Abstract
Low temperature is one of the most important environmental stimuli that control gene transcription programs and development in plants. In Arabidopsis thaliana, the HOS1 locus is a key negative regulator of low temperature-responsive gene transcription. The recessive hos1 mutation causes enhanced induction of the CBF transcription factors by low temperature as well as of their downstream cold-responsive genes. The hos1 mutant plants flower early, and this correlates with a low level of Flowering Locus C gene expression. The HOS1 gene was isolated through positional cloning. HOS1 encodes a novel protein with a RING finger motif near the amino terminus. HOS1 is ubiquitously expressed in all plant tissues. HOS1–GFP translational fusion studies reveal that HOS1 protein resides in the cytoplasm at normal growth temperatures. However, in response to low temperature treatments, HOS1 accumulates in the nucleus. Ectopic expression of HOS1 in wild-type plants causes cosuppression of HOS1 expression and mimics the hos1 mutant phenotypes.
Keywords: Low temperature, signal transduction, HOS1, FLC, RING finger
Plants and other poikilothermic organisms adapt to ambient temperature by adjusting membrane fluidity, metabolism, and gene expression profiles (Murata and Los 1997; Thomashow 1999). Low temperature is one of the most important environmental factors influencing plant distribution, growth, development, and survival. Plants from temperate regions can increase their freezing tolerance by being exposed to low nonfreezing temperatures, a process known as cold acclimation (Guy 1990). Low temperature-induced gene expression is key to cold acclimation (Guy et al. 1985; Thomashow 1999). For example, induction of lipid desaturase genes may contribute to freezing tolerance by altering lipid composition (Gibson et al. 1994). Cold-induced molecular chaperones may help prevent protein denaturation during freezing (Anderson et al. 1994). Many cold-induced genes encode novel proteins, some of which have been shown to discourage the formation of deleterious membrane structures during freezing (Steponkus et al. 1998). Ectopic expression of CBF transcription factors has been shown to induce a set of cold-responsive genes at warm temperatures and to mimic cold acclimation in terms of increases in freezing tolerance (Jaglo-Ottosen et al. 1998; Liu et al. 1998).
Many of the cold-regulated genes can also be induced by the phytohormone abscisic acid (ABA) or by osmotic stress treatment (Gilmour et al. 1992; Kurkela and Borg-Franck 1992; Nordin et al. 1993). Low temperature and osmotic stresses appear to have several features in common. Freezing injury is largely a consequence of freezing-induced dehydration stress. Low nonfreezing temperatures can also lead to water stress by impairing water absorption by the roots and water transport to the shoot (Levitt 1980). ABA is known to play an important role in cold acclimation, and treatment with ABA at normal growth temperatures increases the freezing tolerance of a wide range of plants (Chen et al. 1983). Furthermore, exposure to low temperature transiently increases ABA levels (Chen et al. 1983).
Cold-responsive genes such as RD29A (also known as COR78 or LTI78) and COR15A are known to contain two cis-regulatory elements in their promoters, that is, the ABRE and DRE/CRT elements (Yamaguchi-Shinozaki and Shinozaki 1994; Stockinger et al. 1997). The ABRE element is responsive to ABA, whereas the DRE/CRT element mediates ABA-independent regulation of gene expression by cold or hyperosmotic stress. Transcription factors termed CBFs or DREBs that bind to DRE/CRT are induced early by cold or osmotic stress (Stockinger et al. 1997; Gilmour et al. 1998; Liu et al. 1998). Constitutive or inducible overexpression of some of the transcription factors leads to enhanced expression of downstream cold responsive genes and increased plant tolerance to freezing, drought, and salt stresses (Jaglo-Ottosen et al. 1998; Liu et al. 1998; Kasuga et al. 1999).
Besides the CBF transcription factors, other potential cold signal transduction components in plants include SFR6, ESK1, HOS1, and HOS2, which are defined by Arabidopsis mutations. The molecular identities of these genetic loci await molecular cloning. The sfr6 mutation suppresses cold and ABA induction of genes with the CRT/DRE promoter element (Knight et al. 1999). The esk1 mutants are constitutively freezing-tolerant but are apparently not affected in the expression of genes with the CRT/DRE element, suggesting that ESK1 may be involved in a distinct pathway related to freezing tolerance (Xin and Browse 1998). HOS1 and HOS2 are both negative regulators of cold signaling pathways (Ishitani et al. 1998; Lee et al. 1999). Mutations in HOS1 or HOS2 lead to enhanced cold induction of genes such as RD29A, COR15A, KIN1, and ADH. Without cold acclimation, hos1 mutant plants have reduced capacity for freezing tolerance. After cold acclimation, hos1 plants acquire as much freezing tolerance as do wild-type plants (Ishitani et al. 1998).
hos1 mutant plants flower considerably earlier, which may be related to vernalization, another important plant process regulated by low temperature (Ishitani et al. 1998). Vernalization refers to the promotion of flowering by prolonged periods of low temperature exposure. The early flowering phenotype of hos1 mutant plants appears to be a manifestation of constitutive vernalization (Ishitani et al. 1998). This is because (1) hos1 plants have a reduced threshold of low temperature activation of cold-responsive genes, that is, cold signaling happens even at normal growth temperatures in the mutant plants (Ishitani et al. 1998); and (2) nonvernalized hos1 mutant plants flower as early as vernalized wild-type plants (Ishitani et al. 1998). hos1 mutant plants can be further vernalized by low temperature treatment to flower even earlier.
To address the mechanisms of cold-regulated gene expression and plant development, a necessary step is to determine the molecular nature of the key regulatory genes involved. Toward this goal, we report here the map-based cloning and molecular analysis of HOS1, the first such genetic locus involved in cold-regulated plant development and signaling to be characterized at the molecular level. The sequence data reveal that HOS1 encodes a novel RING finger protein. Studies with HOS1–GFP fusion protein show that HOS1 displays cold-regulated nucleo–cytoplasmic partitioning, suggesting an important role of HOS1 may be to communicate cold-generated signals in the cytoplasm to the nucleus to regulate the level of gene transcription.
Results
Expression of CBF transcription factor genes in hos1 mutant plants
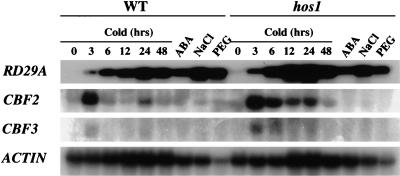
Transcripts for the CBF/DREB1 transcription factors are rapidly induced by low temperature treatment (Gilmour et al. 1998; Liu et al. 1998; Medina et al. 1999). These transcription factors in turn activate the expression of downstream COR/RD/KIN genes such as COR15A, COR47, RD29A, and KIN1. We have shown previously that these COR/RD/KIN genes are superinduced by cold in hos1 mutant plants (Ishitani et al. 1998). Northern analysis was carried out to determine whether CBF genes are also superinduced by cold in the hos1 mutant. Both CBF2 and CBF3 were found to exhibit enhanced cold induction in hos1 mutant plants (Fig. 1). At 3 h after cold treatment, CBF2 expression in hos1 mutants is approximately 50% higher than in wild-type plants. Increased CBF2 and CBF3 expression is more pronounced after longer cold treatment, for example, at 6, 12, 24, and 48 h (Fig. 1). We did not observe significant cold induction of CBF1 in either the wild-type or hos1 mutant plants (not shown).
