Abstract
The migration of neuroblasts along the anteroposterior body axis of C. elegans is controlled by multiple Wnts that act partially redundantly to guide cells to their precisely defined final destinations. How positional information is specified by this system is, however, still largely unknown. Here, we used a novel fluorescent in situ hybridization methods to generate a quantitative spatiotemporal expression map of the C. elegans Wnt genes. We found that the five Wnt genes are expressed in a series of partially overlapping domains along the anteroposterior axis, with a predominant expression in the posterior half of the body. Furthermore, we show that a secreted Frizzled-related protein is expressed at the anterior end of the body axis, where it inhibits Wnt signaling to control neuroblast migration. Our findings reveal that a system of regionalized Wnt gene expression and anterior Wnt inhibition guides the highly stereotypic migration of neuroblasts in C. elegans. Opposing expression of Wnts and Wnt inhibitors has been observed in basal metazoans and in the vertebrate neurectoderm. Our results in C. elegans support the notion that a system of posterior Wnt signaling and anterior Wnt inhibition is an evolutionarily conserved principle of primary body axis specification.
Keywords: C. elegans, Wnt, Neuroblast migration, Secreted Frizzled-related protein
INTRODUCTION
Wnt proteins control many aspects of metazoan development, with prominent functions in cell fate determination, cell proliferation and cell migration (van Amerongen and Nusse, 2009). The activity of Wnt proteins is counteracted by a range of inhibitors, including secreted proteins such as the secreted Frizzled-related proteins (SFRPs) (Leyns et al., 1997; Bovolenta et al., 2008) and Dickkopf (Niehrs, 2006), and intracellular inhibitors such as the TCF/Lef transcription factor Tcf3 (Kim et al., 2000). During neurectoderm development in vertebrates, these inhibitors are expressed in the anterior and counteract the activity of posteriorly expressed Wnts to enable the formation of anterior brain structures and the eyes (Leyns et al., 1997; Kiecker and Niehrs, 2001; Tendeng and Houart, 2006). Opposing expression of Wnts and Wnt inhibitors has also been observed in basal metazoan organisms such as the cnidarians Hydra and Nematostella vectensis (Hobmayer et al., 2000; Kusserow et al., 2005; Guder et al., 2006b; Lee et al., 2006), and in the planarian Schmidtea mediterranea (Petersen and Reddien, 2008), which led to the hypothesis that a system of posterior Wnt signaling and anterior Wnt inhibition is an ancient mechanism that may be used across animal phyla to pattern the primary body axis (Petersen and Reddien, 2009).
The nematode Caenorhabditis elegans expresses five different Wnt proteins that control many aspects of development, including cell fate specification, cell polarity and the highly stereotypic migration of neuroblasts along the anteroposterior body axis (Korswagen, 2002; Silhankova and Korswagen, 2007). Neuroblasts that migrate in a Wnt-dependent manner include: the HSN neurons, which migrate from the posterior to the mid-body region (Sulston et al., 1983; Hedgecock et al., 1987; Pan et al., 2006); the ALM and CAN neurons, which migrate from the anterior to positions in the mid-body region (Sulston et al., 1983; Hedgecock et al., 1987; Zinovyeva and Forrester, 2005); and the Q neuroblast descendants, which migrate in opposite directions on the left and right lateral sides (Sulston and Horvitz, 1977; Harris et al., 1996). With the exception of the left Q cell descendants, the migration of these neuroblasts is controlled through multiple, partially redundantly acting Wnt proteins (Zinovyeva et al., 2008). The expression patterns of the C. elegans Wnt genes have been analyzed using transgenic reporter constructs. These studies revealed a predominantly posterior expression for the Wnt genes lin-44, egl-20 and cwn-1 (Herman et al., 1995; Whangbo and Kenyon, 1999; Pan et al., 2006), whereas mom-2 and cwn-2 were reported to be generally expressed along the anteroposterior axis (Gleason et al., 2006), with a more prominent expression of cwn-2 in the pharynx and anterior muscle cells (Kennerdell et al., 2009; Song et al., 2010). Although there is a large degree of overlap between the reported expression patterns, there are also important differences in the extent of expression along the anteroposterior axis and in the specific cell types that are involved, complicating the analysis of Wnt gene function in neuroblast migration and other aspects of development.
Here, we used single molecule mRNA fluorescent in situ hybridization (smFISH) to determine quantitatively the spatio-temporal expression patterns of the five C. elegans Wnt genes. Our results show that the different Wnt genes are expressed in a series of partially overlapping expression domains, with a predominant expression in the posterior body half and a single Wnt gene with an anterior expression domain. Furthermore, we show that the C. elegans genome contains a single SFRP ortholog that is specifically expressed at the anterior end of the body axis. SFRP-1 functions as an inhibitor of Wnt signaling that represses the most anteriorly expressed Wnts to control the migration of neuroblasts in the anterior body region. Our results demonstrate that opposing Wnt and Wnt inhibitory activities are also key to anteroposterior patterning in C. elegans and provide further support for the evolutionary conservation of this system in primary body axis specification.
MATERIALS AND METHODS
C. elegans strains and culturing
General methods for culture, manipulation and genetics of C. elegans were as described previously (Lewis and Fleming, 1995). Strains were cultured at 20°C. Mutations and transgenes used in this study were: LGI, lin-44(n1792) (Herman et al., 1995), mom-5(gk812), pry-1(mu38) (Maloof et al., 1999; Korswagen et al., 2002), ccIs4251[Pmyo-3::gfp] (Fire et al., 1998); LGII, cwn-1(ok546) (Zinovyeva and Forrester, 2005), mab-5(gk670), mig-14(mu71) (Bänziger et al., 2006), vps-35(hu68) (Coudreuse et al., 2006), muIs32[Pmec-7::gfp] (Ch'ng et al., 2003); LGIV, sfrp-1(gk554), cwn-2(ok895) (Zinovyeva and Forrester, 2005), egl-20(hu105) (Coudreuse et al., 2006); otIs33 (Pkal-1::GFP) (Bulow et al., 2002); ayIs7[Phlh-8::gfp] (Harfe et al., 1998); LGV, mom-2(or309) (Zinovyeva and Forrester, 2005) (note that the balancer for mom-2, nT1, also complements sfrp-1), muIs35[Pmec-7::gfp] (Ch'ng et al., 2003); heIs63[Pwrt-2::ph::gfp] (M. Wildwater and S. van den Heuvel, unpublished); and unassigned, huIs120[Phsp::sfrp-1].
Single molecule mRNA FISH
Probe design and hybridization to perform FISH for single transcript measurement in C. elegans larvae was performed as previously described (Raj et al., 2008) (see also www.singlemoleculefish.com). Animals were collected by washing plates with M9 and were fixed in 4% formaldehyde in 1×PBS for 45 minutes. Fixed animals were permeabilized in 70% ethanol overnight. All probes for hybridization were coupled to either Cy5 (GE Amersham), Alexa594 (Invitrogen) or tetramethylrhodamine (TMR) (Invitrogen), depending on the desired gene combinations for image acquisition. The type of coupled fluorophore did not affect any quantitative results in this study. Images were taken in z-stacks using a Nikon TE2000 epi-fluorescence microscope with a Princeton Instruments CCD camera and appropriate optical filters for DAPI, Cy5, Alexa594 and TMR. All experiments were performed using either wild-type (N2) or wild-type animals expressing cell type specific GFP markers. Three-dimensional positions of bright fluorescent spots in each animal were detected with the aid of a custom program written in MATLAB, as described (Raj et al., 2008), which was later manually corrected for further accuracy. The low background of the smFISH staining procedure is illustrated by the absence of random spots outside the main regions of expression. Examples are the lack of spots for cwn-1, cwn-2, egl-20, lin-44 and sfrp-1 in the early embryo (Fig. 2) and the absence of spots for mom-2, egl-20 and lin-44 around the anterior region of the L1 pharynx (Fig. 1F). Nuclei were visualized with DAPI. We used body length to gauge the developmental age of individual animals. We obtained body lengths of each animal by measuring the distance between the nuclei of hyp4 (the anterior-most cell) and hyp10 (the posterior-most cell) along the anteroposterior (AP) axis. The identity of transcript containing cells was determined using specific GFP markers (body wall muscle cells, seam cells and the undifferentiated M cell descendants), or by determining nuclear positions by DAPI staining (Sulston and Horvitz, 1977; Sulston et al., 1983; Long et al., 2009).
