Abstract
The low level of available iron in vivo is a major obstacle for microbial pathogens and is a stimulus for the expression of virulence genes. In this study, Mycobacterium tuberculosis H37Rv was grown aerobically in the presence of limited iron availability in chemostat culture to determine the physiological response of the organism to iron-limitation. A previously unidentified wax ester accumulated under iron-limited growth, and changes in the abundance of triacylglycerol and menaquinone were also observed between iron-replete and iron-limited chemostat cultures. DNA microarray analysis revealed differential expression of genes involved in glycerolipid metabolism and isoprenoid quinone biosynthesis, providing some insight into the underlying genetic changes that correlate with cell-wall lipid profiles of M. tuberculosis growing in an iron-limited environment.
INTRODUCTION
The process of withholding iron from pathogens is of key importance as a host defence. Iron is not freely available and is bound to glycoproteins such as transferrin. The surplus iron-binding capacity of transferrin ensures that no free iron remains in circulation, and bacteria need to employ specific mechanisms for the acquisition of iron (Wagner et al., 2005). Mycobacterium tuberculosis, which survives in host macrophages, is likely to have to adapt to an iron-limited environment inside the phagosome, particularly as host defences such as interferon-γ decrease the ability of infected macrophages to acquire iron from transferrin (Olakanmi et al., 2002). Mycobacteria have developed sophisticated iron-acquisition systems, a major component of which is the siderophore (Ratledge & Ewing, 1996; Ratledge, 2004). The extracellular carboxymycobactin has a high affinity for ferric iron. It sequesters the iron and returns it to the cell via the unusual, biosynthetically related, intracellular mycobactin. The latter occupies an intracellular location and is thought to act as a short-term iron-storage molecule (De Voss et al., 2000; Ratledge & Dover, 2000; Ratledge, 2004). However, a balance has to be maintained between acquisition and storage of iron. Excess levels can be toxic, as iron serves as a catalyst for oxidative stress. Thus, in an environment where there is excess iron, the metal is stored by iron-binding molecules such as bacterioferritins, of which M. tuberculosis has two (Pessolani et al., 1994; Smith, 2004), and the cell-bound mycobactins.
Part of the response to fluctuating iron levels is controlled in bacteria by two families of iron-dependent regulatory proteins, Fur and DtxR (Kunkle & Schmitt, 2005; Manabe et al., 2005). Mycobacteria possess fur and dtxR homologues (the latter being called ideR). IdeR represses expression of siderophore biosynthetic genes (Gold et al., 2001), whilst activating the expression of genes involved in the storage of iron. Other IdeR-controlled genes have been identified by microarray analysis of M. tuberculosis growing in batch cultures (Rodriguez et al., 1999, 2002; Rodriguez & Smith, 2003), and have been found to encode PE/PPE proteins and enzymes involved in the biosynthesis, metabolism and transport of lipids. The set of genes induced under low iron in the study of Rodriguez et al. (2002) partially overlaps with genes induced upon infection of macrophages (Schnappinger et al., 2003), providing further supporting evidence that the macrophage is an iron-limited environment.
The previous observation that growth in low iron induces genes involved in lipid biosynthesis and metabolism led us to investigate the complement of cell lipids made under these conditions. The mycobacterial cell envelope is a unique structure. The core is an interconnected peptidoglycan/arabinogalactan/mycolic acid, which interacts with complex free lipids, ranging in polarity from apolar triacylglycerols (TAGs) and phthiocerol dimycocerosates (PDIMs) to polar glycopeptides and lipo-oligosaccharides (Minnikin et al., 2002). Some of these lipids, such as phenolic glycolipid and PDIM, have been directly implicated in virulence (Cox et al., 1999; Reed et al., 2004).
The aims of our study were to grow M. tuberculosis under iron-replete and iron-limited conditions in vitro, and to explore the effect of iron availability on the lipid composition of the cell. We describe the accumulation of a novel wax ester (WE) and other lipid changes, and investigate the transcriptional changes that occur under iron-limitation.
METHODS
Growth of M. tuberculosis under iron-replete and iron-limited conditions
Four independent iron-replete cultures (IR1, IR2, IR3 and IR4) were previously grown and used in studies to determine the effects of oxygen availability on gene expression and pathogenesis (Bacon et al., 2004). These cultures represent standard growth conditions (aerobic, carbon-limited and iron-replete) in both the previous and the current studies. The gene expression data for iron-replete growth used in the current study were generated previously (Bacon et al., 2004) from all four of these cultures (IR1–IR4), and the lipids were extracted from cultures IR3 and IR4.
Three further independent, iron-replete steady-state cultures (IR5, IR6 and IR7) were established as described previously (Bacon et al., 2004) under aerobic conditions, in CAMR mycobacterium medium (CMM) (James et al., 2000) at a dissolved oxygen tension (DOT) of 10 % (previously described as 50 % air saturation) (Bacon et al., 2004) at 37 °C, pH 6.9, and a dilution rate of 0.03 h−1 (mean generation time of 23 h). Samples were collected from the culture system to monitor culture turbidity (OD540), viability and nutrient utilization, as described previously (Bacon et al., 2004). Cultures were maintained for 8 days (equivalent to eight culture generations) after the turbidity stabilized, before commencing steady-state sample collection. Replicate samples were taken daily over the following week. The CMM medium was then modified by removing the iron source, ferric sulphate (FeSO4.7H2O). The culture was transferred to a new fermentation vessel, and allowed to adjust to the new medium and to re-establish a steady-state in the iron-limited environment. These cultures were named IL1, IL2 and IL3, respectively. The sampling regimen described for iron-replete cultures was repeated for the iron-limited cultures. Cell samples were removed during steady-state growth, for RNA and lipid extraction. For lipid extraction, the overnight effluents from two of the iron-limited cultures, labelled IL1 and IL2, were diverted to Duran bottles at 4 °C and harvested by centrifugation for 20 min at 15 000 r.p.m. The cell pellets were combined and resuspended in 10 ml water. The cells were spun again by centrifugation at 3500 r.p.m. for 10 min at 4 °C, and frozen at −70 °C. Prior to dispatch for lipid analysis, cells were autoclaved for 20 min at 121 °C and dried overnight in a SpeedVac Plus SC110A concentrator (Thermo Savant).
