Abstract
Receptor-independent G-protein regulators provide diverse mechanisms for signal input to G-protein-based signaling systems, revealing unexpected functional roles for G-proteins. As part of a broader effort to identify disease-specific regulators for heterotrimeric G-proteins, we screened for such proteins in cardiac hypertrophy using a yeast-based functional screen of mammalian cDNAs as a discovery platform. We report the identification of three transcription factors belonging to the same family, transcription factor E3 (TFE3), microphthalmia-associated transcription factor, and transcription factor EB, as novel receptor-independent activators of G-protein signaling selective for Gα16. TFE3 and Gα16 were both up-regulated in cardiac hypertrophy initiated by transverse aortic constriction. In protein interaction studies in vitro, TFE3 formed a complex with Gα16 but not with Gαi3 or Gαs. Although increased expression of TFE3 in heterologous systems had no influence on receptor-mediated Gα16 signaling at the plasma membrane, TFE3 actually translocated Gα16 to the nucleus, leading to the induction of claudin 14 expression, a key component of membrane structure in cardiomyocytes. The induction of claudin 14 was dependent on both the accumulation and activation of Gα16 by TFE3 in the nucleus. These findings indicate that TFE3 and Gα16 are up-regulated under pathologic conditions and are involved in a novel mechanism of transcriptional regulation via the relocalization and activation of Gα16.
Keywords: Cardiac Hypertrophy, G-proteins, Signal Transduction, Tight Junction, Transcription Regulation
Introduction
Heterotrimeric G-proteins play key roles in transducing cell surface stimuli to intracellular signaling events (1, 2). Activation of G-protein coupled receptors (GPCRs)3 at the cell surface initiates nucleotide exchange on Gα subunits, leading to a conformational change in Gαβγ and subsequent transduction of signals to various intracellular effector molecules. In addition to the basic components of the G-protein signaling system (i.e. GPCRs, heterotrimeric G-proteins, and effector molecules), there is a novel class of regulatory proteins for heterotrimeric G-proteins that directly regulate the activation status of heterotrimeric G-proteins independently of GPCRs (3–10).
Such receptor-independent G-protein regulators are involved in unexpected and important functional roles of heterotrimeric G-proteins in multiple cellular events. For example, LGN (activator of G-protein signaling 5 (AGS5)) and AGS3 are involved in the regulation of mitotic spindle dynamics and cell division (11–14). The GTPase-activating protein RGS14 also translocates between the nucleus and the cytoplasm and is associated with centrosomes influencing mitosis (15). Another RGS protein, RGS7, interacts with Gβ5 and migrates into the nucleus as an RGS7-Gβ5 complex (16). Furthermore, signal alteration by G-protein and their various types of regulators is involved in adaptation of cells to maintain homeostasis under pathologic conditions (17–21). In fact, the expression of such regulatory proteins is altered with the development of cardiac hypertrophy in hypertension or in response to pressure overload stress (22, 23).
As part of a broader approach to identify adaptation-specific regulatory proteins for heterotrimeric G-proteins, we previously identified AGS8 from a cDNA library of rat hearts subjected to repetitive transient ischemia (18). AGS8 was up-regulated in cardiomyocytes in response to transient hypoxia and regulated Gβγ signaling. Indeed, AGS8 played a key role in apoptosis of cardiomyocytes induced by hypoxic stress via Gβγ and the channel protein connexin 43 (24). These findings prompted us to investigate the presence of putative AGS proteins in other models of cardiovascular diseases.
We first screened for regulatory proteins for heterotrimeric G-proteins involved in the development of cardiac hypertrophy. Cardiac hypertrophy is a gateway to cardiac dysfunction and acts as an independent risk factor for cardiovascular events. GPCR-mediated signaling pathways, in particular those involving β-adrenergic or angiotensin II receptors, influence gene expression involved in cardiac hypertrophy. Overexpression of Gαs or Gαq in the mouse heart actually results in the development of cardiac hypertrophy and dysfunction.
We report the identification of three Gα16-selective AGS proteins using a yeast-based discovery platform for receptor-independent activators of G-protein signaling to screen cDNA libraries from mouse models of cardiac hypertrophy induced by transverse aortic constriction (TAC) or continuous infusion of the β-adrenergic agonist isoproterenol. Of importance, the three new AGS proteins are microphthalmia-associated transcription factor (MITF)/TFE transcription factors. Although increased expression of TFE3 in heterologous systems had no influence on receptor-mediated Gα16 signaling at the plasma membrane, TFE3 actually translocated Gα16 to the nucleus, leading to the induction of claudin 14, a key component of membrane structure in cardiomyocytes. These findings indicate that Gα16-selective AGS proteins are up-regulated under pathologic conditions and are involved in a novel mechanism of transcriptional regulation via the relocalization and activation of Gα16.
EXPERIMENTAL PROCEDURES
Materials
Anti-Gαi3, anti-Gαs, and anti-phospholipase C (PLC)-β2 antibodies and anti-β2-adrenergic receptor were purchased from Santa Cruz Biotechnology. IGEPAL CA-630 and anti-β-actin antibody were obtained from Sigma. Anti-Gα16 and anti-claudin 14 antibodies were purchased from Medical and Biological Laboratories, Co., Ltd. (Nagoya, Japan) and Abcam, respectively. Anti-Xpress antibody and Lipofectamine 2000 reagent were obtained from Invitrogen. pcDNA3.1::Gα16 and pcDNA3.1::Gα16Q212L were obtained from the Missouri S&T cDNA Resource Center. Gα16G211A was generated by site-direct mutagenesis (PrimeSTAR Mutagenesis Basal kit, Takara, Otsu, Japan). Full-length mouse TFE3, human transcription factor EB (TFEB), and mouse MITF were subcloned into the pYES2 vector (Invitrogen) or pcDNAHis vector (Invitrogen) from cDNA clones (Open Biosystems) (TFE3, MMM1013-98478992; TFEB, MHS1010-7508073; MITF, EMM1002-97035453).
Animal Models
All animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee at Yokohama City University.
TAC
Constriction of the transverse thoracic aorta was performed on 14 male mice (C57BL/6; age, 14–17 weeks; 24–29 g; Charles River Laboratories, Gilroy, CA) as described previously (25). In brief, mice were anesthetized, intubated, and placed on a respirator. The transverse aorta was visualized following midline sternotomy. A 5-0 nylon suture was placed around the aorta distal to the brachiocephalic artery. The suture was tightened around a blunt 27-gauge needle placed adjacent to the aorta to produce −70% constriction. The needle was then removed, and the chest and overlying skin were closed. Six age-matched animals underwent the same surgical procedure but without TAC (sham). Seven days after surgery, the mice were sacrificed for tissue extraction. The left ventricles were quickly separated, frozen in liquid nitrogen, and stored at −70 °C until use.
Cardiac Hypertrophy and Tachycardia
Nineteen male mice (C57BL/6; age, 16–19 weeks; 25–30 g; Charles River Laboratories) were anesthetized, and an osmotic minipump (model 2002, ALZET Osmotic Pumps, Cupertino, CA) was implanted subcutaneously (18). After 7 days of continuous infusion of isoproterenol (60 μg/g of body weight/day), mice were anesthetized, and the hearts were rapidly excised. The left ventricles were rapidly frozen in liquid nitrogen and stored at −70 °C until use.
