Abstract
To shed light on the structure of the basal backbone of the human Y chromosome phylogeny, we sequenced about 200 kb of the male-specific region of the human Y chromosome (MSY) from each of seven Y chromosomes belonging to clades A1, A2, A3, and BT. We detected 146 biallelic variant sites through this analysis. We used these variants to construct a patrilineal tree, without taking into account any previously reported information regarding the phylogenetic relationships among the seven Y chromosomes here analyzed. There are several key changes at the basal nodes as compared with the most recent reference Y chromosome tree. A different position of the root was determined, with important implications for the origin of human Y chromosome diversity. An estimate of 142 KY was obtained for the coalescence time of the revised MSY tree, which is earlier than that obtained in previous studies and easier to reconcile with plausible scenarios of modern human origin. The number of deep branchings leading to African-specific clades has doubled, further strengthening the MSY-based evidence for a modern human origin in the African continent. An analysis of 2204 African DNA samples showed that the deepest clades of the revised MSY phylogeny are currently found in central and northwest Africa, opening new perspectives on early human presence in the continent.
Main Text
The male-specific region of the human Y chromosome (MSY) is characterized by the lowest level of sequence diversity in the human genome (compared to autosomal and X-linked regions), which is probably a consequence of its recent origin and low mutation rate (compared to mtDNA). These features have slowed down identification of polymorphisms in the MSY compared with other genomic regions. In the last decade, however, the number of phylogenetically characterized MSY SNPs has rapidly increased from a dozen to several hundreds. Improved mutation-discovery methods have substantially contributed to this progress, and the Y chromosome has become a powerful resource not only for reconstructing past human demographic events but also for studying relevant aspects of genome evolution, such as intra- and interchromosomal gene conversion,1–5 structural variation,6–8 and MSY-coding-gene evolution.9
The first well-resolved phylogenetic tree of the MSY included 116 binary haplogroups identified by 167 SNPs.10 From then on, the MSY tree has been progressively refined through the discovery and mapping of a large number of SNPs,11,12 the most recently published MSY phylogeny incorporating 599 mutations in 311 distinct binary haplogroups.13
Although the level of resolution of the MSY tree has been significantly increased in the last decade, its basal backbone has remained substantially unchanged. The first branching in the MSY tree has been reported to be the one that separates the African-specific clade A (called clade I in 10) from clade BT (clade II-X in 10), whereas the second branching determines the subdivision of BT in clades B, mostly African, and CT, which comprises the majority of African and all non-African chromosomes.13,14 This branching pattern, along with the geographical distribution of the major clades A, B, and CT, has been interpreted as supporting an African origin for anatomically modern humans,10 with Khoisan from south Africa and Ethiopians from east Africa sharing the deepest lineages of the phylogeny.15,16
The robustness of a branch in a phylogenetic tree relies both on the number of characters supporting the branch and the rate of their recurrent evolution on the tree.14 In the currently accepted phylogeny of the human Y chromosome, clade A appears to be defined by only two markers, M91 and P97,10,13 the former being a length polymorphism at a homopolymer-associated tract, which shows at least one signal of mutational instability.10 Thus, it is conceivable that major haplogroups within clade A (haplogroups A1, A2, and A3) do not represent a monophyletic group of lineages. This point is relevant for the correct placement of the root in the tree, and this, in turn, is crucial for making inferences about early paternal genetic structure among modern humans.
To test the robustness of the backbone and the root of current Y chromosome phylogeny, we searched for SNPs that might be informative in this respect. To this aim, a resequencing analysis of a 205.9 kb MSY portion (183.5 kb in the X-degenerate and 22.4 kb in the X-transposed region) was performed for each of seven chromosomes that are representative of clade A (four chromosomes belonging to haplogroups A1a, A1b, A2, and A3), clade B, and clade CT (two chromosomes belonging to haplogroups C and R) (Table S1 available online).
