Abstract
A favored hypothesis to explain the pathology underlying nuclear envelopathies is that mutations in nuclear envelope proteins alter genome/chromatin organization and thus gene expression. To identify nuclear envelope proteins that play roles in genome organization, we analyzed nuclear envelopes from resting and phytohemagglutinin-activated leukocytes because leukocytes have a particularly high density of peripheral chromatin that undergoes significant reorganization upon such activation. Thus, nuclear envelopes were isolated from leukocytes in the two states and analyzed by multidimensional protein identification technology using an approach that used expected contaminating membranes as subtractive fractions. A total of 3351 proteins were identified between both nuclear envelope data sets among which were 87 putative nuclear envelope transmembrane proteins (NETs) that were not identified in a previous proteomics analysis of liver nuclear envelopes. Nuclear envelope localization was confirmed for 11 new NETs using tagged fusion proteins and antibodies on spleen cryosections. 27% of the new proteins identified were unique to one or the other of the two leukocyte states. Differences in expression between activated and resting leukocytes were confirmed for some NETs by RT-PCR, and most of these proteins appear to only be expressed in certain types of blood cells. Several known proteins identified in both data sets have functions in chromatin organization and gene regulation. To test whether the novel NETs identified might include those that also regulate chromatin, nine were run through two screens for different chromatin effects. One screen found two NETs that can recruit a specific gene locus to the nuclear periphery, and the second found a different NET that promotes chromatin condensation. The variation in the protein milieu with pharmacological activation of the same cell population and consequences for gene regulation suggest that the nuclear envelope is a complex regulatory system with significant influences on genome organization.
The nuclear envelope (NE)1 is a double membrane system consisting of the intermediate filament nuclear lamin polymer and associated proteins attached to the inner nuclear membrane (INM) (1), nuclear pore complexes (NPCs) that direct transport of soluble macromolecules in and out of the nucleus (2), and the outer nuclear membrane (ONM) and associated proteins. Structurally, the ONM is continuous with the endoplasmic reticulum (ER) and is studded with ribosomes (3), yet it also contains unique proteins, many of which connect the cytoskeleton to the NE (4). On the other side, lamins and many INM proteins directly connect chromatin to the NE. Lamins and an increasing number of nuclear envelope transmembrane proteins (NETs) have been linked to a similarly increasing number of diseases ranging from muscular dystrophy to neuropathy, dermopathy, lipodystrophy, bone disorders, and progeroid aging syndromes (5, 6).
A favored hypothesis to explain how different NE proteins can produce such a wide range of disease pathologies is that chromatin-NE connections are disrupted with NE protein mutations, yielding changes in gene regulation. This hypothesis is supported by observations that the distribution of dense peripheral chromatin is affected in fibroblasts from patients with NE-linked muscular dystrophy, cardiomyopathy, mandibuloacral dysplasia, and progeria (7–10). Furthermore, many binding partners have been identified for NETs that are either chromatin proteins, enzymes that modify chromatin proteins, or regulators of gene expression (1, 11). These include markers of silent chromatin such as heterochromatin protein 1 (12) and proteins that modify chromatin to a silent conformation such as histone deacetylase 3 (13). The importance of the NE to global genome organization has been underscored by several recent studies that showed that affinity-based recruitment of a specific chromosome locus by the NE both pulled entire chromosomes to the periphery and affected gene regulation in complex ways (14–16).
To identify NE proteins likely to be involved in genome organization, we turned to lymphocytes as a model system. Lymphocytes in the resting state tend to have massive amounts of dense peripheral chromatin as determined by electron microscopy studies. Upon activation with phytohemagglutinin, this dense chromatin largely dissipates as the cells actively express genes (17–20). Thus, to identify proteins that might be involved in tethering heterochromatin to the NE or in changing its organization, we analyzed the NE proteomes of leukocyte populations (∼70% lymphocytes) in both the resting and phytohemagglutinin (PHA)-activated states. The previously validated subtractive approach was applied (21) using microsomes and mitochondria, the principal membrane contaminants expected, as subtractive fractions.
Many new NE proteins were identified that had not been identified in previous NE proteomics investigations using liver and neuroblastoma cells (21, 22). NE residence was confirmed for 12 novel NETs by expression of epitope-tagged versions and using antibodies on tissue cryosections.
Roughly one-quarter of the proteins identified varied between the resting and activated states. Some NET differences between the two data sets were confirmed by RT-PCR. Among the known proteins identified were several that suggest that changes in NE composition associated with PHA activation contribute to gene regulation. Novel NETs identified also appear to play significant roles in genome organization/regulation as we found that several can either recruit a specific locus to the nuclear periphery or promote chromatin condensation. As several studies have implicated misregulation of chromatin organization in NE diseases (7, 8), these newly identified NETs may contribute to the diverse pathologies associated with NE diseases.
EXPERIMENTAL PROCEDURES
Preparation and Activation of Leukocytes
Human buffy coats from healthy donors were obtained anonymously from the Scottish National Blood Transfusion Service according to approved ethics protocols. Buffy coats were diluted with phosphate-buffered saline (PBS), and a peripheral blood mononuclear cell (PBMC) fraction was isolated by centrifugation on Ficoll-Histopaque-1077 (Sigma) at 250 × g for 30 min. Cells were diluted in PBS, pelleted, and resuspended in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 100 μg/μl penicillin, and 100 μg/μl streptomycin sulfate (Invitrogen). PBMCs were activated by addition of 2.5 μg/ml PHA (Sigma) for 7 days. To determine the composition of PBMC preparations, cells were analyzed by FACS using antibodies from Caltag Medsystems to CD19 for resting B-cells and CD19/CD4 for activated B-cells, CD3 for resting T-cells and CD3/CD45 for activated T-cells, CD3/CD4 for resting helper T-cells and CD4/CD45 for activated helper T-cells, CD3/CD8 for resting cytotoxic T-cells and CD8/CD45 for activated cytotoxic T-cells, and CD11b/CD14 for myeloid cells.
Preparation of Fractions
NEs were isolated using procedures specifically modified for blood leukocytes and detailed previously (23). In brief, nuclei were first isolated from PBMCs by hypotonic lysis in 10 mm HEPES, pH 7.4, 1.5 mm MgCl2, 10 mm KCl, 2 mm DTT using a “loose” Dounce homogenizer (Wheaton). Nuclei were pelleted at 1000 × g for 10 min to separate them from small vesicles and mitochondria that require higher speeds to pellet. To float/remove contaminating membranes, nuclei were resuspended in SHKM (50 mm HEPES, pH 7.4, 25 mm KCl, 5 mm MgCl2, 1 mm DTT and 1.8 m sucrose) and pelleted through a 5 ml 2.1 m sucrose cushion in an SW28 swinging bucket rotor (Beckman) at 4 °C for 2 h at 82,000 × g. NEs were then prepared from isolated nuclei by two rounds of digestion with DNase and RNase in 0.3 m sucrose, 10 mm HEPES, pH 7.4, 2 mm MgCl2, 0.5 mm CaCl2, 2 mm DTT for 20 min followed by layering onto the same buffer with 0.9 m sucrose and centrifugation at 6000 × g for 10 min at 4 °C.
The PBMC cell populations had very little ER, so microsomes representing contaminant membrane fractions were instead isolated from rat liver using well established procedures (24, 25). In brief, liver was diced and homogenized in SHKM using a motorized Dounce homogenizer. After removing nuclei as for NE preparations, 0.5 mm EDTA was added to inhibit metalloproteinases, and mitochondria and other debris from postnuclear supernatants were also removed by pelleting at 10,000 × g for 15 min. The supernatant was made to 2 m sucrose with SHKM and then overlaid with 1.86 and 0.25 m sucrose layers. This was then subjected to centrifugation in an SW28 swinging bucket rotor (Beckman) at 4 °C for 4 h at 57,000 × g to float microsomes. The material between the 1.86 and 0.25 m layers was then diluted 4-fold with 0.25 m SHKM and pelleted at 152,000 × g in a type 45 Ti rotor (Beckman) for 1 h.
NE preparations were divided, and one portion was further extracted on ice with 400 mm NaCl, 25 mm HEPES, pH 8.0, 5 mm MgCl2, 10 mm DTT, 1% β-d-octyl glucoside, and material used for mass spectrometry analysis was pelleted at 15,000 × g for 30 min and washed in sterile H2O. Another portion of NEs and separate microsomes were further extracted with 0.1 n NaOH, 10 mm DTT, and the material used for mass spectrometry was pelleted by centrifugation at 150,000 × g for 30 min and washed three times in sterile H2O.
MudPIT
Two types of digestion were applied to NE and microsome pellets (supplemental Table S1). For the first, dried membrane pellets were incubated overnight in 90% formic acid and 500 mg/ml cyanogen bromide (CNBr). Ammonium hydroxide (NH4OH) was then added to pH 8.5, solid urea was added to 8 m, and tris(2-carboxylethyl)phosphine hydrochloride (TCEP) was added to 5 m and incubated for 30 min. Next free cysteines were alkylated with 20 mm iodoacetamide (IAM) for 30 min prior to addition of endoproteinase Lys-C (Roche Applied Science) at a 1:100 (w/w) enzyme to protein ratio for 6 h at 37 °C. Urea was diluted to 2 m with 0.1 m Tris-HCl, pH 8.5, and CaCl2 (0.5 mm) and modified trypsin (1:100, w/w) were added for 12 h at 37 °C. Digestions were quenched with 5% formic acid (25). For the second digestion (Ti), pellets were solubilized in 0.1 m Tris-HCl, pH 8.5, 8 m urea, 5 mm TCEP. IAM was added to 10 mm for 30 min, and endoproteinase Lys-C and trypsin digestion was performed as above. Samples were centrifuged for 30 min at 17,500 × g. Supernatants were analyzed by MudPIT, whereas pellets were resuspended in 0.1 m sodium carbonate (Na2CO3), pH 11.5, 8 m urea, 5 mm TCEP for 30 min and then in 10 mm IAM for 30 min and then further digested with proteinase K for 4 h at 37 °C and also analyzed by MudPIT.