Generation of cDNA Libraries and Functional Screen in Saccharomyces cerevisiae
mRNA isolated from the left ventricle in the TAC or tachycardia models was used to synthesize cDNAs using a cDNA Synthesis kit (Takara); cDNAs were cloned into the pYES2 yeast expression vector. The cDNA library from the TAC model contained 1.1 × 106 cfu with an average insert size of 1.5 kb, and the library from tachycardia model contained 2.8 × 106 cfu with an average insert size of 1.2 kb. Functional screens and growth assays in the modified strains of S. cerevisiae were conducted as described previously (26–28).
Quantitative Polymerase Chain Reaction (PCR)
RNA isolation, cDNA synthesis, and real time PCR analysis were performed as described previously (24). The primers for RT-PCR were as follows: mouse MITF: forward, 5′-ACTTTCCCTTATCCCATCCACC-3′; reverse, 5′-TGAGATCCAGAGTTGTCGTACA-3′; mouse TFE3: forward, 5′-TGCGTCAGCAGCTTATGAGG-3′; reverse, 5′-AGACACGCCAATCACAGAGAT-3′; mouse TFEB: forward, 5′-CCACCCCAGCCATCAACAC-3′; reverse, 5′-CAGACAGATACTCCCGAACCTT-3′; mouse GNA15: forward, 5′-CGCCAGAATCGACCAGGAG-3′; reverse, 5′-GTAGCCCACACCGTGAATGA-3′; mouse claudin 14: forward, 5′-GCATGGTGGGAACGCTCAT-3′; reverse, 5′-CCACAGTCCCTTCAGGTAGGA-3′; human claudin 14: forward, 5′-CAAACACCGCACCTGCCTA-3′; reverse, 5′-CACGTAGTCGTTCAGCCTGT-3′; rat GNA15: forward, 5′-CAGGAGAACCGTATGAAGGAGAGTC-3′; reverse, 5′-CAGGATGTCTGTCTTGTTGAGGAAG-3′; rat TFE3: forward, 5′-TGTTCGTGCTGTTGGAAGAGC-3′; reverse, 5′-GGGATAGAGGCTGGCTTTTGAG-3′; rat claudin 14: forward, 5′-TCATCACTACTATCCTGCCGCAC-3′; reverse, 5′-ACACACTCCATCCACAGTCCCTTC-3′; and 18 S: forward, 5′-GTAACCCGTTGAACCCCATT-3′; reverse, 5′-CCATCCAATCGGTAGTAGCG-3′. All PCRs were performed in duplicate or triplicate at 95 °C for 2 min followed by 40 cycles at 95 °C for 30 s and 60 or 62 °C for 45 s. The cycle threshold values corresponding to the PCR cycle number at which fluorescence emission in real time reaches a threshold above the base-line emission were determined. 18 S ribosomal RNA was used as a control for the amount of target mRNA in each sample.
Generation of Glutathione S-Transferase (GST) Fusion Protein, Protein Interaction Assays, and Immunoblotting
The coding sequence of TFE3 (amino acids Leu40–Ser572; cDNA1-8) was amplified by PCR and fused in-frame to GST in the pGEX-6T vector (Amersham Biosciences). The GST-TFE3 fusion protein was expressed in bacteria (Escherichia coli BL21; Amersham Biosciences) and purified on a glutathione affinity matrix. The GST fusion protein was eluted from the resin, and glutathione was removed by desalting to allow a solution-phase interaction assay (17). Protein interaction assays and immunoblotting were performed as described previously (17, 29).
Cell Culture and Transfection
COS7 or HEK293 cells were cultured and transfected as described previously (17). In brief, cells were suspended at 0.5–1.0 × 105 cells/ml, and 1.0 (12-well plate), 2.0 (35-mm dish), or 10 ml (100-mm dish) was plated. After 18 h, cells were transfected with 2 (12-well plate), 4–5 (35-mm dish), or 12 μg (100-mm dish) of cDNA with Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. For each experiment, transfection efficiency was monitored by pEGFP vector transfection to generate a fluorescent signal and immunoblotting. The transfection efficiency was 60–80%. Cell lysis and fractionation were performed as described previously (17, 30).
Transfection of Small Interfering RNA (siRNA) to Cultured Cardiomyocytes
Double strand siRNA oligonucleotides to rat GNA15 (Gα16; NCBI Reference Sequence NM_053542) and TFE3 (NCBI Reference Sequence XM_228760) were synthesized (Stealth siRNA, Invitrogen) as follows: GNA15 siRNA: sense, 5′-CCAUGCAGGCCAUGAUUGAAGCAAU-3′; TFE3: sense, 5′-CAGAAGAAAGACAAUCACAACCUAA-3′. The conditions and duplex eliciting the most effective reduction in GNA15 and TFE3 were determined in a series of preliminary experiments. Cardiomyocytes were prepared from the hearts of 1–3-day-old Wistar rats as described previously (24). Approximately 24 h after preparation, neonatal cardiomyocytes at 4.0 × 105 cells in 35-mm plates were transfected with siRNA using Lipofectamine 2000 according to the manufacturer's instructions. Briefly, GNA15siRNA and TFE3siRNA individually in 50 μl of Opti-MEM I medium (Invitrogen) and 2.5 μl of Lipofectamine 2000 in 50 μl of Opti-MEM I medium were mixed, and then the mixture was added to cardiomyocytes. The final concentrations of GNA15siRNA and TFE3siRNA were 50 and 100 nm, respectively. The transfection efficiency of FITC-labeled oligonucleotide was 70–80%. The decrease of mRNA of GNA15 or TFE3 was confirmed by real time PCR following transfection of siRNAs.
Immunoprecipitation
Cell lysates were prepared in 250–500 μl of immunoprecipitation buffer (50 mm Tris, pH 7.4, 70 mm NaCl, 5 mm EDTA, 1% IGEPAL CA-630 (Sigma), and a protease inhibitor mixture (Complete Mini, Roche Applied Science)). The lysates were incubated with 1.0–3.5 μg of antibody for 18 h after preclearing with 25 μl of 50% Sepharose-G for 1 h at 4 °C. The samples were incubated with 25 μl of 50% Sepharose-G for 1 h at 4 °C, and the pellets were washed three times with immunoprecipitation buffer. Proteins were eluted in 30 μl of 2× Laemmli buffer and resolved by SDS-PAGE (24).
Measurements of Inositol Phosphates
COS7 cells were seeded in 12-well plates at 0.5–1.0 × 105 cell/well. Next, 40 h after transfection, the cells were washed three times with phosphate-buffered saline (PBS) and incubated with serum-free Dulbecco's modified Eagle's medium for 4 h. The amount of cellular inositol monophosphate was determined by IP-One ELISA (Cisbio) according to the manufacturer's protocol.
Immunocytochemistry
Tissue Sections
Mouse heart was fixed in 4% paraformaldehyde and embedded in paraffin. Sections (4 μm thick) were prepared after being deparaffinized with xylene and graded ethanol. Sections were incubated in 0.3% H2O2 in methanol for 30 min to inactivate endogenous peroxidases and then rinsed three times for 5 min each with PBS. Tissues were incubated in citrate buffer (pH 6.0) at 100 °C for 10 min. Tissues were blocked in 5% skim milk for 30 min at room temperature and then incubated overnight with goat anti-claudin 14 (ab19035, Abcam; 1:100) antibodies at 4 °C in a humidified chamber. After washing three times for 5 min each in PBS, tissues were processed by the avidin-biotin complex method using a commercially available kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's instructions. Immunocomplexes were visualized with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Dako, Glostrup, Denmark) or with the Liquid DAB-Black Substrate kit (Zymed Laboratories Inc., San Francisco, CA).
