 500–1000 bp (oligos are listed in Table S2). PCR reactions were cleaned (PerfectPrep 96 kit; Eppendorf) and used as templates for T3 and T7 transcription reactions (Megascript T3 and T7 kits, Ambion). Transcription reactions were mixed, cleaned (MegaClear96 kit, Ambion), and annealed by adding 3x Soaking buffer (32.7 mM Na2HPO4, 16.5 mM KH2PO4, 6.3 mM NaCl, 14.1 mM NH4Cl) to a final concentration of 1X and incubating the reactions at 68°C for 10 minutes followed by 37°C for 30 minutes.
RNA Interference
Larval (L4 stage) worms from the strain OD95 were rinsed with M9 and soaked in 5 μl of dsRNA (supplemented with 0.5 μl of a 50/50 mixture of 63 mM spermidine and 1.1 % gelatin) for 24 hrs at 20 °C inside a humid chamber. Worms were transferred to NGM plates seeded with OP50 E. coli and allowed to recover at 20°C for 48 hrs prior to imaging.
Live Imaging
Six worms were anesthetized in a fresh mixture of 1 mg/ml Tricane (ethyl 3-aminobenzoate methanesulfonate salt) and 0.1 mg/ml of tetramisole hydrochloride (TMHC) dissolved in M9 for 15–30 minutes before transferring them to an agarose pad under a coverslip for imaging. Gonads were imaged by collecting an 80 x 0.5μm z-series including DIC, GFP and RFP images for every z-plane. Microscopy was performed with a spinning disk confocal mounted on a Nikon TE2000-E inverted microscope equipped with a 60x 1.4 NA PlanApochromat lens, a krypton-argon 2.5 W water-cooled laser (Spectra-Physics, Mountain View, CA) and a Hamamatsu Orca ER CCD camera. Acquisition parameters, shutters, and focus were controlled by MetaMorph software (Molecular Devices, Downington, PA).
Sterility Assessment
At the time when the other worms in the cohort were anesthetized and imaged, 3 worms were moved to a fresh plate. After 24 hours at 20°C, the number of embryos and L1 progeny on the plate was counted. A gene was “sterile” if no embryos or L1 larvae were observed, “partially sterile” if <50 embryos/L1 worms were present, and “wild-type” if >50 embryos/L1 worms were present. After an additional 24 hours, we assessed whether the knockdown resulted in embryonic lethality. If >50 L1-L2 larvae were present, they were considered “viable”, if <50 L1-L2 larvae were present and there were >5 dead embryos, it was considered “embryonic lethal”.
Antibody Production
Antibodies against K10D2.4 were generated by using the oligos cgcgcgggatccgctttgatgtacccattcca and gcgcgcgaattctcaaggatttgcaggcatattt to amplify a region encoding amino acids 2–81 from a cDNA library. The product was digested with Bam HI/EcoRI and cloned into pGEX6P-1 digested with the same enzymes (GE Healthcare Life Sciences). The purified GST fusion protein was outsourced for injection into rabbits (Covance). Antibodies were affinity purified by binding to columns of the same antigen after removal of the GST tag as described previously (Desai et al., 2003).
Immunoprecipitation and Mass Spectrometry
K10D2.4 was immunoprecipiated from extracts prepared from frozen worm pellets (Desai et al., 2003) and mass spectrometry was performed (Cheeseman et al., 2004) as described, using the most recent version of the predicted C. elegans proteins (Wormprep111).
Hypodermal Cell Experiments, MAP Kinase Experiments, and Membrane Trafficking Assay
For Figure 2C, embryos produced by adult hermaphrodites from the strain UD299 were imaged as described (Fridolfson and Starr, 2010), 20 hours after injection of dsRNA against T09E8.1 into their gonad. Nuclear migration was scored in L1 larvae using DIC optics as described (Starr et al., 2001). The attenuated RNAi, gonad dissection, immunofluorescence, and gonad analysis in Figure 2E and F (Arur et al., 2009) and the GFP-SNB-1 trafficking assay (Shi et al., 2010) were performed as described.
Phenobank and N-Browse
Movies can be accessed via the Phenobank website (http://worm.mpi-cbg.de/phenobank_gonad) or through RNAiDB (http://www.rnai.org). Written instructions and a demo video explaining how to search Phenobank can be found at http://worm.mpi-cbg.de/phenobank_gonad/project. Written instructions and a demo video describing how to visualize the gene network in N-Browse2 can be found at http://worm.mpi-cbg.de/phenobank_gonad/nbrowse. As described in the instructions, gonad and embryo data can be viewed in N-Browse2 by entering the URL http://gnetbrowse.org/gonad.jnlp into your browser and providing the password “phenotypes” at the prompt. Additional information about N-Browse, including system requirements and tutorials, can be found at http://www.gnetbrowse.org.
