Abstract
DNA double-strand breaks (DSB), particularly those induced by ionizing radiation (IR), are complex lesions that can be cytotoxic if not properly repaired. IR-induced DSB often have DNA termini modifications, including thymine glycols, ring fragmentation, 3′-phosphoglycolates, 5′-hydroxyl groups, and abasic sites. Nonhomologous end joining (NHEJ) is a major pathway responsible for the repair of these complex breaks. Proteins involved in NHEJ include the Ku 70/80 heterodimer, DNA–PKcs, processing proteins including Artemis and DNA polymerases μ and λ, XRCC4, DNA ligase IV, and XLF. We will discuss the role of the physical and functional interactions of DNA–PK as a result of activation, with an emphasis on DNA structure, chemistry, and sequence. With the diversity of IR induced DSB, it is becoming increasingly clear that multiple DNA processing enzymes are likely necessary for effective repair of a break. We will explore the roles of several important processing enzymes, with a focus on the nuclease Artemis and its role in processing diverse DSB. The effect of DNA termini on both DNA–PK and Artemis activity will be analyzed from a structural and biochemical view. Antioxid. Redox Signal. 14, 2531–2543.
DNA Double-Strand Breaks
DNA double-strand breaks (DSB) and their contribution to genomic instability, chromosomal translocations, carcinogenesis, and cell death have been extensively studied over the years. These breaks can arise from both endogenous and exogenous sources. Endogenous DSB can occur from reactive oxygen species that create dual single-strand break lesions in close proximity to each other (Fig. 1A). DSB may also arise from replication fork stalling that leads to fork collapse or attempts to replicate past a nick in a leading strand template (Fig. 1B) (47). In addition, certain genomic recombination events, including V(D)J recombination, induce DSB through endonuclease processing (24). Finally, endogenous DSB can result from physical stress that occurs during separation of chromosomes in mitosis (22). DSB can also be produced from a variety of exogenous DNA damaging agents, such as ionizing radiation (IR) and certain chemotherapeutic agents like bleomycin and camptothecin (Fig. 1C) (75).
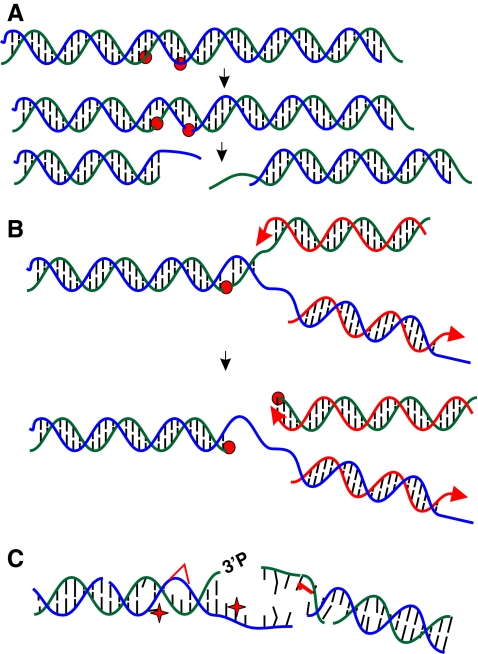
FIG. 1.
DNA double-strand breaks (DSB). (A) Single-strand breaks (SSB) arising from reactive oxygen species (ROS) that are located in close proximity to each other in the genome can lead to a DNA DSB. SSB locations are indicated by red circles. (B) DNA polymerase-driven attempts to replicate past a nick in the leading strand template of DNA can result in a DNA DSB. Nicks are indicated by red circles and newly replicated DNA is indicated by red strands. (C) Exogenous DNA damaging agents like ionizing radiation (IR) produce DNA DSB with a variety of end modifications and various forms of DNA damage around the DNA terminus. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Ionizing Radiation-Induced DNA Double-Strand Breaks
DNA double-strand breaks produced by ionizing radiation typically do not have blunt unmodified termini. Instead, the DNA termini can have a variety of end modifications and base damages. It has been suggested that many of these DNA modifications can occur in a clustered region, potentially near the site of the initial break, and the presence of these multiple lesions increases the mutation rate that arise from IR (32, 85). Identified DNA modifications include thymine glycols, ring fragmentation, 3′-phosphoglycolates, 5′-hydroxyl groups, and abasic sites. Regions of single-strand DNA that arise from strand breakage can also occur at a DSB, leaving a single-strand overhang region at the site of the break. This diversity of damage and structures at the site of DNA DSB are likely to impact rate and accuracy of repair. As the structural complexity found at the site of DSB increases, the ability of repair decreases (74). It is becoming increasingly apparent that the assortment of secondary DNA lesions found at the site of an IR-induced break presents challenges for their repair. Due to the complexities in the DNA lesions produced by IR, one could imagine that different enzymes or pathways would be required to process different types of DNA lesions found at termini towards the joining or resolution of DNA DSB.
Repairing DNA DSBs
Two major pathways have evolved that catalyze the repair of DNA DSB, homology directed repair (HDR) and nonhomologous end joining (NHEJ). HDR only occurs in S or G2 phase of the cell cycle in mammalian cells, as it requires a homologous template. Importantly, specific DNA damage may be retained in HDR and require further repair or processing following initial HDR. NHEJ repairs DSB by simply joining the two DNA ends together in the absence of a homologous template, and is therefore capable of occurring throughout the cell cycle. While in theory a simple mechanism, continuing research is showing that the joining of two nonhomologous DNA ends by NHEJ is, in fact, a sophisticated and complex mechanism of DNA repair.
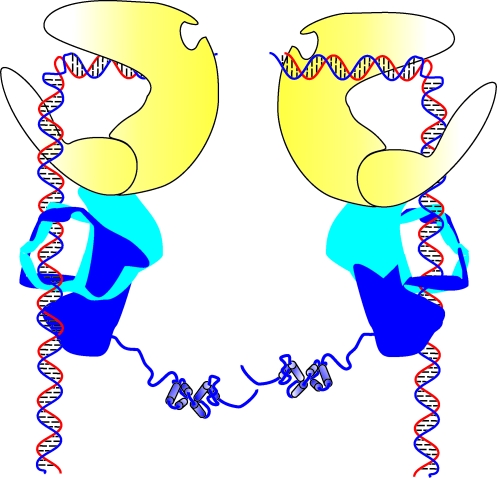
NHEJ can be divided into four specific steps; DNA termini recognition, bridging of the two DNA ends to be joined, also known as formation of the synaptic complex, DNA end processing, and finally DNA ligation (Fig. 2). After a DSB occurs, Ku, a heterodimer composed of Ku70 and Ku80, binds to the end of the break. Once Ku is bound, it recruits the 465 kDa DNA–PK catalytic subunit (DNA–PKcs). Together, these proteins make up a heterotrimeric complex called the DNA-dependent protein kinase, or DNA–PK. The formation of this complex may aid in stabilizing the two DNA ends at the site of the break, forming a synaptic complex that secures the two DNA termini together. The catalytic activity of DNA–PK is activated once bound to DNA, and this unique serine/threonine protein kinase phosphorylates downstream target proteins needed for completion of the pathway.
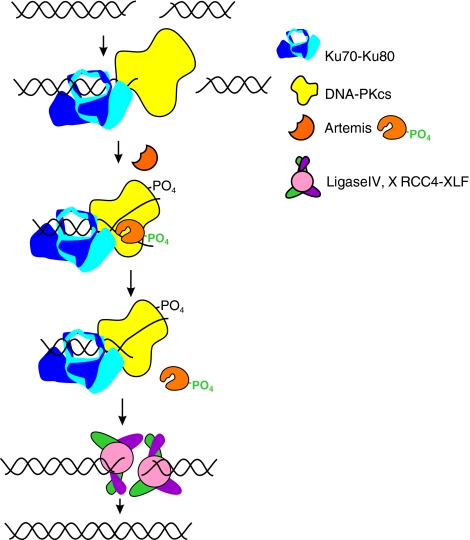
FIG. 2.
