Abstract
Cattle comprise a main reservoir of Shiga toxin-producing Escherichia coli O157:H7 (STEC). The significant differences in host prevalence, transmissibility, and virulence phenotypes among strains from bovine and human sources are of major interest to the public health community and livestock industry. Genomic analysis revealed divergence into three lineages: lineage I and lineage I/II strains are commonly associated with human disease, while lineage II strains are overrepresented in the asymptomatic bovine host reservoir. Growing evidence suggests that genotypic differences between these lineages, such as polymorphisms in Shiga toxin subtypes and synergistically acting virulence factors, are correlated with phenotypic differences in virulence, host ecology, and epidemiology. To assess the genomic plasticity on a genome-wide scale, we have sequenced the whole genome of strain EC869, a bovine-associated E. coli O157:H7 isolate. Comparative phylogenomic analysis of this key isolate enabled us to place accurately bovine lineage II strains within the genetically homogenous E. coli O157:H7 clade. Identification of polymorphic loci that are anchored both in the chromosomal backbone and horizontally acquired regions allowed us to associate bovine genotypes with altered virulence phenotypes and host prevalence. This study catalogued numerous novel lineage II-specific genome signatures, some of which appear to be associated intimately with the altered pathogenic potential and niche adaptation within the bovine rumen. The presented extended list of polymorphic markers is valuable in the development of a robust typing system critical for forensic, diagnostic, and epidemiological studies of this emerging human pathogen.
INTRODUCTION
Shiga toxin-producing, non-sorbitol-fermenting, and β-glucuronidase-negative Escherichia coli (STEC) O157:H7 has evolved from an O55:H7-like progenitor (24, 50, 65) into an emerging human pathogen, with cattle as the main asymptomatic reservoir (12, 54). E. coli O157:H7 is transmitted from cattle to human by means of contaminated food products, such as undercooked meat, unpasteurized milk, fruit and vegetables, or tainted water. As seasonal changes can influence the prevalence and load of E. coli O157:H7 in cattle and super shedders exist within the bovine population (30, 33, 39, 49, 67), physical contact of humans with cattle and their environment introduces added risk of E. coli infection (29). Human infection manifests in various ways, ranging from mild to more severe bloody diarrhea. In some cases, infection can lead to renal dysfunction, i.e., hemolytic uremic syndrome (HUS), and central nervous system (CNS) failure (10, 17, 73). Epidemiological data have demonstrated a high prevalence of E. coli O157:H7 in cattle and their environment but a comparatively low incidence of human infection. This supports the notion that a subset of STEC O157:H7 strains harbored in cattle causes the majority of human disease (18). Genetic heterogeneity among STEC O157:H7 strains has been established using a broad panel of typing methodologies, such as multilocus sequence tagging (2), octamer- and PCR-based genome scanning (41, 66), phage typing (4, 72), multiple-locus variable-number tandem repeat analysis (40, 64), microarrays (37), nucleotide polymorphism assays (56, 87) (M. Eppinger, M. K. Mammel, J. E. LeClerc, T. A. Cebula, and J. Ravel, unpublished data), pulsed-field gel electrophoresis (PFGE) (28), subtractive hybridization (79, 80), and optical mapping (43, 44). High-resolution genotyping studies on E. coli O157:H7 strains utilizing octamer-based genome scanning (OBGS) first demonstrated that the E. coli O157:H7 clonal complex has diverged into two lineages, designated lineages I and II, that were disproportionately represented among human and cattle isolates, respectively (41). Further analyses led to a refined classification system, termed the lineage-specific polymorphism assay (LSPA), that partitioned E. coli O157:H7 strains ultimately into three groupings—lineages I, I/II, and II—based on sequence length polymorphisms at six distinct loci within the O157:H7 genome (84). Whereas E. coli O157:H7 strains of lineages I and I/II were isolated from human clinical cases, lineage II strains were more commonly derived from the bovine reservoir (84). Lineage II isolates indeed are thought to be less pathogenic and possibly impaired in their transmissibility to humans (50). For example, branch-specific genotypes differ substantially in the frequencies with which they are associated with isolates from clinical or bovine settings (8, 88), and testing of STEC infection in the gnotobiotic pig model suggests that the virulence of cattle-derived strains may differ from that of strains isolated from humans (7). The mobilome is a major factor in the genome evolution and differentiation of these two lineages (11, 48, 88). Shiga toxin (stx)-converting phages of toxin subtypes stx2 and stx2c are more frequently carried in clinical STEC isolates, while stx1 is more frequent in O157 and non-O157 STEC isolates of bovine origin (13, 32) (Eppinger, Mammel, LeClerc, Cebula, and Ravel, unpublished). Lineages I, I/II, and II are further distinguished by polymorphisms in toxin production and expression of other synergistic key virulence factors (3, 10). STEC strains produce characteristic attaching and effacing (A/E) lesions mediated by the pathogenicity locus of enterocyte effacement (LEE) (57). The LEE pathogenicity island codes the adhesion and effacement molecule intimin (eae) and its translocated intimin receptor (tir) (ECH7EC869_1708), a type III secretion system, and the LEE effector molecules (espABD) and the EHEC hemolysin (ehxA) (74). The attachment of the bacterium is mediated through intimin and Tir, which is secreted by the type III system through an EspA filament and EspB-EspD pore (58). This mechanism results in host cell actin rearrangement and the formation of A/E lesions. Intimin subtypes in E. coli O157:H7 may alter adhesion capabilities and virulence in bovine and human settings (1, 19, 55, 70). Here, we report the genome and phylogenetic analyses of the bovine strain EC869, a strain originally isolated from a ground-beef sample. The sequencing and phylogenomic analyses of this strain were key in elucidating the genomic plasticity among the E. coli O157:H7 lineages on a genome-wide scale. The approach identified multiple branch-specific polymorphisms that appear to be associated intimately with altered virulence and physiological capabilities of isolates resident in cattle populations.
MATERIALS AND METHODS
Biological material.
The sequenced E. coli O157:H7 strain EC869 was isolated in 2002 by the U. S. Department of Agriculture (USDA) in Wyndmoor, PA, from ground beef. This strain is available from the E. coli strain collection of the Food and Drug Administration (FDA), Center for Food Safety and Applied Nutrition, Office of Applied Research and Safety Assessment, Division of Molecular Biology. Genome features and other associated metadata of the compared E. coli O157:H7 strains are listed in Table S1A in the supplemental material.
Sanger DNA sequencing and genome annotation.
Genomic DNA of strain EC869 was subjected to random shotgun sequencing and closure strategies using a combination of Sanger and 454 sequencing as previously described (23). Two random insert pHOS2 libraries with insert sizes of 3 to 5 kb and 10 to 12 kb and a fosmid library of 35 to 40 kb were constructed. Draft genome sequences were assembled using the Celera assembler (36). The draft contigs were manually annotated using the MANATEE system (http://manatee.sourceforge.net/).