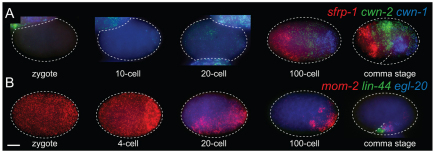
Fig. 2.
Single molecule mRNA FISH analyses of the C. elegans Wnt genes and sfrp-1 during embryonic development. Images are maximum intensity projections of lateral z-stacks. Detection of (A) sfrp-1, cwn-1 and cwn-2, and (B) mom-2, lin-44 and egl-20 transcripts. Embryos were staged using DIC microscopy and DAPI staining of nuclei. Scale bar: 10 μm.
Fig. 1.
Single molecule mRNA FISH analyses of the C. elegans Wnt genes and sfrp-1 in staged L1 animals. Images are maximum intensity projections of lateral z-stacks. (A) Detection of cwn-1, egl-20 and lin-44 transcripts in the L1 larval tail of six individual animals. Broken circles indicate: top, the anal depressor muscle; from bottom left to right, P11/P12, Y, B and the body wall muscle cells VL23/VL24. The rectum is indicated by a solid line. Scale bar: 10 μm. (B) Transcript identification using a custom program written in MATLAB. A sagittal view of cwn-1 smFISH spots shows predominant expression in the four body wall muscle quadrants. (C) Magnification of the boxed area in A. smFISH spot counts of cwn-1, egl-20 and mom-2 are indicated. (D) Expression of cwn-2, egl-20 and mom-2 in the posterior half of the animal. The asterisk indicates the Z2 and Z3 germ line precursor cells, the arrow indicates the position of the tail cells that transiently express mom-2. Scale bar: 10 μm. (E) Expression of sfrp-1 and cwn-2 in an L1 larva. The nuclei of body wall muscle cells are highlighted by nuclear GFP. The posterior ventral nerve cord neuron expressing sfrp-1 was identified as DA7 (indicated by an arrow). Scale bar: 10 μm. (F) Quantification of Wnt and sfrp-1 smFISH spots along the anteroposterior axis of early L1 stage larvae. A DAPI stained animal is included for orientation. For all images, anterior is towards the left and posterior is towards the right. Error bars indicate s.e.m. (cwn-1, n=10; cwn-2, egl-20 and lin-44, n=13; mom-2, n=6 and sfrp-1, n=9).
C. elegans phenotypes, expression constructs and transgenesis
The final positions of the CAN, ALM, HSN and Q descendants and the polarity of the V5 seam cell division were scored in L1 larvae by Nomarski microscopy (Harris et al., 1996; Whangbo et al., 2000). Statistical significance of differences in the final positions of the Q.d, CAN, ALM and HSN neurons was determined using Fisher's exact test in SPSS (version 19). The polarity of the ALM and PLM neurons, dye filling, mab-5::lacZ reporter transgene activation and P12 to P11 fate transformation were analyzed as described (Salser and Kenyon, 1992; Herman and Horvitz, 1994; Prasad and Clark, 2006). To generate a heat shock inducible sfpr-1 expression plasmid, the sfrp-1 cDNA was cloned into the pPD49.78 vector. The plasmid was injected with a Pmyo-2::Tomato co-injection marker and integrated as described (Mello and Fire, 1995).
RESULTS
The five C. elegans Wnt genes are expressed in a series of partially overlapping domains along the anteroposterior axis
To quantitatively determine the spatio-temporal expression patterns of the five C. elegans Wnt genes, we performed single molecule mRNA FISH (smFISH) to measure endogenous transcript levels in staged L1 larvae and during embryonic development (Raj et al., 2008). Using this technique, we were able simultaneously to label and visualize individual transcripts of up to three Wnt genes as bright diffraction-limited fluorescent spots in animals with preserved shape (Fig. 1A). Counting of these spots using a custom program written in MATLAB allowed us to measure transcript levels in any three-dimensional region of the animal (Fig. 1B,C,F). These measurements revealed that the expression patterns of the five Wnt genes are strikingly reproducible in wild-type animals of the same developmental stage (Fig. 1A,F). In general, the overall transcript expression profile of the different Wnt genes agreed with previous expression patterns obtained with transgenes expressing transcriptional or translational reporter constructs; but, as detailed below, there were a number of important differences.
We found that of the five Wnt transcripts, three (lin-44, egl-20 and cwn-1) were mostly localized to the posterior half of L1 larvae (Fig. 1A,F), in a pattern that was already present at the comma stage of embryonic development (Fig. 2A,B). lin-44 transcripts were present in the tail hypodermal cells hyp8, hyp9, hyp10 and hyp11, and at later larval stages in the phasmid socket cells PHso1 and PHso2 (see Fig. S1A in the supplementary material), as previously reported (Herman et al., 1995). In addition, we found that lin-44 is expressed in the rectal epithelial cells B and Y, demonstrating that lin-44 has a more anterior expression domain than has been observed using reporter transgenes. egl-20 was expressed in the rectal epithelial cells K, F, U and B, in the anal depressor muscle and in P11/12, which is in agreement with previous reporter studies (Whangbo and Kenyon, 1999). However, we found that in L1 larvae, egl-20 was also expressed in the posterior ventral body wall muscle quadrants VL23 and VR24 and the rectal epithelial cell Y. cwn-1 was mainly expressed in posterior body wall muscle cells (Fig. 1A,B) and in the M cell descendants that give rise to body wall muscle cells and the vulva and uterine muscle cells (Fig. 1F; see Fig. S1A in the supplementary material). In addition, several cells were found to co-express cwn-1 and egl-20, including the anal depressor muscle, the body wall muscle quadrants VL23 and VR24 and P11/12. Interestingly, we observed that the two lateral canal associated neurons (CANs) simultaneously induce cwn-1 expression during late L1 (see Fig. S1A,B in the supplementary material), an expression that persists throughout larval development.
mom-2 has previously been reported to be widely expressed along the anteroposterior axis of developing larvae, with expression in body wall muscle cells, ventral cord neurons, intestinal cells and seam cells (Gleason et al., 2006). By contrast, we found that mom-2 shows a restricted expression pattern, with mom-2 transcripts localizing only to the germ cell precursors Z2 and Z3, their descendants and a few unidentified cells in the tail (Fig. 1D). mom-2 expression in the germ cells continued throughout larval development, whereas the tail expression reached a maximum at the mid-L1 stage and disappeared before the L1 to L2 molt (see Fig. S1D in the supplementary material). In addition, one or two mom-2 transcripts were occasionally detected in posterior seam cells in early L1 larvae. Consistent with the early embryonic function of mom-2 (Thorpe et al., 1997), we found that mom-2 transcripts were already present in the zygote (Fig. 2B). At the four-cell stage, mom-2 transcripts were enriched in the P2 blastomere. During later stages of embryonic development, mom-2 transcripts were restricted to the posterior, with expression remaining in the tail and in the region of the Z2 and Z3 germ line precursors in comma stage embryos.
The larval expression of cwn-2 has been described using different reporter transgenes, showing either a general expression in body wall muscle cells and ventral nerve cord neurons along the whole body axis (Gleason et al., 2006), or a more restricted expression in the pharynx, anterior muscle cells and the intestine (Kennerdell et al., 2009; Song et al., 2010). We found that cwn-2 transcripts mainly localized to head neurons, anterior body wall muscle cells, anterior P.n cells and the intestine (Fig. 1E,F; see Fig. S1A in the supplementary material). The highest cwn-2 transcript count was observed around the terminal bulb of the pharynx, with a gradual decline in expression levels in more posterior cells. The mostly anterior expression of cwn-2 and posterior expression of cwn-1 was already observed at the 100-cell stage of embryonic development (Fig. 2A).
