Abstract
Background
Alfalfa, [Medicago sativa (L.) sativa], a widely-grown perennial forage has potential for development as a cellulosic ethanol feedstock. However, the genomics of alfalfa, a non-model species, is still in its infancy. The recent advent of RNA-Seq, a massively parallel sequencing method for transcriptome analysis, provides an opportunity to expand the identification of alfalfa genes and polymorphisms, and conduct in-depth transcript profiling.
Results
Cell walls in stems of alfalfa genotype 708 have higher cellulose and lower lignin concentrations compared to cell walls in stems of genotype 773. Using the Illumina GA-II platform, a total of 198,861,304 expression sequence tags (ESTs, 76 bp in length) were generated from cDNA libraries derived from elongating stem (ES) and post-elongation stem (PES) internodes of 708 and 773. In addition, 341,984 ESTs were generated from ES and PES internodes of genotype 773 using the GS FLX Titanium platform. The first alfalfa (Medicago sativa) gene index (MSGI 1.0) was assembled using the Sanger ESTs available from GenBank, the GS FLX Titanium EST sequences, and the de novo assembled Illumina sequences. MSGI 1.0 contains 124,025 unique sequences including 22,729 tentative consensus sequences (TCs), 22,315 singletons and 78,981 pseudo-singletons. We identified a total of 1,294 simple sequence repeats (SSR) among the sequences in MSGI 1.0. In addition, a total of 10,826 single nucleotide polymorphisms (SNPs) were predicted between the two genotypes. Out of 55 SNPs randomly selected for experimental validation, 47 (85%) were polymorphic between the two genotypes. We also identified numerous allelic variations within each genotype. Digital gene expression analysis identified numerous candidate genes that may play a role in stem development as well as candidate genes that may contribute to the differences in cell wall composition in stems of the two genotypes.
Conclusions
Our results demonstrate that RNA-Seq can be successfully used for gene identification, polymorphism detection and transcript profiling in alfalfa, a non-model, allogamous, autotetraploid species. The alfalfa gene index assembled in this study, and the SNPs, SSRs and candidate genes identified can be used to improve alfalfa as a forage crop and cellulosic feedstock.
Background
The advent of next generation high-throughput sequencing has revolutionized the analysis of genomes and transcriptomes [1-5]. When applied to the transcriptome, this methodology is referred to as RNA-Seq (RNA sequencing). RNA-Seq has been used for gene annotation, expression analysis and SNP discovery [6,7]. This methodology has also proven useful for discovery of novel transcripts (coding and non-coding) and identification of alternative splice variants [5,8]. It is expected that RNA-Seq methodologies will supersede microarrays for transcript profiling because of higher sensitivity, base-pair resolution and the larger range of expression values that can be detected [3,5,9]. Furthermore, in contrast to microarrays, RNA-Seq does not require prior knowledge of gene sequences. However, RNA-Seq presents bioinformatic challenges because of the required assembly of millions of short sequence reads that are generated by the methodology.
RNA-Seq has been successfully used for annotation, transcript profiling and/or SNP discovery in a number of plant species. For model plant species with sequenced genomes, sequence reads can be mapped to the reference genome. The model species where RNA-Seq analysis has been applied include Arabidopsis [10,11], soybean [12,13], rice [14], maize [15] and Medicago truncatula [16]. There are also examples of the application of RNA-Seq to non-model plant species that lack a reference genome. In the absence of a reference genome, de novo assembly of sequence reads into contigs is required. RNA-Seq has been used for transcript profiling in Eucalyptus grandis [17], grape (Vitis vinifera L.) [18], California poppy (Eschschlozia califonica) [11], avocado (Persea americana) [11], Pachycladon enysii [19] and Artemisia annua [20]. In Eucalyptus grandis and rape (Brassica napus), RNA-Seq was used for SNP discovery [17,21].
Alfalfa is the most widely cultivated forage legume in the world and the fourth most widely grown crop in the US [22,23]. In addition to its value as a livestock feed, alfalfa also has potential as a cellulosic ethanol feedstock [24,25]. Alfalfa is an allogamous autotetraploid with complex polysomic inheritance [26-28]. Slow progress has been made in improving the agronomic traits of this species using traditional breeding approaches based on phenotypic selection. For the most part, genomic approaches for crop improvement (e.g., molecular breeding) have not been applied to this legume because of limited genomic resources. As of February 2010, there were 12,371 alfalfa ESTs available in the public database. A few SSRs have been detected but SNPs have not yet been identified [28-30]. Recently, we reported on the results of transcript profiling and single feature polymorphism (SFP) detection in alfalfa using the Medicago GeneChip as a cross-species platform [25,31]. The Medicago GeneChip contains probe sets designed for the model plant, Medicago truncatula, a diploid relative of alfalfa. Using a method based on probe affinity differences and affinity shape power, we identified over 10,000s SFPs in the stem internodes of alfalfa genotypes 252 and 1283 that differed in cellulose and lignin concentrations in cell walls [31]. In a subsequent study using the Medicago GeneChip for transcript profiling of alfalfa genotypes 252 and 1283, interspecies variable regions and SFPs were masked prior to data analysis resulting in a 2-fold increase in the number of differentially expressed genes detected in stem internodes of the two genotypes [25]. Although the research of Yang et al. [25,31] significantly advanced alfalfa genomics, the use of a cross-species platform for microarray analysis limits the sensitivity and specificity of transcriptome analysis and polymorphism detection.
The stem tissue of alfalfa is important in determining the value of this forage as a livestock feed and cellulosic feedstock. Increasing the cellulose and decreasing the lignin content in cell walls in stems would improve alfalfa for both uses. In this study, we applied RNA-Seq to gene identification, polymorphism detection and transcript profiling of two alfalfa clonal lines (708, 773) that differ in cell wall composition in stems. The results were used to assemble the first gene atlas for alfalfa (MSGI 1.0). Our research also provides the first report of high-throughput SNP detection and digital gene expression analysis in the alfalfa transcriptome.
Results and discussion
Cell wall composition of stems of genotypes 708 and 773
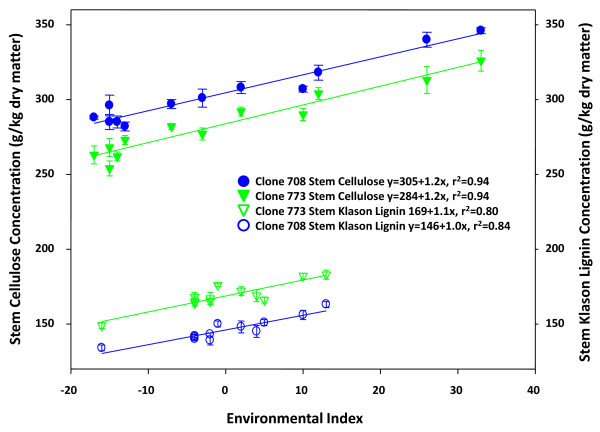
The alfalfa genotypes 708 and 773 used in this study were selected for divergent cell wall composition in stems under field conditions (see Methods for details). Cell wall composition of greenhouse grown stems used for RNA sampling in the current study is shown in Table 1. Cell wall concentration in stems of the two clones did not differ. In contrast, cellulose content (defined as glucose) in the stems of genotype 708 was 5.2% greater compared to genotype 773 (p < 0.05) (Table 1). In addition, galactose and mannose concentrations were 14.2% (p < 0.05) and 8.5% (p < 0.01) greater, respectively, in stems of genotype 708 compared to genotype 773 (Table 1). Klason lignin concentration in the cell wall was 8.0% greater in stems of 773 compared to stems of 708 (p < 0.05) (Table 1). These genotypes consistently displayed differences in cell wall cellulose and lignin content in stems when plants were grown under different field environments (Figure 1) and in the greenhouse (Table 1).
Table 1.
Comparison of cell wall components in stems of genotypes 708 and 773 on a cell wall basis.
| Component | Genotype 708 | Genotype 773 | SEM | p-value |
|---|---|---|---|---|
| ---------- g kg-1 cell wall ---------- | ||||
| Klason lignin | 162 | 175 | 2 | p < 0.05 |
| Glucose | 443 | 421 | 2 | p < 0.05 |
| Xylose | 137 | 149 | 3 | NS |
| Arabinose | 39 | 39 | 1 | NS |
| Galactose | 32 | 28 | 1 | p < 0.05 |
| Mannose | 33.1 | 30.5 | 0.1 | p < 0.01 |
| Rhamnose | 11.5 | 11.4 | 0.4 | NS |
| Fucose | 3.01 | 3.1 | 0.03 | NS |
| Uronic acids | 139 | 142 | 6 | NS |
Values are least square means based on an analysis of variance with three biological replicates for each clone arranged in a randomized complete block design (see Methods for details). SEM = Standard error of mean, NS = Non-significant (p > 0.05).
Figure 1.
Regression analyses of cellulose and Klason lignin concentrations in stems of two alfalfa genotypes. The stems of genotype 708 were consistently higher in cellulose and lower in Klason lignin compared to stems of genotype 773 across twelve environmental indexes (field environments). The high r2 values for all regression lines suggest that genotypic differences in stem cellulose and Klason lignin concentrations were environmentally stable.
RNA-Seq using the Illumina GA-II platform
For RNA-Seq analysis, we developed a total of four cDNA libraries derived from elongating stem (ES) and post-elongation stem (PES) internodes of alfalfa genotypes 708 and 773 (see Methods for details). In alfalfa stems, genes associated with primary cell wall development are preferentially expressed in ES internodes while genes associated with secondary xylem development are enriched in PES internodes [25]. For sequencing by synthesis using the Illumina GA-II platform, cDNA libraries 708ES, 708PES and 773ES were run on two lanes per library while the 773PES library was run on one lane. A total of 234,908,899 EST reads were generated by a single run of 76 cycles. After filtering low quality reads, a total of 198,861,304 reads (76-bp in size) were selected for further analysis (see Methods for details). The Illumina reads generated in this study are available at the NCBI SRA browser (accession number GSE26757; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26757.
de novo assembly of short RNA-Seq reads without a known reference is a challenging task especially for alfalfa, an allogamous autotetraploid with complex polysomic inheritance. In this study, we used the Velvet algorithm [32] for de novo assembly of the 198,861,304 Illumina reads (76 bp) into a total of 132,153 unique sequences with an average length of 284 bp (Additional file 1). The Velvet algorithm has also been used successfully for de novo transcriptome assembly in previous studies [33,34]. The Velvet algorithm was originally developed for de novo assembly of genome sequences where the coverage is expected to be homogeneous throughout the genome. However, the coverage of transcripts is highly heterogeneous due to difference in gene expression. Previous studies showed that de novo assembly using the Velvet program with longer k-mers results in a more contiguous transcript assembly but lower transcript diversity compared to shorter k-mers [32,33]. Although several recent studies introduced new algorithms and methodologies developed for de novo transcriptome assembly [35-38], a consensus standard protocol has not yet emerged for de novo transcriptome assembly. In this study, we optimized our Velvet de novo transcriptome assembly to favor transcript contiguity with high specificity as opposed to increased transcript diversity (see Methods for details). To complement the limitation of the high k-mer that we selected for the Velvet assembly in this study (lower diversity and probably biased toward highly expressed genes), we generated additional ESTs using the GS FLX Titanium platform.
RNA-Seq using the GS FLX Titanium platform
We generated a total of 341,984 additional ESTs (average length 243 bp, minimum length 40 bp, maximum length 792 bp) using the GS FLX Titanium platform http://www.454.com. The additional EST sequences were generated from the cDNA libraries derived from ES (124,533 ESTs, average length 230 bp) and PES (217,451 ESTs, average length 256 bp) internodes of the genotype 773. The additional ESTs obtained using the GS FLX Titanium platform increased the diversity of transcripts discovered and hence provided broader coverage of the alfalfa transcriptome than would have been achieved based on the de novo assembly of the Illumina reads alone. The additional ESTs are also available at the NCBI SRA browser (accession number GSE26757; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26757.