The Non-Homologous End Joining (NHEJ) pathway. Following a DNA DSB induced by IR, the heterodimeric Ku (Ku 70 depicted in light blue and Ku 80 depicted in dark blue) complex is recruited to the DNA terminus, binds to the DNA, and recruits the DNA-dependent protein kinase catalytic subunit (DNA–PKcs). DNA–PKcs forms a heterotrimeric complex with Ku and its serine/theonine protein kinase activity is activated once bound to the DNA terminus. Autophosphorylation and phosphorylation of other target proteins occurs. Artemis, in the presence of DNA–PK and ATP, becomes active and is able to endonucleolytically cleave DNA termini that require processing. Ligase IV/XRCC4/XLF complex is recruited to DNA termini and catalyze ligation of the DNA DSB. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
As mentioned earlier, IR does not frequently produce clean blunt-end breaks, and in fact regularly produces a number of complex breaks that contain DNA discontinuities at the terminus that require processing before proper ligation can occur. Artemis is the main nuclease known to process DNA termini in NHEJ by degrading DNA single-strand overhangs with its 5′-exonuclease and 5′- or 3′-endonuclease activity, and when defective, cells have been shown to possess some radiation sensitivity (49, 52, 73). Polymerases responsible for adding bases at the termini include pol β, μ, and λ. Pol μ is of particular interest, as its level is increased after IR and it is found to be in a complex with Ku and the Ligase IV/XRCC4 complex (56). The discussion of polymerases in NHEJ will be covered in another article in this Forum (70).
After processing of the DNA termini, DNA ligase IV is responsible for ligating the DSB that contain incompatible or compatible overhangs, as well as blunt-ends (27, 51), making it the perfect ligase for a repair pathway that does not require homology. DNA ligase IV is found in a complex with XRCC4, and the flexibility of this complex is apparent by the fact that the complex can ligate one strand even if the second strand cannot be ligated (perhaps because of a 5′-OH) (28). XLF (Cernunnos), a recently discovered protein found to be involved in NHEJ (2, 8), was determined to interact with Ligase IV/XRCC4 (LIV/X4) and is also required for optimal NHEJ. More recent evidence has shown XLF in a complex with LIV/X4, and is believed to be required for stimulating the catalytic activity of LIV/X4 (26). PNKP has also been found to associate with XRCC4, and may be responsible for phosphate replacement at damaged DNA termini (40, 42).
Ku 70/80
Ku, initially discovered as an autoantigen and found to be extremely abundant in the cell, is one of the first proteins to bind to DNA at a double-strand break (62). This DNA binding protein exists predominantly as a stable heterodimer made up of 70 and 86 kDa subunits (3, 29). Ku can also form a heterotrimeric complex with the 465 kDa DNA–PKcs when bound to DNA, forming the ∼610 kDa DNA–PK complex. Overall, Ku has been implicated in other cellular pathways, including telomere regulation and apoptosis (20). Very recent work from the Ramsden laboratory has also shown that Ku has 5′-AP lyase activity, although interestingly, it is suggested that this activity is still used in NHEJ (not BER) to remove AP sites near DSB (72). Overall, the ultimate role of Ku has been shown to be crucial to NHEJ-mediated DNA repair in eukaryotes.
Ku and DNA Binding
Ku binds with a high affinity to double-stranded DNA termini in a sequence-independent fashion and can translocate inward along the length of DNA in an ATP-independent manner (50). This movement is thought to coincide with the recruitment of DNA–PKcs to the site of the break, and is required for DNA–PK to gain access to the end of the DNA substrate (53, 93). Interestingly, discontinuities in the DNA structure, such as bulky cisplatin lesions, do not diminish Ku binding capacity significantly, but can inhibit translocation of Ku along the length of DNA (81). This impairment of Ku movement along the DNA was also found to inhibit LIV/X4 stimulated ligation, presumably because without translocation of Ku along the DNA, the ligase complex is unable to efficiently bind to the DNA (45). A more recent study has addressed the issue of Ku translocating on DNA when the DNA is coated in histones and other DNA binding proteins. Such bulky proteins could prevent the ring-like structure of Ku from sliding onto the end of DNA and moving along the length of it, as suggested by numerous in vitro experiments over the years. Roberts and Ramsden provide data demonstrating that Ku is capable of peeling away as much as 50 base-pairs of DNA from around the histone octamer structure found at the terminus of a double-strand break, thus allowing for DNA–PK to slide along the DNA without the need for chromatin remodeling (71).
Ku Structure
The structural features of Ku as revealed by various methods support much of the biochemical evidence gathered about Ku over the years. The two Ku subunits have a great deal of sequence similarity and both contain regions that contribute to the main DNA binding domain of the heterodimer (23) (Fig. 3). High resolution structural analysis of Ku reveals a ring-like shape that does not appear to undergo any major change in conformation after binding DNA (83). This ring-like structure allows for Ku to slide onto the end of a DNA duplex and it is shown that 1.5–2 turns of DNA can fit through the channel in this ring-like structure, but the shape and relative rigidity of the molecule renders it difficult if not impossible to bind to and interact with DNA in the absence of a free end. The nonspecific interactions between the sugar-phosphate backbone of DNA and the amino acids of the Ku ring structure support binding in a sequence-independent manner (83). Ku 70 and Ku 80 also share homology in that each contain a von Willebrand Factor A (vWA) domain N-terminal of the DNA binding domain (Fig. 3). In addition, each contains C-terminal extensions, a SAP domain in the case of Ku 70 and a less well-conserved extension in the case of Ku 80. The structure of the C-terminal region (CTR) of Ku 80, too flexible for X-ray crystallography and so left out of the heterodimer solved structure, has been solved by solution-based NMR (36). This work has revealed a long flexible linker region with a cluster of six alpha helixes, with the final 12 amino acids largely disordered. This region is important for interaction with DNA–PKcs, and is discussed in detail later in this review.
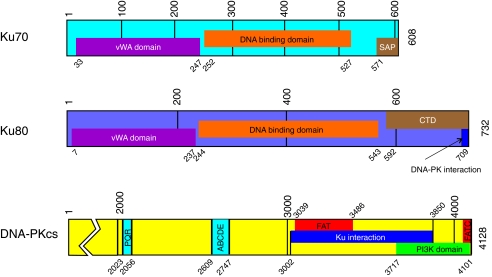
FIG. 3.
Map depicting Ku 70, Ku 80, and DNA–PKcs genetic structure. The conserved vWA domains in the N-terminal regions of Ku 70 and Ku 80 are depicted in purple and the conserved DNA binding domains are depicted in orange. The Ku 70 and 80 C-terminal domains are depicted in brown. Located at the extreme C-terminus of Ku 80 are the 12 amino acids implicated in the Ku-DNA–PKcs interaction and kinase activation that are depicted in blue. The DNA–PKcs map depicts the structurally and functionally important portion of the gene structure. The autophosphorylation clusters, PQR and ABCDE, are depicted in blue. The Ku interaction domain is shown in dark blue and highly conserved FAT and FACT domains are depicted in red. The conserved catalytic site for kinase activity is depicted in green. These conserved structural domains were determined by NCBI conserved domain analysis for human XRCC5, XRCC6 and PRKDC. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
DNA–PK
The 469 kDa DNA-dependent protein kinase catalytic subunit (DNA–PKcs) is the largest protein kinase in the cell reported to date. Sequence analysis places DNA–PKcs as a member of the phosphatidylinositol-3 (PI-3) kinase-like-kinase (PIKK) super-family (along with ATM, ATR, mTOR, SMG-1, and TRRAP). Grouped together because of their similar catalytic domains, the PIKKs catalytic domains share homology with the phosphoinositide (PI) 3-kinase catalytic domains (Fig. 3). However, the PIKKs phosphorylate proteins on serine or threonine residues rather than lipids. DNA–PKcs, like its family members, has a C-terminus kinase domain that is relatively small compared to the rest of the polypeptide (5%–10%), and is flanked by a FAT and FAT-C domain, whose roles are not yet clearly understood (Fig. 3). The N-terminal region is not well conserved, but is predicted to have multiple alpha-helical HEAT repeats (1).
