Abstract
We describe Abacus, a computational tool for extracting spectral counts from tandem mass spectrometry based proteomic datasets. The program aggregates data from multiple experiments, adjusts spectral counts to accurately account for peptides shared across multiple proteins, and performs common normalization steps. It can also output the spectral count data at the gene level, thus simplifying the integration and comparison between gene and protein expression data. Abacus is compatible with the widely used Trans-Proteomic Pipeline suite of tools and comes with a graphical user interface making it easy to interact with the program. The main aim of Abacus is to streamline the analysis of spectral count data by providing an automated, easy to use solution for extracting this information from proteomic datasets for subsequent, more sophisticated statistical analysis.
Keywords: Label free quantification, spectral counts, software, tandem mass spectrometry, protein inference, shared peptides
In the last several years, label-free mass spectrometry (MS) based protein quantification methods have received significant attention and become commonly used [1–3]. Label-free approaches offer several practical advantages over generally more accurate labeling-based methods. They often offer savings in terms of costs of the analysis, are easier to implement, and allow for complex experimental designs unlike labeling-based methods where comparisons can be made for a limited number of samples.
One of the most commonly used label-free quantitation methods is spectral counting. In this approach, the number of tandem mass (MS/MS) spectra assigned to the peptides of a protein, after a proper normalization, is used to measure the protein’s abundance in the sample [4, 5], for a recent review see e.g. [3]. A number of statistical approaches and software tools have been described for assessing the significance of differential protein expression based on spectral count data [6–13], including our previously described program Qspec [7]. These programs take as input a spectral counts matrix (i.e., a table listing all proteins identified with high confidence and the corresponding spectral counts in each of the experiments) extracted from the MS data. While conceptually simple, accurate extraction of spectral counts and their use as a measure of the protein abundance nevertheless requires addressing several challenges. First, the analysis naturally involves processing of multi-sample datasets (biological and technical replicates, multiple cell lines or patient samples, etc). The accession numbers of proteins identified in different samples need to be aligned across the experiments to create a single protein summary list. This task is complicated due to the ambiguities in inferring protein identifies from shotgun proteomic data - the protein inference problem [14]. Directly related to this issue is the uncertainty with the contribution from shared peptides, i.e. peptides present in multiple different proteins, to the spectral count of each their corresponding proteins [7, 15, 16]. Furthermore, protein-level data often need to be matched to genomic data, which requires mapping of protein to gene accession numbers.
In this work we describe “Abacus” – a software tool for extracting spectral counts that is compatible with the Trans-Proteomic Pipeline suite of computational tools [17]. The overview of Abacus is shown in Figure 1. Abacus takes as input PeptideProphet [18] and ProteinProphet [19] output files (in pepXML and protXML format, respectively). The program aggregates data from multiple experiments, adjusts spectral counts based upon how the peptides are shared among the proteins reported in the experimental results, and performs common normalization steps. In order to use Abacus it is necessary to generate a list of all the proteins that were identified from all of the individual mass spectrometry experiments. This is achieved by using ProteinProphet (by the user and prior to running Abacus) to create a combined (“merged”) protXML file from the peptide-level results of each independent experiment. The resulting combined file more accurately represents the protein-level identifications across all experiments. After a combined protXML file is obtained, Abacus parses the individual pepXML and protXML result files storing their respective data into an internal database. Abacus then performs the following steps to arrive at the final results it reports:
Filter out low scoring proteins from the combined file.
Select representative protein identifiers from the combined file.
Collect summary information about the representative proteins from among independent experiments.
Calculate peptide/spectral counts for each protein.