Cultured Cells
Cells were seeded on 24 × 24-mm polylysine-coated coverslips. Cells were fixed with PBS containing 4% paraformaldehyde and 4% sucrose for 15 min and then incubated with 0.2% Triton X-100 in PBS for 5 min. After three washes with PBS, cells were incubated with 5% normal donkey serum in PBS for 1 h. Cells were incubated with primary antibodies for 18 h at 4 °C followed by incubation for 1 h with secondary antibody (goat anti-mouse Alexa Fluor 488 or goat anti-rabbit Alexa Fluor 594, highly cross-absorbed; Molecular Probes) diluted to 1:2000 in PBS. All antibody dilutions were centrifuged at 12,000 × g for 15 min prior to use. In some cases, cells were incubated with 1 μg/ml 4′,6′-diamidino-2-phenylindole, dihydrochloride (DAPI) (Molecular Probes) in PBS for 5 min after incubation with secondary antibodies. Slides were then mounted with glass coverslips with ProLong Gold antifade reagent (Invitrogen). Images were analyzed by deconvolution microscopy (TE2000-E, Nikon, Tokyo, Japan). Obtained images were deconvoluted using NIS-Elements 3.0 software (Nikon) with a “no neighbors” deconvolution algorithm. All images were obtained from approximately the middle plane of the cells.
Miscellaneous Procedures and Statistical Analysis
Immunoblotting and data analysis were performed as described previously (18, 24). The luminescence images captured with an image analyzer (LAS-3000, Fujifilm, Tokyo, Japan) were quantified using Image Gauge 3.4 (Fujifilm). Data are expressed as mean ± S.E. from independent experiments as described in the figure legends. Statistical analyses were performed using the unpaired t test, F-test, and one-way analysis of variance followed by Tukey's multiple comparison post hoc test. All statistical analyses were performed with Prism 4 (GraphPad Software).
RESULTS
Identification of Activators of G-protein Signaling from Hypertrophied Hearts
We utilized an expression cloning system in S. cerevisiae to identify receptor-independent activators of G-protein signaling involved in the development of cardiac hypertrophy (18, 26). The yeast strains used in this screen system lacked the pheromone receptor but expressed mammalian Gα (Gαi3, Gαs, or Gα16) in place of the yeast Gα subunit and provided a readout of growth upon activation of the G-protein-regulated pheromone signaling pathway. cDNA libraries from the left ventricle of the hypertrophy models were constructed in a galactose-inducible vector and introduced into these yeast strains. Functional screening for receptor-independent AGS proteins was then facilitated by selection of colonies growing in a galactose-specific manner.
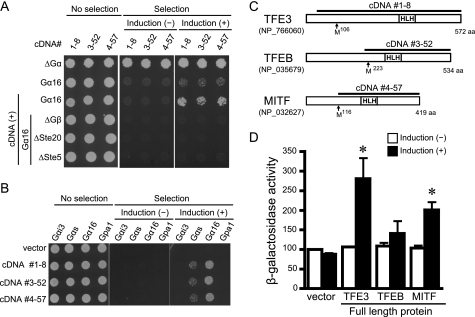
We used two models of cardiac hypertrophy: the TAC-induced pressure overload model and the isoproterenol-induced tachycardiac hypertrophic model (supplemental Fig. 1). cDNA libraries from each model were introduced into the yeast strains expressing mammalian Gαi3, Gαs, or Gα16 (Table 1). Twenty-nine cDNA clones encoding six distinct proteins were isolated from the two cDNA libraries (Gαs strain, 0; Gαi3 strain, 20; Gα16 strain, 9). Each clone was retransformed into yeast to confirm plasmid-dependent growth, and then epistasis analysis was performed to identify the site of action within the pheromone pathway. Epistasis analysis demonstrated that six of these cDNA clones required G-protein to activate the growth-linked G-protein pathway, and thus these clones satisfied the definition of AGS (3, 27) (Table 1 and Fig. 1A).
TABLE 1.
AGS cDNAs isolated from cardiac hypertrophy model of mouse
AGSs are numbered according to the order in which they were isolated from a functional screen in yeast. GPR, G-protein-regulatory motif. The number of transformants screened for each cDNA library of the heart is as follows: transverse aortic constriction, 1.6 × 107; isoproterenol infusion, 2.0 × 107.
| Gene in database | AGS | Cardiac dysfunction model used to generate cDNA libraries for functional screena |
|
|---|---|---|---|
| Transverse aortic constriction | Isoproterenol infusion | ||
| Dynlt1b (the entire coding sequence) | AGS2 | + | − |
| GPSM1 (C-terminal 178 amino acids with 3 GPR motifs) | AGS3 | + | − |
| RGS12 (C-terminal 206 amino acids with GPR motif) | AGS6 | + | + |
| TFE3 (C-terminal 533 amino acids) | AGS11 | + | + |
| TFEB (C-terminal 320 amino acids) | AGS12 | − | + |
| MITF (C-terminal 304 amino acids) | AGS13 | + | + |
a cDNA libraries were screened in yeast strains CY1141 (Gαi3), CY8342 (Gαs), and CY9603 (Gα16).
FIGURE 1.
Bioactivity and diagram of AGSs isolated from mouse hypertrophic heart. In A and B, data are presented in three panels to illustrate the viability of the transformed yeast and the galactose-dependent growth under the selective pressure of exclusion of histidine from the medium. Galactose promotes the expression of each cDNA in the pYES2-containing GAL1 promoter. About 2000 cells were suspended in H2O and spotted on medium with glucose plus histidine (left; no selection), glucose minus histidine (center; selection without induction), or galactose plus histidine (right; selection plus induction). A, epistasis analysis of isolated clones. Transformants in a yeast strain expressing human Gα16 (Gpa1(1–41)) and yeast lacking Gα, Gβ, or downstream signaling molecules (ΔGα, yeast lacking Gα; ΔGβ, yeast lacking Gβ; ΔSte20, yeast lacking p21-activated kinase; ΔSte5, yeast lacking the kinase scaffold protein). B, effect of isolated cDNAs in yeast expressing various types of Gα. C, schematic diagram of the sequences of TFE3, TFEB, and MITF in mouse. The line above the sequence refers to cDNA isolated by the yeast-based functional screen. HLH, helix-loop-helix. D, bioactivity of full-length TFE3, TFEB, and MITF. The full-length clones were transformed into yeast expressing Gα16. The magnitude of activation of G-protein signaling pathway was monitored by β-galactosidase activity. Data are presented as the mean S.E. of five experiments with duplicate determinations. *, p < 0.05 versus non-induction group.
Three clones isolated from yeast expressing Gαi3 encoded the previously characterized proteins AGS2 (Dynlt1b, NCBI Reference Sequence NM_033368), AGS3 (GPSM1, NCBI Reference Sequence NM_700459), and AGS6 (RGS12, NCBI Reference Sequence NM_001156984). The cDNAs encoding AGS3 and AGS6 contained the G-protein-regulatory motif(s) that stabilizes the GDP-bound conformation of Gαi, transducin, and Gαο. An additional three cDNAs (1-8, 3-52, and 4-57) were isolated from yeast expressing Gα16. These three cDNAs exhibited bioactivity in yeast strains expressing Gα16 but not in yeast expressing Gαi3, Gαs, or Gpa1 (yeast Gα), indicating Gα selectivity (Fig. 1B and supplemental Text 1). We therefore focused on these Gα16-specific AGS cDNAs.