DNA–PKcs binds to the site of a DSB following binding of Ku, and binding affinity of the catalytic subunit is increased 100-fold in the presence of Ku (91). The C-terminal region of Ku 80 is thought to participate in this Ku-DNA–PKcs interaction, discussed in greater detail later in this review. Once bound to DNA, DNA–PKcs is able to phosphorylate substrate proteins, targeting serines and threonines that are often followed by a glutamine, although as other sequences have been demonstrated to be phosphorylated by DNA–PK this calls into question the definition of a true consensus sequence. (57). This DNA-dependent kinase activity has been shown to be required for efficient DNA end joining (43). In addition, DNA–PK kinase activity plays a significant role in the DNA damage checkpoint signaling pathway and/or damage signaling to the apoptosis pathway (38).
DNA–PK Structure and Activation: The Role of DNA
One of the biggest challenges in the field remains the determination of the molecular mechanism that drives activation of DNA–PK by DNA. Biochemical and structural studies support the hypothesis that DNA–PK activation is driven by direct contact with DNA, including a study showing the importance of a leucine-rich region of DNA–PKcs that appears to be at least partially responsible for the direct interaction between the kinase and the DNA (31). Significant structural and biochemical advances with DNA–PK are aiding our understanding of kinase activation.
The DNA terminus resulting from an IR-induced DSB can vary in structure, size, and chemistry, each of which may play an important role in DNA–PK activation. Like the DNA binding activity of Ku, it has been shown that DNA–PK kinase activity is strongest with full duplex DNA, while hairpin structures or supercoiled plasmids result in little or no kinase activity (80). Furthermore, DNA–PK is preferentially activated by DNA with 3′-pyrimidine-rich termini, but activity is severely inhibited by terminal cisplatin-DNA adducts (65). These results are attributed to the catalytic subunit of DNA–PK, as there is no strong evidence for sequence-bias or strand bias of Ku DNA binding activity alone, nor are cisplatin lesions thought to inhibit Ku binding (see above in Ku section).
Interestingly, chemical structures attached to DNA, such as biotin, do not inhibit kinase activity (16, 65). A large amount of information regarding the activation of the catalytic subunit has previously been determined in vitro in the absence of Ku. Such studies were made possible because DNA–PKcs can bind to DNA in vitro, specifically in low salt and high magnesium buffers (33). Under these conditions in a Ku-independent reaction, this assay technique results in interesting data, indicating that DNA–PKcs is preferentially activated by single-strand DNA ends (34). These Ku-independent results were also observed in another lab. Furthermore, studies have suggested that melting of the DNA terminus may be necessary for DNA–PK to bind in a stable complex with DNA, and lead to optimal activation of the kinase (39). More recently, similar studies with the heterotrimeric complex, DNA–PK, have been performed with the more physiologically relevant form of DNA–PK, as there is no evidence that DNA–PKcs binds to or is activated by DNA in the cell in the absence of Ku. Studies from our lab have revealed that DNA–PK is preferentially activated by 3′-pyrimidine-rich sequences, and this supports the theory that single-strand ends of DNA may play an important and differential role in NHEJ catalyzed repair of DSBs (65). Building upon this work we have discovered that DNA–PK is preferentially activated by substrates containing 5′-single-strand overhangs compared to 3′-single-strand overhangs (66). These combined results suggest that individual DNA termini play an important role in activation of DNA–PK.
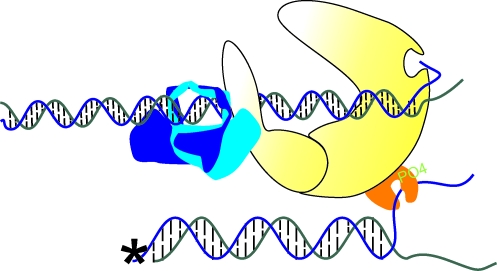
Advances in structural studies have complemented and extended our understanding of DNA–PK with respect to the role of DNA in activation of the kinase. Electron crystallographic studies have shown an open channel in the DNA–PKcs structure that can plausibly interact with double-stranded DNA (48). More recently, higher resolution reconstruction (7Å) of DNA–PKcs was achieved and work shows that DNA–PKcs displays handedness, with a head and base region containing two side connections that create a tunnel-like hollow channel within the protein that is the proposed binding site for DNA (35). Docking experiments revealed that the kinase domain could fit in either the head or base regions, although homology docking work done with PI3Kγ as a model indicate that the base region is the more plausible position. Within the central opening is an alpha-helical like protrusion, a likely candidate for direct interaction with DNA. The authors propose that approximately 1 turn or 10 base-pairs of the dsDNA would need to enter this channel to interact with this alpha-helical region. Interestingly, what appears to be a smaller cavity, only large enough to fit single-strand DNA, is located above the larger central channel (89). A crystal structure of DNA–PKcs has resulted in a structure of DNA–PKcs with the highest resolution yet accomplished, at 6.6Å, where the overall fold of the catalytic subunit is discernable. This reveals that an alpha helical region of HEAT repeats result in a bending of the protein structure into a hollow circular structure, like that described by the cryo-EM data discussed above. Interestingly, these authors place the catalytic domain in the top portion, or head region, of this circular structure, and show that there is a small HEAT repeat region inside the structure that is thought to bind DNA. This higher resolution structure supports the model of DNA threading through the kinase (76). In fact, data showing that the kinase has alpha-helical HEAT repeats scattered throughout the polypeptide and distributed around the structure indicate that the DNA could also interact in various positions on the periphery of the kinase, which could result in activation of DNA–PKcs by DNA. Additional research has suggested that once a double-strand DNA is threaded through the kinase, it can fray to expose a certain length of single-strand DNA, each of which can then be inserted into what are potentially seen in the structural data as two cavities (16, 48). Alternatively, there may be only one cavity on the perimeter of the molecule that may be an active site with dimensions to accommodate single-strand DNA (89). Activation of DNA–PK by a DNA terminus could be happening in either a cis or trans fashion, meaning the strand necessary for activation could interact with the same DNA–PK molecule through which it is threaded (Fig. 4, cis activation) or could interact with a separate DNA–PK molecule bound to the other DNA terminus (Fig. 4, trans activation). All of this structural and biochemical data suggest that DNA is in fact important for activating the kinase, probably through a direct interaction that involves threading of the DNA through the circular structure of DNA–PK followed by insertion of a DNA terminal strand into an active site within the kinase. (Fig. 4).
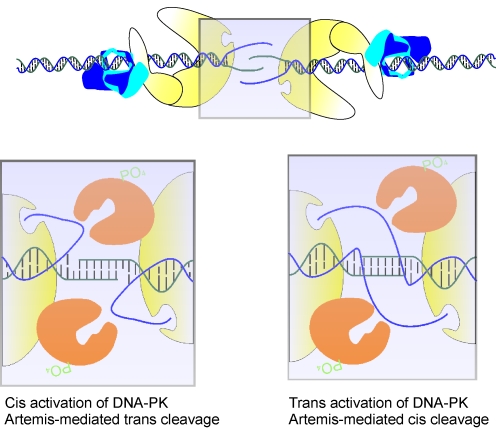
FIG. 4.
DNA–PK activation and Artemis-mediated cleavage. DNA threads through the ring-like structure of the Ku heterodimer and through a channel in DNA–PKcs. Following threading of the DNA, strand separation occurs and the 3′-strand (depicted in green) drives homology-mediated interactions with the opposite 3′-strand, aiding in formation of a synaptic complex between the two DNA termini. The purple shadow in the top panel indicates the region of magnification in the bottom panels. Cis activation of DNA–PK occurs by the 5′-strand (depicted in blue) interacting with an active site that can accommodate single-strand DNA on the periphery of the same DNA–PK molecule that is bound to the DNA terminus. Artemis interacts with DNA–PK and cleaves DNA in a trans fashion by cleaving a single-strand overhang on the opposite DNA terminus. Trans activation occurs by 5′-single-strand DNA interacting with the active on the DNA–PK molecule bound to the opposite DNA terminus. Artemis interacts with DNA–PK and cleaves DNA in a cis fashion by cleaving a single-strand overhang on the same terminus with which DNA–PK and Artemis are interacting. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
DNA–PK Activation: The Role of Protein–Protein Interactions
While the role of DNA is crucial as the name implies, protein–protein interactions are also necessary for DNA–PK activation and are predominantly provided by the Ku heterodimer. Two regions of Ku were not observed in the crystal structure, the C-terminal regions (CTR) of both Ku 80 and Ku 70. Ku 80 CTR has been shown to play an important role in DNA–PK activation. As noted above, the favored model is that Ku binds first to a DNA DSB and then recruits the DNA–PKcs, which becomes active once bound to DNA. Kinetic analysis has shown that DNA–PKcs undergoes an extreme increase in activity when bound to a Ku-DNA complex as compared to just DNA alone, suggesting that a Ku-DNA–PKcs interaction is necessary for optimal activation of the enzyme. Interestingly, incubating DNA–PKcs with Ku molecules devoid of the C-terminal region of Ku 80 results in significantly reduced DNA–PK kinase activity, and it has been shown that only the final 12 amino acids of Ku80 are required for an interaction with DNA–PKcs (23, 77).
