Abstract
In mammals, the transcriptional activity of signal transducer and activator of transcription 3 (STAT3) is regulated by the deacetylase SIRT1. However, whether the newly described nongenomic actions of STAT3 toward mitochondrial oxidative phosphorylation are dependent on SIRT1 is unclear. In this study, Sirt1 gene knock-out murine embryonic fibroblast (MEF) cells were used to delineate the role of SIRT1 in the regulation of STAT3 mitochondrial function. Here, we show that STAT3 mRNA and protein levels and the accumulation of serine-phosphorylated STAT3 in mitochondria were increased significantly in Sirt1-KO cells as compared with wild-type MEFs. Various mitochondrial bioenergetic parameters, such as the oxygen consumption rate in cell cultures, enzyme activities of the electron transport chain complexes in isolated mitochondria, and production of ATP and lactate, indicated that Sirt1-KO cells exhibited higher mitochondrial respiration as compared with wild-type MEFs. Two independent approaches, including ectopic expression of SIRT1 and siRNA-mediated knockdown of STAT3, led to reduction in intracellular ATP levels and increased lactate production in Sirt1-KO cells that were approaching those of wild-type controls. Comparison of profiles of phospho-antibody array data indicated that the deletion of SirT1 was accompanied by constitutive activation of the pro-inflammatory NF-κB pathway, which is key for STAT3 induction and increased cellular respiration in Sirt1-KO cells. Thus, SIRT1 appears to be a functional regulator of NF-κB-dependent STAT3 expression that induces mitochondrial biogenesis. These results have implications for understanding the interplay between STAT3 and SIRT1 in pro-inflammatory conditions.
Keywords: Mitochondrial Metabolism, NF-kappa B, Respiratory Chain, SIRT, STAT Transcription Factor, Protein Phosphorylation, siRNA, Signal Transduction
Introduction
Sirtuins, or silent information regulator proteins, are a conserved family of seven proteins that regulate distinct biological pathways in mammals (1). The sirtuin, SIRT1, a NAD-dependent deacetylase, is located in the nucleus and plays crucial roles in genome organization and stability, stress response, glucose homeostasis, cell differentiation, DNA repair, and cell cycle control (2, 3). It also regulates processes such as cell survival, inflammation, mitochondrial biogenesis, and oxidative damage (4). Although the promotion of mitochondrial biogenesis by SIRT1 has been ascribed to deacetylation of the master regulator PGC-1α and FOXO1 (5, 6), little is known about how SIRT1 controls the energy production of the cell. Proper mitochondrial function is crucial for maintenance of metabolic homeostasis and activation of appropriate stress responses that have been implicated in life span extension and aging (7). The process of oxidative phosphorylation to generate ATP represents one of the principal bioenergetic functions of mitochondria, and yet the molecular mechanisms by which deregulation of SIRT1 expression and/or activity alters the capacity of mitochondria to meet energy demands have only begun to be elucidated.
The signal transducer and activator of transcription 3 (STAT3) transcription factor is a key antiapoptotic factor involved in the inducible expression of target genes responsible for acute-phase response, cell cycle progression, and cell transformation in response to the interleukin-6 family of cytokines and growth factors. The paralogs of STAT3 include STAT1, STAT2, STAT4, STAT5a, STAT5b, and STAT6. The STAT3 protein is a target for post-translational modifications such as phosphorylation, glutathionylation, and acetylation, all of which add layers of STAT3 regulation by extracellular signals (8–11). There is, however, persistent activation of STAT3 in many cancers and transformed cell lines (12). STAT3 is involved in the gp130-mediated signaling pathway due to the JAK-mediated phosphorylation on tyrosine 705 (Tyr(P)-STAT3) in response to interleukin-6 and related cytokines, including oncostatin M (OSM).5 Additional phosphorylation on serine 727 (Ser(P)-STAT3) has been associated with the formation of competent DNA-binding STAT3 homodimers and association with coactivators for maximal transcriptional activity. Of interest, STAT3 is subject to reversible acetylation that occurs both in the cytosol and the nucleus by multiple enzymes. In response to cytokine stimulation, the histone acetyltransferase p300/CREB-binding protein associates with STAT3 and promotes its acetylation (9, 11, 13). Conversely, several histone deacetylases, including SIRT1, interact with STAT3 and negatively regulate its transcriptional activity, through deacetylation of key STAT3 lysine residues (14–16). It is unclear whether other cellular functions of STAT3 involve SIRT1.
Recent data suggest that STAT3 can function both as a transcription factor and as a positive modulator of nongenomic cellular functions. For instance, STAT3 has been found to interact with GRIM-19, a known component of mitochondrial complex I (17, 18) and to regulate mitochondrial complex function of the respiratory chain in primary pro-B lymphocytes and the heart (19, 20). More recently, Jeong et al. (21) reported that catecholamine-induced hypertrophy modulates the expression and activation of STAT3 in mitochondria of cardiomyocytes. In light of these and related observations, an understanding of the role for SIRT1-STAT3 connection in mitochondria respiration is important. To address this issue in more detail, we here investigated the expression and subcellular localization of STAT3 and various markers of mitochondrial function in wild-type and Sirt1-null MEFs. We demonstrate that Sirt1-null cells have significantly higher serine-phosphorylated STAT3 levels in mitochondria that correlated with an increase in the mitochondrial bioenergetics and formation of ATP. Induction of STAT3 expression was linked to accumulation of active NF-κB in the nuclei of Sirt1-null cells, resulting from an increase in the phosphorylation of the p65 subunit and of its cytoplasmic inhibitor IκBα. Correspondingly, suppression of NF-κB activity by p65Rel siRNA led to a concomitant reduction of the mitochondrial function and of STAT3 expression. These results suggest that the activation of NF-κB may play an important role in triggering a pro-inflammatory program of increased cellular respiration in Sirt1-null cells via higher expression and activation of mitochondrial STAT3.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary MEFs in which Sirt1 had been inactivated via exon deletion were a kind gift from Raul Mostoslavsky. MEFs were immortalized in culture using a standard 3T3 protocol (22). The cells were maintained at 37 °C under humidified 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (high glucose) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. The medium was replenished every 3 days, and cells were subcultured after reaching confluence.
Microarray Analysis
Total RNA was isolated from wild-type (WT) and Sirt1-KO MEFs using RNeasy mini kit following the manufacturer's instructions (Qiagen, Valencia, CA) and was hybridized to BeadChips (catalog no. BD-202-0202) from Illumina (San Diego). Raw data were subjected to Z normalization as described previously (23). Parameterized significant analysis was completed according to the significance analysis of microarray protocol (24) with analysis of variance filtering (analysis of variance p < 0.05). Significant STAT genes (STAT1, STAT2, STAT3, STAT5a, and STAT6) were selected by comparison between WT and Sirt1-KO samples taking WT as control with Z test p ≤ 0.05 and Z ratio ≥1.5 in both directions and a false discovery rate of ≤0.30.
Reverse Transcription and Real Time PCR Analysis
Total RNA was isolated from frozen MEFs using the RNeasy mini kit (Qiagen), and first strand cDNA was synthesized using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) following the manufacturer's instructions. Real time PCRs were carried out with SYBR® Green PCR master mix to observe expression of STAT3 on an ABI Prism 7300 sequence detection system (Applied Biosystems). The primer sequences used for STAT3 amplification were 5′-GACCCGCCAACAAATTAAGA (sense) and 5′-TCGTGGTAAACTGGACACCA (antisense). Differences in mRNA expression were calculated according to the 2−ΔΔCT method after normalization to the housekeeping gene Gapdh (25). Each analyzed sample was performed in three biological replicates, and at least three reactions were used to calculate the expression. Fidelity of the PCR was determined by melting temperature analysis.
