Abstract
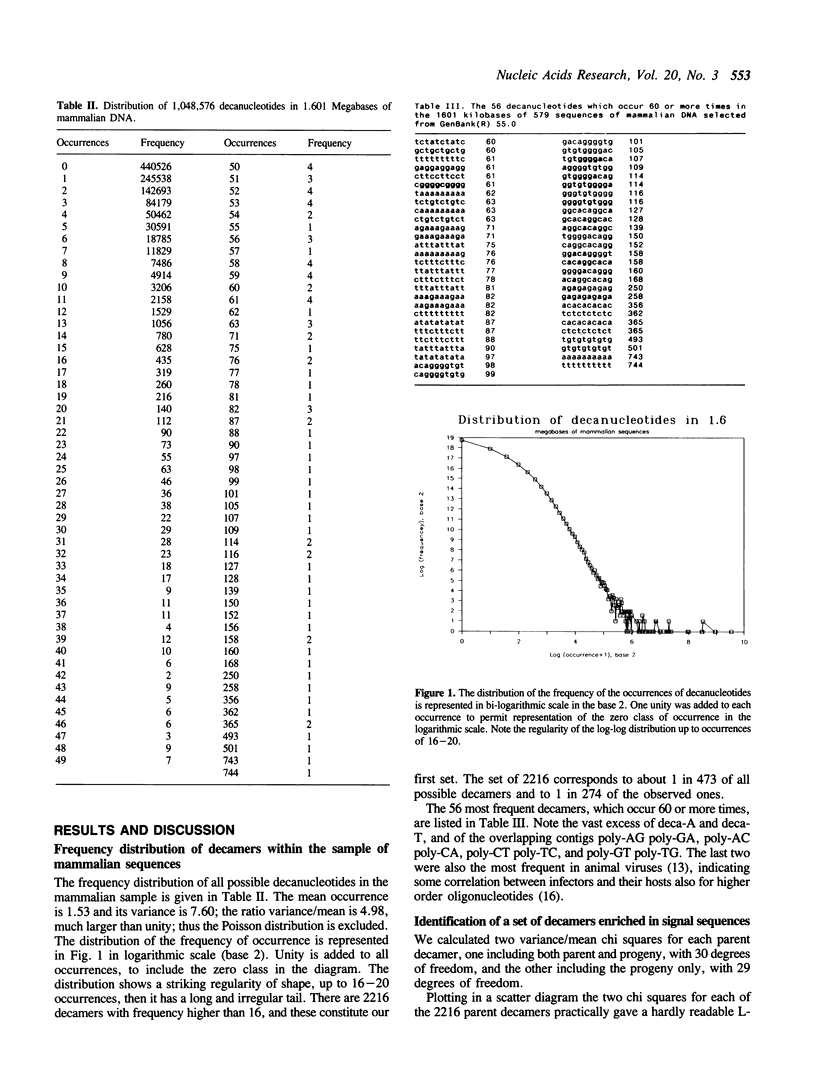
We studied the frequency distribution of oligonucleotides 10 bp long in a sample of 1.6 Mb of mammalian genes, containing 579 sequences from GenBank(R) 55.0, with the aim of detecting transcription control signals. 2216 decamers had a frequency higher than 10 times the mean and were subjected to further statistical analysis. For each of the 2216 decamers (parents), we counted the individual frequencies of the 30 decamers differing from the parent by one base mutation (progeny) and then calculated two variance/mean chi squares for the progeny, with and without the parent. We then studied the distribution of the ratio between the two chi squares. Out of 2216 decamers, 346 had a chi square ratio of 1.9 or larger. In this final set, which corresponds to less than 0.033 per cent of all possible decamers, 18 were found to contain 23 eukaryotic transcription control elements 5-10 bp of length, such as Sp1 and others. Furthermore, when compared to 210 random sets containing 346 decamers, this set contains a highly significant excess of the longer signals.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almagor H. A Markov analysis of DNA sequences. J Theor Biol. 1983 Oct 21;104(4):633–645. doi: 10.1016/0022-5193(83)90251-5. [DOI] [PubMed] [Google Scholar]
- Barrai I., Scapoli C., Barale R., Volinia S. Oligonucleotide correlations between infector and host genomes hint at evolutionary relationships. Nucleic Acids Res. 1990 May 25;18(10):3021–3025. doi: 10.1093/nar/18.10.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J. M., Bougueleret L. Heuristic informational analysis of sequences. Nucleic Acids Res. 1986 Jan 10;14(1):179–196. doi: 10.1093/nar/14.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980 Jan 11;8(1):r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Ikeuchi K., Groopman J., Baltimore D. Factors affecting cellular tropism of human immunodeficiency virus. J Virol. 1990 Nov;64(11):5600–5604. doi: 10.1128/jvi.64.11.5600-5604.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J. R., Waterman M. S., Smith T. F. Regulatory pattern identification in nucleic acid sequences. Nucleic Acids Res. 1983 Apr 11;11(7):2221–2231. doi: 10.1093/nar/11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto M. H., Brunck T. K., Bernstein R. L. Overlapping redundant septuplets identical with regulatory elements of HIV-1 and SV40. Nucleic Acids Res. 1989 Apr 11;17(7):2783–2800. doi: 10.1093/nar/17.7.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S., Sadler J. R. Statistical characterization of nucleic acid sequence functional domains. Nucleic Acids Res. 1983 Apr 11;11(7):2205–2220. doi: 10.1093/nar/11.7.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Bernardi F., Gambari R., Barrai I. Co-localization of rare oligonucleotides and regulatory elements in mammalian upstream gene regions. J Mol Biol. 1988 Sep 20;203(2):385–390. doi: 10.1016/0022-2836(88)90006-x. [DOI] [PubMed] [Google Scholar]
- Volinia S., Gambari R., Bernardi F., Barrai I. The frequency of oligonucleotides in mammalian genic regions. Comput Appl Biosci. 1989 Feb;5(1):33–40. doi: 10.1093/bioinformatics/5.1.33. [DOI] [PubMed] [Google Scholar]
- Volinia S., Scapoli C., Gambari R., Barale R., Barrai I. A set of viral DNA decamers enriched in transcription control signals. Nucleic Acids Res. 1991 Jul 11;19(13):3733–3740. doi: 10.1093/nar/19.13.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. L., Garcia J., Harrich D., Pearson L., Wu F., Gaynor R. Lymphoid specific gene expression of the adenovirus early region 3 promoter is mediated by NF-kappa B binding motifs. EMBO J. 1990 Dec;9(13):4435–4442. doi: 10.1002/j.1460-2075.1990.tb07894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E. Compilation of transcription regulating proteins. Nucleic Acids Res. 1988 Mar 25;16(5):1879–1902. doi: 10.1093/nar/16.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]


