Abstract
Tandem PDZ domains have been suggested to form structurally-independent supramodules. However, dissimilarity between crystallography and NMR models emphasize their malleable conformation. Studies in full length scaffold proteins are needed to examine the effect of tertiary interactions within their native context. Using single molecule fluorescence to characterize the N-terminal PDZ tandem in PSD-95, we provide the first direct evidence that PDZ tandems can be structurally-independent within a full-length scaffold protein. Molecular refinement using our data converged on a single structure with an antiparallel alignment of the ligand binding sites. Devoid of interaction partners, single molecule conditions captured PSD-95 in its unbound, ground state. Interactions between PDZ domains could not be detected while fluctuation correlation spectroscopy showed that other conformations are dynamically sampled. We conclude that ultra-weak interactions stabilize the conformation providing a “low-relief” energy landscape that allows the domain orientation to be flipped by environmental interactions.
Keywords: PSD-95, PDZ tandem, FRET, single molecule fluorescence, Isothermal Titration Calorimetry, Fluctuation Correlation Spectroscopy, PDZ domain
INTRODUCTION
Multidomain scaffold proteins are critical organizers of signal transduction and junctional communication (Pawson and Scott, 1997). Often these proteins contain a series of archetypal protein interaction domains connected by flexible linkers into a larger protein. By physically colocalizing proteins, scaffolds increase the efficiency of signal transduction reactions (Garrington and Johnson, 1999). It is convenient to try and understand scaffold proteins by studying each individual domain as an isolated unit. However, dimerization and incorporation into larger proteins can alter the structure as well as the binding specificity relative to the isolated domain (Irie et al., 1997; Zhang et al., 2001; Peterson et al., 2004; Mishra et al., 2007; van den Berk et al., 2007; Iskenderian-Epps and Imperiali, 2010). Thus, studies of isolated domains can be only partially successful at explaining function in their biological context.
PDZ (PSD-95/Dlg/ZO-1) domains are the most common protein-interaction domains in the human genome (Bhattacharyya et al., 2006) and have been extensively reviewed (Harris and Lim, 2001). PDZ domains typically form part of a larger multidomain protein and often appear in tandem with instances of up to 13 PDZ domains in a single protein (Hung and Sheng, 2002). The structure of PDZ domains, with and without ligand, has been well characterized with over 200 PDZ domain structures in the Protein Data Bank.
PSD-95 was one of the first PDZ-containing proteins identified (Cho et al., 1992). PSD-95 contains three PDZ domains followed by an SH3 domain and an enzymatically inactive guanylate kinase-like domain. PSD-95 is one of four members of the membrane associated guanylate kinase (MAGUK) family of proteins that act as scaffolds for signal transduction in the postsynaptic density of excitatory synapses (Kim and Sheng, 2004). The first two PDZ domains are connected by a five residue linker. Conjoined PDZ domains are often connected to each other by relatively short polypeptide linkers. Such tandem domain arrays are often conceptualized as folded beads on a disordered string. Most structural studies of PDZ tandems find few specific interdomain contacts but nonetheless conclude that interdomain mobility is limited (Kang et al., 2003; Long et al., 2003; Cierpicki et al., 2005; Long et al., 2005; Goult et al., 2007; Sainlos et al., 2010). In contrast, the tandem PDZ domains from GRIP have extensive interdomain interactions that result in a fixed interdomain orientation (Feng et al., 2003; Long et al., 2008). The PDZ tandems of PSD-95 and syntenin have been examined separately with NMR and crystallography (Kang et al., 2003; Long et al., 2003; Cierpicki et al., 2005; Sainlos et al., 2010). In both cases, despite evidence for limited interdomain mobility in solution, the crystal structure differs substantially from the NMR model.
NMR studies suggested that the first two PDZ domains from PSD-95 form a “supramodule” with a limited freedom of rotation relative to each other (Long et al., 2003). NMR investigations of peptide binding to the PDZ tandem of PSD-95 found that interdomain orientation was altered by protein interactions (Wang et al., 2009). Most studies of PDZ tandems to date have examined fragments of much larger proteins. As domain orientation has been shown to be sensitive to protein interactions, the degree to which the structure of the PDZ tandem depends on the remainder of the protein is therefore unknown. Negative-stain electron microscopy of full-length PSD-95 revealed a molecule with the N and C termini “curled back on itself” (Nakagawa et al., 2004), in which the PDZ tandem could interact with other domains. In contrast, immunogold labeling suggested that PSD-95 adopts an extended conformation (Chen et al., 2008), in which intramolecular contacts could be limited.
The crystallization of flexible, multidomain scaffold proteins is challenging. The large molecular weight of such proteins often necessitates the use of truncated constructs. The limited interdomain contact provides few unambiguous NMR restraints. In contrast, fluorescence resonance energy transfer (FRET) allows for the accurate measurement of long intramolecular distances (3–8 nm) so it is particularly useful for studying the orientation of domains that are not in intimate contact. In addition, fluorescence measurements are not subject to molecular weight restrictions, which makes measurements in full-length scaffold proteins possible.
In this paper, we used single molecule fluorescence and other techniques to characterize the interdomain orientation of the first two PDZ domains in full-length PSD-95. We found that while the tandem PDZ domains have a limited range of relative motion, they are not fixed and dynamically sample other conformations on the 10−5 second timescale. By comparing measurements in full-length PSD-95 and truncated fragments, we show that domain orientation of the tandem fragment was identical to full-length PSD-95. This study offers the first structural model for tandem PDZ domains based on measurements made in a full-length scaffold protein and suggests a lack of tertiary interactions with other domains in PSD-95.
RESULTS
Orientation of the first two PDZ Domains in Full Length PSD-95
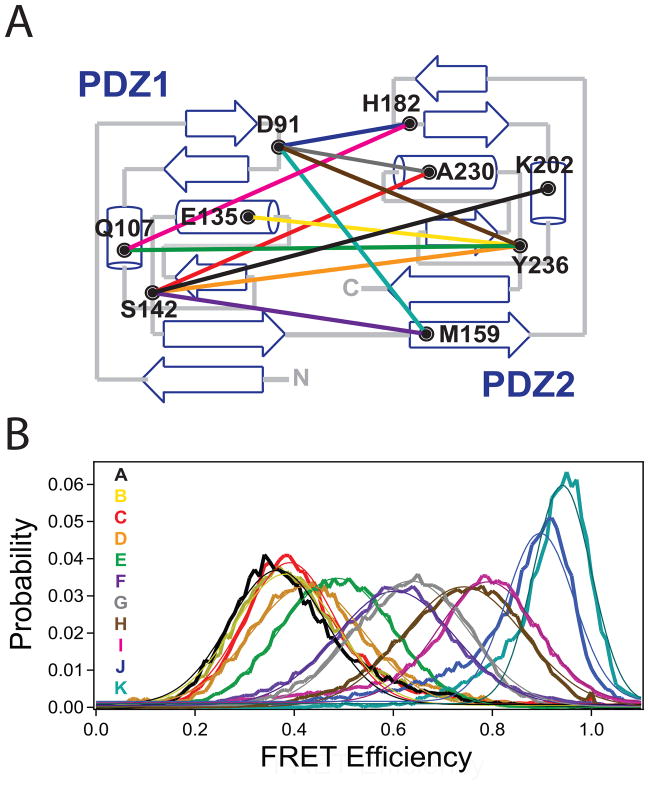
Using the high-resolution structures of the first two PDZ domains (Tochio et al., 2000; Piserchio et al., 2002; Long et al., 2003) four surface-exposed sites in PDZ1 and five in PDZ2 were selected for fluorescent labeling and mutated to cysteine (Figure 1A, Table 1). Accepted labeling sites showed maximal solvent exposure and minimal near neighbor interactions, which should minimize any positional dependence of the photophysical properties. We confirmed this expectation by measuring the anisotropy and quantum yield of the attached dyes, which showed minimal environmental impacts on fluorescence emission (Table 1). The donor anisotropy was similar for all samples and was minimally higher than that measured for unconjugated Alexa 555. Using the nine single labeling sites, eleven pairwise combinations were generated in full-length PSD-95 to create eleven double-cysteine mutants for FRET measurements (Figure 1A). This ensemble of labeling combinations was chosen to sample any possible orientation between these two domains.