Figure 1.
The CBF transcription factor genes are expressed at higher levels in hos1 mutant plants in response to cold treatments. Total RNA (20 μg per lane) was isolated from wild-type and hos1 mutant seedlings treated with cold (0°C for the indicated time periods), ABA (100 μM, 3 h), NaCl (300 mM, 5 h), or PEG (30%, 5 h). The RNA blot was probed with 32P-labeled RD29A, CBF2, CBF3, and actin cDNA probes, with the actin used as a loading control.
The hos1 mutation reduces expression of the Flowering Locus C gene
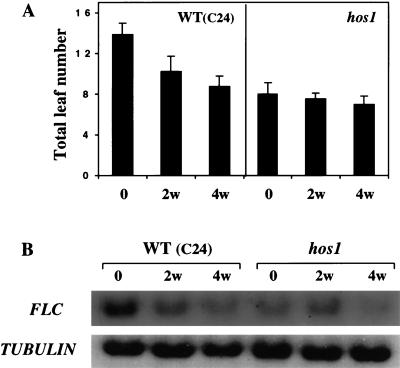
The Flowering Locus C (FLC) gene that encodes a MADS-box protein is a central regulator of flowering induction by vernalization (Michaels and Amasino 1999; Sheldon et al. 2000). The level of FLC determines the extent of the vernalization response in the promotion of flowering: The duration of cold treatment correlates with the extent of down-regulation of FLC expression (Sheldon et al. 2000). Because the hos1 mutant plants flower early (Ishitani et al. 1998) and appear to be constitutively vernalized (Fig. 2A), we carried out Northern analysis to determine whether FLC expression is altered in the mutant. Consistent with previous studies (Sheldon et al. 2000), the FLC transcript level is high in wild-type C24 plants, but it decreases significantly as the plants become vernalized by cold treatments (Fig. 2B). Interestingly, hos1 mutant plants have a substantially lower level of FLC expression even in the absence of vernalization treatment (Fig. 2B). This lower level of FLC expression correlates with the early flowering phenotype of the hos1 mutant plants (Fig. 2A). After 2 wk of vernalization treatment, the FLC level in hos1 mutant plants is not reduced. After 4 wk of vernalization, the FLC level decreases slightly (Fig. 2B), as does the flowering time (Fig. 2A). As a control, the tubulin gene transcript levels are not different between hos1 and the wild type, and are not changed by vernalization (Fig. 2B). These results suggest that wild-type HOS1 is a negative regulator of vernalization and that it is required for a high level of FLC expression.
Figure 2.
The early flowering phenotype and reduced expression of the vernalization regulator FLC in hos1 mutant plants. (A) Flowering time of C24 wild-type and hos1 mutant plants with and without 2 or 4 wk of vernalization treatment at 0°C. Flowering time is indicated by the total number of rosette leaves at time of flower bud emergence. Shown are the average and standard deviation (n = 15). (B) FLC expression. Total RNA (20 μg per lane) was isolated from wild-type and hos1 mutant seedlings grown in agar plates that were incubated at 0°C for 0, 2, and 4 wk. The tubulin gene was used as a loading control.
Positional cloning of HOS1
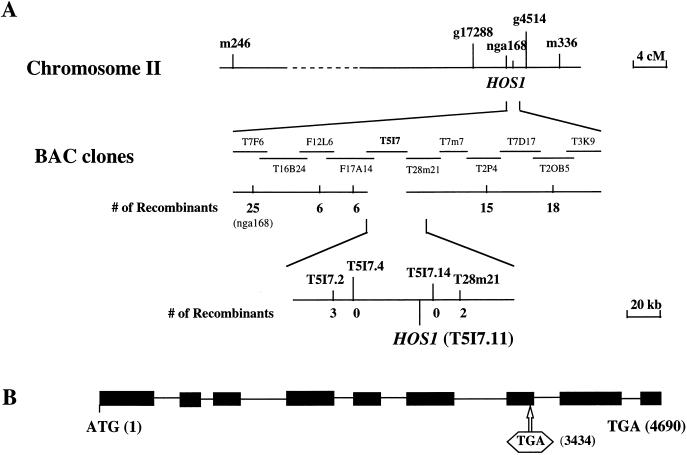
To map hos1 genetically, homozygous hos1 mutant plants in the C24 ecotype were crossed with wild-type plants of the Columbia ecotype. The resulting F1 plants were selfed, and homozygous hos1 mutant plants were selected from the segregating F2 population by luciferase imaging after 2 d of treatment at 0°C. Surveying of representative molecular markers from each of the five Arabidopsis chromosomes located hos1 to chromosome II. Further analysis showed that hos1 is closely linked with the SSLP (simple sequence length polymorphism) marker nga168. Several new SSLP markers were developed based on the genomic sequences of BAC clones in the nga168 region. Fine mapping using these new markers delimited HOS1 to the BAC clone T5I7 (Fig. 3). Candidate open reading frames on T5I7 were sequenced from wild-type as well as hos1 mutant plants. The sequence analysis revealed a single nucleotide substitution in the hypothetical T5I7.11 gene in the hos1 mutant. This mutation is predicted to create a premature stop codon, resulting in a truncated protein.
Figure 3.
Positional cloning of the HOS1 gene. (A) Physical mapping of HOS1. Genetic mapping delimited HOS1 to the BAC clone T5I7. The hos1 mutation was identified by sequencing and comparing all predicted genes on this BAC from hos1 mutant and wild-type plants. (B) Structure of HOS1 and the position of the hos1 mutation. Positions are relative to the initiation codon. Filled boxes indicate the open reading frame, and lines between boxes indicate introns. The premature stop codon created by the hos1 mutation is boxed.