Alfalfa Gene Index 1.0 (MSGI 1.0)
We used the Gene Index Assembly protocol [39,40] for reference transcriptome assembly in alfalfa. This protocol has been used for over a decade to build unigene assemblies for numerous species of animals, plants and microorganisms http://compbio.dfci.harvard.edu/tgi/plant.html. However, no gene index is currently available for alfalfa. In this study, the first alfalfa (Medicago sativa) gene index (MSGI 1.0) was built by combining the de novo assembled Illumina reads using the Velvet program (132,153 sequences), the 341,984 ESTs obtained using the GS FLX Titanium platform, and 12,371 Sanger ESTs for alfalfa available in the public database http://www.ncbi.nlm.nih.gov following the Gene Index Assembly protocol previously described [39,40].
MSGI 1.0 contains a total of 124,025 unique sequences including 22,729 tentative consensus sequences (TCs), 22,315 singletons and 78,981 pseudo-singletons (Additional file 2). Pseudo-singletons refer to the de novo assembled Illumina sequences that were not assembled into contigs during the Gene Index Assembly process. The average length of the unique sequences in MSGI 1.0 is 384 bp. Unique sequence lengths ranged from 100 to 6,956 bp with more than 10,000 sequences larger than 800 bp. The total base count of the sequences in MSGI 1.0 is 47,628,953 bp. The newly built alfalfa gene index increases the number of alfalfa sequences publicly available by about 10-fold.
Gene annotation and functional classification
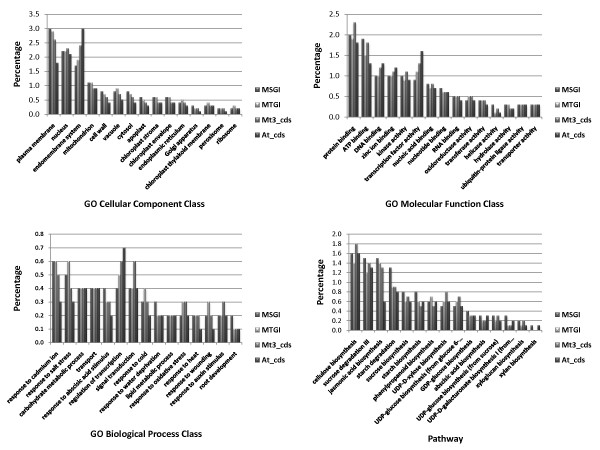
We assigned putative functions for the unique sequences in MSGI 1.0 by conducting BlastX searches against the non-redundant (NR) protein database (e-value cutoff of 1e-10) (Additional file 3). Putative functions could be assigned for about 83% of the sequences. We also assigned gene ontology (GO) functional classes and MapMan functional classifications [41] to the unique sequences in MSGI 1.0 (Additional file 3) (see Methods for details). To examine whether bias occurs among the functional classes represented in MSGI 1.0, we compared the percentages of each GO functional class and pathway in MSGI 1.0 with the percentages found in the M. (Medicago) truncatula Gene Index (MTGI 9.0), the M. truncatula coding sequences (Mt3.0 cds) and the Arabidopsis coding sequences (At cds) (Figure 2). Although most of the sequences in MSGI 1.0 were derived from stem tissues, similar levels of representation of most functional classes were found in MSGI 1.0 and the other databases (MTGI 9.0, Mt3.0 cds, and At cds). These results suggest that MSGI 1.0 can serve as a reference sequence database for genomic analysis in alfalfa.
Figure 2.
Comparison of percentage distribution of gene ontology and pathway classifications using four reference databases. The percentage distributions of gene ontology (GO) classes and pathways are shown for the following reference databases: (1) the Medicago sativa Gene Index (MSGI 1.0) assembled in this study, (2) the Medicago truncatula Gene Index (MTGI 9.0), (3) the M. truncatula coding sequences (Mt3.0 cds), and (4) the Arabidopsis coding sequences (At cds).
SSR detection
We detected simple sequence repeats (SSRs) among sequences in MSGI 1.0 using the MISA program [42] (see Methods for details). A total of 1,294 SSRs were identified among 1,245 sequences which represents about 1.7% of the total unique sequences in MSGI 1.0 (Additional file 4). The estimated frequency of SSRs among the expressed sequences was one SSR per 37 kb. SSR detection frequency is dependent on the SSR detection parameter [43]. The SSR frequency measured in this study is significantly lower than that detected in other species (one SSR per 11 kb) where the same SSR detection parameter was used [40]. The significantly reduced SSR detection frequency found in MSGI 1.0 sequences may be due to the reduced detection efficiency of short length sequences (384 bp on average for MSGI1.0). Alternatively, the SSR frequency among expressed sequences may be lower in alfalfa compared to other species. SSRs with mono-, di-, tri-, tetra-, penta- and hexanucleotide repeats composed about 5.4%, 30.4%, 47.2%, 10.6%, 3.9% and 2.5% of the SSRs in MSGI 1.0, respectively. Using the default parameter of the Primer3 program [44], we designed SSR primers spanning a total of 664 SSRs (Additional file 4).
SNP detection
To identify SNPs between alfalfa genotypes 708 and 773, Illumina EST reads from ES and PES internode libraries were combined for each genotype. The combined ES and PES reads for each genotype were independently aligned to the MSGI 1.0 sequences using the Maq program [45]. From the alignment output of each genotype, we summarized the depth (frequency) of each nucleotide (A, G, C, or T) at each base position in each reference sequence. Next, to reduce the identification of false positive SNPs, we filtered potential SNPs using a stringent nucleotide depth cutoff of 10 [e.g., at least 10 adenines (A) in one genotype vs. at least 10 guanines (G) in the other genotype] for each genotype (see Methods for details). Using this protocol, we identified 10,826 SNPs between genotypes 708 and 773 in 7,282 sequences in MSGI 1.0 (Additional file 5). About 74% of these sequences contained a single SNP while about 2.3% contained 5 or more SNPs.
To validate the SNPs that were predicted using the RNA-Seq data generated in this study, we randomly selected 55 SNPs. Genomic DNAs purified from genotypes 708 and 773 were genotyped by MALDI-TOF mass spectrometry using the iPLEX Gold spectrometry system http://www.sequenom.com. Out of 55 SNPs tested, 47 (85%) were polymorphic between the two genotypes (Additional file 6). In addition to genotypes 708 and 773, we also genotyped 51 additional alfalfa (M. sativa) genotypes selected from different populations of M. sativa ssp. sativa or M. sativa ssp. falcata. The 47 validated SNPs between 708 and 773 also showed polymorphism among the other Medicago genotypes tested (Additional file 6). This suggests that the SNPs predicted in this study can also be used for genotyping in other alfalfa genotypes.
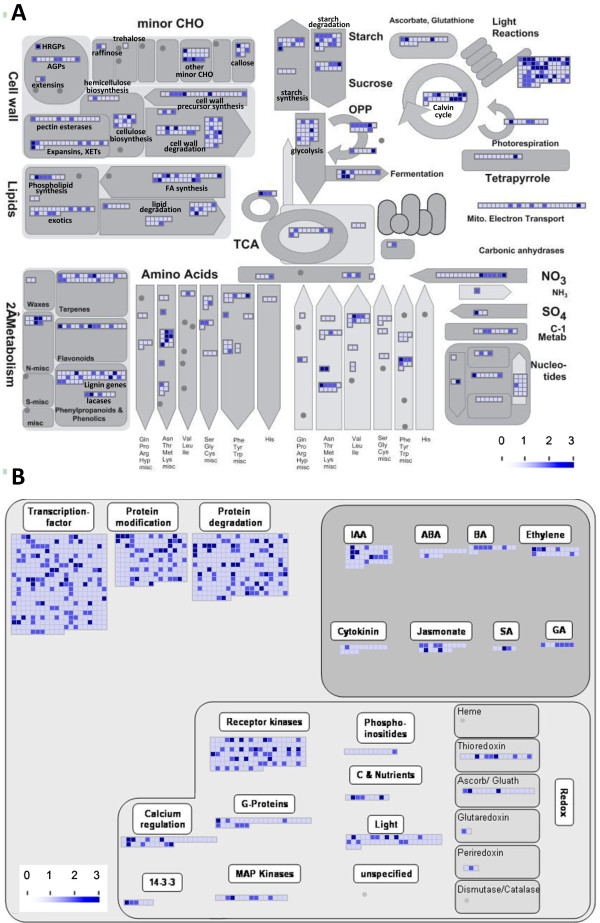
In a previous study that described single-feature polymorphism (SFP) discovery in alfalfa using the Medicago GeneChip as a cross-species platform [31], we proposed candidate gene-based association mapping for selecting alfalfa germplasm with modified cell wall composition in stems. In this study, SNPs were also identified in genes with various functional classes including numerous cell wall-related genes (Figure 3A). For example, SNPs were identified in 14 genes involved in cellulose biosynthesis including 11 cellulose synthase and three COBRA genes [46] (Figure 3A). In addition, SNPs were identified in 21 lignin pathway genes, 20 genes involved in cell wall precursor pathways (Figure 3A) and in numerous regulatory genes including various transcription factor families, signalling genes and hormone genes (Figure 3B).
Figure 3.
MapMan overview of cellular metabolism (A) and regulation (B) showing SNP-harboring genes and SNP frequencies. Individual genes are represented by small squares. The SNP frequency for each gene is indicated by the intensity of the blue color on a 0 to 3 scale. Dark blue (scale intensity 3) indicates genes with three or more SNPs. A complete list of SNP-harboring genes, corresponding MapMan functional categories and SNP frequencies are provided in Additional file 5.
To detect functional classes over- or under-represented among the SNP-harboring genes, we performed Fisher's exact test with Bonferroni correction (z-value cutoff = 1) as previously described [31] (Additional file 7). The functional classes over-represented among SNP-harboring genes included photosynthesis, cell wall, amino acid metabolism, stress response (biotic and abiotic), nodulin-like, protein synthesis and WRKY transcription factor classes (Additional file 7). The SNPs developed in this study can be used for either candidate gene-based or whole genome scanning association mapping studies to identify SNPs associated with cell wall traits in alfalfa stems. With further development, the SNPs identified in this study may prove to be useful in molecular breeding programs focused on improving alfalfa as a forage crop and biomass feedstock via marker-assisted selection.
In this study, we also identified allelic variations (SNPs) within genotypes. Using a minimum SNP depth cutoff of 10, we detected 287,555 and 168,966 allelic variations (SNPs) within genotypes 708 and 773, respectively (Additional files 8 and 9). These SNPs within genotype were detected in 55,320 and 33,406 sequences for genotypes 708 and 773, respectively. Detection of allelic variations (SNPs) within genotypes is equally important as detecting SNPs between genotypes for understanding phenotypic differences (e.g. cell wall composition) and for future applications such as marker-assisted selection.
Comparison of MSGI 1.0 and Mt3.0 cds as reference sequences for digital transcript profiling
The alfalfa gene index (MSGI 1.0) developed in this study provides a reference sequence database that can be used for digital gene expression analysis in alfalfa. However, another option for RNA-Seq analysis in alfalfa is to use Mt3.0 cds as a reference sequence because M. truncatula and alfalfa share significant coding sequence homology [25]. Furthermore, sequences in Mt3.0 cds are full-length sequences (predicted gene models) with better coverage than sequences in MSGI 1.0 where the majority are partial sequences. As an initial step to evaluate the utility of MSGI 1.0 and Mt3.0 cds as reference sequences for transcript profiling of alfalfa, the Illumina EST reads generated in this study were mapped to MSGI 1.0 and Mt3.0 cds sequences using the bowtie program [47] (see Methods for details). On average, about 70% of the EST reads in each library (708 ES, 773 ES, 708 PES, and 773 PES) could be mapped to the MSGI 1.0 sequences. In contrast, only about 30% of the EST reads could be mapped to the Mt3.0 cds sequences (data not shown). We measured the raw digital expression counts for each gene by quantifying the number of EST reads that were mapped to each reference sequence. The raw digital gene expression counts were normalized using the RPKM (reads/Kb/Million) method [1,48] to correct the digital gene expression counts for bias caused by reference sequence size and total EST numbers per library (see Methods for details).
