Abstract
Plants maintain cytosine methylation at CG and non-CG residues to control gene expression and genome stability. In a screen for Arabidopsis mutants that alter methylation and silencing of a densely methylated endogenous reporter gene, we recovered 11 loss-of-function alleles in the CMT3 chromomethylase gene. The cmt3 mutants displayed enhanced expression and reduced methylation of the reporter, particularly at non-CG cytosines. CNG methylation was also reduced at repetitive centromeric sequences. Thus, CMT3 is a key determinant for non-CG methylation. The lack of CMT homologs in animal genomes could account for the observation that in contrast to plants, animals maintain primarily CG methylation.
Keywords: Cytosine methyltransferase, gene silencing, Arabidopsis thaliana
In many eukaryotes, including mammals, higher plants, and some species of fungi, cytosine methylation plays an important role in genome stability and development by altering chromatin structure and patterns of gene expression. In mammalian genomes, methylation is found primarily at cytosines in the symmetric context 5′-CG-3′ (CG), whereas in plant and fungal genomes methylation is found on both CG and non-CG residues (Yoder et al. 1997; Colot and Rossignol 1999; Finnegan and Kovac 2000). Mammals and higher plants carry related cytosine methyltransferases of the Dnmt1/MET1 class that have been implicated by mutational analysis as enzymes that maintain the bulk of genomic methylation (Li et al. 1992; Finnegan et al. 1996; Ronemus et al. 1996). Another class of chromomethylases (CMTs) has been identified by analysis of Arabidopsis thaliana genomic sequences (Henikoff and Comai 1998; McCallum et al. 2000). The CMT class is characterized by the presence of a chromodomain amino acid motif between the cytosine methyltransferase catalytic motifs I and IV. There are three CMT genes encoded in Arabidopsis: CMT1, CMT2, and CMT3 (Henikoff and Comai 1998; Finnegan and Kovac 2000; McCallum et al. 2000). In the Wassilewskija (WS) strain background used for this study, CMT2 and CMT3 are predicted to encode functional proteins, whereas the CMT1 coding sequence is disrupted by an Eve1 (Henikoff and Comai 1998) retroelement insertion (J. Bender, unpubl.). CMT genes have also been identified in several other plant species including Brassica and maize, but not in fungal or animal systems (Rose et al. 1998; Finnegan and Kovac 2000). Recently, Arabidopsis CMT3 (Lindroth et al. 2001) and the maize CMT homolog ZMET2 (Papa et al. 2001) have been implicated in the maintenance of CNG methylation.
In the genome of Arabidopsis, duplicated genes encoding the tryptophan pathway enzyme phosphoribosylanthranilate isomerase (PAI) provide a well-characterized example of endogenous genes that are densely methylated with both CG and non-CG methylation (Luff et al. 1999). In the Arabidopsis strain WS, there are four methylated PAI genes at three unlinked loci: a singlet PAI2 gene that encodes functional enzyme, a singlet PAI3 gene that does not encode functional enzyme, and a tail-to-tail inverted repeat of the PAI1 and PAI4 genes (PAI1-PAI4) in which the PAI1 gene encodes functional enzyme and the PAI4 gene does not (Bender and Fink 1995; Melquist et al. 1999). The functional singlet PAI2 gene is silenced by methylation (Bender and Fink 1995; Jeddeloh et al. 1998; Melquist et al. 1999). In contrast, the functional PAI1 gene in the inverted repeat is expressed despite dense methylation in the body of the gene, providing sufficient PAI enzyme for a wild-type plant phenotype (Melquist et al. 1999). It is likely that the WS PAI1 gene eludes silencing by methylation because of novel promoter sequences lying upstream of the methylated region (Melquist et al. 1999; J. Bender, unpubl.).
Here we describe the isolation and characterization of mutations in the CMT3 chromomethylase gene from a genetic screen for reduced PAI methylation. Southern blot analysis and bisulfite genomic methylation sequencing indicate that cmt3 mutations confer a partial loss of CG methylation and a strong loss of non-CG methylation (both CNG and asymmetric cytosines) from the PAI genes. Southern blot analysis of repetitive methylated genomic sequences indicates that cmt3 mutations also confer reduced CNG methylation on these regions. In contrast to characterized Arabidopsis mutations that confer globally decreased methylation (Finnegan et al. 1996; Kakutani et al. 1996; Ronemus et al. 1996), the cmt3 mutations do not lead to pleiotropic effects upon inbreeding, suggesting that CMT3 function is specialized for only a subset of methylated regions in the genome.
