Abstract
RecA is a key protein in homologous recombination. During recombination, one single-stranded DNA (ssDNA) bound to site I in RecA exchanges Watson–Crick pairing with a sequence-matched ssDNA that was part of a double-stranded DNA molecule (dsDNA) bound to site II in RecA. After strand exchange, heteroduplex dsDNA is bound to site I. In vivo, direct polymerization of RecA on dsDNA through site I does not occur, though it does in vitro. The mechanisms underlying the difference have been unclear. We use single-molecule experiments to decouple the two steps involved in polymerization: nucleation and elongation. We find that elongation is governed by a fundamental clock that is insensitive to force and RecA concentration from 0.2 and 6 µM, though rates depend on ionic conditions. Thus, we can probe nucleation site stability by creating nucleation sites at high force and then measuring elongation as a function of applied force. We find that in the presence of ATP hydrolysis a minimum force is required for polymerization. The minimum force decreases with increasing RecA or ATP concentrations. We propose that force reduces the off-rate for nucleation site binding and that nucleation site stability is the stringency factor that prevents in vivo polymerization.
INTRODUCTION
During homologous recombination initiated by double-strand breaks (DSBs) or single-strand gaps, one DNA strand makes contact with RecA protein in order to form a protein/DNA filament where single-stranded DNA (ssDNA) is bound to the primary site of RecA (site I) (1–4). The filament rapidly scans for a homologous double-stranded DNA (dsDNA) partner that eventually binds to the secondary site of RecA (site II) to allow for strand exchange to take place. During this step, one strand of the duplex DNA in site II unbinds from its partner and pairs instead with the ssDNA in site I thus leaving ssDNA in site II and the newly formed heteroduplex dsDNA in site I. In the absence of hydrolysis, the heteroduplex dsDNA remains bound to site I in RecA, but in the presence of hydrolysis the RecA unbinds after a time of the order of a minute as the strand exchange window proceeds along the dsDNA (5). In vivo, RecA does not polymerize directly on dsDNA (Bishop,D.K., personal communication), whereas in vitro experiments have shown that dsDNA can polymerize directly on dsDNA under certain experimental conditions (6,7). In this article, we will study the factors that allow for RecA polymerization on dsDNA with particular focus on the role of tension exerted on the dsDNA substrate. Given that forces are known to play a role in vivo (8), these results will shed light on the stringency factors that prevent direct polymerization of RecA on dsDNA inside the cell.
The binding of RecA protein to dsDNA has been described as a multi-step process that depends on pH, salts, nucleotide co-factor, temperature and DNA topology and length (6,7). A proposed binding mechanism is characterized by an initial pre-nucleation step that is concentration dependent followed by a slow nucleation step, and subsequent fast polymerization (6,7). Recent visualization of single filaments have provided evidence that 4–5 RecA molecules are required to form a stable nucleation site that can extend into the growth phase (9). Given that nucleation is strongly dependent on solution variables, it has been proposed that nucleation is the limiting step for RecA assembly in vivo (6).
In vitro experiments have shown direct RecA polymerization on dsDNA in the absence of any external force (6,7); however, several experiments demonstrated that force or torque applied to the ends of the dsDNA can play a key role in RecA polymerization (10–12). Some experiments did not separate nucleation and elongation. One experiment measured the force-induced onset of nucleation by probing the lag time between the application of force and the appearance of polymerization, which cannot occur in the absence of a stable nucleation site. Those experiments showed that the lag time between the application of an external force to the ends of the dsDNA and the onset of RecA polymerization decreases with increasing force (11) suggesting that force enhances the rate at which stable nucleation sites are produced; however, the effect of force on the stability of the nucleation sites or the elongation rates was not considered.
Other experiments considered the force dependence of elongation, without explicitly considering nucleation. Such experiments measured elongation rate as a function of force. Those studies showed that the elongation rate is insensitive to force: no change in average growth rates for forces between 0.5 and 9 pN was observed (13) and force changes from 65 to 20 pN resulted in an average growth rate reduction of ∼50% (10). Thus, earlier single-molecule work suggested that force influences the nucleation step more strongly than the elongation step, though force was observed to increase the total polymerization rate in a manner that was not in agreement with the accompanying theory (10). Bulk work had suggested that more than one step might be involved in the creation of stable nucleation sites (6,7); therefore, in what follows, we will try to determine whether applied force plays multiple roles in nucleation and elongation where we consider the possibility that nucleation might include more than one step.
Applying an external force to the ends of a naked dsDNA molecule can affect the dsDNA in several ways including the following: (i) increasing the average extension/base pair; (ii) increasing the actual spacing between phosphates in the backbone; (iii) decreasing the entropy of dsDNA; (iv) decreasing the hydrogen bonding between base pairs; (v) increasing the internal tension in dsDNA; (vi) changing the conformation of dsDNA, including ‘overstretching’ and/or ‘melting’ (14–20). In addition, if an external force is applied to a dsDNA molecule, then for proteins whose binding extends dsDNA, the free-energy changes and thermal fluctuations that normally occur when the protein unbinds are altered. This mechanism was proposed as an explanation for experiments using Rad51, a eukaryotic homolog of RecA (21). In those experiments, direct polymerization of Rad51 on dsDNA under tension took place in a solution in which free Rad51 was present and hydrolysis was suppressed. Flow was used to change to a solution in which hydrolysis was not suppressed and no free Rad51 was present, though tension on the dsDNA was maintained. Without free Rad51, polymerization is not possible, but hydrolysis driven depolymerization can occur. The experiments showed that the depolymerization rate for Rad51 increases if a force that is applied to the ends of the dsDNA decreases (21). It was suggested that depolymerization may originate at the ends of the filament; however, it was not clear whether the ends where depolymerization occurred corresponded to nucleation sites. Given the many effects of force on dsDNA, it is not trivial to determine which effects enhance direct RecA polymerization on dsDNA.
In this work, we will suggest that all of the in vitro results as well as the in vivo results can be unified by describing RecA polymerization as a process that will occur as long as there is stable nucleation, where nucleation stability requires that RecA molecules in the active conformation remain bound at the ends of the RecA filament. We will show that the growth rate/nucleation site occurs at a fundamental clock rate that is insensitive to applied force and RecA concentration, though increasing the applied force can create additional nucleation sites and possibly also allow the binding of higher order RecA oligomers from solution. The latter two effects can result in dramatic force-dependent increases in the total average elongation rate even though the clock rate is force insensitive.