Western Blotting
Unless otherwise indicated, cells were lysed in RIPA buffer supplemented with EDTA and EGTA (Boston BioProducts, Ashland, MA). Insoluble materials were removed by centrifugation, and the Bradford assay method (Bio-Rad) was used to determine the protein concentrations. Proteins were separated by SDS-PAGE under reducing conditions and then transferred onto nitrocellulose membranes. Western blots were performed according to standard methods, which involved blocking in 5% nonfat milk and incubating with the antibody of interest, followed by incubating with a secondary antibody conjugated with the enzyme horseradish peroxidase. The visualization of immunoreactive bands was performed using the ECL Plus Western blotting detection system (GE Healthcare). The quantitation was done by volume densitometry using ImageJ software and normalization to β-actin. In this study, the primary antibodies were directed against STAT1, SIRT1, Brg-1, ERK 1/2, p65Rel, IκBα, phospho-IκBα (Ser-32/36), and α-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA); STAT3, Ser(P)-STAT3 (Ser-727), Tyr(P)-STAT3 (Tyr-705), phospho-NF-κB p65 (Ser-536), acetyl-p53 (Cell Signaling Technology, Inc., Danvers, MA); and complex V (MitoSciences Inc., Eugene, OR); β-actin (Abcam, Cambridge, MA), and were used generally at a dilution recommended by the manufacturer.
Subcellular Fractionation
Fractionation of cytoplasmic and nuclear protein extracts was carried out using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. Mitochondrial, cytosolic, and 100,000 × g vesicular membrane (P100) fractions were isolated using the mitochondria isolation kit for cultured cells (MitoSciences Inc.) following the manufacturer's protocol. The cytosolic fractions were concentrated using Microcon YM10 concentrators (Millipore-Amicon, Billerica, MA).
Oxygen Consumption
Oxygen consumption in WT and Sirt1-KO MEFs was measured using the Seahorse 24XF instrument (Seahorse Biosciences, North Billerica, MA) according to the manufacturer's protocols. Cells were seeded into a Seahorse tissue culture plate at a density of 30,000 cells per well, and 16 h later, medium was changed to unbuffered XF assay media at pH 7.4 (Seahorse Biosciences) supplemented with 25 mm glucose (Sigma) and 1 mm sodium pyruvate and 1 mm GlutaMAX (Invitrogen). Cells were incubated for 1 h at 37 °C at ambient air before measurements were taken. Respiration was measured in four blocks of three for 3 min. The first block measured the basal respiration rate. Next 1 μm oligomycin (EMD Chemicals, Gibbstown, NJ) was added to inhibit complex V, and the second block was measured. Then 0.3 μm carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma) was added to uncouple respiration, and the third block was measured. Finally, 2 μm antimycin A (Sigma) was added to inhibit complex III, and the last measurements were performed. Immediately after finishing the measurements, cells were counted using a Beckman Z1 Coulter counter (Beckman Coulter Inc., Brea, CA).
Mitochondrial Electron Transport Activities
Activities of NADH:coenzyme Q1 oxidoreductase (complex I), succinate deshydrogenase (complex II), cytochrome c oxidase (complex IV), ubiquinol:cytochrome c oxidoreductase (complex III), succinate:cytochrome c reductase (complex II-complex III), NADH:cytochrome c reductase (complex I–III), and citrate synthase (CS) were determined in isolated mitochondria. Specific activities were determined by spectrophotometric methods, as described previously (26, 27). Results were expressed in nmol/mg of protein/min and normalized to CS activity. The activity of CS and aconitase was also measured in total cell lysates.
Plasmid Transfection and RNA Interference
The SIRT1 expression vector and the empty vector were described previously (28). Sirt1-KO MEFs were plated at a density of 3 × 105 cells/well in 6-well plates and grown in DMEM supplemented with 10% FBS. The plasmids were transfected at a final amount of 4.0 μg/well using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. SIRT1 expression was confirmed by immunoblotting. Transfection of Sirt1-KO MEFs with 40 nm SMARTpool siRNA targeted against mouse STAT3 or NFκB p65 (Santa Cruz Biotechnology) was carried out using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's protocol. Transfected cells were assayed 48 h after reverse transfection. The negative control siRNA was “AllStars Neg. Control siRNA” (Qiagen) with no known target gene. Specific target gene silencing was confirmed by immunoblotting.
ATP and Lactate Measurements
Cells were lysed with ATP-releasing buffer (100 mm potassium phosphate buffer at pH 7.8, 1% Triton X-100, 2 mm EDTA, and 1 mm DTT) for the determination of ATP levels using a luciferin-based ATP determination kit (Invitrogen). Values were expressed as pmol/μg of protein. Culture media were collected for the determination of lactate production; the assay was performed on a 96-well microplate using an enzyme-based lactate reagent kit (Trinity Biotech, Berkeley Heights, NJ). Lactate concentrations were expressed as μmol/mg of protein.
Phosphoprotein Profiling by the Phospho Explorer Antibody Microarray
The Phospho Explorer antibody microarray, which was designed and manufactured by Full Moon Biosystems, Inc. (Sunnyvale, CA), contains 1318 antibodies. Each of the antibodies has two replicates that are printed on a coated glass microscope slide, along with multiple positive and negative controls. The antibody array experiment was performed by Full Moon Biosystems, according to their established protocol (29, 30). In brief, cell lysates obtained from Sirt1-KO and WT MEFs were biotinylated with the antibody array assay kit (Full Moon Biosystems, Inc.). The antibody microarray slides were first blocked in a blocking solution (Full Moon Biosystems, Inc.) for 30 min at room temperature, rinsed with Milli-Q grade water for 3–5 min, and dried with compressed nitrogen. The slides were then incubated with the biotin-labeled cell lysates (∼80 μg of protein) in coupling solution (Full Moon Biosystems, Inc.) at room temperature for 2 h. The array slides were washed 4 to 5 times with 1× Wash Solution (Full Moon Biosystems, Inc.) and rinsed extensively with Milli-Q grade water before detection of bound biotinylated proteins using Cy3-conjugated streptavidin. The slides were scanned on a GenePix 4000 scanner, and the images were analyzed with GenePix Pro 6.0 (Molecular Devices, Sunnyvale, CA). The fluorescence signal (I) of each antibody was obtained from the fluorescence intensity of this antibody spot after subtraction of the blank signal (spot in the absence of antibody). For each phospho-specific antibody, a phosphorylation signal ratio change (Δ) was calculated based on Equation 1 (29),
where Ip and Inp are fluorescence signals of the phosphorylated and matching nonphosphorylated/total protein from the experimental sample, respectively. Parameters I0p and I0np are the respective fluorescence signals of the phosphorylated and matching nonphosphorylated/total protein from the control sample. A 95% confidence interval was used to quantify the precision of the phosphorylation signal ratio change based on the analysis of the replicates (data analysis was performed based on the method of an on-line program.
Statistical Analysis
All results were expressed as relative to the control value. Experiments were performed in at least two to three different culture preparations, and two dishes for each experimental condition were plated in each preparation. Results are expressed as means ± S.D. Statistical comparisons between groups were made by t test. Analyses were performed using the Microsoft® Office Excel, 2003 (Microsoft Corp., Redmond, WA), with p values ≤0.05 considered significant.
