Abstract
The adenovirus type 5 (Ad5) E1B-55K and E4orf6 (E1B-55K/E4orf6) proteins are multifunctional regulators of Ad5 replication, participating in many processes required for virus growth. A complex containing the two proteins mediates the degradation of cellular proteins through assembly of an E3 ubiquitin ligase and induces shutoff of host cell protein synthesis through selective nucleocytoplasmic viral late mRNA export. Both proteins shuttle between the nuclear and cytoplasmic compartments via leucine-rich nuclear export signals (NES). However, the role of their NES-dependent export in viral replication has not been established. It was initially shown that mutations in the E4orf6 NES negatively affect viral late gene expression in transfection/infection complementation assays, suggesting that E1B-55K/E4orf6-dependent viral late mRNA export involves a CRM1 export pathway. However, a different conclusion was drawn from similar studies showing that E1B-55K/E4orf6 promote late gene expression without active CRM1 or functional NES. To evaluate the role of the E1B-55K/E4orf6 NES in viral replication in the context of Ad-infected cells and in the presence of functional CRM1, we generated virus mutants carrying amino acid exchanges in the NES of either or both proteins. Phenotypic analyses revealed that mutations in the NES of E1B-55K and/or E4orf6 had no or only moderate effects on viral DNA replication, viral late protein synthesis, or viral late mRNA export. Significantly, such mutations also did not interfere with the degradation of cellular substrates, indicating that the NES of E1B-55K or E4orf6 is dispensable both for late gene expression and for the activity associated with the E3 ubiquitin ligase.
INTRODUCTION
Two early gene products of human adenovirus type 5 (Ad5), E4orf6 and E1B-55K, are known to fulfill multiple functions during productive infection to ensure efficient production of viral progeny (reviewed in references 5, 19, and 26). A complex consisting of these two proteins is known to assemble a Cullin 5 (Cul5)-based E3 ubiquitin ligase to induce proteasomal degradation of cellular substrates, including the tumor suppressor p53; Mre11 and DNA ligase IV, both involved in DNA double-strand break repair; integrin α3 (3, 14, 16, 32, 53, 54, 62); and, most recently, Daxx, whose degradation seems to be independent of E4orf6 (60). It is well established that during the late phase of infection, both early viral proteins are also necessary for the preferential export of viral late mRNAs from the nuclear compartment to the cytoplasm (2, 11, 31, 43, 52). Nevertheless, it is still not understood how the E1B-55K/E4orf6 complex mediates the exclusive nuclear export of viral late mRNAs or, indeed, how export of the complex impacts the activity of the Cul5 ubiquitin ligase, which requires these two early proteins for assembly (8, 66). Extensive investigations have revealed functional nuclear export signals (NES) of the HIV-1 Rev type within both the E4orf6 and the E1B-55K protein (18, 20, 39, 65). This leucine-rich sequence mediates the nuclear export of proteins by the cellular exportin 1 protein, also known as CRM1 (40). The E1B-55K and E4orf6 proteins exhibit nucleocytoplasmic shuttling activity, and both proteins have been reported to exit the nucleus via CRM1-dependent and -independent mechanisms (13, 20, 37, 39, 55, 65). The cellular mechanism for the import of these proteins into the nucleus has not been determined, although it was recently found that nuclear import and localization of E1B-55K may be regulated by SUMOylation (23, 37). Both E1B-55K and E4orf6 have been shown to enter the nucleus in the absence of other viral proteins (18, 20, 39), but the nuclear localization of E1B-55K seems to depend on the E4orf6 protein (51), and it is proposed that the interaction of E4orf6 with E1B-55K leads to the localization of E1B-55K to viral replication centers, promoting selective viral late mRNA export via an unknown mechanism (28, 51).
Since both E1B-55K and E4orf6 can shuttle through a NES-dependent pathway, the role of CRM1-dependent export in viral replication has been examined using the drug leptomycin B (LMB), which irreversibly modifies CRM1 (13, 55), as well as a specific peptide inhibitor of CRM1 (27). The utilization of these compounds successfully blocked NES-dependent export of E4orf6 (55) or E1B-55K (13, 27). In every case, viral late mRNA export (27) or late protein synthesis were not inhibited, indicating that CRM1 does not participate in selective viral mRNA export (13, 27, 55). Nevertheless, the contribution(s) of the E1B-55K or E4orf6 NES, or indeed that of CRM1, to the viral replication cycle has not been characterized in detail. To address this issue, we constructed a set of adenoviral mutants harboring amino acid substitutions within the NES of E1B-55K, E4orf6, or both. We examined the different effects of the functional inactivation of the NES-mediated export of E4orf6 and/or E1B-55K during adenoviral infection in the presence of functional CRM1 on viral progeny production, viral late mRNA export, and E1B-55K/E4orf6-mediated proteasomal degradation of cellular target proteins. Our findings show that neither viral late mRNA export nor the E3 ubiquitin ligase-associated activity requires NES-dependent export of the E1B-55K and E4orf6 proteins.
MATERIALS AND METHODS
Cell culture.
HeLa cells (DSMZ ACC 57; Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany), A549 cells (DSMZ ACC 107), 911 cells (25), 293 cells (29), and W162 (65a) and HEK-293T cells (67) were grown as monolayer cultures in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U of penicillin, and 100 μg of streptomycin per ml under a 5% CO2 atmosphere at 37°C.
To generate polyclonal E4orf6 NES cells, A549 cells were first transduced with lentiviral particles containing the cDNA of the E4orf6 NES and were afterwards selected and maintained in a medium additionally containing neomycin (250 μg/ml).
Construction of lentiviral particles.
Lentiviral particles were generated based on the system previously described by Weber and coworkers (63). The coding sequence of E4orf6 was amplified by PCR introducing a 5′ BamHI site and a 3′ EcoRI site, purified by agarose gel electrophoresis, sequentially digested using the appropriate enzymes, and ligated into LeGO-iVLN2/iBLB2 (63, 64). Mutations within the E4orf6 coding sequence were introduced subsequently via QuikChange site-directed mutagenesis (Agilent) using oligonucleotides 390 and 391 (Table 1). To generate lentiviral particles, HEK-293T cells were transfected with the LeGO construct pCMV-VSV-G (6), pRSV-Rev, and pMDLg/pPRE (22) and were treated as described previously (60). After transfection, the medium was replaced by a standard culture medium additionally supplemented with 20 mM sterile filtered HEPES buffer. Two days after transfection, the supernatant containing the lentiviral particles was collected.
Table 1.
Primers used in this work
| Primer no. | Description | Sequence |
|---|---|---|
| 390 | E4orf6 NES forward | 5′-CGCGGGAGGAGGCTGTAGCCCTGAGGAAGTGTATGC-3′ |
| 391 | E4orf6 NES reverse | 5′-GCATACACTTCCTCAGGGCTACAGCCTCCTCCCGCG-3′ |
| 1110 | L4-100K forward | 5′-AAACTAATGATGGCCGCAGTG-3′ |
| 1111 | L4-100K reverse | 5′-CGTCTGCCAGGTGTAGCATAG-3′ |
Construction of Ad5 recombinants.
To generate Ad5 mutants carrying defined amino acid changes in the NES of E4orf6 and E4orf6 plus E1B-55K (see Fig. 1A), point mutations were first introduced into the E4orf6 gene at nucleotides (nt) 31926 (33805 in plasmid pE4-1155), 31925 (33804 in pE4-1155), 31920 (33799 in pE4-1155), and 31919 (33798 in pE4-1155) by site-directed mutagenesis with oligonucleotide primers 390 and 391 (Table 1), resulting in pE4-1170 (30), which carries two alanine substitutions at amino acid positions 90 and 92 (L90A I92A). The mutations were verified by DNA sequencing. Next, the 5.1-kb BstBI fragments from pH5pg4100 (wild type [wt]) and pH5pm4101 (E1B NES mutant) were replaced with the corresponding fragment from plasmid pE4-1170 to generate adenoviral bacmids pH5pm4116 and pH5pm4119, respectively. Recombinant bacmids were partially sequenced to confirm the mutations in the E1B-55K and E4orf6 genes. Virus mutants were generated as described previously (38). For transfection of viral DNA and propagation of the viruses, complementing W162 cells were used.
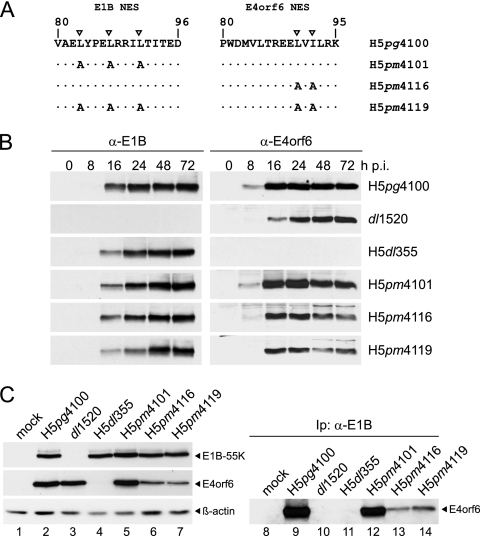
Fig. 1.