Quantification of Wnt transcripts along the anteroposterior axis of staged L1 larvae revealed that the five Wnt genes are expressed in a series of partially overlapping expression domains (Fig. 1F). At the posterior end of the animal, only lin-44 is expressed. Around the rectum, the most abundantly expressed Wnt gene is egl-20. In the posterior region between the gonad premordium and the rectum, cwn-1 is the dominant Wnt, whereas the anterior half of the animal is the domain of cwn-2 expression. This overall anteroposterior expression profile was already present at the comma stage of embryonic development (Fig. 2A,B) and remained essentially unchanged during the remainder of L1 larval development, although quantification of total Wnt transcript numbers revealed changes in the expression levels of the five Wnt genes (see Fig. S1A,C in the supplementary material). Thus, whereas mom-2 expression remained mostly unchanged during early larval development and there was only a gradual increase in the expression of lin-44 and egl-20, there was a sharp increase in the expression of cwn-1 and cwn-2.
The anteriorly expressed secreted Frizzled-related protein gene sfrp-1 controls neuronal migration along the anteroposterior axis
The activity of Wnt proteins is modulated by secreted Wnt-binding proteins such as members of the secreted Frizzled-related proteins (SFRPs), an ancient family of Wnt regulators that are present in organism ranging from sponges to vertebrates (Bovolenta et al., 2008) (see Fig. S2A in the supplementary material). Sequence similarity searches revealed that the C. elegans genome contains a single SFRP ortholog encoded by the predicted gene Y73B6BL.21, which we renamed sfrp-1. Similar to other SFRP family members, SFRP-1 contains a cysteine-rich Frizzled-related domain (CRD) and a netrin-related (NTR) domain (see Fig. S2B in the supplementary material), which is characterized by positively charged residues and six conserved cysteines (Chong et al., 2002).
To determine the expression pattern of sfrp-1, we analyzed sfrp-1 mRNA localization using smFISH. As shown in Fig. 1E, sfrp-1 is expressed in four stripes of cells in the head region, an anterior specific expression that is already present at the 100-cell stage of embryonic development (Fig. 2A). Using a muscle-specific marker, these cells were identified as head body wall muscle cells. In addition, we found that sfrp-1 is expressed at low levels in a single posterior ventral nerve cord neuron and occasionally in one or more cells around the rectum (Fig. 1E; see Fig. S1E in the supplementary material). The predominantly anterior expression of sfrp-1 indicates that SFRP-1 and the posteriorly expressed Wnts form opposing gradients. Although we have not been able to visualize directly such an SFRP-1 concentration gradient, the genetic analysis of sfrp-1 function described below demonstrates that SFRP-1 has both short- and long-range functions in modulating Wnt activity.
To investigate the function of sfrp-1, we used the deletion allele gk554, which truncates the sfrp-1 gene upstream of the CRD and NTR domains and probably represents the null phenotype (see Fig. S2B in the supplementary material). sfrp-1(gk554) is viable and does not induce obvious morphological defects. However, sfrp-1 mutants show clear alterations in the Wnt-dependent anteroposterior positioning of migrating neuroblasts.
One group of neuroblasts that migrates along the anteroposterior axis are the Q neuroblast descendants (Sulston and Horvitz, 1977; Hedgecock et al., 1987). At the end of embryogenesis, two Q neuroblasts are generated at equivalent positions on the left (QL) and right (QR) lateral side of the animal (Fig. 3A). During the first stage of larval development, the two Q neuroblasts each generate three descendants that migrate in opposite directions: on the left side, the QL descendants (QL.d) migrate towards the posterior, whereas on the right side, the QR.d migrate towards the anterior. Both anterior and posterior migration is controlled by Wnt signaling. The posterior migration of the QL.d is mediated by EGL-20, which triggers a canonical Wnt/β-catenin pathway to induce expression of the target gene mab-5 and to direct migration towards the posterior (Harris et al., 1996; Maloof et al., 1999; Whangbo and Kenyon, 1999). The anterior migration of the QR.d is also dependent on EGL-20, but here EGL-20 functions together with CWN-1 to activate a β-catenin independent Wnt signaling pathway that is required for anterior directed migration (Zinovyeva et al., 2008). Although the mechanism remains to be established, current models suggest that a difference in response threshold to EGL-20 determines which pathway is activated (Whangbo and Kenyon, 1999). Thus, QL is primed to activate canonical Wnt/β-catenin signaling in response to EGL-20, whereas QR will only activate this pathway when EGL-20 is overexpressed. At intermediate levels, overexpression of EGL-20 induces overmigration of the QR.d, indicating that Wnt signaling activity not only specifies the direction of migration, but also influences the position at which the cells terminate their migration (Whangbo and Kenyon, 1999). To investigate whether sfrp-1 regulates the Wnt dependent migration of the Q descendants, we determined the final positions of the Q descendants Q.paa and Q.pap relative to the hypodermal seam cells V1 to V6. We found that the QL.d localized around their normal positions in sfrp-1 mutants (Fig. 3B). There was, however, a clear change in the final position of the QR.d, with the QR.d migrating significantly further into the anterior than in wild-type animals (P<0.001, Fisher's exact test). As this phenotype is similar to the extended migration induced by EGL-20 overexpression, these data are consistent with a negative regulatory role for sfrp-1 in QR.d migration. Loss of this negative regulatory activity is, however, insufficient to trigger canonical Wnt/β-catenin signaling and mab-5 expression in QR.
Fig. 3.
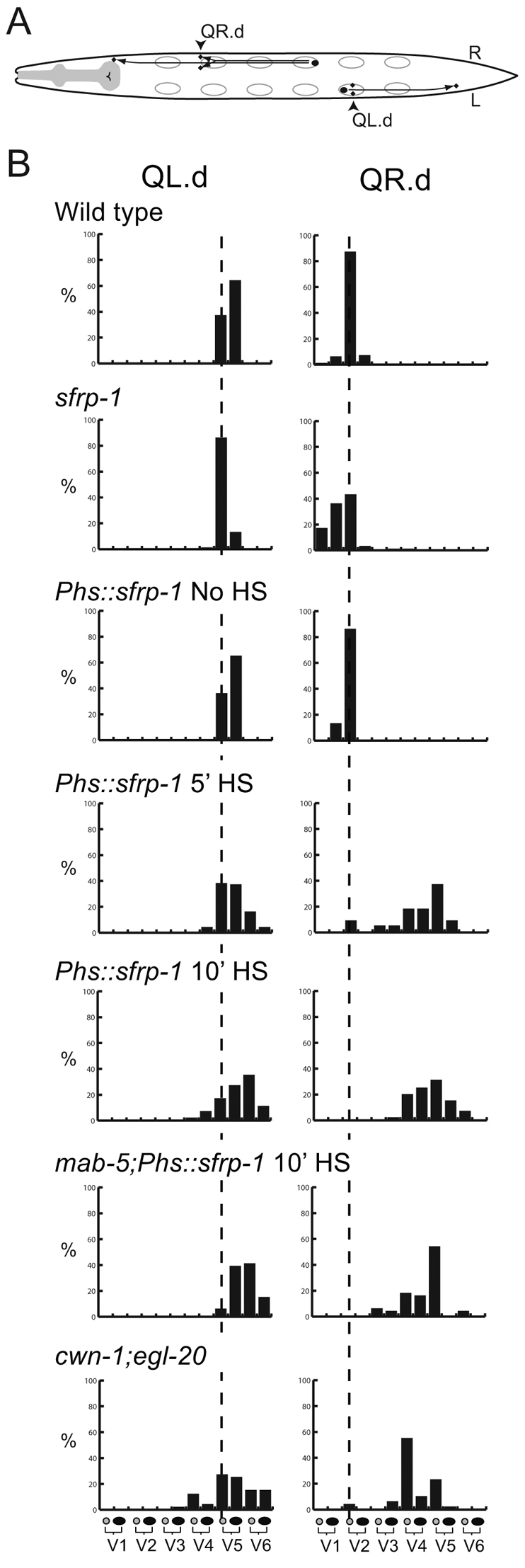
sfrp-1 is required for the migration of the QR descendants. (A) Schematic representation of Q neuroblast descendant migration. Arrowheads indicate the final positions of the Q.paa and Q.pap cells. (B) The final positions of the left and right Q.paa and Q.pap cells are indicated relative to the invariant positions of the seam cells V1 to V6 (n>100 for wild type and sfrp-1; for the other conditions, n>50). The broken lines indicates the wild-type position.