RESULTS
Levels of Serine-phosphorylated STAT3 Increase within the Mitochondria of SirT1-null MEFs
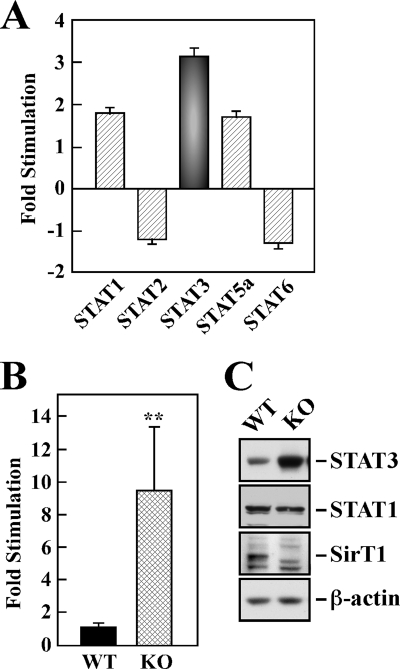
Using cDNA microarray technology, changes in expression profiles of the STAT family of transcription factors were analyzed as a feature of SIRT1 deletion in MEF cells (supplemental Table 1). Among the five STAT members identified, STAT3 was the most highly expressed gene with a 3.2 ± 0.2-fold increase in Sirt1-KO versus wild-type (WT) MEFs (p < 0.05, Fig. 1A). However, SIRT1 depletion was associated with 1.9- and 1.8-fold increase in STAT1 and STAT5a mRNA expression, respectively, and a reduction in the levels of STAT2 and STAT6 mRNA (1.2- and 1.3-fold reduction, respectively). We then analyzed the expression of STAT3 mRNA by real time PCR and found that it was increased nearly 10-fold in Sirt1-KO MEFs, consistent with the cDNA microarray data (p < 0.01; Fig. 1B). Western blot analysis showed a significant 3.1 ± 0.3-fold up-regulation in STAT3 protein levels in Sirt1-null cells when compared with WT MEFs (Fig. 1C). There was no appreciable difference in STAT1 expression (Fig. 1C). These results indicate a contribution of SIRT1 in modulating the transcriptional induction of STAT3.
FIGURE 1.
Up-regulated expression of STAT3 in SirT1-KO MEF cells. A, cDNA microarray expression data from wild-type (WT) and Sirt1-KO (KO) MEF cells. The bars indicate the relative abundance of STAT1, STAT2, STAT3, STAT5a, and STAT6 (ratio of the signal intensities of KO cells relative to WT cells) and are represented as average ± S.D. B, relative mRNA expression of STAT3 was assessed by quantitative PCR, and the values were normalized to GAPDH mRNA levels. The data represent the average ± S.D. of three independent experiments. **, p < 0.01 versus WT group. C, cellular expression of STAT3, STAT1, and SIRT1 proteins was assessed by Western blot analysis. Blot was reprobed for β-actin, which was used as a loading control.
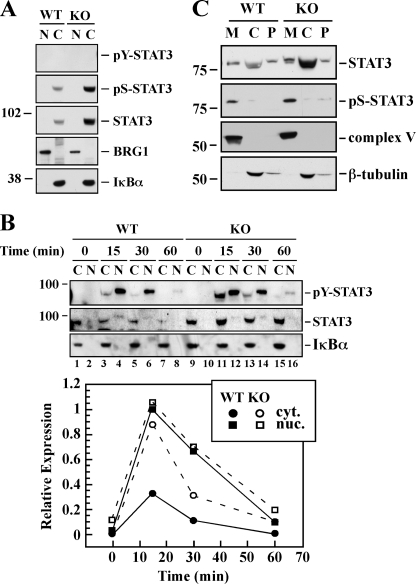
Recent data have demonstrated that STAT3 participates in specific gene regulation after being activated by tyrosine phosphorylation in response to receptor and nonreceptor tyrosine kinases (31). Moreover, the binding of OSM, a member of the interleukin-6 family cytokines, to the transmembrane gp130 subunit triggers JAK-mediated tyrosine phosphorylation of STAT3 (11). To investigate the role of SIRT1 in STAT3 activation and compartmentalization, we performed subcellular fractionation of Sirt1-KO and WT MEFs according to established protocols. Higher levels of total and Ser(P)-STAT3 were present in the cytosol of untreated Sirt1-null cells in the absence of a detectable pool of Tyr(P)-STAT3 (Fig. 2A). Exposure to OSM resulted in a time-dependent increase in Tyr(P)-STAT3 levels in the nuclear fractions of both cell lines, peaking at 15 min before returning to basal levels by 60 min (Fig. 2B). When compared with the cytosolic fraction, the total nuclear STAT3 pool was small but detectable after OSM treatment, indicative of a weak STAT3 nucleocytoplasmic shuttling (Fig. 2B). Nevertheless, the ratio of Tyr(P)-STAT3 to total STAT3 was comparable between Sirt1-KO and WT MEFs, consistent with the ability of OSM to elicit JAK-mediated phosphorylation of STAT3 in a SIRT1-independent manner.
FIGURE 2.
Subcellular distribution of STAT3. The WT and Sirt1-KO MEFs were incubated in the absence (A) or presence (B) of 20 ng/ml murine OSM for 15, 30, and 60 min. Nuclear (N) and cytosolic (C) fractions were prepared and processed for Western blot analysis using antibodies specific for phosphotyrosine (pY), phosphoserine (pS), and unphosphorylated/total forms of STAT3. The membranes were reprobed with antibodies specific for the nuclear marker BRG-1 and the cytoplasmic IκBα. The relative subcellular distribution of Tyr(P)-STAT3 levels in response to OSM treatment was determined by densitometry. Similar results were obtained in a second independent experiment. C, cytosolic (C), mitochondrial (M) and P100 membrane (P) fractions from the WT and Sirt1-KO MEFs were processed for the detection of Ser(P)-STAT3 and STAT3 by Western blot. The membranes were reprobed with the mitochondrial marker complex V and the cytosolic marker β-tubulin. The migration of molecular mass markers (values in kilodaltons) is shown on the left of immunoblots.
STAT3 has also been shown to participate in a number of nongenomic actions and accumulate in mitochondria (32). Although the bulk of STAT3 protein was observed in the cytosolic fraction of both cell lines, mitochondria of untreated Sirt1-KO cells showed significantly higher Ser(P)-STAT3 levels than WT MEFs (Fig. 2C). It is tempting to speculate that the mitochondrial pool of Ser(P)-STAT3 interacts closely with protein complexes that mediate cellular respiration.
Changes in Cellular Respiration in SirT1-null Cells
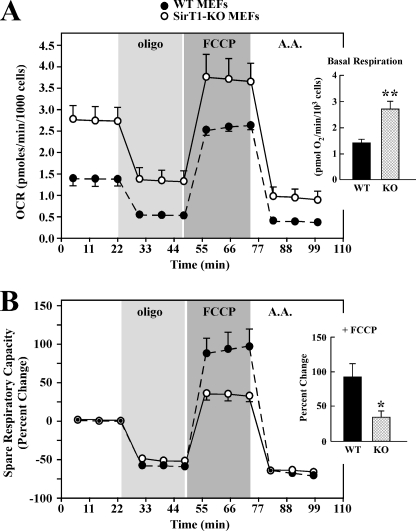
To measure cellular respiration as an index of mitochondrial function, we used the Seahorse Bioscience XF analyzer. Sirt1-KO MEFs cultured under standard conditions had basal oxygen consumption rates (OCRs) that were more than 2-fold higher than those for WT MEFs (Fig. 3A). Inhibition of the F1F0-ATPase with oligomycin is a good measure of the fraction of the OCR that is coupled to ATP production, as it typically induces an ∼70% decrease in basal OCR levels (33). The basal OCRs decreased approximately by 47 and 67% following the addition of 1 μm oligomycin to the Sirt1-null cells and WT MEFs, respectively (Fig. 3A). As a further test of their differential respiratory function, MEFs were treated with FCCP, which uncouples the mitochondrial membrane potential to cause an increase in the OCR. The FCCP-induced increase in the OCR relative to the oligomycin-treated rate corresponds to the maximum oxidative phosphorylation capacity, which is an approximate measure of the Vmax for the electron transport chain in isolated mitochondria (33, 34). As shown in Fig. 3A, FCCP produced an ∼2.6-fold increase in OCRs in Sirt1-KO cells compared with oligomycin-treated OCRs. The increase in OCRs produced by FCCP in WT MEF cultures was 5.1-fold. Finally, when the complex III inhibitor antimycin A was used, OCR was markedly blocked in the presence of FCCP, with an ∼71 and 86% reduction in Sirt1-KO and WT MEFs, respectively (Fig. 3A).