Effects of NES amino acid changes on the stability of the E1B-55K and/or E4orf6 protein and coimmunoprecipitation of E4orf6 with E1B-55K. (A) Amino acid substitutions in E1B-55K and/or E4orf6 mutant viruses. The NES-specific residues in E1B-55K and E4orf6 are indicated by inverted triangles. Numbers refer to amino acid residues in the wt E1B-55K and E4orf6 proteins from H5pg4100. Amino acid changes in the E1B and E4orf6 proteins of H5pm4101, H5pm4116, and H5pm4119 are shown. (B) Steady-state expression levels of E1B-55K and E4orf6 proteins. A549 cells were infected with wt and mutant viruses at a multiplicity of 20 FFU per cell. Cells were harvested at the indicated times p.i., and total-cell extracts were prepared. Proteins (20-μg samples for E1B-55K; 100-μg samples for E4orf6) from each time point were separated by SDS-12% PAGE and were subjected to immunoblotting using anti-E1B-55K (α-E1B) mouse MAb 2A6 or anti-E4orf6 mouse MAb RSA3. (C) Coimmunoprecipitation of E4orf6 with E1B-55K. Whole-cell extracts from infected A549 cells were prepared at 16 h p.i., and samples containing 800 μg of protein (or 2 mg for H5pm4116 and H5pm4119) were coimmunoprecipitated (Ip) with MAb 2A6 and separated by SDS-12% PAGE, followed by immunoblotting with anti-E4orf6 rabbit polyclonal antibody 1807 (lanes 8 to 14). For analysis of steady-state levels of E1B-55K, E4orf6, and β-actin, proteins (20-μg samples for E1B-55K and β-actin; 100-μg samples for E4orf6) were separated by SDS-12% PAGE and were subjected to immunoblotting using anti-E1B-55K mouse MAb 2A6, anti-E4orf6 rabbit polyclonal antibody 1807, or anti-β-actin mouse MAb AC-15 (lanes 1 to 7).
Viruses.
H5pg4100 served as the wt Ad5 parent virus in these studies (37). H5pm4101 has been described recently (37) and carries three leucine-to-alanine substitutions at amino acid positions 83, 87, and 91 (L83A L87A L91A) in the NES of the E1B-55K product. The E1B mutant virus dl1520 is a chimeric group C adenovirus (Ad2 and Ad5) that contains an 827-bp deletion within the 55K open reading frame in combination with a nonsense mutation at the third codon (4). The Ad5 mutant H5dl355 contains a 14-bp deletion in E4 open reading frame 6 (E4orf6), which prevents expression of the E4orf6 protein (31). H5pm4149 has been described recently (16) and carries four stop codons in the 55K reading frame. H5pm4154 expresses normal E4orf6/7 protein but an unstable or nonfunctional product of E4orf6 containing only the first 66 residues (8). The construction of mutant viruses H5pm4116 and H5pm4119 is described above. H5pg4100, dl1520, H5pm4149, and H5pm4101 were propagated in 911 monolayer cultures, whereas H5dl355, H5pm4154, H5pm4116, and H5pm4119 were propagated in W162 cells. The titers of all viruses were determined by fluorescent focus assays. To measure virus growth, infected cells were harvested 72 h postinfection (p.i.). The cell lysates were serially diluted in DMEM for the infection of 911 or 293 cells, and the virus yield was determined by quantitative E2A-72K immunofluorescence staining at 24 h p.i. (30). Viral DNA replication was determined by quantitative PCR as described previously (58). PCR products were analyzed by agarose gel electrophoresis on a 1% agarose gel and were quantitated using the ChemiDoc system and Quantity One software (Bio-Rad).
Antibodies and inhibitors.
Primary antibodies specific for the Ad proteins used in this study included mouse monoclonal antibody (MAb) 2A6 against E1B-55K (59), rat MAb 7C11 against E1B-55K (37), mouse MAb B6-8 against E2A-72K (DBP) (56), rabbit polyclonal antibody 1807 against E4orf6 (9), mouse MAb RSA3 against E4orf6 (46), rat MAb 6B10 against L4-100K (41), and rabbit polyclonal serum L133 against Ad5, generated by immunizing New Zealand White rabbits (Charles River) with purified and inactivated viral particles. Primary antibodies specific for cellular proteins included mouse MAb AC-15 against β-actin (Sigma-Aldrich, Inc.), mouse MAb DO-1 against p53 (Santa Cruz), and rabbit polyclonal antibody pNB-100-142 against Mre11 (Novus Biologicals, Inc.). Secondary antibodies conjugated to horseradish peroxidase (HRP) for detection of proteins by immunoblotting were donkey anti-rabbit immunoglobulin G (IgG), goat anti-mouse IgG, and sheep anti-rat IgG (Amersham, Biosciences). Fluorescent secondary antibodies were affinity-purified fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG and Texas Red-conjugated donkey anti-mouse IgG (Dianova). These were used at a 1:100 dilution in all immunofluorescence experiments, and images were acquired on a DMRB fluorescence microscope (Leica). For fluorescence analysis with an LSM 510 confocal laser scanning microscope (Zeiss), affinity-purified Alexa Fluor 488-conjugated goat anti-rat IgG, Alexa Fluor 555-conjugated goat anti-rabbit IgG, and Alexa Fluor 633-conjugated goat anti-mouse IgG (Invitrogen) were used at a 1:400 dilution.
Leptomycin B (LMB) was purchased from Enzo Life Sciences and was used at concentrations of 20 nM to block the CRM1-dependent nuclear export pathway.
Protein analysis.
Preparation of cell lysates for protein analysis and immunoblotting were conducted exactly as described previously (38). For immunoprecipitation, protein A-Sepharose (30 mg/ml) was incubated with 1 ml hybridoma supernatant 2A6 for 2 h at 4°C, and the mixture was washed twice in lysis buffer. The antibodies bound to protein A-Sepharose were added to the protein A-Sepharose precleared extracts and were rotated overnight at 4°C. The immune complexes were washed three times with lysis buffer (38), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by immunoblotting, and band intensities were measured densitometrically using GeneTools (Syngene).
Indirect immunofluorescence.
For indirect immunofluorescence, A549 cells were grown on glass coverslips and were infected as described previously (38). For immunofluorescence analysis with a DMRB fluorescence microscope (Leica), cells were fixed in freshly prepared phosphate-buffered saline (PBS)–2% paraformaldehyde (PFA) for 10 min at room temperature at the times indicated in the corresponding figure legends and were permeabilized in PBS–0.5% Triton X-100 for 15 min. Cells were then incubated for 10 min in PBS–4% PFA and were washed three times with PBS. After 1 h of blocking in Tris-buffered sulfate-BG buffer (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 0.05% Tween 20, 0.05% sodium azide, 5 mg/ml glycerol, and 5 mg/ml bovine serum albumin), the cells were reacted for 1 h with the primary antibody diluted in PBS, washed three times in PBS, and incubated for 1 h with the corresponding FITC- or Texas Red-conjugated secondary antibody (Invitrogen). Coverslips were washed three times in PBS and were mounted in Glow medium (Energene), and digital images were acquired on a DMRB fluorescence microscope (Leica) with a charge-coupled device camera (Diagnostic Instruments). Images were cropped using Adobe Photoshop CS4 and were assembled with Adobe Illustrator CS4.
For immunofluorescence analysis with an LSM 510 confocal laser scanning microscope (Zeiss), cells were fixed with ice-cold methanol for 15 min at −20°C and were permeabilized in PBS–0.5% Triton X-100 for 15 min at room temperature. After 1 h of blocking in TBS-BG buffer, cells were reacted with the primary antibody diluted in PBS, washed three times in PBS, and then incubated for 1 h with the corresponding Alexa Fluor 488-, 555-, and 633-conjugated secondary antibodies (Invitrogen). Coverslips were washed three times in PBS and were mounted in Glow medium (Energene), and digital images were acquired in multitrack mode on an LSM 510 Meta confocal laser scanning microscope with a 63× (numerical aperture, 1.40) oil objective (Zeiss). Images were prepared using Adobe Photoshop CS4 and were assembled with Adobe Illustrator CS4.
To visualize nuclei, fluorescence-coupled secondary antibodies were supplemented with 0.5 μg/ml 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI).
Quantitative PCR analysis.
Ad-infected cells were harvested 48 h p.i. and were washed in ice-cold PBS. Cells were resuspended in lysis buffer (10 mM HEPES [pH 7,8], 10 mM KCl, 20% glycerol, 1 mM dithiothreitol [DTT], 0.25% Nonidet P-40) and were lysed on ice for a maximum of 2 min. Centrifugation for 5 min at 470 × g and 4°C separated the cytoplasmic from the nuclear fraction. The pellet containing the nuclei was washed in lysis buffer once.
RNA was isolated from the supernatant (cytoplasmic fraction), pelleted nuclei, or total-cell pellets (total RNA) with 1 ml Trizol reagent (Invitrogen) as described by the manufacturer. The RNA pellets were dissolved in diethyl pyrocarbonate-treated water.
One microgram of RNA was used for transcription with the Reverse Transcription system (Promega). Reverse transcription (RT) was primed with oligo(dT) to select for processed mRNA. RT was performed as described by the manufacturer.