If, as suggested above, the elongation rate is insensitive to both force and RecA concentration, then elongation should be observed whenever a stable nucleation site is formed as long as the force and RecA concentration are in the range over which the elongation rate is insensitive to them. Thus, within this range of conditions, the absence of polymerization would indicate the absence of stable nucleation sites. Under conditions where the elongation rate is insensitive to force and RecA concentration, in buffers where RecA hydrolyses ATP, we observe that there is a RecA concentration-dependent lower bound on the force at which polymerization takes place, even in molecules where we have created a nucleation site that has shown polymerization at higher forces. This result suggests that nucleation is not stable at low forces. In what follows we will present results that indicate that external force stabilizes the nucleation site binding, where the force required to stabilize the binding increases with decreasing RecA or ATP concentration.
MATERIALS AND METHODS
Bulk fluorescence experiments
RecA protein labeled with Alexa 568 dye (Invitrogen) was prepared following a standard protocol included in the labeling kit, with the following modification: 50 μl of 2 mg/ml RecA dialyzed overnight in PBS were reacted with 1/500 of the provided solid dye dissolved in 100 mM sodium bicarbonate. A solution containing 2 ng/ml lambda-phage dsDNA (annealed and ligated to 25-mer oligonucleotides containing 12 complementary bases and a stretch of 13 bases with several biotin labels that yielded 3′5′ biotinylated ssDNA tails), 1 µM fluorescently labeled RecA, and 1 mM ATP or ATPγS in RecA buffer was incubated at 37°C for several hours. Aliquots were taken and mixed with YOYO-1 final concentration 0.1 µM and diluted 1:5 in RecA buffer, and immediately placed on a glass slide for observation. Images were taken on a Nikon Eclipse TE2000-U inverted microscope with a Plan Apo TIRF 60×-oil objective, Roper Scientific Coolsnap HQ2 Firewire digital camera, and G-2E/C TRITC (red) and FITC HYQ (green) fluorescence filter sets. Red-emission channel images were taken with a 10 s exposure time, followed by green-emission images, with a 1–4 s exposure time, where the exposure times were chosen to approximately equate the total signal intensities from both channels. Intensities in each channel were then thresholded and contrast adjusted to reveal overlapping regions of green and red signal. At least five independent images were taken for each sample.
Single-molecule experiments
In single-molecule experiments, we used the 3'5'-biotin-labeled dsDNA to attach to extravidin molecules adsorbed on a glass surface and superparamagnetic beads (4.5 µm in diameter, Invitrogen). The ssDNA tails allow free rotation of the bonds in the phosphate backbone so no steady-state torque can be maintained in the dsDNA molecules. After an aliquot of dsDNA in RecA buffer (70 mM Tris–HCl, 10 mM MgCl2 and 5 mM dithiothreitol, pH 7.6) was mixed with RecA (New England Biolabs), ATP or ATPγS, and the beads, it was placed in a square micro-cell with cross-section of 0.8 mm × 0.8 mm containing a round inner capillary, 0.55 mm in diameter and closed at its ends. When necessary, MgCl2 was replaced by CaCl2; the regeneration system contained 10 U/ml of pyruvate kinase (Sigma-Aldrich), 2 mM phosphoenolpyruvate and 1 mM KCl. The inner capillary was modified by adsorption of 1 mg/ml extravidin in PBS (phosphate-buffered saline) pH 7.4 overnight at room temperature. After an initial incubation of 10 min, the DNA molecules became tethered between the glass capillary surface and the extravidin-coated beads. The micro-cell was then placed in a magnetic tweezers apparatus consisting of a stack of permanent magnets held at a variable distance from the sample. The 2–200 pN force on the beads was controlled by the distance between the magnets and the micro-cell. The position of each bead was tracked by bead-tracking software and recorded as described in previous work (22). The observation time was limited to 1 h.
Automated calculation of the slopes of bead position as a function of time was performed using custom-written scripts running under Matlab software (The Mathworks). The algorithm assumes that when the applied force is constant, the extension increases linearly with time; however, one single growth rate does not characterize the entire observation time. We find that the growth is piecewise linear, with abrupt stochastic changes in growth rate separating one linear growth region from the next. Thus, we model the extension as a function of time as a continuous set of linear regions. Linear least-squares fitting is performed on all possible lines that are at least 2 s long. The line with the minimum absolute value of the sum of its residuals per unit length multiplied by the sum of the autocorrelation of its residuals is chosen as the best fit line. The algorithm continues with a new starting point at the time point where the previous fit ended, until the entire curve is exhausted. The algorithm is allowed to skip short regions (<1 s) of high noise at the transitions between regions with different slope. Because the precise transition point may not be known, the fitter allows a 0.5 s overlap between bordering sloping regions. For histograms, sloped regions that remain at the same slope for longer than 20 s and that occur more than 10 s after a change in force are retained in the analysis.
RESULTS
Observation of nucleation
In this work, we have studied how externally applied tension affects RecA polymerization on dsDNA under various experimental conditions in order to separate the effect of tension on nucleation from the effect of tension on growth and to shed light on some of the discrepancies found in previous reports on the influence of tension on RecA polymerization. We performed two types of experiments: (i) bulk experiments, where the DNA is not under any external tension, were performed using fluorescently labeled RecA; and (ii) single-molecule experiments, where the dsDNA is under external tension were performed using unlabeled RecA.
In the bulk experiments, the ionic conditions and the temperature of the sample could easily be varied and observation times in excess of 24 h were readily obtained. In some cases, full filament formation was observed in the absence of tension; however, even under the most favorable conditions full filament formation required >3 h.
In single-molecule experiments, external tension was applied using magnetic tweezers and the observation time was limited to ∼1 h. In agreement with the bulk results, no filament formation was observed during the 1-h observation time if the tension on the filament was maintained at a low constant value as long as the DNA had not been incubated with ligands that intercalate between bases. This lack of growth could be due to a failure of nucleation or of polymerization. In order to separate the two effects, we induced nucleation by briefly applying high forces up to ∼75 pN for about 1 min which is short enough to create one or two nucleation sites. If polymerization is observed, we lower the force. After lowering the force, we observe strong polymerization at forces where no polymerization was observed after an hour in samples that had never been exposed to high forces. These results suggest that the long lag time required to observe growth at low forces is a consequence of the time required for nucleation.