FIGURE 3.
SirT1-KO MEF cells exhibit increased basal mitochondrial respiration. Eighteen h after plating, WT and Sirt1-KO MEFs were incubated in unbuffered XF assay media in ambient air for 1 h. OCR was determined using Seahorse XF-24 metabolic flux analyzer. A, OCRs from WT cells (black) have lower basal rates of mitochondrial respiration than Sirt1-KO MEFs (open symbols). Vertical lines indicate time of addition of mitochondrial inhibitors (i) oligomycin (1 μm), (ii) FCCP (0.3 μm), or (iii) antimycin A (A.A.) (2 μm). The maximal respiratory capacity (FCCP) is significantly lower in WT than Sirt1-KO cells. Inset, average basal respiration between WT and Sirt1-KO cells. Data represent the average ± S.D. of five independent experiments. B, percent change in spare respiratory capacity in response to mitochondrial inhibitors. Spare respiratory capacity is significantly higher in WT than Sirt1-KO cells (*, p < 0.05 as compared with WT group; n = 5). Inset, average values of the spare respiratory capacity of the WT and Sirt1-KO cells after exposure to FCCP. Data represents the average ± S.D. of five independent experiments. * and **, p < 0.05 and 0.01 as compared with WT groups.
The spare respiratory capacity, described by Fern (35) as a measure of the ATP-generating reserve available to cope with an increase in ATP demand, is key in the ability of cells to survive stressful conditions. The spare respiratory capacity is defined as the increase in respiration above basal OCR levels. Despite the increase in OCR, SirT1-null cells exhibited a 2.7-fold reduction in spare respiratory capacity in the presence of FCCP as compared with WT MEFs (p < 0.05, see Fig. 3B).
Up-regulation of the Complexes I and III in Cultured SirT1-KO MEF Cells
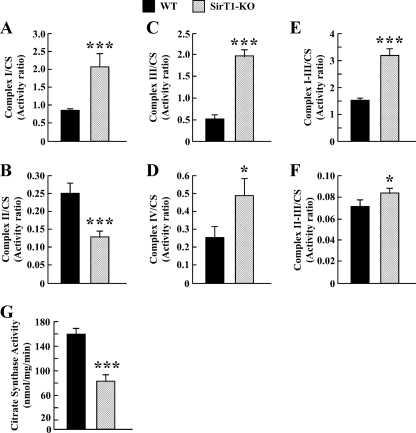
Enzymatic measurements of the respiratory chain complex activities revealed that, in Sirt1-null cells, the activities of complex I and III and complex I plus III (NADH:cytochrome c oxidoreductase) were significantly increased (supplemental Fig. 1, A, C and E), whereas the activity of complex II and complex II plus III (succinate:cytochrome c oxidoreductase) were decreased (p < 0.001) and that of complex IV remained unchanged as compared with WT controls (supplemental Fig. 1, B and D). CS is a marker of mitochondrial mass (36), whose activity is not related to oxidative phosphorylation. There was a 2-fold reduction in mitochondrial CS activity in Sirt1-KO cells versus WT cells (p < 0.001, see Fig. 4G). Following normalization for CS activity, the analyses showed a true complex II deficiency but a marked up-regulation of complex I, which translated into higher activities of the different complexes (III, I plus III, and IV) in Sirt1-null cells (Fig. 4, A–F). This may reflect some metabolic adaptation to sustain increased oxygen consumption as a consequence of SIRT1 deficiency (see Fig. 3A).
FIGURE 4.
Enzyme activities of the mitochondrial electron transport chain complexes. Enzymatic activities of complex I (A), complex II (B), complex III (C), complex IV (D), complex I plus III (E), and complex II plus III (F) were measured in isolated mitochondria from WT (black bars) and Sirt1-KO MEFs (hatched bars) as described under “Experimental Procedures.” Values are expressed as activity ratio, whereby nanomoles of substrate (donor or acceptor) consumed by min per mg of protein were normalized to the mitochondrial CS activity (G). The data represent the average ± S.D. of three independent experiments. * and ***, p < 0.05 and 0.001 versus WT groups.
We have also measured CS and aconitase activities in total cell lysates. The results showed a significant ∼40 and 23% reduction in CS (supplemental Fig. 1G) and aconitase (data not shown) activities, respectively, in the Sirt1-null cells as compared with the WT controls. These data are consistent with SirT1-KO MEFs having lower mitochondrial mass.
SIRT1 Regulates Lactate/ATP Production via STAT3 Expression in MEF Cells
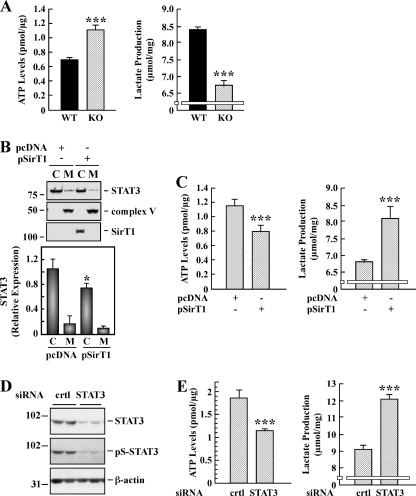
To examine whether increases in cellular respiration were accompanied by alteration in glycolysis, WT and Sirt1-KO MEFs were processed for the assessment of intracellular ATP levels and production of lactate. There was a significant inverse relationship between lactate and ATP levels in the two cell lines studied, such that a greater increase in ATP was accompanied by a lower production of lactate in Sirt1-null cells (Fig. 5A). The lactate/ATP ratio decreased from 12.1 ± 0.5 to 6.1 ± 0.4 (p < 0.001, n = 3) in WT and Sirt1-KO cells, respectively.
FIGURE 5.
Effect of SIRT1 and STAT3 on intracellular ATP levels and lactate production. A, WT and Sirt1-KO MEFs were processed for the determination of intracellular ATP (left panel) and lactate production (right panel). Bars represent the average ± S.D. of three independent experiments. B, Sirt1-KO MEF cells were transfected with pcDNA control and SIRT1 expressing plasmid for 48 h followed by isolation of mitochondrial (M) and cytosolic (C) fractions. Representative Western blot shows the signal associated with STAT3, complex V, and SIRT1. Bars represent the average ± S.D. (n = 3). *, p < 0.05 versus pcDNA. C, Sirt1-KO MEF cells were transiently transfected as in B and processed for the determination of intracellular ATP (left panel) and lactate production (right panel). Bars represent the average ± S.D. of two independent experiments performed in triplicate dishes. D, Sirt1-KO MEF cells were transfected with 40 nm of control (crtl) and STAT3 siRNAs. Following 48 h of siRNA knockdown, cells were subjected to Western blot analysis for STAT3 and Ser(P)-STAT3 (pS-STAT3) expression levels. The blot was reprobed with anti-β-actin as a loading control. The migration of molecular mass markers (values in kilodaltons) is shown on the left of immunoblots. E, control and STAT3-depleted Sirt1-KO cells were processed for the determination of intracellular ATP (left panel) and lactate production (right panel). Bars represent the average ± S.D. of two independent experiments performed in triplicate. ***, p < 0.001 versus WT or control cells.