Figure 1. Single Molecule FRET Measurements between PDZ1 and PDZ2 in Full Length PSD-95.
(A) Topology diagram showing the position of the cysteine labeling sites in each domain and the eleven combinations of labeling sites used for fluorescence studies. A representation of the labeling sites on the ribbon diagram of the two domains is shown in Figure S1. (B) smFRET histograms for all eleven FRET pairs made in full-length PSD-95. Letter codes (indicated within the panel) were assigned to each mutant in order of increasing FRET efficiency. The coloring of the FRET distributions corresponds to the colored lines denoting labeling combinations in panel A. Thin lines indicate the fit to a single Gaussian function. See also Figure S1.
Maleimide labeling of cysteine is well established but results in random incorporation of the donor and acceptor dyes at both sites. This is problematic for ensemble studies because the ratio of different species is not known. Here single molecule analysis was used to distinguish molecules containing two instances of the donor or acceptor dye from molecules containing one of each. The two double labeled species (DA and AD) are indistinguishable and were treated equally in the analysis. Thus, we used single molecule methods to tackle the experimental heterogeneity associated with sample production.
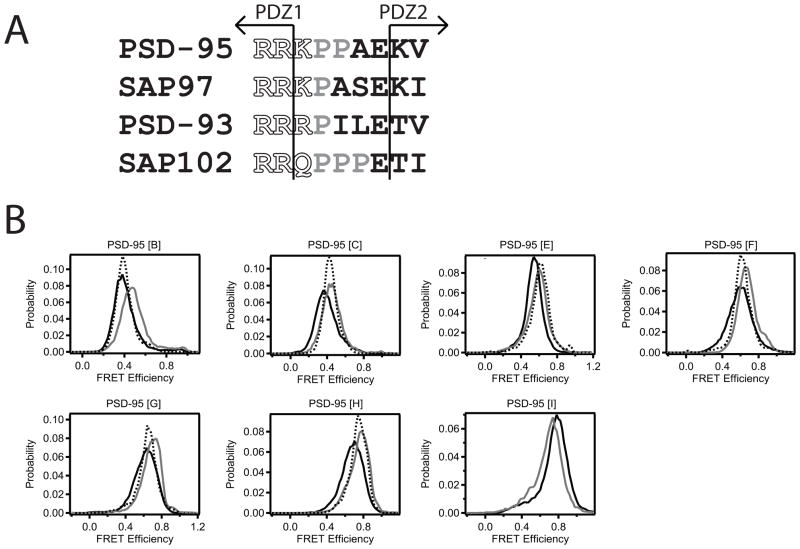
All eleven labeling combinations produced smFRET histograms containing a single peak of narrow width that was well described by a single Gaussian curve (Figure 1B, Table 2). The narrow widths are similar to those observed in duplex DNA and could either arise from a fixed interdomain orientation or time averaging of rapid motions. Paradoxically, these two extremes are indistinguishable in this assay. The different labeling site combinations showed a wide dispersion in their mean smFRET efficiencies (from 0.37 to 0.94), which would not be expected if the two domains are undergoing completely isotropic motion.
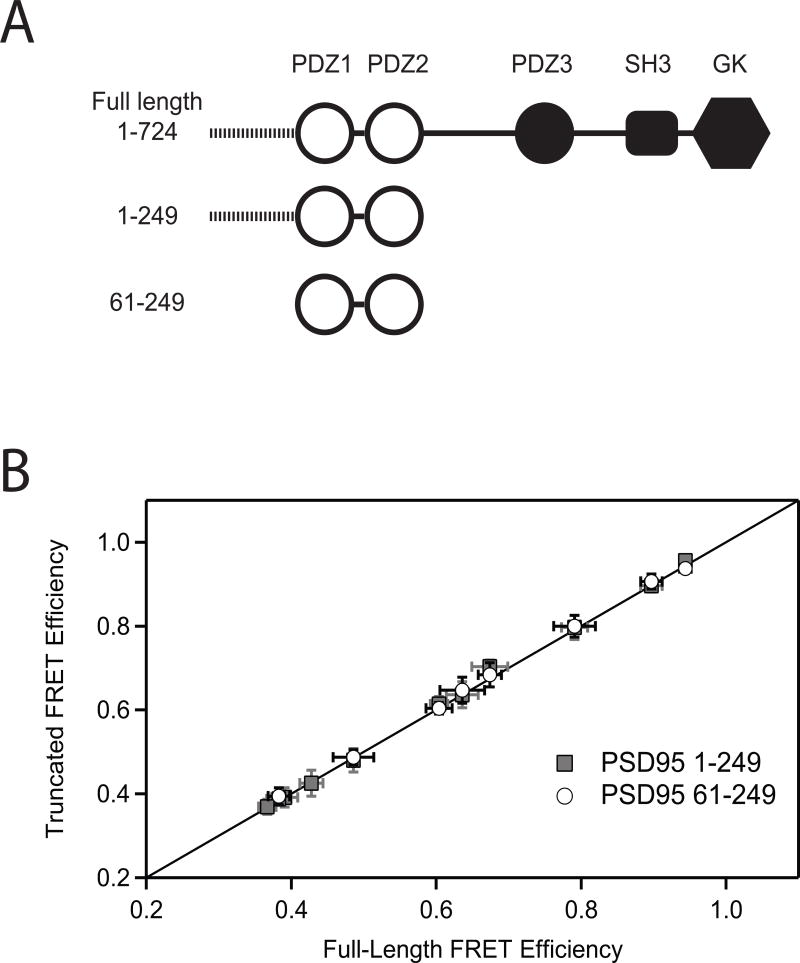
These are the first measurements made on the PDZ tandem in full-length PSD-95, which contains three other domains and a 60 residue disordered N-terminal region. To see if domain orientation is dependent on interdomain contacts within the full-length protein, we measured smFRET efficiencies in truncated PSD-95 constructs lacking the additional domains (Figure 2A). None of the labeling site combinations showed significant changes in mean FRET efficiency or the width of the FRET distribution relative to full length PSD-95 (Figure 2B, Table 2). Thus, domain orientation in the PDZ tandem was identical in the full length protein and truncated fragments. Given the sensitivity of domain orientation to protein interactions (Wang et al., 2009), this suggests a lack of intramolecular interactions with other domains present in full-length PSD-95.
Figure 2. Truncation of the PDZ Tandem Does Not Alter Domain Orientation.
(A) Cartoon representation of the constructs used in this experiment. Construct 1-724 is full length PSD-95. Construct 1-249 is truncated after PDZ2 but retains the 60 residue disordered N-terminal domain. Construct 61-249 encompasses only the PDZ tandem. (B) The mean smFRET efficiency measured in the two truncated fragments (y-axis) is plotted against the mean FRET efficiency between the same labeling sites in the full-length protein (x-axis). Mean smFRET for the 1-249 truncations are shown in gray while those for the 61-249 truncation are colored white. The solid line represents equality between mean smFRET measured in the full length protein and measured in the truncations. Error bars indicate the standard deviation in mean FRET from three or more replicate histograms.