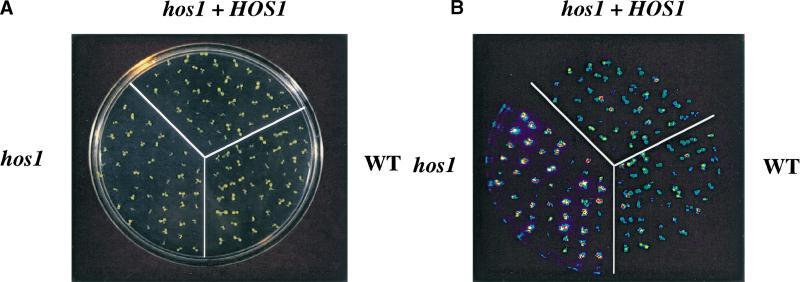
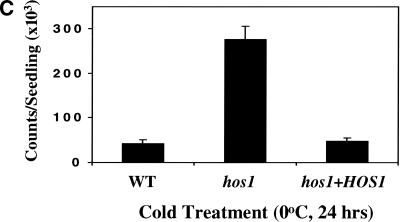
To confirm that we have cloned the correct gene, the T5I7.11 sequence including 2807 bp upstream of the predicted ATG start codon was amplified from wild-type plants and cloned into the binary vector pCIT20. The construct was introduced into hos1 mutant plants via Agrobacterium-mediated transformation. Transformants were selected based on hygromycin resistance; the progenies of two transformants were tested by luciferase imaging after 2 d of cold treatment. Both hygromycin resistance and cold-induced RD29A–LUC expression segregated in the T2 generation. All hygromycin-resistant plants exhibited the wild-type phenotype in terms of cold-induced RD29A–LUC expression and time to flower. Figure 4A–C shows an example of complemented hos1 mutant plants in the T3 generation. In response to cold treatment, hos1 mutant plants exhibited brighter luminescence, indicating a higher level of RD29A–LUC expression compared with the wild type (Fig. 4B). In contrast, luminescence intensities from hos1 mutant seedlings containing the introduced wild-type HOS1 gene were similar to those in wild-type plants (Fig. 4B). Quantification of the luminescence intensities confirmed that the mutant was fully complemented by the wild-type HOS1 gene (Fig. 4C). The wild-type HOS1 transgene also rescued all the other phenotypes exhibited by hos1 mutant plants, such as pale, small leaves and early flowering (data not shown).
Figure 4.
Complementation of hos1 mutant by the wild-type HOS1 gene. Ten-day-old wild-type seedlings, hos1, and homozygous T3 progenies of a hos1 plant containing the wild-type HOS1 transgene (designated as hos1 + HOS1) grown in agar medium were incubated at 0°C for 24 h before the bioluminescence image was taken. (A) A picture of wild-type, hos1, and T3 progenies of hos1 plants containing the HOS1 complementation construct. (B) Bioluminescence image of plants shown in A. (C) Quantification of the luminescence intensities in B.
HOS1 encodes a novel RING finger protein
The transcribed sequence of the HOS1 gene was determined by sequencing cDNAs obtained by the reverse transcriptase polymerase chain reaction (RT–PCR). Comparison of the cDNA with the genomic sequence showed that the HOS1 gene contains nine exons and eight introns (Fig. 3B). HOS1 is predicted to encode a polypeptide of 915 amino acids, with an estimated molecular mass of 104 kD (Fig. 5). Database searches revealed that HOS1 contains a short stretch of amino acids near the amino terminus that are similar to the RING finger domain found in a group of proteins known as IAPs (inhibitor of apoptosis) (Fig. 6; Miller 1999). The rest of HOS1 does not show significant homology to any other sequences in the GenBank database. The hos1 mutation, a G → A substitution, is predicted to create a premature stop codon that truncates the protein after His 643. This suggests that the carboxy-terminal one-third of HOS1 is functionally essential.
Figure 5.
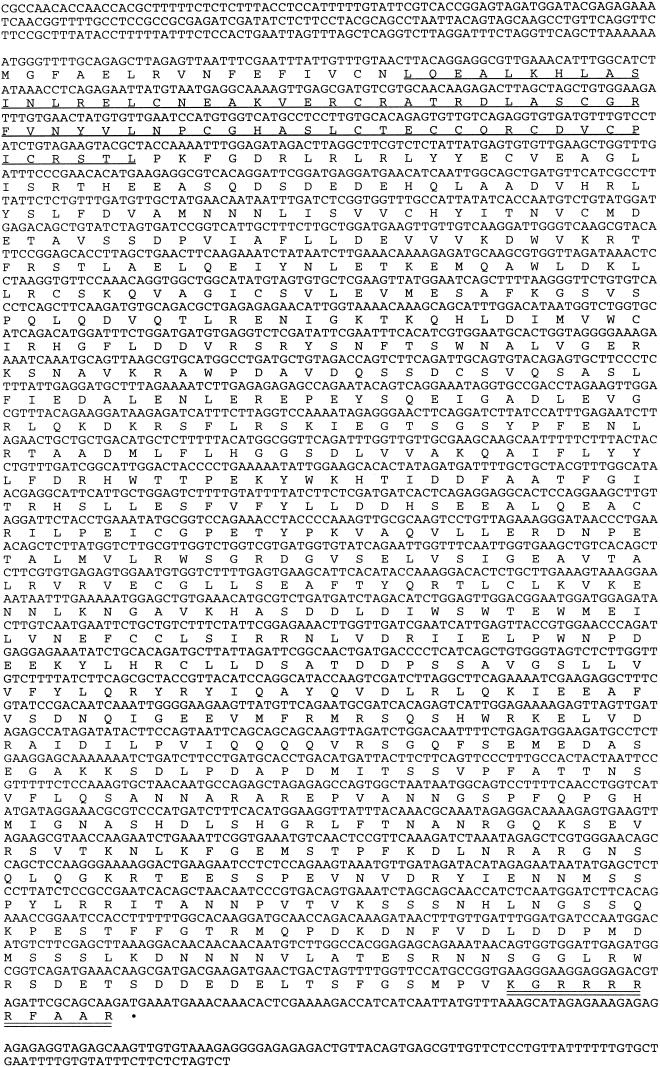
. The HOS1 cDNA sequence and the conceptual translational product of its longest ORF (GenBank accession no. AAB87130). The RING finger domain is underlined, and a putative nuclear localization signal is double-underlined.
Figure 6.
Alignment of the RING finger domain of HOS1 with those of animal IAPs. Amino acid residues in black indicate identical matches, and those in gray indicate conserved substitutions. The conserved Cys and His residues of the RING finger are indicated by asterisk marks. A missing Cys residue in HOS1 is indicated by a question mark. GeneBank accession numbers for various IAPs are as follows: Human IAP, AAC83232; Pig IAP, AAC39271; Mouse IAP, AAC42078; Rat IAP, AAG22971; HOS1, AAB87130.