A Rotor-Gene 6000 real-time analyzer (Corbett Research) and the SensiMix Plus SYBR kit (Quantace) were used for quantitative RT-PCR. A 5 μM concentration of each primer was added to 12.5 μl of reaction mixture, followed by 1 μl of diluted template in a total volume of 25 μl. Quantitative PCR was done in triplicate for each sample by denaturing at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C (L5 and 18S rRNA) or 68°C (L4) for 30 s, and 72°C for 15 s. Raw numbers were normalized to those for the internal control, 18S rRNA. Relative RNA concentrations were determined by the standard curve method. The subcellular distributions of L4 and L5 transcripts are presented as the ratios of cytoplasmic to nuclear L4 and L5 mRNAs. The relative levels of total L4 and L5 mRNAs resulted from normalization to total-mRNA levels of H5pg4100. Standard deviations were calculated by the best value of sum or quotient. The primers used for the amplification of viral L5 RNA were described by Woo and Berk (66). Oligonucleotide primers 1110 and 1111 (Table 1), used for amplification of viral L4 RNA, were located in the L4-100K region, generating a product with a length of 199 bp. The primers used for the amplification of cellular 18S rRNA have been described recently (60).
Statistics and calculations.
Statistical analyses were performed using Dunnett's multiple comparison test. P values of <0.05 were considered statistically significant.
RESULTS
Construction of viruses harboring mutations within the E1B-55K and/or E4orf6 NES.
To study the function of the nuclear export signals of the adenoviral early proteins E1B-55K and E4orf6, we generated virus mutants with amino acid substitutions in either or both NES sequences, following the procedure described in Materials and Methods. The previously established wild-type (wt) Ad5 genome bacmid pH5pg4100 (30) was used as a template to modify the coding sequence of E4orf6 by site-directed mutagenesis replacing leucine-90 and isoleucine-92 within the nuclear export signal with alanines, resulting in H5pm4116 (Fig. 1A). To generate the E1B-55K/E4orf6 double-NES (DNES) mutant virus H5pm4119, we used the previously constructed bacmid pH5pm4101, which contains three amino acid substitutions (L83A L87A L91A) within the E1B-55K NES (37), and introduced the same L90A and I92A mutations into E4orf6.
Effect of mutational inactivation of the E1B-55K and/or E4orf6 NES on the proteins' stability and intracellular distribution.
Alanine substitution at key leucine or isoleucine residues within the E1B-55K or E4orf6 NES has been used previously in efforts to elucidate the contribution of this sequence to the proteins' ability to shuttle and to virus replication (see the introduction). The proteins with altered NES, either transiently synthesized during transfection assays (13, 18, 20, 39, 55, 65) or expressed from the viral genome, in the case of E1B-55K (37), are specifically impaired in their ability to shuttle but are otherwise functional, indicating that such amino acid substitutions do not alter the overall conformation or stability of either protein. Nevertheless, since the simultaneous mutation of both proteins' NES has not been studied previously, we first tested whether the mutations introduced interfered with protein stability. A549 cells infected at a multiplicity of 20 FFU per cell were harvested at different time points, and Western blot analyses using the E1B-55K-specific antibody 2A6 (59) and the E4orf6-specific antibody RSA3 (46) were performed. Figure 1B shows representative results. Infection with H5pg4100 showed high-level expression of both the E4orf6 and E1B-55K proteins as early as 8 and 16 h postinfection, respectively. The previously described virus mutant dl1520 (ONYX-015 [4]), which harbors a large deletion within the E1B coding sequence, showed no expression of the adenoviral E1B-55K protein, but despite a delay of approximately 8 h compared to wt virus expression, E4orf6 accumulated to levels comparable to those of the wt. The deletion mutant H5dl355 (31) displayed no difference in E1B-55K protein levels from H5pg4100. In agreement with our recent findings (37), functional inactivation of the E1B-55K NES did not lead to a decrease in E1B-55K protein expression and/or stability (H5pm4101). Similar results with regard to the level of E1B-55K were obtained for the E1B-55K/E4orf6 double-NES mutant H5pm4119 and the E4orf6 NES mutant H5pm4116. Surprisingly, in contrast to the E1B NES mutant (H5pm4101), comparison of the E4orf6 protein levels synthesized by either of the E4orf6 NES mutants (H5pm4116 or H5pm4119) with those for the wt showed that at very late times postinfection (48 and 72 h), E4orf6 protein levels were clearly lower in the mutants, suggesting that mutation of the E4orf6 NES induces a reduction in the expression or stability of the protein very late during infection. However, the E4orf6 protein attained levels comparable to those of the wt during both the early and late phases (16 and 24 h, respectively), indicating that the protein was stable (Fig. 1B).
We next tested whether the NES mutations inhibited formation of the E1B-55K/E4orf6 complex by using coimmunoprecipitation assays of the E4orf6 protein with the E1B-55K-specific MAb, 2A6. For this purpose, we first determined the steady-state concentration levels of both proteins. While E1B-55K is expressed at similar levels, densitometry measurement revealed that E4orf6 protein levels are 1.96-fold lower in H5pm4116 and 3.03-fold lower in H5pm4119 than those in wt-infected cells. Representative results of the immunoprecipitation experiments are shown in Fig. 1C, where, as expected, high levels of the E4orf6 protein could be coimmunoprecipitated by E1B-55K in both H5pg4100- and H5pm4101-infected cells (Fig. 1C, lanes 9 and 12) confirming that the E1B-55K NES mutation does not interfere with E4orf6 binding. In contrast, the levels of E4orf6 coimmunoprecipitated by E1B-55K in cells infected by mutants carrying E4orf6 NES mutations were 3- to 5-fold lower than those in the wt (Fig. 1C, lanes 13 and 14). In these assays, a specific band corresponding to E4orf6 could be detected, suggesting that although clearly reduced, E1B-55K/E4orf6 binding was not completely inhibited by mutation of either protein's NES. It has been suggested that E1B-55K and E4orf6 associate through an indirect interaction that requires assembly of the Cul5 E3 ubiquitin ligase (7), and it should be interesting to determine whether the E4orf6 NES mutation interferes with the protein′s ability to interact with the Cul5 E3 ubiquitin ligase complex.
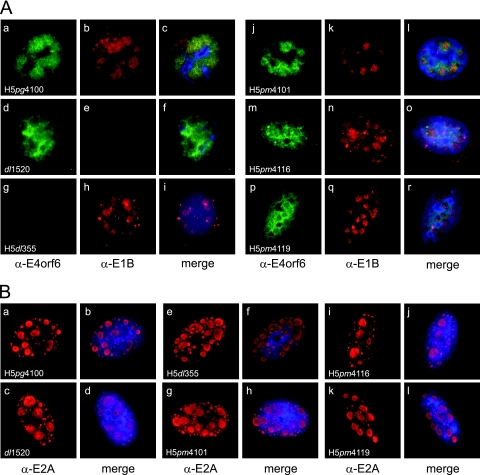
The intracellular localization of E1B-55K and E4orf6 to replication centers and to cytoplasmic foci has been related to their activities in selective viral late mRNA metabolism and in the assembly of the Cul5-containing E3 ubiquitin ligase, respectively (28, 45, 51, 61). To determine the effect of functional inactivation of the NES on the intracellular distribution of both proteins, we performed double-label immunofluorescence analysis of infected cells at 20 h postinfection, a time at which both the E1B-55K and E4orf6 proteins with altered NES accumulated to levels comparable to those for the wt (Fig. 1B). We probed for E1B-55K and E4orf6 to visualize the intracellular localization of both proteins. As has been reported previously, E1B-55K showed a complex distribution pattern that correlated closely with the presence or absence of E4orf6 (28, 44, 51) and was clearly altered in NES mutants. Infection with the H5pg4100 virus led to a nuclear distribution that is characteristic for E1B-55K: the protein localized in spherical bodies that most likely correspond to viral replication centers (37). In the cytoplasm, the protein accumulated in numerous brightly stained cytoplasmic bodies (Fig. 2Ab). Elimination of NES-dependent nuclear export of E1B-55K induced its complete relocalization to the nucleus, where the protein displayed a closer association with brightly stained nuclear bodies (Fig. 2Ak) similar to the viral replication centers but displaying a signal with higher density (37). Interestingly, the same phenotype could be observed in H5pm4119-infected cells, indicating that the nuclear localization of E1B-55K in the absence of its functional NES is not influenced by the status of the E4orf6 NES (Fig. 2Aq). In the E4orf6 deletion virus H5dl355, the nuclear population of E1B-55K was not as clearly associated with these spherical bodies; rather, it appeared more closely and densely packed in these structures, displaying some dense foci (Fig. 2Ah). In the cytoplasm the protein accumulated in punctate aggregates, indicating that the cytoplasmic localization of E1B-55K was not affected by the absence of E4orf6 (Fig. 2Ag to i). The effect of NES mutation on the intracellular localization of E4orf6 was difficult to assess at the level of resolution that can be obtained by these assays. The NES-dependent nucleocytoplasmic shuttling of this protein has been demonstrated using transfected heterokaryon assays (18), and its export has been shown by use of leptomycin B treatment to be mediated by CRM1 (55). However, E4orf6 displays an essentially nuclear distribution in wt Ad-infected cells by immunofluorescence, indicating that the import of the protein into this organelle is more efficient than its export (20). In our experiments, E4orf6 was detected exclusively in the nucleus, where the protein was distributed throughout the nucleoplasm but displayed a higher density in close association with spherical bodies in cells infected with H5pg4100 or the E1B mutant dl1520 or H5pm4101 (Fig. 2Aa, d, and j), suggesting that the status of E1B-55K does not have a major impact on the nuclear distribution of E4orf6, in agreement with previous reports (28, 50). The characteristic localization pattern that E1B and E4orf6 showed in the different mutant virus-infected cells was detectable in nearly all of the cells examined (n, >50), and the single selected cells show representative stainings of both proteins. Interestingly, mutation of the E4orf6 NES resulted in subtle but clear differences in the nuclear distribution of the protein, where it localized homogeneously and was seemingly excluded from the nuclear structures where E1B-55K could be detected (Fig. 2Am and p). These observations suggest that although the NES mutation does not interfere with E4orf6 transport into the nucleus, it does perturb its proper localization in this organelle. The effect of NES mutation on the intranuclear localization of other viral or cellular proteins has not been studied in detail. However, altered intranuclear patterns of distribution have been shown for other NES-bearing proteins upon either NES mutation or LMB treatment (21, 35). It should be interesting to determine whether the intranuclear distribution of E1B-55K or E4orf6 may be functionally connected to nucleocytoplasmic shuttling.