Structural Dynamics in the PDZ Tandem
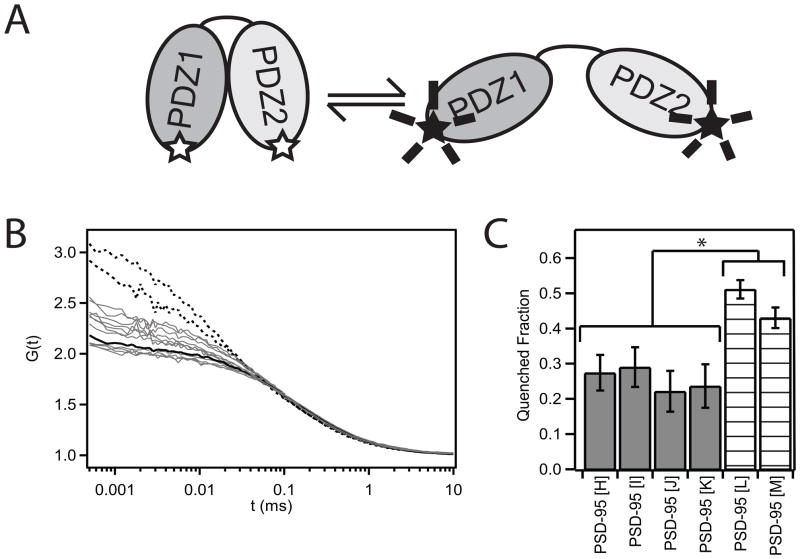
Our time resolution in smFRET experiments (0.1 sec/frame) renders our measurements insensitive to rapid structural dynamics. Protein domain motions on faster timescales would be time averaged. To probe the extent of conformational dynamics, we used fluctuation correlation spectroscopy (FCS) (Hess et al., 2002; Medina and Schwille, 2002), which has been successfully used to detect structural changes in both DNA (Bonnet et al., 1998; Kim et al., 2006) and proteins (Chattopadhyay et al., 2002; Mukhopadhyay et al., 2007; Neuweiler et al., 2009). We monitored tetramethylrhodamine (TMR) self-quenching, which has been used to characterize dynamics in chemically denatured proteins (Chattopadhyay et al., 2005). Similar to FRET, TMR self-quenching is distance dependent (Bernacchi and Mély, 2001), so the brightness of a doubly-labeled molecule will depend on the fluorophore separation (Figure 3A). The timescale of conformational fluctuations and the accessibility of the quenched state can be obtained from the autocorrelation curve of TMR emission (Bonnet et al., 1998).
Figure 3. Fluorescence Correlation Spectroscopy Measurement of Structural Dynamics.
(A) Cartoon illustrating the tetramethylrhodamine (TMR) self-quenching assay. The fluorescence intensity for a protein doubly-labeled with TMR dyes will decrease as the dyes are brought into close proximity. (B) Autocorrelation curves for TMR-labeled PSD-95. Singly-labeled PSD-95 is shown as a black solid line. PDZ1-PDZ2 mutants used in the FRET studies are all colored gray. Mutants labeled in PDZ1 and the disordered N-terminal region are shown as dashed lines. All experiments were performed using the 1-249 construct. (C) Amplitude of the relaxation term required to fit the FCS autocorrelation. Samples were only included if the F-ratio test showed a statistical improvement in the fit by incorporating the additional terms. The amplitude is reported in terms of the fraction in the dark (quenched) state. (* P < 0.01 based on standard deviation from replicate measurements) See also Figure S2.
The orientation and dynamics of the remaining domains in PSD-95 are unknown and could give rise to variable diffusion times or fluorescence quenching by endogenous tryptophan residues. To avoid these complications, we used the truncated PDZ tandem, which has the same conformation as in the full length protein. In the presence of triplet state quenchers, the autocorrelation curve for singly-labeled PSD-95 could be fit using a 3D diffusion model (Figure 3B). In contrast, PSD-95 doubly-labeled in PDZ1 and the disordered N-terminus showed clear evidence of dynamic quenching (Figure 3B). The F-ratio test showed that fitting these curves required an additional relaxation component in addition to the diffusion model (P=0.05). The disordered N-terminus is expected to be dynamic, so this finding confirms that our assay detects intramolecular dynamics in PSD-95.
When similar quenching measurements were made on the eleven constructs used in FRET studies, four showed statistically significant improvement when the fit incorporated the additional relaxation term (P=0.1). Two showed a trend towards dynamics but failed the F-ratio test while five showed little difference relative to the single labeled control. The timescale for domain motion was ~10−5 seconds while the amplitude of the relaxation component varied as a function of the labeling positions (Figure 3C). All had significantly lower amplitude relaxation terms than the N-terminal sites.
The amplitude from the FCS fit has been related to the accessibility of the quenched state (Bonnet et al., 1998; Chattopadhyay et al., 2002; Kim et al., 2006). The disordered N-terminal region readily allows for the large-scale motions necessary to bring the dye pairs into close proximity, while dye quenching is less accessible for labeling site combinations between the PDZ domains. Isotropic domain motions should allow quenching for all labeling site combinations so the low and variable extent of quenching is consistent with limited domain motions. However, the FCS data confirms that the conformation is not rigid; other orientations are sampled on a timescale much faster than the smFRET measurements.
To support the finding of limited conformational dynamics we also utilized lysine-crosslinking. If the domains frequently sample alternate conformations then crosslinking should capture and stabilize discrete static structures from within this ensemble. This would change the shape of the smFRET distribution (Choi et al., 2010). In agreement with this expectation, the presence of crosslinker changes the smFRET distribution for two labeling site combinations between the disordered N-terminus and PDZ1 (Figure S2A). In contrast, crosslinking had no effect on the mean FRET value or peak width in the smFRET distribution for two labeling site combinations between PDZ1 and PDZ2 (Figure S2B). To confirm the presence of intramolecular crosslinking, we used SDS-PAGE (Figure S2C), which showed altered mobility.
Modeling of Domain Orientation Using FRET Distance Restraints
When combined with known structures for individual components, smFRET-derived distances can be used as restraints to generate 3D structural models (Muschielok et al., 2008; Brunger et al., 2010; Choi et al., 2010; Vrljic et al., 2010). Following this idea, we created a model for the PDZ tandem based on our smFRET-derived distances. We used molecular dynamics simulations to calculate the mean dye position relative to the protein backbone for each of the nine labeling sites (Choi et al., 2010; Vrljic et al., 2010). The mean FRET efficiencies were converted to distances using a calibrated Förster’s radius.
We performed extensive torsion angle/rigid body molecular dynamics simulations with atoms in PDZ1 and PDZ2 fixed while atoms in the interdomain linker were unrestrained. FRET-derived distance restraints were applied to guide domain docking. This type of rigid body refinement has long been used to generate models from NMR data (Clore and Bewley, 2002; Clore and Schwieters, 2003) and more recently from EPR data (Bhatnagar et al., 2007).