Genomic DNA extracted from blood or saliva was used for the analysis. The study was approved by the “Policlinico Umberto I, Sapienza Università di Roma” ethical committee (document no. 1016/10), and informed consent was obtained from all participants. MSY regions known to be involved in gene conversion events1–5 were excluded from the analysis. Sequencing templates were obtained through PCR. After DNA amplification, the PCR products were purified with the QIAquick PCR Purification Kit (QIAGEN). Cycle sequencing was performed with the use of the BigDye Terminator Cycle Sequencing Kit with Amplitaq DNA polymerase (Applied Biosystems). Sequencing products were purified by ethanol precipitation and run on an ABI Prism 3730XL DNA sequencer (Applied Biosystems). Sequencing chromatograms were aligned and analyzed for mutations with the use of Sequencher 4.8 (Gene Codes). Length variations at both microsatellites and homopolymeric tracts were not considered for further analysis.
We detected a total of 146 biallelic MSY variants, including eight indel polymorphisms 1–12 bases long and 138 single-nucleotide substitutions (Table S2). Similar to previous reports,17,18 transitions were about twice as frequent as transversions (93 versus 45). The observed nucleotide diversity was high (π = 2.1 × 10−4 ± 0.3 × 10−4) as compared to the values reported for regions not involved in gene conversion in previous MSY resequencing studies (π in the range of 0.9 × 10−4 to 1.5 × 10−4).5,9,19
We used the 146 identified DNA variants and the MEGA 5 software to infer a maximum parsimony (MP) phylogeny of the human MSY. Previously reported phylogenetic relationships among the seven Y chromosomes resequenced in the present study were not taken into account. We obtained a strict-consensus MP tree, which was rooted with respect to either orthologous chimp MSY sequence (131 sites on the X-degenerated MSY) or paralogous human X chromosome sequence (15 sites on the X-transposed MSY), as reported in the UCSC Genome Browser (March 2006 chimp assembly and February 2009 human assembly, respectively). The resulting MSY tree is shown in Figure 1. The deepest branching separates A1b from a monophyletic clade whose members (A1a, A2, A3, B, C, and R) all share seven mutually reinforcing derived mutations (five transitions and two transversions, all at non-CpG sites). To retain the information from the reference MSY tree13 as much as possible, we named this clade A1a-T (Figure 1). Within A1a-T, the transversion V221 separates A1a from a monophyletic clade (called A2-T) consisting of three branches: A2, A3, and BT, the latter being supported by ten mutations (Figure 1).
Figure 1.
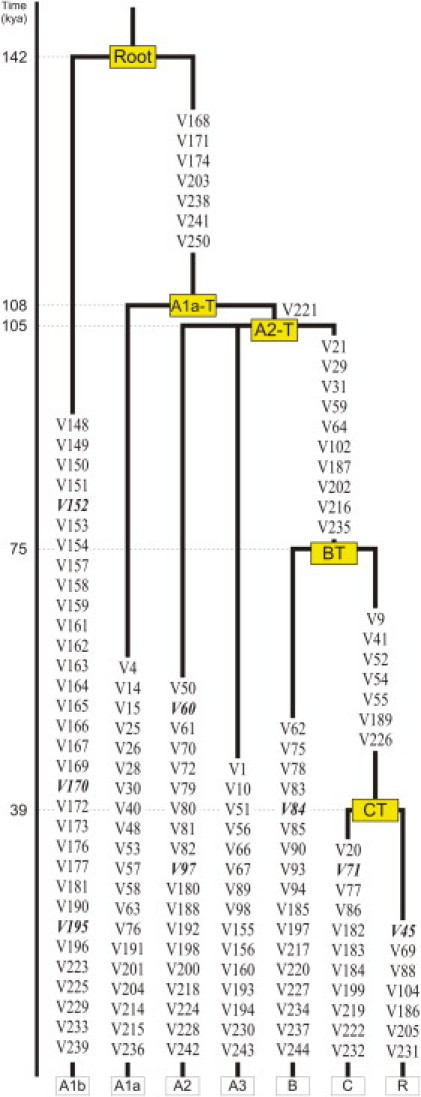
Revised Human Y Chromosome Phylogeny
The tree is based on the 146 mutations detected in the present study. Mutations are shown along the branches (indels in italics). Branch lengths are proportional to the rho estimates of the time to the MRCA (TMRCA; estimates to the left). The nodes are shown in yellow. The sequenced chromosomes are indicated at the bottom (see also Table S1). Haplogroup nomenclature follows Karafet et al.13 However, the revised tree required the introduction of new names for higher-order haplogroups (A1a-T and A2-T; see main text).