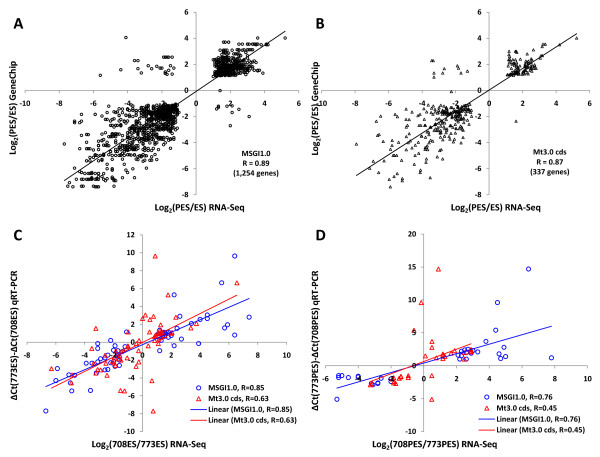
Further evaluation of MSGI 1.0 and Mt3.0 cds as reference sequence databases for alfalfa was conducted by comparing RNA-Seq data with the previously generated GeneChip data for the same stem tissues but in different alfalfa genotypes [25] (see Methods for details). The RNA-Seq data generated using MSGI 1.0 or Mt3.0 cds showed a linear relationship with GeneChip data with similar Pearson correlation coefficients (R = 0.89 and R = 0.87, respectively) (Figure 4A and 4B). A total of 1,254 genes were commonly-selected between RNA-Seq and GeneChip data when MSGI 1.0 was used as reference sequences (Figure 4A). However, when Mt3.0 cds was used as reference sequences, the number of genes commonly-selected between RNA-Seq and GeneChip data decreased to 337 reflecting a significant decrease in detection sensitivity (Figure 4B). This is not surprising because, as described above, only about 30% of the EST reads could be mapped to the Mt3.0 cds while about 70% of the EST reads could be mapped to the MSGI 1.0 (data not shown).
Figure 4.
Comparison of MSGI 1.0 and Mt3.0 cds as reference sequences for digital gene expression analysis. For a subset of genes involved in stem development independent of genotypic variation, Log2(PES/ES) values from the RNA-Seq data (x-axis) generated using (A) the Medicago sativa Gene Index (MSGI1.0) or (B) the Medicago truncatula coding sequences (Mt3.0 cds) as reference sequences were plotted against Log2(PES/ES) values from the GeneChip data (y-axis) previously generated [25]. For 63 randomly selected genes (C) and 34 selected cell wall genes (D), Log ratio values from the RNA-Seq data (x-axis) generated using MSGI1.0 (O) and Mt3.0 cds (Δ) as reference sequences were plotted against ΔΔCT values obtained from the qRT-PCR data (y-axis).
As a final evaluation of MSGI 1.0 and Mt3.0 cds as reference sequences for digital gene expression analysis in alfalfa, we compared the digital gene expression data generated using MSGI 1.0 and Mt3.0 cds sequences with real -time quantitative RT-PCR (qRT-PCR) data obtained from 97 genes (63 randomly selected, 34 cell wall genes) (Additional file 10) (see Methods for details). Previous studies showed a linear relationship between ΔΔCT values from qRT-PCR and the log gene expression ratio obtained in microarray analysis [25,49,50]. We plotted ΔΔCT values obtained from the qRT-PCR data for randomly selected genes against Log2(708ES/773ES) values from the RNA-Seq data with MSGI 1.0 or Mt3.0 cds as reference sequences. The results showed a linear relationship between qRT-PCR data and the RNA-Seq data using both reference sequences. However, using MSGI 1.0 increased the Pearson correlation coefficient (R) from 0.63 to 0.85 (Figure 4C). Next, we plotted ΔΔCT values obtained from the qRT-PCR data for selected cell wall genes against Log2(708PES/773PES) values from the RNA-Seq data. Using MSGI 1.0 as the reference sequence database also increased the Pearson correlation coefficient (R) for selected cell wall genes from 0.45 to 0.76 (Figure 4D). On the basis of these results, we chose to use MSGI 1.0 as reference sequences for digital gene expression analysis of stems of alfalfa genotypes 708 and 773.
Transcript profiling of stems of alfalfa genotypes 708 and 773
For transcript profiling of stems of alfalfa genotypes 708 and 773, we analyzed the RPKM-normalized digital gene expression counts for each sequence in MSGI 1.0 for cDNA libraries derived from ES and PES internodes of each genotype (Additional file 11). Among the 124,025 sequences in MSGI 1.0, about 94.7% were transcriptionally active (RPKM > 0) in at least one library while about 5.3% (6,629 sequences) were silent in all four libraries examined (RPKM = 0 in all 4 libraries) (Additional file 11).
Among the transcriptionally-active genes in each library, we identified the top 500 most abundant transcripts (Additional file 12). The Fisher's exact test with Bonferroni correction (z-value cutoff = 1) revealed that genes belonging to photosynthesis, amino acid metabolism and transport classes were significantly over-represented among the most abundantly expressed transcripts in all 4 libraries which suggests roles as housekeeping genes in alfalfa stems (Additional file 13). We also identified functional classes over-represented among the most abundant genes expressed in a genotype- or tissue-specific manner suggesting their role in determining genotype or tissue identity (Additional file 13). Interestingly, genes involved in lignin biosynthesis were significantly over-represented among the most abundant genes. The lignin genes over-represented in one or more libraries include CCoAOMT (caffeoyl-CoA O-methyltransferase), CCR1 (cinnamoyl-CoA reductase1) and COMT (caffeic acid O-methyltransferase) genes (Additional file 13). On the other hand, the transcription factor family class was significantly under-represented among the most abundant transcripts in three libraries (Additional file 13). Table 2 shows the top 10 most abundant protein-coding transcripts identified in each alfalfa stem internode library. Interestingly, a putative COMT gene (MSGI1_1270) was among the top 10 most abundant protein-coding transcripts and it was up-regulated in 773 (high lignin genotype) in both ES and PES internodes compared to 708 (low lignin genotype). The promoters of these highly expressed genes, including strong constitutive and tissue-specific promoters, may be useful for transgenic studies in alfalfa.
Table 2.
Top 10 most abundant protein-coding transcripts identified in each alfalfa stem internodes library.
| Unique_ID | Libraries | Putative Functions | |||
|---|---|---|---|---|---|
| 708_ES | 708_PES | 773_ES | 773_PES | ||
| ------------ RPKM-normalized expression counts ------------ | |||||
| MSGI1_2417 | 6068 (1)† | 6754 (1) | 3271 (2) | 6034 (1) | Leucine-rich repeat family protein |
| MSGI1_8746 | 4812 (2) | 4697 (3) | 3850 (1) | 3586 (2) | Chlorophyll a/b binding protein |
| MSGI1_523 | 4213 (3) | 5428 (2) | 2719 (6) | 3552 (3) | Beta ketoacyl CoA synthase |
| MSGI1_18145 | 3859 (4) | 979 | 2555 (8) | 3400 (4) | Rubisco small chain |
| MSGI1_27309 | 748 | 2453 (7) | 1317 | 3336 (5) | Metallothionein |
| MSGI1_11989 | 2315 (6) | 2574 (5) | 1171 | 2387 (6) | Uncharacterized protein |
| MSGI1_6529 | 265 | 328 | 2393 (9) | 2350 (7) | Glycine rich protein |
| MSGI1_1166 | 1458 | 1486 | 901 | 2182 (8) | AAA ATPase |
| MSGI1_62398 | 160 | 225 | 1267 | 2168 (9) | Stress (ABA)-inducible protein |
| MSGI1_21335 | 2012 (8) | 2465 (6) | 1275 | 2155 (10) | Cytochrome P450-like |
| MSGI1_8707 | 2762 (5) | 2632 (4) | 1854 | 1833 | Chlorophyll a/b binding protein |
| MSGI1_4749 | 1425 | 1682 (10) | 1298 | 1693 | Polyubiquitin |
| MSGI1_5229 | 2287 (7) | 2140 (8) | 1507 | 1561 | Chlorophyll a/b binding protein |
| MSGI1_1270 | 744 | 969 | 3145 (3) | 1470 | Caffeic acid O-methyltransferase |
| MSGI1_1415 | 1723 (9) | 1215 | 764 | 1468 | Elongation factor 1-alpha |
| MSGI1_36219 | 1633 | 1777 (9) | 723 | 1256 | Uncharacterized protein |
| MSGI1_5153 | 1705 (10) | 1531 | 1058 | 1090 | Chlorophyll a/b binding protein |
| MSGI1_29285 | 324 | 513 | 2861 (4) | 1035 | Stress (ABA)-inducible protein |
| MSGI1_13276 | 86 | 77 | 2750 (5) | 423 | Cold acclimation responsive protein |
| MSGI1_7576 | 274 | 114 | 2357 (10) | 284 | Cold-acclimation-specific protein (CAS) |
| MSGI1_96533 | 11 | 5 | 2580 (7) | 16 | Cold acclimation-specific protein CAS) |
† Top 10 most abundant protein-coding transcripts selected from each library are highlighted in bold. Numbers enclosed in parenthesis represent rank based on transcript frequency for the top 10 most abundant protein-coding transcripts in each library.
We also identified putative housekeeping genes (HKGs) that showed little variation in expression but were expressed at relatively high levels. To identify HKGs, we first selected genes with an average RPKM-normalized transcript count greater than 10. Next, we selected the top 300 genes with the lowest coefficient of variation (CV = standard deviation/mean) (Additional file 14) [13]. These HKGs may be useful as reference genes in qRT-PCR or other experiments to normalize gene expression levels across different conditions [51].
Identification of differentially expressed genes
We used a MA-plot-based method with a random sampling model in a DEGSeq program [52] to identify genes differentially expressed between stems of alfalfa genotypes 708 and 773. A total of 3,838 and 4,428 genes were differentially expressed between ES and PES tissues of genotypes 708 and 773, respectively (p < 0.001, FDR < 0.025, ≥ 2-fold difference) (Additional files 15 and 16). Among the genes that were differentially expressed between ES and PES internodes, 849 genes were detected in internodes of both genotypes. In addition, a total of 8,883 and 4,799 genes were differentially expressed between genotypes 708 and 773 within ES and PES internodes, respectively (p < 0.001, FDR < 0.025, ≥ 2-fold difference) (Additional files 17 and 18). Of the genes that were differentially expressed between the two genotypes, 2,422 were detected in both ES and PES internodes. Among the 13,797 differentially expressed genes identified in four pair-wise comparisons of ES and PES internodes of the two genotypes, about 85% were ubiquitously expressed in all four libraries (RPKM-normalized transcript count > 0 in all 4 libraries), about 5.5% were expressed in three libraries, about 9.6% were expressed in two libraries, and 16 genes were expressed in only one library (Additional file 19). These results suggest that stem tissue internodes in alfalfa may be characterized on the basis of differential expression of ubiquitous genes or tissue/genotype-specific expression of selected genes as shown in previous studies with other species [12,13,40]. SNPs were detected in 700 differentially expressed genes. Interestingly, about 14% of these SNP-harboring differentially expressed genes were cell wall-related genes.
