Abstract
Psoriasis is well known as a chronic inflammatory dermatosis. The disease affects persons of all ages and is a burden worldwide. Psoriasis is associated with various diseases such as arthritis. The disease is characterized by well-demarcated lesions on the skin of the elbows and knees. Various genetic and environmental factors are related to the pathogenesis of psoriasis. In order to identify enzymes that are potential therapeutic targets for psoriasis, we utilized a computational approach, combining microarray analysis and protein interaction prediction. We found 6,437 genes (3,264 upregulated and 3,173 downregulated) that have significant differences in expression between regions with and without lesions in psoriasis patients. We identified potential candidates through protein-protein interaction predictions made using various protein interaction resources. By analyzing the hub protein of the networks with metrics such as degree and centrality, we detected 32 potential therapeutic candidates. After filtering these candidates through the ENZYME nomenclature database, we selected 5 enzymes: DNA helicase (RUVBL2), proteasome endopeptidase complex (PSMA2), nonspecific protein-tyrosine kinase (ZAP70), I-kappa-B kinase (IKBKE), and receptor protein-tyrosine kinase (EGFR). We adopted a computational approach to detect potential therapeutic targets; this approach may become an effective strategy for the discovery of new drug targets for psoriasis.
1. Introduction
Psoriasis is a common inflammatory disease affecting more than 25 million people in North America and Europe. It is associated with arthritis, myopathy, enteropathy, spondylotic joint disease, and atopic dermatitis. This disease is characterized by well-demarcated lesions on the skin of the elbows, knees, and scalp. It is an autoimmune disease triggered by an activated cellular immune system resulting from a combination of genetic and environmental factors. It is also frequently inherited and is passed from one generation to the next [1].
Many factors trigger psoriasis, including bacterial pharyngitis, stress, and various medications (e.g., lithium and β-blockers). Perturbation of epidermal keratinocytes is considered an activating signal in psoriasis [2], and in regions with psoriasis lesions, keratinocyte proliferation is increased along with inflammation and angiogenesis [3]. Recent studies have reported that the interaction between T-cells and keratinocytes gives rise to a cytokine soup dominated by Th1-type and Th17-type cytokines such as interleukin- (IL-) 12, IL-17, interferon- (IFN-) γ, and tumor necrosis factor (TNF) [4]. In addition, keratinocytes stimulated with IL-20 upregulate a variety of inflammatory genes, including monocyte chemotactic protein-1 (MCP-1) and myeloid-related protein-14 (MRP-14) [5, 6].
Some genetic studies have reported a strong association between psoriasis and human leukocyte antigen- (HLA-) C, particularly with the HLA-Cw0602 allele. Individuals who are homozygous have a 2.5-fold higher risk of developing psoriasis than those who are heterozygous [7]. Moreover, a genomewide association study revealed that polymorphisms in genes related to IL-23 and nuclear transcription factor κB (NFκB) signaling are associated with psoriasis [8]. TNF is currently considered a major target in psoriasis pathogenesis, because much higher levels of TNF are found in lesional skin than in normal skin [9]. Treatment with TNF antagonists elicits a response in patients with psoriasis. However, despite the success of anti-TNF-α therapies, the involvement of TNF-α in disease pathogenesis is not yet fully understood. Furthermore, these drugs have clinical nonresponse rates that range from 20% to 50% in patients with psoriasis [10]. Therefore, there is a need for new and effective drug targets and compounds.
New research initiatives have been undertaken to collect high-throughput mRNA expression and protein-protein interaction (PPI) data from different organisms. This important source of biological information has been effectively employed in the search for new drugs [11]. Systematic analysis using bioinformatics has enabled researchers to extract and manipulate biological information with the goal of understanding the pathogenesis of disease. In particular, the combined analysis of gene expression and PPI may help identify candidates that are potential therapeutic targets. Recent studies analyzing protein interaction networks have been carried out in Saccharomyces cerevisiae and Caenorhabditis elegans [12, 13]; such studies have confirmed that topological metrics of protein interaction networks are useful for predicting essential target proteins. These studies have also been expanded to organisms of medical importance, such as the malaria parasite [14], as a starting point for the discovery of new drug targets. In humans, the analysis of PPIs has also been useful in detecting important proteins, such as hub proteins, when the interactions were predicted using a homologous approach [15].