In addition to the defect in the anterior migration of the QR.d, we found that sfrp-1 mutants show misplacement of the ALM and CAN neurons (Fig. 6A; see Fig. S5A in the supplementary material). Both neurons migrate during the end of embryogenesis from the anterior to final positions in the mid-body region (Sulston et al., 1983; Hedgecock et al., 1987). In both cases, posterior migration depends on the combined activity of CWN-1 and CWN-2 (Zinovyeva et al., 2008). In sfrp-1 mutants, the posterior migration of the ALM neurons was significantly truncated (P<0.001) (Fig. 6B). Also, in the case of the CAN neurons, mutation of sfrp-1 induced undermigration (P<0.001), although this effect was less pronounced than observed with the ALM neurons (see Fig. S5B in the supplementary material).
Fig. 6.
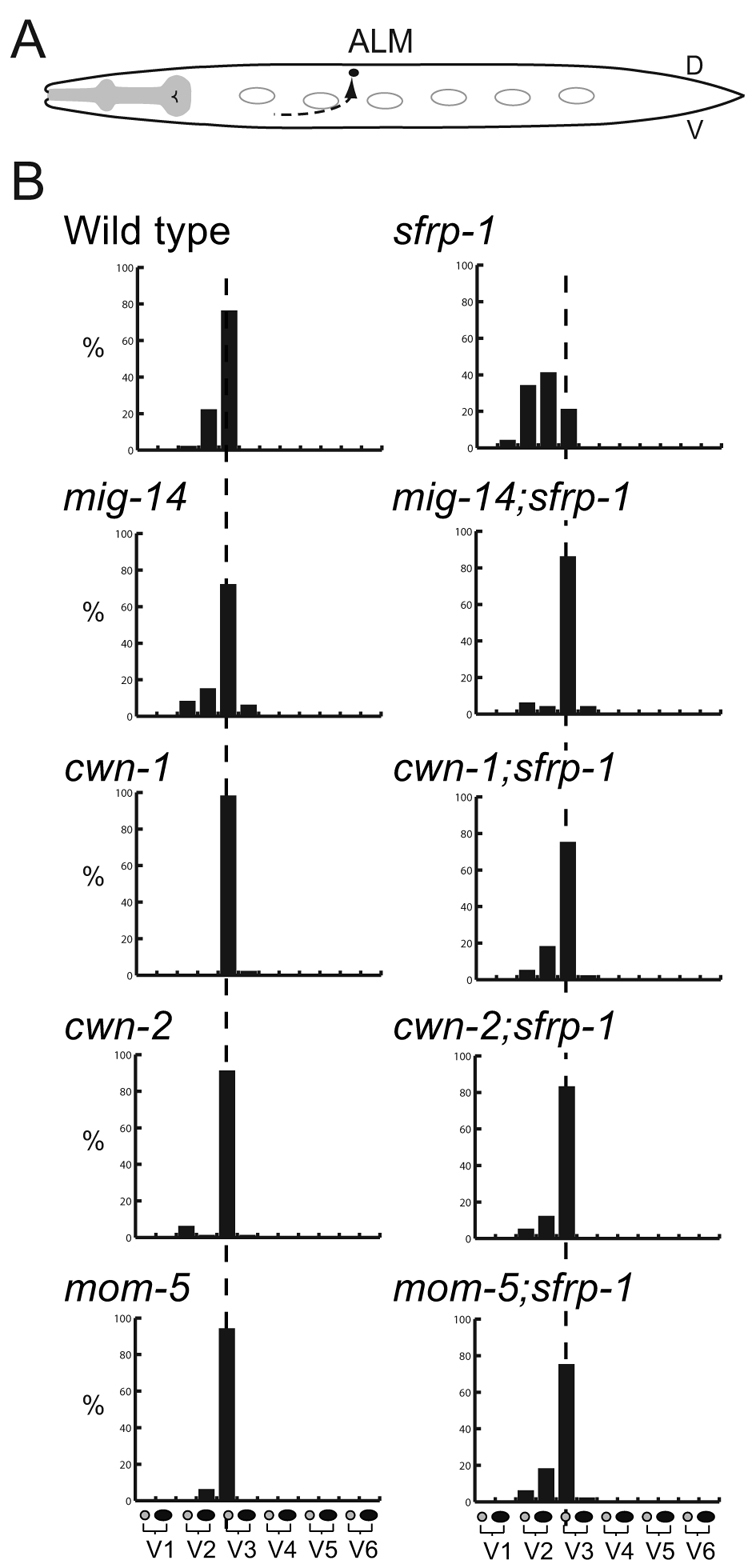
The sfrp-1-induced undermigration of the ALM neurons is suppressed by mutation of cwn-1 or cwn-2. (A) Schematic representation of the ALM migration. Note that the migration takes place at the end of embryogenesis. (B) The final positions of the ALML and ALMR neurons are indicated relative to the seam cells V1 to V6 (n >50). The broken lines indicate the wild-type position.
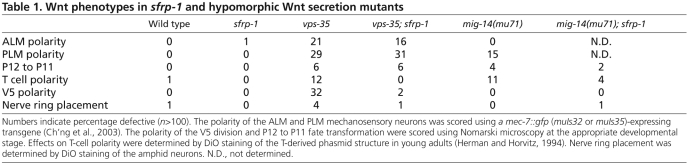
Mutation of sfrp-1 did not significantly affect other Wnt-dependent processes (Table 1; data not shown). Thus, there were no defects in the anterior migration of the HSN neurons (Pan et al., 2006), in the polarization of the mechanosensory neurons ALM and PLM (Prasad and Clark, 2006), in the polarization of the division of the hypodermal seam cells V5 and T (Herman et al., 1995; Whangbo et al., 2000), in the positioning of the nerve ring (Kennerdell et al., 2009), or in the specification of P12 fate (Jiang and Sternberg, 1998). With the exception of nerve ring positioning, all of these processes control cells in the posterior or mid-body region, whereas the migration of the QR.d, ALM and CAN neurons takes place in the anterior. These results are therefore consistent with the anterior-specific expression of sfrp-1 and a function of SFRP-1 in modulating Wnt activity in the anterior body region.
Table 1.
Wnt phenotypes in sfrp-1 and hypomorphic Wnt secretion mutants
SFRP-1 is a global inhibitor of Wnt signaling
One of the main functions of SFRPs is to negatively regulate Wnt signaling (Kawano and Kypta, 2003). There are, however, also examples of SFRPs functioning as facilitators or direct mediators of Wnt signaling; for example, by assisting the spreading of Wnt in the tissue or by directly interacting with the Wnt receptor Frizzled (Rodriguez et al., 2005; Mii and Taira, 2009). To investigate potential Wnt inhibitory or stimulatory functions of sfrp-1, we overexpressed sfrp-1 using a heat-shock-inducible promoter (Fig. 3B). A short induction of sfrp-1 expression before the Wnt-dependent migration of the Q descendants resulted in an almost complete loss of the anterior migration of the QR.d, a phenotype that is also observed in double mutants of egl-20 and cwn-1, or in mutants in which all five Wnt genes have been deleted (Zinovyeva et al., 2008). Also, the effect of sfrp-1 overexpression on the QL.d was similar to egl-20; cwn-1 double mutants, with a more variable and posterior localization of the cells (Fig. 3B). Taken together, these results show that the migration phenotype induced by overexpression of sfrp-1 closely resembles that of mutants defective in multiple Wnts, consistent with a negative regulatory role for SFRP-1 in the Wnt-dependent control of Q.d migration. This conclusion is further supported by the observation that overexpression of sfrp-1 induces a similar loss of anterior QR.d and QL.d migration in a mab-5 null mutant background (Fig. 3B), demonstrating that the posterior localization of the QR.d and QL.d does not result from activation of the EGL-20 target gene mab-5. We found that sfrp-1 overexpression also inhibited the EGL-20 dependent anterior migration of the HSN neurons as well as other Wnt-dependent processes, such as the polarized division of the seam cell V5 (data not shown), indicating that SFRP-1 can inhibit the activity of most if not all of the Wnt proteins of C. elegans. Importantly, no phenotypes were observed that suggest a stimulatory function of SFRP-1 in Wnt signaling.