RNA was prepared from cultures IL1, IL2 and IL3. A 20 ml aliquot of culture was collected daily from the chemostat. This sample was immediately added to four volumes of guanidium thiocyanate, and the RNA was extracted as described previously (Bacon et al., 2004). The level of iron (p.p.m.) used by two of the iron-replete cultures (IR5 and IR6) and two iron-limited cultures (IL1 and IL2) was determined by flame atomic absorption spectrophotometry (Eurotest) (Table 1). Iron was confirmed as the limiting nutrient by the addition of a pulse (directly to the vessel) of 20 ml iron-replete medium containing 2.04 mg FeSO4.7H2O, and monitoring the optical density of the culture over time. This was performed on both iron-replete and iron-limited cultures.
Table 1.
Iron levels in iron-replete and iron-limited culture
| Culture | Medium (m) or spent supernatant (s) |
Iron level (p.p.m.) |
|---|---|---|
| Iron-replete | ||
| IR5 | m | 0.58 |
| IR5 | s | 0.04 |
| IR6 | m | 0.31 |
| IR6 | s | 0.06 |
| Iron-limited | ||
| IL1 | m | 0.06 |
| IL1 | s | 0.02 |
| IL2 | m | 0.05 |
| IL2 | s | 0.03 |
Lipid extraction and analysis
Dry biomass (50 mg) was extracted according to Dobson et al. (1985) to provide apolar and polar lipid fractions. The apolar and polar lipid fractions were resuspended in petroleum ether or chloroform/methanol (2 : 1, v/v), respectively, and 50 μg was applied to 6.6×6.6 cm Merck 5554 aluminium-backed TLC plates. Plates were developed using several solvent systems, designed to cover the whole range of lipid polarities (Dobson et al., 1985) (see below for summary of solvent systems). Charred TLC plates were scanned using a Bio-Rad GS-700 Imaging Densitometer, and the image was captured using PDQuest (Bio-Rad) software. Quantitative analyses were carried out on these images using QuantityOne (Bio-Rad) software, global background subtraction being employed over a large representative area of a plate image. Relative, but not absolute, proportions of the lipids were calculated. Since these lipids occur naturally as complex, uncharacterized homologous mixtures, it is not practical to obtain standards to allow absolute quantification.
Summary of solvent systems
The solvent systems used were as follows. System A: (1) petroleum ether (boiling point 60–80 °C)/ethyl acetate (98 : 2, three times); (2) petroleum ether/acetone (98 : 2). System B: (1) petroleum ether/acetone (92 : 8, three times); (2) toluene/acetone (95 : 5). System C: (1) chloroform/methanol (96 : 4); (2) toluene/acetone (80 : 20). System D: (1) chloroform/methanol/water (100 : 14 : 0.8); (2) chloroform/acetone/methanol/water (50 : 60 : 2.5 : 3). System E: (1) chloroform/methanol/water (60 : 30 : 6); (2) chloroform/acetic acid/methanol/water (40 : 25 : 3 : 6).
Isolation, purification and analysis of putative WEs
A portion of the combined apolar fraction (2 mg) from iron-limited cultures IL1 and IL2 was loaded onto a 10×10 cm Merck 5735 plastic-backed TLC plate. The plate was developed in solvent system A (Fig. 1) and visualized by spraying with 0.01 % ethanolic rhodamine 6G spray, and separated components were identified by fluorescence under 366 nm long-wave UV light. The lipids of interest were isolated by scraping the silica gel from the plate and extracting them in glass tubes, employing three 20 min washes with 3 ml diethyl ether per wash. After each wash, the tubes were centrifuged for 5 min at 3500 r.p.m., and the combined supernatants were filtered into pre-weighed tubes and dried under nitrogen. Yields of apolar and polar lipids from 50 mg dried biomass were ~8 and ~4 mg, respectively, for the iron-limited cultures, and ~4 mg each for iron-replete cultures. 1H-NMR spectra were obtained in deuterated trichloromethane (CDCl3) on a Bruker WP300 spectrometer, using stearyl oleate and stearyl stearate as standards. MALDI-TOF MS was performed using a Bruker Biflex IV mass spectrometer.
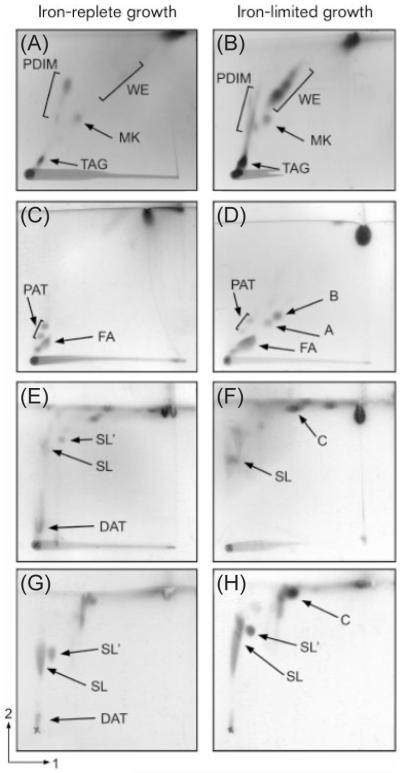
Fig. 1.
2D non-phospholipid patterns of representative chemostat-grown M. tuberculosis H37Rv. (A), (C), (E) and (G) are TLC images for iron-replete culture IR3. (B), (D), (F) and (H) are TLC images for iron-limited culture IL1. (A–F) are apolar lipids. (G) and (H) are polar lipids. Profiles in (A) and (B) were resolved using solvent system A. Profiles (C) and (D) were resolved using solvent system B. Profiles (E–H) were resolved using solvent system D. FA, fatty acids; A, B and C, unknowns; SL and SL′, sulfolipids.