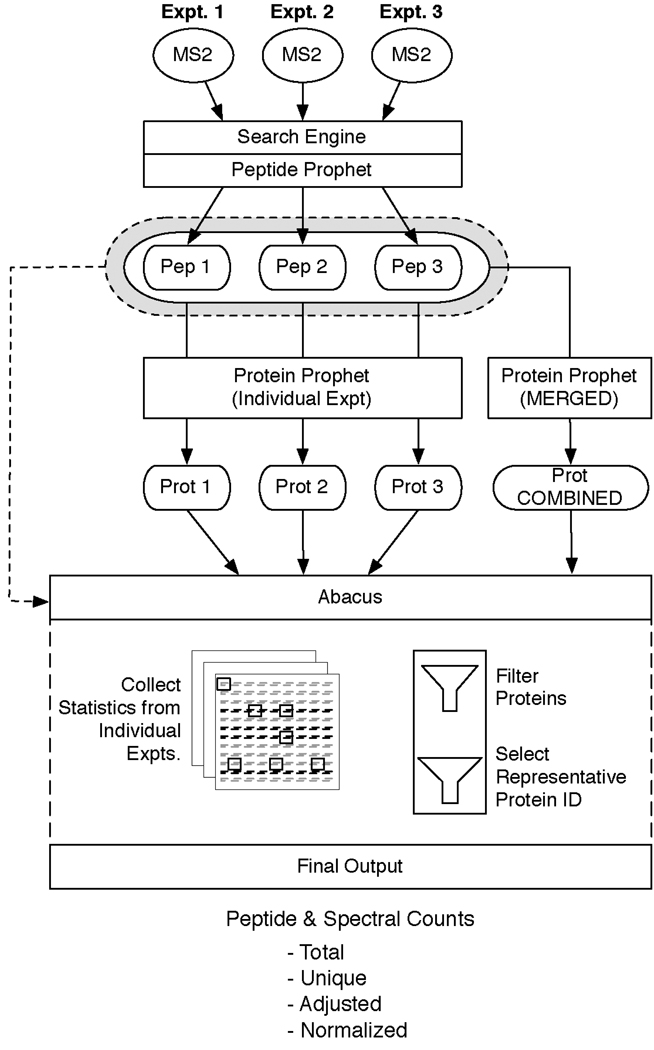
Figure 1. Overview of Abacus.
Schematic representation of the workflow including processing of the data through Abacus. In this illustration three individual MS experimental data sets were searched with a standard protein search engine (i.e.: SEQUEST, Mascot, etc.) and then post processed using PeptideProphet. The resulting pepXML files (Pep1, Pep2, and Pep3) were then processed through ProteinProphet to create three individual protXML files (Prot1, Prot2, and Prot3). The pepXML files were also processed together through ProteinProphet to merge all of their results into a single, combined protXML file (Prot COMBINED). All of the pepXML and protXML files were then parsed into Abacus and filters were applied to the merged results to filter false positives and select a representative protein for each protein group. Statistics for the representative protein were then extracted from each of the individual protXML files and used to generate the final output that is produced by Abacus.
The first step in the Abacus algorithm is to remove low scoring protein identifications recorded in the combined file. Abacus allows filtering of protein-level identifications based upon several features: the ProteinProphet posterior protein probability from the combined file, the maximum protein probability observed across the individual experiments, as well as the maximum peptide probability observed for the protein. These parameters are adjustable allowing for precise control over what proteins are retained for subsequent analysis. It should be noted that in the context of this work merging of multiple MS runs implies that all of the individual results from each MS run are combined together regardless of how often a particular protein is identified across all the replicates. This merging of multiple MS run results tends to increase the number of false positives [20]. Since the probability-based estimates provided by ProteinProphet may not be accurate in the case of very large multi-replicate experiments, filtering can be performed in such a way as to achieve a desired false discovery rate (FDR) based on the target-decoy strategy [20, 21].