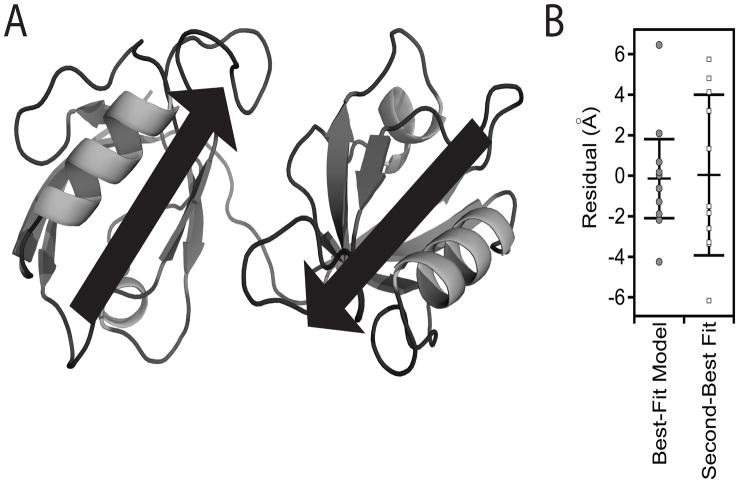
From 500 trials with randomly assigned starting orientations, a single model was found that best satisfied the FRET distance restraints (Figure 4A). Our best-fit model for the PDZ tandem showed a relatively closed configuration but with the distance between domains greater than a single bond. The orientation in our model shows an antiparallel alignment of the two ligand binding pockets (Figure 4A). To insure that we have not overfit or misfit the FRET data, docking simulations were repeated with each of the FRET restraints omitted from the refinement. The final results from all 11 simulations were highly similar structures, indicating that FRET restraints oversampled the domain orientation in the PDZ tandem (data not shown).
Figure 4. FRET Model for the Tandem PDZ Domains in Full-length PSD-95.
(A) Cartoon representation of the best fit model to the eleven FRET distance restraints. The model shows a relatively compact orientation without interdomain contacts. The position and direction of canonical PDZ ligand binding sites are represented as arrows. (B) Goodness of fit for the first and second best-fit models based on the smFRET distance restraints. Each model was assessed by plotting the residual (xFRET − xModel) for each of the 11 labeling-site pairs. The structure of the second best model is shown in Figure S3. See also Figure S3.
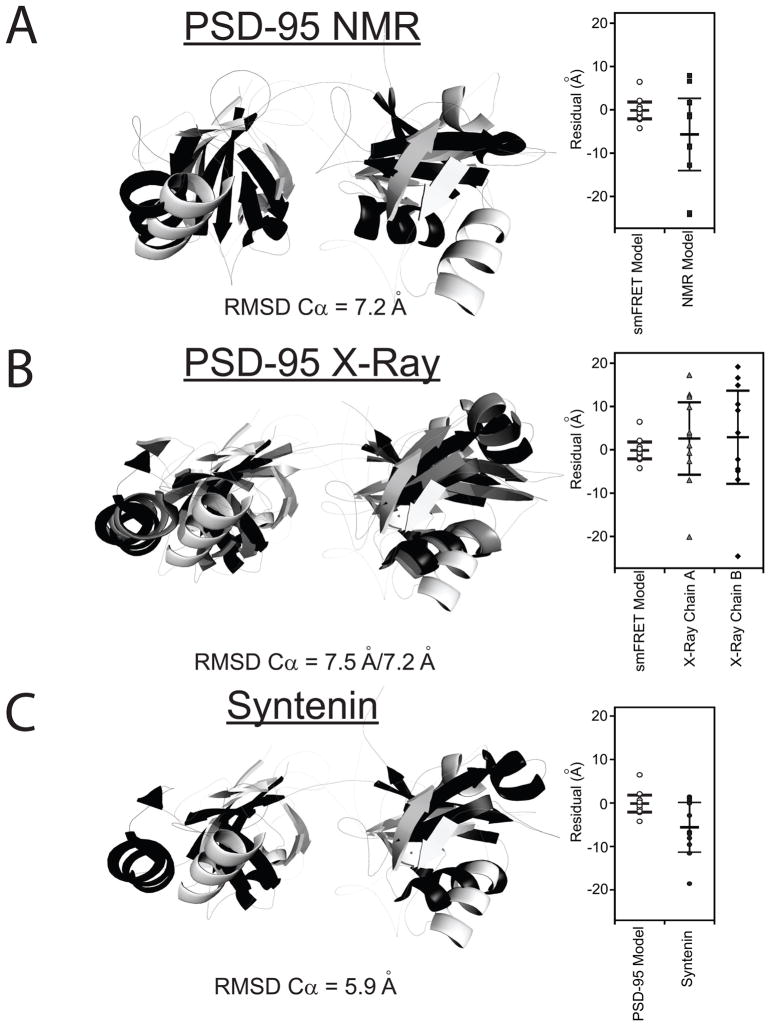
To illustrate the agreement between models and the entire data set, we plotted the residual (in Å) between the distance measured with FRET and the fluorophore separation in the model for each FRET pair (Figure 4B). The root mean square error (ERMS) for the best-fit model was 2.54 Å, while in the second model ERMS was 5.21 Å. The best-fit model was more compact, with the centers of mass for the two domains positioned 7.7 Å closer (Figure S3). The second model had the relative orientation flipped with PDZ2 positioned on the opposite face of PDZ1.
The antiparallel alignment of the ligand binding pockets in our model for the PDZ tandem differs from the orientation suggested by NMR experiments (Long et al., 2003). NMR experiments detected no interdomain distance restraints (i.e. NOEs), so the relative orientation of the two domains was examined through chemical shift mapping and a single measurement of residual dipolar couplings in the presence of bacteriophage. In their selected representative model, the binding pockets are co-aligned although the family of NMR models shows a visible distribution of domain orientations (Figure 2B in Long et al., 2003). Distances resulting from simulations using the representative NMR model were poorly fit by our FRET restraints (ERMS=11.97 Å, Figure 5A). This suggests that our smFRET measurements would distinguish between the two models.
Figure 5. Comparison the smFRET Model to Other Tandem PDZ Structures.
Only residues 61-249 are shown. The best-fit smFRET model is shown in white. (A) Alignment with the representative NMR model of the PDZ tandem from PSD-95 (black). This representative model was kindly provided by Dr. Mingjie Zhang but other models for domain orientation were also compatible with their data (Long et al., 2003). (B) Alignment with the two conformers observed in the crystal structure for the PDZ tandem from PSD-95 (PDB ID: 3GSL) Chains A and B are colored gray and black, respectively. (C) Alignment with the crystal structure for the PDZ tandem of syntenin (black) (PDB ID: 1N99). Dye positions were simulated for the other models and the goodness of fit with the FRET efficiency data is plotted next to the alignment. RMSD values are for the best overall alignment. See also Figure S4.
The crystal structure of the PDZ tandem from PSD-95 used a self-interacting PDZ ligand sequence to induce protein interactions necessary for crystal formation (Sainlos et al., 2010). They observed two copies of the PDZ tandem within the asymmetric unit that adopted different interdomain orientations. Both conformers have an antiparallel alignment of the ligand binding pockets akin to our smFRET model (Sainlos et al., 2010). However, neither conformation was identical to our smFRET model (Figure 5B) nor the representative NMR model (Figure S4). Thus, four completely different conformations have been observed for the same PDZ tandem (Figure S4).
A comparison of our model to the other known tandem PDZ domain structures found a similar conformation in the crystal structure of the PDZ tandem from syntenin (Figure 5C), which showed a comparable domain separation and orientation of the binding pockets. Simulations using homologous positions in the syntenin structure showed that, although a better fit to our restraints (ERMS = 8.24 Å), these two structures could be resolved using FRET.
What Stabilizes the Interdomain Orientation in the PDZ Tandem?
Amino acids within the interdomain linker could play a role in limiting domain mobility. The synaptic MAGUK proteins each contain between one and three sequential prolines in the linker that could restrict domain motions (Figure 6A). In addition, positively charged residues that begin the linker are also highly conserved (Figure 6A). Salt bridges between the PDZ domain and the linker were observed in the syntenin structure that could stabilize the orientation (Kang et al., 2003).
