Abstract
To assess the diversity of AbaR genomic resistance islands in Acinetobacter baumannii European clone I (MLST clonal complex 1), we investigated 26 multidrug-resistant strains of this major clone isolated from hospitals in 21 cities of 10 European countries between 1984 and 2005. Each strain harbored an AbaR structure integrated at the same position in the chromosomal ATPase gene. AbaR3, including four subtypes based on variations in class 1 integron cassettes, and AbaR10 were found in 15 and 2 strains, respectively, whereas a new, unique AbaR variant was discovered in each of the other 9 strains. These new variants, designated AbaR11 to AbaR19 (19.8 kb to 57.5 kb), seem to be truncated derivatives of AbaR3, likely resulting from the deletions of its internal parts mediated by either IS26 elements (AbaR12 to AbaR19) or homologous recombination (AbaR11). AbaR3 was detected in all 10 strains isolated in 1984 to 1991, while AbaR11 to AbaR19 were carried only by strains isolated since 1997. Our results and those from previous publications suggest that AbaR3 is the original form of AbaR in European clone I, which may have provided strains of the lineage with a selective advantage facilitating their spread in European hospitals in the 1980s or before.
INTRODUCTION
Nosocomial infections caused by multidrug-resistant (MDR) strains of Acinetobacter baumannii have become a serious therapeutic and epidemiological problem worldwide (9). Resistance to multiple antimicrobials in the species has been associated with several international lineages, particularly with the so-called European (EU) clones I and II (7, 9). The earliest known MDR strain of EU clone I was isolated in a Swiss hospital in 1977 (16), and the lineage prevailed among outbreak and MDR Acinetobacter strains in some European countries in the 1980s and 1990s (8, 18). Further studies in these countries have demonstrated a shift toward the predominance of EU clone II among hospital A. baumannii strains in the early 2000s (19, 25), considered to be a result of the spread of carbapenem-resistant strains (subclones) of this clone (19). Although strains belonging to or related to EU clone II seem to dominate in the current global population of MDR A. baumannii strains (5, 13, 14), EU clone I and other MDR lineages can be common or even prevail in some regions (10, 26).
Recent studies have revealed that the chromosomes of some A. baumannii strains harbor large clusters of horizontally transferred genes conferring resistance to multiple antibiotics and heavy metals, which are integrated at a specific site in a particular ATPase gene (12, 22). Nine such genomic resistance islands (A. baumannii resistance islands [AbaRs]) have been fully characterized, eight of which were found in strains of EU clone I, i.e., AbaR1 (12), AbaR3 (2), AbaR5 (20), AbaR6, AbaR7 (22), AbaR8 (21), AbaR9, and AbaR10 (1). These AbaRs share a structure represented by a 16.3-kb backbone transposon (Tn6019) interrupted by a large compound transposon that contains a variable-resistance region bounded by directly oriented copies of Tn6018 (Fig. 1) (22). Exceptions are AbaR6 and AbaR7, each with a large deleted region that includes the left-hand copy of Tn6018 and part of the Tn6019 backbone (22). Some strains of EU clone I were also found to harbor a blaOXA-23 gene-carrying island termed AbaR4, which was integrated at a chromosomal site different from that of the ATPase gene (2, 24) and the backbone of which is formed by a Tn6021 transposon instead of Tn6019 (22). Much less is known about AbaRs in EU clone II. The resistance islands harbored by this clone are integrated at the same site of the ATPase gene as is known for AbaRs in EU clone I. Apart from AbaR2, a largely truncated AbaR island found in a fully sequenced strain (15), AbaRs from two EU clone II strains were partially characterized and shown to contain a transposon related to Tn6021 (22). In addition, an epidemic strain of EU clone II from the United Kingdom was shown to carry an AbaR4-type island integrated in the ATPase gene (24). These findings suggest that the genomic islands associated with the ATPase gene evolved independently in two main A. baumannii clones.