Gα16-specific AGS Proteins
Sequence analysis of the Gα16-specific cDNAs indicated that all encoded MITF/TFE transcription factors (31–33). cDNA1-8 encoded the C-terminal 533 amino acids of TFE3 (NCBI Reference Sequence NP_766060), cDNA3-52 encoded the C-terminal 320 amino acids of TFEB (NCBI Reference Sequence NP_035679), and cDNA4-57 encoded the C-terminal 304 amino acids of MITF (NCBI Reference Sequence NP_032627) (Fig. 1C). In accordance with the numbering of previously discovered AGS proteins (18), cDNA1-8, cDNA3-52, and cDNA4-57 were termed AGS11, AGS12, and AGS13, respectively (Table 1).
Full-length TFE3, TFEB, and MITF were cloned into a yeast expression vector, and the bioactivity for the G-protein signaling pathway was determined by β-galactosidase reporter assays (Fig. 1D). Full-length TFE3 and MITF, but not TFEB, activated the G-protein pathway in Gα16-expressing cells. Full-length TFE3, MITF, and TFEB did not activate growth of yeast expressing Gαs (supplemental Text 2). Immunoblot analysis indicated that the full-length proteins were expressed at the expected size and that their expression did not alter the levels of Gα16. These findings suggest that TFE3, MITF, and TFEB are transcription factors that act as receptor-independent G-protein activators. AGSs with various functions have been identified; however, no transcription factors have previously been described as AGS proteins.
Expression of TFE3, TFEB, and MITF in Cardiac Hypertrophy Models
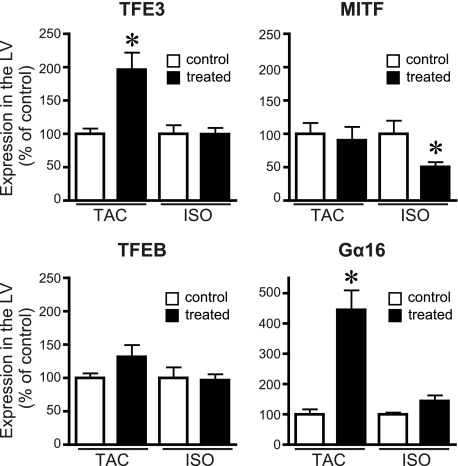
It was reported previously that the expression level of MITF was associated with development of cardiac hypertrophy in mouse (34). We sought to determine whether the three Gα16-specific AGS proteins were up-regulated in cardiac hypertrophy or were constitutively expressed in the myocardium. RNA expression of TFE3, MITF, TFEB, and the target Gα16 subunit was determined in the hypertrophied myocardium (Fig. 2). TFE3 mRNA expression was up-regulated in the left ventricle in the TAC model but not in the isoproterenol model. MITF was unchanged in the TAC model but reduced in the isoproterenol model. TFEB did not show any significant changes of expression in either model. Notably, Gα16 mRNA expression was also increased in the TAC model in which TFE3 was up-regulated. As TFE3 and Gα16 were both significantly up-regulated in the TAC model, we focused on the characterization of TFE3.
FIGURE 2.
Expression of MITF/TFE transcription factors and Gα16 in mouse cardiac hypertrophy model. The expression of mRNA of each gene was analyzed by real time PCR as described under “Experimental Procedures.” Control refers to the sham-operated or saline-infused mouse. Data are expressed as the -fold change in level compared with the control group. ISO, continuous infusion of isoproterenol; LV, left ventricle. Data are presented as the mean ± S.E. of five experiments with duplicate determinations. *, p < 0.05 versus control group.
Formation of TFE3-Gα16 Complex in Cells
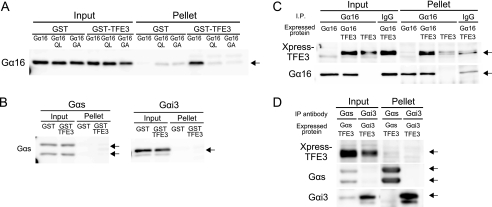
The above findings suggested that TFE3 plays an important role via Gα16 in the development of cardiac hypertrophy. We thus examined whether TFE3 indeed was able to form a complex with Gα16. As a first approach, the interaction of GST-tagged TFE3 (GST-TFE3) with Gα16 was examined in vitro. GST-TFE3 successfully pulled down transfected Gα16 from cell lysates. However, neither a constitutively active mutant of Gα16 (Gα16Q212L) nor an inactive mutant of Gα16 (Gα16G211A) was pulled down, suggesting that the interaction of Gα16 and TFE3 was dependent upon the conformation of Gα16 and regulated by guanine nucleotide binding (Fig. 3A) (35, 36). In contrast, GST-TFE3 did not pull down transfected Gαs or Gαi3 from cell lysates (Fig. 3B). We also examined whether TFE3 interacted with Gα16 in mammalian cells. Expressed TFE3 was co-immunoprecipitated with Gα16 from COS7 cell lysates, suggesting that TFE3 and Gα16 formed a stable complex within these cells (Fig. 3C). In contrast, TFE3 did not co-immunoprecipitate with Gαs or Gαi3 (Fig. 3D). We next examined the role of this interaction in Gα16-mediated signaling events.
FIGURE 3.
Interaction of TFE3 with Gα16in vitro and in cell. A and B, GST pulldown assay of TFE3 with COS7 lysate expressing various Gα subunits. The C-terminal 533-amino acid fragment of TFE3 was expressed as a GST fusion protein (GST-TFE3). GST-TFE3 (300 nm) was incubated with 1 mg of cell lysate in a total volume of 500 μl at 4 °C. Lysates of COS7 cells were prepared as described under “Experimental Procedures” following transfection of 10 μg of the Gα subunit in pcDNA3. C and D, COS7 cells in a 100-mm dish were transfected with a combination of pcDNA3, pcDNA3::Gα16 (5 μg/dish), and pcDNA3.1-His::TFE3 (5 μg/dish). The amount of DNA transfected was adjusted to 10 μg/well with the pcDNA3 vector. The preparation of a whole-cell lysate including the nuclear fraction and immunoprecipitation (IP) were performed as described under “Experimental Procedures.” The Gα subunit was immunoprecipitated with a specific antibody for each Gα subunit. QL, Gα16Q212L; GA, Gα16G211A.
TFE3 Is Not Involved in Receptor-mediated Gα16 Signaling
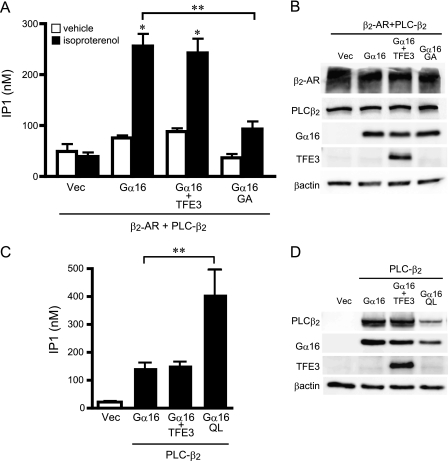
Gα16 is coupled to multiple GPCRs including β2-adrenergic receptors mediating signal transfer to the effector molecule PLC-β (37, 38). Thus, we examined whether TFE3 regulated β2-adrenergic receptor-mediated PLC-β2 activation as a representative of Gα16-mediated signaling (39). In a transient expression system in COS7 cells, Gα16 activated PLC-β2 following β2-adrenergic receptor stimulation as determined by inositol monophosphate production (Fig. 4). The magnitude of PLC-β2 activation was reduced in the presence of an inactive Gα16 mutant (Gα16G211A), indicating that PLC-β2 activation was mediated by Gα16 (Fig. 4, A and B). However, TFE3 overexpression did not alter this receptor-mediated Gα16 signaling. We also examined the effect of TFE3 overexpression on the basal activity of PLC-β2/Gα16 in the absence of receptor stimulation. TFE3 overexpression did not alter PLC-β2 activity, whereas a constitutively active mutant of Gα16 (Gα16Q212L) increased the activity even in the absence of receptor stimulation (Fig. 4, C and D). These data are consistent with a lack of TFE3 involvement in regulating the conventional GPCR-mediated Gα16 signaling pathway.