To better understand the pathogenesis of psoriasis and to identify potential therapeutic targets, we performed a microarray analysis comparing lesional and nonlesional psoriatic skin and a protein interaction network analysis that was constructed using differentially expressed genes obtained from the microarray data. We identified potential therapeutic or drug target candidates by analyzing the protein interaction network with the metrics of degree and centrality. We then selected the enzymes from the candidates and detected nonsynonymous single-nucleotide polymorphisms (SNPs) in the enzyme genes that could cause structural changes in the proteins. These putative enzyme targets are a starting point for the discovery of new psoriasis drugs.
2. Materials and Methods
2.1. Microarray Analysis Related to Psoriasis
Microarray data from psoriasis patients were downloaded from Gene Expression Omnibus (GEO), which is a public database of centrally archived raw microarray data [16]. We used 2 microarray datasets (GDS2518 and GDS3539) generated using Affymetrix human genome microarrays, which have more than 4 million gene expression measurements. The GDS2518 dataset contained transcriptome data of lesional and nonlesional skin from 13 patients with plaque-type psoriasis [17]. The GDS3539 dataset contained similar data from 33 patients [18]. In order to identify genes that are differentially expressed in psoriasis patients, we compared lesional and nonlesional skin data to microarray datasets.
2.2. Identification of Differentially Expressed Genes from Transcriptome
We removed probe redundancy because 1 gene has several probes on a single microarray chip. After removing the redundancy, the average expression profiles were calculated for the probe clusters having multiple expression profiles. From each of the given microarray datasets, we obtained differentially expressed genes (DEGs) by unpaired two class analysis (Sigma = 2.4, Q-value = 0.0001) by using significance analysis for microarray (SAM) [19]. We then combined the DEGs obtained from GDS2518 and GDS3539.
2.3. PPI Resources
Predictions of PPI have been applied in various studies in order to understand the mechanism of disease development, find key proteins related to species lethality, and search for drug targets between a host and pathogen [20]. PPI resources were assembled from a combination of several experimental protein interaction databases. The protein interaction resources include 6 databases: DIP [21], BIND [22], IntAct [23], MINT [24], HPRD [25], and BioGrid [26]. We performed a redundancy test to remove identical protein sequences from the interaction databases. The databases contain 116,773 proteins and 229,799 interactions.
2.4. Protein Network Prediction from the DEGs
There are computational methods for predicting PPIs such as gene neighborhood [27], gene fusion [28], phylogenetic profile [29], and interolog [30]. In particular, the interolog approach is widely used to predict PPIs when the sequences of target proteins are known. In the interolog approach, the interaction of 2 query proteins is predicted when both have homologousproteins that are already known to interact [30]. A protein network of DEGs selected from the microarrays was predicted from homologous interactions. To find homologous interactions among the DEGs, we converted the DEGs to proteins, and we aligned these with proteins from the interaction resources using PSI-BLAST [31] with a minimal cutoff of 40% sequence identity, 70% length coverage, and an E-value of 0.0001.
2.5. Detection of Essential Proteins with Topological Metrics and Selecting for Enzymes
The protein network analysis has been applied to find essential proteins, such as hubs, that are related to a disease or biological pathway [11]. To find essential proteins, we performed a protein network analysis with topological metrics such as degree and betweenness centrality using Perl program. Proteins with a high number of interaction partners in the network were regarded as degree-based hubs. Proteins with many shortest paths between other proteins were regarded as betweenness-based hubs. Proteins that have high betweenness scores mainly exist in the middle of the protein network. If the protein is removed from the network, the proportion of unreachable protein pairs and the mean shortest path length between all pairs of reachable nodes are increased [32]. We selected the essential proteins that have the top 1% of total degree and 1% of total betweenness in the protein network as hubs. Finally, the proteins were filtered through the list of enzyme proteins stored in the ENZYME nomenclature database (http://expasy.org/enzyme/).