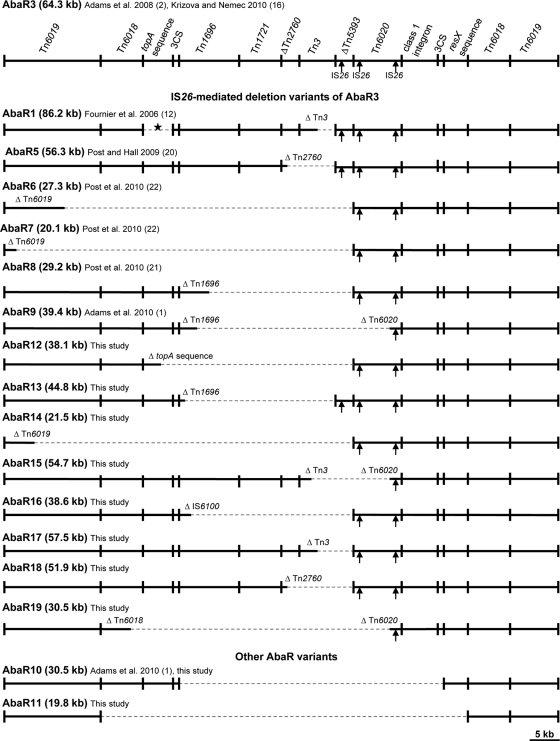
Fig. 1.
Schematic overview of AbaR variants in European clone I. A diagram of AbaR3 is shown at the top. Only AbaR3 components discussed in the text are shown (demarcated by vertical lines); for a more detailed structure of AbaR3, see reference 16. The other AbaRs are lined up below AbaR3 according to the presence of the regions homologous to those of AbaR3. Unbroken and dashed horizontal lines indicate, respectively, theAbaR3 regions present or absent in each of the AbaRs. Partial deletion of an AbaR3 component is indicated by the absence of a vertical line. The arrows indicate the positions of three copies of IS26 sequences present in AbaR3. The location of the 29-kb specific sequence in AbaR1 is denoted by an asterisk. The sizes of the individual AbaRs are proportional to the scale, except for the regions with class 1 integrons (Table 1). The sequences of AbaR1, AbaR3, AbaR5, AbaR6, AbaR7, AbaR8, AbaR9, and AbaR10 are available from the GenBank database under accession numbers CT025832, CP001182, FJ172370, GQ406245, GQ406246, HM590877, ADGZ00000000, ADHA00000000, respectively.
The EU clone I strains harboring the completely characterized AbaRs were isolated in Europe (AbaR1), the United States (AbaR3, AbaR9, and AbaR10), or Australia (AbaR5 to AbaR8) between 1997 and 2009 (1, 2, 12, 20, 21, 22). Comparative analysis of these AbaRs has suggested that AbaR1 and AbaR5 to AbaR10 are truncated variants of AbaR3, while AbaR1 is the only known AbaR structure that contains additional DNA compared to AbaR3 (1, 22). Recently, we identified an intact AbaR3 island in an outbreak MDR strain of EU clone I from 1977 (16), indicating that AbaR variants already existed in A. baumannii in the late 1970s. The aim of the present study was to obtain more insight into the structural diversity of AbaR islands in the European population of EU clone I. To this end, we investigated 26 strains isolated in 10 countries over a period of 21 years.
MATERIALS AND METHODS
Selection and properties of bacterial strains.
Twenty-six A. baumannii strains isolated from hospitals in 21 cities of 10 European countries between 1984 and 2005 were investigated (Table 1). The strains known to belong to EU clone I were selected from published (7, 8, 17, 18, 19) or unpublished (strains NIPH 783 and NIPH 827) studies to be as diverse as possible in time and place of isolation without any known epidemiological link, with the emphasis on isolates from the 1980s and 1990s. Strains from the same city were included only if they differed in molecular-typing characteristics and/or in the content of genes associated with the known AbaRs. The strains had been previously assigned to EU clone I by multilocus sequence typing (MLST) as developed at the Institut Pasteur (7) and by amplified fragment length polymorphism (AFLP) analysis (9). By MLST, all strains had either sequence type (ST) 1 or one of its single-locus variants (Table 1), which together form clonal complex 1, corresponding to EU clone I (7). By AFLP analysis, the isolates were further classified into 16 types at a similarity level of 90% (Table 1). The minimal inhibitory concentrations of 10 antimicrobial agents known to be primarily effective against A. baumannii were determined by the agar dilution method using CLSI guidelines (4) and are shown in Table 2.