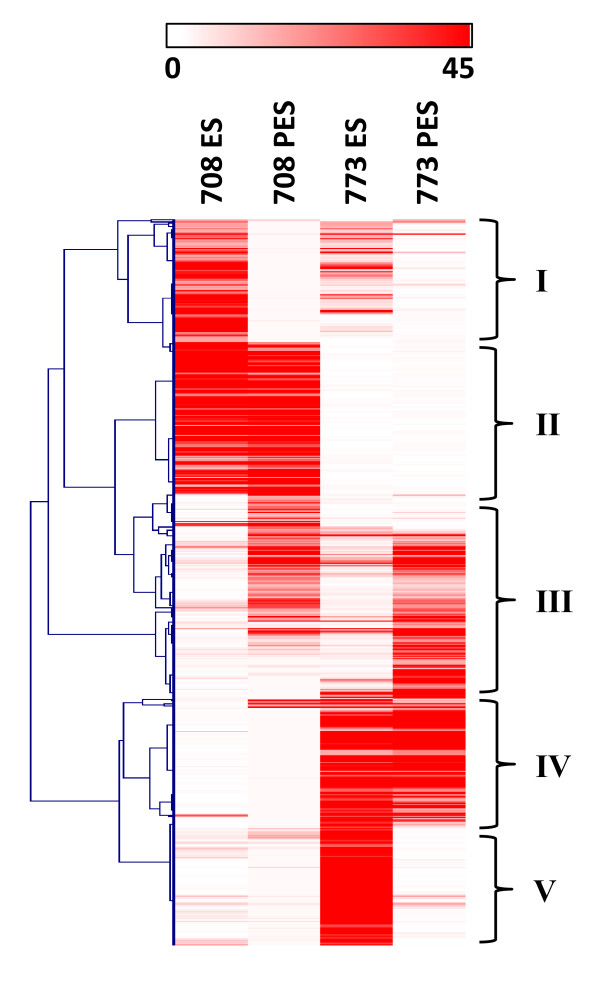
To illustrate the differential expression of genes detected in the stem internodes of 708 and 773, we generated a heatmap of RPKM-normalized transcript counts for the top 200 most differentially expressed genes in each pair-wise comparison (Figure 5, Additional file 20). Groups I and III in Figure 5 contain genes that were differentially expressed in a tissue-specific manner which suggests their role in alfalfa stem development. For example, one expansin and four pectin esterase genes included in group I were up-regulated in ES compared to PES internodes in both genotypes. These genes are involved in cell wall loosening and cell elongation [53,54]. On the other hand, a putative alfalfa cellulose synthase gene, IRREGULAR XYLEM 3 (IRX3), included in group III (Figure 5) was up-regulated in PES internodes compared to ES in both genotypes. Several previous studies demonstrated xylem specific expression of IRX3 and its role in secondary cell wall development in Arabidopsis [55-57]. Groups II and IV in Figure 5 contain genes differentially expressed in a genotype-specific manner suggesting possible roles in the genotypic variation between stems of 708 and 773. For example, two extensin genes and a cellulose synthase gene (CESA4) included in group II were up-regulated in genotype 708 compared to 773 in both ES and PES internodes. These genes may be responsible for the higher cellulose content in stem internodes of genotype 708 compared to 773. Group V in Figure 5 contains genes differentially expressed in both a genotype- and tissue-specific manner.
Figure 5.
Hierarchical clustering analysis of the top 200 most differentially expressed genes selected from pair-wise comparisons. Pair-wise comparisons of gene expression were made between stem tissues (ES, PES) in alfalfa genotypes 708 and 773. The RPKM-normalized expression counts for each gene in each library are represented by intensity of the red color on a 0 to 45 scale. Dark red (scale intensity 45) indicates genes with RPKM-normalized expression counts ≥ 45. See Methods for details. Groups I and III, genes differentially expressed in a tissue-specific manner; Groups II and IV, genes differentially expressed in a genotype-specific manner; and Group V, genes differentially expressed in both a genotype- and tissue-specific manner. A complete list of the genes, RPKM-normalized expression counts, and corresponding MapMan functional categories are provided in Additional file 20.
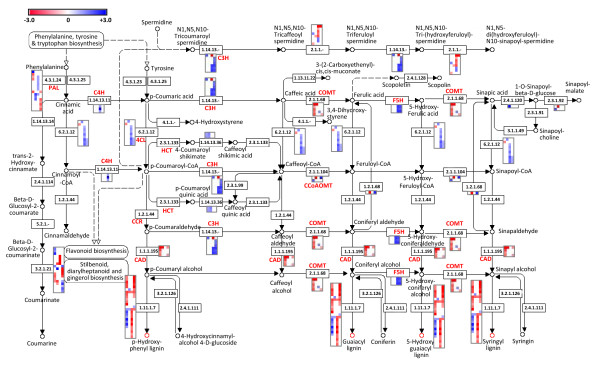
Lignin content in alfalfa stems affects the quality of alfalfa as a forage crop and biomass feedstock. Lignin is indigestible and reduces cell wall digestibility in ruminants [58-60]. In addition, the pre-treatment process to remove lignin is one the most costly steps of cellulosic ethanol production [61-64]. Over multiple environments, alfalfa genotype 773 consistently showed higher cell wall lignin content in stems compared to genotype 708 (Figure 1) suggesting differences in the genetics of lignin biosynthesis. In an effort to identify key genes responsible for differences in cell wall properties in stems of genotypes 708 and 773, we identified lignin (phenylpropanoid) pathway genes among the 13,797 genes detected (Additional file 21). Next, we generated a heatmap of gene expression ratios for each selected lignin pathway gene for each pair-wise comparison (see Methods for details). The heatmaps generated were inserted into the lignin biosynthetic pathway (Figure 6). As expected, numerous lignin pathway genes were up-regulated in PES compared to ES internodes (Figure 6, Additional file 21). We also identified lignin genes differentially expressed between the two alfalfa genotypes. For example, several CAD and COMT genes were up-regulated in 773 compared to 708 especially in ES internodes (Figure 6, Additional file 21). These genes may contribute to difference in lignin content in cell walls of stems of genotypes 708 and 773.
Figure 6.
Lignin pathway genes differentially expressed in stem tissues of two alfalfa genotypes. Pair-wise comparisons were made between stem tissues (ES, PES) of genotypes 708 and 773. Columns in each heatmap from left to right: Log2(708ES/773ES), Log2(708PES/773PES), Log2(708PES/708ES), and Log2(773PES/773ES). The rows in each heatmap represent lignin gene sequences identified in MSGI 1.0. The Log2 expression ratio values were false color-coded using a scale of -3 to 3. The intensity of blue and red indicates the degree of up- and down-regulation of the corresponding lignin gene in the denominator in each column mentioned above. The red and blue color saturates at -3 and 3, respectively. See Methods for details. The heatmaps generated were inserted next to the corresponding lignin gene in the lignin biosynthetic pathway diagram downloaded from the KEGG pathway database http://www.genome.jp/kegg/pathway/map/map00940.html. PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumarate-CoA ligase; HCT, hydroxycinnamoyltransferase; C3H, p-coumarate 3-hydroxylase; CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; CCR1, cinnamoyl-CoA reductase 1; F5H, ferulate 5-hydroxylase; COMT, caffeic acid O-methyltransferase; CAD, cinnamyl-alcohol dehydrogenase.
A previous study [25] and the current study both suggest significant genotypic variation for gene expression in alfalfa stem internodes. To identify genes involved in general stem development (ES vs. PES internodes) independent of genotypic variation in gene expression, we selected a subset of alfalfa genes differentially expressed between ES and PES internodes in both genotype 708 and genotype 773 (p < 0.001, FDR < 0.025, ≥ 2-fold difference). A total of 594 genes were identified by further selecting genes with similar differential expression patterns in both genotypes [Log2(PES/ES) ≥ 1 or ≤ -1 in both genotypes] (Additional file 22). Among these genes, about 19% were cell wall-related genes. These genes included 5 cellulose synthase genes (a putative IRX3, two CesA8s, and two COBRAs) and six lignin pathway genes (three 4CLs and three F5Hs) that were up-regulated in PES compared to ES internodes in both genotypes (Additional file 22). In Arabidopsis, IRX3, CesA8 (IRX1) and COBRA genes are involved in cellulose biosynthesis during secondary cell wall development [46,55-57,65,66]. The gene families that were significantly over-represented among genes up-regulated in PES compared to ES internodes in both genotypes (Fisher's exact test with Bonfferoni correction with z-value cutoff of 1) included arabinogalactan protein (AGP), arginosuccinate synthase, metal handling, and transporter (sucrose, amino acids, and phosphate) families (Additional file 23). The gene families significantly over-represented among genes up-regulated in ES compared to PES internodes in both genotypes included invertase, pectin esterase, simple phenol, gibberellin-responsive, cold-responsive, lipid transfer protein (LTP), and GDSL-motif lipase families (Additional file 23). Cell wall family genes were over-represented among genes up-regulated in both ES and PES Internodes.
Assimilated photosynthetic carbon is translocated primarily as sucrose in higher plants [67]. Membrane-bound, energy dependent, H+-symporting sucrose transporters (SUC or SUT proteins) play an essential role in sucrose uptake in sink tissues and sucrose release in source tissues [67]. In this study, members of the sucrose transporter gene family were over-represented among genes up-regulated in PES compared to ES internodes in both genotypes (Additional file 23). Previous studies showed that the expression of sucrose transporter genes was developmentally regulated in plants [68-73]. For example, sucrose transporter genes were up-regulated during secondary cell wall synthesis in developing cotton fibers [73]. In this study, we identified five putative sucrose transporters (MsSUCs) that were up-regulated in PES compared to ES internodes in both genotypes (Additional file 22, Additional file 24). As stem development progresses from ES to PES, sink strength may also increase due to secondary cell wall formation in secondary xylem. The up-regulation of MsSUCs in PES internodes may be in response to increased demand for sucrose and UDP-glucose to support cellulose synthesis during secondary cell wall formation. Consistent with this explanation is our finding that three sucrose synthase (MsSuSy) genes were up-regulated in PES compared to ES internodes in both genotypes. Sucrose synthase provides the UDP-glucose needed for cellulose synthesis [74,75]. In addition to their roles in providing sucrose and UDP-glucose for cellulose synthesis in secondary cell walls, MsSUCs and MsSuSy genes, respectively, may play important roles in modulating sugar sensing and signal transduction pathways during stem development in alfalfa [76].
In addition to the SUC transporter gene family, we also found that the phosphate (Pi) transporter gene family was over-represented among genes up-regulated in PES compared to ES internodes in both genotypes (Additional file 23). We identified six putative PHOSPHATE1 (PHO1) genes up-regulated in PES compared to ES internodes in both genotypes (Additional file 22, Additional file 24). In Arabidopsis root epidermal and cortical cells, PHO1 is involved in Pi loading into the xylem [77,78]. A recessive mutation in PHO1 in Arabidopsis resulted in reduced Pi loading into xylem [77,78]. PHO1 is expressed predominantly in roots and up-regulated under conditions of Pi starvation [78-80]. A recent study in Arabidopsis showed that the expression of PHO1 was modulated by WRKY6 and WRKY42 transcription factors in response to low Pi [81]. Up-regulation of PHO1 genes in PES may be needed to meet the requirements of Pi uptake and redistribution during cellulose synthesis in secondary cell walls. For example, the fructose released by SuSy (sucrose -> UDP-glucose + fructose) needs to be phosphorylated to be recycled by sucrose phosphate synthase (SPS).
The plant hormone auxin is a key regulator of plant growth and development [82]. In addition to its role in cell wall loosening and cell elongation [82], auxin also regulates vascular tissue differentiation and patterning in plants [82-85], secondary xylem development in trees [86,87], and fiber development in cotton [88]. Indole-3-acetic acid (IAA), the major auxin species, is made in the shoot apex and transported to the root apex [82]. Directional auxin transport is mainly controlled by the coordinated action of auxin influx (AUX1) and efflux (PIN) carrier complexes [82]. AUX1, an amino acid permease-like membrane protein, was originally identified after screening for auxin resistant mutants [89]. In Arabidopsis, AUX1 was preferentially expressed in xylem compared to phloem and nonvascular tissues of the root-hypocotyl [90]. Arabidopsis AUX1 mutants showed a reduction in lateral root formation [91] but enhanced root generation in shoot regeneration media [92]. In addition, disruption of polar auxin transport in Arabidopsis resulted in ectopic vascular differentiation in leaves [93]. Polarized auxin transport is essential for providing directional and positional signals for various developmental processes such as apical dominance, organ development, tropic growth, embryogenesis and vascular development [82-85,94-98]. In this study, the amino acid transporter gene families, which include AUX1 genes, were over-represented among genes up-regulated in PES compared to ES internodes in both genotypes (Additional file 23). A total of 5 putative AUX1 genes were up-regulated in PES (Additional file 22, Additional file 24).The up-regulation of AUX1 in PES internodes of alfalfa and the resultant increase in auxin uptake may play an important role in the formation of secondary xylem. A recent study in trees suggested that the radial auxin concentration gradient in cell types of secondary xylem modulates the expression of a small number of key genes that regulate secondary xylem development [87].