To further investigate the function of sfrp-1 in modulating Wnt activity, we analyzed double mutants between sfrp-1 and mutants in which Wnt secretion is reduced: a hypomorphic allele (mu71) of the Wnt sorting receptor mig-14/Wls (Bänziger et al., 2006; Yang et al., 2008) and a null allele of the retromer subunit gene vps-35 (Coudreuse et al., 2006). By assaying whether specific Wnt phenotypes are suppressed or enhanced, these mutants provide a sensitive assay to test negative or positive effects of sfrp-1 on Wnt signaling. In mig-14(mu71) and vps-35 single mutants, a reduction in EGL-20 levels leads to a loss of mab-5 expression in QL (Harris et al., 1996) and anterior migration of the QL.d (Fig. 4). We found that in double mutants with sfrp-1, posterior migration was almost fully restored in mig-14(mu71) and significantly rescued in vps-35. Importantly, the rescue of posterior migration depended on activation of mab-5 expression. Thus, mutation of sfrp-1 failed to restore posterior QL.d localization in a mig-14(mu71); mab-5 double mutant background (Fig. 4). Furthermore, mab-5 expression was markedly increased in the QL.d of mig-14(mu71); sfrp-1 double mutants [20% of mig-14(mu71) versus 75% of mig-14(mu71); sfrp-1 animals showed activation of a mab-5::lacZ reporter, whereas the reporter was expressed in 86% and 83% of wild type and sfrp-1 single mutants, respectively; in each case n>50]. These results suggest that the reduction in EGL-20 signaling can be overcome by removal of SFRP-1, consistent with a negative regulatory role of SFRP-1 in the EGL-20 dependent activation of canonical Wnt/β-catenin signaling in QL. We observed a similar inhibitory role for sfrp-1 in QR.d migration. Thus, whereas the QR.d showed reduced anterior migration in mig-14(mu71) and vps-35 mutants, the QR.d localized at their correct positions in double mutants with sfrp-1 (see Fig. S3 in the supplementary material). A comprehensive analysis of other Wnt phenotypes in mig-14(mu71) and vps-35 mutants showed that loss of sfrp-1 also suppressed defects in the migration of the ALM neurons and the polarity of the seam cells V5 and T (Fig. 6B and Table 1). Importantly, there were no instances in which the vps-35 or mig-14(mu71) phenotype was enhanced by loss of sfrp-1. Taken together with the strong Wnt inhibitory activity of sfrp-1 overexpression, these results support the conclusion that SFRP-1 functions as a global inhibitor of Wnt signaling in C. elegans.
Fig. 4.
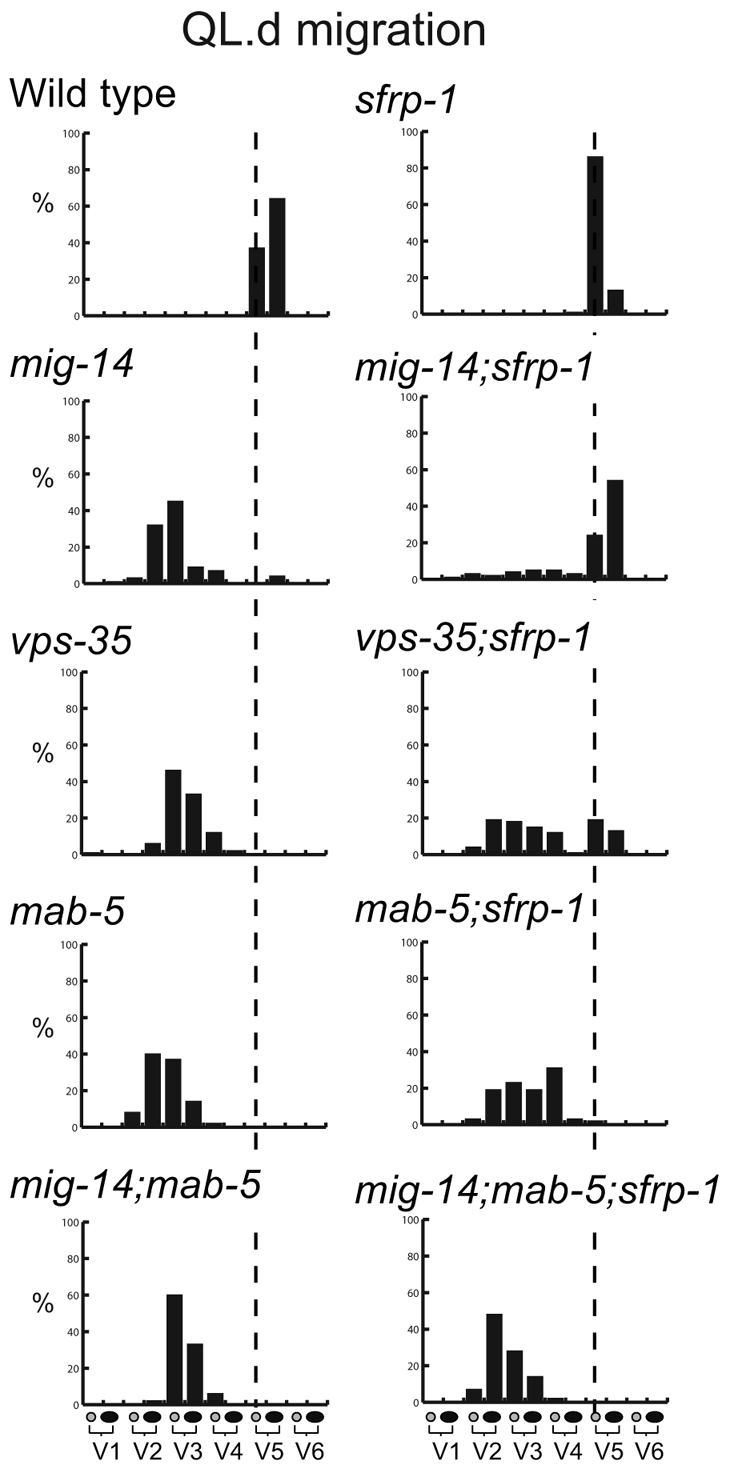
sfrp-1 rescues QL.d migration in hypomorphic Wnt secretion mutants. The final positions of QL.paa and QL.pap cells are indicated relative to the seam cells V1 to V6 (n>50). The broken lines indicate the wild-type position.
The sfrp-1-induced overmigration of the QR descendants is suppressed by mutation of cwn-2
The overmigration of the QR.d in sfrp-1 mutants is similar to the extended migration induced by ubiquitous EGL-20 expression (Whangbo and Kenyon, 1999), indicating that the overmigration is a result of a gain in Wnt signaling activity. To investigate which Wnts mediate the sfrp-1-induced overmigration, we constructed double mutants between sfrp-1 and null mutants of the different Wnt genes. We found that mutation of lin-44 or mom-2 did not suppress the sfrp-1-induced overmigration (Fig. 5). In double mutants between sfrp-1 and egl-20, there was a partial suppression of the overmigration, but also a clear undermigration of the QR.d, an effect that was even more pronounced in double mutants with cwn-1. EGL-20 and CWN-1 function partially redundantly in specifying anterior QR.d migration and loss of either egl-20 or cwn-1 results in a distinct undermigration of the QR.d (Zinovyeva et al., 2008). The intermediate phenotype of the egl-20; sfrp-1 and cwn-1; sfrp-1 double mutants therefore suggests that egl-20 and cwn-1 either function in parallel to sfrp-1 or play only a minor role in the sfrp-1-induced overmigration of the QR.d. By contrast, we found that the sfrp-1-induced overmigration was fully rescued by a null mutation in cwn-2 (Fig. 5). Thus, whereas cwn-2 has no significant effect on QR.d positioning on its own, the QR.d localized at their wild-type position in sfrp-1; cwn-2 double mutants. Taken together, these results indicate that derepression of CWN-2 signaling is primarily responsible for the QR.d overmigration phenotype of sfrp-1 mutants.