Despite results revealing the Ku 80 CTR both physically interacts with DNA–PKcs and is needed for optimal kinase activity, it is still unclear if this region promotes activation through recruitment of DNA–PKcs to the DSB or through direct contact with the DNA–PKcs polypeptide to activate the kinase. Initial in vivo studies showed that DNA–PKcs does not accumulate at DNA DSB in cells that are Ku 80 null, indicating that perhaps the CTR of Ku 80 is needed for recruitment of DNA–PKcs to the DSB (82). Interestingly, studies also revealed that Ku 80 CTR truncations result in a level of radiosensitivity in cells that is similar to that seen with DNA–PKcs null cells, and this Ku 80 truncation mutant phenotype is postulated to be from lack of recruitment of DNA–PKcs to the site of a break (77). However, controversial and more recent in vitro and in vivo studies show that DNA–PKcs is recruited to the site of a DSB in the absence of the CTR of Ku 80 (but with the remaining heterotrimeric protein intact) (87). These results indicate that deletion of the Ku 80 CTR does not disrupt recruitment of DNA–PKcs to a DNA terminus. Although this same group reported only a 50% decrease in kinase activation with mutant Ku that was missing the CTR of Ku 80, our laboratory has data suggesting that DNA–PK kinase activity is severely inhibited by loss of the Ku 80 CTR, with kinase activity near background level when incubated with truncated Ku. This would indicate that while recruitment of DNA–PKcs to the site of DSB is not dependent on Ku 80 CTR, kinase activation, and therefore repair of DSB, is Ku 80 CTR dependent. Interestingly, new structural data using small angle X-ray scattering (SAXS) suggests that the Ku 80 CTR can extend considerably from the remainder of the Ku heterodimer, and is attached to the core by the disordered, flexible region. The considerable flexibility coupled with the distance of the C-terminal region from the core suggests this region could easily recruit and then retain interaction with DNA–PKcs at the site of a DSB. It is also possible that this disordered region interacts with other parts of the Ku molecule to induce a conformational change that could in turn activate the kinase. These possibilities raise the question of whether the C-terminal region of Ku 80 is responsible for DNA–PKcs recruitment, retention, and activation, or potentially plays a role in all three (35). Clearly, further studies need to be done to determine the exact role of Ku 80 CTR.
Protein–Protein and Protein–DNA Interactions: Juxtaposition of a DNA DSB
Successful repair of a double-strand break requires that the two ends of the DSB are brought together to allow for ligation by Ligase 4/XRCC4/XLF. Emerging data over the years have indicated that NHEJ proteins must bind to DNA in order to bring the two ends together into a synaptic complex to allow for the ligation reaction. Ku and DNA–PKcs are recognized as the first proteins to bind to the site of the damage and may be responsible for the recruitment of other proteins in the pathway. This leads to the hypothesis that this protein complex could be responsible for bringing the two ends of DNA together and maintaining them in a synaptic complex at the site of a DSB.
A large amount of data exists to suggest that Ku, DNA–PKcs, or the heterotrimeric complex play a crucial role in synapses of DNA ends. Atomic force microscopy revealed a complex of Ku and the two DNA ends of a linearized plasmid, suggesting that Ku holds the two termini together in a synaptic complex (63). Data showing that Ku can transfer between two strands of DNA, whether they contain homologous or nonhomologous sequence regions, also suggest that Ku is responsible for the juxtaposition of DNA ends (4, 13) More recent electron microscopy, as well as two-photon fluorescence cross-correlation spectroscopy, have revealed that two DNA ends are in fact brought together by two DNA–PKcs molecules into a synaptic complex and biochemical analysis revealed that kinase activation occurs following formation of the complex (16, 60). Higher resolution structural work suggests that dimeric complexes made up of Ku/-DNA–PKcs/DNA interact within the HEAT repeat regions (79).
SAXS structural data suggest that both the DNA–PKcs and Ku molecules play a role in stabilizing two DNA termini in a synaptic complex, as the Ku 80 CTR is made up of a dynamic arm that is significantly extended from the core of the molecule such that it could interact with a DNA–PKcs molecule, and furthermore shows that DNA–PKcs can form head-to-head dimers (35). The SAXS work shows that the dimensions coupled with the extreme flexibility of the C-terminal region of Ku 80 are ample enough to allow interactions with both the DNA–PKcs bound at the same DSB terminus, as well as across the DSB to a DNA–PKcs molecule bound to the opposing terminus, contacting the molecule in a trans fashion. This would suggest that the CTR of Ku 80 indeed is responsible in some way for retaining DNA–PKcs at the site of a break, perhaps through a tethering mechanism that retains the kinase in a synaptic complex at the DSB site. Recent biochemical data out of our lab and structural SAXS data also suggest that the CTR of Ku 80 can form a dimer. Our laboratory generated the 16 kDa Ku 80 CTR fragment indicated to be important for DNA–PK activation, and cross-linking data show a homodimer being formed with this mutant protein. SAXS data suggests that the Ku 80 CTR can interact with the remainder of the Ku polypeptide in a total of five possible positions (35). This Ku80 CTR homodimerization may facilitate tethering of the two DNA termini at a DSB to enhance synaptic complex formation and end-joining activity (Fig. 5).
FIG. 5.
Ku 80 C-term interactions. The Ku 80 CTR (depicted in dark blue and extending out from the Ku molecule) is a highly flexible region with a cluster of six α-helical repeats at the terminus. Ku 80 CTR can interact with regions of the Ku heterodimer, including itself, and this CTR–CTR interaction may promote synaptic complex formation. Interactions between Ku 80 CTR and DNA–PKcs can also occur and may also play a role in synapsis of the DNA termini. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
As discussed above, protein–protein interactions play an important role in formation and stabilization of a synaptic complex. Protein–DNA and DNA–DNA interactions may also play a role in synaptic complexes at the site of a DSB. Recent work analyzing the Mycobacterium tuberculosis DNA–PK suggests that the 3′-ends of DNA at each terminus of a double-strand break are important in maintaining a synaptic complex consisting of the NHEJ machinery. The authors propose that the 3′-ends protruding from each LigD molecule can pair together, thus bringing each LigD-bound DNA termini into proximity of each other in a synaptic complex (7). This work is consistent with observations in our laboratory, where a greater than additive level of DNA–PK activation was observed when mixing two substrates containing regions of microhomology in a 3′-overhang region, while substrates with complementary 5′-overhangs did not display this synergistic activity (66). This work suggests that the 3′-end of DNA is important for searching for an opposing strand on the opposite side of the break, particularly for a DNA end with microhomology, thus assisting in synaptic complex formation (66) (Fig. 4). It is becoming increasingly clear that the synaptic complex is a crucial step in NHEJ, and that Ku, DNA–PKcs, and the DNA strands all play an important role in the formation.
Impact of DNA–PK Autophosphorylation on Protein–Protein and Protein–DNA interactions
As it appears that the main role of active DNA–PK is in NHEJ, considerable work has been done to map DNA–PK dependent phosphorylation sites of the core NHEJ protein machinery and uncover the in vivo relevance of these phosphorylation events. While several of the key NHEJ players are phosphorylated in vitro by DNA–PK (18, 41, 84, 94) only two proteins have been demonstrated to be regulated by phosphorylation, DNA–PKcs and Artemis (58). Artemis, a DNA nuclease implicated in end processing in the NHEJ pathway, gains endonucleolytic activity on DNA substrates following DNA–PK phosphorylation (54, 55). The other DNA–PK dependent phosphorylation substrate that has been shown to have in vivo relevance is DNA–PK itself through autophosphorylation.