Control of ATP hydrolysis
In the absence of ATP hydrolysis, dsDNA bound to site I in RecA does not unbind even in the absence of force (6); however, in the presence of hydrolysis unbinding can occur (12). In order to simplify our interpretation of the experimental results, we will first consider interactions between dsDNA and free RecA in the presence of ATPγS, a poorly hydrolysable analog of ATP, so that the results will not include significant unbinding. We will then consider the analogous results in solutions containing ATP with MgCl2, where both binding and unbinding can occur and ATP with CaCl2 where the ATPase activity of RecA is known to be negligible.
Polymerization in the absence of external tension
A solution containing lambda-phage dsDNA, fluorescently labeled RecA and 1 mM ATP or ATPγS in RecA buffer was incubated at 37°C for several hours; after adding YOYO, the samples were immediately observed under a fluorescence microscope. Images were taken after excitation with green light to excite Alexa 568-labeled RecA and blue light to excite YOYO-labeled DNA. The color channels were then merged and analyzed for coincidence of dsDNA and RecA molecules, which appear yellow in the images. We found that our samples consistently produced either positive (coincident colors) or negative (no coincident colors) results, demonstrating that the presence of coincident fluorescence can readily be used as an indicator of RecA binding and polymerization at zero-force.
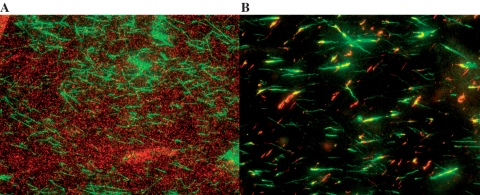
Figure 1A and B show the results obtained in RecA buffer after a 4-h incubation period in ATPγS and a 24-h incubation period in ATP, respectively. In ATPγS, no polymerization was observed after 3 h; however, after 4 h RecA covers approximately one-fourth of the molecule. This result is consistent with earlier results that have shown that nucleation is the rate limiting step and once nucleation takes place elongation proceeds rapidly. If RecA polymerized over approximately one-fourth of the lambda-phage molecule in 1 h, and each nucleation site grew at a constant rate, then the observed growth rate would be of the order of 2 µ/h/N = 0.5 nm/s/N, where N is the number of nucleation sites, or 1 RecA molecule/s if N = 1, consistent with previously observed growth rates (9,10,12) as well as the results presented in this work (see below). We note that ATPγS is not present in vivo, so these results do not imply that RecA can directly polymerize under in vivo conditions.
Figure 1.
Fluorescent images of samples incubated in the presence of 1 µM-labeled RecA for 4 h at 37°C and 0.1 µM YOYO for 2 min. (A) 1 mM ATP; (B) 1 mM ATPγS.
Consistent with in vivo results, we find that in the absence of external tension in 10 mM MgCl2, 1 mM ATP and 1 µM RecA at pH 7.6, RecA does not polymerize on intact dsDNA even after 24 h at 37°C. Similarly, if the ATP concentration is increased by 10-fold no polymerization is observed after 24 h.
Polymerization in the presence of external tension
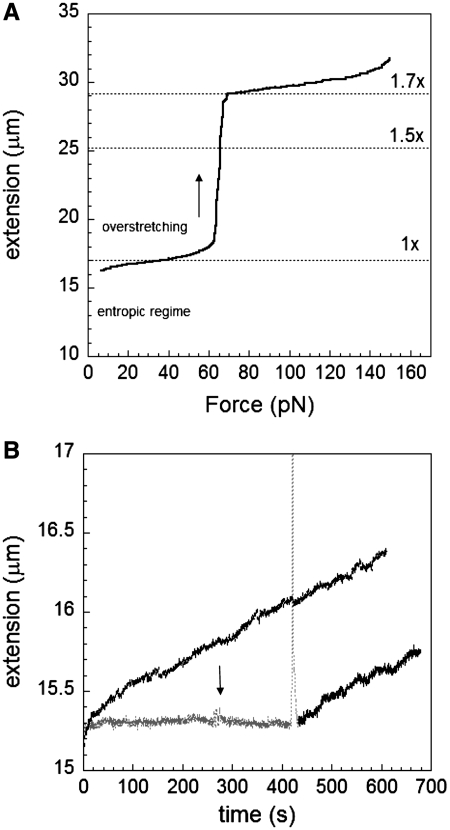
In the absence of RecA, only very low forces (∼0.5 pN) are required to extend linear dsDNA out of its natural random coil configuration (the ‘entropic regime’). No significant increase in DNA length occurs for forces from 2 to 55 pN as shown in Figure 2A. At forces below 60 pN, most of the structure is believed to remain in the B-form configuration. Starting at around 60 pN, dsDNA undergoes an overstretching transition in which this structural change results in a length increase to almost 1.7× that of B-DNA. If the force is kept constant at a value significantly below the overstretching transition, then in the absence of RecA the bead-to-surface distance remains constant as a function of time. In contrast, if the same experiment is done in the presence of RecA, then we observe an increase in the bead-to-surface distance as a function of time due to 1.5× elongation of DNA upon RecA binding (4,23). Thus, if the constant applied force is well below the overstretching transition, then we can monitor RecA binding by measuring the bead-to-surface distance as a function of time. The number of RecA monomers bound to a dsDNA molecule under force can be determined by measuring the change in DNA extension versus time and using the additional fact that there is one RecA per three bases and bases are extended on average by about 1.5 times (4), yielding an increase of about 0.5 nm per bound RecA.
Figure 2.
Nucleation through overstretching. (A) Overstretching curve for lambda-phage dsDNA. (B) Absence of growth without overstretching and constant force of 50 pN (black curve); however, a fast increase in force through the overstretching transition (sharp peak) can start RecA polymerization showing linear growth at 50 pN on dsDNA at pH 7.6, 10 mM MgCl2, 1 mM ATP and 1 µM RecA (gray and black curve). A rapid change in force between 47 and 55 pN is indicated by the arrow.
It is possible to trigger nucleation by applying mechanical force to the dsDNA substrate, as demonstrated by previous work (10–12). A typical experiment begins with an overstretching cycle during which the force is increased from 2 to 75 pN, and then reduced back to some value, Fc, that is then held constant for a time ΔT. During ΔT, the extension of each dsDNA molecule is measured. Figure 2B shows a molecule that displays no growth when a constant force of 50 pN is applied (gray trace), suggesting that no nucleation sites are present. Even when the force is oscillated around 50 pN (arrow) no growth is observed; however after a 12-s interval during which the force was increased to 75 pN and quickly decreased back to 50 pN (peak), the molecule length as a function of time clearly increases while the force is held fixed at 50 pN. This result demonstrates that overstretching can induce nucleation, and in what follows we use this technique for generating nucleation sites to investigate the force dependence of the elongation rate once at least one nucleation site has been established.
