Abstract
The emerging role of single-nucleotide polymorphisms (SNPs) in clinical association and pharmacogenetic studies has created a need for high-throughput genotyping technologies. We describe a novel method for multiplexed genotyping of SNPs that employs PCR amplification on microspheres. Oligonucleotide PCR primers were designed for each polymorphic locus such that one of the primers contained a recognition site for BbvI (a type IIS restriction enzyme), followed by 11 nucleotides of locus-specific sequence, which reside immediately upstream of the polymorphic site. Following amplification, this configuration allows for any SNP to be exposed by BbvI digestion and interrogated via primer extension, four-color minisequencing. Primers containing 5′ acrylamide groups were attached covalently to the solid support through copolymerization into acrylamide beads. Highly multiplexed solid-phase amplification using human genomic DNA was demonstrated with 57 beads in a single reaction. Multiplexed amplification and minisequencing reactions using bead sets representing eight polymorphic loci were carried out with genomic DNA from eight individuals. Sixty-three of 64 genotypes were accurately determined by this method when compared to genotypes determined by restriction-enzyme digestion of PCR products. This method provides an accurate, robust approach toward multiplexed genotyping that may facilitate the use of SNPs in such diverse applications as pharmacogenetics and genome-wide association studies for complex genetic diseases.
The capability to discern genetic variation among individuals is fundamental to the further understanding of genetic predisposition to complex diseases such as cancer, mental illness, and diabetes. The multifactorial and heterogeneous nature of these complex diseases has highlighted the need for genetic association studies, as opposed to linkage mapping, as the preferred method to determine the underlying genetic cause of these diseases (Risch and Merikangas 1996; Gray et al. 2000; Schork et al. 2000). Coupled with this focus on genetic association studies has been the reemergence of single-nucleotide polymorphisms (SNPs) as the marker of choice in these types of studies based on their abundance, stability, and adaptability to automation (Kruglyak 1999). Major efforts in both the public (Human Genome Project) as well as the private (The SNP Consortium) sectors are underway to generate high-density, evenly spaced SNP maps, the first of which has recently been published (Sachidanandam et al. 2001). These polymorphisms, and the maps derived from them, will provide the framework for powerful new studies to identify genes involved in the pathophysiology of polygenic diseases, as well as diagnostic markers and predictors of differential drug response. Clearly, the wide-ranging applications of SNPs in pharmacogenetics and clinical association studies will necessitate further development of robust, flexible, cost-effective technology platforms for scoring genotypes in large numbers of samples.
A variety of schemes useful in the molecular genotyping of SNPs have been described over the years (for review, see Landegren et al. 1998 and Shi 2001) including those that discriminate alleles via hybridization (allele-specific PCR [Liu et al. 1997], DNA microarrays [Wang et al. 1998], Taqman [Livak 1999]) and those that discriminate alleles via enzymatic means (RFLP [Kan and Dozy 1978], oligonucleotide ligation assay [OLA; Grossman et al. 1994; Iannone et al. 2000], single-base chain extension [Pastinen et al. 1997; Cai et al. 2000] and pyrosequencing [Alderborn et al. 2000]). These methods offer the capability of accurate genotyping, but they all rely on standard PCR amplification of target sequences as the initial front-end step in generating material. Because of the technically demanding nature of standard solution-based multiplexed PCR, which often can require extensive optimization of primers and reaction conditions (Edwards and Gibbs 1994), the inherent amplification requirement effectively limits the extent to which these varied platforms can be modified for highly multiplexed genotyping. The ability to convert hundreds of PCR primer-pairs into a single-tube, multiplexed reaction producing specific, robust products from a complex genomic DNA template would greatly reduce the requirements for large-scale, population-based SNP genotyping.
In this report, we describe a novel SNP genotyping platform that combines PCR amplification and four-color minisequencing (Pastinen et al. 1997) on acrylamide microspheres, with fragment detection and analysis using an automated sequencer. This system is capable of highly multiplexed PCR in a single reaction tube, as demonstrated by the simultaneous amplification of 57 human genetic loci. Multiplexed PCR and four-color minisequencing with eight individuals were carried out using bead sets representing eight polymorphic loci. 63 of the 64 genotypes were accurately determined with this approach. This system combines the sensitivity of PCR and a universal fluorescent read-out step with the capability of high multiplexing, creating an accurate, robust approach to SNP genotyping.
RESULTS
Solid-Phase PCR from Human Genomic DNA
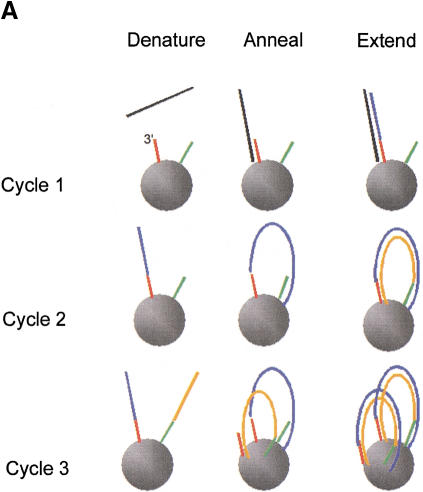
Figure 1A is a schematic diagram illustrating solid-phase PCR. During the first cycle, denatured target DNA anneals to primers that are covalently bound on their 5′ ends to beads. Primers are extended on their free 3′ OH ends during the extension phase. During the second PCR cycle, covalently bound primer extension products hybridize to complementary primers that reside in close proximity on the bead surface. After the second extension cycle, double-stranded PCR product is covalently bound to the beads at both of its 5′ ends. During the third and all subsequent denaturation, annealing, and extension cycles, additional PCR product is generated on the solid phase until primers are consumed. High concentrations of localized primers, coupled with the lack of solution-phase primers, provide each template with a minimal-complexity oligonucleotide probe set and decreases the likelihood of nonspecific priming and product cross-reactivity that normally can occur during multiplexed amplification.
Figure 1.