Fig. 2.
Intracellular localization of E1B-55K, E4orf6, and E2A-72K in wt- and NES mutant-infected cells. A549 cells were infected with wt or E1B-55K/E4orf6 mutant viruses at a multiplicity of 20 FFU per cell and were fixed at 20 h p.i. (A) Cells were double labeled with anti-E4orf6 rabbit polyclonal antibody 1807 (α-E4orf6) and anti-E1B-55K mouse MAb 2A6 (α-E1B) as primary antibodies and with FITC- and Texas Red-conjugated secondary antibodies, respectively. Representative anti-E4orf6 (green) (a, d, g, j, m, and p) and anti-E1B (red) (b, e, h, k, n, and q) staining patterns are shown. Overlays of DAPI staining (blue) with the green and red images (merge) are also shown (c, f, i, l, o, and r). Nuclei were visualized using DAPI. Magnification, ×7,600. (B) Cells were labeled with anti-E2A-72K mouse MAb B6-8 and Texas Red-conjugated secondary antibodies (red) (a, c, e, g, i, and k). Representative anti-E2A staining patterns are shown. Overlays of DAPI staining (blue) with the red images (merge) are also shown (b, d, f, h, j, and l). Nuclei were visualized using DAPI. Magnification, ×7,600.
In parallel, we also probed for the DNA-binding protein E2A in order to visualize the adenoviral DNA replication centers (Fig. 2B). All of the mutants examined in this study showed similar distributions of the E2A protein within the nucleus, where the protein localized in brightly stained, spherical structures, indicating that the mutations introduced did not interfere either with the expression of E2A or with the formation of DNA replication centers during infection of human A549 cells.
Effects of elimination of NES-mediated nuclear export of E1B-55K and/or E4orf6 on viral production and late gene expression.
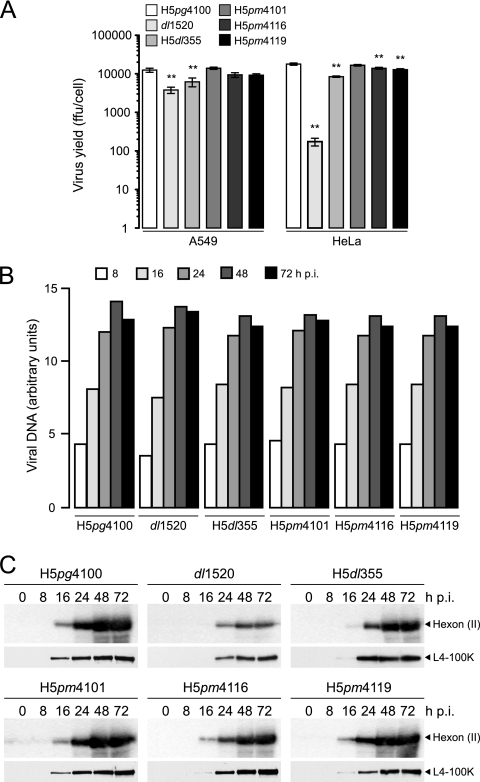
Although the effects of CRM1 inhibition and the role of NES-dependent transport of E1B-55K and E4orf6 in viral late gene expression were reported some time ago (13, 18, 20, 27, 39, 55, 65), the effect of mutation of the proteins' NES has never been examined in adenovirus recombinants and in the presence of active CRM1. We therefore decided to investigate whether mutations in the E1B-55K and/or E4orf6 NES lead to defects in adenoviral replication due to functional inactivation of the CRM1-dependent nucleocytoplasmic shuttling of either or both proteins. We initially analyzed viral progeny production in both A549 and HeLa cells. Figure 3A shows representative results for the efficiency of virus production by all virus mutants examined in this study. Virus production was determined in at least two independent experiments as described in Materials and Methods. As expected, the E1B-55K-null mutant, dl1520, showed a defect of about 4-fold in A549 cells and approximately 15-fold in HeLa cells compared to H5pg4100 (4, 57). Also as expected, the reduction of progeny virus production in H5dl355-infected cells was less prominent than that in dl1520-infected cells; about half the number of viral particles produced by H5pg4100 were produced by H5dl355 in either cell type. As we reported previously, the E1B-55K NES mutation did not impair progeny virus production in either cell line examined, confirming that CRM1-mediated nuclear export of E1B-55K is not required for efficient viral replication (37). Functional inactivation of the E4orf6 NES led to a slight (in HeLa cells) to moderate (in A549 cells) decrease in viral production, which was not significantly reduced further by concurrent inactivation of the E1B-55K NES (Fig. 3A). The values for the E4orf6 NES mutant viruses are statistically significant (P, <0.05) only in infected HeLa cells. The moderately lower efficiency of virus production induced by mutation of the E4orf6 NES could not be accounted for by defects in early E2 gene expression, since the accumulation of the E2A protein and the formation of E2A-containing replication centers were similar for the NES mutants and the wt (Fig. 2B). We therefore decided to measure the effects of mutation of the E4orf6 and E1B-55K NES on viral DNA replication and on viral late protein synthesis. All viruses produced comparable amounts of DNA and with similar kinetics, indicating that early gene expression, progression into the late phase, and viral DNA replication were not affected either by the absence of E1B-55K or E4orf6 or by mutation of the proteins' NES (Fig. 3B). As expected, the absence of E1B-55K in dl1520-infected cells produced delayed and lower levels of both the hexon and L4-100K proteins, and in H5dl355-infected cells a clear, albeit less severe reduction of late protein synthesis was observed. In contrast, mutation of either or both of the E1B-55K and E4orf6 NES (H5pm4101, H5pm4116, and H5pm4119) resulted in hexon protein levels that were comparable to those produced in wt-infected cells. However, in H5pm4116- and H5pm4119-infected cells, the expression of L4-100K was delayed and reduced to the same extent as in the E4orf6-null mutant (Fig. 3C). These data indicate that the effects of E4orf6 NES mutation in the H5pm4116 or H5pm4119 virus (Fig. 1B) might result from defects in the late phase of adenoviral replication, since the delayed accumulation of L4-100K (Fig. 3C) could impact viral late protein synthesis as well as assembly of the viral capsid and could explain reduced virus production (Fig. 3A).
Fig. 3.
Effects of amino acid substitutions in the E1B-55K and E4orf6 NES on virus growth, viral DNA accumulation, and viral late protein synthesis. (A) Virus growth. A549 and HeLa cells were infected with wt or mutant viruses at a multiplicity of 10 FFU per cell and were harvested 72 h p.i., and virus yield was determined by quantitative E2A-72K immunofluorescence staining on 911 cells. The results are reported with Dunnett′s multiple comparison test applied to the log-transformed virus yield (double asterisks indicate significant differences [P, <0.05] between H5pg4100 and mutant viruses). Each result represents the average for three independent infections in two independent experiments. Error bars indicate the standard errors of the means. (B) Viral DNA synthesis. A549 cells were infected with wt or mutant viruses at a multiplicity of 20 FFU per cell. Total nuclear DNA was isolated at the indicated times after infection and was subjected to quantitative PCR. PCR products were analyzed and quantitated using the Quantity One software of the ChemiDoc system (Bio-Rad). (C) Viral late protein synthesis. A549 cells were infected with wt or mutant viruses at a multiplicity of 20 FFU per cell. Total-cell extracts were prepared at the indicated times postinfection. Proteins (20-μg samples) were separated by SDS-12% PAGE, transferred to nitrocellulose membranes, and probed with anti-Ad5 rabbit polyclonal serum L133 or anti-L4-100K rat MAb 6B10. The bands corresponding to the viral late protein hexon (II) are indicated on the right.
