Abstract
5-Fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine (SCH 66712) is a potent mechanism-based inactivator of human cytochrome P450 2D6 that displays type I binding spectra with a Ks of 0.39 ± 0.10 μM. The partition ratio is ∼3, indicating potent inactivation that addition of exogenous nucleophiles does not prevent. Within 15 min of incubation with SCH 66712 and NADPH, ∼90% of CYP2D6 activity is lost with only ∼20% loss in ability to bind CO and ∼25% loss of native heme over the same time. The stoichiometry of binding to the protein was 1.2:1. SDS-polyacrylamide gel electrophoresis with Western blotting and autoradiography analyses of CYP2D6 after incubations with radiolabeled SCH 66712 further support the presence of a protein adduct. Metabolites of SCH 66712 detected by mass spectrometry indicate that the phenyl group on the imidazole ring of SCH 66712 is one site of oxidation by CYP2D6 and could lead to methylene quinone formation. Three other metabolites were also observed. For understanding the metabolic pathway that leads to CYP2D6 inactivation, metabolism studies with CYP2C9 and CYP2C19 were performed because neither of these enzymes is significantly inhibited by SCH 66712. The metabolites formed by CYP2C9 and CYP2C19 are the same as those seen with CYP2D6, although in different abundance. Modeling studies with CYP2D6 revealed potential roles of various active site residues in the oxidation of SCH 66712 and inactivation of CYP2D6 and showed that the phenyl group of SCH 66712 is positioned at 2.2 Å from the heme iron.
Introduction
Cytochromes P450 (P450s) are a family of heme-containing enzymes that are ubiquitous in nature. In humans there are 57 P450 enzymes responsible for the metabolism of a variety of substrates including lipids, hormones, vitamins, and exogenous small molecules including pharmaceuticals (Guengerich, 2003). The basic reaction of P450 enzymes is a mono-oxygenation of substrate through activation of molecular oxygen with electrons provided by P450 NADPH reductase, although a variety of products are possible due to various rearrangements (Guengerich, 2003).
A direct consequence of the capacity of P450s to accommodate a wide range of drug substrates is that they are susceptible to inhibition by many agents (Ortiz de Montellano and Correia, 1983; Correia and Ortiz de Montellano, 2005). A fairly common mechanism of enzyme inhibition related to drug-drug interactions is mechanism-based inhibition (Ortiz de Montellano and Correia, 1983; Correia and Ortiz de Montellano, 2005). In the strict definition of a mechanism-based inhibitor, a substrate (inhibitor) is transformed by the enzyme in the usual way to a reactive intermediate (usually electrophilic) that has a finite half-life (Ortiz de Montellano and Correia, 1983; Guengerich and Shimada, 1991; Correia and Ortiz de Montellano, 2005). The inhibition of the P450 is NADPH-, time-, and concentration-dependent. The reactive intermediate can partition between reaction with the enzyme (at the active site) to inactivate the enzyme or undergo a different transformation to yield a stable (released) product (Ortiz de Montellano and Correia, 1983; Silverman, 1988; Guengerich and Shimada, 1991; Correia and Ortiz de Montellano, 2005). Mechanism-based inactivation of P450s has been shown to occur by several mechanisms including covalent binding of the inhibitor to the heme or to the protein at specific amino acids (or both) or covalent linking of modified heme to the protein (Ortiz de Montellano and Correia, 1983; Guengerich and Shimada, 1991; Correia and Ortiz de Montellano, 2005).
CYP2D6 is responsible for the metabolism of approximately 25 to 30% of commercially available pharmaceutical drugs including several antidepressants (Guengerich, 2003). The only P450 that metabolizes more clinically relevant drugs is CYP3A4, an enzyme that accounts for 30% of the P450 content in liver; CYP2D6 accounts for on average 5% of total P450 in liver (Shimada et al., 1994; Guengerich, 2005). CYP2D6 notably metabolizes drugs containing a basic nitrogen and a planar aromatic ring (Wolff et al., 1985; Islam et al., 1991; Koymans et al., 1992; Rowland et al., 2006). These moieties are common in drugs with narrow therapeutic indexes targeted to central nervous system or cardiovascular disorders (Rowland et al., 2006).
CYP2D6 is an important drug-metabolizing enzyme of wide interest because of its ability to metabolize a large number of drugs and its multiple polymorphic states. Individuals with poor metabolizer phenotype are susceptible to a variety of drug-induced side effects, some of which have severe clinical outcomes. Irreversible inactivation of CYP2D6 by mechanism-based inhibitors can lead to drug-induced loss of P450 activity and thus produce a poor metabolizer type of response in individuals who are not poor metabolizers. Thus, understanding of the inactivation of CYP2D6 by mechanism-based inhibitors is of clinical interest. Beyond clinical understanding of inactivation of CYP2D6, mechanism-based inhibitors also can be used to understand the basic biochemistry of the CYP2D6 enzyme mechanism and catalysis.
5-Fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine (SCH 66712) was the first mechanism-based inhibitor of CYP2D6 to be described previously (Palamanda et al., 2001). It was under development as an antagonist of human dopamine receptor D4 but was later abandoned (Palamanda et al., 2001). Although previously SCH 66712 was shown to be a mechanism-based inhibitor of CYP2D6, the mechanism for inactivation was not determined (Palamanda et al., 2001). Therefore, the current study was initiated to investigate the mechanism of this inhibition.
Materials and Methods
Chemicals.
SCH 66712 was obtained from Schering-Plough Research Institute (Kenilworth, NJ) and was reconstituted in water for use in assays described below. Radiolabeled SCH 66712 (3H and 14C) was synthesized by Schering-Plough Research Institute. [14C]SCH 66712 had a specific activity of 55 mCi/mmol and a purity of ≥99% as determined by high-performance liquid chromatography (HPLC). [3H]SCH 66712 had a specific activity of 25.5 Ci/mmol. Ultra-pure solvents (water, ACN, and methanol) for MS were purchased from EMD Chemicals, Inc. (Gibbstown, NJ). All other solvents were HPLC-grade and were purchased from Sigma-Aldrich (St. Louis, MO). Glutathione was purchased from Cayman Chemical (Ann Arbor, MI). Potassium phosphate, N-acetylcysteine, NADPH, l-α-dilauroyl-phosphatidylcholine phospholipids, ACN, glucose 6-phosphate, glucose-6-phosphate dehydrogenase, NADP+, bufuralol, 1′-hydroxybufuralol, catalase, and all other reagents were purchased from Sigma-Aldrich.
Enzymes.
Human CYP2D6 with P450 reductase (Supersomes) were purchased from BD Gentest (Woburn, MA) and used for all experiments except spectral analysis and binding titrations and hemochrome assays. For spectral analysis and binding titrations and the hemochrome assays described below, recombinant human CYP2D6 and recombinant P450 NADPH reductase were purified from Escherichia coli as described previously and were a generous gift from Dr. F. P. Guengerich (Vanderbilt University, Nashville, TN) (Gillam et al., 1995).
Spectral Binding Titrations.
Spectral binding titration studies were performed with recombinant purified CYP2D6 (1 μM) in 100 mM potassium phosphate buffer, pH 7.4. The enzyme was evenly divided between two cuvettes, and the experiments were performed at room temperature by titrating in aliquots of SCH 66712 (0.05–16 μM) to the sample cuvette with the solvent control added to the reference cuvette. A baseline of the reference cuvette was recorded (350–500 nm) on a Cary 300 dual-beam spectrophotometer (Varian, Inc., Walnut Creek, CA). Ligand was subsequently added, and the spectra were recorded (350–500 nm) after each addition. The solvent was added to the reference cuvette. The difference in absorbance between the wavelength maximum and minimum was plotted versus the concentration of SCH 66712, and the data were analyzed by nonlinear regression methods with KaleidaGraph (Synergy Software, Reading, PA). The dissociation constant, Ks, was determined using the following quadratic velocity equation or tight-binding equation: [CYP2D6 · SCH 66712] = 0.5 (Ks + Et + St) − [0.25 (Ks + Et + St)2 − EtSt]1/2, where S represents substrate concentration, E is the total enzyme concentration, and Ks is the spectral dissociation constant for the reaction CYP2D6 + SCH 66712 ⇆ CYP2D6 · SCH 66712.