FIGURE 4.
Effect of TFE3 on activation of phospholipase C-β2. A, effect of TFE3 on the generation of inositol phosphate (IP1) following receptor stimulation. COS7 cells were transfected in 12-well plates with control vectors (Vec) or cDNAs as indicated (0.4 μg of pcDNA::PLC-β2, 0.5 μg of pcDNA::TFE3, 0.5 μg of pcDNA::Gα16, and 0.6 μg of pEGFP::β2-adrenergic receptor (AR)). The amount of transfected DNA was adjusted to 2 μg/well with the pcDNA vector. Cells were stimulated with 10 μm isoproterenol for 30 min and assayed immediately. Data are expressed as the mean ± S.E. of five experiments with duplicate determinations. B, expression of transfected proteins of A. The expression of each protein was determined by immunoblotting of 10 μg of whole-cell lysates. C, effect of TFE3 on the generation of inositol phosphate. COS7 cells were transfected in 12-well plates with control vectors or cDNAs as indicated (0.5 μg of pcDNA::PLC-β2, 0.75 μg of pcDNA::TFE3, and 0.75 μg of pcDNA::Gα16). The amount of transfected DNA was adjusted to 2 μg/well with the pcDNA vector. Data are expressed as the mean ± S.E. of five experiments with duplicate determinations. D, expression of transfected cDNA of C. The expression of each protein was determined by immunoblotting of 10 μg of whole-cell lysates. *, p < 0.05 versus control group; **, p < 0.05 between two groups. QL, Gα16Q212L; GA, Gα16G211A.
TFE3 Induces Accumulation of Gα16 in Nucleus
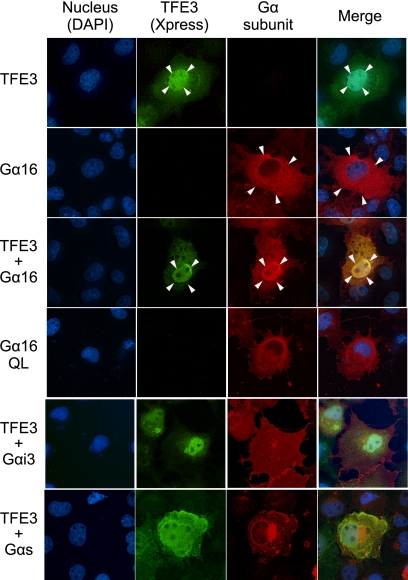
The identification of transcription factors as Gα16-specific AGS proteins suggested that MITF/TFE transcription factors may interact with a subpopulation of Gα16 distinct from that involved in the conventional G-protein signaling at the plasma membrane. To address this issue, we first examined the subcellular distribution of Gα16 and TFE3 when each was independently overexpressed in the cell. Overexpressed TFE3 was predominantly found in the nucleus as expected, whereas Gα16 was found in the plasma membrane and cytoplasm but not in the nucleus (Fig. 5, arrow, and supplemental Fig. 2, A, B, and D). However, when Gα16 and TFE3 were overexpressed together, Gα16 predominantly accumulated in the nucleus (Fig. 5, arrow). This novel nuclear translocation of Gα16 was not due to Gα16 activation because the constitutively active mutant of Gα16 (Gα16Q212L) was not found in the nucleus when it was overexpressed by itself. These data suggested that Gα16 forms a complex with TFE3 and translocates to the nucleus. Nuclear accumulation of G-protein by TFE3 was not observed for Gαi3 or Gαs.
FIGURE 5.
Localization of expressed Gα subunits and TFE3 in COS7 cells. COS7 cells were transfected in a 35-mm dish with 2.0 μg of Gα subunits in pcDNA3 and/or 2.0 μg of pcDNA3.1-His::TFE3. The amount of transfected DNA was adjusted to 4 μg/well with the pcDNA3 vector. The Gα subunit and TFE3 were determined using a specific antibody for each Gα (red) or Xpress antibody (green), respectively. QL, Gα16Q212L.
Up-regulation of Claudin 14 mRNA by TFE3-Gα16 Complex
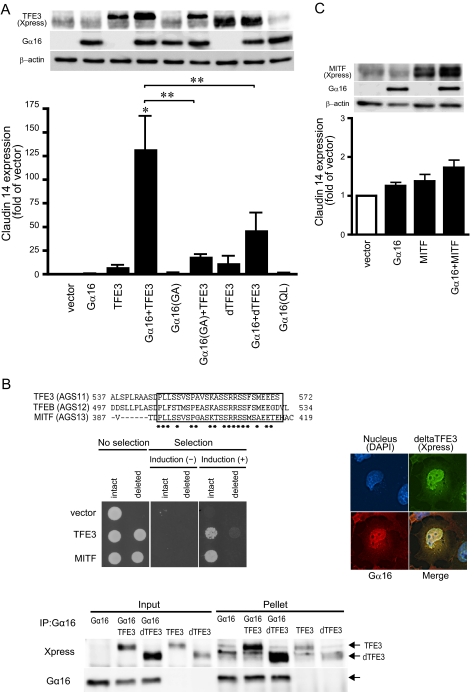
The co-localization of TFE3 and Gα16 suggested an involvement of a nuclear TFE3-Gα16 complex in regulating the expression of particular genes. To address this issue, genes regulated by TFE3 and Gα16 were screened by microarray analysis of mRNA of HEK293 cells transfected with TFE3 and/or Gα16. In the screening of more than 40,000 human genes, we found that claudin 14 mRNA was highly up-regulated by the simultaneous transfection of TFE3 and Gα16. Parallel experiments indicated that the co-overexpression of TFE3 and Gα16 in HEK293 cells increased claudin 14 mRNA by 133-fold, whereas independent overexpression of TFE3 (8.3-fold) or Gα16 (1.0-fold) had minimal effect on the induction of claudin 14 (Fig. 6A). The induction of claudin 14 was significantly decreased in the presence of the inactive mutant of Gα16 (Gα16G211A) compared with wild type Gα16, suggesting that Gα16 activation was also required for the induction of this gene.
FIGURE 6.