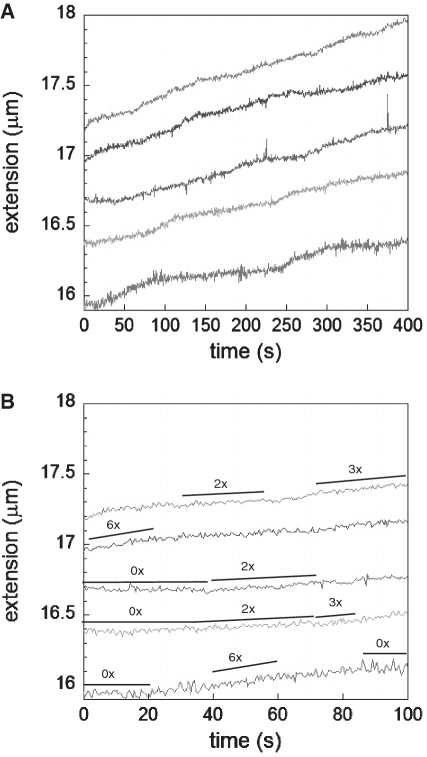
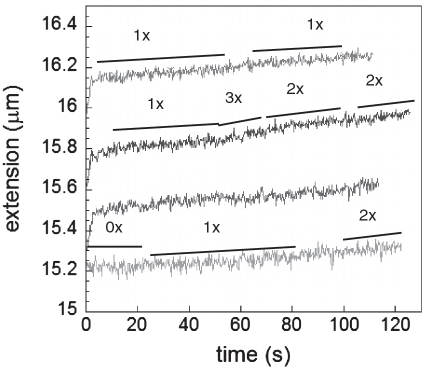
Figure 3A shows plots of extension as a function of time for molecules subjected to a series of constant forces after they are initially overstretched to produce nucleation sites for RecA binding in ATPγS. Between 10 and 50 pN RecA polymerizes at the same average rate suggesting that once nucleation takes place, extension is not strongly affected by force; however, it has been shown that nucleation itself is force dependent and can be favored by going to forces above 60 pN where nucleation and extension occur simultaneously. In addition, it should be noted that over long time intervals the elongation is not constant but characterized by growth periods alternated by pauses or growth periods with different slopes. A more detailed analysis of a narrower time window shows these pauses and changes in slope for different molecules and forces where some characteristic elongation rates are shown next to the curves (Figure 3B, black lines). The changes in slope may indicate a change in the number of nucleation sites participating in the polymerization process or a change in the growth rate/nucleation site. For cases where the previous slope was zero, a change in slope either indicates the growth rate for a single nucleation site or it shows that the growth between different nucleation sites is coordinated and the change indicates the sum of the changes for the individual sites.
Figure 3.
Extension versus time for RecA growth on dsDNA at pH 7.6, 10 mM MgCl2 and 1 mM ATPgS at 10 pN, 20 pN, 30 pN, 40 pN and 50 pN (bottom curve to top curve, respectively). (A) Full-time range. (B) Replotting the first 100 s where a few growth rates that are twice, three and six times the average rate, are indicated with the black lines next to each curve. In addition, periods where no growth is observed are indicated as 0×.
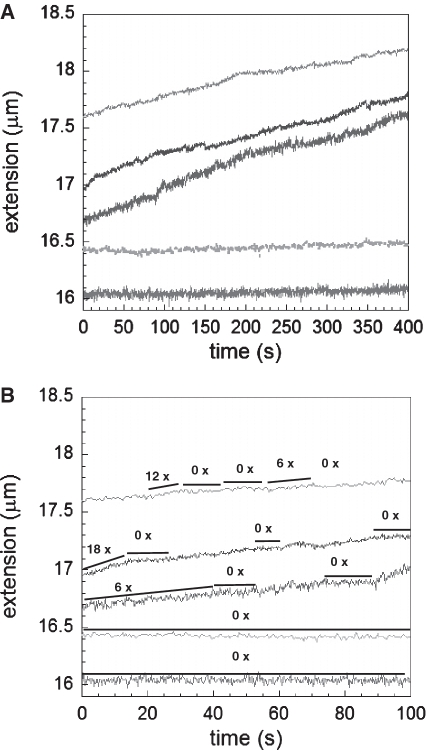
Additional experiments were performed in the presence of ATP and MgCl2. Polymerization results in ATP may be harder to interpret than similar results in ATPγS since in ATP both polymerization and depolymerization are possible (12); however, earlier work demonstrated that if linear growth was observed when a constant force was applied to dsDNA, then depolymerization played a negligible role in the absence of substantial concentrations of ADP (12). Previous experiments have also shown that the affinity of RecA for dsDNA strongly depends on the nucleotide cofactor (9,24) which affects the nucleation step whereas the growth rates in solutions containing ATP are similar to the growth rates in solutions containing ATPγS (9), which is consistent with the stringency governing direct RecA polymerization on dsDNA being determined by nucleation.
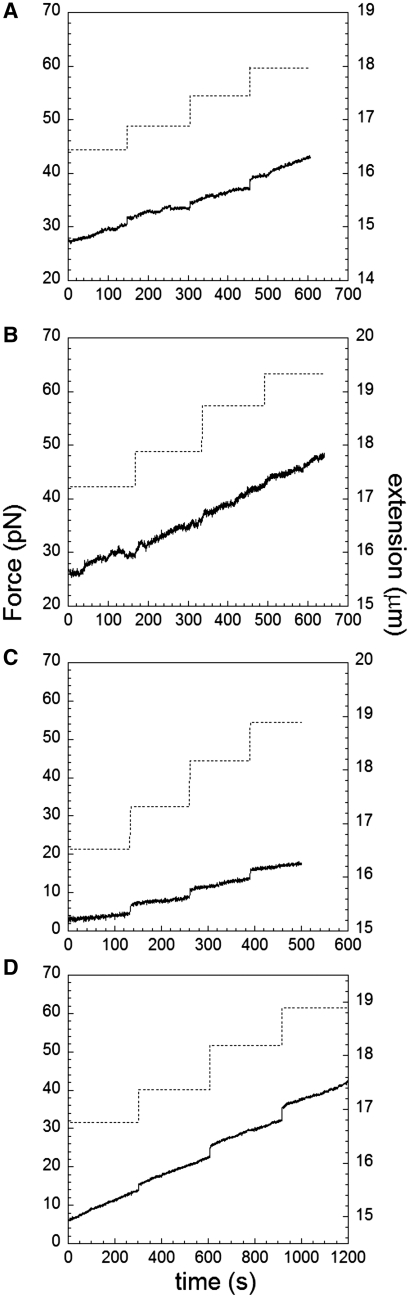
At forces around 60 pN, both nucleation and elongation take place simultaneously typically exhibiting increases in extension with time that are not always clearly linear. At forces below 20 pN, we do not observe polymerization in the standard RecA buffer containing ATP at 25°C. As discussed above, we create nucleation sites by briefly applying high forces and then lowering the force to a value that is held constant for some time. For forces between 30 and 50 pN RecA polymerizes with piecewise linear rates independent of force as can be seen in Figure 4A and B. Within large periods of time the elongation exhibits some changes in slope and occasionally small pauses yielding a lower average rate if slope changes are neglected. Some typical slopes are indicated next to the curves (black lines). We note that previous work in ATP observed characteristic growth rates of 10 nm/s (13) and 6 nm/s where the latter was attributed to growth starting from the two single-stranded ends of lambda-phage dsDNA yielding 3 nm/s per site (12). Similarly, Ref. (9) reported average growth rates between 3 and 10 nm/s in ATP and a distribution of rates of 3.6 ± 1.1 nm/s measured from individual nucleation sites (n = 34) in ATPγS.