The RING finger is a small zinc-binding domain found in many proteins with diverse functions. Recent studies have shown that proteins containing a RING finger may act as E3 ubiquitin protein ligases, and that the RING finger can interact with E2 ubiquitin conjugating enzymes, thereby promoting ubiquitination of specific target proteins (for review, see Joazeiro and Weissman 2000). The RING finger motif is defined as a series of conserved cysteine and histidine residues: Cys-X2-Cys-X9–39-Cys-X1–3-His-X2–3-Cys/His-X2-Cys-X4–48-Cys-X2-Cys, where X can be any amino acid (Saurin et al. 1996). The RING finger of HOS1 is most similar to that of IAPs, which can be classified as a C3HC4-type RING (Miller 1999). Interestingly, the first Cys in the RING finger of HOS1 is replaced with Leu (Fig. 6). RING variants have also been seen in other well-known RING finger proteins (Saurin et al. 1996). For example, in mouse MDM2, Cys3 is replaced by a Thr residue, whereas in CART1, Cys7 is substituted for an Asp residue (Saurin et al. 1996).
HOS1 expression in Arabidopsis seedlings
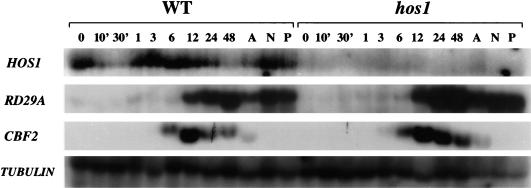
Northern analysis was carried out to examine HOS1 transcript abundance in Arabidopsis seedlings subjected to various treatments. In wild-type plants, HOS1 mRNA was detected in unstressed control as well as cold-, ABA-, NaCl-, or PEG-treated samples (Fig. 7). HOS1 transcript abundance was not substantially influenced by ABA, NaCl, or PEG treatment. In response to cold treatment, HOS1 expression declined quickly and transiently. At 30 min after cold treatment, HOS1 expression was almost undetectable (Fig. 7). However, by 1 h after cold treatment, HOS1 expression recovered to pretreatment levels. This rapid and transient reduction of HOS1 message in response to brief cold treatment was consistently observed in independent experiments. After recovery from the decline, HOS1 expression was then maintained at this level until 2 d after cold treatment, before declining again (Fig. 7).
Figure 7.
HOS1 transcript levels in wild-type and hos1 mutant plants subjected to various stress treatments. Wild-type and hos1 mutant 3-week-old seedlings were incubated at 0°C for 0 min, 10 min, 30 min, 1 h, 3 h, 6 h, 12 h, 24 h, or 48 h. Seedlings were also treated with ABA (A: 100 μM, 3 h), NaCl (N: 300 mM, 5 h), and PEG (P: 30%, 5 h). Total RNA was extracted, and 20 μg RNA was loaded into each lane. The blots were hybridized with 32P-labeled HOS1, RD29A, CBF2, and β-tubulin cDNA probes. The β-tubulin gene was used as a loading control.
Very low levels of HOS1 transcript were detected in hos1 mutant plants subjected to the same treatments as were applied to wild-type plants (Fig. 7). The low level of HOS1 expression is probably attributable to active degradation of the mutant mRNA, since it is known that mRNA surveillance mechanisms degrade transcripts with premature stop codons (Hilleren and Parker 1999). To verify that the stress treatments were appropriate, the RNA blot was also probed with RD29A and CBF2. The results show that the two genes were up-regulated with the appropriate kinetics, that is, CBF2 induction occurred earlier than RD29A (Fig. 7), although not as early as the CBF2 induction seen in some other experiments (Fig. 1). The results also confirm that CBF2 as well as RD29A are expressed at higher levels in hos1 mutant plants under cold treatment. Hybridization with a β-tubulin cDNA probe showed RNA loading in stressed and in control unstressed sample lanes (Fig. 7).
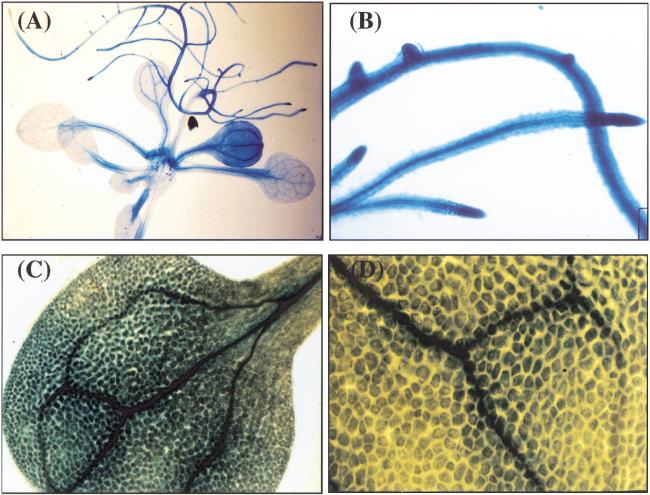
To examine HOS1 expression in various plant tissues, 2.78 kb of the genomic sequence upstream of the HOS1 open reading frame (−2807 to −23 from ATG) was fused with the β-glucuronidase reporter gene (GUS), and the resulting construct was introduced into wild-type Arabidopsis plants. GUS expression was observed in all plant tissues examined (Fig. 8). In leaves, the expression was stronger in the vasculature (Fig. 8C), but could be seen in mesophyll tissues as well (Fig. 8D). Strong expression was observed in roots (Fig. 8A), with root tips having especially high levels (Fig. 8B).
Figure 8.
Ubiquitous expression of HOS1 in Arabidopsis seedlings. Fifteen-day-old transgenic plants containing the HOS1 promoter–GUS construct were analyzed for GUS expression. (A) Whole seedling, (B) roots, (C) leaf, and (D) close-up of part of a leaf.
HOS1 accumulates in the nucleus in response to cold treatment
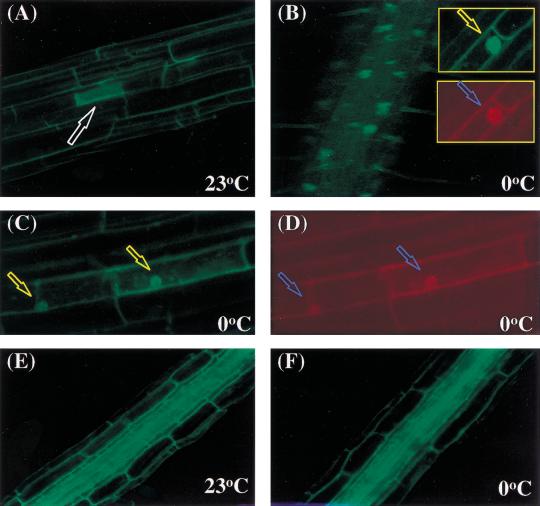
Knowledge of the subcellular localization of a protein is crucial to the understanding of its cellular function. To determine the subcellular localization of HOS1 gene product, HOS1 cDNA was fused in-frame at its C terminus with the GFP (green fluorescence protein) marker. HOS1–GFP fusion protein was expressed under the CaMV 35S promoter in transgenic Arabidopsis plants. The subcellular localization of HOS1–GFP was determined by green fluorescence imaging under a confocal microscope. Without cold treatment, only very faint green fluorescence signals slightly above background (i.e., nontransformed plants) could be detected in the majority of cells (Fig. 9A). This suggests that HOS1–GFP was either expressed at a very low level or was being rapidly degraded at normal growth temperatures. Occasionally, a few cells were found to express a high level of HOS1–GFP. In these cells, the green fluorescence was entirely cytoplasmically localized (Fig. 9A, indicated by arrow). Amazingly, when the HOS1–GFP transgenic plants were placed at 0°C for 1–2 d, green fluorescence could be seen in structures consistent with the nuclei (Fig. 9B). To verify that the structures emitting green fluorescence were nuclei, the seedling roots were stained with propidium iodide. The results show that the green fluorescence-emitting organelles were stained with propidium iodide, suggesting that the organelles are, indeed, nuclei (Fig. 9B, inserts). The occasional cells expressing high levels of cytoplasmic HOS1–GFP at normal temperatures also showed strong nuclear HOS1–GFP expression after cold treatment (Fig. 9C,D).