Table 1.
Characteristics of the A. baumannii strains of European clone I used in the present study
| Strain | City, country,a yr of isolation | STb (allelic profile) | AFLP typec | AbaR islandd | Gene cassettes of the AbaR-associated class 1 integron | Reference(s) |
|---|---|---|---|---|---|---|
| RUH 875 | Dordrecht, NL, 1984 | 1 (1111511) | 1 | AbaR3 | dfrA1 | 7, 8, 17 |
| RUH 510 | Nijmegen, NL, 1984 | 8 (1111111) | 2 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 8, 17 |
| RUH 3239 | London, UK, 1985-8 | 1 (1111511) | 3 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 8, 17 |
| RUH 2037 | Venlö, NL, 1986 | 1 (1111511) | 3 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 8, 17 |
| RUH 3238 | Sheffield, UK, 1987 | 1 (1111511) | 4 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 8, 17 |
| RUH 3242 | Basildon, UK, 1989 | 1 (1111511) | 2 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 8, 17 |
| RUH 3247 | Leuven, BE, 1990 | 1 (1111511) | 4 | AbaR3 | aacA4 | 7, 8, 17 |
| RUH 3282 | Salford, UK, 1990 | 1 (1111511) | 5 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 8, 17 |
| NIPH 7 | Praha, CZ, 1991 | 7 (1112511) | 6 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 17, 18 |
| NIPH 10 | Praha, CZ, 1991 | 1 (1111511) | 6 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 17, 18 |
| NIPH 56 | Praha, CZ, 1992 | 1 (1111511) | 7 | AbaR10 | None | 7, 17, 18 |
| NIPH 321 | Tábor, CZ, 1994 | 1 (1111511) | 4 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 7, 17, 18 |
| NIPH 783 | Debrecen, HU, 1995 | 1 (1111511) | 8 | AbaR10 | None | |
| NIPH 827 | Sofia, BG, 1997 | 1 (1111511) | 9 | AbaR3 | aacC1-orfP-orfQ-aadA1 | |
| NIPH 470 | Č. Budějovice, CZ, 1997 | 1 (1111511) | 10 | AbaR11 | None | 17, 18 |
| LUH 6013 | Rome, IT, 1997 | 1 (1111511) | 11 | AbaR12 | aacC1-(orfP)2-orfQ-aadA1 | 7 |
| LUH 6015 | Rome, IT, 1998 | 1 (1111511) | 12 | AbaR13 | aacC1-orfP-orfQ-aadA1 | 7 |
| LUH 5881 | Madrid, ES, 1998 | 1 (1111511) | 13 | AbaR14 | aacA4 | 7 |
| LUH 6125 | Krakow, PL, 1998 | 1 (1111511) | 10 | AbaR15 | aacC1-(orfP)2-orfQ-aadA1 | 7 |
| LUH 7140 | London, UK, 2000 | 1 (1111511) | 10 | AbaR16 | aacC1-orfP-orfQ-aadA1 | 7 |
| NIPH 1605 | Sedlčany, CZ, 2001 | 1 (1111511) | 14 | AbaR3 | aacC1-orfP-orfQ-aadA1 | 17 |
| LUH 8592 | Sofia, BG, 2001 | 1 (1111511) | 10 | AbaR17 | aacC1-orfP-orfQ-aadA1 | 7 |
| LUH 9668 | Dublin, IE, 2003 | 1 (1111511) | 2 | AbaR3 | aacC1-orfP-orfQ-aadA1 | 7 |
| NIPH 2605 | Most, CZ, 2005 | 1 (1111511) | 15 | AbaR3 | aacC1-(orfP)2-orfQ-aadA1 | 19 |
| NIPH 2713 | Kladno, CZ, 2005 | 1 (1111511) | 16 | AbaR18 | aacC1-orfP-orfQ-aadA1 | 19 |
| NIPH 2554 | Praha, CZ, 2005 | 1 (1111511) | 16 | AbaR19 | aacC1-(orfP)2-orfQ-aadA1 | 19 |
Country abbreviations: BE, Belgium; BG, Bulgaria; CZ, Czech Republic; ES, Spain; HU, Hungary; IE, Ireland; IT, Italy; NL, Netherlands; PL, Poland; UK, United Kingdom.
