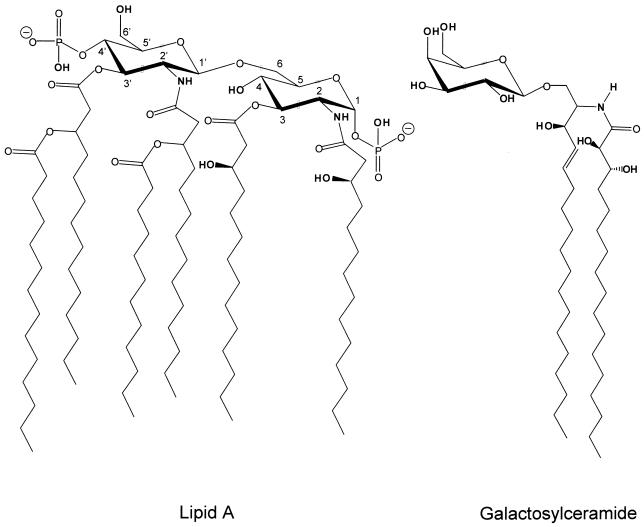
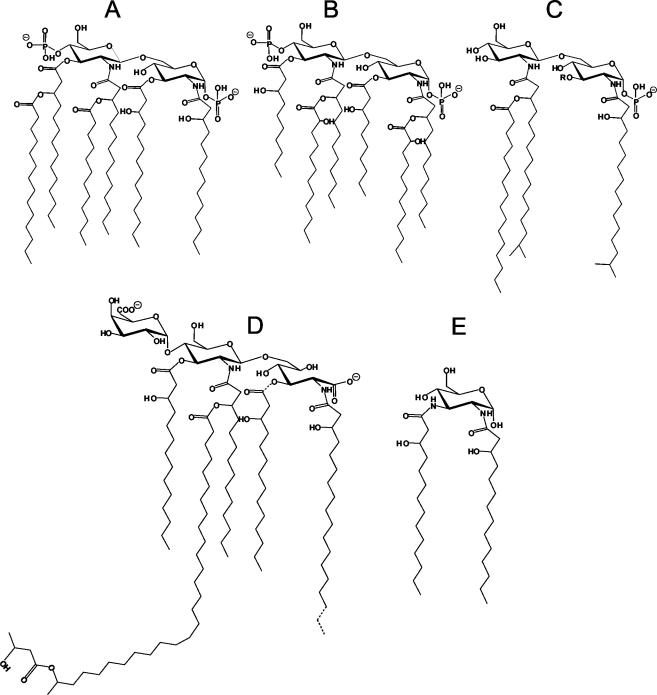
Abstract
Gram-negative bacteria characteristically are surrounded by an additional membrane layer, the outer membrane. Although outer membrane components often play important roles in the interaction of symbiotic or pathogenic bacteria with their host organisms, the major role of this membrane must usually be to serve as a permeability barrier to prevent the entry of noxious compounds and at the same time to allow the influx of nutrient molecules. This review summarizes the development in the field since our previous review (H. Nikaido and M. Vaara, Microbiol. Rev. 49:1-32, 1985) was published. With the discovery of protein channels, structural knowledge enables us to understand in molecular detail how porins, specific channels, TonB-linked receptors, and other proteins function. We are now beginning to see how the export of large proteins occurs across the outer membrane. With our knowledge of the lipopolysaccharide-phospholipid asymmetric bilayer of the outer membrane, we are finally beginning to understand how this bilayer can retard the entry of lipophilic compounds, owing to our increasing knowledge about the chemistry of lipopolysaccharide from diverse organisms and the way in which lipopolysaccharide structure is modified by environmental conditions.
INTRODUCTION
Gram-negative bacteria characteristically are surrounded by an outer membrane (OM). Analysis of prokaryote phylogeny using signature sequences in proteins even led to the notion that a major phylogenetic division exists between organisms with double-membrane envelopes (diderms) and those with only a simple cytoplasmic membrane (monoderms) (240). It is likely that the most important function for the additional membrane layer, the OM, in gram-negative bacteria is to serve as a selective permeation barrier. My earlier review of this subject, written with Martti Vaara, appeared in 1985 (464). It was an opportune time for such a review because the fundamental properties of the Escherichia coli porins and the specific maltodextrin channel, LamB, were characterized a few years earlier (see, for example, references 388 and 460-462) and the asymmetric structure of the OM bilayer, with lipopolysaccharides (LPS) located exclusively in the outer leaflet (316), offered an explanation for the unusually slow influx of lipophilic solutes. In the intervening 18 years, explosive progress has occurred in the field. Diffusion through the nonspecific porin channels and the specific channels are now understood in molecular detail, thanks to the elucidation of the crystal structure of these proteins. Channel proteins were identified in many nonenteric bacteria as a result of the rapid growth of our knowledge of the genome sequences, and in some cases this has offered insights into the physiology of various organisms in their native habitat. Studies of LPS not only showed us the impressive diversity of LPS structures in the bacterial kingdom but also led to the realization that the LPS structure may be modified in response to the conditions prevailing in the environment.
The present review attempts to summarize the development in the field since 1985. The major problem in presenting the results has been the information explosion. For example, more than 650 articles with the word “porin” in the title have been published during this period. If the databases are searched with “porin” or “lipopolysaccharide” as keywords, literally thousands of references are retrieved. Therefore, I had to be severely restrictive with citations in order to keep the review within a reasonable (and perhaps useful) size. I tried to limit the discussion strictly to selective permeability, eliminating most papers dealing with the biosynthesis and assembly of the OM or with the role of the OM in the interaction of bacteria with the environment, including higher animals and plants. I also apologize at the outset for the omission of many references which were not cited because the main message could be found in other articles or reviews or, probably most frequently, because of my oversight.
PROTEIN CHANNELS
OMs, like other biological membranes, are fundamentally built as a bilayer of lipids. As such, lipid bilayers show little permeability for hydrophilic solutes, including most nutrients. Therefore, they contain channel-forming proteins for the purpose of allowing the influx of nutrients and perhaps for the extrusion of waste products. One such nonspecific channel-forming protein, porin, was discovered in 1976 (438), and the word “porin” was proposed specifically for this class of proteins forming nonspecific diffusion channels. As predicted, porins were found in every species of gram-negative bacteria investigated and even in a group of “gram-positive” bacteria, the Corynebacterium-Nocardia-Mycobacterium complex, which produces a lipid-rich, bilayer-like “cell wall” (see below). (Interestingly, the Corynebacterium-Nocardia-Mycobacterium complex appears to be related to gram-negative bacteria on the basis of signature sequence analysis, since both of these groups have a 12-residue deletion in the S12 ribosomal protein, unlike archaebacteria and other gram-positive bacteria [240]).
At about the same time as bacterial porin was discovered, porins in mitochondria were discovered (584); this was followed a few years later by a study in my own laboratory using different approaches (773). The mitochondrial porins (voltage-dependent anion channels [VDAC]) are not discussed further, except to note that the channels are apparently large (up to 3 nm in diameter), as expected, because mitochondria have no need to exclude toxic molecules in the environment, and that the overall structure is thought to be similar to the β-barrel structure (to be discussed below) of bacterial porins. Many reviews of mitochondrial porins exist (see, for example, references 65 and 397). Another eukaryotic organelle in which porins were found is peroxisome (540-542).
The word “porin” has suffered from its popularity. As stated above, it was defined to mean only proteins that form nonspecific channels. Workers often concocted fanciful names like “maltoporin” for the LamB channel, which is specific, or “phosphoporin” for the Escherichia coli PhoE porin, which prefers anions in general and has no specificity for phosphate. These abuses of the term cause confusion and misunderstanding, and I present a plea, once again, to limit the word “porin” to references to nonspecific channels only.
Classical Porins
Families of classical porins.
The OM must allow transmembrane passage of nutrient molecules, which are usually small and hydrophilic. At the time of the first version of this review (464), E. coli was known to produce three trimeric porins (OmpF, OmpC, and PhoE). Since the studies of these porins formed the basis of our current knowledge of many other porins, they (and their homologs) are called “classical porins” in the present review. They show general preferences for charge and size of the solute, with OmpF and OmpC preferring cations slightly over anions and PhoE preferring anions and with OmpF allowing the permeation of slightly larger solutes than OmpC does. Furthermore, the diffusion rates of sugars of various sizes led to the estimate that the OmpF porin channel had a diameter of about 12 Å. Since that time, a great deal of progress has occurred in the field, some of which is described in minireviews (343, 453, 587, 598, 599).
X-ray crystallographic analysis showed that porins exist as transmembrane β-barrels (see below). However, crystal structures are available for only a few porins, and therefore it becomes highly desirable to derive as much information from primary sequence. Unfortunately, comparison of primary sequences of porins is extremely difficult. This is because the external loops between the transmembrane β-strands undergo very rapid mutational alterations as they interact with elements of the external world, such as antibodies, components of the innate immune system, bacteriocins, and phages. Because of this, simple alignment programs such as BLAST are “fooled” and create gaps at improper places. It is therefore important to first identify the transmembrane β-strands and compare different sequences solely on the basis of the sequences of these strands. However, none of the simple algorithms proposed for the detection of transmembrane strands work in a satisfactory manner, as mentioned by Ferenci (199). The proposal made by Ferenci was to look at the alignment of related porin sequences and assume that the less variant regions correspond to transmembrane strands. This works very nicely when many sequences from isolates of the same species, for example, exist (362, 482). However, in other cases one has to start from an alignment of sequences of distantly related porins, and this requires the prior identification of transmembrane strands.
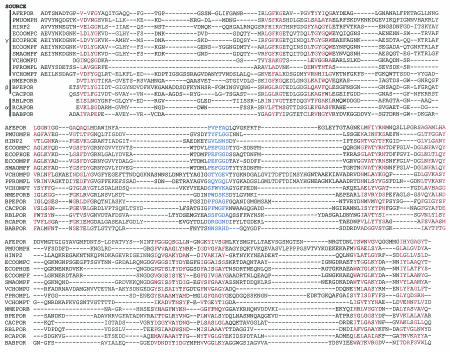
One method that seems to work reasonably well for the detection of transmembrane strands is that of Jeanteur et al. (303, 304), which uses the sum of the hydrophobicity and hydrophobic moment of each 9- to 10-residue segments. Their alignment, together with the addition of some newer sequences aligned by myself, is shown in Fig. 1. In this way, we can see that porins from α-, β-, and γ-protcobacteria are indeed related to each other, a conclusion that BLAST, for example, completely fails to show. Figure 1 also shows some characteristic features of porins from various groups of bacteria. (i) Porins from γ-proteobacteria show strong similarity to each other and are characterized by the presence of short (10-residue) extensions at their N termini (Vibrio cholerae OmpU is an exception). If the alignment presented is correct, porins from the Vibrio-Photobacterium group are unusual in containing exceptionally long L3 loops. Enterobacterial porins contain the characteristic PEFGGD signature sequence in L3, as noted previously (303). (ii) Porins from β-proteobacteria have sequences that are similar to each other but quite divergent from those of the γ-proteobacteria. Although Fig. 1 shows only the sequences of Neisseria meningitidis PorB and Bordetella pertussis porin, several other sequences were aligned earlier (303, 304). (iii) Porins from α-proteobacteria again seem to form a group of their own.
FIG. 1.
Sequence alignment of porins from α-, β-, and γ-proteobacteria. The classification of the source organisms into γ-, β-, and α-subdivisions is shown at the beginning. Transmembrane β-strands are indicated by highlighting, in red, the presence of hydrophobic amino acid residues at alternate positions. The PEFGGD motif of loop 3 in the Enterobacteriaceae and corresponding sequences are colored in blue. The alignment of E. coli OmpF (ECOOMPF), E. coli OmpC (ECOOMPC), E. coli PhoE (ECOPHOE), Haemophilus influenzae Rd P2 porin (HINP2), Neisseria meningitidis PorB (NMEPORB), Bordetella pertussis porin (BPEPOR), Comamonas acidovorans Omp32 (CACPOR), Rhodopseudomonas blastica porin (RBLPOR), and Rhodobacter capsulatus porin (RCAPOR) is basically that of Jeanteur et al. (303, 304), with minor adjustments. The rest of the sequences (Acidithiobacillus ferrooxidans porin [AFEPOR], Pasteurella multocida OmpH [PMUOMPH], Serratia marcescens OmpF [SMAOMPF], Vibrio cholerae OmpU [VCHOMPU], Photobacterium profundus OmpL [PPROMPL], V. cholerae OmpT [VCHOMPT], and Brucella abortus porin [BABPOR]) were aligned by me. The alignment relied mostly on the plot of (average hydrophobicity + average hydrophobic moment) as specified by Jeanteur et al. (303, 304) and took into account the prediction of turns (485). The Gibbs motif sampling program (442) was also utilized (http://bayesweb.wadsworth.org/gibbs/gibbs.html), although this program predicted only the β-strands facing the lipid bilayer. When multiple sequences were available, deletions and insertions were assumed to have occurred in loops (199); this approach was useful in the analysis of P. multocida OmpH (129, 390) and B. abortus porin (424, 482). The alignment of V. cholerae OmpU, V. cholerae OmpT, and P. profundum OmpL was difficult but was helped by the comparison among these three, as well as with Vibrio fischeri OmpU and VCH1008 from the V. cholerae genome-sequencing project (both sequences retrieved from GenBank). No attempt was made to align the variable-loop sequences carefully.
Porins from the organisms in the ɛ-proteobacteria, which forms a very deep branch almost reaching the bottom of the whole Proteobacteria group, are indeed quite different in sequence, and I have not been able to align porins from Helicobacter pylori (60, 178, 196) or Campylobacter jejuni (78, 79, 362, 782) with the porin sequences shown in Fig. 1.
Functional assays.
Nonspecific diffusion of hydrophilic solutes across the OM usually occurs through porin channels, and thus the activity of these channels in intact cells is most conveniently assayed by determining the flux of hydrophilic solutes or ions. For isolated porin proteins, reconstitution into planar bilayer lipids allows the measurement of flux of ions through single channels. Although the single-channel conductance values have been used to calculate the sizes of the channel by assuming that ions within the channel behave identically to those in bulk solutions, this procedure often leads to misleading results (452, 453). A remarkable example is found in some mutants of E. coli OmpF porin, where both crystallography and the proteoliposome swelling assay (see below) showed enlargement of the channel whereas single-channel conductance showed a significant decrease (571). Another problem in the use of single-channel conductance is that it is often a result of insertion of a porin trimer, containing three open channels, rather than that of a “single” channel. Thus, the data that E. coli OmpF gave a single-channel conductance of about 2 nS in 1 M KCl led to the calculated pore diameter of 9 Å (52); this was thought to validate the calculation because of the agreement of calculated diameter with that found later by crystallography. However, the conductance of 2 nS was actually the result of insertion of three channels in a trimer, and the true single-channel conductance of OmpF is around 0.7 nS, which gives an unreasonable prediction for the pore diameter (452). Single-channel conductance is a reasonable indicator of pore size for large channels, but it must be used with the utmost caution for small channels. This problem has also been discussed in a recent review (693). Nevertheless, the planar bilayer study is the only functional assay performed with many porins; therefore, the single-channel conductance value for OmpF, 0.7 nS, is used as the reference in the discussion of many such porins (see “Other porins” below).
The bilayer domain of the OM is asymmetric, at least in Enterobacteriaceae, with the outer and inner leaflets containing nearly exclusively LPS and phospholipid molecules, respectively (see “Lipid bilayer as a diffusion barrier” below). Therefore, there is concern about the use of the phospholipid bilayer, or even nonphysiological barriers such as oxidized cholesterol, for planar film studies. In fact, some authors have argued that porin channels behave quite differently when they are in a natural, asymmetric bilayer of the OM (168, 169). In this connection, it is important that an asymmetric planar bilayer, containing on one side a deep-rough LPS exclusively, was used for experiments involving a trimeric porin of Paracoccus denitrificans (732). There was no difference in single-channel conductance regardless of whether the bilayer was asymmetric or symmetric (i.e., containing phospholipids in both leaflets). However, the spontaneous insertion of porin from the phospholipid side was accelerated more than an order of magnitude if LPS was present on the other side, a result that may have important implications for the mechanism of assembly of OM proteins in intact cells. The voltage-gating behavior was affected somewhat, in a way that was expected if the total potential sensed by the porin molecules was the sum of external voltage and the internal potential arising from the presence of excess negative charges on the LPS-containing outer leaflet.
Another approach is the reconstitution of porins into multilayered proteoliposomes and the measurement of solute diffusion rates that are reflected in the rates of osmotic swelling of these vesicles in media containing the test solutes (460). Comparison of diffusion rates of solutes of various sizes gave remarkably reliable values of the channel size with respect to the crystallographic structure (see below). However, because swelling occurs in response to the movement of any solute, including components of the buffer, extreme care is needed when this method is used to study the diffusion of charged solutes (461).
Finally, diffusion rates through porin channels can be measured in intact cells by coupling the influx of hydrophilic solutes with a “sink” process. A convenient assay is to examine the influx of cephalosporins by coupling it to their hydrolysis by periplasmic β-lactamase (462); cephalosporins are especially useful because a diverse collection of cephalosporins has been synthesized and because the hydrolysis can be monitored easily by recording changes in the optical density at 260 nm.
Crystallographic structure of porins.
Undoubtedly the most important progress in the study of porins was the elucidation of the three-dimensional structures of trimeric porins by electron diffraction (297-300, 705) and X-ray crystallography. The latter approach had its first success with a Rhodobacter capsulatus trimeric porin (714-718), an achievement that was quickly followed by the elucidation of the structure of the E. coli OmpF and PhoE porins (152). Important conclusions from these studies include the following (there are recent minireviews on the structure of porins [343, 598, 599]).
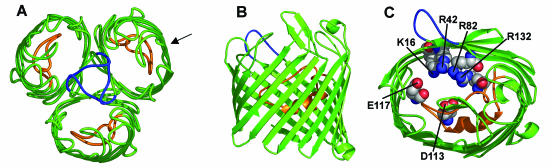
(i) As predicted from earlier studies, porin monomers were shown to cross the lipid bilayer as a β-barrel or a series of 16 β-strands. The strands are tilted rather strongly (by 30 to 60°) in relation to the barrel axis, as shown earlier by Fourier transform infrared spectroscopy (436), and this tilting increases the diameter of the barrel. (For the concept of sheer number, which is related to the degree of tilt, see reference 599.) The length of each transmembrane strand spans the range from only 7 (in strand 5) to 16 (in strand 1) residues in OmpF. Contact among the monomers is stabilized by hydrophobic and polar interactions, and loop 2 tends to bend over the wall of the barrel of the neighboring subunit, playing a significant role in stabilization (Fig. 2A).
FIG. 2.
Structure of the OmpF porin of E. coli. (A) View of the trimer from the top, that is, in a direction perpendicular to the plane of the membrane. Loop 2, colored blue, plays a role in interaction of the monomer with its neighboring unit. Loop 3, colored orange, narrows the channel. (B) View of the monomeric unit from the side, roughly in the direction of the arrow in panel A. Loops 2 and 3 are colored as in panel A. (C) View of the monomeric unit from the top, showing the “eyelet” or the constricted region of the channel. The eyelet is formed by Glu117 and Asp113 from the L3 loop, as well as four basic residues from the opposing barrel wall, Lys16, Arg42, Arg82, and Arg132, all shown as spheres. The diagrams are based on PDB file 2OMF. This figure and Fig. 4 and 6 were drawn by using the program PyMol (Warren L. DeLano, DeLano Scientific LLC, San Carlos, Calif. [http://www.pymol.org]).
(ii) The external surface of the barrel is occupied by lipophilic side chains. One striking observation about the R. capsulatus porin was the presence of many aromatic amino acid residues at both the outer and inner interfaces between the bilayer and the aqueous medium (714). The presence of these “aromatic girdles” has since been observed in the structure of E. coli OmpF and PhoE (152) and many other OM proteins. They are now known to exist, to a somewhat lesser extent, also in the structure of integral inner membrane proteins (see also reference 755).
(iii) It was observed, just before the elucidation of the structures of OmpF and PhoE, that porin sequences almost invariably end with a C-terminal phenylalanine (635). This feature was confirmed with practically all OM channel proteins, although in rare cases the C-terminal residue is tryptophan. This C-terminal residue is located at the OM/periplasm interface in OmpF and PhoE (152), and its conservation is at least partially explained by this location.
(iv) Transmembrane strands are connected by short “turns” on the periplasmic side, but the “loops” that connect the strands on the external sides are often long. With both R. capsulatus porin and OmpF, loop 2 folds back outward and contributes to the connection with the neighboring monomer. Loop 3, connecting strand 5 with strand 6, is especially long (33 residues in OmpF) and folds into the barrel to produce the narrowing of the channel (often called the “eyelet”) (Fig. 2B and C). It was impossible to predict from simple folding prediction algorithms that the eyelet region, which has the strongest effects on the function of porin, is made in this manner, which falls outside of the regular succession of β-strands. The size of the eyelet region of OmpF was 7 by 11 Å (152), very close to the estimate of a diameter of 12 Å from sugar diffusion studies (see above).
(v) The nature of the residues lining the channel wall provided a reasonable explanation of the diffusion characteristics through porins. Thus, OmpF and PhoE prefer cations and anions, respectively, despite having a 72% similarity in the mature-protein sequence; this difference in charge preference was shown to be due mainly to the replacement of Gly131 in the eyelet region of OmpF with the positively charged Lys125 in PhoE (42, 152). The electrostatic properties of the OmpF and PhoE channels were calculated and compared (322).
The OmpC channel appears to be slightly smaller than the OmpF channel on the basis of diffusion rates of organic molecules (461, 462). Although the crystal structure of E. coli OmpC is not yet known, the structure of an OmpC homolog from Klebsiella pneumoniae has been determined (187). However, the size of the constriction region and the arrangement of charged residues there are almost exactly the same as in OmpF, and it is not easy to explain the difference in diffusion rates. Schulz has pointed out that more charged residues are pointing toward the pore lumen in OmpC (599). Perhaps this may decrease the functional radius of the solute diffusion pathway. Molecular dynamics simulation (see below) will be valuable in solving this question.
Lipophilicity in the solute molecule strongly retards its diffusion through the porin channel (461, 462). Schulz explained this effect by assuming that the structure of eyelet, in which the cationic and anionic amino acid residues are located in the opposite sides of the channel (Fig. 2C), orients the water molecules in the channels in a highly directional manner, making the disruption of this ordered structure by hydrophobic solutes energetically unfavorable (597).
OmpF, OmpC, and PhoE are so strongly similar in sequence and also in their three-dimensional structure (as shown experimentally for OmpF and PhoE [152]) that they apparently form mixed trimers almost at random (217).
Regulation of porin expression.
The regulation of expression of nonspecific porins in E. coli is briefly summarized. PhoE is expressed only under phosphate starvation, since the phoE gene is a member of the phosphate regulon (664). The expression of the two major porins, OmpF and OmpC, is exquisitely regulated. The apparent purpose of this regulation became clear when it was discovered that OmpF produces a slightly larger channel than OmpC (461, 462). Thus, noxious agents such as antibiotics and bile acids diffuse far better through the larger OmpF channel, as seen clearly from the observation that low concentrations of antibiotics select for ompF mutants but never for ompC mutants (255) (see “Porins and antibiotic resistance” below). In its natural habitat, the intestinal tract, E. coli encounters 4 to 16 mM bile salts (84), and it is most important to minimize their influx. The conditions prevailing in the intestinal tract, high osmotic strength and high temperature, both favor the production of OmpC (with its narrower channel) and repress the production of OmpF (259). On the other hand, the increased production of OmpF under low-temperature, low-osmolarity conditions (for example, in lake water) will benefit E. coli by facilitating the influx of scarce nutrients. The molecular mechanisms of this regulation have been studied extensively and reviewed in a clear and concise manner (507). Thus, environmental osmotic activity is sensed by the sensor component EnvZ of the archetypal two-component system, EnvZ-OmpR, and high osmolarity results in the phosphorylation of OmpR. The ompF gene, with its high affinity OmpR-binding sites, is transcribed even when the phosphorylated OmpR is scarce (i.e., under low-osmolarity conditions). However, when the concentration of activated OmpR increases, additional binding of these molecules results in increased transcription of ompC and repression of ompF. High temperature, on the other hand, increases the transcription of an antisense RNA, micF (172) (A putative regulatory protein, EnvY, has been reported to affect the temperature regulation process [389], but its effect on micF transcription is not known.) This RNA binds to the 5′-region of the ompF mRNA and inhibits its translation. More recently, another twist was added to this complex regulatory network (383). When E. coli is starved for carbon sources, OmpF production responds strongly to the growth rate (or the concentration of glucose in the medium). Finally, as described below, oxidative stress and the presence of salicylate also increase micF transcription and prevent the production of OmpF porin posttranscriptionally.
The intestinal tract, the normal environment of E. coli, is thought to be mostly anaerobic. Interestingly, anaerobiosis was found to modify the osmoregulation of OmpF and OmpC (406). Thus, under anaerobiosis, OmpC is expressed at a rather high level even in fairly low-osmolarity media, and the repression of OmpF by osmotic activity occurs more strongly than under aerobic conditions. This modification of the regulatory response, which is expected to favor the survival of E. coli in the intestinal tract, occurs through the cross talk activation of OmpR by the ArcB sensor, which senses the anaerobic condition.
Another environmental factor that acts in some cases through the EnvZ-OmpR system is the medium pH (270). At an acidic pH, such as 5.2, the production of OmpF porin becomes strongly repressed and the expression of OmpC becomes increased. This acid induction phenomenon is quite complex (658) and is affected additionally by the nature of the carbon source. A partial explanation of this effect may be the direct phosphorylation of OmpR by acetyl phosphate (269). What advantage would the increased synthesis of OmpC confer to the bacterium trying to survive in an acidic environment? In terms of proton influx, the small difference in the channel size between OmpF and OmpC is unlikely to produce any significant difference in influx. Perhaps a critical factor for survival under acid-stressed conditions is the neutralization of the periplasm, achieved by the decarboxylation of glutamate in the cytoplasm followed by the export of γ-aminobutyrate into the periplasm, where it may act as a buffer (83). In this scenario, the narrower channel of OmpC may contribute to the retention of these buffer molecules in the periplasm. The closure of porin channels by low pH and by endogenously synthesized polyamines produced under acid stress (see “Regulation of porin function” below) may also help in the same way. The acid response involving OmpR also involves, in Salmonella enterica serovar Typhimurium, the downstream two-component regulatory system SsrAB, encoded by the genes in pathogenicity island 2, and is essential for the survival of this species inside macrophages (34, 368).
Regulation of porin expression also occurs in response to chemicals in the environment. It was found in 1991 that salicylate in the medium decreased OmpF synthesis (560). This is now known to be a part of the global regulation of porins mediated by three XylS-AraC family regulatory proteins, MarA, SoxS, and Rob (9, 174). Thus, the increased production of MarA (caused by some environmental chemicals, such as salicylate, inactivating its cognate repressor, MarR) or SoxS (caused by the inactivation of its repressor SoxR via its oxidation) or the binding of coregulators such as dipyridyl (561) or some bile salts (558) to Rob activates the transcription of micF antisense RNA, decreasing OmpF synthesis. Interestingly, all these environmental signals also result in the increased production of the main multidrug efflux pump, AcrAB. Together, these responses result in prevention of the influx of toxic molecules, a reasonable response for E. coli. The benefit is clear from the observation that resistance to several antibiotics is moderately increased in the presence of bile salts (558), a normal component of the environment of E. coli.
A very interesting possibility is that R plasmids could bring in a new regulatory element that would repress porin synthesis, making the bacteria generally more resistant to drugs. An early example is the finding by Iyer and coworkers (289, 290) that N compatibility group R plasmids decrease drastically the synthesis of OmpF porin when introduced into E. coli. Although Iyer favored the interpretation that the preexisting ompF mutants acted as a better recipient for the plasmids, it seems more likely that the plasmid contains a gene that represses OmpF synthesis. I am not aware of other studies in this potentially important area, except that Rossouw and Rowbury (562) showed that the presence of F compatibility group plasmid R124 resulted in the repression of OmpF, causing increased resistance to many agents.
Porin mutants.
Benson et al. (49) used an ingenious approach to isolate porin mutants that allowed the diffusion of large maltodextrins, which cannot diffuse through the wild-type OmpF channel at significant rates. These mutations changed the large, charged residues (Arg82 or Asp113 in the pore constriction [Fig. 2C]) into residues with smaller side chains, such as serine, cysteine, or glycine, or resulted in short, in-frame deletions of the L3 loop (152). The crystal structure of these mutant proteins confirmed the enlargement of the eyelet (387) as expected. Surprisingly, the single-channel conductance in 1 M NaCl was not increased in any of the mutant porins (571). This observation emphasizes the danger of using single-channel conductance uncritically as the indicator for channel size (described in “Functional assays” above). Conductance is affected by the selectivity of the channel for cations versus anions, and the total conductance is the sum of complex factors. Indeed, a liposome-swelling assay with disaccharides unequivocally showed that the mutant channels were larger than the wild-type channels (571). Similar, larger-channel mutants were also isolated in OmpC (419), and the enlargement of the channels was confirmed by biochemical studies (555).
In an interesting case, selection of colicin-N-resistant mutants of E. coli produced a strain in which the Gly119 residue of OmpF was mutated to Asp (305). This change essentially divided the constriction zone of the channel into two smaller channels, and drastically reduced the permeation rates of both ions and sugars.
Site-directed mutagenesis studies were carried out with samples containing residues lining the eyelet region. However, the results are not always easy to interpret. In one example in which the channel assay was accompanied by a structural determination by X-ray crystallography (590), an attempt was made to narrow the eyelet of the Rhodopseudomonas blastica porin (351) by replacing the residues surrounding the eyelet with bulky tryptophan. The introduction of two or three tryptophan residues narrowed the eyelet and indeed decreased single-channel conductance, as expected. However, the introduction of four or six tryptophan residues produced unstable proteins. Similarly, replacement of four eyelet-surrounding residues with alanine produced only a marginal (17%) increase in eyelet cross-section in the X-ray structure, apparently because the remaining arginine side chain assumed a more expanded conformation and narrowed the eyelet. The study by Benson et al. of OmpF mutants (49), mentioned above, was recently complemented by that of site-directed mutants (494). Thus, converting the three arginine residues at the eyelet into a much smaller alanine and converting the glutamate and aspartate residues nearby to uncharged glutamine and asparagine (see Fig. 2C for the location of these residues) produced a 77% enlargement in the eyelet opening, as judged by crystallography. However, the single-channel conductance of this OmpF mutant was decreased by 30% in comparison to that of the wild-type protein (494). Liposome swelling assay with disaccharides, however, showed that the permeation rates of these large sugars was about eight times higher in the mutant.
Functional studies with new approaches.
Computer simulation of Brownian movement of cations and anions through channels (589) has become a valuable tool in the study of wild-type and mutant porins. This approach rather accurately predicted the conductance behavior of various mutants with site-directed mutations at the eyelet region of OmpF (494). These analyses do have restrictions, however: only a single ion was usually observed, so that ion-to-ion interaction was neglected, the protein was assumed to have a rigid structure, and the simulation still assumed that each ion behaves as an unhydrated, naked ion. In the most recent studies by Im and Roux (286, 287), most of these limitations were overcome. Thus, a molecular dynamics simulation involving more than 70,000 atoms, including an OmpF trimer, more than 100 phospholipid molecules, 13,470 water molecules, and 231 K+ ions, and 201 Cl− ions was carried out. Observations of great interest from this simulation include the following. (i) The channel is remarkably efficient in taking up monovalent cations even from micromolar solutions. (ii) The constriction “eyelet” of the channel is very stable and shows no sign of closing during simulation. (iii) Water molecules are strongly ordered at the eyelet region, as predicted earlier (597). A recent analysis combining molecular dynamics simulation with planar bilayer studies suggests also that cations bind to an anionic side chain(s) on the eyelet, most importantly Asp113, and that this binding is significant in the cation penetration across the channel (158). Therefore, the diffusion of preferred solutes through the nonspecific porin channels occurs by a mechanism similar to that through the specific channels (see “Specific channels” below), in the sense that both involve solute (ligand) binding to the channel wall. The difference between nonspecific porins and specific channels has now become quantitative rather than qualitative.
Atomic force microscopy has proven to be a useful tool to examine the porin surface (583). Recently, observation in low ionic strength solution detected the electrostatic potential on the surface of porin trimers with very good resolution (496). This may be a valuable method of determining the electrostatic potentials on the surface of various proteins.
Single-channel experiments can now be carried out with much refinement, so that it is possible to observe the clogging caused by the partitioning of polyethylene glycol into the OmpF channel (563). Here polyethylene glycol 1360 was the cutoff size for the OmpF porin, in comparison with polyethylene glycol 2200, which was the cutoff size for a larger, staphylococcal alpha-toxin channel, a result showing excellent agreement with the known diameters of these channels. Even more impressively, transient clogging of the channel accompanying the diffusion of antibiotic ampicillin through the OmpF channel could be observed and analyzed in single-channel experiments (440).
Voltage gating.
Planar lipid film reconstitution of porins showed from the earliest days that the channel can be closed (“gated”) at high voltage, typically 100 mV or more (see reference 464 and references therein). Much effort has thus been spent in understanding the structural basis of the gating phenomenon. According to one hypothesis (103), transmembrane voltages make the channel narrower by bringing the cationic and anionic amino acid side chains closer to each other within the channel. As mentioned above, the constriction zone, or eyelet, of OmpF and PhoE contain acidic residues on the L3 loop and basic residues on the facing barrel wall (Fig. 2C). Therefore, the movement of the loop containing the acidic residues in response to voltage is in principle possible, as shown by theoretical analysis (710). However, when the loop was fixed to the barrel wall by a disulfide bond, the gating still occurred (29, 194, 492), a result that rules out at least a large-scale movement of the loop against the barrel wall. Small movements in parts of the loop are still possible, and some molecular dynamics simulation studies (628, 659) indeed seem to support this idea. Another possibility is the movement of external loops that are located outside the channel in the crystal structure. This mechanism was supported by an atomic force microscopy study, in which the application of voltage was shown to cause the movement of external loops, resulting in the closure of channel entrance (433). With Haemophilus influenzae porin, which shows significant sequence homology to E. coli porins (Fig. 1), this idea was supported by site-directed mutagenesis as well as chemical modification of basic amino acid residues in the external loops (20, 21, 157).
When the results did not favor the movement of loop 3 in gating, some workers took a broader look and considered the fact that even the channel made by the α-toxin of Staphylococcus aureus, which is an empty 14-member β-barrel with no constrictions or infolding loops, shows typical gating behavior in planar bilayers (28). They refer to the studies in which even holes in a plastic film (polyethylene terephthalate) were shown to exhibit the gating phenomenon, presumably as a result of the fluctuation in the ionization of the charged groups either within or near the channel (347). This is an extreme position, but the possibility that gating can occur in the total absence of conformational changes in channel proteins should be considered more seriously.
Regardless of the mechanism of the gating, we must ask if the phenomenon has any physiological significance. It is difficult to imagine the presence of such a high membrane potential across the OM, and distribution of ions across the OM, which measures directly the potential, show that only low (less than 30 mV) values of Donnan potential exist here (601). This Donnan potential was of the size expected from the presence, in the periplasm, of polyanionic oligosaccharides (membrane-derived oligosaccharides [MDO]) (333) that cannot diffuse through the porin channels. Therefore, clearly there is no large electrical potential across the OM. In the days when the structure of porins was completely unknown, the observation of gating encouraged some workers to propose that porins might serve as “models” of voltage-gated ion channels in nerve cells, for example. However, we now know that real voltage-gated ion channels are constructed in a totally different manner, and the porin gating observed under these high-voltage conditions may be no more than an interesting laboratory artifact. At lower voltages, the open states often last for many seconds (or even minutes) (168), and this also gives an impression that porin channels are normally open at least most of the time. In retrospect, the emphasis on voltage gating could have produced more confusion rather than enlightenment. The most obvious possible benefit of the voltage gating of porins may be to prevent the formation of open channels when porins are misincorporated into the cytoplasmic membrane. A similar benefit may also apply to the (voltage-gated) mitochondrial porin, which apparently becomes inserted occasionally into other membranes of the eukaryotic cell (657).
On the other hand, it was argued that the gating occurs in the OM of living cells. This argument was supported most strongly by early studies involving patch clamping of the membranes of E. coli spheroplasts. The open state was much less stable, and the closure occurred at lower voltages (periplasmic side negative). This behavior is ascribed to the more “natural” environment occupied by the porins here (168, 169). A patch of the size used in these early studies should have contained close to 105 porin monomers if it came from the OM. However, in reports of patch clamping of E. coli “OM,” only a dozen or so open channels were observed in a patch (559). These data seemed to imply that an overwhelming majority of porin channels were closed in intact cells and that they opened only under specific conditions such as starvation (168, 169).
Although a microscopic observation suggested that the OM was intact in the spheroplasts (107), the same paper reported that the patches also contained stretch-activated channels (107), now known to be a component of the inner membrane (642). Therefore, it seems possible, even likely, that these early patch clamp studies using spheroplasts observed the behavior of the inner membrane, containing perhaps a very few porin channels misincorporated into the wrong membrane, or that patches were made on mixed membrane fragments. This view is now shared by one of the authors of these early papers (A. Delcour, personal communication). Consistent with this interpretation, we can calculate the permeability coefficient of the OM in intact cells to uncharged or zwitterionic solutes of a known size on the basis of the cross-section of the always-open porin channels in the OM (460). This calculation yields the predicted permeability coefficient, P, of 2.5 μm/s for lactose (342 Da), a value quite close to the measured value of P of 1 μm/s (601) for a zwitterionic antibiotic of a comparable size (415 Da), cephaloridine. This agreement again reinforces the notion that most of the porin channels are open in intact E. coli cells.
Indeed, the porin channels were open most of the time, even when investigated by the patch clamp method, if OM fragments were diluted into a large excess of phospholipid bilayers (171). Using this latter system, functional studies were carried out. The addition of MDO produced prolonged closure of E. coli porin channels (170). The physiological significance of this observation is not clear. First, the closure required about 10 mM MDO. This is not an excessively high concentration in cells grown in low-osmolarity media, but once the cells are grown in more physiologically relevant media of moderate osmolarity (for example, containing 0.3 M NaCl) the MDO concentration decreases almost 20-fold (601). Second, the closure could be seen only when the voltage across the membrane was periplasmic-side positive, opposite of the direction of the Donnan potential. Delcour argues that inside-positive potential could be created by the influx of cations when bacteria are diluted into a high-salt medium (168), but this has not been demonstrated experimentally. Perhaps the most interesting observation of this group was that polyamines increased the closure of porin channels in patch clamp experiments; since it seems likely that this represents closure, rather than intensified voltage gating, these data are discussed in the next section.
Regulation of porin function.
Delcour's group observed that polyamines affected the permeability of porin channels not only in artificial membranes (166) but also in intact cells when measured with cephalorididine influx (167). The effect was modest when the polyamines were added to the medium; a 40% decrease in OmpF permeability required the addition of 100 mM putrescine, 60% inhibition required 30 mM spermidine, and 70% required 1 mM spermine. These polyamine concentrations can be compared to the total intracellular concentrations of putrescine and spermidine, 20 and 6 mM, respectively (which are mostly tied up by polyanionic macromoles such as ribosomes and LPS), and the inability of E. coli to synthesize spermine (643). However, Samartzidou and Delcour (574) made an important observation that when E. coli is synthesizing and excreting cadaverine, the OM permeability decreases to about 30% of the normal level. In an experiment in which acid stress was used to induce cadaverine synthesis, the decrease in OM permeability could be interpreted as the result of increase in OmpC and decrease in OmpF, a part of the acid pH response (see “Regulation of porin expression” above). However, in the experiment where cadaverine synthesis was controlled by a lactose promoter, the decrease is unlikely to be the result of anything other than the cadaverine synthesis and excretion. The concentration of cadaverine in the medium, after 1 h of induction, was only 0.2 mM. Thus, surprisingly low concentrations of endogenously produced cadaverine exerted this profound effect in intact cells, although cadaverine needed to be present at 300 mM for 50 to 60% inhibition of porin activity when added to the external medium. Perhaps the constant efflux of cadaverine from the periplasm tends to close the porin channel more effectively (574). (It should be noted that the channel closing is not the result of voltage gating but appears to be the result of direct interaction between the polyamines and the interior of the channel [291]). It is also likely that polyamine synthesis and export, as a part of the acid stress response, are meant to lead to the neutralization of the periplasm and that closing of the porin channels tends to build up high concentrations of polyamines in the periplasm by retarding the release of polyamines into the medium (see “Regulation of porin expression” above). Thus, the polyamine effect seems to be truly significant in bacterial physiology. Indeed, this point was proven in a recent study in which a strain producing polyamine-resistant OmpC (with the Gly195 changed to Asp) was shown to be more susceptible to acid shock (575).
A single-channel conductance assay of OmpF and OmpC by Todt et al. (662) showed that the conductance value was lowered to almost one-half by going from pH 8.1 to 5.4. A liposome-swelling assay also was reported to show a similar decrease in permeability. However, the authors' finding of very high permeation rate for maltose (about 60% of the rate for glucose at pH 9.4) is quite unexpected, because in our hands disaccharides usually penetrate through the OmpF channel with rates that are about 2 orders of magnitude lower than that of glucose (460, 461). The same group suspected the participation of the sole histidine residue of OmpF or OmpC in this switching of channel size and reported that diethylpyrocarbonate modification of His21 abolished this pH-induced alteration of pore function (661). However, another laboratory could not reproduce the acidic-pH-induced alteration of pore size and saw no effect of a His21-to-Thr substitution (572). The latter authors suggest that the different results were possibly obtained because of the presence of LPS in the porin preparations used by the earlier workers. This explanation seems reasonable because Todt et al. (660) showed that the alteration of channel size can be seen in whole E. coli cells. This important issue needs renewed attention.
Evolution of porins.
In the real world, the exposed external loops of porins interact with antibodies, phages, and colicins. For example, OmpF (together with LPS) is the receptor for phage K20 (616, 666) and for colicins A and N (210). The specific maltodextrin channel LamB was initially known only as the receptor for phage lambda. Therefore, it is understandable that these loops underwent rapid evolutionary alterations in structure, as seen clearly by comparisons of sequences of the same (orthologous) porin from different strains of the same species or from strains belonging to the related species (the latter is seen in Fig. 1). In contrast, comparison of porins of Rhodobacter capsulatus strains kept for more than 30 years in separate laboratories (that is, in the absence of external selective agents) revealed that changes occurred nearly exclusively in the transmembrane strands (677).
“Evolution” experiments can be carried out in the laboratory. When E. coli was grown for a few hundred generations in chemostat under glucose-limited conditions, mutations that resulted in the overproduction of the specific LamB channel (which also facilitates the diffusion of glucose [see “Specific channels” below]) were regularly observed (465). In contrast, when a disaccharide lactose was the sole carbon source in a minimal medium, “evolved” strains overproduced the larger-channel porin OmpF over OmpC and, moreover, contained ompF mutations that altered the residues in the constriction region (Arg82 and Asp113) (Fig. 2C) or resulted in a short deletion in the channel-constricting loop L3 (780). These are precisely the mutations found by Benson et al. (49), who looked for larger channel mutants of OmpF; indeed, the presence of the larger channel in the evolved strains was confirmed by their hypersusceptibility to large antibiotics such as cloxacillin. This experiment shows that the porin channel can become limiting for the diffusion of even disaccharides when their concentration is low. Furthermore, the experiment is important in two other areas. (i) Such “evolution” obviously does not take place in nature, in spite of the obvious advantage of the larger channel. This is consistent with the idea that in its natural habitat, E. coli must balance the desirability of more efficient uptake of nutrients against the danger of more rapid influx of toxic compounds, especially bile salts. It would be interesting to repeat the in vitro evolution experiment in the presence of bile salts. (ii) If the porin channels are mostly closed, as claimed by some workers, the mutations will occur in the regions of porin protein that affect the voltage-induced closing. The constriction zone is not such an area, as described above. Therefore, the results do not favor the idea of normally closed porin channels.
Slow Porins
OprF is the major porin in P. aeruginosa.
The study of what I now propose to call “slow porins” has had a long, tortuous, and sometimes acrimonious history. It began with the identification of protein OprF as the major porin of Pseudomonas aeruginosa (246). This porin is very different from the trimeric, classical porins. (i) There is no strong evidence that it exists as a stable oligomer. (ii) On reconstitution into proteoliposomes, it allows a much slower diffusion of small solutes such as monosaccharides (in one experiment, the influx of arabinose was 50 times slower than through the E. coli OmpF channel [767]) but allows the diffusion of much larger solutes that cannot penetrate through the OmpF channel (246). Although the low permeation rates were in agreement with the very low permeability of intact P. aeruginosa OM, measured by using hydrophilic solutes such as cephaloridine (19, 766), the apparent contradiction between the large pore size and the lower penetration rates was unexpected, and some simplistic “solutions” were proposed (see below). (iii) When the oprF gene was sequenced, it was found to be a homolog of E. coli OmpA protein, which at that time was not known to have a channel function. This finding further added skepticism about the porin function of OprF. (d) The mobility of OprF and OmpA in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) decreases when the samples are heated at 100°C, a property often called heat modifiability.
Nakae and coworkers criticized almost every aspect of the concept of OprF as the major P. aeruginosa porin (references are cited in reference 459). The argument was based on results that purportedly showed that the porin channels in the intact P. aeruginosa OM was much narrower, instead of wider, than the E. coli porin channels and that the purified OprF lacked pore-forming activity. This group also claimed that the major porins in P. aeruginosa were OM proteins other than OprF. Careful reexamination of the data published by this group showed, however, that their claims could not be substantiated (459). The claim for the narrower channel was based on an improper and arbitrary way of plotting the permeation rate versus the solute size. Purification of OprF following their procedure produced an active pore-forming protein, with a wide channel and low penetration rates. Finally comparison of various P. aeruginosa OM proteins for nonspecific porin activity showed that OprF was the major porin in this organism.
Even more decisive was the study from the laboratory of Hancock (46). They showed, by expressing genes for the metabolism of raffinose (MW 505) in P. aeruginosa, that oprF+ cells, but not an oprF mutant, can grow readily by utilizing this sugar. Since raffinose cannot diffuse through E. coli general porin channels at sufficient rates, this is the ultimate proof that the larger pore size of OprF channel is not an in vitro artifact and that the major porin of P. aeruginosa is indeed OprF.
The apparent dilemma of low permeability through a large channel.
Although it has been established that OprF is the major porin, it was difficult to explain why solutes penetrated the P. aeruginosa OM at rates about 2 orders of magnitude lower than the rates at which they penetrated the E. coli OM (19, 766), because the number of OprF molecules per cell was similar to that of the major porins in E. coli and because the larger channel in OprF porin was expected to result in a faster, not slower, diffusion of solutes. The answer to this question came through the studies of E. coli OmpA, a homolog of OprF. Like OprF, OmpA also produces permeability on reconstitution into proteoliposomes, but the diffusion rates of solutes are about 2 orders of magnitude lower than in proteoliposomes containing comparable amounts of classical porins (638). (The channel-forming activity of OmpA was also confirmed by a planar lipid bilayer assay [24, 570]). When unilamellar proteoliposomes each containing only a few molecules of OmpA were fractionated on the basis of permeability to sucrose, only a small percentage of the OmpA molecules were found to have the ability to form open channels (636). A similar experiment using OprF also showed that the OprF population was heterogeneous, with only a minority containing the open channel (E. Sugawara, E. K. Nestrovich, S. M. Bezrukov, and H. Nikaido, unpublished data).
These data explain the slow permeation of solutes mediated by OmpA and OprF but leave open the question of the nature of the difference between the “open” and “closed” proteins. OmpA has always been hypothesized to fold as a two-domain protein (545), with its N-terminal half spanning the OM as an eight-strand β-barrel and its C-terminal segment located in the periplasm and interacting with the peptidoglycan layer (173, 342) (Fig. 3, Majority Conformer). This folding model is supported by a large amount of convincing data, beginning with the location of phage resistance mutations in the predicted external loops (429) and culminating in the determination of the crystal structure of the N-terminal domain of OmpA as an eight-stranded β-barrel (488, 489). Since the eight-stranded β-barrel has little room inside for the passage of even small molecules, this conformer cannot serve as an open channel for the passage of organic solutes. (However, N-terminal domains of both OmpA and OprF were shown to produce very small conductance steps [24, 100], and these observations have been rationalized by the recent molecular dynamics simulation study [81]).
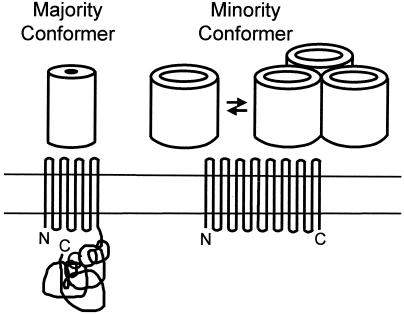
FIG. 3.
Folding model of OmpA-OprF family slow porins. The major fraction of the population folds as a two-domain protein (left) and is important in binding the OM to the underlying peptidoglycan, since the C-terminal globular domain contains a peptidoglycan-binding motif (165, 342). A minor fraction of the population, however, folds differently to produce an open β-barrel (right). In E. coli, which produces trimeric, high-permeability porins, the presence of this fraction has no functional consequence. However, in fluorescent pseudomonads, which lack the high-permeability porin, this fraction functions as the major nonspecific porin. This fraction also tends to form a loosely associated oligomeric structure, as shown. The oligomer is shown as a trimer only for illustrative purposes. Modified from reference 455a with permission of the publisher.