To independently ascertain the role of SIRT1 in cellular energy metabolism, Sirt1-KO MEFs were transiently transfected with control or SIRT1-expressing plasmid for 48 h. Control experiment indicated that overexpression of SIRT1 resulted in a 29 ± 5% reduction in STAT3 levels (Fig. 5B). Moreover, ectopic expression of SIRT1 resulted in lower ATP levels but higher production of lactate when compared with cells transfected with plasmid control (Fig. 5C). Consequently, the lactate/ATP ratio increased significantly, passing from 4.9 ± 0.6 to 12.1 ± 1.7 after pSIRT1 transfection (p < 0.001, n = 5), a ratio similar in WT MEFs.
Because of the up-regulation of mitochondrial STAT3 levels in Sirt1-null cells, we investigated the role of STAT3 in modulating cellular energy metabolism. siRNA-mediated ablation of STAT3 in Sirt1-KO cells resulted in an increase in lactate production with a concomitant decrease in ATP levels when compared with KO cells transfected with a control nontargeting siRNA (Fig. 5E). The lactate/ATP ratio rose from 4.9 ± 0.5 to 10.4 ± 0.4 upon STAT3 knockdown, approaching that of WT MEFs. Western blot analysis confirmed the silencing of STAT3 (Fig. 5D) in Sirt1-KO MEFs. Taken together, these results indicate that SIRT1-dependent alteration in STAT3 expression contributes to cell energy metabolism.
Activation of NF-κB Leads to STAT3 Mitochondrial Signaling in SirT1-KO MEFs
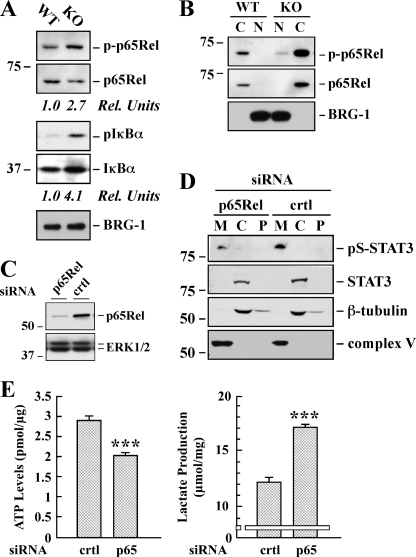
To explore the molecular mechanism(s) underlying STAT3 induction in Sirt1-KO MEFs, a comprehensive phospho-proteomic-based study was performed using a commercial phospho-antibody microarray, which provides a high throughput platform for efficient protein phosphorylation status profiling. This method facilitates the detection and analysis of phosphorylation events at specific sites (29), allowing the survey of potential links between SIRT1 and some signaling molecules of known relevance to the transcriptional network controlling STAT3 expression. The experiment was performed by applying cell lysates obtained from Sirt1-KO and WT MEFs onto the Phospho Explorer antibody microarray. We identified a spectrum of proteins whose phosphorylation levels were increased more than 2.5-fold with low values of 95% confidence interval in Sirt1-null cells. Many of these proteins are transcription factors that, when phosphorylated, modulate expression of target genes. These transcription factors included NF-κB, Elk1, Smad2/3, GATA1, FOXO1A/3A, and CREB (Table 1 and supplemental Table 2). Among these proteins, the p65Rel subunit of NF-κB has been reported to bind to the Stat3 promoter, and SIRT1 induces transcriptional repression of NF-κB via deacetylation of p65Rel (37, 38). The NF-κB activity is tightly regulated by the inhibitory IκBα protein, whose phosphorylation on Ser-32/36 allows the release of NF-κB from the inhibitory cytosolic complex and its translocation into the nucleus to affect gene transcription (39). We confirmed by immunoblotting the increased phosphorylation (Fig. 6A) and nuclear accumulation (Fig. 6B) of p65Rel in Sirt1-KO cells versus WT cells. Moreover, the levels of phosphorylated IκBα (p-Ser32/36) were 4.1-fold higher in Sirt1-null cells (Fig. 6A), consistent with NF-κB activation in these cells.
TABLE 1.
Partial list of transcription factors whose phosphorylation increased in Sirt1-KO MEF cells
The phospho-antibody microarray identified a list of transcription factors whose phosphorylation states increased in MEFs derived from Sirt1-KO mice. The signal intensities of phosphorylated and total forms of each protein were determined, and the ratio (phospho/total) of each protein was calculated between Sirt1-KO and WT MEFs. 95% confidence interval (CI) was determined to demonstrate the significance of the signal alteration for each transcription factor. The impact of site-specific phosphorylation on the biological function of these transcription factors is shown. This information can be found on line.
| Phosphorylation sites | Ratio (Sirt1-KO/WT) | 95% CI | Function |
|---|---|---|---|
| NF-κB-p65 (Ser(P)-529) | 6.54 | 5.39–7.68 | Activation |
| Elk1 (Thr(P)-417) | 5.25 | 3.50–7.00 | Activation |
| NF-κB-p65 (Ser(P)-468) | 4.03 | 3.04–5.03 | Slight inhibition |
| NF-κB-p65 (Ser(P)-536) | 3.68 | 2.15–5.20 | Activation |
| Smad2/3 (Thr(P)-8) | 3.67 | 3.09–4.24 | Activation |
| GATA1 (Ser(P)-310) | 3.62 | 3.35–3.88 | Activation |
| NF-κB-p105/p50 (Ser(P)-932) | 3.28 | 1.27–5.30 | Activation |
| FOXO1A/3A (Ser(P)-322/325) | 3.28 | 2.79–3.78 | Inactivation |
| CREB (Ser(P)-129) | 2.64 | 2.26–3.03 | Activation |
FIGURE 6.
Effect of p65Rel silencing on STAT3 expression and ATP/lactate ratio in SirT1-KO MEFs. A, total lysates of WT and Sirt1-KO MEFs were prepared and analyzed by Western blot for total and phosphorylated p65Rel, as well as total and phosphorylated IκBα. The blot was reprobed for BRG1 as a loading control. Rel Units, for each phospho-specific antibody, a phosphorylation signal ratio change (phospho/total) was calculated and normalized to the WT sample. The migration of molecular-mass markers (values in kilodaltons) is shown on the left of immunoblots. B, cytosolic (C) and nuclear (N) fractions were prepared from the WT and Sirt1-KO MEFs and processed for immunoblot analysis. The blot was reprobed with anti-BRG1 antibody to demonstrate the quality of our nuclear fractionation. C, Sirt1-KO MEFs were transfected with 40 nm of either control (crtl) or a pool of four siRNAs targeting p65Rel. Following 48 h of siRNA knockdown, cell lysates were subjected to Western blot analysis for p65Rel and ERK1/2 expression levels. D, mitochondrial (M), cytosolic (C), and P100 membrane (P) fractions were prepared from Sirt1-null MEFs transfected with either control or p65-Rel siRNA. Western blot analysis was performed with the indicated antibodies. Similar results were obtained in two independent experiments performed in triplicate dishes. Ser(P)-STAT3 (pS-STAT3). E, control and p65Rel-depleted Sirt1-KO cells were processed for lactate determination and ATP levels. The data represents the average ± S.D. of three independent experiments. ***, p < 0.001 versus control siRNA-transfected cells.