Microarray procedures
RNA was extracted from three independent chemostat cultures grown under iron-limitation. Four separate labellings were carried out with each RNA sample, giving a total of 12 labelled products. For each array, 8 μg total RNA was used as a template for reverse transcriptase (200 U Superscript II RNase H μl−1; Life Technologies) in the presence of random primers and cyanine (Cy)5-labelled dCTP. Each aliquot of Cy5-labelled cDNA generated from RNA (test sample) was co-hybridized with Cy3-labelled DNA generated from genomic DNA (control sample). The DNA (1 μg) was used as a template for DNA polymerase (5 U Klenow μl−1; Life Technologies) in the presence of random primers and Cy3-labelled dCTP. The genomic DNA used in this work had been extracted previously from a cell pellet of M. tuberculosis H37Rv harvested from an aerobic steady-state culture. The same batch of genomic DNA was used in the present and previously published array experiments (Bacon et al., 2004). We used whole-genome arrays of M. tuberculosis H37Rv, containing 3924 gene-specific PCR-amplified products, prepared by the Bacterial Microarray Group at St George’s University (http://bugs.sgul.ac.uk/). The hybridization method has been described previously (Bacon et al., 2004). The arrays were scanned for the Cy5 channel at more than one gain level to capture the wide range of intensity values.
Microarray data analysis
Semilinear high-dimensional model (SLIM) normalization (Fan, 2005) followed by quantile normalization (Bacon et al., 2004; Wernisch et al., 2003) were used across iron-replete and iron-limited data. These normalizations generated a common distribution of log ratios across all arrays and conditions. Two scans for the Cy5 channel were integrated as follows, in order to extrapolate intensity values with poor quality incurred from saturation and quantization. A regression coefficient was estimated using the low scan as an independent variable and the higher scan as a response variable. Intensity values below the fifth quantile were excluded from the low-intensity data, and values >40 000 (approximately the fifteenth percentile) were excluded from the high-intensity data. The Cy5 intensity values were transformed by taking the square root. Regression was then performed using the robust regression technique, MM estimator (Venables & Ripley, 2002). Outliers with residuals higher than the ninetieth percentile of all the residuals were excluded before a second regression was performed. The coefficients from the second regression were used to extrapolate values below the fifth quantile and above the high-intensity threshold. The two expression profiles were then integrated. To achieve this, the mean of the two intensity values for each spot was calculated and squared to recover values comparable to the original data, before the above square root transformation.
Following the normalization procedures, genes were identified that were differentially regulated under iron-replete and iron-limited conditions, using Limma (http://bioinf.wehi.edu.au/limma/index.html), a microarray analysis software that uses a linear model within an empirical Bayes framework. The moderated t test in Limma uses a variance based on the combined information across genes to compensate underestimated sample variances. This is similar to other test statistics with a compensated SD, such as significance analysis of microarray (SAM) (Tusher et al., 2001). With a small number of replicates available per gene, the outlier replicates distorted the statistical result significantly. We identified which genes had outlying values that caused high coefficients of variation. These outliers were subsequently down-weighted using weighted least-square regression in Limma, before significance analysis was applied. Differentially regulated genes were then selected on the basis of a positive B score; the log odds for differential expression were calculated. Consequently, genes with a positive B value were selected.
Real-time quantitative PCR (RT-qPCR)
Reverse transcription took place in a total volume of 20 μl containing 100 ng total RNA, 300 ng random primers (Invitrogen Life Technologies), 10 mM DTT, 0.5 mM each of dCTP, dATP, dGTP and dTTP, and 200 U Superscript III (Invitrogen Life Technologies). For primer annealing, RNA and random primers were heated to 65 °C for 10 min in a volume of 13 μl, and snap-cooled on ice prior to the addition of the remaining components. For reverse transcription, the reactions were incubated at 55 °C for 50 min. One microlitre (equivalent to 5 ng RNA) of cDNA was used in the RT-qPCR reactions. RT-qPCR reactions were set up using the DyNAmo SYBR Green qPCR kit (MJ Research), and RT-qPCR was performed using the DNA Engine Opticon 2 System (GRI). Twenty-microlitre reactions were set up on ice containing 1× DNA Master SYBR Green I mix, 1 μl cDNA product and 0.3 μM each primer. Reactions were heated to 95 °C for 10 min, before 35 cycles of 95 °C for 30 s, 62 °C for 20 s, and 72 °C for 20 s. Fluorescence was captured at the end of each cycle after heating to 80 °C to ensure the denaturation of primer–dimers. At the end of the PCR reaction, melting-curve analysis was performed, and PCR products were analysed on an agarose gel to ensure product specificity.
RESULTS
Continuous growth of M. tuberculosis under iron-replete and iron-limited conditions
M. tuberculosis was cultured for 14 generations in a steady-state under either iron-replete [IR1, IR2, IR3, IR4 (Bacon et al., 2004), IR5, IR6 and IR7] or iron-limited growth conditions (IL1, IL2, IL3), at a dilution rate of 0.03 h−1, which corresponded to a constant mean generation time of 23 h. Steady-state growth was confirmed in all cultures by monitoring the culture OD540, which remained constant at a mean of 2.4±0.2 and 0.969±0.1 for iron-replete and iron-limited growth, respectively. The higher optical density for the iron-replete growth was reflected by higher viable counts, which were 8.8±0.1 log10(c.f.u. ml−1) compared to 8.2±0.1 log10(c.f.u. ml−1) for iron-limited growth. Iron was confirmed as the limiting nutrient in the iron-limited cultures IL1, IL2 and IL3 by a twofold increase in the optical density over a 24 h period, following the addition of a pulse of medium containing iron. No increase in optical density was observed following the addition of iron to the iron-replete cultures. The limiting nutrient in the iron-replete cultures was glycerol (Bacon et al., 2004).
The levels of iron were measured in iron-replete (IR5 and IR6) and iron-limited cultures (IL1 and IL2) (Table 1) to determine the amount of iron being used in these two growth environments. The quantity of iron used by the former was found to be ~10-fold higher than that of the latter.