To estimate the age of ancestral nodes in the tree, we used the rho statistic,20,21 considering a germline MSY mutation rate of 1.0 × 10−9 single-nucleotide substitutions (SNS) per base per year.22 Indel variants were excluded from this calculation. We obtained a time estimate for the root of the MSY tree of 141.5 ± 15.6 KY, with an age in mutations (rho) of 29.1 ± 3.2 and values of 107.6 ± 12.2 KY, 104.9 ± 13.1 KY, 74.5 ± 12.5 KY, and 38.8 ± 9.7 KY for the coalescence age of A1a-T, A2-T, BT, and CT, respectively (Figure 1 and Table S3). We compared the rho estimates with the maximum likelihood (ML) estimates obtained with PAML 4.4. The differences between the rho and ML estimators of the TMRCA were minor (<8%; Table S3) for the root and major clades (A1a-T and A2-T).
How does the present MSY tree compare with the backbone of the recently published “reference” MSY phylogeny?13 The phylogenetic relationships we observed among chromosomes belonging to haplogroups B, C, and R are reminiscent of those reported in the tree by Karafet et al.13 These chromosomes belong to a clade (haplogroup BT) in which chromosomes C and R share a common ancestor (Figure 2). However, when the deepest branches of the two phylogenies are considered, important differences emerge. Chromosomes A1a and A1b appear to be separated at the root of the present tree, rather than being connected as in the reference phylogeny (Figure 2). Moreover, haplogroups A2 and A3 form a trifurcating clade together with BT in the present phylogeny, implying that “haplogroup” A can no longer be recognized as a monophyletic group of lineages.
Figure 2.
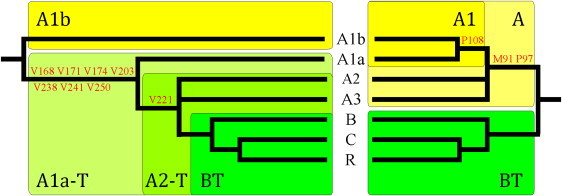
Comparison of Human Y Chromosome Phylogenies
Backbones of the MSY phylogeny as reported in the present study (to the left) and by Karafet et al.13 (to the right). Only mutations that identify “conflicting” clades between phylogenies are shown (in red).
Three key mutations define haplogroups A (M91 and P97) and A1 (P108) (Figure 2) in the phylogeny by Karafet et al.13 Regarding the poly-T marker M91, the chimp-sequence-based inferred polarity of the mutation is formally correct (T9 is the ancestral allele). However, as shown by a comparative analysis of human and chimp sequences, homopolymeric stretches are prone to recurrent mutation,23 suggesting some caution when the ancestral or derived state of a length polymorphism is inferred by interspecies comparison. By comparing a human sequence of 500 kb MSY flanking the M91 site to the orthologous sequence in the chimp, we have observed that less than one-third of the 9-Ts homopolymeric stretches have retained the same length in the two species (15/56 human versus chimp, 15/53 chimp versus human, see Table S4), which is consistent with the hypothesis that this marker might be evolutionarily unstable. Concerning SNPs P97 and P108, the inspection of the chimp reference sequence (March 2006 chimp assembly, UCSC Genome Browser) would indicate that the derived allele of these markers should be placed at the root of the BT and A2-T clades, respectively, rather than A and A1. We confirmed this to be the case by resequencing the relevant orthologous regions in three unrelated chimps. Thus, P108 now supports the second branching in the tree, being phylogenetically equivalent to V221, whereas P97 and, possibly, M91 should be considered to be phylogenetically equivalent to the markers that define haplogroup BT.