To substantiate these observations, we looked for genes that significantly changed with Sirt1 deletion and known to be involved in biological pathways modulated by NF-κB, such as inflammation, migration, and cell survival. A comprehensive list of NF-κB target genes compiled by T. D. Gilmore can be found on line. Using the cDNA microarray technology, changes in expression profiles of select target genes were detected when comparing Sirt1-null cells with WT MEFs (supplemental Table 3). One of the genes identified was Nfkbia, whose transcript encodes IκBα, a protein inhibitor of the NF-κB complex.
Finally, the role of p65Rel in modulating STAT3 expression and cellular energy metabolism was investigated in Sirt1-null MEFs. siRNA-mediated ablation of Rela resulted in 6.3 ± 1.9-fold reduction in p65Rel transcript levels (Fig. 6C) and an ∼68% decrease in mitochondrial Ser(P)-STAT3 levels in p65Rel knockdown cells as compared with cells transfected with a control nontargeting siRNA (Fig. 6D; p < 0.01, n = 4). Moreover, the silencing of p65Rel was associated with an increase in lactate production with a concomitant decrease in ATP levels (Fig. 6E). The lactate/ATP ratio rose from 4.2 ± 0.2 to 8.4 ± 0.1 upon p65Rel knockdown. These results demonstrate the cooperation between NF-κB and STAT3 mitochondrial signaling in SirT1-null MEFs.
DISCUSSION
SIRT1 has attracted great interest as a key player in the control of mitochondrial bioenergetics through activation of gene expression by the deacetylation of important signaling proteins, such as transcription factors and cotranscriptional regulators. This study extends these observations, identifying STAT3, which is a contributor to cellular respiration, as an additional SIRT1 target. This study shows a selective induction of STAT3 expression in cells with targeted gene deletion of Sirt1. Transcriptional induction of Stat3 in Sirt1-null MEFs was associated with an increase in the constitutive activation of STAT3, identified by Ser-727 phosphorylation in the absence of Tyr(P)-STAT3. Additionally, there was accumulation of Ser(P)-STAT3 in the mitochondria of Sirt1-null MEFs, which correlated with higher cellular respiration and ATP formation due in large part to the up-regulation of complex I activity. Further documenting the importance of SIRT1 in STAT3 mitochondrial signaling, ectopic expression of SIRT1 reduced STAT3 expression and ATP formation in Sirt1-null MEFs. There was constitutive activation of the transcription factor NF-κB and induction of target genes in SirT1-null cells. Also gene knockdown of NF-κB p65Rel subunit lowered STAT3 expression and consequently down-modulated mitochondrial respiration. These findings suggest that the control of mitochondrial bioenergetics by STAT3 may represent a novel NF-κB-mediated pathway that is dependent on SIRT1 regulation.
Several studies have shown that non-tyrosine-phosphorylated STAT3 is signaling competent and has been detected in the nuclear fraction of tumors and primary cells (40, 41). Immunohistochemical analysis showed that nuclear Ser(P)-STAT3 staining is associated with advanced stage breast cancer and increases with tumor size (42). Furthermore, treatment of cells with insulin, interleukin 3, or GM-CSF elicits STAT3 phosphorylation on Ser-727 in the absence of Tyr-705 phosphorylation (43, 44). Additionally, Notch signaling is accompanied by phosphorylation of STAT3 on serine but not tyrosine (45). However, STAT3 can also localize to the mitochondria where it stimulates respiration in normal and transformed cells (20), while contributing to cardioprotection by inhibition of mitochondrial permeability transition pore opening (46). The ability of STAT3 to promote ATP synthesis requires Ser-727 phosphorylation but does not depend on its phosphorylation-mediated dimerization and function as a transcriptional activator. Depending on the cellular and environmental context, the presence of Ser(P)-STAT3 in mitochondria can support transformation of cells by the Ras oncogene, which is responsible for many human cancers (47, 48). A significant pool of STAT3 is mainly present in the matrix of subsarcolemmal and interfibrillar cardiomyocyte mitochondria under physiological conditions, where it interacts with cyclophilin D (46) as well as complexes I/II (20). However, the mechanism by which STAT3 alters mitochondrial respiration is controversial. The observations made by Phillips et al. (49) indicated a largely unfavorable ratio of complexes I/II and STAT3 in cardiac tissue, implying the existence of an additional mechanism of STAT3 regulation of ATP production in vivo.
A number of cotranscriptional regulators, molecular chaperones, and signaling proteins interact with STAT3, leading to the modulation of its transcriptional activity. For example, a recent report shows the direct physical interaction of SIRT1 with the STAT3 DNA-binding domain (15), and type 1 histone deacetylase also binds with the acetylated form of STAT3 (11, 14). A large fraction of mitochondrial proteins are acetylated (50), including cyclophilin D whose deacetylation by SIRT3 induces dissociation of hexokinase II from the mitochondria that is necessary for stimulation of oxidative phosphorylation (51). Therefore, it is possible that the mitochondrial localization of STAT3 serves as a nodal point for the formation of functional multiprotein complexes encompassing mitochondrion-specific deacetylases (e.g. SIRT3, -4, and -5), STAT3, and interacting proteins involved in mitochondrial function. Regardless of the responsible mechanism, the activation of non-tyrosine-phosphorylated STAT3 in mitochondria may be critical to the pathogenesis of chronic inflammatory conditions such as infection and in aging (see below).
A number of serine kinase activities have been reported to elicit STAT3 Ser-727 phosphorylation and transactivation. Enhanced Ser(P)-STAT3 levels were observed in response to ataxia telangiectasia-mutated, MAPKs, mitogen- and stress-activated protein kinase 1, and p90 ribosomal protein S6 kinases (52–55). Moreover, PKCδ, TAK1·Nemo-like kinase, and ZIP kinase have all been found to promote STAT3 serine phosphorylation in cells treated with interleukin-6 (56–58). In contrast, neither Akt-1 nor MAPKs regulate STAT3 serine phosphorylation in primary human macrophages (41). Other data indicate that mTOR promotes STAT3 signaling via Ser-727 phosphorylation (59). Additionally, the viability and maintenance of breast cancer stem-like cells appear to require activation of the PTEN/mTOR/STAT3 pathway (60). Conversely, resveratrol inhibits mitogenic signaling by down-regulating the PI3K/Akt/mTOR pathway, and it leads also to the reduction in STAT3 expression and lowering of the constitutive and IL-6-induced activation of STAT3 (61, 62). However, the mechanism of resveratrol signaling is controversial. In fact, resveratrol has been proposed to reduce mTOR activity in a SIRT1-dependent manner (63), whereas other recent reports have questioned the notion that resveratrol is a direct activator of SIRT1 (64–66). Thus, it is unclear whether SIRT1-mediated inhibition of mTOR signaling is accompanied by STAT3 inactivation through impairment of Ser-727 phosphorylation. Our phospho-antibody array data revealed that many of these putative STAT3 kinases were activated in Sirt1-null cells as compared with WT MEFs (supplemental Table 2). Moreover, we noted that SIRT1 deficiency correlated with an elevated mTOR pathway, as assessed by phosphorylation of its downstream targets p70 ribosomal protein S6 kinase and 4E-BP1 (supplemental Table 2). Because of the elevation of mTOR activity in Sirt1-null cells, we speculate that it directly phosphorylates STAT3 on Ser-727. Clearly, more work is needed to identify the protein serine kinase(s) responsible for the constitutive phosphorylation of STAT3.