In the second step of the algorithm, Abacus chooses a single protein identifier to represent each of the remaining entries in the combined file. ProteinProphet collects proteins that share peptide evidence into protein groups [14]. When there is an ambiguity, Abacus selects a single representative identifier from within each of the protein groups of the combined file to report in the final output. This representative protein is chosen based upon the following hierarchy. The first two heuristics are applied across the independent experimental results. The last four are executed on the data within the protein group of the combined file. These heuristics are followed sequentially until any ties are broken or the last rule is reached: (1) The protein identified the most often among independent experimental results; (2) The protein with the highest protein probability among the independent experiments; (3) The protein with the highest scoring peptide assigned to it; (4) The protein with the most number of distinct peptide sequences matched to it; (5) The protein with the highest spectral count; (6) The top protein identifier after alphanumeric sorting of all remaining identifiers.
Having selected a representative protein accession number, the third step in the algorithm is to collect basic statistics about this protein from the results of the independent experiments. In addition to computing the total (i.e. regardless of whether the peptides are shared across multiple proteins) peptide and spectral counts for each protein, adjusted spectral counts are computed for the representative protein across each of the individual experiments. ProteinProphet already addresses the problem of shared peptides in the context of protein identification [14, 19] (and other approaches, e.g. [22–24]). In ProteinProphet, peptides shared across multiple proteins provide varying contributions (i.e. weight) to the protein’s final score. Peptides that are highly redundant contribute less, whereas peptides unique to a single protein are given more importance. In Abacus, the same framework is applied to the calculation of spectral counts, leading to a more realistic quantitative measurement of a protein’s abundance. A similar spectral count adjustment approach was investigated in [15], which showed that adjusting spectral counts based upon how shared peptides are distributed gave the best agreement between computationally and experimentally derived measurements of protein abundance.
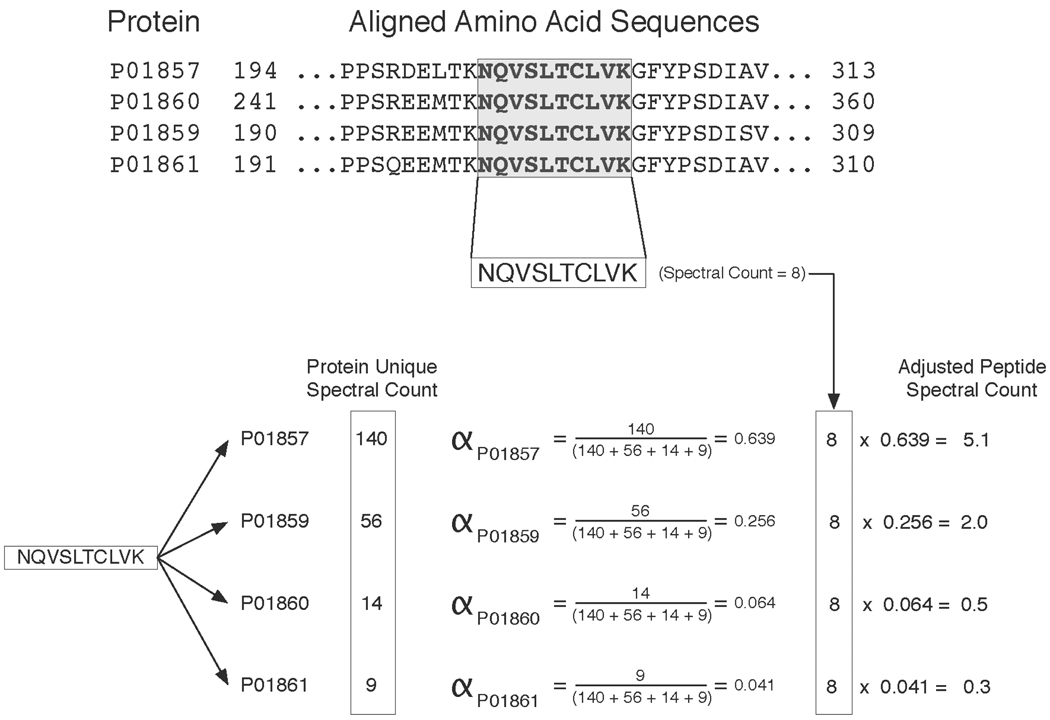