Each digested sample was analyzed independently by MudPIT as described previously (26–28) using a flow rate of 200–300 nl/min. 12–15 cycles (supplemental Table S1) of 120 min each of increasing salt concentrations followed by organic gradients (5–80% acetonitrile with 0.1% formic acid) slowly released peptides directly into the mass spectrometer (26). The last two or last five (of 12 or 15) chromatography steps consisted of a high salt wash with 5% acetonitrile, 0.1% formic acid, 500 mm ammonium acetate followed by the acetonitrile gradient. The distal application of a 2.5-kV voltage electrosprayed the eluting peptides directly into ion trap mass spectrometers equipped with a nano-LC electrospray ionization source (ThermoFinnigan). Each full MS scan (from 400 to 1600 m/z) was followed by three (XP-Deca) or five (LTQ) MS/MS events (supplemental Table S1) using data-dependent acquisition where the first most intense ion was isolated and fragmented by collision-induced dissociation (at 35% collision energy) followed by the second to third or fifth most intense ions. The raw mass spectrometric data may be downloaded from ProteomeCommons.org by going to the URL indicated in Table I and then inserting the hash identifiers from Table I.
Table I. Hash identifiers to access raw mass spectrometry files for each MudPIT run.
Individual runs are organized by cell fraction, extraction, and protease to generate peptides for analysis. MM, microsomal membrane.
| Cell fraction | Extraction/digestion | Sample name used in supplemental Tables S1–S5 | At https://www.proteomecommons.org/tranche/downloads.jsp, the raw mass spectrometry files can be downloaded using the following hash identifiers. In all cases, there are two files ranging from 100 MB to 1.5 GB that are archived and compressed tar.gz files: use tar −xzf Linux command to uncompress to *.RAW files (1 per MudPIT step) |
|---|---|---|---|
| PBMC NEs | NaOH/CNBr-trypsin | HsNaOH-NE_NT_lymphocytes_CNBrTi_1 | 4mWjs+LvJQb4FUwQD8O3X5MkkLit6/3bHJfJZK3owfOX4jwMtCUQFhRv95ORUTT8gT5HKx6RdVxGlCjy/nOHw/pjLeMAAAAAAAAGUg== |
| PBMC NEs | NaOH/trypsin | HsNaOH-NE_NT_lymphocytes_Ti_1 | 5aTYg17TJJuGrxJ/wra09yMtKGoVwtlgYWi+EK7Np92HD73ed5417f2/x0ndBFzSjLxwFldZheB74gYe829QI8CDDiwAAAAAAAAFrQ== |
| PBMC NEs | NaOH/proteinase K | HsNaOH-NE_NT_lymphocytes_PK_1 | MBcaJI2EOh07R/EEOLUVt85OXiVbebs7/PiRkN3HvoazDKMmEZxw53RLG96Vx9pgGhIxM0Dxk2O05V0QiEiQi0+oUqoAAAAAAAAGlQ== |
| PBMC NEs | Salt-detergent/trypsin | HsSD-NE_NT_lymphocytes_Ti_1 | rgMMQvG4dw8tJ9WeX0DHi0k6vvEclU34F8mzsnExggk9AbENEtfaBPxvTuBlhJ/iAnDU7XQLLNr8fJV2W72mziU6KrMAAAAAAAAFsQ== |
| PBMC NEs | Salt-detergent/proteinase K | HsSD-NE_NT_lymphocytes_PK_1 | bQnUrYCBdxRh8wM32qka9Jg80U4ow7Z0HK+SmdbPbxPVo3TzLgeTqCWgzKPDYAkUltDGXybehz2ZFVq2EJ+G/et3AiwAAAAAAAAGmQ== |
| PHA-treated PBMC NEs | NaOH/CNBr-trypsin | HsNaOH-NE_PHA_lymphocytes_CNBrTi_1 | /0iTLp3cH9L60CR441Dxg/REq/GKoYRz9Kd/QloEd+3h265EsD1S213sb8ic267lWS5gGDdQHV3xR+T7qZ36K4+O/ZUAAAAAAAAGXA== |
| PHA-treated PBMC NEs | NaOH/trypsin | HsNaOH-NE_PHA_lymphocytes_Ti_1 | gKqv8i+5tPSdMhZM9IZW93nSIXTeQUpYhgyc6SEXUgYJAj+AqeF49+VneqJ5RTQQrEnOM9t2ANPZQJWRZAJaKY0uMBAAAAAAAAAFtQ== |
| PHA-treated PBMC NEs | NaOH/proteinase K | HsNaOH-NE_PHA_lymphocytes_PK_1 | pi3VbyTD3uo8WvUrRgmJn0phN7KpMp5yNInoWpvXSInLu/N0cNKjTY79b3okjGy6+e5zSkQATmN2dQzQ4giyJMdVmvMAAAAAAAAGnw== |
| PHA-treated PBMC NEs | Salt-detergent/trypsin | HsSD-NE_PHA_lymphocytes_Ti_1 | 5nw33XkNPBMdAuQNg6HRD2MPyTa7osxDObK2cxSdAWtVHsRV+sC3kW6omUK3PrvBNa4WCarVz3Hnskjrbn4e7JuU6+sAAAAAAAAFuw== |
| PHA-treated PBMC NEs | Salt-detergent/proteinase K | HsSD-NE_PHA_lymphocytes_PK_1 | QPGdJRqWff6OE9isf9e9zSnB9D0xmV0rQzyR7+Jy5Z6hZZNQDzuGYKNO5+vZJFLxasQ7/Tx4lWHp/Jd5yztAJ095NuUAAAAAAAAGpA== |
| Liver MMs | NaOH/CNBr-trypsin | MmNaOH-MM_liver_CNBrTi_1 | Y+ITtCT8NMlD0geT1lJiVcUIdaOHJmM08umEd9yhyGOq/rqjAkuft0Txw1yeznfC8iXNbkfR/0v35Y0qLBhVyvrgaSwAAAAAAAAGBg== |
| Liver MMs | NaOH/CNBr-trypsin | MmNaOH-MM_liver_CNBrTi_2 | 2KBwq4C/GrCKjyrUMRoRdZ55hzMbJr3wc+O3kZqwnFLV/90uaYuK64FxNneldmnQ7HO1dRNBaNG51EGUnz3EV18OrusAAAAAAAAGAA== |
| Liver MMs | NaOH/trypsin | RnNaOH-MM_liver_Ti_1 | bpp6s9dcwf3lzclX0cPDJCJ01f97QesmXl6anF4PVaUNuNmfdIRrtqkELKcBM6l2X08B/d69Hys6o4vUq66TYHBc4CQAAAAAAAAFaQ== |
| Liver MMs | NaOH/trypsin | RnNaOH-MM_liver_Ti_2 | 7wO8Wz/6iv2rZU5WACjlm1sY5sIkiMWTC5X2+Tjyt5SN+fPoCR/LxuWiD3DetTh+oTWNkU7wd6gp2c3XKx8Oybxl6HMAAAAAAAAFaQ== |
| Liver MMs | NaOH/trypsin | RnNaOH-MM_liver_Ti_3 | ElAxLpoQLOHTnIp0FpP1FeRrEckCbdz0VHUt4h9ISX8v8sMEoutFOarYhBK3+d+jUv1Ihe5rRfXhhR7UcXqx0Bicw4sAAAAAAAAFaQ== |
| Liver MMs | NaOH/trypsin | RnNaOH-MM_liver_Ti_4 | cnJ7dLVqF73eGbFqgZyTn3Egb/XL2RzMWrP7gXG41VfvbRri1eHa1uaHWjvMjbXGdvpq4XCQoz1u+gU//rR/MO7/hckAAAAAAAAFaQ== |
| Liver MMs | NaOH/proteinase K | RnNaOH-MM_liver_PK_1 | ENorMHs0SFhkyD9Y5w4gbEh2bVCdl7gT5SF6dnp0zebaxoFG7kYpPTwYFQXGmhPBjKR/ZG8ewsnnSZZcEzmJZvYq8ecAAAAAAAAGVA== |
| Liver MMs | NaOH/proteinase K | RnNaOH-MM_liver_PK_2 | H36mWYfElnzq6ISRTgc6v6VJVs4WxrUkCNa0LTmufeP+OoMPQaxkkBxn6nHlEv8iCgcw8ghryUbgIY9i+Udjk4k8XnsAAAAAAAAGUg== |
RAW files were extracted into ms2 file format (29) using RAW_Xtract v.1.0 (30). For the human PBMC data set (supplemental Table S2), MS/MS spectra were queried for peptide sequence information using SEQUEST v.27 (revision 9) (31) against 30,552 human proteins (non-redundant NCBI sequences on March 4, 2008) plus 162 sequences from usual contaminants (e.g. human keratins etc.) and 313 previously identified NETs (21). To estimate false discovery rates, each non-redundant protein entry was randomized and added to the database bringing the total search space to 61,864 sequences (supplemental Table S1). For the mouse (supplemental Table S3) (21) and rat (supplemental Table S4) liver microsomal membrane data sets, the species-specific databases consisted of 29,998 mouse or 28,400 rat proteins (non-redundant NCBI sequences on June 22, 2007 and July 10, 2006, respectively) complemented as described above with sequences for contaminants and previously identified NETs as well as their corresponding randomized sequences (supplemental Table S1). MS/MS spectra were searched without specifying differential modifications, but +57 Da were added statically to cysteine residues to account for carboxamidomethylation. No enzyme specificity was imposed during searches, setting a mass tolerance of 3 amu for precursor ions and of ±0.5 amu for fragment ions.
Results from different runs were compared using DTASelect and Contrast (32). Spectrum/peptide matches were retained if at least 7 amino acids long with ends complying with proteolytic specificity, e.g. Met, Lys, or Arg before the amino terminus and at the carboxyl terminus of peptides for those cleaved with CNBr prior to trypsin digestion. Other criterion were ΔCn ≥ 0.08, XCorr ≥ 1.8 for singly, 2.0 for doubly, and 3.0 for triply charged spectra, and a maximum Sp rank of 10. For proteinase K-digested samples, no specific peptide ends were imposed, but the ΔCn cutoff was increased to 0.15 (33), and XCorr minima were increased to 2.5 for doubly and 3.5 for triply charged spectra (SEQUEST parameters for the spectrum to peptide matches leading to the identification of proteins from human PBMC NEs, mouse liver microsomes, and rat liver microsomes are provided in supplemental Tables S2A, S3A, and S4A, respectively). Peptide hits from all analyses were merged to establish a master list of proteins identified by at least two peptides or one peptide with two independent spectra (see supplemental Tables S2B, S3B, and S4B for the detailed peptide and spectral counts of proteins detected by MudPIT analysis of human PBMC NEs, mouse liver microsomes, and rat liver microsomes, respectively). Based on the merged detected peptides, proteins could fall into three categories following the parsimony principle. (i) Proteins detected by the exact same set of peptides were grouped together because they could not be distinguished based on the available peptide data (see column named “Proteins in Group” in supplemental Tables S2–S4), and only one arbitrarily selected representative protein entry is reported for such groups of proteins (see column labeled “Locus” in supplemental Tables S2–S4). (ii) Proteins with at least one peptide uniquely mapping to them were considered unique entries. (iii) Subset proteins for which no unique peptides were detected were removed from the final list of identified proteins.