Enzyme Assays and Inactivation.
A primary reaction mixture (inhibition assay) containing 16 μM SCH 66712 and 20 pmol of CYP2D6 Supersomes in 100 mM potassium phosphate buffer, pH 7.4 (final volume 200 μl), was preincubated in a 37°C shaking water bath. After 3 min, reactions were initiated by the addition of NADPH (1 mM final). Aliquots of the primary reaction (10 μl) were removed at 0, 2, 5, 10, 15, 30, and 60 min and added to a secondary reaction (activity assay) containing 1 mM NADPH and 100 μM bufuralol in 100 mM potassium phosphate buffer, pH 7.4, in a final volume of 200 μl. Each secondary reaction was incubated for 10 min at 37°C in a shaking water bath before being quenched with 15 μl of 70% perchloric acid. Reaction mixtures were centrifuged (2000g, 5 min) to remove the precipitated enzyme, and the recovered supernatants were directly injected onto HPLC for analysis. The formation of the 1′-hydroxybufuralol product was quantified by HPLC with a Symmetry 300 C18 column (5 μm, 4.6 × 250 mm) on a 515 HPLC pump system with a 474 fluorescence detector controlled with Empower software (Waters, Milford, MA). The detector was set at an excitation wavelength of 252 nm and an emission wavelength of 302 nm. The mobile phase consisted of 30% acetonitrile and 70% 1 mM perchloric acid in water. All analyses were performed at ambient temperature with a flow rate of 1 ml/min (Hanna et al., 2001a,b).
Hemochrome Assays.
Native heme in CYP2D6 was determined using the methods of Omura and Sato (1964). Primary reactions containing 1 μM recombinant purified CYP2D6, 2 μM recombinant purified P450 NADPH reductase, 30 μM freshly sonicated l-α-dilauroyl-phosphatidylcholine phospholipids, 100 mM potassium phosphate buffer (pH 7.4), 400 units/ml catalase (8 μg/ml), 80 units/ml superoxide dismutase (0.18 μM), and 16 μM SCH 66712 were incubated for 0 to 40 min with an NADPH-generating system (5 mM glucose 6-phosphate, 0.5 mM NADP+, and 0.5 units/ml glucose-6-phosphate dehydrogenase). Controls lacked either SCH 66712 or the NADPH-generating system. Incubations were quenched by addition of solubilizing buffer [100 mM potassium phosphate (pH 7.4) containing 1.0 mM EDTA, 20% glycerol (v/v), 0.50% sodium cholate (w/v), and 0.40% Emulgen 913 (w/v)] and placed on ice. To quenched samples were added 22% pyridine (v/v) and 0.01 N NaOH (final concentrations). The samples were immediately mixed by inversion and divided between two cuvettes. A baseline spectrum was recorded from 520 to 620 nm. After the addition of 1 to 2 mg of NaS2O4 to the sample cuvette, the spectrum was measured between 520 and 620 nm. The change in absorbance at 557 nm relative to that at 575 nm was used to determine the heme content using 32.4 mM−1cm−1 as the molar absorptivity.
Native Heme Analysis by HPLC.
Reaction mixtures containing 16 μM SCH 66712 and 50 pmol of CYP2D6 Supersomes in 100 mM potassium phosphate buffer, pH 7.4, (final volume 200 μl) were preincubated in a 37°C shaking water bath. After 3 min, the reactions were initiated by the addition of NADPH (1 mM final); an equivalent volume of water was added to the control. After 0, 2, 5, 10, 15, and 40 min, the reactions were quenched by the addition of 10 μl of acetonitrile, and samples were placed on ice. For heme adduct analysis, incubation mixtures were injected onto a PROTO 300 C4 column (5 μm, 2.1 × 250 mm) connected to a Waters Alliance e2695 HPLC system, and the mobile phase was a gradient elution with initial conditions of 70% A (0.1% TFA in H2O):30% B (0.05% TFA in ACN) that was ramped linearly to 20% A over 30 min and then returned to the initial conditions. Heme was monitored using a Waters model 2487 dual wavelength UV/visible detector at 405 nm. Heme eluted at approximately 22.5 min.
Partition Ratio.
Primary reaction mixtures containing 0 to 15 μM SCH 66712 and 20 pmol of CYP2D6 Supersomes in 100 mM potassium phosphate buffer, pH 7.4 (final volume of 100 μl), were preincubated in a 37°C shaking water bath. After 5 min, the primary reactions were initiated with the addition of NADPH (1 mM, final) and incubated at 37°C. To allow the inactivation to go to completion (Silverman, 1988), inactivation essays were incubated for 60 min. Then, aliquots of 10 μl of were removed and added to secondary reaction mixtures containing 100 μM bufuralol and 1 mM NADPH in 100 mM potassium phosphate buffer, pH 7.4, in a final volume of 200 μl. The secondary mixtures (in triplicate) were incubated for 10 min at 37°C and quenched with 15 μl of 70% perchloric acid. Reaction mixtures were centrifuged (2000g, 5 min) to remove the precipitated enzyme, and aliquots of the recovered supernatants were directly injected onto HPLC for analysis as described above.
Spectral Analysis.
P450-reduced CO difference spectra were measured using the methods of Omura and Sato (1964). Primary reactions containing 1 μM recombinant purified CYP2D6, 2 μM recombinant purified P450 NADPH reductase, 30 μM freshly sonicated l-α-dilauroyl-phosphatidylcholine phospholipids, 100 mM potassium phosphate buffer, pH 7.4, 400 units/ml catalase (8 μg/ml), 80 units/ml superoxide dismutase (0.18 μM), and 16 μM SCH 66712 were incubated for 0 to 25 min with an NADPH-generating system. Incubations were terminated by the addition of ice-cold quenching buffer [100 mM potassium phosphate buffer (pH 7.4) containing 1.0 mM EDTA, 20% glycerol (v/v), 0.50% sodium cholate (w/v), and 0.40% Emulgen 913 (w/v)]. Incubations without SCH 66712 or without the NADPH-generating system were performed as controls. Quenched samples were divided into two cuvettes, and a baseline spectrum from 400 to 500 nm was recorded using a Cary 300 dual-beam spectrophotometer (Varian, Inc.). To the sample cuvette only, CO was bubbled for 1 min and then ∼1 to 2 mg of Na2S2O4 was added to both the reference and sample cuvettes. The spectrum between 400 and 500 nm was recorded. The change in absorbance at 450 nm relative to that at 475 nm was used to determine the P450 content using 91 mM−1cm−1 as the molar absorptivity (Omura and Sato, 1964).
LC-ESI-MS Analysis of CYP2D6.
Reactions containing CYP2D6 Supersomes (50 pmol) and SCH 66712 (100 μM) in 100 mM potassium phosphate buffer (pH 7.4) were preincubated for 3 min at 30°C in a shaking water bath. Reactions were initiated by the addition of NADPH (1 mM); control incubations received an equal volume of water (final reaction volume 200 μl). Incubations were continued at 30°C for 10, 20, and 40 min. Reactions were terminated by placing samples on ice. The same experiments were also done at 37°C. An aliquot of each reaction (20 μl, 5 pmol) was directly injected onto a reverse-phase PROTO 300 C4 column (5 μm, 2.1 × 250 mm) and chromatographic separation was performed using a Waters Alliance 2690 HPLC system. The solvent system consisted of A (0.1% formic acid in water) and B (0.1% formic acid in ACN). A flow rate of 0.2 ml/min was used. After an initial 20-min hold at 90% A, a linear gradient of 90% A to 5% A over 35 min was applied for resolving of protein components followed by a 10-min hold at 5% A. The column effluent starting at 20 min was directed into an LXQ mass analyzer (Thermo Fisher Scientific, Waltham, MA) operated in the positive ion mode using the Xcalibur software package. The system had been optimized with horse heart myoglobin. The ESI conditions were the following: sheath gas, 20 arbitrary units; auxiliary gas, 9 arbitrary units; spray voltage, 5 kV; capillary temperature, 275°C; capillary voltage, 48 V; and tube lens offset, 120 V. The molecular masses of CYP2D6 were determined by deconvolution of the apoprotein charge envelopes using ProMass (Novatia, LLC, Mammouth Junction, NJ).