Figure 4.
Extension versus time for RecA growth on dsDNA at pH 7.6, 10 mM MgCl2, and 1 mM ATP at 10 pN, 30 pN, 40 pN and 50 pN (bottom curve to top curve, respectively). (A) Full-time range. (B) Replotting the first 100 s where a few growth rates that are several times higher than the minimum rate are indicated with the black lines next to each curve. A few lines labeled as 0× show absence of growth at the lowest forces or pauses at higher forces.
In the standard RecA buffer at 25°C, after initial nucleation at high forces, we observed growth down to 5 pN in ATPγS, close to the 2 pN lower limit of the force calibration in our apparatus. Forces as low as 10 pN are enough to achieve growth rates of ∼3 nm/s in ATPγS, whereas growth in 1 mM ATP and MgCl2 requires forces of about 30 pN or higher after nucleation. The observation that the growth rates in ATP and ATPγS are similar is consistent with earlier results, and supports the contention that depolymerization does not play a significant role in the force range where linear growth is observed.
ATP hydrolysis can be also suppressed if MgCl2 is replaced by CaCl2. As shown in Figure 5, in the presence of CaCl2 polymerization takes place at low forces similar to the result obtained in ATPγS. Interestingly, the elongation rates in CaCl2 after nucleation: 0.3, 0.7 and 1 nm/s (represented by black lines labeled 1×, 2× and 3×, respectively), are different from those obtained in the presence of MgCl2; however, these values change in integer units and again support the idea that there is either a change in nucleation sites or a change in the fundamental RecA growth rates. All the curves shown in the figure correspond to the extension versus time for the same molecule. The force was increased in successive steps from 21 to 54 pN, but the elongation rate remained the same. Similar results were obtained for individual single molecules in buffers containing ATP or ATPγS and MgCl2.
Figure 5.
RecA polymerization in the absence of hydrolysis or negligible hydrolysis: elongation of one single dsDNA molecule in 1 mM ATP and 10 mM CaCl2, pH 7.6 at 21 pN, 32 pN, 44 pN and 54 pN (bottom curve to top curve, respectively).
The insensitivity of the growth rate to the applied force is dramatically illustrated in Figure 6, which shows the extension versus time curves for typical individual single molecules stretched by a series of constant applied forces that are increased with time. Results are shown for four different buffer conditions: ATP/MgCl2, ATP and a regeneration system, ATPγS and ATP/CaCl2. Figure 6 also explores the effect of ATP hydrolysis since it includes results in two conditions (ATPγS and ATP/ CaCl2) where little or no ATP hydrolysis occurs, as well as results in the presence of an ATP regeneration system that rapidly converts ADP back into ATP (6). Figure 6A and B demonstrate that during the initial RecA polymerization, ATP hydrolysis does not play a relevant role and growth rates with and without the ATP regeneration system are indeed similar; however, the force at which the extension starts to decrease is lower in the presence of the regeneration system in agreement with the results obtained at higher ATP concentrations (see below).
Figure 6.
Changes in force and measured extension versus time for typical single molecules. (A) 1 mM ATP, 10 mM MgCl2, pH 7.6. (B) Same as (a) in the presence of 10 U/ml pyruvate kinase, 2 mM phosphoenolpyruvate and 1 mM KCl; (C) 1 mM ATPγS, 10 mM MgCl2, pH 7.6; (D) 1 mM ATP and 10 mM CaCl2, pH 7.6.
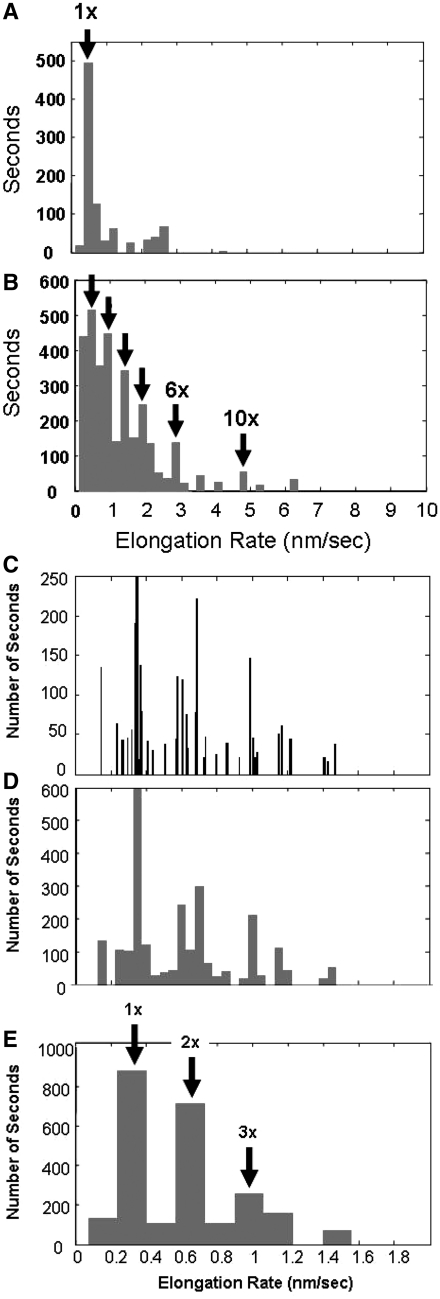
In order to characterize the rates of RecA polymerization on dsDNA, the data obtained for 21 single-molecule extension curves were analyzed; Figure 7A shows a histogram of the slopes of the extension versus time for ATPγS data at 20–40 pN, at times between 10 and 400 s at constant force, where the slopes were determined using an automated algorithm. Figure 7B shows the same curves at all times after 1000 s at constant force. The distribution shows a peak at 0.48 ± 0.05 nm/s = 1×. This rate probably corresponds to the characteristic growth rate/nucleation site for a RecA binding in its monomeric form.