Fig. 5.
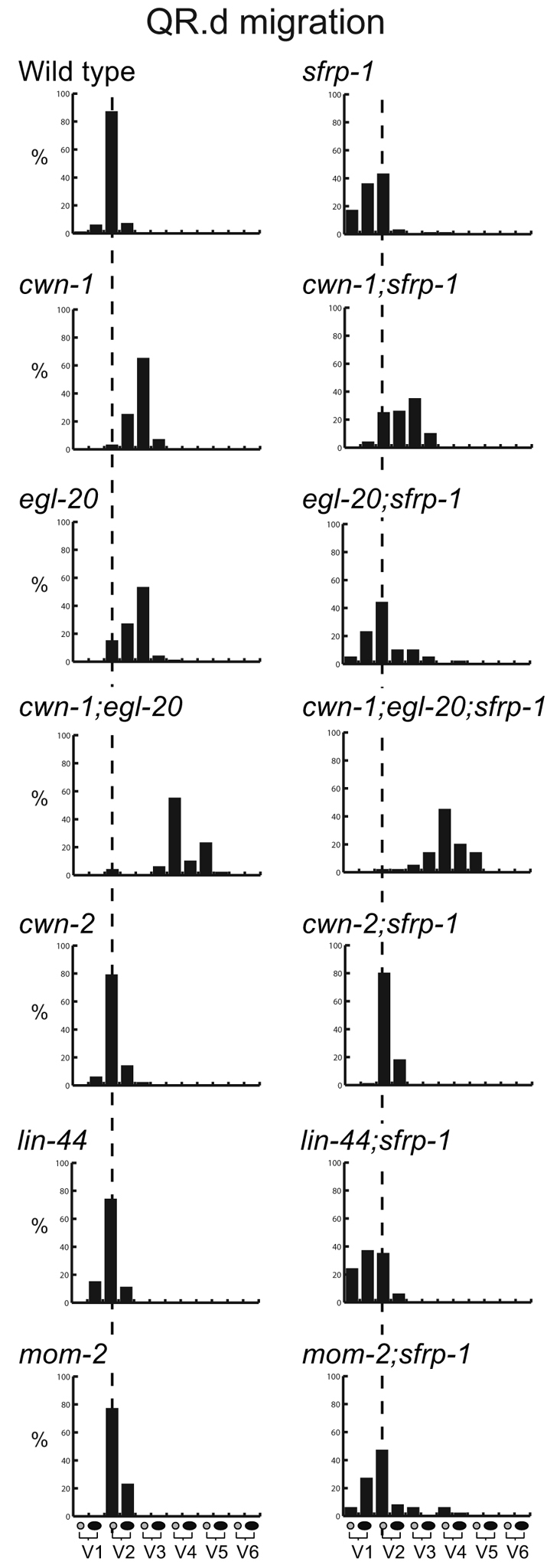
The sfrp-1-Induced overmigration of the OR.d is suppressed by mutation of cwn-2. The final positions of QR.paa and QR.pap cells are indicated relative to the seam cells V1 to V6 (n>50). The broken lines indicate the wild-type position.
The sfrp-1-induced undermigration of the ALM and CAN neurons is suppressed by mutation of cwn-1 or cwn-2
The posterior migration of the ALM neurons is dependent on the redundant activity of CWN-1 and CWN-2, with a strong inhibition of migration in cwn-1; cwn-2 double mutants (Zinovyeva et al., 2008). A similar undermigration is observed in sfrp-1 mutants, indicating that Wnt overactivity also interferes with the correct posterior migration of the ALM neurons. This conclusion is supported by the observation that the ALM undermigration phenotype of sfrp-1 is suppressed by reducing Wnt secretion through mutation of the Wnt sorting receptor mig-14/Wls (Fig. 6B). To investigate which Wnts are required for the sfrp-1-induced undermigration of the ALM neurons, we analyzed double mutants with null alleles of each of the five different Wnt genes and tested which combination could suppress the ALM undermigration phenotype. Whereas lin-44, egl-20 and mom-2 did not affect the sfrp-1-induced ALM undermigration, mutation of cwn-1 or cwn-2 fully restored the migration of the ALM neurons to their wild-type positions (Fig. 6B; see Fig. S4B in the supplementary material). These results suggest that in the absence of SFRP-1, overactivity of CWN-1 and CWN-2 interferes with the correct positioning of the ALM neurons, and that normal migration can be restored by removing either of the two Wnt genes. Furthermore, the sfrp-1-induced undermigration of the ALM neurons was fully suppressed in double mutants between sfrp-1 and the Frizzled mom-5 (Fig. 6B), indicating that CWN-1 and CWN-2 control ALM positioning through the MOM-5/Frizzled receptor.
Similar results were obtained for the sfrp-1-induced undermigration of the CAN neurons, which was also dependent on CWN-1 and CWN-2. Thus, CAN undermigration was not suppressed in lin-44, egl-20 or mom-2 mutants, but was rescued in either cwn-1 or cwn-2 mutants (see Fig. S5B in the supplementary material).
The function of SFRP-1 in suppressing CWN-1 and CWN-2 activity is consistent with the anterior expression of sfrp-1 and is in agreement with a role for SFRP-1 in modulating Wnt activity in the anterior body region. Our results show that this inhibitory activity is particularly important for controlling the Wnt-dependent migration of neuroblasts along the primary body axis of C. elegans.
DISCUSSION
During C. elegans development, the migration of neuroblasts along the anteroposterior axis is controlled through a complex network of partially redundantly acting Wnt proteins. Here, we used smFISH to map quantitatively the spatio-temporal expression pattern of the C. elegans Wnt genes. We show that the five Wnt genes are expressed in partially overlapping expression domains along the anteroposterior axis, with the most prominent Wnt expression in the posterior body region. Furthermore, we demonstrate that Wnt signaling in the anterior body region is repressed by the secreted Frizzled-related protein SFRP-1. These results show that the anteroposterior positioning of neuroblasts is controlled by opposing Wnt and Wnt inhibitory activities and provide further evidence for the evolutionary conservation of this system in patterning of the primary body axis.
A quantitative gene expression map of the C. elegans Wnt family
Conventional methods for gene expression analysis in C. elegans are mostly based on transgenic reporter constructs (Mello et al., 1991). In this study we used for the first time smFISH (Raj et al., 2008) to determine the spatio-temporal expression pattern of a gene family in C. elegans. By visualizing single transcripts as bright diffraction-limited spots, we could directly measure endogenous gene expression in vivo by counting the number of spots in a three-dimensional area of interest, such as a specific cell or tissue. Importantly, this method also allowed us to quantify dynamic changes in gene expression, such as the transient expression of mom-2 in cells of the tail and the activation of cwn-1 expression in the CAN neurons during the first stage of larval development. We found that the expression level and spatial distribution of transcripts detected by smFISH was highly reproducible between staged animals, indicating that this method accurately visualizes endogenous gene expression patterns. We conclude that smFISH can be used to produce quantitative spatio-temporal maps of endogenous gene expression patterns. It should be noted, however, that the expression pattern of the protein product may be influenced by post-transcriptional regulation.