Covalent Binding of SCH 66712 to CYP2D6.
For SDS-PAGE analysis, CYP2D6 Supersomes (100 pmol/ml), control and inactivated samples, in the presence of 14C- or 3H-labeled SCH 66712 were resolved on a 10% polyacrylamide gel and then transferred to a nitrocellulose membrane. Blots were blocked and then treated with anti-human CYP2D6 antibodies. CYP2D6 was detected by chemiluminescence using a secondary antibody with horseradish peroxidase activity. After Western blot analysis, the nitrocellulose membrane was subjected to autoradiography.
Stoichiometry of Binding.
Stoichiometry of binding was determined using the method of Chan et al. (1993). In brief, reactions containing CYP2D6 Supersomes (0.1 nmol, 0.2 μM final) and 14C-labeled-SCH 66712 (16 μM) in 100 mM potassium phosphate buffer (pH 7.4) were preincubated for 3 min at 37°C in a shaking water bath. Reactions were initiated by the addition of NADPH (1 mM) or the addition of an equal volume of water for the control (final reaction volume 500 μl). Incubations were continued at 37°C for 60 min. A 5-fold volume (2.5 ml) of 5% sulfuric acid in methanol was added to quench the reactions, and the mixtures were vortexed for 10 s. The protein pellet obtained after centrifugation at 1800g for 10 min was washed a total of 4 times or until the count was less than 300 cpm/0.5 ml of for the supernatant. The final protein pellet was dissolved in 0.6 ml of 1 N NaOH, and 0.1 ml of the resulting sample was analyzed using a Beckman LS 6000 scintillation counter.
Metabolism of SCH 66712.
A mixture of CYP2D6 Supersomes (500 pmol), 100 mM potassium phosphate buffer (pH 7.4), SCH 66712 (100 μM), freshly prepared glutathione (10 mM), or freshly prepared N-acetylcysteine (10 mM) was preincubated for 3 min at 37°C before addition of 150 μl of the NADPH-generating system. The total reaction volume was 1 ml. After 40 min of incubation in a water bath, all samples were placed on ice and quenched with 60 μl of 1% TFA. All samples were then applied to a 1-ml C18 solid-phase extraction column (Agilent Technologies, Santa Clara, CA), preconditioned by washing with 1 ml of methanol followed by 1 ml of water. After the sample was loaded, the column was washed with 1 ml of water then 2 ml of methanol and 300 μl of acetonitrile. All fractions were collected. The methanol and acetonitrile fractions were combined and dried under a stream of nitrogen to a final volume of approximately 50 μl. The sample was then resuspended in 200 μl of solvent A that was composed of 90% water, 10% methanol, and 0.05% TFA for LC-MS analysis. A 20-μl aliquot of the sample was injected onto a Kinetex C18 (2.6 μm, 2.1 × 100 mm) column (Phenomenex, Torrance, CA) for chromatographic separation using a Waters Alliance 2690 HPLC system with a solvent system composed of solvent mixtures A (90% water, 10% methanol, and 0.05% TFA) and B (90% acetonitrile, 10% methanol, and 0.05% TFA). After 5 min at the initial conditions (5% B), separation of metabolites was achieved by a linear gradient of 5% B to 50% B over 20 min and then holding at 50% B for 1 min before returning to the initial conditions. The flow rate was 0.1 ml/min. The column was allowed to re-equilibrate for 15 min at the initial conditions (95% A:5% B) before the next injection. Mass spectrometry was performed using an LXQ mass spectrometer (Thermo Fisher Scientific) with an ESI source in a positive ion mode. The ESI conditions were the following: sheath gas, 27 arbitrary units; auxiliary gas, 5 arbitrary units; spray voltage, 5 kV; and capillary temperature, 250°C.
Molecular Modeling and Docking Simulations.
AutoDock 4.0 was used to perform docking simulations and molecular models (http://autodock.scripps.edu) (Morris et al., 1998; Huey et al., 2007). The protein structures used in these studies were CYP2D6 (PDB: 2F9Q) and CYP2C9 (PDB: 1R9O). Ligands and solvent molecules were removed, but the heme was retained. The Fe atom of the heme was assigned a charge of +3. The PDB structure of 2D6 was modified in Swiss DeepView at position 374 to the reference amino acid (M374V). Residues within 5 Å of the heme iron were identified and set as flexible residues for computations. For each protein structure, charges were calculated by the Gasteiger-Marsili method. The three-dimensional structures of the ligands for docking studies were built in Spartan 4.0 (Wavefunction, Inc., Irvine, CA) with all hydrogen atoms added and energy minimization. The grid maps were calculated using AutoGrid. The dimensions of the grid box were set to 40 × 40 × 40 Å, and the grid spacing was set to 0.375 Å. Docking was performed using the Lamarckian genetic algorithm. Each docking experiment was performed 100 times, yielding 100 docked conformations. The consensus binding postures of the molecules were obtained by visual inspection and docking scores.
Results
Inactivation of CYP2D6 by SCH 66712.
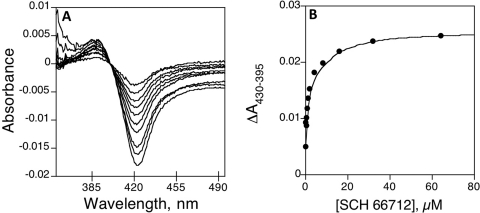
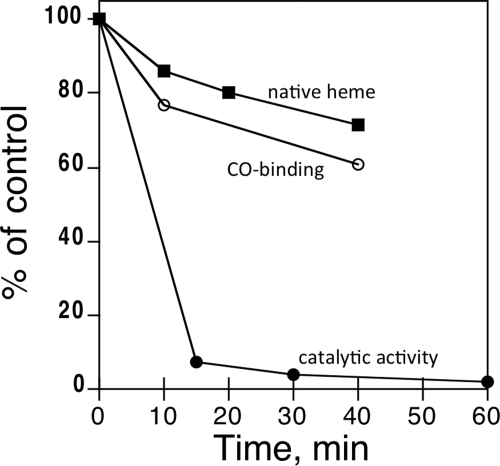
Binding analysis with SCH 66712 (Fig. 1) and CYP2D6 showed type I binding spectra characteristic of substrate binding. The Ks was determined to be 0.39 ± 0.10 μM (Fig. 2). Treatment of CYP2D6 with SCH 66712 in the presence of NADPH led to a ∼90% loss of enzyme activity as evidenced by reduced bufuralol hydroxylation within less than 15 min (Fig. 3). The same results were found when dextromethorphan was used as a substrate (data not shown). At the same time, the loss of ability to bind CO in reduced-CO difference assays was ∼25%, and the loss of native heme in hemochrome assays was ∼15% (Fig. 3; Supplemental Fig. 1). Loss of native heme and CO-binding ability correlated fairly well, although the extent of the loss of P450 activity was much higher (Fig. 3). Furthermore, the control hemochrome assays that lacked SCH 66712 or NADPH showed nearly the same degree of loss of hemochrome formation over time as the reaction containing both SCH 66712 and NADPH (Supplemental Fig. 2). Inactivation reactions with 2 mM potassium cyanide as an iminium scavenger showed the same rate of inactivation as reactions without cyanide (Supplemental Fig. 3). In concordance with this finding, previous studies have shown that other exogenous nucleophiles including glutathione and reactive oxygen species scavengers including superoxide dismutase and mannitol had no affect on the inactivation (Palamanda et al., 2001).