Figure 6. Effect of Linker Sequence on Interdomain Positioning.
(A) Sequence alignment of the PDZ1-2 linker in the four synaptic MAGUK homologues in Rattus novegicus. The last structured residue of each PDZ domain is indicated by the arrows. The well-conserved positively charged tripeptide at the beginning of the linker was mutated to alanines (RRK150-152AAA). The diproline sequence in the center of the PSD-95 linker was mutated to glycines (PP153,154GG). (B) smFRET measurements between identical sites on PSD-95 (1-249) containing mutations within interdomain linker were compared to wildtype (solid black). RRK150-152AAA (dashed black) and PP153,154GG (gray) mutants. See also Figure S5.
To test the role of linker sequence in domain orientation, we introduced mutations to disrupt these elements by changing charged residues to alanine or proline residues to glycine. Analytical size exclusion on the truncated PDZ tandem (61-249) showed no difference in the radius of hydration (RH) between the RRK150-152AAA mutant (RH = 2.19 ± 0.04 nm) and the wildtype linker (2.18 ± 0.03 nm). The PP152,153GG mutants showed a slight increase in RH (2.23 ± 0.01 nm). The domain orientation was also examined using smFRET, which showed small changes in mean FRET efficiency relative to measurements in wild type PSD-95 (Figure 6B). The largest shift in FRET efficiency corresponds to less than a 4 Å change in dye separation. The limited effects of these drastic amino acid substitutions suggest that the linker sequence plays a minimal role in the domain orientation.
Unpublished NMR titrations were reported to show chemical shift differences compared to the free domains (Long et al., 2003). To estimate the affinity of interdomain interactions, we expressed and purified PDZ1 and PDZ2 separately and used isothermal titration calorimetry (ITC) to measure the thermodynamics of their interaction. Injection of PDZ1 into PDZ2 was similar to injection of PDZ1 into buffer alone (Figures S5A, B). Thus, we were unable to detect binding by ITC at 10−4 M protein concentrations.
Some binding events or ultra-weak interactions would be missed by ITC so we also used smFRET to detect binding events between the two separately expressed domains. Donor-labeled PDZ2 was coencapsulated within 100 nm liposomes with acceptor-labeled PDZ1 (μM effective protein concentration). This approach is useful to study weak interactions because single molecule observations can capture rare binding events at concentrations below the KD (Benítez et al., 2010). Alternating laser excitation confirmed the presence of both domains within a liposome before the time traces were examined for binding events. Instances of FRET were observed, but the histograms were dominated by the peak at zero FRET indicating a lack of observable binding at this concentration (Figure S5C). Thus, the interdomain affinity is greater than mM.
Discussion
PSD-95 is implicated in several overlapping functions. There are more than ten different protein ligands identified as binding directly to the PDZ tandem in PSD-95 (Kim and Sheng, 2004). At mature excitatory synapses, PSD-95 controls localization of three different glutamate receptor subtypes; all have been shown to be dependent on an interaction with the PDZ tandem. How PSD-95 selects between ligands and recruits the appropriate signaling machinery remains a mystery.
Structural studies of PDZ tandems have all found evidence for limited interdomain motions, which has led to the suggestion that they form independent supramodules. These studies highlight the geometrical constraints on ligand selectivity when domains are conjoined, but none have examined domain orientation in a full-length scaffold protein. Protein interactions have been shown to affect domain orientation so tertiary interactions within a scaffold protein could be a determinant of conformation in tandem PDZ domains.
To address this, we used eleven FRET pairs to probe domain orientation for the first two PDZ domains in full length PSD-95 (Figure 1A). Each FRET pair was characterized to rule out photophysical artifacts. These constructs showed a dispersion of observed FRET values that is completely incompatible with isotropic rotation. Such narrow Gaussian smFRET peaks are consistent with either a fixed structure or rapid domain motions. Isotropic motions would result in similar FRET values for all measured pairs, while slower domain motions would broaden the histograms.
We used FCS measurement of TMR self-quenching to probe the extent of domain motions in the PDZ tandem. Four of the eleven labeling combinations showed statistically significant (P=0.1) evidence of dynamic quenching on the 10−5 second timescale, a reasonable timescale for domain motions of this type. The low amplitude of the relaxation and the reduced statistical significance compared to the disordered N-terminus (P=0.05) suggest that motions within the tandem provide limited or infrequent accessibility to domain orientations necessary for quenching (Figure 3).
The lysine crosslinking results support the interpretation that domain motions in the PDZ tandem are limited or infrequent when compared to the disordered N-terminus (Figure S2). However, the distribution of lysine residues is not uniform so not all orientations would be equally captured by crosslinking, a limitation of this assay. Our results are consistent with the PDZ tandem having a unique lowest energy conformation but that the domains do sample orientations different from those observed in our model. Effective quenching would imply large domain movements since TMR quenching, like FRET, is sensitive to changes on the nanometer length scale.
None of the eleven FRET measurements showed differences in PDZ domain orientation between full-length PSD-95 and truncated constructs (Figure 2B). Thus, in PSD-95 the PDZ tandem is not affected by interactions within the full length protein, which was not predictable from previous studies. Negative-stain electron microscopy showed a compact arrangement of the domains in PSD-95 (Nakagawa et al., 2004). The conformation of the PDZ tandem from PSD-95 has been demonstrated to be sensitive to protein interactions (Wang et al., 2009; Sainlos et al., 2010), so it was surprising to find no difference in the domain orientation in full-length PSD-95. Unless the PDZ tandem is internally stabilized, this would suggest a lack of tertiary interactions between the PDZ tandem and the other domains. Thus our results confirm the independence of the PDZ tandem in full-length PSD-95.
To obtain a model for the domain orientation, we used rigid body molecular dynamics simulations incorporating our smFRET derived restraints, which converged on a unique solution that best fit the data. The best-fit model was robust to the omission of any of the smFRET restraints from the refinement. This study presents the first structural model for the PDZ tandem of PSD-95 in the context of the full-length protein (Figure 4).
It remains unclear what limits the interdomain mobility. The linker is an obvious constraint that prevents full isotropic rotation. We introduced mutations to examine the role of the linker sequence in domain orientation. Changing a diproline sequence to glycines or a positively charged tripeptide to alanines affected domain spacing but did not change the overall orientation (Figure 6). Many linker conformations are compatible with the domain orientation observed in our model. Linker length has been shown to be important for domain orientation and function (Long et al., 2003; Dyson and Wright, 2005) but our results suggest that the amino acid sequence plays a minimal role.
PDZ domains conjoined by a short linker co-occupy a small volume. We estimate ~10−23 L from our SEC measurement of the radius of hydration (RH = 2.2 nm). In the absence of stabilization, this would ensure non-specific collisional interactions, which are distinct from specific binding interactions. Steric repulsion may serve as a major determinant of domain orientation. The lowest energy orientation minimizes steric clashes. While not completely rigid (in agreement with our FCS data and the distribution of NMR models), the degree to which the domains are free to move without encountering negative van der Waals forces is limited. At the high effective concentration (~10−2 M based on our measured RH), even a very low affinity interactions could be significant for stabilization. Such, ultra-weak protein interactions would not have been detected by our interdomain binding assays (Figure S5). For example, although we used no attractive potential and kept loops rigid, our model places the backbone Cα atoms of two solvent exposed isoleucine residues within surface loops (I88 in PDZ1 and I208 in PDZ2) within 8 Å. A lone hydrophobic pair could provide stability in excess of thermal fluctuations at room temperature. The linker length sets the effective concentration and domain surface features control the final orientation. This implies a relatively featureless, “low-relief” energy landscape, where the PDZ tandem achieves a lowest energy conformation but can be displaced.