Optical mapping.
Optical maps for strain EC869 were generated, which facilitated assembly and allowed for a detailed study of the prophage dynamics and their respective genome localization in strain EC869. Optical maps were prepared by OpGen, Madison, WI. Following gentle lysis and dilution, high-molecular-mass genomic DNA molecules were spread and immobilized onto derivatized glass slides and digested with BamHI. The DNA digests were stained with YOYO-1 fluorescent dye and photographed using a fluorescent microscope interfaced with a digital camera. Automated image analysis software located and sized fragments and assembled multiple scans into whole-chromosome optical maps.
Biolog phenotype microarray.
The Biolog system was used for phenotype microarray (PM) studies of strains EC869 and EC508. Strains were plated on Biolog universal growth medium and incubated overnight at 37°C. Cells were swabbed from the plates after overnight growth and suspended in the appropriate medium containing dye mix C; 100 μl of a 1:200 dilution of an 85% transmittance suspension of cells was added to each well of the PM plates. Plates 1 to 8, which test for catabolic pathways for carbon, nitrogen, phosphorus, sulfur, and biosynthetic pathways, and plates 9 and 10, which test for osmotic/ion and pH effects, were utilized in this study. IF-0 GN base was used for PM plates 1 and 2. IF-0 GN base plus 20 mM sodium succinate, pH 7.1, and 2 μM ferric citrate was used for plates 3 to 8. IF-10 base was used for plates 9 and 10. Plates were incubated in the OmniLog for 48 h, with readings taken every 15 min. Data analysis was performed using kinetic and parametric software (Biolog). Phenotypes were determined based on the area difference under the kinetic curve of dye formation between the mutant and wild type. Data points for the entire 48 h were used for PM1 through PM8, and area differences were mean centered by plate.
Bovine-specific SNPs and phylogenetic analysis.
Bovine lineage II-specific single nucleotide polymorphisms (SNPs) that are shared among strains EC869, FRIK2000, and FRIK966 but not present in tested human clinical lineage I and I/II strains were extracted. Concatenated SNP data were analyzed by the HKY93 method (34) with 500 bootstrap replicates, and the results were used to generate a phylogenetic tree according to the PhyLM algorithms (31) using the Geneious software package and SplitsTree4 (35).
Nucleotide sequence accession number.
The EC869 genome has been deposited in GenBank under accession no. ABHU00000000 (54,466 reads, 147 contigs, 8.66×). The respective genome assemblies have been deposited in the NCBI Assembly Archive, and the electropherogram data of the sequencing traces are available from the NCBI Trace Archive.
RESULTS AND DISCUSSION
LSPA typing.
In silico analyses of lineage-specific polymorphism assay (LSPA) sites (45, 84) revealed that EC869, FRIK966, and FRIK2000 are lineage II-derived (LSPA222222) strains (see Table S1B in the supplemental material). Recall that the LSPA typing schema originally described is based on PCR amplicon length polymorphisms. Sequence comparisons thus permitted closer inspection of these sites and revealed previously undetected polymorphisms in the LSPA-6 and LSPA-1 loci of E. coli O157:H7 strains (see Table S1B). Within the LSPA-6 region, a 9-bp sequence (AGTGTATGA) is tandemly repeated (TR) once in lineage I and lineage I/II strains (TR2), three times in lineage II strains FRIK2000 and FRIK966 (LSPA-2; TR4), and twice in strain EC869 (TR3). TR3, found in EC869, is thus termed LSPA-6-2b. Strains are further distinguished by a length polymorphism within the LSPA-1 locus. Whereas lineage I strains harbor a 9-bp deletion (CTGAGGTCG) in this region, lineage I/II and II strains do not. Closer inspection of the sequences shows that a base substitution (G22A) at the LSPA-1 locus (EC4115 position 653,503) further distinguishes these lineages. Whereas both lineage I and lineage II strains harbor the G allele, both the G and A alleles are distributed among lineage I/II strains (see Table S2 in the supplemental material).
Phylogenomic analysis of the bovine isolates. (i) Comparative analysis of protein sequences.
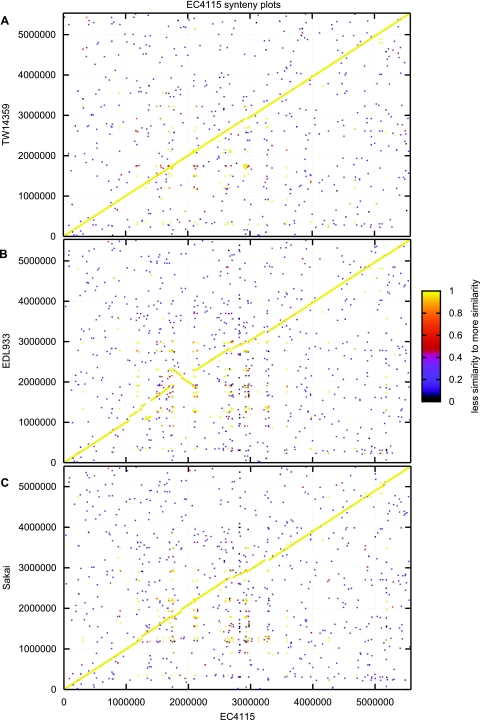
The close genetic relatedness within the E. coli O157:H7 clade is reflected in a high degree of overall protein sequence conservation, as evidenced by a BLAST score ratio (BSR) analysis that is evident for isolates derived from both humans and cattle (see Table S3 in the supplemental material) (71). Bioinformatic comparisons classified 748 divergent protein sequences (BSR ≤ 0.8) and 4,567 conserved protein sequences (BSR > 0.8), which is about 86% of the coding capacity of the reference strain, EC4115. Such a high degree of protein sequence conservation is comparable to data obtained for genetically highly homogenous bacterial species, like Yersinia pestis (23). For E. coli O157:H7, however, this data set is not sufficient to deduce a monomorphic population structure. Due to the genome dynamics driven by the highly homologous prophages, such comparative analysis must consider the overall genomic architecture and phage prevalence, which disrupts the E. coli O157:H7 genome synteny (Fig. 1).
Fig. 1.
Genome organization in completed E. coli O157:H7 genomes. Genome synteny in strains EC4115, TW14359 (A), EDL933 (B), and Sakai (C) is disrupted by isolate-specific lateral acquired genomic regions introduced by prophages and mobile genetic elements, such as transposases and insertion sequence elements. Each protein of the reference genome EC4115 is plotted on the x axis and was queried for its presence in the y axis query genome using BLASTP. For a match, the N-terminal coordinates of both proteins were plotted as x and y coordinates. The color represents the level of similarity of the match expressed by the BLAST score ratio (71).