Our smFISH analysis of the Wnt gene family showed similarities, but also important differences with expression patterns based on transgenic reporter constructs. We found that cwn-2 is mainly expressed in head neurons and anterior body wall muscle cells, resolving a conflict in the previously reported expression patterns for cwn-2 (Gleason et al., 2006; Kennerdell et al., 2009; Song et al., 2010). Furthermore, we observed that mom-2 is not generally expressed along the anteroposterior body axis (Gleason et al., 2006), but is restricted to the germ line precursor cells and transiently to a group of cells in the tail. Another important difference is the expression of lin-44 outside of the tail hypodermal cells (Herman et al., 1995). The more anterior expression of lin-44 in the B and Y rectal epithelial cells is particularly interesting for the function of LIN-44 as a directional signal in T-cell polarity (Goldstein et al., 2006) and for the inhibition of presynaptic assemblies in the DA9 neuron (Klassen and Shen, 2007).
The smFISH analysis revealed that the five Wnt genes are expressed in a series of partially overlapping expression domains, with expression of three of the five Wnt genes in the posterior and one in the anterior half of the body. The expression of the different Wnt genes in serial domains correlates with their function in controlling the migration of neuroblasts along the anteroposterior axis. Thus, the posteriorly expressed Wnt EGL-20 controls migration in the posterior and mid-body region, while CWN-1 and CWN-2 are particularly important for the migration of the QR descendants and the CAN and ALM neurons in the anterior half of the animal (Harris et al., 1996; Pan et al., 2006; Zinovyeva et al., 2008). We propose that the staggered series of Wnt expression domains provides a system for positional information along the anteroposterior body axis of C. elegans.
The secreted Frizzled-related protein SFRP-1 is an inhibitor of Wnt signaling
SFRP proteins are characterized by an N-terminal cysteine-rich domain (CRD) that is similar to the Wnt binding CRD domain of Frizzled (Bovolenta et al., 2008). SFRPs are secreted proteins that have been shown to act as inhibitors of Wnt signaling, most probably by competing with Wnt receptors for Wnt binding. However, SFRPs have also been reported to promote Wnt signaling, for example, by facilitating the spreading of Wnt in the tissue (Mii and Taira, 2009) or by directly interacting with Frizzleds to stimulate signaling in a Wnt-independent manner (Rodriguez et al., 2005). Phylogenetic analysis has shown that the SFRP family appeared very early in metazoan evolution, as clear SFRP orthologs are already present in the cnidarians Hydra and Nematostella vectensis (Guder et al., 2006a). Their function has, however, not been studied in any of the genetically tractable invertebrate model systems. The Drosophila genome does not contain SFRP orthologs, indicating that this gene family may have been lost in insects (but not in all arthropods, as the genome of the tick Ixodes scapularis contains an SFRP ortholog). In this study, we show that the C. elegans genome contains a single SFRP ortholog, sfrp-1, which has enabled us to study potential Wnt inhibitory or stimulatory functions of SFRPs in a well-defined model system. We found that SFRP-1 functions exclusively as an inhibitor of Wnt signaling: first, overexpression of sfrp-1 induced a strong defect in Wnt signaling, similar to the phenotype observed in mutants in which all five Wnt genes have been mutated (Zinovyeva et al., 2008). Second, loss of sfrp-1 suppressed the Wnt signaling defect of mutants that induce a reduction in Wnt secretion and, finally, all the phenotypes observed in sfrp-1 mutants could be suppressed by removing specific Wnts, indicating that mutation of sfrp-1 leads to derepression of Wnt signaling. These results suggest that the stimulatory function of SFRPs in Wnt signaling has either been lost in the nematode lineage, or is a more recent invention of organisms of higher complexity. Studies on the cnidarian SFRPs may shed light on this issue.
An anterior SFRP-1 inhibitory gradient controls the positioning of neuroblasts in the anterior body region
The predominant anterior expression of sfrp-1 suggests that it counteracts the more posteriorly expressed Wnts. Loss of this inhibitory activity leads to defects in the migration of neuroblasts in the anterior body region. Thus, the QR.d migrate too far into the anterior, whereas the extent of the posterior migration of the CAN and ALM neurons is reduced. In each of these cases, the final position of the cells is shifted anteriorly, indicating that SFRP-1 counteracts a Wnt activity that promotes anterior localization. We found that the sfrp-1-induced anterior displacement of the QR.d could be suppressed by mutation of cwn-2, whereas ALM and CAN migration could be restored by deletion of either cwn-2 or cwn-1. These results are consistent with a local inhibitory function of SFRP-1 in controlling the activity of the two most anteriorly expressed Wnts.
In addition to this short-range function in the anterior body region, our experiments in Wnt secretion mutants showed that SFRP-1 also has a long-range inhibitory activity. Thus, mutation of sfrp-1 rescued the posterior migration of the QL.d and the polarity of the V5 and T cell divisions in hypomorphic Wnt secretion mutants, consistent with a function of SFRP-1 in modulating Wnt activity in the mid to posterior body region. This long-range inhibition may fine-tune the activity gradients of the posteriorly expressed Wnt genes.
An evolutionarily conserved function of Wnts and Wnt inhibitors in patterning the primary body axis
We found that four out of the five C. elegans Wnt genes are expressed in a series of partially overlapping domains along the anteroposterior axis. This staggered expression is remarkably similar to the expression of Wnt genes in the cnidarian Nematostella vectensis (Kusserow et al., 2005) and in the planarian Schmidtea mediterranea (Petersen and Reddien, 2008). It has been proposed that the staggered expression of Wnt genes provides an ancestral mechanism for positional information along the primary body axis (Guder et al., 2006a) and our results suggest that C. elegans has retained such a system.
Another important similarity is the anterior-specific expression of sfrp-1 and the mostly posterior expression of the Wnt genes. This opposite expression of Wnts and Wnt inhibitors is already present in cnidarians, where Wnt inhibitors are expressed at the aboral side and Wnts at the oral side of the primary body axis (Hobmayer et al., 2000; Kusserow et al., 2005; Guder et al., 2006b; Lee et al., 2006). Posterior Wnt signaling and anterior Wnt inhibition is also a central feature of vertebrate neurectodermal patterning, with the formation of the eyes and anterior brain structures depending on the anterior activity of both intracellular and secreted Wnt inhibitory factors (Kim et al., 2000; Kiecker and Niehrs, 2001; Niehrs, 2006). In protostomes, anterior specific expression of an SFRP has been observed in Schmidtea mediterranea (Petersen and Reddien, 2008), but Wnt inhibitors have not been studied in any of the other protostome model organisms. Our studies in C. elegans show that the opposite expression of Wnts and Wnt inhibitors is also an important feature of nematode development, supporting the notion that a system of posterior Wnt activity and anterior Wnt inhibition is a unifying principle of primary body axis specification in animals (Petersen and Reddien, 2009).