Identifications mapping to shuffled peptides were used to estimate false discovery rates. Spectral FDR was calculated as follows.
 |
Protein level FDR was calculated as follows.
Under these criteria, the final FDRs at the protein and peptide levels were on average 1.5 and 0.5%, respectively (supplemental Tables S2B–S4B, bottom).
To estimate relative protein levels, normalized spectral counts were calculated for each non-redundant protein as described (34–36).
 |
We implemented a new algorithm on this data set to refine spectral counts to deal with peptides shared between multiple proteins (37). For each run, dNSAFs were calculated based on distributed spectral counts in which shared spectral counts were distributed based on spectral counts unique to each isoform (supplemental Tables S2B–S4B).
Bioinformatics Analysis
To be able to directly compare the lists of proteins detected in the human PBMC NEs and rodent liver microsome data sets, proteins were mapped to an Ensembl gene to remove redundancy from differences in protein annotations. Human gene orthologous groups were identified with Ensembl release 48 (38) to remove redundancy and false variation that might have resulted from differences in human and rodent gene assignments. Orthologous group IDs were sorted according to frequency of detection in the NE and microsome runs, relative levels (determined by averaged dNSAF values), membrane helix prediction (determined using TMHMM 2.0; http://www.cbs.dtu.dk/services/TMHMM-2.0/ (39)), and dNSAF ratios between values measured in NEs versus microsomes (supplemental Table S5).
Biologically interesting GO terms and their corresponding child terms were retrieved from the mySQL database AmiGO (http://amigo.geneontology.org (40)). To ensure a fair comparison for term enrichment, only human mapped genes in our data set were considered. These were compared with the genomics data set of human Ensembl genes as well as those GO defined as having nuclear localization. For a given GO term, the fraction of genes containing that term or any of the child terms was calculated for all data sets. The -fold difference was calculated by dividing this fractional value from our data set of interest by the value from the reference group.
Comparison of expression levels in different blood cell lineages was done by downloading numbers for each NET and blood cell type from BioGPS (http://biogps.gnf.org/#goto=welcome (41)) and calculating the -fold expression over the median value from all cell types tested in this transcriptome database. Thus, the numbers not only reflect differences among the blood cell lineages but also differences with other tissues.
Antibodies and Western Blotting
Antibodies used were GAPDH (Enogene, E1C604), Calreticulin (Cell Signaling Technology, 2891S), Calnexin (Stressgen, SPA-860), lamins A and B1 (3262 and 3931; Ref. 42), Nup153 (Covance), and Nup358 (raised against recombinant human protein amino acids 2595–2881; a kind gift from F. Melchior). NET antibodies were rabbit polyclonal antibodies generated to peptides from human sequences by Millipore, Tmem126 (06-1037), C17orf62 (06-1033), C17orf32 (06-1035), and MARCH5 (06-1036), and the previously characterized NETs NET31/Tmem209 (06-1020), SUN2 (06-1038). All fluorophore-conjugated secondary antibodies were minimally cross-reactivity from donkey (Jackson ImmunoResearch Laboratories) or goat (Molecular Probes).
To increase lamina solubility, PBMC or liver NEs were incubated on ice in 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm MgCl2, 0.2% Nonidet P-40 in the presence of protease inhibitor mixture (Roche Applied Science, 11 873 580 001) for 15 min and then sonicated in a 4 °C sonic bath. Microsomes were treated similarly. Protein concentrations of liver NEs and microsomes (see Fig. 5) were determined by Bradford assay before adding sample buffer (100 mm Tris, pH 6.8, 4 m urea, 2% SDS, 50 mm DTT, 15% sucrose), heating at 65 °C, and then sonicating again in a 4 °C sonic bath. PBMC NEs and microsomes were adjusted for equal loading based on Coomassie Blue staining on polyacrylamide gels (see Fig. 3A). Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane (LI-COR Biosciences). Membranes were blocked in PBS, 5% milk, 0.2% Tween. Primary antibodies were diluted in this buffer (1:200 for Millipore NET peptide antibodies, 1:500 for calreticulin, 1:200 for calnexin, and 1:2000 for lamin A) and allowed to incubate overnight at 4 °C. Secondary IR800-conjugated goat anti-rabbit antibodies (LI-COR Biosciences) were added at 1:5000 dilution at RT for 2 h. Visualization and quantification were performed using a LI-COR Odyssey system and software (Odyssey 3.0.16) using median background subtraction. Three independent blots were run for each NET and control, and averages from all three are presented in Fig. 5B.
Fig. 5.
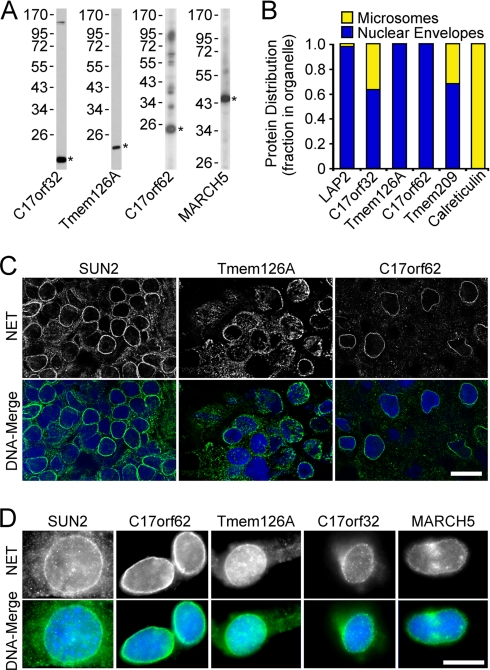
Antibody staining confirms novel NET identifications. A, NET antibody validation by Western blotting. C17orf32 and Tmem126A antibodies were tested on human PBMC lysates, whereas C17orf62 antibodies were tested on a C2C12 lysate to check background in the cell line used to test pre-extraction in D. MARCH5 antibodies were tested using the Jurkat human blood cell line. Asterisks indicate expected molecular weight. B, comparison of relative NET amounts in ER (microsome) and NE fractions by quantitative Western blotting. LAP2 and Tmem209 antibodies are NET controls, whereas calreticulin antibodies are an ER control. Blots were quantified using a LI-COR Odyssey system (three repeats averaged). C, cryosections of rat spleen stained with antibodies to new NETs and a SUN2 control. Nuclear rim staining was clearly observed for all NETs tested. Scale bars, 10 μm. D, antibody staining on Triton-pre-extracted C2C12 cells. A nuclear rim staining was observed for the control NET SUN2 and all novel NETs tested.
Fig. 3.
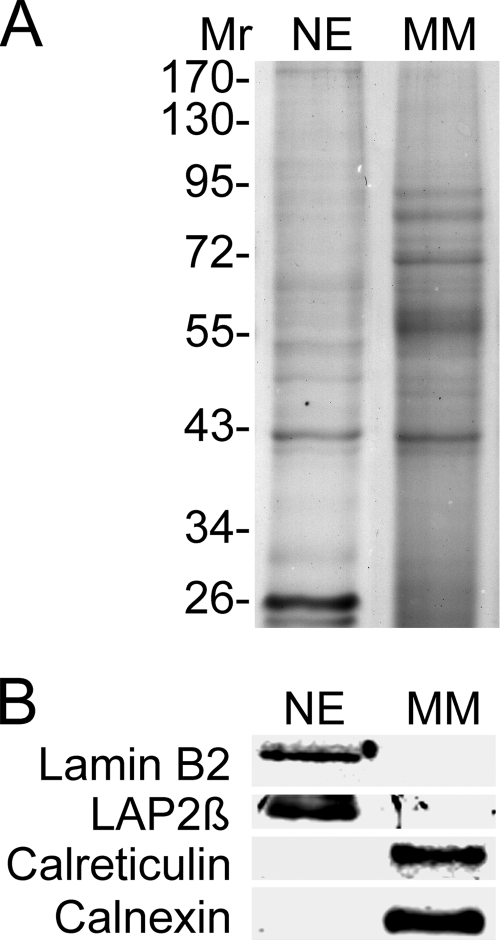
Fraction purity. A, Coomassie-stained gel of NE and microsome fractions analyzed. B, Western blot of the above fractions stained with organelle markers. ER markers calreticulin and calnexin were absent from NEs, whereas NE markers lamin B2 and the NET LAP2β were absent from microsomes. Similar amounts of total protein were loaded. MM, microsomal membranes.
Plasmid Construction
IMAGE clones for human NETs were obtained from RZPD and Geneservice. NET gene names are followed by IMAGE numbers or gene IDs in parentheses: STT3A (3891543), TAPBPL (3916213), METTL7A (2900478), C17orf62 (2822930), IAG2 (3834858), Tmem109 (3453616), C20orf3 (3530962), C17orf32 (5429060), Sec11C (3895819), Tmem126A (1667589), AADACL1 (4815834), and Tmem41A (3838280). Coding sequences were amplified by PCR with added 5′ and 3′ restriction sites, sequenced from both ends in intermediate cloning vectors, and then inserted into the mammalian expression vector pmRFP with a carboxyl-terminal tag (derived from Clontech pEGFP-N2 by replacing the GFP coding sequence with that of monomeric red fluorescent protein). Lamin A-GFP was obtained from Anne Straube (Marie Curie Research Institute, Oxted, UK), and calreticulin-GFP was obtained from Tom Rapoport (Harvard, Boston, MA).
Cell Culture and Transient Transfection
HT1080, HT1080 5.1 (carrying a lac operator (lacO) array and lac repressor-GFP), HeLa cells stably transfected with H2B-GFP, COS-7, and C2C12 cells were maintained in high glucose DMEM with 4.5 g/liter glucose (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 μg/μl penicillin, and 100 μg/μl streptomycin sulfate (Invitrogen). PBMCs were maintained in RPMI 1640 medium with 10% FBS and antibiotics. PBMCs were activated with 2.5 μg/ml PHA (Sigma) for 7 days. Adherent cells destined for microscopy were plated on coverslips, and DNA was transfected the next day using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions.