The adjustment procedure in Abacus is performed as follows. First, the number of unique spectra assigned to each protein, s, is calculated. For each peptide p present in multiple proteins j = 1…N, its contribution to the spectral count of protein i is weighted by an adjustment factor, αp,i:
Given this definition, α weights range from zero to one. In essence, α determines what proportion of the spectral counts from peptide p should be ascribed to protein i. The sum of the peptides’ adjusted counts constitutes the each protein’s adjusted spectral count. The calculation of alpha and how it is applied to a single peptide case is illustrated in Figure 2 using a subset of the prostatic secretion (EPS) dataset from 9 prostate cancer patients [25] (this dataset is also provided as a sample data along with the Abacus software). After analyzing the X! Tandem search results with PeptideProphet and ProteinProphet, Abacus was used to extract adjusted spectral counts for one of the nine patient samples. Figure 2 focuses on four related immunoglobulin proteins (IGHG1 through IGHG4) that were identified with high confidence in the sample. These homologous proteins share numerous peptides in common. Figure 2 demonstrates how the spectral count of a single one of these common peptides is distributed among these proteins. Following spectral count adjustment, IGHG1 protein is assigned 5 of the peptide’s 8 spectral counts. The remaining three spectra are assigned to IGHG2 and IGHG3 respectively. IGHG4 is not assigned any of the spectra from this peptide. These adjusted counts are based upon the unique spectral evidence ascribed to each of the proteins independently as described above. Without spectral adjustment, all 8 of the peptide’s spectra would have contributed equally to each protein’s final spectral count.
Figure 2. Illustration of the spectral count adjustment procedure.
Example of how the alpha factor is calculated for a single peptide shared among 4 immunoglobulins (IGHG1 through IGHG4) identified in the EPS data set. The upper panel shows the protein sequence alignment between the 4 immunoglobulins and highlights a peptide common to all of them. This specific peptide has 8 unique spectra assigned to it making its spectral count 8. The lower panel shows how alpha is computed for each of the proteins that share this peptide. The alpha factor is based upon the unique spectral count of each protein. In this example PO1857, PO1859, PO1860, PO1861 each have unique spectral counts of 140, 56, 14, and 9 respectively. These unique spectral counts are derived from the peptides that are exclusive to each of the proteins in the example. The peptide’s total spectral count of 8 is multiplied by the alpha factor of each protein. The resulting values indicate how the peptide’s 8 spectral counts are to be distributed among each of the 4 proteins.