(ii) SNP discovery and typing.
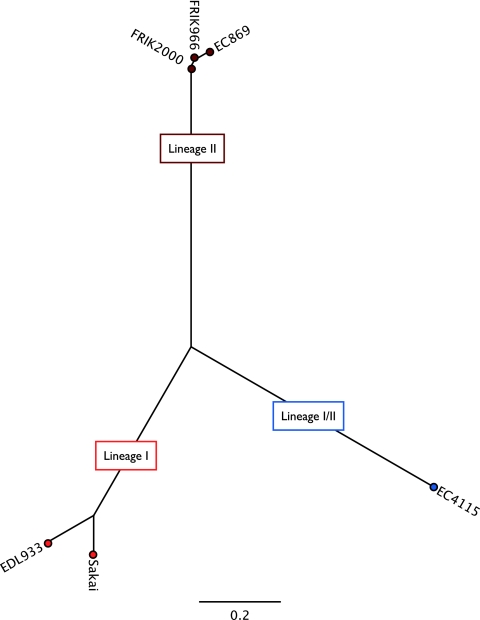
Detection of genetic diversity between STEC strains is an important component of outbreak investigations. Nucleotide polymorphisms are key in determining relatedness of genetically homogenous pathogens and enable one to discriminate between and among environmental and outbreak strains (23) (Eppinger, Mammel, LeClerc, Cebula, and Ravel, unpublished). To achieve the necessary high phylogenetic resolution and discover branch-specific genome signatures, we applied a multitier approach of SNP-derived genotyping coupled with an analysis of phage content and polymorphisms. SNPs have been identified previously in different subsets of STEC genomes (14, 50, 56, 87); however, the identified SNPs are biased toward strains originating from human outbreaks and thus may be limited in their ability to distinguish genetic STEC subtypes present in cattle (56). We applied a phylogenomic SNP discovery and SNP validation pipeline that was specifically developed for the high-resolution typing of E. coli O157:H7 strains. Genotyping identified 298 SNPs that separate strain EC869 from the remainder of the tested isolates. All 298 SNPs were confirmed in two other completed lineage II genomes, FRIK2000 and FRIK966 (20) (see Table S2 in the supplemental material). Thus, detected SNPs are considered to represent lineage II branch-specific SNPs. The SNP-derived high-resolution phylogenetic analysis places the bovine isolates on a distinct branch (Fig. 2) and identified the lineage I/II strains EC508 as a close phylogenetic relative.
Fig. 2.
Genetic relatedness of human and bovine lineages. To deduce the genetic relatedness of the lineage II and lineage I and I/II outbreak isolates, the phylogeny was reconstructed utilizing an SNP library specifically developed for the high-resolution typing of E. coli O157:H7 (Eppinger, Mammel, LeClerc, Cebula, and Ravel, unpublished). SNP discovery detected 298 lineage II-specific SNPs unique to the analyzed bovine strains. The colors blue, brown, and red represent the lineage associations of the analyzed strains.
Association of polymorphisms with altered physiology and virulence.
Such detected fine polymorphisms are key in understanding the genetic makeup and altered physiology of the bovine lineage II isolates. The panel of SNPs is comprised of 81 synonymous SNPs (sSNPs), 153 nonsynonymous SNPs (nsSNPs), and 64 intergenic SNPs (see Table S2 in the supplemental material). A 16-bp deletion within the fim-switching region of fimA, shown to underlie the lack of expression of type I fimbriae in E. coli O157:H7 strains, was found in both human- and bovine-derived strains analyzed in the present studies (51, 75, 78). These data are consistent with the hypothesis that the fim switch mutation occurred early on in E. coli O157:H7 evolution, before the divergence into three sublineages (78). A C→A substitution in fimH results in the replacement of an asparagine residue by lysine in the mannose-binding pocket of the FimH protein (see Table S2); however, because of the fim mutation, type I fimbriae are not expressed. Although the N135K polymorphism has been previously noted (78), we show here that the C→A transversion in the fimH gene is distributed nonrandomly within the three lineages analyzed. That is, while the C allele is found both in lineage I/II and lineage II strains, the A allele is found in each of the four lineage I strains, but only one of 18 lineage I/II strains, examined in this study (see Table S2).
A nonrandom distribution of a T255A base substitution within the intimin receptor has been reported, with isolates containing the A allele, to be 34 times more likely to be of bovine rather than human origin, although the physiological relevance of this base substitution that leads to an Asp→Glu substitution in LII strains is not known (14, 18). Indeed, the three analyzed bovine strains harbor the A allele, while the human lineage I/II isolate, strain EC508, contains the T allele. LII isolates are also distinguished by an 18-bp sequence (CAAAAGGCGTTGGGGAGT), which is located within the N-terminal Tir receptor domain (11) (see Fig. S1 in the supplemental material). This sequence is perfectly duplicated to form an 18-bp tandem repeat in the three analyzed lineage II isolates. Though the underlying cause for this host restriction or its impact on pathogenicity is unknown, these novel alleles provide valuable genetic markers for the grouping of STEC isolates based on their likely human or bovine origin.
SNP discovery detected eight lineage II genes with branch-specific premature stop codons (see Table S2 in the supplemental material), and some of these are implicated in STEC pathogenicity and might play a role in the host adaption to the bovine environment. For example, one such stop occurs in luxR, a transcription regulator that governs expression of proteins of ETTSS2, a type III secretion system (see Fig. S2 in the supplemental material). The genome of E. coli O157:H7 harbors two type III secretion systems. In strain EC4115, the loci are ETTSS1, located within the LEE island (4691936 bp→4725720 bp), and ETTSS2, localized between ECH74115_4114 and ECH74115_4154. In contrast to the ETTSS1 system, the ETTSS2 system is not directly involved in the injection of virulence factors. Recently, however, the expression of three genes within the ETTSS2 locus has been shown to negatively impact the expression of ETTSS1 genes (86). In the absence of luxR, it is expected that overexpression of ETTSS2 proteins would subvert injection of virulence factors by the ETTSS1 system (59, 86).