The open conformers, in contrast, are likely to contain more β-strands like the classical porins (Fig. 3, Minority Conformer). In fact, a model assuming that OprF folds essentially as a continuous β-barrel, containing up to 16 transmembrane β-strands as in the classical porins, was proposed (534, 747). The surface exposure of residues in the C-terminal region was indeed confirmed, for example, by inserting malarial antigen epitopes at various places in the OprF sequence (748). Such an OmpF-OmpC-like folding model, however, cannot explain the low permeability of the OprF porin. Furthermore, a circular dichroism study of the OprF protein isolated without the use of denaturing detergent showed clearly that at least the majority of OprF molecules, just like OmpA, contained a substantial fraction of helical structures (presumably coming from the periplasmic, globular domain of the two-domain conformation) (639), in contrast to the essentially all β-structure predicted from the earlier model. The heat modifiability of OprF (and OmpA) is also likely to be explained by the two-domain conformation of the majority conformer, since the N-terminal β-barrel will not be denatured in SDS unless the protein is heated (639). However, if we assume that only the minority, open-channel conformer of OprF takes the one-domain conformation, as with OmpA (636), then we can explain the low permeability of OprF and all other available data on this protein. Thus, the epitopes in the C-terminal half of OprF will be exposed on the surface of intact cells, if they exist in the loop region of the continuous β-barrel conformer. However, because this conformer represents a minority fraction of OprF, the reactivity of such epitopes will be substantially lower than that of the same epitopes located in the loop region of the N-terminal half. In retrospect, this was precisely the result obtained (748), although it was not interpreted in this manner at that time.
Recently, the current model of OmpA and OprF (Fig. 3) has received strong experimental support. The model predicts that the N-terminal fragment (alone) of these proteins will not produce significant permeability because it contains only the eight-stranded β-barrel and that the entire protein sequence is needed to produce an approximately 16-stranded β-barrel that would allow the permeation of large molecules. Indeed, Arora et al. (24) obtained such a result by using our OmpA preparation that was enriched for open conformers (see below), and this was soon followed by similar papers dealing with OprF (100, 190).
The two-conformer model of OmpA-OprF is thus quite useful and is likely to be correct. The model has received further support. The C-terminal domain of OmpA (as well as that of OprF) contains the immunodominant epitope, which is present on the surface of intact bacteria on the basis of studies using fluorescent-labeled antibodies. However, these data appeared to be inconsistent with the two-domain model of these proteins. A recent study showed that the reaction, with S. enterica serovar Typhimurium cells, of monoclonal antibodies directed to the C-terminal domain of OmpA was dramatically enhanced when the OM was made permeable to these antibodies, confirming that the C-terminal domain was exposed only in minority conformers (621). If residues in the C-terminal domain of OprF are exposed on the cell surface only in the minority conformer containing open channels, one should be able to enrich for this conformer by inserting an additional cysteine residue in one of the predicted external loops, by labeling the residue with a biotinylation reagent in intact cells, and by capturing this conformer (but not the majority conformer, in which the cysteine residue is hidden in the periplasm) via binding to avidin. We have indeed been able to enrich for the biotin-labeled species and to show that this species has higher specific activity in terms of pore formation (E. Sugawara, E. K. Nestrovich, S. M. Bezrukov, and H. Nikaido, unpublished data). Even more intriguing is the observation that the open-channel conformers tend to have a larger size (637), presumably in an oligomeric form. We have again been able to utilize this property and enrich the OmpA and OprF preparation for their open conformers (Sugawara et al., unpublished data).
There is therefore no longer any controversy about the porin functions of OprF and OmpA, in spite of several reviews that argue that their porin function is controversial or unsubstantiated (see, for example, reference 343). Presumably the C-terminal portion of their majority conformer stabilizes the cell envelope structure through their interaction with peptidoglycan (173, 342, 533), and this is their primary function. However, in organisms that lack the classical trimeric porin, such as fluorescent pseudomonads, the protein of this family functions as the major porin and contributes to the high levels of intrinsic resistance to toxic agents through their low permeability. At least in one strain of P. fluorescens, the deletion of oprF gene is followed by compensatory suppressor mutations resulting in the overexpression of OprD family channel proteins (described in “Specific channels in bacteria other than the Enterobacteriaceae” below), a result that convincingly demonstrates that OprF functions as the major porin (130). It is remarkable that OprF-OmpA family proteins are found in almost every gram-negative bacterial genome sequenced; they may serve as the major porin also in some organisms other than the pseudomonads.
One remaining question, though, is whether the two folding pathways of these proteins are regulated. In this connection, P. fluorescens was found to produce OprF of lower single-channel conductance when grown at low temperature (163). Although low ionic conductance cannot be immediately equated with smaller channels (see “Classical porins” above), the low-temperature conformer is more susceptible to protease hydrolysis (190), as expected for the two-domain conformer, and it seems likely that the growth temperature affects the folding of OprF, at least in this organism.
Other Porins
Other porins in E. coli and Salmonella.
The genome of E. coli K-12 codes for several general-purpose porins other than OmpF-OmpC-PhoE. The NmpC porin, which belongs to the OmpF-OmpC-PhoE cluster and which is not expressed in K-12 because of the insertion of the IS 5 element close to the distal end of the gene, is essentially identical to the lc porin coded by the genome of a lambdoid phage, PA-2 (71). Indeed, the nmpC gene is a part of the genome of a defective phage inserted into the E. coli chromosome; this finding suggests that phages sometimes code for a new porin gene, which would help in the rapid biosynthesis of phage material by allowing a rapid influx of nutrients. The NmpC sequence is more similar to OmpF-PhoE than to OmpC in that it lacks the long insertion that is found in OmpC in loop 3. It is more similar to PhoE than to OmpF, in that it lacks the small insertion of OmpF in loop 6, but it retains Gly131 (OmpF numbering), which is crucial in the cation selectivity of OmpF (see above). The transcription of nmpC is apparently up-regulated by growth in a slightly alkaline media (140).
A recent survey of the K-12 genome using a program detecting β-barrel proteins indicated that the product of gene b1377 is a homolog of OmpC belonging to the classical porin family (778). Actually this trimeric porin, called OmpN, was originally found in E. coli B and then in K-12 and was expressed and purified (510). Its channel property was also reported to be very similar to that of OmpC. This protein is apparently expressed at a very low level in wild-type strains; I am not aware of any report on the effect of environmental conditions on the production of this porin.
S. enterica serovar Typhimurium produces OmpD, an additional member of the classical porin family. It has channel properties comparable to those of OmpF-OmpC as judged from single-channel conductance data (51). Liposome-swelling studies apparently have not been carried out with this porin. Interestingly, the synthesis of this porin is dependent on cyclic AMP (B. Rotman and G. F.-L. Ames, personal communication). The gene ompD is located downstream of the putative transporter gene yddG, and recently the inactivation of either of these genes was reported to cause hypersusceptibility to methyl viologen (577). The authors propose that the OmpD porin forms a multiprotein complex with the YddG pump, playing the role of the exit channel like TolC. However, OmpD does not contain the periplasmic “tunnel” found in TolC (see “export channels of the TolC family” below). Furthermore, this reviewer found that homologs of yddG gene in other organisms occur without the neighboring ompD-homologous gene. A porin not corresponding to OmpF and OmpC was reported in S. enterica serovar Enteritidis and was called OmpE (126). The nomenclature suggests that the authors were aware of the presence of OmpD, yet they did not compare their porin with OmpD. The production pattern of this porin on complex agar media and in synthetic liquid media seems to fit with the idea that it is actually OmpD that is synthesized in a cyclic AMP-dependent manner.
Mutants expressing OmpG porin were isolated in E. coli K-12 by Misra and Benson (420) by using a selection procedure that favors mutant cells capable of taking up large nutrients. OmpG is a porin with unusual properties (197). First, it appears to lack, on the basis of its sequence, the large loop 3 that is ubiquitous in classical trimeric porins. Second, it produces an unusually large channel, as expected from this structure. Third, it appears to exist as a monomer, unlike members of the classical porin family. The large channel size and the monomeric nature of OmpG were confirmed by single-channel conductance and folding studies in another laboratory (143, 144). The protein was made into a two-dimensional crystal, and its study also confirms the monomeric nature of this porin (45). The ompG gene appears to be the last gene in a putative 11-gene operon, which contains genes needed for the ATP-binding cassette (ABC) transporter-catalyzed uptake of oligosaccharides, as well as various genes presumably involved in the degradation of such compounds (197). Therefore, it seems likely that it is a large-channel porin needed for the uptake of larger oligosaccharides. OmpG is expressed, at a low level, in Salmonella and Shigella, but only trace levels are seen in wild-type E. coli K-12.
The ompL gene of K-12 was identified by transposon insertion mutagenesis as a gene whose inactivation made a dsbA mutant more dithiothreitol resistant (159). Its sequence suggests that it may exist as a β-barrel in the OM, and the purified OmpL showed porin activity. One could explain the phenotype of ompL mutants if it were assumed that OmpL facilitated the outward diffusion of hypothetical, low-molecular-weight oxidizing agents in the periplasm, but recent attempts to reproduce the reported phenotypes of ompL and ompL dsbA mutant strains have failed (578). OmpL is not expressed at a high level in wild-type cells and presumably plays only a minor role in the flux of nutrients in general. Recently, OmpL was found to be a homolog of the Erwinia chrysanthemi oligogalacturonate channel protein, KdgM (490) (see “Other specific channels” below).
E. coli OmpW, a small (191 residues in the mature form) OM protein, is used by a colicin as a receptor (498); otherwise, its function is unknown. Interestingly, this protein belongs to a homology group that includes P. putida AlkL, which is hypothesized to be an alkane channel (690), and NahQ (SwissProt accession no. Q51498), which is part of the cluster coding for naphthalene utilization and is also likely to code for an OM channel. This AlkL-OmpW group also includes Omp21 of Comamonas acidovorans, which was studied as a two-dimensional crystal (31) and showed patterns similar to other eight-strand β-barrel proteins. With slow porins like OmpA and OprF, there was a possibility of alternative folding to produce large channels. However, it is currently unclear how these small proteins of the AlkL-OmpW group would produce sizable channels, if they indeed function as channels.
Many other small OM proteins are known (701) or suspected (31) to produce eight-stranded β-barrels. These include OmpX of E. coli (701), opacity proteins of Neisseria, and Ail protein of Yersinia. More recently, PagP, the enzyme that transfers a palmitoyl residue from phospholipid to lipid A (discussed in “Physiological adaptation in LPS structure” below), was shown to be an eight-stranded β-barrel protein by solution nuclear magnetic resonance spectroscopy (NMR) (285). One example of the opacity protein family protein was actually shown by X-ray crystallography to have a 10-strand β-barrel structure (511), and another example from this family, NspA, was recently characterized to be an 8-strand β-barrel protein (691). As Baldermann and Engelhardt argue (31), what is important in these proteins are the external loops that may be needed for interaction with the external structure, such as the surface of the host cell membrane or the substrates of enzyme reaction, and the 8- or 10-strand β-barrel may simply serve as the OM anchor. None of these proteins has been shown to form channels, as far as I am aware. Other examples where OM-spanning β-barrels are thought to function as simple anchors include OmpT and OmpLA. The crystal structure of OmpT, an OM-associated protease, shows that its 10-strand β-barrel protrudes far beyond the plane of the OM bilayer and that the membrane-external portion of the barrel and the loops apparently function in proteolysis (692). In OmpLA, the OM-associated phospholipase A, the structure shows a 12-strand β-barrel (625), yet the interior of the barrel appears to be occluded by a dense hydrogen-bonding network, and the protein is not thought to function as a channel.
Most E. coli strains, including K-12 and B, are incapable of metabolizing raffinose, a trisaccharide. However, some strains contain plasmids that contain genes for degradation and transport of this sugar. One of the genes is rafY, which produces an OM channel (353). When RafY, a fairly large (50.7-kDa) protein that exists as a trimer, was inserted into a planar lipid bilayer, it produced a high single-channel conductance of 2.9 nS in 1 M KCl (16). Interestingly, however, there was no blocking of the channel by several sugars, including raffinose. Although negative results must be interpreted with caution, it seems likely that RafY, unlike such specific channels as LamB or ScrY (which shows low single-channel conductance that is blocked by sugars), simply produces a pore large enough for the influx of trisaccharides. RafY does not appear to be strongly similar to other known porins or channel proteins.
Porins in members of the Enterobacteriaceae other than E. coli and Salmonella.
Among porins of members of the Enterobacteriaceae other than the E. coli-Shigella-Salmonella cluster, those from Serratia have been studied in several laboratories. In an early study (518), S. marcescens was found to produce two major (and one minor) porin-like proteins. The ompF and ompC genes were characterized (283, 284), but the properties of the channels were not studied. The upstream operator region of the ompF gene in this species appears truncated in comparison with that of E. coli, in such a way that would make the repression of OmpF by high osmotic activity much less efficient. This seems to fit with the natural habitat of this saprophytic organism, which would derive little advantage by shutting down larger channels under high-osmolarity conditions. An early study (396) utilizing a clinical isolate showed the predominant production of the OmpC-like porin, with OM permeability to cephaloridine that was similar to that of E. coli (462). Interestingly, a study using a clinical isolate with high-level resistance to a wide range of cephalosporins showed OM permeability that was about an order of magnitude lower than that found in E. coli (530); it is possible that this strain contained mutations in a porin gene(s). Perhaps the low general permeability, due to lowered porin expression, mutations in porin genes, or both, is a general hallmark of many strains of clinical origin (see also “porins and antibiotic resistance” below); in this respect, it is interesting that enterohemorrhagic E. coli O157:H7 was recently shown to have much lower OM permeability to a hydrophilic, anionic compound than the laboratory strain K-12 (401).
Enterobacter cloacae also produces homologs of OmpF and OmpC (319) (GenBank accession numbers CAC48382 and CAC48383). Interestingly, the OM permeability of a clinical strain of E. cloacae was more than an order of magnitude lower than that of E. coli K-12 when the monoanionic cephalosporin cefazolin (704) or the zwitterionic compound cephaloridine (458) was used as the probe. An especially large fraction of β-lactam-resistant clinical isolates of Enterobacter aerogenes were found to have alterations in porins (125). Since this species contains an inducible chromosomal cephalosporinase (509), this result is not easy to explain. Perhaps the modest extent of induction of this enzyme, seen in a reference strain (125), may be relevant. An example of alteration of porin structure is described in “Porins and antibiotic resistance” (below).
Klebsiella pneumoniae is unusual among the Enterobacteriaceae in lacking the ability to produce a chromosomally encoded, inducible class C β-lactamase and instead produces a chromosomally encoded, weak class A enzyme (491). Thus, they become resistant to extended-spectrum cephalosporins such as cefotaxime and ceftazidime primarily through the acquisition of plasmids with genes coding for mutated class A enzymes with wider specificity (495) rather than by the constitutive production of class C chromosomal enzymes (295). This situation also suggests that the loss of porin might play a larger role in the resistance of K. pneumoniae to β-lactams. This species produces two porins (319, 580). Genes coding for these two porins were later cloned, and their sequences were confirmed to correspond to E. coli ompF (coding for OmpK35) and ompC (coding for OmpK36), respectively (267). As mentioned above, the crystal structure of the OmpC homolog OmpK36 is known (187). In an intriguing study, it was found that a K. pneumoniae strain from a patient lacked one of these porins. A later, more cephalosporin-resistant isolate from the same patient was deficient even in this remaining porin, and introduction of a plasmid coding for OmpK36 resulted in the regaining of drug susceptibility (402) (see, however, “Porins and antibiotic resistance” [below] for more on these results). In carbapenem-resistant strains lacking both of the porins (404), the decreased OM permeability is likely to play a role in resistance.
Biochemical studies of porins from Proteus vulgaris, Proteus mirabilis, Morganella morganii, Providencia rettgeri, and Providencia alcalifaciens suggested that each of these species produced predominantly a single porin with a subunit size of around 35 to 37 kDa (422). Proteoliposome swelling studies showed pore sizes similar to that in E. coli porins, and the channels favored more hydrophilic members among cephalosporins, just as in E. coli (462). All porins, just like E. coli porins, were cation selective, as indicated by the preference for zwitterionic cephaloridine over monoanionic cephalosporins, with the interesting exception of the P. vulgaris porin, which allowed influx, at equal rates, of these two classes of compounds.
The OmpC homolog in Yersinia enterocolitica was shown to be a trimeric porin with the expected single channel conductance (104). Disruption of ompF and ompC in this organism led to decreases in OM permeability (105).
A nematode symbiont, Xenorhabdus nematophilus, which is required for the nematode to kill the host insects, predictably produced a trimeric porin, OpnP, that resembled the E. coli porins (209). Also, in this case the gene lacked the regulatory region needed for its repression under high-osmolarity conditions.
The OmpC porin from Rahnella aquatilis, a rhizosphere bacterium, shows strong homology to other enterobacterial OmpC proteins and also acts as a root adhesin (4). This is one of many examples where surface-exposed OM proteins were shown to play an additional role in the interaction with the environment.
Photobacterium and Vibrio are phylogenetically close to the Enterobacteriaceae, and their porins are discussed here. A major OM protein (35 to 42 kDa) of Vibrio (now Listonella) anguillarum was purified and made into a two-dimensional crystal (617). The protein was a trimer and showed conductivity properties similar to those of E. coli OmpF. The sequence of this protein is unknown. The deep-sea Photobacterium sp. strain SS9 (Photobacterium profundum) has been studied extensively. At atmospheric pressure, it produces predominantly an “OmpL” protein (34.7 kDa), which shows homology to enterobacterial porins (719). This protein is also an oligomeric porin, and its complete denaturation requires boiling in 2% SDS. At higher pressures, however, another porin, “OmpH,” (31.1 kDa) (37), is preferentially produced (132). This regulation apparently involves an extracytoplasmic function sigma factor, RpoE (131), and proteins that are expected to regulate the RpoE activity (36). We do not yet know, however, the properties of OmpL and OmpH that might make them suitable for environments with different pressures. Vibrio vulnificus was shown to express, depending on the growth medium, a few porin-like proteins (345); with the recent sequencing of the genome of this species (GenBank accession numbers NC004459 and NC004460), a more detailed study may appear in the future. Vibrio cholerae is known to produce two porins: OmpU (120), whose transcription is activated by the regulatory protein ToxR (155), and OmpT, which is repressed by ToxR (374). Cells expressing OmpU exclusively are more resistant to bile salts than are those expressing OmpT alone (513, 514). E. coli cells expressing only the narrower-channel porin OmpC are also more resistant to bile salts than are those expressing the wider-channel porin OmpF (653). These results are consistent with the assumption that OmpU produces a narrower channel than does OmpT (although a contrary conclusion was drawn earlier on the basis of liposome-swelling data [120]). This notion is also supported by the observation that OmpT-expressing cells allow faster penetration of a zwitterionic cephalosporin, cephaloridine, across the OM than do OmpU-expressing cells (727). Recent patch clamp results are also consistent with this, since the single-channel conductance values for OmpU and OmpT were, respectively, about 50 and 350 pS in 150 mM NaCl (618); the authors, however, favor the interpretation that these conductance values represent the insertion of (open) monomers and trimers, respectively.
Porins in γ-proteobacteria outside the Enterobacteriaceae.
γ-Proteobacteria are divided into three groups by 16S rRNA analysis (739). The γ-3 group contains the Enterobacteriaceae, vibrios, Haemophilus, fluorescent pseudomonads, and Xanthomonas. The fluorescent pseudomonads are characterized by the absence of genes coding for classical porins, as noted above. The trimeric porin P2 from H. influenzae type b has been extensively characterized in the laboratory of Coulton in terms of both its exclusion limit (689) and single-channel conductance (688). The OM permeability to cephaloridine, when calculated from the data given (150), seems to be around 1 to 2 μm s−1, that is, similar to what was found in E. coli (462). Recently this porin was found to bind specifically NAD+ and nicotinamide mononucleotide with the half-saturation constant of 4 to 8 mM, apparently an adaptation to the growth requirement of this organism for nicotinamide nucleotides (17). This again seems to be an example showing that the distinction between the general, nonspecific porins and specific channels is far from absolute. A major trimeric porin was sequenced (234) from an acidophilic, obligate lithotroph, Thiobacillus (now Acidithiobacillus [332]) ferrooxidans, which belongs to the γ branch according to the 16S rRNA sequence (332). The porin sequence shows a strong similarity to enterobacterial porins (Fig. 1). This porin has one of the lowest single-channel conductance values reported for porins, 30 pS in KCl (614). Guiliani and Jerez (234) note that in this porin from an extreme acidophile (with an optimum growth pH of 2.5) the pore-narrowing L3 loop, which contains many more acidic residues than basic residues in E. coli porins, appears to be nearly neutral. The structure-function relationship of these extremophile porins will be an interesting topic for further study.
The γ-2 branch is rather distant from the γ-3 group and contains only Legionella and related genera. The porin in Legionella pneumophila was identified, by extraction in the presence of 2-mercaptoethanol and reconstitution into planar bilayer, as a major OM protein of about 28 kDa (212). The protein could not be extracted in the absence of 2-mercaptoethanol, and the native structure of this porin turned out to be a disulfide-linked oligomer (probably a trimer) containing one subunit that was slightly larger (31 kDa) (275). The two kinds of subunits turned out to be the same protein, OmpS, but the 31-kDa monomer is larger because it has a fragment of peptidoglycan attached covalently near its C terminus (274). This covalent attachment to peptidoglycan is unusual, and the sequence of this oligomeric porin is not strongly related to that of any other classical porins (274), a feature consistent with the phylogenetic distance of Legionella from the γ-3 group.
A porin from Coxiella burnetii has been purified, and its activity was ascertained by the liposome-swelling assay (33). An unusual feature of this porin is that it dissociated, in the presence of mercaptoethanol, into subunits of 29.5 and 31 kDa, both relatively small for classical porins. Since both reacted with the same monoclonal antibody, they probably were derived from an identical precursor. GenBank contains a recent entry, AAM03442, which is annotated as the porin sequence from C. burnetii. This protein, when processed, would indeed have a rather low molecular weight of 24,500. Interestingly, the predicted sequence does not contain any cysteine residues, and mercaptoethanol must therefore contribute to subunit dissociation by a mechanism other than the reduction of disulfide bonds. Other features of this porin, its small size and the apparent presence of two types of subunits differing slightly in molecular weight, are reminiscent of the Legionella porin. C. burnetii is indeed closely related to Legionella according to their 16S rRNA sequences (476).
The γ-1 branch contains some purple sulfur bacteria such as Chromatium and Ectothiorhodospira. A porin gene from Ectothiorhodospira, a halophilic photosynthetic organism, has been sequenced, but the sequence does not appear to resemble the sequence of genes encoding enterobacterial porins (742), again a reasonable result in view of the distance between the two branches. Porins from two other species, Ectothiorhodospira shaposhnikovii and E. vacuolata, were shown to be trimeric, anion-preferring porins with single-channel conductance values typical for classical trimeric porins (743).
Porins in β-proteobacteria.
Comamonas (now Delftia [720]; previously Pseudomonas) acidovorans produces its major trimeric porin, Omp32, as a crystalline array in the OM (192). Its structure was determined by X-ray crystallography (774). It is a trimeric porin, and the monomer folds as a 16-strand β-barrel, as in other classical porins. However, a 6-residue segment in the middle of the second β-strand protrudes into the barrel lumen and constricts the eyelet region together with the ubiquitous infolding of the L3 loop. Consequently, the opening of the constricted region is narrower (5 by 7 Å) than in the E. coli OmpF porin. Furthermore, there are many basic amino acid residues (especially arginine) throughout the channel, and their charges are not neutralized by acidic residues, which are very few. This feature is consistent with the strong anion selectivity (the flux of Cl− was 17 times higher than the flux of K+ [405]) of this porin and with the preference of this organism for organic acids as carbon sources (720). At the constriction zone, three arginine residues are clustered, yet apparently all of them remain positively charged. A large factor here is the presence of a glutamate residue, whose side chain does not protrude into the channel yet stabilizes the positive charges of the two arginine residues protruding into the channel (770). Inspection of three-dimensional structures as well as the primary sequences of many porins from β- and γ-proteobacteria suggests that this arrangement is rather common in the trimeric, classical porins (770). Omp32 is copurified and cocrystallized with a small protein of 54 amino acid residues.
The sequence of Omp32 is similar to the sequences of the porins of other bacteria belonging to the β-proteobacteria, such as Bordetella pertussis, Neisseria gonorrhoeae, Neisseria meningitidis, and Acidovorax (previously Pseudomonas) delafieldii (220, 303, 774) (Fig. 1). The large excess of basic amino acids in the channel interior seems to be a common feature in these porins, on the basis of primary sequence alignment (774). This fits with the strong anion selectivity of the Omp34 porin from A. delafieldii (103). Also, the anion selectivity was already noted in one of the first papers describing the N. gonorrhoeae porin (408). The porin of B. pertussis also produces an anion-selective channel (22). This protein appears to occur as a two-dimensional crystalline array in the OM (334, 341). (A crystalline arrangement of a trimeric porin was also reported from the stalk region of an unidentified prosthecate bacterium [121]). Another related porin was reported to be induced by the virulence regulatory system bvg (206); the functional properties of this porin have not been investigated.
Neisserial porins have been studied extensively, in part due to their potential use in vaccines. The N. gonorrhoeae porin gene was first cloned and sequenced in 1987 (230). N. meningitidis expresses two porins, PorA and PorB. The porB locus is occupied in any given strain by either of the two alleles, each expressing either class 2 or class 3 PorB protein, which are only 60 to 70% identical. The class 3 porin, which has extensive alterations in the external-loop sequence (including many short deletions), exhibits the interesting property that its trimers are unusually labile in the presence of SDS even at room temperature (418), producing monomers that apparently still retain the β-barrel structure but are altered (denatured) in the loop structure. In this connection, we note that with the OmpF porin of E. coli, modification of loop 2, which connects the neighboring monomers, can make even this porin susceptible to SDS-induced dissociation at lower temperatures (493).
Burkholderia cepacia shows strong intrinsic resistance to a number of antibiotics, and the permeability of its OM to nitrocefin (a cephalosporin) is an order of magnitude lower than that in E. coli (483). When the major porin species was isolated, it dissociated into a major 36-kDa species and a minor 27-kDa species on being heated in SDS. The authors speculate that the active porin is a trimer of the former, a conclusion that seems to be consistent with a later study from another laboratory (229). It produced also a low conductance (0.23 nS) channel in 1 M KCl (483). The gene coding for the 37-kDa species, opcP, was cloned; the sequence is most similar to that of the B. pertussis porin (680).
Interestingly, a 37-kDa OM protein of Vibrio furnissii, which is induced by the presence of chitooligosaccharides and facilitates the specific diffusion of such oligosaccharides (335), shows homology to the neisserial porins.
Porins in α-proteobacteria.
The α-proteobacteria have been divided, by the use of 16S rRNA signature sequences, into three groups, α-1, α-2, and α-3 (738). The porin for which the first three-dimensional structure was determined by crystallography (714, 717, 718) was from Rhodobacter capsulatus, a member of the α-3 group. The monomer produces a 16-strand β-barrel, which is assembled into a now classical trimer structure (see “Crystallographic structure of porins” above). In planar lipid bilayers, the porin produces a high single-channel conductance of 3.2 nS in 1 M KCl (741), presumably corresponding to the insertion of a trimer with three open channels. A similar structure was shown for Rhodopseudomonas blastica (now Rhodobacter blasticus) (351).
The structure of the trimeric porin from Paracoccus denitrificans, also belonging to the α-3 branch (531), was reported at 3.1-Å resolution (273). Although this organism was earlier reported to produce a dimeric porin (772), this conclusion was obviously incorrect, presumably a result of undue reliance on the mobility of undenatured oligomers in SDS-PAGE. Unlike Rhodobacter and enterobacterial porins, which contain an excess of acidic amino acid residues in the channel (although the eyelet itself has only two acidic residues [Fig. 2C], there are many more acidic residues on the channel wall, including Glu62, Asp97, Asp107, Glu296, and Asp312 in OmpF approximately at the same level as the eyelet), and unlike the Omp32 porin, which contains mostly basic residues in the channel, the P. denitrificans porin channel contains about equal numbers of acidic and basic residues (273). This is apparently reflected in the ion selectivity of this porin, which is reported to be almost nonselective or slightly anion selective (581, 582). Isolation of an oligomeric (most probably trimeric) porin from another member of the α-3 branch, a phototrophic, marine organism, Roseobacter denitrificans, was reported (441); this porin, like other trimeric porins from the α-3 branch, is dissociated into monomeric units at low temperature in the presence of EDTA, apparently a hallmark of porins in this group. An oligomeric porin from Thiobacillus (now Paracoccus) versutus, with rather high single-channel conductance, was reported (740).
Besides these organisms, Brucella, which belongs to the α-1 branch, appears to be the only organism whose porins were studied in some detail. Following the demonstration by the proteoliposome-swelling assay that the Omp2 protein of Brucella abortus forms a channel approximately equal in size to the E. coli OmpF channel (180), the gene for this protein was identified as omp2b (400). Interestingly, there is another homologous gene, omp2a, located right next to it but read in the opposite direction. Although Omp2a was apparently not expressed in B. abortus, it allowed the permeation of a large tetrasaccharide when expressed in E. coli (400), suggesting that it produces a larger channel. Indeed, comparison of the sequence suggests that Omp2a has a large deletion in the putative channel-narrowing L3 loop. However, in a planar lipid bilayer study, Omp2a behaved in a rather anomalous manner, in contrast to Omp2b, which produced conductance steps expected for stable, trimeric porins (424). In any case, the juxtaposition of two very homologous genes apparently produces constant variation in gene sequences via gene conversion mechanism (482), and the effect of this variation on the physiology and evolution of Brucella species remains an interesting topic. OM protein IIIa of Rhizobium leguminosarum is homologous to Brucella porins (128), but no further study of the Rhizobium porin seems to exist.
Porins in δ- and ɛ-proteobacteria.
The δ-proteobacteria consists of organisms including myxobacteria, Bdellovibrio, and some sulfate reducers (476, 737). I am not aware of any detailed studies of the porins from these organisms.
More recently, Campylobacter and Helicobacter were recognized to form another deep branch in the Proteobacteria, with only very remote relationships to the other four subdivisions (365, 678); this group is now called the ɛ-proteobacteria. Because of the clinical significance of these organisms, their porins have been investigated. Campylobacter jejuni produces a trimeric porin called the major OM protein (MOMP) (79), with a monomer molecular weight of about 45,000. The trimeric structure is unstable and is easily dissociated by SDS even at room temperature (79). The trimer conformation, however, has been confirmed in two-dimensional crystallization studies (12, 787). Primary sequence analysis utilizing local hydrophobicity and sequence divergence among isolates suggested the presence of 18, rather than 16, transmembrane β-strands (362, 782), and comparison with specific channels LamB and ScrY (described below) showed potential conservation of some short signature motifs (362). On this basis, MOMP will be a specific channel rather than a general porin. However, its high single-channel conductance (0.4 nS per monomer) (162) is unexpected for a specific channel. Unfortunately, no further functional study has been carried out on this protein. Another, slightly larger, putative porin, Omp50, was identified in C. jejuni (78). This protein shows a low single-channel conductance of 50 pS, consistent with a specific channel, but no other functional information is available.
Helicobacter pylori, another member of the ɛ-branch, was found to produce at least four “heat-modifiable” envelope proteins, a characteristic that is suggestive of the β-barrel structure (196). These proteins (48 to 67 kDa), which are called HopA, HopB, HopC, and HopD, are related to each other, and all produced ion channels in planar lipid bilayers. In a separate study, a 30-kDa protein was suspected of being a porin (681); indeed, this protein, called HopE, was shown to produce 1.5-nS channels in 1 M KCl (178). It also appears to exist as oligomers (most probably as trimers). More than 30 paralogs of this protein were found in H. pylori (663). Analysis of the HopE sequence and linker insertion experiments suggested that it is a 16-member β-barrel (60), but the folding model, with a small L3 loop and a huge L6 loop, looks very different from the enterobacterial porins.
Porins in the Planctomyces-Chlamydia group.
The MOMP of Chlamydia trachomatis, an obligate intracellular pathogen, was suggested to be the porin by liposome-swelling assay (43). Since then, a somewhat truncated version of the gene (750) and a full version (309) were cloned and the protein was expressed in E. coli. The pore-forming activity of this protein was confirmed by ion conductance after planar bilayer film reconstitution and liposome swelling, respectively. The genome sequence, however, indicated the existence of another homolog, porB, and this gene was shown to be a minor porin (352). Interestingly, PorB appears to prefer, by a wide margin, intermediates of tricarboxylic acid cycles such as 2-ketoglutarate to simple neutral sugars (353). This preference might be important for Chlamydia, which lives within animal cells.
Porins in the Cytophaga-Flexibacter-Bacteroides group.
In 1992, Wexler's group reported the identification of an OmpA-like major protein from the OM of Bacteroides distasonis, a protein that showed pore-forming activity in the proteoliposome-swelling assay (723). The protein was characterized subsequently (725). More recently, the same group identified a heterodimeric protein complex of approximately 200 kDa (Omp200) from Bacteroides fragilis (722). This complex also shows porin activity and is thought to be the major porin in this organism. The genes coding for the subunits (omp121 and omp71) have been identified (724). The larger protein appears to be rich in β-strands, and the smaller protein appears more hydrophobic. This seems to be a porin of a totally new type, and more studies of its structure and function should be carried out. (OM proteins involved in the degradation of polysaccharides are discussed in “Specific channels” below).
Another group noted that β-lactam-resistant clinical isolates of B. fragilis tended to lack a 45-kDa envelope protein and reported that this protein showed pore-forming activity after electroelution from SDS-PAGE (472). The relative contribution of this protein, as well as the OmpA-like protein and Omp200, to the overall permeability of this organism remains to be determined. In another study, 51-, 92-, and 125-kDa proteins were isolated from the B. fragilis OM; although all proteins were active in pore formation, the specific activity was orders of magnitude lower than that of E. coli OmpF (318).
Porins in spirochetes.
The Treponema pallidum OM contains an almost 100-fold-lower level of proteins than does the E. coli OM (525). The identification of the major porin in this organism has therefore been difficult and controversial. One group has identified a 31-kDa OM protein, Tromp1, as the major porin, based primarily on the ability of this protein to form channels in planar-bilayer assays (70). Another group (8), however, found that this protein did not produce permeability on reconstitution into liposomes. The protein lacked the extensive β-structure expected for porins. Furthermore, the protein sequence was found to be homologous to that of the solute-binding proteins, and the gene was a part of an operon coding for components of an ABC transport system. These results suggest strongly that Tromp1 is a solute-binding protein, anchored to the cytoplasmic membrane. The final proof of this interpretation came with the solution of the crystal structures of Tromp1 (or TroA) with or without the ligand, Zn2+ (369, 370), which showed a fold typical of periplasmic solute-binding proteins rich in α-helices. The first group presented a rebuttal, reporting that the urea denaturation of the protein followed by renaturation through dialysis produced a trimeric conformer with porin activity (781). However, the circular dichroism spectrum of the renatured protein showed no evidence of a predominant β-conformation. Furthermore, the argument of these authors that genes coding for proteins unrelated to solute-binding activity are sometimes present in ABC transport operons is incorrect. The authors cite the case of “adhesins” in gram-positive bacteria, but these are metal-binding proteins anchored to the plasma membrane, which also function as adhesins. Although I do not understand why the protein produces channels, the case for Tromp1 as the major porin is not convincing at present.
The Borrelia burgdorferi OM showed two kinds of channels, one small and one large (622). The 25-kDa protein, Oms28, was shown to produce the channel with the lower conductance of about 1 nS in 1 M KCl (622). A larger protein, Oms66 (SwissProt accession number Q44841) was shown to produce channels of very high conductance, about 10 nS (623). In Leptospira, a 31-kDa protein, OmpL1, produces channels with a conductance of 1 nS and also to form a rather unstable trimer (605). The C-terminal aromatic amino acid residue (usually phenylalanine), which characteristically occurs in classical porins of the Enterobacteriaceae (635), does not occur in any of these spirochetal porins.
Porins in cyanobacteria.
Envelopes from cyanobacteria were shown to produce ion-permeable channels on reconstitution into planar lipid bilayers (50, 311). The responsible porins were identified only fairly recently (252). The proteins travel as a 140-kDa complex if subjected to SDS-PAGE without heat denaturation, but they collapse into about 50-kDa monomers if heated in SDS; this result suggests that they exist as a tight trimer. On insertion into a planar lipid bilayer, they produce a single-channel conductance of approximately 0.5 nS in 1 M KCl. (However the same laboratory reported an order of magnitude higher value of 5 nS earlier [251], and the authors were unable to give reasons for this difference.) This value has been compared with the “much higher” single-channel conductance values of 2 to 3 nS for enterobacterial porins, and it has been argued that cyanobacteria does not need large channels because they require, as autotrophs, the uptake of only inorganic ions (276). However, this comparison unfortunately disregards the fact that the high “single-channel” conductance values of enterobacterial porins actually correspond to the insertion of three open channels of a trimer. Since it is not known whether the conductance of cyanobacterial porin corresponds to one or three channels, such an argument seems premature. More convincing assays, such as liposome swelling, are sorely needed for cyanobacterial porins.
Determination of the partial amino acid sequence of these porins led to the realization that one of them was SomA, a major OM protein identified earlier (683). There are six homologs in the genome of Synechocystis strain PCC 6803 (276). All these proteins are much larger than the classical porins of the Enterobacteriaceae, and the additional mass exists at the N terminus as an S-layer homology domain, which is likely to interact strongly with the peptidoglycan layer (252, 276).
Porins in Fusobacterium.
Fusobacterium, a genus of gram-negative bacteria which seems to be related, surprisingly, to the gram-positive Corynebacterium-Nocardia-Mycobacterium complex on the basis of 16S rRNA sequence (476), has been studied in several laboratories because of its potential role as a periodontal pathogen. An OM protein of about 40 kDa was identified as a porin by the liposome-swelling assay and was shown to exhibit heat modifiability (see “Slow porins” above) (644). The fomA gene was cloned and sequenced, and a 16-strand β-barrel model was proposed mainly on the basis of comparison of protein sequences from different strains (80), a procedure that has produced reliable predictions in the past (452). The predicted pattern is interesting in that loop 3, which is always large in the classical enterobacterial porins, is small and, instead, loop 6 is the largest. The protein was shown to exist as a trimer by cross-linking and to form channels of very weak ion selectivity (338). Introduction of epitope insertions and short in-frame deletions confirmed the original folding model proposed and showed that partial deletion in loop 6 indeed led to the production of channels with much higher permeation rates for cephalosporins (339, 519). Recent results (520), however, suggest that the N terminus of this protein is located in the periplasm and that the β-barrel actually contains 14 strands.
Porins in the Deinococcus-Thermus group.
A porin of exceptionally large apparent size (185 kDa) was found in the Thermus thermophilus OM. This porin is also unusual in producing a channel of exceptionally high conductivity, 20 nS in 1 M KCl (394), some 30 times higher than that of E. coli OmpF.
Porin in Thermotoga.
Thermotoga maritima, an extremely thermophilic eubacterium, produces a “sheath” with a crystalline array of trimeric proteins with triple indentations (524). This protein is also porin-like in terms of the high content of β-structure. This protein, called Omp-beta, has since been shown to produce cation-selective channels in planar bilayers (191).
Porins in the Corynebacterium-Nocardia-Mycobacterium group.
Bacteria in the Corynebacterium-Nocardia-Mycobacterium group belong to the gram-positive bacteria, yet their peptidoglycan layer is covered by a lipid-rich layer. They produce long-chain mycolic acids, covalently linked to arabinogalactan, which in turn is covalently linked to peptidoglycan. Mycolic acids present in mycobacteria may contain as many as 90 carbons and may contain no or very few double bonds. In 1993, it was shown that the chains of mycolic acids are arranged in a direction perpendicular to the cell surface (457), and it was proposed that these bacteria produce, outside the peptidoglycan layer, an OM-like structure that is composed of an inner leaflet of covalently linked mycolate residues and an outer leaflet of extractable lipids (99). It was further found that the hydrocarbon domain of this cell wall had exceptionally low fluidity (381, 382). These observations explain the high intrinsic resistance of many mycobacterial species to lipophilic antibiotics that are expected to diffuse through the lipid bilayer regions of the cell wall (301).
Mycobacteria must, however, take up hydrophilic nutrient molecules. Thus porins were expected to be present. Furthermore, the permeability of the mycobacterial cell wall to hydrophilic solutes was shown to be quite low: compared with the permeability of the E. coli OM, it was about 1,000-fold lower in M. chelonae (302) and about 100-fold lower in M. tuberculosis (122). Therefore, the property or the amount of the porins should explain this extremely low permeability.
The first report on mycobacterial porins was that of Trias et al. (670, 673). These workers assayed detergent-solubilized proteins of the cell wall of M. chelonae for channel formation in planar bilayers and discovered a fraction that produced a rather high single-channel conductance of 2.7 nS in 1 M KCl. The protein appeared to be about 59 kDa on the basis of its mobility on SDS-PAGE, and the paucity of this protein in the cell wall appeared to explain the low permeability of the intact cell wall.
A similar study with M. smegmatis showed the presence of porins producing 3-nS channels in 1 M NaCl (671). The use of ions with different radii led to the prediction that the channel diameter was about 3 nm. When the protein was purified, it behaved as though it had a molecular mass of about 100 kDa in SDS-PAGE (444). (An M. smegmatis porin producing a large channel was also reported by another laboratory [432]. The N-terminal sequence of this protein is different from that of MspA [discussed below], and its role in cell wall permeability is not clear.) Importantly, heating of the sample in 80% dimethyl sulfoxide (but not in SDS) resulted in the conversion of the protein into a 20-kDa band. N-terminal sequencing of the protein led to the cloning of the mspA gene (444). In a crucial experiment (630), a deletion of this gene caused a large (four- to ninefold) decrease in the cell wall permeability, as assessed by the diffusion of cephaloridine and glucose, a result indicating that MspA is indeed the major porin for M. smegmatis. The permeability did not go to zero. Indeed, the use of degenerate primers resulted in the identification of other close homologs of MspA in this organism, MspB, MspC, and MspD, whose sequences are essentially identical to that of MspA except for the region close to the N terminus (630). In a recent study (193), Engelhardt et al. showed that the isolated MspA has a negative-stain-filled channel of 2.5 nm in diameter and that such channels were also present on the surface of intact cell wall. The size is in good agreement with the original estimate of Trias and Benz (671). Furthermore, the native MspA oligomer was shown, by cross-linking, to be a tetramer, and the end-on view of the oligomer showed a fourfold symmetry. Since the length of the channel appeared to be unusually long, about 10 nm, in the side view, each of the monomers probably extends for this entire length. This length is consistent with the thickness of the mycobacterial cell wall, made thicker primarily because of the extraordinary length of the mycolic acid chains (99). This length will also slow the diffusion of solutes. The surface density of the channel appears to be about 50-fold lower than that of porin channels in the E. coli OM, and both of these factors are probably sufficient to explain the low permeability of the M. smegmatis cell wall. The remarkable stability of the tetrameric β-sheet porin, MspA, which requires heating to 92 to 112°C for denaturation (even in the presence of SDS), was described recently (263).
A porin from M. phlei migrates in SDS-PAGE as a 135-kDa protein, which collapses into a 22-kDa monomer on heating in dimethyl sulfoxide (547); it is likely that this porin is a homolog of MspA. Cell wall extracts of M. tuberculosis was shown to contain porins behaving like MspA, as well as another porin that produces a much lower single-channel conductance (323). The situation here seems similar to the coexistence of at least two porin channels in M. bovis BCG (377).
Studies of porins in other members of the Corynebacterium-Nocardia-Mycobacterium complex have also been done. Three species of Nocardia were found to produce porins that had a large aggregate size (>80 kDa) and formed channels with high single-channel conductance values (3 to 6 nS) (546, 549, 550). On heating in SDS, these proteins collapsed into monomers of approximately 20 kDa (when identified), and thus the nocardial porins are probably similar to the MspA-type porins of mycobacteria. C. glutamicum was shown to contain a channel producing a high single-channel conductance of 6 nS in 1 M KCl (445). The porin was isolated following its initial extraction into organic solvents (376). It turned out to be an extraordinarily small protein of only 45 amino acid residues, encoded by the porA gene, and PorA was confirmed to be the major porin by deletion of the gene (148). Information about the three-dimensional structure of this protein is not yet available. Since corynemycolic acids have much shorter chains (32 to 38 carbons) than mycolic acids, it is perhaps not impossible for such a small protein to cover the entire thickness of the corynebacterial cell wall, if the MspA model is applicable here. Porins have been found in the cell wall of Rhodococcus erythropolis (378) and R. equi (548).
The M. tuberculosis genome contains a homolog of E. coli OmpA. The product of this gene (OmpAtb) was shown to generate pores in reconstituted systems (603). Although the low permeability of OmpA-like porins (see “Slow porins” above) is attractive in explaining the generally low permeability of the mycobacterial cell wall, it is rather difficult to imagine that a protein similar to those that traverse the thickness of E. coli OM could span the much thicker mycobacterial cell wall. A deletion mutant containing a mutation of this gene was constructed recently (535). It had difficulty in growing in acidic media, and at pH 5.5 the uptake of serine was nearly totally abolished in the mutant, suggesting that this protein is a major porin under these conditions.
Putative porins in archaea.
Cell envelopes of Haloferax volcanii were found to contain ion channels that are mostly open, showing a behavior resembling that of porin (55). However, OMs are not known in archaebacteria. The authors speculate that the region of the envelope hitherto thought to correspond to “periplasm” may actually be membranous.
Porins and Antibiotic Resistance
The properties of constitutively expressed porins obviously exert a profound influence on the intrinsic level of antibiotic resistance in gram-negative bacteria. Thus, P. aeruginosa (and most probably other members of the fluorescent pseudomonads, although their resistance to antibiotics is rarely assessed) shows a well-known intrinsic resistance to a wide range of antibiotics, in comparison with the members of the Enterobacteriaceae, in large part due to the use of a slow porin, rather than a classical trimeric porin, as the major channel. The low-permeability OM becomes even more effective in preventing the influx of antibiotics, thanks to the synergistic effect of the multidrug efflux complex, which extrudes drugs directly into the medium (450, 454).
Route of antibiotic influx.
In many cases it is not easy to determine the pathway of influx of antibiotics. Agents that contain a strong acid group, a quaternary ammonium group, or multiple charged groups are expected to have difficulty in crossing the bilayer regions of the OM. This is especially so because the LPS-containing bilayer of the OM shows permeability to lipophilic probes that is about 2 orders of magnitude lower than that of the conventional phospholipid bilayer membranes (504) (see “Lipid bilayer as a diffusion barrier” below). As an experimental criterion, one can compare the antibiotic susceptibility of the wild-type strain with that of an LPS mutant strain (either a “deep-rough”-type rfa mutant or a leaky lpxA mutant) (685). Alternatively, one can add polymyxin B nonapeptide (PMBN) to perturb the LPS-containing bilayer (684). A greatly increased susceptibility in the mutant or in the PMBN-treated bacterium indicates strongly that the major pathway of diffusion is through the OM bilayer. It must be realized, however, that this change in susceptibility occurs mainly because the drug influx is counteracted by the active drug efflux; this is clear from the observation that inactivation of the major multidrug efflux pump AcrAB in E. coli produces drug hypersusceptibility almost exactly like that seen on permeabilization of the OM (455). Therefore, the observation of increased susceptibility is significant, but negative results may not mean much because they could be caused by intrinsically high influx, poor efflux, or both. The same caution must be exercised in interpreting drug susceptibility data from porin-deficient mutants. For example, in an ompR mutant of E. coli in which the expression of both OmpF and OmpC was decreased, there was no gross changes in the fluoroquinolone MICs (497). However, because of the consideration just described, these results do not rule out the diffusion of quinolones through the porin channels.
It would be ideal if one could measure the flux of drugs through the porin channels. This is possible for β-lactams by using intact cells or unilamellar liposomes, as mentioned above (see “Functional assays” in the section “Classical porins”). Unfortunately, for most other drugs the assay is difficult. The liposome-swelling assay is easy for uncharged or zwitterionic agents and possible for monoanionic hydrophilic drugs after some compromise (461). However, it is currently impossible for other classes of charged drugs.
If we take the theoretical considerations together with the experimental data, it is likely that small agents (β-lactams, tetracycline, chloramphenicol, and fluoroquinolones) use mainly the porin channels for penetration, at least in the Enterobacteriaceae, with their high-permeability porins. In contrast, large, lipophilic agents, such as macrolides, rifamycins, novobiocin, and fusidic acid, have difficulty in diffusing through the porin channels, and therefore even the slow diffusion across the lipid bilayer becomes significant.
Aminoglycosides have been proposed to diffuse across the OM of P. aeruginosa by first perturbing the LPS leaflet (“self-promoted uptake”) just like polymyxin does (242, 249). This hypothesis is discussed below (see “Alterations of the OM bilayer barrier”).
Resistance caused by loss or modification of porins.