ST of the Institut Pasteur MLST scheme available at http://www.pasteur.fr/mlst.
AFLP types distinguished by comparative cluster analysis of the current set of strains at 90% similarity level and numbered for the purpose of this study.
The AbaR3 designation is used for all four subtypes of AbaR3 based on variations in class 1 integron cassettes.
Table 2.
MICs of the A. baumannii EU clone I strains included in the study
| Strain | MIC (μg/ml)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CTX | FEP | MEM | AMK | GEN | NET | TOB | CIP | CST | |
| RUH 875 | 8 | 32 | 16 | 1 | 8 | >32 | 32 | >16 | 1 | 0.5 |
| RUH 510 | 8 | 16 | 8 | 0.5 | 8 | >32 | 32 | 4 | 0.5 | 0.5 |
| RUH 3239 | 8 | 32 | 16 | 1 | 8 | >32 | 32 | 4 | 1 | 0.5 |
| RUH 2037 | 128 | >64 | 128 | 2 | 8 | >32 | 16 | 4 | >32 | 0.5 |
| RUH 3238 | 8 | 16 | 8 | 0.5 | 4 | >32 | 8 | 2 | 0.25 | 0.5 |
| RUH 3242 | 8 | 32 | 16 | 0.5 | 4 | >32 | 8 | 2 | 0.5 | 0.5 |
| RUH 3247 | 8 | 16 | 16 | 0.25 | 16 | 32 | >64 | >16 | 8 | 0.5 |
| RUH 3282 | 128 | >64 | 32 | 1 | >128 | >32 | >64 | 4 | 8 | 0.25 |
| NIPH 7 | 8 | 16 | 8 | 0.5 | >128 | >32 | 16 | 4 | 2 | 0.5 |
| NIPH 10 | 16 | 32 | 32 | 1 | >128 | >32 | 64 | 8 | 8 | 0.5 |
| NIPH 56 | 4 | 8 | 4 | 0.25 | 4 | 4 | 8 | 2 | 0.25 | 0.5 |
| NIPH 321 | 8 | 16 | 8 | 0.5 | 128 | >32 | 16 | 4 | 8 | 0.5 |
| NIPH 783 | 8 | 32 | 4 | 0.25 | 4 | >32 | 8 | >16 | 32 | 0.5 |
| NIPH 827 | 64 | >64 | 16 | 0.5 | >128 | >32 | 16 | >16 | 0.5 | 0.5 |
| NIPH 470 | 64 | >64 | 16 | 0.5 | 128 | 8 | 64 | 2 | 4 | 0.5 |
| LUH 6013 | 128 | >64 | 16 | 1 | >128 | >32 | 8 | 2 | >32 | 0.5 |
| LUH 6015 | >128 | >64 | 16 | 0.5 | 4 | >32 | 8 | 2 | >32 | 0.5 |
| LUH 5881 | 16 | 64 | 16 | 0.25 | 32 | >32 | >64 | >16 | >32 | 0.25 |
| LUH 6125 | 16 | >64 | 16 | 0.125 | >128 | >32 | 8 | 2 | 8 | 0.5 |
| LUH 7140 | 64 | >64 | 32 | 1 | 8 | >32 | 32 | 4 | >32 | 0.5 |
| NIPH 1605 | 4 | 16 | 8 | 0.25 | 8 | >32 | 16 | >16 | 32 | 0.5 |
| LUH 8592 | 128 | >64 | 16 | 0.5 | 128 | >32 | 8 | 2 | 32 | 0.5 |
| LUH 9668 | 4 | 16 | 8 | 0.25 | 8 | >32 | 16 | 4 | 4 | 0.5 |
| NIPH 2605 | 8 | 32 | 4 | 0.25 | 128 | >32 | 8 | 2 | 16 | 0.5 |
| NIPH 2713 | >128 | >64 | 16 | 8 | 128 | >32 | 32 | 2 | 4 | 0.5 |
| NIPH 2554 | 4 | 16 | 8 | 0.25 | 4 | >32 | 8 | 2 | 32 | 0.25 |
AMK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; CST, colistin; CTX, cefotaxime; FEP, cefepime; GEN, gentamicin; MEM, meropenem; NET, netilmicin; TOB, tobramycin.