Automated clustering using MINE
MINE (Module Identification in Networks) is a graph clustering algorithm that shows comparable or better performance compared with similar algorithms in identifying functional modules in dense interaction networks (K. Rhrissorrakrai and K.C. Gunsalus, in press). MINE, which is similar to MCODE (Bader and Hogue, 2003) but uses a modified weighting scheme and takes into account network modularity, is available as a Cytoscape plug-in from http://www.cytoscape.org or as a Perl package (upon request from the authors).
Supplementary Material
Acknowledgments
This work was supported by grants from the American Cancer Society (PF-06-254-01-CCG, to RG), the Helen Hay Whitney Foundation (to AA), and the NIH (R01 GM074207 to KO, R01 HD046236, to KCG and FP; R01 GM085503, to KCG; R01 GM085150 to TS; R01 GM088151 to AA) and by funding from the Ludwig Institute for Cancer Research to KO and AD. We thank Kahn Rhrissorrakrai for the MINE algorithm, Michael Volkmer for movie formatting, and the Caenorhabditis Genetics Center, funded by the NIH National Center for Research Resources, for strains.
Footnotes
Author Contributions: Conceived and designed the experiments: KO, AD, RG, AA. Performed the experiments: RG, AA, SN, SA, TS, SW, HF, DS, JM. Scored the data: AA, RG, KO. Manually partitioned the genes into classes: RG, KO. Reworked Phenobank to include the gonad morphology data: SS, RG, KO, AH. Developed N-Browse: HK, MS, FP, KG. Developed CSI: KO, RG, HK, MS, FP, KG. Conceptualized and performed network analysis: HK, FP, KG. Contributed reagents/materials/analysis tools: KO, RG, AA, SN, KL, SA, TS, AD, HK, MS, KG, FP, HF, DS.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc Natl Acad Sci U S A. 2009;106:4776–4781. doi: 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu Rev Biophys Biomol Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods. 2008;5:711–718. doi: 10.1038/nmeth.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Weissman JS, Krogan NJ. From information to knowledge: new technologies for defining gene function. Nat Methods. 2009;6:721–723. doi: 10.1038/nmeth1009-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C, Gerlich DW. Automated microscopy for high-content RNAi screening. J Cell Biol. 2010;188:453–461. doi: 10.1083/jcb.200910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A, Hickson GR, Foley E, O’Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essex A, Dammermann A, Lewellyn L, Oegema K, Desai A. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell. 2009;20:1252–1267. doi: 10.1091/mbc.E08-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus KC. A Caenorhabditis elegans genetic-interaction map wiggles into view. J Biol. 2008;7:8. doi: 10.1186/jbiol70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus KC, Ge H, Schetter AJ, Goldberg DS, Han JD, Hao T, Berriz GF, Bertin N, Huang J, Chuang LS, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic Analysis of Human Protein Complexes Identifies Chromosome Segregation Proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur TM, Audhya A, Pilgrim DB. UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev. Biol. 2008;314:287–299. doi: 10.1016/j.ydbio.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL WormBook. Germline proliferation and its control. The C. elegans Research Community, WormBook. 2005 August 15; doi: 10.1895/wormbook.1.13.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Kops GJ, Voet MV, Manak MS, van Osch MH, Naini SM, Brear A, McLeod IX, Hentschel DM, Yates JR, 3rd, van den Heuvel S, et al. APC16 is a conserved subunit of the anaphase-promoting complex/cyclosome. J Cell Sci. 2010;123:1623–1633. doi: 10.1242/jcs.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Ohmachi M, Arur S, Nayak S, Francis R, Church D, Lambie E, Schedl T. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics. 2007;177:2039–2062. doi: 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Sims D, Baum B. Parallel RNAi screens across different cell lines identify generic and cell type-specific regulators of actin organization and cell morphology. Genome Biol. 2009;10:R26. doi: 10.1186/gb-2009-10-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathey-Prevot B, Perrimon N. Drosophila genome-wide RNAi screens: are they delivering the promise? Cold Spring Harb Symp Quant Biol. 2006;71:141–148. doi: 10.1101/sqb.2006.71.027. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Hyman AA WormBook. Cell division. The C. elegans Research Community, WormBook. 2006 January 19; doi: 10.1895/wormbook.1.72.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Piano F, Gunsalus KC, Hill DE, Vidal M. WormBook. 2006. C. elegans network biology: a beginning; pp. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol. 2000;10:1619–1622. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Morton DG, Gunsalus KC, Reinke V, Kim SK, Kemphues KJ. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr Biol. 2002;12:1959–1964. doi: 10.1016/s0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhrissorrakrai K, Gunsalus KC. MINE: Module Identification in NEtworks. BMC Bioinformatics. doi: 10.1186/1471-2105-12-192. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shi A, Chen CC, Banerjee R, Glodowski D, Audhya A, Rongo C, Grant BD. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol. Biol. Cell. 2010;21:2930–2943. doi: 10.1091/mbc.E10-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol. 2009;186:229–241. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperlen P, Fraser AG, Kamath RS, Martinez-Campos M, Ahringer J. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 2001;20:3984–3992. doi: 10.1093/emboj/20.15.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.