For agents that cross the OM via porin channels, a decrease in porin-mediated permeability may increase the level of resistance. Loss of the porin that produces the largest channel is predicted to increase resistance to hydrophilic antibiotics, since these molecules are usually larger than the common nutrients and are barely able to pass through these channels (for example, see reference 765). Examples of increased resistance due to porin loss are too numerous to mention in an exhaustive manner, but I will give some early examples here. Mutants with mutations of the ompF gene are easily selected in E. coli by the use of bulky, relatively β-lactamase-resistant, β-lactam compounds (255). One of the earliest examples of porin-deficient clinical isolates was the report of an S. marcescens strain that became simultaneously resistant to both aminoglycosides and β-lactams (228). When the broad-spectrum cephalosporins such as cefotaxime were introduced, it was found that the derepressed production of β-lactamase, accompanied by the decreased production of porins, produced elevated resistance most frequently in E. cloacae (111, 721). With E. coli, which cannot respond to cephalosporins by inducing its chromosomally encoded β-lactamase, the decrease in the production of porins was the only way to gain resistance (30). A clinical isolate of S. enterica serovar Typhi showed chloramphenicol resistance because it could not express the wider-channel porin, OmpF (665) (undoubtedly with the synergism with the multidrug efflux system [see below]). In another example of a strain from clinical sources, an S. enterica serovar Typhimurium strain originally expressed only OmpC porin and became more resistant by losing its only porin species (412). A drug-resistant B. cepacia strain with a reduced expression of porin was reported (23). Several H. influenzae strains with reduced levels of porin were more resistant to several antibiotics (110).
Several studies reported the alteration of porin patterns in drug-resistant K. pneumoniae. Loss of porin accompanied by the production of β-lactamase produced cephamycin resistance in vivo in K. pneumoniae (480). This organism normally expresses OmpK35 (an OmpF homolog) and OmpK36 (an OmpC homolog). In one study, a clinical isolate did not express one of the two porins from the beginning but became more strongly antibiotic resistant after the apparent loss of the remaining porin (402). OmpK35 may not always play a predominant role in drug influx, because mutants with single mutations in either OmpK35 or OmpK36, produced in the laboratory, were essentially unaltered in drug susceptibility, although deletion of both porins increased resistance, as expected (268). A study using clinical isolates was repeated by the same investigators (267). In strains that are highly resistant to bulky broad-spectrum cephalosporins, the larger-channel porin, OmpK35, is not usually expressed, leaving behind only the narrower-channel porin, OmpK36. In some strains, both of the porins appear to be absent, but these bacteria apparently survive by expressing, at a low level, a normally quiescent porin, OmpK37, which produces a channel that is even more restrictive than OmpK36 (179).
Loss of the major porins, however, is expected to have deleterious effects on the survival of the organism in its natural habitat. More recently, antibiotic-resistant mutants of E. aerogenes were reported in which the properties of their porins were altered (395). In a detailed study, alteration of the channel-constricting loop L3, resulting in the strongly decreased influx of anionic compounds, was identified (161). It is likely that the influx of particular drugs used in therapy (usually cephalosporins of the most recent “generation” with large substituents that make the drugs inert to the action of commonly occurring β-lactamases) is drastically decreased without, presumably, decreasing significantly the influx of small, common nutrients. Detailed comparison of the permeability of these relevant compounds, however, is not available.
A mutation, penB, in N. gonorrhoeae has been known for a long time to increase the resistance level to penicillin and tetracycline, when the strain also has the mtr mutation that leads to the overexpression of the major multidrug efflux system, MtrCDE. The penB mutation was also known to be linked to the porin gene, por. A genetic study by Gill et al. (222) showed that penB is an allele of por, with alterations in the channel-constricting loop 3 of the protein, so that two additional negative charges are introduced into the channel. The influx rate of penicillin was shown to be drastically reduced in strains expressing this variant porin.
Imipenem crosses the P. aeruginosa outer membrane primarily through the OprD specific channel (674) (see “Specific channels” below). Mutants with decreased expression of this protein become predominant easily during imipenem therapy, since OprD is not important for the uptake of most nutrients. Martinez-Martinez and coworkers found that growth of P. aeruginosa in the presence of siliconized latex urinary catheters, a clinically relevant condition, induces non-hereditary resistance to imipenem (403), an effect now traced to the elution of zinc, which represses OprD expression (142).
Specific Channels
Specific channels catalyze the downhill (spontaneous) diffusion of specific classes of nutrients. A prototypic member of this class is the LamB protein, or phage lambda receptor protein of E. coli, which catalyzes the influx of maltose and higher oligosaccharides of the maltose series. It may sound strange that E. coli needs this specific channel, when the classical porins OmpF and OmpC allow the ready influx of at least maltose. However, diffusion rates through the porin channels are proportional to the concentration difference across the membrane, and therefore the rates become minuscule when the external concentration of the solute drops far below the millimolar level (464). This effect is especially severe for a fairly large solute such as maltose, which diffuses through the OmpF porin channel at a rate about 2 orders of magnitude lower than the diffusion rate of a pentose, arabinose (see Fig. 5 of reference 464). Since starch is degraded primarily to maltose by intestinal amylase, E. coli is thought to need a specific channel for this most important carbon source in the natural habitat. However, more recent studies have shown that LamB is actually important for the uptake of other sugars as well (see below).
FIG. 5.
Hypothetical models of the ExbBD-TonB (left) and TolQR-TolB-PAL (right) systems. As seen, most proteins in these two systems are similar in their membrane topology, except for the periplasmic protein To1B and the OM lipoprotein PAL, which do not have counterparts in the Ton system. TonB is drawn as though it will interact directly with the OM receptors while still associated with the inner membrane (IM). However, this may be an oversimplification, and there are pieces of evidence that favor the shuttling of TonB between the OM and the inner membrane (372).
LamB or maltose channel.
Early studies of the LamB channel, which suggested that the channel favored the diffusion of maltose and maltodextrins, were discussed in the previous version of this review (464). The first significant advance since then was the planar lipid bilayer study (53). The single-channel conductance had a rather low value of 0.16 nS in 1 M KCl. However, this ion conductance was inhibited by the oligosaccharides of the maltose series, and this information not only showed the specific binding of these ligands to the interior of the channel but also allowed the determination of their binding affinity. Expressed as KD values, the affinity ranged from quite poor (10 mM for maltose) to reasonable (about 60 μM for maltopentaose, hexaose, and heptaose). This approach was further refined by analysis of the current “noise” that is generated by the binding and dissociation of sugars (15). The study showed that there was not much difference among sugars in the “on” rate but that the “off” rates differed widely from 2,000 s−1 for maltotriose to 180 s−1 for maltopentaose. More recently, this line of approach was further refined to the observation of the transport of individual sugar molecules through a single channel (355); such studies showed that each of the channels within a trimer functioned independently and that the entry of sugar was easier from one end of the channel but that once the sugar was bound to the interior of the channel, the direction of its initial entry did not produce any difference in the behavior of the sugar molecule.
Perhaps the work that had the strongest impact was the solution of the X-ray crystallographic structure of LamB in 1995 (588) (Fig. 4). Each subunit within the LamB trimer was shown to contain a β-barrel consisting of 18 transmembrane β-strands, in contrast to the 16 found in classical porins. As in the porins, the external loop L3 folds back into the channel and causes a constriction, which is much narrower (0.5 to 0.6 nm in diameter) in LamB. The outer half of the pore is also more constricted than in the porin channel, owing to the infolding of the additional loops L1 and L6. In spite of the narrowness of the channel, the path for maltodextrins could be traced in the crystal soaked with maltotetraose, since the entire length of the channel wall is lined with a succession of six aromatic residues, called the greasy slide, with which the rings in sugar residues interact (Fig. 4B and C). The interaction between the sugars and the greasy slide was elucidated in greater detail by soaking the crystal with maltose, maltotriose, and maltohexaose (188). Thus the sugars enter in the natural, left-handed helical conformation into the channel, with the nonreducing end first. The aromatic residues of the greasy slide are arranged in a helical manner, and they interact via van der Waals forces with the less polar side of the sugar rings (Fig. 4C). There is a slight difference in the helix curvature between the longer maltooligosaccharides and the greasy slide, so that maltohexaose strongly interacts only with the central three aromatic residues of the slide. Especially at the constriction region, there are also many charged residues that are predicted to form hydrogen bonds with the hydroxyl groups of the sugars (188); the role of these “polar tracks” was confirmed by site-directed mutagenesis (186). Finally, soaking of the crystal with sucrose, trehalose, and melibiose showed that the last two could bind to the greasy slide and could be transported, although the interaction at any given time would involve only one sugar unit of the disaccharide (709). With sucrose, on the other hand, the fructose residue prevents the entry of the sugar into the constricted region of the channel and hence prevents its translocation. These results are in complete agreement with the liposome-swelling study showing that LamB allowed the diffusion of trehalose and melibiose but not sucrose (388). The structure of the greasy slide also predicts that LamB will catalyze the diffusion of glucose, and an in vitro reconstitution confirmed this prediction (388). It is interesting in this connection that a chemostat study showed that under carbohydrate-limited conditions, LamB production by E. coli became strongly derepressed and that the presence of LamB conferred advantages to growth at limiting concentrations of not only glucose but also lactose, arabinose, and even glycerol (164). Thus, the LamB channel is not really a maltose-specific channel (or maltoporin), but its function in E. coli physiology is to facilitate the influx of a wide variety of carbohydrates when they exist in low concentrations in the environment.
FIG. 4.
X-ray crystallographic structure of LamB. (A) Side view of the monomeric unit. The β-barrel contains 18 strands in this protein, in contrast to the 16 strands seen in the trimeric porins. In addition to loop 3 (orange), loop 1 (red) folds deeply into the channel. Loop 2 (blue) folds outward and interacts with the neighboring subunit in the trimer, as in OmpF (Fig. 2A). Other loops also are often large and tend to cover the entrance of the channel from the outside. (B) View of the monomeric unit from the top. The greasy slides (Tyr41, Tyr6, Trp420, Trp358, and Phe227) are shown as blue stick diagrams, and Tyr118, which constricts the diffusion channel from the opposite side, is shown as a yellow stick diagram. (C) View of the greasy slide and its interaction with maltotriose. This is a side view with the front of the β-barrel cut out for a better view of the slide. The aromatic residues that comprise the greasy slide and Tyr118 are shown as stick diagrams colored as in panel B. The maltotriose molecule (Triose) is shown as a stick diagram colored orange. The coloring of the loops is the same as in panel A. The diagrams are based on PDB coordinate files 1MAL and 1MPN.
ScrY or sucrose channel.
A specific channel that is homologous to LamB (25% identity at the amino acid level), ScrY, is encoded by a plasmid that allows some strains of E. coli and Salmonella to utilize sucrose as a carbon source (256). This channel allows the rapid diffusion of a large variety of sugars, such as glucose, fructose, arabinose, maltose, lactose, raffinose, and maltodextrins, in addition to sucrose (13, 256, 591, 595). The N-terminal segment of ScrY, about 70 residues long, has no equivalent part in LamB, and in the crystal structure of ScrY this portion was disordered. The structure of the C-terminal 413-residue portion of this protein (208) provides an interesting contrast to that of LamB. As in LamB, the protein forms a trimer, each subunit consisting of an 18-stranded β-barrel. The channel is constricted by the infolding of loop 3, but the constriction is significantly wider (0.85 by 1.1 nm) than in LamB. The channel contains a greasy-slide structure very similar to that in LamB. Sucrose, however, was seen to interact with the aromatic residues of the slide only through the pyranose ring of the glucosyl residue; the fructosyl residue pointed away into the lumen of the channel. Furthermore, the residues in LamB that stick out into the lumen and prevent the diffusion of the bent sucrose molecules are absent in ScrY. In fact, conversion of Asp201 of ScrY into Tyr, to produce a constriction structure resembling that in LamB, prevented the diffusion of sucrose without impairing that of maltose (682). In a converse experiment, changing the residues that present obstruction to sucrose permeation in LamB, Arg109 and Tyr118, into alanine produced a mutant channel that now allowed sucrose to enter (694).
The function of the N-terminal 70-residue extension in ScrY is somewhat controversial. When this extension was deleted, there was not much change in the binding of various sugars to the channel, but the diffusion of maltopentaose was greatly enhanced in intact cells for unexplained reasons (596). In a more recent study (185), the peptide comprising this domain was shown to form a parallel, three-stranded coiled coil, which was shown to bind to sucrose in equilibrium dialysis with an affinity 2 orders of magnitude higher than the binding to the greasy slide within the channel. (The binding of sucrose to this peptide was also observed by NMR [415].) This appeared to support the idea that the N-terminal extension forms an extension of the greasy slide to facilitate the movement of sucrose into the periplasm. When the N-terminal sequence was added to LamB, the diffusion of higher maltodextrins was greatly enhanced, a result that is not inconsistent with the idea of the periplasmic extension of the transport pathway. The authors argue that such an extension may not be needed for LamB in intact E. coli cells because maltodextrins will be captured immediately by the periplasmic maltose-binding protein. However, these data do not fit well with the better transport of maltopentaose by the N-terminus-truncated version of ScrY.
“BglH,” an aryl-β-d-glucoside channel.
A gene immediately downstream from the bglGFB operon, yieC, was found to code for a sequence homologous to LamB (708). It was thought to be a part of the operon that functions in the degradation of aryl-β-glucosides in E. coli and was named bglH and overexpressed (18). The BglH protein (58 kDa) was a trimeric OM protein, and when inserted into the planar lipid bilayer it produced ion channels that were blocked most effectively by aryl-β-glucosides such as arbutin (4-hydroxyphenyl-β-glucoside) or salicin (2-hydroxymethylphenyl-β-glucoside), with an apparent KD values of 1 to 3 mM (18). The protein showed strong sequence similarity to both LamB and ScrY.
Other specific channels.
The role of E. coli Tsx (receptor of phage T6[six]) protein in nucleoside transport has been known since the 1970s (253). The expression of the tsx gene is increased in the presence of nucleosides through the action of the DeoR and CytR repressors (98). This relatively small protein (31.4 kDa) has been purified and shown to produce channels that can be blocked by nucleosides (393). Comparison of sequences of homologs from other members of Enterobacteriaceae confirms a 14-strand β-barrel structure (447). Interestingly, loop 3, which is normally very large in classical porins, is minute here and loops 6 and 7 are relatively large. The Tsx channel functions in the uptake of an antibiotic albicidin, and albicidin-resistant mutants contain alterations in this protein (61). Identification of mutational alterations showed that residues close to the loop-barrel junctions in loops 1, 6, and 7 are important in substrate recognition (211). As far as I am aware, Tsx is not known to form stable oligomers, and this fact, together with its small size, makes Tsx very different from LamB or ScrY and somewhat reminiscent of OmpG (discussed in “Other porins in E. coli and Salmonella” above) and CymA (see below).
The CymA protein of Klebsiella oxytoca was identified as a product of a large cluster of cyclodextrin utilization genes (205). It is a relatively small protein (322 residues) and seems to exist as monomers (479). However, the channel in this protein binds cyclodextrins at high affinity, as judged from the KS of 28 μM for the blocking of the ion conductance produced by this protein. Several features of this protein, including the presence of the gene within a large cluster of oligosaccharide utilization genes, its monomeric nature, and its relatively small size, are reminiscent of Tsx and OmpG of E. coli described above.
FadL of E. coli, a somewhat larger protein (46.0 kDa), was identified as an OM channel necessary for the efficient utilization of long-chain fatty acids as a carbon source (66, 68). This protein, which apparently exists as monomers (67), binds its substrates with an affinity that is extraordinary among specific OM channels (for example, the KD for oleate is 2 × 10−7 M) (360). A 20-strand β-barrel structure has been proposed (156). Mutagenesis experiments showed that residues close to the C terminus are important in substrate binding (360). However, long-chain alcohol cannot displace the bound fatty acids, and this suggests that the carboxyl group of fatty acids also plays a role in binding. A search for basic amino acid residues involved in binding led to the identification of His110 as an important factor in the process (69). Interestingly, homologs apparently facilitating the diffusion of aromatic compounds have recently been identified. These include the TbuX toluene channel in Ralstonia picketii (313), the m-xylene channel XylN encoded by the TOL plasmid pWW0 of P. putida (324), the putative salicylate ester channel SalD of Acinetobacter (310), the putative p-cymene channel CymD of P. putida (189), and the putative cumene channel CumH of P. fluorescens (241).
Recently, a study of a small (236-residue) OM protein, KdgM, of Erwinia chrysanthemi was published (75). This protein acted as oligogalacturonate channel in intact cells, and its conductance in planar lipid bilayer assay was inhibited by the addition of oligogalacturonates. There was no evidence that this small protein formed an oligomer. Experimental evidence shows that KdgM produces a 14-strand β-barrel and that loop 6, rather than loop 3, may cause constriction of the channel (490).
Specific channels in bacteria other than the Enterobacteriaceae.
Specific channels have been found in other bacteria. Fluorescent pseudomonads, which do not produce high-permeability classical porins (see “Slow porins” above), appear to be especially rich in these proteins, which are presumably needed for the uptake of various nutrients. A recent analysis of the P. aeruginosa genome (244) suggests that for the OprD family alone, there are 19 members in this organism.
One P. aeruginosa OM protein, OprB (earlier called OprD1), was assumed to be a specific channel for glucose (for references, see reference 464). Detailed study with various sugars showed that although OprB allowed the slow, nonspecific penetration of solutes of <300 Da, it significantly accelerated the diffusion of d-glucose and d-xylose (676). Structure prediction by using LamB as a model showed that a similar greasy slide with aromatic residues may exist in this protein. Mutants lacking this protein were shown to be reduced in their capacity to utilize several monosaccharides (749). By using the oprB promoter fusion plasmid, it was confirmed that glucose was the most effective inducer of OprB (6). Acidic pH produced a strong additional increase in oprB transcription, but bacteria grown at acidic pH on glucose paradoxically took up glucose more slowly than did those grown at pH 7. Unfortunately, the authors did not study the effect of pH on the channel properties of OprB, and thus the results remain rather confusing. The P. aeruginosa genome contains one close and one distant relative of the oprB gene (244).
OprD (earlier called OprD2), a protein of 45.9 kDa, came to attention because imipenem-resistant mutants of P. aeruginosa were found to lack it (523). Such mutants were indeed defective in the permeation of imipenem across the OM (672), and liposome-swelling studies with purified OprD showed that this protein allowed a much faster diffusion of imipenem than expected from its molecular mass (674). Finally, the OprD channel was shown to facilitate the diffusion of basic amino acids and peptides, and imipenem was proposed to mimic these compounds (675). Expression of OprD is specifically induced by arginine via the arginine repressor ArgR (471).
One laboratory claimed that OprD was the main nonspecific porin of P. aeruginosa (763). The finding of extremely low single-channel conductance (20 pS in 1 M KCl) was incompatible with this notion, and this laboratory emphasized the occasional conductance steps that were much larger (>400 pS) (288). However, the smaller conductance steps are clogged by micromolar concentrations of imipenem and thus represent the imipenem-conducting pathway (279). If OprD is a specific channel, the size of the conductance for inorganic ions, which are not the substrates for the channel, is expected to be small. Therefore, these results suggest that OprD behaves as a specific channel, contrary to the claim.
Analysis of the primary sequence of OprD suggested that it could be modeled as a homolog of E. coli OmpF-OmpC (280). It is interesting that in addition to loops 2 and 3, the OprD model has a large loop 7, and that deletion of many of the external loops was shown to affect the permeability of the channel (280). Eight-residue deletions in loops 2 and 3 were found to produce proteins with channels that not only are incapable of facilitating the diffusion of imipenem but also are not blocked by added imipenem (279, 470).
OprD was reported to exist as a trimer in nonionic detergents (764). This oligomeric structure, however, is said to be dissociated by all of the ionic detergents tried, including such mild and nondenaturing detergents as cholate and deoxycholate. OprD appeared monomeric when the OM was solubilized in SDS at low temperature (245). It was claimed to be a protease (762). However, the kcat reported was insignificant (about 10−5 s−1), indicating that one cleavage occurs after tens of generations of the bacterium; clearly, this does not have any physiological relevance. A recent review (244) indicated that there are many homologs of OprD in the P. aeruginosa genome, as stated above, and that those more closely related to OprD play roles in amino acid or peptide transport. However, I am not aware of any functional studies of proteins other than OprD in this family. An interesting homolog of OprD, PhaK of P. putida, is involved in the influx of phenylacetic acid, a carbon source for this organism (475), a result suggesting that many homologs of OprD in P. aeruginosa are likely to be specific channels for a wide range of nutrients.
OprE (earlier named protein E1) was found to be decreased in some P. aeruginosa mutants resistant to cephalosporins carrying double negative charges (752) and was thus thought to be a specific channel. It was then cloned and sequenced (751). OprE appears to be a homolog of OprD. OprE was induced when P. aeruginosa was grown anaerobically with nitrate as the electron acceptor.
P. aeruginosa also produces protein E2 (formerly protein E). The level of this protein was found to be decreased in a mutant showing a wide range of increased resistance to cephalosporins and fluoroquinolones (752). Otherwise, its properties are not known (244).
OprP of P. aeruginosa was first identified as an OM protein induced under phosphate starvation conditions (248). Unlike E. coli PhoE, which is a general porin with an anion preference, OprP appears to be a true specific channel since it has a much higher affinity for phosphate than for other anions (243). The gene was sequenced (612), and the significance of various lysine residues in the protein was assessed by site-directed mutagenesis (641). Epitope insertion mutagenesis gave results compatible with a 16-strand β-barrel model (640).
OprO, a 45.3-kDa protein of P. aeruginosa, was identified as a pyrophosphate-selective channel (247). It is a trimeric porin like OprP, and its sequence is 76% identical to that of OprP. It shows a low single-channel conductance of 0.26 nS in 1 M KCl (613).
A methylotroph (an organism that can grow on one-carbon compounds such as methanol or formamide), Methylophilus methylotrophus, was found to contain an fmdC gene in its formamide utilization gene cluster (416). The FmdC protein (39 kDa) is induced by formamide, acetamide, and urea and is hypothesized to form a specific channel for these compounds, but experimental evidence is still lacking. The sequence of FmdC is not similar to that of any other OM proteins studied.
TonB-Dependent Receptors or Gated Channels
Siderophores (iron chelators of microbial origin) and vitamin B12 are too large to pass through the classical porin channels of E. coli, and it would be difficult to design specific channels for them without making them nonspecifically permeable to many large solutes, thus compromising the bacterial resistance to environmental toxic compounds. These compounds are transported by utilizing TonB-dependent receptors in the OM. Examples of such receptors in E. coli K-12 include BtuB for vitamin B12 and six receptors for Fe3+-siderophore complexes (FepA for Fe3+ enterobactin, FhuA for ferrichrome, FecA for Fe3+ citrate, FhuE for Fe3+ coprogen, and Cir and Fiu for Fe3+ catecholates and Fe3+ dihydroxybenzoylserine [95]). (A recent addition to the list of siderophore receptors in S. enterica and enteropathogenic E. coli is IroN, which transports Fe3+ salmochelin [254].) These receptors bind to the ligands with high affinity (submicromolar or even nanomolar Kd), and their function requires an interaction with the protein TonB, which is thought by some workers to span the thickness of periplasm (425). The action of TonB requires that the cytoplasmic membrane is energized and that the energy derived from the proton motive force is somehow transferred to the receptors with the assistance of two cytoplasmic membrane proteins, ExbB and ExbD (94, 271, 425) (Fig. 5), so that an active transport results across the OM (505). At least for vitamin B12, evidence has been presented that the periplasmic concentration can far exceed the external concentration (543).
That the receptor functions as a gated channel was demonstrated by Rutz et al. (567). These workers hypothesized that the receptor for ferric enterobactin, FepA, folds as a 29-strand β-barrel and deleted large internal segments. Some of these deletion mutant proteins not only allowed the passive diffusion of enterobactin but also apparently functioned as large, open channels through which relatively large antibiotics could penetrate. These results led to the hypothesis that the surface loops of this β-barrel acted to close the large diffusion channel. When the crystal structures of several siderophore receptors, FepA (106), ferrichrome receptor FhuA (202, 385), ferric citrate receptor FecA (200), and, most recently, the vitamin B12 receptor BtuB (133), were solved, the overall structure of these receptors was found to be quite different from what was anticipated by Rutz et al. (567). (Crystallographic structures of various receptors have recently been reviewed [201].) The unliganded and liganded structures of FecA are shown in Fig. 6. The β-barrel in all these monomeric receptor proteins is composed of only 22 strands, and the N-terminal portion, consisting of about 150 to 200 residues, is found as a globular domain that is inserted into the barrel from the periplasmic side, forming a “plug” or “cork.” Some of these receptors were crystallized with and without ligands. The ligands bind to a site that is close to the external entrance of the barrel and is composed of both plug and barrel sequences (Fig. 6B). It was gratifying to see that ligand binding resulted in the propagation of conformational changes within the plug, so that the most N-terminal portion containing a short motif (TonB box) (shown in light blue in Fig. 6A), which was shown to interact with the TonB on the basis of its disulfide cross-linking with BtuB (113) and, more recently, with FecA (474), appears to undergo a large conformational change. Interestingly, conversion of some residues of the TonB box of BtuB into proline or glycine nearly abolishes the transport activity without impairing the binding of the ligand (112). Mutagenesis of various residues of the TonB box into cysteine, followed by labeling with a spin label, showed that some of these residues are immobile, presumably because they are docked within the β-barrel. When the ligand, vitamin B12, binds to BtuB, all of the residues become mobile, a result thought at that time to be due to the release of the N-terminal sequence of the protein, containing this box, into the periplasm, so that the box sequence could interact with TonB (138). These conclusions were also consistent with the site-directed labeling of residues in the N-terminal domain of BtuB (198, 414). Recently, the crystallization of unliganded and liganded BtuB (133) showed for the first time the details of the conformational change in the TonB box on ligand binding. The TonB box does not become extruded into the periplasm, but there is a flipping of a portion of the box. The interaction of TonB with the receptors was recently reviewed by Postle and Kadner (506).
FIG. 6.
X-ray crystallographic structures of the ferric citrate receptor, FecA, of E. coli. (A) Side view of the unliganded FecA. The “plug” domain inside the β-barrel is shown in orange. At its N-terminal end, the short sequence comprising the Ton box is shown in light blue. Loops 7 and 8 are shown in deep blue and mauve, respectively. (B) Liganded FecA. On binding of the ferric citrate (with two large blue balls near the top indicating the two iron atoms, and citrate molecules in stick diagrams), large displacements are seen at the N-terminal end of the plug domain, where residues 80 through 95 (including the Ton box of residues 80 to 84) become disordered and invisible. Loops 7 and 8 also undergo large conformational changes, with the loss of part of the helical structure in loop 7. The diagrams are based on PDB files 1KMO and 1KMP.
These results explain the sequence of events up to the binding of ligands and the conformational alteration of the Ton box so that it can now interact with TonB. However, inspection of structures with and without the ligand shows no massive movement of the plug domain relative to the barrel to create a large channel that may be necessary for the influx of the ligands (385). Two lines of evidence suggest that the next stage of transport is initiated by the movement of external loops. First, the structures of FecA with and without ferric citrate showed that the ligand binding causes extensive movement of some of the loops, so that the high-affinity binding site now becomes shielded from the external medium (200) (Fig. 6). Although this movement itself does not create a large channel, it can be viewed as the first step, which may then be followed by a large-scale conformational change caused by the energized TonB. Second, chemical cross-linking of Lys483 (close to the tip of L7) in FepA to the neighboring OmpA and OmpF proteins becomes impossible on binding of the ligand, enterobactin (600); this shows that the loops come together and close the top of the FepA protein on ligand binding, as shown crystallographically with FecA (200). (Conformational alteration in surface loops on ligand binding is also suggested by site-specific fluorescence labeling [115].) Currently, the urgent question in this area is to understand the nature of the expected TonB-dependent conformational alterations of the receptors. In this connection, it is interesting that the spin label at residue 280 of FepA, now known to be near the base of L3, showed large TonB-dependent changes in mobility during enterobactin transport in intact cells (306). ExbB and ExbD are related to MotA and MotB, respectively (346; see also reference 94), which are thought to form a stator in the flagellar motor; this homology suggests that perhaps the proton flux through the interface between ExbB and ExbD may generate a rotation or conformational change of another protein, such as TonB. A hypothetical model of the proton pathway through the ExbB-ExbD-TonB complex has been proposed (779).
A surprising observation, in view of the general consensus about the role of the TonB box, was that the deletion of the entire “plug” from FhuA still produced a TonB-dependent transport of ferrichrome (92, 336). (A much earlier study of the deletion between residues 21 and 128 showed a similar, unimpaired transport phenotype [116], but the mutant protein still contained the Ton box.) Thus, the β-barrel domain alone of FhuA is still capable of energy-dependent transport and many other functions of this protein described in the references cited. The structure of wild-type FhuA suggests that deletion of the plug would produce a wide-open channel. However, the deletion made E. coli only very slightly more susceptible to various antibiotics (92), and the mutant protein did not produce stable channels after reconstitution into planar lipid bilayers (93). These observations suggest that the channel might be still closed by the external loops. Indeed, deletion of part of the L4 loop (or the region from the end of the L4 loop and the external half of β-strand 8 that protrudes out from the surface of the OM), when combined with the plug deletion, leads to a protein producing large, stable channels (93). The movement of the external loops accompanying ligand binding is described above.
The utilization of polysaccharides by Bacteroides has been studied in the laboratory of A. A. Salyers. Unlike many bacteria, which degrade polymers in the external medium by secreting enzymes, Bacteroides appears to contain polysaccharide-degrading enzymes within the cells, probably in the periplasm (573), or associated with the envelope (609). Thus, polysaccharides, or at least their large degradation products, must be transported across the outer membrane. A large OM protein of 1,065 residues, CsuF, which is essential for growth of B. thetaiotaomicron on chondroitin sulfate and which binds this polysaccharide, was discovered in 1995 (127). Its N terminus, about a 60-residue domain, had a striking similarity to the similar region of TonB-dependent receptors of E. coli (127), now known to correspond to the plug domain. A close homolog of CsuF, SusC (again a large protein [115 kDa]), is encoded by an operon for starch utilization, is necessary for growth on starch, is located in the OM, and binds starch (536). Furthermore, my analysis of the CsuF and SusC sequences with the program PSI-BLAST (11) shows that these proteins belong to the family of TonB-dependent receptors. Therefore, it is likely that CsuF and SusC are also energy-dependent transporters utilizing a TonB homolog, although little evidence is available for their precise mechanisms of action. SusC apparently forms a complex with another OM protein, SusD (134).
Export Channels of the TolC Family
The OM is expected to act as a major barrier when proteins have to be exported out of gram-negative cells. Currently at least five different mechanisms are known to be utilized to overcome this barrier, and three of them are schematically shown in Fig. 7. In the type I machinery, the protein is exported in a single step through a multisubunit export assembly that does not depend on the conventional Sec pathway. The type I pathways export, for example, hemolysin and colicin V in E. coli, metalloprotease in E. chrysanthemi, alkaline protease in P. aeruginosa, and cyclolysin in B. pertussis (706). Type I export assembly is composed of an ABC-type transporter in the cytoplasmic membrane, a periplasmic helper protein now classified as the membrane fusion protein (175), and an OM channel, now known to comprise a family (14), called OM factor (OMF) (757). In E. coli, a single OMF protein, TolC, functions in the export of various proteins (707); in contrast, in other species of bacteria, the three component proteins of any given type I system tend to be located on the same operon, resulting in the requirement of a specific OMF for a specific substrate protein (14).
FIG. 7.
Major export pathways of proteins across the OM. Components of type I, type II, and type III secretion pathways are shown schematically. The type I pathway is composed of an ABC transporter (here HlyB for E. coli hemolysin), a membrane fusion protein (MFP) family protein (HlyD), and an OM channel (TolC). In the type II pathway, the proteins reach the periplasmic space via the Sec pathway and are then secreted across the OM by a machinery with many components, including pilin-like proteins (pseudopilins). The proteins are labeled according to the universal Gsp (general secretory pathway) nomenclature, and the scheme owes most to a recent review by Thanassi (652). The energy is apparently supplied by ATP hydrolysis by the GspE protein. The type III system is involved in the secretion (or perhaps injection) of virulence-related proteins into animal and plant host cells. Many of the components have homologies to the proteins of the flagellar hook and basal plate system. The figure is based mainly on a recent proposal that relies heavily on this similarity (73), as well as experimental studies of the “needle complex” (354, 648). The names of the proteins are those from the type III pathway in S. enterica serovar Typhimurium, with those from that of Y. enterocolitica shown in parentheses. The energy for export is thought to be supplied by the InvC (YscN) ATPase. IM, inner membrane.
One of the first protein export complexes of this type studied was that for the export of hemolysin in E. coli, an assembly containing the HlyB exporter (an ABC transporter), the HlyD periplasmic protein, and the OM channel TolC (706, 707) (Fig. 7). More recently, exporters of a very wide range of small molecules, ranging from solvents and dyes to most amphiphilic and lipophilic antibiotics, were also shown to be constructed as a tripartite complex that also contained members of the OMF family. A major multidrug efflux transport complex of E. coli is thus composed of the AcrB antiporter protein, the AcrA periplasmic protein of the membrane fusion protein family, and, again, the TolC OM channel (450). Similarly, the major multidrug efflux pump of P. aeruginosa is composed of the MexB antiporter, the MexA periplasmic protein, and the OprM OM channel, and there are now many more examples (for reviews, see references 448 and 450). The P. aeruginosa genome codes for 18 homologs of OprM, and the function of some of the members in drug efflux is beginning to be identified (307).
Some information is available about the interaction between the components of these systems. Using an affinity matrix that binds the protein substrates of the type I secretion machinery of E. chrysanthemi, the protein substrates were shown to bind to the ABC transporter first, which then forms a complex with the periplasmic component and finally forms a complex with the OM channel protein (373). More recently, chemical cross-linking in intact cells showed that HlyB and a trimer of HlyD form a complex in the absence of substrate and TolC but that the formation of the complex containing a trimeric TolC required the presence of the substrate (651). Similar cross-linking experiments showed that the AcrB antiporter forms a complex with a trimer of the periplasmic protein AcrA even in the absence of TolC (777), and AcrA was shown to be a highly elongated protein (up to 20 nm long) (26, 775). Finally, the crystal structure of the AcrB transporter was elucidated (434), revealing a homotrimeric protein with huge periplasmic domains. The structure of AcrB with bound drugs, elucidated recently (769), suggests the way in which drugs become captured by this pump of extraordinary wide specificity (768). The molecular details about how these proteins interact and form an export complex, however, are not clear.
An important discovery has been the solution of the crystal structure of the TolC trimer (348). The trimer was found to consist of two barrel-like structures joined together (Fig. 8A). At one end is a β-barrel structure that is typical of OM proteins, such as porins, specific channels, and high-affinity siderophore receptors. The interior of the 12-strand β-barrel, however, is not constricted or obstructed in any way by the infolding of loops and thus has a wide diameter (up to 3.5 nm). Furthermore, a single barrel is made of three monomeric units, each contributing four β-strands. This barrel is connected to a long (about 100-Å) α-tunnel structure consisting essentially of 12 α-helices. The α-tunnel contains an internal cavity, which has a diameter in the range of 20 Å in the part close to the β-barrel but is closed at the end facing the cytoplasmic membrane. This is because of the way in which neighboring helices interact as a coiled coil (607). However, unlike the rigid β-barrel structure, the α-tunnel can be opened up widely (by 30 Å) by a small uncoiling movement of the component α-helices (348, 607), and this dynamic nature of the α-tunnel is likely to have important implications for how the protein functions. It is not known what causes the tunnel opening—one proposal is that the periplasmic proteins, such as AcrA or HlyD, which themselves contain a coiled-coil structure (308, 499), may interact with the α-tunnel domain of TolC, stabilizing the inherently unstable open conformation of this tunnel (607). In any case, it is intriguing that the outer end of the periplasmic domain of the AcrB trimer seems to fit nicely to the inner end of the α-tunnel domain of TolC (434). Recently, TolC in S. enterica serovar Typhimurium was shown to allow some influx of methyl viologen from the external medium (577); this may not be totally surprising because the closure of the end of the α-tunnel is thought to be a dynamic phenomenon.
FIG. 8.
X-ray crystallographic structures of the TolC trimer and the OM-puncturing needle apparatus of phage T4. (A) TolC trimer. Each subunit is shown in a different color. At the top, a 12-strand β-barrel is formed by each subunit contributing four strands each. Below the barrel, there is a long periplasmic tunnel composed of 12 α-helices. Structure Based on PDB coordinate file 1EK9. (B) OM-puncturing needle apparatus of phage T4. This structure, produced by protein gp5, is also trimeric, with each monomer shown in a different color. The extended needle structure, which traverses the OM and the periplasm just like the β-barrel and the α-helical tunnel of TolC, is constructed in a totally different manner, as a trimeric β-helix. The structure rich in α-helix, surrounding the upper middle part of the needle, is the lysozyme domain, which is thought to play a role in puncturing a hole in the peptidoglycan. The domain at the very top interacts with another protein, gp27, which was omitted for clarity. Structure based on PDB coordinate file 1K28. This figure was drawn by using program VMD (282).
OprM, one of the TolC homologs of P. aeruginosa, was studied by creating short deletions within the gene (375). Those that prevented protein expression occurred mostly in the region predicted to be in the α-tunnel domain in the TolC structure, a result that fits with the importance of this domain in the overall folding of the trimer. A similar result was reported from another laboratory on the basis of both insertion and deletion mutations (745). The surface accessibility of inserted epitopes of the same series of mutants, however, resulted in a different, porin-like folding model in an earlier study (746); these results appear to remain unexplained in the newer study.
Secretins, Ushers, and Autotransporters Producing Oligomeric Rings
Secretins.
Many proteins are secreted by gram-negative bacteria via the type II secretion pathway or the main terminal branch of the general secretory pathway (515, 516), where they are first secreted into the periplasm by the Sec-dependent process and then cross the OM by a process requiring many components (Fig. 7). The more intensively studied examples include the Pul (pullulanase) system of K. oxytoca, the Out system of E. chrysanthemi, which secretes pectate lyase and other enzymes; and the Xcp system of P. aeruginosa, which secretes toxin A and various enzymes (564, 576). The features of these systems include the large number (14 or more) of protein components required for secretion and the observation that the secreted protein apparently folds into its mature conformation in the periplasm (517). From the early days, PulD and its homologs (or GspD in the universal nomenclature and also in Fig. 7), among these large number of exporter components, were thought to form channels of some kind across the OM, where they are located (516). Based on the knowledge (219) that protein pIV of filamentous phage f1, a protein that is located in the OM and is essential for the extrusion of the 6.5-nm wide phage into the environment, is homologous to PulD and its homologs in the type II secretion system (and also to the OM component of type III secretion systems [see below]), Russel proposed the seminal idea (565, 566) that oligomers of these proteins may form OM channels for export of proteins and filamentous phages. (Recent reviews [652, 654] summarize our current knowledge of these OM channels.) Furthermore, this group showed that pIV occurs as multimers composed of 10 to 12 subunits (331), an observation suggesting that pIV homologs function by forming a ring-like oligomeric complex, which was soon visualized by electron microscopy (379). PulD and its homologs (or GspDs) in type II secretion systems, often called secretins, were also later shown to occur as oligomeric complexes containing 12 to 14 subunits (64, 257, 466). (Secretins of P. aeruginosa have been reviewed [63].) Homologs of PulD and pIV are also involved in the extrusion of type IV pili, and one such component, PilQ, was also shown to form an oligomer (64). A recent study presents the description of the ring structure of PilQ from N. meningitidis (141).
The C-terminal halves of various secretins show strong similarity and were predicted to contain 10 to 13 β-strands (64, 235). In agreement with the hypothesis that the N-terminal halves reside in the periplasm and the C-terminal halves cross the membrane, limited protease digestion of PulD (467) and P. aeruginosa XcpQ (101) left the C-terminal domains undigested, and these domains still formed a ring-like structure capable of forming ion channels upon insertion into lipid bilayers. In intact cells, the isolated C-terminal domain of PulD still formed multimeric channels if the N-terminal domain was coexpressed in trans; the function of the N-terminal domain here is not clearly understood (235). Interestingly, electron micrographs of intact PulD suggested that the N-terminal portion may fold back into the central channel and close it (467). However, the situation may be more complex, since the ring-like structure in pullulanase secretion system apparently is composed of 1:1 dimers of PulD and PulS (467). The PulD-PulS oligomer is unable to form stable open channels in planar-bilayer assays (466). An informative study of the channel and its gating again comes from that of pIV (398). Thus, the expression of unaltered pIV did not make E. coli hypersusceptible to a large, hydrophilic, and normally OM-impermeant antibiotic, vancomycin, whereas that of a single-amino-acid substitution mutant, S324G, did. Insertion of oligomers into planar bilayers showed that the pIV channels were closed (unless extraordinarily high voltage was applied) whereas the mutant channels were frequently open and that the single-channel conductance was about 1 order of magnitude larger than that of E. coli porins, suggesting that the diameter of the channel was large. Finally, this mutant pIV allows the entry of maltodextrins into the periplasm in the absence of the specific channel LamB. Marciano et al. (399) showed that this entry of maltodextrin is blocked by the assembly and extrusion of f1 phage, confirming that the channel indeed serves as the exit for the phage particles.
Plant and animal pathogens often possess extracellular secretion systems of a different type, type III, which delivers various effector proteins directly into the cytosol of the host cells (for reviews, see references 145-147, 213, 281, and 443) (Fig. 7). The type III complex resembles a syringe needle or the basal body of flagella (7). Remarkably, this complex also contains one OM component, InvG in S. enterica serovar Typhimurium or YscC in Yersinia (and its homologs), which shows homology to PulD and its homologs in the type II secretion system, as well as pIV. These InvG and YscC homologs were also shown to produce ring-like multimers containing 12 to 14 subunits (154, 349).
Ushers.
Components of P pili and type I pili of E. coli are secreted into the periplasm by the Sec machinery, just like the substrates of the type II export machinery. However, they are secreted across the OM by a pathway completely different from the type II pathway described above (652, 654). The pathway here, called the chaperone-usher pathway, requires essentially only two protein species, a periplasmic chaperone (PapD for P pili) and an OM channel or usher that may also help in the folding and polymerization of pili subunits (PapC for P pili), in contrast to a dozen or so proteins required in the type II secretion pathway. The ushers (80 to 90 kDa) are somewhat larger than secretins. PapC, as well as its counterpart in type I pili secretion, FimD, forms ring-like structures, as shown by electron microscopy (579, 655). These rings are 15 nm across and contain central pores 2 to 3 nm in diameter. Osmotic swelling assays with PapC-containing proteoliposomes showed the diffusion rates of various sugars to be relatively insensitive to their size, confirming the large pore size (655). It is still unclear how many monomers are needed to form the ring. The PapC oligomer was estimated to contain at least six monomers, but no evidence for oligomerization has been found so far for FaeD, an usher for the export of K88 pili in enterotoxigenic E. coli (258). The monomeric subunit of PapC and FaeD each seems to contain many β-strands, at least 22 (258, 586, 652), most of which are present in the central domain.
It is curious that the “terminal branch” (i.e., the part that causes secretion across the OM) of the general secretion pathway is so different between the chaperone-usher pathway and the type II secretion pathway. Possibly the cause of this difference is the requirement for energy. In the chaperone-usher pathway, the transported proteins polymerize into the pilus structure during secretion, and the stabilization through polymerization is thought to provide the energy for transport (294). In contrast, the type II secretion pathway exports monomeric proteins (usually enzymes) and thus requires a large number of proteins to provide the scaffolding and transduction of energy.
Autotransporters.
Some proteins contain the information needed for their transport in their own sequences. These proteins, called autotransporters, are thought to use their C-terminal domains to form OM channels for transport of their N-terminal domains (for a review, see reference 266). This hypothesis was proposed in a concrete form by Klauser et al. (337), in part on the basis of the identification of putative 15 β-strands in the C-terminal domain of the Neisseria immunoglobulin A protease precursor, a prototype autotransporter. This idea has since been supported by the fusion of numerous “passenger” peptides and proteins to the C-terminal domains of various autotransporters. For example, the secretion of the passenger, cholera toxin B subunit, by the C-terminal domain of an autotransporter, AIDA-I, was shown to require all of the presumed β-strands and the intervening linker sequence (407). Furthermore, the C-terminal domain of another autotransporter, BrkA of B. pertussis, was found to produce channels in planar lipid bilayers (606). However, in all these studies, each autotransporter molecule was hypothesized to act independently, its C-terminal domain facilitating the export of its own N-terminal cargo domain (for example, see reference 407).
In a recent study of the C-terminal domain of the immunoglobulin A protease precursor, this domain was again found to produce presumably nonspecific pores, detected by the osmotic swelling of the reconstituted proteoliposomes (698). However, more importantly, electron microscopy and size exclusion chromatography showed that this domain produced a ring-like oligomeric complex containing at least six, and perhaps more, monomers. Thus autotransporters apparently function by building a secretin-like oligomeric structure with a 2-nm channel in the center. This explains why proteins that are folded and sometimes disulfide stabilized in the periplasm are exported by the C-terminal domains of autotransporters (91, 697).
Other Export Channels
Type IV secretion pathway.
The type IV secretion pathway carries out the export of proteins or protein-DNA complexes, and participates not only in the secretion of proteins but also in the transfer of plasmids between bacterial cells and the injection into plant cells of tumor-generating T-DNA by Agrobacterium tumefaciens. This is a complex system requiring many gene products, and the way in which the exported macromolecules cross the OM is still not clear in spite of the recent crystallization of ATPase molecules that form ring-like structures (for reviews, see references 35 and 135).
Two-partner secretion pathway.
A protein secretion pathway somewhat reminiscent of the autotransporters has attracted attention in recent years. In this two-partner secretion pathway, reviewed recently (293), both the exoprotein to be secreted and the separate helper or channel protein are moved into the periplasm via the usual Sec-dependent pathway. Then the N-terminal domain of the passenger protein is recognized by the channel protein, which is thought to form a β-barrel spanning the OM, and secretion results. Passenger proteins are large (100 to 500 kDa) and apparently have a characteristic folding pattern, like that of B. pertussis filamentous hemagglutinin, FhaB. In this example, electron microscopy and modeling suggest that the protein folds in an unusual β-helix conformation, eventually attaining a length of 50 nm (314). The best studied transporters, ShlB of S. marcescens (344) and FhaC of B. pertussis (233), were shown to fold as β-barrels by epitope insertion and protease cleavage studies. Both transporters were shown to produce channels in planar lipid bilayers, but such channels were unstable, suggesting the presence of some form of gating (292, 344). FhaC allowed the permeation of sugars, but again the specific activity was low. When a secreted exoprotein is accumulated in the periplasm, it is extremely sensitive to proteolysis, a result suggesting that it is in an extended conformation (585). Furthermore, chimeric proteins containing a C-terminal addition of easily folded globular domains cannot be translocated in this system (293). Therefore, the secreted exoprotein is likely to cross the OM in an extended conformation and become folded once it reaches the cell surface. This folding can provide the energy for secretion. FhaB hemagglutinin is synthesized as a 357-kDa giant protein, but its C-terminal 137-kDa portion apparently becomes cleaved by a subtilisin-like protease, SphB1, which gets to the cell surface by using its own autotransporter function (151). It is tempting to imagine that at least one function of this cleaved C-terminal sequence is to act as a chaperone, in order to keep the FhaB protein in an unfolded conformation within the periplasm.
Both ShlB and FhaC migrate as monomers in SDS-PAGE, even when extraction is done at low temperature, although a small amount of a higher-molecular-weight fraction was observed for the latter transporter (292, 344). If the passenger protein is exported in its extended conformation, perhaps the lumen of a single β-barrel is sufficiently large for this purpose and a large channel formed by an oligomeric complex of secretin-like proteins may not be necessary (293).
Export channels for polysaccharides.
Gram-negative bacteria often export capsular polysaccharides or exopolysaccharides. Such export processes have been studied in depth, especially in E. coli and related organisms. The most important classes of capsular polysaccharides in E. coli are the group 1 and group 2 polysaccharides (296, 726). The former type of polysaccharide is made up of oligosaccharide repeating units (just like many O-antigen chains of LPS) and is synthesized in a manner analogous to such O-antigen chains of LPS, that is, by the addition of prefabricated repeating units to the reducing end of the growing undecaprenol-pyrophosphate-linked polymer, presumably at the external surface of the inner membrane. The transport of the undecaprenol-linked repeating unit across the cytoplasmic membrane is assumed to be carried out by a polysaccharide transport (PST) family transporter, presumably utilizing the proton motive force as energy (633). In contrast, the group 2 capsule is usually made up of one or perhaps two component sugars, is synthesized in the cytoplasm by the successive addition of new sugar units at the nonreducing end of the growing polymer, and is then exported across the envelope by an ABC transporter. In both cases, however, the export across the OM is thought to occur through the channels of a class of homologous proteins in the OMA (OM auxiliary) protein family (487). Furthermore, the export requires periplasmic proteins anchored to the cytoplasmic membrane, belonging to either the MPA-1 family (for “cytoplasmic membrane-periplasmic auxiliary protein 1”) for group 2 capsules or the MPA-2 family for group 1 capsules. MPA-1 family proteins are especially interesting, because their cytosolic domain contains an ATPase-like structure, which is thought to regulate the length of the polysaccharides exported via its protein tyrosine kinase activity (44, 446, 699, 700).