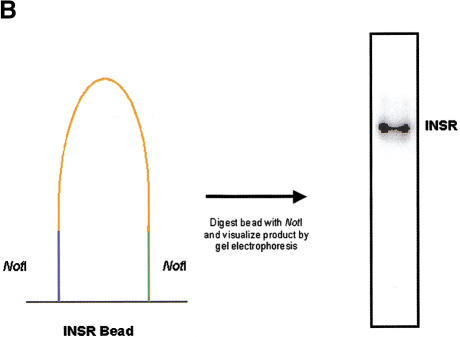
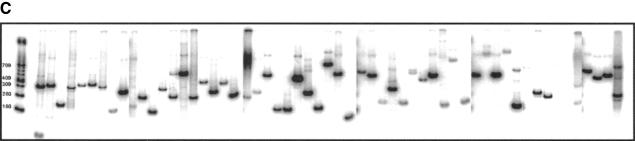
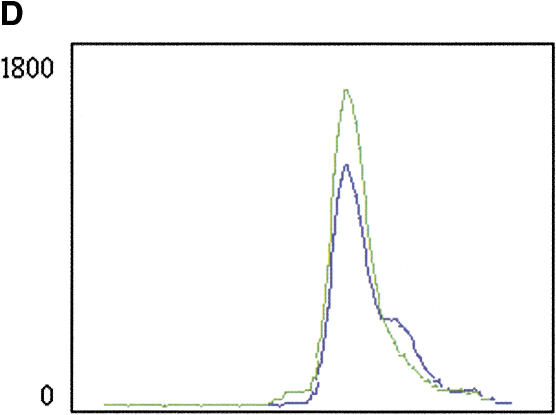
(A) Solid-phase amplification. Acrydite-containing primers are incorporated into an acrylamide bead or spot. Multiple cycles of denaturation, annealing, and extension result in a product that is covalently attached to the surface at both ends. (B) Solid-phase amplification from genomic DNA. Digested human genomic DNA was amplified using an acrylamide derivatized polyester sheet containing the INSR RsaI-specific primers. Amplification products were released from the beads with NotI and run on a 10% acrylamide TBE gel. (C) Multiplex, solid-phase PCR with human genomic DNA. Shown is a composite of six 10% Acrylamide TBE gels. Fifty-eight beads (57 beads containing PCR primers and one blank bead) were multiplexed and the resulting radiolabeled products released from the beads are shown in random order. No amplification is observed with beads lacking oligonucleotide primers. Amplification products range in size from ∼70 bp to ∼1300 bp.
Solid-phase amplification of preamplified DNA templates previously has been demonstrated (Adams and Kron 1997; Mitra and Church 1999). In an attempt to determine if solid-phase amplification using locus-specific primers was sensitive enough to amplify from genomic DNA, we polymerized Acrydite-containing primers (Rehman et al. 1999) for the insulin receptor (INSR) RsaI polymorphism into spots immobilized on an acrylamide derivatized polyester sheet. Both the forward and reverse primer contained a NotI restriction site 5′ to the locus-specific sequence as a means to release the amplification product from the support (Fig. 1B). Following amplification and digestion with NotI, the correct size fragment was seen using as little as 8 ng/μL of sheared human genomic DNA. Furthermore, the removal of the genomic DNA template after a target hybridization step, but prior to amplification, indicates that the product is a result of amplification on the solid phase (data not shown). The PCR process can be described by the equation N = N0 (1 + E)n, where E defines the amplification efficiency, N0 is the initial number of target molecules, and N is the amount of product synthesized. Amplification reactions at early and intermediate cycle numbers were used to determine that the efficiency of solid-phase PCR was near 0.3 (data not shown), whereas typical solution-phase amplification generally has an efficiency of 0.8. Additionally, the effect of primer concentration on solid-phase product yield also has been examined. No product is detected when the primer concentration is 1.0 μM. Product is detectable at a primer concentration of 10 μM, but increases with 100 μM primer concentration. These results are in general agreement with calculations that at 100 μM primer concentration, there is one primer molecule per 10 Å on or at the bead surface. Thus, at 1 μM primer concentration, the spacing of primers (1 primer per 1000 Å) can no longer support formation of a solid-phase product of average length, which is 250 bp.
Multiplexed, Solid-Phase PCR
To evaluate if solid-phase PCR could be multiplexed, we performed a single PCR reaction containing 57 1.0 μL acrylamide beads representing 53 different genes from 16 chromosomes. The primer sequences were obtained from the Genome Database (www.gdb.org) and were not chosen or optimized for their ability to enable solid- or solution-phase amplification. The primers also contained unique restriction sites for NotI that allow product release following amplification. The reaction products range in size from ∼70 bp to 1300 bp. Following amplification en masse, the beads were digested individually with NotI, and supernatants were loaded onto acrylamide gels. Because the beads cannot be readily deconvoluted prior to digestion of product, the gel contains a random order of amplification products (Fig. 1C). Fifty-three of the 57 loci exhibit a predominant amplification product. We also have demonstrated multiplexing of up to 102 beads (17 distinct amplicons each represented six times, data not shown), which confirmed the reproducibility of product yield and purity of each amplification product.
Single-Color SNP Minisequencing
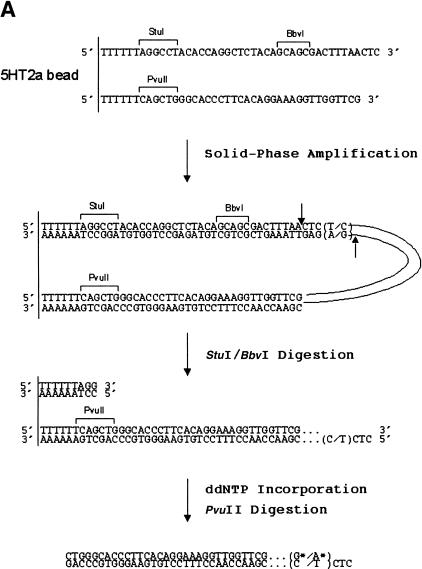
There are several general features of the primer design used for solid-phase amplification and genotyping. Each primer pair designed for a specific polymorphic locus contained the locus-specific sequence and introduced a BbvI, a StuI site, and a PvuII restriction site into the resulting PCR product. Figure 2A illustrates the primers used to amplify the T102C polymorphism in the gene for human 5-hydroxytryptamine type 2a (5-HT2a) receptor (Warren et al. 1993) and the product expected following amplification and digestion. The forward primer contains, after the T6 spacer and the StuI site, 15 bases of locus-specific sequence followed by the artificially introduced BbvI site, and then 11 additional bases of locus-specific sequence at the 3′ end. The reverse primer contains 28 bases of locus-specific sequence following the T6 spacer and PvuII site. The purpose of the Type IIS BbvI restriction site, which is precisely positioned 11 bases 5′ to the base with genetic variation, is to expose the polymorphic site upon digestion. The StuI site (always positioned on the same primer as the BbvI site) is used to remove one of the BbvI-generated 5′ overhangs and ensure that each bead utilizes only the 5′ overhang containing the polymorphic site immediately adjacent to the 3′ hydroxyl group of the recessed nucleotide as a template for ddNTP incorporation. PvuII digestion releases the amplification product after ddNTP incorporation.