Effect of TFE3 or MITF on expression of claudin 14. A, expression of claudin 14 in transfected HEK293 cells. HEK293 cells were transfected in 6-well plates with a combination of control vectors or cDNAs as indicated (2.0 μg of Gα subunits in pcDNA3 and 2.0 μg of TFE3 or delTFE3 in pcDNA3.1-His). The amount of transfected DNA was adjusted to 4 μg/well with the pcDNA3 vector. The expression of claudin 14 mRNA was analyzed by real time PCR. Data are expressed as the -fold change from the level of claudin 14 expression in control cells transfected with the vector alone. Data are expressed as the mean ± S.E. of five experiments with duplicate determinations. Upper inset, expression of proteins determined by immunoblotting (∼10 μg of whole-cell lysate). Data are representative of five experiments. *, p < 0.05 versus control group; **, p < 0.05 between two groups. B, effect of delTFE3. Upper panel, amino acid sequence of C-terminal MITF/TFE transcription factors. The square indicates conserved amino acid sequence. *, consensus amino acid. Middle left panel, bioactivity of intact or deleted TFE3 in yeast expressing Gα16. The assay was performed as described under “Experimental Procedures.” Middle right panel, localization of transfected Gα16 and TFE3 in COS7 cells. COS7 cells were transfected in a 35-mm dish with 2.0 μg of pcDNA3::Gα16 and 2.0 μg of pcDNA3.1-His::TFE3. Gα16 and TFE3 were determined using Gα16 antibody (red) or Xpress antibody (green), respectively. Lower panel, interaction of Gα16 with TFE3 or delTFE3. COS7 cells in a 100-mm dish were transfected with a combination of pcDNA3, pcDNA3::Gα16 (5 μg/dish), pcDNA3.1-His::TFE3 (5 μg/dish), and pcDNA3.1-His::delTFE3 (5 μg/dish). The amount of transfected DNA was adjusted to 10 μg/well with the pcDNA3 vector. The preparation of the cell lysate and immunoprecipitation (IP) were performed as described under “Experimental Procedures”. C, effect of MITF on claudin 14 expression in transfected HEK293 cells. HEK293 cells were transfected in 6-well plates with a combination of control vectors or cDNAs as indicated (2.0 μg of pcDNA3::Gα16 and 2.0 μg of pcDNA3.1-His::MITF). The amount of transfected DNA was adjusted to 4 μg/well with the pcDNA3 vector. The expression of claudin 14 mRNA was analyzed by real time PCR. Data are expressed as the -fold change in the level of claudin 14 in control cells transfected with the vector alone. Data are expressed as the mean ± S.E. of five experiments with duplicate determinations. Upper inset, expression of proteins determined by immunoblotting (∼10 μg of whole-cell lysate). Data are representative of five experiments. QL, Gα16Q212L; GA, Gα16G211A.
Requirement of Gα16 Activation for Gene Induction by TFE3
The requirement of Gα16 activation for this gene induction was further characterized utilizing a truncated mutant of TFE3 (delTFE3), which showed less bioactivity for Gα16 activation in the yeast system. Analysis of the amino acid sequences of the MITF/TFE family indicated that the C-terminal 27 acids were conserved among the Gα16-selective AGS proteins (Fig. 6B, upper panel). Deletion of the C-terminal 27 amino acids resulted in the loss of bioactivity of TFE3 and MITF for G-protein activation (Fig. 6B, left middle panel, and supplemental Text 3). Despite the loss of bioactivity for Gα16 activation, delTFE3 was still able to form a complex with Gα16 and induce the translocation of Gα16 to the nucleus (Fig. 6B, left lower and right panels, and supplemental Fig. 2, C and D). Thus, nuclear translocation by itself did not require Gα16 activation as long as TFE3 and Gα16 formed a complex (Fig. 6A).
Although the delTFE3-Gα16 complex was found in the nucleus, the subsequent up-regulation of claudin 14 was blunted, suggesting that Gα16 activation is critical for this gene induction (Fig. 6A). Furthermore, the constitutively active mutant of Gα16 (Gα16Q212L), which was not expressed in the nucleus (Fig. 5), failed to induce claudin 14. MITF, which had a similar ability to activate Gα16 (Fig. 1D), failed to induce claudin 14 (Fig. 6C). Taken together, the results suggest that in addition to the nuclear translocation of a TFE3-Gα16 complex activation of Gα16 in the nucleus was required for the induction of claudin 14.
Regulation of Claudin 14 Expression in Cardiomyocytes
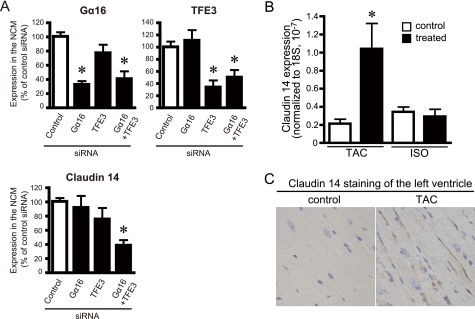
The influence of Gα16 and TFE3 on the expression of claudin 14 was also examined in neonatal cardiomyocytes following knockdown of Gα16 and/or TFE3 by siRNA. Gα16siRNA or TFE3siRNA successfully suppressed the level of target molecules to 33–34% of the level of cardiomyocytes treated with negative control siRNA (Fig. 7A). The level of claudin 14 in cardiomyocytes was not influenced by Gα16siRNA or TFE3siRNA itself when they were separately introduced (Fig. 7A, lower panel). However, interestingly, the simultaneous knockdown of Gα16 and TFE3 by siRNAs significantly reduced the claudin 14 mRNA (38.9 ± 7.4%, p < 0.05 versus control siRNA), indicating that both Gα16 and TFE3 were required for the regulation claudin 14 expression. These results are consistent with the data observed in HEK293 cells.
FIGURE 7.
Expression of claudin 14 in cultured cardiomyocytes and hypertrophied heart. A, effect of knockdown of Gα16 and TFE3 on the level of claudin 14 mRNA in cultured cardiomyocytes. Neonatal cardiomyocytes (NCM) were transfected with each siRNA and/or universal control siRNA (Stealth RNAi Negative Control, Invitrogen). Forty-eight hours after transfection, the level of mRNA of Gα16 (A), TFE3 (B), and claudin 14 (C) were analyzed by real time PCR. Transfection efficiency of siRNA was estimated at 70–80% using FITC-labeled double strand RNA (Block It Fluorescent Oligo, Invitrogen) (right panel). *, p < 0.05 versus negative siRNA. Data are expressed as the mean ± S.E. of seven to eight independent experiments. B and C, expression of claudin 14 in the mouse cardiac hypertrophy model. B, the left ventricular expression of claudin 14 mRNA was analyzed by real time PCR. Control refers to the sham-operated or saline-infused mouse. Data are expressed as the -fold change in claudin 14 level from that in control group. Data are expressed as the mean ± S.E. of five experiments with duplicate determinations. C, immunohistochemical staining for claudin 14 (1:100; brown) of the left ventricle of sham- or TAC-operated mouse. A frozen section (8 μm) of the mouse heart was subjected to immunohistochemical staining as described under “Experimental Procedures”. Blue, nucleus. ISO, continuous infusion of isoproterenol. *, p < 0.05 versus control group.
Up-regulation of Claudin 14 in Mouse Heart upon Pressure Overload Stress
As TFE3 and Gα16 were simultaneously up-regulated in the left ventricle in the TAC model (Fig. 2), we examined whether ventricular claudin 14 was also up-regulated in the models of cardiac hypertrophy. Quantitative PCR analysis indicated that claudin 14 mRNA was increased 5-fold in the left ventricle in the TAC model but not in the isoproterenol model of cardiac hypertrophy (Fig. 7B), consistent with the expression profile of TFE3 and Gα16 in these stimulated models (Fig. 2). Immunocytochemical analysis indicated that expression of claudin 14 was increased in the lateral membrane of cardiomyocytes rather than the intercalated disks (Fig. 7C). Thus, similar to our findings in cultured cells, the simultaneous up-regulation of TFE3 and Gα16 was associated with gene induction of claudin 14 in vivo under pathologic conditions. Gene induction by Gα16 and TFE3 is therefore postulated to be part of the cardiac adaptation process to pressure overload stress.
DISCUSSION
We report the identification of three MITF/TFE transcription factors, TFE3, MITF, and TFEB, as new AGS proteins selective for the Gα16 subunit. These factors belong to the Myc supergene family of basic helix-loop-helix leucine zipper transcription factors that act either as a homo- or heterodimer within the family members (31–33). TFE3 formed a complex with and activated Gα16 in cells. Formation of TFE3-Gα16 complex resulted in the translocation of Gα16 to the nucleus and up-regulation of the cell junction protein claudin 14. Expression of claudin 14 was also induced in vivo in the hypertrophied ventricle, and this was associated with the up-regulation of Gα16 and TFE3. Thus, the transcription factor TFE3 is postulated to act as a G-protein activator for the Gα16 subunit and regulate gene induction in response to pathophysiologic stress.