An important observation on the OM channels of OMA family proteins is that of Drummelsmith and Whitfield (182). They showed that an OMA member, WzaK30, a 40-kDa lipoprotein involved in the export of group 1 capsulare polysaccharide K30 of E. coli, occurs as secretin-like multimers that are 11 nm in diameter and contain a 3-nm central cavity (or hole). Among members of OMA family, all those involved in the group 1 polysaccharide export are quite similar to WzaK30 in sequence, including the wza gene in the cps cluster in E. coli K-12, involved in the biosynthesis of colanic acid (a nonspecific “mucus” exopolysaccharide composed of hexasaccharide repeat units) (633). The same is true also for OMA members that are involved in the export of group 2 polysaccharides in H. influenzae (BexD for the export of polyribitol phosphate in type b) and N. meningitidis (CtrA for the export of polysialic acid). It is therefore reasonable to expect that these proteins also function in the same manner.
The mechanism of export of the group 2 capsule in E. coli has been studied extensively by using K1 antigen (polysialic acid) (72) and K5 antigen (552, 726), and it appears more complex than the export of group 1 polysaccharides. The KpsD protein, which is a distantly related member of the OMA family (487), has several unusual features. Unlike other members of the OMA family, it does not contain the lipoprotein signal sequence. It is much larger (about 64 kDa) than the other OMA family members, and the C-terminal half of KpsD shows no homology to the other members. Indeed, KpsD was seen as a periplasmic protein (615). In a recent study, KpsD was found in the OM, periplasm, and cytoplasmic membrane depending on the genetic constitution of E. coli (25); it is found exclusively in the OM only when all other transport components are present but the capsular polysaccharide is not made because of the defect in one of the polysaccharide biosynthesis genes. The authors suggest that KpsD, together with the MPA-1 protein KpsE, may drive the transperiplasmic transport of the polysaccharide. It has also been suggested that the polysaccharide is extruded through the porin channels (72); this idea was supported by the observation that all naturally occurring capsule-producing E. coli strains produced an extra porin-like protein in the OM, called protein K (478). The sequence of protein K has not been deposited in databases, and it is not known if this protein is related to conventional porins.
A family of putative polysaccharide export channels unrelated to the OMA family is known. These channels include AlgE of P. aeruginosa (539) and AlgJ of Azotobacter vinelandii (538), both of which are thought to be involved in the export of alginate (a polysaccharide containing mannuronate and iduronate). AlgE was purified and shown to produce an anion channel, whose conductivity is inhibited by the addition of compounds containing mannuronate. These proteins (about 52 kDa) are larger than classical porins and were predicted to span the OM as a 18-strand β-barrel (539).
Entry of Colicins and Phage Nucleic Acids
Gram-negative bacteria are vulnerable to infection by phages and killing by colicins. The A group colicins use the TolA system (see below), and the B group colicins use the TonB system, (see above) for penetration across the OM. Table 1 shows examples of A group colicins, which use various OM proteins as receptors and additionally may require another OM protein, often a channel protein, for penetration. Although the structures of several colicins have been determined by X-ray crystallography (for a review, see reference 771) and the domains involved in receptor binding, translocation, and final killing activity have been localized by mostly genetic approaches, the pathway by which colicin molecules cross the OM is still somewhat controversial (114). Briefly, some scientists favor the idea that the initial receptor serves as the translocation channel whereas others propose that the translocation occurs at a site separate from the initial binding site, perhaps through a porin or through the lipid bilayer. Cao and Klebba (114) argue, mainly on the basis of the data from Letellier and coworkers (47, 85, 183), that at least colicins A and N (Table 1) are likely to pass through the porin channels in a more or less denatured form. Zakharov and Cramer (771) also conclude that colicin E1 (Table 1) will pass through the TolC channel tunnel in a largely denatured form, despite its initial interaction with the receptor, BtuB. The inward movement of the long, unwound colicin molecules through the narrow channels of the OM proteins, however, cannot occur without being driven by an input of energy. This is where the Tol (or Ton) system is thought to function.
TABLE 1.
Representative members of group A colicinsa
| Colicin | Receptor | Translocation | Cytotoxic activity |
|---|---|---|---|
| A | BtuB | OmpF, TolQRAB | Pore formation |
| E1 | BtuB | TolC, TolQRA | Pore formation |
| E2, E7, E8, E9 | BtuB | OmpF, TolQRAB | DNase |
| E3, E4, E6 | BtuB | OmpF, TolQRAB | RNase hydrolyzing 16S rRNA |
| E5 | BtuB | OmpF, TolRAB | RNase hydrolyzing the anticodon loop of tRNA |
| N | OmpF | TolQRA | Pore formation |
| K | Tsx | OmpF, OmpA, TolQRAB | Pore formation |
| U, 28b | OmpA | OmpF, LPS, TolQRAB | Pore formation |
| DF13 | IutA | TolQRA | RNase hydrolyzing 16S rRNA |
Based on data in reference 366.
As described above, energization of transport through the gated receptor channels by TonB requires two associated proteins in the inner membrane, ExbB and ExbD. The Tol system is very similar in having the TolA protein (which lies mostly in the periplasm with its N-terminus inserted in the inner membrane), which is surrounded by TolQ and TolR (which correspond to ExbB and ExbD, respectively, in terms of topology) (384) (Fig. 5). There is an extra member in the Tol system, TolB, which is a periplasmic protein. The Tol system, which “stabilizes” the OM structure through a poorly understood mechanism (see “Alterations of the OM bilayer barrier” below), somehow supplies the energy (or even movement) needed for colicin uptake. However, despite the tantalizing similarity of TolQR proteins to MotAB stator proteins of the flagellar motor (119, 346), the mechanistic details remain unknown.
The injection of bacteriophage nucleic acids does not usually require the Tol system and, hence, energy. Most of the studies in this area have been done with tailed double-stranded DNA phages. Phages of the family Myoviridae (for classification of tailed phages, see reference 5), including T4, P1, and P2, have contractile tails, in which the contraction of the sheath is thought to bring the central hollow tube through the OM (431). Some morphological studies suggest that this insertion of the tail core occurs at, or induces the formation of, the OM-inner membrane fusion structure (649). The structure of the central, needle-like device which is used by T4 to puncture the OM was solved recently (317). In this structure (Fig. 8B), three molecules of gp5 protein come together so that its C-terminal domain forms a three-strand β-helix with a triangular cross-section. The width of this tube-like structure becomes smaller as it reaches the tip, and thus it seems to function as a sharp puncturing needle at the time of sheath contraction. Unlike the T4 short tail fiber made of a similar triple β-helix of gp12 (421), the central core of the helix of gp5 is not strongly hydrophobic. However, the central channel is not wide enough for the conduction of DNA, and this presumably occurs by the subsequent juxtaposition of the end of the tail tube onto the cytoplasmic membrane (317).
Injection of DNA by members of the Siphoviridae (including phages T5, λ, and T1) has been studied in detail in recent years. Phage λ was found to inject its DNA into proteoliposomes containing its receptor, LamB, in 1986 (556). This observation has been confirmed by the appearance of high-conductance channels (0.7 nS), but it is not clear if this is the result of an expansion of the LamB channel or if it is formed by the insertion of phage proteins into the bilayer (54). Phage T5 uses the ferrichrome receptor FhuA as the receptor. Binding to FhuA is mediated by protein pb5 (502), but protein pb2, which forms the long tail fiber and has a high molecular mass (108 kDa), is not needed for adsorption. In 1990, pb2 was purified, inserted into the black lipid membrane, and shown to produce channels of an exceptionally high conductance of 4.6 nS in the absence of FhuA (204). When T5 was later shown to inject its DNA into FhuA-containing proteoliposomes (501), the results with pb2 were somehow forgotten and it was proposed that T5 DNA would diffuse through the opened FhuA channel (82). A more recent study by using cryoelectron tomography (77) shows beautifully that FhuA simply acts as an anchor and that the tail fiber made of pb2 traverses the lipid bilayer nearby, possibly allowing DNA transfer. Interestingly, the tail tip undergoes a major conformational change, and its length is shortened from 50 to 23 nm, with a concomitant increase in diameter. In T7, which belongs to the family Podoviridae (also including P22), the tail is very short. Here the proteins that form DNA channels are ejected from the phage particle core (rather than being a part of the preexisting tail). One of the proteins, gp14 (21 kDa), is localized in the OM, and a remarkably large protein, gp16 (144 kDa), becomes located in the inner membrane. Together with gp15, these proteins are hypothesized to form the channel for DNA injection, but details are not yet known (426). Data on phage DNA injection up to 1994 have been reviewed by Dreiseikelmann (181).
In contrast to the systems so far described, the Tol system (and therefore energy) is required for infection of F+ E. coli by the filamentous phage f1 (136) and of V. cholerae by the cholera toxin-encoding filamentous phage CTXφ (261). Although the Tol system is required for the insertion of the major capsid protein into the inner membrane, presumably simultaneously with the disassembly of the phage and the translocation of DNA into the cytoplasm (137), this does not necessarily rule out the possibility that it is needed for an earlier event, such as the translocation of phage tip protein pIII to the surface of the inner membrane, a process that may be analogous to the entry of colicin into the periplasmic space. In fact, for the related phage fd, the N-terminal domain of pIII (or g3p) was shown to interact with the C-terminal domain of TolA (presumably after penetrating across the OM barrier), once the block on this interaction is lifted by the interaction of the central domain of g3p with the pilus (544). Since g3p is known to produce a channel of rather high conductance (0.82 nS) in planar bilayer (223), this interaction with TolA is hypothesized to bring down the C-terminal domain of g3p into the inner membrane, where it is thought to form a DNA transfer channel (544).
In an exceptional case (the double-stranded RNA phage ϕ6 of P. syringae), the early stage of infection involves the fusion of the viral capsid membrane with that of the host OM (32). This unusual membrane fusion event is catalyzed by a fusogenic protein, P6 (32, 417). For another unusual double-stranded DNA phage containing an internal membrane, PRD1, genetic evidence suggests that a viral protein, P11, produces a channel across the OM (231).
THE LIPID BILAYER AS A DIFFUSION BARRIER
The Asymmetric Bilayer
The location of LPS in the OM was studied in our laboratory many years ago. First, quantitation of LPS and phospholipids led to the prediction that each species may cover an equal area, corresponding to the surface of the OM (excluding the area occupied by OM proteins) (624). We then suggested that LPS comprised all of the outer leaflet of the OM, because chemical labeling of amino groups with intact cells of S. enterica serovar Typhimurium, by the use of an OM-impermeable macromolecular reagent, failed to label any of the phosphatidylethanolamine molecules (316). The latter paper was rejected twice by Biochemistry because a reviewer insisted that such an asymmetric distribution of bilayer components was “thermodynamically impossible.” It amuses the present author that this concept is mentioned in many recent publications without any reference, as though it is self-evident.
I mention this ancient history because the concept of an asymmetric bilayer does not appear to have been tested many times with similar rigor. Labeling intact cells of Erwinia carotovora with trinitrobenzenesulfonic acid showed that 3% of the total phosphatidylethanolamine was exposed on the cell surface (611); it was not tested if the reagent penetrated through the OM. The generally high susceptibility of N. gonorrhoeae to lipophilic agents may be consistent with the presence of a mixed LPS-phospholipid leaflet in the OM, but phospholipase C digestion of intact cells suggested that the outer leaflet did not contain much phospholipid (391). In contrast, the presence of phospholipids on the outer surface of OM was detected by using a macromolecular labeling reagent with V. cholerae (486), and a similar situation may be found with some other gram-negative bacteria.
General Structure of LPS
Many reviews are available on the structure, biosynthesis, and genetic determination of LPS (262, 526-528, 594), including an extensive review with 810 references, emphasizing structure (735). A recent and comprehensive review discusses LPS structure and biosynthesis in detail (529). LPS typically is composed of lipid A, a short core oligosaccharide, and an O antigen that may be a long polysaccharide. As seen in Fig. 9 (left), E. coli K-12 lipid A consists of a β-1′,6-linked glucosamine disaccharide backbone, with its 2, 3, 2′, and 3′ positions acylated by the characteristic (R)-3-hydroxymyristic acid residues. The fatty acids linked to the nonreducing sugar residue are further acylated on their hydroxyl groups, producing characteristic piggyback (or, more correctly, acyloxyacyl) structures. Furthermore, both “ends” of the disaccharide structure, i.e., the 1 and 4′ positions, are typically phosphorylated. The core domain of LPS usually contains a few KDO (3-deoxy-d-manno-oct-2-ulosonic acid) residues at its inner end, and this is extended by a few heptose residues, some of which are phosphorylated or carry phosphoethanolamine or pyrophosphoethanolamine substituents. The outermost part of the core consists mostly of hexoses. The structure of the core oligosaccharides is shown in Fig. 10, and it can be seen that this region contains many anionic groups, which appear to be more evenly distributed in E. coli (Fig. 10A) than in P. aeruginosa (Fig. 10B). The core oligosaccharide can be divided into the outer core and inner core, the latter usually consisting of the region composed of heptose and KDO residues (Fig. 10A). Although E. coli K-12 cannot synthesize the O chain because of an insertion mutation (634) and thus produces a “rough” or R-type LPS devoid of O chains, most wild-type strains of members of the Enterobacteriaceae synthesize this outermost structure of LPS, which often consists of oligosaccharide repeating units (735).
FIG. 9.
Lipid A (from E. coli K-12) and galactosylceramide from intestinal epithelial cells. Groups that may act as H-bond donors are shown in boldface.
FIG. 10.
Structure of the R-core oligosaccharide in E. coli K-12 (A) (735) (the Hep residue indicated by the asterisk is replaced by GlcNAc in S. enterica serovar Typhimurium) and in P. aeruginosa (B) (568). Individual LPS molecules may not necessarily contain all of the residues shown. Furthermore, the core structure in the O-antigen-containing LPS of P. aeruginosa is modified from the structure in the R-mutant LPS shown (569). Abbreviations: Glc, d-glucose; Gal, d-galactose; Hep, l-glycero-d-manno-heptose; Kdo, 3-deoxy-d-manno-oct-2-ulosonic acid; EtN, ethanolamine; Rha, l-rhamnose; GalN, d-galactosamine. The anomeric configurations of Hep and Kdo are always α. In the P. aeruginosa core, there are more formal negative charges (2 from Kdo residues plus about 4.5 from monophosphates, assuming roughly 1.5 negative charges per phosphatomonoester, i.e., a total of 6.5), especially concentrated in the trisubstituted heptose residue (although the negative charge is partially compensated by the presence of one positive charge in N-alanylgalactosamine). The E. coli core carries about 5.5 formal negative charges, but they are present in a more dispersed manner.
Because LPS biosynthesis is covered well in other reviews, especially in the review by Raetz and Whitfield (529), this topic is not discussed here. However, it should be pointed out that an ABC transporter, MsbA, located in the cytoplasmic membrane, whose structure was solved by X-ray crystallography (124), is involved in the flipping of lipid A (and possibly the R-type LPS containing the core) across the membrane (176, 177, 786). It is not clear how the completed LPS reaches the outer membrane. A recent paper claims that an OM protein, Omp85, in Neisseria performs this function but is not involved in the export of OM proteins (218). However, the results rely entirely on the phenotype of the strain in which the expression of this protein was hindered, and the conclusion is not yet totally convincing. The controversial nature of the conclusion can be also seen from the report of another group concluding that the same protein is involved in the export of OM proteins (703). A more direct biochemical study is necessary in this area.
There is no question that the LPS leaflet in the OM produces a very effective barrier for the spontaneous diffusion of lipophilic compounds. Earlier, we thought that the Salmonella OM containing the intact LPS leaflet was essentially impermeable to lipophilic solutes (451). This apparent impermeability later turned out to be the result of slow influx across the OM counteracted by active efflux (456). Nevertheless, when the diffusion rates of very hydrophobic test solutes, i.e., steroids, were measured experimentally by coupling their influx with oxidation in the cytoplasm (503, 504), their diffusion across the OM was found to be about 2 orders of magnitude slower than the diffusion through typical cell membranes containing phospholipid bilayers, showing that, indeed, the lipid bilayer domain of OM is an effective barrier.
Mutants that synthesize severely truncated cores, commonly called deep rough mutants, cannot insert many OM proteins and consequently have to fill the “void” in the OM with phospholipids (464). Cells of these mutants, containing patches of phospholipid bilayers, unlike in the wild type (316), are hypersensitive to lipophilic inhibitors (464, 685), a result consistent with the fact that phospholipid bilayers allow a much more rapid penetration of these compounds. With these data alone, we cannot eliminate the possibility that the deep rough LPS itself produces a more permeable leaflet, a hypothesis advocated by some workers on the basis of minor differences in the melting temperature of isolated LPS preparations (90). However, such differences are more likely to be due to variations in divalent cation content among LPS preparations (see below) (735). Lipophilic probes indeed partition with equal efficiency into the lipid domains of normal as well as deep rough LPS (686). Furthermore, reconstitution of vesicles surrounded by pure LPS bilayers (627) and measurement of the bilayer permeability showed unequivocally that bilayers of deep rough LPS and wild-type LPS have comparable permeability (626).
Some recent data, however, are not easily explained by this concept. Although a defect in the phosphorylation of the distal core heptose (HepII in Fig. 10A) used to be thought to result in the truncated core and therefore to lead to a deep rough phenotype (265), disruption of the waaP gene, responsible for this phosphorylation, did not produce a complete truncation of the core (758). Therefore, the mutant used in the early study, isolated by chemical mutagenesis followed by selection for core-truncated mutants (265), must have contained multiple mutations. Even more importantly, the “clean” waaP mutant did not show decreased levels of OM proteins yet was hypersensitive to hydrophobic agents (759, 760). It is difficult to reconcile this result with a large body of evidence supporting the phospholipid bilayer hypothesis mentioned above, and further study is obviously needed.
The O chains define the properties of the cell surface in species that do not produce capsules and are important in evading immune attacks from the host, for example by inventing novel and unusual structures (for example, 3,6-dideoxyhexoses in Salmonella) or by mimicking the structures on host cell surface (for example, mimicking gangliosides in C. jejuni [428]). Some wild-type organisms produce LPS containing very short or no O chains. A well-known example is N. gonorrhoeae, and some workers advocate the use of term “lipooligosaccharide” in order to emphasize this feature. For some organisms (e.g., N. gonorrhoeae), the production of short chains may be beneficial because they allow intimate interaction with the host cell membrane. For others (e.g., N. meningitidis), an O chain would not be needed because the cell surface is covered by a thick layer of capsular polysaccharide.
It is not impossible that interactions between neighboring O side chains contribute to the stabilization of the LPS leaflet, but not many papers have been published on this point since the intriguing study by Morrison and Leive (430). One important observation is that the melting temperature of the hydrocarbon domain of LPS is the lowest in Re LPS and increases in S LPS (90, 439); this is likely to reflect the increased interaction between neighboring LPS molecules through the O chain (and maybe the outer core region [439]). Yeh and Jacobs (756) found that when added to phosphatidylcholine liposomes, the LPS fraction containing long O side chains decreased the fluidity of the phospholipid bilayer more effectively than did the fraction with short or no side chains, suggesting that O side chains affect the behavior of lipid A domain. Furthermore, the lateral diffusion of an OM protein, IcsA, becomes somewhat faster (perhaps twofold) in a mutant that cannot synthesize the LPS O chain (551). A recent report shows that deletion of the O side chain increases the weak-acid sensitivity of both E. coli and S. enterica serovar Typhimurium (38), but the effect is very slight. It should be noted that in most strains, only a small fraction of LPS molecules appear to contain long O side chains, a feature regulated by a gene called rol (reference 41 and references therein), cld, or wzz (537).
What Makes the LPS Leaflet an Effective Permeability Barrier?
Low fluidity of the LPS hydrocarbon domain.
The fatty acid substituents in LPS from enteric bacteria, grown under the usual conditions, are all saturated. Further, increased numbers of fatty acyl substituents per molecule of lipid usually decrease the fluidity of the hydrocarbon region (see below). Therefore, it is reasonable to expect that the hydrocarbon regions of LPS are in a gel-like state of very low fluidity under the usual conditions of growth for bacteria. Indeed, differential scanning calorimetry using intact E. coli cells detected no thermal transition caused by the melting of LPS, up to about 60°C, where proteins begin to become denatured (413), and isolated LPS in the presence of 10 mM Mg2+ showed no thermal melting up to 75°C (463), clearly suggesting that the lipid interior of LPS remains in a gel-like state even at a high temperature. In addition, X-ray diffraction studies of isolated LPS showed that the hydrocarbon chains are highly ordered and in a nearly crystalline arrangement, with a characteristic, sharp 4.2-Å reflection (the measured value was actually 4.25 Å) rather than the diffuse 4.5-Å reflection seen in liquid-crystalline lipids (363). Furthermore, this gel-like structure did not melt at least up to 50°C. This rigidity of the lipid interior explains the low permeability of small hydrophobic solutes across the OM (504).
This concept of the gel-like interior of LPS has, however, been challenged by some studies. To understand the nature of this controversy, it is important to realize that LPS is a polyanionic lipid. Thus, there are usually two monophosphate substitutents in lipid A (at the 1 and 4′ positions)(Fig. 9, left) and in the inner core region there are a few carboxyl groups of KDO residues and monophosphate (or pyrophosphate [diphosphate]) substituents on the heptose residue(s) (Fig. 10). Considering that lipid A usually contains six hydrocarbon chains, the density of formal negative charges is more than 1 per hydrocarbon chain. This is far higher than in the usual acidic phospholipids, where the ratio is about 0.5 per hydrocarbon chain. With acidic lipids, the neutralization and bridging of negative charges by divalent cations is a major factor in determining the lateral interaction between lipids and hence the fluidity and melting behavior of the bilayer. As an example, with the acidic lipid phosphatidylserine, the melting temperature increases by more than 50°C when 1 mM Ca2+ is added to bridge the negative charges (481). Effects of similar magnitude are expected with LPS. In fact, an early study (214) showed that removing most of the monovalent cations from LPS by electrodialysis and replacing them with various cations produced huge changes in the aggregation state of LPS. Thus, a bulky organic cation, triethylamine, produced a clear solution with the smallest aggregate size whereas the divalent cations Mg2+ and especially Ca2+ produced huge, insoluble aggregates. In fact, E. coli LPS is known to contain, even after extensive dialysis, about 4 mol of Mg2+ and 1 mol of Ca2+ per mol (149). Electron probe microanalysis showed that in thin sections of washed E. coli cells, the content of divalent cations in the envelope region is about 10 mol of Mg2+ and 2 mol of Ca2+ per mol of LPS (data from reference 123 recalculated according to the LPS content per cell [624]). When Wiese et al. (732) calculated the intrinsic transmembrane potential in the LPS-phospholipid asymmetric bilayer by assuming that all phosphate and carboxylate groups were dissociated, they found a value of −74 mV. In contrast, when the actual potential was measured under the conditions where the LPS molecules were facing 10 mM Mg2+, it was only −38 mV (731), suggesting that at least half of these formal negative charges are neutralized by divalent cations. A molecular dynamics simulation showed, quite predictably, that lateral association of LPS molecules was not possible unless divalent cations were present (350); this study was marred, however, by the “visualization,” by atomic force microscopy, of O-antigen side chains that should be absent in the E. coli JM109 strain, a derivative of K-12.
In spite of these data, early studies of melting behavior paid little attention to the divalent-cation content of LPS. Thus, many studies claiming that LPS hydrocarbon “melts” between 30 and 37°C (see, for example, reference 90) were done in the absence of any added divalent cations and seem to have little relevance to the situation in real bacterial cell. Unfortunately, the thermal transition in LPS has not been studied in intact cells since the early work of Melchior and Steim (admittedly carried out with a low-sensitivity instrument) that detected no cooperative melting up to about 50°C (413). I nevertheless feel that the cooperative melting of LPS around 37°C, i.e., the optimum growth temperature of E. coli, is unlikely to occur in intact cells. This is because lipid bilayers are known to become highly leaky, at the thermal transition temperature, to solutes of up to 900 Da (74, 695). (This finding was confirmed in several other laboratories [reference 260 and references therein].) Since the major function of the bacterial OM is to serve as a permeability barrier, it is highly unlikely that the structure of LPS has evolved to defeat this purpose precisely at the optimal growth temperature.
LPS in intact cells is therefore likely to melt at a much higher temperature than the temperatures observed with purified LPS. Nevertheless, the study of the latter system gives us some interesting pieces of information. In more recent studies (89, 604), it was shown that Mg2+ raises the melting temperature of lipid A. Furthermore, the melting temperature of lipid A measured by Fourier transform infrared spectroscopy was about 57°C, in comparison with that of dioleoylglycerophosphate (about 3°C) (89), showing clearly that lipid A hydrocarbons are in a much more strongly ordered state. It should also be emphasized that simply adding divalent cations to LPS preparations may not be enough; the huge effect of cations (described above) could be seen only when most of the resident cations were first removed by electrodialysis. One study (439), however, included a few electrodialyzed and then neutralized LPS samples and found that the Ca2+ form of Salmonella S-LPS melted at a temperature 6°C higher than did the Na+ form of Citrobacter S-LPS, although it is unclear if such a comparison of LPS of different origin can be justified.
There were differences in the melting temperature of mutant LPS molecules from the same species. Thus, the complete S-LPS had a relatively high transition temperature, which decreased in Re LPS but increased again in lipid A (90, 439). In an X-ray diffraction study using different types of LPS with high water content (88), the molecular cross-section of lipid A and S-LPS did not change much when the temperature was raised from 20 to 40°C, but under the same conditions, that of deep rough LPS increased drastically (about twofold). Presumably, neighboring lipid A molecules may undergo strong intermolecular interactions because only a few phosphate moieties are present, but the interaction is weakened in Re LPS owing to the additional presence of two negatively charged KDO residues. These considerations then suggest that further increases in negative charges, due to the presence of phosphate groups on heptose residues, must be more than balanced by intermolecular interactions between the distal parts of the core and/or the O side chains.
In view of their importance in intermolecular interactions, the anionic groups in lipid A and the deep core region of LPS merit some consideration. It seems most likely that phosphomonoester groups on lipid A as well as on the heptose residue are dissociated in the LPS leaflet of the outer membrane, in view of the very low pKa values of these groups. However, I think it is debatable whether the carboxylate groups of KDO are always dissociated in this highly acidic local environment, since the simple carboxyl groups are not strong acids. Perhaps they are there to increase lateral interactions between LPS molecules through divalent cation bridging if such ions are available, but they become protonated (so as to prevent the increase in intermolecular repulsion) when divalent cations are scarce or the pH becomes more acidic in the environment.
In connection with the important role played by divalent cations, some authors assume that a source of such cations may be an “LPS-associated” calcium exporter (10). However, this seems unlikely. LPS chelates very large amounts of divalent cations (123), as mentioned above. Since the intracellular concentration of Ca2+ in bacterial cells must be very low, at submicromolar or even nanomolar levels, as in any living cells, exporting Ca2+ from cytosol does not seem to make much difference to the supply of this ion for the construction of the LPS layer. Furthermore, there is no evidence that this putative Ca2+ pump has anything to do with LPS assembly, except that its gene is located near the LPS biosynthetic genes (10).
Strong lateral interactions between LPS molecules.
LPS from many species have powerful biological actions when introduced into the bloodstream of higher animals and humans. This “endotoxic” activity of LPS has attracted the attention of many scientists over the years. However, the significance of LPS synthesis for most, nonpathogenic gram-negative bacteria undoubtedly lies in the fact that it can produce a very effective permeability barrier, presumably as a consequence of the gel-like interior of the LPS leaflet. Production of this low-fluidity interior presumably requires a strong interaction between neighboring LPS molecules.
In fact, when patches of LPS were added to phospholipid bilayers in the presence of Mg2+, domains of pure LPS apparently persisted for days without becoming mixed with phospholipids, showing the existence of a very strong lateral interaction between LPS molecules (647). As mentioned above, the ionic interactions, including the bridging action of divalent cations, is probably a major factor in this lateral interaction. Are there any other factors? By comparing the structure of LPS with that of simple phospholipids, we can detect possible additional features of LPS that may contribute to their strong lateral interaction. (i) The first feature is the presence of several (six or seven in E. coli and Salmonella) fatty acid chains per molecule of LPS, in comparison with only two such chains in the usual phospholipids. The presence of more fatty acid chains per lipid molecule is likely to result in stronger lateral interactions because of the increased surface area. In fact, a major lipid in the plasma membrane of a thermophile, Thermus, is a dihexosyl-(N-acylglucosaminyl)-hexosyl-(diacylglycerol), containing three, rather than two, acyl chains (477). It is likely that the presence of one extra acyl group in this lipid contributes to the increased membrane stability at high temperatures, because the fatty acid chains are of normal lengths (the carbohydrate head groups may also contribute to the lateral interaction [see below]). (ii) Some glycerophospholipids, e.g., phosphatidylcholine, do not contain any groups that can act as H-bond donors. In contrast, many such groups are present in LPS. Lipid A is a glycolipid with the β-1,6-linked glucosaminyl-glucosamine as its backbone, and the 4-OH group of the reducing glucosamine residue is free to act as an H-bond donor. Furthermore, the two NH groups at the 2 and 2′ positions can also act in this way. Finally, the core oligosaccharide linked to the 6′ position of the nonreducing glucosamine provides numerous groups that could act as both donors and acceptors of H bonds (Fig. 10A). (iii) In contrast to phospholipids, lipid A contains a number of hydroxy fatty acids. The characteristic 3-OH-myristic acid residues occur at the 2, 3, 2′, and 3′ positions, as mentioned above. The residues at the 2 and 3 positions do not usually carry the piggyback substituuents, and thus there are two hydroxyl groups within the membrane interior that can act as H-bond donors (Fig. 9, left).
Features (ii) and (iii) above suggest that H bonding between neighboring LPS molecules may be important in stabilizing the structure of the LPS leaflet. These features are strikingly similar to the properties of glycosphingolipids (Fig. 9, right), whose head groups also contain sugars with many hydroxyl groups. In addition, in sphingolipids the sphingosine base itself contains the NH group and either one (sphingosine) or two (phytosphingosine) free OH groups. Finally, the fatty acid residue that is amide linked to the base is often hydroxylated. Interestingly, the number of OH groups in sphingolipids seems to parallel the degree of stress that the cells must endure (321). Thus, sphingolipids from muscle cells and erythrocytes contain sphingosine (with only one free OH group) substituted with nonhydroxy fatty acyl residues. In contrast, those from cells of the intestinal epithelium contain mostly 2-hydroxy fatty acyl residues attached to the base containing two OH groups. In unicellular organisms (yeast and amoebae), which are often exposed to toxic compounds in the environment, the acyl moiety can even be dihydroxylated, increasing the number of free OH groups in the substituted ceramide to four (321). These features suggest that glycosphingolipids may produce unusually stable membranes through their strong lateral interactions mediated by H bonds, and in recent years “rafts” composed of sphingolipids and cholesterol have indeed been isolated as Triton X-100-insoluble patches of eukaryotic cell membranes (386). Furthermore, the lateral interaction via numerous H bonds has been demonstrated by X-ray crystallographic analysis of a galactosyl ceramide (484). Intermolecular H-bonds between lipids persist even in aqueous media, because the lipids are placed in close proximity by the hydrophobic effect and the van der Waals interactions (76); this is similar to the formation of an H-bonded α-helix in aqueous solution of some short peptides. Triton X-100 does not disrupt H-bonded structures, and this may be one of the reasons why “rafts” remain insoluble in this detergent. Similarly, the OM is also insoluble in Triton X-100 (unless the LPS leaflet is destablized by the removal of divalent cations, as reported originally by Schnaitman in 1971 [592, 593; see also reference 449]). These observations strongly suggest that LPS and glycosphingolipids are similar in producing stable domains in the membrane as a result of the extensive H bonding between neighboring molecules. Interestingly, the structure proposed for a deep rough LPS from E. coli (328) suggests that the free OH group of 3-OH-myristoyl moieties at the 2 and 3 positions of lipid A are located very close to the inner core sugar moiety at the 6′ position of the neighboring LPS molecule.
Do glycosphingolipid leaflets also act as permeability barrier? The observation that the sphingolipid-rich rafts are found in the apical membranes of cells of the intestinal epithelium or of the epithelium of bladder and kidney tubules (620) suggests that this is indeed the case. The intermolecular H bonding, as well as the saturated nature of hydrocarbon chains in sphingolipids, will produce an ordered rather than liquid crystalline lipid interior (386), and this will make the spontaneous transmembrane diffusion of solutes difficult. However, just as the research on LPS function was dominated by concerns about their endotoxic activity, studies of rafts have concentrated on their roles in the signaling pathways (619); surprisingly few inquiries were made about the permeability of the sphingolipid domains. Recently, Hill and Zeidel (272) showed that bilayers composed of lipids with the calculated composition of the outer leaflet of the apical membrane of MDCK kidney cells, containing mostly sphingolipids and cholesterol, were nearly 100-fold less permeable to small solutes, such as glycerol, than were bilayers containing mostly glycerophospholipids. It therefore turns out that both E. coli and our own intestinal epithelial cells, facing the same milieu, employ the same strategy to build membrane barriers that are unusually resistant to the solubilizing activity of bile salts and that prevent the permeation of such noxious solute molecules. Similarity between the two systems is even more pronounced, since toxic molecules that slowly leak in through these membranes are actively pumped out by multidrug efflux pumps (such as AcrAB-TolC in E. coli [776] and MDR1, which is highly expressed in the apical membranes of intestinal cells [656]).
Conformation of LPS in bilayers.
The first model of lipid A and LPS, derived largely via an energy minimization approach, appeared in 1985 (363). The model used the heptaacylated lipid A from Salmonella, with the PhoPQ-regulated addition of palmitate residue on the hydroxyl group of 3-OH-myristate amide-linked to the 2 position of the reducing glucosamine residue (see “Physiological adaptation in LPS structure” below). The seven fatty acid chains occupy the nodes of a hexagonal lattice with an edge of 4.9 Å, a finding that explains the presence of a sharp, 4.25-Å (=sin60° × 4.9) meridional reflection, corresponding to the ordered lipid phase, in X-ray diffraction experiments (363). Importantly, the GlcN-(β1′6)-GlcN disaccharide backbone was predicted to be tilted in relation to the plane of the membrane, with the nonreducing glucosamine residue farther away from the membrane. This is precisely the conformation found for LPS bound to FhuA (described in the next section). The quasi-crystalline, ordered arrangement of hydrocarbon chains was also confirmed by using synthetic lipid A with the correct structure, whereas the synthetic compounds representing the earlier, incorrect structure showed a liquid crystalline, disordered structure (364).
The modeling was extended to hexa-acylated lipid A from E. coli, and the prediction was similar in that the six hydrocarbon chains were optimally located in the nodes of a hexagonal lattice (326, 439) (Fig. 11). It was also predicted that the bridging by divalent cations would bring the carboxylate group of KDOII close to the 1-phosphate on the reducing GlcN residue, a feature that is found in the experimentally determined structure of LPS bound to FhuA (203), with the distance between KDOII carboxyl and the phosphodiester oxygens being only 5 Å (see below). Again, the axis of glucosamine disaccharide is predicted to be tilted by about 40° in relation to the plane of the membrane. (The conformation of the polysaccharide chain of LPS was also estimated by energy minimization, with the result that O chains were predicted to be bent strongly against the membrane normal [325].) The energy minimization approach predicted several different conformers of Re LPS (326), and molecular dynamics simulation of single LPS molecules preserved the initial conformation (469). The next important step, the simulation of the LPS aggregate, an effort that required 3 months of computer time, was performed by Kotra et al. (350). The paper, unfortunately, does not supply much information, aside from the necessity for divalent cations to form a monolayer/leaflet structure. Thus, although the abstract of that paper indicates that LPS molecules are tightly assembled next to each other, there is no detailed description of the packing between LPS molecules. The equilibrium positions of individual fatty acyl chains are not described. Although simulation at a single-LPS level did not convert one conformer to the other, as described above, Kotra et al. (350) did not specify which conformer was used for simulation nor give information about interconversion between different conformers. The structures of the R core and O antigen (which should not exist in this strain) chosen for the modeling were not described either. Some of these deficiencies are not present in the molecular dynamic simulation of the P. aeruginosa OM bilayer (380, 610). Thus, the GlcN-GlcN backbone of all LPS molecules become oriented in a similar direction, and, most importantly, the divalent cation (Ca2+ was used in this simulation) binds four neighboring LPS molecules by coordinating with phosphate groups at the 1 and 4′ positions. Unfortunately, the P. aeruginosa LPS structure used by the authors was incorrect in the distribution of acyloxyacyl substituents in lipid A (see “Physiological adaptation in LPS structure” below and Fig. 14B).
FIG. 11.
Positions of fatty acyl chains in an energy-minimized model of LPS. The positions of fatty acids linked at the 2, 3, 2′, and 3′ positions, as well as those linked as piggybacked residues on the 2′- and 3′-linked 3-OH-myristoyl residues [these are shown as (2′) and (3′)] of E. coli Re LPS are shown by overlaying the structure of the energy-minimized conformer A described by Obst et al. (469) onto the two-dimensional hexagonal lattice.
FIG. 14.
Lipid A with nonclassical structures. (A) E. coli lipid A (as reference). (B) Lipid A from P. aeruginosa (the fatty acid substituents are shorter than in E. coli and are distributed symmetrically over the two glucosamine residues; the “piggyback” fatty acid residues are 2-hydroxylated, so that the lipid A contains more free hydroxyl groups than does the E. coli lipid A; the acyl residue shown at the 3-position may be absent). (C) Lipid A from P. gingivalis (R is H in one strain and an acyl group in another [see the text]). (D) Lipid A from R. leguminosarum (the “reducing” glucosamine residue has been converted into 2-amino-2-deoxygluconic acid; the usual phosphate group at the 4′-position is replaced by a galacturonic acid residue [58], and the characteristic 27-OH-C28:0 acid was found most recently to occur as a piggyback substitutent [40]). (E) Lipid A from Rhodopseudomonas viridis, based on a 2,3-diaminoglucose monomer and totally lacking phosphate substituents (409) (not many details are known about this structure, and the stoichiometry of acyl substituents has not been determined; although the 27-OH-C28:0 acid is known to be present, its position is not known).
An X-ray crystallographic analysis of LPS would be highly desirable. Kato's laboratory has crystallized numerous natural LPS species and synthetic lipid A (for reference, see reference 328). However, none of the crystals diffracted to a high resolution. The atomic force microscopy of Re LPS films gave interesting results. The surface representing the methyl ends (omega carbons) of the acyl chains had a hexagonal pattern, with a lozenge-shaped unit cell with sides of 6.5 and 5.5 Å. Each fatty acyl chain is thought to be located at the nodes of this hexagonal lattice. In contrast, the hydrophilic surface produced a somewhat less ordered pattern, thought to reflect the presence of two KDO residues. On the basis of these results, a model was proposed in which the carboxyl group of KDOII is located rather close to the 1-phosphate substituent on the reducing GlcN (328). This model is quite different from the earlier energy minimization models (439) in that the hydrocarbon chains are much farther apart (5.5 and 6.5 Å) than 4.9 Å. One likely reason is that Re LPS has the lowest melting point and has the loose packing of hydrocarbon chains (439). Another is that this crystal was made in triethylamine, a bulky organic cation, which presumably expanded the structure by competing with divalent cations. Therefore, the tilt of the diglucosamine axis, which is the consequence of the tight packing of the hydrocarbon chains against each other, was rather small in this model. Interestingly, the distance between various hydrocarbon chains in the FhuA-bound LPS is quite short (in the range of 4.2 to 4.8 Å), and in this sense again this structure is closer to the tightly packed model (439) than to the loosely packed, expanded model of Kato et al. (328).
Conformation of LPS in the LPS-FhuA complex.
In a recent X-ray crystallographic structure of OM protein FhuA, one molecule of LPS was cocrystallized (202). (Tight association of LPS to E. coli porins was reported [554], but the crystal structure of the classical porins does not show LPS molecules.) Although this may not necessarily tell us everything about the nature of LPS-LPS interactions, the structure reinforces the importance of anionic groups. The interaction between LPS and FhuA was studied in detail; this study led to the conclusion that the basic amino acid side chains playing important roles in this interaction are also conserved in other LPS-binding proteins, such as the serum LPS-binding protein, neutrophil bactericidal/permeability-increasing protein (BPI), lactoferrin, lysozyme, and Limulus antibacterial and anti-LPS factor (203). Furthermore, a similar putative LPS-binding site was recently found on the external surface of the 10-strand β-barrel of the OM-associated protease, OmpT (692). The study of the FhuA-LPS complex (203) revealed the proximity of various acidic groups of LPS to basic groups on the surface of FhuA. However, these interactions, which are interpreted as hydrogen bonding in that paper, are more likely to be ionic interactions (or salt bridges) in my opinion. My analysis of ionic interactions, based on the Protein Data Bank (PDB) structure 1QFG, is presented here (Fig. 12). The terminal phosphate and the secondary phosphate groups in the 1-pyrophosphate moiety of lipid A interact strongly with the ɛ-amino groups of Lys441 (2.5 Å) (the numbering of the residues here and in Fig. 12 is that of the engineered FhuA, which contains a 11-residue insertion after Pro405) and Lys439 (3.1 Å). The 4′-phosphate group interacts strongly with the ɛ-amino group of Lys351 (2.5 Å). The carboxylate group of KDOII interacts tightly with the guanidinium N of Arg384 (3.0 Å). Somewhat longer-range electrostatic interactions are listed in the legend to Fig. 12.
FIG. 12.
Electrostatic interactions between the E. coli LPS and the FhuA protein. The LPS structure is shown in a ball-and-stick diagram, with the oxygen atoms in the acidic moieties indicated by large red balls. A, B, C, D, E, and F refer to the 1-pyrophosphate of GlcNI (the reducing glucosamine), the 4′-phosphate of GlcNII (the nonreducing glucosamine), KDOI carboxylate, KDOII carboxylate, the HepI 4-pyrophosphate, and the HepII 4-phosphate, respectively. The amino acid residues apparently interacting with these anionic groups are shown as thin green sticks, with the basic nitrogen atoms indicated by blue balls. The amino acid residues making tight interactions are discussed in the text. The numbering of residues here is that of the engineered FhuA, which contains an 11-residue insertion after Pro405. Thus, Lys439 and Lys441 here correspond to Lys428 and Lys430 of the native FhuA, the numbering used in Table 1 of reference 203. In addition, the following residues make somewhat longer-range interactions: Arg382 with the secondary phosphate of 1-pyrophosphate (4.6 Å); Lys306 with 4′-phosphate (6 to 7 Å); Lys351 and Arg384 with KDOI carboxylate (4.7 and 7 Å); Lys439 with KDOII carboxylate (4.9 Å); and Arg474 and Arg435 with HepI 4-pyrophosphate (4.7 and about 10 Å). The figure was drawn with DS Viewer Pro (Accelrys, San Diego Calif.) using PDB file 1QFG. In addition to these electrostatic interactions, some amino acid residues may be involved in H-bonding interactions, which are not shown.
It seems remarkable to me that a weak base (lysine) is used for tight interactions with the strongly acidic groups of phosphate and pyrophosphate whereas a much stronger base (arginine) is used for weakly acidic groups of KDO. This is likely to ensure that the nature of the interaction will be ionic, by protonating the weak base and by deprotonating the weak acid. Perhaps Mg2+ and Ca2+ similarly deprotonate KDO and produce divalent cation bridges in the structure of the LPS leaflet, as was described in the preceding section.
Another interesting feature of this LPS structure is that the piggybacked fatty acid chains are in a conformation that allows their methyl ends to be aligned almost exactly with the ends of the 3-OH-myristic acid chains. Also, the plane of GlcN-(β-1′6)-GlcN disaccharide is strongly tilted in relation to the plane perpendicular to the direction of the fatty acid chain, as was predicted by the modeling of the LPS (discussed above). It has been pointed out that this tilting results from the tight packing of ordered fatty acid chains. Indeed, the LPS in the FhuA structure (PDB: 1QFG) contains fatty acid chains in a very tight association with each other (the distances between the neighboring hydrocarbon chains are all somewhat below 5 Å, as was predicted in the “frozen,” ordered structure); this is surprising because the LPS hydrocarbons here are not packed tightly by the presence of neighboring LPS molecules. One unexpected feature is that the hydrocarbons of this single LPS molecule are nearly in a single plane (although the 3-OH-myristate linked to the 3 position was disordered and could not be modeled), in contrast to the models in which they were thought to occupy successive planes in a hexagonal lattice, in a zig-zag fashion (Fig. 11).
Physiological Adaptation in LPS Structure
Especially in bacteria that have complex life cycles, LPS structures are modified in response to the requirement of different environment. One of these adaptation phenomena illustrates the importance of the factors presumed to be involved in the stabilization of LPS leaflet (described above). During infection, Salmonella must survive inside mammalian cells, more specifically in phagosomes. As we have seen, the OM containing the “normal” LPS molecules must be stabilized by the bridging action of divalent cations, Mg2+ and Ca2+. The concentrations of these ions are quite high (millimolar levels) in serum but are apparently very low (micromolar levels) in phagocytic vacuoles (215), and this will destabilize the OM. The low Mg2+ concentration may be sensed by the PhoQ sensor in the cytoplasmic membrane and may cause the activation, by phosphorylation, of the PhoP regulator (232). The phosphorylated PhoP (PhoP-P) then activates the transcription of a number of genes involved in modifying the structure of LPS (238) (Fig. 13). Recently, cationic antimicrobial peptides were shown to activate the PhoPQ system (and consequently the PmrAB system [see below]) in S. enterica serovar Typhimurium (27) and also to activate the PmrAB system directly in P. aeruginosa (411). (The PhoPQ system up-regulates the OM protease hydrolyzing some cationic peptides [236]; this observation now appears even more significant with the inducing capacity of these peptides.) (i) PhoP-P activates another two-component system, pmrAB (see reference 327 for the mechanism), the genes originally identified as being responsible for causing moderate resistance to a polycationic antibiotic, polymyxin (687). Some of the PmrAB-activated genes (ugd and the pbgPE operon) code for enzymes needed for the addition of 4-aminoarabinose to the 4′-phosphate group on lipid A (96, 97, 237, 668, 669) (Fig. 13). This modification decreases the net charge at this position from about −1.5 to 0 and reduces electrostatic repulsion between the neighboring LPS molecules. Interestingly, growth of E. coli in the presence of metavanadate (VO3−) produces modifications in lipid A similar to the PmrAB-mediated modification seen in S. enterica serovar Typhimurium (783). In some of the lipid A species produced under these conditions, aminoarabinose is attached to the 1-phosphate rather than to the 4′-phosphate (785). (ii) The PmrAB system also causes the addition of the phosphoethanolamine moiety to lipid A (784), confirming the earlier data obtained with a polymyxin-resistant mutant (468). An enzyme catalyzing the transfer of the phosphoethanolamine group has recently been identified in N. meningitidis (153). On the basis of the structure of lipid A produced by the disruption of the lptA gene (NMB1638), the enzyme appears to add the phosphoethanolamine moiety on top of the 4′-phosphate and 1-phosphate (and possibly even the 1-pyrophosphate) moieties of lipid A. If we assume that the primary phosphate ester group has a formal charge of −1.5 at a neutral pH, as postulated above, the addition of phosphoethanolamine will decrease the net charge to −1. In addition, the presence of the positively charged ethanolamine may increase the lateral interaction between LPS in a more specific manner. (iii) PhoP-P activates the transcription of pagP, which codes for an acyl transferase (239). Although at that time PagP was thought to be an enzyme located in the inner membrane, subsequent studies (62) showed it to be an outer membrane protein oriented so that the active site is close to the interface of the outer leaflet (62, 285). This enzyme uses phosphatidylethanolamine as a donor and transfers palmitate (C16:0) to the OH group of 3-hydroxymyristic acid residue linked to position 2 of lipid A (62) (Fig. 13). This increases the number of acyl groups per lipid A molecule from six to seven. It also is probably important that the chain of this newly added fatty acid is longer than those that existed earlier in lipid A. These features are expected to stabilize the LPS leaflet by increasing the hydrophobic and van der Waals interactions between neighboring LPS molecules. Perhaps the reported increased resistance to basic peptides mediated by PagP (239) is an indication of this effect. PagP homologs from other bacteria have also been studied (508, 553). (iv) Finally, PhoP-P induces the production of LPS containing the 2-OH-myristic acid residue as a piggyback fatty acyl chain connected to the 3-OH-myristic acid residue at the 3′-position (238) (Fig. 13). This modification is done by converting the myristic acid residue in the LPS into the 2-OH acid by an oxygenase encoded by lpxO (221). This compensates for the decrease in the number of OH groups caused by the PagP-catalyzed reaction and again maintains the LPS leaflet in a stable state by maintaining the number of H-bond donors, such as the OH group, at the same level. The authors also speculate that 2-OH-myristate, released inside the host cell, may inhibit the signal transduction pathway by acting as an inhibitor of protein myristoyl transferase (221). In any case, we can see that all four modifications of lipid A are in the direction of increased stabilization of LPS leaflet in the Mg2+-poor environment of phagosomes. In fact, by measuring the permeability of the OM to a large, lipophilic probe, we have shown that in a low-Mg2+ medium, the PhoP-constitutive strain producing fully modified lipid A is more successful in preventing the entry of the probe than is the PhoP-defective S. enterica serovar Typhimurium producing unmodified lipid A (W. W. Tseng, T. Guina, S. H. Miller, and H. Nikaido, unpublished data). Another PhoPQ-regulated gene product, PagL, was shown to remove the 3-OH-myristic acid residue at the 3 position (667). However, this modification was observed only under conditions of PagL overproduction from the cloned gene and has not been observed in normal cells. Possibly this enzyme is needed for minor adjustments of the structure of the LPS leaflet after the addition of the seventh fatty acid residue.