Inactivation of Ser(P)-STAT3 has to be regulated to terminate STAT3 signaling. It is interesting that HDAC3 influences phosphorylation of STAT3 at Ser-727 by acting as a scaffold protein for the protein phosphatase 2A (16). Expression of the small protein phosphatase PTP-1B is decreased whereas that of SIRT1 is increased in the muscle of exercised old rats (67). Therefore, inducible interaction between deacetylases and protein-serine phosphatases may play a role in the constitutive phosphorylation/dephosphorylation cycle of STAT3 at Ser-727 that can impact on the intensity and duration of the cellular respiratory quotient.
A physiological balance between ATP and lactate production exists in living organisms depending on the demand for oxygen consumption and oxygen supply (68). Conversion of pyruvate from glycolysis to acetyl-CoA as compared with lactate is determined in part by the activity of the mitochondrial pyruvate dehydrogenase complex, whose activity is negatively regulated by phosphorylation involving pyruvate dehydrogenase kinase 4 (PDK4). Inhibition of pyruvate dehydrogenase prevents the entry of pyruvate into the citric acid cycle and activation of the electron transport chain, ultimately altering cellular energy balance (69). Gerhart-Hines et al. (70) showed lower PDK4 expression in Sirt1-null MEFs, and therefore, efficient mitochondrial entry of pyruvate as the main supply of acetyl-CoA that makes ATP must occur at the expense of lactate formation in these cells. Our data support this notion and add evidence of greater oxygen consumption (Fig. 3) and a shift in the ATP/lactate ratio (Fig. 5A) when comparing Sirt1-null versus WT MEFs. Remarkably, the ATP/lactate ratio in Sirt1-null cells significantly returned to that in WT MEFs by knocking down STAT3 or p65Rel, implying that the observed rate in cellular respiration can be accounted for, at least in part, by NF-κB-induced STAT3 transcription.
Our data indicate that genetic depletion of SIRT1 significantly increased the transcription of known NF-κB target genes (supplemental Table 3). It has been established that reactive oxygen species elicit NF-κB activation and inflammatory gene expression both in cultured cells and in vivo (71). Moreover, SIRT1 regulates cellular reactive oxygen species levels by inducing expression of antioxidant proteins while down-modulating NF-κB activity (72). We cannot rule out the existence of additional mechanisms of SIRT1 regulation of NF-κB involving interaction with transcription coregulators.
In summary, genetic deletion of Sirt1 contributes to enhance cellular respiration through at least two mechanisms as follows: induction and activation of STAT3, via Ser-727 phosphorylation, and subcellular localization of STAT3 in mitochondria. Our study provides a better understanding of the molecular mechanisms by which SIRT1 controls mitochondrial oxidative function through an NF-κB-STAT3 connection. These results have important implications for our understanding of how these pathways might be altered in metabolic diseases, cancer, and aging.
Supplementary Material
Acknowledgments
We thank Drs. Yongqing Zhang, William H. Wood III, and Kevin G. Becker from the Gene Expression and Genomics Unit, Research Resources Branch, NIA, National Institutes of Health, for support with the cDNA microarray analysis; Dr. Xiong Li from the Dept. of Mathematics and Computer Science, Emory University (Atlanta, GA), for invaluable help with the analysis of the phospho-antibody array data; and Sutapa Kole for technical assistance. We also thank Dr. Raul Mostoslavsky (Harvard Medical School, Boston) for providing us with the Sirt1-KO and WT MEFs.
This work was supported, in whole or in part, by National Institutes of Health NIA Intramural Research Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1–3.
- OSM
- oncostatin M
- NF-κB
- nuclear factor κ-light-chain-enhancer of activated B cells
- MEF
- murine embryonic fibroblast
- FCCP
- carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone
- OCR
- oxygen consumption rate
- CS
- citrate synthase
- mTOR
- mammalian target of rapamycin.
REFERENCES
- 1. Haigis M. C., Sinclair D. A. (2010) Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gorospe M., de Cabo R. (2008) Trends Cell Biol. 18, 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vaquero A., Reinberg D. (2009) Genes Dev. 23, 1849–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michan S., Sinclair D. (2007) Biochem. J. 404, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cantó C., Auwerx J. (2009) Curr. Opin. Lipidol. 20, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nemoto S., Fergusson M. M., Finkel T. (2004) Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 7. Guarente L. (2007) Cold Spring Harbor Symp. Quant. Biol. 72, 483–488 [DOI] [PubMed] [Google Scholar]
- 8. Lee J. L., Wang M. J., Chen J. Y. (2009) J. Cell Biol. 185, 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ray S., Boldogh I., Brasier A. R. (2005) Gastroenterology 129, 1616–1632 [DOI] [PubMed] [Google Scholar]
- 10. Xie Y., Kole S., Precht P., Pazin M. J., Bernier M. (2009) Endocrinology 150, 1122–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuan Z. L., Guan Y. J., Chatterjee D., Chin Y. E. (2005) Science 307, 269–273 [DOI] [PubMed] [Google Scholar]
- 12. Grivennikov S. I., Karin M. (2010) Curr. Opin. Genet. Dev. 20, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang R., Cherukuri P., Luo J. (2005) J. Biol. Chem. 280, 11528–11534 [DOI] [PubMed] [Google Scholar]
- 14. Ray S., Lee C., Hou T., Boldogh I., Brasier A. R. (2008) Nucleic Acids Res. 36, 4510–4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nie Y., Erion D. M., Yuan Z., Dietrich M., Shulman G. I., Horvath T. L., Gao Q. (2009) Nat. Cell Biol. 11, 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Togi S., Kamitani S., Kawakami S., Ikeda O., Muromoto R., Nanbo A., Matsuda T. (2009) Biochem. Biophys. Res. Commun. 379, 616–620 [DOI] [PubMed] [Google Scholar]
- 17. Lufei C., Ma J., Huang G., Zhang T., Novotny-Diermayr V., Ong C. T., Cao X. (2003) EMBO J. 22, 1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J., Yang J., Roy S. K., Tininini S., Hu J., Bromberg J. F., Poli V., Stark G. R., Kalvakolanu D. V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9342–9347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Potla R., Koeck T., Wegrzyn J., Cherukuri S., Shimoda K., Baker D. P., Wolfman J., Planchon S. M., Esposito C., Hoit B., Dulak J., Wolfman A., Stuehr D., Larner A. C. (2006) Mol. Cell. Biol. 26, 8562–8571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., Dulak J., Baker D. P., Wolfman A., Stuehr D., Hassan M. O., Fu X. Y., Avadhani N., Drake J. I., Fawcett P., Lesnefsky E. J., Larner A. C. (2009) Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeong K., Kwon H., Min C., Pak Y. (2009) Exp. Mol. Med. 41, 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Todaro G. J., Green H. (1963) J. Cell Biol. 17, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheadle C., Vawter M. P., Freed W. J., Becker K. G. (2003) J. Mol. Diagn. 5, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quinzii C., Naini A., Salviati L., Trevisson E., Navas P., Dimauro S., Hirano M. (2006) Am. J. Hum. Genet. 78, 345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rustin P., Chretien D., Bourgeron T., Gérard B., Rötig A., Saudubray J. M., Munnich A. (1994) Clin. Chim. Acta 228, 35–51 [DOI] [PubMed] [Google Scholar]