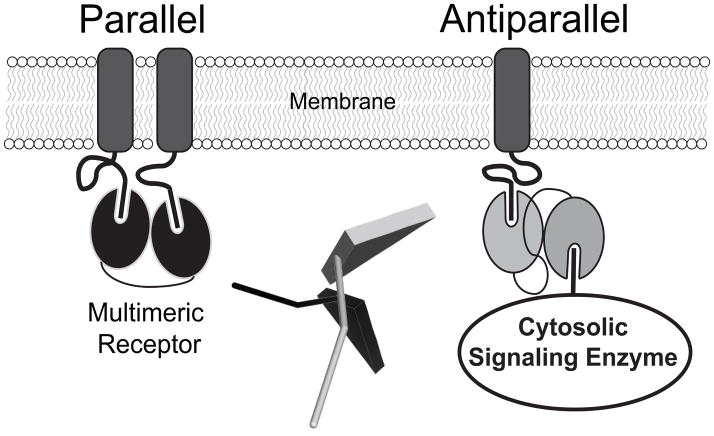
Our model and the two crystal structures for the PSD-95 PDZ tandem show an antiparallel alignment of the binding pockets (Sainlos et al., 2010). The representative NMR model showed co-alignment of the ligand binding pockets (Long et al., 2003). However, none of the four models show the same conformation (Figure S4). The NMR data was collected in the presence of 20 mg/mL of the negatively-charged, proteinaceous Pf1 bacteriophage (Long et al., 2003). Pf1 partially orients the PDZ tandem in solution through steric and electrostatic interactions between PSD-95 and the bacteriophage otherwise the residual dipolar coupling measurement is not possible (Zweckstetter and Bax, 2000; Prestegard et al., 2004). The crystallization experiments included a self-interacting sequence to force protein interactions (Sainlos et al., 2010). In contrast, we encapsulated PSD-95 with neutral phospholipids for which PSD-95 has no affinity (data not shown), so our measurements captured PSD-95 in an isolated state devoid of interactions, which can be thought of as its ground state. In light of the strong effect of peptide binding on domain orientation (Wang et al., 2009), the four different conformations likely reflect the response of the PDZ tandem to these different environments. This notion is supported by the dissimilarity of the solution and crystal structures of the syntenin PDZ tandem, which were attributed to crystal-packing forces (Kang et al., 2003; Cierpicki et al., 2005). The observed conformation likely reflects the lowest energy configuration for a particular environment so the different experimental conditions captured different conformations. Thus, although the PDZ tandem is independent in PSD-95 it clearly lacks a single, rigid structure.This has implications for PSD-95, which functions in the crowded environment of the post synaptic density (Sheng and Hoogenraad, 2007). The conformation determines the relative orientation of the PDZ ligand binding sites, which has implications for ligand selection. Although the PDZ domains recognize short linear peptide sequences, these are part of much larger proteins that must sterically fit together to co-occupy PSD-95. For example, the C-terminus of the 2B-NMDAR is ~75 kDa while neuronal nitric oxide synthase is ~160 kDa and these proteins are thought to co-occupy the PDZ tandem of PSD-95. Co-aligned binding sites would favor the selection of ligands coming from the same direction, such as multimeric transmembrane receptors (Figure 7). Such an orientation could support recognition of multivalent ligands like the glutamate receptors. Indeed, it has been suggested that the rigid, supramodular structure observed in the NMR model favors the selection of bivalent ligands. In contrast, an antiparallel orientation, as observed by FRET and crystallography, would favor selection of ligands from opposite directions, such as recruitment of cytosolic signaling factors to transmembrane receptors (Figure 7). This orientation would not favor binding of bivalent ligands. The 2A- and 2B-NMDA subtype glutamate receptors are differentially trafficked and connected to different signaling pathways by PSD-95 despite having the exact same C-terminal PDZ ligand sequence. Such selectivity has not been explained by the numerous single domain structures. Switching domain orientation may allow differential interaction with proteins containing identical PDZ ligand sequences.
Figure 7. Domain Orientation of Tandem PDZ Domains Can Influence Function.
PDZ domains are depicted as ovals attached to C-terminal ligand peptides at the cell surface. The relative alignment of tandem PDZ binding sites determines the geometry of higher order complexes at the scaffold. Left: Parallel orientation (black) selects for proteins originating from the same side of the scaffold, such as multimeric transmembrane receptors. Right: The antiparallel configuration (grey) would allow coordinate binding of membrane receptors and cytosolic signaling enzymes. Domain positioning could be a key element in understanding the functions of scaffold proteins such as PSD-95. Center: Relative orientation of the canonical peptide ligand in the antiparallel (grey) and co-aligned (black) PDZ configurations in the smFRET and representative NMR models. The ligand site in PDZ1 was aligned as shown in Figure S2A. The peptide was modeled in PDZ2 based on the peptide-bound NMR structure (PDB ID: 2KA9).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The cDNA for PSD-95 from Rattus norvegicus was expressed as a 6 His fusion in the Rosetta bacterial strain (Novagen, Madison WI) from the vector pet28a (Novagen) or pPROEX (Invitrogen, Carlsbad, CA). The native cysteine residues were mutated to serines and cysteines for labeling were introduced by site-directed mutagenesis. Proteins were purified using a combination of Ni-NTA (Qiagen Inc, Valencia, CA), anion exchange and size exclusion chromatography (GE Healthcare, Piscataway, NJ) under reducing conditions. Purified proteins were labeled using a 5-10 fold molar excess of maleimide-conjugated Alexa 555 and Alexa 647 dyes (Invitrogen). Unconjugated dye was removed by desalting with Sephadex G-50 (GE Healthcare). Labeling efficiency was greater than 90% as determined from absorbance measurements. Proteins were randomly labeled with both dyes. Analytical size exclusion showed that neither the introduction of cysteines nor fluorescent labeling altered the retention time for any mutant, supporting the contention that mutations and fluorescent dyes did not alter domain folding (Data not shown).
Data Acquisition and Analysis
Fluorescently-labeled protein samples were encapsulated at 0.5 μM by extrusion into 100 nm liposomes composed of egg phosphatidylcholine with 0.1% biotinylated phosphatidylethanolamine (Avanti Polar Lipids, Alabaster, AL). Unencapsulated protein was removed by desalting on Sepharose CL-4B (GE Healthcare). At these concentrations, the vast majority of liposomes are empty or contain a single protein. Liposomes were immobilized via streptavidin to a quartz slide coated with biotinylated-BSA. Data was recorded with a prism-type total internal reflection microscope, detected by an Andor iXon EMCCD camera (Andor Technologies, Belfast, UK) as previously described (McCann et al., 2010).
Data analysis was as described previously (McCann et al., 2010). Single molecules containing both donor and acceptor dye were confirmed by single step photobleaching to baseline. Briefly, FRET efficiency is calculated from the background subtracted intensity as:
where IA and ID are the acceptor and donor intensities respectively and γ is the normalization factor used to correct for differences in quantum yield and detection efficiency between dyes. We applied per molecule γ correction determined from single molecule photobleaching (McCann et al., 2010). Histograms of FRET efficiency for hundreds of molecules were characterized by non-linear least squared fitting with Gaussian functions.