FIG. 13.
Adaptative changes in S. enterica serovar Typhimurium lipid A structure in response to a low-Mg2+ environment. Alterations are highlighted in boldface, and the enzymes involved (LpxO, PagP, and PmrAB-regulated enzymes of 5-aminoarabinose addition) are indicated by large type. LpxO generates the (S)-2-hydroxymyristate moiety, as shown. Stereochemistry was not indicated for most of the other asymmetric centers shown in Fig. 9. An additional modification of lipid A induced by PmrAB is the addition of phosphoethanolamine on 1-phosphate, as discussed in the text.
There are probably additional PhoPQ-regulated modifications of the LPS structure that contributes to the stabilization of OM. For example, the LPS of pmrAB-constitutive mutants of S. enterica serovar Typhimurium and E. coli contains more of its heptose-pyrophosphate moiety capped by ethanolamine (264, 468). The gene (lpt-3) responsible for the addition of the phosphoethanolamine moiety to a heptose residue in the core region of N. meningitidis was identified (392); it is a homolog of the lptA gene involved in the addition of the phosphoethanolamine to lipid A, described above. Schnaitman and Klena (594) reported that a tolC mutant of E. coli produced a similar LPS, with an increased content of pyrophosphorylethanolamine. The connection suggested between this LPS alteration and the drug hypersensitivity of the tolC mutants (594) is unlikely to be correct, now that we know the contribution of TolC to active drug efflux (450). However, there is an interesting possibility that E. coli cells suffering from the accumulation of toxic chemicals due to the absence of TolC produce this modified LPS through a regulatory pathway that could include PhoPQ-PmrAB.
A somewhat similar modification of lipid A occurs in P. aeruginosa strains infecting the airways of cystic fibrosis patients (195). First, as an environmental microorganism, P. aeruginosa has a lipid A that contains somewhat shorter fatty acids than does E. coli or Salmonella lipid A (see below); thus, at positions 2 and 2′, the 3-OH-myristate (C14:0) of the latter species are replaced by 3-OH-laurate (C12:0), positions 3 and 3′ are substituted by 3-OH-decanoate (C10:0), and, finally 2-OH-laurate, rather than myristate and laurate, is present as the piggybacking fatty acyl residues (357). Interestingly, these piggyback substitutions occur symmetrically on fatty acids linked to the 2 and 2′ positions (357) rather than on those linked to the 2′ and 3′ positions as in Salmonella lipid A (compare Fig. 14B with Fig. 14A). The substituent at the 3 position is often deacylated, producing a penta-acyl species (357). Therefore, as if to compensate for the decreased van der Waals intermolecular interaction due to the shorter (and sometimes smaller number of) fatty acid chains, the total number of free OH groups is increased from two or three in the E. coli and Salmonella lipid A to five (or even six in the penta-acyl form) in P. aeruginosa, greatly increasing the possibility of stabilization of the LPS leaflet through H bonding. Interestingly, in P. aeruginosa isolated from infants suffering from cystic fibrosis (i.e., in the early phase of the disease), the lipid A structure is modified (i) by the addition of a 5-aminoarabinose residue(s) and (ii) by the addition of C16:0 as a piggyback residue to the 3-OH-decanoate residue linked to the 3′ position (195). (Also, one of the piggyback fatty acids was laurate instead of 2-OH-laurate.) These modifications are reminiscent of the PhoPQ-induced modification in Salmonella discussed above, although the palmitoylation occurs at a different position in the latter system. (The P. aeruginosa genome does not appear to contain a close homolog of pagP.) In fact, the PhoPQ system was shown to cause this palmitoylation reaction in P. aeruginosa in response to low Mg2+ concentrations in the medium (195). Since P. aeruginosa strains from cystic fibrosis patients produced lipid A of this PhoPQ-modified type even when cultured on Mg2+-sufficient laboratory media, it is likely that these isolates have accumulated a mutation(s) leading to the production of such a lipid A, which is likely to be beneficial to the bacteria in this environment. It is difficult to imagine that the airway exudate contains only micromolar concentrations of Mg2+; possible advantages might involve resistance to cationic peptides, for example.
What would happen under conditions that are the opposite of those inducing the PhoPQ response, that is, an excess of divalent cations, especially Ca2+? Ca2+, because of its dehydrating effect, is known to strongly increase the rigidity of the lipid bilayer. In bilayers containing large amounts of acidic lipids, millimolar levels of Ca2+ raises the melting temperature of the membrane interior by many tens of degrees (481). LPS is obviously a strongly acidic lipid, and OM “freezes” in high concentrations of Ca2+, apparently producing cracks that allow the entry of DNA in transformation (139) and that of large proteins in the periplasmic reconstitution procedure (108). Very interestingly, the laboratory of Raetz discovered that millimolar levels of Ca2+ in the medium induce an enzyme that transfers the phosphoethanolamine group from phosphatidylethanolamine to one of the KDO residues in LPS (320). Possibly this zwitterionic group prevents the binding of Ca2+ to the high-affinity binding sites nearby, but the cation-binding properties of such modified LPS remain to be studied.
A recent study illustrates the effect of the total number of acyl chains in lipid A on the stability of the OM (435). Lipid A is synthesized in E. coli and Salmonella by the transfer of four 3-OH-myristoyl residues to the 2, 3, 2′, and 3′ positions to produce lipid IVA, then by the transfer of two KDO residues, and finally by the transfer of two piggyback acyl chains, C12:0 to the 2′-linked 3-OH-myristoyl residue by HtrB and C14:0 to the 3′-linked 3-OH-myristoyl residue by MsbB (528). Mutants with mutations of msbB gene were hitherto thought to be without phenotype, but Murray et al. (435) found that the mutant could not grow in Luria-Bertani broth without NaCl even though it grew slowly in the same medium with NaCl and grew almost as fast as the parent strain in Luria-Bertani broth containing 2 mM Mg2+ and Ca2+. Apparently, with one fewer acyl chain, the van der Waals interaction between the neighboring LPS molecules cannot compensate for the electrostatic repulsion, which becomes more pronounced in the lower-ionic-strength environment of the NaCl-free medium. The repulsion, obviously, can be minimized by an excess of divalent cations.
In a different system, pathogenic Y. enterocolitica, growth in an acidic pH or in EGTA-containing media decreased the degree of acylation of LPS, thereby increasing the fluidity of the OM (48). The molecular mechanism of this regulation is not known.
As if to avoid the excessive “freezing” of lipids, the acyl substitutents in lipid A are usually shorter than in phospholipids. In E. coli or Salmonella, most chains are C14, with one C12 acid. That the chain length is relevant to the regulation of viscosity is seen from the observation that when Salmonella cells are grown at low temperatures (12°C), there is now an incorporation of monounsaturated fatty acid (cis-Δ9-C16:1), apparently at the expense of C12:0 (744). The molecular mechanism of this modification is now known (117); low temperature induces the synthesis of a new acyl transferase, LpxP, catalyzing the transfer of palmitoleate (cis-Δ9-C16:1). The mutant in which the lpxP gene was inactivated grows at 12° C but is hypersusceptible to a large (nearly 1,500-Das); rather hydrophilic antibiotic, vancomycin (702), a result suggesting that an excessively rigid LPS interior produces a brittle OM that is unstable and undergoes transient breakage events. In a psychrophilic Pseudomonas syringae strain that was grown at 4°C, lipid A somewhat unexpectedly showed a decrease in the content of unsaturated palmitoleate, and this was compensated by strong increases in 3-OH-C10 and 3-OH-C12 acids (361). This could be because hydrophobic interactions become weak at these low temperatures and therefore must be replaced by intermolecular H bonding to maintain the LPS leaflet in a stable form.
Interaction with the host cells is also expected to produce modifications of the more highly exposed O-chain portion of the LPS. These changes are presumably more closely related to the interaction of bacterial cell surface with host components and are outside the scope of this review. One particularly well-characterized example is cited, however. When Rhizobium etli infects the roots of its host plants, it alters the structure of the O-antigen chain of its LPS. Outside the host plants, the chains are “capped” by an exceedingly lipophilic sugar residue, a tri-O-methyl-6-deoxyhexose, which must play a major role in the contact of bacterial cell with the environment; remarkably, this capping residue appears to become lost in the plant environment (184).
Different Lipid A Structures in Various Organisms
Length, number, and position of fatty acyl groups.
In the prototype lipid A of the Enterobacteriaceae, the 2, 3, 2′, and 3′ positions of the backbone disaccharide are all substituted by 3-hydroxymyristic acid (3-OH-C14:0). In many other bacteria, however, fatty acyl groups at these positions are either shorter or longer (see Table 4 of reference 735). In many cases, these differences can be rationalized as a feature of adaptation to the particular habitat of the species. For example, all four of these positions are substituted by a much shorter acid, 3-OH-C10:0 in both Rhodocyclus gelatinosus and Sphaerotilus natans, both of which are aquatic organisms presumably adapted for life at lower temperatures. As described above, P. aeruginosa, which is basically an environmental saprophyte, also contains shorter fatty acids in its lipid A than does E. coli. It was also mentioned that, as if to compensate for the decreased intermolecular hydrophobic interactions due to the shorter fatty acid chains, P. aeruginosa lipid A contains larger numbers of free OH groups (Fig. 14B), which should increase H-bonding interactions between neighboring lipid A molecules. In contrast to these environmental organisms, in H. pylori, which lives symbiotically in the stomach of higher animals (and thus exists at temperatures around 37°C), these positions contain two 3-OH-C16:0 and two 3-OH-C18:0 residues. Temperature adaptation therefore seems to play a major role in the length of fatty acids.
In the classical E. coli lipid A, the substitution of the 3-OH groups of 3-OH-C14:0 occurs in an asymmetric manner, with only the residues linked to the nonreducing glucosamine residue becoming substituted in the piggyback manner; therefore, two and four hydrocarbon chains are connected to the reducing and nonreducing sugar residues, respectively (Fig. 14A). In contrast, lipid A from N. gonorrhoeae is acylated in a symmetrical manner, with both sugar residues carrying three hydrocarbon chains (645). This difference in the positions of the acyl chains, however, cannot be the cause of the presumed high permeability of the neisserial OM, because P. aeruginosa, whose OM bilayer is at least as impermeable as that of Salmonella (503), produces lipid A that also has the symmetrical pattern of acylation (735) (Fig. 14B). The lipid A of N. meningitidis is known to carry pyrophosphorylethanolamine substitutents at both the 1 and 4′ positions (356), and the possibility remains that this may be related to the production of a highly permeable bilayer.
An unusual acyl group, 27-OH-C28:0, was first found in Rhizobium leguminosarum lipid A in 1991 (59). The enzymatic mechanism of the addition of this fatty acid has been clarified (39, 102). From its length and the presence of the hydroxyl group at the other end of the chain, it has been hypothesized that this fatty acid traverses the entire thickness of OM. Obviously this substituent is likely to stabilize the OM structure. Its presence is sometimes thought to be related to the life within eukaryotic cells, since it occurs in bacteria belonging to the α-2 subgroup of proteobacteria, which includes many such symbionts and pathogens such as Brucella and Bartonella, in addition to Rhizobium (57). However, this fatty acid is also found in many free-living members of the α-2 subgroup, including Rhodopseudomonas and Nitrobacter (57). When the gene coding for the special acyl carrier protein used for the synthesis of this long-chain fatty acid is inactivated, the mutant cells grow normally in the laboratory but are hypersensitive to deoxycholate (608) and to hyperosmotic or acidic conditions (696).
The average number of fatty acid substituents per molecule of lipid A is usually close to the number observed in the Enterobacteriaceae, i.e., about six. Significant deviation is seen in the lipid A of Porphyromonas gingivalis (Fig. 14C). In one strain, both the 3 and 3′ positions were unsubstituted and the lipid contained only three fatty acid residues (473). This was rather surprising, but in a later study a single strain was shown to produce a heterogeneous mixture of lipid A, ranging from diacyl to penta-acyl compounds (358). Furthermore, the acyl chains are rather long, with the amide-linked chains, for example, being terminally branched 3-OH-C17, a feature that may help in stabilizing the bilayer in spite of the smaller numbers of acyl chains. Interestingly, in both studies phosphate substitution at the 4′ position was found to be absent or very incomplete (see also below); the presence of fewer hydrocarbon chains may be compensated for by the decreased electrostatic repulsion between neighboring lipid A molecules.
Groups at the ends of the disaccharide backbone.
As mentioned above, both ends of the disaccharide (1 and 4′ positions) are usually phosphorylated (and the 1 position is often pyrophosphorylated) in E. coli and Salmonella; this contributes several negative charges to lipid A. In Mg2+-poor environments, PmrAB-regulated genes add a positively charged 4-aminoarabinose to the 4′-phosphate, thus decreasing the net charge (Fig. 13), as described above. This modification is apparently constitutive in Proteus spp., for example (735). This is the reason why Proteus is intrinsically resistant to the polycationic agent polymyxin B, but it can probably be better understood as an adaptation of this organism to the aquatic habitat (with low divalent-cation levels).
Among organisms outside the Enterobacteriaceae, many organisms, especially aquatic ones like R. capsulatus, Rhodocyclus gelatinosus, and S. natans, contain lower net negative charges in lipid A (Table 2). This would decrease the electrostatic repulsion between neighboring LPS molecules in a divalent-cation-poor environment, a useful feature for aquatic dwellers. Interestingly, N. meningitidis lipid A also has a lower net negative charge (Table 2), and this is probably the cause of its exceptionally high intrinsic resistance to polymyxin B (728).
TABLE 2.
Charged substituents of the classical lipid Aa
| Species | Substituent at position 1 | Substituent at position 4′ | Formal net chargeb |
|---|---|---|---|
| Escherichia coli | PP | P | −4 |
| Proteus mirabilis | P | PEtN | −1.5 |
| Rhizobium meliloti | P | P | −3 |
| Rhodobacter capsulatus | PPEtN | PEtN | −1 |
| Rhodocyclus gelatinosus | PEtN | PPEtN | −1 |
| Sphaerotilus natans | P | PPEtN | −2.5 |
| Neisseria meningitidis | PPEtN | PPEtN | −2 |
For references, see reference 735. Abbreviations: PP, pyrophosphate; P, phosphate; PEtN, phosphoethanolamine; PPEtN, pyrophosphoethanolamine.
For simplicity, phosphomonoesters were assumed to have a 1.5 negative charge at neutral pH.
B. fragilis and Porphyromonas LPS lack phosphorylation at the 4′ position, as mentioned above. In Rhodomicrobium vanielli, another aquatic organism, the 1 and 4′ positions are not phosphorylated, so that the backbone is totally devoid of negative charge. In addition, the 4′ position carries a mannose substituent in most cases (409). It will be of interest to study the structure of the inner core of this LPS, particularly the presence and distribution of charged residues. Similar, nonphosphorylated lipid A has also been isolated from Selenomonas ruminantium cells pretreated with hot 5% trichloroacetic acid (315); in this case, however, at least the 1-phosphate group could have been split off by the acid pretreatment.
Interestingly, lipid A from R. leguminosarum does not contain any phosphoryl substituents, and 2-amino-2-deoxygluconic acid and galacturonic acid now supply the negative charges contributed by the 1- and 4′-phosphates in the classical structure (Fig. 14D) (58). (2-Amino-2-deoxygluconic acid is generated by an oxidase located in the outer membrane [521, 522].) In the closely related R. etli, even the core region is devoid of phosphoryl substituents and the negative charges are provided by uronic acids and KDO (207). This may give an ecological advantage to this organism in phosphorus-poor environment. Carboxylates could also become protonated more easily and may help the survival of these organisms in an environment poor in the bridging divalent cations by decreasing the repulsion between neighboring LPS molecules.
Backbones containing 2,3-diamino-2,3-dideoxyglucose (GlcN3N).
In 1975, Roppel et al. (557) found that some Rhodopseudomonas species produced lipid A containing GlcN3N rather than glucosamine. The list of bacteria producing lipid A with this feature has grown significantly (735) since then and now includes not only α-2 proteobacteria (Rhodopseudomonas, Nitrobacter, Thiobacillus novellus, Phenylobacterium, Bradyrhizobium, Brucella, etc.) but also some γ-proteobacteria (Chromatium, Thiocapsa, Thiocystis, and Legionella) and ɛ-proteobacteria (Campylobacter jejuni). In α-2 proteobacteria, GlcN3N appears to be essentially the only sugar component of lipid A, but in others glucosamine and GlcN3N often seem to coexist. An example of the latter type, LPS from Rhodospirillum salinarium was studied in some detail (532). Conversion of the usual ester linkage at the 3 and 3′ positions into amide linkages creates two NH groups, which may function as additional H-bond donors to increase the lateral interaction between LPS molecules. Many of these bacteria are indeed environmental aquatic organisms that may need this feature in divalent-cation-poor environment. Phosphate groups are absent in many species, including Rhodopseudomonas palustris, R. viridis, Nitrobacter, Phenylobacterium immobile, Bradyrhizobium, Chromatium, Thiocapsa, and Thiocystis (see Table 7 of reference 735), and this feature will increase the lateral interaction further by decreasing electrostatic repulsion. Another characteristic feature of the α-2 proteobacteria is that their lipid A very often contains 27-OH-C28:0 (mentioned above), another feature that would stabilize the OM structure.
In R. viridis and P. immobile (and possibly in some others), the backbone is a GlcN3N monosaccharide and the phosphate groups are entirely absent (409) (Fig. 14E). This decreases drastically the number of hydrocarbon chains per molecule and the hydrophobic lateral interaction between LPS molecules: in these cases, the increased intermolecular interaction through H bonding and the drastically decreased electrostatic repulsion due to the absence of phosphate groups may be needed for the construction of an effective permeability barrier.
Extractable Lipids Substituting for or Supplementing LPS
LPS is known to be present in most gram-negative bacteria. However, among spirochetes, T. pallidum and B. burgdorferi lack LPS, in contrast to Leptospira, which produces LPS with O antigens (109). The absence of LPS in Sphingomonas is described below. Furthermore, in contrast to enteric bacteria, mutations abolishing lipid A biosynthesis are not lethal in Neisseria species (631, 632), as long as capsular polysaccharide is produced.
In some groups of bacteria, there are characteristic lipids that occur in the OM either in addition to or in place of LPS. (Reference 736 provides a comprehensive review of lipids of gram-negative bacteria, with more than 1,000 references up to 1987.) These special lipids include sphingolipids, sulfonolipids, and ornithine lipids (Fig. 15).
FIG. 15.
Extractable lipids that replace or supplement LPS in various bacteria. (Left) A sphingolipid, d-glucuronosylceramide, from Sphingomonas yanoikuyae (437). (Center) A sulfonolipid from surface-grown F. johnsoniae (1). (Right) An ornithine lipid from Paracoccus denitrificans (734). In the last lipid, the positions of the double bonds have not been determined and a biosynthetic pathway via cis-vaccenic acid has been assumed.
Sphingolipids.
This review has already mentioned that glycosphingolipids have many features in common with LPS. Indeed, glycosphingolipids replace all of the LPS in Sphingomonas paucimobilis (330). In all strains of Sphingomonas and related genera examined, glycosphingolipids were abundant (646); these lipids contain not only the OH group in the sphingosine base but also 2-OH or 3-OH fatty acid substituents, and these are expected to form an extensive H-bonding network with neighboring lipid molecules, especially with the OH groups in the sugar head groups. The head groups of Sphingomonas sphingolipids usually contain sugar acids, such as d-glucuronic acid (330) (Fig. 15, left) or d-galacturonic acid (437), although in some cases the negative charge is neutralized by the presence of an amino sugar (330). The presence of sphingolipids instead of LPS in the OM may produce a more flexible and pliable structure, and this may be related to the utilization of polymers, such as polyethylene glycol and alginate, by these bacteria (329, 427). In fact, unusual structures (called mouth-like pits) have been reported on the surface of alginate-utilizing Sphingomonas spp. (427). Asymmetric bilayers were reconstituted in vitro in which one leaflet consisted exclusively of a sphingolipid from S. paucimobilis, containing a monosaccharide as the carbohydrate component (731). The hydrocarbon chains appeared to be more rigidly packed, and the phase transition occurred in the range of 10 to 40°C, far higher than the temperature at which phospholipid transition is observed. Thus, the experimental data are in agreement with the hypothesis that glycosphingolipid and LPS leaflets behave in a similar manner in terms of physical chemistry.
Sphingolipids are also known to be a major component of lipids in Bacteroides, Porphyromonas, and Sphingobacterium species (736). In these cases also, the amide-linked fatty acids are often 3-OH acids, and in some species the hydroxy fatty acids constitute up to 43% of the fatty acids of extractable lipids. However, these lipids usually occur either as ceramides (N-acylsphingosines) (423, 753) or as sphingophospholipids (736), and because of the absence of sugars in the head group the lateral interaction through H bonding is expected to be weaker than in Sphingomonas. In fact, in contrast to Sphingomonas, the Bacteroides OM does contain LPS (87, 359, 473, 712, 713), and the observation that sphingolipids can comprise up to one-half of the total lipids in Bacteroides (736) suggests that they also form a significant fraction of the inner membrane lipids.
Sulfonolipids.
Sulfonolipids are found characteristically in the Cytophaga-Flexibacter group (224) and are located mainly in the OM (225). These lipids are N-acyl derivatives of 2-amino-3-hydroxy-15-methyl-hexadecane-1-sulfonic acid, and most of the acyl groups are also hydroxylated at the 3 position (sometimes at the 2 position) (226) (Fig. 15, center). This is reminiscent of the structures of lipid A and sphingolipids, in that they all contain hydroxyl groups as well as an amide NH group(s), all of which are potential donors of H bonds, near the hydrophilic-hydrophobic interface. As with LPS and hexuronic acid-containing glycosphingolipids, these lipids also add negative charges to the external surface of the OM.
The more fluid bilayer produced by the presence of sulfonolipids may be essential for the rapidly gliding motility of these organisms, which appears to involve the rapid lateral movement of OM components (410). Sulfonolipid-deficient mutants of Flavobacterium johnsoniae (formerly Cytophaga johnsonae) indeed lack gliding motility (2). Interestingly, when F. johnsoniae is grown on a solid surface, the composition of the acyl substituent in the sulfonolipid becomes altered. Instead of the mixture of 3-OH acids and nonhydroxylated acids found in cells grown in liquid media, cells grown on solid surfaces contain either 2-OH acids or 2,3-dihydroxy acids (1) (Fig. 15, center). This may be an adaptation for better gliding motility, but it is possible that the increased H bonding through the use of these substituents is needed on solid media, where negatively charged surfaces of neighboring cells are in close contact with each other and a better stabilization of OM becomes necessary. The latter interpretation is supported by a similar observation on the modification of S. enterica serovar Typhimurium LPS. Defects in the ugd gene and the pbgPE operon, which are needed for the decrease of the net negative charge on lipid A (through the addition of 4-aminoarabinose [237]; see “Physiological adaptation in LPS structure” above), hinders the growth of mutants on solid, low-Mg2+ medium but not in liquid medium (629); presumably the increased net negative charge of LPS in the absence of divalent cations hinders growth more strongly in colonies, where the surfaces of neighboring cells contact each other.
Ornithine lipids.
Three different classes of ornithine-containing lipids are known to exist in bacteria (227). Here we discuss only one type, in which one 3- (or 2-)hydroxy fatty acid is linked through an amide linkage to the α-amino group of ornithine and the second fatty acid is linked to the hydroxyl group of the first fatty acid via an ester linkage (Fig. 15, right), the piggyback configuration found in the lipid A. These lipids occur in many bacteria (736) and were shown to be present predominantly in the OM in Paracoccus denitrificans (734). In relation to the possible lateral interaction of these lipids, it is interesting that the majority of the ester-linked fatty acids become hydroxy fatty acids in B. cepacia when grown at a higher temperature (40°C) (650). Furthermore, F. johnsoniae contains ornithine lipids in addition to the OM sulfonolipids, and in a sulfonolipid-deficient mutant strain the content of ornithine lipid increases (500).
These observations suggest that the OM of relatively low permeability can be constructed by using lipids with a high capacity for lateral H-bond interactions, including LPS, glycosphingolipids, sulfonolipids, and ornithine lipids. It is not clear what advantages could be gained by the use of the smaller lipids, but perhaps it is reasonable to assume that one can obtain higher flexibility at the possible cost of somewhat lowered barrier capacity. The extremely high susceptibility of P. denitrificans to lipophilic agents (733) is indeed consistent with the latter assumption, although we do not know anything about the active efflux of drugs in this organism. Perhaps the availability of these “LPS substitutes” also gives bacteria the ability to survive in an environment that is unfavorable for the production and stabilization of LPS monolayers. Thus, P. denitrificans, when grown in complex media containing a low concentration of divalent cations, a condition that would destabilize the OM due to electrostatic repulsion between the highly acidic LPS molecules, produces more of the zwitterionic ornithine lipids, possibly to substitute for LPS (711).
Alterations of the OM Bilayer Barrier
Effect of energy state.
In the previous version of this review (464), a section (“Potential dynamic fluctuations in structure”) was devoted to the curious observation that the deenergization of the inner membrane somehow drastically increased the apparent permeability of the outer membrane. This mystery has now been completely solved. We now know that the influx of lipophilic compounds into gram-negative bacterial cells is limited not only by the OM permeability barrier but also by their active efflux by ubiquitous, “multidrug” efflux pumps, which are usually energized by the proton motive force (450). Thus, in these earlier experiments, the deenergization was inhibiting the active-efflux process. Entry of the lipophilic compounds across the OM is a slow process (504) but is much faster than was previously thought (see, for example, reference 451).
However, there is one area of OM organization that appears to require an energized inner membrane. I have already mentioned that TolQ-TolR-TolA (Fig. 5), a system homologous to ExbB-ExbD-TonB, is involved in the import of the A group colicins. TolQ (with three transmembrane helices like ExbB) and TolR (with one transmembrane helix like ExbD) are hypothesized to energize the largely periplasmic TolA protein for this process. Unlike TonB, which interacts with the Ton-box-containing OM receptors, the C-terminal domain of TolA interacts with a periplasmic protein, TolB, or the OM lipoprotein PAL (384). Defects in any of the TolQR-TolB system components result in profound functional problems with the OM barrier, increasing drastically the formation of OM vesicles (see below), causing periplasmic proteins (such as RNase and β-lactamase) to leak out of the cell, and making the cells hypersusceptible to various noxious agents (367). The hypersusceptibility pattern is interesting in that it includes not only lipophilic agents expected to traverse the OM bilayer (such as SDS and cholate) (367) but also vancomycin (119), which is large but hydrophilic, so that it does not cross the OM unless large, supramolecular defects open up. The known interaction between the TolB periplasmic protein and PAL (86) and between PAL and peptidoglycan through the peptidoglycan-binding motif of the PAL (118, 173) suggests that the TolQR-TolB system, through PAL and other OM proteins, somehow stabilizes the OM organization in an energy-dependent manner (367, 384). Indeed, overproduction of PAL stabilizes the OM in mutants lacking the Braun lipoprotein (118), suppressing their OM-leaky phenotype, which is very reminiscent of that of the mutants with mutations of the TolQR-TolB system. However, 7 × 105 copies of the Braun lipoprotein are supposed to exist in a single cell, in contrast to a few thousand copies of PAL (118). It is unclear why so many fewer PAL and the associated TolQR-TolB system proteins are capable of stabilizing the OM-peptidoglycan complex. The X-ray crystallographic structure of TolB (3), which includes an N-terminal α/β domain connected to a C-terminal β-propeller domain, gives an interesting hint. Here it was suggested that the homology of the C-terminal domain to oligopeptide peptidases and also metallo-β-lactamase may mean that TolB, together with PAL (which, unlike the Braun lipoprotein, interacts noncovalently with the peptidoglycan and therefore can move to different places in the peptidoglycan network), reorganizes the OM-peptidoglycan complex by local hydrolysis of the peptidoglycan and religation of the Braun lipoprotein with a new site in the peptidoglycan. The TolA and TolQ proteins were also reported to be needed for the export of the O-antigen portion of LPS (216); it is not known if this is a direct effect or a consequence of the destabilization of the OM in mutants defective in the Tol system.
Perturbation of the OM permeability barrier.
As discussed in detail above, the LPS-containing outer leaflet of OM slows considerably the spontaneous permeation of lipophilic solutes across the asymmetric bilayer of the OM. However, this relatively impermeable bilayer can be perturbed to produce a higher permeability. There are two ways to achieve this perturbation of the barrier: mutational alteration of the LPS structure and the use of drugs that bind to the bilayer and perturb its organization. The LPS mutants include the so-called deep rough mutants, which produce a very defective R core and have been studied extensively. The mutants with a complete R core (Ra LPS) (see Fig. 10 for the structure of the R core) or the mutants with a core deficient only in the galactose and N-acetylglucosamine moieties (Rb LPS) produce an essentially unaltered barrier. Those with a core containing only one glucose residue (Rc LPS) may or may not produce a permeable OM, depending on the species. Defects in any earlier steps of biosynthesis, resulting in LPS containing only KDO residues or KDO plus one or two heptose residues, show a typical deep rough phenotype with increased permeability to lipophilic antibiotics and inhibitors. In the previous version of this review (464), we concluded that the cause of this phenotype was not the increased permeability of the deep rough LPS per se but was the decreased incorporation of OM proteins in the presence of these defective LPS molecules, a situation that produces an OM bilayer with patches of phospholipid bilayers. Such glycerophospholipid bilayers have been experimentally observed in the OM of deep rough mutants (316). Furthermore, deep rough LPS was shown to be less capable of assisting the folding, insertion, and trimerization of porins (165, 602). The crucial structure for this function of LPS appears to be the presence of the phosphate substituent on HepII (Fig. 10) (464). It is interesting that this is the only anionic group that does not interact with basic groups of the FhuA protein in the structure of FhuA-LPS complex (Fig. 12). Other lines of evidence that either support or contradict the phospholipid bilayer concept are described above (see “General structure of LPS”). The literature on deep rough mutants up to 1993 has been lucidly summarized by the coauthor of the previous version of this review, Martti Vaara (685). One of the important observations cited in that review is that a leaky mutation in lpxA, which codes for the first enzyme of lipid A biosynthesis, produces a phenotype that is far more hypersusceptible to many agents than are the classical deep rough strains. I am not aware of studies of the quantitative composition of the OM outer leaflet in the lpxA strain.
The second method for perturbing the OM bilayer to increase its permeability has again been discussed critically by Vaara (684). To summarize, polycationic agents are the classical compounds that permeabilize the OM bilayer. One point that should be emphasized is that the initial electrostatic interaction between a polycationic agent and the polyanionic surface of the LPS leaflet shows an enormous dependence on the ionic strength. Using the Debye-Hückel theory, one can predict that the interaction between a pentavalent cation and a pentavalent anionic (small) section of the OM will increase 2,000-fold if the ionic environment is changed from 0.1 M NaCl (mimicking physiological conditions in the human body) to 10 mM HEPES-Na buffer (pH 7.5) (455). Thus, one should always ask if an agent loses its activity as a result of the addition of 0.1 M NaCl. If it does, then it is very unlikely to increase the permeability of OM in the human bloodstream or tissue fluids.
As summarized by Vaara (684), polymyxin B (a cyclic peptide with a net positive charge of 4 and a fatty acid tail) is an exceptionally powerful OM perturbant, presumably because of the optimal spatial distribution of the charges. Polymyxin B produces a large defect in the LPS/phospholipid asymmetric bilayer assembled in vitro (730), partly through its detergent-like capacity mediated by the fatty acid tail, and then damages the inner membrane. A recent study using intact cells (160) also confirmed that modest concentrations of polymyxin B rapidly increase the OM permeability to such lipophilic compounds as tetraphenylphosphonium and phenyldicarbaundecaborane. PMBN is devoid of the fatty acid tail and produces OM permeabilization without killing gram-negative cells (684). The involvement of anionic groups of LPS in the action of polymyxin B is clear from the fact that lipid A of polymyxin B-resistant mutants of Salmonella contains 5-aminoarabinose substituents, which neutralize one of the negative charges on lipid A (see “Physiological adaptation in LPS structure” above). The interaction between polymyxin B and lipid A was characterized by NMR (512). The solution structure of polymyxin B is altered significantly on binding of polymyxin B to lipid A, and modeling suggests that one antibiotic molecule may bind to one lipid A head group primarily through electrostatic interactions. Interestingly, the side chains of the two hydrophobic amino acid residues in polymyxin B (Phe6 and Leu7) are inserted into the hydrocarbon interior of lipid A in this model; this would explain the OM-permeabilizing effect of PMBN, which lacks the fatty acyl tail. A recent study showed that replacing Phe6 with tyrosine or replacing Leu7 with alanine resulted in a compound with weaker OM-permealizing activity (679), a result consistent with this hypothesis. We should not, however, rely too heavily on the one-to-one stoichiometry model of the polymyxin B-lipid A interaction, because the use of synthetic lipid A analogs with shorter or no acyl chains showed the stoichiometry of less than 1 (average, 0.64) polymyxin B molecule per lipid A molecule (761); the authors suggest that the predominant binding mode of polymyxin B may be the one that straddles two lipid A molecules. One note of caution is that the synthetic lipid A preparations used lacked the piggyback acyl residues or 3-hydroxyl groups on acyl substituents, and these features could have affected the results. In any case, this study provides a welcome new direction, because most other studies concentrated on polymyxin B, and what happens to the conformation of lipid A has not been pursued so far, although one reference indicates that its hydrocarbon interior becomes much more fluid, without giving detailed data (340).
Many cationic peptides of animal origin (such as defensins) have been claimed, at low concentrations, to produce permeabilization of the outer membrane. However, many of these experiments were carried out by using buffers of exceptionally low ionic strength. Indeed, some of these compounds work presumably within specialized animal cells and were optimized to exert their action only at very high concentrations. In contrast, BPI from the azurophilic granules of polymorphonuclear leukocytes binds LPS very tightly, with KD values in the micromolar range (684). BPI has since been shown to bind to the in vitro-reconstituted LPS-phospholipid bilayer and to cause rigidification of LPS followed by membrane rupture (729). The structural aspect of interaction between LPS and proteins including BPI is discussed in “Conformation of LPS in the LPS-FhuA complex” above.
Aminoglycosides, which are polycationic, were proposed to traverse the OM of P. aeruginosa by perturbing the bilayer just as polymyxin does (“self-promoted uptake” hypothesis). This pathway is likely to be significant in P. aeruginosa, which has a porin of exceptionally low permeability (see “Slow porins” above). Aminoglycosides were also reported to cause general permeabilization of the OM bilayer (250). However, these experiments were done under the extremely artificial conditions of using 5 mM HEPES buffer, and the addition of 0.15 M NaCl completely abolished the permeabilization. It is therefore necessary to conclude that the effect would be insignificant in human body fluids.
Because the LPS-containing outer leaflet is stabilized by the bridging actions of divalent cations (see “Strong lateral interactions between LPS molecules” above), removal of these cations by chelators such as EDTA destabilizes this leaflet, resulting in the release of a large fraction of LPS into the medium, as shown by the pioneering study by Leive (371). This presumably results in the consequent migration of glycerophospholipids from the inner leaflet to the outer leaflet, producing, in the OM, glycerophospholipid bilayer domains, which allow the rapid permeation of lipophilic inhibitors (464). As discussed by Vaara (684), divalent cations can also be removed by other agents.
OM Vesicles
Gram-negative cells have been known since the 1960s to “bleb off” vesicles of the OM (for a review, see reference 56). Because strong electrostatic repulsion may develop between neighboring LPS molecules and because LPS molecules may assemble in nonbilayer forms under certain conditions (for example, see reference 604), the formation of blebs with strong curvature is not a complete surprise. These vesicles contain LPS and some periplasmic enzymes (56). More surprisingly, vesicles from some species contain DNA, and vesicle-mediated transformation has been reported (754).
The vesicles, however, have not been carefully characterized in detail in most studies. For example, the reported protein composition of vesicles from P. aeruginosa (312) is strikingly different from that of the OM, yet little attention has been paid to this observation. An exception is studies by Horstman and Kuehn (277, 278), which suggest a significant role for OM vesicles for the secretion of E. coli heat-labile toxin. Here the vesicles were shown to be enriched in monomeric OM proteins, OmpA, OmpX, and OmpW, but to contain less of the trimeric porins OmpF and OmpC. The two major protein bands that are strongly enriched in vesicles, at about 45,000 and 30,000 Da, remain unidentified, however. Despite much work, it is still unclear whether the production of these vesicles is a catalyzed, active process or the simple consequence of OM instability.
EPILOGUE
When I began writing this review almost 2 years ago, I intended to use, as a motto, a fragment of a poem by the earliest known Greek lyric poet Archilochus (early seventh century B.C.), which says, “A fox knows many things, but a hedgehog one great thing.” Since this is a fragment, it is not absolutely clear what the poet intended to say, but most people think that the poet was comparing himself to the hedgehog. Having worked in the field of OM for 30 years, I thought I knew a lot about this one field, just like the hedgehog of Archilochus. But during the writing of this review, I learned how little I knew in this area, and this was a humbling and sobering experience. I am certain that I have made many mistakes due to my ignorance, and I hope that the review will be useful despite its many faults.
ADDENDUM IN PROOF
The cAMP-dependent expression of OmpD in S. enterica serovar Typhimurium (mentioned in “Other porins”) was recently rediscovered (C. A. Santiviago, C. S. Toro, A. A. Hidalgo, P. Youderian, and G. C. Mora, J. Bacteriol. 185:5901-5905, 2003).
As regards the structure of LPS (discussed in “General structure of LPS”), additional and fairly recent reviews exist on the structure of lipid A (U. Zähringer, B. Lindner, and E. Rietschel, p. 93-114, in H. Brade, S. M. Opal, S. N. Vogel, and D. C. Morrison, ed., Endotoxin in Health and Disease, 1999), on the structure of core (O. Holst, p. 115-154, in Endotoxin in Health and Disease), and on the structure of O-polysaccharide chains (P.-E. Jansson, p. 155-178, in Endotoxin in Health and Disease).
Acknowledgments
I am indebted to many colleagues, too numerous to mention, who provided invaluable help by answering my questions or by reading and correcting parts of the manuscript. I thank my coworkers, especially for their patience during the long period needed to write this review. I also thank the anonymous reviewers of this article for many constructive and useful suggestions. I benefited greatly from the information in Milton Saier's website (http://www.biology.ucsd.edu/∼msaier/transport/), which presents an up-to-date classification of transport proteins.
The studies in my laboratory have been supported by U.S. Public Health Service grant AI-09644.