Haplogroup A1b was first reported by Karafet et al.13 as being defined by marker P114. However, no information is available on the geographic distribution of this haplogroup, recognized as one of the two deepest clades in the present phylogeny. To fill this gap in our knowledge and shed light on the phylogeography of the deepest branches (haplogroups A1b, A1a, and A2-T) of the revised tree, we considered our sample of 2204 African subjects (Figure S1 and Table S5). Chromosomes that were not previously assigned to a specific Y haplogroup24–28 were analyzed by DHPLC for the mutation defining haplogroup A2-T (V221; Figure 1 and Table S2). All but eight chromosomes were shown to carry the derived allele at V221 and were therefore classified as A2-T. The eight chromosomes carrying the ancestral allele at V221 were further analyzed for markers defining haplogroups A1a (V4, V14, V25, and M31) and A1b (V164, V166, V196, and P114). Four subjects (two Berbers from northwest Africa, one Tuareg and one Fulbe from Niger) were confirmed as belonging to clade A1a.24,29 It is worth noting that this clade was previously detected in west Africa, although at low frequencies.10,30–32 Three chromosomes from the Bakola pygmy group from southern Cameroon (central Africa) were found to carry the derived allele at V164, V166, V196, and P114 and were classified as A1b. Interestingly, one chromosome from an Algerian Berber group (northwest Africa) was found to carry the derived allele at V164, V166, and V196 but carried the ancestral one at P114, implying a bipartite structure for A1b, where P114 defines an internal node.
Three features of our data are of particular interest. First, the branching order at the deepest points of the tree is different from that previously recognized. The root of the tree now falls between A1b and A1a-T, and the number of deep branchings leading to African-specific clades has doubled (Figure 2), providing further strong support for the MSY-based evidence of a modern human origin in the African continent.
Second, the MSY tree is deeper than previously believed. The present figure of about 140 KY for the inferred most recent common ancestor (MRCA) of the MSY phylogeny is older than previous estimates (about 100 KY or below)33–35 and easier to reconcile with plausible scenarios of modern human origin.36 Clearly, calculation of the precise age of the tree largely depends on the accuracy of the assumed mutation rate. In any case, an antiquity of the root greater than that previously estimated is evident from the present tree structure. It is worth noting that A1b, long neglected in previous large-scale resequencing studies of the MSY, contributes to the older TMRCA and high nucleotide diversity values that we observe, highlighting the importance of targeted studies on rare haplogroups.
Third, contrary to previous phylogeny-based conclusions,15,16 the deepest clades of the revised MSY phylogeny are currently found in central and northwest Africa. MSY lineages from these regions coalesce at an older time (142 KY) than do those from east and south Africa (105 KY), opening new perspectives concerning early modern human evolution. A scenario of a Y chromosome “Adam” living in central-northwest Africa about 140 KY ago would provide a good fit to the present data. However, we also note that, because of the still largely incomplete geographic coverage of the African MSY diversity and unknown consequences of past population processes such as growth, extinction, and migration, any phylogeny-based inference on the geographical origin of human MSY diversity in Africa should be made with caution. Additional Y chromosome data and future discoveries in other disciplines are required in order to provide crucial information in support of the proposed scenario. Interestingly, there is an accumulation of a growing body of evidence that indicates that African regions that have been long neglected in studies on the origin of Homo sapiens may have been important early sites of modern human occupation, possibly connected to other areas of the continent by routes that are hidden today (see 37 and references therein).
In conclusion, we present here a Y chromosome phylogenetic tree deeply revised in its root and earliest branches. Our data do not uphold previous models of Y chromosomal emergence15,16 and demand a reevaluation of some fundamental ideas concerning the early history of the human genetic diversity we find today.38–40 Our phylogeny shows a root in central-northwest Africa. Although this point requires further attention, we think that it offers a new prospect from which to view the initial development of our species in Africa.
Acknowledgments
We thank Gabriella Spedini for collecting the Bakola Pygmy samples from Cameroon and A. Achilli for helpful advice on maximum likelihood analysis. This research received support from the Italian Ministry of Education, University and Research (grant “Progetti di Ricerca di Interesse Nazionale”), the Sapienza Università di Roma (Ricerche Universitarie) (both to R.S.), and the Ateneo Federato della Scienza e della Tecnologia (grant “Progetti AST”) (to F.C.).
Contributor Information
Fulvio Cruciani, Email: fulvio.cruciani@uniroma1.it.
Rosaria Scozzari, Email: rosaria.scozzari@uniroma1.it.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
UCSC Genome Browser, http://genome.ucsc.edu/
Accession Numbers
The dbSNP accession numbers for the 146 biallelic variants reported in this paper are ss410064561–ss410064706.