2.6. Structural Changes Induced by Nonsynonymous SNPs
Nonsynonymous single-nucleotide polymorphisms (nsSNPs) have been implicated in various diseases; nsSNPs can alter protein function [33], destabilize core protein structure, and reduce protein solubility [34]. Because polymorphisms are an important genetic factor in psoriasis [7], we checked for nsSNPs in the selected enzymes using the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/), and we predicted the structural changes in the enzymes caused by the nsSNPs through structure modeling and stability analysis. On the basis of homology modeling, three-dimensional structural models of the target enzymes were built using SWISS-MODEL [35]. The SWISS-MODEL program automatically provides an all-atom model using alignments between the query sequence and known homologous structures. For homology modeling, the known homologous structures of enzymes from the Protein Data Bank (PDB) (http://www.pdb.org/) were used as the structural templates. Further, the stability of the enzyme structure was checked using the CPUSAT program [36].
3. Results and Discussion
3.1. Psoriasis Microarray Coupled with SAM Analysis
We compared the microarray datasets GDS2518 and GDS3539 from regions with and without lesions in psoriasis patients. Using SAM (Sigma 2.6 cutoff value with a 0.0001 Q-value), 6,437 candidate genes were found, and 3,264 genes were upregulated while 3,173 genes were downregulated. Among the listed candidates were several distinct genes that have been known to be associated with psoriasis; these genes coded for TNF-α, IFN-γ receptor 1, IL-8, and IL-20 (data not shown). In this study, we used bioinformatic analyses in which several methods were integrated, including PPI and structure prediction, to detect novel candidate proteins except well-known genes.
3.2. Identification of Putative Therapeutic Candidates
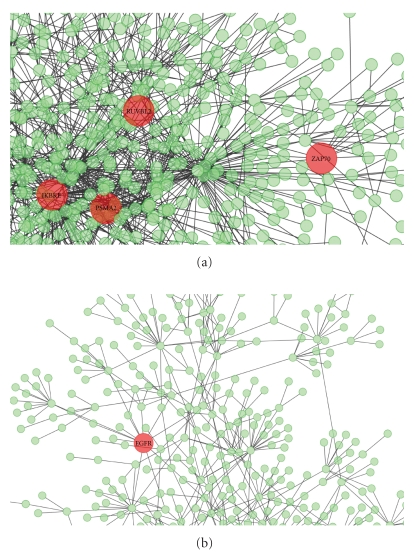
There are several ways to obtain candidate genes for disease analysis, including cDNA microarray and proteomics approaches such as mass spectrometry. Although we could obtain many upregulated or downregulated genes in psoriasis using high-throughput omics approaches, it was very difficult to choose the important genes. However, PPI mapping can help find potential targets among these candidates. Recent studies have successfully applied this approach to discover drug targets using computational predictions of protein networks in the bacterium Mycobacterium tuberculosis and in humans [37–39]. Therefore, we predicted the protein interaction network of psoriasis-related proteins using homologous interaction prediction. We separately constructed 2 protein interaction networks using 3,264 upregulated genes and 3,173 downregulated genes in psoriasis. The protein interaction network constructed from the upregulated genes contained 1,310 proteins and 1,934 interactions, and the network constructed from the downregulated genes contained 985 proteins and 1,205 interactions, and they are shown in Figure 1. In order to find potential therapeutic candidates, we calculated the number of interacting partners (degree) [40] and betweenness centrality [41] for each protein. These 2 topological metrics have been shown to improve the detection of essential proteins in protein networks [42]. Betweenness centrality correlates more closely with essentiality than degree, thereby exposing critical nodes that usually belong to the group of scaffold proteins or proteins involved in cross-talk between signaling pathways. This metric has also been proposed in the new paradigm of network pharmacology as a good method for investigating potential drug targets [43]. In the protein network, we selected the top 1% of the degree and centrality rank as hub proteins because this cutoff value identifies the group with the highest probability to be an essential group of proteins. We chose 17 proteins in the upregulated gene protein interaction network and 15 proteins in the downregulated gene protein interaction network. The protein list and network properties are described in Tables 1 and 2. We found the 5 enzymes by filtering the group of detected 32 proteins through the ENZYME nomenclature database. Among the upregulated genes, 4 enzymes were selected as putative targets: DNA helicase (RUVBL2), proteasome endopeptidase complex (PSMA2), non-specific protein-tyrosine kinase (ZAP70), and I-kappa-B kinase (IKBKE). From the downregulated genes, EGF receptor (EGFR) was selected.