- 28. Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 29. He H. J., Zong Y., Bernier M., Wang L. (2009) Proteomics Clin. Appl. 3, 1440–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang S., Elf S., Lythgoe K., Hitosugi T., Taunton J., Zhou W., Xiong L., Wang D., Muller S., Fan S., Sun S. Y., Marcus A. I., Gu T. L., Polakiewicz R. D., Chen Z. G., Khuri F. R., Shin D. M., Chen J. (2010) J. Clin. Invest. 120, 1165–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reich N. C., Liu L. (2006) Nat. Rev. Immunol. 6, 602–612 [DOI] [PubMed] [Google Scholar]
- 32. Myers M. G., Jr. (2009) Science 323, 723–724 [DOI] [PubMed] [Google Scholar]
- 33. Affourtit C., Brand M. D. (2009) Methods Enzymol. 457, 405–424 [DOI] [PubMed] [Google Scholar]
- 34. Gnaiger E. (2003) Adv. Exp. Med. Biol. 543, 39–55 [DOI] [PubMed] [Google Scholar]
- 35. Fern R. (2003) J. Neurosci. Res. 71, 759–762 [DOI] [PubMed] [Google Scholar]
- 36. López-Lluch G., Hunt N., Jones B., Zhu M., Jamieson H., Hilmer S., Cascajo M. V., Allard J., Ingram D. K., Navas P., de Cabo R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1768–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salminen A., Kauppinen A., Suuronen T., Kaarniranta K. (2008) BioEssays 30, 939–942 [DOI] [PubMed] [Google Scholar]
- 38. Schug T. T., Xu Q., Gao H., Peres-da-Silva A., Draper D. W., Fessler M. B., Purushotham A., Li X. (2010) Mol. Cell. Biol. 30, 4712–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karin M. (2006) Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 40. Meyer T., Gavenis K., Vinkemeier U. (2002) Exp. Cell Res. 272, 45–55 [DOI] [PubMed] [Google Scholar]
- 41. Liu H., Ma Y., Cole S. M., Zander C., Chen K. H., Karras J., Pope R. M. (2003) Blood 102, 344–352 [DOI] [PubMed] [Google Scholar]
- 42. Yeh Y. T., Ou-Yang F., Chen I. F., Yang S. F., Wang Y. Y., Chuang H. Y., Su J. H., Hou M. F., Yuan S. S. (2006) Int. J. Cancer 118, 2943–2947 [DOI] [PubMed] [Google Scholar]
- 43. Ceresa B. P., Pessin J. E. (1996) J. Biol. Chem. 271, 12121–12124 [DOI] [PubMed] [Google Scholar]
- 44. Decker T., Kovarik P. (2000) Oncogene 19, 2628–2637 [DOI] [PubMed] [Google Scholar]
- 45. Androutsellis-Theotokis A., Leker R. R., Soldner F., Hoeppner D. J., Ravin R., Poser S. W., Rueger M. A., Bae S. K., Kittappa R., McKay R. D. (2006) Nature 442, 823–826 [DOI] [PubMed] [Google Scholar]
- 46. Boengler K., Hilfiker-Kleiner D., Heusch G., Schulz R. (2010) Basic Res. Cardiol. 105, 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gough D. J., Corlett A., Schlessinger K., Wegrzyn J., Larner A. C., Levy D. E. (2009) Science 324, 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leslie K., Gao S. P., Berishaj M., Podsypanina K., Ho H., Ivashkiv L., Bromberg J. (2010) Breast Cancer Res. 12, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Phillips D., Reilley M. J., Aponte A. M., Wang G., Boja E., Gucek M., Balaban R. S. (2010) J. Biol. Chem. 285, 23532–23536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang J. Y., Hirschey M. D., Shimazu T., Ho L., Verdin E. (2010) Biochim. Biophys. Acta 1804, 1645–1651 [DOI] [PubMed] [Google Scholar]
- 51. Shulga N., Wilson-Smith R., Pastorino J. G. (2010) J. Cell Sci. 123, 894–902 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52. Ceresa B. P., Horvath C. M., Pessin J. E. (1997) Endocrinology 138, 4131–4137 [DOI] [PubMed] [Google Scholar]
- 53. Zhang Y., Liu G., Dong Z. (2001) J. Biol. Chem. 276, 42534–42542 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y., Cho Y. Y., Petersen B. L., Bode A. M., Zhu F., Dong Z. (2003) J. Biol. Chem. 278, 12650–12659 [DOI] [PubMed] [Google Scholar]
- 55. Wierenga A. T., Vogelzang I., Eggen B. J., Vellenga E. (2003) Exp. Hematol. 31, 398–405 [DOI] [PubMed] [Google Scholar]
- 56. Schuringa J. J., Dekker L. V., Vellenga E., Kruijer W. (2001) J. Biol. Chem. 276, 27709–27715 [DOI] [PubMed] [Google Scholar]
- 57. Kojima H., Sasaki T., Ishitani T., Iemura S., Zhao H., Kaneko S., Kunimoto H., Natsume T., Matsumoto K., Nakajima K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4524–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sato N., Kawai T., Sugiyama K., Muromoto R., Imoto S., Sekine Y., Ishida M., Akira S., Matsuda T. (2005) Int. Immunol. 17, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 59. Yokogami K., Wakisaka S., Avruch J., Reeves S. A. (2000) Curr. Biol. 10, 47–50 [DOI] [PubMed] [Google Scholar]
- 60. Zhou J., Wulfkuhle J., Zhang H., Gu P., Yang Y., Deng J., Margolick J. B., Liotta L. A., Petricoin E., 3rd, Zhang Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16158–16163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bhardwaj A., Sethi G., Vadhan-Raj S., Bueso-Ramos C., Takada Y., Gaur U., Nair A. S., Shishodia S., Aggarwal B. B. (2007) Blood 109, 2293–2302 [DOI] [PubMed] [Google Scholar]
- 62. Yu L. J., Wu M. L., Li H., Chen X. Y., Wang Q., Sun Y., Kong Q. Y., Liu J. (2008) Neoplasia 10, 736–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ghosh H. S., McBurney M., Robbins P. D. (2010) PLoS ONE 5, e9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beher D., Wu J., Cumine S., Kim K. W., Lu S. C., Atangan L., Wang M. (2009) Chem. Biol. Drug Des. 74, 619–624 [DOI] [PubMed] [Google Scholar]
- 65. Tang B. L. (2010) Brain Res. Bull. 81, 359–361 [DOI] [PubMed] [Google Scholar]
- 66. Pacholec M., Bleasdale J. E., Chrunyk B., Cunningham D., Flynn D., Garofalo R. S., Griffith D., Griffor M., Loulakis P., Pabst B., Qiu X., Stockman B., Thanabal V., Varghese A., Ward J., Withka J., Ahn K. (2010) J. Biol. Chem. 285, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pauli J. R., Ropelle E. R., Cintra D. E., De Souza C. T., da Silva A. S., Moraes J. C., Prada P. O., de Almeida Leme J. A., Luciano E., Velloso L. A., Carvalheira J. B., Saad M. J. (2010) Mech. Ageing Dev. 131, 323–329 [DOI] [PubMed] [Google Scholar]
- 68. Valenza F., Aletti G., Fossali T., Chevallard G., Sacconi F., Irace M., Gattinoni L. (2005) Crit. Care 9, 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Holness M. J., Sugden M. C. (2003) Biochem. Soc. Trans. 31, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 70. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Csiszar A., Labinskyy N., Jimenez R., Pinto J. T., Ballabh P., Losonczy G., Pearson K. J., de Cabo R., Ungvari Z. (2009) Mech. Ageing Dev. 130, 518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pfluger P. T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9793–9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.