Fluorescence Correlation Spectroscopy (FCS)
FCS measurements were performed on a Zeiss ConfoCor 2 LSM-510 Meta (Carl Zeiss AG, Oberkochen, Germany). Labeling with tetramethylrhodamine-5-maleimide (Sigma-Aldrich) was done as described above. Samples were excited with a 543 nm laser, and detected through a 560 nm long pass filter. The auto-correlation curve for singly-labeled protein could be fit using a single component three-dimensional Brownian diffusion model (τD = 148 μsec). Triplet state quencher and low laser power eliminated the need to incorporate a triplet state contribution to the fit. We make the simplifying assumption that TMR quenching can be approximated by a two state system (bright or dark). The autocorrelation curves for doubly labeled samples were fit with a diffusion model plus an additional component to describe the dynamics (Bonnet et al., 1998).
where p denotes the bright fraction, N is the number of molecules in the focal volume and τR is the time scale of the relaxation. τD was fixed to the value obtained for singly-labeled protein.
Lysine Crosslinking
An ~1.3 x ratio (relative to lysine residues) of BS3 crosslinking reagent (Thermo Scientific, Rockford, IL) was reacted with PSD-95 1-249. This ratio achieved maximal intramolecular crosslinking with minimal intermolecular crosslinking as assessed by SDS-PAGE (Figure S3C). Unreacted BS3 was inactivated with Tris and removed by desalting using Sephadex G-50 (GE Healthcare).
Isothermal Titration Calorimetry (ITC)
Thermodynamic measurements were made using a MicroCal VP-ITC MicroCalometer (MicroCal, LLC, Northampton, MA). Individual domains were expressed and purified separately as described above. In 20 mM Tris 150 mM NaCl and 0.5 mM TCEP, PDZ1 at 1.2 mM was injected (in 10 μL injections) into a sample cell containing either 100 μM PDZ2 or protein-free buffer . Data was analyzed and figures generated using the MicroCal Origin software package.
Analytic Size Exclusion Chromatography
PSD-95 (residues 61-249) was labeled with Alexa 647 and injected onto a Shodex KW 802.5 column (Showa Denko America, Inc., New York, NY) in 20 mM Tris 300 mM NaCl pH 7.5. The elution profile was monitored by UV absorbance at 651 nm.
Fluorescence Spectroscopy
Ensemble fluorescence measurements were made on a SLM AMINCO-Bowman Series 2 spectrofluorimeter (Thermo Fisher Scientific, Waltham, MA). FRET samples were dual-labeled with Alexa 555. The absorbance value at the excitation wavelength (532 nm) was adjusted to 0.05 AU for all samples. The average donor quantum yield for each FRET construct was determined by ratiometric measurements relative to the quantum yield standard of Rhodamine 101 in ethanol (QY=1.0) (Karstens and Kobs, 1980). Fluorescence anisotropy was measured in the L configuration using film polarizers. The average values were used because the dataset contains a random distribution of the two dyes across two labeling sites.
Modeling Using FRET Derived Distance Restraints
Structural refinement of the PDZ tandem was performed using CNS version 1.3 (Brunger et al., 1998). To estimate the location of the dye relative to the protein backbone, we performed simulations using an atomic model of the dyes attached to the protein (Vrljic et al., 2010). The dye and linker were allowed free rotation while the protein structure was fixed. 200 separate simulations were performed for each labeling site used in the experiments (100 of each dye) and the mean position of the central carbon atom relative to the backbone was taken as a fixed locus for applying the FRET-derived restraints.
smFRET measurements were converted to distance using a Förster radius of 5.5 nm as calibrated using smFRET measurements on known structures (Choi et al., 2010). Distances defined harmonic square well potentials associated with the fluorophore positions. Variable bounds were used for the square well potential depending on how close the expected distance is to R0 (Choi et al., 2010). The individual domains, PDZ1(61-149) and PDZ2(160-244), were treated as independent rigid bodies. The torsion angles for amino acids in the linker (150-159) were unconstrained but bond lengths and angles were fixed (Vrljic et al., 2010). The energy function included a repulsive term for non-bonded interactions and the distance restraints but lacked electrostatics and the attractive van der Waals terms (Brunger et al., 2010). Cluster analysis of 500 simulations with randomly generated starting positions was used to isolate similar structures. An exponential scoring function based upon the difference between FRET distant restraints and the model results was used to identify a best-fit model (Vrljic et al., 2010). Figures were generated using PyMol (Schrödinger LLC).
Supplementary Material
Figure S1 (related to Figure 1): Ribbon diagram showing the position of labeling sites in the PDZ domains.
Figure S2 (related to Figure 3): Effect of crosslinking on the smFRET distribution and conformation of crosslink formation.
Figure S3 (related to Figure 4): Second best fit model from molecular refinement.
Figure S4 (related to Figure 5): Comparison of domain organization in the different structural models for the PDZ tandem.
Figure S5 (related to Figure 6): Estimation of the affinity of interdomain interactions in the absence of the linker.
Acknowledgments
We benefited from conversations with Axel Brunger, Stephen E. Kaiser, Nicolas Nassar, Anne Hinderliter and Ucheor B. Choi. We thank: Stuart McLaughlin for access to the FCS and W. Todd Miller for assistance with ITC. The authors acknowledge the National Institutes of Health for funding to MEB (MH081923).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benítez JJ, Keller AM, Chen P, Nils GW. Methods in Enzymology. Academic Press; 2010. Nanovesicle Trapping for Studying Weak Protein Interactions by Single-Molecule FRET; pp. 41–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernacchi S, Mély Y. Exciton interaction in molecular beacons: a sensitive sensor for short range modifications of the nucleic acid structure. Nucleic Acids Research. 2001;29:e62. doi: 10.1093/nar/29.13.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar J, Freed JH, Crane BR. Rigid body refinement of protein complexes with long-range distance restraints from pulsed dipolar ESR. Methods Enzymol. 2007;423:117–133. doi: 10.1016/S0076-6879(07)23004-6. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc Natl Acad Sci U S A. 1998;95:8602–8606. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Strop P, Vrljic M, Chu S, Weninger KR. Three-dimensional molecular modeling with single molecule FRET. J Struct Biol. 2010 doi: 10.1016/j.jsb.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay K, Elson EL, Frieden C. The kinetics of conformational fluctuations in an unfolded protein measured by fluorescence methods. Proc Natl Acad Sci U S A. 2005;102:2385–2389. doi: 10.1073/pnas.0500127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay K, Saffarian S, Elson EL, Frieden C. Measurement of microsecond dynamic motion in the intestinal fatty acid binding protein by using fluorescence correlation spectroscopy. Proc Natl Acad Sci U S A. 2002;99:14171–14176. doi: 10.1073/pnas.172524899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Winters C, Azzam R, Li X, Galbraith JA, Leapman RD, Reese TS. Organization of the core structure of the postsynaptic density. Proc Natl Acad Sci U S A. 2008;105:4453–4458. doi: 10.1073/pnas.0800897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- Choi UB, Strop P, Vrljic M, Chu S, Brunger AT, Weninger KR. Single-molecule FRET-derived model of the synaptotagmin 1-SNARE fusion complex. Nat Struct Mol Biol. 2010;17:318–324. doi: 10.1038/nsmb.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cierpicki T, Bushweller JH, Derewenda ZS. Probing the Supramodular Architecture of a Multidomain Protein: The Structure of Syntenin in Solution. Structure. 2005;13:319–327. doi: 10.1016/j.str.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Clore GM, Bewley CA. Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings. J Magn Reson. 2002;154:329–335. doi: 10.1006/jmre.2001.2489. [DOI] [PubMed] [Google Scholar]