In addition to transporter family genes that were differentially expressed between ES and PES internodes of both genotypes, we also identified transporter family genes that were differentially expressed between genotypes. For example, several sugar (glucose, hexose, and sucrose) transporters and AUX1 genes were up-regulated in 708 compared to 773 in both ES and PES internodes (Additional file 24, Additional file 25). These transporters may play a role in the higher cellulose and sugar (galactose and mannose) content in stem internodes of genotype 708 compared to 773 (Table 1). We also identified numerous transporter families that were up-regulated in both ES and PES internodes of 773 compared to 708. Among these up-regulated transporter families were the multi-drug toxic efflux carrier (MATE) and ATP-binding cassette (ABC) transporter families (Additional file 24, Additional file 25). Recent studies suggest that monolignols synthesized in the cytoplasm are transported across the plasma membrane into the cell wall matrix where they are polymerized into lignin [99,100]. However, little is known about the transport mechanism. Previous studies have suggested that monolignol transport across the plasma membrane may involve passive diffusion [101] or may be mediated by membrane-bound transporters [102]. Genes in the MATE transporter family may be good candidates for monolignol transporters because they are involved in transport of proanthocyanidin precursors across the tonoplast in Arabidopsis and M. truncatula [103,104]. A role for ABC transporters in monolignol transport across the plasma membrane has been postulated because of their known role in transporting various secondary metabolites in plants [99,100,105,106]. Additional research will be required to determine whether the up-regulation of the MATE efflux carrier and ABC transporter families in stems of 773 (high-lignin) compared to 708 (low lignin) (Additional file 24, Additional file 25) contributes to the higher lignin content in cell walls of 773 (Table 1). The up-regulated MATE efflux carrier and ABC transport genes that we identified provide a list of candidate genes that will be useful in future research to evaluate the involvement of these gene families in monolignol transport.
Conclusion
This study represents the first application of RNA-Seq technology for genomic studies in alfalfa. Our results demonstrate that RNA-Seq can be successfully used for gene identification, polymorphism detection and transcript profiling in alfalfa. Using RNA-Seq has several advantages over other technologies, especially for non-model species with few genomic resources such as alfalfa. Unlike hybridization-based technologies such as microarrays, RNA-Seq does not require pre-existing sequence information and, as shown in this study, RNA-Seq can integrate multiple tasks in a single pipeline saving time and money. The integrated approach used in this study can be applied to other non-model species. The newly built alfalfa gene index (MSGI 1.0), and the SNPs, SSRs and candidate genes identified in this study will be a valuable resource for advancing genetic/genomic research in alfalfa and eventually for improving alfalfa as a forage crop and cellulosic ethanol feedstock.
Methods
Plant materials and cell wall analysis
Alfalfa [Medicago sativa (L) subsp. sativa] genotypes 708 and 773 were selected from a population (UMN 3097) created by mixing seeds from six commercial alfalfa cultivars (5312, Rushmore, Magnagraze, Wintergreen, Windstar and WL 325HQ) as previously described [25]. The alfalfa clonal lines 708 and 773 were propagated from cuttings and grown in the greenhouse. The greenhouse experiments consisted of three replicates arranged in a randomized complete block design. For each replicate, there were eight plants of each clone in individual pots. For cell wall analysis, stem internodes tissues were harvested at full bloom and plant material for analysis was composited within each replicate (2 blocks × 3 reps = 6 data points per genotype). Cell wall analysis was performed in duplicate as previously described [25]. An analysis of variance was done to test if the means (g kg-1 cell wall) for cell wall components of the two genotypes were equal (Table 1). For RNA-Seq, ES and PES internodes were harvested as previously described [25].
RNA extraction, cDNA library preparation and sequencing
Total RNA was purified from three replicates of elongating and post-elongation stem internodes of genotypes 708 and 773 using the CTAB based protocol previously described [40]. Contaminating genomic DNA was removed from each RNA sample using the DNA-free™ kit following the manufacturer's recommendations http://www.ambion.com. An equal amount of total RNA was pooled from each replicate for each stem tissue sample. RNA samples were quantified using Quant-iT™ RiboGreen® RNA Reagent http://www.invitrogen.com and the RNA integrity was checked with RNA6000 Nano Assay using the Agilent 2100 Bioanalyzer™ (Agilent Technologies, Palo Alto, CA). cDNA library preparation and sequencing reactions were conducted in the Biomedical Genomics Center, University of Minnesota. Illumina library prep, clustering and sequencing reagents were used throughout the process following the manufacturer's recommendations http://www.illumina.com. Briefly, mRNAs were purified using poly-T oligo-attached magnetic beads and then fragmented. The first and the second strand cDNAs were synthesized and end repaired. Adaptors were ligated after adenylation at the 3'-ends. After gel purification, cDNA templates were enriched by PCR. cDNA libraries were validated using a High Sensitivity Chip on the Agilent2100 Bioanalyzer™ (Agilent Technologies, Palo Alto, CA). The cDNA library was quantified using PicoGreen Assay and by qPCR. The samples were clustered on a flow cell using the cBOT. After clustering, the samples were loaded on the Illumina GA-II machine. The samples were sequenced using a single read with 76 cycles. Initial base calling and quality filtering of the Illumina GA-II image data were performed using the default parameters of the Illumina GA Pipeline GERALD stage http://www.illumina.com. Additional filtering for homopolymers and read size (< 75 bp) was performed using custom written code.
For RNA-Seq using the GS FLX Titanium platform http://www.454.com, mRNA was reverse transcribed with SuperScript III reverse transcriptase http://www.invitrogen.com using dT15VN2 primer. cDNA was synthesized using E. coli DNA Ligase, E. coli DNA polymerase I and E coli RNaseH. cDNA was then fragmented by sonication. The cDNA was then used for 454 sstDNA preparation in the "GS20 DNA Library Preparation" step2 http://www.454.com. The rest of the library preparation and the 454 sequencing procedures were performed following the manufacturer's recommendations http://www.454.com. Standard post-run and bioinformatics processing on the 454 platform to determine reads that passed various quality filters were also performed following the manufacturer's recommendations http://www.454.com.
de novo transcriptome assembly
The Velvet algorithm [32] was used for de novo assembly of the 198,861,304 Illumina reads (76 bp). During the de novo assembly using the Velvet program, short EST reads were first hashed based on a predefined hash length in base pairs (k-mer length). Next, the contigs were built based on a series of overlapping k-mers using de Brujin graphs [32]. In general, longer k-mers increase transcript contiguity (longer transcript length) and specificity (less spurious overlaps) but decrease diversity (smaller number of contigs) compared to shorter k-mers [32]. To optimize our Velvet assembly toward higher transcript contiguity and specificity, we tested a series of k-mers (31, 37, 41, 47, 51, 57, 61, 63, 65) for de novo assembly of short EST reads (Additional file 26). We used the median contig length (N50) generated for each k-mer as an indicator of the transcript contiguity of de novo assembly. As k-mer values increased from 31 to 61, N50 values increased to a value of 289 reflecting increased efficiency of de novo assembly. The N50 values declined significantly at k-mer values above 61 (Additional file 26). On the basis of these results, we used a k-mer value of 61 for de novo assembly of alfalfa EST reads.
Alfalfa Gene Index assembly
The alfalfa gene index (MSGI 1.0) was built following the Gene Index Assembly protocol previously described [39,40]. The gene ontology (GO) functional classes and pathways for each sequence in MSGI 1.0 were assigned based on Arabidopsis GO SLIM and pathway annotation ftp://ftp.arabidopsis.org/home/tair/Ontologies/. For GO characterization, the unique sequences in MSGI 1.0 were compared with the Arabidopsis proteome using the BlastX program with e-value cutoff of 1e-10. Top protein matches from Arabidopsis sequences were assigned to each of the MSGI 1.0 sequences. The MapMan gene functional classification system [41] was assigned to each sequence in MSGI 1.0 following the method previously described [31]. The functional class over-representation analysis was performed using PageMan [107] as previously described [25,31].
Polymorphism detection
The MISA program [42] was used to detect simple sequence repeats (SSRs) among sequences in MSGI 1.0. The minimum number of nucleotide repeats specified during SSR analysis was 20, 10, 7, 5, 5, and 5 for mono-, di-, tri-, tetra-, penta-, and hexanucleotide repeats, respectively. The maximum number of bases interrupting 2 SSRs in a compound microsatellite was set at 100 bp. The primers spanning each SSR were designed using the default parameter of the Primer3 program [44].
For SNP detection, the Illumina GA-II reads were mapped to the sequences in MSGI 1.0 using the Maq program [45]. Next, the coverage and nucleotide differences were extracted using the pileup command of the Maq program. The pileup output was further compiled for genotypes 708 and 773 with custom written script using filtering based on coverage and quality scores. Custom written script was used for additional sorting and filtering of the pileup output based on a nucleotide depth cutoff of 10 for each SNP.
Digital gene expression analysis
For digital gene expression analysis, the raw digital gene expression counts were measured by quantifying the number of Illumina GA-II reads that were mapped to the reference sequences (MSGI 1.0 or Mt3.0 cds) using the bowtie program [47]. The best-match option with a maximum of 3 nucleotide mismatches was used (-v 3 --best). The raw digital gene expression counts were normalized using the RPKM (reads/Kb/Million) method [1,48]. Custom written scripts were used to summarize the bowtie output from the raw digital expression counts and the RPKM-normalized expression counts. To identify differentially expressed genes, an expression profile matrix was built representing the digital gene expression count for each gene in each library, then imported into the DEGSeq program [52]. A DEGSeq program that utilized a MA-plot-based method with random sampling model was used to identify differentially expressed genes in each pair-wise comparison (p < 0.001, FDR < 0.025, ≥ 2-fold difference). Heatmaps based on hierarchical cluster analysis [108] of RPKM-normalized expression counts (Figure 5, Additional file 25) and expression ratios (Figure 6) were generated using MultiExperiment Viewer http://www.tm4.org/mev/.
In a previous study, we generated GeneChip data for ES and PES internodes of alfalfa genotypes 252 and 1283 [25]http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13602. To compare the digital gene expression data generated using MSGI 1.0 and Mt3.0 cds sequences with the previously generated GeneChip data [25], we first compared two Medicago reference sequences (MSGI 1.0 and Mt3.0 cds) with Medicago GeneChip probe set consensus sequences using the Blastn program (e-value cutoff of 1e-10). Top sequence matches from the Medicago GeneChip probe sets were assigned to each RNA-Seq reference sequence. Next, we selected from GeneChip data and RNA-Seq data a subset of genes involved in general stem development independent of genotypic variation in gene expression (Log2(PES/ES) ≥ 1 or ≤ -1 in both genotypes). Genes that were commonly selected between RNA-Seq and GeneChip data were identified based on sequence homology. Log2(PES/ES) values from the RNA-Seq data generated using MSGI 1.0 and Mt3.0 cds as reference sequences were compared with Log2(PES/ES) values from the GeneChip data (Figure 4A, 4B).
To compare the digital gene expression data generated using MSGI 1.0 and Mt3.0 cds sequences with the qRT-PCR data, we first compared two Medicago reference sequences (MSGI 1.0 and Mt3.0 cds) using the the Blastn program (e-value cutoff of 1e-10). Top sequence matches from the Mt3.0 cds were assigned to each MSGI 1.0 sequence. Primers for qRT-PCR were designed based on the MSGI 1.0 sequences (Additional file 10). Log ratio values from the RNA-Seq data generated using MSGI and Mt3.0 cds as reference sequences were compared with ΔΔCT values obtained from the qRT-PCR data (Figure 4C, 4D).
SNP genotyping
The SNP genotyping was conducted in the Biomedical Genomics Center, University of Minnesota. Briefly, a total of 55 SNPs predicted between genotypes 708 and 773 were randomly selected for validation by MALDI-TOF mass spectrometry using the iPLEX Gold spectrometry system http://www.sequenom.com. Genomic DNAs were purified from young leaves of genotypes 708 and 773 using DNeasy Plant Mini Kit http://www.qiagen.com. The multiplex assays were designed using Mass-ARRAY Assay Design 3.0 software and primers were obtained from IDT (Coralville, Iowa). Reactions (PCR, shrimp alkaline phosphatase treatment followed by extension) were performed according to iPLEX Gold method http://www.sequenom.com. Mass ARRAY workstation software (v. 3.3) was used to analyze the SNP genotyping results.