Another premature stop occurs with the ethanolamine utilization (eut) operon. As recent studies have shown that E. coli O157:H7 incubated in bovine rumen under aerobic conditions can utilize ethanolamine present in the rumen as an efficient nitrogen source for growth (9), our finding of a lineage II-specific premature stop in eutA, encoding reactivating factor, is especially noteworthy (see Table S2 in the supplemental material). Although the eutA gene product is essential for ethanolamine utilization under aerobic conditions, as it rescues ethanolamine ammonia lyase from suicide inactivation by toxic by-products of exogenously added B12, only eutB and eutC gene products are necessary under anaerobic conditions for the utilization of ethanolamine as a nitrogen source (76). Thus, the mutation that we localized in the bovine lineage does not compromise its ability to grow and compete within the animal reservoir. Rather, as ethanolamine catabolism has been implicated in bacterial pathogenicity, i.e., linked to intestinal host colonization, impaired gut functioning, and immune evasion in a diverse range of enteric pathogens (27), we theorize that the nonsense mutation attenuates virulence in eutA strains. That is, host-pathogen interactions result in the upregulation of eut genes, which is accompanied, at least in Salmonella enterica serovar Typhimurium (47), by the activation of global virulence regulators. If the E. coli O157:H7 eutA mutation behaves as it does in S. Typhimurium, strains harboring this mutation, like the bovine isolates described here, may be impaired in their abilities for human transmission and pathogenicity.
STEC isolates produce cellulose as a component of their extracellular matrix that is involved in virulence, colonization, and biofilm formation (90). Bacterial cellulose biosynthesis (bcs) is conferred by the constitutively transcribed bcsABZC operon (ECH7EC869_1529, ECH7EC869_1534). This locus was initially described in Acetobacter xylinum and shares high protein conservation and syntenic arrangement with corresponding loci of E. coli and S. enterica (83, 90). Within E. coli O157:H7, we found four distinct genotypes, a 10-bp deletion (GTTACAACAA), and three independent nsSNPs (see Table S2 in the supplemental material) within this operon (see Fig. S3 in the supplemental material). One of these SNPs, at position 1,674, leads to a truncated cellulose synthase C gene (ECH7EC869_1530, ECH7EC869_1531) (see Table S2). Previous research demonstrated that the four-gene synthase operon is essential for maximal cellulose synthesis in A. xylinum (83), and we therefore suspect reduced cellulose production in the genetic background of lineage II isolates. This novel polymorphism may underlie reported differences in cellulose production in STEC isolates (85). As cellulose is abundant in the bovine rumen, we speculate that the discovered lineage II cellulose phenotype is not disadvantageous within the bovine environment, but within the human milieu, bcs mutants may manifest with impaired transmissibility to humans and reduced pathogenicity (90).
The lsrACDBFGtam operon is an integral part of the AI-2 quorum-sensing (QS) system monitoring cell density in biofilms (5, 52). We detected a lineage II-specific 1,339-bp deletion that leads to a C-terminal-truncated version of the S-adenosyl-l-methionine-dependent methyltransferase (tam) (ECH7EC869_4857, 88 amino acids [aa]) compared to those of the other STEC isolates, such as EC4115 (ECH74115_2132, 252 aa) (see Fig. S4 in the supplemental material). The lineage II genomes lack the neighboring conserved hypothetical protein (ECH74115_2133) with no assigned function. Tam catalyzes the methylesterification of trans-aconitate from the citric acid cycle intermediate cis-aconitate. The physiological role of aconitate conversion in E. coli cells is not clear. However, it is known that further conversion of trans-aconitate to tricarballylic acid by the rumen microbiome can induce hypomagnesemia and grass tetany in cattle (77). Thus, a tam mutant of E. coli O157:H7 might be a welcomed resident within the cattle host.
Prophage content and dynamics.
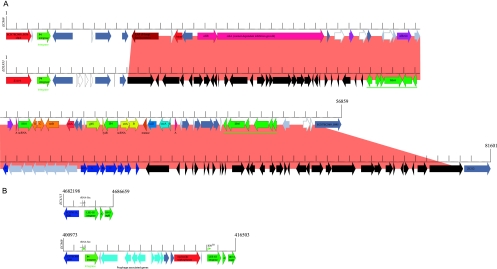
E. coli O157:H7 has become increasingly more resistant to streptomycin, sulfisoxazole, and tetracycline, likely resulting from the antibiotic treatment of livestock (10) with noted differences in resistance phenotypes between the two major lineages (89). This finding is supported by the discovery of a novel strain-specific phage that underlies the multidrug resistance (MDR) phenotype in strain EC869. The human strain EDL933 and bovine strain EC869 carry phylogenetically unrelated P4-type prophages that, in both cases, are located within the clpA locus (68) (Fig. 3A). The EC869 prophage is an entirely novel P4 prophage. This 56,859-bp phage introduces many loci that are potentially associated with the pathogenic potential and niche adaptation to the bovine rumen. The novel phage carries three predicted adhesins (ECH7EC869_5853, _5857, _5859). Phylogenetic analyses showed that one of the three predicted phage-borne adhesins (ECH7EC869_5857) is related to adhesins found in the bovine E. coli strain RW1374 (see Fig. S5A in the supplemental material), which suggests a role in bovine niche adaptation (38). Delineated from its domain of unknown function (DUF638), this adhesin may act as hemagglutinin. We identified a Tn10-like transposon as an integral part of this phage. This transposon introduces resistance loci for streptomycin (strAB), tetracycline (tetDBAR), sulfonamide (sul2), and cobalt-zinc-cadmium (czcAB) (53). Predictions made for the EC869 MDR genotype were validated and confirmed utilizing Biolog-derived phenotypic microarrays as well as antibiotic susceptibility assays (data not shown) compared to the P4 prophage-deficient strain EC508. The bovine tetDCBA and strB resistance loci are phage-borne, while analysis of the genome draft of EC4192 suggests a plasmid-borne origin of the tetRA and strB genes (82). The EC869 phage carries two heavy-metal resistance genes that mediate the efflux of cadmium, zinc, and cobalt (czcAB). This two-gene system is highly homologous and partly syntenic to the czcABCD resistance locus extensively studied in Alcaligenes eutrophus (63). The CzcA, CzcB, and CzcC transporters mediate Co2+, Zn2+, and Cd2+ efflux, while CzcC acts as a modifier protein required to change substrate specificity. To our knowledge, this is the first report of czc transporter loci in E. coli. The phylogenetic analysis of the cadmium/zinc antiporters is presented in Fig. S5B and C in the supplemental material.
Fig. 3.