Immunofluorescence Microscopy
After 30 h, cells were either directly fixed for 7 min in 3.7% formaldehyde or washed with PBS; then extracted for 1 min with 1% Triton X-100, 25 mm Tris, pH 8.0, 150 mm KOAc, 15 mm NaCl, 5 mm MgCl2; washed again with PBS; and then fixed with formaldehyde. For antibody staining, cells that were not pre-extracted were permeabilized for 6 min in 0.2% Triton X-100 after fixation. Cells were then blocked with 10% FBS, 200 mm glycine in PBS and incubated for 40 min at RT with relevant antibodies. DNA was visualized with Hoechst 33342 or 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) and coverslips mounted in Fluoromount-G (EM Sciences). For structured illumination (OMX) microscopy, Alexa Fluor secondary antibodies (Molecular Probes) were used. Most images were obtained using a Nikon TE-2000 microscope equipped with a 1.45 numerical aperture 100× objective, Sedat quad filter set, and CoolSnapHQ High Speed Monochrome charge-coupled device camera (Photometrics). Structured illumination images (Fig. 4C) were taken on the OMX system at the University of Dundee microscopy facility (details described at http://microscopy.lifesci.dundee.ac.uk/omx/).
Fig. 4.
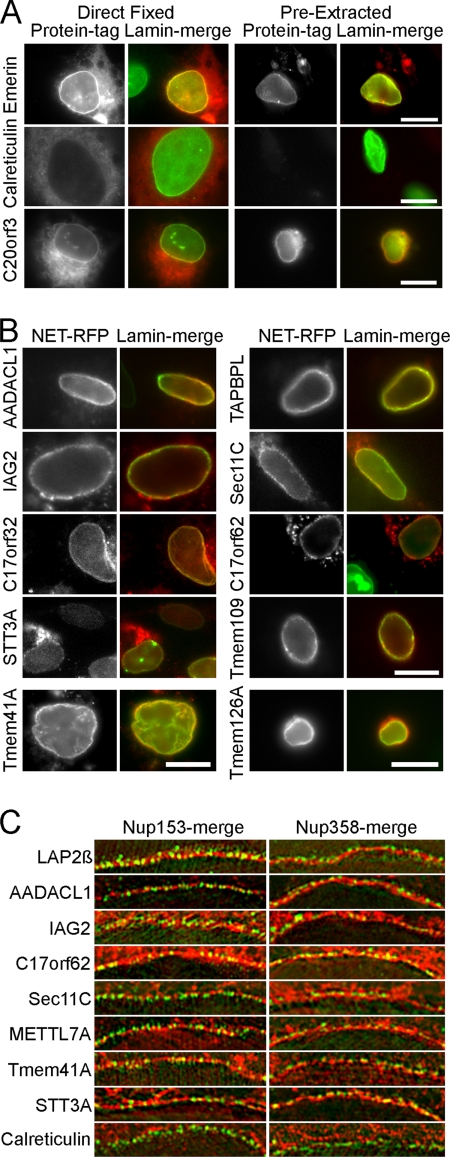
mRFP fusions confirm NE targeting for several novel PBMC NETs. A, HT1080 cells expressing NETs fused to mRFP were directly fixed (left) or extracted with Triton X-100 prior to fixation (right). The NE is marked by lamin A in green so that yellow indicates co-localization of the NET at the NE. The directly fixed emerin image has part of an untransfected cell, confirming that none of the NET staining at the nuclear rim is due to bleed-through from the lamin A channel. Note that prefixation extraction affects morphology and sometimes leaves aggregated protein in the cytoplasm. The emerin control and new NET C20orf3 are retained after extraction, whereas the ER protein calreticulin is not. Scale bar, 10 μm. B, other NET-mRFP fusions were similarly pre-extracted and retained at the NE. Tmem41A is shown in COS-7 cells because it was not expressed in HT1080 cells, and Tmem126A is shown in Jurkat cells because it failed to be expressed in either HT1080 or COS-7 cells. Scale bar, 10 μm. C, inner versus outer nuclear membrane targeting. If a NET (red) is in the INM it should appear in the same plane as the nuclear basket protein Nup153 (green, left) and internal to the cytoplasmic filament protein Nup358 (green, right) using structured illumination microscopy. Characterized NET LAP2β and most new NETs tested appeared in the INM. One INM NET, METTL7A, did not resist Triton pre-extraction. Scale bars, 5 μm.
For cryosections, fresh rat spleen cut into 2–3-mm cubes were embedded in optimal cutting temperature compound (Tissue-Tek), snap frozen in liquid nitrogen, and maintained at −80 °C. Sections were cut on a Leica CM 1900 cryostat at 6–8-μm thickness and fixed in −20 °C methanol. After rehydration, sections were incubated with NET antibodies overnight at 4 °C followed by secondary antibodies as above. Cryosection images were recorded using an SP5 confocal system (Leica) with 63× oil 1.4 numerical aperture objective using argon and UV lasers. Micrographs were saved from source programs as .tif files and prepared for figures using Photoshop 8.0.
The positional distribution of the lacO array was determined using a macro (available on request) written in Visual Basic within Image Pro Plus. In brief, the total nuclear area was automatically measured on DAPI images, and the area was then divided into five equal regions of interest through eroding 20% of the total area from the outer limits of the DAPI-defined nucleus. The nuclear sector containing the lacO spot was determined and exported to Microsoft Excel where the results from each cell were summed.
Images of chromatin condensation in Fig. 8 were generated from 60-h post-transfection cells fixed with formaldehyde using a wide field DeltaVision microscope (Applied Precision) using a 60× Plan Apo oil objective. Image stacks (0.2-μm steps) were deconvolved using DeconQ and processed using SoftWorks.
Fig. 8.
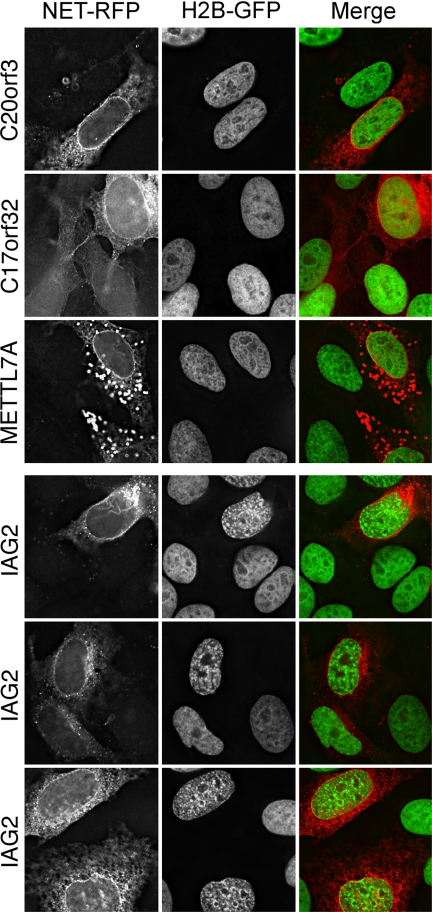
Screen for PBMC NETs that promote chromatin compaction. HeLa cells stably expressing H2B-GFP were transfected with the same set of nine NETs used in Fig. 7. Only IAG2 displayed strong chromatin condensation, whereas all other NETs (METTL7A, C17orf32, and C20orf3 are shown) exhibited no differences in chromatin compaction or distribution compared with adjacent untransfected cells. It is noteworthy that IAG2 had no effect in the lacO screen and that NETs STT3A and TAPBPL, which altered lacO positioning, had no effect on chromatin compaction.
Electron Microscopy
For transmission electron microscopy, whole PBMCs or isolated NEs were fixed in 3% glutaraldehyde in 0.1 m sodium cacodylate, pH 7.3 for 2 h; washed in 0.1 m sodium cacodylate; postfixed in 1% osmium tetroxide for 45 min; washed again; and dehydrated in 50, 70, 90, and 100% normal grade acetones for 10 min each and then for a further two 10-min changes in analar acetone. Samples were then embedded in Araldite resin. 60-nm-thick sections were cut on a Reichert OMU4 ultramicrotome (Leica), stained in uranyl acetate and lead citrate, and then viewed in a Phillips CM120 transmission electron microscope. Images were taken at 100 kV at 11,000× magnification using a Gatan Orius charge-coupled device camera.
RESULTS
Generation of Lymphocyte-enriched Populations and NE Fractions
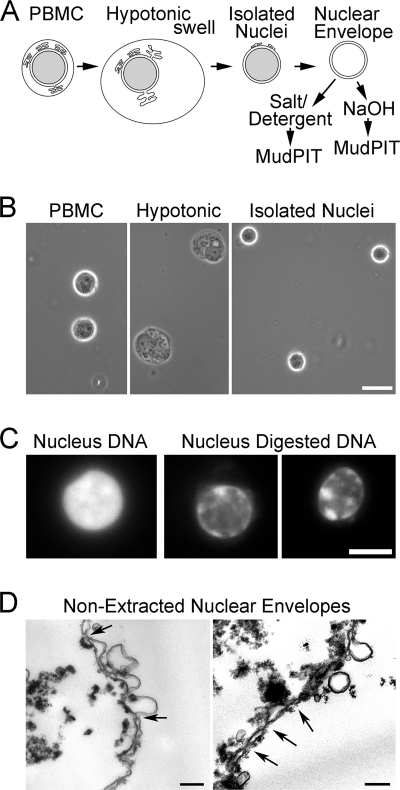
NEs were prepared from PBMCs using methods specially optimized for blood cells (23) (Fig. 1A). Central to these methods is isolating clean nuclei from the rest of the cell (Fig. 1B) that are subsequently digested with DNase and RNase and salt-washed to remove most of the nuclear contents (Fig. 1C). Even after such isolation, NEs maintain strong connections to partners such as peripheral chromatin (Fig. 1D). Prior to mass spectrometry, NEs were further extracted with 1% β-octyl glucoside, 400 mm NaCl or 0.1 n NaOH that enriched for proteins associated with the insoluble intermediate filament lamin polymer or integral membrane proteins, respectively. Some well characterized NETs distribute to one or the other fraction (21, 22); hence, both detergent/salt- and NaOH-extracted fractions were analyzed to be comprehensive. Both extractions remove most contaminants so that only strongly bound and relevant partners should remain.
Fig. 1.