- Clore GM, Schwieters CD. Docking of protein-protein complexes on the basis of highly ambiguous intermolecular distance restraints derived from 1H/15N chemical shift mapping and backbone 15N-1H residual dipolar couplings using conjoined rigid body/torsion angle dynamics. J Am Chem Soc. 2003;125:2902–2912. doi: 10.1021/ja028893d. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Feng W, Shi Y, Li M, Zhang M. Tandem PDZ repeats in glutamate receptor-interacting proteins have a novel mode of PDZ domain-mediated target binding. Nat Struct Biol. 2003;10:972–978. doi: 10.1038/nsb992. [DOI] [PubMed] [Google Scholar]
- Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- Goult BT, Rapley JD, Dart C, Kitmitto A, Grossmann JG, Leyland ML, Lian LY. Small-angle X-ray scattering and NMR studies of the conformation of the PDZ region of SAP97 and its interactions with Kir2.1. Biochemistry. 2007;46:14117–14128. doi: 10.1021/bi701257z. [DOI] [PubMed] [Google Scholar]
- Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- Hess ST, Huang S, Heikal AA, Webb WW. Biological and chemical applications of fluorescence correlation spectroscopy: a review. Biochemistry. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- Hung AY, Sheng M. PDZ domains: structural modules for protein complex assembly. J Biol Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Iskenderian-Epps WS, Imperiali B. Modulation of Shank3 PDZ Domain Ligand-Binding Affinity by Dimerization. Chembiochem. 2010 doi: 10.1002/cbic.201000246. [DOI] [PubMed] [Google Scholar]
- Kang BS, Cooper DR, Jelen F, Devedjiev Y, Derewenda U, Dauter Z, Otlewski J, Derewenda ZS. PDZ tandem of human syntenin: crystal structure and functional properties. Structure. 2003;11:459–468. doi: 10.1016/s0969-2126(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Karstens T, Kobs K. Rhodamine-B and Rhodamine-101 as Reference Substances for Fluorescence Quantum Yield Measurements. Journal of Physical Chemistry. 1980;84:1871–1872. [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kim J, Doose S, Neuweiler H, Sauer M. The initial step of DNA hairpin folding: a kinetic analysis using fluorescence correlation spectroscopy. Nucleic Acids Res. 2006;34:2516–2527. doi: 10.1093/nar/gkl221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Wei Z, Feng W, Yu C, Zhao YX, Zhang M. Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1. J Mol Biol. 2008;375:1457–1468. doi: 10.1016/j.jmb.2007.11.088. [DOI] [PubMed] [Google Scholar]
- Long JF, Feng W, Wang R, Chan LN, Ip FC, Xia J, Ip NY, Zhang M. Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem. Nat Struct Mol Biol. 2005;12:722–728. doi: 10.1038/nsmb958. [DOI] [PubMed] [Google Scholar]
- Long JF, Tochio H, Wang P, Fan JS, Sala C, Niethammer M, Sheng M, Zhang M. Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD–95. J Mol Biol. 2003;327:203–214. doi: 10.1016/s0022-2836(03)00113-x. [DOI] [PubMed] [Google Scholar]
- McCann JJ, Choi UB, Zheng L, Weninger K, Bowen ME. Optimizing methods to recover absolute FRET efficiency from immobilized single molecules. Biophys J. 2010;99:961–970. doi: 10.1016/j.bpj.2010.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MA, Schwille P. Fluorescence correlation spectroscopy for the detection and study of single molecules in biology. Bioessays. 2002;24:758–764. doi: 10.1002/bies.10118. [DOI] [PubMed] [Google Scholar]
- Mishra P, Socolich M, Wall MA, Graves J, Wang Z, Ranganathan R. Dynamic scaffolding in a G protein-coupled signaling system. Cell. 2007;131:80–92. doi: 10.1016/j.cell.2007.07.037. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschielok A, Andrecka J, Jawhari A, Bruckner F, Cramer P, Michaelis J. A nano-positioning system for macromolecular structural analysis. Nat Methods. 2008;5:965–971. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Futai K, Lashuel HA, Lo I, Okamoto K, Walz T, Hayashi Y, Sheng M. Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron. 2004;44:453–467. doi: 10.1016/j.neuron.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Neuweiler H, Johnson CM, Fersht AR. Direct observation of ultrafast folding and denatured state dynamics in single protein molecules. Proc Natl Acad Sci U S A. 2009;106:18569–18574. doi: 10.1073/pnas.0910860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Peterson FC, Penkert RR, Volkman BF, Prehoda KE. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- Piserchio A, Pellegrini M, Mehta S, Blackman SM, Garcia EP, Marshall J, Mierke DF. The PDZ1 domain of SAP90. Characterization of structure and binding. J Biol Chem. 2002;277:6967–6973. doi: 10.1074/jbc.M109453200. [DOI] [PubMed] [Google Scholar]
- Prestegard JH, Bougault CM, Kishore AI. Residual Dipolar Couplings in Structure Determination of Biomolecules. Chemical Reviews. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- Sainlos M, Tigaret C, Poujol C, Olivier NB, Bard L, Breillat C, Thiolon K, Choquet D, Imperiali B. Biomimetic divalent ligands for the acute disruption of synaptic AMPAR stabilization. Nat Chem Biol. 2010;7:81–91. doi: 10.1038/nchembio.498. [DOI] [PubMed] [Google Scholar]
- Sheng M, Hoogenraad CC. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev Biochem. 2007;76:823–847. doi: 10.1146/annurev.biochem.76.060805.160029. [DOI] [PubMed] [Google Scholar]
- Tochio H, Hung F, Li M, Bredt DS, Zhang M. Solution structure and backbone dynamics of the second PDZ domain of postsynaptic density-95. J Mol Biol. 2000;295:225–237. doi: 10.1006/jmbi.1999.3350. [DOI] [PubMed] [Google Scholar]
- van den Berk LC, Landi E, Walma T, Vuister GW, Dente L, Hendriks WJ. An allosteric intramolecular PDZ-PDZ interaction modulates PTP-BL PDZ2 binding specificity. Biochemistry. 2007;46:13629–13637. doi: 10.1021/bi700954e. [DOI] [PubMed] [Google Scholar]
- Vrljic M, Strop P, Ernst JA, Sutton RB, Chu S, Brunger AT. Molecular mechanism of the synaptotagmin-SNARE interaction in Ca2+-triggered vesicle fusion. Nat Struct Mol Biol. 2010;17:325–331. doi: 10.1038/nsmb.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Weng J, Zhang X, Liu M, Zhang M. Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95. J Am Chem Soc. 2009;131:787–796. doi: 10.1021/ja8076022. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Fan JS, Zhang M. Interdomain chaperoning between PSD-95, Dlg, and Zo-1 (PDZ) domains of glutamate receptor-interacting proteins. J Biol Chem. 2001;276:43216–43220. doi: 10.1074/jbc.M105996200. [DOI] [PubMed] [Google Scholar]
- Zweckstetter M, Bax A. Prediction of Sterically Induced Alignment in a Dilute Liquid Crystalline Phase: Aid to Protein Structure Determination by NMR. Journal of the American Chemical Society. 2000;122:3791–3792. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (related to Figure 1): Ribbon diagram showing the position of labeling sites in the PDZ domains.
Figure S2 (related to Figure 3): Effect of crosslinking on the smFRET distribution and conformation of crosslink formation.
Figure S3 (related to Figure 4): Second best fit model from molecular refinement.
Figure S4 (related to Figure 5): Comparison of domain organization in the different structural models for the PDZ tandem.
Figure S5 (related to Figure 6): Estimation of the affinity of interdomain interactions in the absence of the linker.