Prophage polymorphisms. (A) Strains EC869 and EDL933 share parts of P4 prophages targeting the clpA locus, which in the case of EC869 introduces loci that are intimately associated with host prevalence and pathogenic potential. The phage architecture in EC869 is distinguished by a 56,859-kb insertion, featuring a Tn10-type transposon. This lateral acquired region carries 57 genes (ECH7EC869_5838 to _5894), mediating contact-dependent inhibition of growth (cdiAB), adhesion, and resistance to streptomycin (strAB), tetracyclines (tetDBAR), sulfonamides (sul) (dark blue), and cobalt-zinc-cadmium (czcAB) (light blue). To our knowledge this is the first report of czc resistance in E. coli. Corresponding loci are colored accordingly, with hypothetical proteins (light gray), conserved hypothetical proteins (dark gray), pseudogenes (white), mobile elements (dark green), and adhesins (purple). In the phage of strain EDL933, dark blue marks the tetracycline resistance operon. (B) Strain EC869 shows polymorphisms within the borders of the LEE pathogenicity island caused by the integration of an 11,098-bp P4 prophage remnant targeting the selC locus (ECH74115_5031). This novel phage remnant was also identified in the draft contigs of the FRIK2000 and FRIK966 genomes, supporting their SNP-derived phylogenetic placement. The integration side is a likely target for lateral acquisition, as evidenced by its deviating GC content linked to the AT-rich LEE-borne phage integrase and neighboring IS911 element. Integration results in a 30-bp imperfect direct repeat (IDR; arrows) and introduces 12 genes (ECH7EC869_1671 to _1682). Corresponding loci are colored accordingly, with hypothetical proteins (light gray) and conserved hypothetical proteins (dark gray).
Contact-dependent inhibition of growth (cdi), initially described in E. coli strain EC93, is advantageous to the microbe in competing for certain ecological niches (6). We discovered a P4 prophage-borne cdiAB locus that displays high homology to the corresponding locus in EC93, with the notable absence of cdiI (Fig. 3A; see also Fig. S5D and E in the supplemental material). We speculate that this locus enables strain EC869 to compete successfully with phylogenetically diverse and abundant microorganisms that are known to colonize the bovine rumen.
Polymorphisms in the LEE island.
Strain EC869 is distinguished by LEE polymorphisms, which are caused by the integration of an 11,098-bp P4 prophage remnant within the borders of the pathogenicity island. The integration site features a deviating G+C content neighboring a transposable element and is thus a likely target for lateral acquisition. Its insertion at the tRNA-Sec locus (ECH74115_5031) resulted in a 30-bp imperfect direct repeat (IDR) (Fig. 3B). We note that both the EC869-carried P4 prophage and smaller P4 prophage remnant are present in the draft genomes of lineage II strains FRIK2000 and FRIK966, which supports their SNP-derived phylogenetic placement (20).
Mutator phage content and localization.
Both bovine genomes FRIK2000 and FRIK966 feature a mutator (MU) phage insertion that is not present in strain EC869. The mutator phages in strain Sakai and the 2006 Taco John outbreak isolates (EC4501, TW14588) are inserted within the ECs4942 and fbpC loci, respectively (68) (Eppinger, Mammel, LeClerc, Cebula, and Ravel, unpublished data), while this phage has yet another integration in FRIK2000, disrupting a predicted DEAD/DEAH box helicase domain gene (Escherichia coli O157_19017; introduced at the N-terminal end of a pseudogene, at position 5174) (see Fig. S6 in the supplemental material). We noted that all bovine isolates show a T deletion at position 1831, generating a premature stop (TAA) that results in a truncated variant of 389 aa compared to the full-length protein of 2014 aa (ECs5259). The Mu prophage is inserted at position 5174 of the helicase gene. The sequence data do not allow us to localize the Mu-like insertion for FRIK966 because of contig size and fragmentation, though from our sequence analyses, it is at a site distinct from those found for FRIK2000, Sakai, and Taco John. We detected length polymorphism in the mutator gene Mu-gp35 (ECH7EC4501_3951) due to variable numbers of a characteristic 6-bp perfect repeat (AGCCGA)5–15 (5–15 represents the range of repeat [AGCCGA] copy numbers in the analyzed Mu-gp35 mutator gene.). The analyzed mutator phage proteins (AE)5–15 within E. coli O157:H7 range from 128 to 140 aa, while we note that this prophage is otherwise highly conserved and syntenically organized (60). The affected protein is annotated as conserved hypothetical with no assigned physiological function. However, this novel identified MU prophage polymorphism may serve as an additional genomic marker.
Polymorphisms in the Enterobacteria P2 prophage.
Comprehensive analyses of an Enterobacteria P2-like prophage inserted at the yegQ locus within E. coli O157:H7 revealed another polymorphic region unique to the analyzed lineage II isolates (16, 44). It was noted that the deletion of 4,210 bp encompasses four genes that code hypothetical proteins with no assigned physiological function. This region may serve as an additional marker for lineage II (ECH74115_3055 to _3058), as this region is absent in the lineage I/II strain EC508 or the lineage I/II strains from the 2006 spinach (SO) and Taco Bell (TB) outbreaks.
Association of Shiga toxin subtypes and bacterial habitat.
Carriage of both stx2 and stx2c rather than stx1 and altered expression ratios of stx2/stx1 have been implicated in the greater virulence of human outbreak isolates and reduced virulence and impaired transmissibility in the lineage II clade (11, 22). Moreover, stx2c was identified as a key factor in HUS manifestation (56) (Eppinger, Mammel, LeClerc, Cebula, and Ravel, unpublished). An analysis of the prophage insertion sites demonstrates differences of the bovine STEC genome architecture (11). However, our findings do not support epidemiological data that show a nonrandom distribution of stx subtypes among bovine and human isolates, with stx2 typically biased toward human isolates (15, 18, 25, 26). The analyzed bovine lineage isolates contained stx1 and stx2c, much like a number of clinical isolates do, but lack the stx2 subtype. Draft genomes of the bovine isolates allowed us to locate the stx1 prophage at the yehV locus, as in clinical isolates, such as strain EDL933 (ECH7EC869_329, Escherichia coli O157_010100002259, Escherichia coli O157EcO_010100024977). Both the wrbA and argW loci that were previously identified as potential sites for stx2 prophage integrations are intact in the analyzed bovine isolates (20, 44) (Eppinger, Mammel, LeClerc, Cebula, and Ravel, unpublished). Unlike the stx1- and stx2-converting phages that are found integrated at more than one chromosomal loci in the E. coli O157:H7 lineage, the stx2c type preferentially targets the sbcB locus (ECH7EC869_3164). The overall stx2c prophage architecture of the bovine isolates is organized highly syntenic to other stx2c prophages compared to the SO strain EC4115 or P1717 (NC_011357; unpublished data). Phage insertion resulted in 13-bp (TTTCACGATTACG) perfect direct repeats that could be detected in all analyzed bovine isolates.
Antiterminator Q polymorphisms may relate to toxicity.