Figure 2.
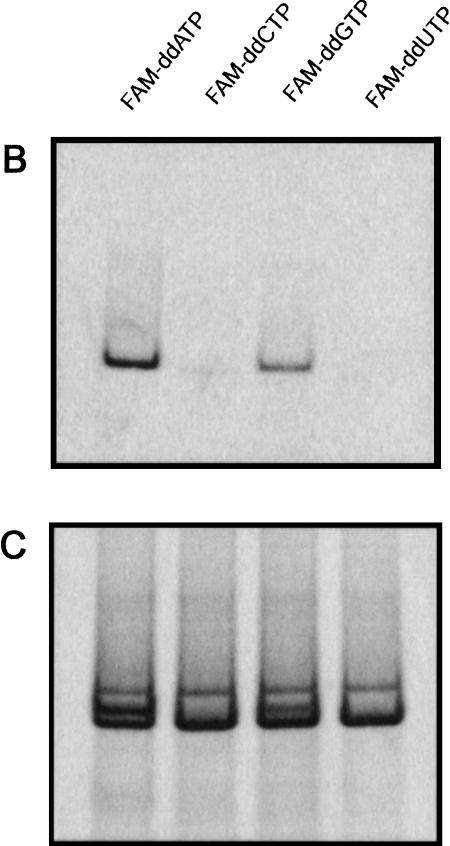
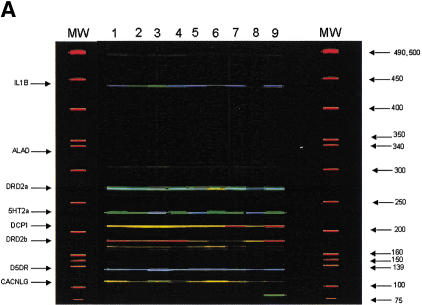
Single-nucleotide polymorphism (SNP) minisequencing. (A) Diagram of the components and products of solid-phase amplification and SNP minisequencing using 5HT2A-specific primers. The polymorphic site (C/T) is shown in parentheses. The fluorescently labeled nucleotides (G* or A*) are depicted following BbvI digestion and nucleotide incorporation. (B) Fluorescent image of 10% acrylamide gel with single-color FAM-ddNTP minisequencing of 5-HT2A solid-phase PCR product. (C) Sybr green I staining of same gel from panel B. (D) Four-color minisequencing of 5HT2A locus.
To assess the specificity of the genotyping scheme, a model 135 bp amplicon containing the 5-HT2aT102C polymorphism was evaluated using a single-color SNP minisequencing format. Individual beads containing a 5-HT2a primer pair were used for target hybridization and solid-phase amplification with genomic DNA template from a single individual who had previously been genotyped as a C/T heterozygote using a PCR-RFLP protocol (data not shown). Following StuI and BbvI cleavage, beads were used in minisequencing reactions with individual FAM-ddNTPs (Fig. 2B). Fluorescent signal is observed only from the FAM-ddATP and FAM-ddGTP labeling reactions, corresponding to incorporation into the opposite strand of the C/T polymorphism. Sybr Green I staining of the gel after fluorescent imaging (Fig. 2C) shows that equal amounts of 5-HT2a PCR product were synthesized and released from each bead. The results show that BbvI can accurately cleave a solid-phase amplification product, which then can serve as a template for ddNTP incorporation.
Four-Color SNP Minisequencing
Single-color minisequencing reactions were converted to four-color minisequencing reactions by using a cocktail of all four ddNTPs that were individually labeled with distinct rhodamine dyes. Fluorescent-ddNTP incorporation reactions were carried out directly on bead-bound products. Fluorescent products were enzymatically released from beads, separated by denaturing gel electrophoresis, and data collection and analysis was via an automated sequencer with Genescan software. This platform therefore permitted quantitative detection of any color combination in an amplification product, including SNPs that are not biallelic. Figure 2D shows the four-color minisequencing of the 5HT2A locus amplified from the same heterozygous individual as used in Figure 2B. Incorporation of the rhodamine dye-labeled ddNTPs resulted in a gel image that was blue for R110-ddGTP, red for ROX-ddUTP, green for R6G-ddATP, and yellow for TAMRA-ddCTP. Near equal green and blue peaks, indicating a C/T heterozygote, shows that the technique is able to accurately incorporate the correct base on a solid-phase template.