Figure 1.
Protein interaction networks constructed by DEGs in psoriasis. The node indicates protein and the edge indicates protein-protein interaction. The red nodes are essential enzymes predicted by topological metrics such as degree and betweenness centrality. (a) Protein interaction network constructed from genes upregulated in microarray data from regions with and without lesions in psoriasis patients. The network consisted of 1,310 proteins and 1,934 interactions. (b) Protein interaction network constructed from downregulated genes. The network consisted of 985 proteins and 1,205 interactions.
Table 1.
Selected hub proteins in the protein interaction network constructed from upregulated genes.
| Gene symbol | Refseq ID | Full name (enzyme number) | Topological metrics |
|---|---|---|---|
| CDKN1A | NP_000380.1 | Cyclin dependent kinase inhibitor 1A | centrality: 64419 |
| RUVBL2 | NP_006657.1 | RuvB like 2 (3.6.4.12: DNA helicase) | centrality: 50925 |
| COPS6 | NP_006824.2 | COP9 constitutive photomorphogenic homolog subunit 6 | centrality:60596 |
| EWSR1 | NP_001156757.1 | Ewing sarcoma breakpoint region 1 | centrality: 62682 |
| FTSJ1 | NP_036412.1 | FtsJ homolog 1 | degree: 27 |
| SFN | NP_006133.1 | 14-3-3 sigma | centrality: 50372 |
| GRB2 | NP_002077.1 | Grb2 | centrality: 397096 |
| CCDC85C | NP_001138467.1 | C14orf65 protein | centrality: 76862 |
| EIF6 | NP_002203.1 | Eukaryotic translation initiation factor 6 | centrality: 61909 |
| ARF6 | NP_001654.1 | ADP ribosylation factor 6 | centrality: 45220 |
| NFKB1 | NP_001158884.1 | NFKB1 | degree: 18 |
| PCNA | NP_002583.1 | Proliferating cell nuclear antigen | centrality: 46316 |
| PINX1 | NP_060354.4 | Pin2 interacting protein X1 | centrality: 51027 |
| PSMA2 | NP_002778.1 | Proteasome subunit alpha type 2 (3.4.25.1: Proteasome endopeptidase complex) | degree: 17 |
| VDAC1 | NP_003365.1 | VDAC1 | degree: 18 |
| ZAP70 | NP_001070.2 | ZAP70 (2.7.10.2: Nonspecific protein-tyrosine kinase) | centrality: 50140 |
| IKBKE | NP_001180250.1 | IKKE (2.7.11.10: I-kappa-B kinase) | centrality: 412760 |
Table 2.
Selected hub proteins in the protein interaction network constructed from downregulatedgenes.