Supplementary Material
Acknowledgements
We thank Dr Bert Hobmayer for critically reading the manuscript, Remco Mentink for assistance with statistical analysis, Dr Andrew Fire for expression vectors and the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis) for strains. This work was funded by a NWO VIDI fellowship (016.076.317) to H.C.K., a Boehringer Ingelheim Foundation fellowship to M.H. and a NIH Pioneer award (1DP1OD003936) to A.v.O. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.064733/-/DC1
References
- Bänziger C., Soldini D., Schütt C., Zipperlen P., Hausmann G., Basler K. (2006). Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125, 509-522 [DOI] [PubMed] [Google Scholar]
- Bovolenta P., Esteve P., Ruiz J. M., Cisneros E., Lopez-Rios J. (2008). Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci. 121, 737-746 [DOI] [PubMed] [Google Scholar]
- Bulow H. E., Berry K. L., Topper L. H., Peles E., Hobert O. (2002). Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc. Natl. Acad. Sci. USA 99, 6346-6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng Q., Williams L., Lie Y. S., Sym M., Whangbo J., Kenyon C. (2003). Identification of genes that regulate a left-right asymmetric neuronal migration in Caenorhabditis elegans. Genetics 164, 1355-1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J. M., Uren A., Rubin J. S., Speicher D. W. (2002). Disulfide bond assignments of secreted Frizzled-related protein-1 provide insights about Frizzled homology and netrin modules. J. Biol. Chem. 277, 5134-5144 [DOI] [PubMed] [Google Scholar]
- Coudreuse D. Y. M., Roël G., Betist M. C., Destrée O., Korswagen H. C. (2006). Wnt gradient formation requires retromer function in Wnt-producing cells. Science 312, 921-924 16645052 [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811 [DOI] [PubMed] [Google Scholar]
- Gleason J. E., Szyleyko E. A., Eisenmann D. M. (2006). Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Dev. Biol. 298, 442-457 [DOI] [PubMed] [Google Scholar]
- Goldstein B., Takeshita H., Mizumoto K., Sawa H. (2006). Wnt signals can function as positional cues in establishing cell polarity. Dev. Cell 10, 391-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guder C., Philipp I., Lengfeld T., Watanabe H., Hobmayer B., Holstein T. W. (2006a). The Wnt code: cnidarians signal the way. Oncogene 25, 7450-7460 [DOI] [PubMed] [Google Scholar]
- Guder C., Pinho S., Nacak T. G., Schmidt H. A., Hobmayer B., Niehrs C., Holstein T. W. (2006b). An ancient Wnt-Dickkopf antagonism in Hydra. Development 133, 901-911 [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Vaz Gomes A., Kenyon C., Liu J., Krause M., Fire A. (1998). Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 12, 2623-2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J., Honigberg L., Robinson N., Kenyon C. (1996). Neuronal cell migration in C. elegans: regulation of Hox gene expression and cell position. Development 122, 3117-3131 [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Culotti J. G., Hall D. H., Stern B. D. (1987). Genetics of cell and axon migrations in Caenorhabditis elegans. Development 100, 365-382 [DOI] [PubMed] [Google Scholar]
- Herman M. A., Horvitz H. R. (1994). The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120, 1035-1047 [DOI] [PubMed] [Google Scholar]
- Herman M. A., Vassilieva L. L., Horvitz H. R., Shaw J. E., Herman R. K. (1995). The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell 83, 101-110 [DOI] [PubMed] [Google Scholar]
- Hobmayer B., Rentzsch F., Kuhn K., Happel C. M., von Laue C. C., Snyder P., Rothbacher U., Holstein T. W. (2000). WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407, 186-189 [DOI] [PubMed] [Google Scholar]
- Jiang L. I., Sternberg P. W. (1998). Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development 125, 2337-2347 [DOI] [PubMed] [Google Scholar]
- Kawano Y., Kypta R. (2003). Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 116, 2627-2634 [DOI] [PubMed] [Google Scholar]
- Kennerdell J. R., Fetter R. D., Bargmann C. I. (2009). Wnt-Ror signaling to SIA and SIB neurons directs anterior axon guidance and nerve ring placement in C. elegans. Development 136, 3801-3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker C., Niehrs C. (2001). A morphogen gradient of Wnt/β-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 128, 4189-4201 [DOI] [PubMed] [Google Scholar]
- Kim C. H., Oda T., Itoh M., Jiang D., Artinger K. B., Chandrasekharappa S. C., Driever W., Chitnis A. B. (2000). Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 407, 913-916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen M. P., Shen K. (2007). Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell 130, 704-716 [DOI] [PubMed] [Google Scholar]
- Korswagen H. C. (2002). Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: variations on a common signaling theme. BioEssays 24, 801-810 [DOI] [PubMed] [Google Scholar]
- Korswagen H. C., Coudreuse D. Y. M., Betist M. C., van de Water S., Zivkovic D., Clevers H. C. (2002). The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans. Genes Dev. 16, 1291-1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A., Pang K., Sturm C., Hrouda M., Lentfer J., Schmidt H. A., Technau U., von Haeseler A., Hobmayer B., Martindale M. Q., et al. (2005). Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156-160 [DOI] [PubMed] [Google Scholar]
- Lee P. N., Pang K., Matus D. Q., Martindale M. Q. (2006). A WNT of things to come: evolution of Wnt signaling and polarity in cnidarians. Semin. Cell Dev. Biol. 17, 157-167 [DOI] [PubMed] [Google Scholar]
- Lewis J. A., Fleming J. T. (1995). Basic culture methods. Methods Cell Biol. 48, 3-29 [PubMed] [Google Scholar]
- Leyns L., Bouwmeester T., Kim S. H., Piccolo S., De Robertis E. M. (1997). Frzb-1 is a secreted antagonist of Wnt signaling expressed in the Spemann organizer. Cell 88, 747-756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F., Peng H., Liu X., Kim S. K., Myers E. (2009). A 3D digital atlas of C. elegans and its application to single-cell analyses. Nat. Methods 6, 667-672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof J. N., Whangbo J., Harris J. M., Jongeward G. D., Kenyon C. (1999). A Wnt signaling pathway controls Hox gene expression and neuroblast migration in C. elegans. Development 126, 37-49 [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A. (1995). DNA transformation. Methods Cell Biol. 48, 451-482 [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mii Y., Taira M. (2009). Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083-4088 [DOI] [PubMed] [Google Scholar]
- Niehrs C. (2006). Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 25, 7469-7481 [DOI] [PubMed] [Google Scholar]
- Pan C. L., Howell J. E., Clark S. G., Hilliard M., Cordes S., Bargmann C. I., Garriga G. (2006). Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10, 367-377 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2008). Smed-β-catenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327-330 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2009). Wnt signaling and the polarity of the primary body axis. Cell 139, 1056-1068 [DOI] [PubMed] [Google Scholar]
- Prasad B. C., Clark S. G. (2006). Wnt signaling establishes anteroposterior neuronal polarity and requires retromer in C. elegans. Development 133, 1757-1766 [DOI] [PubMed] [Google Scholar]
- Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Esteve P., Weinl C., Ruiz J. M., Fermin Y., Trousse F., Dwivedy A., Holt C., Bovolenta P. (2005). SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat. Neurosci. 8, 1301-1309 [DOI] [PubMed] [Google Scholar]
- Salser S. J., Kenyon C. (1992). Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature 355, 255-258 [DOI] [PubMed] [Google Scholar]
- Silhankova M., Korswagen H. C. (2007). Migration of neuronal cells along the anterior-posterior body axis of C. elegans: Wnts are in control. Curr. Opin. Genet. Dev. 17, 320-325 [DOI] [PubMed] [Google Scholar]
- Song S., Zhang B., Sun H., Li X., Xiang Y., Liu Z., Huang X., Ding M. (2010). A Wnt-Frz/Ror-Dsh pathway regulates neurite outgrowth in Caenorhabditis elegans. PLoS Genet. 6, e10010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev. Biol. 56, 110-156 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119 [DOI] [PubMed] [Google Scholar]
- Tendeng C., Houart C. (2006). Cloning and embryonic expression of five distinct sfrp genes in the zebrafish Danio rerio. Gene Expr. Patterns 6, 761-771 [DOI] [PubMed] [Google Scholar]
- Thorpe C. J., Schlesinger A., Carter J. C., Bowerman B. (1997). Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90, 695-705 [DOI] [PubMed] [Google Scholar]
- van Amerongen R., Nusse R. (2009). Towards an integrated view of Wnt signaling in development. Development 136, 3205-3214 [DOI] [PubMed] [Google Scholar]
- Whangbo J., Kenyon C. (1999). A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol. Cell 4, 851-858 [DOI] [PubMed] [Google Scholar]
- Whangbo J., Harris J., Kenyon C. (2000). Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans. Development 127, 4587-4598 [DOI] [PubMed] [Google Scholar]
- Yang P. T., Lorenowicz M. J., Silhankova M., Coudreuse D. Y., Betist M. C., Korswagen H. C. (2008). Wnt signaling requires retromer-dependent recycling of MIG-14/Wntless in Wnt-producing cells. Dev. Cell 14, 140-147 [DOI] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Forrester W. C. (2005). The C. elegans Frizzled CFZ-2 is required for cell migration and interacts with multiple Wnt signaling pathways. Dev. Biol. 285, 447-461 [DOI] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Yamamoto Y., Sawa H., Forrester W. C. (2008). Complex network of Wnt signaling regulates neuronal migrations during Caenorhabditis elegans development. Genetics 179, 1357-1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.