Early studies with DNA–PK revealed that autophosphorylation of DNA–PKcs resulted in decreased protein kinase activity (12). This loss of activity was linked with dissociation of the DNA–PK catalytic subunit from Ku and DNA bound complex, or the “active” complex. These studies demonstrate the dynamic complex that forms at a DNA DSB, and reveals a physiological role for DNA–PK autophosphorylation (61). Further experiments mapped the autophosphorylation sites within DNA–PKcs and initially identified a cluster of six sites within residues 2609–2647, dubbed the ABCDE cluster (19). Interestingly, mutational analysis of these residues revealed that autophosphorylation of any one of the sites alone is not necessary for NHEJ, as determined by radiosensitivity assays. However, when all six of the serine or threonine residues were mutated to alanine, cells displayed similar high levels of radiosensitivity as cells that were DNA–PKcs null (61). When mutated, this cluster was also unable to efficiently repair IR-induced DSB (5). Several more autophosphorylation sites have been identified, and to date the ABCDE mutant and the PQR mutant (6, 14) are the clusters that appear to be the most functionally relevant. Interestingly, each of these mutant clusters display severe radiosensitivity, but still maintain full phosphorylation activity and can associate with and dissociate from DNA DSB. The major defect exhibited by the two mutants described is a disruption in DNA end processing during NHEJ. Further studies have shown that autophosphorylation at the ABCDCE sites renders DNA termini accessible for end processing, but not by DNA–PK dissociation. Instead, it is proposed that following autophosphorylation of ABCDE, DNA–PK remains bound but undergoes a conformational change so that DNA ends are now made accessible to end processing factors and the ligase complex, and DNA–PK remains in position on the DNA termini to support a synaptic complex This model is further supported by recent evidence showing that autophosphorylation of DNA–PKcs occurs in trans, both in vitro and in vivo (59). This is consistent with the formation of a synaptic complex, thus allowing for trans autophosphorylation between two DNA–PK molecules located at each termini of the DNA DSB. Furthermore, this synaptic complex could protect ends from processing until autophosphorylation occurs and alters the complex formation, liberating ends for processing (58). Recent in vivo photobleaching studies (82) and in vitro structural work support the model that autophosphorylation does render DNA termini accessible for processing, and this occurs because of a conformational change in DNA–PKcs following autophosphorylation. SAXS data revealed dramatic conformational changes throughout the phosphorylated DNA–PKcs structure, including an opening up of the head and palm regions that have previously been postulated to encircle the DNA that most likely results in dissociation of DNA–PKcs from DNA. While additional work needs to be done to understand more clearly the mechanism of DNA–PKcs dissociation and define the residues involved, it is clear that DNA–PK autophosphorylation plays two important roles, one involved in DNA termini accessibility and the other in dissociation (35).
End Processing Events
Once DNA termini have been recognized, bound, and stabilized by DNA–PK, processing of DNA to create ligatable ends occurs. While a single I-SCE1 break is extremely useful for analysis of DNA DSB signaling response, the processing necessary for IR-induced breaks is likely to be considerable more complex. In attempts to induce more complex events, asymmetric I-SCE1 has been used to show that processing can occur in these systems, though the specific enzymes and mechanisms required to process these types of ends remains to be determined (86). To recapitulate these events more accurately will require complex lesions that more closely mimic damage induced by IR. Depending on the complexity of the DNA discontinuity, an assortment of different processing enzymes may be required to remove DNA damage at the site of the break to allow for ligation. Numerous enzymes have been implicated in DNA processing and include but are not limited to, FEN-1 (90), polynucleotide kinase-phosphatase (PNKP) (40), Werner protein (44, 68), MRN (46), DNA polymerase μ and λ (52), and the nuclease Artemis (55). While all of these enzymes have been implicated in NHEJ, specific roles of certain enzymes like MRN remain unclear. Further research may reveal that a subset of these enzymes, along with yet unknown molecules, play a role in DNA DSB signaling.
NHEJ is proficient in joining blunt termini or two DNA ends that have nonhomologous overhang regions. Frequently, joining of overhangs requires some sort of removal or addition of nucleotides at the site of the break (9). Despite the term “nonhomologous” end joining, it has been shown that there can be a greater tendency to join two broken ends that contain sequences with 1–4 nucleotides that are complementary (88), dubbed more recently as areas of microhomology. It is suggested that to align these ends of DNA at regions of microhomology, processing that results in the loss or addition of nucleotides must occur. While often characterized as error-prone, NHEJ when presented with a break that does not require processing can efficiently join the termini with high fidelity (9; 30). When necessitated by the complexity of the breaks being joined, additions are carried out by family X polymerases (70) and we will focus on the loss of nucleotides, which are carried out by nucleases, including Artemis.
Artemis
The nuclease Artemis is involved in cleaving hairpins generated during V(D)J recombination and has been shown to play a role in NHEJ repair of IR-induced DNA DSB. The importance of Artemis participation in NHEJ is based on in vivo data demonstrating that Artemis null cells are more sensitive to IR than wild-type counterparts (73). Artemis has been found to possess 5′-3-exonucleolytic activity on single-strand DNA (this activity is explored further in this section), DNA–PK, and ATP dependent endonuclease activity on DNA hairpin structures, and DNA–PK ATP dependent endonuclease processing of 3′- and 5′-single-strand overhangs, with preferential cleavage at the dsDNA/ssDNA junction (55). Artemis has also been shown to remove 3′-phosphoglycolate groups from DNA termini, like those generated by IR (69).
Artemis is a member of the β-CASP family, a relatively new group of the metallo-β-lactamase fold superfamily made up of enzymes acting on nucleic acids (10). All β-CASP family members to date are enzymes that process nucleic acids and have two major structural domains, the metallo-β-lactamase domain and the β-CASP domain (which is embedded in the metallo-β-lactamse domain). Artemis also has an extended C-terminal region following the β-CASP domain, which appears to be fairly disordered (data not shown). The metallo-β-lactamase domain is made up of a four-layered β-sandwich that is flanked by α-helices, with zinc coordination sites located in the sandwich region (11). Artemis hydrolyses the phosphodiester backbone of DNA, and recent work has revealed a histidine residue within the β-CASP domain that is critical for the catalytic activity of Artemis (in vitro and in vivo) (15). Higher resolution structural data is needed for a greater understanding of how this enzyme is activated and regulated, but extensive biochemical analysis has answered some questions regarding Artemis nuclease activity.
Artemis has been shown to be phosphorylated by DNA–PK in vitro (54) and in vivo (78). It has been suggested that the stimulation of endonuclease activity of Artemis requires binding and phosphorylation by DNA–PK that causes a conformational change in the C-terminal region of Artemis, resulting in relief of Artemis autoinihibition of the endonuclease active site (54). DNA–PKcs activation has also been suggested to be necessary for Artemis to retain association with DNA–PK/DNA complex at the site of the break (21). Other research has suggested that autophosphorylation of DNA–PK results in a conformational change in the DNA-bound kinase which in turn alters the conformation of DNA such that it can be easily recognized and cleaved by Artemis (25, 92). While each model differs slightly in mechanism, both models suggest that Artemis endonuclease activity is DNA–PK and ATP dependent.