Toxin production is regulated by the induction of the stx-converting phages resulting in multiplication of toxin gene copies and the transcription activator proteins Q and Q′ located upstream of the Shiga toxins (61). These antiterminator proteins control gene expression by recognizing control signals near the promoter and prevent transcriptional termination. In the stx-converting lambdoid prophages, stx expression has been genetically linked to the late antiterminator antQ and the prV promoter and associated qut site, with the trV terminator located directly upstream of the stx coding sequences (42, 62). Two major subtypes of this regulator, antQ and antQ′, have been previously identified in stx1 and stx2(c) converting phages (46) (Eppinger, Mammel, LeClerc, Cebula, and Ravel, unpublished). We tried to locate the stx subtypes and investigate their association colocalization to stx-inducing antiterminator antQ among the bovine draft genomes (EC869, FRIK2000, FRIK966) and completed clinical genomes (EC4115, EDL933, and Sakai). Our comprehensive analysis found polymorphisms in the stx1 and stx2c antiterminator genes, in their respective prophage associations, and further in phage-carried insertion element insertions, all of which might be intimately associated with altered Shiga toxin production levels (21).
Stx-converting prophage polymorphism in clinical and bovine isolates.
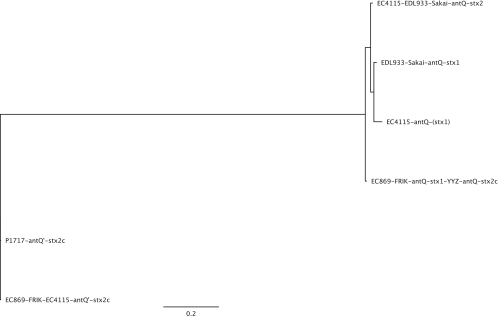
The stx2c-antQ′ terminator gene (ECH74115_2910, ECH7EC869_2226, Escherichia coli O157_010100022153, Escherichia coli O157EcO_010100023776) and the promoter terminator region between stx2c and antQ are genetically identical among the studied bovine lineage I and clinical lineage I/II isolates (Fig. 4). We further noted that the stx2c-converting prophage YYZ-2008 features antiterminator Q in the context of the stx2c prophage and not the typical stx2c-antQ′ combination (Fig. 4). Thus, these terminators are in general useful but not sufficient molecular markers to distinguish among Stx2-, Stx1-, and Stx2c-converting phages in the E. coli O157:H7 lineage. This finding is indicative of a potential antiterminator shuffling of this key regulator within the stx-converting prophage pool. Our comprehensive analysis of the toxin-producing prophages identified the transposable insertion sequence elements IS629 as a driver of stx1 and stx2c prophage microevolution in lineage II. The bovine and clinical isolates are distinguished by stx prophage-carried insertions of the IS629 element. The IS629 (ECH74115_3231, ECH74115_3232) insertion within the potentially stx1-converting phage at the yehV locus is absent in the analyzed bovine lineage II isolates (EC869, FRIK2000, FRIK966) and the closely related strain EC508 but present in all other analyzed human lineage I isolates, such as the 2006 SO isolates and the phylogenetically more distant human outbreak STEC isolates. Vice versa, the sbcB-occupying bovine stx2c phage variant carries an IS629 insertion element in strain EC869 (ECH7EC869_2207, ECH7EC869_2208), while this insertion is absent in lineage I isolates, such as in the SO- and TB-carried stx2c prophages. This insertion is present in the 62,147-bp Stx2c-converting phage PP1717 (NC_011357, unpublished; Stx2-1717_gp55/56) but is absent in the 54,896-bp phage YYZ2008 (NC_011356, unpublished). In strains FRIK2000 and FRIK966, the IS629 insertions are not traceable due to contig sizes and fragmentation. It was discovered that both stx prophage insertion element polymorphisms may serve as additional genomic markers for lineage II isolates. We note that in both cases the IS629 insertion disrupts the prophage terminase and replication machinery, creating fragmented and truncated pseudogenes. Affected are the large prophage terminase subunit (stx2c, ECH7EC869_2206, 338 aa; ECH74115_2893, 553 aa) and replication gene repP (stx1, ECH7EC869_3230/3233), which may result in prophage immobilization. The detected polymorphisms might relate to altered Stx production levels and asymptomatic manifestation (21). However, the underlying molecular mechanisms remain unclear from the current research and literature. As the alleged regulatory genes and sequences show no polymorphisms, we hypothesize that alterations in bacteriophage architectures and gene content may have altered the Stx2 toxin expression system in the bovine strains.
Fig. 4.
Phylogeny of the Shiga toxin antiterminator Q regulators in E. coli O157:H7. The prototypical SO strain EC4115 carries three distinct transcription antiterminators regulating toxin production in the stx2-, stx2c-, and potentially stx1-converting prophages. The antiterminator antQ colocalizing with stx1 (stx1-antQ) and antiterminator stx2-antQ of strain EC4115 (ECH74115_3220, ECH74115_3538, 144 aa) share 95.4% nucleotide identity, featuring 23 SNPs, and 97.2% protein identity, while stx1-antQ and stx2-antQ show no identity with stx2c-antQ′ (EC74115_2910, 163 aa) at the nucleotide sequence level and only weak protein identities of 22.8% and 23.4%, respectively (81).
Understanding the genomic plasticity among the bovine and human lineages provides insights into differences in the pathogenic potential, physiology, and ecology. Here, we present multitier high-resolution typing approaches based on comparative genomic data that provide the basis for population-based epidemiological studies and surveillance of this genetically homogenous pathogen. Identifying genomic polymorphisms in virulence content and regulatory networks is key in understanding why cattle prevalence and incidence of human illness are of a nonlinear relationship, and likely only a subset of the STEC isolates residing in cattle may cause the majority of human disease (18). Key in studying the polymorphisms of these two STEC lineages is the association of lineage-specific genotypes with disease, strain prevalence, and source. The identified signatures primarily reside in the conserved chromosomal STEC backbone, biased by the applied SNP methodology, but also within mobile elements. Identified signatures can help to detect strain profiles that enable transmissibility from the bovine reservoir and human infectivity and monitor their prevalence in the bovine reservoir (49). The environmental conditions in bovine and human settings are also of major importance, such as the host-specific expression levels of the stx target receptors in cattle and human (69). This study could identify two parameters that might be intimately associated with Shiga toxin production in the E. coli O157:H7 lineage: first the prevalence, exchange, and polymorphism in the antiterminator Q and Q′ phage regulators and second the potential position effects due to insertion of the Stx2-converting phage at the argW and wrbA loci in clinical isolates. Next-generation sequencing will become increasingly important and enable us to include and compare in silico results with sequence data gathered from ongoing outbreaks. However, association studies in broader panels of isolates from cattle and other animal host species and environmental isolates are necessary to get insights into the genetically distinct STEC genotypes associated with differences in disease manifestation and host reservoir. The set of novel polymorphisms presented in this study complements current techniques used to classify strains and provides a basis for the phylogenomic analysis of this emerging pathogen.