Multiplexed, Solid-Phase Amplification and Four-Color SNP Minisequencing
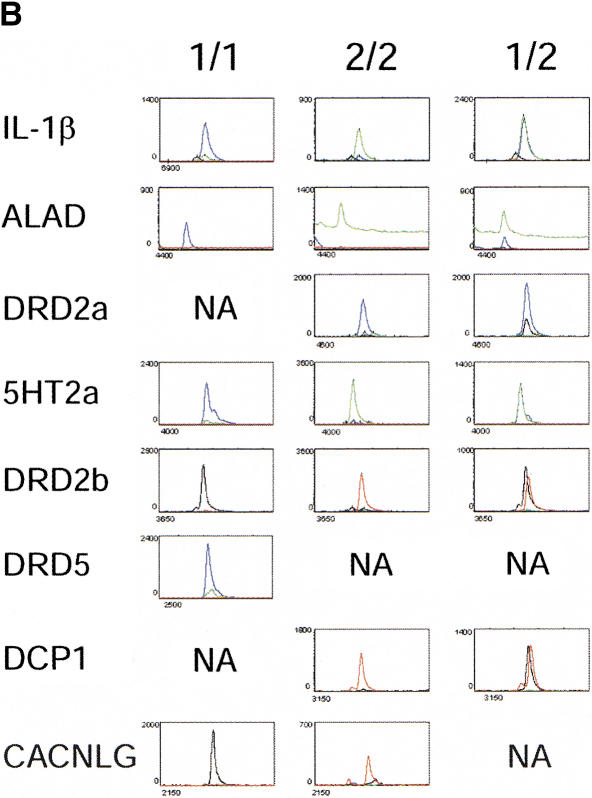
Multiplexing of the solid-phase amplification and minisequencing reactions was achieved by designing each released product to have a specific, discrete length. Thus, the length of the detected product identifies the polymorphic locus. Eight polymorphic loci were used to assess the accuracy of the multiplexed genotyping scheme. Bead sets for the eight polymorphic loci were used for target hybridization and multiplexed solid-phase amplification with genomic DNA from eight different individuals. A single reaction tube was used for each individual to genotype the eight loci. These individuals previously had been genotyped at each of the polymorphic loci by restriction enzyme digestion of PCR products or by direct DNA sequencing of PCR products. Following multiplexed solid-phase PCR, each bead set was used for four-color SNP minisequencing. Labeled products were released by PvuII digestion, and separated on a 6%, 8M urea acrylamide gel. A representative Genescan gel image is shown in Figure 3A. The eight displayed products for each individual span a range of sizes from ∼420 b (IL1B) to ∼110 b (CACNLG). Each of the eight PCR products from the eight individuals was assigned a genotype after analysis of the electropherograms. Representative electropherograms for each genotype are shown in Figure 3B. Peak height ratios for the relevant colors were determined for all electropherograms. The results are summarized for each locus in Table 1. This collective data set shows that 63 of 64 genotypes determined by the multiplexed solid-phase amplification and four-color minisequencing method were accurately determined. The IL1B genotype for DNA sample No. 4 determined by AvaI enzyme digests was CC. When genotyped by the current method, this sample showed an elevated green peak (T allele) that prevented a definitive genotype from being assigned. Whereas the peak height ratio was twofold higher than the CT heterozygote (DNA sample No. 3), it was not as high as the five additional CC homozygotes. Although peak height ratios differed between amplicons, they generally were consistent within a given locus.
Figure 3.
(A) An image of released solid-phase PCR/four-color minisequencing products separated on a 6%, 8M urea acrylamide gel. Lanes 1–8 contain products from eight unrelated individuals. Lane 9 is an internal reference standard, and lanes labeled MW contain the GENESCAN-500 ROX size standards. The identity of each amplified locus is shown on the left, and the sizes of the ROX-labeled standards are shown on the right. (B) Electropherograms for representative genotypes of each locus are shown. The Y axis denotes relative fluorescent intensity, and the X axis denotes scan number. The numerical genotype designations are indicated in Table 1.
Table 1.
Genotype Scoring
| Locus | Peak height comparison (Allele 1/Allele2) | DNA 1 | DNA 2 | DNA 3 | DNA 4 | DNA 5 | DNA 6 | DNA 7 | DNA 8 |
|---|---|---|---|---|---|---|---|---|---|
| IL-1β | C/T | CC;CC | TT;TT | CT;CT | CC;CT | CC;CC | CC;CC | CC;CC | CC;CC |
| 6.5 | 0.2 | 0.9 | 1.8 | 7.3 | 10.1 | 11.4 | 9.3 | ||
| ALAD | C/T | TT;TT | CC;CC | CC;CC | TT;TT | TT;TT | TC;TC | TT;TT | TT;TT |
| 0.1* | 22* | 46* | 0.05* | 0.04* | 0.6 | 0.04* | 0.1* | ||
| DRD2a | G/C | GG;GG | GG;GG | GG;GG | GG;GG | GG;GG | GG;GC | GG;GG | GG;GG |
| 89* | 90* | 152* | 89* | 8.0 | 3.0 | 115* | 5.6 | ||
| 5-HT2a | T/C | TT;TT | CT;CT | CC;CC | TT;TT | CC;CC | TT;TT | CT;CT | CC;CC |
| 13.8 | 1.5 | 0.3 | 5.0 | 0.5 | 4.5 | 1.0 | 0.1 | ||
| DCP1 | C/T | CC;CC | CC;CC | CC;CC | CC;CC | CC;CC | CC;CC | TT;TT | CT;CT |
| 113* | 170* | 131* | 120* | 133* | 118* | 0.1 | 1.3 | ||
| DRD5 | C/T | CC;CC | CC;CC | CC;CC | CC;CC | CC;CC | CC;CC | CC;CC | CC;CC |
| 3.2 | 3.4 | 3.5 | 5.6 | 6.1 | 4.1 | 4.2 | 5.4 | ||
| DRD2b | T/C | TT;TT | TT;TT | TT;TT | TT;TT | TC;TC | TC;TC | TC;TC | TC;TC |
| 21.3 | 39* | 21 | 18 | 0.8 | 0.7 | 0.7 | 1.0 | ||
| CACNLG | G/A | GG;GG | GG;GG | GG;GG | AA;AA | GG;GG | GG;GG | GG;GG | GG;GG |
| 98* | 31* | 125* | 0.2 | 62* | 102* | 48* | 19* |
The genotype determined by PCR-RFLP is shown, followed by the genotype determined by multiplexed solid-phase amplification and four-color minisequencing. Note that the only discrepancy is seen with the IL-1β polymorphism in DNA Sample #4.
Peak height ratios were determined for each individual and are shown below the genotypes. The ALAD amplicon generated the weakest overall fluorescent signal, which may have contributed to the higher baseline values for the green tracings in the electropherograms. Nonetheless, blue peak height (C allele) to green peak height (T allele) ratios for the TT and CC homozygotes and the CT heterozygote were determined by subtracting the green baseline signal from actual samples. An asterisk (*) indicates that the default threshold value of 20 was used for the minor peak.
A value shown in bold indicates that the electropherogram is shown in Figure 3B.
DISCUSSION
We have enabled a high throughput genotyping platform that takes advantage of both the ability to simultaneously amplify multiple loci in a single reaction and the coupling of these reaction products to a generic minisequencing method that does not require additional locus-specific primers. The multiplexed amplification was carried out on a solid phase, thereby greatly reducing the local complexity of primers and eliminating artifactual priming events and primer-primer interactions. Furthermore, one of the primers is designed such that digestion of the amplification product with a type IIS restriction enzyme will expose the site of polymorphism in a form that is directly amenable to interrogation via incorporation of a fluorescently labeled dideoxynucleotide. This methodology overcomes two critical bottlenecks faced in high-throughput genotyping; namely, the ability to perform highly multiplexed amplifications from genomic DNA and the ability to genotype multiple loci without having to add a complex mixture of primers.