REFERENCES
- 1.Abbanat, D. R., W. Godchaux III, and E. R. Leadbetter. 1988. Surface-induced synthesis of new sulfonolipids in the gliding bacterium Cytophaga johnsonae. Arch. Microbiol. 149:358-364. [Google Scholar]
- 2.Abbanat, D. R., E. R. Leadbetter, W. Godchaux III, and A. Escher. 1986. Sulfonolipids are molecular determinants of gliding motility. Nature 324:367-369. [Google Scholar]
- 3.Abergel, C., E. Bouveret, J. M. Claverie, K. Brown, A. Rigal, C. Lazdunski, and H. Benedetti. 1999. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 Å resolution. Struct. Fold Des. 7:1291-1300. [DOI] [PubMed] [Google Scholar]
- 4.Achouak, W., J. M. Pages, R. De Mot, G. Molle, and T. Heulin. 1998. A major outer membrane protein of Rahnella aquatilis functions as a porin and root adhesion. J. Bacteriol. 180:909-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ackermann, H.-W. 1999. Tailed bacteriophages: The order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adewoye, L. O., and E. A. Worobec. 1999. Multiple environmental factors regulate the expression of the carbohydrate-selective OprB porin of Pseudomonas aeruginosa. Can. J. Microbiol. 45:1033-1042. [PubMed] [Google Scholar]
- 7.Aizawa, S. I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 8.Akins, D. R., E. Robinson, D. Shevchenko, C. Elkins, D. L. Cox, and J. D. Radolf. 1997. Tromp1, a putative rare outer membrane protein, is anchored by an uncleaved signal sequence to the Treponema pallidum cytoplasmic membrane. J. Bacteriol. 179:5076-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alekshun, M. N., and S. B. Levy. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allaway, D., L. Cavalca, S. Saini, P. Hocking, E. M. Lodwig, M. E. Leonard, P. S. Poole, and L. Calvaco. 2000. Identification of a putative LPS-associated cation exporter from Rhizobium leguminosarum bv. viciae. FEMS Microbiol. Lett. 186:47-53. [DOI] [PubMed] [Google Scholar]
- 11.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amako, K., N. Baba, N. Suzuki, S. N. Wai, and A. Umeda. 1997. A structural analysis of the regularly arranged porin on the outer member of Campylobacter jejuni based on correlation averaging. Microbiol. Immunol. 41:855-859. [DOI] [PubMed] [Google Scholar]
- 13.Andersen, C., R. Cseh, K. Schulein, and R. Benz. 1998. Study of sugar binding to the sucrose-specific ScrY channel of enteric bacteria using current noise analysis. J. Membr. Biol. 164:263-274. [DOI] [PubMed] [Google Scholar]
- 14.Andersen, C., C. Hughes, and V. Koronakis. 2000. Chunnel vision. Export and efflux through bacterial channel-tunnels. EMBO Rep. 1:313-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen, C., M. Jordy, and R. Benz. 1995. Evaluation of the rate constants of sugar transport through maltoporin (LamB) of Escherichia coli from the sugar-induced current noise. J. Gen. Physiol. 105:385-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen, C., D. Krones, C. Ulmke, K. Schmid, and R. Benz. 1998. The porin RafY encoded by the raffinose plasmid pRSD2 of Escherichia coli forms a general diffusion pore and not a carbohydrate-specific porin. Eur. J. Biochem. 254:679-684. [DOI] [PubMed] [Google Scholar]
- 17.Andersen, C., E. Maier, G. Kemmer, J. Blass, A. K. Hilpert, R. Benz, and J. Reidl. 2003. Porin OmpP2 of Haemophilus influenzae shows specificity for nicotinamide-derived nucleotide substrates. J. Biol. Chem. 278:24269-24276. [DOI] [PubMed] [Google Scholar]
- 18.Andersen, C., B. Rak, and R. Benz. 1999. The gene bgIH present in the bgl operon of Escherichia coli, responsible for uptake and fermentation of beta-glucosides encodes for a carbohydrate-specific outer membrane porin. Mol. Microbiol. 31:499-510. [DOI] [PubMed] [Google Scholar]
- 19.Angus, B. L., A. M. Carey, D. A. Caron, A. M. Kropinski, and R. E. Hancock. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob. Agents Chemother. 21:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arbing, M. A., D. Dahan, D. Boismenu, O. A. Mamer, J. W. Hanrahan, and J. W. Coulton. 2000. Charged residues in surface-located loops influence voltage gating of porin from Haemophilus influenzae type b. J. Membr. Biol. 178:185-193. [DOI] [PubMed] [Google Scholar]
- 21.Arbing, M. A., J. W. Hanrahan, and J. W. Coulton. 2001. Mutagenesis identifies amino acid residues in extracellular loops and within the barrel lumen that determine voltage gating of porin from Haemophilus influenzae type b. Biochemistry 40:14621-14628. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong, S. K., T. R. Parr, Jr., C. D. Parker, and R. E. W. Hancock. 1986. Bordetella pertussis major outer membrane porin protein forms small, anion-selective channels in lipid bilayer membranes. J. Bacteriol. 166:212-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronoff, S. C. 1988. Outer membrane permeability in Pseudomonas cepacia: diminished porin content in a beta-lactam-resistant mutant and in resistant cystic fibrosis isolates. Antimicrob. Agents Chemother. 32:1636-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arora, A., D. Rinehart, G. Szabo, and L. K. Tamm. 2000. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J. Biol. Chem. 275:1594-600. [DOI] [PubMed] [Google Scholar]
- 25.Arrecubieta, C., T. C. Hammarton, B. Barrett, S. Chareonsudjai, N. Hodson, D. Rainey, and I. S. Roberts. 2001. The transport of group 2 capsular polysaccharides across the periplasmic space in Escherichia coli: roles for the KpsE and KpsD proteins. J. Biol. Chem. 276:4245-4250. [DOI] [PubMed] [Google Scholar]
- 26.Avila-Sakar, A. J., S. Misaghi, E. M. Wilson-Kubalek, K. H. Downing, H. Zgurskaya, H. Nikaido, and E. Nogales. 2001. Lipid-layer crystallization and preliminary three-dimensional structural analysis of AcrA, the periplasmic component of a bacterial multidrug efflux pump. J. Struct. Biol. 136:81-88. [DOI] [PubMed] [Google Scholar]
- 27.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. [DOI] [PubMed] [Google Scholar]
- 28.Bainbridge, G., I. Gokce, and J. H. Lakey. 1998. Voltage gating is a fundamental feature of porin and toxin beta-barrel membrane channels. FEBS Lett. 431:305-308. [DOI] [PubMed] [Google Scholar]
- 29.Bainbridge, G., H. Mobasheri, G. A. Armstrong, E. J. A. Lea, and J. H. Lakey. 1998. Voltage-gating of Escherichia coli porin: A cystine-scanning mutagenesis study of loop 3. J. Mol. Biol. 275:171-176. [DOI] [PubMed] [Google Scholar]
- 30.Bakken, J. S., C. C. Sanders, and K. S. Thomson. 1987. Selective ceftazidime resistance in Escherichia coli: association with changes in outer membrane protein. J. Infect. Dis. 155:1220-1225. [DOI] [PubMed] [Google Scholar]
- 31.Baldermann, C., and H. Engelhardt. 2000. Expression, two-dimensional crystallization, and three-dimensional reconstruction of the β8 outer membrane protein Omp21 from Comamonas acidovorans. J. Struct. Biol. 131:96-107. [DOI] [PubMed] [Google Scholar]
- 32.Bamford, D. H., M. Romantschuk, and P. J. Somerharju. 1987. Membrane fusion in prokaryotes: bacteriophage phi 6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 6:1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee-Bhatnagar, N., C. R. Bolt, and J. C. Williams. 1996. Pore-forming activity of Coxiella burnetii outer membrane protein oligomer comprised of 29.5- and 31-kDa polypeptides. Inhibition of porin activity by monoclonal antibodies 4E8 and 4D6. Ann. N. Y. Acad. Sci. 791:378-401. [DOI] [PubMed] [Google Scholar]
- 34.Bang, I. S., J. P. Audia, Y. K. Park, and J. W. Foster. 2002. Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol. Microbiol. 44:1235-1250. [DOI] [PubMed] [Google Scholar]
- 35.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 36.Bartlett, D. H. 2002. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595:367-381. [DOI] [PubMed] [Google Scholar]
- 37.Bartlett, D. H., E. Chi, and M. E. Wright. 1993. Sequence of the ompH gene from the deep-sea bacterium Photobacterium SS9. Gene 131:125-128. [DOI] [PubMed] [Google Scholar]
- 38.Barua, S., T. Yamashino, T. Hasegawa, K. Yokoyama, K. Torii, and M. Ohta. 2002. Involvement of surface polysaccharides in organic acid resistance of Shiga toxin-producing Escherichia coli O157:H7. Mol. Microbiol. 43:629-640. [DOI] [PubMed] [Google Scholar]
- 39.Basu, S. S., M. J. Karbarz, and C. R. Raetz. 2002. Expression cloning and characterization of the C28 acyltransferase of lipid A biosynthesis in Rhizobium leguminosarum. J. Biol. Chem. 277:28959-28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basu, S. S., K. A. White, N. L. S. Que, and C. R. H. Raetz. 1999. A deacylase in Rhizobium leguminosarum membranes that cleaves the 3-O-linked beta-hydroxymyristoyl moiety of lipid A precursors. J. Biol. Chem. 274:11150-11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batchelor, R. A., P. Alifano, E. Biffali, S. I. Hull, and R. A. Hull. 1992. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J. Bacteriol. 174:5228-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer, K., M. Struyve, D. Bosch, R. Benz, and J. Tommassen. 1989. One single lysine residue is responsible for the special interaction between polyphosphate and the outer membrane porin PhoE of Escherichia coli. J. Biol. Chem. 264:16393-16398. [PubMed] [Google Scholar]
- 43.Bavoil, P., A. Ohlin, and J. Schachter. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44:479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker, A., and A. Puehler. 1998. Specific amino acid substitutions in the proline-rich motif of the Rhizobium meliloti ExoP protein result in enhanced production of low-molecular-weight succinoglycan at the expense of high-molecular-weight succinoglycan. J. Bacteriol. 180:395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behlau, M., D. J. Mills, H. Quader, W. Kuhlbrandt, and J. Vonck. 2001. Projection structure of the monomeric porin OmpG at 6 Å resolution. J. Mol. Biol. 305:71-77. [DOI] [PubMed] [Google Scholar]
- 46.Bellido, F., N. L. Martin, R. J. Siehnel, and R. E. W. Hancock. 1992. Reevaluation, using intact cells, of the exclusion limit and role of porin OprF in Pseudomonas aeruginosa outer membrane permeability. J. Bacteriol. 174:5196-5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benedetti, H., R. Lloubes, C. Lazdunski, and L. Letellier. 1992. Colicin A unfolds during its translocation in Escherichia coli cells and spans the whole cell envelope when its pore has formed. EMBO J. 11:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bengoechea, J. A., K. Brandenburg, M. D. Arraiza, U. Seydel, M. Skurnik, and I. Moriyon. 2003. Pathogenic Yersinia enterocolitica strains increase the outer membrane permeability in response to environmental stimuli by modulating lipopolysaccharide fluidity and lipid A structure. Infect. Immun. 71:2014-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benson, S. A., J. L. Occi, and B. A. Sampson. 1988. Mutations that alter the pore function of the OmpF porin of Escherichia coli K12. J. Mol. Biol. 203:961-970. [DOI] [PubMed] [Google Scholar]
- 50.Benz, R., and H. Bohme. 1985. Pore formation by an outer membrane protein of the cyanobacterium, Anabaena variabilis. Biochim. Biophys. Acta 812:286-292. [Google Scholar]
- 51.Benz, R., J. Ishii, and T. Nakae. 1980. Determination of ion permeability through the channels made of porins from the outer membrane of Salmonella typhimurium in lipid bilayer membranes. J. Membr. Biol. 56:19-29. [DOI] [PubMed] [Google Scholar]
- 52.Benz, R., K. Janko, W. Boos, and P. Lauger. 1978. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim. Biophys. Acta 511:305-319. [DOI] [PubMed] [Google Scholar]
- 53.Benz, R., A. Schmid, and G. H. Vos-Scheperkeuter. 1987. Mechanism of sugar transport through the sugar-specific LamB channel of Escherichia coli outer membrane. J. Membr. Biol. 100:21-29. [DOI] [PubMed] [Google Scholar]
- 54.Berrier, C., M. Bonhivers, L. Letellier, and A. Ghazi. 2000. High-conductance channel induced by the interaction of phage lambda with its receptor maltoporin. FEBS Lett. 476:129-133. [DOI] [PubMed] [Google Scholar]
- 55.Besnard, M., B. Martinac, and A. Ghazi. 1997. Voltage-dependent porin-like ion channels in the Archaeon Haloferax volcanii. J. Biol. Chem. 272:992-995. [DOI] [PubMed] [Google Scholar]
- 56.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhat, U. R., R. W. Carlson, M. Busch, and H. Mayer. 1991. Distribution and phylogenetic significance of 27-hydroxy-octacosanoic acid in lipopolysaccharides from bacteria belonging to the alpha-2 subgroup of Proteobacteria. Int. J. Syst. Bacteriol. 41:213-217. [DOI] [PubMed] [Google Scholar]
- 58.Bhat, U. R., L. S. Forsberg, and R. W. Carlson. 1994. Structure of lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide: unique nonphosphorylated lipid A containing 2-amino-2-deoxygluconate, galacturonate and glucosamine. J. Biol. Chem. 269:14402-14410. [PubMed] [Google Scholar]
- 59.Bhat, U. R., H. Mayer, A. Yokota, R. I. Hollingsworth, and R. W. Carlson. 1991. Occurrence of lipid A variants with 27-hydroxyoctacosanoic acid in lipopolysaccharides from members of the family Rhizobiaceae. J. Bacteriol. 173:2155-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bina, J., M. Bains, and R. E. W. Hancock. 2000. Functional expression in Escherichia coli and membrane topology of porin HopE, a member of a large family of conserved proteins in Helicobacter pylori. J. Bacteriol. 182:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Birch, R. G., J. M. Pemberton, and W. V. S. Basnayake. 1990. Stable albicidin resistance in Escherichia coli involves in altered outer-membrane nucleoside uptake system. J. Gen. Microbiol. 136:51-58. [DOI] [PubMed] [Google Scholar]
- 62.Bishop, R. E., H. S. Gibbons, T. Guina, M. S. Trent, S. I. Miller, and C. R. H. Raetz. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 19:5071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bitter, W. 2003. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch. Microbiol. 179:307-314. [DOI] [PubMed] [Google Scholar]
- 64.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 65.Blachly-Dyson, E., and M. Forte. 2001. VDAC channels. IUBMB Life 52:113-118. [DOI] [PubMed] [Google Scholar]
- 66.Black, P. N. 1991. Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J. Bacteriol. 173:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Black, P. N., and C. C. Dirusso. 1994. Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim. Biophys. Acta 1210:123-145. [DOI] [PubMed] [Google Scholar]
- 68.Black, P. N., B. Said, C. R. Ghosn, J. V. Beach, and W. D. Nunn. 1987. Purification and characterization of an outer membrane-bound protein involved in long-chain fatty acid transport in Escherichia coli. J. Biol. Chem. 262:1412-1419. [PubMed] [Google Scholar]
- 69.Black, P. N., and Q. Zhang. 1995. Evidence that His-110 of the protein FadL in the outer membrane of Escherichia coli is involved in the binding and uptake of long-chain fatty acids: possible role of this residue in carboxylate binding. Biochem. J. 310:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanco, D. R., C. I. Champion, M. M. Exner, H. Erdjument-Bromage, R. E. W. Hancock, P. Tempst, J. N. Miller, and M. A. Lovett. 1995. Porin activity and sequence analysis of a 31-kilodalton Treponema pallidum subsp. Pallidum rare outer membrane protein (Tromp1). J. Bacteriol. 177:3556-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blasband, A. J., W. R. Marcotte, Jr., and C. A. Schnaitman. 1986. Structure of the 1c and nmpC outer membrane porin protein genes of lambdoid bacteriophage. J. Biol. Chem. 261:12723-12732. [PubMed] [Google Scholar]
- 72.Bliss, J. M., and R. P. Silver. 1996. Coating the surface: A model for expression of capsular polysialic acid in Escherichia coli K1. Mol. Microbiol. 21:221-231. [DOI] [PubMed] [Google Scholar]
- 73.Blocker, A., K. Komoriya, and S. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blok, M. C., L. L. van Deenen, and J. De Gier. 1976. Effect of the gel to liquid crystalline phase transition on the osmotic behaviour of phosphatidylcholine liposomes. Biochim. Biophys. Acta 433:1-12. [DOI] [PubMed] [Google Scholar]
- 75.Blot, N., C. Berrier, N. Hugouvieux-Cotte-Pattat, A. Ghazi, and G. Condemine. 2002. The oligogalacturonate-specific porin KdgM of Erwinia chrysanthemi belongs to a new porin family. J. Biol. Chem. 277:7936-7944. [DOI] [PubMed] [Google Scholar]
- 76.Boggs, J. M. 1987. Lipid intermolecular hydrogen bonding influence on structural organization and membrane function. Biochim. Biophys. Acta 906:353-404. [DOI] [PubMed] [Google Scholar]
- 77.Bohm, J., O. Lambert, A. S. Frangakis, L. Letellier, W. Baumeister, and J. L. Rigaud. 2001. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr. Biol. 11:1168-1175. [DOI] [PubMed] [Google Scholar]
- 78.Bolla, J. M., E. De, A. Dorez, and J. M. Pages. 2000. Purification, characterization and sequence analysis of Omp50, a new porin isolated from Campylobacter jejuni. Biochem. J. 352:637-643. [PMC free article] [PubMed] [Google Scholar]
- 79.Bolla, J.-M., E. Loret, M. Zalewski, and J.-M. Pages. 1995. Conformational analysis of the Campylobacter jejuni porin. J. Bacteriol. 177:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bolstad, A. I., J. Tommassen, and H. B. Jensen. 1994. Sequence variability of the 40-kDa outer membrane proteins of Fusobacterium nucleatum strains and a model for the topology of the proteins. Mol. Gen. Genet. 244:104-110. [DOI] [PubMed] [Google Scholar]
- 81.Bond, P. J., J. D. Faraldo-Gomez, and M. S. P. Sansom. 2002. OmpA: a pore or not a pore? Simulation and modeling studies. Biophys. J. 83:763-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bonhivers, M., A. Ghazi, P. Boulanger, and L. Letellier. 1996. FhuA, a transporter of the Escherichia coli outer membrane, is converted into a channel upon binding of bacteriophage T5. EMBO J. 15:1850-1856. [PMC free article] [PubMed] [Google Scholar]
- 83.Booth, I. R., P. Cash, and C. O'Byrne. 2002. Sensing and adapting to acid stress. Antonie Leeuwenhoek 81:33-42. [DOI] [PubMed] [Google Scholar]
- 84.Borgstrom, B. 1974. Bile salts—their physiological functions in the gastrointestinal tract. Acta Med. Scand. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 85.Bourdineaud, J. P., P. Boulanger, C. Lazdunski, and L. Letellier. 1990. In vivo properties of colicin A: channel activity is voltage dependent but translocation may be voltage independent. Proc. Natl. Acad. Sci. USA 87:1037-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bouveret, E., H. Benedetti, A. Rigal, E. Loret, and C. Lazdunski. 1999. In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J. Bacteriol. 181:6306-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bramanti, T. E., G. G. Wong, S. T. Weintraub, and S. C. Holt. 1989. Chemical characterization and biologic properties of lipopolysaccharide from Bacteroides gingivalis strains W50, W83, and ATCC 33277. Oral Microbiol. Immunol. 4:183-192. [DOI] [PubMed] [Google Scholar]
- 88.Brandenburg, K., S. S. Funari, M. H. Koch, and U. Seydel. 1999. Investigation into the acyl chain packing of endotoxins and phospholipids under near physiological conditions by WAXS and FTIR spectroscopy. J. Struct. Biol. 128:175-186. [DOI] [PubMed] [Google Scholar]
- 89.Brandenburg, K., and U. Seydel. 1990. Investigation into the fluidity of lipopolysaccharide and free lipid A membrane systems by Fourier-transform infrared spectroscopy and differential scanning calorimetry. Eur. J. Biochem. 191:229-236. [DOI] [PubMed] [Google Scholar]
- 90.Brandenburg, K., and U. Seydel. 1984. Physical aspects of structure and function of membranes made from lipopolysaccharides and free lipid A. Biochim. Biophys. Acta 775:225-238. [Google Scholar]
- 91.Brandon, L. D., and M. B. Goldberg. 2001. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J. Bacteriol. 183:951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Braun, M., H. Killmann, and V. Braun. 1999. The beta-barrel domain of FhuAΔ5-160 is sufficient for TonB-dependent FhuA activities of Escherichia coli. Mol. Microbiol. 33:1037-1049. [DOI] [PubMed] [Google Scholar]
- 93.Braun, M., H. Killmann, E. Maier, R. Benz, and V. Braun. 2002. Diffusion through channel derivatives of the Escherichia coli FhuA transport protein. Eur. J. Biochem. 269:4948-4959. [DOI] [PubMed] [Google Scholar]
- 94.Braun, V., and M. Braun. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 529:78-85. [DOI] [PubMed] [Google Scholar]
- 95.Braun, V., K. Hantke, and W. Koster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Metal Ions Biol. Syst. 35:67-145. [PubMed] [Google Scholar]
- 96.Breazeale, S. D., A. A. Ribeiro, and C. R. Raetz. 2003. Origin of lipid A species modified with 4-amino-4-deoxy-l-arabinose in polymyxin-resistant mutants of Escherichia coli. An aminotransferase (ArnB) that generates UDP-4-deoxyl-l-arabinose. J. Biol. Chem. 278:24731-24739. [DOI] [PubMed] [Google Scholar]
- 97.Breazeale, S. D., A. A. Ribeiro, and C. R. Raetz. 2002. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid a species modified with 4-amino-4-deoxy-l-arabinose. J. Biol. Chem. 277:2886-2896. [DOI] [PubMed] [Google Scholar]
- 98.Bremer, E., A. Middendorf, J. Martinussen, and P. Valentin-Hansen. 1990. Analysis of the tsx gene, which encodes a nucleoside-specific channel-forming protein (Tsx) in the outer membrane of Escherichia coli. Gene 96:59-66. [DOI] [PubMed] [Google Scholar]
- 99.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 100.Brinkman, F. S., M. Bains, and R. E. Hancock. 2000. The amino terminus of Pseudomonas aeruginosa outer membrane protein OprF forms channels in lipid bilayer membranes: correlation with a three-dimensional model. J. Bacteriol. 182:5251-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294:1169-1179. [DOI] [PubMed] [Google Scholar]
- 102.Brozek, K. A., R. W. Carlson, and C. R. Raetz. 1996. A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium. J. Biol. Chem. 271:32126-32136. [DOI] [PubMed] [Google Scholar]
- 103.Brunen, M., and H. Engelhardt. 1993. Asymmetry of orientation and voltage gating of the Acidovorax delafieldii porin Omp34 in lipid bilayers. Eur. J. Biochem. 212:129-135. [DOI] [PubMed] [Google Scholar]
- 104.Brzostek, K., J. Hrebenda, R. Benz, and W. Boos. 1989. The OmpC protein of Yersinia enterocolitica: purification and properties. Res. Microbiol. 140:599-614. [DOI] [PubMed] [Google Scholar]
- 105.Brzostek, K., and W. W. Nichols. 1990. Outer membrane permeability and porin proteins of Yersinia enterocolitica. FEMS Microbiol. Lett. 58:275-277. [DOI] [PubMed] [Google Scholar]
- 106.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitakar, R. Chakraborty, D. Van Der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 107.Buechner, M., A. H. Delcour, B. Martinac, J. Adler, and C. Kung. 1990. Ion channel activities in the Escherichia coli outer membrane. Biochim. Biophys. Acta 1024:111-121. [DOI] [PubMed] [Google Scholar]
- 108.Bukau, B., J. M. Brass, and W. Boos. 1985. Calcium-induced permeabilization of the Escherichia coli outer membrane: comparison of transformation and reconstitution of binding-protein-dependent transport. J. Bacteriol. 163:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bulach, D. M., T. Kalambaheti, A. de la Pena-Moctezuma, and B. Adler. 2000. Lipopolysaccharide biosynthesis in Leptospira. J. Mol. Microbiol. Biotechnol. 2:375-380. [PubMed] [Google Scholar]
- 110.Burns, J. L., and A. L. Smith. 1987. A major outer-membrane protein functions as a porin in Haemophilus influenzae. J. Gen. Microbiol. 133:1273-1278. [DOI] [PubMed] [Google Scholar]
- 111.Bush, K., S. K. Tanaka, D. P. Bonner, and R. B. Sykes. 1985. Resistance caused by decreased penetration of beta-lactam antibiotics into Enterobacter cloacae. Antimicrob. Agents Chemother. 27:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cadieux, N., C. Bradbeer, and R. J. Kadner. 2000. Sequence changes in the Ton box region of BtuB affect its transport activities and interaction with TonB protein. J. Bacteriol. 182:5954-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cao, Z., and P. E. Klebba. 2002. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie 84:399-412. [DOI] [PubMed] [Google Scholar]
- 115.Cao, Z., P. Warfel, S. M. Newton, and P. E. Klebba. 2003. Spectroscopic observations of ferric enterobactin transport. J. Biol. Chem. 278:1022-1028. [DOI] [PubMed] [Google Scholar]
- 116.Carmel, G., and J. W. Coulton. 1991. Internal deletions in the FhuA receptor of Escherichia coli K-12 define domains of ligand interactions. J. Bacteriol. 173:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carty, S. M., K. R. Sreekumar, and C. R. H. Raetz. 1999. Effect of cold shock on lipid A biosynthesis in Escherichia coli: Induction at 12°C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 274:9677-9685. [DOI] [PubMed] [Google Scholar]
- 118.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cascales, E., R. Lloubes, and J. N. Sturgis. 2001. The To1Q-To1R proteins energize To1A and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795-807. [DOI] [PubMed] [Google Scholar]
- 120.Chakrabarti, S. R., K. Chaudhuri, K. Sen, and J. Das. 1996. Porins of Vibrio cholerae: purification and characterization of OmpU. J. Bacteriol. 178:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chalcroft, J. P., H. Engelhardt, and W. Baumeister. 1987. Structure of the porin from a bacterial stalk. FEBS Lett. 211:53-58. [DOI] [PubMed] [Google Scholar]
- 122.Chambers, H. F., D. Moreau, D. Yajko, C. Miick, C. Wagner, C. Hackbarth, S. Kocagoz, E. Rosenberg, W. K. Hadley, and H. Nikaido. 1995. Can penicillins and other beta-lactam antibiotics be used to treat tuberculosis? Antimicrob. Agents Chemother. 39:2620-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chang, C. F., H. Shuman, and A. P. Somlyo. 1986. Electron probe analysis, X-ray mapping, and electron energy-loss spectroscopy of calcium, magnesium, and monovalent ions in log-phase and in dividing Escherichia coli B cells. J. Bacteriol. 167:935-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 125.Charrel, R. N., J.-M. Pages, P. De Micco, and M. Mallea. 1996. Prevalence of outer membrane porin alteration in beta-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob. Agents Chemother. 40:2854-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chart, H., J. A. Frost, and B. Rowe. 1993. Expression of a new porin “OmpE” by strains of Salmonella enteritidis. FEMS Microbiol. Lett. 109:185-188. [DOI] [PubMed] [Google Scholar]
- 127.Cheng, Q., M. C. Yu, A. R. Reeves, and A. A. Salyers. 1995. Identification and characterization of a Bacteroides gene, csuF, which encodes an outer membrane protein that is essential for growth on chondroitin sulfate. J. Bacteriol. 177:3721-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chevalier, G., and C. Delamarche. 1992. Protein IIIa of Rhizobium leguminosarum is probably a porin. Biochimie 74:1121-1123. [DOI] [PubMed] [Google Scholar]
- 129.Chevalier, G., H. Duclohier, D. Thomas, E. Shechter, and H. Wroblewski. 1993. Purification and characterization of protein H, the major porin of Pasteurella multocida. J. Bacteriol. 175:266-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chevalier, S., J. F. Burini, M. A. Freulet-Marriere, C. Regeard, G. Schools, J. Guespin-Michel, R. De Mot, and N. Orange. 2000. Characterization of an OprF-deficient mutant suggests that OprF is an essential protein for Pseudomonas fluorescens strain MF0. Res. Microbiol. 151:619-627. [DOI] [PubMed] [Google Scholar]
- 131.Chi, E., and D. H. Bartlett. 1995. An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium sp. strain SS9. Mol. Microbiol. 17:713-726. [DOI] [PubMed] [Google Scholar]
- 132.Chi, E., and D. H. Bartlett. 1993. Use of a reporter gene to follow high-pressure signal transduction in the deep-sea bacterium Photobacterium sp. strain SS9. J. Bacteriol. 175:7533-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394-401. [DOI] [PubMed] [Google Scholar]
- 134.Cho, K. H., and A. A. Salyers. 2001. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 183:7224-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Click, E. M., and R. E. Webster. 1997. Filamentous phage infection: required interactions with the To1A protein. J. Bacteriol. 179:6464-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Click, E. M., and R. E. Webster. 1998. The To1QRA proteins are required for membrane insertion of the major capsid protein of the filamentous phage f1 during infection. J. Bacteriol. 180:1723-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coggshall, K. A., N. Cadieux, C. Piedmont, R. J. Kadner, and D. S. Cafiso. 2001. Transport-defective mutations alter the conformation of the energy-coupling motif of an outer membrane transporter. Biochemistry 40:13964-13971. [DOI] [PubMed] [Google Scholar]
- 139.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Coll, J. L., M. Heyde, and R. Portalier. 1994. Expression of the nmpC gene of Escherichia coli K-12 is modulated by external pH. Identification of cis-acting regulatory sequences involved in this regulation. Mol. Microbiol. 12:83-93. [DOI] [PubMed] [Google Scholar]
- 141.Collins, R. F., R. C. Ford, A. Kitmitto, R. O. Olsen, T. Tonjum, and J. P. Derrick. 2003. Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J. Bacteriol. 185:2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Conejo, M. C., I. Garcia, L. Martinez-Martinez, L. Picabea, and A. Pascual. 2003. Zinc eluted from siliconized latex urinary catheters decreases OprD expression, causing carbapenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:2313-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Conlan, S., and H. Bayley. 2003. Folding of a monomeric porin, OmpG, in detergent solution. Biochemistry 42:9453-9465. [DOI] [PubMed] [Google Scholar]
- 144.Conlan, S., Y. Zhang, S. Cheley, and H. Bayley. 2000. Biochemical and biophysical characterization of OmpG: a monomeric porin. Biochemistry 39:11845-11854. [DOI] [PubMed] [Google Scholar]
- 145.Cornelis, G. R. 2000. Type III secretion: a bacterial device for close combat with cells of their eukaryotic host. Philos. Trans. R. Soc. London Ser. B 355:681-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cornelis, G. R. 1998. The Yersinia deadly kiss. J. Bacteriol. 180:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 148.Costa-Riu, N., A. Burkovski, R. Kramer, and R. Benz. 2003. PorA represents the major cell wall channel of the Gram-positive bacterium Corynebacterium glutamicum. J. Bacteriol. 185:4779-4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Coughlin, R. T., S. Tonsager, and E. J. McGroarty. 1983. Quantitation of metal cations bound to membranes and extracted lipopolysaccharide of Escherichia coli. Biochemistry 22:2002-2007. [DOI] [PubMed] [Google Scholar]
- 150.Coulton, J. W., P. Mason, and D. Dorrance. 1983. The permeability barrier of Haemophilus influenzae type b against beta-lactam antibiotics. J. Antimicrob. Chemother. 12:435-449. [DOI] [PubMed] [Google Scholar]
- 151.Coutte, L., R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 20:5040-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghosh, R. A. Pauptit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two Escherichia coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 153.Cox, A. D., J. C. Wright, J. Li, D. W. Hood, E. R. Moxon, and J. C. Richards. 2003. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J. Bacteriol. 185:3270-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Crago, A. M., and V. Koronakis. 1998. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol. Microbiol. 30:47-56. [DOI] [PubMed] [Google Scholar]
- 155.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 156.Cristalli, G., C. C. DiRusso, and P. N. Black. 2000. The amino-terminal region of the long-chain fatty acid transport protein FadL contains an externally exposed domain required for bacteriophage T2 binding. Arch. Biochem. Biophys. 377:324-333. [DOI] [PubMed] [Google Scholar]
- 157.Dahan, D., V. Vachon, R. Laprade, and J. W. Coulton. 1994. Voltage gating of porins from Haemophilus influenzae type b. Biochim. Biophys. Acta 1189:204-211. [DOI] [PubMed] [Google Scholar]
- 158.Danclon, C., A. Suenaga, M. Winterhalter, and I. Yamato. 2003. Molecular origin of the cation selectivity in OmpF porin: single channel conductances vs. free energy calculation. Biophys. Chem. 104:591-603. [DOI] [PubMed] [Google Scholar]
- 159.Dartigalongue, C., H. Nikaido, and S. Raina. 2000. Protein folding in the periplasm in the absence of primary oxidant DsbA: modulation of redox potential in periplasmic space via OmpL porin. EMBO J. 19:5980-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Daugelavicius, R., E. Bakiene, and D. H. Bamford. 2000. Stages of polymyxin B interaction with the Escherichia coli cell envelope. Antimicrob. Agents Chemother. 44:2969-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.De, E., A. Basle, M. Jaquinod, N. Saint, M. Mallea, G. Molle, and J. M. Pages. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 162.De, E., M. Jullien, G. Labesse, J. M. Pages, G. Molle, and J. M. Bolla. 2000. MOMP (major outer membrane protein) of Campylobacter jejuni; a versatile pore-forming protein. FEBS Lett. 469:93-97. [DOI] [PubMed] [Google Scholar]
- 163.De, E., N. Orange, N. Saint, J. Guerillon, R. De Mot, and G. Molle. 1997. Growth temperature dependence of channel size of the major outer-membrane protein (OprF) in psychrotrophic Pseudomonas fluorescens strains. Microbiology 143:1029-1035. [DOI] [PubMed] [Google Scholar]
- 164.Death, A., L. Notley, and T. Ferenci. 1993. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J. Bacteriol. 175:1475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.de Cock, H., K. Brandenburg, A. Wiese, O. Holst, and U. Seydel. 1999. Non-lamellar structure and negative charges of lipopolysaccharides required for efficient folding of outer membrane protein PhoE of Escherichia coli. J. Biol. Chem. 274:5114-5119. [DOI] [PubMed] [Google Scholar]
- 166.Delavega, A. L., and A. H. Delcour. 1995. Cadaverine induces closing of E. coli porins. EMBO J. 14:6058-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Delavega, A. L., and A. H. Delcour. 1996. Polyamines decrease Escherichia coli outer membrane permeability. J. Bacteriol. 178:3715-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Delcour, A. H. 1997. Function and modulation of bacterial porins: insights from electrophysiology. FEMS Microbiol. Lett. 151:115-123. [DOI] [PubMed] [Google Scholar]
- 169.Delcour, A. H. 2002. Structure and function of pore-forming beta-barrels from bacteria. J. Mol. Microbiol. Biotechnol. 4:1-10. [PubMed] [Google Scholar]
- 170.Delcour, A. H., J. Adler, C. Kung, and B. Martinac. 1992. Membrane-derived oligosaccharides (MDO's) promote closing of an Escherichia coli porin channel. FEBS Lett. 304:216-220. [DOI] [PubMed] [Google Scholar]
- 171.Delcour, A. H., B. Martinac, J. Adler, and C. Kung. 1989. Voltage-sensitive ion channel of Escherichia coli. J. Membr. Biol. 112:267-276. [DOI] [PubMed] [Google Scholar]
- 172.Delihas, N. 1995. Regulation of gene expression by trans-encoded antisense RNAs. Mol. Microbiol. 15:411-414. [DOI] [PubMed] [Google Scholar]
- 173.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 174.Demple, B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon: a review. Gene 179:53-57. [DOI] [PubMed] [Google Scholar]
- 175.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr. 1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 176:3825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Doerrler, W. T., and C. R. Raetz. 2002. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277:36697-36705. [DOI] [PubMed] [Google Scholar]
- 177.Doerrler, W. T., M. C. Reedy, and C. R. Raetz. 2001. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 276:11461-11464. [DOI] [PubMed] [Google Scholar]
- 178.Doig, P., M. M. Exner, R. E. W. Hancock, and T. J. Trust. 1995. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 177:5447-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Domenech-Sanchez, A., S. Hernandez-Alles, L. Martinez-Martinez, V. J. Benedi, and S. Alberti. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: Its role in beta-lactam antibiotic resistance. J. Bacteriol. 181:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Douglas, J. T., E. Y. Rosenberg, H. Nikaido, D. R. Verstreate, and A. J. Winter. 1984. Porins of Brucella species. Infect. Immun. 44:16-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dreiseikelmann, B. 1994. Translocation of DNA across bacterial membranes. Microbiol. Rev. 58:293-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Drummelsmith, J., and C. Whitfield. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 19:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Duche, D., D. Baty, M. Chartier, and L. Letellier. 1994. Unfolding of colicin A during its translocation through the Escherichia coli envelope as demonstrated by disulfide bond engineering. J. Biol. Chem. 269:24820-24825. [PubMed] [Google Scholar]
- 184.Duelli, D. M., A. Tobin, J. M. Box, V. S. K. Kolli, R. W. Carlson, and K. D. Noel. 2001. Genetic locus required for antigenic maturation of Rhizobium etli CE3 lipopolysaccharide. J. Bacteriol. 183:6054-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dumas, F., S. Frank, R. Koebnik, E. Maillet, A. Lustig, and P. Van Gelder. 2000. Extended sugar slide function for the periplasmic coiled coil domain of ScrY. J. Mol. Biol. 300:687-695. [DOI] [PubMed] [Google Scholar]
- 186.Dumas, F., R. Koebnik, M. Winterhalter, and P. Van Gelder. 2000. Sugar transport through maltoporin of Escherichia coli. Role of polar tracks. J. Biol. Chem. 275:19747-19751. [DOI] [PubMed] [Google Scholar]
- 187.Dutzler, R., G. Rummel, S. Alberti, S. Hernandez-Alles, P. S. Phale, J. P. Rosenbusch, V. J. Benedi, and T. Schirmer. 1999. Crystal structure and functional characterization of OmpK36, the osmoporin of Klebsiella pneumoniae. Structure 7:425-434. [DOI] [PubMed] [Google Scholar]
- 188.Dutzler, R., Y. F. Wang, P. J. Rizkallah, J. P. Rosenbusch, and T. Schirmer. 1996. Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway. Structure 4:127-134. [DOI] [PubMed] [Google Scholar]
- 189.Eaton, R. W. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-Cymene to p-cumate. J. Bacteriol. 179:3171-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.El Hamel, C., M. A. Freulet, M. Jaquinod, E. De, G. Molle, and N. Orange. 2000. Involvement of the C-terminal part of Pseudomonas fluorescens OprF in the modulation of its pore-forming properties. Biochim. Biophys. Acta 1509:237-244. [DOI] [PubMed] [Google Scholar]
- 191.Engel, A. M., M. Brunen, and W. Baumeister. 1993. The functional properties of Omp-beta, the regularly arrayed porin of the hyperthermophilic bacterium Thermotoga maritima. FEMS Microbiol. Lett. 109:231-236. [Google Scholar]
- 192.Engelhardt, H., S. Gerbl-Rieger, D. Krezmar, S. Schneider-Voss, A. Engel, and W. Baumeister. 1990. Structural properties of the outer membrane and the regular surface protein of Comamonas acidovorans. J. Struct. Biol. 105:92-102. [Google Scholar]
- 193.Engelhardt, H., C. Heinz, and M. Niederweis. 2002. A tetrameric porin limits the cell wall permeability of Mycobacterium smegmatis. J. Biol. Chem. 277:37567-37572. [DOI] [PubMed] [Google Scholar]
- 194.Eppens, E. F., N. Saint, P. Van Gelder, R. Van Boxtel, and J. Tommassen. 1997. Role of the constriction loop in the gating of outer membrane porin PhoE of Escherichia coli. FEBS Lett. 415:317-320. [DOI] [PubMed] [Google Scholar]
- 195.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 196.Exner, M. M., P. Doig, T. J. Trust, and R. E. W. Hancock. 1995. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 63:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fajardo, D. A., J. Cheung, C. Ito, E. Sugawara, H. Nikaido, and R. Misra. 1998. Biochemistry and regulation of a novel Escherichia coli K-12 porin protein, OmpG, which produces unusually large channels. J. Bacteriol. 180:4452-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fanucci, G. E., K. A. Coggshall, N. Cadieux, M. Kim, R. J. Kadner, and D. S. Cafiso. 2003. Substrate-induced conformational changes of the perplasmic N-terminus of an outer-membrane transporter by site-directed spin labeling. Biochemistry 42:1391-400. [DOI] [PubMed] [Google Scholar]
- 199.Ferenci, T. 1994. From sequence alignment to structure prediction: the case of the OmpF porin family. Mol. Microbiol. 14:188-189. [DOI] [PubMed] [Google Scholar]
- 200.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. Van Der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 201.Ferguson, A. D., and J. Deisenhofer. 2002. TonB-dependent receptors-structural perspectives. Biochim. Biophys. Acta 1565:318-332. [DOI] [PubMed] [Google Scholar]
- 202.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: Crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 203.Ferguson, A. D., W. Welte, E. Hofmann, B. Lindner, O. Holst, J. W. Coulton, and K. Diederichs. 2000. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Struct. Fold. Des. 8:585-592. [DOI] [PubMed] [Google Scholar]
- 204.Feucht, A., A. Schmid, R. Benz, H. Schwarz, and K. J. Heller. 1990. Pore formation associated with the tail-tip protein pb2 of bacteriophage T5. J. Biol. Chem. 265:18561-18567. [PubMed] [Google Scholar]
- 205.Fiedler, G., M. Pajatsch, and A. Boeck. 1996. Genetics of a novel starch utilisation pathway present in Klebsiella oxytoca. J. Mol. Biol. 256:279-291. [DOI] [PubMed] [Google Scholar]
- 206.Finn, T. M., Z. Li, and E. Kocsis. 1995. Identification of a Bordetella pertussis bvg-regulated porin-like protein. J. Bacteriol. 177:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Forsberg, L. S., and R. W. Carlson. 1998. The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359: the complete structure of the core region of R. etli lipopolysaccharides. J. Biol. Chem. 273:2747-2757. [DOI] [PubMed] [Google Scholar]
- 208.Forst, D., W. Welte, T. Wacker, and K. Diederichs. 1998. Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose. Nat. Struct. Biol. 5:37-46. [DOI] [PubMed] [Google Scholar]
- 209.Forst, S., J. Waukau, G. Leisman, M. Exner, and R. Hancock. 1995. Functional and regulatory analysis of the OmpF-like porin, OpnP, of the symbiotic bacterium Xenorhabdus nematophilus. Mol. Microbiol. 18:779-789. [DOI] [PubMed] [Google Scholar]
- 210.Fourel, D., S. Mizushima, A. Bernadac, and J.-M. Pages. 1993. Specific regions of Escherichia coli OmpF protein involved in antigenic and colicin receptor sites and in stable trimerization. J. Bacteriol. 175:2754-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fsihi, H., B. Kottwitz, and E. Bremer. 1993. Single amino acid substitutions affecting the substrate specificity of the Escherichia coli K-12 nucleoside-specific Tsx channel. J. Biol. Chem. 268:17495-17503. [PubMed] [Google Scholar]
- 212.Gabay, J. E., M. Blake, W. D. Niles, and M. A. Horwitz. 1985. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J. Bacteriol. 162:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 214.Galanos, C., and O. Luderitz. 1975. Electrodialysis of lipopolysaccharides and their conversion to uniform salt forms. Eur. J. Biochem. 54:603-610. [DOI] [PubMed] [Google Scholar]
- 215.Garcia-del Portillo, F., J. W. Foster, M. E. Maguire, and B. B. Finlay. 1992. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol. Microbiol. 6:3289-3297. [DOI] [PubMed] [Google Scholar]
- 216.Gaspar, J. A., J. A. Thomas, C. L. Marolda, and M. A. Valvano. 2000. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol. Microbiol. 38:262-275. [DOI] [PubMed] [Google Scholar]
- 217.Gehring, K. B., and H. Nikaido. 1989. Existence and purification of porin heterotrimers of Escherichia coli K12 OmpC, OmpF, and PhoE proteins. J. Biol. Chem. 264:2810-2815. [PubMed] [Google Scholar]
- 218.Genevrois, S., L. Steeghs, P. Roholl, J. J. Letesson, and P. van der Ley. 2003. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 22:1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Genin, S., and C. A. Boucher. 1994. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol. Gen. Genet. 243:112-118. [DOI] [PubMed] [Google Scholar]
- 220.Gerbl-Rieger, S., J. Peters, J. Kellermann, F. Lottspeich, and W. Baumeister. 1991. Nucleotide and derived amino acid sequences of the major porin of Comamonas acidovorans and comparison of porin primary structures. J. Bacteriol. 173:2196-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gibbons, H. S., S. Lin, R. J. Cotter, and C. R. H. Raetz. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, a new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J. Biol. Chem. 275:32940-32949. [DOI] [PubMed] [Google Scholar]
- 222.Gill, M. J., S. Simjee, K. Al-Hattawi, B. D. Robertson, C. S. F. Easmon, and C. A. Ison. 1998. Gonococcal resistance to beta-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob. Agents Chemother. 42:2799-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Glaser-Wuttke, G., J. Keppner, and I. Rasched. 1989. Pore-forming properties of the adsorption protein of filamentous phage fd. Biochim. Biophys. Acta 985:239-247. [DOI] [PubMed] [Google Scholar]
- 224.Godchaux, W., III, and E. R. Leadbetter. 1983. Unusual sulfolipids are characteristic of the Cytophaga-Flexibacter group. J. Bacteriol. 153:1238-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Godchaux, W., III, and E. R. Leadbetter. 1988. Sulfonolipids are localized in the outer membrane of the gliding bacterium Cytophaga johnsonae. Arch. Microbiol. 150:42-47. [Google Scholar]
- 226.Godchaux, W., III, and E. R. Leadbetter. 1984. Sulfonolipids of gliding bacteria. Structure of the N-acylaminosulfonates. J. Biol. Chem. 259:2982-2990. [PubMed] [Google Scholar]
- 227.Goldfine, H. 1982. Lipids of prokaryotes—structure and distribution. Curr. Top. Membr. Transp. 17:1-43. [Google Scholar]
- 228.Goldstein, F. W., L. Gutmann, R. Williamson, E. Collatz, and J. F. Acar. 1983. In vivo and in vitro emergence of simultaneous resistance to both beta-lactam and aminoglycoside antibiotics in a strain of Serratia marcescens. Ann. Microbiol. (Paris) 134A:329-337. [PubMed] [Google Scholar]
- 229.Gotoh, N., K. Nagino, K. Wada, H. Tsujimoto, and T. Nishino. 1994. Burkholderia (formerly Pseudomonas) cepacia porin is an oligomer composed of two component proteins. Microbiology 140:3285-3291. [DOI] [PubMed] [Google Scholar]
- 230.Gotschlich, E. C., M. E. Seiff, M. S. Blake, and M. Koomey. 1987. Porin protein of Neisseria gonorrhoeae. Cloning and gene structure. Proc. Natl. Acad. Sci. USA 84:8135-8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Grahn, A. M., R. Daugelavicius, and D. H. Bamford. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46:1199-1209. [DOI] [PubMed] [Google Scholar]
- 232.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Guedin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 275:30202-30210. [DOI] [PubMed] [Google Scholar]
- 234.Guiliani, N., and C. A. Jerez. 2000. Molecular cloning, sequencing, and expression of omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl. Environm. Microbiol. 66:2318-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Guilvout, I., K. R. Hardie, N. Sauvonnet, and A. P. Pugsley. 1999. Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J. Bacteriol. 181:7212-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 238.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 239.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 240.Gupta, R. S. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62:1435-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Habe, H., K. Kasuga, H. Nojiri, H. Yamane, and T. Omori. 1996. Analysis of cumene (isopropylbenzene) degradation genes from Pseudomonas fluorescens IPO1. Appl. Environ. Microbiol. 62:4471-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hancock, R. E. W. 1981. Aminoglycoside uptake and mode of action—with special reference to streptomycin and gentamicin. II. Effects of aminoglycosides on cells. J. Antimicrob. Chemother. 8:429-445. [DOI] [PubMed] [Google Scholar]
- 243.Hancock, R. E. W., and R. Benz. 1986. Demonstration and chemical modification of a specific phosphate binding site in the phosphate-starvation-inducible outer membrane porin protein P of Pseudomonas aeruginosa. Biochim. Biophys. Acta 860:699-707. [DOI] [PubMed] [Google Scholar]
- 244.Hancock, R. E. W., and F. S. Brinkman. 2002. Function of pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17-38. [DOI] [PubMed] [Google Scholar]
- 245.Hancock, R. E. W., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hancock, R. E. W., G. M. Decad, and H. Nikaido. 1979. Identification of the protein producing transmembrane diffusion pores in the outer membrane of Pseudomonas aeruginosa PA01. Biochim. Biophys. Acta 554:323-331. [DOI] [PubMed] [Google Scholar]
- 247.Hancock, R. E. W., C. Egli, R. Benz, and R. J. Siehnel. 1992. Overexpression in Escherichia coli and functional analysis of a novel PPi-selective porin, OprO, from Pseudomonas aeruginosa. J. Bacteriol. 174:471-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hancock, R. E. W., K. Poole, and R. Benz. 1982. Outer membrane protein P of Pseudomonas aeruginosa: regulation by phosphate deficiency and formation of small anion-specific channels in lipid bilayer membranes. J. Bacteriol. 150:730-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hancock, R. E. W., V. J. Raffle, and T. I. Nicas. 1981. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 19:777-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hancock, R. E. W., and P. G. Wong. 1984. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 26:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hansel, A., A. Schmid, M. H. Tadros, and U. J. Juergens. 1994. Isolation and characterization of porin from the outer membrane of Synechococcus PCC 6301. Arch. Microbiol. 161:163-167. [DOI] [PubMed] [Google Scholar]
- 252.Hansel, A., and M. H. Tadros. 1998. Characterization of two pore-forming proteins isolated from the outer membrane of Synechococcus PCC 6301. Curr. Microbiol. 36:321-326. [DOI] [PubMed] [Google Scholar]
- 253.Hantke, K. 1976. Phage T6—colicin K receptor and nucleoside transport in Escherichia coli. FEBS Lett. 70:109-112. [DOI] [PubMed] [Google Scholar]
- 254.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Harder, K. J., H. Nikaido, and M. Matsuhashi. 1981. Mutants of Escherichia coli that are resistant to certain beta-lactam compounds lack the ompF porin. Antimicrob. Agents Chemother. 20:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hardesty, C., C. Ferran, and J. M. DiRienzo. 1991. Plasmid-mediated sucrose metabolism in Escherichia coli: characterization of scrY, the structural gene for a phosphoenolpyruvate-dependent sucrose phosphotransferase system outer membrane porin. J. Bacteriol. 173:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 258.Harms, N., W. C. Oudhuis, E. A. Eppens, Q. A. Valent, M. Koster, J. Luirink, and B. Oudega. 1999. Epitope tagging analysis of the outer membrane folding of the molecular usher FaeD involved in K88 fimbriae biosynthesis in Escherichia coli. J. Mol. Micribiol. Biotechnol. 1:319-325. [PubMed] [Google Scholar]
- 259.Hasegawa, Y., H. Yamada, and S. Mizushima. 1976. Interactions of outer membrane proteins O-8 and O-9 with peptidoglycan sacculus of Escherichia coli K-12. J. Biochem. 80:1401-1409. [DOI] [PubMed] [Google Scholar]
- 260.Hays, L. M., J. H. Crowe, W. Wolkers, and S. Rudenko. 2001. Factors affecting leakage of trapped solutes from phospholipid vesicles during thermotropic phase transitions. Cryobiology 42:88-102. [DOI] [PubMed] [Google Scholar]
- 261.Heilpern, A. J., and M. K. Waldor. 2000. CTXφ infection of Vibrio cholerae requires the tolQRA gene products. J. Bacteriol. 182:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Heinrichs, D. E., J. A. Yethon, and C. Whitefield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 263.Heinz, C., H. Engelhardt, and M. Niederweis. 2003. The core of the tetrameric mycobacterial porin MspA is an extremely stable beta-sheet domain. J. Biol. Chem. 278:8678-8685. [DOI] [PubMed] [Google Scholar]
- 264.Helander, I. M., I. Kilpelainen, and M. Vaara. 1994. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: A31P-NMR study. Mol. Microbiol. 11:481-487. [DOI] [PubMed] [Google Scholar]
- 265.Helander, I. M., M. Vaara, S. Sukupolvi, M. Rhen, S. Saarela, U. Zahringer, and P. H. Makela. 1989. rfaP mutants of Salmonella typhimurium. Eur. J. Biochem. 185:541-546. [DOI] [PubMed] [Google Scholar]
- 266.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hernandez-Alles, S., S. Alberti, D. Alvarez, A. Domenech-Sanchez, L. Martinez-Martinez, J. Gil, J. M. Tomas, and V. J. Benedi. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 268.Hernandez-Alles, S., M. Conejo, A. Pascual, J. M. Tomas, V. J. Benedi, and L. Martinez-Martinez. 2000. Relationship between outer membrane alterations and susceptibility to antimicrobial agents in isogenic strains of Klebsiella pneumoniae. J. Antimicrob. Chemother. 46:273-277. [DOI] [PubMed] [Google Scholar]
- 269.Heyde, M., P. Laloi, and R. Portalier. 2000. Involvement of carbon source and acetyl phosphate in the external-pH-dependent expression of porin genes in Escherichia coli. J. Bacteriol. 182:198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Heyde, M., and R. Portalier. 1987. Regulation of major outer membrane porin proteins of Escherichia coli K12 by pH. Mol. Gen. Genet. 208:511-517. [DOI] [PubMed] [Google Scholar]
- 271.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44:271-281. [DOI] [PubMed] [Google Scholar]
- 272.Hill, W. G., and M. L. Zeidel. 2000. Reconstituting the barrier properties of a water-tight epithelial membrane by design of leaflet-specific liposomes. J. Biol. Chem. 275:30176-30185. [DOI] [PubMed] [Google Scholar]
- 273.Hirsch, A., J. Breed, K. Saxena, O. M. H. Richter, B. Ludwig, K. Diederichs, and W. Welte. 1997. The structure of porin from Paracoccus denitrificans at 3.1 A resolution. FEBS Lett. 404:208-210. [DOI] [PubMed] [Google Scholar]
- 274.Hoffman, P. S., M. Ripley, and R. Weeratna. 1992. Cloning and nucleotide sequence of a gene (ompS) encoding the major outer membrane protein of Legionella pneumophila. J. Bacteriol. 174:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hoffman, P. S., J. H. Seyer, and C. A. Butler. 1992. Molecular characterization of the 28- and 31-kilodalton subunits of the Legionella pneumophila major outer membrane protein. J. Bacteriol. 174:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hoiczyk, E., and A. Hansel. 2000. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J. Bacteriol. 182:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Horstman, A. L., and M. J. Kuehn. 2002. Bacterial surface association of heat-labile enterotoxin through lipopolysaccharide after secretion via the general secretory pathway. J. Biol. Chem. 277:32538-32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Horstman, A. L., and M. J. Kuehn. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489-12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Huang, H., and R. E. W. Hancock. 1996. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J. Bacteriol. 178:3085-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Huang, H., D. Jeanteur, F. Pattus, and R. E. W. Hancock. 1995. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol. Microbiol. 16:931-941. [DOI] [PubMed] [Google Scholar]
- 281.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27-28, 33-38. [DOI] [PubMed] [Google Scholar]
- 283.Hutsul, J.-A., and E. Worobec. 1994. Molecular characterization of a 40 kDa OmpC-like porin from Serratia marcescens. Microbiology 140:379-387. [DOI] [PubMed] [Google Scholar]
- 284.Hutsul, J.-A., and E. Worobec. 1997. Molecular characterization of the Serratia marcescens OmpF porin, and analysis of S. marcescens OmpF and OmpC osmoregulation. Microbiology 143:2797-2806. [DOI] [PubMed] [Google Scholar]
- 285.Hwang, P. M., W. Y. Choy, E. I. Lo, L. Chen, J. D. Forman-Kay, C. R. Raetz, G. G. Prive, R. E. Bishop, and L. E. Kay. 2002. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. USA 99:13560-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Im, W., and B. Roux. 2002. Ion permeation and selectivity of OmpF porin: a theoretical study based on molecular dynamics, Brownian dynamics, and continuum electrodiffusion theory. J. Mol. Biol. 322:851-869. [DOI] [PubMed] [Google Scholar]
- 287.Im, W., and B. Roux. 2002. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J. Mol. Biol. 319:1177-1197. [DOI] [PubMed] [Google Scholar]
- 288.Ishii, J., and T. Nakae. 1993. Lipopolysaccharide promoted opening of the porin channel. FEBS Lett. 320:251-255. [DOI] [PubMed] [Google Scholar]
- 289.Iyer, R. 1977. Plasmid mediated alterations in composition and structure of envelopes of Escherichia coli B/r. Biochim. Biophys. Acta 470:258-272. [DOI] [PubMed] [Google Scholar]
- 290.Iyer, R., V. Darby, and I. B. Holland. 1978. Alterations in the outer membrane proteins of Escherichia coli B/r associated with the presence of the R plasmid rRM98. FEBS Lett. 85:127-132. [DOI] [PubMed] [Google Scholar]
- 291.Iyer, R., Z. Wu, P. M. Woster, and A. H. Delcour. 2000. Molecular basis for the polyamine-OmpF porin interactions: Inhibitor and mutant studies. J. Mol. Biol. 297:933-945. [DOI] [PubMed] [Google Scholar]
- 292.Jacob-Dubuisson, F., C. El-Hamel, N. Saint, S. Guedin, E. Willery, G. Molle, and C. Locht. 1999. Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 274:37731-37735. [DOI] [PubMed] [Google Scholar]
- 293.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 294.Jacob-Dubuisson, F., R. Striker, and S. J. Hultgren. 1994. Chaperone-assisted self-assembly of pili independent of cellular energy. J. Biol. Chem. 269:12447-12455. [PubMed] [Google Scholar]
- 295.Jacoby, G. A. 1997. Extended-spectrum beta-lactamases and other enzymes providing resistance to oxyimino-beta-lactams. Infect. Dis. Clin. North Am. 11:875-887. [DOI] [PubMed] [Google Scholar]
- 296.Jann, B., and K. Jann. 1997. Capsules of Escherichia coli, p. 113-143. In M. Sussman (ed.), Escherichia coli: mechanisms of virulence. Cambridge University Press, Cambridge, United Kingdom.