Figure 7.
Histogrammed slopes of extension versus time obtained for several experiments where the elongation at constant forces between 20 and 40 pN was measured in 1 µM RecA, 1 mM ATPγS and 10 mM MgCl2 at pH 7.6. (A) Slopes at times >1000 s at constant force, where the dominant peak at 0.48 nm/s (1×) differs from the average by 2 SDs. (B) Slopes at times 10–400 s at constant force, where arrows mark slopes at 1×, 2×, 3×, 4×, 6× and 10× integer multiples of the dominant rate. (C) Slopes of extension versus time measured in 1 mM ATP and 10 mM CaCl2, showing the 1×, 2× and 3× multiples of the dominant rate of 0.33 nm/s, 0.010 nm/s bin size; (D) 0.050 nm/s bin size; (E) 0.165 nm/s bin size, where the arrows show the integer multiples of the 1× rate.
Figure 7C–E show a histogram of the slope distribution for all ATP and CaCl2 data at 20–40 pN, at 10–400 s and at constant force as determined by the same automated algorithm used for the data in Figure 7A. The bin widths in the histograms are 0.010 nm/s, 0.050 nm/s and 0.165 nm/s, respectively. The last bin size was chosen to represent a half integer multiple of the 1× growth rate. The three quantized peaks are prominent for all histogram bin widths, indicating that they are not an artifact of the binning. A measure of the statistical significance of this result can be obtained by calculating the difference between the total number of points in the integer bins minus the total number of points in the half integer bins and dividing the result by the square root of the total number of points. The result is 23, indicating that the quantization is highly statistically significant. The growth rate histogram shows a very strong peak at ∼0.33 ± 0.05 nm/s, which we believe corresponds to monomeric growth at the clock rate. Additional peaks are also seen at 2 × 0.33 nm/s and 3 × 0.33 nm/s. These peaks could correspond to multiple nucleation sites growing at the monomeric rate.
In general, the elongation rates decrease at long times, possibly because nucleation sites collide or reach the end of the molecule. Long-term growth rates are often dominated by monomeric growth of a single nucleation site. At earlier times we observed much larger elongation rates as shown in Figures 3B and 4B. The same figures also show large sudden changes in elongation rates. These large slope changes could represent simultaneous changes in the growth rates of many nucleation sites; however, they are often separated by regions of zero slope where all elongation ceases. Thus, if they represent growth from many nucleation sites, there must be a mechanism that allows the growth from different sites to couple so that they all start or stop simultaneously. Another plausible explanation for such large changes in elongation rates is that the RecA is binding in a multimeric form, where the clock rate for the multimer binding is the same as the clock rate for the monomer. We note that if RecA hexamers bind at the clock rate shown in this paper, then the growth rate/nucleation site would be 2.9 ± 0.1 nm/s, which is consistent with the rate/nucleation site observed in Ref (9).
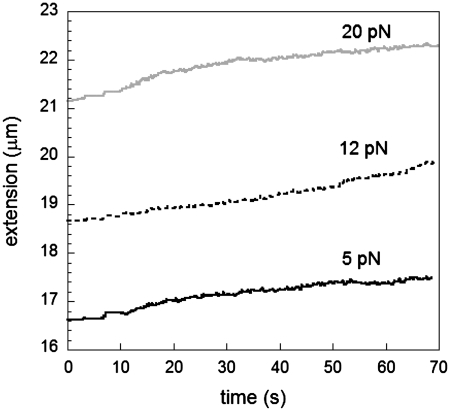
In all the results presented above, growth was initiated by applying high forces during an overstretching cycle; however, previous work has shown that RecA polymerization can take place at 6 pN on lambda phage under significantly different experimental conditions such as pH 6.8, 37°C and 10 µM RecA (12). In our current experimental conditions, an increase in stability of bound RecA was achieved by adding an intercalator such as YOYO that changes the elongation of dsDNA to a significant extent (25), thus allowing for growth at forces as low as about 5 pN in ATP at pH 7.6, as can be seen in Figure 8. The characteristic growth rates in this case are significantly higher and above 10 nm/s suggesting that nucleation and growth occur simultaneously even at very low forces; therefore, we are unable to extract the elongation rate independently of the nucleation rate.
Figure 8.
Elongation versus time in the presence of an intercalator in 1 mM ATP, 10 mM MgCl2 and 0.1 µM YOYO, pH 7.6.
Analysis of the effect of force, RecA concentration and ATP concentration on the stability of the nucleation sites
As shown above, the elongation rate/nucleation site is insensitive to force and RecA concentration for a force range from 5 to 50 pN and a RecA concentration range from 0.2 to 6 µM. Thus, if a nucleation site is present we should observe elongation, which can be used to probe for the presence of nucleation sites. In order to investigate the stability of the nucleation sites, we performed experiments where we overstretched a dsDNA molecule to create nucleation sites and then lowered the force to ∼40 pN and observed elongation. We then lowered the force again. In the absence of hydrolysis, we continued to observe growth down to the lower force limit of our apparatus which is ∼ 2 pN. In the presence of hydrolysis, we found that there was a force, Fmin, below which we did not observe elongation. In fact, below that force we observed a net decrease in extension. Such a decrease in extension may be associated with the unbinding of RecA due to a finite koff such as that observed in previous work (26).