It is widely held that PCR is the rate-limiting step in genome-wide analyses of sequence variation and that amplification efficiency, product specificity, and yield suffer when PCR amplifications are multiplexed (Edwards and Gibbs 1994; Landegren et al. 1998). Methods aimed at alleviating the requirement for amplification (Lizardi et al. 1998; Hall et al. 2000) or making the amplification of 100s–1000s of markers more efficient (Westin et al. 2000; Broude et al. 2001) have gained in popularity. The strength of our method stems from the ability to perform highly multiplexed amplifications from genomic DNA on acrylamide beads, thereby allowing us to combine the sensitivity of PCR with the capability to multiplex beads with distinct primer pairs in a single reaction. Because the primers remain immobilized to the bead surface through the 5′ end during amplification, PCR product is synthesized on the bead itself, thereby reducing the total primer complexity that would occur if primers were present free in solution. This minimizes the interactions between primers from different loci during multiplexed reactions, which can contribute to unwanted products such as primer-dimers or nonspecific amplification products.
Immobilization of the primers to the solid phase also allows for the removal of any unbound genomic DNA prior to the start of thermal cycling. Once prebinding of the target DNA to primers is complete, nonhybridized DNA is removed prior to the addition of enzymes for the extension and amplification reactions. This prevents production of solution-phase amplification products by trace amounts of primer that may leach from the beads during thermal cycling and minimizes formation of any artifactual products (mis-primed, primer dimers) commonly associated with multiplexed, solution-phase amplification. We have observed the maximum amount of product that can be generated on a 1-μL bead via solid-phase amplification to be ∼100 ng. This yield is a function of multiple parameters including primer length, sequence composition, and surface density, which all function in concert to define an overall efficiency of solid-phase amplification. In addition, the sequence and length of the amplicon may affect the efficiency of the reaction, whereas the efficiency of amplification can range theoretically from zero to one, the value can vary experimentally from 0.46 to 0.99 for different genes in solution PCR (Chelly et al. 1988; Choi et al. 1989; Wang et al. 1998). Furthermore, the value has varied from 0.8 to 0.99 when the same gene was amplified in independent reactions under the same conditions (Wiesner 1992). We ascribe the decreased efficiency of solid-phase PCR (0.3) to the fact that the complementary DNA strands are held in close proximity to each other and can reanneal, thus decreasing the amount of DNA that can act as template in the next round of amplification. Experiments designed to shift the equilibrium away from reannealing of PCR product strands toward annealing of product strands to amplification primers (i.e., use of amplification primers with higher Tm and decreasing the amplicon length) are a means to increase the efficiency of solid-phase amplification.
The use of type IIS enzymes in genetic analysis has been described successfully in a variety of applications, such as gene expression analysis (Velculescu et al. 1995; Brenner et al. 2000) and mass spectrometry-based genotyping (Laken et al. 1998). The current method takes advantage of a type IIS restriction enzyme site positioned in one of the amplification primers, and digestion with the type IIS enzyme exposes the polymorphic site in such a way that it can be interrogated directly via fluorescent dye-labeled nucleotide incorporation. No additional synthesis of locus-specific oligonucleotides is required for the detection component of this genotyping system. This method not only utilizes the fidelity of the DNA polymerase in allele determination but also alleviates any need for separate genotyping probes and minimizes artifacts based on primer heterodimerization and false priming.
A potential drawback in this scheme is the requirement that the recognition site for the type IIS restriction enzyme is present only once in the amplification product and it is introduced via the primer. The impact of this requirement is minimized by having the freedom to choose which amplification primer (forward or reverse strand) will contain the type IIS recognition site and the need to amplify a minimum of one nucleotide, the polymorphic site. The requirement then is limited to only the nucleotides between the recognition site and the cut site (the “throw” of the enzyme). in the case of BbvI, with a throw of 12 nucleotides, there are only nine opportunities for the GCAGC recognition site to appear, out of a total of ∼1.7 × 107 (412) different sequence combinations that can be present. Therefore, <1 in 2 × 106 polymorphisms will not be amenable to this method because a second BbvI site will be present in the amplification product. Another potential drawback in the current method is the use of gel electrophoresis as a means to deconvolute the multiplexed amplification products. Although this method has proved useful in genotyping of SNPs via minisequencing (Krook et al. 1992; Pastinen et al. 1996) and OLA (Grossman et al. 1994), the number of loci that can be discriminated by size is well short of the theoretical capacity of the multiplexed amplification reaction. Several solutions exist that could be applied to the current method. First, solid-phase amplification could be performed on spatially addressable arrays rather than beads, as demonstrated by Westin et al. (2000). Second, it is conceivable to replace the product-release step with one in which the SNP genotype is read out directly on the bead itself. To this end, current efforts are underway to use an optical encoding scheme in which specific fluorophores distinct from the rhodamine dye-labeled ddNTPs are entrapped within the beads (Chen et al. 2000; Steemers et al. 2000). Lastly, incorporation of a unique DNA tag into the amplification product, followed by hybridization to a “tag array” containing spatially addressed sequences complimentary to each unique tag (Fan et al. 2000) will convert our solution phase, released products to a deconvoluted solid-phase array.
In conclusion, we have developed a high-throughput genotyping system that takes advantage of the ability to perform highly multiplexed amplification reactions on the solid phase and couples these products with a generic genotyping scheme that utilizes information contained within the amplification product and does not require additional nucleic-acid primers. The modular nature of this method should prove beneficial to existing methodologies that require either highly multiplexed amplification or the need for a generic, rapid readout. We believe the current method will prove useful as the number of SNPs and their use in genetic association studies increases.
METHODS
Oligonucleotide Synthesis
Oligonucleotides with a 5′ acrylamide group (Acrydite, Mosaic Technologies) (Rehman et al. 1999) were obtained from Operon Technologies, Inc. Lyophilized oligonucleotides were resuspended at a concentration of 1 mM with 1X TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) and were stored frozen at −20°C.
Each human locus that was analyzed is listed in Table 2 along with the Acrydite oligonucleotides used for solid-phase amplification.
Table 2.