The moderate defects in both L4-100K protein and viral progeny synthesis observed in H5pm4116- and H5pm4119-infected cells could be due to lower levels of E4orf6 protein expression in these mutants. Therefore, a semistable cell line expressing the E4orf6 NES mutant protein was generated using a lentiviral construct as described in Materials and Methods. This cell line was infected with the wt (H5pg4100), the E1B-55K- or E4orf6-null mutant (H5pm4149 or H5pm4154, respectively), or the single or double E1B-55K or E4orf6 NES mutant, as described for Fig. 1B. Time course analyses were performed for E1B-55K and E4orf6, viral late proteins, and viral progeny production. Figure 4A shows the expression levels of E4orf6 in mock-infected cells and in cells infected with the wt or the different mutants. The level of expression of E4orf6 protein obtained upon infection of this cell line with each virus mutant was similar to that observed in wt-infected cells (compare Fig. 1B and 4A). Although E4orf6 protein levels were reduced in H5pm4154-infected cells at late times postinfection, similar levels were produced in these cells by the E1B-55K- and E4orf6-null mutants (H5pm4149 and H5pm4154, respectively) and by the single NES mutants (H5pm4101 and H5pm4116), indicating that exogenously expressed E4orf6 NES was stable. Surprisingly, when the cell line expressing the recombinant E4orf6 NES was infected with the double-NES mutant, both the E1B-55K and E4orf6 protein levels were significantly reduced.
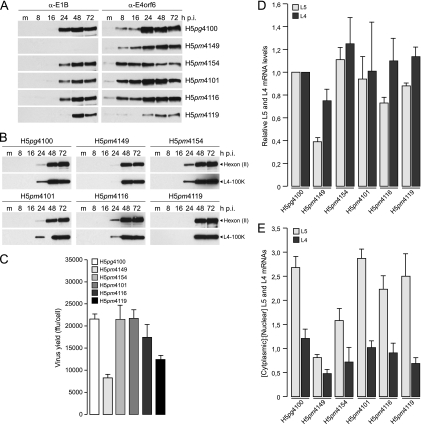
Fig. 4.
Effect of E4orf6 NES expression from a lentiviral construct and concomitant infection of A549 cells with wt and mutant viruses on E1B-55K and/or E4orf6 protein stability, viral late protein synthesis, virus growth, viral late mRNA expression, and the nuclear export efficiency of viral late mRNAs. (A) Steady-state expression levels of E1B-55K and E4orf6. E4orf6 NES-expressing cells were infected with wt and mutant viruses at a multiplicity of 20 FFU per cell. Noninfected (m, mock) and infected cells were harvested at the indicated times p.i., and total-cell extracts were prepared. Proteins (25-μg samples) from each time point were separated by SDS-10% PAGE and were subjected to immunoblotting using anti-E1B-55K mouse MAb 2A6 or anti-E4orf6 rabbit polyclonal antibody 1807. (B) Viral late protein synthesis. Total-cell extracts were prepared at the indicated times postinfection. Proteins (10-μg samples) were separated by SDS-10% PAGE, transferred to nitrocellulose membranes, and probed with anti-Ad5 rabbit polyclonal serum L133 or anti-L4-100K rat MAb 6B10. The bands corresponding to the viral late protein hexon (II) are indicated on the right. (C) Virus growth. E4orf6 NES-expressing cells were infected with wt or mutant viruses at a multiplicity of 10 FFU per cell and were harvested 72 h p.i., and virus yield was determined by quantitative E2A-72K immunofluorescence staining on 293 cells. The results represent averages for three independent experiments. Error bars indicate the standard deviations of the means. (D) Total amounts of viral late mRNAs L5 and L4. E4orf6 NES-expressing cells were infected with wt or mutant viruses at a multiplicity of 10 FFU per cell. Steady-state concentrations of both RNAs were determined by real-time PCR at 48 h postinfection. Total L5 and L4 values were corrected using 18S rRNA as an internal control and are expressed relative to the wild-type value. The results represent averages for three independent experiments, each performed in duplicate. Error bars indicate the standard deviations of the means. (E) Nuclear export efficiency of viral late mRNAs L5 and L4. E4orf6 NES-expressing A549 cells were infected with wt or mutant viruses at a multiplicity of 10 FFU per cell. Steady-state concentrations of both RNAs in the cytoplasm and nucleus were determined by real-time PCR at 48 h postinfection. Raw numbers were corrected using 18S rRNA as an internal control, and the normalized numbers were used to calculate the ratios of cytoplasmic to nuclear L5/L4 mRNAs. The results represent averages for three independent experiments, each performed in duplicate. Error bars indicate the standard deviations of the means.
The E4orf6 NES-expressing cell line was then used to evaluate levels of viral late proteins and virus production (Fig. 4B and C). Interestingly, expression of the E4orf6 NES rescued L4-100K protein levels and virus growth in cells infected with the E4orf6-null virus (H5pm4154) to essentially wt levels, indicating that mutation of the protein's NES has no effect on either viral late proteins or virus reproduction. Significantly, these observations also indicate that the NES-mutated protein is not functionally defective. The levels of E4orf6 were also similar to wt levels in cells infected with the E1B-55K-null mutant (H5pm4149) or either the E1B-55K (H5pm4101) or the E4orf6 (H5pm4116) NES mutant. Expression of exogenous E4orf6 NES in cells infected with these mutants rescued normal levels of late proteins and virus production, although the latter did not attain wt levels in the H5pm4116 mutant. These properties were more defective in H5pm4119, suggesting that they result from lower E4orf6 and E1B-55K protein levels (Fig. 4A to C).
We next investigated whether the E1B-55K NES and/or the E4orf6 NES is required for late mRNA synthesis and preferential export during the late phase of infection. For these experiments, we used the NES virus mutants to measure total-mRNA levels and analyzed their nucleocytoplasmic distribution by fractionation and quantification of RNA, performing quantitative RT-PCR on E4orf6 NES-expressing A549 cells (Fig. 4D and E). Using the E1B-null mutant H5pm4149, we could confirm that E1B-55K is required for efficient synthesis of viral late mRNAs (2, 31, 49). The absence of this protein led to a 20 to 60% drop in total viral late mRNA levels from those in H5pg4100-infected cells when L4 or L5 mRNA levels were measured, respectively (Fig. 4D). As was seen for virus growth, viral late mRNA production was completely rescued by exogenously expressed E4orf6 NES in H5pm4154-infected cells (Fig. 4D). A similar rescue was achieved for the NES mutants, since none of the NES mutations in either E1B-55K, E4orf6, or both led to reductions in mRNA production of more than 30% from wt levels. These results indicate that the presence of a functional NES in either or both of these early adenoviral proteins is not essential for the synthesis or stability of late mRNA (Fig. 4D). To evaluate efficient nuclear export of late adenoviral mRNAs, we extracted the L4 and L5 mRNAs after fractionation of the nuclear and cytoplasmic compartments, and we analyzed them by quantitative PCR (Fig. 4E). As expected, both viral null mutants were significantly impaired in nuclear export of L4 and L5 mRNA compared to H5pg4100. As we have shown previously, functional inactivation of the E1B-55K NES does not decrease late mRNA accumulation within the cytoplasm, suggesting that mRNA transport does not depend on CRM1-mediated export of E1B-55K (37). Interestingly, concurrent inactivation of the E4orf6 NES in the H5pm4119 virus, as well as the mutation of E4orf6 alone in H5pm4116, led to a cytoplasmic accumulation of viral late mRNA that was comparable to that in the wt (Fig. 4E). These data indicate that functional inactivation of E1B-55K and/or E4orf6 NES-dependent export does not abrogate viral late mRNA accumulation in the cytoplasm, and they are in perfect agreement with previous reports in which inhibition of CRM1 was shown not to interfere with viral late gene expression (13, 27, 55). These findings are significant because they allow us to extend this to conclude that the NES sequences that mediate the export of both E1B-55K and E4orf6 in the normal context of adenovirus-infected cells and in the presence of active CRM1 do not contribute to viral late gene expression.
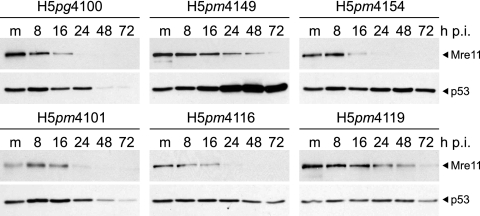
Mutation of E1B-55K and E4orf6 NES does not preclude degradation of p53 or Mre11 during infection of A549 cells.
Since CRM1-dependent export of E1B-55K and E4orf6 is not required for viral late gene expression, we decided to examine the effects of mutations of both the E1B-55K and E4orf6 NES on the degradation of p53 and Mre11, two well-known cellular substrates of the E3 ubiquitin ligase assembled by E1B-55K/E4orf6. Lysates of infected E4orf6 NES-expressing A549 cells were obtained at various times postinfection, and p53 and Mre11 protein levels were analyzed by Western blotting. As expected, infection of A549 cells with the H5pg4100 virus led to the degradation of p53 and Mre11 (7, 32, 53, 54, 62), since p53 levels declined after 8 h p.i. and Mre11 levels were clearly reduced by 16 h and no longer detectable at 72 h after infection (Fig. 5). In contrast, Mre11 protein levels were not efficiently reduced, and p53 accumulated, in the course of infection with the E1B-55K-null mutant, H5pm4149. Also as expected, mutation of the E1B-55K NES (H5pm4101) did not interfere with the degradation of either cellular protein (Fig. 5) (37).
Fig. 5.