| Gene symbol | Refseq ID | Full name (enzyme number) | Topological metrics |
|---|---|---|---|
| EIF1B | NP_005866.1 | Translation factor sui1 homolog | centrality: 41540 |
| RNPS1 | NP_006702.1 | RNA binding protein S1, serine-rich domain | degree: 21 |
| EGFR | NP_005219.2 | EGF receptor (2.7.10.1: Receptor protein-tyrosine kinase) | centrality: 83945 |
| NINL | NP_079452.3 | KIAA0980 protein | centrality: 53502 |
| HTT | NP_002102.4 | Huntingtin | centrality: 99972 |
| HMGB1 | NP_002119.1 | High mobility group box 1 | centrality: 43032 |
| APP | NP_000475.1 | Amyloid beta A4 protein | centrality: 51716 |
| RIF1 | NP_001171134.1 | Rap1 interacting factor 1 | degree: 17 |
| PLSCR4 | NP_001121778.1 | Phospholipid scramblase 4 | centrality: 60954 |
| BCL6 | NP_001124317.1 | B cell lymphoma 6 protein | degree: 18 |
| TBP | NP_001165556.1 | TATA box binding protein | centrality: 40738 |
| SUMO1 | NP_001005781.1 | SMT3 suppressor of mif two 3 homolog 1 | degree: 21 |
| UNC119B | NP_001074002.1 | Unc-119 homolog B | centrality: 42366 |
| NCOR1 | NP_001177367.1 | Nuclear receptor corepressor 1 | degree: 16 |
| UTP14C | NP_067677.4 | UTP14, U3 small nucleolarribonucleoprotein, homolog C | centrality: 60679 |
3.3. Experimental Evidence of the Predicted Targets Related to Psoriasis
The predicted enzymes were directly or indirectly related to psoriasis. The predicted enzymes have several significant features. IKBKE was already known as a psoriasis-related protein that is essential for the regulation of antiviral signaling and inflammatory pathways. TNF-α, which exists on upstream of IKBKE signaling pathway, has been used as a psoriasis drug target. TNF-α inhibitors such as etanercept are used for treating rheumatoid and psoriatic arthritis. The abnormal activation of T-cells is known to be a factor in the development of psoriasis. Therefore, T-cell infiltration and the inhibition of cytokines are the major modalities for the treatment of psoriasis [1].
ZAP70 plays a role in T-cell development and lymphocyte activation [44]. This enzyme, which is phosphorylated at tyrosine residues upon T-cell antigen receptor (TCR) stimulation, functions in the initial step of TCR-mediated signal transduction in combination with the Src family kinases Lck and Fyn [45]. Mutations in this enzyme cause selective T-cell defects and a severe combined immunodeficiency disease characterized by a selective absence of CD8-positive T-cells [46]. Furthermore, a significant change in ZAP70 expression was reported during the course of chronic lymphocytic leukemia [47].
We found EGFR, which plays a role in morphogenesis by modulating cell adhesion, as a downregulated target. EGFR promotes keratinocyte antimicrobial defenses in a differentiation-dependent manner. Ligands of the EGF family regulate autocrine keratinocyte proliferation, and IL-1 family cytokines have epithelial defense responses [48].
Two proteasome-related enzymes were also found among the upregulated target candidates. RUVBL2 is a DNA helicase that is essential for homologous recombination and DNA double-strand break repair. Recent research reported that the depletion of RUVBL2 leads to tumor growth arrest and that it is overexpressed in a majority of hepatocellular carcinomas [49]. Moreover, the enzyme levels are strictly controlled by a posttranslational mechanism involving proteasomal degradation of newly synthesized proteins [50]. PSMA2 is a proteasome subunit alpha type 2 and is a member of the peptidase T1A family. Proteasome-related enzymes may be valuable as therapeutic candidates since a proteasome blockade causes the overexpression of the suppressor of cytokine signaling (SOCS) 3protein, which inhibits the IFN-α response which are overexpressed in psoriasis [51].
This may be reasonable because these enzymes are important regulators in the mechanism of development of psoriasis. Although more information on the biological functions of these interesting enzymes is needed, they are potential candidates for future drug screening and therapeuticor drug target development.
3.4. Structural Stability Analysis of Enzymes Using nsSNPs
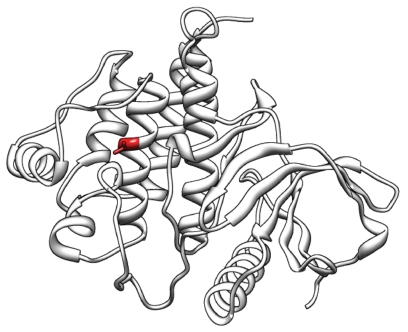
We predicted the structural changes in ZAP70 caused by nsSNPs with structural modeling and stability analysis because mutations can cause selective T-cell defects [46]. Known homologous structures of ZAP70, PDB entries 1M61 and 1U59 (100% and 98.9% sequence identity), were used as structural templates. Currently, 19 nsSNPs in the dbSNP database have been reported to cause nonsense or missense changes in ZAP70. Using the CUPSAT program, we found 10 nsSNPs that cause unstable structural changes. The nsSNPs are shown in Table 3. In particular, Val527Gly had the lowest unstable energy value. The native structure and changed structures are shown in Figure 2. Although there is no experimental evidence which shows that nsSNPs regulate the expression level in T-cells, stability analysis of the target structure can give us more biological information to understand the enzymatic mechanism and to evaluate the drug effect.