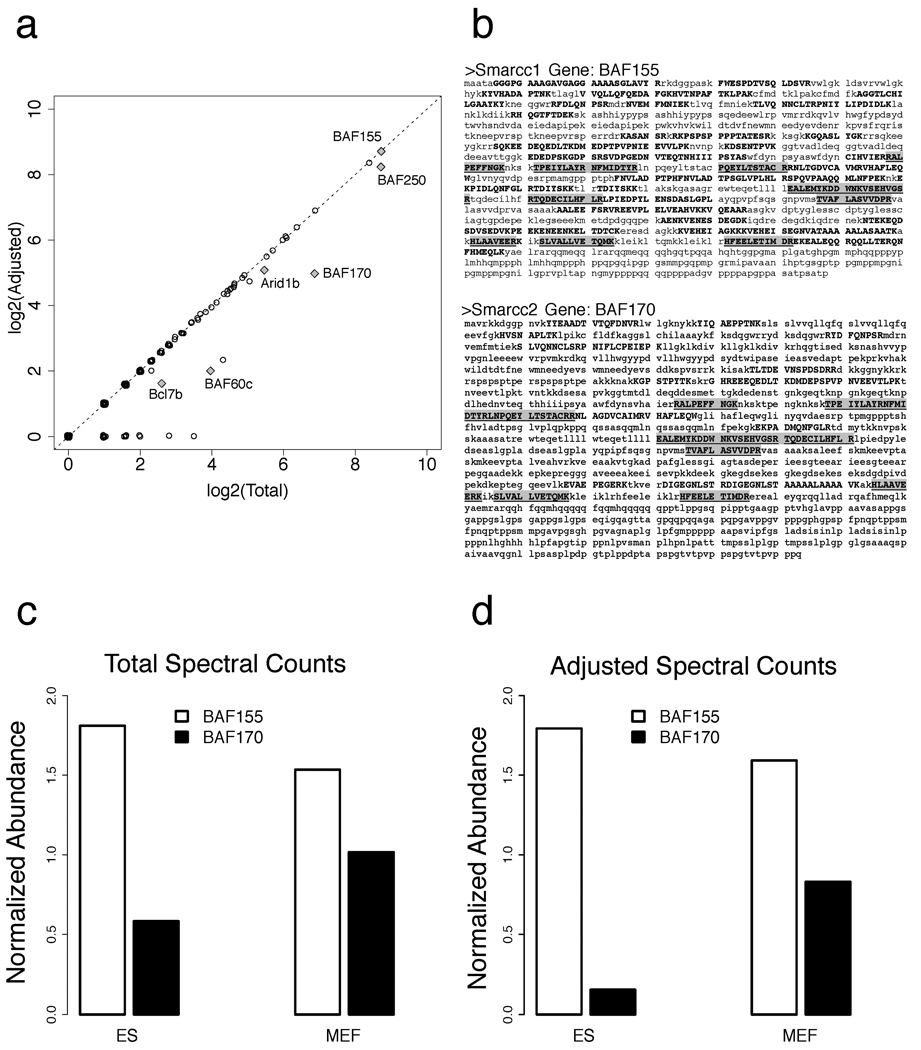
In some cases, the use of adjusted counts is helpful for achieving accurate biological interpretation. This is illustrated in Figure 3 using data from a recent study of an embryonic stem cell chromatin remodeling complex, esBAF [26]. In [26], the analysis was performed using a semi-manual spectral count adjustment procedure, for which Abacus now provides an automated software solution. The mammalian BAF (Brg/Brahma-associated factors) chromatin remodeling complexes play a key role in establishing and maintaining pluripotency. These complexes contain 11 core subunits, several of which are encoded by gene families. The diversity of BAF complexes is derived from the combinatorial assembly of alternative family members, some of which have a high degree of sequence homology to each other. As a result, accurate characterization of the composition of this protein complex requires appropriate adjustment of the spectral counts to account for the high number of shared peptides. Figure 3a shows the difference between the total and adjusted spectral counts for proteins identified in the analysis of the BAF complex in the mouse embryonic stem cells (ES). A number of core complex components, notably BAF170 (protein Smarcc2), have substantially reduced protein abundance after adjustment. This is further illustrated in Figure 3b, which shows the sequences of BAF170 and BAF155 (Smarcc1, 61.7% sequence homology with Smarcc2), and the peptides mapping to these two proteins identified from MS data. The comparison of the normalized abundances of these two proteins (NSAF factors) within the complex at two different stages of differentiations (ES and mouse embryonic fibroblast, MEF), computed using the spectral adjustment procedure is shown in Figure 3d. Of the most biological importance is the significant reduction of BAF170 in the BAF complex purified from ES cells, in agreement with quantitative immunobloting data and other evidence [26]. At the same time, the abundance of BAF170 in ES cells based on the total (unadjusted) counts is overestimated (Figure 3c). For example, without the adjustment the ratio of estimated BAF 155 vs. BAF 170 protein abundances in ES cells is ~ 3:1, compared with ~ 12:1 after the adjustment for shared peptides (the latter shows a much better agreement with quantitative immunobloting data shown in Figure 3c in [26]).
Figure 3. Comparison of total and adjusted spectral counts: application to BAF chromatin remodeling complex [26].