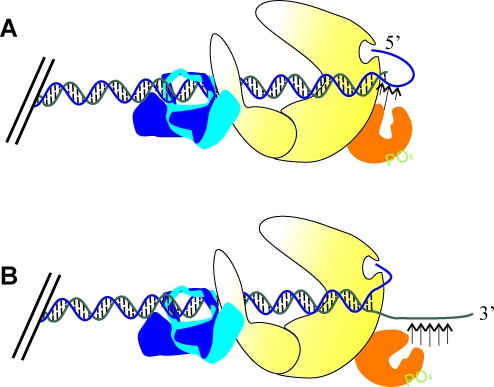
Despite the biochemical data gathered, many questions remain regarding DNA–PK-dependent Artemis endonuclease activity. It is clear that Artemis requires DNA–PK and ATP for endonuclease activity, but it is unclear if this activation by DNA–PK is through a cis or trans mechanism (Fig. 6). Several laboratories have performed in vitro nuclease assays with a radiolabeled overhang substrate, but include unlableled double-strand DNA to increase DNA–PK activity, arguing that this increases Artemis nuclease activity. However, it is hard to imagine an in vivo scenario at the site of a DSB where DNA–PK is acting in trans to activate Artemis (Fig. 6). In fact, work by Povirk's group has shown that Artemis nuclease activity is not increased in the presence of a double-strand DNA stimulus (69), and Lees–Miller's group sees robust endonuclease activity in the absence of stimulating DNA (25). Rather, an alternative is that DNA–PK is bound to the site of a DSB, Artemis is recruited to this same DNA end, and Artemis is activated in cis by this DNA–PK molecule. Once DNA–PK dependent Artemis-catalyzed endonuclease cleavage is activated, cleavage events could also be happening through a cis or trans mechanism. The DNA/DNA–PK/Artemis complex described could be envisioned to stimulate trans endonucleolytic cleavage. That is, just as trans autophosphorylation of DNA–PK occurs, Artemis too could be bound to one terminus in a complex with DNA–PK and act catalytically across the synaptic complex to cleave DNA on the opposite terminus (Fig. 4). As a DNA DSB induced by IR can be very complex, with a variety of DNA discontinuities on either terminus at the site of the DSB, it is possible that Artemis cleaves a DNA terminus in trans that needs to be processed, even if the side it is bound to doesn't require processing. It is also possible that Artemis cleaves a DNA strand in cis (Fig. 4), similar to DNA–PK being activated in cis.
FIG. 6.
Activation of Artemis endonuclease activity. Artemis endonuclease activity is dependent on DNA–PK and ATP, and this activation event could occur in trans by a DNA–PK molecule bound to a different DNA terminus than the terminus Artemis is cleaving. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
The specifics of DNA cleavage by Artemis have yet to be fully elucidated. It appears that in vitro, DNA–PK dependent Artemis endonuclease activity preferentially cleaves single-strand overhang DNA around the SS/DS junction on synthetic oligonucleotide substrate. Biochemical studies performed with DNA substrates containing a 21 basepair double-strand region with a 15 base 5′-single-strand overhang result in endonuclease activity at the SS/DS DNA junction, generating a 15 base single-strand cleavage product (and a blunt double-strand product). Paradoxically, work from the Lees–Miller laboratory with a similar substrate (25 basepair double-strand substrate with a 15 base 5′-single-strand overhang) resulted in 24 and 26 nucleotide excision fragments, indicating that DNA–PK dependent Artemis activity can cleave DNA at the n+1 and n-1 positions at the SS/DS junction (25). 3′-Overhang substrates (21 basepairs of double-strand DNA with a 15 base single-strand overhang) are preferentially cleaved in the SS region, reducing the 15 base overhang to a 4–5 single-strand overhang (55). According to Yannone and Povirk, single-strand overhang length impacts the cleavage position on 3′-overhangs, with longer single-strand overhangs (15 base and 13 base) trimmed to leave a 5-base overhang while a 9-base overhang was trimmed to a 4-base overhang (69). This group also observed endonuclease activity on plasmid substrates, albeit somewhat decreased compared to longer overhang substrates, with 4-, 5-, and 6-base overhangs, resulting in 2–4 base cleavage products. Detailed biochemical studies with purified Artemis and DNA–PK further suggest that in the presence of active DNA–PK, Artemis can make small endonuclease cleavage products, or “trim”, DNA termini on both the 3′ and 5′ strand. The authors point out that this versatile activity of Artemis would allow the enzyme to process a variety of DNA structures and chemistries that might exist at a DSB, thus setting the break site up for polymerase-mediated extension and ultimate ligation of the DSB (92). Artemis has also recently been shown to possess DNA–PK dependent endonuclease activity on single-strand DNA, with a sequence preference favoring cleavage of thymidines.
As mentioned earlier, it has been suggested that Artemis has 5′–3′-exonuclease activity on single-strand DNA. However, our lab has recently separated the 5′–3′-exonuclease activity of Artemis from the DNA–PK dependent endonuclease activity with little to no loss of overall protein or endo activity, which strongly suggests that the exonuclease activity is not an intrinsic component of the Artemis polypeptide (67). This work is supported by studies done by several other laboratories in an attempt to map the active site of Artemis. While the Artemis endonuclease active site has been successfully mapped, mutational analysis throughout the protein has been unable to disrupt the exonuclease activity of Artemis (15, 37, 64). Furthermore, metallo-β-lactamase fold enzymes have been classified as only having one active site that has been shown to be the functional catalytic site for all activity (17). While it appears that the exonuclease activity originally associated with Artemis is a contaminant of some sort, it is still possible that this exonuclease is endogenously associated with Artemis and plays a significant role in NHEJ. Additional work needs to be done to identify this exonuclease and define its role in NHEJ.
While the biochemical studies of Artemis to date clearly indicate that more research needs to be done to understand the molecular mechanism of DNA–PK and Artemis-mediated DNA cleavage in NHEJ, combined results from the field suggest a model for DNA–PK and Artemis interactions with DNA ends. DNA strand polarity clearly plays a role, as 3′ and 5′ cleavage of single-strand overhangs varies slightly, with a 3′-overhang getting cleaved to within 4 nt of the double-strand junction while 5′-overhangs are cleaved predominantly at the double-strand junction with minor cleavage events at n+1, n-1. Our laboratory and others have also shown that a 3′-overhang results in a more heterogeneous cleavage set, with the n+4 position being cleaved slightly more predominantly but additional cleavage of surrounding nucleotides up to two in either direction as well. This differs from the homogenous cleavage pattern of a 5′-overhang resulting in predominant cleavage at the double-strand junction. It is possible that this variation can in part be explained by altered geometry of loading DNA–PK and Artemis, where the Ku70/80 complex loads from the end with the same geometry, 80 distal to the end and 70 proximal while DNA–PKcs displays overhang orientation dependent on loading to the terminus. It is more likely that this variation is a product of DNA positioning as driven by DNA–DNA–PK interactions. Data demonstrate that 5′-overhangs may be crucial for activation and thus may possess more defined interactions with the DNA–PKcs subunit, which creates a uniform alteration in the DNA structure to allow more precise cleavage by Artemis at the single-strand-double-strand DNA junction (Fig. 7A). 3′ overhangs do not necessarily have such a defined interaction with DNA–PKcs, as Artemis is forced to interact with DNA overhangs in a stochastic manner, creating a more random cleavage pattern that results in cleavage products of assorted sizes (Fig. 7B).
FIG. 7.
Artemis endonuclease activity on single-strand overhangs. DNA–PK dependent Artemis-mediated cleavage of overhangs varies based on strand polarity. (A) Artemis cleaves 5′-overhangs in a homogeneous, with cleavage mainly occurring at the single-strand/double-strand junction and a few cleavage events occurring at the n+1 or n-1 positions. (B) Artemis cleaves 3′-overhangs in a heterogenous manner, with cleavage products occurring at the n+6, n+5, n+4, n+3, and n+2 positions. All of these potential cleavage products result in a 3′-overhang remaining at the DNA terminus. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Conclusion
IR-induced DNA DSB are complicated and can vary in many different ways at each break site, with the potential for one terminus of the DSB to have a different DNA structure than the opposite terminus. As research progresses in the NHEJ field, it becomes more evident that there are many proteins involved in the processing of this large variety of damage found at the DNA DSB. With the variety of proteins, including polymerases, nucleases, and kinases, it is becoming apparent that the processing steps are not adequately accomplished by just one protein. Rather, it is likely that not every DNA DSB is processed the same way, and that only a subset of the processing proteins are required to repair any individual break. It is not known how the correct protein needed to process a specific DNA discontinuity at the terminus is recruited. However, it is clear that this is a dynamic and complicated process that is likely to involve a variety of proteins, some of which may not yet have been identified.