Gene detection and AbaR mapping.
The genes associated with AbaR islands were detected by PCR using primers derived from the sequences of AbaR3 (GenBank accession no. CP001182) and AbaR1 (GenBank accession no. CT025832). The 25 genes studied, primers, and annealing temperatures are listed in Table S1 in the supplemental material. To investigate the structures of AbaRs, PCR mapping experiments were performed as described previously (16). DNA fragments (up to 12 kb in size) corresponding to overlapping internal segments of AbaR3-like islands were amplified using a Long Range PCR Kit (Qiagen, Hilden, Germany) and primers listed in Table S2 in the supplemental material. The resulting amplicons were analyzed for restriction fragment length polymorphism (RFLP) using AccI, ApaI, BfaI, BclI, BglI, BsmAI, ClaI, EcoRI, EcoRV, HincII, HindIII, or StyI restriction enzymes (Fermentas, Vilnius, Lithuania). EU clone I strains AYE (12) and HK302 (16), carrying AbaR1 and AbaR3, respectively, were used as positive controls for PCR detection of the AbaR-associated genes, while strain HK302 was a positive control for PCR-RFLP mapping experiments.
DNA sequence analysis.
Sequence analysis was performed on the regions different from those of AbaR3 using a primer-walking strategy. PCR amplicons were purified with a QIAquick PCR Purification Kit (Qiagen) or, after visualization in agarose gel, with a High Pure PCR Product Purification Kit (Roche Diagnostic GmbH, Mannheim, Germany). DNA sequencing was performed using a BigDye Terminator v3.1 Cycle Sequencing Kit and an ABI 3130 XL genetic analyzer (Applied Biosystems). DNA sequences were assembled using the software KODON (Applied Maths, St-Martens-Latem, Belgium) and annotated using BLASTN (http://www.ncbi.nlm.nih.gov/blast) and the sequence annotation tools integrated into the Sequin program (available at http://www.ncbi.nlm.nih.gov/Sequin).
Nucleotide sequence accession numbers.
The partial nucleotide sequences of the AbaR structures have been deposited in the GenBank database under accession numbers JF262165 (AbaR10 from NIPH 56), JF262166 (AbaR10 from NIPH 783), JF262167 (AbaR11), JF262168 (AbaR12), JF262169 (AbaR13), JF262170 (AbaR14), JF262171 (AbaR15), JF262172 (AbaR16), JF262173 (AbaR17), JF262174 (AbaR18), and JF262175 (AbaR19).
RESULTS AND DISCUSSION
Structural analysis of AbaRs.
To analyze the structure of AbaR resistance islands, we used a strategy based on the structural homology of the AbaR-type islands in strains of EU clone I (16). This strategy included (i) PCR determination of the presence and interruption of the ATPase gene, (ii) PCR analysis of the junctions between the inserted DNA and the ATPase gene, (iii) PCR detection of the genes corresponding to those found in the known AbaR in EU clone I, (iv) PCR-RFLP mapping based on AbaR3 and on the content of the AbaR-associated genes in a given strain, and (v) sequence analysis of the amplicons which differ in size and/or RFLP pattern from those of AbaR3.
All 26 strains carried the ATPase gene interrupted by a DNA insert at the same position as in the known AbaRs (16, 22). PCR analysis of the junctions using primers designed to distinguish between Tn6019- and Tn6021-like transposons revealed that the ends of the inserted sequence corresponded to Tn6019, known to be associated with AbaRs in EU clone I (22). PCR screening for the presence of 25 genes harbored by AbaR1 or AbaR3 showed that individual strains carried from 5 to 20 of these genes in 10 different combinations (data not shown). Thus, at least nine variants of AbaRs different from AbaR1 and AbaR3 occurred in the strains of the study.