(a) The difference between the total and adjusted spectral counts for proteins identified in the analysis of the BAF complex in the mouse embryonic stem cells (ES). Selected core components of the complex most affected by the adjustment procedure are marked. (b) The sequences of two homologous proteins, BAF170 and BAF155, and the peptides mapping to these two proteins identified from MS data in the ES cells (identified peptide sequences are in bold, and those that are shared between the proteins are in grey boxes). (c) The normalized abundances of these two proteins (NSAF factors, additionally normalized to levels of Brg protein [26] in each cell line) within the complex at two different stages of differentiations (ES, MEF). (d) Same as (c), but using adjusted spectral counts.
Adjusted spectral counts allowed more accurate quantification in the study described above, and more accurate reconstruction of protein complexes from affinity purification-mass spectrometry (AP-MS) protein interaction data in another of our recent works [27]. At the same time, our experience with label-free quantification data suggests that different research questions warrant the use of different counts (or simultaneous use of multiple measures). While adjusted counts can be more informative than total counts for relative quantification of highly homologous proteins, they may underestimate the absolute protein abundances. For example, when using spectral count-based quantification measures as a basis for separating between true and false protein interactions using the SAINT statistical model [28, 29], we routinely utilize total peptide or spectral counts (instead of adjusted counts) in order to perform more conservative assessments and to eliminate non-specific binders. Thus, Abacus reports a number of different abundance metrics, including adjusted, total, and unique counts, for spectra and peptides, as well as normalized spectral abundance factors, NSAF [30]. It is worth noting that similar challenges of dealing with shared sequence counts are present in other data, most notably RNA-Seq data (‘multiread’ counts), see e.g. [31, 32].
Abacus can also provide counts (both spectral and peptide) at the gene level. The members of a protein group are often related isoforms derived from the same gene locus. For gene-centric consolidation, proteins are mapped to their parent gene. Once proteins have been mapped, their peptides are then associated with the parent gene. Spectral counts are still adjusted as described above except that alpha factors are computed based upon genes not proteins. The genes are assigned the maximum protein probability reported from among their observed protein products in the combined file. This gene-centric output is often useful for performing quantitative comparisons between protein and gene expression data. As next-generation sequencing methods become more established, such comparisons will become more popular [33–35]. It must be emphasized that mapping proteins back to their parent gene loci is not a trivial task and is an on-going challenge. For this reason, Abacus requires the user to provide a gene-to-protein mapping file. Such files can be easily generated for the public databases and we provide example programs in the Supplementary Materials.
A key aim in the development of Abacus was making it user friendly. Abacus comes with a graphical user interface making it easy to interact with the program (See Supplementary Materials for a detailed description of the software). This interface allows users to easily apply filters and choose what information is reported. The flexibility provided by Abacus is one of its strongest features. The user is given a great deal of control in deciding what to report as well as how the data is filtered. Currently there are few other open source tools that provide a platform independent method to extract spectral counts from proteomics data sets in a user-friendly manner. In its simplest usage, Abacus can provide summary statistics for a large collection of experiments that would otherwise be too complicated to manage. An option for creating a QSpec-compatible [7] output format is available, which simplifies the subsequent statistical analysis of differential expression. Should the existing options be insufficient, users can directly query the database that holds all the information for their data. Abacus uses the HyperSQL database as a back end to store and query the information it extracts from the pepXML and protXML files [36]. Having this database distributed with Abacus allows users to directly access their data in a robust relational database should the default output of Abacus not fulfill their needs. Abacus is written in JAVA and has been tested to verify reproducible results on Windows, Linux, and MacOS X platforms. The software is open-source and distributed under the Apache License 2.0. The software, source code, documentation and sample data are available at http://abacustpp.sourceforge.net.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants R01-CA-126239 and R01-GM-094231. We would like to thank Lena Ho, Jeff Ranish, Hyungwon Choi, and Dattatreya Mellacheruvu for helpful discussions.