Table 3.
Nonsynonymous SNPs of ZAP70.
| rsNumber | DNA substitution | Amino-acid substitution | Overall stability | Predicted ΔΔG (kcal/mol) |
|---|---|---|---|---|
| rs56077145 | 106C > G | Leu36Val | Destabilising | −1.03 |
| rs113994172 | 239C > A | Pro80Gln | Stabilising | 0.82 |
| rs56264206 | 308A > T | Asn103Ile | Stabilising | 0.71 |
| rs55845489 | 309C > A | Asn103Lys | Destabilising | −0.36 |
| rs55679020 | 311G > C | Arg104Pro | Stabilising | 0.11 |
| rs111771234 | 550T > A | Phe115Ile | Stabilising | 0.67 |
| rs55964305 | 524G > T | Arg175Leu | Destabilising | −0.67 |
| rs56403250 | 572C > T | Pro191Leu | Stabilising | 0.08 |
| rs76059124 | 653C > T | Ala218Val | Destabilising | −1.86 |
| rs113310375 | 1274T > C | Val356Ala | Destabilising | −2.01 |
| rs113994174 | 1393C > T | Arg465Cys | Destabilising | −0.81 |
| rs56059280 | 1468G > A | Asp490Asn | Destabilising | −0.29 |
| rs104893674 | 1554C > A | Ser518Arg | Destabilising | −0.56 |
| rs56189815 | 1568G > T | Trp523Leu | Stabilising | 3.36 |
| rs56146954 | 1580T > G | Val527Gly | Destabilising | −4.09 |
| rs113994175 | 1714A >T | Met572Leu | Stabilising | 0.17 |
| rs55803111 | 1781G > A | Arg594Gln | Stabilising | 1.11 |
| rs56326640 | 1783G > T | Ala595Ser | Destabilising | −0.36 |
| rs56250717 | 1826G > A | Gly609Asp | — | — |
Figure 2.
Protein structure of ZAP70 and structural variations by nsSNPs. We predicted the structural changes in ZAP70 caused by nsSNPs through structural modeling and stability analysis because mutations can cause selective T-cell defects [46]. In order to predict the structure of ZAP70, we used PDB entries 1M61 and 1U59 as the structural templates, which have 100% and 98.9% sequence identity, respectively, which were found to be suitable structural templates. We found 19 nsSNPs from the dbSNP database that cause nonsense or missense changes in ZAP70. From these nsSNPs, 10 nsSNPs were found to lead to unstable structural changes by using CUPSAT. In particular, Val527Gly had the lowest unstable energy value (Predicted ΔΔG: −4.09). The red region shows the location of Val527Gly in the middle of the alpha-helix structure.
4. Conclusion
In this study, we attempted to explore potential therapeutic candidates for psoriasis by utilizing bioinformatic methods. We have presented essential proteins from upregulated and downregulated genes in psoriasis; some of these findings have been supported by experimental evidence reported in the literature. Of particular interest are the predicted essential enzymes that are important proteins related to the pathogenesis of psoriasis; these enzymes can be explored as therapeutic or drug target candidates. Further studies should be conducted to determine the role of these candidate enzymes in psoriasis and to explore agonists of the upregulated candidates or antagonists of downregulated candidates as drug targets by exploiting the property of multiple targeting. These results will aid future drug discovery efforts, enabling drug development in a more timely and cost-effective manner.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (MOST) (no. 20090083538). The authors thank Aging Tissue Bank for providing research information.