Results and Discussion
To identify factors that control methylation and silencing of the WS PAI genes, we isolated a mutant variant of WS, pai1C251Y, in which silencing of the methylated singlet PAI2 gene can be visualized by the intensity of a blue fluorescent plant phenotype under ultra-violet (UV) light (Bartee and Bender 2001). In the pai1C251Y strain, the only potential sources of PAI enzyme activity are the PAI1 gene, which is crippled by a missense mutation, and the PAI2 gene, which is silenced. Because of this PAI deficiency, the strain accumulates fluorescent tryptophan pathway intermediates, as well as displaying yellow-green leaf pigmentation, reduced size, increased bushiness, and reduced fertility. However, second-site mutations that relieve PAI2 silencing will suppress the PAI-deficient phenotypes (Bartee and Bender 2001). Thus, we mutagenized the pai1C251Y strain and screened for seedlings with suppressed weak fluorescent phenotypes. As a secondary screen, we tested PAI methylation by Southern blot analysis with methylation-sensitive restriction enzymes. Specifically, we assayed methylation with the isoschizomers HpaII and MspI, which recognize the sequence 5′-CCGG-3′. HpaII is sensitive to methylation of both the inner (CG) and the outer (CNG) cytosines, whereas MspI is only sensitive to methylation of the outer cytosines. These enzymes cleave once in each WS PAI locus and reveal both the density and the pattern of methylation for each gene (Bender and Fink 1995; Luff et al. 1999; Melquist et al. 1999).
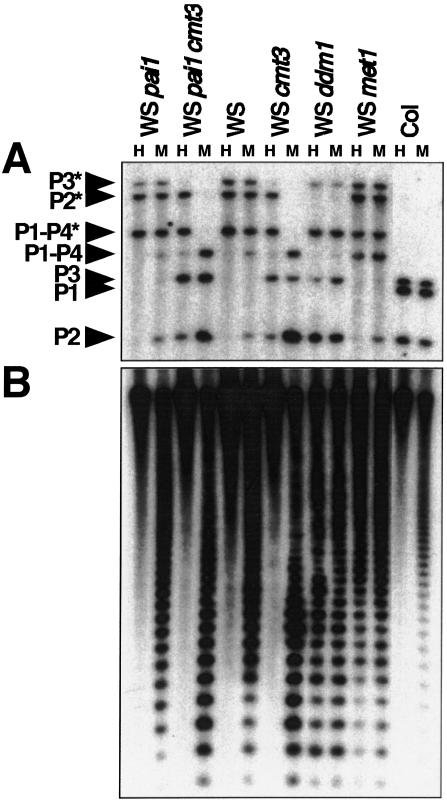
From this screening strategy we isolated 11 loss-of-function alleles in the CMT3 gene (see below and Materials and Methods). The cmt3 mutants in the pai1C251Y background displayed strongly reduced fluorescence in early seedling development and partially reduced fluorescence in adult plants, with increased size, decreased bushiness, and increased fertility (Fig. 1). These intermediate fluorescent cmt3 isolates did not revert to nonfluorescence, which is diagnostic of loss of residual PAI2 methylation (Bender and Fink 1995), at a detectable frequency. They displayed partially increased cleavage with HpaII and strongly increased cleavage with MspI for the PAI genes relative to parental pai1C251Y (Fig. 2A). The cleavage pattern suggested that the cmt3 mutants were most affected in maintenance of CNG methylation of the PAI genes. To determine whether the cmt3 mutants also affected methylation of a highly repeated genomic sequence, we reprobed the HpaII/MspI Southern blot with a probe to the 180-bp centromere-associated repeat (CEN) sequences (Vongs et al. 1993). This probe revealed little effect on HpaII cleavage but increased MspI cleavage, consistent with the pattern observed for the PAI genes (Fig. 2B). A similar pattern of increased MspI cleavage was also observed at the repeated rDNA (data not shown). All of 11 cmt3 alleles tested had identical methylation patterns in these assays. Moreover, when the cmt3 alleles were segregated away from the pai1C251Y allele into a wild-type WS background, they also displayed identical methylation patterns in these assays (Fig. 2). The PAI and CEN methylation patterns were distinct from the patterns induced by the characterized ddm1 and met1 methylation-deficient mutations (Fig. 2; Bartee and Bender 2001).
Figure 1.
Wassilewskija (WS) pai1C251Y cmt3 plant phenotypes. (A) Two-week-old seedlings of the indicated genotypes grown on agar medium are shown under visible (top) and UV (bottom) light. (B) Four-week-old adult plants of the indicated genotypes grown in soil are shown under visible (top) and UV (bottom) light. (C) Representative 2-week-old T2 generation transgenic seedlings of the indicated genotypes grown on agar medium are shown under visible (top) and UV (bottom) light. The phenotypes of the cmt3G456D allele shown are representative of the phenotypes observed with 10 other cmt3 alleles.