Lipid profiling for M. tuberculosis grown under different iron availabilities
Apolar and polar lipids were sequentially extracted from cells collected during steady-state continuous growth under iron-replete and iron-limited conditions (IR3, IR4, IL1 and IL2). Lipid extracts were analysed by TLC, using five solvent systems (A–E) of increasing polarity (Fig. 1) (Dobson et al., 1985). Individual lipid components were quantified by densitometry, and the relative proportions of the non-phospholipid fractions are shown in Table 2. While the phospholipid patterns previously obtained from batch cultures were clearly resolved (unpublished data), those from the chemostat samples were not well separated, so it was not possible to obtain chromatograms suitable for quantification.
Table 2.
Proportion of non-phospholipids from chemostat cultures of M. tuberculosis H37Rv grown under iron-replete (IR3, IR4) and iron-limited (IL1, IL2) conditions
| Lipid | Proportion (%) |
|||
|---|---|---|---|---|
| Iron-replete |
Iron-limited |
|||
| IR3 | IR4 | IL1 | IL2 | |
| TAG | 2.7 | 5.1 | 11.9 | 11.4 |
| PDIM | 17.0 | 7.6 | 9.1 | 4.1 |
| WE | 2.9 | 3.3 | 30.8 | 16.4 |
| MK | 1.7 | 1.2 | 0.8 | 0.6 |
| FA | 8.8 | 11.5 | 8.3 | 20.4 |
| A, B | ND | ND | 6.4 | ND |
| C | ND | 9.6 | 7.4 | 8.7 |
| ST* | 66.5 | 61.7 | 25.3 | 38.4 |
The sum of all trehalose-based lipids (DATs, PATs, SL and SL′).
FA, fatty acids; A, B and C, unknowns; SL and SL′, sulfolipids.
ND, Not detected.
Both subtle and major differences in apolar lipids were found between cells grown under iron-replete and iron-limited conditions, when separated using solvent system A. PDIMs, menaquinones (MKs), TAGs and WEs were identified under both growth conditions (Fig. 1A, B, Table 2). We observed a decrease in the abundance of an MK in the iron-limited profiles compared with the iron-replete profiles shown in Fig. 1. Pronounced increases in the abundance of two classes of lipid were observed in iron-limitation; these were TAGs and the accumulation of a large amount of an unidentified lipid series, consisting of three distinct spots with the character of WEs. No clear trends were observed for PDIMs.
Examples of TLC system D, applied to both apolar and polar lipid fractions, are shown in Fig. 1E–H. Diacyl trehaloses (DATs) were found in cells from both growth conditions (Fig. 1D, F). High proportions of sulfolipids (SL, SL′) were observed in all the lipid profiles from continuous cultures. The proportions of trehalose-based lipids [DATs, pentaacyl trehaloses (PATs) and sulfolipids] differed under the two growth conditions (Table 2, ST). The iron-replete cultures (IR3 and IR4) had >60 % of their non-phospholipids as various acylated trehaloses, but under iron-limitation (IL1 and IL2), the proportion was reduced to ~30 %.
In TLC system B, two unknown components (A and B), with the chromatographic behaviour of diacylglycerols (DAGs) were observed under iron-limitation. A further unidentified lipid C was also observed. These findings need to be investigated further, as they were not seen in replicate cultures.
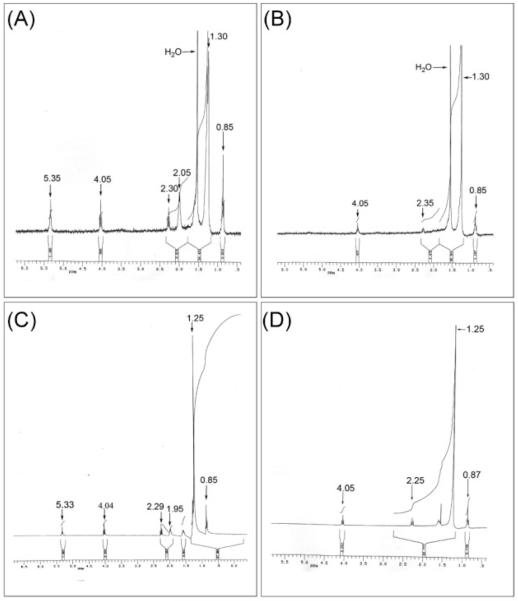
The two major components of the putative WEs identified in system A (Fig. 1B) were isolated and purified by 2D preparative TLC. These compounds were then subjected to NMR analysis (Fig. 2), in order to elucidate their structures. The least mobile component (see Fig. 1A) of this unidentified lipid series is postulated to be an unsaturated WE, as indicated by the similarity of its 1H-NMR results (Fig. 2A) to those of stearyl oleate (Fig. 2C). Signals at 5.35 and 2.05 p.p.m. corresponded to a cis -CH2CH=CHCH2- unit. Similarly, triplets at 4.05 and 2.30 p.p.m. indicated the presence of a -CH2-OCO-CH2- unit. A large signal at 1.30 p.p.m. corresponded to long-chain -(CH2)x- units, with the terminal -CH3 triplet located at 0.85 p.p.m. In comparison with the spectrum of stearyl stearate (Fig. 2D), the 1H-NMR spectrum of the more mobile component (Fig. 1B) appeared to indicate a saturated WE, based on the lack of 1H-NMR resonances at 5.35 and 2.05 p.p.m. that corresponded to the cis -CH2CH=CHCH2- unit. The small amount of available material did not allow the recording of informative 13C-NMR spectra of these WEs.
Fig. 2.
1H-NMR spectra of the two major components of the putative WE (Fig. 1B, WE) that accumulated under iron-limitation. (A) Least mobile component, (B) more mobile component, (C) stearyl oleate, and (D) stearyl stearate. x axes of plots show p.p.m.
MALDI-TOF MS of the standard stearyl stearate and stearyl oleate gave the expected molecular ions (M+Na+) at m/z 560 and 558, respectively. The two natural esters did not produce any recognizable molecular ions or mass spectrometric fragments.