Figure 9.
Cold treatment alters HOS1–GFP localization in Arabidopsis seedlings. (A) Cytoplasmic localization of HOS1–GFP fusion protein in root cells without cold treatment. T2 10-day-old seedlings containing the HOS1–GFP translational fusion construct grown on MS agar plates in the dark were analyzed for GFP expression under confocal microscope. The arrow points to a cell with strong cytoplasmic green fluorescence. (B) Nuclear localization of HOS1–GFP after cold treatment. Seedlings were incubated at 0°C for 48 h before GFP analysis. The spots emitting green fluorescence correspond to nuclei as confirmed by propidium iodide staining (red stain in insert). Arrows point to nuclei. (C) Those cells expressing strong cytoplasmic GFP at normal growth temperatures show strong nuclear GFP after cold treatment. Arrows point to nuclei. (D) Cells in C were stained with propidium iodide. Arrows point to nuclei. (E,F) Cytoplasmic GFP localization in the roots of transgenic Arabidopsis expressing an acidic ribosomal protein–GFP fusion (Cutler et al. 2000) either before (E) or after 48 h of cold treatment (F).
The nuclear accumulation of HOS1–GFP fusion protein was clearly visible after plants were subjected to 0°C treatment for 20 h. Longer cold treatment did not change the intensity of the nuclear green fluorescence. After the cold-treated plants were returned to normal growth temperatures (18°–23°C) for more than 12 h, the nuclear green fluorescence disappeared, suggesting that the fusion protein moved back to the cytoplasm and/or was rapidly degraded.
To determine whether the cold-induced nuclear accumulation is specific to HOS1–GFP, we treated transgenic Arabidopsis plants containing a known cytoplasmically localized small acidic ribosomal protein–GFP fusion (line Q1; Cutler et al. 2000) at 0°C for 48 h. Only cytoplasmically localized green fluorescence was detected in these control plants either at 23°C or 0°C (Fig. 9E,F). These results show that cold treatment does not induce a general nonspecific nuclear accumulation of GFP fusion proteins.
Cosuppression of HOS1 expression in wild-type plants mimics hos1 mutant phenotypes
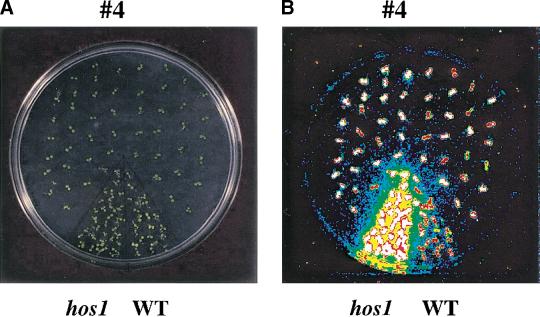
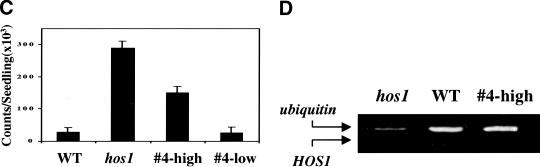
The HOS1 open reading frame was overexpressed in wild-type plants under the CaMV 35S promoter to determine whether increased HOS1 expression would down-regulate RD29A–LUC regulation and affect plant tolerance to chilling or freezing stress. Among 25 independent transgenic plant lines tested in the T2 and T3 generations, none showed reduced RD29A–LUC induction by cold treatments compared to wild type (data not shown). We also did not find any transgenic lines with altered chilling or freezing tolerance. Interestingly, there were two transgenic lines that showed higher than wild-type levels of RD29A–LUC expression in response to cold treatment. This phenotype segregated in the T2 generation, with an ∼3:1 ratio of the high expression to regular expression plants. The RD29A–LUC phenotype of one of the lines, #4, is shown in Figure 10A–C. The high expression plants were all found to carry the 35S–HOS1 transgene according to their hygromycin resistance.
Figure 10.
Ectopic expression of HOS1 causes silencing of HOS1 expression and mimics the hos1 mutant phenotypes. Ten-day-old wild-type seedlings (WT), hos1, and T2 progenies of a wild-type plant containing the 35S–HOS1 construct (designated as line #4) were incubated at 0°C for 24 h before the bioluminescence image was taken. (A) Picture of wild-type, hos1, and T2 progenies of transgenic line #4. (B) Bioluminescence image of plants in A. (C) Quantification of the luminescence intensities in B. Shown are the averages and standard deviations (n = 15). T2 progenies of the #4 line were segregated into two groups, which are designated #4-high and #4-low, corresponding to those exhibiting high expression and low expression of RD29A–LUC, respectively. (D) Reduced HOS1 transcript abundance in the #4 cosuppression plants, as determined by quantitative RT–PCR.
The increased RD29A–LUC expression in these 35S–HOS1 transgenic lines mimics the phenotype of the hos1 mutant plants, although the luciferase expression level was not as high as in the mutant (Fig. 10C). The strong luminescence phenotype of these transgenic lines is probably a result of posttranscriptional cosuppression of HOS1 expression, because semiquantitative RT–PCR tests revealed a decreased amount of HOS1 transcript in the high expression plants derived from the #4 line (Fig. 10D).
Discussion
Low temperature is an important environmental signal for all living organisms. Two well-known plant physiological processes are associated with the low temperature cue, namely, cold acclimation and vernalization. Cold acclimation refers to the phenomenon that plants from temperate regions gain freezing tolerance by preexposure to low nonfreezing temperatures (Guy 1990; Thomashow 1994). Vernalization is a plant-specific process whereby prolonged exposure to low temperature accelerates flowering (Chandler et al. 1996). HOS1 is the only gene known to regulate both cold acclimation and vernalization (Ishitani et al. 1998). For cold acclimation, HOS1 negatively regulates the expression of cold-responsive genes that are known to contribute to cold acclimation. CBF/DREB transcription factor genes are induced by cold at higher levels in the hos1 mutant (Figs. 1 and 7). This is probably responsible for higher levels of cold induction of RD29A, COR47, COR15A, and KIN1 in the mutant because these later genes are controlled by the CBF/DREB transcription factors (Stockinger et al. 1997; Liu et al. 1998). However, the hos1 mutation seems to also affect genes outside the CBF regulon. This is suggested by the effect of the hos1 mutation on cold induction of the alcohol dehydrogenase gene (Ishitani et al. 1998), which does not appear to be under control of CBF transcription factors because its promoter is not known to contain the DRE/CRT element. These observations indicate that HOS1 functions early in cold signal transduction. An early role of HOS1 in cold signaling is consistent with its involvement in vernalization, that is, FLC expression and flowering time control.