Abbreviations
- SAM:
Significance analysis of microarray
- PPI:
Protein-protein interactions
- SNP:
Single-nucleotide polymorphism.
References
- 1.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445(7130):866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 2.Nickoloff BJ. The immunologic and genetic basis of psoriasis. Archives of Dermatology. 1999;135(9):1104–1110. doi: 10.1001/archderm.135.9.1104. [DOI] [PubMed] [Google Scholar]
- 3.Gudjonsson JE, Johnston A, Sigmundsdottir H, Valdimarsson H. Immunopathogenic mechanisms in psoriasis. Clinical and Experimental Immunology. 2004;135(1):1–8. doi: 10.1111/j.1365-2249.2004.02310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kupper TS, Fuhlbrigge RC. Immune surveillance in the skin: mechanisms and clinical consequences. Nature Reviews Immunology. 2004;4(3):211–222. doi: 10.1038/nri1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg H, Conklin D, Xu W, et al. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104(1):9–19. doi: 10.1016/s0092-8674(01)00187-8. [DOI] [PubMed] [Google Scholar]
- 6.Rich BE, Kupper TS. Cytokines: IL-20—a new effector in skin inflammation. Current Biology. 2001;11(13):R531–R534. doi: 10.1016/s0960-9822(01)00312-8. [DOI] [PubMed] [Google Scholar]
- 7.Bowcock AM. Understanding the pathogenesis of psoriasis, psoriatic arthritis, and autoimmunity via a fusion of molecular genetics and immunology. Immunologic Research. 2005;32(1–3):45–56. doi: 10.1385/IR:32:1-3:045. [DOI] [PubMed] [Google Scholar]
- 8.Nair RP, Duffin KC, Helms C, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nature Genetics. 2009;41(2):199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krueger G, Callis K. Potential of tumor necrosis factor inhibitors in psoriasis and psoriatic arthritis. Archives of Dermatology. 2004;140(2):218–225. doi: 10.1001/archderm.140.2.218. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Valladares I, Cuchacovich R, Espinoza LR. Comparative assessment of biologics in treatment of psoriasis: drug design and clinical effectiveness of ustekinumab. Drug Design, Development and Therapy. 2011;(5):41–49. doi: 10.2147/DDDT.S10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature Reviews Genetics. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 13.Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nature Genetics. 2008;40(2):181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 14.LaCount DJ, Vignali M, Chettier R, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438(7064):103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22(18):2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets-10 years on. Nucleic Acids Research. 2011;39(supplement 1):D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reischl J, Schwenke S, Beekman JM, Mrowietz U, Stürzebecher S, Heubach JF. Increased expression of Wnt5a in psoriatic plaques. Journal of Investigative Dermatology. 2007;127(1):163–169. doi: 10.1038/sj.jid.5700488. [DOI] [PubMed] [Google Scholar]
- 18.Yao Y, Richman L, Morehouse C, et al. Type I interferon: potential therapeutic target for psoriasis? PLoS ONE. 2008;3(7, article e2737) doi: 10.1371/journal.pone.0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JG, Park D, Kim BC, et al. Predicting the interactome of Xanthomonas oryzae pathovar oryzae for target selection and DB service. BMC Bioinformatics. 2008;9, article 41 doi: 10.1186/1471-2105-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Research. 2000;28(1):289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader GD, Donaldson I, Wolting C, Ouellette BFF, Pawson T, Hogue CWV. BIND—the biomolecular interaction network database. Nucleic Acids Research. 2001;29(1):242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermjakob H, Montecchi-Palazzi L, Lewington C, et al. IntAct: an open source molecular interaction database. Nucleic Acids Research. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT: a molecular INTeraction database. FEBS Letters. 2002;513(1):135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 25.Peri S, Navarro JD, Kristiansen TZ, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Research. 2004;32:D497–D501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Research. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends in Biochemical Sciences. 1998;23(9):324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 28.Enright AJ, Illopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402(6757):86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 29.