Abbreviations Used
- DNA–PK
DNA dependent protein kinase
- DNA–PKcs
DNA dependent protein kinase catalytic subunit
- DSB
double-strand break
- dsDNA
double-strand DNA
- HDR
homology directed repair
- IR
ionizing radiation
- Ku 80 CTR
Ku 80 C-terminal region
- LIV/X4
ligase IV / XRCC4
- NHEJ
non-homologous end joining
- nt
nucleotides
- PIKK
(PI-3) kinase-like kinases
- PNKP
human polynucleotide kinase-phosphatase
- Pol
polymerase
- SAXS
small angle X-ray scattering
- ssDNA
single-strand DNA
- SS/DS junction
single-strand/double-strand junction
- vWA
von Willebrand Factor A
- XLF
XRCC4-like factor
Acknowledgments
We thank the members of the Turchi Lab for their helpful discussion. Research in the Turchi lab is supported by NIH awards R01CA82741 and R21CA128628.
References
- 1.Abraham RT. PI 3-kinase related kinases: 'Big' players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Ahnesorg P. Smith P. Jackson SP. XLF interacts with the XRCC4-DNA ligase IV omplex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Badie C. Goodhardt M. Waugh A. Doyen N. Foray N. Calsou P. Singleton B. Gell D. Salles B. Jeggo P. Arlett CF. Malaise EP. A DNA double-strand break defective fibroblast cell line (180BR) derived from a radiosensitive patient represents a new mutant phenotype. Cancer Res. 1997;57:4600–4607. [PubMed] [Google Scholar]
- 4.Bliss TM. Lane DP. Ku selectively transfers between DNA molecules with homologous ends. J Biol Chem. 1997;272:5765–5773. doi: 10.1074/jbc.272.9.5765. [DOI] [PubMed] [Google Scholar]
- 5.Block WD. Merkle D. Meek K. Lees–Miller SP. Selective inhibition of the DNA-dependent protein kinase (DNA- PK) by the radiosensitizing agent caffeine. Nucleic Acids Res. 2004;32:1967–1972. doi: 10.1093/nar/gkh508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Block WD. Yu Y. Merkle D. Gifford JL. Ding Q. Meek K. Lees–Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brissett NC. Pitcher RS. Juarez R. Picher AJ. Green AJ. Dafforn TR. Fox GC. Blanco L. Doherty AJ. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 8.Buck D. Malivert L. de CR. Barraud A. Fondaneche MC. Sanal O. Plebani A. Stephan JL. Hufnagel M. le DF. Fischer A. Durandy A. de Villartay JP. Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 9.Budman J. Chu G. Processing of DNA for nonhomologous end-joining by cell-free extract. EMBO J. 2005;24:849–860. doi: 10.1038/sj.emboj.7600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callebaut I. Moshous D. Mornon JP. de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: The beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattell E. Sengerova B. McHugh PJ. The SNM1/Pso2 family of ICL repair nucleases: From yeast to man. Environ Mol Mutagen. 2010;51:635–645. doi: 10.1002/em.20556. [DOI] [PubMed] [Google Scholar]
- 12.Chan DW. Lees–Miller SP. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 13.Chiu CF. Lin TY. Chou WG. Direct transfer of Ku between DNA molecules with nonhomologous ends. Mutat Res. 2001;486:185–194. doi: 10.1016/s0921-8777(01)00080-5. [DOI] [PubMed] [Google Scholar]
- 14.Cui X. Yu Y. Gupta S. Cho YM. Lees–Miller SP. Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Villartay JP. Shimazaki N. Charbonnier JB. Fischer A. Mornon JP. Lieber MR. Callebaut I. A histidine in the beta-CASP domain of Artemis is critical for its full in vitro and in vivo functions. DNA Repair (Amst) 2009;8:202–208. doi: 10.1016/j.dnarep.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 16.DeFazio LG. Stansel RM. Griffith JD. Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- 18.Douglas P. Gupta S. Morrice N. Meek K. Lees–Miller SP. DNA–PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. Dna Repair. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Douglas P. Sapkota GP. Morrice N. Yu YP. Goodarzi AA. Merkle D. Meek K. Alessi DR. Lees–Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downs JA. Jackson SP. A means to a DNA end: The many roles of Ku. Nature Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 21.Drouet J. Frit P. Delteil C. de Villartay JP. Salles B. Calsou P. Interplay between Ku, artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends. J Biol Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 22.Erenpreisa J. Kalejs M. Ianzini F. Kosmacek EA. Mackey MA. Emzinsh D. Cragg MS. Ivanov A. Illidge TM. Segregation of genomes in polyploid tumour cells following mitotic catastrophe. Cell Biol Int. 2005;29:1005–1011. doi: 10.1016/j.cellbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Gell D. Jackson SP. Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Ann Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 25.Goodarzi AA. Yu Y. Riballo E. Douglas P. Walker SA. Ye R. Harer C. Marchetti C. Morrice N. Jeggo PA. Lees–Miller SP. DNA–PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu J. Li S. Zhang X. Wang LC. Niewolik D. Schwarz K. Legerski RJ. Zandi E. Lieber MR. DNA–PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair (Amst) 2010;9:429–437. doi: 10.1016/j.dnarep.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu J. Lu H. Tippin B. Shimazaki N. Goodman MF. Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu J. Lu H. Tsai AG. Schwarz K. Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: Influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y. Jin S. Gao Y. Weaver DT. Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guirouilh–Barbat J. Huck S. Bertrand P. Pirzio L. Desmaze C. Sabatier L. Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Gupta S. Meek K. The leucine rich region of DNA–PKcs contributes to its innate DNA affinity. Nucleic Acids Res. 2005;33:6972–6981. doi: 10.1093/nar/gki990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hada M. Georgakilas AG. Formation of clustered DNA damage after high-LET irradiation: a review. J Radiat Res (Tokyo) 2008;49:203–210. doi: 10.1269/jrr.07123. [DOI] [PubMed] [Google Scholar]
- 33.Hammarsten O. Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammarsten O. DeFazio LG. Chu G. Activation of DNA-dependent protein kinase by single-stranded DNA ends. J Biol Chem. 2000;275:1541–1550. doi: 10.1074/jbc.275.3.1541. [DOI] [PubMed] [Google Scholar]
- 35.Hammel M. Yu Y. Mahaney BL. Cai B. Ye R. Phipps BM. Rambo RP. Hura GL. Pelikan M. So S. Abolfath RM. Chen DJ. Lees–Miller SP. Tainer JA. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J Biol Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris R. Esposito D. Sankar A. Maman JD. Hinks JA. Pearl LH. Driscoll PC. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR) J Mol Biol. 2004;335:573–582. doi: 10.1016/j.jmb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y. Giblin W. Kubec M. Westfield G. St CJ. Chadde L. Kraftson S. Sekiguchi J. Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability. J Exp Med. 2009;206:893–908. doi: 10.1084/jem.20082396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson SP. DNA-dependent protein kinase. [Review] Intl J Biochem Cell Biol. 1997;29:935–938. doi: 10.1016/s1357-2725(97)00006-x. [DOI] [PubMed] [Google Scholar]
- 39.Jovanovic M. Dynan WS. Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res. 2006;34:1112–1120. doi: 10.1093/nar/gkj504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karimi–Busheri F. Rasouli–Nia A. lalunis–Turner J. Weinfeld M. Human polynucleotide kinase participates in repair of DNA double-strand breaks by nonhomologous end joining but not homologous recombination. Cancer Res. 2007;67:6619–6625. doi: 10.1158/0008-5472.CAN-07-0480. [DOI] [PubMed] [Google Scholar]
- 41.Kim DK. Stigger E. Lee SH. Role of the 70-kDa subunit of human replication protein A (I). Single-stranded DNA binding activity, but not polymerase stimulatory activity, is required for DNA replication. J Biol Chem. 1996;271:15124–15129. doi: 10.1074/jbc.271.25.15124. [DOI] [PubMed] [Google Scholar]
- 42.Koch CA. Agyei R. Galicia S. Metalnikov P. O'Donnell P. Starostine A. Weinfeld M. Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurimasa A. Kumano S. Boubnov NV. Story MD. Tung CS. Peterson SR. Chen DJ. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusumoto R. Dawut L. Marchetti C. Wan LJ. Vindigni A. Ramsden D. Bohr VA. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry. 2008;47:7548–7556. doi: 10.1021/bi702325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kysela B. Doherty AJ. Chovanec M. Stiff T. Ameer-Beg SM. Vojnovic B. Girard PM. Jeggo PA. Ku stimulation of DNA ligase IV-dependent ligation requires inward movement along the DNA molecule. J Biol Chem. 2003;278:22466–22474. doi: 10.1074/jbc.M303273200. [DOI] [PubMed] [Google Scholar]