Molecular cloning of HOS1 revealed that it encodes a novel protein with a modified RING finger domain. The RING finger motif was first identified in the human Really Interesting New Gene 1 (RING1) product (for review, see Freemont 2000). Several hundred proteins from diverse eukaryotes have been found to contain this motif or its variants. RING finger proteins include some of the most important regulators of human diseases and various cellular signaling pathways, for example, BRCA1, a product of a breast cancer-associated gene; the protooncogene products Cbl, BMI-1, and PML; and the p53-regulator MDM2 (for reviews, see Freemont 2000; Jackson et al. 2000; Joazeiro and Weissman 2000). However, no specific function was ascribed to the RING finger until recently. In the past two years, a number of RING finger proteins have been found to have E3 ubiquitin ligase activities, that is, they mediate the transfer of ubiquitin to heterologous substrates and/or to the RING finger proteins themselves. The RING finger sequence of HOS1 is most similar to those found in the family of antiapoptotic molecules in animals known as IAPs (Fig. 6). IAPs were recently shown to catalyze their own ubiquitination, and this ubiquitination activity is dependent on the RING domain (Yang et al. 2000).
It is now becoming abundantly clear that the stability of critical regulatory proteins in the cell is dynamically controlled in response to environmental or developmental stimuli (Joazeiro and Weissman 2000). Because protein degradation must be highly selective in order for the cell not to cannibalize itself, the substrate-recognition step mediated by the RING finger E3 ubiquitin ligases is of critical importance. The existence of a large number of RING finger proteins may thus reflect the need to selectively target many different substrate proteins for degradation.
No RING finger protein has been previously reported to function in low temperature signal transduction. Thus, our results provide the first evidence that a RING finger protein is also important in regulating gene transcription and plant development in response to cold environments. Future studies will determine whether HOS1 can serve as an E3 ubiquitin ligase and what types of substrate proteins might be specifically targeted by HOS1 for degradation. Because HOS1 negatively regulates CBF transcript levels, it is tempting to speculate that HOS1 might control the turnover of ICE, the proposed inducer of CBF expression (Thomashow 1999). The Arabidopsis COP1 is a well-known RING finger protein that negatively regulates light-responsive gene expression and photomorphogenesis (Deng et al. 1992). It has been shown recently that COP1 functions by controlling the degradation of HY5, a transcription factor that activates light-regulated gene expression (Osterlund et al. 2000).
The analogy with COP1 does not stop at the RING finger motif or negative regulation of gene transcription. The HOS1 and COP1 RING finger proteins both exhibit nucleo–cytoplasmic partitioning in response to environmental stimuli. COP1 is localized in the nucleus in the dark and translocates to the cytoplasm in response to light signals (von Arnim and Deng 1994). HOS1 accumulates in the nucleus in response to cold environments (Fig. 9). To our knowledge, HOS1 represents the first example of cold-induced nucleo–cytoplasmic partitioning of proteins. Our findings provide a connection between cold-triggered cytoplasmic signaling and nuclear gene transcription. Two histidine kinases and a response regulator were recently shown to contribute to cold perception and signaling in the cyanobacterium Synechocyctis (Suzuki et al. 2000). It is known that the second-messenger calcium and protein phosphorylation are involved in early events of cold signaling in plant cells (Monroy and Dhindsa 1995; Jonak et al. 1996; Knight et al. 1996; Monroy et al. 1998). How these cytoplasmic events relay the cold signal to the nucleus, where cold-induced gene transcription occurs, is unknown. Perhaps the nucleo–cytoplasmic movement of HOS1 contributes to the communication between the cytoplasm and the nucleus under cold conditions.
It is a matter of speculation as to the mechanism of the nuclear accumulation of HOS1 in response to cold treatment. There is a putative nuclear localization sequence in HOS1 (Fig. 5). Whether this sequence is required or sufficient for HOS1 accumulation in the nucleus remains to be determined. It is possible that the nuclear translocation of HOS1 is triggered by phosphorylation changes in this protein. Alternatively, HOS1 may be cotranslocated to the nucleus by binding to another protein that is destined to accumulate in the nuclear compartment in response to cold treatments.
It is interesting that HOS1 transcript abundance is transiently down-regulated by cold shock (Fig. 7). Because the genetic role of HOS1 is to attenuate cold signaling, perhaps the transient down-regulation of HOS1 transcript is important to allow cellular cold signals to be amplified. Our observation that cosuppression of HOS1 mimics hos1 mutant phenotypes (Fig. 10) indicates an inverse correlation between HOS1 transcript level and the expression of cold-responsive RD29A. It will be of interest to determine whether natural plant variations in the extent of cold acclimation and vernalization might be correlated with their expression levels of HOS1 or its orthologs.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana plants of the C24 ecotype expressing the RD29A promoter–Luciferase (RD29A–LUC) transgene (referred as wild type herein) were obtained by Agrobacterium transformation (Ishitani et al. 1997). The methodology of mutant screening and characterization were described previously (Ishitani et al. 1997, 1998). For bioluminescence image analysis, wild-type and hos1 mutant seeds were surface-sterilized and sown onto Petri plates containing Murashige and Skoog salts (MS salts) supplemented with 3% sucrose and solidified with 0.6% agar. Seedlings were grown at 23°C with cycles of 16 h light and 8 h darkness before stress treatments. For cold treatment, 1-wk-old seedlings in agar plate were incubated at 0°C (±0.1°C) for 48 h in the dark. For ABA treatment, 100 μM ABA (mixed isomers) in water was sprayed on leaves, and the plants were incubated at room temperature (∼23°C) under light. For NaCl or PEG treatment, seedlings were placed on 3M filter paper saturated with MS salt solution supplemented with 300 mM NaCl or 30% PEG6000 and incubated under light at room temperature. After indicated times of incubation, bioluminescence images were taken as described (Ishitani et al. 1997).
For measuring flowering time in hos1 and the wild-type, plants were grown in pots in a growth chamber under the conditions as described above. At the emergence of the first flower, the total number of rosette leaves was counted (n = 15).