Cellular fractionation of PBMCs. A, method schematic. Crude nuclei prepared from hypotonic lysis of PBMCs were cleaned of contaminating cellular structures by gradient centrifugation to float contaminating membranes while pelleting the denser nuclei. Crude NEs were prepared by digesting/extracting nuclear contents from isolated nuclei. Before MudPIT analysis, these were further extracted with 1% β-octyl glucoside, 400 mm NaCl or 0.1 n NaOH to enrich for proteins associated with the insoluble lamin polymer or integral membrane proteins, respectively. B, buffy coats from human blood were separated on Ficoll-Hypaque density gradients to enrich for mononuclear leukocytes (left panel). The cells were swollen hypotonically (middle panel) and Dounce homogenized to release nuclei (right panel), which were then further purified on sucrose gradients. Phase-contrast light microscope images are shown. Scale bar, 10 μm. C, enrichment for NEs by chromatin digestion. DAPI staining for DNA visualizes significant nuclear chromatin content in an isolated PBMC nucleus (left panel) and the loss of most of this material after two rounds of digestion with DNase and RNase, each followed by salt washes (two right panels). A fluorescence microscope image is shown. Scale bar, 5 μm. D, ultrastructure of isolated NEs. Electron micrographs of PBMC NEs show that most membranes in the population are the characteristic double membrane with little contamination from single membrane vesicles. Arrows point to positions of NPCs. In some places, the hypotonic treatment used to swell the NEs while digesting/extracting most of the chromatin resulted in membrane blebbing. These NEs were further extracted with salt and detergent to enrich for proteins associated with the intermediate filament lamin polymer or with an alkaline treatment to enrich for transmembrane proteins prior to analysis by MudPIT. After such treatment, no structure remains that can be readily discerned by EM with the characteristics of NEs. Scale bar, 0.2 μm.
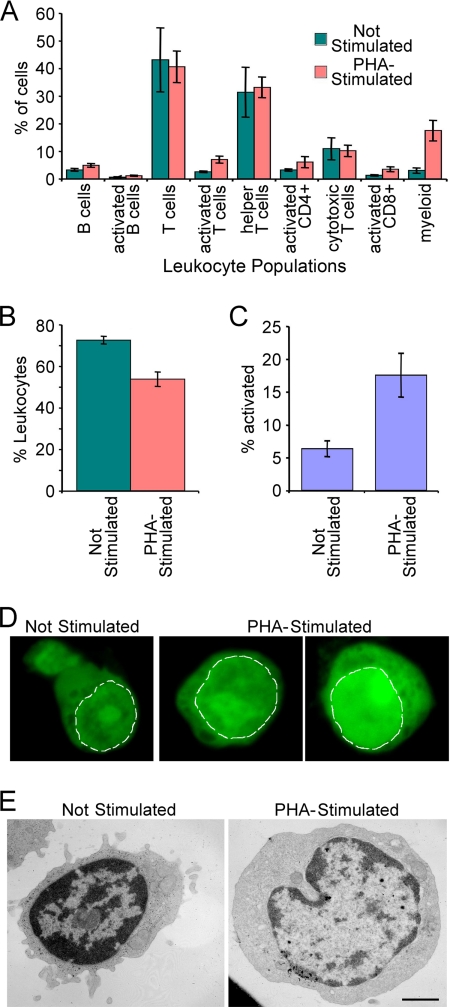
PBMCs were ∼70% lymphocytes as determined by FACS and antibody staining for CD antigens (Fig. 2A). PBMC NEs were isolated with or without PHA stimulation, which mimics immune responses (43) and increased activated cells >3-fold (Fig. 2, B and C). PHA activation also results in dissolution of dense chromatin at the nuclear periphery (17–20). Staining of cells with the DNA-binding dye acridine orange confirmed the reduction of denser chromatin at the periphery. Acridine orange has been reported to bind more strongly to single-stranded DNA and thus gives a brighter signal in actively transcribed chromatin regions with condensed chromatin not allowing efficient intercalation (44). Thus, a weaker signal concentrated largely at the periphery was observed in unstimulated lymphocytes, whereas a brighter signal that was more diffuse through the nucleoplasm was observed for PHA-activated cells (Fig. 2D). This was further confirmed at the ultrastructural level as electron microscope images of the cells revealed significant differences in the chromatin distribution. Although patches could still be observed at the periphery in PHA-activated cells, they were less dense and broken (Fig. 2E). Proteins identified in the PBMC data sets would thus likely include proteins involved in chromatin remodeling, and some of these proteins might be highlighted by differences between proteins identified in the unstimulated and PHA-activated PBMC NE fractions.
Fig. 2.
PBMC composition. A, cells treated as for NE purification were incubated with CD markers for different blood cell types and analyzed by flow cytometry. The percentage of each cell type for PBMC composition in leukocytes is graphed. B, percentage of leukocytes in the total population. C, percentage of lymphocyte activation in the different PBMC populations measured by appearance of surface markers. Error bars in A, B, and C indicate standard deviation. D, PHA activation gauged by changes in nucleotide distribution using acridine orange dye. Acridine orange intercalates more strongly with single-stranded DNA than double-stranded DNA, so intensity to some degree measures “open” DNA that is being actively transcribed. A dashed white line delineates the periphery of the nucleus. The unstimulated PBMCs have weak staining at the periphery of the nucleus and no staining in some central areas. PHA-activated lymphocytes have more uniform distributions and brighter staining, indicating more open DNA. E, PHA activation effects on chromatin organization assessed by electron microscopy. The unstimulated cell is smaller and has more compacted chromatin concentrated at the nuclear periphery. In the PHA-activated cell, much of this dense peripheral chromatin has become less compact, and both the cell and nucleus have increased in volume as the activated cells are now rapidly transcribing DNA and making protein. Images were taken at 11,000×. Scale bar, 1 μm.
Microsomes were prepared separately as a comparative/subtractive fraction as they are rich in the main expected transmembrane protein contaminants from the ER, which is continuous with the NE. Thus, they provide a mechanism to distinguish INM proteins and ONM-enriched proteins from those such as ribosomes that inhabit both the ONM and ER. Because isolated lymphocytes have very little ER, we used rodent liver for production of microsomes. Actual contamination of NE fractions with ER was minimal because, although some single membrane vesicles can be observed by electron microscopy (see Fig. 1D), ER markers such as calreticulin and calnexin were undetectable in the crude PBMC NE fractions (Fig. 3). Conversely, NE proteins such as lamin A and the NET LAP2β were not detectable in microsome fractions (Fig. 3), indicating that the hypotonic lysis step only disrupted the plasma membrane and did not contribute NE membranes to the microsome fraction.
To remove donor variation as a factor in potential differences measured between unstimulated and PHA-activated PBMCs, each PBMC preparation was divided in two: half was treated with PHA, and the other half was used as unstimulated. Some degree of variation between blood donors was expected because of the wide variation in human immune responses, so donor variation was averaged out within each condition by combining NEs isolated from at least 15 separate preparations for analysis. Combining this large number of samples was necessary for the independent reason that the vast majority of material was lost in isolating clean NEs from PBMCs. In this study, it was not possible to further control for variation due to factors such as age or racial differences as all blood donor information was anonymous, but all buffy coats were obtained within 2 days of donation. The combined NE preparations were divided and extracted with either 0.1 m NaOH or 1% β-octyl glucoside, 400 mm NaCl, and insoluble material was pelleted. The alkali extraction should have enriched for transmembrane proteins, and so vesicles were pelleted at 150,000 × g for 30 min, whereas the detergent/salt extraction should have removed membranes and enriched for both soluble and transmembrane proteins strongly associated with the higher order intermediate filament lamin polymer; these proteins were pelleted at 15,000 × g for 30 min.
MudPIT Analysis
After extraction, pellets of isolated NE fractions were digested with either a combined cyanogen bromide-endoproteinase Lys-C-trypsin treatment or with a combined endoproteinase Lys-C-trypsin treatment. For the latter, treatment material that could be pelleted at 17,500 × g for 30 min was subsequently digested with proteinase K. The multiple and processive digestions were favored over direct replicates on the limiting material to better access material tightly associated with the intermediate filament lamin polymer that is resistant to 2% Triton X-100, 2 m NaCl, and even 4 m urea. Indeed, additional identifications were made from the proteinase K-digested material (supplemental Table S2B). Five separate MudPIT (27, 28) runs were performed for each of the unstimulated and PHA-activated PBMC NE fractions, three for the NaOH-extracted and two for the salt/detergent-extracted NEs. Thus, using different extraction conditions and different digestions and performing multiple runs were expected to provide comprehensive coverage to identify all proteins in the PBMC NE. After removing redundancy by converting protein IDs to orthologous gene groups, 3351 proteins were identified between the unstimulated and PHA-activated PBMC NE data sets, 2923 from the unstimulated and 2839 from the PHA-activated cells, 2411 of which were in both (supplemental Table S2B).
To complement previously published microsome data sets isolated from mouse liver (21), microsome data sets were separately generated from rat liver as a subtractive fraction (supplemental Table S4). These newly acquired microsome factions were treated similarly to the NE fractions, i.e. digested with trypsin and proteinase K and analyzed on LTQ ion trap mass spectrometers (supplemental Table S1). Another potential contaminant is mitochondria, but as the mitochondrial proteome has been extensively studied, lists of mitochondrial proteins were generated from a previous mass spectrometry study of that organelle (45) for subtraction instead of reanalyzing mitochondria (supplemental Table S5). After subtracting 177 proteins that were previously identified in mitochondria, 2756 proteins remained in unstimulated PBMC NEs, 2760 remained in PHA-activated PBMC NEs, and 3174 remained in total. Using microsomes strictly as an absolute subtractive fraction would have resulted in loss of several well characterized NETs that have been demonstrated to inhabit both the NE and ER. For example, emerin, one of the most established INM NETs (46), was detected in both NE and microsome data sets (supplemental Table S5) in agreement with the fact that emerin also functions in the ONM, ER, and interstitial discs (47–49). Moreover, as a recent elegant proteomics study suggests that at least a third of all proteins have multiple cellular localizations (50), we reasoned that absolute subtraction would also remove relevant identifications. Thus, proteins were ordered according to an estimate of abundance based on normalized spectral counts (dNSAF) (35), and NE data sets were restricted to those proteins either not detected in the microsomes or at least 5× more abundant than in the microsome fractions based on their NSAF values, leaving 2183 proteins in unstimulated PBMC NEs, 2141 in PHA-activated, and 2542 in total (supplemental Table S5).