Amplification Primers
| Primer | Oligonucleotide sequence | Reference |
|---|---|---|
| IL1Bf | QTTTTTTAGGCCTTCAGAGGCTCCTGCCGCAGCCAGAGAGCTCC | di Giovine et al. 1992 |
| IL1Br | QTTTTTTCAGCTGGAATACCTGATTTCACAATCAAGTTAAAGG | |
| ALADf | QTTTTTTAGGCCTTTCAACCCCTCTACCGCAGCCCACACAGGTA | Astrin et al. 1991 |
| ALADr | QTTTTTTCAGCTGCCTCCCACCTCTCCACCTCCCGAGTAGC | |
| DRD2af | QTTTTTTCAGCTGATGGAAATCACACAGTCACAAAGGAGCAGA | Hauge et al. 1991 |
| DRD2ar | QTTTTTTAGGCCTTGGACTCACGAAGGCGCAGCCGGTGACCATT | |
| 5-HT2Af | QTTTTTTAGGCCTACACCAGGCTCTACAGCAGCGACTTTAACTC | Warren et al. 1993 |
| 5-HT2Ar | QTTTTTTCAGCTGGTTGGTGGCATTCTGCGGCTTTTTCTCTAG | |
| DCP1f | QTTTTTTCAGCTGAGGGCCGCTCCCTCCTCATTCCTGTCTTTC | Rieder et al. 1999 |
| DCP1r | QTTTTTTAGGCCTAGCCGGGGTTGGCCCGCAGCCGCAGGGAGAC | |
| DRD5f | QTTTTTTAGGCCTACTGCATGGTCCCTTGCAGCAGTGGACACCC | Sommer et al. 1993 |
| DRD5r | QTTTTTTCAGCTGGGCAAACACCTTCTGAAAGTCGGCGTTG | |
| DRD2bf | QTTTTTTCAGCTGATAAGCATCAAGTGTTTGGAACAGTGCC | Hauge et al. 1991 |
| DRD2br | QTTTTTTAGGCCTAGAGGAAGGAGTGGCGCAGCGTTCCCTAGTC | |
| CACNLGf | QTTTTTTAGGCCTCTGTGCCGCCTTCATGCAGCTCTTTCTCGGC | Olckers et al. 1993 |
| CACNLGr | QTTTTTTCAGCTGGAGGGTCGCTAGGGCCGCAGGAGGGTTA |
The sequence of the oligonucleotides (5′ to 3′) for each locus are shown with the BbvI site (GCAGC) depicted in bold type, the PvuII site (CAGCTG) in italics, and the StuI site (AGGCCT) is underlined.
(Q) Designates the acrylamide group; (f) forward primer; (r) reverse primer.
Preparation of Acrylamide Beads
Gel solutions contained 10% acrylamide (29:1 w/w acrylamide: bis-acrylamide), 10mM sodium borate buffer, 0.2% ammonium persulfate, and 100 μM of each Acrydite primer. Nitrogen-saturated mineral oil containing 0.4% TEMED (N, N, N‘, N‘-tetramethylenediamine) was placed into a small polyethylene dish (weighboat), and 1.0 or 0.5 μL aliquots of the gel solution were pipetted under the mineral oil. Beads were polymerized at room temperature for 1 h. The mineral oil was decanted and the beads were recovered in TE. Beads were loaded into large wells of a 2% Agarose gel in 0.5× TBE, and unpolymerized primer was removed by electrophoresis at 130V for 1 h. Beads were removed from the wells with a large bore pipet tip and washed in TE. Beads were stored in TE at 4°C for up to 4 wk. Alternatively, acrylamide solutions containing Acrydite primers and both polymerization catalysts were spotted onto acrylamide-derivatized polyester sheets (GelBond PAG, FMC BioPolymer), and polymerized under nitrogen for a few minutes. Spots were treated to remove unpolymerized primer as described for beads.
Beads or spots were pre-cycled to decrease nonspecific amplification (94°C for 10 min, then 94°C for 45 sec, 45°C for 1 min, 72°C for 1 min for 15 cycles) in 1× GeneAmp PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.001% [w/v] gelatin) containing 0.25 ng/μL Escherichia coli genomic DNA (Sigma). Beads were washed three times in STE (TE containing 50 mM NaCl), and twice in 1× GeneAmp PCR buffer prior to target hybridization.
Target Hybridization and Solid-Phase Amplification
For target hybridization, beads were incubated in 1× GeneAmp or 1× Taq Extender (Stratagene) buffer (20 mM Tris-HCl pH 8.8, 10 mM KCl, 10 mM [NH4]2SO4, 2 mM MgSO4, 0.1% Triton X-100, 0.1 mg/mL bovine serum albumin) containing 100–125 ng/μL denatured, human genomic DNA that had been either sheared or digested with StuI. Target hybridization volumes were adjusted to just cover the beads (10 μL for a single 1-μL bead, 80–100 μL for 50 1-μL or 100 0.5-μL beads). Beads were hybridized with human genomic DNA for 12–24 h in an Eppendorf thermomixer at 45°C and 850 rpm. After target hybridization, reactions were washed twice in STE, and twice in 1× Taq Extender buffer. Solid-phase amplification reactions contained 1× Taq Extender buffer, 200 μM of each dNTP, 2.5U Amplitaq, 5U AmplitaqTaq Gold, and 5U Taq Extender. In cases where PCR products were visualized directly, 1 μL (10 μCi) of 32P-α-dCTP (3000 Ci/mmole) was also included. Reactions underwent an initial extension protocol (60°C for 5 min, 68°C for 5 min, 72°C for 10 min) prior to cycling (94°C for 10 min, then 94°C for 45 sec, 65°C (−1°C/cycle) for 1 min, 72°C for 1 min for 20 cycles, followed by 94°C for 45 sec, 45°C for 1 min, 72°C for 1 min for 70 cycles). After cycling, beads were washed five times in STE.
Release of 32P-Labeled, Solid-Phase PCR Product
Single beads were manually recovered by using wide-bore pipette tips with a diameter greater than an individual bead. Beads were individually pipetted into tubes and incubated for 12–16 h in 10 μL of solution containing appropriate restriction enzymes. Supernatants were loaded onto a 10% Acrylamide TBE (89 mM Tris-borate, 89 mM boric acid, 2 mM EDTA) gel and run at 200V for 1 h. Gels were dried and exposed to phosphoimaging cassettes for 1–3 h. Screens were scanned with a Storm 860 instrument (Molecular Dynamics) at 200 μm resolution.