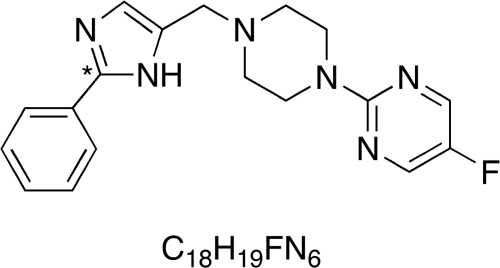
Fig. 1.
Structure of SCH 66712. SCH 66712 contains functional groups typically associated with substrates for CYP2D6 including basic nitrogens and planar aromatic rings. *, location of the 14C radiolabel.
Fig. 2.
Spectral binding titration of SCH 66712. A, purified CYP2D6 (1 μM) was divided into each of two cuvettes, and a baseline was set. Aliquots of SCH 66712 in H2O were added to the sample cuvette, and equal volumes of H2O were added to the reference cuvette. B, plot of ΔA430–395 (from A) versus concentration of SCH 66712. Ks was determined to be 0.39 ± 0.10 μM.
Fig. 3.
Effect of SCH 66712 on CYP2D6 catalytic activity, reduced CO difference spectra, and native heme content. CYP2D6 was treated with SCH 66712 (16 μM) in the presence of NADPH. Residual hydroxybufuralol activity (●) was determined at the times indicated as described under Materials and Methods. Reduced CO difference spectra (○) were collected at 0, 10, and 40 min for inactivation of CYP2D6 by SCH 66712. Residual native heme (■) was determined by a hemochrome assay at 0, 10, 20, and 40 min for the inactivation of CYP2D6 by SCH 66712. The percentage of control indicates relative catalytic activity, CO binding, or native heme remaining compared with time 0.
Analysis of Heme.
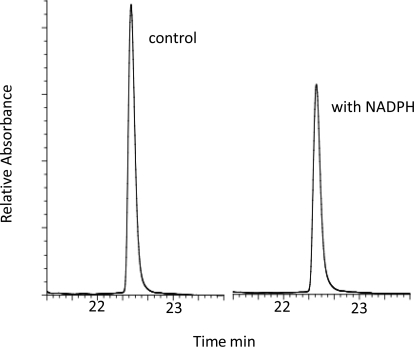
Hemochrome and spectral assays (Fig. 3) suggested that heme adduction was not a significant contributor to CYP2D6 inactivation. However, because of the nature of hemochrome colorimetric assays, it is possible that a heme adduct could be formed but still detected as native heme in the hemochrome assay above. To further examine possible heme adduction, HPLC analysis of heme using UV/visible detection was performed. The results show an ∼25% decrease in native heme content upon incubation with SCH 66712 and NADPH compared with the control reaction that did not contain NADPH (Fig. 4). This finding is consistent with the loss of CO-binding ability (Fig. 3) and a slightly greater loss of native heme than found in hemochrome assays (Fig. 4). Furthermore, MS analysis of the heme showed only m/z 616 with no peaks at potential adducted masses (data not shown). Taken together, these findings suggest that heme adduct formation is unlikely.
Fig. 4.
Analysis of native heme. CYP2D6 was treated with SCH 66712 (16 μM) in the presence or absence of NADPH for 0, 2, 5, 10, 15, and 40 min (only data for the 40-min incubation are shown). Heme content was analyzed by HPLC using absorbance at 405 nm. Native heme eluted at 22.5 min. After a 40-min incubation with SCH 66712 and NADPH (right panel), there was a 25% decrease in native heme compared with time 0 (left panel).
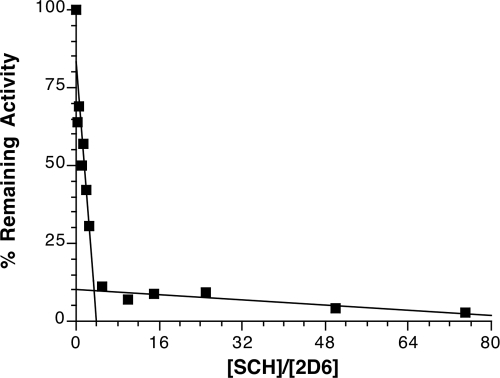
Determination of the Partition Ratio.
The number of molecules of SCH 66712 metabolized per molecule of inactivated CYP2D6, i.e., the partition ratio, was determined by incubation of CYP2D6 with various concentrations of SCH 66712 over 60 min to allow the inactivation to progress until essentially complete. The percentage of the activity remaining was plotted as a function of the molar ratio of SCH 66712 to CYP2D6. The turnover number (partition ratio + 1) was estimated from the intercept of the linear regression line obtained from the lower ratios of SCH 66712 to CYP2D6 with the straight line derived from the higher ratios of SCH 66712 to 2D6 as described previously (Silverman, 1988). With this method, the turnover number was 4, and consequently the partition ratio was 3 (Fig. 5).
Fig. 5.
Loss of CYP2D6 activity as a function of the ratio of SCH 66712 to CYP2D6. CYP2D6 was incubated with various concentrations of SCH 66712 for 60 min to allow for complete inactivation. The partition ratio was estimated to be ∼3.
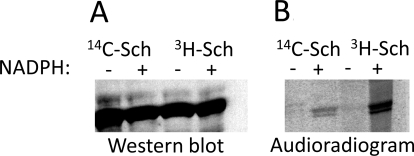
Covalent Binding of SCH 66712 to CYP2D6.
SDS-PAGE with Western blotting and autoradiography analyses of CYP2D6 from Supersomes after incubation with radiolabeled SCH 66712 suggested the presence of a protein adduct (Fig. 6). Both 3H- and 14C-labeled SCH 66712 bound to CYP2D6 in the presence of NADPH but not in the absence of NADPH (Fig. 6).
Fig. 6.
Covalent binding of SCH 66712 to CYP2D6. Labeled SCH 66712 was incubated with Supersomes (100 pmol/ml) for 10 min at 37°C in the presence or absence of 1 mM NADPH. Samples were chilled on ice after the incubation. For SDS-PAGE, 20-μl aliquots were removed, mixed with 20 μl of gel loading buffer, and boiled for 3 min before loading on the 10% acrylamide gel. For analysis, 2 pmol of 2D6 protein were loaded on the gel. Protein was transferred from gel to nitrocellulose paper for Western blotting. A, Western blot with anti-CYP2D6 antibodies. Chemiluminescence was used for detection. B, audioradiogram of gel in A.
Attempts to identify adducted CYP2D6 by LC-ESI-MS were inconclusive because of lowered ionization of CYP2D6 upon inactivation. CYP2D6 (from Supersomes) in the presence of SCH 66712 but absence of NADPH produced reasonable mass spectra that allowed for deconvolution of the parent protein with mass of 55,781 Da (Supplemental Fig. 4). However, upon incubation with NADPH, ionization was greatly reduced and no protein adduct was identified (Supplemental Fig. 4). The normalized level in the mass spectra of CYP2D6 after inactivation by SCH 66712 was decreased by more than 1 order of magnitude (Supplemental Fig. 4), consistent with protein adduction and the loss of enzyme ionization. Varying temperature (37 or 30°C) and duration of incubation (10, 20, or 40 min) had no affect on improving ionization. We confirmed our experimental method with the use of CYP2B4 inactivation by 4-tert-butylphenylacetylene (tBPA) as a positive control because tBPA is an inactivator known to form an adduct with CYP2B4 apoprotein. LC-ESI-MS analysis and deconvolution of CYP2B4 inactivation showed a characteristic increase in protein mass by 174 amu, consistent with protein adduct formation and previous findings (Zhang et al., 2009) (Supplemental Fig. 5).
Stoichiometry of Binding.