This large number of proteins associated with the NE includes chromatin proteins and other factors that are tethered directly or indirectly to both INM and ONM proteins and so, although they can associate with the NE, are not necessarily primarily resident at the NE. The subset of NETs is more likely to be enriched at the NE and constitutes the surest NE core components in PBMCs. These proteins include all 13 of the previously well characterized NETs, 39 NETs previously detected in the liver NE data sets (21), and 423 proteins predicted to contain at least one transmembrane domain by TMHMM or membrane anchor by SignalP. Based on available GO terms and annotations, we selected 87 of these proteins as putative blood NETs (supplemental Table S5). It is of note that five of 12 proteins we further characterized for NE targeting (see below) did not pass the criterion of 5-fold enrichment over microsomes once the new rat liver microsome data set was acquired. However, NE residence was confirmed for all five, indicating that our selection criteria were very stringent and that additional NETs are likely to be found in this data set.
Confirmation of NE Residence
The putative NETs identified could be in the INM and/or in the ONM that is continuous with the ER or be contaminants. To directly test the validity of novel protein identifications, 12 putative NETs from PBMC data sets were cloned as tagged fusions and expressed in cells to determine whether they target to the NE. Adherent tissue culture cells were used instead of freshly isolated PBMCs because lymphocytes are very difficult to transfect, and their small cytoplasm made it difficult to distinguish the NE from the ER. To be able to clearly distinguish the NE from the ER and other cell membranes, adherent flat HT1080 cells were used for this analysis because lymphocytes are round suspension cells with little ER that is packed closely around the nucleus. Another issue is that, when overexpressed, NETs tend to saturate binding sites at the NE and accumulate in the ER, making clear confirmation of NE targeting difficult (Fig. 4A, left panels). To get around this problem, most of this extraneous material was removed with a prefixation detergent extraction. Most previously characterized NETs resist a prefixation extraction with 1% Triton X-100 and 400 mm KCl presumably because of tight associations with the intermediate filament lamin polymer (e.g. emerin; Fig. 4A, top panels), whereas most ER proteins (e.g. calreticulin; Fig. 4A, middle panels) are extracted with this treatment. Staining for lamins is also shown as co-localization (yellow) with this well defined NE protein, indicating NE localization. One of the novel NETs identified here, C20orf3, behaved similarly to the well characterized NET emerin in this assay (Fig. 4A, bottom panels). In all, ten of the 12 putative NETs tested were retained at the NE after a prefixation detergent extraction (Fig. 4; all except Tmem126A and METTL7A). The putative NETs Tmem41A and Tmem126A both failed to be expressed in the HT1080 cells first tested, but Tmem41A was expressed and targeted to the NE in COS-7 cells and resisted the detergent pre-extraction (Fig. 4B, bottom left). Tmem126A also failed to be expressed in COS-7 cells but was expressed and targeted in lymphocytes, which due to their small size and being suspension cells could not be properly tested for resistance to detergent pre-extraction (Fig. 4B, bottom right). METTL7A exhibited a concentration at the nuclear periphery that co-localized with lamins but was never observed after a pre-fixation detergent extraction (data not shown).
As the NE is comprised equally of INM and ONM, some NETs may preferentially accumulate in one or the other subcompartment. A recent study used three-dimensional structured illumination microscopy (OMX) to distinguish INM from ONM localization of lamins in cells when co-stained for proteins from the nuclear pore nucleoplasmic (Nup153) or cytoplasmic (Nup358) face (51). We have previously used this system to test several NETs identified in the earlier liver proteomics data sets (52). Control NET LAP2β and all the new NETs tested here accumulated in the INM as both the NET (red) and Nup153 (green) localize to the same ring, whereas an inner NET ring was observed compared with Nup358 (Fig. 4C). In contrast to the NETs, a separable inner Nup153 ring was observed against the ER protein calreticulin. Yellow co-localization was not generally observed in this system because the resolution of OMX microscopy is sufficient to distinguish the NPC proteins that are at ∼120-nm gaps in the membrane from NETs that are inserted in the membrane itself. Even METTL7A, the putative NET that was not resistant to the detergent pre-extraction, appeared in the INM.
To test localization of endogenous proteins, polyclonal antibodies were generated to three novel NETs (C17orf32, Tmem126A, and C17orf62). Each recognized a protein of the correct size in immunoblots (Fig. 5A). First, to test whether NETs were principally in the NE or inhabited multiple cellular localizations as has been indicated for roughly 40% of the human proteome (50), antibodies were used on Western blots comparing NE and microsome fractions. Tmem126A and C17orf62 were exclusively in the NE fraction, but about a third of C17orf32 occurred in the microsome fraction (Fig. 5B). This is similar to the previously characterized NET31/Tmem209 that was detected in both the PBMC NE data sets and the earlier liver NE data set (52).
The Tmem126A and C17orf62 antibodies yielded strong and distinctive staining at the nuclear periphery when used on cryosections from rat spleen (Fig. 5C). The nuclei are clearly visualized with the DAPI DNA stain, and the antibody staining is concentrated in a crisp rim just surrounding the nucleus that is very similar to that observed with antibodies to the well characterized NET SUN2. C17orf62 and C17orf32 both resisted pre-extraction with Triton X-100 when expressed as fusions in cell lines, and similarly, the antibodies to these NETs yielded a rim stain in cultured cells pre-extracted with detergent. Adherent tissue culture cells were used again because lymphocytes have little cytoplasm and do not withstand the prefixation extraction well. However, C2C12 cells were used instead of the HT1080 cells used earlier for targeting of fusion proteins. According to available transcriptome data, all the NETs for which we had antibodies were at least weakly expressed in C2C12 cells, whereas some were not expressed at all in HT1080 cells. This allowed confirmation that Tmem126A also resists detergent pre-extraction (Fig. 5D). Several proteins identified in these data sets were previously characterized proteins that had other reported cellular localizations. Among these was MARCH5, previously reported to be a mitochondrial protein that possibly contributes to perinuclear localization of mitochondria (53). Although widely staining throughout the cytoplasm in non-extracted cells (data not shown), antibodies to MARCH5 (Fig. 5A) also yielded nuclear rim staining after the detergent pre-extraction (Fig. 5D, right panels), indicating that the endogenous protein has a separate function at the NE. Thus MARCH5 is among the estimated 40% of cellular proteins that have multiple cellular localizations (50).
Changes in PBMC NE Composition following PHA Activation
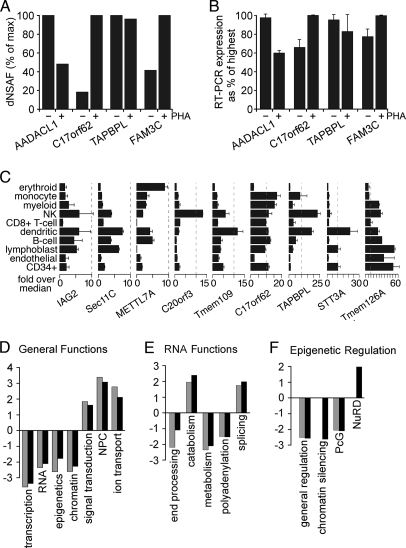
Of the 87 core putative new PBMC NETs (supplemental Table S5), 14% uniquely appeared in the unstimulated PBMC NE fraction, and 14% uniquely appeared in the PHA-activated PBMC NE fractions with 72% shared between both conditions. These numbers were similar when considering all transmembrane proteins identified (73% shared) or both transmembrane and soluble proteins identified (72% shared). Differences between the unstimulated and PHA-stimulated fractions were also observed in RNA expression levels for NETs that appeared in both fractions that correlated with peptide recovery. For example, four peptides were recovered for AADACL1 in the PHA-activated versus seven in the unstimulated lymphocytes (Fig. 6A and supplemental Table S2B), and correspondingly, AADACL1 transcript levels were reduced in PHA-activated lymphocytes to 60% of the level in unstimulated lymphocytes (Fig. 6B). The trends for expression changes also agreed with estimated protein levels for C17orf62, TAPBPL, and FAM3C.
Fig. 6.
Differences in NE composition with PHA activation and effects on genome organization. A, differences in abundance estimated from dNSAF values for some NETs with or without PHA activation. dNSAF values were taken from the two most equivalent runs based on total protein coverage and identifications. B, RT-PCR of the NETs in A revealed reproducible differences in expression that are consistent with their abundance estimates based on peptide recoveries. C, transcript levels for several of the NETs identified differ among blood cell lineages. Data from the BioGPS transcriptome study comparing 84 different tissues/cell types are plotted relative to the median value over all 84 tissues sampled. Ticks along the bottom are for increments of 5-fold above the median. Each NET had its own unique expression pattern. Surface markers defining cell populations were CD71 for early erythroid cells, CD14 for monocytes, CD33 for myeloid cells, CD56 for natural killer (NK) cells, CD8 for T-cells, BDCA4 for myeloid dendritic cells, CD19 for B-cells, 721 for B-lymphoblasts, CD105 for endothelial cells, and CD34 for polyploidy progenitor cells. Error bars in B and C indicate standard deviation. D, within the subset of proteins in NE data sets with GO annotations, the fraction with a particular functional annotation was calculated. Similar fractions were calculated against all “nuclear”-annotated proteins in the GO database. The ratio of NE/nuclear fractions was then calculated, setting a 1:1 ratio to 0 so that positive values are -fold enrichment and negative values are -fold deficiency at the NE compared with the whole nucleus. Resting and activated PBMC data sets are represented by gray and black bars, respectively. E, the same analysis applied to the specific subset associated with RNA functions. F, the same analysis applied to the specific subset associated with functions in epigenetic regulation. PcG, Polycomb group.
Many of the novel NETs identified that were directly tested here varied not only between the unstimulated and PHA-activated leukocytes but appeared to be uniquely expressed in different blood cell lineages. Extracting relative transcript levels from a large scale transcriptome data set that compared RNA transcript levels across multiple tissues (41), most of the novel NETs had completely distinct patterns of expression among 10 different blood cell types ranging from erythroid cells to B-cells (Fig. 6C). Only IAG2 and Sec11C of those shown had similar general patterns of expression, and only Tmem41A and C17orf32 (not shown) were expressed similarly in all lineages. The data are expressed relative to a median value that was generated from a much wider range of tissues, and these NETs ranged from ∼10× the median value to nearly 60× (Fig. 6C). Thus, many of the novel NETs identified from the PBMC proteomics are reasonably restricted in the tissues and cell types in which they are expressed. The interpretation of these results is tempered by the observation that some changes in the population composition occur upon PHA activation (Fig. 2A). Myeloid cells were found to increase, and this is consistent with some of the NETs being more specific to myeloid dendritic cells (Fig. 6C); however, the percentage of myeloid cell-associated proteins at the NE did not increase. This could be attributed to having already made a full identification of myeloid cell-associated proteins in the unstimulated cells. Interestingly, when considered as a percentage of the total myeloid cell-associated proteins, roughly 10% occur at the NE, indicating this organelle as a dynamic player in cell type specification.