Effects of E4orf6 NES expression from a lentiviral construct and concomitant infection of A549 cells with wt or mutant viruses on steady-state protein concentrations of p53 and Mre11. E4orf6 NES-expressing A549 cells were infected with wt or mutant viruses at a multiplicity of 20 FFU per cell. Noninfected (m, mock) and infected cells were harvested at the indicated times p.i., and whole-cell extracts were prepared. Proteins (40-μg samples) from each time point were separated on SDS-10% polyacrylamide gels and were subjected to immunoblotting using anti-Mre11 rabbit polyclonal antibody pNB-100-142 and anti-p53 mouse MAb DO-1.
Interestingly, for H5pm4154 or H5pm4116 infections, exogenous expression of the E4orf6 NES did not preclude the degradation of Mre11, and p53 did not accumulate to the levels observed in H5pm4149-infected cells. As with other properties examined (Fig. 4), the double-NES mutant, H5pm4119, displayed reduced efficiency at Mre11 and p53 degradation in E4orf6 NES-expressing cells (Fig. 5). Taken together, these findings suggest that neither CRM1-mediated export of E1B-55K nor CRM1-mediated export of E4orf6 is required for the degradation of Mre11 or p53.
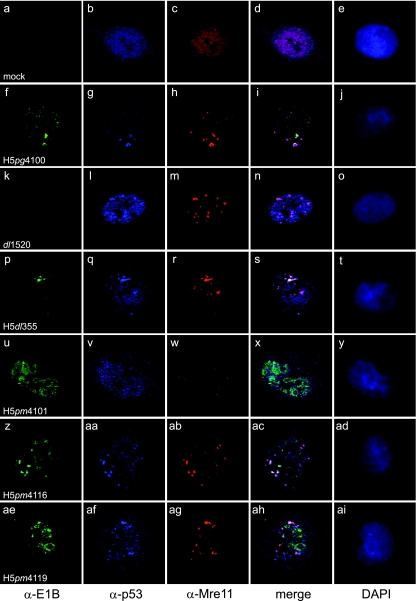
Since our findings showed that degradation of p53 and Mre11 does not require CRM1-dependent export of E1B-55K and E4orf6, we decided to examine the effects of NES mutations on the intracellular localization of p53 and Mre11 using triple-label immunofluorescence and confocal microscopy analysis of Ad5-infected cells. It has been proposed that the E1B-55K/E4orf6-dependent Cul5 ubiquitin ligase assembles in juxtanuclear cytoplasmic aggregates, where the complex colocalizes and induces polyubiquitylation of p53, and presumably of Mre11 and other cellular targets (45, 61). Analysis of the localization of E1B-55K, p53, and Mre11 in A549 cells infected with the wt virus H5pg4100 showed lower levels of the nuclear populations of both p53 and Mre11 (Fig. 6g and h) than in mock-infected cells (Fig. 6b and c) and the colocalization of both cellular proteins with E1B-55K in cytoplasmic aggregates (Fig. 6f to j). In cells infected with the dl1520 mutant, in the absence of E1B-55K, neither degradation nor relocalization of p53 was induced: the protein clearly accumulated in the nuclear compartment and was not relocalized to cytoplasmic aggregates (Fig. 6l). The nuclear population of Mre11 was relocalized from a homogeneous and diffuse distribution (Fig. 6c) to numerous nuclear specks; however, the protein was not detected in the cytoplasm (Fig. 6m), indicating that although the nuclear relocalization of Mre11 is independent of E1B-55K, export to the cytoplasm requires this viral protein. No such requirement was observed for E4orf6; both p53 and Mre11 could be detected in cytoplasmic perinuclear aggregates in H5dl355-infected cells (Fig. 6q and r). In H5pm4101-infected cells, reduced p53 protein levels were observed in the nuclear compartment, and no formation of perinuclear cytoplasmic aggregates could be detected (Fig. 6v). In contrast, reduced levels of Mre11 could be detected in cytoplasmic bodies, indicating that Mre11 can be exported in the absence of an E1B-55K NES, although with lower efficiency (Fig. 6w). It has been shown that E4orf6 is required for the degradation of both cellular targets (14, 53, 54, 62); however, the absence of E4orf6 (in H5dl355) did not preclude the export of E1B-55K to the cytoplasm, nor was the E4orf6 protein necessary for the cytoplasmic localization of either p53 or Mre11 (Fig. 6p to t). Significantly, mutation of the E4orf6 NES in H5pm4116-infected cells did not block the relocalization of p53 or Mre11 to E1B-55K-containing cytoplasmic aggregates (Fig. 6z to ad), indicating that NES-dependent export of E4orf6 does not contribute to the cytoplasmic export of either p53 or Mre11.
Fig. 6.
Intracellular localization of E1B-55K, p53, and Mre11 in wt and mutant virus-infected cells. A549 cells were either left uninfected or infected with wt or E1B-55K/E4orf6 mutant viruses at a multiplicity of 20 FFU/cell and were fixed at 24 h p.i. Cells were triple labeled with anti-E1B rat MAb 7C11 (α-E1B), anti-p53 mouse MAb DO-1 (α-p53), and anti-Mre11 rabbit polyclonal antibody pNB-100-142 (α-Mre11) along with Alexa Fluor 488-, 633-, and 555-conjugated secondary antibodies, respectively. Representative anti-E1B (green) (a, f, k, p, u, z, and ae), anti-p53 (blue) (b, g, l, q, v, aa, and af), and anti-Mre11 (red) (c, h, m, r, w, ab, and ag) staining patterns are shown. Overlays of the green, blue, and red images (merge) are also shown (d, i, n, s, x, ac, and ah). Nuclei were visualized using DAPI (blue) (e, j, o, t, y, ad, and ai). Magnification, ×6,300.
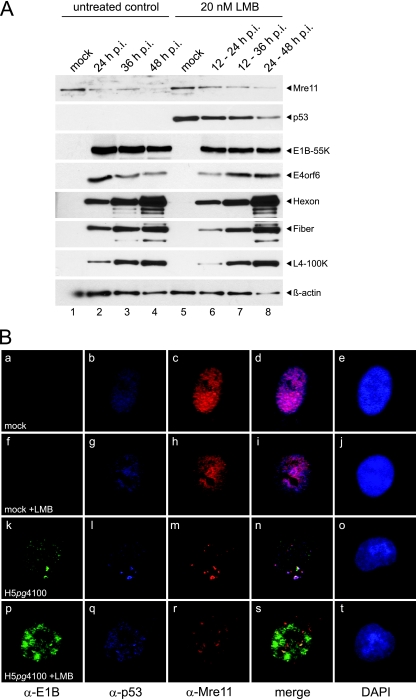
To confirm that the properties exhibited by the E1B-55K and E4orf6 NES mutants correlated with impaired nucleocytoplasmic export, we next examined the impact of inhibition of CRM1 on the degradation and localization of p53 and Mre11 in mock- and Ad-infected cells. The LMB concentration used for these assays has been shown not to have significant toxic effects for as long as 24 h (13). Ad-infected cells were either left untreated or treated with LMB and were harvested at the times postinfection indicated in Fig. 7, and protein levels were compared by Western blotting. Representative results are shown in Fig. 7 A. As expected, treatment of Ad-infected cells with LMB at different times postinfection had no effect on late protein synthesis: the steady-state levels of the hexon and L4-100K proteins were comparable to those in untreated cells. Furthermore Mre11 degradation occurred with essentially the same efficiency in LMB-treated or untreated cells, indicating that in agreement with our observations using the NES virus mutants (Fig. 5 and 6), CRM1-dependent export is not required for the degradation of Mre11 (Fig. 7A). Consistent with published observations (42, 47), LMB treatment of mock-infected cells for 24 h led to the accumulation of p53 (Fig. 7A, lane 5) compared to levels in the untreated control (Fig. 7A, lane 1). p53 levels were efficiently reduced in the course of adenoviral infection (Fig. 7A, lanes 5 to 8), indicating that degradation of p53 occurs in the absence of active CRM1, consistent with the reduction of p53 levels observed in cells infected with NES virus mutants (Fig. 5). Furthermore, analysis by immunofluorescence of the intracellular distribution of p53 and Mre11 in Ad-infected cells in the presence of LMB (Fig. 7B) also confirmed our results for the effects of E1B-55K and E4orf6 NES mutations on the relocalization of the cellular proteins (Fig. 6). In mock-infected cells, LMB induced no change in p53 or Mre11 distribution; both proteins displayed an essentially nuclear localization (Fig. 7B, compare panels a to e with panels f to j). The characteristic colocalization pattern that E1B, p53, and Mre11 exhibit in Fig. 6 and 7 was detectable in approximately 90 to 100% of the cells examined (n, >50). Treatment of Ad-infected cells with LMB completely inhibited the relocalization of both E1B-55K and p53 to cytoplasmic aggregates (Fig. 7B, compare panels k and l with panels p and q), and as expected, E1B-55K and E4orf6 displayed the same nucleocytoplasmic distribution as that observed in cells infected with the NES mutants (data not shown). Interestingly, Mre11 levels were reduced (Fig. 7A), an effect similar to that observed with the E1B-55K NES mutant, and the protein was relocalized both to numerous nuclear specks and to cytoplasmic aggregates (Fig. 7Br), indicating that export of Mre11 is not completely blocked by inhibition of CRM1. Taken together, our data confirm that CRM1-dependent export of neither E1B-55K nor E4orf6 is required for efficient synthesis of viral late proteins, and they indicate that while efficient export of p53 depends on the E1B NES and active CRM1, Mre11 requires neither the E1B nor the E4 NES and may use a CRM1-independent export pathway.