One-Color SNP Minisequencing
Individual beads containing primers for a polymorphism in the human 5-hydroxytryptamine type 2a receptor (5-HT2a) were used for solid-phase amplification. The primers were 5′-QT12AGGCCTACACCAGGCTCTACAGCAGCGACTTTAACT-3′ and 5′-QT12CAGCTGGGCACCCTTCACAGGAAAGG TTGGTTCG-3′ where Q is the Acrydite group. After amplification, beads were equilibrated into 1× NEBuffer 2 (New England Biolabs; 50 mM NaCl, 10 mM Tris-HCl pH 7.9 @25°C, 10 mM MgCl2, 1 mM DTT, and digested at 37°C for 16 h with 10 U StuI per bead. Beads then were rinsed with 1× NEBuffer 2, and incubated for 4 h at 37°C with 1 U BbvI per bead. Beads were rinsed and equilibrated into 1× Amplitaq FS buffer for 10 min at room temperature. One-color sequencing reactions contained 1× Amplitaq FS buffer, 1.5 μM of the single FAM (5-carboxyfluorescein)-labeled ddNTP (ddATP, ddCTP, ddGTP, or ddUTP) as well as 1.5 μM of each of the three unlabeled ddNTPs. Reactions were incubated at 68°C for 30 min, and then beads were rinsed twice with TE buffer and unincorporated fluorescent ddNTP was removed by electrophoresis of the beads at 130 V for 1 h. Beads were equilibrated into 1× NEBuffer 2, and digested overnight at 37°C with 10 U PvuII enzyme. Released products were analyzed by electrophoresis on 10% Acrylamide 1× TBE gels and run at 200V for 1 h. Gels were scanned with a Storm 860 instrument (blue fluorescence filter, PMT 900 V) at 200 μm resolution to detect the fluorescent signal. Gels then were stained with Sybr Green I (Molecular Probes) diluted 1:10,000 in 1× TBE for 30 min and reimaged using the same instrument setup to detect total released PCR product in all lanes.
Four-Color SNP Sequencing
Following solid-phase amplification reactions, beads were equilibrated into 1× NEBuffer 2 and digested overnight at 37°C with 40 U StuI restriction enzyme. Beads were washed once with fresh 1× NEBuffer 2, and then digested overnight at 37°C with 10 U of the type IIS enzyme BbvI. Reactions then were equilibrated into 1× Amplitaq FS buffer. Four-color sequencing reactions (100 μL) contained 1× Amplitaq FS buffer, 2 mM manganese citrate, 1.5 μM R6G-ddATP, 1.5 μM R110-ddGTP, 1.5 μM ROX (6-carboxy-X-rhodamine)-ddUTP, 1.5 μM TAMRA (N, N, N‘, N‘-tetramethyl-6-carboxyrhodamine)-ddCTP, and 5 U Amplitaq FS. All fluorescently labeled ddNTPs were from NEN Life Science Products. Reactions were incubated at 68°C for 30 min, rinsed twice with TE buffer, and unincorporated fluorescent ddNTPs were removed by electrophoresis of the beads at 100V for 1 h. Individual beads were separated (if the solid-phase amplification reaction had been multiplexed) and equilibrated into 1× NEBuffer 2, and digested overnight at 37°C with 40 U PvuII enzyme. Reaction supernatants were precipitated with NaOAc/EtOH and resuspended in 10 μL TE buffer. Aliquots were mixed with an equal volume of loading solution (5:1 deionized Formamide: 50 mM EDTA), heated at 90°C for 2 min followed by an ice quench, and run on 6% acrylamide (19:1 acrylamide: bis-acrylamide), 8.3 M urea, 1× TBE gels on an ABI 373 DNA Sequencer. Data was analyzed using the ABI 672 Genescan Collection Software (version 1.1) and Genescan PCR Analysis Software (version 1.2.2–1). The BbvI recognition site was placed into the forward primer in the ALAD, 5-HT2a, IL1B, DRD5, and CACNLG amplicons resulting in fluorescent labeling of the noncoding strand such that blue electropherogram tracings indicated a C allele, red tracings indicated an A allele, green tracings indicated a T allele, and black (yellow) tracings indicated a G allele. The BbvI recognition site was placed into the reverse primer for the DRD2a, DRD2b, and DCP1 amplicons resulting in fluorescent labeling of the coding strand such that blue electropherogram tracings indicated a G allele, red tracings indicated a T allele, green tracings indicated an A allele, and black (yellow) tracings indicated a C allele.
Acknowledgments
We thank Drs. Chris Boles, Ezra Abrams, Chris Adams, and Bill Dower for scientific discussions and Michelle Boatwright for technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL keith_jones@affymetrix.com; FAX (408) 481-0435.
Article published on-line before print: Genome Res., 10.1101/gr. 205001.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.205001.
REFERENCES
- Adams, C. and Kron, S. 1997. Method for performing amplification of nucleic acid with two primers bound to a single solid support. US Patent No. 5641658.