PHA-induced changes include physiologically relevant functions such as re-entering the active cell cycle (43) and altering peripheral chromatin organization (17). Individual NETs could contribute to chromatin organization and transcriptional regulation to facilitate one of the individual lineages observed in Fig. 6C. To gauge the likelihood of NE contributions to genome functions, the known proteins in the NE data sets (both NETs and soluble proteins) that had functional annotations in the GO database were extracted. A separate data set was extracted for all proteins in the GO database associated with the nucleus. GO functional annotations were then applied to these data sets, and the fraction of proteins within each data set with a particular GO functional annotation was calculated. These fractions were compared with one another to give the -fold enrichment of a particular function at the NE compared with that function as a fraction of activity within the nucleus in general (Fig. 6, D–F). For example, transcription takes place throughout the nucleus, and the nuclear periphery is considered to be a generally transcriptionally repressive environment (54): the fraction of proteins identified at the NE involved in transcription, as thus expected, is much lower than that for the nucleus, yielding a negative -fold value of −3.5 (Fig. 6D). In contrast, proteins involved in nucleocytoplasmic transport would be expected to be enriched at the NE compared with the nucleus as a whole, and the value obtained for NPC proteins showed enrichment of 3.5-fold at the NE (Fig. 6D).
The general functional categories varied little between unstimulated and PHA-activated PBMC NEs; however, analysis of more specific subcategories revealed interesting differences. RNA functions, as might be expected, varied little upon PHA activation except that a few more end-processing proteins were found at the NE (Fig. 6E), consistent with an increased burden of RNAs to be exported from the nucleus with the significant gene activation resulting from PHA treatment. Functions in epigenetic regulation yielded more dynamic changes upon leukocyte activation (Fig. 6F). Chromatin silencing functions were less enriched at the NE with PHA activation, clearly indicating that changes at the NE play a role in the major chromatin reorganization that takes place. NuRD complex proteins in contrast were specifically enriched at the NE upon PHA activation.
Novel NET Functions in Chromatin/Genome Organization
The finding of so many known proteins with chromatin regulatory functions in these data sets that change between the data sets in ways that match the physiological changes observed in chromatin upon PHA activation argues that some novel proteins identified might also have chromatin functions. To screen for such NETs, we tested nine PBMC NETs in assays for effects on genome organization.
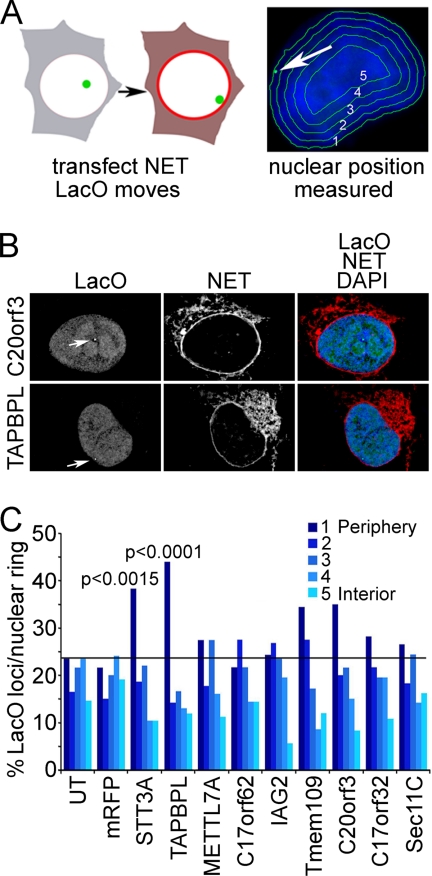
The first assay utilized an HT1080-derived cell line carrying a lacO repeat insertion in chromosome 5, the position of which is visualized with a lac repressor-GFP fusion that binds to the array. This lacO array tends to be in the nuclear interior. These cells were transfected with expression constructs for NET-mRFP fusions and assayed for a change in the distribution of the lacO array position with respect to the nuclear periphery. If a NET altered the position of the lacO array by pulling it to the nuclear periphery it could be considered to function in genome organization (Fig. 7A, left). To determine lacO positioning, a previously published algorithm was utilized that erodes the nucleus based on DAPI staining for DNA into five concentric rings of roughly equal area (55). The ring containing the lacO array was marked in each cell and recorded (Fig. 7A, right). The NET C20orf3 had no effect on the position of the lacO array; however, TAPBPL expression increased the occurrence of the array at the periphery (Fig. 7B). The ring containing the lacO array was marked in ∼100 cells, and the distribution was plotted (Fig. 7C). The lacO array tended to be in the nuclear interior, occurring in the peripheral ring only ∼20% of the time. Most blood NETs tested yielded no significant differences, but in cells expressing STT3A and TAPBPL, the occurrence at the periphery roughly doubled in multiple separate experiments with p values <0.0015 and 0.0001, respectively (Fig. 7C).
Fig. 7.
Cell-based screen for PBMC NETs that promote recruitment of chromosome loci to nuclear periphery. A, schematic representation of a screen to determine whether overexpression of a particular NET can recruit a specific chromatin locus to the NE. NETs were transfected into cells containing a lacO repeat integration that is typically in the interior. The lacO position was visualized with GFP-lacI (green) and measured using an algorithm that partitions the nucleus based on DAPI staining (blue) into five concentric circles of roughly equal area. B, example of NET-transfected cells. The position of the lacO locus is highlighted by the white arrows. The lacO locus position is unaffected by C20orf3 expression but moves to the periphery with TAPBPL expression. DAPI staining added to the merged image confirms that the movement of the locus is not an artifact of generalized chromatin condensation at the periphery. Deconvolved images are shown. C, the ring containing the locus was recorded in roughly 100 transfected cells. p values were calculated for NETs that increased the locus at the periphery in comparison with untransfected (UT) or mRFP-transfected control cells using a χ2 test.
For the second assay, the same set of blood NETs was transfected into a HeLa cell line stably expressing H2B-GFP, and cells were observed for general changes in overall chromatin distribution. The two NETs that had strong effects in the lacO screen had no effect in this screen, further indicating that the interactions pulling the locus to the periphery are specific. A different NET, IAG2, had a strong effect on chromatin condensation (Fig. 8; compare with cells transfected with C20orf3, C17orf32, and METTL7A in upper panels), and this effect was repeated in all of three independent experiments. A ring of condensed chromatin is noticeable at the periphery, although the effect has clearly propagated throughout the nucleus. Distribution of the NET alone in the left panels clearly indicates that, whereas some of the overexpressed protein has accumulated in the ER, none is in the nucleoplasm: thus, the effect must be propagated from the NE.
DISCUSSION
This analysis of the leukocyte NE proteome identified many more NE-associated proteins and NETs than an earlier study of liver NEs (21). Moreover, some proteome differences were observed between unstimulated and PHA-activated leukocytes, many of which correspond to proteins that are expressed in only specific blood cell lineages. Many of these new NETs appear to be tissue-specific, and our results further show that NE protein composition changes in response to chemical stimulation. These findings indicate the potential of other organelle proteomes to vary with chemical treatments or between tissues.
Large scale changes have been reported in the organization of leukocyte peripheral chromatin upon PHA stimulation (17–20). These data sets should contain both NETs and tightly bound chromatin proteins involved in making these changes. As several NETs have known associations with chromatin-silencing proteins (12, 13), it is not surprising that several of the proteins identified here in association with the NE had functions in gene regulation and RNA processing. The PHA-induced general decrease in epigenetic silencing factors is consistent with the observed reduction in dense peripheral chromatin. Contrasting with this, the specific relative increase in NuRD complex proteins at the NE is interesting as this chromatin remodeling complex plays roles in development and was recently shown to function in progeria defects caused by NE mutations (56).
NET developmental roles are consistent with the blood cell type specificity of the two NETs that affected positioning of the lacO array (∼25-fold higher in blood cells over other tissues). Several tissue-specific genes move from the NE to the nuclear interior during differentiation (e.g. IgH locus, Mash1, and CFTR) (57–59); however, the NE proteins that regulate NE tethering and release have yet to be identified. Recent studies showed that tethering lac repressor to an NE protein could recruit the lacO array to the periphery by an affinity-based mechanism (14–16). As lac repressor was not fused to the NETs used in our screen, the specific effects of TAPBPL and STT3A likely operate through affinity for other chromosome regions on the same chromosome as the lacO array. This is consistent with the earlier studies showing that the whole chromosome accompanied the array to the periphery (14, 16). The interaction cannot be due to a more general affinity for chromatin because if so it would increase chromatin density at the periphery, and these two NETs had no effect in the second screen for chromatin compaction. Thus, our identification here of endogenous NETs that can alter the positioning of genes/chromosomes in the nucleus appears to reflect the unique chromatin characteristics of lymphocytes. As several studies have implicated misregulation of chromatin organization in NE diseases (7, 8), these new blood NETs may contribute to the diverse pathologies associated with NE diseases.
Supplementary Material
Acknowledgments
We thank D. Tollervey and W. C. Earnshaw for comments; W. Bickmore, F. Melchior, T. Rapoport, J. Stewart, M. Waterfall, E. King, D. Kelly, T. Guan, and S. Mitchell for reagents/technical assistance; and the Scottish National Blood Transfusion Service.
Footnotes
* This work was supported in part by the Wellcome Trust (a senior research fellowship to E. C. S.) and the Stowers Institute for Medical Research (to L. F.). Use of the OMX microscope was supported by the Scottish University Life Sciences Alliance.
 This article contains supplemental Tables S1–S5.
This article contains supplemental Tables S1–S5.
1 The abbreviations used are:
- NE
- nuclear envelope
- NET
- nuclear envelope transmembrane protein
- INM
- inner nuclear membrane
- ONM
- outer nuclear membrane
- PBMC
- peripheral blood mononuclear cell
- PHA
- phytohemagglutinin
- MudPIT
- multidimensional protein identification technology
- NPC
- nuclear pore complex
- ER
- endoplasmic reticulum
- TCEP
- tris(2-carboxylethyl)phosphine hydrochloride
- IAM
- iodoacetamide
- FDR
- false discovery rate
- dNSAF
- distributed normalized spectral abundance factor
- ID
- identity
- GO
- gene ontology
- mRFP
- monomeric red fluorescent protein
- lacO
- lac operator
- NuRD
- nucleosome remodeling and deacetylase.