The stoichiometry of binding of SCH 66712 to CYP2D6 was determined using scintillation counting and 14C-radiolabeled SCH 66712. Inactivation reactions were treated with 5% sulfuric acid in methanol and protein-precipitated as described under Materials and Methods and as described previously (Chan et al., 1993). Although there was some nonNADPH-dependent binding apparent in samples, the difference between samples that received NADPH and those that did not was ∼1.2 nmol of SCH 66712 bound/nmol CP2D6 (Table 1). Therefore, NADPH-dependent binding indicated a roughly 1.2:1 stoichiometry between SCH 66712 and CYP2D6 (Table 1). Because heme was not present in the protein precipitate after acid washes, this stoichiometry is indicative of protein adduction.
TABLE 1.
Covalent binding of SCH 66712 to CYP2D6
| Radiolabeled Protein |
|||
|---|---|---|---|
| +NADPH | −NADPH | NADPH-Dependent | |
| nmol SCH 66712/nmol of total CYP2D6 | |||
| Precipitation assay (n = 3) | 3.15 | 1.95 | 1.20 |
Metabolism of SCH 66712.
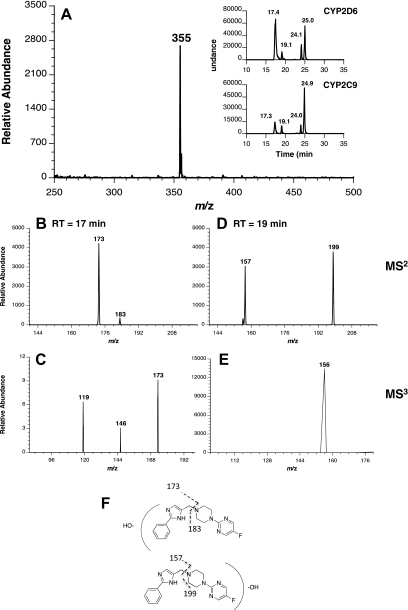
Four SCH 66712 metabolites were observed with a molecular ion at m/z 355 in samples incubated with CYP2D6 Supersomes and NADPH (Fig. 7). The elution times for these products were 17, 19, 24, and 25 min. The metabolites were present in the same abundance when GSH or NAC was included in the reactions, although no GSH or NAC conjugates were observed (both GSH and NAC in reduced forms were observed; data not shown). The m/z 355 products were not observed in control (no NADPH) reactions. The masses of these products are all consistent with mono-oxygenation of SCH 66712. Collision-induced dissociation (CID) fragmentation (MS2) of the mono-oxygenated product at 17 min gave primarily two peaks, m/z 173 and m/z 183 (splitting of the molecule at the methylene group) (Fig. 7; Supplemental Fig. 6). This result is consistent with addition of oxygen to the side of the molecule containing the phenyl and imidazole rings. Further CID fragmentation (MS3) of m/z 173 gave m/z 119 and m/z 146, and CID of m/z 146 (MS4) gave m/z 119. The m/z 146 fragment is consistent with loss of an HCN fragment from the imidazole ring, whereas the m/z 119 fragment is consistent with loss of C3H5N from the imidazole ring and methyl substituent (Fig. 7; Supplemental Fig. 6). Both results suggest mono-oxygenation of the phenyl ring; this is consistent with predictions from molecular modeling experiments (vide infra). Because of the low abundance of m/z 183, we were unable to observe fragmentation of that peak although m/z 183 is consistent with unmodified SCH 66712 (Fig. 8). The presence of dioxygenated, dehydrogenated, ring-opened, or N-dealkylated SCH 66712 metabolites was not observed or supported by MSn data.
Fig. 7.
Analysis of SCH 66712 metabolites formed by CYP2D6 and CYP2C9 in the presence of NADPH. CYP2D6 or CYP2C9 was incubated with SCH 66712 and NADPH, and the resulting reaction mixtures were prepared for MS as described under Materials and Methods. A, mass spectrum of the molecular ion eluting at 17 min and extracted ion chromatogram of the molecular ion [M + H]+ with m/z of 355 (inset). B, tandem mass spectrometry (MS/MS) of the monooxygenated SCH 66712 metabolite eluting at 17 min with m/z 355. C, MS3 of the m/z 173 ion from B. D, MS/MS of the monooxygenated SCH 66712 metabolite eluting at 19 min with m/z 355. E, MS3 of the m/z 199 ion from D. F, MS2 fragmentation patterns giving rise to observed mass spectra in B and D. Further analysis of fragmentation patterns is given in Supplemental Fig. 6. The data shown are representative of multiple analyses, and results with CYP2C9 were identical to those with CYP2C19 (data not shown).
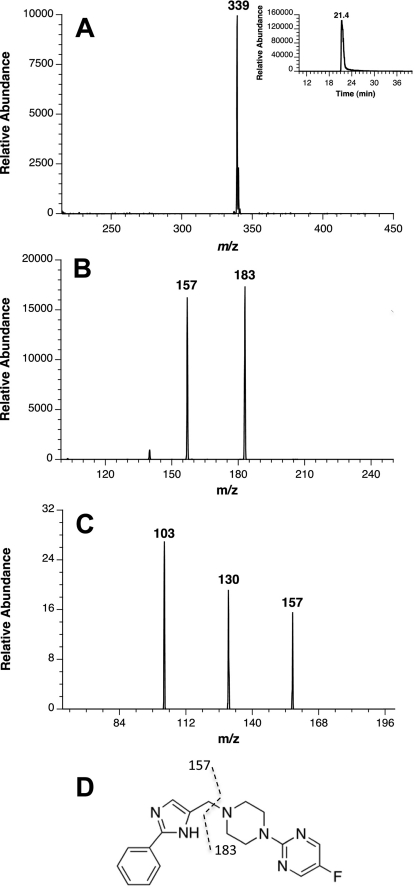
Fig. 8.
Mass spectral analysis of SCH 66712. A, mass spectrum of the molecular ion eluting at 21 min and extracted ion chromatogram of the molecular [M + H]+ ion with m/z of 339 (inset). B, tandem mass spectrometry of SCH 66712. C, MS3 of the m/z 157 ion from B. D, MS2 fragmentation pattern of SCH 66712.
The m/z 355 peaks eluting at 19, 24, and 25 min (Fig. 7, D and E) probably represent monooxygenation at different sites of the SCH 66712 molecule. The major fragments observed were at m/z 157 and m/z 199 (Fig. 7D) consistent with monooxygenation on the side of the molecule with the piperazine and fluorinated heteroaromatic rings and might include hydroxylation, N-oxide formation, or piperazine ring oxidation consistent with what has been reported in the literature for other piperazine-containing compounds (Kamel et al., 2010; Miraglia et al., 2010). CID analysis (MS3) did not confirm specific site modifications but suggested that the modification is on the piperazine ring (Supplemental Fig. 6). Because of the low abundance of fragments, further fragmentation experiments were not possible.
In comparison, fragmentation of the parent SCH 66712 ([M + H]+ with m/z 339) yielded primarily two peaks, m/z 157 (16 amu less than the m/z 173 observed in the metabolite peak at 17 min) and m/z 183 (16 amu less than the m/z 199 observed in the metabolite peak at 19, 24, and 25 min) (Fig. 8). Further fragmentation (MS3) of m/z 157 gave m/z 103 and m/z 130, both 16 amu less than that of the SCH 66712 metabolite.
When CYP2C9 and CYP2C19 were used in metabolism studies with SCH 66712, the same metabolites with a molecular ion of m/z 355 were observed (Fig. 7A, inset; data for CYP2C19 are not shown but were identical to those for CYP2C9). In addition, the fragmentation patterns noted for each metabolite were the same as those observed with CYP2D6 (Fig. 7). The metabolites were also present when GSH or NAC was included in the reactions, although no GSH or NAC conjugates were observed. However, the distribution of each of the metabolites is different with much less formation of the peak at 17 min with CYP2C9 and CYP2C19. The peak at 17 min represents monooxygenation of SCH 66712, most likely on the phenyl ring. The other three metabolites observed, all of which represent monooxygenation on SCH 66712 at the piperazine or fluorinated heteroaromatic ring, are present in similar abundance in reactions with CYP2C9 and CYP2C19 as was observed with CYP2D6 (Fig. 7A, inset). This result of oxygenation by CYP2C9 and CYP2C19 on SCH 66712 at the piperazine or fluorinated heterocyclic aromatic rings is also consistent with molecular docking experiments (vide infra). No metabolites of SCH 66712 were observed from incubations with CYP3A4.