Fig. 7.
Effects of leptomycin B treatment on steady-state concentrations and intracellular localization of p53 and Mre11. (A) Steady-state protein levels of p53 and Mre11 in wt-infected and leptomycin B (LMB)-treated cells. A549 cells were either mock infected or infected with the wt virus at a multiplicity of 20 FFU per cell, treated with 20 nM LMB from 12 to 24 (lane 6), 12 to 36 (lane 7), or 24 to 48 (lanes 5 and 8) h p.i., and harvested at the indicated times p.i., and total-cell extracts were prepared. LMB-treated cell lysates (lanes 5 to 8) were compared to untreated controls harvested at the same time points after infection (lanes 1 to 4). Proteins (30-μg samples for p53 and Mre11; 20-μg samples for E1B-55K and E4orf6; 10-μg samples for hexon, fiber, and L4-100K) from each time point were separated by SDS-10% PAGE and were subjected to immunoblotting using anti-p53 mouse MAb DO-1, anti-Mre11 rabbit polyclonal antibody pNB-100-142, anti-E1B-55K mouse MAb 2A6, anti-E4orf6 rabbit polyclonal antibody 1807, anti-Ad5 rabbit polyclonal serum L133, or anti-L4-100K rat MAb 6B10. (B) Intracellular localization of E1B-55K, p53, and Mre11 in wt-infected and LMB-treated cells. A549 cells were either left uninfected or infected with the wt virus at a multiplicity of 20 FFU per cell; either left untreated or treated with 20 nM LMB from 12 to 24 h p.i.; and fixed at 24 h p.i. Cells were triple labeled with anti-E1B rat MAb 7C11 (α-E1B), anti-p53 mouse MAb DO-1 (α-p53), and anti-Mre11 rabbit polyclonal antibody pNB-100-142 (α-Mre11), along with Alexa Fluor 488-, 633-, and 555-conjugated secondary antibodies, respectively. Representative anti-E1B (green) (a, f, k, and p), anti-p53 (blue) (b, g, l, and q), and anti-Mre11 (red) (c, h, m, and r) staining patterns are shown. Overlays of the green, blue, and red images (merge) are also shown (d, i, n, and s). Nuclei were visualized using DAPI (blue) (e, j, o, and t). Magnification, ×6,300.
DISCUSSION
The ability of the E1B-55K and E4orf6 proteins to shuttle between the nucleus and the cytoplasm has been extensively studied using different approaches and a variety of established cell lines (13, 18, 20, 27, 37, 39, 55, 65). Both early proteins possess bona fide nuclear export signals (NES) that can direct their nuclear export via a CRM1-dependent pathway (18, 20, 39, 65), and both can also exit the nucleus by other, less characterized pathways (20, 37, 55). However, the cellular mechanism(s) exploited for nucleocytoplasmic transport by E1B-55K/E4orf6 to induce the almost exclusive export of viral late mRNA or the assembly and activity of the adenovirus-infected cell-specific E3 ubiquitin ligase has not been determined. Indeed, the contribution(s) made by the shuttling of E1B-55K/E4orf6 to these activities or to viral replication is not fully understood.
In this report we have used adenovirus recombinants with specific substitutions in the E1B-55K and E4orf6 NES sequences to investigate the role of their CRM1-dependent transport in the adenoviral replication cycle. The properties exhibited by adenovirus recombinants with altered NES sequences confirm that neither the E1B-55K nor the E4orf6 NES is necessary for normal levels of late gene expression (13, 27, 37, 55). Interestingly, nucleocytoplasmic export and degradation of p53 and Mre11 displayed different requirements for the E1B-55K and E4orf6 NES.
The NES mutants directed the synthesis of stable E1B-55K and E4orf6, as evidenced by the fact that protein levels were similar to those produced by the wild-type virus, during both the early and late phases. A clear reduction in steady-state levels of E4orf6 was induced by the NES mutation at very late time points postinfection (Fig. 1B). The reason(s) for this reduction is not clear, since the substitutions introduced to generate the altered NES have previously resulted in seemingly stable proteins in transfected cells (13, 18, 55, 65), and they do not perturb any known regulatory sequence in E4. E4orf6 is known to regulate alternative splicing of the late viral mRNA tripartite leader (49), and a temporal pattern for the production of E4 mRNA splicing has been shown to be influenced by the E1B-55K/E4orf6 proteins (17). Regulation of E4orf6 mRNA was not implicated in these studies. However, the role of E4orf6 NES-dependent export or the impact of CRM1 on the export or stability of early mRNAs has not been examined. Nevertheless, transcription of E4 genes declines rapidly upon entry into the late phase (reviewed in reference 48), and the E4orf6 protein half-life is reduced late after infection (15). Irrespective of the endogenous E4orf6 NES protein levels, exogenous expression of E4orf6 NES completely rescued the defective phenotypes of an E4orf6-null mutant, H5pm4154, in viral late protein and virus production, indicating that the E4orf6 NES protein was functional (Fig. 3C). However, for NES virus mutants, exogenously expressed E4orf6 NES resulted in lower accumulation or expression of both the E1B-55K and E4orf6 NES proteins (Fig. 4A) only in cells infected with the double-NES (DNES) mutant virus (H5pm4119). Such an effect was unexpected and cannot be explained by these experiments. However, the nuclear distribution of E1B-55K and its binding with E4orf6 were affected by mutation of the E4orf6 NES in single and double mutants, leading us to speculate that the lower stability of the E4orf6 NES could have a dominant negative effect, in which its overexpression could lead to the formation of a defective and thereby less stable E1B-55K NES/E4orf6 NES complex. This is supported by the fact that overexpression of the E4orf6 NES protein in DNES mutant virus-infected cells could not rescue the defective properties of the virus.
The efficiency of L4-100K protein synthesis and viral progeny production was decreased in viruses harboring a mutated E4orf6 NES, although to a lower extent than that shown by the dl1520 or H5dl355 viruses, and this decrease could be rescued by expression of exogenous E4orf6 NES, except for the DNES mutant virus. Since viral DNA replication was unaffected, those properties are likely to result from defects in the late phase of infection (Fig. 3). E4orf6 is known to stimulate exon skipping of the i-leader within the tripartite leader of viral late mRNA and to have a similar effect on alternatively spliced chimeric β-globin transcripts (49). Consequently, it is possible that lower levels of E4orf6 may impact the posttranscriptional processing and accumulation of viral late mRNAs. Moreover, the inadequate activation of the E3 ubiquitin ligase by the E1B-55K/Eorf6 complex (Fig. 1C) could result in such phenotypes as inhibition of cellular substrate degradation or decreased production of encapsidation-competent genomes (10, 34, 36, 62).
In agreement with the previously reported effect of E1B-55K NES mutation on the ability of this protein to induce the degradation of p53 or Mre11 (37), our data indicate that degradation of either p53 or Mre11 does not require E1B-55K or E4orf6 NES-dependent transport (Fig. 5). Furthermore, while nuclear export of these cellular proteins is unaffected by inactivation of the E4orf6 NES, the E1B-55K NES is required for p53 nuclear export only (Fig. 6). Interestingly, in mock-infected cells, inactivation of CRM1 by LMB did not alter either p53 or Mre11 localization. In contrast, LMB treatment of Ad-infected cells precluded export of p53, but Mre11 could still relocalize to cytoplasmic aggregates (Fig. 7B), suggesting that upon infection, relocalization of Mre11 to the cytoplasm is achieved by a CRM1-independent mechanism. These findings indicate that separate and distinct requirements exist for CRM1-mediated export of E1B-55K and E4orf6 with regard to the induced export and degradation of p53 and Mre11. This conclusion is consistent with reports that have shown that degradation of different cellular proteins can occur independently and that individual targets require separate substrate recognition events by E1B-55K (12, 61). It has been proposed that activation of the E3 ubiquitin ligase requires the binding of E4orf6 to Cullin 5, Rbx1, and elongins B and C and that E1B-55K provides substrate specificity, facilitating the recruitment of p53, and presumably that of Mre11 (7, 33, 53, 54). Since relocalization of p53 to the cytoplasm depends on CRM1-dependent E1B-55K export, E1B-55K might also function as a shuttling factor for the cellular target protein. Mre11 is known to colocalize in the nucleus with E4orf3, which then targets the cellular protein to cytoplasmic aggregates (1, 24, 36). Interestingly, our findings have shown that efficient export of Mre11 requires neither active CRM1 nor the E1B-55K NES, suggesting that Mre11 can access a CRM1- and E1B-55K NES-independent mechanism to exit the nucleus.