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(8):4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews LR, Vaglio P, Reboul J, et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Research. 2001;11(12):2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albert R, Jeong H, Barabási AL. Error and attack tolerance of complex networks. Nature. 2000;406(6794):378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(1 I):258–261. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proia RL, Neufeld EF. Synthesis of beta-hexosaminidase in cell-free translation and in intact fibroblasts: an insoluble precursor alpha chain in a rare form of Tay-Sachs disease. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(20):6360–6364. doi: 10.1073/pnas.79.20.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 36.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Research. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He F, Zhang Y, Chen H, Zhang Z, Peng YL. The prediction of protein-protein interaction networks in rice blast fungus. BMC Genomics. 2008;9, article 519 doi: 10.1186/1471-2164-9-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raman K, Yeturu K, Chandra N. targetTB: a target identification pipeline for Mycobacterium tuberculosis through an interactome, reactome and genome-scale structural analysis. BMC Systems Biology. 2008;2(1, article 109) doi: 10.1186/1752-0509-2-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang S, Son SW, Kim SC, Kim YJ, Jeong H, Lee D. A protein interaction network associated with asthma. Journal of Theoretical Biology. 2008;252(4):722–731. doi: 10.1016/j.jtbi.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Batada NN, Hurst LD, Tyers M. Evolutionary and physiological importance of hub proteins. PLoS Computational Biology. 2006;2(7, article e88):0748–0756. doi: 10.1371/journal.pcbi.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joy MP, Brock A, Ingber DE, Huang S. High-betweenness proteins in the yeast protein interaction network. Journal of Biomedicine and Biotechnology. 2005;2005(2):96–103. doi: 10.1155/JBB.2005.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Computational Biology. 2007;3(4, article e59):713–720. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature Chemical Biology. 2008;4(11):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Kadlecek TA, Au-Yeung BB, et al. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harbor Perspectives in Biology. 2010;2(5) doi: 10.1101/cshperspect.a002279. Article ID a002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo XJ, Li J, Ni PH, Ren LP, Xu WG. The transcription levels of linker for activation of T cell and its upstream regulatory factors in T cells of asthmatic patients. Chinese Journal of Tuberculosis and Respiratory Diseases. 2008;31(2):125–128. [PubMed] [Google Scholar]
- 46.Arpaia E, Shahar M, Dadi H, Cohen A, Rolfman CM. Defective T cell receptor signaling and CD8 thymic selection in humans lacking zap-70 kinase. Cell. 1994;76(5):947–958. doi: 10.1016/0092-8674(94)90368-9. [DOI] [PubMed] [Google Scholar]
- 47.Zanotti R, Frattini F, Ghia P, et al. ZAP-70 expression is associated with increased risk of autoimmune cytopenias in CLL patients. American Journal of Hematology. 2010;85(7):494–498. doi: 10.1002/ajh.21737. [DOI] [PubMed] [Google Scholar]
- 48.Johnston A, Gudjonsson JE, Aphale A, Guzman AM, Stoll SW, Elder JT. EGFR and IL-1 signaling synergistically promote keratinocyte antimicrobial defenses in a differentiation-dependent manner. Journal of Investigative Dermatology. 2011;131(2):329–337. doi: 10.1038/jid.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rousseau B, Ménard L, Haurie V, et al. Overexpression and role of the ATPase and putative DNA helicase RuvB-like 2 in human hepatocellular carcinoma. Hepatology. 2007;46(4):1108–1118. doi: 10.1002/hep.21770. [DOI] [PubMed] [Google Scholar]
- 50.Haurie V, Ménard L, Nicou A, et al. Adenosine triphosphatase pontin is overexpressed in hepatocellular carcinoma and coregulated with Reptin through a new posttranslational mechanism. Hepatology. 2009;50(6):1871–1883. doi: 10.1002/hep.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eriksen KW, Woetmann A, Skov L, et al. Deficient SOCS3 and SHP-1 expression in psoriatic T cells. Journal of Investigative Dermatology. 2010;130(6):1590–1597. doi: 10.1038/jid.2010.6. [DOI] [PubMed] [Google Scholar]