Real-time quantitative RT-PCR (qRT-PCR)
A portion of the pooled total RNA used for the RNA-Seq analysis was used to make cDNAs for qRT-PCR. The first strand cDNA for each sample was made using random hexamers and Taqman Reverse Transcription Reagents (Applied Biosystems, CA) following the manufacturer's recommendations. Gene specific primers based on MSGI 1.0 sequences were subsequently designed using Primer Express (Applied Biosystems, CA) (Additional file 10). Samples and standards were run in triplicate on each plate and repeated on two plates using SYBR-Green PCR Master Mix (Applied Biosystems, CA) on a StepOnePlus™ Real-Time PCR System (Applied Biosystems, CA) following the manufacturer's recommendations. qRT-PCR was performed in a 20 μl reaction containing 4 μl ddH2O, 10 μl 2× PCR mix, 1 μl forward primer (1 μM), 1 μl reverse primer (1 μM), and 4 μl of template cDNA (5 ng/μl). The PCR conditions were as follows: two minutes of pre-incubation at 50°C, 10 minutes of pre-denaturation at 94 °C, 40 cycles of 15 seconds at 95 °C and one min at 60 °C, followed by steps for dissociation curve generation (30 seconds at 95 °C, 60 seconds at 60 °C and 30 seconds at 95 °C). The StepOnePlus software (Applied Biosystems, CA) was used for data collection and analysis. Dissociation curves for each amplicon were carefully examined to confirm lack of multiple amplicons at different melting temperatures (Tms). Relative transcript levels for each sample were obtained using the "comparative CT method" [109] using the CT value of the 18S rRNA for each sample as a normaliser.
Abbreviations
ES: elongating stem; PES: post-elongation stem; SNP: single nucleotide polymorphism; EST: expressed sequence tag; GO: gene ontology; cds: coding sequence; SSR: simple sequence repeat; RPKM: reads/Kb/Million; q-RT PCR: real-time quantitative RT-PCR; HKG: housekeeping gene; CV: coefficient of variation; CesA: cellulose synthase; PAL: phenylalanine ammonia-lyase; C4H: cinnamate-4-hydroxylase; 4CL: 4-coumarate-CoA ligase; HCT: hydroxycinnamoyl transferase; C3H: p-coumarate 3-hydroxylase; CCoAOMT: caffeoyl-CoA 3-O-methyltransferase; CCR1: cinnamoyl-CoA reductase 1; F5H: ferulate 5-hydroxylase; COMT: caffeic acid O-methyltransferase; CAD: cinnamyl-alcohol dehydrogenase; AGP: arabinogalactan protein; LTP: lipid transfer protein; LHB1B1: Photosystem II light harvesting complex gene; RBCS-1A: rubisco small subunit 1; SUC: sucrose transporter; SuSy: sucrose synthase; PHO1: PHOSPHATE 1; IAA: Indole-3-acetic acid; AUX1: auxin influx carrier; MATE: multi-drug toxic efflux carrier; ABC: ATP-binding cassette.
Authors' contributions
SY and ZT performed the computational analysis involved in the de novo assembly, digital gene expression and SNP detection. CF performed the computational analysis involved in MSGI 1.0 assembly and SSR detection. SY and WX performed the computational analysis involved in the identification of differentially expressed genes. SY performed the computational analysis involved in the functional classification and over-representation analysis. SY conducted the qRT-PCR. JL identified the genotypes used in the study. HJ conducted the cell wall analysis of the alfalfa genotypes. All authors contributed to the analysis of results and writing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
de novo assembly of alfalfa Illumina GA-II EST reads. A fasta file containing a total of 132,153 unique sequences generated after de novo assembly of Illumina GA-II EST reads derived from 4 cDNA libraries developed in this study. The Velvet program [32] with k-mer 61 was used for de novo assembly.
Alfalfa Gene Index 1.0 (MSGI 1.0). A fasta file containing Alfalfa Gene Index 1.0 (MSGI 1.0) sequences. MSGI 1.0 contains a total of 124,025 unique sequences including 22,729 tentative consensus sequences (TCs), 22,315 singletons and 78,981 pseudo-singletons. The average length of the unique sequences in MSGI 1.0 is 384 bp (100 bp minimum and 6,956 bp maximum) with more than 10,000 sequences larger than 800 bp. The total base count of the sequences in MSGI 1.0 is 47,628,953 bp. Unfortunately, the current pipe line of the DFCI gene index database http://compbio.dfci.harvard.edu/tgi/ is not suited for short reads (personal communication with a DFCI Gene Index staff). The Gene Index Project team has indicated that it plans to address this issue soon. When a gene index database is established for alfalfa, MSGI1.0 will be uploaded to the DFCI gene index database.
Functional classification and annotation of sequences in the Alfalfa Gene Index 1.0 (MSGI 1.0). A table listing Gene ontology (GO), pathway, MapMan functional classes and gene annotation for sequences in the Alfalfa Gene Index 1.0 (MSGI 1.0).
Simple sequence repeats (SSRs) detected in MSGI 1.0. A table listing SSR-containing sequence IDs, SSR types and position, and primers spanning each SSR for the sequences in the Alfalfa Gene Index 1.0 (MSGI 1.0).
Single nucleotide polymorphisms (SNPs) predicted between alfalfa genotypes 708 and 773. A table listing SNPs predicted between alfalfa genotypes 708 and 773 including SNP-containing sequence ID, SNP type, SNP position and depth in each genotype.
Validation of SNPs predicted between alfalfa genotypes 708 and 773 using RNA-Seq data. A table showing SNP validation results. A total of 55 SNPs were randomly selected to genotype genomic DNAs purified from the genotypes 708 and 773 by MALDI-TOF mass spectrometry using the iPLEX Gold spectrometry system http://www.sequenom.com. In addition to genotypes 708 and 773, we also genotyped 51 additional alfalfa (M. sativa) genotypes selected from different populations of M. sativa ssp. sativa or M. sativa ssp. falcata.
Functional classes over- or under-represented among SNP-harboring genes. A figure showing the functional class over-representation analysis conducted for SNP-harboring genes. Functional classes that are over- or under-represented among SNP-harboring genes were identified using the PageMan over-representation analysis module. The z-vlaues for significant classes identified after Fisher's exact test with Bonferroni correction (z-value cutoff of 1) were false color coded using a scale of -5 to +5. The intensity of blue and red indicate the degree of over- and under-representation of the corresponding class, respectively.
Allelic variations (SNPs) detected within genotype 708. A table listing a total of 287,555 allelic variations (SNPs) detected within genotype 708 using minimum SNP depth cutoff of 10.
Allelic variations (SNPs) detected within genotype 773. A table listing a total of 168,966 allelic variations (SNPs) detected within genotype 773 using minimum SNP depth cutoff of 10.
qRT-PCR validation of RNA-Seq data generated by two reference sequences (MSGI 1.0 and Mt3.0 cds). A table showing the source data used to generate Figure 4. The table contains MSGI 1.0 and Mt3.0 cds IDs of genes used for qRT-PCR, qRT-PCR and RNA-Seq data generated by two reference sequences (MSGI 1.0 and Mt3.0 cds), and primers used for qRT-PCR.
An expression profile matrix for each library showing digital gene expression count of each gene in MSGI 1.0. A table showing the digital gene expression counts of each gene in MSGI 1.0 for ES and PES internodes of alfalfa genotypes 708 and 773. The raw expression counts generated by bowtie program were normalized using the RPKM method [1,48].
Top 500 most abundant transcripts in each library. A table showing the RPKM-normalized digital gene expression counts and MapMan functional classes for the top 500 most abundant transcripts selected in each library.
Functional classes over- or under-represented among the top 500 most abundant transcripts in each library. A figure showing the results from functional class over-representation analysis for the top 500 most abundant transcripts in ES and PES internodes of alfalfa genotypes 708 and 773. For details, see the description for additional file 7.
300 housekeeping genes selected. A table listing 300 housekeeping genes (HKGs) with relatively high levels of expression. To identify these HKGs, we first selected genes with an average RPKM-normalized transcript count greater than 10. Next, we selected the top 300 genes from the list with the lowest coefficient of variation (CV = standard deviation/mean). The RPKM-normalized expression counts, MapMan functional class and description for each HKG selected are also presented in the table.
Genes differentially expressed between ES and PES internodes of alfalfa genotype 708.A table listing 3,838 genes differentially expressed between ES and PES internodes of alfalfa genotype 708 in MSGI 1.0. We used a MA-plot-based method with random sampling model in a DEGSeq program to select these genes (p-value < 0.001, FDR < 0.025, ≥ 2-fold difference). RPKM-normalized expression counts, log ratios, z-scores, p-values, and q-values for each gene selected are also presented in the table.
Genes differentially expressed between ES and PES internodes of alfalfa genotype 773. A table listing 4,428 genes differentially expressed between ES and PES internodes of alfalfa genotype 708 in MSGI 1.0. For details, see the description for additional file 11.
Genes differentially expressed between alfalfa genotypes 708 and 773 in ES internodes.A table listing 8,883 genes differentially expressed between alfalfa genotypes 708 and 773 in ES internodes in MSGI 1.0. For details, see the description for additional file 11.
Genes differentially expressed between alfalfa genotypes 708 and 773 in PES internodes. A table listing 4,799 genes differentially expressed between alfalfa genotypes 708 and 773 in PES internodes in MSGI 1.0. For details, see the description for additional file 11.
Genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773. A table listing 13,797 genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773 in MSGI 1.0. Genes selected in additional files 15, 16, 17 and 18 were combined together to produce this table. The RPKM-normalized expression counts, MapMan functional class and description for each gene selected are also presented in the table.
Top 200 most differentially expressed genes in each pair-wise comparison. A table that lists 657 genes that were generated after combining the top 200 most differentially expressed genes selected in each pair-wise comparison of gene expression between ES and PES internodes of genotypes 708 and 773. This table is a data source for Figure 5. The RPKM-normalized expression counts, MapMan functional class and description for each gene selected are also presented in the table.
Phenylpropanoid (lignin) pathway genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773. A table listing phenylpropanoid (lignin) pathway genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773 (p-value < 0.001, FDR,0.025, ≥ 2-fold difference). This table is a data source for Figure 6. The log ratios from each pair-wise comparison, EC number, and enzyme ID for each gene selected are also presented in the table.
Candidate genes identified in 708 and 773 that may be involved in general stem development independent of genotypic variation in gene expression. A table listing 594 genes potentially involved in general stem development independent of genotypic variation in gene expression in alfalfa (Log2(PES/ES)≥1 or ≤-1 in both genotypes 708 and 773). The RPKM-normalized expression counts, log ratios, MapMan functional class and description for each gene selected are also presented in the table.
Functional classes over- or under-represented among genes involved in general stem development independent of genotypic variation in alfalfa. A figure showing the functional class over-representation analysis for genes involved in general stem development independent of genotypic variation in alfalfa (Log2(PES/ES)≥1 or ≤-1 in both genotypes 708 and 773). "Up in PES" and "Up in ES" indicate genes up-regulated in PES and ES internodes in both genotypes, respectively. For details, see the description for additional file 7.
Putative transporter genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773. A table listing 478 transporter genes in ES and PES internodes of alfalfa genotypes 708 and 773 in MSGI 1.0. The RPKM-normalized expression counts, log ratios from each pair-wise comparison, MapMan functional class and description for each transporter gene selected are also presented in the table.
Hierarchical clustering analysis of selected transporter genes differentially expressed between 708 and 773 in both ES and PES internodes. A figure showing a heatmap for 42 transporter genes differentially expressed between 708 and 773 in both ES and PES internodes (p < 0.001, FDR < 0.025, ≥ 2-fold difference). The RPKM-normalized expression counts for each gene in each library are represented by the intensity of the red color on a 0 to 22 scale. Dark red (scale intensity 22) indicates genes with RPKM-normalized expression counts ≥ 22. See Methods for details. A complete list of the transporter genes selected, RPKM-normalized expression counts, and corresponding MapMan functional categories are provided in Additional file 24.
Optimization of de novo assembly of Illumina GA-II EST reads with a series of k-mers using the Velvet program [32]. A figure showing the median sequence length of the contigs (y-axis) for a series of k-mers (31, 37, 41, 47, 51, 57, 61, 63, 65) tested using the Velvet program. k-mer 61 produced the longest median sequence length.
Contributor Information
S Samuel Yang, Email: sam.yang@ars.usda.gov.
Zheng Jin Tu, Email: ztu@msi.umn.edu.
Foo Cheung, Email: foo.cheung@nih.gov.