Molecular Modeling.
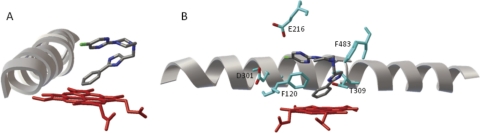
A series of molecular modeling studies were performed to better understand the metabolism of SCH 66712 that would lead to inactivation of CYP2D6. Initial docking experiments showed that the phenyl group of SCH 66712 is positioned closest to the heme iron of CYP2D6 (Fig. 9A). This finding is consistent with MS metabolite studies, which indicated that monooxygenation on the phenyl end of SCH 66712 by CYP2D6 produces the most abundant metabolite (Fig. 7A, inset). The phenyl group is ∼2.1 Å from the heme iron, within reasonable distance for metabolism.
Fig. 9.
Molecular modeling of SCH 66712 bound to CYP2D6. AutoDock was used to model binding of SCH 66712 to the active site of CYP2D6 as described under Materials and Methods. A, docking simulation of SCH 66712 with CYP2D6 is shown. The phenyl moiety of SCH 66712 faces the heme iron. Helix I is also shown. B, active site amino acids Phe120, Phe483, Asp301, Glu216, and Thr309 are shown. The distance from Thr309 oxygen to the phenyl ring of SCH 66712 is 2.6 Å. The heme is shown in red.
Previous studies have identified the active site residues of CYP2D6 associated with ligand binding and orientation as Asp301, Glu216, Phe483, and Phe120 (McLaughlin et al., 2005; Rowland et al., 2006; Ito et al., 2008; Marechal et al., 2008). In the docking model, Phe120 showed π-π stacking with the fluorinated heterocyclic aromatic ring as well as edge-to-edge interaction with the phenyl group of SCH 66712 (distances of 3.2 and 2.7 Å, respectively). Glu216 is within hydrogen bonding distance to a nitrogen in the fluorinated heterocyclic ring of SCH 66712 (∼3.5 Å). The closest nucleophile identified as a potential target for inactivation by an electrophilic SCH 66712 metabolic intermediate was Thr309, an amino acid positioned at the juncture of the phenyl and imidazole rings of SCH 66712 and ∼2.6 Å from the phenyl ring (Fig. 9B).
In contrast, when SCH 66712 was modeled with CYP2C9 (structure by Wester et al., 2004) the binding orientation was seen most commonly in opposite orientation, namely the fluorinated heterocyclic aromatic ring was pointing toward the heme with a distance of ∼5 Å to the heme iron from the ring center (Supplemental Fig. 7). However, a horizontal binding orientation was also frequently observed in simulations with CYP2C9, an orientation not observed with CYP2D6 (Supplemental Fig. 7). The horizontal binding orientation may be more consistent with observed SCH 66712 metabolites that result from monooxygenation of either end of SCH 66712 (Fig. 7). In the metabolism studies both CYP2C9 and CYP2C19 produced metabolites of SCH 66712 that are more commonly monooxygenated on the piperazine or fluorinated heterocyclic aromatic ring end of the molecule (Fig. 8), consistent with the molecular simulation models.
Discussion
SCH 66712 was the first reported mechanism-based inhibitor of CYP2D6 (Palamanda et al., 2001). Inactivation of CYP2D6 is potent with ∼90% loss of enzyme activity within ∼15 min and a low partition ratio of ∼3 (Figs. 3 and 5). Within the same incubation period, the loss of native heme and CO-binding ability are not reduced as effectively (Figs. 3 and 4). These results support the role of SCH 66712 as an inactivator of CYP2D6 by adduct formation with the apoprotein rather than with the heme.
LC-ESI-MS of microsomal proteins (Supersomes) yielded a deconvoluted mass of CYP2D6 of 55,781 Da, consistent with reported values (Supplemental Fig. 4). However, when CYP2D6 was inactivated by SCH 66712 in the presence of NADPH, we were unable to determine a definitive mass increase for the apoprotein because of loss of ionizable P450 or aggregation after adduction or both. The inability to detect the adducted mass of CYP2D6 after inactivation is not uncommon, and there are other reports in the literature that have shown similar results with CYP2B6, CYP3A4, and others (Bateman et al., 2004; Kent et al., 2006). The use of radiolabeled SCH 66712 in the inactivation assays allowed for the detection of radiolabeled CYP2D6 using SDS-PAGE with audioradiography and Western blotting supporting adduct formation (Fig. 6). Using radiochemical analysis and scintillation counting, the stoichiometry of binding was determined to be ∼1.2, further supporting protein adduction (Table 1).
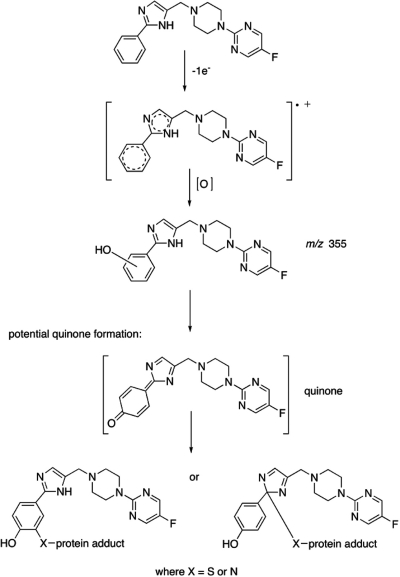
Metabolism of SCH 66712 by CYP2D6 produced monooxygenated metabolites (m/z 355) (Fig. 7). No dioxygenated, dehydrogenated, ring-opened, or N-dealkylated metabolites were observed from reactions with CYP2D6, CYP2C9, or CYP2C19. The m/z 355 metabolite eluting at 17 min is present in more abundant quantities in reactions with CYP2D6 than in those with CYP2C9 or CYP2C19, whereas the other three metabolites are present roughly in the same amounts (Fig. 7A, inset). Furthermore, the metabolite at 17 min is the only one that represents modification of SCH 66712 at the end of the molecule with the phenyl and imidazole groups (Fig. 7, B and C). Thus, the metabolite eluting at 17 min could be the one that leads to the inactivation of CYP2D6. CID analysis of this metabolite compared with that of the parent SCH 66712 supports monooxygenation of the phenyl ring, most certainly a hydroxylation on the aromatic ring. If the hydroxylation were to occur in the para position relative to the imidazole ring, a methylene quinone-derived electrophile capable of inactivating the protein via the adduction on the imidazole or the phenyl ring could arise (Fig. 10). Quinones are known to be involved in enzyme inactivation as reported for CYP3A4 by raloxifene (Chen et al., 2002).
Fig. 10.
Proposed metabolism and inactivation pathway for SCH 66712 by CYP2D6. p-Hydroxylation of the phenyl ring can lead to the formation of a methylene quinone. Collapse of the quinone with the capture of a nucleophile on the phenyl or the imidazole ring may lead to protein adduct formation.
Given the very low partition ratio of the inactivation and the fact that previous studies indicated no protection from inactivation of CYP2D6 by SCH 66712 upon addition of exogenous nucleophiles (Palamanda et al., 2001), it is perhaps not surprising that we were unable in the current study to trap any reactive intermediates of SCH 66712 by use of GSH or NAC. GSH and NAC both have soft nucleophile cysteine residues. Because there was no detection of adduction of these nucleophiles in the incubations of SCH 66712 with CYP2D6 and NADPH, a different type of nucleophile (a hard nucleophile) might be the target for inactivation by SCH 66712 or these nucleophiles may simply not have access to the inactivator.