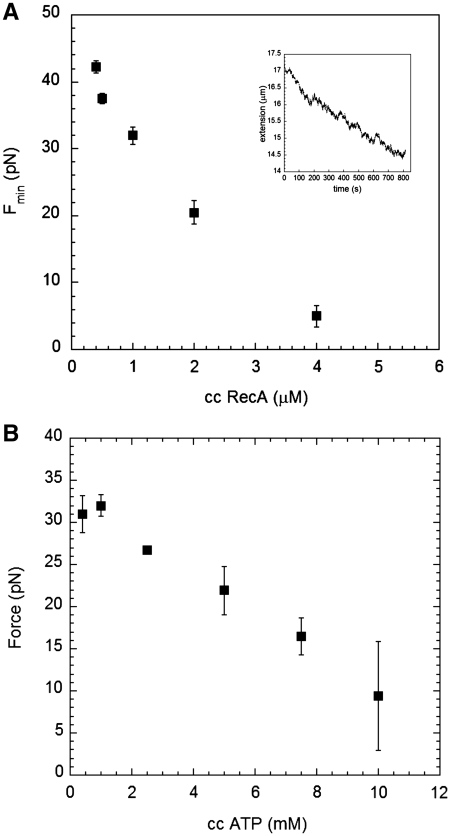
Figure 9A shows a plot of Fmin versus RecA concentration, where there is a clear decrease in Fmin as a function of RecA concentration at all RecA concentrations shown in the figure. Moreover, as observed in Rad51 experiments, changes in RecA filaments are characterized by bursts and pauses (21) in agreement with unbinding events of ATP monomers at a disassembly end (inset). Similarly, Figure 9B shows a plot of Fmin as a function of ATP concentration, where there is a clear decrease in Fmin with ATP concentrations above 1 mM. At lower concentrations, Fmin may become insensitive to ATP concentration.
Figure 9.
Minimum force required to observe a decrease in length versus time. (A) 1 mM ATP at pH 7.6 and several RecA concentrations; the inset shows the change in length after changing force to 5 pN in 4 µM RecA, 1 mM ATP and pH 7.6. (B) 1 µM RecA at pH 7.6, and several ATP concentrations.
DISCUSSION
We have studied different polymerization conditions for RecA on dsDNA. The direct polymerization of RecA onto dsDNA is not known to occur in vivo. It has been proposed that the nucleation step provides the stringency factor that prevents direct RecA polymerization on dsDNA in vivo (6); however, previous work had shown that direct polymerization can occur in vitro where changes in pH, salt concentration and composition, temperature, RecA concentration and DNA length can dramatically affect the nucleation step (6,7,11). More recent single-molecule experiments demonstrated that the mechanical distortion of the dsDNA substrate either by pulling or unwinding can induce polymerization, which requires nucleation (10–12). In the presence of intercalators that are known to increase the base pair separation and partially unwind the dsDNA, nucleation and growth can occur simultaneously at forces as low as 5 pN in ATP. Thus, nucleation can be rapidly induced at applied forces where the measured extension/bp is smaller than the average extension for B-form dsDNA due to the contribution of entropic coiling at such low forces.
If the creation of nucleation sites were simply due to sequence-independent thermally driven increases in length, then the number of nucleation sites should continue to increase as long as the high force is applied; however, in our experiments the number of nucleation sites saturates at longer times where the maximum number of nucleation sites is less than 10 even though lambda phage has ∼50 000 bp. Other works also showed fewer than 10 nucleation sites for lambda phage. (9,12), and there is some evidence that the nucleation sites occur preferentially in AT rich regions (9) suggesting that nucleation occurs at non-B-form conformations rather than thermally extended B-form DNA, where particular sequences may be much more susceptible to conversion to the alternate conformation. The presence of these alternate conformation regions is not sufficient to guarantee nucleation: it is also necessary that the on-rate for RecA binding to these regions dominates over the off-rate.
There is general agreement that each nucleation site requires 4–5 RecA molecules, whereas elongation has been suggested to be monomeric (27), bidirectional (9) and exhibits a characteristic rate of 3–10 nm/s (9,10,12). It is important to note that the work by Joo et al. (27) was done using very short DNA molecules. More recent work on RecA bound to long ssDNA has concluded that both nucleation and elongation involve multimeric forms of RecA (28) that are known to exist in solution (29,30).
In the absence of force at pH 7.6 and 10 mM MgCl2, we find that polymerization does not take place using ATP as nucleotide cofactor but it does occur if ATPγS is used instead and the sample is incubated for at least 3 h at 37°C which is in agreement with the fact that RecA does not bind to dsDNA in vivo. In our current conditions, which are comparable to in vivo conditions, the stringency of RecA polymerization on dsDNA is thus determined by nucleation.
In single-molecule experiments, we find that the growth rate/nucleation site occurs at a fundamental rate that is insensitive to the applied force and RecA concentration for a force range from 10 to 50 pN and a RecA concentration range from 0.2 to 6 µM. At low RecA concentrations (<0.5 µM), low forces (<40 pN) and long times after nucleation (>240 s), we observe that in buffers containing ATPγS or ATP and CaCl2, one growth rate dominates over all others. The dominant growth rate is 0.48 ± 0.05 nm/s in the presence of ATPγS, whereas in ATP and CaCl2 the dominant growth rate is 0.33 ± 0.05 nm/s. At high forces, high RecA concentrations, and short times after nucleation we observe changes in total polymerization rates (nucleation plus elongation).
Earlier work had suggested that during net polymerization there is no significant koff for RecA in regimes where linear growth is observed even in the presence of ATP hydrolysis (12). We believe that once growth is initiated at a nucleation site, the elongation process can proceed for long times (>1000 s) without any significant depolymerization, where the rate is independent of force at all applied forces where elongation is observed. In addition, we find that in the presence of an ATP regeneration system that can rapidly reconvert ADP into ATP, the growth rates are independent of force and equal to the growth rates measured in ATP and MgCl2. Thus, there is no evidence for a force dependent off-rate for the elongation process. Once depolymerization is triggered by lowering the force until the nucleation site becomes unstable, the situation becomes more complex and the depolymerization process may display a force dependent off-rate consistent with earlier results observed in Rad51 where the off-rate was studied in a regime where only depolymerization was present (21).
In ATP, we find that there is a minimum force, Fmin that decreases as RecA concentration increases, below which the RecA–dsDNA interaction is dominated by depolymerization. We believe this is the result of instability in the nucleation sites that unbind if the off-rate for RecA bound to the nucleation site exceeds the on-rate where the off-rate is decreased by increasing force and the on-rate is increased by increasing RecA concentration. Similarly, since RecA can only bind if it makes the hydrolysis driven transition from the active conformation with bound ATP to the inactive conformation with bound ADP, the stability of the nucleation sites might be enhanced by adding free ATP or an ATP regeneration system in solution so that the ADP can be replaced by ATP before the RecA unbinds. This prediction is consistent with our observation that Fmin decreases with increasing ATP concentration for concentrations above 1 mM, though it may become sensitive at lower concentrations where the characteristic time required to bind is longer than the time that the inactive RecA conformation remains bound to the dsDNA.