References
- 1.Rozen S., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 2.Bosch E., Hurles M.E., Navarro A., Jobling M.A. Dynamics of a human interparalog gene conversion hotspot. Genome Res. 2004;14:835–844. doi: 10.1101/gr.2177404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosser Z.H., Balaresque P., Jobling M.A. Gene conversion between the X chromosome and the male-specific region of the Y chromosome at a translocation hotspot. Am. J. Hum. Genet. 2009;85:130–134. doi: 10.1016/j.ajhg.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruciani F., Trombetta B., Macaulay V., Scozzari R. About the X-to-Y gene conversion rate. Am. J. Hum. Genet. 2010;86:495–497. doi: 10.1016/j.ajhg.2010.01.033. author reply 497–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trombetta B., Cruciani F., Underhill P.A., Sellitto D., Scozzari R. Footprints of X-to-Y gene conversion in recent human evolution. Mol. Biol. Evol. 2010;27:714–725. doi: 10.1093/molbev/msp231. [DOI] [PubMed] [Google Scholar]
- 6.Repping S., Korver C.M., Oates R.D., Silber S., van der Veen F., Page D.C., Rozen S. Are sequence family variants useful for identifying deletions in the human Y chromosome? Am. J. Hum. Genet. 2004;75:514–517. doi: 10.1086/423394. author reply 517–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Repping S., van Daalen S.K., Brown L.G., Korver C.M., Lange J., Marszalek J.D., Pyntikova T., van der Veen F., Skaletsky H., Page D.C., Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat. Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 8.Jobling M.A. Copy number variation on the human Y chromosome. Cytogenet. Genome Res. 2008;123:253–262. doi: 10.1159/000184715. [DOI] [PubMed] [Google Scholar]
- 9.Rozen S., Marszalek J.D., Alagappan R.K., Skaletsky H., Page D.C. Remarkably little variation in proteins encoded by the Y chromosome's single-copy genes, implying effective purifying selection. Am. J. Hum. Genet. 2009;85:923–928. doi: 10.1016/j.ajhg.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Underhill P.A., Shen P., Lin A.A., Jin L., Passarino G., Yang W.H., Kauffman E., Bonné-Tamir B., Bertranpetit J., Francalacci P. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 11.Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jobling M.A., Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 13.Karafet T.M., Mendez F.L., Meilerman M.B., Underhill P.A., Zegura S.L., Hammer M.F. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Underhill P.A., Kivisild T. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu. Rev. Genet. 2007;41:539–564. doi: 10.1146/annurev.genet.41.110306.130407. [DOI] [PubMed] [Google Scholar]
- 15.Hammer M.F., Karafet T.M., Redd A.J., Jarjanazi H., Santachiara-Benerecetti S., Soodyall H., Zegura S.L. Hierarchical patterns of global human Y-chromosome diversity. Mol. Biol. Evol. 2001;18:1189–1203. doi: 10.1093/oxfordjournals.molbev.a003906. [DOI] [PubMed] [Google Scholar]
- 16.Semino O., Santachiara-Benerecetti A.S., Falaschi F., Cavalli-Sforza L.L., Underhill P.A. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am. J. Hum. Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nachman M.W., Crowell S.L. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebersberger I., Metzler D., Schwarz C., Pääbo S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen P., Wang F., Underhill P.A., Franco C., Yang W.H., Roxas A., Sung R., Lin A.A., Hyman R.W., Vollrath D. Population genetic implications from sequence variation in four Y chromosome genes. Proc. Natl. Acad. Sci. USA. 2000;97:7354–7359. doi: 10.1073/pnas.97.13.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forster P., Harding R., Torroni A., Bandelt H.J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- 21.Saillard J., Forster P., Lynnerup N., Bandelt H.J., Nørby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am. J. Hum. Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y., Wang Q., Long Q., Ng B.L., Swerdlow H., Burton J., Skuce C., Taylor R., Abdellah Z., Zhao Y. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr. Biol. 2009;19:1453–1457. doi: 10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelkar Y.D., Tyekucheva S., Chiaromonte F., Makova K.D. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruciani F., Santolamazza P., Shen P., Macaulay V., Moral P., Olckers A., Modiano D., Holmes S., Destro-Bisol G., Coia V. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am. J. Hum. Genet. 2002;70:1197–1214. doi: 10.1086/340257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruciani F., La Fratta R., Santolamazza P., Sellitto D., Pascone R., Moral P., Watson E., Guida V., Beraud Colomb E., Zaharova B. Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am. J. Hum. Genet. 2004;74:1014–1022. doi: 10.1086/386294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruciani F., La Fratta R., Trombetta B., Santolamazza P., Sellitto D., Beraud Colomb E., Dugoujon J.M., Crivellaro F., Benincasa T., Pascone R. Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol. Biol. Evol. 2007;24:1300–1311. doi: 10.1093/molbev/msm049. [DOI] [PubMed] [Google Scholar]
- 27.Cruciani F., Trombetta B., Sellitto D., Massaia A., Destro-Bisol G., Watson E., Beraud Colomb E., Dugoujon J.M., Moral P., Scozzari R. Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur. J. Hum. Genet. 2010;18:800–807. doi: 10.1038/ejhg.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruciani F., Trombetta B., Sellitto D., Massaia A., Destro-Bisol G., Watson E., Beraud Colomb E., Dugoujon J.M., Moral P., Scozzari R. Chadic languages and Y haplogroups. Reply to Lancaster. Eur. J. Hum. Genet. 2010;18:1186–1187. doi: 10.1038/ejhg.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King T.E., Parkin E.J., Swinfield G., Cruciani F., Scozzari R., Rosa A., Lim S.K., Xue Y., Tyler-Smith C., Jobling M.A. Africans in Yorkshire? The deepest-rooting clade of the Y phylogeny within an English genealogy. Eur. J. Hum. Genet. 2007;15:288–293. doi: 10.1038/sj.ejhg.5201771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonçalves R., Rosa A., Freitas A., Fernandes A., Kivisild T., Villems R., Brehm A. Y-chromosome lineages in Cabo Verde Islands witness the diverse geographic origin of its first male settlers. Hum. Genet. 2003;113:467–472. doi: 10.1007/s00439-003-1007-4. [DOI] [PubMed] [Google Scholar]
- 31.Wood E.T., Stover D.A., Ehret C., Destro-Bisol G., Spedini G., McLeod H., Louie L., Bamshad M., Strassmann B.I., Soodyall H., Hammer M.F. Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes. Eur. J. Hum. Genet. 2005;13:867–876. doi: 10.1038/sj.ejhg.5201408. [DOI] [PubMed] [Google Scholar]
- 32.Rosa A., Ornelas C., Jobling M.A., Brehm A., Villems R. Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective. BMC Evol. Biol. 2007;7:124. doi: 10.1186/1471-2148-7-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson R., Pritchard J.K., Shen P., Oefner P.J., Feldman M.W. Recent common ancestry of human Y chromosomes: evidence from DNA sequence data. Proc. Natl. Acad. Sci. USA. 2000;97:7360–7365. doi: 10.1073/pnas.97.13.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H., Siegmund D.O., Shen P., Oefner P.J., Feldman M.W. Frequentist estimation of coalescence times from nucleotide sequence data using a tree-based partition. Genetics. 2002;161:447–459. doi: 10.1093/genetics/161.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilder J.A., Mobasher Z., Hammer M.F. Genetic evidence for unequal effective population sizes of human females and males. Mol. Biol. Evol. 2004;21:2047–2057. doi: 10.1093/molbev/msh214. [DOI] [PubMed] [Google Scholar]
- 36.Blum M.G., Jakobsson M. Deep divergences of human gene trees and models of human origins. Mol. Biol. Evol. 2011;28:889–898. doi: 10.1093/molbev/msq265. [DOI] [PubMed] [Google Scholar]
- 37.Balter M. Was North Africa the launch pad for modern human migrations? Science. 2011;331:20–23. doi: 10.1126/science.331.6013.20. [DOI] [PubMed] [Google Scholar]
- 38.Behar D.M., Villems R., Soodyall H., Blue-Smith J., Pereira L., Metspalu E., Scozzari R., Makkan H., Tzur S., Comas D. The dawn of human matrilineal diversity. Am. J. Hum. Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.M., Doumbo O. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henn B.M., Gignoux C.R., Jobin M., Granka J.M., Macpherson J.M., Kidd J.M., Rodríguez-Botigué L., Ramachandran S., Hon L., Brisbin A. From the Cover: Feature Article: Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