Molecular docking experiments were used to predict the modes of SCH 66712 binding to CYP2D6 and CYP2C9 and to rationalize experimental observations of inhibition in one case (with CYP2D6) versus little inhibition in the other (with CYP2C9), and the more abundant formation of a metabolite representing hydroxylation of the phenyl ring (with CYP2D6). SCH 66712 is positioned in the active site of CYP2D6 such that the phenyl group is 2.2 Å from the heme iron. Conversely, with CYP2C9, the fluorinated heterocyclic aromatic ring of SCH 66712 is closest to the heme iron as well as a low energy binding conformation that is more horizontal (Supplemental Fig. 7). The active site of CYP2D6 contains Thr309, a possible nucleophilic target of a reactive electrophile (Fig. 9). Use of a Thr309 mutant of CYP2D6 would allow for further investigation of this potential site of inactivation. At this time, the identity of amino acid targets for the any of the mechanism-based inhibitors of CYP2D6 is unknown.
Interactions between aromatic moieties, i.e., π-π interactions, play a crucial role in binding and substrate conformation in enzyme active sites and lead to increased inhibitory potential of mechanism-based inhibitors (Sridhar et al., 2010). In particular, modeling research with polyaromatic hydrocarbon inhibitors of P450s has shown that face-to-face and edge-to-edge orientations are unfavorable and may be preliminary steps leading to mechanism-based inhibition (versus edge-to-face or misaligned stacking orientations that are favorable orientations not as strongly associated with mechanism-based inhibition) (Sridhar et al., 2010). Our modeling simulations of binding show SCH 66712 interacting with Phe120 via π face-to-face and edge-to-edge interactions at distances of 3.2 and 2.7 Å, respectively (Fig. 9). These distances are well within the range of previously reported binding distances for π-π interactions that can be associated with potent inhibition (Sridhar et al., 2010). Thus, the modeling studies shown here are consistent with the hypothesis that face-to-face and edge-to-edge interactions could be preliminary steps in mechanism-based inhibition.
Because SCH 66712 was first reported as a mechanism-based inhibitor of CYP2D6, a few other compounds have been reported to be mechanism-based inhibitors, including a related compound, EMTPP, that was intentionally studied on the basis of its similarity to SCH 66712 (Hutzler et al., 2004). For EMTPP, the methylene carbon of the ethyl substituent on the imidazole group is the site of oxidation (see structure of EMTPP in Supplemental Fig. 8) (Hutzler et al., 2004). For SCH 66712, there is a phenyl group at this location that is the site of oxidation. The Ki values for inhibition of CYP2D6 (from Supersomes) by SCH 66712 and by EMTPP are 0.55 and 5.5 μM, respectively, consistent with much more potent inhibition of CYP2D6 by SCH 66712 (Palamanda et al., 2001; Hutzler et al., 2004). Likewise, the ki for inhibition by SCH 66712 is 0.32 min−1, whereas it is lower for inhibition by EMTPP at 0.09 min−1. The partition ratios further differentiate these compounds with a value of ∼3 for SCH 66712 and of ∼99 for EMTPP. The difference in partition ratio values might also explain the fact that few metabolites of SCH 66712 could be found upon inactivation, whereas reactions with EMTPP produced at least four abundant metabolites in addition to the adducted CYP2D6 (Hutzler et al., 2004).
Molecular modeling studies shown here with CYP2D6 and EMTPP show that EMTPP, like SCH 66712, binds with the fluorinated ring away from the heme iron with a distance of 2.9 Å from the site of oxidation to the heme iron (Supplemental Fig. 8). The distance between the heme iron atom and the proposed site of oxidation in SCH 66712 is 2.1 Å. For other known P450 mechanism-based inhibitors, modeling studies show the moiety of the inhibitor that is activated for protein adduction pointing toward the heme iron. This includes inhibitors such as tBPA with CYP2B1 and CYP2B4 (Zhang et al., 2009; Lin et al., 2010) and mifepristone and raloxifene with CYP3A4 (Zhang et al., 2009; Moore et al., 2010), as well as inhibitors that inactivate P450s by heme alkylation such as gemfibrozil glucuronide with CYP2C8 (Baer et al., 2009) and others (Sridhar et al., 2010).
In conclusion, the inactivation of CYP2D6 by SCH 66712 is very potent with production of few metabolites. SCH 66712 causes inactivation probably by apoprotein adduction that we postulate would be at Thr309 on the basis of modeling studies. Finally, the current study supports the notion that some substituted imidazole groups may tend to give rise to inactivation of CYP2D6 and should be examined for mechanism-based inactivation early in the drug discovery process.
Supplementary Material
Acknowledgments
We thank Dr. F. P. Guengerich for making this collaboration possible. We also thank Drs. P. Hollenberg, H. Zhang, Y. Osawa, A. Venter, and G. Slough and G. Cavey for helpful comments. We thank Dr. M. V. Martin for preparing purified recombinant CYP2D6 and P450 NADPH reductase. We are indebted to Dr. Mark Hail of Novatia for assistance with ProMass software and to Drs. D. Hachey and W. Calcutt of Vanderbilt University for training in use and maintenance of MS.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants 1R15-GM086767-01, 3R15-GM086767-01S1] (to L.L.F.); Kalamazoo College [Grant 52006304 from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program]; and the Hutchcroft Fund of Kalamazoo College.
Parts of this work were previously presented at the following conference: Mocny CS, Arthur EJ, Butler BF, Diffenderfer LE, Palamanda JR, Guengerich FP, Nomeir AA, and Furge LL (2010) Mechanism-based inhibition of human cytochrome P450 2D6 by Schering 66712. 2010 American Society for Biochemistry and Molecular Biology Annual Meeting; 2010 Apr 24–38; Anaheim, CA. American Society for Biochemistry and Molecular Biology, Bethesda, MD.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.037630.
- P450
- cytochrome P450
- SCH 66712
- 5-fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine
- HPLC
- high-performance liquid chromatography
- ACN
- acetonitrile
- MS
- mass spectrometry
- TFA
- trifluoroacetic acid
- LC
- liquid chromatography
- ESI
- electrospray ionization
- PAGE
- polyacrylamide gel electrophoresis
- PDB
- Protein Data Bank
- tBPA
- 4-tert-butylphenylacetylene
- amu
- atomic mass units
- NAC
- N-acetylcysteine
- EMTPP
- (1-[(2-ethyl-4-methyl-1H-imidazol-5-yl)-methyl]-4-[4-(trifluoromethyl)-2-pyridinyl]piperazine
- CID
- collision-induced dissociation.
Authorship Contributions
Participated in research design: Nagy, Mocny, Diffenderfer, Hsi, Palamanda, Nomeir, and Furge.
Conducted experiments: Nagy, Mocny, Diffenderfer, Hsi, Butler, Arthur, Fletke, Palamanda, and Furge.
Contributed new reagents or analytic tools: Palamanda and Nomeir.
Performed data analysis: Nagy, Mocny, Diffenderfer, Hsi, Arthur, and Furge.
Wrote or contributed to the writing of the manuscript: Nagy, Diffenderfer, Nomeir, and Furge.
Other: Furge acquired funding for the research.