Transcriptional profiling by DNA microarray analysis
Microarray analysis was performed in order to identify changes in gene expression that underlie the altered lipid profiles during growth under different iron availabilities. The gene expression data for iron-replete growth used in the current study were generated previously from the four cultures IR1, IR2, IR3 and IR4 (Bacon et al., 2004), while iron-limited microarray data were generated from cultures IL1, IL2 and IL3.
Following the normalization procedures, genes were identified that were differentially regulated under iron-replete and iron-limited conditions, using Limma micro-array analysis software. With a small number of replicates available per gene, the outlier replicates significantly distorted the results of statistical analysis. However, Limma allowed us to perform weighted least-square regression to explicitly down-weight outlier replicates in the data. A comparison of Limma, with or without down-weighting outlier replicates to detect differentially expressed genes, showed that there were fewer significantly differentially expressed genes without the down-weighting. Differentially regulated genes were then selected on the basis of a positive B score. Fully annotated microarray data containing information for the fold change, M, B and t values for all genes have been deposited in BμG@Sbase (accession no. E-BUGS-35; http://bugs.sgul.ac.uk/) and also Array Express (http://www.ebi.ac.uk/arrayexpress/; accession no. E-BUGS-35).
Differential expression of genes under iron-limitation
We identified a total of 316 genes expressed at significantly higher levels under iron-limited growth (Supplementary Table S1). Fifty-four of the genes shown in our study to be induced by iron-limitation have been identified elsewhere as being induced under low-iron growth (Rodriguez et al., 2002) (see Supplementary Table S1). That study has presented evidence that 100 genes from M. tuberculosis H37Rv are induced by low iron. Our data support some of these, do not confirm others, and identify additional genes induced by iron-limitation in the chemostat. We identified a total of 304 genes expressed at significantly higher levels under iron-replete growth (Supplementary Table S2). Twelve of the genes shown in our study to be induced by high levels of iron have been identified elsewhere as being induced under high-iron growth (Rodriguez et al., 2002) (see Supplementary Table S2).
There are some differences in the differential expression of certain genes in response to iron availability between the current study and that of Rodriguez et al. (2002); some of the genes identified as upregulated by Rodriguez et al. were down-regulated in our study, and vice versa. These are also indicated in Supplementary Tables S1 and S2. The putative function of genes upregulated under iron-limitation was established using annotations within Tuberculist (http://genolist.pasteur.fr/TubercuList/index.html), MycoDB (http://myco.bham.ac.uk) and National Center for Biotechnology Information BLAST (http://www.ncbi.nlm.nih.gov/blast/).
Some of the genes that were differentially expressed in our study are associated with lipid biosynthesis (Table 3), fatty acid metabolism and cell wall biosynthesis/partition. Four genes involved in biosynthesis of isoprenoid lipids, Rv3582c (ispD), Rv3581c (ispF), Rv3383c (idsB) and Rv0560c, that have been identified previously as being upregulated in macrophages, were also induced under iron-limitation. Supplementary Table S3 highlights those genes upregulated under iron-limitation in the chemostat and in macrophages (Schnappinger et al., 2003). None of these lipid genes was upregulated under low oxygen in the chemostat, suggesting a response that was specific to iron-limitation rather than a generalized response to stress. A role related to the biosynthesis of the isoprenoid lipid MK can be envisaged for Rv0560c, which encodes a putative benzoquinone methyl-transferase (Sun et al., 2001), the induction of which has been observed in connection with the biosynthesis of isoprenoid quinones (Garbe et al., 1996). We observed upregulation of Rv2182c, Rv3842c, Rv3837c and Rv3097c (lipY), which are putatively involved in glycerophospholipid metabolism (Bono et al., 1998; Karp et al., 1999). These genes could be associated with the biosynthesis of the WE; in particular, Rv3097c (lipY), which is a triacylglycerol lipase/esterase (Deb et al., 2006).
Table 3.
Differential expression of genes encoding lipid biosynthetic proteins in M. tuberculosis grown under different iron availability
| Rv no. | Gene name | Fold change | B score | Putative function |
|---|---|---|---|---|
|
Induction under
iron-limited growth |
||||
| Rv0450c | mmpl4 | 2.97 | 2.05 | Transmembrane transport protein |
| Rv0560c | 9.29 | 0.50 | Benzoquinone methyltransferase | |
| Rv0676c | mmpl5 | 3.61 | 5.18 | Transmembrane transport protein |
| Rv0753c | mmsa | 4.06 | 3.90 | Methylmalonate semialdehyde dehydrogenase |
| Rv1342c | pks14 | 1.96 | 0.40 | Membrane protein |
| Rv2182c | 3.97 | 4.77 | 1-Acylglycerol-3-phosphate O-acyltransferase | |
| Rv2933 | ppsC | 2.75 | 2.82 | Phenolphthiocerol synthesis type-1 polyketide synthase |
| Rv2934 | ppsD | 4.30 | 0.36 | Phenolphthiocerol synthesis type-1 polyketide synthase |
| Rv3097c | lipY | 2.25 | 2.38 | Triacylglycerol lipase/esterase |
| Rv3383c | idsB | 2.88 | 0.59 | Polyprenyl synthetase |
| Rv3581c | ispF | 2.87 | 2.22 | Biphenyl-2,3-diol 1,2-dioxygenase |
| Rv3582c | ispD | 3.64 | 3.44 | 4-Diphosphocytidyl-2C-methyl-D-erythritol |
| Rv3825c | pks2 | 3.02 | 3.42 | Polyketide synthase |
| Rv3837c | 2.36 | 1.24 | Phosphoglycerate mutase | |
| Rv3842c | glpQ1 | 2.16 | 1.78 | Glycerolphosphoryl diester phosphodiesterase |
|
Induction under
iron-replete growth |
||||
| Rv0403c | mmps1 | 0.45 | 0.38 | Conserved membrane protein |
| Rv0506 | mmps2 | 0.41 | 3.43 | Conserved membrane protein |
| Rv1372 | pks18 | 0.34 | 1.57 | Conserved hypothetical protein |
| Rv1660 | pks10 | 0.50 | 0.89 | Chalcone synthase |
| Rv1665 | pks11 | 0.21 | 7.02 | Chalcone synthase |
| Rv2198c | mmps3 | 0.34 | 0.16 | Conserved membrane protein |
| Rv2339 | mmps9 | 0.34 | 4.05 | Conserved membrane transport protein |
| Rv2349c | plcC | 0.46 | 0.25 | Phospholipase |
| Rv2350c | plcB | 0.36 | 0.75 | Phospholipase |
Some genes associated with lipid biosynthesis were induced during iron-replete growth (Table 3), including Rv1665 (pks11), Rv2350 and Rv2356 (plcB and plcC, phospholipases), and Rv1660 (pks10), which has been implicated in phthiocerol synthesis. However, upregulation of Rv1660 (pks10) under iron-replete growth seems to conflict with the upregulation of Rv2350 (ppsC) and Rv2356 (ppsD) under iron-limitation. Neither of these changes was reflected in our lipid analyses, in which the PDIM content was similar under both culture conditions.