AbaR3 and its integron-based subtypes.
The structures of AbaRs were further investigated using an array of 14 overlapping long-range PCRs designed to cover the whole sequence of AbaR3 (16) (see Table S2 in the supplemental material), followed by RFLP analysis of the resulting amplicons. Between 3 and 14 PCRs were performed for each strain, depending on the presence of AbaR1- or AbaR3-associated genes. In 10 strains, all 14 amplicons corresponded in size and RFLP pattern to those of AbaR3, which indicated that these strains carried AbaRs structurally congruent with AbaR3 (Table 1). Each of five other strains yielded 12 amplicons identical to those of AbaR3, but the two amplicons spanning the variable region of a class 1 integron were different. Sequence analysis revealed that these five strains carried gene cassette arrays different from that originally identified in AbaR3 (aacC1-orfP-orfP-orfQ-aadA1). While one copy of orfP was absent from this cassette array in three strains (NIPH 827, NIPH 1605, and LUH 9668), the variable region in two other strains (RUH 875 and RUH 3247) included only a single gene cassette, either dfrA1 or aacA4 (Table 1). These results indicate that the already established AbaR3 islands may have undergone diversification through changes in the structure of class 1 integrons, possibly via loss, acquisition, or replacement of integron gene cassettes. For reasons of simplicity, we do not propose new designations for these integron-based subtypes of the complete AbaR3 island.
Truncated AbaR3 variants.
Structural analysis using PCR-RFLP and sequencing revealed that all 11 remaining strains carried AbaR structures that can be considered truncated versions of the AbaR3-type islands. Except for AbaR10, found in two strains, each of these structures was detected in a single strain and represented a unique, yet-undescribed variant of AbaR. The nine new AbaRs have been termed AbaR11 to AbaR19, and their nearly complete or partial sequences are available from GenBank under accession numbers JF262167 to JF262175.
Compared to AbaR3, all of these new AbaRs lacked internal regions of different lengths, most likely as a result of deletions mediated by an IS26 element, except for AbaR11 (Fig. 1). IS26 is known to create deletions of adjacent regions, especially those containing antibiotic resistance genes (6, 11, 22). Three copies of IS26 are located in the right half of AbaR3, and as depicted in Fig. 1, each of them could be associated with 5′-ward deletions in AbaRs. In AbaR13, the IS26 located next to the right end of Tn3 is likely to have caused deletions reaching to the left part of Tn1696, and the same IS26 copy seems to have been responsible for deletions in AbaR1 and AbaR5 (20). The IS26 located at the left end of Tn6020 has most likely created deletions of different lengths in AbaR12 (reaching to the topA sequence and partially removing orfY in this sequence), AbaR14 (reaching to the left part of Tn6019 and partially deleting the tniB gene), AbaR16 (leaving only part of IS6100 in Tn1696), AbaR17 (completely removing blaTEM-1 carried by Tn3), and AbaR18 (partially deleting the catA1 gene located in ΔTn2760). It may have also been involved in deletions in AbaR6, AbaR7 (22), and AbaR8 (21). Finally, the IS26 located at the right end of Tn6020 has probably removed internal regions in AbaR15 (partially deleting the tpnR gene of Tn3), AbaR19 (reaching to the left-hand copy of Tn6018 and partially deleting the tpnA gene), and AbaR9 (1). Although multiple genetic events might have been responsible for the formation of the AbaRs discussed above, it is also possible that, except for AbaR1 (Fig. 1), all these structures arose from the complete AbaR3-type islands through a single deletion event mediated by IS26. Given the nonspecificity of IS26-mediated deletions, a number of additional AbaR variants can be expected in the current EU clone I population. Therefore, it should be considered whether to continue numbering IS26-based variants, especially when confronted with tiny and genetically irrelevant differences.