- 46.Lee JH. Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 47.Lehmann AR. Fuchs RP. Gaps and forks in DNA replication: Rediscovering old models. DNA Repair (Amst) 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Leuther KK. Hammarsten O. Kornberg RD. Chu G. Structure of DNA-dependent protein kinase: Implications for its regulation by DNA. EMBO J. 1999;18:1114–1123. doi: 10.1093/emboj/18.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 50.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lieber MR. Grawunder U. Wu X. Yaneva M. Tying loose ends: Roles of Ku and DNA-dependent protein kinase in the repair of double-strand breaks. [Review] Curr Opinion Genet Devel. 1997;7:99–104. doi: 10.1016/s0959-437x(97)80116-5. [DOI] [PubMed] [Google Scholar]
- 52.Lieber MR. Lu H. Gu J. Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: Relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 53.Ma Y. Lieber MR. DNA length-dependent cooperative interactions in the binding of Ku to DNA. Biochemistry. 2001;40:9638–9646. doi: 10.1021/bi010932v. [DOI] [PubMed] [Google Scholar]
- 54.Ma YM. Pannicke U. Lu HH. Niewolik D. Schwarz K. Lieber MR. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human artemis. J Biol Chem. 2005;280:33839–33846. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 55.Ma YM. Pannicke U. Schwarz K. Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 56.Mahajan KN. Nick McElhinny SA. Mitchell BS. Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: Role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahaney BL. Meek K. Lees–Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meek K. Dang V. Lees–Miller SP. DNA–PK: The means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 59.Meek K. Douglas P. Cui X. Ding Q. Lees–Miller SP. Trans-autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol. 2007;27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merkle D. Block WD. Yu Y. Lees–Miller SP. Cramb DT. Analysis of DNA-dependent protein kinase-mediated DNA end joining by two-photon fluorescence cross-correlation spectroscopy. Biochemistry. 2006;45:4164–4172. doi: 10.1021/bi0524060. [DOI] [PubMed] [Google Scholar]
- 61.Merkle D. Douglas P. Moorhead GBG. Leonenko Z. Yu YP. Cramb D. Bazett–Jones DP. Lees–Miller SP. The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry. 2002;41:12706–12714. doi: 10.1021/bi0263558. [DOI] [PubMed] [Google Scholar]
- 62.Mimori T. Hardin JA. Steitz JA. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J Biol Chem. 1986;261:2274–2278. [PubMed] [Google Scholar]
- 63.Pang D. Yoo S. Dynan WS. Jung M. Dritschilo A. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 1997;57:1412–1415. [PubMed] [Google Scholar]
- 64.Pannicke U. Ma YM. Hopfner KP. Niewolik D. Lieber MR. Schwarz K. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J. 2004;23:1987–1997. doi: 10.1038/sj.emboj.7600206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pawelczak KS. Andrews BJ. Turchi JJ. Differential activation of DNA–PK based on DNA strand orientation and sequence bias. Nucleic Acids Res. 2005;33:152–161. doi: 10.1093/nar/gki157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pawelczak KS. Turchi JJ. A mechanism for DNA–PK activation requiring unique contributions from each strand of a DNA terminus and implications for microhomology-mediated nonhomologous DNA end joining. Nucleic Acids Res. 2008;36:4022–4031. doi: 10.1093/nar/gkn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pawelczak KS. Turchi JJ. Purification and characterization of exonuclease-free Artemis: Implications for DNA–PK-dependent processing of DNA termini in NHEJ-catalyzed DSB repair. DNA Repair (Amst) 2010;9:670–677. doi: 10.1016/j.dnarep.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perry JJ. Yannone SM. Holden LG. Hitomi C. Asaithamby A. Han S. Cooper PK. Chen DJ. Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 69.Povirk LF. Zhou T. Zhou R. Cowan MJ. Yannone SM. Processing of 3'-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J Biol Chem. 2007;282:3547–3558. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- 70.Ramsden DA. Polymerases in nonhomologous end joining: Building a bridge over broken chromosomes. AntioxidRedox Signal. 2011;14:2509–2519. doi: 10.1089/ars.2010.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts SA. Ramsden DA. Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA ends. J Biol Chem. 2007;282:10605–10613. doi: 10.1074/jbc.M611125200. [DOI] [PubMed] [Google Scholar]
- 72.Roberts SA. Strande N. Burkhalter MD. Strom C. Havener JM. Hasty P. Ramsden DA. Ku is a 5'-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rooney S. Alt FW. Lombard D. Whitlow S. Eckersdorff M. Fleming J. Fugmann S. Ferguson DO. Schatz DG. Sekiguchi J. Defective DNA repair and increased genomic instability in artemis-deficient murine cells. J Exp Med. 2003;197:553–565. doi: 10.1084/jem.20021891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shikazono N. Noguchi M. Fujii K. Urushibara A. Yokoya A. The yield, processing, and biological consequences of clustered DNA damage induced by ionizing radiation. J Radiat Res (Tokyo) 2009;50:27–36. doi: 10.1269/jrr.08086. [DOI] [PubMed] [Google Scholar]
- 75.Shrivastav M. De Haro LP. Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 76.Sibanda BL. Chirgadze DY. Blundell TL. Crystal structure of DNA–PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singleton BK. Torres–Arzayus MI. Rottinghaus ST. Taccioli GE. Jeggo PA. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soubeyrand S. Pope L. de CR. Gosselin D. Dong F. de Villartay JP. Hache RJ. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J Mol Biol. 2006;358:1200–1211. doi: 10.1016/j.jmb.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 79.Spagnolo L. Rivera–Calzada A. Pearl LH. Llorca O. Three-dimensional structure of the human DNA–PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 80.Suwa A. Hirakata M. Takeda Y. Jesch SA. Mimori T. Hardin JA. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci USA. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turchi JJ. Henkels KM. Zhou Y. Cisplatin-DNA adducts inhibit translocation of the Ku subunits of DNA–PK. Nucleic Acids Res. 2000;28:4634–4641. doi: 10.1093/nar/28.23.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uematsu N. Weterings E. Yano K. Morotomi–Yano K. Jakob B. Taucher–Scholz G. Mari PO. van G. Chen BP. Chen DJ. Autophosphorylation of DNA–PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walker JR. Corpina RA. Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 84.Wang YG. Nnakwe C. Lane WS. Modesti M. Frank KM. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J Biol Chem. 2004;279:37282–37290. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 85.Ward JF. Radiation mutagenesis: The initial DNA lesions responsible. Radiat Res. 1995;142:362–368. [PubMed] [Google Scholar]
- 86.Weinstock DM. Brunet E. Jasin M. Induction of chromosomal translocations in mouse and human cells using site-specific endonucleases. J Natl Cancer Inst Monogr. 2008:20–24. doi: 10.1093/jncimonographs/lgn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weterings E. Verkaik NS. Keijzers G. Florea BI. Wang SY. Ortega LG. Uematsu N. Chen DJ. van G. The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol Cell Biol. 2009;29:1134–1142. doi: 10.1128/MCB.00971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Willers H. Husson J. Lee LW. Hubbe P. Gazemeier F. Powell SN. hm-Daphi J. Distinct mechanisms of nonhomologous end joining in the repair of site-directed chromosomal breaks with noncomplementary and complementary ends. Radiat Res. 2006;166:567–574. doi: 10.1667/RR0524.1. [DOI] [PubMed] [Google Scholar]
- 89.Williams DR. Lee KJ. Shi J. Chen DJ. Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008;16:468–477. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu X. Wilson TE. Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: The order of strand annealing and nucleolytic processing events. Proc Natl Acad Sci USA. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yaneva M. Kowalewski T. Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: Biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yannone SM. Khan IS. Zhou RZ. Zhou T. Valerie K. Povirk LF. Coordinate 5' and 3' endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:3354–3365. doi: 10.1093/nar/gkn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoo S. Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: Recruitment of DNA–PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu Y. Mahaney BL. Yano K. Ye R. Fang S. Douglas P. Chen DJ. Lees–Miller SP. DNA–PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst) 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]