Validation of the microarray data was performed using RT-qPCR. We examined Rv0560c, Rv3825c (pks2), Rv3765 (tcrX) and Rv2703 (sigA); sigA has traditionally been used to normalize data (Fig. 3). Under these conditions, there was no significant change in sigA expression during iron-limitation. However, all the other genes showed significant upregulation with Rv0560c, Rv3825c (pks2) and Rv3765 (tcrX) showing fold changes that were concordant with the microarray data.
Fig. 3.
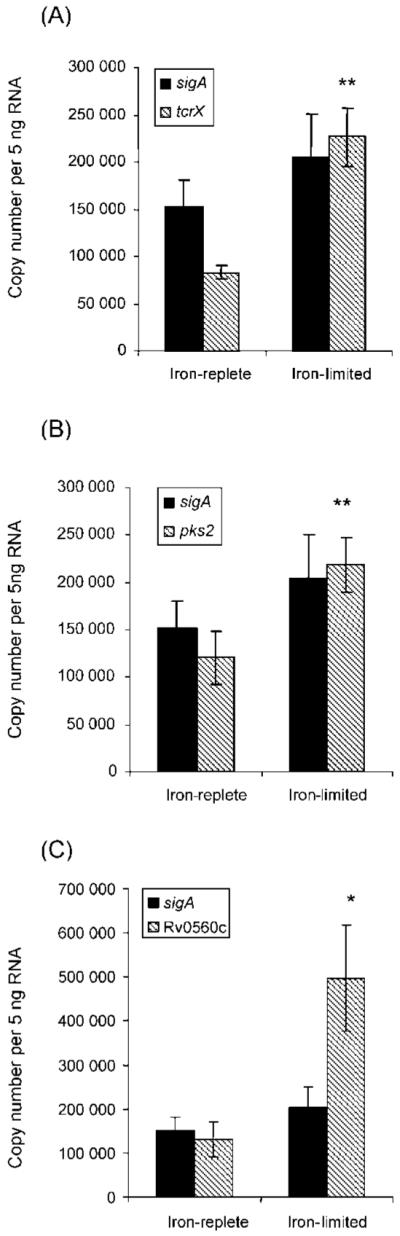
RT-qPCR analysis of (A) tcrX, (B) pks2 and (C) Rv0560c from M. tuberculosis grown under iron-replete or iron-limited conditions. Levels of sigA were not significantly different under iron-limitation. The other genes were significantly induced under iron-limitation (unpaired Student’s t test); *P<0.01, **P<0.001.
DISCUSSION
Complex cell-wall lipids in M. tuberculosis play an important role in the interaction with the immune system during the early stages of infection (Korf et al., 2005). Some components of the cell envelope, such as the mycolic acids, have a structural role, essential for survival in the host. Other lipids, such as the PDIMs, PATs, DATs and sulfolipids, are considered to be expressed on the cell surface and possibly to interact with host cells (Minnikin et al., 2002). Earlier transcriptome studies (Rodriguez et al., 2002) have shown changes in the expression of lipid biosynthetic and transport genes under low-iron growth, which led us to investigate how the lipid profiles of M. tuberculosis were modified under iron-limited growth in a chemostat, and how these changes correlated with the differential gene expression identified through microarray analysis.
The most notable finding from the lipid analysis of cells growing continuously under iron-limitation was the appearance of a family of novel apolar lipids, later indicated to be WEs. These were very minor components in iron-replete, phosphate-limited and low-oxygen cultures (unpublished data), and they had never been observed previously in batch cultures of M. tuberculosis grown to different optical densities (unpublished data). However, they increased markedly during iron-limited growth (Fig. 1A, B, Table 2, IL1 and IL2), indicating that the wax accumulation is specific to iron-limited growth conditions. The two main components of the WEs were isolated and shown by 1H-NMR analyses to have spectra (Fig. 2) clearly indicative of saturated and unsaturated WEs. Detailed characterization of these molecules will be performed following isolation of larger cell samples.
An increase in the proportion of TAGs was also seen in iron-limited cultures (Fig. 1A, B, Table 2, cultures IL1 and IL2). TAGs clearly have a storage role, probably residing in mycobacterial globular lipid inclusions (Garton et al., 2002). WEs are considered to have a similar function in other bacteria (Alvarez et al., 2000; Waltermann et al., 2005; Waltermann & Steinbuchel, 2005). Consistent with the concept that mycobacterial growth is iron-limited in vivo during the early stages of disease, inclusion bodies containing TAG have been detected in M. tuberculosis recovered from sputum (Garton et al., 2002). The relatively small increase in TAG in this study, as compared to that of WE, is intriguing. The location of WE in mycobacteria has not been determined. However, in Acinetobacter species, intracellular WE is located in inclusions, which are likely to be flat, disk-like or rectangular, the shape being correlated to some extent with the nature of the substrates incorporated by the bacterium (Waltermann & Steinbuchel, 2005). In the lipid-rich mycobacterial cell envelope (Minnikin et al., 2002), the outer membrane organelle may offer a haven for linear molecules, such as WEs, that interact with the long chains of mycolic acids.