Supplementary Material
ACKNOWLEDGMENT
This work was supported with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under NIAID contract N01 AI-30071.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 18 March 2011.
REFERENCES
- 1. Abu-Ali G. S., et al. 2010. Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One 5:e10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Afset J. E., et al. 2008. Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis. J. Clin. Microbiol. 46:2280–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmad A., Zurek L. 2006. Evaluation of the anti-terminator Q933 gene as a marker for Escherichia coli O157:H7 with high Shiga toxin production. Curr. Microbiol. 53:324–328 [DOI] [PubMed] [Google Scholar]
- 4. Ahmed R., Bopp C., Borczyk A., Kasatiya S. 1987. Phage-typing scheme for Escherichia coli O157:H7. J. Infect. Dis. 155:806–809 [DOI] [PubMed] [Google Scholar]
- 5. Anand S. K., Griffiths M. W. 2003. Quorum sensing and expression of virulence in Escherichia coli O157:H7. Int. J. Food Microbiol. 85:1–9 [DOI] [PubMed] [Google Scholar]
- 6. Aoki S. K., et al. 2005. Contact-dependent inhibition of growth in Escherichia coli. Science 309:1245–1248 [DOI] [PubMed] [Google Scholar]
- 7. Baker D. R., Moxley R. A., Francis D. H. 1997. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv. Exp. Med. Biol. 412:53–58 [DOI] [PubMed] [Google Scholar]
- 8. Baker D. R., et al. 2007. Differences in virulence among Escherichia coli O157:H7 strains isolated from humans during disease outbreaks and from healthy cattle. Appl. Environ. Microbiol. 73:7338–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bertin Y., et al. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13:365–377 [DOI] [PubMed] [Google Scholar]
- 10. Besser R. E., Griffin P. M., Slutsker L. 1999. Escherichia coli O157:H7 gastroenteritis and the hemolytic uremic syndrome: an emerging infectious disease. Annu. Rev. Med. 50:355–367 [DOI] [PubMed] [Google Scholar]
- 11. Besser T. E., et al. 2007. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl. Environ. Microbiol. 73:671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beutin L. 2006. Emerging enterohaemorrhagic Escherichia coli, causes and effects of the rise of a human pathogen. J. Vet. Med. B Infect. Dis. Vet. Public Health 53:299–305 [DOI] [PubMed] [Google Scholar]
- 13. Boerlin P., et al. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bono J. L., et al. 2007. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect. Dis. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cherla R. P., Lee S. Y., Tesh V. L. 2003. Shiga toxins and apoptosis. FEMS Microbiol. Lett. 228:159–166 [DOI] [PubMed] [Google Scholar]
- 16. Christie G. E., Temple L. M., Bartlett B. A., Goodwin T. S. 2002. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J. Bacteriol. 184:6522–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cimolai N., Morrison B. J., Carter J. E. 1992. Risk factors for the central nervous system manifestations of gastroenteritis-associated hemolytic-uremic syndrome. Pediatrics 90:616–621 [PubMed] [Google Scholar]
- 18. Clawson M. L., et al. 2009. Phylogenetic classification of Escherichia coli O157:H7 strains of human and bovine origin using a novel set of nucleotide polymorphisms. Genome Biol. 10:R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cookson A. L., Woodward M. J. 2003. The role of intimin in the adherence of enterohaemorrhagic Escherichia coli (EHEC) O157:H7 to HEp-2 tissue culture cells and to bovine gut explant tissues. Int. J. Med. Microbiol. 292:547–553 [DOI] [PubMed] [Google Scholar]
- 20. Dowd S. E., et al. 2010. Microarray analysis and draft genomes of two Escherichia coli O157:H7 lineage II cattle isolates FRIK966 and FRIK2000 investigating lack of Shiga toxin expression. Foodborne Pathog. Dis. 7:763–773 [DOI] [PubMed] [Google Scholar]
- 21. Dowd S. E., Ishizaki H. 2006. Microarray based comparison of two Escherichia coli O157:H7 lineages. BMC Microbiol. 6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dowd S. E., Williams J. B. 2008. Comparison of Shiga-like toxin II expression between two genetically diverse lineages of Escherichia coli O157:H7. J. Food Prot. 71:1673–1678 [DOI] [PubMed] [Google Scholar]
- 23. Eppinger M., et al. 2010. Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. J. Bacteriol. 192:1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng P., Lampel K. A., Karch H., Whittam T. S. 1998. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750–1753 [DOI] [PubMed] [Google Scholar]
- 25. Friedrich A. W., et al. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 26. Friedrich A. W., et al. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garsin D. A. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8:290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerner-Smidt P., et al. 2005. Molecular surveillance of Shiga toxigenic Escherichia coli O157 by PulseNet U. S. A. J. Food Prot. 68:1926–1931 [DOI] [PubMed] [Google Scholar]
- 29. Goode B., et al. 2009. Outbreak of Escherichia coli O157:H7 infections after petting zoo visits, North Carolina State Fair, October-November 2004. Arch. Pediatr. Adolesc. Med. 163:42–48 [DOI] [PubMed] [Google Scholar]
- 30. Grauke L. J., et al. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guindon S., Lethiec F., Duroux P., Gascuel O. 2005. PHYML online—a Web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gyles C., et al. 1998. Association of enterohemorrhagic Escherichia coli hemolysin with serotypes of Shiga-like-toxin-producing Escherichia coli of human and bovine origins. Appl. Environ. Microbiol. 64:4134–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hancock D. D., et al. 1994. The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol. Infect. 113:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hasegawa M., Kishino H., Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174 [DOI] [PubMed] [Google Scholar]
- 35. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 36. Huson D. H., et al. 2001. Design of a compartmentalized shotgun assembler for the human genome. Bioinformatics 17(Suppl. 1):S132–S139 [DOI] [PubMed] [Google Scholar]
- 37. Jackson S. A., et al. 2007. Interrogating genomic diversity of E. coli O157:H7 using DNA tiling arrays. Forensic Sci. Int. 168:183–199 [DOI] [PubMed] [Google Scholar]
- 38. Jores J., Rumer L., Kiessling S., Kaper J. B., Wieler L. H. 2001. A novel locus of enterocyte effacement (LEE) pathogenicity island inserted at pheV in bovine Shiga toxin-producing Escherichia coli strain O103:H2. FEMS Microbiol. Lett. 204:75–79 [DOI] [PubMed] [Google Scholar]
- 39. Kenny B., et al. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520 [DOI] [PubMed] [Google Scholar]
- 40. Keys C., Kemper S., Keim P. 2005. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J. Appl. Microbiol. 98:928–940 [DOI] [PubMed] [Google Scholar]
- 41. Kim J., Nietfeldt J., Benson A. K. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. U. S. A. 96:13288–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koitabashi T., et al. 2006. Genetic characterization of Escherichia coli O157: H7/− strains carrying the stx2 gene but not producing Shiga toxin 2. Microbiol. Immunol. 50:135–148 [DOI] [PubMed] [Google Scholar]
- 43. Kotewicz M. L., Jackson S. A., LeClerc J. E., Cebula T. A. 2007. Optical maps distinguish individual strains of Escherichia coli O157:H7. Microbiology 153:1720–1733 [DOI] [PubMed] [Google Scholar]