REFERENCES
- 1.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44:1637–1660. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 2.Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lundgren DH, Hwang SI, Wu LF, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 4.Liu HB, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical Chemistry. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 5.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Molecular & Cellular Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho PC, Fischer JS, Chen EI, Yates JR, 3rd, Barbosa VC. PatternLab for proteomics: a tool for differential shotgun proteomics. BMC Bioinformatics. 2008;9:316. doi: 10.1186/1471-2105-9-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinecke NL, Pratt BS, Vaisar T, Becker L. PepC: Proteomics software for identifying differentially expressed proteins based on spectral counting. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Gray W, Zhang HX, Chung CH, Billheimer D, Yarbrough WG, Liebler DC, Shyr Y, Slebos RJC. Comparative Shotgun Proteomics Using Spectral Count Data and Quasi-Likelihood Modeling. J. Proteome Res. 2010;9:4295–4305. doi: 10.1021/pr100527g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little KM, Lee JK, Ley K. ReSASC: A resampling-based algorithm to determine differential protein expression from spectral count data. Proteomics. 2010;10:1212–1222. doi: 10.1002/pmic.200900328. [DOI] [PubMed] [Google Scholar]
- 11.Pham TV, Piersma SR, Warmoes M, Jimenez CR. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics. 2010;26:363–369. doi: 10.1093/bioinformatics/btp677. [DOI] [PubMed] [Google Scholar]
- 12.Searle BC. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. PROTEOMICS. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 13.Pavelka N, Fournier ML, Swanson SK, Pelizzola M, Ricciardi-Castagnoli P, Florens L, Washburn MP. Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Molecular & Cellular Proteomics. 2008;7:631–644. doi: 10.1074/mcp.M700240-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 16.Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting Signals, N-Terminal Modifications and Abundance of the Chloroplast Proteome. PLoS One. 2008;3:19. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. A guided tour of the Trans-Proteomic Pipeline. PROTEOMICS. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 19.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. Journal of Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 22.Li YF, Arnold RJ, Li Y, Radivojac P, Sheng Q, Tang H. A bayesian approach to protein inference problem in shotgun proteomics. J Comput Biol. 2009;16:1183–1193. doi: 10.1089/cmb.2009.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Zimmerman LJ, Park BH, Tabb DL, Liebler DC, Zhang B. Network-assisted protein identification and data interpretation in shotgun proteomics. Mol Syst Biol. 2009;5:303. doi: 10.1038/msb.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerster S, Qeli E, Ahrens CH, Buhlmann P. Protein and gene model inference based on statistical modeling in k-partite graphs. Proc Natl Acad Sci U S A. 2010;107:12101–12106. doi: 10.1073/pnas.0907654107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake RR, Elschenbroich S, Lopez-Perez O, Kim Y, Ignatchenko V, Ignatchenko A, Nyalwidhe JO, Basu G, Wilkins CE, Gjurich B, Lance RS, Semmes OJ, Medin JA, Kislinger T. In-depth proteomic analyses of direct expressed prostatic secretions. J Proteome Res. 2010;9:2109–2116. doi: 10.1021/pr1001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi H, Kim S, Gingras AC, Nesvizhskii AI. Analysis of protein complexes through model-based biclustering of label-free quantitative AP-MS data. Mol. Syst. Biol. 2010;6:11. doi: 10.1038/msb.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi H, Larsen B, Lin Z-Y, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras A-C, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Meth. 2010 doi: 10.1038/nmeth.1541. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin Z-Y, Breitkreutz B-J, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras A-C, Nesvizhskii AI, Tyers M. A Global Protein Kinase and Phosphatase Interaction Network in Yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 31.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenbaum D, Jansen R, Gerstein M. Analysis of mRNA expression and protein abundance data: an approach for the comparison of the enrichment of features in the cellular population of proteins and transcripts. Bioinformatics. 2002;18:585–596. doi: 10.1093/bioinformatics/18.4.585. [DOI] [PubMed] [Google Scholar]
- 34.Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR., 3rd Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3107–3112. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, Khaitovich P. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.HyperSQ. HyperSQL Database Engine. 2010 ed.)^eds.). p. http://hsqldb.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.