Taken together, our findings suggest that the activity associated with the E3 ubiquitin ligase requires a complex pattern of CRM1-dependent and -independent activities of E1B-55K and E4orf6. Although CRM1-dependent export of E1B-55K or E4orf6 makes no contribution to viral late mRNA export, the relocalization and degradation of cellular targets that depend on these viral proteins seem to have distinct requirements. Even though E1B-55K may function as a shuttling factor for at least some cellular substrates through the CRM1 pathway, others may use a separate route. Furthermore, the degradation of different cellular substrates has separate requirements for E1B-55K or E4orf6. This conclusion is consistent with a recent observation suggesting that a more complex mechanism than previously anticipated is required to regulate the activities of E1B-55K and E4orf6, since the E1B-55K-dependent degradation of Daxx does not require E4orf6 (60). Our findings suggest that a complex molecular interplay exists between E1B-55K and E4orf6 to induce selective export of viral late mRNA or relocalization and degradation of cellular targets and that these mechanisms involve more than one cellular export pathway.
ACKNOWLEDGMENTS
We thank Kristoffer Weber for plasmid LeGO-iVLN2/iBLB2.
This work was supported by the Erich and Gertrud Roggenbuck Foundation, Hamburg, Germany. P.W. was supported by a grant from the Studienstiftung des Deutschen Volkes e.V., Ahrstrasse 41, 53175 Bonn, Germany. R.A.G. received a grant from CONACyT-Mexico (SEP-2004-C01-84582) and support from the Alexander Von Humboldt Foundation. The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
Footnotes
Published ahead of print on 11 May 2011.
REFERENCES
- 1. Araujo F. D., Stracker T. H., Carson C. T., Lee D. V., Weitzman M. D. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382–11391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Babiss L. E., Ginsberg H. S., Darnell J. E., Jr 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker A., Rohleder K. J., Hanakahi L. A., Ketner G. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barker D. D., Berk A. J. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107–121 [DOI] [PubMed] [Google Scholar]
- 5. Berk A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685 [DOI] [PubMed] [Google Scholar]
- 6. Beyer W. R., Westphal M., Ostertag W., von Laer D. 2002. Oncoretrovirus and lentivirus vectors pseudotyped with lymphocytic choriomeningitis virus glycoprotein: generation, concentration, and broad host range. J. Virol. 76:1488–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanchette P., et al. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanchette P., et al. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boivin D., Morrison M. R., Marcellus R. C., Querido E., Branton P. E. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyer J., Rohleder K., Ketner G. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307–312 [DOI] [PubMed] [Google Scholar]
- 11. Bridge E., Ketner G. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345–353 [DOI] [PubMed] [Google Scholar]
- 12. Carson C. T., et al. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carter C. C., Izadpanah R., Bridge E. 2003. Evaluating the role of CRM1-mediated export for adenovirus gene expression. Virology 315:224–233 [DOI] [PubMed] [Google Scholar]
- 14. Cheng C. Y., Blanchette P., Branton P. E. 2007. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology 364:36–44 [DOI] [PubMed] [Google Scholar]
- 15. Cutt J. R., Shenk T., Hearing P. 1987. Analysis of adenovirus early region 4-encoded polypeptides synthesized in productively infected cells. J. Virol. 61:543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dallaire F., Blanchette P., Groitl P., Dobner T., Branton P. E. 2009. Identification of integrin α3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dix I., Leppard K. N. 1993. Regulated splicing of adenovirus type 5 E4 transcripts and regulated cytoplasmic accumulation of E4 mRNA. J. Virol. 67:3226–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dobbelstein M., Roth J., Kimberly W. T., Levine A. J., Shenk T. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobner T., Kzhyshkowska J. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25–54 [DOI] [PubMed] [Google Scholar]
- 20. Dosch T., et al. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du J. X., Bialkowska A. B., McConnell B. B., Yang V. W. 2008. SUMOylation regulates nuclear localization of Kruppel-like factor 5. J. Biol. Chem. 283:31991–32002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dull T., et al. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Endter C., Kzhyshkowska J., Stauber R., Dobner T. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. U. S. A. 98:11312–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans J. D., Hearing P. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fallaux F. J., et al. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215–222 [DOI] [PubMed] [Google Scholar]
- 26. Flint S. J., Gonzalez R. A. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287–330 [DOI] [PubMed] [Google Scholar]
- 27. Flint S. J., Huang W., Goodhouse J., Kyin S. 2005. A peptide inhibitor of exportin1 blocks shuttling of the adenoviral E1B 55 kDa protein but not export of viral late mRNAs. Virology 337:7–17 [DOI] [PubMed] [Google Scholar]
- 28. Gonzalez R. A., Flint S. J. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Graham F. L., Smiley J., Russell W. C., Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 30. Groitl P., Dobner T. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol. Med. 130:29–39 [DOI] [PubMed] [Google Scholar]
- 31. Halbert D. N., Cutt J. R., Shenk T. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harada J. N., Shevchenko A., Shevchenko A., Pallas D. C., Berk A. J. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harada N., et al. 2005. E1B-deleted adenovirus replicates in p53-deficient lung cancer cells due to the absence of apoptosis. Oncol. Rep. 14:1155–1163 [PubMed] [Google Scholar]
- 34. Jayaram S., Bridge E. 2005. Genome concatenation contributes to the late gene expression defect of an adenovirus E4 mutant. Virology 342:286–296 [DOI] [PubMed] [Google Scholar]
- 35. Jiang H., et al. 2006. A novel CRM1-dependent nuclear export signal in adenoviral E1A protein regulated by phosphorylation. FASEB J. 20:2603–2605 [DOI] [PubMed] [Google Scholar]
- 36. Karen K. A., Hoey P. J., Young C. S., Hearing P. 2009. Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection. J. Virol. 83:4565–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kindsmüller K., et al. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U. S. A. 104:6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kindsmuller K., et al. 2009. A 49-kilodalton isoform of the adenovirus type 5 early region 1B 55-kilodalton protein is sufficient to support virus replication. J. Virol. 83:9045–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krätzer F., et al. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850–857 [DOI] [PubMed] [Google Scholar]
- 40. Kutay U., Guttinger S. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15:121–124 [DOI] [PubMed] [Google Scholar]
- 41. Kzhyshkowska J., Kremmer E., Hofmann M., Wolf H., Dobner T. 2004. Protein arginine methylation during lytic adenovirus infection. Biochem. J. 383:259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lecane P. S., Kiviharju T. M., Sellers R. G., Peehl D. M. 2003. Leptomycin B stabilizes and activates p53 in primary prostatic epithelial cells and induces apoptosis in the LNCaP cell line. Prostate 54:258–267 [DOI] [PubMed] [Google Scholar]
- 43. Leppard K. N., Shenk T. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lethbridge K. J., Scott G. E., Leppard K. N. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84:259–268 [DOI] [PubMed] [Google Scholar]
- 45. Liu Y., Shevchenko A., Shevchenko A., Berk A. J. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79:14004–14016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marton M. J., Baim S. B., Ornelles D. A., Shenk T. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 64:2345–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naniwa J., et al. 2003. Leptomycin B enhances CDDP-sensitivity via nuclear accumulation of p53 protein in HPV-positive cells. Cancer Sci. 94:1099–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nevins J. R. 1987. Regulation of early adenovirus gene expression. Microbiol. Rev. 51:419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nordqvist K., Ohman K., Akusjarvi G. 1994. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Orlando J. S., Ornelles D. A. 2002. E4orf6 variants with separate abilities to augment adenovirus replication and direct nuclear localization of the E1B 55-kilodalton protein. J. Virol. 76:1475–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ornelles D. A., Shenk T. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pilder S., Moore M., Logan J., Shenk T. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Querido E., et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Querido E., et al. 2001. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rabino C., Aspegren A., Corbin-Lickfett K., Bridge E. 2000. Adenovirus late gene expression does not require a Rev-like nuclear RNA export pathway. J. Virol. 74:6684–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reich N. C., Sarnow P., Duprey E., Levine A. J. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480–484 [DOI] [PubMed] [Google Scholar]
- 57. Rothmann T., Hengstermann A., Whitaker N. J., Scheffner M., zur Hausen H. 1998. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J. Virol. 72:9470–9478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rubenwolf S., Schutt H., Nevels M., Wolf H., Dobner T. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 71:1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sarnow P., Sullivan C. A., Levine A. J. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510–517 [DOI] [PubMed] [Google Scholar]
- 60. Schreiner S., et al. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwartz R. A., et al. 2008. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 82:9043–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stracker T. H., Carson C. T., Weitzman M. D. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352 [DOI] [PubMed] [Google Scholar]
- 63. Weber K., Bartsch U., Stocking C., Fehse B. 2008. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol. Ther. 16:698–706 [DOI] [PubMed] [Google Scholar]
- 64. Weber K., Mock U., Petrowitz B., Bartsch U., Fehse B. 2010. Lentiviral gene ontology (LeGO) vectors equipped with novel drug-selectable fluorescent proteins: new building blocks for cell marking and multi-gene analysis. Gene Ther. 17:511–520 [DOI] [PubMed] [Google Scholar]
- 65. Weigel S., Dobbelstein M. 2000. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J. Virol. 74:764–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65a. Weinberg D. H., Ketner G. 1983. A cell line that supports the growth of a defective early region 4 deletion mutant of human adenovirus type 2. Proc. Natl. Acad. Sci. U. S. A. 80:5383–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Woo J. L., Berk A. J. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamashita T., et al. 1996. Competition between Mg2+ and spermine for a cloned IRK2 channel expressed in a human cell line. J. Physiol. 493 (Pt 1):143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]