AbaR10 and AbaR11 represent AbaRs with no apparent deletions mediated by IS26. In the present study, two strains carried AbaRs, the partial sequences of which corresponded to that of the recently described AbaR10 (1). Compared to AbaR3, AbaR10 lacks a 31.5-kb region, which is located between the left-hand copy of the class 1 integron 3′ conserved segment (3′CS) and the resX sequence, while AbaR11 is missing several additional segments, including one copy of Tn6018, the topA and resX sequences, and the second copy of the 3′CS (Fig. 1). Even though AbaR10 and AbaR11 might also be less developed progenitors of the AbaR3-type islands, it is likely that each of these AbaRs has arisen from a larger AbaR structure through a deletion resulting from recombination between homologous sequences present in AbaR3. In AbaR10 and AbaR11, such deletions could be caused by recombination between two copies of 3′CS (1) and two copies of Tn6018, respectively. Thus, similar to the vast majority of the AbaRs with IS26-mediated deletions, both AbaR10 and AbaR11 may have arisen from an AbaR3-type structure via a single genetic event.
AbaRs in space and time.
The available data indicate that AbaR3 is the most widespread of the AbaRs in space and time among EU clone I strains. So far, AbaR3 has been found in two strains isolated at the Walter Reed Army Medical Center in 2004 (1, 2) and in a Swiss isolate from 1977 (16). In the present study, strains with AbaR3 (including all its integron-based subtypes) were isolated in six European countries between 1984 and 2005 (Table 1). In addition, we identified AbaR3 (carrying an integron with aacC1-orfP-orfP-orfQ-aadA1) in an EU clone I strain isolated in Australia in 1995 (L. Krizova and A. Nemec, unpublished data). In contrast, each of the non-AbaR3 variants was found in a single strain isolated not earlier than 1997. The only exception was AbaR10, found in two European strains from 1992 and 1995 (NIPH 56 and NIPH 783, respectively), and in a U.S. isolate from 2003 (1).
Concluding remarks.
The results of the present study and previously published data suggest that AbaR3 is the original genomic structure from which the hitherto known AbaRs in EU clone I have been derived. This assumption is supported by the distribution of different AbaRs in time and space and by the observation that nearly all non-AbaR3 islands are most likely truncated derivatives of AbaR3 that might have arisen from AbaR3 through a single genetic event. Notably, AbaR3 and its integron-based subtypes harbor a number of genes conferring resistance to antimicrobials used in the 1970s and 1980s, including early generations of β-lactams (the blaTEM-1 gene), aminoglycosides (aphA1, aacC1, aadA1, and aacA4), tetracyclines (tetA), chloramphenicol (catA1), sulfonamides (sul1), and trimethoprim (dfrA1). Therefore, it is conceivable that these structures provided strains of EU clone I with a selective advantage, which facilitated their dissemination in European hospitals in the 1980s or before (8, 16). Reportedly, genomic islands integrated into the ATPase gene of A. baumannii represent a hot spot that could explain the rapid acquisition of resistance markers under antimicrobial pressure (12). However, AbaR1, found in an epidemic strain in France, is the only known AbaR in EU clone I that has incorporated additional genes encoding antibiotic resistance compared to AbaR3. Although recent isolates of EU clone I can harbor horizontally acquired genes encoding resistance to modern antibiotics, such as carbapenems, these genes (e.g., blaOXA-23) have not been located on any of the AbaRs integrated in the ATPase gene in this clone (1, 24). In addition, many mechanisms conferring resistance to antimicrobials currently used against MDR strains of A. baumannii result from the upregulation or mutational modification of intrinsic chromosomal genes rather than from horizontal gene transfer (3, 9, 23). Consequently, the accumulation of truncated derivatives of AbaR3 in the recent population of EU clone I may reflect the diminishing role of these structures in resistance to antimicrobial therapy. In light of these considerations, AbaRs may be seen as footprints left behind by an important phase in the evolution of EU clone I, but currently they do not seem to play a substantial role in the ongoing development of antimicrobial resistance in A. baumannii.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brigitte Berger-Bächi, Sylvain Brisse, Rossi Dobrewski, Séamus Fanning, László Kiss, Tyrone Pitt, Laurent Poirel, Encho Savov, and Richard Spence for providing strains. Martin Musílek and Martina Maixnerová are acknowledged for technical assistance and Eva Kodytková for linguistic revision of the manuscript.