Wayne Wenzhong Xu, Email: wxu@msi.umn.edu.
JoAnn FS Lamb, Email: joann.lamb@ars.usda.gov.
Hans-Joachim G Jung, Email: hans.jung@ars.usda.gov.
Carroll P Vance, Email: carroll.vance@ars.usda.gov.
John W Gronwald, Email: john.gronwald@ars.usda.gov.
Acknowledgements
This work was carried out in part using computing resources at the University of Minnesota Supercomputing Institute for Advance Computational Research. Funding for this research was provided by USDA-ARS CRIS Project 3640-12210-001-00D. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. We thank Dr. David Garvin, Dr. Jamie O'Rourke, and Dr. Deborah Samac for critical review of the manuscript.
References
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Lister R, Gregory BD, Ecker JR. Next is now:new technologies for sequencing of genomes, trancriptomes, and beyond. Curr Opin Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Bähler J. RNA-seq: from technology to biology. Cell Mol Life Sci. 2010;67:569–579. doi: 10.1007/s00018-009-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Landry J-R. RNA-Seq-quantitative measurement of expression through massively parallel RNA-Sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. Comprehensive annotation of the trancriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Last RL. Shotguns and SNPS: how fast and cheap sequencing is revolutionizing plant biology. Plant J. 2010;61:922–927. doi: 10.1111/j.1365-313X.2009.04030.x. [DOI] [PubMed] [Google Scholar]
- Howard BE, Heber S. Towards reliable isoform quantification using RNA-SEQ data. BMC Bioinformatics. 2010;11(Suppl 3):56. doi: 10.1186/1471-2105-11-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pryrosequencing. Plant Physiol. 2007;144(1):32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall PK, Leebens-Mack J, Chanderbali AS, Barakat A, Wolcott E, Liang H, Landherr L, Tomsho LP, Hu Y, Carlson JE, Ma H, Schuster SC, Soltis DE, Soltis PS, Altman N, dePamphilis CW. Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics. 2009;10:347. doi: 10.1186/1471-2164-10-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;10:1111. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, Graham MA, Cannon SB, May GD, Vance CP, Shoemaker RC. RNA-Seq atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biology. 2010;10:160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Lu G, Fan D, Zhu C, Li W, Zhao Q, Feng Q, Zhao Y, Guo Y, Li W, Huang X, Han B. Functional annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010;20:1238–1249. doi: 10.1101/gr.106120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS. SNP discovery via 454 transcriptome sequencing. Plant J. 2007;51(5):910–918. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F, Haas BJ, Goldberg SMD, May GD, Xiao Y, Town CD. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7:272. doi: 10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novaes E, Drost DR, Farmerie WG, Pappas GJ Jr, Grattapaglia D, Sederoff RR, Kirst M. High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics. 2008;9:312. doi: 10.1186/1471-2164-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin D, Ferrarini A, Chimento A, Kaiser O, Levenkova N, Bouffard P, Delledonne M. Combining next-generation pyrosequencing with microarray for large scale expression analysis in non-model species. BMC Genomics. 2009;10:555. doi: 10.1186/1471-2164-10-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LJ, Biggs PJ, Voelckel C, Joly S. An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Informatics. 2008;21:3–14. [PubMed] [Google Scholar]
- Wang W, Wang Y, Zhang Q, Qi Y, Guo D. Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genomics. 2009;10:465. doi: 10.1186/1471-2164-10-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M, Long Y, Meng J, Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploidy Brassica napus using Solexa transcriptome sequencing. Plant Biotech J. 2009;7:334–346. doi: 10.1111/j.1467-7652.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- Michaud R, Lehman WF, Rumbaugh MD. In: Alfalfa and alfalfa improvement - Agronomy Monograph no. 29. Hanson AA, Barnes DK, Hill RR Jr, editor. Madison, WI: ASA-CSSA-SSSA; 1988. World Distribution and Historical Development; pp. 25–91. [Google Scholar]
- National Agricultural Statistics Service. 2009. http://www.nass.usda.gov On-line resource.
- Samac DA, Jung H-JG, Lamb JFS. In: Alcoholic Fuels. Minteer S, editor. Boca Raton, FL: CRC Press; 2006. Development of alfalfa (Medicago sativa L.) as a feedstock for production of ethanol and other bioproducts; pp. 79–98. [Google Scholar]
- Yang SS, Xu WW, Tesfaye M, Lamb JFS, Jung H-JG, VandenBosch KA, Vance CP, Gronwald JW. Transcript profiling of two alfalfa genotypes with contrasting cell wall composition in stems using a cross-species platform: optimizing analysis by masking biased probes. BMC Genomics. 2010;11:323. doi: 10.1186/1471-2164-11-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh MD, Caddel JL, Rowe E. Alfalfa and Alfalfa Improvement. ASA Monograph 29. Madison, WI: American Society of Agronomy; 1988. Breeding and Quantitative Genetics; pp. 777–808. [Google Scholar]
- Brummer EC, Sledge MK, Bouton JH, Kochert G. In: DNA-based markers in plants. Phillips RL, Vasil IK, editor. The Netherlands: Kluwer Academic; 2001. Molecular Marker Analyses in Alfalfa and Related Species; pp. 169–180. [Google Scholar]
- Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol. 2003;3:9. doi: 10.1186/1471-2229-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan N, Bhagwat AA, Bauchan GB, Cregan PB. Simple sequence repeat DNA markers in alfalfa and perennial and annual Medicago species. Genome. 1997;40:887–895. doi: 10.1139/g97-115. [DOI] [PubMed] [Google Scholar]
- Sledge MK, Ray IM, Jiang G. An expressed sequence tag SSR map of tetrapolid alfalfa (Medicago sativa L.) Theor Appl Genet. 2005;111:980–992. doi: 10.1007/s00122-005-0038-8. [DOI] [PubMed] [Google Scholar]
- Yang SS, Xu WW, Tesfaye M, Lamb JFS, Jung H-JG, Samac DA, Vance CP, Gronwald JW. Single-feature polymorphism discovery in the transcriptome of tetraploid alfalfa. Plant Genome. 2009;2:224–232. doi: 10.3835/plantgenome2009.03.0014. [DOI] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JG, Janson EM, Hittinger CT, Johnston M, Abbot P, Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol Biol Evol. 2009;26(12):2731–2744. doi: 10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- Mizrachi E, Hefer CA, Ranik M, Joubert F, Myburg AA. de novo assembled expressed gene catalog of a fast-growing Eucalyptus tree produced by Illumina mRNA-Seq. BMC Genomics. 2010;11:681. doi: 10.1186/1471-2164-11-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Patel RK, Tyagi AK, Jain M. de novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Research. 2011. [DOI] [PMC free article] [PubMed]
- Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, Stazyk G, Morin RD, Zhao Y, Hirst M, Schein JE. et al. de novo transcriptome assembly with ABySS. Bioinformatics. 2009;25(21):2872–2877. doi: 10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- Surget-Groba Y, Montoya-Burgos JI. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Research. 2010;20:1432–1440. doi: 10.1101/gr.103846.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oases. http://www.ebi.ac.uk/~zerbino/oases (Jan 31st, website last accessed)
- Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- Yang SS, Cheung F, Lee JJ, Ha M, Wei NE, Sze SH, Stelly DM, Thaxton P, Triplett B, Town CD, Chen ZJ. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 2006;47:761–775. doi: 10.1111/j.1365-313X.2006.02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: a user driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor Appl Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- Ellis JR, Burke JM. EST-SSRs as a resource for population genetic analyses. Heredity. 2007;99:125–132. doi: 10.1038/sj.hdy.6801001. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S, Misener S, editor. Totowa, NJ: Humana Press; 2000. Primer3 on the www for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18(11):1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnidelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001;15(9):1115–1127. doi: 10.1101/gad.879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human gemone. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Zhou W, Gregg K, Yu N, Davis S, Davis S. A method for cross-species gene expression analysis with high-density oligonucleotide arrays. Nucl Acids Res. 2004;32:e93. doi: 10.1093/nar/gnh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SS, Valdés-López O, Xu WW, Bucciarelli B, Gronwald JW, Hernández G, Vance CP. Transcript profiling of common bean (Phaseolus vulgaris L.) using the GeneChip® Soybean Genome Array: optimizing analysis by masking biased probes. BMC Plant Biol. 2010;10:85. doi: 10.1186/1471-2229-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005;6:242. doi: 10.1186/gb-2005-6-12-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007;12:267–277. doi: 10.1016/j.tplants.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Morrison WH III, Freshour GD, Hahn MG, Ye ZH. Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 2003;132:786–795. doi: 10.1104/pp.102.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosca S, Barton CJ, Taylor NG, Ryden P, Neumetzler L, Pauly M, Roberts K, Seifert GJ. Interactions between MUR10/CesA7 dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol. 2006;142:1353–1363. doi: 10.1104/pp.106.087700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Bernard-Vailhé MA, Chabbert B, Besle JM, Opsomer C, Van Montagu M, Botterman J. Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol Biol. 1999;39:437–447. doi: 10.1023/A:1006182925584. [DOI] [PubMed] [Google Scholar]
- Guo D, Chen F, Inoue K, Blount JW, Dixon RA. Down-regulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa (Medicago sativa L.): impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell. 2001;13:73–88. doi: 10.1105/tpc.13.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MSS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.) Proc Natl Acad Sci USA. 2005;102:16573–16578. doi: 10.1073/pnas.0505749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley R, Ruth M, Glassner D, Sheehan J. Process design and costing of bioethanol technology: a tool for determining the status and direction of research and development. Biotechnol Prog. 1999;15:794–803. doi: 10.1021/bp990107u. [DOI] [PubMed] [Google Scholar]
- Wooley R, Ruth M, Sheehan J, Ibsen K, Majdeski H, Galvez A. Lignocellulosic biomass to ethanol process design and economics utilizing co-current dilute acid prehydrolysis and enzymatic hydrolysis: current and futuristic scenarios. National Renewable Energy Laboratory, Golden CO, NREL/TP-580-26157; 1999. [Google Scholar]
- Aden A, Ruth M, Ibsen K, Jechura J, Neeves K, Sheehan J, Wallace B, Montague L, Slayton A, Lukas J. Lignocellulosic biomass to ethanol process design and economics utilizing co-current dilute acid prehydrolysis and enzymatic hydrolysis for corn stover. National Renewable Energy Laboratory, Golden CO, NREL/TP-510-32438; 2002. [Google Scholar]
- Yang B Wyman CE Pretreatment: the key to unlocking low cost cellulosic ethanol Biofuels Bioproducts & Biorefining 2008226–40. 10.1002/bbb.4921324176 [DOI] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12:2529–2539. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Lett. 2007;581:2309–2317. doi: 10.1016/j.febslet.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Riesmeier JW, Hirner B, Frommer WB. Potato sucrose transporter expression in minor veins indicates a role in phloem loading. Plant Cell. 1993;5:1591–1598. doi: 10.1105/tpc.5.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta. 1995;196:564–570. doi: 10.1007/BF00203657. [DOI] [PubMed] [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J. 1999;19:269–278. doi: 10.1046/j.1365-313X.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- Barth I, Meyer S, Sauer N. PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell. 2003;15:1375–1385. doi: 10.1105/tpc.010967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N. Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol. 2004;134:684–693. doi: 10.1104/pp.103.033399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Singh B, Wang G, Zhang D. In: Genetics and Genomics of Cotton. Plant Genetics and Genomics: Crops and Models 3. Paterson AH, editor. New York, USA: Springer Science Business Media; 2009. Genomics of cotton fiber secondary wall deposition and cellulose biogenesis; pp. 385–417. [Google Scholar]
- Somerville CR. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Fujii S, Hayashi T, Mizuno K. Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant Cell Physiol. 2010;51:294–301. doi: 10.1093/pcp/pcp190. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007;50:982–994. doi: 10.1111/j.1365-313X.2007.03108.x. [DOI] [PubMed] [Google Scholar]
- Ribot C, Wang Y, Poirier Y. Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta. 2008;227:1025–1036. doi: 10.1007/s00425-007-0677-x. [DOI] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell. 2009;21:3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Ye ZH. Vascular tissue differentiation and pattern formation in plants. Annu Rev Plant Biol. 2002;53:183–202. doi: 10.1146/annurev.arplant.53.100301.135245. [DOI] [PubMed] [Google Scholar]
- De Smet I, Jürgens G. Patterning the axis in plants - auxin in control. Curr Opin Genet Dev. 2007;17:337–343. doi: 10.1016/j.gde.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Lucas M, Godin C, Jay-Allemand C, Laplaze L. Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot. 2008;59:55–66. doi: 10.1093/jxb/erm171. [DOI] [PubMed] [Google Scholar]
- Sundberg B, Uggla C, Tuominen H. In: Cell and Molecular Biology of Wood Formation. Savidge R, Barnett J, Napier R, editor. Oxford, UK: BIOS Scientific Publishers; 2000. Cambial growth and auxin gradients; pp. 169–188. [Google Scholar]
- Nilsson J, Karlberg A, Antti H, Lopez-Vernaza M, Mellerowicz E, Perrot-Rechenmann C, Sandberg G, Bhalerao RP. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell. 2008;20:843–855. doi: 10.1105/tpc.107.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001;127:1361–1366. doi: 10.1104/pp.010724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 2005;138(2):803–818. doi: 10.1104/pp.105.060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G. AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell. 2002;14:589–597. doi: 10.1105/tpc.010354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakani A, Li G, Peng Z. Role of AUX1 in the control of organ identity during in vitro organogenesis and in mediating tissue-specific auxin and cytokinin interaction in Arabidopsis. Planta. 2009;229:645–657. doi: 10.1007/s00425-008-0846-6. [DOI] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berleth T. Responses of plant vascular systems to auxin transport inhibition. Development. 1999;126:2979–2991. doi: 10.1242/dev.126.13.2979. [DOI] [PubMed] [Google Scholar]
- Klee H, Estelle M. Molecular genetic approaches to plant hormone biology. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:529–551. doi: 10.1146/annurev.pp.42.060191.002525. [DOI] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18:2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/S0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Masson PH. Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol. 2003;133:1677–1690. doi: 10.1104/pp.103.032169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Li X, Chapple C. Understanding lignification: challenges beyond monolignol biosynthesis. Plant Physiol. 2010;154:449–452. doi: 10.1104/pp.110.162842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Ann Rev Genet. 2010;44:337–363. doi: 10.1146/annurev-genet-102209-163508. [DOI] [PubMed] [Google Scholar]
- Boija E, Johansson G. Interactions between model membranes and lignin-related compounds studied by immobilized liposome chromatography. Biochim Biophys Acta. 2006;1758:620–626. doi: 10.1016/j.bbamem.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Rensing KH, Wong JCT, Banno B, Mansfield SD, Samuels AL. Tracking monolignols during wood development in lodgepole pine. Plant Physiol. 2008;147:1750–1760. doi: 10.1104/pp.108.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weber B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell. 2007;19(6):2023–2038. doi: 10.1105/tpc.106.046029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. MATE transporters facilitate vacuolar uptake of epicatechin 3'-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell. 2009;21:2323–2340. doi: 10.1105/tpc.109.067819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2006;580:1183–1191. doi: 10.1016/j.febslet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Rea PA. Plant ATP-binding cassette transporters. Annu Rev Plant Biol. 2007;58:347–375. doi: 10.1146/annurev.arplant.57.032905.105406. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, Fernie AR, Stitt M. PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
de novo assembly of alfalfa Illumina GA-II EST reads. A fasta file containing a total of 132,153 unique sequences generated after de novo assembly of Illumina GA-II EST reads derived from 4 cDNA libraries developed in this study. The Velvet program [32] with k-mer 61 was used for de novo assembly.