- 44. Kotewicz M. L., Mammel M. K., LeClerc J. E., Cebula T. A. 2008. Optical mapping and 454 sequencing of Escherichia coli O157:H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology 154:3518–3528 [DOI] [PubMed] [Google Scholar]
- 45. Laing C., et al. 2008. Rapid determination of Escherichia coli O157:H7 lineage types and molecular subtypes by using comparative genomic fingerprinting. Appl. Environ. Microbiol. 74:6606–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laing C. R., et al. 2009. In silico genomic analyses reveal three distinct lineages of Escherichia coli O157:H7, one of which is associated with hyper-virulence. BMC Genomics 10:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lawhon S. D., et al. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633–1645 [DOI] [PubMed] [Google Scholar]
- 48. LeJeune J. T., Abedon S. T., Takemura K., Christie N. P., Sreevatsan S. 2004. Human Escherichia coli O157:H7 genetic marker in isolates of bovine origin. Emerg. Infect. Dis. 10:1482–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. LeJeune J. T., et al. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leopold S. R., et al. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc. Natl. Acad. Sci. U. S. A. 106:8713–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li B., Koch W. H., Cebula T. A. 1997. Detection and characterization of the fimA gene of Escherichia coli O157:H7. Mol. Cell Probes 11:397–406 [DOI] [PubMed] [Google Scholar]
- 52. Li J., et al. 2007. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 189:6011–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liebert C. A., Hall R. M., Summers A. O. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lowe R. M., et al. 2009. Escherichia coli O157:H7 strain origin, lineage, and Shiga toxin 2 expression affect colonization of cattle. Appl. Environ. Microbiol. 75:5074–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Malik A., et al. 2006. Serotypes and intimin types of intestinal and faecal strains of eae+ Escherichia coli from weaned pigs. Vet. Microbiol. 114:82–93 [DOI] [PubMed] [Google Scholar]
- 56. Manning S. D., et al. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U. S. A. 105:4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McDaniel T. K., Jarvis K. G., Donnenberg M. S., Kaper J. B. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McNally A., et al. 2001. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect. Immun. 69:5107–5114 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. Monday S. R., Minnich S. A., Feng P. C. 2004. A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H− strains. J. Bacteriol. 186:2319–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morgan G. J., Hatfull G. F., Casjens S., Hendrix R. W. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337–359 [DOI] [PubMed] [Google Scholar]
- 61. Muhldorfer I., et al. 1996. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect. Immun. 64:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neely M. N., Friedman D. I. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255–1267 [DOI] [PubMed] [Google Scholar]
- 63. Nies D. H. 1992. Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid 27:17–28 [DOI] [PubMed] [Google Scholar]
- 64. Noller A. C., McEllistrem M. C., Pacheco A. G., Boxrud D. J., Harrison L. H. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ogura Y., et al. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ohnishi M., et al. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. U. S. A. 99:17043–17048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Omisakin F., MacRae M., Ogden I. D., Strachan N. J. 2003. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 69:2444–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Perna N. T., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 69. Pruimboom-Brees I. M., et al. 2000. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc. Natl. Acad. Sci. U. S. A. 97:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ramachandran V., et al. 2003. Distribution of intimin subtypes among Escherichia coli isolates from ruminant and human sources. J. Clin. Microbiol. 41:5022–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rasko D. A., Myers G. S., Ravel J. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ratnam S., March S. B., Ahmed R., Bezanson G. S., Kasatiya S. 1988. Characterization of Escherichia coli serotype O157:H7. J. Clin. Microbiol. 26:2006–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Riley D. G., Gray J. T., Loneragan G. H., Barling K. S., Chase C. C., Jr 2003. Escherichia coli O157:H7 prevalence in fecal samples of cattle from a southeastern beef cow-calf herd. J. Food Prot. 66:1778–1782 [DOI] [PubMed] [Google Scholar]
- 74. Ritchie J. M., Thorpe C. M., Rogers A. B., Waldor M. K. 2003. Critical roles for stx2, eae, and tir in enterohemorrhagic Escherichia coli-induced diarrhea and intestinal inflammation in infant rabbits. Infect. Immun. 71:7129–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roe A. J., Currie C., Smith D. G., Gally D. L. 2001. Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: a comparison between serotypes O157 and O26. Microbiology 147:145–152 [DOI] [PubMed] [Google Scholar]
- 76. Roof D. M., Roth J. R. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Russell J. B. 1985. Enrichment and isolation of rumen bacteria that reduce trans-aconitic acid to tricarballylic acid. Appl. Environ. Microbiol. 49:120–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shaikh N., Holt N. J., Johnson J. R., Tarr P. I. 2007. Fim operon variation in the emergence of enterohemorrhagic Escherichia coli: an evolutionary and functional analysis. FEMS Microbiol. Lett. 273:58–63 [DOI] [PubMed] [Google Scholar]
- 79. Steele M., et al. 2009. Genomic regions conserved in lineage II Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 75:3271–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Steele M., et al. 2007. Identification of Escherichia coli O157:H7 genomic regions conserved in strains with a genotype associated with human infection. Appl. Environ. Microbiol. 73:22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Strauch E., et al. 2008. Bacteriophage 2851 is a prototype phage for dissemination of the Shiga toxin variant gene 2c in Escherichia coli O157:H7. Infect. Immun. 76:5466–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Williams L. E., Detter C., Barry K., Lapidus A., Summers A. O. 2006. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 72:4899–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wong H. C., et al. 1990. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. U. S. A. 87:8130–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yang Z., et al. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl. Environ. Microbiol. 70:6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yoo B. K., et al. 2010. Selection and characterization of cellulose-deficient derivates of Shiga toxin-producing Escherichia coli. J. Food Prot. 73:1038–1046 [DOI] [PubMed] [Google Scholar]
- 86. Zhang L., et al. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang W., et al. 2006. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 16:757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang Y., et al. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ziebell K., et al. 2008. Genotypic characterization and prevalence of virulence factors among Canadian Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 74:4314–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zogaj X., Nimtz M., Rohde M., Bokranz W., Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.