A bifunctional wax ester synthase/diacylglycerol acyltransferase (WS/DGAT) has recently been discovered in Acinetobacter calcoaceticus (Kalscheuer & Steinbuchel, 2003), to which 15 M. tuberculosis genes (tgs) have homology. One of these genes is Rv3130c, which has been shown to be expressed by M. tuberculosis in sputum (Garton et al., 2002), and to be involved in biosynthesis of TAG (Daniel et al., 2004). The WS activity of these tgs gene products has been demonstrated by expression of a homologue from Mycobacterium smegmatis mc2155 in recombinant Escherichia coli lysates (Daniel et al., 2004; Kalscheuer & Steinbuchel, 2003).
The accumulation of the WEs, seen in our lipid analyses, is unlikely to be attributed to the WS/DGAT class of enzymes, however, as the microarray data clearly showed a lack of induction of these genes. In contrast, we observed induction of genes involved in glycerophospholipid metabolism: Rv2182c (1-acylglycerol-3-phosphate O-acyltransferase), Rv3842c (glpQ1, glycerophosphoryl diester phosphodiesterase), Rv3837c (phosphoglycerate mutase) and Rv3097c (PE-PGRS family, triacylglycerol lipase/esterase). Triacylglycerol lipases generate DAG and fatty acid from TAG, and recently the product of Rv3097c (lipY) has been shown to hydrolyse long-chain TAGs with high specific activity. It has been demonstrated that hypoxic cultures of M. tuberculosis, which accumulate TAG, hydrolyse the stored TAG when subjected to nutrient starvation (Deb et al., 2006). Under these conditions, lipY is induced more than all the other lipase homologues in M. tuberculosis, suggesting that it has a central role in the utilization of stored TAG. It has also been shown that TAG utilization is drastically decreased in a lipY-deficient mutant under nutrient-deprived conditions (Deb et al., 2006). Combined with our observation that TAGs and WEs accumulate under iron-limitation, the transcriptional profiling indicates that the WEs are formed from fatty acids that have been hydrolysed from TAG by Rv3097c, as it has previously been shown that lipases can catalyse the synthesis of WEs either from free fatty acids or through degradation of TAG (Tsujita et al., 1999). Spherical lipid bodies in Rhodococcus opacus have been found to include significant percentages of DAG and free fatty acids, in addition to the predominant TAGs (Alvarez et al., 2000; Waltermann & Steinbuchel, 2005). In one of the iron-limited cultures (Table 2, IL1), possible DAGs were noted as components A and B (Fig. 2D). The other iron-limited culture (Table 2, IL2) lacked A and B, but had increased proportions of free fatty acids, perhaps compensating for the lack of putative DAGs. This suggests that the well-documented presence of free fatty acids in mycobacteria (Dobson et al., 1985; Minnikin et al., 1985) may not be incidental, and they may have a positive storage or structural role.
To conclude, our studies showed that M. tuberculosis adapted and grew in an iron-limited environment by altering the composition and relative abundance of its lipids. Of particular interest is the substantial induction of a novel WE in M. tuberculosis. The microarray analysis identified differential expression in a number of genes involved in lipid biosynthesis and metabolism, and some of these changes may form part of the genetic network underlying production of TAG and the WE. Inactivation of these genes, particularly Rv3097c, will provide further evidence for a link between the transcriptomic and lipid changes observed.
The function of the family of WEs also comes into question. The discovery of such novel lipids is of interest, due to the increasing focus on unique cell-wall lipid biosynthetic pathways as targets for the development of new antitubercular drugs. The WE was induced under in vivo-like conditions in the chemostat, and a number of genes were induced that were previously found to be expressed in macrophages. This reinforces the view that the chemostat can provide useful information about global responses to relevant host stimuli.
Supplementary Material
ACKNOWLEDGEMENTS
This study was funded by the Department of Health and the Health Protection Agency UK. The views expressed in the publication are those of the authors and not necessarily those of the Department of Health and Health Protection Agency. The authors acknowledge the multi-collaborative bacterial microarray group at St George’s, University of London (BuG@S: http://www.sgul.ac.uk/depts/medmicro/), which is supported by the Wellcome Trust Functional Genomics Resource Initiative. G. S. B., a Lister–Jenner Research Fellow, acknowledges support from the UK Medical Research Council. S. L. K. was funded by the UK Biotechnology and Biological Sciences Research Council grant 48/P18545.
Abbreviations
- Cy
cyanine
- DAG
diacylglycerol
- DAT
diacyl trehalose
- MK
menaquinone
- PAT
pentaacyl trehalose
- PDIM
phthiocerol dimycocerosate
- RT-qPCR
real-time quantitative PCR
- TAG
triacylglycerol
- WE
wax ester
- WS/DGAT
wax ester synthase/diacylglycerol acyltransferase
Footnotes
Tables showing genes upregulated under iron-limitation, genes upregulated under iron-replete growth, and genes induced under iron-limitation in the chemostat and 24 h post-infection in the macrophage, are available as supplementary data with the online version of this paper.
