Abstract
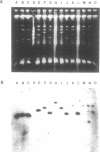
In order to construct a human chromosome 4-specific YAC library, we have utilized pYAC4 and a mouse/human hybrid cell line HA(4)A in which the only human chromosome present is chromosome 4. From this cell line, approximately 8Mb of chromosome 4 have been cloned. The library includes 65 human-specific clones that range in size from 30kb to 290kb, the average size being 108kb. In order to optimize the manipulation of YAC libraries, we have begun to investigate the stability of YACs containing human DNA in yeast cells; these studies will also determine if there are intrinsic differences in the properties of chromosomes containing higher eukaryotic DNAs. We are examining two kinds of stability: 1] mitotic stability, the ability of the YAC to replicate and segregate properly during mitosis, and 2] structural stability, the tendency of the YAC to rearrange. We have found that the majority of YACs examined are one to two orders of magnitude less stable than authentic yeast chromosomes. Interestingly, the largest YAC analyzed displayed a loss rate typical for natural yeast chromosomes. Our results also suggest that increasing the length of an artificial chromosome improves its mitotic stability. One YAC that showed a very high frequency of rearrangement by mitotic recombination proved to be a mouse/human chimera. In contrast to studies using total human DNA, the frequency of chimeras (i.e., mouse/human) in the YAC pool appeared to be low.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertsen H. M., Abderrahim H., Cann H. M., Dausset J., Le Paslier D., Cohen D. Construction and characterization of a yeast artificial chromosome library containing seven haploid human genome equivalents. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4256–4260. doi: 10.1073/pnas.87.11.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. Autonomously replicating sequences in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6329–6333. doi: 10.1073/pnas.77.11.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Dani G. M., Zakian V. A. Mitotic and meiotic stability of linear plasmids in yeast. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3406–3410. doi: 10.1073/pnas.80.11.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E. D., Olson M. V. Chromosomal region of the cystic fibrosis gene in yeast artificial chromosomes: a model for human genome mapping. Science. 1990 Oct 5;250(4977):94–98. doi: 10.1126/science.2218515. [DOI] [PubMed] [Google Scholar]
- Green E. D., Riethman H. C., Dutchik J. E., Olson M. V. Detection and characterization of chimeric yeast artificial-chromosome clones. Genomics. 1991 Nov;11(3):658–669. doi: 10.1016/0888-7543(91)90073-n. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst M. C., Rack K., Nakahori Y., Roche A., Bell M. V., Flynn G., Christadoulou Z., MacKinnon R. N., Francis M., Littler A. J. A YAC contig across the fragile X site defines the region of fragility. Nucleic Acids Res. 1991 Jun 25;19(12):3283–3288. doi: 10.1093/nar/19.12.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. S., Ripps M. E., Korman S. H., Deckelbaum R. J., Breslow J. L. Hypobetalipoproteinemia due to an apolipoprotein B gene exon 21 deletion derived by Alu-Alu recombination. J Biol Chem. 1989 Jul 5;264(19):11394–11400. [PubMed] [Google Scholar]
- Lehrman M. A., Russell D. W., Goldstein J. L., Brown M. S. Exon-Alu recombination deletes 5 kilobases from the low density lipoprotein receptor gene, producing a null phenotype in familial hypercholesterolemia. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3679–3683. doi: 10.1073/pnas.83.11.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little R. D., Porta G., Carle G. F., Schlessinger D., D'Urso M. Yeast artificial chromosomes with 200- to 800-kilobase inserts of human DNA containing HLA, V kappa, 5S, and Xq24-Xq28 sequences. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1598–1602. doi: 10.1073/pnas.86.5.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Hoekstra M. F. Relationships between a hyper-rec mutation (REM1) and other recombination and repair genes in yeast. Genetics. 1984 May;107(1):33–48. doi: 10.1093/genetics/107.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D., Hartwell L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986 Jan 17;44(1):43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Schultes N. P., Szostak J. W. Chromosome length controls mitotic chromosome segregation in yeast. Cell. 1986 May 23;45(4):529–536. doi: 10.1016/0092-8674(86)90284-9. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Construction of artificial chromosomes in yeast. Nature. 1983 Sep 15;305(5931):189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Neil D. L., Villasante A., Fisher R. B., Vetrie D., Cox B., Tyler-Smith C. Structural instability of human tandemly repeated DNA sequences cloned in yeast artificial chromosome vectors. Nucleic Acids Res. 1990 Mar 25;18(6):1421–1428. doi: 10.1093/nar/18.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y., Weber J. L., Killary A. M., Ledbetter D. H., Smith J. R., Pereira-Smith O. M. Genetic analysis of indefinite division in human cells: evidence for a cell senescence-related gene(s) on human chromosome 4. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5635–5639. doi: 10.1073/pnas.88.13.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolenghi S., Giglioni B. The deletion in a type of delta 0-beta 0-thalassaemia begins in an inverted AluI repeat. Nature. 1982 Dec 23;300(5894):770–771. doi: 10.1038/300770a0. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Hill C. W. Recombination between repeated genes in microorganisms. Annu Rev Genet. 1988;22:147–168. doi: 10.1146/annurev.ge.22.120188.001051. [DOI] [PubMed] [Google Scholar]
- Rouyer F., Simmler M. C., Page D. C., Weissenbach J. A sex chromosome rearrangement in a human XX male caused by Alu-Alu recombination. Cell. 1987 Nov 6;51(3):417–425. doi: 10.1016/0092-8674(87)90637-4. [DOI] [PubMed] [Google Scholar]
- Runge K. W., Wellinger R. J., Zakian V. A. Effects of excess centromeres and excess telomeres on chromosome loss rates. Mol Cell Biol. 1991 Jun;11(6):2919–2928. doi: 10.1128/mcb.11.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J., Szostak J. W., Blackburn E. H. DNA sequences of telomeres maintained in yeast. Nature. 1984 Jul 12;310(5973):154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Blackburn E. H. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982 May;29(1):245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989 Aug 25;17(16):6463–6471. doi: 10.1093/nar/17.16.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984 May 25;12(10):4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Copy number control by a yeast centromere. Gene. 1983 Aug;23(2):221–232. doi: 10.1016/0378-1119(83)90054-9. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A. Structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]