REFERENCES
- 1. Schirmer E. C., Foisner R. (2007) Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 313, 2167–2179 [DOI] [PubMed] [Google Scholar]
- 2. Suntharalingam M., Wente S. R. (2003) Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4, 775–789 [DOI] [PubMed] [Google Scholar]
- 3. Callan H. G., Tomlin S. G. (1950) Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc. R. Soc. Lond. B Biol. Sci. 137, 367–378 [DOI] [PubMed] [Google Scholar]
- 4. Starr D. A., Fischer J. A. (2005) KASH 'n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. BioEssays 27, 1136–1146 [DOI] [PubMed] [Google Scholar]
- 5. Stewart C. L., Roux K. J., Burke B. (2007) Blurring the boundary: the nuclear envelope extends its reach. Science 318, 1408–1412 [DOI] [PubMed] [Google Scholar]
- 6. Worman H. J., Bonne G. (2007) “Laminopathies”: a wide spectrum of human diseases. Exp. Cell Res. 313, 2121–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fidziañska A., Toniolo D., Hausmanowa-Petrusewicz I. (1998) Ultrastructural abnormality of sarcolemmal nuclei in Emery-Dreifuss muscular dystrophy (EDMD). J. Neurol. Sci. 159, 88–93 [DOI] [PubMed] [Google Scholar]
- 8. Goldman R. D., Shumaker D. K., Erdos M. R., Eriksson M., Goldman A. E., Gordon L. B., Gruenbaum Y., Khuon S., Mendez M., Varga R., Collins F. S. (2004) Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 101, 8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maraldi N. M., Squarzoni S., Sabatelli P., Lattanzi G., Ognibene A., Manzoli F. A. (2002) Emery-Dreifuss muscular dystrophy, nuclear cell signaling and chromatin remodeling. Adv. Enzyme Regul. 42, 1–18 [DOI] [PubMed] [Google Scholar]
- 10. Ognibene A., Sabatelli P., Petrini S., Squarzoni S., Riccio M., Santi S., Villanova M., Palmeri S., Merlini L., Maraldi N. M. (1999) Nuclear changes in a case of X-linked Emery-Dreifuss muscular dystrophy. Muscle Nerve 22, 864–869 [DOI] [PubMed] [Google Scholar]
- 11. Bengtsson L., Wilson K. L. (2004) Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr. Opin. Cell Biol. 16, 73–79 [DOI] [PubMed] [Google Scholar]
- 12. Ye Q., Worman H. J. (1996) Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem. 271, 14653–14656 [DOI] [PubMed] [Google Scholar]
- 13. Somech R., Shaklai S., Geller O., Amariglio N., Simon A. J., Rechavi G., Gal-Yam E. N. (2005) The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 118, 4017–4025 [DOI] [PubMed] [Google Scholar]
- 14. Finlan L. E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J. R., Bickmore W. A. (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4, e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumaran R. I., Spector D. L. (2008) A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reddy K. L., Zullo J. M., Bertolino E., Singh H. (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452, 243–247 [DOI] [PubMed] [Google Scholar]
- 17. Drings P., Sonnemann E. (1974) Phytohemagglutinin-induced increase of euchromatin contents in human lymphocytes. Res. Exp. Med. 164, 63–76 [DOI] [PubMed] [Google Scholar]
- 18. Hirschhorn R., Decsy M. I., Troll W. (1971) The effect of PHA stimulation of human peripheral blood lymphocytes upon cellular content of euchromatin and heterochromatin. Cell. Immunol. 2, 696–701 [DOI] [PubMed] [Google Scholar]
- 19. Manteĭfel' V. M., Andreĭchuk T. N., Karu T. I. (1992) A comparative study of chromatin from lymphocyte nuclei upon activation of transcription by irradiation from an He-Ne-laser or phytohemagglutinin. Mol. Biol. 26, 1054–1062 [PubMed] [Google Scholar]
- 20. Pompidou A., Rousset S., Macé B., Michel P., Esnous D., Renard N. (1984) Chromatin structure and nucleic acid synthesis in human lymphocyte activation by phytohemagglutinin. Exp. Cell Res. 150, 213–225 [DOI] [PubMed] [Google Scholar]
- 21. Schirmer E. C., Florens L., Guan T., Yates J. R., 3rd, Gerace L. (2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382 [DOI] [PubMed] [Google Scholar]
- 22. Dreger M., Bengtsson L., Schöneberg T., Otto H., Hucho F. (2001) Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc. Natl. Acad. Sci. U.S.A. 98, 11943–11948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korfali N., Fairley E. A., Swanson S. K., Florens L., Schirmer E. C. (2009) Use of sequential chemical extractions to purify nuclear membrane proteins for proteomics identification. Methods Mol. Biol. 528, 201–225 [DOI] [PubMed] [Google Scholar]
- 24. Walter P., Blobel G. (1983) Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 96, 84–93 [DOI] [PubMed] [Google Scholar]
- 25. Florens L., Korfali N., Schirmer E. C. (2008) Subcellular fractionation and proteomics of nuclear envelopes. Methods Mol. Biol. 432, 117–137 [DOI] [PubMed] [Google Scholar]
- 26. Florens L., Washburn M. P. (2006) Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 328, 159–175 [DOI] [PubMed] [Google Scholar]
- 27. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 28. Wolters D. A., Washburn M. P., Yates J. R., 3rd (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690 [DOI] [PubMed] [Google Scholar]
- 29. McDonald W. H., Tabb D. L., Sadygov R. G., MacCoss M. J., Venable J., Graumann J., Johnson J. R., Cociorva D., Yates J. R., 3rd (2004) MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 30. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 31. Eng J., McCormack A., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 32. Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zybailov B. L., Florens L., Washburn M. P. (2007) Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol. Biosyst. 3, 354–360 [DOI] [PubMed] [Google Scholar]
- 34. Florens L., Carozza M. J., Swanson S. K., Fournier M., Coleman M. K., Workman J. L., Washburn M. P. (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paoletti A. C., Parmely T. J., Tomomori-Sato C., Sato S., Zhu D., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U.S.A. 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y., Wen Z., Washburn M. P., Florens L. (2010) Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272–2281 [DOI] [PubMed] [Google Scholar]
- 38. Flicek P., Aken B. L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Down T., Dyer S. C., Eyre T., Fitzgerald S., Fernandez-Banet J., Gräf S., Haider S., Hammond M., Holland R., Howe K. L., Howe K., Johnson N., Jenkinson A., Kähäri A., Keefe D., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Prlic A., Rice S., Rios D., Schuster M., Sealy I., Slater G., Smedley D., Spudich G., Trevanion S., Vilella A. J., Vogel J., White S., Wood M., Birney E., Cox T., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Hubbard T. J., Kasprzyk A., Proctor G., Smith J., Ureta-Vidal A., Searle S. (2008) Ensembl 2008. Nucleic Acids Res. 36, D707–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 40. Carbon S., Ireland A., Mungall C. J., Shu S., Marshall B., Lewis S. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Su A. I., Cooke M. P., Ching K. A., Hakak Y., Walker J. R., Wiltshire T., Orth A. P., Vega R. G., Sapinoso L. M., Moqrich A., Patapoutian A., Hampton G. M., Schultz P. G., Hogenesch J. B. (2002) Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 99, 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schirmer E. C., Guan T., Gerace L. (2001) Involvement of the lamin rod domain in heterotypic lamin interactions important for nuclear organization. J. Cell Biol. 153, 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hirschhorn K., Bach F., Kolodny R. L., Firschein I. L., Hashem N. (1963) Immune Response and Mitosis of Human Peripheral Blood Lymphocytes in Vitro. Science 142, 1185–1187 [DOI] [PubMed] [Google Scholar]
- 44. Ichimura S., Zama M., Fujita H. (1971) Quantitative determination of single-stranded sections in DNA using the fluorescent probe acridine orange. Biochim. Biophys. Acta 240, 485–495 [DOI] [PubMed] [Google Scholar]
- 45. Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 46. Manilal S., Nguyen T. M., Sewry C. A., Morris G. E. (1996) The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 5, 801–808 [DOI] [PubMed] [Google Scholar]
- 47. Cartegni L., di Barletta M. R., Barresi R., Squarzoni S., Sabatelli P., Maraldi N., Mora M., Di Blasi C., Cornelio F., Merlini L., Villa A., Cobianchi F., Toniolo D. (1997) Heart-specific localization of emerin: new insights into Emery-Dreifuss muscular dystrophy. Hum. Mol. Genet. 6, 2257–2264 [DOI] [PubMed] [Google Scholar]
- 48. Lattanzi G., Ognibene A., Sabatelli P., Capanni C., Toniolo D., Columbaro M., Santi S., Riccio M., Merlini L., Maraldi N. M., Squarzoni S. (2000) Emerin expression at the early stages of myogenic differentiation. Differentiation 66, 208–217 [DOI] [PubMed] [Google Scholar]
- 49. Salpingidou G., Smertenko A., Hausmanowa-Petrucewicz I., Hussey P. J., Hutchison C. J. (2007) A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 178, 897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 51. Schermelleh L., Carlton P. M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M. C., Agard D. A., Gustafsson M. G., Leonhardt H., Sedat J. W. (2008) Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Malik P., Korfali N., Srsen V., Lazou V., Batrakou D. G., Zuleger N., Kavanagh D. M., Wilkie G. S., Goldberg M. W., Schirmer E. C. (2010) Cell-specific and lamin-dependent targeting of novel transmembrane proteins in the nuclear envelope. Cell Mol. Life Sci. 67, 1353–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karbowski M., Neutzner A., Youle R. J. (2007) The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 178, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pickersgill H., Kalverda B., de Wit E., Talhout W., Fornerod M., van Steensel B. (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 55. Croft J. A., Bridger J. M., Boyle S., Perry P., Teague P., Bickmore W. A. (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145, 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pegoraro G., Kubben N., Wickert U., Göhler H., Hoffmann K., Misteli T. (2009) Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol. 11, 1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kosak S. T., Skok J. A., Medina K. L., Riblet R., Le Beau M. M., Fisher A. G., Singh H. (2002) Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296, 158–162 [DOI] [PubMed] [Google Scholar]
- 58. Williams R. R., Azuara V., Perry P., Sauer S., Dvorkina M., Jørgensen H., Roix J., McQueen P., Misteli T., Merkenschlager M., Fisher A. G. (2006) Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 119, 132–140 [DOI] [PubMed] [Google Scholar]
- 59. Zink D., Amaral M. D., Englmann A., Lang S., Clarke L. A., Rudolph C., Alt F., Luther K., Braz C., Sadoni N., Rosenecker J., Schindelhauer D. (2004) Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 166, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.