Positional cloning
To generate the mapping population for HOS1, hos1 mutant plants were crossed to wild-type Arabidopsis plants of the Columbia ecotype. A total of 586 hos1 mutant plants were selected from the segregating F2 population based on high luminescence expression of RD29A–LUC after cold treatment. Genomic DNA from these plants was extracted and analyzed for cosegregation with respect to SSLP (simple sequence length polymorphism) markers. These markers were developed by searching for simple repeat sequences with the RepeatMasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). Primer pairs covering the repeat regions were designed and used for PCR amplification of the genomic fragments from both C24 and Col ecotypes. Those that exhibited PCR fragment length polymorphisms were used as markers for mapping. Primer pairs for fine mapping of HOS1 are as follows: F12L6-F, 5′-CACGCT TATTCTTAGCCTAG-3′; F12L6-R, 5′-TTGAAAGATCACGC CGGCGA-3′; F17A14-F, 5′-GGAGAATGAAAGACCGAATC-3′; F17A14-R, 5′-TGTAGAGACATGAGAATCT-3′; T28M21-F, 5′-TAGGGGGATATTCAAGTTTG-3′; T28M21-R, 5′-AAT TTCCCCACTCCACTCCT-3′; T2P4-F, 5′-GTATTCGATCTT CTTTAGGT-3′; T2P4-R, 5′-AGGAGTATACGTCCAATTGA-3′; T20B5-F, 5′-GAAGAGCAACGCATTACAGT-3′; T20B5-R, 5′-CTTCATAAATAACGAGGCCT-3′. In addition, nucleotide differences between C24 and Col ecotypes were also identified by direct sequencing of the open reading frames of T5I7.2, T5I7.4, and T5I7.14, and were used for fine mapping.
RNA analysis
Seedlings grown for 15 d on MS agar plates under continuous light were treated with low temperature, ABA, NaCl, or PEG as described above. Total RNA from control or stressed plants was extracted and analyzed as described by Liu and Zhu (1997). The RD29A gene-specific probe was from the 3′ noncoding region (Liu and Zhu 1997). The CBF2 and CBF3 gene-specific probes were generated by PCR with the following primer pairs: CBF2-F, 5′-TTCGATTTTTATTTCCATTTTTGG-3′; CBF2-R, 5′-CCA AACGTCCTTGAGTCTTGAT-3′; CBF3-F, 5′-TAAAACTCA GATTATTATTTCCATTT-3′; CBF3-R, 5′-GAGGAGCCACG TAGAGGGCC-3′. The FLC gene-specific probe was obtained by PCR with the following primer pair: FLC-F, 5′-CCGCTC GAGCTTAGTATCTCCGGCG-3′; FLC-R, 5′-GGACTAGTC GCCCTTATCAGCGGA-3′. The β-tubulin gene was amplified by PCR with the following primer pair: Tubulin-5′, 5′CGTG GATCACAGCAATACAGAGCC-3′; Tubulin-3′, 5′-CCTCCT GCACTTCCACTTCGTCTTC-3′. It was used for the loading control.
hos1 mutant complementation
For complementation of hos1 mutant plants, T5I7 BAC (bacterial artificial chromosome) DNA was digested with XhoI, and the fragment corresponding to HOS1 was cloned into the binary vector pCIT20 (Ma 1992). This and all other constructs described in this paper were completely sequenced to ensure that they did not contain PCR errors. The construct was transformed into Agrobacterium strain GV3101 by electroporation. hos1 mutant plants were vacuum-infiltrated with the Agrobacteria containing the HOS1 complementation construct. Hygromycin-resistant transgenic plants were selected, and their T2 and T3 progenies were subjected to luminescence imaging analysis for RD29A–LUC expression in response to cold, ABA, and NaCl treatments as described above.
Analysis of HOS1 promoter–GUS expression
The promoter region (2.78 kb upstream from the initiation codon) of the HOS1 gene was PCR-amplified with the following primer pair: HOS1–GUS-F1, 5′-CGCAAGCTTCTCGAGGTAA GACCGTAGATTCA-3′; HOS1–GUS-R1, 5′-CGGTCTAGAC CTAAGACCTGAGCTAAACTAAT-3′. The resulting fragment was digested with HindIII and XbaI and inserted into the pIG121 binary vector. This HOS1 promoter–GUS construct was introduced into Agrobacterium strain GV3101 and transformed into wild-type Arabidopsis, and 20 independent lines of hygromycin-resistant transgenic plants were obtained. For GUS staining, T2 seedlings grown on MS agar plate were incubated with X-Gluc for 12 h at 37°C and then washed 5 times with 70% (v/v) ethanol at 70°C to remove chlorophyll.
Expression and localization of HOS1–GFP fusion protein
The coding region of the HOS1 gene was obtained by RT-PCR using the following primers: HOS1BAMHF, 5′-CGCGGATC CATGGGTTTTGCAGAGCTTAGAGTTAA-3′; HOS1BAMHR, 5′-CGCGGATCCGCGTTCTTGCTGCGAATCTACGTCTCC3′. The resulting PCR fragment was digested with BamH1 and inserted into the binary vector pBIN35S-mGFP4 downstream from the CaMV 35S promoter. Agrobacterium strain GV3101 containing this HOS1–GFP translational fusion was introduced into Arabidopsis (Col ecotype), 40 lines of kanamycin-resistant transgenic plants were selected, and their T2 were analyzed for GFP expression. To visualize the nucleus, root tissues were stained with propidium iodide (1 μg/mL). Green fluorescence (GFP expression) and red fluorescence (propidium iodide staining) analysis of transgenic plants were performed with a 1024 confocal laser-scanning microscope (Grebenok et al. 1997).
HOS1 cosuppression and analysis
HOS1 cDNA was isolated by RT-PCR using primers HOS1RTF1, 5‘-GATCCCGGGGCTCAGGTCTTAGGATTTCTAG-3‘; HOS1RTR1, 5′-GCTCCCGGGTACCTCTCTCTCTCTTTCT CTATGC-3′. The PCR fragment was cloned into the pCR-BluntII-TOPO vector (Invitrogen) and sequenced. The coding sequence of the HOS1 gene was PCR-amplified from the cDNA with primers HOS1OVER-F1, 5′-CGAGCTCGGCTCAGGTCT TAGGATTTCTAG-3′; HOS1OVER-R1, 5′-CCCCCGGGGGT ACCTCTCTCTCTCTTTCTCTATGC-3′. The PCR fragment was digested with SstI and SmaI and inserted into a pRTL104 vector downstream from the CaMV 35S promoter. The HOS1 promoter–GUS fusion was removed from the construct by digestion with SphI and cloned into pCAMBIA 1200. After transformation of wild-type Arabidopsis plants via Agrobacterium strain GV3101, 25 lines of hygromycin-resistant transgenic plants were obtained. Luciferase imaging was carried out on the T2 plants. For analysis of the transcription level of HOS1 in cosuppression lines, the following primer pair was used in quantitative RT–PCR: 5′-ACGGAATGGATGGAGATACT-3′; 5′-AAGAGACTACCCACAGCTGA-3′ (resulting in a 170-bp PCR fragment). Total RNA from wild-type, hos1, and #4-high plants was extracted using the TRIzol (Gibco BRL) reagent system, and 1 μg of RNA from each sample was used for RT–PCR reaction with the EZ rTth RNA PCR Kit (PERKIN ELMER, part no. N808-0179) according to the manufacturer’s instructions. PCR products were then separated in a 4% agarose gel. As an internal control, the ubiquitin gene was amplified along with HOS1 using the following primer pair: UBQ11-F, 5′-GAGGTATGCA GATCTTCGTG-3′; UBQ11-R, 5′-GTAGACTCCTTCTGGAT GTTG-3′.