- 297.Jap, B. K. 1988. High-resolution electron diffraction of reconstituted PhoE porin. J. Mol. Biol. 199:229-232. [DOI] [PubMed] [Google Scholar]
- 298.Jap, B. K. 1989. Molecular design of PhoE porin and its functional consequences. J. Mol. Biol. 205:407-420. [DOI] [PubMed] [Google Scholar]
- 299.Jap, B. K., K. H. Downing, and P. J. Walian. 1990. Structure of PhoE porin in projection at 3.5 Å resolution. J. Struct. Biol. 103:57-63. [DOI] [PubMed] [Google Scholar]
- 300.Jap, B. K., and P. J. Walian. 1990. Biophysics of the structure and function of porins. Q. Rev. Biophys. 23:367-403. [DOI] [PubMed] [Google Scholar]
- 301.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 302.Jarlier, V., and H. Nikaido. 1990. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J. Bacteriol. 172:1418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Jeanteur, D., J. H. Lakey, and F. Pattus. 1991. The bacterial porin superfamily: sequence alignment and structure prediction. Mol. Microbiol. 5:2153-2164. [DOI] [PubMed] [Google Scholar]
- 304.Jeanteur, D., J. H. Lakey, and F. Pattus. 1994. The porin superfamily: diversity and common features, p. 363-380. In J.-M. Ghuysen and R. Hakenbeck (ed.), New comprehensive biochemistry, vol. 27. Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 305.Jeanteur, D., T. Schirmer, D. Fourel, V. Simonet, G. Rummel, C. Widmer, J. P. Rosenbusch, F. Pattus, and J.-M. Pages. 1994. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc. Natl. Acad. Sci. USA 91:10675-10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Jiang, X., M. A. Payne, Z. Cao, S. B. Foster, J. B. Feix, S. M. C. Newton, and P. E. Klebba. 1997. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science 276:1261-1264. [DOI] [PubMed] [Google Scholar]
- 307.Jo, J. T., F. S. Brinkman, and R. E. Hancock. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Johnson, J. M., and G. M. Church. 1999. Alignment and structure prediction of divergent protein families: Periplasmic and outer membrane proteins of bacterial efflux pumps. J. Mol. Biol. 287:695-715. [DOI] [PubMed] [Google Scholar]
- 309.Jones, H. M., A. Kubo, and R. S. Stephens. 2000. Design, expression and functional characterization of a synthetic gene encoding the Chlamydia trachomatis major outer membrane protein. Gene 258:173-181. [DOI] [PubMed] [Google Scholar]
- 310.Jones, R. M., V. Pagmantidis, and P. A. Williams. 2000. sal genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobacter sp. strain ADP1. J. Bacteriol. 182:2018-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Juergens, U. J. 1989. Pore-forming activity of outer membrane extracts from the unicellular cyanobacterium Synechocystis PC 6714. Z. Naturforsch. Sect. C. 44:165-169. [Google Scholar]
- 312.Kadurugamuwa, J. L., and T. J. Beveridge. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177:3998-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kahng, H.-Y., A. M. Byrne, R. H. Olsen, and J. J. Kukor. 2000. Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. J. Bacteriol. 182:1232-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kajava, A. V., N. Cheng, R. Cleaver, M. Kessel, M. N. Simon, E. Willery, F. Jacob-Dubuisson, C. Locht, and A. C. Steven. 2001. Beta-helix model for the filamentous haemagglutinin adhesion of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42:279-292. [DOI] [PubMed] [Google Scholar]
- 315.Kamio, Y., K. C. Kim, and H. Takahashi. 1972. Identification of the basic structure of a glycolipid from Selenomonas ruminantium as β-glucosaminyl-1,6-glucosamine. Agric. Biol. Chem. 36:2195-2201. [Google Scholar]
- 316.Kamio, Y., and H. Nikaido. 1976. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase C and cyanogen bromide activated dextran in the external medium. Biochemistry 15:2561-2570. [DOI] [PubMed] [Google Scholar]
- 317.Kanamaru, S., P. G. Leiman, V. A. Kostyuchenko, P. R. Chipman, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossmann. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553-557. [DOI] [PubMed] [Google Scholar]
- 318.Kanazawa, K., Y. Kobayashi, M. Nakano, M. Sakurai, N. Gotoh, and T. Nishino. 1995. Identification of three porins in the outer membrane of Bacteroides fragilis. FEMS Microbiol. Lett. 127:181-186. [Google Scholar]
- 319.Kaneko, M., A. Yamaguchi, and T. Sawai. 1984. Purification and characterization of two kinds of porins from the Enterobacter cloacae outer membrane. J. Bacteriol. 158:1179-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Kanipes, M. I., S. Lin, R. J. Cotter, and C. R. H. Raetz. 2001. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-d-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide: a novel membrane enzyme dependent upon phosphatidylethanolamine. J. Biol. Chem. 276:1156-1163. [DOI] [PubMed] [Google Scholar]
- 321.Karlsson, K.-A. 1982. Glycolipids and surface membranes, p. 1-74. In D. Chapman (ed.), Biological membranes, vol. 4. Academic Press, Inc., New York, N.Y.
- 322.Karshikoff, A., V. Spassov, S. W. Cowan, R. Ladenstein, and T. Schirmer. 1994. Electrostatic properties of two porin channels from Escherichia coli. J. Mol. Biol. 240:372-384. [DOI] [PubMed] [Google Scholar]
- 323.Kartmann, B., S. Stenger, M. Niederweis, and S. Stengler. 1999. Porins in the cell wall of Mycobacterium tuberculosis. J. Bacteriol. 181:6543-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Kasai, Y., J. Inoue, and S. Harayama. 2001. The TOL plasmid pWW0 xylN gene product from Pseudomonas putida is involved in m-xylene uptake. J. Bacteriol. 183:6662-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Kastowsky, M., T. Gutberlet, and H. Bradaczek. 1992. Molecular modelling of the three-dimensional structure and conformational flexibility of bacterial lipopolysaccharide. J. Bacteriol. 174:4798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Kastowsky, M., A. Sabisch, T. Gutberlet, and H. Labischinski. 1991. Molecular modelling of bacterial deep rough mutant lipopolysaccharide of Escherichia coli. Eur. J. Biochem. 197:707-716. [DOI] [PubMed] [Google Scholar]
- 327.Kato, A., T. Latifi, and E. A. Groisman. 2003. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc. Natl. Acad. Sci. USA 100:4706-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kato, N., T. Sugiyama, S. Naito, Y. Arakawa, H. Ito, N. Kido, M. Ohta, and K. Sasaki. 2000. Molecular structure of bacterial endotoxin (Escherichia coli Re lipopolysaccharide): implications for formation of a novel heterogeneous lattice structure. Mol. Microbiol. 36:796-805. [DOI] [PubMed] [Google Scholar]
- 329.Kawai, F. 1999. Sphingomonads involved in the biodegradation of xenobiotic polymers. J. Ind. Microbiol. Biotechnol. 23:400-407. [DOI] [PubMed] [Google Scholar]
- 330.Kawasaki, S., R. Moriguchi, K. Sekiya, T. Nakai, E. Ono, K. Kume, and K. Kawahara. 1994. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J. Bacteriol. 176:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Kazmierczak, B. I., D. L. Mielke, M. Russel, and P. Model. 1994. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J. Mol. Biol. 238:187-198. [DOI] [PubMed] [Google Scholar]
- 332.Kelly, D. P., and A. P. Wood. 2000. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. E vol. Microbiol. 50:511-516. [DOI] [PubMed] [Google Scholar]
- 333.Kennedy, E. P. 1982. Osmotic regulation and the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc. Natl. Acad. Sci. USA 79:1092-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kessel, M., M. J. Brennan, B. L. Trus, M. E. Bisher, and A. C. Steven. 1988. Naturally crystalline porin in the outer membrane of Bordetella pertussis. J. Mol. Biol. 203:275-278. [DOI] [PubMed] [Google Scholar]
- 335.Keyhani, N. O., X.-B. Li, and S. Roseman. 2000. Chitin catabolism in the marine bacterium Vibrio furnissii. Identification and molecular cloning of a chitoporin. J. Biol. Chem. 275:33068-33076. [DOI] [PubMed] [Google Scholar]
- 336.Killmann, H., C. Herrmann, A. Torun, G. Jung, and V. Braun. 2002. TonB of Escherichia coli activates FhuA through interaction with the beta-barrel. Microbiology 148:3497-509. [DOI] [PubMed] [Google Scholar]
- 337.Klauser, T., J. Kramer, K. Otzelberger, J. Pohlner, and T. F. Meyer. 1993. Characterization of the Neisseria Iga beta-core. The essential unit for outer membrane targeting and extracellular protein secretion. J. Mol. Biol. 234:579-593. [DOI] [PubMed] [Google Scholar]
- 338.Kleivdal, H., R. Benz, and H. B. Jensen. 1995. The Fusobacterium nucleatum major outer-membrane protein (FomA) forms trimeric, water-filled channels in lipid bilayer membranes. Eur. J. Biochem. 233:310-316. [DOI] [PubMed] [Google Scholar]
- 339.Kleivdal, H., P. Puntervoll, and H. B. Jensen. 2001. Topological investigations of the FomA porin from Fusobacterium nucleatum and identification of the constriction loop L6. Microbiology 147:1059-1067. [DOI] [PubMed] [Google Scholar]
- 340.Koch, P. J., J. Frank, J. Schuler, C. Kahle, and H. Bradaczek. 1999. Thermodynamics and structural studies of the interaction of polymyxin B with deep rough mutant lipopolysaccharides. J. Colloid Interface Sci. 213:557-564. [DOI] [PubMed] [Google Scholar]
- 341.Kocsis, E., B. L. Trus, A. C. Steven, P. R. Smith, J. H. Hannah, M. J. Brennan, and M. Kessel. 1993. Orientation of porin channels in the outer membrane of Bordetella pertussis. Mol. Microbiol. 9:469-476. [DOI] [PubMed] [Google Scholar]
- 342.Koebnik, R. 1995. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol. Microbiol. 16:1269-1270. [DOI] [PubMed] [Google Scholar]
- 343.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 344.Koenninger, U. W., S. Hobbie, R. Benz, and V. Braun. 1999. The haemolysin-secreting ShlB protein of the outer membrane of Serratia marcescens: determination of surface-exposed residues and formation of ion-permeable pores by ShlB mutants in artificial lipid bilayer membranes. Mol. Microbiol. 32:1212-1225. [DOI] [PubMed] [Google Scholar]
- 345.Koga, T., and K. Takumi. 1994. Isolation and characterization of three porin-like proteins from Vibrio vulnificus: effect of different growth media on their production. Microbiol. Immunol. 38:931-936. [DOI] [PubMed] [Google Scholar]
- 346.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 347.Korchev, Y. E., C. L. Bashford, G. M. Alder, P. Y. Apel, D. T. Edmonds, A. A. Lev, K. Nandi, A. V. Zima, and C. A. Pasternak. 1997. A novel explanation for fluctuations of ion current through narrow pores. FASEB J. 11:600-608. [DOI] [PubMed] [Google Scholar]
- 348.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 349.Koster, M., W. Bitter, H. de Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789-797. [DOI] [PubMed] [Google Scholar]
- 350.Kotra, L. P., D. Golemi, N. A. Amro, G.-Y. Liu, and S. Mobashery. 1999. Dynamics of the lipopolysaccharide assembly on the surface of Escherichia coli. J. Am. Chem. Soc. 121:8707-8711. [Google Scholar]
- 351.Kreusch, A., and G. E. Schulz. 1994. Refined structure of the porin from Rhodopseudomonas blastica comparison with the porin from Rhodobacter capsulatus. J. Mol. Biol. 243:891-905. [DOI] [PubMed] [Google Scholar]
- 352.Kubo, A., and R. S. Stephens. 2000. Characterization and functional analysis of PorB, a Chlamydia porin and neutralizing target. Mol. Microbiol. 38:772-780. [DOI] [PubMed] [Google Scholar]
- 353.Kubo, A., and R. S. Stephens. 2001. Substrate-specific diffusion of select dicarboxylates through Chlamydia trachomatis PorB. Microbiology 147:3135-3140. [DOI] [PubMed] [Google Scholar]
- 354.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 97:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Kullman, L., M. Winterhalter, and S. M. Bezrukov. 2002. Transport of maltodextrins through maltoporin: a single-channel study. Biophys. J. 82:803-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Kulshin, V. A., U. Zaehringer, B. Lindner, C. E. Frasch, C. M. Tsai, B. A. Dmitriev, and E. T. Rietschel. 1992. Structural characterization of the lipid A component of pathogenic Neisseria meningitidis. J. Bacteriol. 174:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kulshin, V. A., U. Zahringer, B. Lindner, K. E. Jager, B. A. Dmitriev, and E. T. Rietschel. 1991. Structural characterization of the lipid A component of Pseudomonas aeruginosa wild-type and rough mutant lipopolysaccharides. Eur. J. Biochem. 198:697-704. [DOI] [PubMed] [Google Scholar]
- 358.Kumada, H., Y. Haishima, T. Umemoto, and K.-I. Tanamoto. 1995. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J. Bacteriol. 177:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Kumada, H., K. Watanabe, T. Umemoto, Y. Haishima, S. Kondo, and K. Hisatsune. 1988. Occurrence of O-phosphorylated 2-keto-3-deoxyoctonate in the lipopolysaccharide of Bacteroides gingivalis. FEMS Microbiol. Lett. 51:77-80. [Google Scholar]
- 360.Kumar, G. B., and P. N. Black. 1993. Bacterial long-chain fatty acid transport: identification of amino acid residues within the outer membrane protein FadL required for activity. J. Biol. Chem. 268:15469-15476. [PubMed] [Google Scholar]
- 361.Kumar, G. S., M. V. Jagannadham, and M. K. Ray. 2002. Low-temperature-induced changes in composition and fluidity of lipopolysaccharides in the antarctic psychrotrophic bacterium Pseudomonas syringae. J. Bacteriol. 184:6746-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Labesse, G., E. Garnotel, S. Bonnel, C. Dumas, J. M. Pages, and J. M. Bolla. 2001. MOMP, a divergent porin from Campylobacter: cloning and primary structural characterization. Biochem. Biophys. Res. Commun. 280:380-387. [DOI] [PubMed] [Google Scholar]
- 363.Labischinski, H., G. Barnickel, H. Bradaczek, D. Naumann, E. T. Rietschel, and P. Giesbrecht. 1985. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer membrane. J. Bacteriol. 162:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Labischinski, H., D. Naumann, C. Schultz, S. Kusumoto, T. Shiba, E. T. Rietschel, and P. Giesbrecht. 1989. Comparative X-ray and Fourier-transform-infrared investigations of conformational properties of bacterial and synthetic lipid A of Escherichia coli and Salmonella minnesota as well as partial structures and analogues thereof. Eur. J. Biochem. 179:659-665. [DOI] [PubMed] [Google Scholar]
- 365.Lane, D. J., A. P. Harrison, Jr., D. Stahl, B. Pace, S. J. Giovannoni, G. J. Olsen, and N. R. Pace. 1992. Evolutionary relationships among sulfur- and iron-oxidizing eubacteria. J. Bacteriol. 174:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lazzaroni, J. C., J. F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84:391-397. [DOI] [PubMed] [Google Scholar]
- 367.Lazzaroni, J. C., P. Germon, M. C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177:191-197. [DOI] [PubMed] [Google Scholar]
- 368.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-ssrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Lee, Y. H., R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 1999. Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat. Struct. Biol. 6:628-633. [DOI] [PubMed] [Google Scholar]
- 370.Lee, Y. H., M. R. Dorwart, K. R. Hazlett, R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 2002. The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation. J. Bacteriol. 184:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Leive, L. 1965. Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem. Biophys. Res. Commun. 21:290-296. [DOI] [PubMed] [Google Scholar]
- 372.Letain, T. E., and K. Postle. 1997. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol. Microbiol. 24:271-283. [DOI] [PubMed] [Google Scholar]
- 373.Letoffe, S., P. Delepelaire, and C. Wandersman. 1996. Protein secretion in Gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 15:5804-5811. [PMC free article] [PubMed] [Google Scholar]
- 374.Li, C. C., J. A. Crawford, V. J. DiRita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 375.Li, X. Z., and K. Poole. 2001. Mutational analysis of the OprM outer membrane component of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 183:12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Lichtinger, T., A. Burkovski, M. Niederwies, R. Kraemer, and R. Benz. 1998. Biochemical and biophysical characterization of the cell wall porin of Corynebacterium glutamicum: the channel is formed by a low molecular mass polypeptide. Biochemistry 37:15024-15032. [DOI] [PubMed] [Google Scholar]
- 377.Lichtinger, T., B. Heym, E. Maier, H. Eichner, S. T. Cole, and R. Benz. 1999. Evidence for a small anion-selective channel in the cell wall of Mycobacterium bovis BCG besides a wide cation-selective pore. FEBS Lett. 454:349-355. [DOI] [PubMed] [Google Scholar]
- 378.Lichtinger, T., G. Reiss, and R. Benz. 2000. Biochemical identification and biophysical characterization of a channel-forming protein from Rhodococcus erythropolis. J. Bacteriol. 182:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Linderoth, N. A., M. N. Simon, and M. Russel. 1997. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science 278:1635-1638. [DOI] [PubMed] [Google Scholar]
- 380.Lins, R. D., and T. P. Straatsma. 2001. Computer simulation of the rough lipopolysaccharide membrane of Pseudomonas aeruginosa. Biophys. J. 81:1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Liu, J., C. E. Barry III, G. S. Besra, and H. Nikaido. 1996. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 271:29545-29551. [DOI] [PubMed] [Google Scholar]
- 382.Liu, J., E. Y. Rosenberg, and H. Nikaido. 1995. Fluidity of the lipid domain of cell wall from Mycobacterium chelonae. Proc. Natl. Acad. Sci. USA 92:11254-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Liu, X., and T. Ferenci. 1998. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J. Bacteriol. 180:3917-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Lloubes, R., E. Cascales, A. Walburger, E. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152:523-529. [DOI] [PubMed] [Google Scholar]
- 385.Locher, K., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 386.London, E., and D. A. Brown. 2000. Insolubility of lipids in Triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim. Biophys. Acta 1508:182-195. [DOI] [PubMed] [Google Scholar]
- 387.Lou, K.-L., N. Saint, A. P. G. Rummel, S. A. Benson, J. P. Rosenbusch, and T. Schirmer. 1996. Structural and functional characterization of OmpF porin mutants selected for larger pore size. I. Crystallographic analysis. J. Biol. Chem. 271:20669-20675. [PubMed] [Google Scholar]
- 388.Luckey, M., and H. Nikaido. 1980. Specificity of diffusion channels produced by lambda phage receptor protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 77:167-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Lundrigan, M. D., and C. F. Earhart. 1984. Gene envY of Escherichia coli K-12 affects thermoregulation of major porin expression. J. Bacteriol. 157:262-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Luo, Y., Q. Zeng, J. R. Glisson, M. W. Jackwood, I. H. N. Cheng, and C. Wang. 1999. Sequence analysis of Pasteurella multocida major outer membrane protein (OmpH) and application of synthetic peptides in vaccination chickens against homologous strain challenge. Vaccine 17:821-831. [DOI] [PubMed] [Google Scholar]
- 391.Lysko, P. G., and S. A. Morse. 1981. Neisseria gonorrhoeae cell envelope: permeability to hydrophobic molecules. J. Bacteriol. 145:946-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Mackinnon, F. G., A. D. Cox, J. S. Plested, C. M. Tang, K. Makepeace, P. A. Coull, J. C. Wright, R. Chalmers, D. W. Hood, J. C. Richards, and E. R. Moxon. 2002. Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol. Microbiol. 43:931-943. [DOI] [PubMed] [Google Scholar]
- 393.Maier, C., E. Bremer, A. Schmid, and R. Benz. 1988. Pore-forming activity of the Tsx protein from the outer membrane of Escherichia coli. Demonstration of a nucleoside-specific binding site. J. Biol. Chem. 263:2493-2499. [PubMed] [Google Scholar]
- 394.Maier, E., G. Polleichtner, B. Boeck, R. Schinzel, and R. Benz. 2001. Identification of the outer membrane porin of Thermus thermophilus HB8: the channel-forming complex has an unusually high molecular mass and an extremely large single-channel conductance. J. Bacteriol. 183:800-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Mallea, M., J. Chevalier, C. Bornet, A. Eyraud, A. Davin-Regli, C. Bollet, and J. M. Pages. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003-3009. [DOI] [PubMed] [Google Scholar]
- 396.Malouin, F., G. D. Campbell, M. Halpenny, G. W. Becker, and T. R. Parr, Jr. 1990. Outer membrane and porin characteristics of Serratia marcescens grown in vitro and in rat intraperitoneal diffusion chambers. Infect. Immun. 58:1247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Mannella, C. A., M. Forte, and M. Colombini. 1992. Toward the molecular structure of the mitochondrial channel, VDAC. J. Bioenerg. Biomembr. 24:7-19. [DOI] [PubMed] [Google Scholar]
- 398.Marciano, D. K., M. Russel, and S. M. Simon. 1999. An aqueous channel for filamentous phage export. Science 284:1516-1519. [DOI] [PubMed] [Google Scholar]
- 399.Marciano, D. K., M. Russel, and S. M. Simon. 2001. Assembling filamentous phage occlude pIV channels. Proc. Natl. Acad. Sci. USA 98:9359-9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Marquis, H., and T. A. Ficht. 1993. The omp2 gene locus of Brucella abortus encodes two homologous outer membrane proteins with properties characteristic of bacterial porins. Infect. Immun. 61:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Martinez, M. B., M. Flickinger, L. Higgins, T. Krick, and G. L. Nelsestuen. 2001. Reduced outer membrane permeability of Escherichia coli O157:H7: suggested role of modified outer membrane porins and theoretical function in resistance to antimicrobial agents. Biochemistry 40:11965-11974. [DOI] [PubMed] [Google Scholar]
- 402.Martinez-Martinez, L., S. Hernandez-Alles, S. Alberti, J. M. Tomas, V. J. Benedi, and G. A. Jacoby. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 40:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Martinez-Martinez, L., A. Pascual, M. C. Conejo, L. Picabea, and E. J. Perea. 1999. Resistance of Pseudomonas aeruginosa to imipenem induced by eluates from siliconized latex urinary catheters is related to outer membrane protein alterations. Antimicrob. Agents Chemother. 43:397-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Martinez-Martinez, L., A. Pascual, S. Hernandez-Alles, D. Alvarez-Diaz, A. I. Suarez, J. Tran, V. J. Benedi, and G. A. Jacoby. 1999. Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Mathes, A., and H. Engelhardt. 1998. Nonlinear and asymmetric open channel characteristics of an ion-selective porin in planar membranes. Biophys. J. 75:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Matsubara, M., S.-I. Kitaoka, S.-I. Takeda, and T. Mizuno. 2000. Tuning of the porin expression under anaerobic growth conditions by His-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes Cells 5:555-569. [DOI] [PubMed] [Google Scholar]
- 407.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Mauro, A., M. Blake, and P. Labarca. 1988. Voltage gating of conductance in lipid bilayers induced by porin from outer membrane of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 85:1071-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Mayer, H., J. H. Krauss, A. Yokota, and J. Weckesser. 1990. Natural variants of lipid A. Adv. Exp. Med. Biol. 256:45-70. [DOI] [PubMed] [Google Scholar]
- 410.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 411.McPhee, J. B., S. Lewenza, and R. E. W. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 412.Medeiros, A. A., T. F. O'Brien, E. Y. Rosenberg, and H. Nikaido. 1987. Loss of Omp-C porin in a strain of Salmonella typhimurium causes increased resistance to cephalosporins during therapy. J. Infect. Dis. 156:751-757. [DOI] [PubMed] [Google Scholar]
- 413.Melchior, D. L., and J. M. Steim. 1976. Thermotropic transitions in biomembranes. Annu. Rev. Biophys. Bioeng. 5:205-238. [DOI] [PubMed] [Google Scholar]
- 414.Merianos, H. J., N. Cadieux, C. H. Lin, R. J. Kadner, and D. S. Cafiso. 2000. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nat. Struct. Biol. 7:205-209. [DOI] [PubMed] [Google Scholar]
- 415.Michels, J., A. Geyer, V. Mocanu, W. Welte, A. L. Burlingame, and M. Przybylski. 2002. Structure and functional characterization of the periplasmic N-terminal polypeptide domain of the sugar-specific ion channel protein (ScrY porin). Protein Sci. 11:1565-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Mills, J., N. R. Wyborn, J. A. Greenwood, S. G. Williams, and C. W. Jones. 1997. An outer-membrane porin inducible by short-chain amides and urea in the methylotrophic bacterium Methylophilus methylotrophus. Microbiology 143:2373-2379. [DOI] [PubMed] [Google Scholar]
- 417.Mindich, L., and D. H. Bamford. 1988. Lipid-containing bacteriophages, p. 475-520. In R. Calendar (ed.), Bacteriophages, vol. 2. Plenum Press, New York, N.Y.
- 418.Minetti, C. A. S. A., M. S. Blake, and D. P. Remeta. 1998. Characterization of the structure, function, and conformational stability of PorB class 3 protein from Neisseria meningitidis. A porin with unusual physicochemical properties. J. Biol. Chem. 273:25329-25338. [DOI] [PubMed] [Google Scholar]
- 419.Misra, R., and S. A. Benson. 1988. Isolation and characterization of OmpC porin mutants with altered pore properties. J. Bacteriol. 170:528-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Misra, R., and S. A. Benson. 1989. A novel mutation, cog, which results in production of a new porin protein (OmpG) of Escherichia coli K-12. J. Bacteriol. 171:4105-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Mitraki, A., S. Miller, and M. J. van Raaij. 2002. Review: conformation and folding of novel beta-structural elements in viral fiber proteins: the triple beta-spiral and triple beta-helix. J. Struct. Biol. 137:236-247. [DOI] [PubMed] [Google Scholar]
- 422.Mitsuyama, J., R. Hiruma, A. Yamaguchi, and T. Sawai. 1987. Identification of porins in outer membrane of Proteus, Morganella, and Providencia spp. and their role in outer membrane permeation of beta-lactams. Antimicrob. Agents Chemother. 31:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Miyagawa, E., R. Azuma, T. Suto, and I. Yano. 1979. Occurrence of free ceramides in Bacteroides fragilis NCTC 9343. J. Biochem. (Tokyo) 86:311-320. [DOI] [PubMed] [Google Scholar]
- 424.Mobasheri, H., T. A. Ficht, H. Marquis, E. J. Lea, and J. H. Lakey. 1997. Brucella Omp2a and Omp2b porins: single channel measurements and topology prediction. FEMS Microbiol. Lett. 155:23-30. [DOI] [PubMed] [Google Scholar]
- 425.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: Mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 426.Molineux, I. J. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1-8. [DOI] [PubMed] [Google Scholar]
- 427.Momma, K., M. Okamoto, Y. Mishima, S. Mori, W. Hashimoto, and K. Murata. 2000. A novel bacterial ATP-binding cassette transporter system that allows uptake of macromolecules. J. Bacteriol. 182:3998-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Moran, A. P., M. M. Prendergast, and B. J. Appelmelk. 1996. Molecular mimicry of host structures by bacteria lipopolysaccharides and its contribution to disease. FEMS Immunol. Med. Microbiol. 16:105-115. [DOI] [PubMed] [Google Scholar]
- 429.Morona, R., M. Klose, and U. Henning. 1984. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J. Bacteriol. 159:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Morrison, D. C., and L. Leive. 1975. Fractions of lipopolysaccharide from Escherichia coli O111:B4 prepared by two extraction procedures. J. Biol. Chem. 250:2911-2919. [PubMed] [Google Scholar]
- 431.Mosig, G., and F. Eiserling. 1988. Phage T4: structure and metabolism, p. 521-606. In R. Calendar (ed.), Bacteriophages, vol. 2. Plenum Press, New York, N.Y.
- 432.Mukhopadhyay, S., D. Basu, and P. Chakrabarti. 1997. Characterization of a porin from Mycobacterium smegmatis. J. Bacteriol. 179:6205-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Muller, D. J., and A. Engel. 1999. Voltage- and pH-induced channel closure of porin OmpF visualized by atomic force microscopy. J. Mol. Biol. 285:1347-1351. [DOI] [PubMed] [Google Scholar]
- 434.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 435.Murray, S. R., D. Bermudes, K. S. de Felipe, and K. B. Low. 2001. Extragenic suppressors of growth defects in msbB Salmonella. J. Bacteriol. 183:5554-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Nabedryk, E., R. M. Garavito, and J. Breton. 1988. The orientation of beta-sheets in porin. A polarized Fourier transform infrared spectroscopic investigation. Biophys. J. 53:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Naka, T., N. Fujiwara, E. Yabuuchi, M. Doe, K. Kobayashi, Y. Kato, and I. Yano. 2000. A novel sphingoglycolipid containing galacturonic acid and 2-hydroxy fatty acid in cellular lipids of Sphingomonas yanoikuyae. J. Bacteriol. 182:2660-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Nakae, T. 1976. Outer membrane of Salmonella. Isolation of protein complex that produces transmembrane channels. J. Biol. Chem. 251:2176-2178. [PubMed] [Google Scholar]
- 439.Naumann, D., C. Schultz, A. Sabisch, M. Kastowsky, and H. Labischinski. 1989. New insights into the phase behaviour of a complex anionic amphiphile: architecture and dynamics of bacterial deep rough lipopolysaccharide membrane as seen by FTIR, X-ray, and molecular modelling techniques. J. Mol. Struct. 214:213-246. [Google Scholar]
- 440.Nestorovich, E. M., C. Danelon, M. Winterhalter, and S. M. Bezrukov. 2002. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl. Acad. Sci. USA 99:9789-9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Neumann, U., E. Maier, E. Schiltz, J. Weckesser, and R. Benz. 1997. Characterization of porin from Roseobacter denitrificans. Antonie Leeuwenhoek 72:135-140. [DOI] [PubMed] [Google Scholar]
- 442.Neuwald, A. F., J. S. Liu, and C. E. Lawrence. 1995. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 4:1618-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Nguyen, L., I. T. Paulsen, J. Tchieu, C. J. Hueck, and M. H. Saier, Jr. 2000. Phylogenetic analyses of the constituents of type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2:125-144. [PubMed] [Google Scholar]
- 444.Niederweis, M., S. Ehrt, C. Heinz, U. Kloecker, S. Karosi, K. M. Swiderek, L. W. Riley, and R. Benz. 1999. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol. Microbiol. 33:933-945. [DOI] [PubMed] [Google Scholar]
- 445.Niederweis, M., E. Maier, T. Lichtinger, R. Benz, and R. Kramer. 1995. Identification of channel-forming activity in the cell wall of Corynebacterium glutamicum. J. Bacteriol. 177:5716-5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Niemeyer, D., and A. Becker. 2001. The molecular weight distribution of succinoglycan produced by Sinorhizobium meliloti is influenced by specific tyrosine phosphorylation and ATPase activity of the cytoplasmic domain of the ExoP protein. J. Bacteriol. 183:5163-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Nieweg, A., and E. Bremer. 1997. The nucleoside-specific Tsx channel from the outer membrane of Salmonella typhimurium, Klebsiella pneumoniae and Enterobacter aerogenes: Functional characterization and DNA sequence analysis of the tsx genes. Microbiology 143:603-615. [DOI] [PubMed] [Google Scholar]
- 448.Nikaido, H. 1999. Antibiotic efflux mechanisms. Curr. Opin. Infect. Dis. 12:529-536. [DOI] [PubMed] [Google Scholar]
- 449.Nikaido, H. 1994. Isolation of outer membranes. Methods Enzymol. 235:225-234. [DOI] [PubMed] [Google Scholar]
- 450.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Nikaido, H. 1976. Outer membrane of Salmonella typhimurium. Transmembrane diffusion of some hydrophobic substances. Biochim. Biophys. Acta 433:118-132. [DOI] [PubMed] [Google Scholar]
- 452.Nikaido, H. 1992. Porins and specific channels of bacterial outer membranes. Mol. Microbiol. 6:435-442. [DOI] [PubMed] [Google Scholar]
- 453.Nikaido, H. 1994. Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 269:3905-3908. [PubMed] [Google Scholar]
- 454.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-388. [DOI] [PubMed] [Google Scholar]
- 455.Nikaido, H. 1998. The role of outer membrane and efflux pumps in the resistance of gram-negative bacteria. Can we improve drug access? Drug Resist. Updates 1:93-98. [DOI] [PubMed] [Google Scholar]
- 455a.Nikaido, H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin. Cell. Dev. Biol. 12:215-223. [DOI] [PubMed]
- 456.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Nikaido, H., S.-H. Kim, and E. Y. Rosenberg. 1993. Physical organization of lipids in the cell wall of Mycobacterium chelonae. Mol. Microbiol. 8:1025-1030. [DOI] [PubMed] [Google Scholar]
- 458.Nikaido, H., W. Liu, and E. Y. Rosenberg. 1990. Outer membrane permeability and beta-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob. Agents Chemother. 34:337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Nikaido, H., K. Nikaido, and S. Harayama. 1991. Identification and characterization of porins in Pseudomonas aeruginosa. J. Biol. Chem. 266:770-779. [PubMed] [Google Scholar]
- 460.Nikaido, H., and E. Y. Rosenberg. 1981. Effect on solute size on diffusion rates through the transmembrane pores of the outer membrane of Escherichia coli. J. Gen. Physiol. 77:121-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Nikaido, H., and E. Y. Rosenberg. 1983. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J. Bacteriol. 153:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Nikaido, H., E. Y. Rosenberg, and J. Foulds. 1983. Porin channels in Escherichia coli: studies with beta-lactams in intact cells. J. Bacteriol. 153:232-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Nikaido, H., Y. Takeuchi, S. I. Ohnishi, and T. Nakae. 1977. Outer membrane of Salmonella typhimurium. Electron spin resonance studies. Biochim. Biophys. Acta 465:152-164. [DOI] [PubMed] [Google Scholar]
- 464.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Notley-McRobb, L., and T. Ferenci. 1999. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1:45-52. [DOI] [PubMed] [Google Scholar]
- 466.Nouwen, N., N. Ranson, H. Saibil, B. Wolpensinger, A. Engel, A. Ghazi, and A. P. Pugsley. 1999. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc. Natl. Acad. Sci. USA 96:8173-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Nouwen, N., H. Stahlberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Nummila, K., I. Kilpelainen, U. Zaehringer, M. Vaara, and I. M. Helander. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271-278. [DOI] [PubMed] [Google Scholar]
- 469.Obst, S., M. Kastowsky, and H. Bradaczek. 1997. Molecular dynamics simulations of six different fully hydrated monomeric conformers of Escherichia coli R-lipopolysaccharide in the presence and absence of Ca2+. Biophys. J. 72:1031-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Ochs, M. M., M. Bains, and R. E. W. Hancock. 2000. Role of putative loops 2 and 3 in imipenem passage through the specific porin OprD of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:1983-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Ochs, M. M., C.-D. Lu, R. E. W. Hancock, and A. T. Abdelal. 1999. Amino acid-mediated induction of the basic amino acid-specific outer membrane porin OprD from Pseudomonas aeruginosa. J. Bacteriol. 181:5426-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Odou, M. F., E. Singer, M. B. Romond, and L. Dubreuil. 1998. Isolation and characterization of a porin-like protein of 45 kilodaltons from Bacteroides fragilis. FEMS Microbiol. Lett. 166:347-354. [DOI] [PubMed] [Google Scholar]
- 473.Ogawa, T. 1993. Chemical structure of lipid A from Porphyromonas (Bacteroides) gingivalis lipopolysaccharide. FEBS Lett. 332:197-201. [DOI] [PubMed] [Google Scholar]
- 474.Ogierman, M., and V. Braun. 2003. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J. Bacteriol. 185:1870-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Olivera, E. R., B. Minambres, B. Garcia, C. Muniz, M. A. Moreno, A. Ferrandez, E. Diaz, J. L. Garcia, and J. M. Luengo. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 95:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Oshima, M., and T. Yamakawa. 1974. Chemical structure of a novel glycolipid from an extreme thermophile, Flavobacterium thermophilum. Biochemistry 13:1140-1146. [DOI] [PubMed] [Google Scholar]
- 478.Paakkanen, J., E. C. Gotschlich, and P. H. Makela. 1979. Protein K: a new major outer membrane protein found in encapsulated Escherichia coli. J. Bacteriol. 139:835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Pajatsch, M., C. Andersen, A. Mathes, A. Boeck, R. Benz, and H. Engelhardt. 1999. Properties of a cyclodextrin-specific, unusual porin from Klebsiella oxytoca. J. Biol. Chem. 274:25159-25166. [DOI] [PubMed] [Google Scholar]
- 480.Pangon, B., C. Bizet, A. Bure, F. Pichon, A. Philippon, B. Regnier, and L. Gutmann. 1989. In vivo selection of a cephamycin-resistant, porin-deficient mutant of Klebsiella pneumoniae producing a TEM-3 beta-lactamase. J. Infect. Dis. 159:1005-1006. [DOI] [PubMed] [Google Scholar]
- 481.Papahadjopoulos, D., A. Portis, and W. Pangborn. 1978. Calcium-induced lipid phase transitions and membrane fusion. Ann. N. Y. Acad. Sci. 308:50-66. [DOI] [PubMed] [Google Scholar]
- 482.Paquet, J. Y., M. A. Diaz, S. Genevrois, M. Grayon, J. M. Verger, X. de Bolle, J. H. Lakey, J. J. Letesson, and A. Cloeckaert. 2001. Molecular, antigenic, and functional analyses of Omp2b porin size variants of Brucella spp. J. Bacteriol. 183:4839-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Parr, T. R., Jr., R. A. Moore, L. V. Moore, and R. E. W. Hancock. 1987. Role of porins in intrinsic antibiotic resistance of Pseudomonas cepacia. Antimicrob. Agents Chemother. 31:121-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Pascher, I., and S. Sundell. 1977. Molecular arrangements in sphingolipids—crystal structure of cerebroside. Chem. Phys. Lipids 20:175-191. [DOI] [PubMed] [Google Scholar]
- 485.Paul, C., and J. P. Rosenbusch. 1985. Folding patterns of porin and bacteriorhodopsin. EMBO J. 4:1593-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Paul, S., K. Chaudhuri, A. N. Chatterjee, and J. Das. 1992. Presence of exposed phospholipids in the outer membrane of Vibrio cholerae. J. Gen. Microbiol. 138:755-761. [DOI] [PubMed] [Google Scholar]
- 487.Paulsen, I. T., A. M. Beness, and M. H. Saier. 1997. Computer-based analyses of the protein constituents of transport systems catalysing export of complex carbohydrates in bacteria. Microbiology 143:2685-2699. [DOI] [PubMed] [Google Scholar]
- 488.Pautsch, A., and G. E. Schulz. 2000. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298:273-282. [DOI] [PubMed] [Google Scholar]
- 489.Pautsch, A., and G. E. Schulz. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5:1013-1017. [DOI] [PubMed] [Google Scholar]
- 490.Pellinen, T., H. Ahlfors, N. Blot, and G. Condemine. 2003. Topology of the Erwinia chrysanthemi oligogalacturonate porin KdgM. Biochem. J. 372:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Petit, A., H. Ben-Yaghlane-Bouslama, L. Sofer, and R. Labia. 1992. Characterization of chromosomally encoded penicillinases in clinical isolates of Klebsiella pneumoniae. J. Antimicrob. Chemother. 29:629-638. [DOI] [PubMed] [Google Scholar]
- 492.Phale, P., T. Schirmer, A. Prilipov, K.-L. Lou, A. Hardmeyer, and J. P. Rosenbusch. 1997. Voltage gating of Escherichia coli porin channels: Role of the constriction loop. Proc. Natl. Acad. Sci. USA 94:6741-6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Phale, P. S., A. Philippsen, T. Kiefhaber, R. Koebnik, V. P. Phale, T. Schirmer, and J. P. Rosenbusch. 1998. Stability of trimeric OmpF porin: the contributions of the latching loop L2. Biochemistry 37:15663-15670. [DOI] [PubMed] [Google Scholar]
- 494.Phale, P. S., A. Philippsen, C. Widmer, V. P. Phale, J. P. Rosenbusch, and T. Schirmer. 2001. Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry 40:6319-6325. [DOI] [PubMed] [Google Scholar]
- 495.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Philippsen, A., W. Im, A. Engel, T. Schirmer, B. Roux, and D. J. Muller. 2002. Imaging the electrostatic potential of transmembrane channels: atomic probe microscopy of OmpF porin. Biophys. J. 82:1667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Piddock, L. J. V., and R. Wise. 1986. The effect of altered porin expression in Escherichia coli upon susceptibility to 4 quinolones. J. Antimicrob. Chemother. 18:547-549. [DOI] [PubMed] [Google Scholar]
- 498.Pilsl, H., D. Smajs, and V. Braun. 1999. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J. Bacteriol. 181:3578-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Pimenta, A., M. Blight, D. Clarke, and I. B. Holland. 1996. The Gram-negative cell envelope ‘springs’ to life; coiled-coil trans-envelope proteins. Mol. Microbiol. 19:643-645. [DOI] [PubMed] [Google Scholar]
- 500.Pitta, T. P., E. R. Leadbetter, and W. Godchaux, III. 1989. Increase of ornithine amino lipid content in a sulfonolipid-deficient mutant of Cytophaga johnsonae. J. Bacteriol. 171:952-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Plancon, L., M. Chami, and L. Letellier. 1997. Reconstitution of FhuA, an Escherichia coli outer membrane protein, into liposomes. Binding of phage T5 to FhuA triggers the transfer of DNA into the proteoliposomes. J. Biol. Chem. 272:16868-16872. [DOI] [PubMed] [Google Scholar]
- 502.Plancon, L., C. Janmot, M. le Maire, M. Desmadril, M. Bonhivers, L. Letellier, and P. Boulanger. 2002. Characterization of a high-affinity complex between the bacterial outer membrane protein FhuA and the phage T5 protein pb5. J. Mol. Biol. 318:557-569. [DOI] [PubMed] [Google Scholar]
- 503.Plesiat, P., J. R. Aires, C. Godard, and T. Kohler. 1997. Use of steroids to monitor alterations in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 179:7004-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Plesiat, P., and H. Nikaido. 1992. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol. Microbiol. 6:1323-1333. [DOI] [PubMed] [Google Scholar]
- 505.Postle, K. 1999. Active transport by customized beta-barrels. Nat. Struct. Biol. 6:3-6. [DOI] [PubMed] [Google Scholar]
- 506.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 507.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 508.Preston, A., E. Maxim, E. Toland, E. J. Pishko, E. T. Harvill, M. Caroff, and D. J. Maskell. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48:725-736. [DOI] [PubMed] [Google Scholar]
- 509.Preston, K. E., and R. A. Venezia. 2002. Chromosomal sequences from Klebsiella pneumoniae flank the SHV-5 extended-spectrum beta-lactamase gene in pACM1. Plasmid 48:73-76. [DOI] [PubMed] [Google Scholar]
- 510.Prilipov, A., P. S. Phale, R. Koebnik, C. Widmer, and J. P. Rosenbusch. 1998. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J. Bacteriol. 180:3388-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Prince, S. M., M. Achtman, and J. P. Derrick. 2002. Crystal structure of the OpcA integral membrane adhesin from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 99:3417-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Pristovsek, P., and J. Kidric. 1999. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J. Med. Chem. 42:4604-4613. [DOI] [PubMed] [Google Scholar]
- 513.Provenzano, D., and K. E. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Provenzano, D., C. M. Lauriano, and K. E. Klose. 2001. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J. Bacteriol. 183:3652-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Pugsley, A. P., C. d'Enfert, I. Reyss, and M. G. Kornacker. 1990. Genetics of extracellular protein secretion by gram-negative bacteria. Annu. Rev. Genet. 24:67-90. [DOI] [PubMed] [Google Scholar]
- 517.Pugsley, A. P., O. Francetic, O. M. Possot, N. Sauvonnet, and K. R. Hardie. 1997. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene 192:13-19. [DOI] [PubMed] [Google Scholar]
- 518.Puig, M., C. Fuste, and M. Vinas. 1993. Outer membrane proteins from Serratia marcescens. Can. J. Microbiol. 39:108-111. [DOI] [PubMed] [Google Scholar]
- 519.Puntervoll, P., H. Kleivdal, K. O. Dahl, W. Bitter, J. Tommassen, and H. B. Jensen. 2000. The Fusobacterium nucleatum porin FomA possesses the general topology of the non-specific porins. Microbiology 146:1437-1445. [DOI] [PubMed] [Google Scholar]
- 520.Puntervoll, P., M. Ruud, L. J. Bruseth, H. Kleivdal, B. T. Hogh, R. Benz, and H. B. Jensen. 2002. Structural characterization of the fusobacterial non-specific porin FomA suggests a 14-stranded topology, unlike the classical porins. Microbiology 148:3395-3403. [DOI] [PubMed] [Google Scholar]
- 521.Que-Gewirth, N. L., M. J. Karbarz, S. R. Kalb, R. J. Cotter, and C. R. Raetz. 2003. Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase LpxQ. J. Biol. Chem. 278:12120-12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Que-Gewirth, N. L., S. Lin, R. J. Cotter, and C. R. Raetz. 2003. An outer membrane enzyme that generates the 2-amino-2-deoxy-gluconate moiety of Rhizobium leguminosarum lipid A. J. Biol. Chem. 278:12109-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Quinn, J. P., E. J. Dudek, C. A. Divincenzo, D. A. Lucks, and S. A. Lerner. 1986. Emergence of resistance to imipenem during therapy for Pseudomonas aeruginosa infections. J. Infect. Dis. 154:289-294. [DOI] [PubMed] [Google Scholar]
- 524.Rachel, R., A. M. Engel, R. Huber, K. O. Stetter, and W. Baumeister. 1990. A porin-type protein is the main constituent of the cell envelope of the ancestral eubacterium Thermotoga maritima. FEBS Lett. 262:64-68. [Google Scholar]
- 525.Radolf, J. D., M. V. Norgard, and W. W. Schulz. 1989. Outer membrane ultrastructure explains the limited antigenicity of virulent Treponema pallidum. Proc. Natl. Acad. Sci. USA 86:2051-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Raetz, C. R. 1990. Biochemistry of endotoxins. Annu. Rev. Biochem. 59:129-170. [DOI] [PubMed] [Google Scholar]
- 527.Raetz, C. R. H. 1993. Bacterial endotoxins: extraordinary lipids that activate eucaryotic signal transduction. J. Bacteriol. 175:5745-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Raetz, C. R. H. 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles, p. 1035-1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 529.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Raimondi, A., F. Sisto, and H. Nikaido. 2001. Mutation in Serratia marcescens AmpC beta-lactamase producing high-level resistance to ceftazidime and cefpirome. Antimicrob. Agents Chemother. 45:2331-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Rainey, F. A., D. P. Kelly, E. Stackebrandt, J. Burghardt, A. Hiraishi, Y. Katayama, and A. P. Wood. 1999. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int. J. Syst. Bacteriol. 49:645-651. [DOI] [PubMed] [Google Scholar]
- 532.Rau, H., U. Seydel, M. Freudenberg, J. Weckesser, and H. Mayer. 1995. Lipopolysaccharide of Rhodospirillum salinarum 40: structural studies on the core and lipid A region. Arch. Microbiol. 164:280-289. [DOI] [PubMed] [Google Scholar]
- 533.Rawling, E. G., F. S. Brinkman, and R. E. Hancock. 1998. Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J. Bacteriol. 180:3556-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Rawling, E. G., N. L. Martin, and R. E. Hancock. 1995. Epitope mapping of the Pseudomonas aeruginosa major outer membrane porin protein OprF. Infect. Immun. 63:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Raynaud, C., K. G. Papavinasasundaram, R. A. Speight, B. Springer, P. Sander, E. C. Bottger, M. J. Colston, and P. Draper. 2002. The functions of OmpATb, a pore-forming protein of Mycobacterium tuberculosis. Mol. Microbiol. 46:191-201. [DOI] [PubMed] [Google Scholar]
- 536.Reeves, A. R., J. N. D'Elia, J. Frias, and A. A. Salyers. 1996. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J. Bacteriol. 178:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 538.Rehm, B. H. A. 1996. The Azotobacter vinelandii gene algJ encodes an outer-membrane protein presumably involved in export of alginate. Microbiology 142:873-880. [DOI] [PubMed] [Google Scholar]
- 539.Rehm, B. H. A., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: Subcellular localization, purification, ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Reumann, S., E. Maier, R. Benz, and H. W. Heldt. 1995. The membrane of leaf peroxisomes contains a porin-like channel. J. Biol. Chem. 270:17559-17565. [DOI] [PubMed] [Google Scholar]
- 541.Reumann, S., E. Maier, R. Benz, and H. W. Heldt. 1996. A specific porin is involved in the malate shuttle of leaf peroxisomes. Biochem. Soc. Trans. 24:754-757. [DOI] [PubMed] [Google Scholar]