Alfalfa Gene Index 1.0 (MSGI 1.0). A fasta file containing Alfalfa Gene Index 1.0 (MSGI 1.0) sequences. MSGI 1.0 contains a total of 124,025 unique sequences including 22,729 tentative consensus sequences (TCs), 22,315 singletons and 78,981 pseudo-singletons. The average length of the unique sequences in MSGI 1.0 is 384 bp (100 bp minimum and 6,956 bp maximum) with more than 10,000 sequences larger than 800 bp. The total base count of the sequences in MSGI 1.0 is 47,628,953 bp. Unfortunately, the current pipe line of the DFCI gene index database http://compbio.dfci.harvard.edu/tgi/ is not suited for short reads (personal communication with a DFCI Gene Index staff). The Gene Index Project team has indicated that it plans to address this issue soon. When a gene index database is established for alfalfa, MSGI1.0 will be uploaded to the DFCI gene index database.
Functional classification and annotation of sequences in the Alfalfa Gene Index 1.0 (MSGI 1.0). A table listing Gene ontology (GO), pathway, MapMan functional classes and gene annotation for sequences in the Alfalfa Gene Index 1.0 (MSGI 1.0).
Simple sequence repeats (SSRs) detected in MSGI 1.0. A table listing SSR-containing sequence IDs, SSR types and position, and primers spanning each SSR for the sequences in the Alfalfa Gene Index 1.0 (MSGI 1.0).
Single nucleotide polymorphisms (SNPs) predicted between alfalfa genotypes 708 and 773. A table listing SNPs predicted between alfalfa genotypes 708 and 773 including SNP-containing sequence ID, SNP type, SNP position and depth in each genotype.
Validation of SNPs predicted between alfalfa genotypes 708 and 773 using RNA-Seq data. A table showing SNP validation results. A total of 55 SNPs were randomly selected to genotype genomic DNAs purified from the genotypes 708 and 773 by MALDI-TOF mass spectrometry using the iPLEX Gold spectrometry system http://www.sequenom.com. In addition to genotypes 708 and 773, we also genotyped 51 additional alfalfa (M. sativa) genotypes selected from different populations of M. sativa ssp. sativa or M. sativa ssp. falcata.
Functional classes over- or under-represented among SNP-harboring genes. A figure showing the functional class over-representation analysis conducted for SNP-harboring genes. Functional classes that are over- or under-represented among SNP-harboring genes were identified using the PageMan over-representation analysis module. The z-vlaues for significant classes identified after Fisher's exact test with Bonferroni correction (z-value cutoff of 1) were false color coded using a scale of -5 to +5. The intensity of blue and red indicate the degree of over- and under-representation of the corresponding class, respectively.
Allelic variations (SNPs) detected within genotype 708. A table listing a total of 287,555 allelic variations (SNPs) detected within genotype 708 using minimum SNP depth cutoff of 10.
Allelic variations (SNPs) detected within genotype 773. A table listing a total of 168,966 allelic variations (SNPs) detected within genotype 773 using minimum SNP depth cutoff of 10.
qRT-PCR validation of RNA-Seq data generated by two reference sequences (MSGI 1.0 and Mt3.0 cds). A table showing the source data used to generate Figure 4. The table contains MSGI 1.0 and Mt3.0 cds IDs of genes used for qRT-PCR, qRT-PCR and RNA-Seq data generated by two reference sequences (MSGI 1.0 and Mt3.0 cds), and primers used for qRT-PCR.
An expression profile matrix for each library showing digital gene expression count of each gene in MSGI 1.0. A table showing the digital gene expression counts of each gene in MSGI 1.0 for ES and PES internodes of alfalfa genotypes 708 and 773. The raw expression counts generated by bowtie program were normalized using the RPKM method [1,48].
Top 500 most abundant transcripts in each library. A table showing the RPKM-normalized digital gene expression counts and MapMan functional classes for the top 500 most abundant transcripts selected in each library.
Functional classes over- or under-represented among the top 500 most abundant transcripts in each library. A figure showing the results from functional class over-representation analysis for the top 500 most abundant transcripts in ES and PES internodes of alfalfa genotypes 708 and 773. For details, see the description for additional file 7.
300 housekeeping genes selected. A table listing 300 housekeeping genes (HKGs) with relatively high levels of expression. To identify these HKGs, we first selected genes with an average RPKM-normalized transcript count greater than 10. Next, we selected the top 300 genes from the list with the lowest coefficient of variation (CV = standard deviation/mean). The RPKM-normalized expression counts, MapMan functional class and description for each HKG selected are also presented in the table.
Genes differentially expressed between ES and PES internodes of alfalfa genotype 708.A table listing 3,838 genes differentially expressed between ES and PES internodes of alfalfa genotype 708 in MSGI 1.0. We used a MA-plot-based method with random sampling model in a DEGSeq program to select these genes (p-value < 0.001, FDR < 0.025, ≥ 2-fold difference). RPKM-normalized expression counts, log ratios, z-scores, p-values, and q-values for each gene selected are also presented in the table.
Genes differentially expressed between ES and PES internodes of alfalfa genotype 773. A table listing 4,428 genes differentially expressed between ES and PES internodes of alfalfa genotype 708 in MSGI 1.0. For details, see the description for additional file 11.
Genes differentially expressed between alfalfa genotypes 708 and 773 in ES internodes.A table listing 8,883 genes differentially expressed between alfalfa genotypes 708 and 773 in ES internodes in MSGI 1.0. For details, see the description for additional file 11.
Genes differentially expressed between alfalfa genotypes 708 and 773 in PES internodes. A table listing 4,799 genes differentially expressed between alfalfa genotypes 708 and 773 in PES internodes in MSGI 1.0. For details, see the description for additional file 11.
Genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773. A table listing 13,797 genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773 in MSGI 1.0. Genes selected in additional files 15, 16, 17 and 18 were combined together to produce this table. The RPKM-normalized expression counts, MapMan functional class and description for each gene selected are also presented in the table.
Top 200 most differentially expressed genes in each pair-wise comparison. A table that lists 657 genes that were generated after combining the top 200 most differentially expressed genes selected in each pair-wise comparison of gene expression between ES and PES internodes of genotypes 708 and 773. This table is a data source for Figure 5. The RPKM-normalized expression counts, MapMan functional class and description for each gene selected are also presented in the table.
Phenylpropanoid (lignin) pathway genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773. A table listing phenylpropanoid (lignin) pathway genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773 (p-value < 0.001, FDR,0.025, ≥ 2-fold difference). This table is a data source for Figure 6. The log ratios from each pair-wise comparison, EC number, and enzyme ID for each gene selected are also presented in the table.
Candidate genes identified in 708 and 773 that may be involved in general stem development independent of genotypic variation in gene expression. A table listing 594 genes potentially involved in general stem development independent of genotypic variation in gene expression in alfalfa (Log2(PES/ES)≥1 or ≤-1 in both genotypes 708 and 773). The RPKM-normalized expression counts, log ratios, MapMan functional class and description for each gene selected are also presented in the table.
Functional classes over- or under-represented among genes involved in general stem development independent of genotypic variation in alfalfa. A figure showing the functional class over-representation analysis for genes involved in general stem development independent of genotypic variation in alfalfa (Log2(PES/ES)≥1 or ≤-1 in both genotypes 708 and 773). "Up in PES" and "Up in ES" indicate genes up-regulated in PES and ES internodes in both genotypes, respectively. For details, see the description for additional file 7.
Putative transporter genes differentially expressed in ES and PES internodes of alfalfa genotypes 708 and 773. A table listing 478 transporter genes in ES and PES internodes of alfalfa genotypes 708 and 773 in MSGI 1.0. The RPKM-normalized expression counts, log ratios from each pair-wise comparison, MapMan functional class and description for each transporter gene selected are also presented in the table.
Hierarchical clustering analysis of selected transporter genes differentially expressed between 708 and 773 in both ES and PES internodes. A figure showing a heatmap for 42 transporter genes differentially expressed between 708 and 773 in both ES and PES internodes (p < 0.001, FDR < 0.025, ≥ 2-fold difference). The RPKM-normalized expression counts for each gene in each library are represented by the intensity of the red color on a 0 to 22 scale. Dark red (scale intensity 22) indicates genes with RPKM-normalized expression counts ≥ 22. See Methods for details. A complete list of the transporter genes selected, RPKM-normalized expression counts, and corresponding MapMan functional categories are provided in Additional file 24.
Optimization of de novo assembly of Illumina GA-II EST reads with a series of k-mers using the Velvet program [32]. A figure showing the median sequence length of the contigs (y-axis) for a series of k-mers (31, 37, 41, 47, 51, 57, 61, 63, 65) tested using the Velvet program. k-mer 61 produced the longest median sequence length.