- Alderborn A, Kristofferson A, Hammerling U. Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res. 2000;10:1249–1258. doi: 10.1101/gr.10.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrin KH, Kaya AH, Wetmur JG, Desnick RJ. RsaI polymorphism in the human delta-aminolevulinate dehydratase gene at 9q34. Nucleic Acids Res. 1991;19:4307. doi: 10.1093/nar/19.15.4307-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Broude NE, Zhang L, Woodward K, Englert D, Cantor CR. Multiplex allele-specific target amplification based on PCR suppression. Proc Natl Acad Sci. 2001;98:206–211. doi: 10.1073/pnas.98.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, White PS, Torney D, Deshpande A, Wang Z, Marrone B, Nolan JP. Flow cytometry-based minisequencing: A new platform for high-throughput single-nucleotide polymorphism scoring. Genomics. 2000;66:135–143. doi: 10.1006/geno.2000.6218. [DOI] [PubMed] [Google Scholar]
- Chelly J, Kaplan JC, Maire P, Gautron S, Kahn A. Transcription of the dystrophin gene in human muscle and non-muscle tissue. Nature. 1988;333:858–860. doi: 10.1038/333858a0. [DOI] [PubMed] [Google Scholar]
- Chen J, Iannone MA, Li MS, Taylor JD, Rivers P, Nelsen AJ, Slentz-Kesler KA, Roses A, Weiner MP. A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension. Genome Res. 2000;10:549–557. doi: 10.1101/gr.10.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YW, Kotzin B, Herron L, Callahan J, Marrack P, Kappler J. Interaction of Staphylococcus aureus toxin “superantigens” with human T cells. Proc Natl Acad Sci. 1989;86:8941–8945. doi: 10.1073/pnas.86.22.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Giovine FS, Takhsh E, Blakemore AI, Duff GW. Single base polymorphism at −511 in the human interleukin-1 beta gene (IL1 beta) Hum Mol Genet. 1992;1:450. doi: 10.1093/hmg/1.6.450. [DOI] [PubMed] [Google Scholar]
- Edwards MC, Gibbs RA. Multiplex PCR: Advantages, development, and applications. PCR Methods Appl. 1994;3:S65–S75. doi: 10.1101/gr.3.4.s65. [DOI] [PubMed] [Google Scholar]
- Fan JB, Chen X, Halushka MK, Berno A, Huang X, Ryder T, Lipshutz RJ, Lockhart DJ, Chakravarti A. Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res. 2000;10:853–860. doi: 10.1101/gr.10.6.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray IC, Campbell DA, Spurr NK. Single nucleotide polymorphisms as tools in human genetics. Hum Mol Genet. 2000;9:2403–2408. doi: 10.1093/hmg/9.16.2403. [DOI] [PubMed] [Google Scholar]
- Grossman PD, Bloch W, Brinson E, Chang CC, Eggerding FA, Fung S, Iovannisci DM, Woo S, Winn-Deen ES, Iovannisci DA. High-density multiplex detection of nucleic acid sequences: Oligonucleotide ligation assay and sequence-coded separation. Nucleic Acids Res. 1994;22:4527–4534. doi: 10.1093/nar/22.21.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JG, Eis PS, Law SM, Reynaldo LP, Prudent JR, Marshall DJ, Allawi HT, Mast AL, Dahlberg JE, Kwiatkowski RW, et al. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc Natl Acad Sci. 2000;97:8272–8277. doi: 10.1073/pnas.140225597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauge XY, Grandy DK, Eubanks JH, Evans GA, Civelli O, Litt M. Detection and characterization of additional DNA polymorphisms in the dopamine D2 receptor gene. Genomics. 1991;10:527–530. doi: 10.1016/0888-7543(91)90431-d. [DOI] [PubMed] [Google Scholar]
- Iannone MA, Taylor JD, Chen J, Li MS, Rivers P, Slentz-Kesler KA, Weiner MP. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry. 2000;39:131–140. [PubMed] [Google Scholar]
- Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human β-globin structural gene: Relationship to sickle mutation. Proc Natl Acad Sci. 1978;75:5631–5635. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook A, Stratton IM, O’Rahilly S. Rapid and simultaneous detection of multiple mutations by pooled and multiplex single nucleotide primer extension: application to the study of insulin-responsive glucose transporter and insulin receptor mutations in non-insulin-dependent diabetes. Hum Mol Genet. 1992;1:391–395. doi: 10.1093/hmg/1.6.391. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Laken SJ, Jackson PE, Kinzler KW, Vogelstein B, Strickland PT, Groopman JD, Friesen MD. Genotyping by mass spectrometric analysis of short DNA fragments. Nat Biotechnol. 1998;16:1352–1356. doi: 10.1038/4333. [DOI] [PubMed] [Google Scholar]
- Landegren U, Nilsson M, Kwok PY. Reading bits of genetic information: Methods for single-nucleotide polymorphism analysis. Genome Res. 1998;8:769–776. doi: 10.1101/gr.8.8.769. [DOI] [PubMed] [Google Scholar]
- Liu Q, Thorland EC, Heit JA, Sommer SS. Overlapping PCR for bidirectional PCR amplification of specific alleles: A rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res. 1997;7:389–398. doi: 10.1101/gr.7.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Mitra RD, Church GM. In situ localized amplification and contact replication of many individual DNA molecules. Nucleic Acids Res. 1999;27:e34. doi: 10.1093/nar/27.24.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olckers A, Jedlicka AE, Powers PA, Hogan K, Gregg RG, Levitt RC. G to A polymorphism in the CACNLG gene. Hum Mol Genet. 1993;2:2198. doi: 10.1093/hmg/2.12.2198-a. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Kurg A, Metspalu A, Peltonen L, Syvanen AC. Minisequencing: A specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 1997;7:606–614. doi: 10.1101/gr.7.6.606. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Partanen J, Syvanen AC. Multiplex, fluorescent, solid-phase minisequencing for efficient screening of DNA sequence variation. Clin Chem. 1996;42:1391–7. [PubMed] [Google Scholar]
- Rehman FN, Audeh M, Abrams ES, Hammond PW, Kenney M, Boles TC. Immobilization of acrylamide-modified oligonucleotides by co-polymerization. Nucleic Acids Res. 1999;27:649–655. doi: 10.1093/nar/27.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA. Sequence variation in the human angiotensin converting enzyme. Nat Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Schork NJ, Fallin D, Lanchbury JS. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet. 2000;58:250–264. doi: 10.1034/j.1399-0004.2000.580402.x. [DOI] [PubMed] [Google Scholar]
- Shi MM. Enabling large-scale pharmacogenetic studies by high-throughput mutation detection and genotyping technologies. Clin Chem. 2001;47:164–172. [PubMed] [Google Scholar]
- Sommer SS, Sobell JL, Heston LL. A common exonic polymorphism in the human D5 dopamine receptor gene. Hum Genet. 1993;92:633–634. doi: 10.1007/BF00420955. [DOI] [PubMed] [Google Scholar]
- Steemers FJ, Ferguson JA, Walt DR. Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nat Biotechnol. 2000;18:91–94. doi: 10.1038/72006. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- Warren JT, Peacock ML, Rodriguez LC, Fink JK. An MspI polymorphism in the human serotonin receptor gene (HTR2): Detection by DGGE and RFLP analysis. Hum Mol Genet. 1993;2:338. doi: 10.1093/hmg/2.3.338. [DOI] [PubMed] [Google Scholar]
- Westin L, Xu X, Miller C, Wang L, Edman CF, Nerenberg M. Anchored multiplex amplification on a microelectronic chip array. Nat Biotechnol. 2000;18:199–204. doi: 10.1038/72658. [DOI] [PubMed] [Google Scholar]
- Wiesner RJ. Direct quantification of picomolar concentrations of mRNAs by mathematical analysis of a reverse transcription/exponential polymerase chain reaction assay. Nucleic Acids Res. 1992;20:5863–5864. doi: 10.1093/nar/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]