CONCLUSION
The direct polymerization of RecA onto dsDNA occurs at a characteristic rate/nucleation site that is insensitive to the applied force or RecA concentration, suggesting that it is a fundamental property of RecA interacting with dsDNA. This characteristic rate is not dependent on thermal fluctuations in dsDNA, on diffusion times for RecA, or on a force-dependent off-rate for elongation. Nucleation sites can be created by spontaneous thermal processes or by applied force, where these nucleation sites may represent non-B-form conformations to which RecA can bind; however, under ionic conditions that are similar to those found in vivo, the binding of RecA to these sites is not stable in the presence of ATP hydrolysis because the off-rate for RecA bound to the nucleation site exceeds the on-rate. Applying an external force reduces the off-rate for the nucleation sites, allowing stable nucleation which then leads to elongation. Similarly, the off-rate can be reduced by increasing RecA or ATP concentration or adding an ATP regenerator system. Thus, our results suggest that the stability of the binding of RecA to the nucleation sites is the stringency factor that prevents RecA from directly polymerizing on dsDNA in vivo.
FUNDING
Harvard University (to M.P.); National Institutes of Health (RO1-GM025326 and R01-GM044794 to N.K.). Funding for open access charge: Harvard University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 2.Cox MM. The bacterial RecA protein as a motor. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 3.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 5.van der Heijden T, Modesti M, Hage S, Kanaar R, Wyman C, Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol. Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Pugh BF, Cox MM. Stable binding of recA protein to duplex DNA. Unraveling a paradox. J. Biol. Chem. 1987;262:1326–1336. [PubMed] [Google Scholar]
- 7.Pugh BF, Cox MM. General mechanism for RecA protein binding to duplex DNA. J. Mol. Biol. 1988;203:479–493. doi: 10.1016/0022-2836(88)90014-9. [DOI] [PubMed] [Google Scholar]
- 8.Fisher JK, Ballenger M, O’Brien ET, Haase J, Superfine R, Bloom K. DNA relaxation dynamics as a probe for the intracellular environment. Proc. Natl Acad. Sci. USA. 2009;106:9250–9255. doi: 10.1073/pnas.0812723106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 10.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl Acad. Sci. USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leger JF, Robert J, Bourdieu L, Chatenay D, Marko JF. RecA binding to a single double-stranded DNA molecule: a possible role of DNA conformational fluctuations. Proc. Natl Acad. Sci. USA. 1998;95:12295–12299. doi: 10.1073/pnas.95.21.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shivashankar GV, Feingold M, Krichevsky O, Libchaber A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc. Natl Acad. Sci. USA. 1999;96:7916–7921. doi: 10.1073/pnas.96.14.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Heijden T, Van Noort J, Van Leest H, Kanaar R, Wyman C, Dekker N, Dekker C. Torque-limited RecA polymerization on dsDNA. Nucleic Acids Res. 2005;33:2099–2105. doi: 10.1093/nar/gki512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 15.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, Caron F. DNA: an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 16.Bensimon D, Simon AJ, Croquette V, Bensimon A. Stretching DNA with a receding meniscus: experiments and models. Phys. Rev. Lett. 1995;74:4754–4757. doi: 10.1103/PhysRevLett.74.4754. [DOI] [PubMed] [Google Scholar]
- 17.Schaumann HC, Rief M, Tolksdorf C, Gaub HE. Mechanical stability of single DNA molecules. Biophys. J. 2000;78:1997–2007. doi: 10.1016/S0006-3495(00)76747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocco S, Yan J, Léger JF, Chatenay D, Marko JF. Overstretching and force-driven strand separation of double-helix DNA. Phys. Rev. E. 2004;70:011910. doi: 10.1103/PhysRevE.70.011910. [DOI] [PubMed] [Google Scholar]
- 19.Hatch K, Danilowicz C, Coljee V, Prentiss M. Demonstration that the shear force required to separate short double-stranded DNA does not increase significantly with sequence length for sequences longer than 25 base pairs. Phys. Rev. E. 2008;78 doi: 10.1103/PhysRevE.78.011920. 011920 1–4. [DOI] [PubMed] [Google Scholar]
- 20.Danilowicz C, Limouse C, Hatch K, Conover A, Coljee VW, Kleckner N, Prentiss M. The structure of DNA overstretched from the 5′5′ ends differs from the structure of DNA overstretched from the 3′3′ ends. Proc. Natl Acad. Sci. USA. 2009;106:13196–13201. doi: 10.1073/pnas.0904729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJG, Wuite GJL. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danilowicz C, Coljee VW, Bouzigues C, Lubensky DK, Nelson DR, Prentiss M. DNA unzipped under a constant force exhibits multiple metastable intermediates. Proc. Natl Acad. Sci. USA. 2003;100:1694–1699. doi: 10.1073/pnas.262789199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stasiak A, DiCapua E. The helicity of DNA in complexes with RecA protein. Nature. 1982;229:185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 24.Zaitsev EN, Kowalczykowski SC. Binding of double-stranded DNA by Escherichia coli RecA protein monitored by a fluorescent dye displacement assay. Nucleic Acids Res. 1998;26:650–654. doi: 10.1093/nar/26.2.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sischka A, Toensing K, Eckel R, Wilking SD, Sewald N, Ros R, Anselmetti D. Molecular mechanisms and kinetics between DNA and DNA binding ligands. Biophys. J. 2005;88:404–411. doi: 10.1529/biophysj.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox JM, Tsodikov OV, Cox MM. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLOS Biol. 2005;3:231–243. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joo C, McKinney SA, Nakamura M, Rasnik I, Myon S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 28.van Loenhout MT, van der Heijden T, Kanaar R, Wyman C, Dekker C. Dynamics of RecA filaments on single-stranded DNA. Nucleic Acids Res. 2009;37:4089–4099. doi: 10.1093/nar/gkp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner SL, Zlotnick A, Griffith JD. RecA protein self-assembly. Multiple discrete aggregation states. J. Mol. Biol. 1988;204:959–972. doi: 10.1016/0022-2836(88)90055-1. [DOI] [PubMed] [Google Scholar]
- 30.Wilson DH, Benight AS. Kinetic analysis of the pre-equilibrium steps in the self-assembly of RecA protein form Escherichia coli. J. Biol. Chem. 1990;265:7351–7359. [PubMed] [Google Scholar]