Although an increasing body of data implicates heterotrimeric G-proteins and their regulators as key regulators in multiple cellular events (40, 41), this is the first demonstration that activation of a Gα subunit by an AGS drives relocalization of Gα to the nucleus and gene transcription in mammalian cells. Previous studies reported that heterotrimeric Gβ5 translocated to the nucleus when complexed with RGS7 (16). However, the effect of RGS7-Gβ5 on gene regulation has not yet been characterized. This study is the first to demonstrate a direct effect of nuclear translocation of a Gα subunit on specific gene regulation.
The magnitude of gene induction by TFE3-Gα16 was clearly dependent on the guanine nucleotide binding status of Gα16 as well as the bioactivity of TFE3 for Gα16 activation. Activation of Gα16 in the cytosol or plasma membrane was not sufficient to induce claudin 14 expression because a constitutively active Gα16 in the cytosol and plasma membrane failed to induce claudin 14 expression. Conversely, translocation of Gα16 to the nucleus by the delTFE3, which lacked the ability to activate Gα16, showed a blunted induction of claudin 14 as compared with intact TFE3. These observations suggest that TFE3-mediated activation of Gα16 within the nucleus is essential to induce claudin 14 expression. TFE3 may serve as a direct guanine nucleotide exchange factor for Gα16 upon complex formation. Alternatively, Gα16 may be activated in the nucleus following removal or addition of a factor to the TFE3 complex when it is translocated into the nucleus.
The up-regulation of claudin 14 reported in this study may be an important event in remodeling of the heart following pressure overload stress. Claudin 14 was expressed in the lateral membrane of cardiomyocytes and was increased upon pressure overload stress. Claudin 14 is a member of the claudin family of more than 20 highly conserved proteins (42–44). It is interesting that the overexpression of claudin 14 induces apoptosis of cells independently of the caspase-mediated pathway (45). Moreover, in addition to its barrier function, claudin is also involved in activating pro-matrix metalloproteinase 2, which plays a role in reorganization of the extracellular matrix (46). Accordingly, the claudin-mediated sealing and/or molecular remodeling of the lateral region where cardiomyocytes are associated with the basal lamina or extracellular matrix is important for adaptation to mechanical stress. Indeed, changes in the expression of claudin 5 have been reported in the lateral membrane of cardiomyocytes in a dystrophic mouse with dilated cardiomyopathy (47, 48).
It is possible that the transcription factor MITF/TFE acts as a heterologous protein complex and binds to promoter regions to regulate the transcription of claudin 14. TFE3-Gα16 may be required to assemble such a transcriptional complex, leading to increased transcription. Alternatively, TFE3-Gα16 may regulate nuclear PLC-β activity and the nuclear phosphoinositide cycle independently of the plasma membrane phosphoinositide cycle influencing cell cycle and cell differentiation (49). Activation of PLC-β in the nucleus is not usually detectable in whole-cell experiments as used in this study (Fig. 4).
This is the first report of a regulatory protein for the Gα16 subunit, which can be coupled to multiple GPCRs in a variety of experimental systems (37, 38). Although Gα16 is enriched in hematopoietic tissue, it is also expressed in other tissues including heart (50, 51) where its expression is increased 4-fold by the cardiac stress induced in the TAC animal model. It is of particular interest to find that this multifunctional Gα16 is translocated into the nucleus by a specific G-protein regulator where it plays a previously unappreciated functional role.
Various AGS proteins are involved in adaptation to various pathologic conditions (3, 20). For example, we previously identified AGS8 as a novel regulatory protein for the Gβγ subunit in a repetitive transient ischemia model in the rat heart (18). AGS8 was up-regulated in the myocardium by ischemic/hypoxic stress and played a critical role in hypoxia-induced apoptosis of cardiomyocytes (18, 24). Our ability to rapidly identify AGS8 and now TFE3 directly from disease-specific mRNA libraries using our yeast-based functional screen highlights its usefulness in discovering disease-specific regulatory proteins for heterotrimeric G-proteins. Such disease-specific or adaptation-specific regulatory proteins represent novel therapeutic targets in treating human diseases.
Supplementary Material
Acknowledgments
We acknowledge Dr. James R. Broach (Molecular Biology, Princeton University, Princeton, NJ) and Cadus Pharmaceutical Corp. (New York, NY) for providing yeast strains used in this study. We thank Dr. Kazuhiro Ogata (Biochemistry and Gene Regulation, Yokohama City University) for helpful comments.
This work was supported, in whole or in part, by National Institutes of Health Grants NS24821 and DA025896 (both to S. M. L.). This work was also supported by Grants-in-aid for Scientific Research (C) 18599006 and 20590212, the Yokohama Foundation for Advancement of Medical Science, and Strategic Research Project Grants K18017 and K19021 from Yokohama City University, Japan (all to M. S.) and by grants from the Ministry of Health Labor and Welfare; Ministry of Education, Culture, Sports, Science and Technology of Japan; Takeda Science Foundation; Cosmetology Research Foundation; and Kitsuen Research Foundation (all to Y. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Text 1–3.
- GPCR
- G-protein coupled receptor
- TFE
- transcription factor E
- MITF
- microphthalmia-associated transcription factor
- TFEB
- transcription factor EB
- AGS
- activators of G-protein signaling
- RGS
- regulator of G protein signaling
- TAC
- transverse aortic constriction
- PLC
- phospholipase C.