REFERENCES
- Alvarez HM, Kalscheuer R, Steinbuchel A. Accumulation and mobilization of storage lipids by Rhodococcus opacus PD630 and Rhodococcus ruber NCIMB 40126. Appl Microbiol Biotechnol. 2000;54:218–223. doi: 10.1007/s002530000395. [DOI] [PubMed] [Google Scholar]
- Bacon J, James BW, Wernisch L, Williams A, Morley KA, Hatch GJ, Mangan JA, Hinds J, Stoker NG, other authors The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84:205–217. doi: 10.1016/j.tube.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Bono H, Ogata H, Goto S, Kanehisa M. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 1998;8:203–210. doi: 10.1101/gr.8.3.203. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M, Jacobs WR. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, Kolattukudy PE. A novel lipase belonging to the hormonesensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J Biol Chem. 2006;281:3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Voss JJ, Rutter K, Schroeder BG, Su H, Zhu Y, Barry CE., III The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc Natl Acad Sci U S A. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson G, Minnikin DE, Minnikin SM, Partlett JH, Goodfellow M, Ridell MMM. Systematic Analysis of Complex Mycobacterial Lipids. Academic Press; London: 1985. [Google Scholar]
- Fan J. Semilinear high-dimensional model for normalization of microarray data: a theoretical analysis and partial consistency. J Am Stat Assoc. 2005;100:781–813. [Google Scholar]
- Garbe TR, Hibler NS, Deretic V. Response of Mycobacterium tuberculosis to reactive oxygen and nitrogen intermediates. Mol Med. 1996;2:134–142. [PMC free article] [PubMed] [Google Scholar]
- Garton NJ, Christensen H, Minnikin DE, Adegbola RA, Barer MR. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology. 2002;148:2951–2958. doi: 10.1099/00221287-148-10-2951. [DOI] [PubMed] [Google Scholar]
- Gold B, Rodriguez GM, Marras SA, Pentecost M, Smith I. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol Microbiol. 2001;42:851–865. doi: 10.1046/j.1365-2958.2001.02684.x. [DOI] [PubMed] [Google Scholar]
- James BW, Williams A, Marsh PD. The physiology and pathogenicity of Mycobacterium tuberculosis grown under controlled conditions in a defined medium. J Appl Microbiol. 2000;88:669–677. doi: 10.1046/j.1365-2672.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R, Steinbuchel A. A novel bifunctional wax ester synthase/acyl-CoA : diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem. 2003;278:8075–8082. doi: 10.1074/jbc.M210533200. [DOI] [PubMed] [Google Scholar]
- Karp PD, Krummenacker M, Paley S, Wagg J. Integrated pathway-genome databases and their role in drug discovery. Trends Biotechnol. 1999;17:275–281. doi: 10.1016/s0167-7799(99)01316-5. [DOI] [PubMed] [Google Scholar]
- Korf J, Stoltz A, Verschoor J, De Baetselier P, Grooten J. The Mycobacterium tuberculosis cell wall component mycolic acid elicits pathogen-associated host innate immune responses. Eur J Immunol. 2005;35:890–900. doi: 10.1002/eji.200425332. [DOI] [PubMed] [Google Scholar]
- Kunkle CA, Schmitt MP. Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J Bacteriol. 2005;187:422–433. doi: 10.1128/JB.187.2.422-433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe YC, Hatem CL, Kesavan AK, Durack J, Murphy JR. Both Corynebacterium diphtheriae DtxR(E175K) and Mycobacterium tuberculosis IdeR(D177K) are dominant positive repressors of IdeR-regulated genes in M. tuberculosis. Infect Immun. 2005;73:5988–5994. doi: 10.1128/IAI.73.9.5988-5994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin DE, Dobson G, Draper P. The free lipids of Mycobacterium leprae harvested from experimentally infected ninebanded armadillos. J Gen Microbiol. 1985;131:2007–2011. doi: 10.1099/00221287-131-8-2007. [DOI] [PubMed] [Google Scholar]
- Minnikin DE, Laurent K, Dover LG, Besra GS. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem Biol. 2002;9:545–553. doi: 10.1016/s1074-5521(02)00142-4. [DOI] [PubMed] [Google Scholar]
- Olakanmi O, Schlesinger LS, Ahmed A, Britigan BE. Intraphagosomal Mycobacterium tuberculosis acquires iron from both extracellular transferrin and intracellular iron pools. Impact of interferon-gamma and hemochromatosis. J Biol Chem. 2002;277:49727–49734. doi: 10.1074/jbc.M209768200. [DOI] [PubMed] [Google Scholar]
- Pessolani MC, Smith DR, Rivoire B, McCormick J, Hefta SA, Cole ST, Brennan PJ. Purification, characterization, gene sequence, and significance of a bacterioferritin from Mycobacterium leprae. J Exp Med. 1994;180:319–327. doi: 10.1084/jem.180.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb) 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- Ratledge C, Ewing M. The occurrence of carboxymycobactin, the siderophore of pathogenic mycobacteria, as a second extracellular siderophore in Mycobacterium smegmatis. Microbiology. 1996;142:2207–2212. doi: 10.1099/13500872-142-8-2207. [DOI] [PubMed] [Google Scholar]
- Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE., 3rd A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Smith I. Mechanisms of iron regulation in mycobacteria: role in physiology and virulence. Mol Microbiol. 2003;47:1485–1494. doi: 10.1046/j.1365-2958.2003.03384.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Gold B, Gomez M, Dussurget O, Smith I. Identification and characterization of two divergently transcribed iron regulated genes in Mycobacterium tuberculosis. Tuber Lung Dis. 1999;79:287–298. doi: 10.1054/tuld.1999.0219. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL. The physiological role of ferritin-like compounds in bacteria. Crit Rev Microbiol. 2004;30:173–185. doi: 10.1080/10408410490435151. [DOI] [PubMed] [Google Scholar]
- Sun Z, Cheng SJ, Zhang H, Zhang Y. Salicylate uniquely induces a 27-kDa protein in tubercle bacillus. FEMS Microbiol Lett. 2001;203:211–216. doi: 10.1111/j.1574-6968.2001.tb10843.x. [DOI] [PubMed] [Google Scholar]
- Tsujita T, Sumiyoshi M, Okuda H. Wax ester-synthesizing activity of lipases. Lipids. 1999;34:1159–1166. doi: 10.1007/s11745-999-0467-4. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD. Modern Applied Statistics with S. 4th edn. Springer; New York: 2002. [Google Scholar]
- Wagner D, Maser J, Lai B, Cai Z, Barry CE, III, Honer zu Bentrup K, Russell DG, Bermudez LE. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- Waltermann M, Steinbuchel A. Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J Bacteriol. 2005;187:3607–3619. doi: 10.1128/JB.187.11.3607-3619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stoveken T. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol. 2005;55:750–763. doi: 10.1111/j.1365-2958.2004.04441.x. other authors. [DOI] [PubMed] [Google Scholar]
- Wernisch L, Kendall SL, Soneji S, Wietzorrek A, Parish T, Hinds J, Butcher PD, Stoker NG. Analysis of whole-genome microarray replicates using mixed models. Bioinformatics. 2003;19:53–61. doi: 10.1093/bioinformatics/19.1.53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.