This work was supported by grant 310/08/1747 from the Czech Science Foundation. L.K. was also supported by grant no. 263206 from the Charles University Grant Agency.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Adams M. D., Chan E. R., Molyneaux N. D., Neil D. M., Bonomo R. A. 2010. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:3569–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams M. D., et al. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adams M. D., et al. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical Laboratory Standards Institute 2008. Performance standards for antimicrobial susceptibility testing, 18th informational supplement. Approved standards M100-S18 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. D'Arezzo S., et al. 2011. Changing carbapenemase gene pattern in an epidemic multidrug-resistant Acinetobacter baumannii lineage causing multiple outbreaks in central Italy. J. Antimicrob. Chemother. 66:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dawes F. E., et al. 2010. Distribution of class 1 integrons with IS26-mediated deletions in their 3′-conserved segments in Escherichia coli of human and animal origin. PLoS One 5:e12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diancourt L., Passet V., Nemec A., Dijkshoorn L., Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dijkshoorn L., et al. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dijkshoorn L., Nemec A., Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951 [DOI] [PubMed] [Google Scholar]
- 10. Di Popolo A., Giannouli M., Triasi M., Brisse S., Zarrilli R. 2011. Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin. Microbiol. Infect. 17:197–201 [DOI] [PubMed] [Google Scholar]
- 11. Doublet B., et al. 2008. Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52:3745–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fournier P. E., et al. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu Y., et al. 2010. Wide dissemination of OXA-23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J. Antimicrob. Chemother. 65:644–650 [DOI] [PubMed] [Google Scholar]
- 14. Higgins P. G., Dammhayn C., Hackel M., Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233–238 [DOI] [PubMed] [Google Scholar]
- 15. Iacono M., et al. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krizova L., Nemec A. 2010. A 63 kb genomic resistance island found in a multidrug-resistant Acinetobacter baumannii isolate of European clone I from 1977. J. Antimicrob. Chemother. 65:1915–1918 [DOI] [PubMed] [Google Scholar]
- 17. Nemec A., Dijkshoorn L., J. van der Reijden T. 2004. Long-term predominance of two pan-European clones among multi-resistant Acinetobacter baumannii strains in the Czech Republic. J. Med. Microbiol. 53:147–153 [DOI] [PubMed] [Google Scholar]
- 18. Nemec A., Janda L., Melter O., Dijkshoorn L. 1999. Genotypic and phenotypic similarity of multiresistant Acinetobacter baumannii isolates in the Czech Republic. J. Med. Microbiol. 48:287–296 [DOI] [PubMed] [Google Scholar]
- 19. Nemec A., et al. 2008. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 62:484–489 [DOI] [PubMed] [Google Scholar]
- 20. Post V., Hall R. M. 2009. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:2667–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Post V., Nigro S. J., Hall R. M. 2010. Multiple antibiotic resistance in Acinetobacter baumannii from an Australian hospital, abstr. 024. Abstr. 8th Int. Symp. Biol. Acinetobacter. [Google Scholar]
- 22. Post V., White P. A., Hall R. M. 2010. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:1162–1170 [DOI] [PubMed] [Google Scholar]
- 23. Ruzin A., Immermann F. W., Bradford P. A. 2010. RT-PCR and statistical analyses of adeABC expression in clinical isolates of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Microb. Drug Resist. 16:87–89 [DOI] [PubMed] [Google Scholar]
- 24. Turton J. F., Baddal B., Perry C. 2011. Use of the accessory genome for characterization and typing of Acinetobacter baumannii. J. Clin. Microbiol. 49:1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turton J. F., Gabriel S. N., Valderrey C., Kaufmann M. E., Pitt T. L. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 8:807–815 [DOI] [PubMed] [Google Scholar]
- 26. Villalón P., et al. 2011. Clonal diversity of nosocomial epidemic Acinetobacter baumannii in Spain. J. Clin. Microbiol. 49:875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.