REFERENCES
- 1. Birnbaumer L. (2007) Biochim. Biophys. Acta 1768, 772–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oldham W. M., Hamm H. E. (2006) Q. Rev. Biophys. 39, 117–166 [DOI] [PubMed] [Google Scholar]
- 3. Sato M., Blumer J. B., Simon V., Lanier S. M. (2006) Annu. Rev. Pharmacol. Toxicol. 46, 151–187 [DOI] [PubMed] [Google Scholar]
- 4. Cismowski M. J. (2006) Semin. Cell Dev. Biol. 17, 334–344 [DOI] [PubMed] [Google Scholar]
- 5. Blumer J. B., Smrcka A. V., Lanier S. M. (2007) Pharmacol. Ther. 113, 488–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Riddle E. L., Schwartzman R. A., Bond M., Insel P. A. (2005) Circ. Res. 96, 401–411 [DOI] [PubMed] [Google Scholar]
- 7. Tall G. G., Gilman A. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16584–16589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee M. J., Dohlman H. G. (2008) Curr. Biol. 18, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Marcos M., Ghosh P., Farquhar M. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3178–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willars G. B. (2006) Semin. Cell Dev. Biol. 17, 363–376 [DOI] [PubMed] [Google Scholar]
- 11. Du Q., Stukenberg P. T., Macara I. G. (2001) Nat. Cell Biol. 3, 1069–1075 [DOI] [PubMed] [Google Scholar]
- 12. Gotta M., Dong Y., Peterson Y. K., Lanier S. M., Ahringer J. (2003) Curr. Biol. 13, 1029–1037 [DOI] [PubMed] [Google Scholar]
- 13. Sanada K., Tsai L. H. (2005) Cell 122, 119–131 [DOI] [PubMed] [Google Scholar]
- 14. Blumer J. B., Kuriyama R., Gettys T. W., Lanier S. M. (2006) Eur. J. Cell Biol. 85, 1233–1240 [DOI] [PubMed] [Google Scholar]
- 15. Shu F. J., Ramineni S., Amyot W., Hepler J. R. (2007) Cell. Signal. 19, 163–176 [DOI] [PubMed] [Google Scholar]
- 16. Hepler J. R. (2005) Sci. STKE 2005, pe38. [DOI] [PubMed] [Google Scholar]
- 17. Sato M., Gettys T. W., Lanier S. M. (2004) J. Biol. Chem. 279, 13375–13382 [DOI] [PubMed] [Google Scholar]
- 18. Sato M., Cismowski M. J., Toyota E., Smrcka A. V., Lucchesi P. A., Chilian W. M., Lanier S. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blumer J. B., Lord K., Saunders T. L., Pacchioni A., Black C., Lazartigues E., Varner K. J., Gettys T. W., Lanier S. M. (2008) Endocrinology 149, 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato M., Ishikawa Y. (2010) Pathophysiology 17, 89–99 [DOI] [PubMed] [Google Scholar]
- 21. Hendriks-Balk M. C., Peters S. L., Michel M. C., Alewijnse A. E. (2008) Eur. J. Pharmacol. 585, 278–291 [DOI] [PubMed] [Google Scholar]
- 22. Heximer S. P., Srinivasa S. P., Bernstein L. S., Bernard J. L., Linder M. E., Hepler J. R., Blumer K. J. (1999) J. Biol. Chem. 274, 34253–34259 [DOI] [PubMed] [Google Scholar]
- 23. Rogers J. H., Tamirisa P., Kovacs A., Weinheimer C., Courtois M., Blumer K. J., Kelly D. P., Muslin A. J. (1999) J. Clin. Investig. 104, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sato M., Jiao Q., Honda T., Kurotani R., Toyota E., Okumura S., Takeya T., Minamisawa S., Lanier S. M., Ishikawa Y. (2009) J. Biol. Chem. 284, 31431–31440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hill J. A., Karimi M., Kutschke W., Davisson R. L., Zimmerman K., Wang Z., Kerber R. E., Weiss R. M. (2000) Circulation 101, 2863–2869 [DOI] [PubMed] [Google Scholar]
- 26. Cismowski M. J., Takesono A., Ma C., Lizano J. S., Xie X., Fuernkranz H., Lanier S. M., Duzic E. (1999) Nat. Biotechnol. 17, 878–883 [DOI] [PubMed] [Google Scholar]
- 27. Takesono A., Cismowski M. J., Ribas C., Bernard M., Chung P., Hazard S., 3rd, Duzic E., Lanier S. M. (1999) J. Biol. Chem. 274, 33202–33205 [DOI] [PubMed] [Google Scholar]
- 28. Cismowski M. J., Takesono A., Ma C., Lanier S. M., Duzic E. (2002) Methods Enzymol. 344, 153–168 [DOI] [PubMed] [Google Scholar]
- 29. Sato M., Ribas C., Hildebrandt J. D., Lanier S. M. (1996) J. Biol. Chem. 271, 30052–30060 [DOI] [PubMed] [Google Scholar]
- 30. Sato M., Kataoka R., Dingus J., Wilcox M., Hildebrandt J. D., Lanier S. M. (1995) J. Biol. Chem. 270, 15269–15276 [DOI] [PubMed] [Google Scholar]
- 31. Hodgkinson C. A., Moore K. J., Nakayama A., Steingrímsson E., Copeland N. G., Jenkins N. A., Arnheiter H. (1993) Cell 74, 395–404 [DOI] [PubMed] [Google Scholar]
- 32. Hughes M. J., Lingrel J. B., Krakowsky J. M., Anderson K. P. (1993) J. Biol. Chem. 268, 20687–20690 [PubMed] [Google Scholar]
- 33. Steingrímsson E., Copeland N. G., Jenkins N. A. (2004) Annu. Rev. Genet. 38, 365–411 [DOI] [PubMed] [Google Scholar]
- 34. Tshori S., Gilon D., Beeri R., Nechushtan H., Kaluzhny D., Pikarsky E., Razin E. (2006) J. Clin. Investig. 116, 2673–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heasley L. E., Storey B., Fanger G. R., Butterfield L., Zamarripa J., Blumberg D., Maue R. A. (1996) Mol. Cell. Biol. 16, 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou J., Stanners J., Kabouridis P., Han H., Tsoukas C. D. (1998) Eur. J. Immunol. 28, 1645–1655 [DOI] [PubMed] [Google Scholar]
- 37. Huang J., Wilkie T. M. (2006) UCSD-Nature Molecule Pages 10.1038/mp.a000972.01 [DOI] [Google Scholar]
- 38. Offermanns S., Simon M. I. (1995) J. Biol. Chem. 270, 15175–15180 [DOI] [PubMed] [Google Scholar]
- 39. Wu D., Kuang Y., Wu Y., Jiang H. (1995) J. Biol. Chem. 270, 16008–16010 [DOI] [PubMed] [Google Scholar]
- 40. Burchett S. A. (2003) J. Neurochem. 87, 551–559 [DOI] [PubMed] [Google Scholar]
- 41. Spiegelberg B. D., Hamm H. E. (2007) Curr. Opin. Genet. Dev. 17, 40–44 [DOI] [PubMed] [Google Scholar]
- 42. Tsukita S., Furuse M. (2002) Curr. Opin. Cell Biol. 14, 531–536 [DOI] [PubMed] [Google Scholar]
- 43. Wilcox E. R., Burton Q. L., Naz S., Riazuddin S., Smith T. N., Ploplis B., Belyantseva I., Ben-Yosef T., Liburd N. A., Morell R. J., Kachar B., Wu D. K., Griffith A. J., Riazuddin S., Friedman T. B. (2001) Cell 104, 165–172 [DOI] [PubMed] [Google Scholar]
- 44. Ben-Yosef T., Belyantseva I. A., Saunders T. L., Hughes E. D., Kawamoto K., Van Itallie C. M., Beyer L. A., Halsey K., Gardner D. J., Wilcox E. R., Rasmussen J., Anderson J. M., Dolan D. F., Forge A., Raphael Y., Camper S. A., Friedman T. B. (2003) Hum. Mol. Genet. 12, 2049–2061 [DOI] [PubMed] [Google Scholar]
- 45. Hu Y., Lehrach H., Janitz M. (2010) Mol. Biol. Rep. 37, 3381–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miyamori H., Takino T., Kobayashi Y., Tokai H., Itoh Y., Seiki M., Sato H. (2001) J. Biol. Chem. 276, 28204–28211 [DOI] [PubMed] [Google Scholar]
- 47. Sanford J. L., Edwards J. D., Mays T. A., Gong B., Merriam A. P., Rafael-Fortney J. A. (2005) J. Mol. Cell. Cardiol. 38, 323–332 [DOI] [PubMed] [Google Scholar]
- 48. Mays T. A., Binkley P. F., Lesinski A., Doshi A. A., Quaile M. P., Margulies K. B., Janssen P. M., Rafael-Fortney J. A. (2008) J. Mol. Cell. Cardiol. 45, 81–87 [DOI] [PubMed] [Google Scholar]
- 49. Irvine R. F. (2002) Sci. STKE 2002, re13. [DOI] [PubMed] [Google Scholar]
- 50. Wilkie T. M., Scherle P. A., Strathmann M. P., Slepak V. Z., Simon M. I. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10049–10053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Giannone F., Malpeli G., Lisi V., Grasso S., Shukla P., Ramarli D., Sartoris S., Monsurró V., Krampera M., Amato E., Tridente G., Colombatti M., Parenti M., Innamorati G. (2010) J. Mol. Endocrinol. 44, 259–269 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.