References
- Baer BR, DeLisle RK, Allen A. (2009) Benzylic oxidation of gemfibrozil-1-O-β-glucuronide by P450 2C8 leads to heme alkylation and irreversible inhibition. Chem Res Toxicol 22:1298–1309 [DOI] [PubMed] [Google Scholar]
- Bateman KP, Baker J, Wilke M, Lee J, Leriche T, Seto C, Day S, Chauret N, Ouellet M, Nicoll-Griffith DA. (2004) Detection of covalent adducts to cytochrome P450 3A4 using liquid chromatography mass spectrometry. Chem Res Toxicol 17:1356–1361 [DOI] [PubMed] [Google Scholar]
- Chan WK, Sui Z, Ortiz de Montellano PR. (1993) Determinants of protein modification versus heme alkylation: inactivation of cytochrome P450 1A1 by 1-ethynylpyrene and phenylacetylene. Chem Res Toxicol 6:38–45 [DOI] [PubMed] [Google Scholar]
- Chen Q, Ngui JS, Doss GA, Wang RW, Cai X, DiNinno FP, Blizzard TA, Hammond ML, Stearns RA, Evans DC, et al. (2002) Cytochrome P450 3A4-mediated bioactivation of raloxifene: irreversible enzyme inhibition and thiol adduct formation. Chem Res Toxicol 15:907–914 [DOI] [PubMed] [Google Scholar]
- Correia MA, Ortiz de Montellano PR. (2005) Inhibition of cytochrome P450 enzymes, in Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano PR. ed) pp 247–322, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- Gillam EM, Guo Z, Martin MV, Jenkins CM, Guengerich FP. (1995) Expression of cytochrome P450 2D6 in Escherichia coli, purification, and spectral and catalytic characterization. Arch Biochem Biophys 319:540–550 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2003) Cytochromes P450, drugs, and diseases. Mol Interv 3:194–204 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2005) Human cytochrome P450 enzymes, in Cytochrome P450: Structure, Mechanism, and Biochemistry (Ortiz de Montellano PR. ed) pp 377–530, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- Guengerich FP, Shimada T. (1991) Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem Res Toxicol 4:391–407 [DOI] [PubMed] [Google Scholar]
- Hanna IH, Kim MS, Guengerich FP. (2001a) Heterologous expression of cytochrome P450 2D6 mutants, electron transfer, and catalysis of bufuralol hydroxylation: the role of aspartate 301 in structural integrity. Arch Biochem Biophys 393:255–261 [DOI] [PubMed] [Google Scholar]
- Hanna IH, Krauser JA, Cai H, Kim MS, Guengerich FP. (2001b) Diversity in mechanisms of substrate oxidation by cytochrome P450 2D6. Lack of an allosteric role of NADPH-cytochrome P450 reductase in catalytic regioselectivity. J Biol Chem 276:39553–39561 [DOI] [PubMed] [Google Scholar]
- Huey R, Morris GM, Olson AJ, Goodsell DS. (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152 [DOI] [PubMed] [Google Scholar]
- Hutzler JM, Steenwyk RC, Smith EB, Walker GS, Wienkers LC. (2004) Mechanism-based inactivation of cytochrome P450 2D6 by 1-[(2-ethyl-4-methyl-1H-imidazol-5-yl)methyl]-4-[4-(trifluoromethyl)-2-pyridinyl]piperazine: kinetic characterization and evidence for apoprotein adduction. Chem Res Toxicol 17:174–184 [DOI] [PubMed] [Google Scholar]
- Islam SA, Wolf CR, Lennard MS, Sternberg MJ. (1991) A three-dimensional molecular template for substrates of human cytochrome P450 involved in debrisoquine 4-hydroxylation. Carcinogenesis 12:2211–2219 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kondo H, Goldfarb PS, Lewis DF. (2008) Analysis of CYP2D6 substrate interactions by computational methods. J Mol Graph Model 26:947–956 [DOI] [PubMed] [Google Scholar]
- Kamel A, Obach RS, Colizza K, Wang W, O'Connell TN, Coelho RV, Jr, Kelley RM, Schildknegt K. (2010) Metabolism, pharmacokinetics, and excretion of the 5-hydroxytryptamine1B receptor antagonist elzasonan in humans. Drug Metab Dispos 38:1984–1999 [DOI] [PubMed] [Google Scholar]
- Kent UM, Lin HL, Mills DE, Regal KA, Hollenberg PF. (2006) Identification of 17-α-ethynylestradiol-modified active site peptides and glutathione conjugates formed during metabolism and inactivation of P450s 2B1 and 2B6. Chem Res Toxicol 19:279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koymans L, Vermeulen NP, van Acker SA, te Koppele JM, Heykants JJ, Lavrijsen K, Meuldermans W, Donné-Op den Kelder GM. (1992) A predictive model for substrates of cytochrome P450-debrisoquine (2D6). Chem Res Toxicol 5:211–219 [DOI] [PubMed] [Google Scholar]
- Lin HL, Zhang H, Jushchyshyn M, Hollenberg PF. (2010) Covalent modification of Thr302 in CYP2B1 by the mechanism-based inactivator 4-tert-butylphenylacetylene. J Pharmacol Exp Ther 333:663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal JD, Kemp CA, Roberts GC, Paine MJ, Wolf CR, Sutcliffe MJ. (2008) Insights into drug metabolism by cytochromes P450 from modelling studies of CYP2D6-drug interactions. Br J Pharmacol 153 (Suppl 1):S82–S89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LA, Paine MJ, Kemp CA, Maréchal JD, Flanagan JU, Ward CJ, Sutcliffe MJ, Roberts GC, Wolf CR. (2005) Why is quinidine an inhibitor of cytochrome P450 2D6? The role of key active-site residues in quinidine binding. J Biol Chem 280:38617–38624 [DOI] [PubMed] [Google Scholar]
- Miraglia L, Pagliarusco S, Bordini E, Martinucci S, Pellegatti M. (2010) Metabolic disposition of casopitant, a potent neurokinin-1 receptor antagonist, in mice, rats, and dogs. Drug Metab Dispos 38:1876–1891 [DOI] [PubMed] [Google Scholar]
- Moore CD, Shahrokh K, Sontum SF, Cheatham TE, 3rd, Yost GS. (2010) Improved cytochrome P450 3A4 molecular models accurately predict the Phe215 requirement for raloxifene dehydrogenation selectivity. Biochemistry 49:9011–9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. (1998) Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem 19:1639–1662 [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239:2370–2378 [PubMed] [Google Scholar]
- Oritz de Montellano PR, Correia MA. (1983) Suicidal destruction of cytochrome P-450 during oxidative drug metabolism. Annu Rev Pharmacol Toxicol 23:481–503 [DOI] [PubMed] [Google Scholar]
- Palamanda JR, Casciano CN, Norton LA, Clement RP, Favreau LV, Lin C, Nomeir AA. (2001) Mechanism-based inactivation of CYP2D6 by 5-fluoro-2-[4-[(2-phenyl-1H-imidazol-5-yl)methyl]-1-piperazinyl]pyrimidine. Drug Metab Dispos 29:863–867 [PubMed] [Google Scholar]
- Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, et al. (2006) Crystal structure of human cytochrome P450 2D6. J Biol Chem 281:7614–7622 [DOI] [PubMed] [Google Scholar]
- Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. (1994) Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther 270:414–423 [PubMed] [Google Scholar]
- Silverman RB. (1988) Mechanism-Based Enzyme Inactivation: Chemistry and Enzymology, CRC Press, Boca Raton, FL [Google Scholar]
- Sridhar J, Jin P, Liu J, Foroozesh M, Stevens CL. (2010) In silico studies of polyaromatic hydrocarbon inhibitors of cytochrome P450 enzymes 1A1, 1A2, 2A6, and 2B1. Chem Res Toxicol 23:600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. J Biol Chem 279:35630–35637 [DOI] [PubMed] [Google Scholar]
- Wolff T, Distlerath LM, Worthington MT, Groopman JD, Hammons GJ, Kadlubar FF, Prough RA, Martin MV, Guengerich FP. (1985) Substrate specificity of human liver cytochrome P-450 debrisoquine 4-hydroxylase probed using immunochemical inhibition and chemical modeling. Cancer Res 45:2116–2122 [PubMed] [Google Scholar]
- Zhang H, Lin HL, Walker VJ, Hamdane D, Hollenberg PF. (2009) tert-Butylphenylacetylene is a potent mechanism-based inactivator of cytochrome P450 2B4: inhibition of cytochrome P450 catalysis by steric hindrance. Mol Pharmacol 76:1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.