Acknowledgments
We sincerely thank Cheol-Soo Kim, Joon-Hyun Park, and Suk-Whan Hong for assistance, and Sean Cutler, David Ehrhardt, and Chris Somerville for their generous gift of the Q1 GFP transgenic plants. This work was supported by National Science Foundation grant IBN-9808398 and USDA NRI grant 2000-00664.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jkzhu@ag.arizona.edu; FAX (520) 621-7186.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.866801.
References
- Anderson JV, Li QB, Haskell DW, Guy CL. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat shock cognate gene and expression of 70-kilodalton heat shock genes during cold acclimation. Plant Physiol. 1994;104:1359–1370. doi: 10.1104/pp.104.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Wilson A, Dean C. Arabidopsismutants showing an altered response to vernalization. Plant J. 1996;10:637–644. doi: 10.1046/j.1365-313x.1996.10040637.x. [DOI] [PubMed] [Google Scholar]
- Chen H-H, Li PH, Brenner ML. Involvement of abscisic acid in potato cold acclimation. Plant Physiol. 1983;71:362–365. doi: 10.1104/pp.71.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsisat a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβhomologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- Freemont PS. Ubiquitination: RING for destruction? Curr Biol. 2000;10:R84–R87. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- Gibson S, Arondel V, Iba K, Somerville C. Cloning of a temperature-regulated gene encoding a chloroplast omega-3 desaturase from Arabidopsis thaliana. Plant Physiol. 1994;106:1615–1621. doi: 10.1104/pp.106.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Artus NN, Thomashow MF. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol Biol. 1992;18:13–32. doi: 10.1007/BF00018452. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zrka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced CORgene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Grebenok RJ, Pierson E, Lambert GM, Gong FC, Afonso CL, Haldeman-Cahill R, Carrington JC, Galbraith DW. Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 1997;11:573–586. doi: 10.1046/j.1365-313x.1997.11030573.x. [DOI] [PubMed] [Google Scholar]
- Guy CL. Cold acclimation and freezing stress tolerance: Role of protein metabolism. Ann Rev Plant Physiol Plant Mol Biol. 1990;41:187–223. [Google Scholar]
- Guy CL, Niemi KL, Brambl R. Altered gene expression during cold acclimation of spinach. Proc Natl Acad Sci USA. 1985;82:3673–3677. doi: 10.1073/pnas.82.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, Parker R. Mechanisms of mRNA surveillance in eukaryotes. Annu Rev Genet. 1999;33:229–260. doi: 10.1146/annurev.genet.33.1.229. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu J-K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu J-K. HOS1, a genetic locus involved in cold responsive gene expression in Arabidopsis thaliana. Plant Cell. 1998;10:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JDR. The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces CORgenes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Joazeiro CAP, Weissman AM. RING finger proteins: Mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Jonak G, Kiegerl S, Lighterink W, Barker PJ, Huskisson NS. Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc Natl Acad Sci USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotech. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell. 1996;8:489–503. doi: 10.1105/tpc.8.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Veale EL, Warren GJ, Knight MR. The sfr6 mutation in Arabidopsissuppresses low temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell. 1999;11:875–886. doi: 10.1105/tpc.11.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Borg-Franck M. Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol Biol. 1992;19:689–692. doi: 10.1007/BF00026794. [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Ishitani M, Zhu J-K. Cold-regulated gene expression and freezing tolerance in an Arabidopsis thalianamutant. Plant J. 1999;17:301–308. doi: 10.1046/j.1365-313x.1999.00375.x. [DOI] [PubMed] [Google Scholar]
- Levitt J. Responses of plants to environmental stress. 1: Chilling, freezing, and high temperature stress. New York: Academic Press; 1980. [Google Scholar]
- Liu J, Zhu J-K. Proline accumulation and salt-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol. 1997;114:591–596. doi: 10.1104/pp.114.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Klee HJ, Bowman JL, Meyerowitz EM. Vectors for plant transformation and cosmid libraries. Gene. 1992;117:161–167. doi: 10.1016/0378-1119(92)90725-5. [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. The Arabidopsis CBFgene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–470. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS Cencodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LK. An exegesis of IAPs: Salvation and surprises from BIR motifs. Trends Cell Biol. 1999;9:323–328. doi: 10.1016/s0962-8924(99)01609-8. [DOI] [PubMed] [Google Scholar]
- Monroy AF, Dhindsa RS. Low-temperature signal transduction: Induction of cold acclimation-specific genes of alfalfa by calcium at 25°C. Plant Cell. 1995;7:321–331. doi: 10.1105/tpc.7.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy AF, Sangwan V, Dhindsa RS. Low temperature signal transduction during cold acclimation: Protein phosphatase 2A as an early target for cold-inactivation. Plant J. 1998;13:653–660. [Google Scholar]
- Murata N, Los DA. Membrane fluidity and temperature perception. Plant Physiol. 1997;115:875–879. doi: 10.1104/pp.115.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin K, Vahala T, Palva ET. Differential expression of two related, low-temperature-induced genes in Arabidopsis thaliana(L.) Heynh. Plant Mol Biol. 1993;21:641–653. doi: 10.1007/BF00014547. [DOI] [PubMed] [Google Scholar]
- Osterlund M, Hardtke C, Wei N, Deng X-W. Targeted destabilization of HY5 during light regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Borden KLB, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci USA. 2000;97:3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:14570–14575. doi: 10.1073/pnas.95.24.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I, Los DA, Kanesaki Y, Mikami K, Murata N. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 2000;19:1327–1334. doi: 10.1093/emboj/19.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Arabidopsis thalianaas a model for studying mechanisms of plant cold tolerance. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 807–834. [Google Scholar]
- Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-W. Light inactivation of Arabidopsisphotomorphogenic COP1 involves a cell-specific regulation of its nucleo–cytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Xin Z, Browse J. eskimo1 mutants of Arabidopsisare constitutively freezing-tolerant. Proc Natl Acad Sci USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsisgene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]