- 542.Reumann, S., E. Maier, H. W. Heldt, and R. Benz. 1998. Permeability properties of the porin of spinach leaf peroxisomes. Eur. J. Biochem. 251:359-366. [DOI] [PubMed] [Google Scholar]
- 543.Reynolds, P. R., G. P. Mottur, and C. Bradbeer. 1980. Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of BtuC and TonB. J. Biol. Chem. 255:4313-4319. [PubMed] [Google Scholar]
- 544.Riechmann, L., and P. Holliger. 1997. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell 90:351-360. [DOI] [PubMed] [Google Scholar]
- 545.Ried, G., R. Koebnik, I. Hindennach, B. Mutschler, and U. Henning. 1994. Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Mol. Gen. Genet. 243:127-135. [DOI] [PubMed] [Google Scholar]
- 546.Riess, F. G., and R. Benz. 2000. Discovery of a novel channel-forming protein in the cell wall of the non-pathogenic Nocardia corynebacteroides. Biochim. Biophys. Acta 1509:485-495. [DOI] [PubMed] [Google Scholar]
- 547.Riess, F. G., U. Dorner, B. Schiffler, and R. Benz. 2001. Study of the properties of a channel-forming protein of the cell wall of the gram-positive bacterium Mycobacterium phlei. J. Membr. Biol. 182:147-157. [DOI] [PubMed] [Google Scholar]
- 548.Riess, F. G., M. Elflein, M. Benk, B. Schiffler, R. Benz, N. Garton, and I. Sutcliffe. 2003. The cell wall of the pathogenic bacterium Rhodococcus equi contains two channel-forming proteins with different properties. J. Bacteriol. 185:2952-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Riess, F. G., T. Lichtinger, R. Cseh, A. F. Yassin, K. P. Schaal, and R. Benz. 1998. The cell wall porin of Nocardia farcinica: biochemical identification of the channel-forming protein and biophysical characterization of the channel properties. Mol. Microbiol. 29:139-150. [DOI] [PubMed] [Google Scholar]
- 550.Riess, F. G., T. Lichtinger, A. F. Yassin, K. P. Schaal, and R. Benz. 1999. The cell wall porin of the gram-positive bacterium Nocardia asteroides forms cation-selective channels that exhibit asymmetric voltage dependence. Arch. Microbiol. 171:173-182. [DOI] [PubMed] [Google Scholar]
- 551.Robbins, J. R., D. Monack, S. J. McCallum, A. Vegas, E. Pham, M. B. Goldberg, and J. A. Theriot. 2001. The making of a gradient: IcsA (VirG) polarity in Shigella flexneri. Mol. Microbiol. 41:861-872. [DOI] [PubMed] [Google Scholar]
- 552.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 553.Robey, M., W. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Rocque, W. J., R. T. Coughlin, and E. J. McGroarty. 1987. Lipopolysaccharide tightly bound to porin monomers and trimers from Escherichia coli K-12. J. Bacteriol. 169:4003-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Rocque, W. J., and E. J. McGroarty. 1990. Structure and function of an OmpC deletion mutant porin from Escherichia coli K-12. Biochemistry 29:5344-5351. [DOI] [PubMed] [Google Scholar]
- 556.Roessner, C. A., and G. M. Ihler. 1986. Formation of transmembrane channels in liposomes during injection of lambda DNA. J. Biol. Chem. 261:386-390. [PubMed] [Google Scholar]
- 557.Roppel, J., H. Mayer, and J. Weckesser. 1975. Identification of a 2,3-diamino-2,3-dideoxyhexose in lipid A component of lipopolysaccharides of Rhodopseudomonas viridis and Rhodopseudomonas palustris. Carbohydr. Res. 40:31-40. [DOI] [PubMed] [Google Scholar]
- 558.Rosenberg, E., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609-1619. [DOI] [PubMed] [Google Scholar]
- 559.Rosenbusch, J. P. 1974. Characterization of the major envelope protein from Escherichia coli. Regular arrangement on the peptidoglycan and unusual dodecyl sulfate binding. J. Biol. Chem. 249:8019-8029. [PubMed] [Google Scholar]
- 560.Rosner, J. L., K. J. Chai, and J. Foulds. 1991. Regulation of OmpF porin expression by salicylate in Escherichia coli. J. Bacteriol. 173:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Rosner, J. L., B. Dangi, A. M. Gronenborn, and R. G. Martin. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Rossouw, F. T., and R. J. Rowbury. 1984. Effects of the resistance plasmid R124 on the level of the OmpF outer membrane protein and on the response of Escherichia coli to environmental agents. J. Appl. Bacteriol. 56:63-79. [DOI] [PubMed] [Google Scholar]
- 563.Rostovtseva, T. K., E. M. Nestorovich, and S. M. Bezrukov. 2002. Partitioning of differently sized poly(ethylene glycol)s into OmpF porin. Biophys. J. 82:160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 565.Russel, M. 1994. Mutants at conserved positions in gene IV, a gene required for assembly and secretion of filamentous phages. Mol. Microbiol. 14:357-369. [DOI] [PubMed] [Google Scholar]
- 566.Russel, M. 1994. Phage assembly: a paradigm for bacterial virulence factor export? Science 265:612-614. [DOI] [PubMed] [Google Scholar]
- 567.Rutz, J. M., J. Liu, J. A. Lyons, J. Goranson, S. K. Armstrong, M. A. McIntosh, J. B. Feix, and P. E. Klebba. 1992. Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science 258:471-475. [DOI] [PubMed] [Google Scholar]
- 568.Sadovskaya, I., J. R. Brisson, J. S. Lam, J. C. Richards, and E. Altman. 1998. Structural elucidation of the lipopolysaccharide core regions of the wild-type strain PAO1 and O-chain-deficient mutant strains AK1401 and AK1012 from Pseudomonas aeruginosa serotype O5. Eur. J. Biochem. 255:673-684. [DOI] [PubMed] [Google Scholar]
- 569.Sadovskaya, I., J. R. Brisson, P. Thibault, J. C. Richards, J. S. Lam, and E. Altman. 2000. Structural characterization of the outer core and the O-chain linkage region of lipopolysaccharide from Pseudomonas aeruginosa serotype O5. Eur. J. Biochem. 267:1640-1650. [DOI] [PubMed] [Google Scholar]
- 570.Saint, N., E. De, S. Julien, N. Orange, and G. Molle. 1993. Ionophore properties of OmpA of Escherichia coli. Biochim. Biophys. Acta 1145:119-123. [DOI] [PubMed] [Google Scholar]
- 571.Saint, N., K.-L. Lou, C. Widmer, M. Luckey, T. Schirmer, and J. P. Rosenbusch. 1996. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J. Biol. Chem. 271:20676-20680. [PubMed] [Google Scholar]
- 572.Saint, N., A. Prilipov, A. Hardmeyer, K.-L. Lou, T. Schrimer, and J. P. Rosenbusch. 1996. Replacement of the sole histidinyl residue in OmpF porin from E. coli by threonine (H21T) does not affect channel structure and function. Biochem. Biophys. Res. Commun. 223:118-122. [DOI] [PubMed] [Google Scholar]
- 573.Salyers, A. A., and M. O'Brien. 1980. Cellular location of enzymes involved in chondroitin sulfate breakdown by Bacteroides thetaiotaomicron. J. Bacteriol. 143:772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Samartzidou, H., and A. H. Delcour. 1999. Excretion of endogenous cadverine leads to a decrease in porin-mediated outer membrane permeability. J. Bacteriol. 181:791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Samartzidou, H., M. Mehrazin, Z. Xu, M. J. Benedik, and A. H. Delcour. 2003. Cadaverine inhibition of porin plays a role in cell survival at acidic pH. J. Bacteriol. 185:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 577.Santiviago, C. A., J. A. Fuentes, S. M. Bueno, A. N. Trombert, A. A. Hildago, L. T. Socias, P. Youderian, and G. C. Mora. 2002. The Salmonella enterica sv. Typhimurium smvA, yddG and ompD (porin) genes are required for the efficient efflux of methyl viologen. Mol. Microbiol. 46:687-698. [DOI] [PubMed] [Google Scholar]
- 578.Sardesai, A. A., P. Genevaux, F. Schwager, D. Ang, and C. Georgopoulos. 2003. The OmpL porin does not modulate redox potential in the periplasmic space of Escherichia coli. EMBO J. 22:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Saulino, E. T., E. Bullitt, and S. J. Hultgren. 2000. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl. Acad. Sci. USA 97:9240-9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Sawai, T., S. Hirano, and A. Yamaguchi. 1987. Repression of porin synthesis by salicylate in Escherichia coli, Klebsiella pneumoniae, and Serratia marcescens. FEMS Microbiol. Lett. 40:233-237. [Google Scholar]
- 581.Saxena, K., V. Drosou, E. Maier, R. Benz, and B. Ludwig. 1999. Ion selectivity reversal and induction of voltage-gating by site-directed mutations in the Paracoccus denitrificans porin. Biochemistry 38:2206-2212. [DOI] [PubMed] [Google Scholar]
- 582.Saxena, K., O.-M. H. Richter, B. Ludwig, and R. Benz. 1997. Molecular cloning and functional characterization of the Paracoccus denitrificans porin. Eur. J. Biochem. 245:300-306. [DOI] [PubMed] [Google Scholar]
- 583.Schabert, F. A., C. Henn, and A. Engel. 1995. Native Escherichia coli OmpF porin surfaces probed by atomic force microscopy. Science 268:92-94. [DOI] [PubMed] [Google Scholar]
- 584.Schein, S. J., M. Colombini, and A. Finkelstein. 1976. Reconstitution in planar lipid bilayers of a voltage-dependent anion-selective channel obtained from paramecium mitochondria. J. Membr. Biol. 30:99-120. [DOI] [PubMed] [Google Scholar]
- 585.Schiebel, E., H. Schwarz, and V. Braun. 1989. Subcellular location and unique secretion of the hemolysin of Serratia marcescens. J. Biol. Chem. 264:16311-16320. [PubMed] [Google Scholar]
- 586.Schifferli, D. M., and M. A. Alrutz. 1994. Permissive linker insertion in the outer membrane protein of 987P fimbriae of Escherichia coli. J. Bacteriol. 176:1099-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Schirmer, T. 1998. General and specific porins from bacterial outer membranes. J. Struct. Biol. 121:101-109. [DOI] [PubMed] [Google Scholar]
- 588.Schirmer, T., T. A. Keller, Y.-F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science 267:512-514. [DOI] [PubMed] [Google Scholar]
- 589.Schirmer, T., and P. S. Phale. 1999. Brownian dynamics simulation of ion flow through porin channels. J. Mol. Biol. 294:1159-1167. [DOI] [PubMed] [Google Scholar]
- 590.Schmid, B., L. Maveyraud, M. Kroemer, and G. E. Schulz. 1998. Porin mutants with new channel properties. Protein Sci. 7:1603-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Schmid, K., R. Ebner, K. Jahreis, J. W. Lengeler, and F. Titgemeyer. 1991. A sugar-specific porin, ScrY, is involved in sucrose uptake in enteric bacteria. Mol. Microbiol. 5:941-950. [DOI] [PubMed] [Google Scholar]
- 592.Schnaitman, C. A. 1971. Effect of ethylenediaminetetraacetic acid, Triton X-100, and lysozyme on the morphology and chemical composition of isolate cell walls of Escherichia coli. J. Bacteriol. 108:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Schnaitman, C. A. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Schuelein, K. 1991. The sugar-specific outer membrane channel ScrY contains functional characteristics of general diffusion pores and substrate-specific porins. Mol. Microbiol. 5:2233-2241. [DOI] [PubMed] [Google Scholar]
- 596.Schuelein, K., C. Andersen, and R. Benz. 1995. The deletion of 70 amino acids near the N-terminal end of the sucrose-specific porin ScrY causes its functional similarity to LamB in vivo and in vitro. Mol. Microbiol. 17:757-767. [DOI] [PubMed] [Google Scholar]
- 597.Schulz, G. E. 1993. Bacterial porins: structure and function. Curr. Opin. Cell Biol. 5:701-707. [DOI] [PubMed] [Google Scholar]
- 598.Schulz, G. E. 2000. β-Barrel membrane proteins. Curr. Opin. Struct. Biol. 10:443-447. [DOI] [PubMed] [Google Scholar]
- 599.Schulz, G. E. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565:308-317. [DOI] [PubMed] [Google Scholar]
- 600.Scott, D. C., S. M. C. Newton, and P. E. Klebba. 2002. Surface loop motion in FepA. J. Bacteriol. 184:4906-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Sen, K., J. Hellman, and H. Nikaido. 1988. Porin channels in intact cells of Escherichia coli are not affected by Donnan potentials across the outer membrane. J. Biol. Chem. 263:1182-1187. [PubMed] [Google Scholar]
- 602.Sen, K., and H. Nikaido. 1991. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J. Bacteriol. 173:926-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Senaratne, R. H., H. Mobasheri, K. G. Papavinasasundaram, P. Jenner, E. J. A. Lea, and P. Draper. 1998. Expression of a gene for a porin-like protein of the OmpA family from Mycobacterium tuberculosis H37Rv. J. Bacteriol. 180:3541-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Seydel, U., M. H. Koch, and K. Brandenburg. 1993. Structural polymorphisms of rough mutant lipopolysaccharides Rd to Ra from Salmonella minnesota. J. Struct. Biol. 110:232-243. [DOI] [PubMed] [Google Scholar]
- 605.Shang, E. S., M. M. Exner, T. A. Sumemrs, C. Martinich, C. I. Champion, R. E. W. Hancock, and D. A. Haake. 1995. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect. Immun. 63:3174-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Shannon, J. L., and R. C. Fernandez. 1999. The C-terminal domain of the Bordetella pertussis autotransporter BrkA forms a pore in lipid bilayer membranes. J. Bacteriol. 181:5838-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Sharff, A., C. Fanutti, J. Shi, C. Calladine, and B. Luisi. 2001. The role of the TolC family in protein transport and multidrug efflux. From stereochemical certainty to mechanistic hypothesis. Eur. J. Biochem. 268:5011-5026. [DOI] [PubMed] [Google Scholar]
- 608.Sharypova, L. A., K. Niehaus, H. Scheidle, O. Holst, and A. Becker. 2003. Sinorhizobium meliloti acpXL mutant lacks the C28 hydroxylated fatty acid moiety of lipid A and does not express a slow migrating form of lipopolysaccharide. J. Biol. Chem. 278:12946-12954. [DOI] [PubMed] [Google Scholar]
- 609.Shipman, J. A., K. H. Cho, H. A. Siegel, and A. A. Salyers. 1999. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 181:7206-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Shroll, R. M., and T. P. Straatsma. 2002. Molecular structure of the outer bacterial membrane of Pseudomonas aeruginosa via classical simulation. Biopolymers 65:395-407. [DOI] [PubMed] [Google Scholar]
- 611.Shukla, S. D., C. Green, and J. M. Turner. 1980. Phosphatidylethanolamine distribution and fluidity in outer and inner membranes of the gram-negative bacterium Erwinia carotovora. Biochem. J. 188:131-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Siehnel, R., N. L. Martin, and R. E. W. Hancock. 1990. Sequence and relatedness in other bacteria of the Pseudomonas aeruginosa oprP gene coding for the phosphate-specific porin P. Mol. Microbiol. 4:831-838. [DOI] [PubMed] [Google Scholar]
- 613.Siehnel, R. J., C. Egli, and R. E. W. Hancock. 1992. Polyphosphate-selective porin OprO of Pseudomonas aeruginosa: Expression, purification and sequence. Mol. Microbiol. 6:2319-2326. [DOI] [PubMed] [Google Scholar]
- 614.Silva, M., A. Ferreira, M. Rodriguez, and D. Wolff. 1992. The major Thiobacillus ferrooxidans outer membrane protein forms low conductance ion channels in planar lipid bilayers. FEBS Lett. 296:169-173. [DOI] [PubMed] [Google Scholar]
- 615.Silver, R. P., W. Aaronson, and W. F. Vann. 1987. Translocation of capsular polysaccharides in pathogenic strains of Escherichia coli requires a 60-kilodalton periplasmic protein. J. Bacteriol. 169:5489-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Silverman, J. A., and S. A. Benson. 1987. Bacteriophage K20 requires both the OmpF porin and lipopolysaccharide for receptor function. J. Bacteriol. 169:4830-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Simon, M., A. Mathes, A. Blanch, and H. Engelhardt. 1996. Characterization of a porin from the outer membrane of Vibrio anguillarum. J. Bacteriol. 178:4182-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Simonet, V. C., A. Basle, K. E. Klose, and A. H. Delcour. 2003. The Vibrio cholerae porins OmpU and OmpT have distinct channel properties. J. Biol. Chem. 278:17539-17545. [DOI] [PubMed] [Google Scholar]
- 619.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell. Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 620.Simons, K., and G. van Meer. 1988. Lipid sorting in epithelial cells. Biochemistry 27:6197-6202. [DOI] [PubMed] [Google Scholar]
- 621.Singh, S. P., Y. U. Williams, S. Miller, and H. Nikaido. 2003. The C-terminal domain of Salmonella enterica serovar Typhimurium OmpA is an immunodominant antigen in mice but appears to be only partially exposed on the bacterial cell surface. Infect. Immun. 71:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Skare, J. T., C. I. Champion, T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1996. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J. Bacteriol. 178:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Skare, J. T., T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, J. Bunikis, S. Bergstrom, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1997. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect. Immun. 65:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Smit, J., Y. Kamio, and H. Nikaido. 1975. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J. Bacteriol. 124:942-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Snijder, H. J., I. Ubarretxena-Belandia, M. Blaauw, K. H. Kalk, H. M. Verheij, M. R. Egmond, N. Dekker, and B. W. Dijkstra. 1999. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401:717-721. [DOI] [PubMed] [Google Scholar]
- 626.Snyder, D. S., and T. J. McIntosh. 2000. The lipopolysaccharide barrier: correlation of antibiotic susceptibility with antibiotic permeability and fluorescent probe binding kinetics. Biochemistry 39:11777-11787. [DOI] [PubMed] [Google Scholar]
- 627.Snyder, S., D. Kim, and T. J. McIntosh. 1999. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry 38:10758-10767. [DOI] [PubMed] [Google Scholar]
- 628.Soares, C. M., J. Bjorksten, and O. Tapia. 1995. L3 loop-mediated mechanisms of pore closing in porin: a molecular dynamics perturbation approach. Protein Eng. 8:5-12. [DOI] [PubMed] [Google Scholar]
- 629.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Stahl, C., S. Kubetzko, I. Kaps, S. Seeber, H. Engelhardt, and M. Niederweis. 2001. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40:451-464. [DOI] [PubMed] [Google Scholar]
- 631.Steeghs, L., H. de Cock, E. Evers, B. Zomer, J. Tommassen, and P. van der Ley. 2001. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 20:6937-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Steeghs, L., R. den Hartog, A. den Boer, B. Zomer, P. Roholl, and P. van der Ley. 1998. Meningitis bacterium is viable without endotoxin. Nature 392:449-450. [DOI] [PubMed] [Google Scholar]
- 633.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634.Stevenson, G., B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist, and P. Reeves. 1994. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176:4144-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 636.Sugawara, E., and H. Nikaido. 1994. OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. J. Biol. Chem. 269:17981-17987. [PubMed] [Google Scholar]
- 637.Sugawara, E., and H. Nikaido. 1997. Only oligomeric forms of low efficiency porins OmpA and OprF appear to contain open channels. Biophys. J. 72:A138. [Google Scholar]
- 638.Sugawara, E., and H. Nikaido. 1992. Pore-forming activity of OmpA protein of Escherichia coli. J. Biol. Chem. 267:2507-2511. [PubMed] [Google Scholar]
- 639.Sugawara, E., M. Steiert, S. Rouhani, and H. Nikaido. 1996. Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. J. Bacteriol. 178:6067-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Sukhan, A., and R. E. W. Hancock. 1995. Insertion mutagenesis of the Pseudomonas aeruginosa phosphate-specific porin OprP. J. Bacteriol. 177:4914-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Sukhan, A., and R. E. W. Hancock. 1996. The role of specific lysine residues in the passage of anions through the Pseudomonas aeruginosa porin OprP. J. Biol. Chem. 271:21239-21242. [DOI] [PubMed] [Google Scholar]
- 642.Sukharev, S. I., P. Blount, B. Martinac, F. R. Blattner, and C. Kung. 1994. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature 368:265-268. [DOI] [PubMed] [Google Scholar]
- 643.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Takada, H., T. Ogawa, F. Yoshimura, K. Otsuka, S. Kokeguchi, K. Kato, T. Umemoto, and S. Kotani. 1988. Immunobiological activities of a porin fraction isolated from Fusobacterium nucleatum ATCC 10953. Infect. Immun. 56:855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Takayama, K., N. Qureshi, K. Hyver, J. Honovich, R. J. Cotter, P. Mascagni, and H. Schneider. 1986. Characterization of a structural series of lipid A obtained from the lipopolysaccharides of Neisseria gonorrhoeae: combined laser desorption and fast atom bombardment mass spectral analysis of high-performance liquid chromatography-purified dimethyl derivatives. J. Biol. Chem. 261:10624-10631. [PubMed] [Google Scholar]
- 646.Takeuchi, M., K. Hamana, and A. Hiraishi. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. E vol. Microbiol. 51:1405-1417. [DOI] [PubMed] [Google Scholar]
- 647.Takeuchi, Y., and H. Nikaido. 1981. Persistence of segregated phospholipid domains in phospholipid-lipopolysaccharide mixed bilayers: studies with spin-labeled phospholipids. Biochemistry 20:523-529. [DOI] [PubMed] [Google Scholar]
- 648.Tamano, K., S. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Tarahovsky, Y. S., A. A. Khusainov, A. A. Deev, and Y. V. Kim. 1991. Membrane fusion during infection of Escherichia coli cells by phage T4. FEBS Lett. 289:18-22. [DOI] [PubMed] [Google Scholar]
- 650.Taylor, C. J., A. J. Anderson, and S. G. Wilkinson. 1998. Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology 144:1737-1745. [DOI] [PubMed] [Google Scholar]
- 651.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Thanassi, D. G. 2002. Ushers and secretins: channels for the secretion of folded proteins across the bacterial outer membrane. J. Mol. Microbiol. Biotechnol. 4:11-20. [PubMed] [Google Scholar]
- 653.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 655.Thanassi, D. G., E. T. Saulino, M.-J. Lombardo, R. Roth, J. Heuser, and S. J. Hultgren. 1998. The PapC usher forms an oligomeric channel: Implications for pilus biogenesis across the outer membrane. Proc. Natl. Acad. Sci. USA 95:3146-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Thiebaut, F., T. Tsuruo, H. Hamada, M. M. Gottesman, I. Pastan, and M. C. Willingham. 1987. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc. Natl. Acad. Sci. USA 84:7735-7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Thinnes, F. P. 1992. Evidence for extra-mitochondrial localization of the VDAC/Porin channel in eucaryotic cells. J. Bioenerg. Biomembr. 24:71-75. [DOI] [PubMed] [Google Scholar]
- 658.Thomas, A. D., and I. R. Booth. 1992. The regulation of expression of the porin gene ompC by acid pH. J. Gen. Microbiol. 138:1829-1835. [DOI] [PubMed] [Google Scholar]
- 659.Tieleman, D. P., and H. J. C. Berendsen. 1998. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 74:2786-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Todt, J. C., and E. J. McGroarty. 1992. Acid pH decreases OmpF and OmpC channel size in vivo. Biochem. Biophys. Res. Commun. 189:1498-1502. [DOI] [PubMed] [Google Scholar]
- 661.Todt, J. C., and E. J. McGroarty. 1992. Involvement of histidine-21 in the pH-induced switch in porin channel size. Biochemistry 31:10479-10482. [DOI] [PubMed] [Google Scholar]
- 662.Todt, J. C., W. J. Rocque, and E. J. McGroarty. 1992. Effects of pH on bacterial porin function. Biochemistry 31:10471-10478. [DOI] [PubMed] [Google Scholar]
- 663.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 664.Tommassen, J., and B. Lugtenberg. 1982. PHO-regulon of Escherichia coli K12: a minireview. Ann. Microbiol. (Paris) 133:243-249. [PubMed] [Google Scholar]
- 665.Toro, C. S., S. R. Lobos, I. Calderon, M. Rodriguez, and G. C. Mora. 1990. Clinical isolate of a porin-less Salmonella typhi resistant to high levels of chloramphenicol. Antimicrob. Agents Chemother. 34:1716-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Traurig, M., and R. Misra. 1999. Identification of bacteriophage K20 binding regions of OmpF and lipopolysaccharide in Escherichia coli K-12. FEMS Microbiol. Lett. 181:101-108. [DOI] [PubMed] [Google Scholar]
- 667.Trent, M. S., W. Pabich, C. R. H. Raetz, and S. I. Miller. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 276:9083-9092. [DOI] [PubMed] [Google Scholar]
- 668.Trent, M. S., A. A. Ribeiro, W. T. Doerrler, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. Accumulation of a polyisoprene-linked amino sugar in polymyxin-resistant Salmonella typhimurium and Escherichia coli: structural characterization and transfer to lipid A in the periplasm. J. Biol. Chem. 276:43132-43144. [DOI] [PubMed] [Google Scholar]
- 669.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 670.Trias, J., and R. Benz. 1993. Characterization of the channel formed by the mycobacterial porin in lipid bilayer membranes: Demonstration of voltage gating and of negative point charges at the channel mouth. J. Biol. Chem. 268:6234-6240. [PubMed] [Google Scholar]
- 671.Trias, J., and R. Benz. 1994. Permeability of the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 14:283-290. [DOI] [PubMed] [Google Scholar]
- 672.Trias, J., J. Dufresne, R. C. Levesque, and H. Nikaido. 1989. Decreased outer membrane permeability in imipenem-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 33:1201-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Trias, J., V. Jarlier, and R. Benz. 1992. Porins in the cell wall of mycobacteria. Science 258:1479-1481. [DOI] [PubMed] [Google Scholar]
- 674.Trias, J., and H. Nikaido. 1990. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 34:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Trias, J., and H. Nikaido. 1990. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265:15680-15684. [PubMed] [Google Scholar]
- 676.Trias, J., E. Y. Rosenberg, and H. Nikaido. 1988. Specificity of the glucose channel formed by protein D1 of Pseudomonas aeruginosa. Biochim. Biophys. Acta 938:493-496. [DOI] [PubMed] [Google Scholar]
- 677.Trieschmann, M. D. A., F. Pattus, and M. H. Tadros. 1996. Molecular characterization and organization of porin from Rhodobacter capsulatus strain 37B4. Gene 183:61-68. [DOI] [PubMed] [Google Scholar]
- 678.Trust, T. J., S. M. Logan, C. E. Gustafson, P. J. Romaniuk, N. W. Kim, V. L. Chan, M. A. Ragan, P. Guerry, and R. R. Gutell. 1994. Phylogenetic and molecular characterization of a 23S rRNA gene positions the genus Campylobacter in the epsilon subdivision of the Proteobacteria and shows that the presence of transcribed spacers is common in Campylobacter spp. J. Bacteriol. 176:4597-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679.Tsubery, H., I. Ofek, S. Cohen, M. Eisenstein, and M. Fridkin. 2002. Modulation of the hydrophobic domain of polymyxin B nonapeptide: effect on outer-membrane permeabilization and lipopolysaccharide neutralization. Mol. Pharmacol. 62:1036-1042. [DOI] [PubMed] [Google Scholar]
- 680.Tsujimoto, H., N. Gotoh, J.-I. Yamagishi, Y. Oyamada, and T. Nishino. 1997. Cloning and expression of the major porin protein gene opcP of Burkholderia (formerly Pseudomonas) cepacia in Escherichia coli. Gene 186:113-118. [DOI] [PubMed] [Google Scholar]
- 681.Tufano, M. A., F. Rossano, P. Catalanotti, G. Liguori, C. Capasso, M. T. Ceccarelli, and P. Marinelli. 1994. Immunobiological activities of Helicobacter pylori porins. Infect. Immun. 62:1392-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Ulmke, C., J. Kreth, J. W. Lengeler, W. Welte, and K. Schmid. 1999. Site-directed mutagenesis of loop L3 of sucrose porin ScrY leads to changes in substrate selectivity. J. Bacteriol. 181:1920-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Umeda, H., H. Aiba, and T. Mizuno. 1996. SomA, a novel gene that encodes a major outer-membrane protein of Synechococcus sp. PCC 7942. Microbiology 142:2121-2128. [DOI] [PubMed] [Google Scholar]
- 684.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.Vaara, M. 1993. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 37:2255-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Vaara, M., W. Z. Plachy, and H. Nikaido. 1990. Partitioning of hydrophobic probes into lipopolysaccharide bilayers. Biochim. Biophys. Acta 1024:152-158. [DOI] [PubMed] [Google Scholar]
- 687.Vaara, M., T. Vaara, and M. Sarvas. 1979. Decreased binding of polymyxin by polymyxin-resistant mutants of Salmonella typhimurium. J. Bacteriol. 139:664-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Vachon, V., R. Laprade, and J. W. Coulton. 1986. Properties of the porin of Haemophilus influenzae type b in planar lipid bilayer membranes. Biochim. Biophys. Acta 861:74-82. [DOI] [PubMed] [Google Scholar]
- 689.Vachon, V., D. J. Lyew, and J. W. Coulton. 1985. Transmembrane permeability channels across the outer membrane of Haemophilus influenzae type b. J. Bacteriol. 162:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 690.van Beilen, J. B., S. Panke, S. Lucchini, A. G. Franchini, M. Rothlisberger, and B. Witholt. 2001. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology 147:1621-1630. [DOI] [PubMed] [Google Scholar]
- 691.Vandeputte-Rutten, L., M. P. Bos, J. Tommassen, and P. Gros. 2003. Crystal structure of Neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J. Biol. Chem. 278:24825-24830. [DOI] [PubMed] [Google Scholar]
- 692.Vandeputte-Rutten, L., R. A. Kramer, J. Kroon, N. Dekker, M. R. Egmond, and P. Gros. 2001. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. 20:5033-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Van Gelder, P., F. Dumas, and M. Winterhalter. 2000. Understanding the function of bacterial outer membrane channels by reconstitution into black lipid membranes. Biophys. Chem. 85:153-167. [DOI] [PubMed] [Google Scholar]
- 694.Van Gelder, P., R. Dutzler, F. Dumas, R. Koebnik, and T. Schirmer. 2001. Sucrose transport through maltoporin mutants of Escherichia coli. Protein Eng. 14:943-948. [DOI] [PubMed] [Google Scholar]
- 695.van Hoogevest, P., J. de Gier, and B. de Kruijff. 1984. Determination of the size of the packing defects in dimyristoylphosphatidylcholine bilayers, present at the phase transition temperature. FEBS Lett. 171:160-164. [Google Scholar]
- 696.Vedam, V., E. L. Kannenberg, J. G. Haynes, D. J. Sherrier, A. Datta, and R. W. Carlson. 2003. A Rhizobium leguminosarum AcpXL mutant produces lipopolysaccharide lacking 27-hydroxyoctacosanoic acid. J. Bacteriol. 185:1841-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Veiga, E., V. de Lorenzo, and L. A. Fernandez. 1999. Probing secretion and translocation of a beta-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol. Microbiol. 33:1232-1243. [DOI] [PubMed] [Google Scholar]
- 698.Veiga, E., E. Sugawara, H. Nikaido, V. de Lorenzo, and L. A. Fernandez. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO. J. 21:2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Vincent, C., P. Doublet, C. Grangeasse, E. Vaganay, A. J. Cozzone, and B. Duclos. 1999. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J. Bacteriol. 181:3472-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Vincent, C., B. Duclos, C. Grangeasse, E. Vaganay, M. Riberty, A. J. Cozzone, and P. Doublet. 2000. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J. Mol. Biol. 304:311-321. [DOI] [PubMed] [Google Scholar]
- 701.Vogt, J., and G. E. Schulz. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Struct. Fold. Des. 7:1301-1309. [DOI] [PubMed] [Google Scholar]
- 702.Vorachek-Warren, M. K., S. M. Carty, S. Lin, R. J. Cotter, and C. R. H. Raetz. 2002. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis. Absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 degreeC. J. Biol. Chem. 277:14186-14193. [DOI] [PubMed] [Google Scholar]
- 703.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 704.Vu, H., and H. Nikaido. 1985. Role of beta-lactam hydrolysis in the mechanism of resistance of a beta-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum beta-lactams. Antimicrob. Agents Chemother. 27:393-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Walian, P. J., and B. K. Jap. 1990. Three-dimensional electron diffraction of PhoE porin to 2.8 Å resolution. J. Mol. Biol. 215:429-438. [DOI] [PubMed] [Google Scholar]
- 706.Wandersman, C. 1992. Secretion across the bacterial outer membrane. Trends Genet. 8:317-322. [DOI] [PubMed] [Google Scholar]
- 707.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 87:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Wang, J., J.-M. Betton, V. Michel, M. Hofnung, and A. Charbit. 1997. Identification of a cryptic porin gene in the Escherichia coli genome: Expression and insertion of a monomeric form of the protein into the outer membrane. Mol. Microbiol. 26:1141-1143. [PubMed] [Google Scholar]
- 709.Wang, Y.-F., R. Dutzler, P. J. Rizkallah, J. P. Rosenbusch, and T. Schirmer. 1997. Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin. J. Mol. Biol. 272:56-63. [DOI] [PubMed] [Google Scholar]
- 710.Watanabe, M., J. Rosenbusch, T. Schirmer, and M. Karplus. 1997. Computer simulations of the OmpF porin from the outer membrane of Escherichia coli. Biophys. J. 72:2094-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Wee, S., and B. J. Wilkinson. 1988. Increased outer membrane ornithine-containing lipid and lysozyme penetrability of Paracoccus denitrificans grown in a complex medium deficient in divalent cations. J. Bacteriol. 170:3283-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Weintraub, A., U. Zaehringer, H. W. Wollenweber, U. Seydel, and E. T. Rietschel. 1989. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur. J. Biochem. 183:425-432. [DOI] [PubMed] [Google Scholar]
- 713.Weintraub, A., U. Zahringer, and A. A. Lindberg. 1985. Structural studies of the polysaccharide part of the cell wall lipopolysaccharide from Bacteroides fragilis NCTC 9343. Eur. J. Biochem. 151:657-662. [DOI] [PubMed] [Google Scholar]
- 714.Weiss, M. S., U. Abele, J. Weckesser, W. Welte, E. Schiltz, and G. E. Schulz. 1991. Molecular architecture and electrostatic properties of a bacterial porin. Science 254:1627-1630. [DOI] [PubMed] [Google Scholar]
- 715.Weiss, M. S., A. Kreusch, E. Schiltz, U. Nestel, W. Welte, J. Weckesser, and G. E. Schulz. 1991. The structure of porin from Rhodobacter capsulatus at 1.8 Å resolution. FEBS Lett. 280:379-382. [DOI] [PubMed] [Google Scholar]
- 716.Weiss, M. S., and G. E. Schulz. 1992. Structure of porin refined at 1.8 Å resolution. J. Mol. Biol. 227:493-509. [DOI] [PubMed] [Google Scholar]
- 717.Weiss, M. S., T. Wacker, U. Nestel, D. Woitzik, J. Weckesser, W. Kreutz, W. Welte, and G. E. Schulz. 1989. The structure of porin from Rhodobacter capsulatus at 0.6 nm resolution. FEBS Lett. 256:143-146. [Google Scholar]
- 718.Weiss, M. S., T. Wacker, J. Weckesser, W. Welte, and G. E. Schulz. 1990. The three-dimensional structure of porin from Rhodobacter capsulatus at 3 Å resolution. FEBS Lett. 267:268-272. [DOI] [PubMed] [Google Scholar]
- 719.Welch, T. J., and D. H. Bartlett. 1996. Isolation and characterization of the structural gene for OmpL, a pressure-regulated porin-like protein for the deep-sea bacterium Photobacterium species strain SS9. J. Bacteriol. 178:5027-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720.Wen, A., M. Fegan, C. Hayward, S. Chakraborty, and L. I. Sly. 1999. Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:567-576. [DOI] [PubMed] [Google Scholar]
- 721.Werner, V., C. C. Sanders, W. E. Sanders, Jr., and R. V. Goering. 1985. Role of beta-lactamases and outer membrane proteins in multiple beta-lactam resistance of Enterobacter cloacae. Antimicrob. Agents Chemother. 27:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Wexler, H. M., and C. Getty. 1996. The isolation and characterization of a porin protein from Bacteroides fragilis. Anaerobe 2:305-312. [Google Scholar]
- 723.Wexler, H. M., C. Getty, and G. Fisher. 1992. The isolation and characterisation of a major outer-membrane protein from Bacteroides distasonis. J. Med. Microbiol. 37:165-175. [DOI] [PubMed] [Google Scholar]
- 724.Wexler, H. M., E. K. Read, and T. J. Tomzynski. 2002. Characterization of omp200, a porin gene complex from Bacteroides fragilis: omp121 and omp71, gene sequence, deduced amino acid sequences and predictions of porin structure. Gene 283:95-105. [DOI] [PubMed] [Google Scholar]
- 725.Wexler, H. M., E. K. Read, and T. J. Tomzynski. 2002. Identification of an OmpA protein from Bacteroides fragilis: ompA gene sequence, OmpA amino acid sequence and prediction of protein structure. Anaerobe 8:180-191. [Google Scholar]
- 726.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 727.Wibbenmeyer, J. A., D. Provenzano, C. F. Landry, K. E. Klose, and A. H. Delcour. 2002. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 70:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Wiedemann, B., and H. Grimm. 1996. Susceptibility to antibiotics: species incidence and trends, p. 900-1168. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, Md.
- 729.Wiese, A., K. Brandenburg, S. F. Carroll, E. T. Rietschel, and U. Seydel. 1997. Mechanisms of action of bactericidal/permeability-increasing protein BPI on reconstituted outer membranes of gram-negative bacteria. Biochemistry 36:10311-10319. [DOI] [PubMed] [Google Scholar]
- 730.Wiese, A., M. Munstermann, T. Gutsmann, B. Lindner, K. Kawahara, U. Zahringer, and U. Seydel. 1998. Molecular mechanisms of polymyxin B-membrane interactions: direct correlation between surface charge density and self-promoted transport. J. Membr. Biol. 162:127-138. [DOI] [PubMed] [Google Scholar]
- 731.Wiese, A., J. O. Reiners, K. Brandenburg, K. Kawahara, U. Zaehringer, and U. Seydel. 1996. Planar asymmetric lipid bilayers of glycosphingolipid or lipopolysaccharide on one side and phospholipids on the other: membrane potential, porin function, and complement activation. Biophys. J. 70:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Wiese, A., G. Schroder, K. Brandenburg, A. Hirsch, W. Welte, and U. Seydel. 1994. Influence of the lipid matrix on incorporation and function of LPS-free porin from Paracoccus denitrificans. Biochim. Biophys. Acta 1190:231-242. [DOI] [PubMed] [Google Scholar]
- 733.Wilkinson, B. J. 1977. Cell envelope of Paracoccus denitrificans: outer membrane permeability to lysozyme and hydrophobic antibiotics. FEMS Microbiol. Lett. 2:285-288. [Google Scholar]
- 734.Wilkinson, B. J., K. A. Sment, and W. R. Mayberry. 1982. Occurrence, localization, and possible significance of an ornithine-containing lipid in Paracoccus denitrificans. Arch. Microbiol. 131:338-343. [Google Scholar]
- 735.Wilkinson, S. G. 1996. Bacterial lipopolysaccharides—themes and variations. Prog. Lipid Res. 35:283-343. [DOI] [PubMed] [Google Scholar]
- 736.Wilkinson, S. G. 1988. Gram-negative bacteria, p. 299-488. In C. Ratledge and S. G. Wilkinson (ed.), Microbial lipids, vol. 1. Academic Press, Ltd., London, United Kingdom.
- 737.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 738.Woese, C. R., E. Stackebrandt, W. G. Weisburg, B. J. Paster, M. T. Madigan, V. J. Fowler, C. M. Hahn, P. Blanz, R. Gupta, K. H. Nealson, and G. E. Fox. 1984. The phylogeny of purple bacteria: the alpha subdivision. Syst. Appl. Microbiol. 5:315-326. [DOI] [PubMed] [Google Scholar]
- 739.Woese, C. R., W. G. Weisburg, C. M. Hahn, B. J. Paster, L. B. Zablen, B. J. Lewis, T. J. Macke, W. Ludwig, and E. Stackebrandt. 1985. The phylogeny of purple bacteria: the gamma subdivision. Syst. Appl. Microbiol. 6:25-33. [Google Scholar]
- 740.Woitzik, D., R. Benz, A. Lustig, and J. Weckesser. 1989. Porin from Thiobacillus versutus. FEMS Microbiol. Lett. 65:319-322. [DOI] [PubMed] [Google Scholar]
- 741.Woitzik, D., J. Weckesser, R. Benz, S. Stevanovic, G. Jung, and J. P. Rosenbusch. 1990. Porin of Rhodobacter capsulatus: biochemical and functional characterization. Z. Naturforsch. Sect. C 45:576-82. [DOI] [PubMed] [Google Scholar]
- 742.Wolf, E., G. Achatz, J. F. Imhoff, E. Schiltz, J. Weckesser, and M. C. Lamers. 1997. Porin from the halophilic species, Ectothiorhodospira vacuolata: cloning; structure of the gene and comparison with other porins. Gene 191:225-232. [DOI] [PubMed] [Google Scholar]
- 743.Wolf, E., M. Zahr, R. Benz, J. F. Imhoff, A. Lustig, E. Schiltz, J. Stahl-Zeng, and J. Weckesser. 1996. The porins from the halophilic species Ectothiorhodospira shaposhnikovii and Ectothiorhodospira vacuolata. Arch. Microbiol. 166:169-175. [DOI] [PubMed] [Google Scholar]
- 744.Wollenweber, H. W., S. Schlecht, O. Luderitz, and E. T. Rietschel. 1983. Fatty acid in lipopolysaccharides of Salmonella species grown at low temperature. Identification and position. Eur. J. Biochem. 130:167-171. [DOI] [PubMed] [Google Scholar]
- 745.Wong, K. K. Y., F. S. L. Brinkman, R. S. Benz, and R. E. W. Hancock. 2001. Evaluation of a structural model of Pseudomonas aeruginosa outer membrane protein OprM, an efflux component involved in intrinsic antibiotic resistance. J. Bacteriol. 183:367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Wong, K. K. Y., and R. E. W. Hancock. 2000. Insertion mutagenesis and membrane topology model of the Pseudomonas aeruginosa outer membrane protein OprM. J. Bacteriol. 182:2402-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 747.Wong, R. S., H. Jost, and R. E. Hancock. 1993. Linker-insertion mutagenesis of Pseudomonas aeruginosa outer membrane protein OprF. Mol. Microbiol. 10:283-292. [PubMed] [Google Scholar]
- 748.Wong, R. S., R. A. Wirtz, and R. E. Hancock. 1995. Pseudomonas aeruginosa outer membrane protein OprF as an expression vector for foreign epitopes: the effects of positioning and length on the antigenicity of the epitope. Gene 158:55-60. [DOI] [PubMed] [Google Scholar]
- 749.Wylie, J. L., and E. A. Worobec. 1995. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J. Bacteriol. 177:3021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Wyllie, S., D. Longbottom, A. J. Herring, and R. H. Ashley. 1999. Single channel analysis of recombinant major outer membrane protein porins from Chlamydia psittaci and Chlamydia pneumoniae. FEBS Lett. 445:192-196. [DOI] [PubMed] [Google Scholar]
- 751.Yamano, Y., T. Nishikawa, and Y. Komatsu. 1993. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosa PAO1. Mol. Microbiol. 8:993-1004. [DOI] [PubMed] [Google Scholar]
- 752.Yamano, Y., T. Nishikawa, and Y. Komatsu. 1990. Outer membrane proteins responsible for the penetration of beta-lactams and quinolones in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 26:175-184. [DOI] [PubMed] [Google Scholar]
- 753.Yano, I., S. Imaizumi, I. Tomiyasu, and E. Yabuuchi. 1983. Separation and analysis of free ceramides containing 2-hydroxy fatty acids in Sphingobacterium species. FEMS Microbiol. Lett. 20:449-453. [Google Scholar]
- 754.Yaron, S., G. L. Kolling, L. Simon, and K. R. Matthews. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66:4414-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Yau, W. M., W. C. Wimley, K. Gawrisch, and S. H. White. 1998. The preference of tryptophan for membrane interfaces. Biochemistry 37:14713-14718. [DOI] [PubMed] [Google Scholar]
- 756.Yeh, H. Y., and D. M. Jacobs. 1992. Characterization of lipopolysaccharide fractions and their interactions with cells and model membranes. J. Bacteriol. 174:336-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Yen, M. R., C. R. Peabody, S. M. Partovi, Y. Zhai, Y. H. Tseng, and M. H. Saier. 2002. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta 1562:6-31. [DOI] [PubMed] [Google Scholar]
- 758.Yethon, J. A., J. S. Gunn, R. K. Ernst, S. I. Miller, L. Laroche, D. Malo, and C. Whitfield. 2000. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence In vivo. Infect. Immun. 68:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Yethon, J. A., D. E. Heinrichs, M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273:26310-26316. [DOI] [PubMed] [Google Scholar]
- 760.Yethon, J. A., E. Vinogradov, M. B. Perry, and C. Whitfield. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 182:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Yin, N., R. L. Marshall, S. Matheson, and P. B. Savage. 2003. Synthesis of lipid A derivatives and their interactions with polymyxin B and polymyxin B nonapeptide. J. Am. Chem. Soc. 125:2426-2435. [DOI] [PubMed] [Google Scholar]
- 762.Yoshihara, E., N. Gotoh, T. Nishino, and T. Nakae. 1996. Protein D2 porin of the Pseudomonas aeruginosa outer membrane bears the protease activity. FEBS Lett. 394:179-182. [DOI] [PubMed] [Google Scholar]
- 763.Yoshihara, E., and T. Nakae. 1989. Identification of porins in the outer membrane of Pseudomonas aeruginosa that form small diffusion pores. J. Biol. Chem. 264:6297-6301. [PubMed] [Google Scholar]
- 764.Yoshihara, E., H. Yoneyama, and T. Nakae. 1991. In vitro assembly of the functional porin trimer from dissociated monomers in Pseudomonas aeruginosa. J. Biol. Chem. 266:952-957. [PubMed] [Google Scholar]
- 765.Yoshimura, F., and H. Nikaido. 1985. Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob. Agents Chemother. 27:84-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 766.Yoshimura, F., and H. Nikaido. 1982. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J. Bacteriol. 152:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 767.Yoshimura, F., L. S. Zalman, and H. Nikaido. 1983. Purification and properties of Pseudomonas aeruginosa porin. J. Biol. Chem. 258:2308-2314. [PubMed] [Google Scholar]
- 768.Yu, E. W., J. R. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185:5657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, and D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 770.Zachariae, U., A. Koumanov, H. Engelhardt, and A. Karshikoff. 2002. Electrostatic properties of the anion selective porin Omp32 from Delftia acidovorans and of the arginine cluster of bacterial porins. Protein Sci. 11:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Zakharov, S. D., and W. A. Cramer. 2002. Colicin crystal structures: pathways and mechanisms for colicin insertion into membranes. Biochim. Biophys. Acta 1565:333-346. [DOI] [PubMed] [Google Scholar]
- 772.Zalman, L. S., and H. Nikaido. 1985. Dimeric porin from Paracoccus denitrificans. J. Bacteriol. 162:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Zalman, L. S., H. Nikaido, and Y. Kagawa. 1980. Mitochondrial outer membrane contains a protein producing nonspecific diffusion channels. J. Biol. Chem. 255:1771-1774. [PubMed] [Google Scholar]
- 774.Zeth, K., K. Diederichs, W. Welte, and H. Engelhardt. 2000. Crystal structure of Omp32, the anion-selective porin from Comamonas acidovorans, in complex with a periplasmic peptide at 2.1 ANG resolution. Structure 8:981-992. [DOI] [PubMed] [Google Scholar]
- 775.Zgurskaya, H. I., and H. Nikaido. 1999. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285:409-420. [DOI] [PubMed] [Google Scholar]
- 776.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Zhai, Y., and M. H. Saier, Jr. 2002. The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes. Protein Sci. 11:2196-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 779.Zhai, Y. F., W. Heijne, and M. H. Saier, Jr. 2003. Molecular modeling of the bacterial outer membrane receptor energizer, ExbBD/TonB, based on homology with the flagellar motor, MotAB. Biochim. Biophys. Acta 1614:201-210. [DOI] [PubMed] [Google Scholar]
- 780.Zhang, E., and T. Ferenci. 1999. OmpF changes and the complexity of Escherichia coli adaptation to prolonged lactose limitation. FEMS Microbiol. Lett. 176:395-401. [DOI] [PubMed] [Google Scholar]
- 781.Zhang, H. H., D. R. Blanco, M. M. Exner, E. S. Shang, C. I. Champion, M. L. Phillips, J. N. Miller, and M. A. Lovett. 1999. Renaturation of recombinant Treponema pallidum rare outer membrane protein 1 into a trimeric, hydrophobic, and porin-active conformation. J. Bacteriol. 181:7168-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Zhang, Q., J. C. Meitzler, S. Huang, and T. Morishita. 2000. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Zhou, Z., S. Lin, R. J. Cotter, and C. R. Raetz. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274:18503-18514. [DOI] [PubMed] [Google Scholar]
- 784.Zhou, Z., A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller, and C. R. H. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PmrA-dependent 4-amino-4-deoxy-l-arabinose and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]
- 785.Zhou, Z., A. A. Ribeiro, and C. R. Raetz. 2000. High-resolution NMR spectroscopy of lipid A molecules containing 4-amino-4-deoxy-l-arabinose and phosphoethanolamine substituents. Different attachment sites on lipid A molecules from NH4VO3-treated Escherichia coli versus kdsA mutants of Salmonella typhimurium. J. Biol. Chem. 275:13542-13551. [DOI] [PubMed] [Google Scholar]
- 786.Zhou, Z., K. A. White, A. Polissi, C. Georgopoulos, and C. R. Raetz. 1998. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 273:12466-12475. [DOI] [PubMed] [Google Scholar]
- 787.Zhuang, J., A. Engel, J.-M. Pages, and J.-M. Bolla. 1997. The Campylobacter jejuni porin trimers pack into different lattice types when reconstituted in the presence of lipid. Eur. J. Biochem. 244:575-579. [DOI] [PubMed] [Google Scholar]