Figure 2.
cmt3 mutations confer reduced PAI and CEN methylation. (A) Genomic DNAs prepared from 4-week-old plants of the indicated genotypes were cleaved with either HpaII (H) or MspI (M) and used for Southern blot analysis with a PAI probe (Bender and Fink 1995). (P1–P4) PAI1–PAI4; (P2) PAI2; (P3) PAI3; (P1) Columbia (Col) strain PAI1; asterisks indicate the positions of species methylated at internal HpaII/MspI sites (Bender and Fink 1995; Luff et al. 1999). The Col strain is included as a control for the positions of unmethylated PAI2 and PAI3 species. (B) The blot shown in A was reprobed with a 180-bp CEN repeat probe. The phenotypes of the cmt3G456D allele shown are representative of the phenotypes observed with 10 other cmt3 alleles.
To more precisely determine methylation patterns in the cmt3 mutant background, we performed genomic sequencing of methylation patterns in the PAI1 and PAI2 promoter regions of a representative cmt3 allele by using sodium bisulfite mutagenesis (Frommer et al. 1992). This analysis revealed that the majority of methylated cytosines (87% in PAI1 and 70% in PAI2) occurred at CG residues (Fig. 3; Table 1). Compared with the wild-type WS PAI1 promoter (Luff et al. 1999; Table 1), CG methylation was reduced 34%, CNG methylation was eliminated, and asymmetric methylation was reduced 93%; in the PAI2 promoter, CG methylation was reduced 8%, CNG methylation was reduced 92%, and non-CG methylation was reduced 75%. Thus, loss of CMT3 function has a strong effect on maintenance of CNG and asymmetric methylation and a weaker effect on maintenance of CG methylation. These results are consistent with reports that Arabidopsis CMT3 and maize ZMET2 are important for maintenance of CNG methylation at various genomic sites (Lindroth et al. 2001; Papa et al. 2001), but they further show that CMT3 is also important for maintenance of asymmetric methylation for the PAI genes. This result implies either that CMT3 directly controls both symmetric and asymmetric methylation or that the reduction in symmetric methylation in the cmt3 mutant background causes reduced asymmetric methylation as a secondary consequence. Because the methylated sequences in the promoter and first exon of the PAI2 reporter gene (∼370 bp) contain only 16 dispersed CG motifs, loss of non-CG methylation significantly hypomethylates this region of the gene (Fig. 3), accounting for enhanced PAI2 expression in the suppressor mutant.
Figure 3.
Sequencing of PAI promoter methylation in the cmt3 mutant. Bisulfite genomic methylation sequencing was performed as described (Jeddeloh et al. 1998; Luff et al. 1999) for the top strands of the PAI1 and PAI2 promoters in Wassilewskija (WS) pai1C251Y cmt3G456D DNA. For each region, eight independent molecules were sequenced. Vertical lines indicate positions of cytosines, with the height of each line representing how many sequenced molecules had 5-methyl-cytosine. (Black) CG cytosines; (blue) CNG cytosines; (red) other cytosines. Asterisks indicate sites with no methylation. The black horizontal line indicates the region of PAI identity, and the gray horizontal line indicates flanking upstream heterologous sequence unique to each gene. The exon and intron structures of PAI1 and PAI2 are shown as open boxes and dashed lines, respectively, under each sequence. These structures are based on full-length cDNA sequences for each gene (Melquist et al. 1999).
Table 1.
Effects of a cmt3 mutation on patterns of PAI promoter cytosine methylationa
| Strain
|
PAI gene
|
CG
|
CNG
|
Other C
|
Total C
|
|---|---|---|---|---|---|
| WS | PAI1 | 115 (100%) | 61 (100%) | 149 (100%) | 325 (100%) |
| cmt3b | PAI1 | 76 (66%) | 0 (0%) | 11 (7%) | 87 (27%) |
| WS | PAI2 | 122 (100%) | 53 (100%) | 184 (100%) | 359 (100%) |
| cmt3 | PAI2 | 112 (92%) | 4 (8%) | 45 (25%) | 161 (45%) |
The numbers of methylated cytosines in the indicated sequence contexts for eight independent top strand bisulfite sequencing clones for the indicated PAI promoter regions of the indicated strains are shown. The wild-type Wassilewskija (WS) data are derived from data published previously in Luff et al. (1999). The cmt3 data are from this work, and are shown in diagram form in Figure 3.
The sequenced DNA was from the WS pai1C251Y cmt3G456D strain.
The cmt3 mutant locus in the pai1C251Y cmt3 suppressor isolates was mapped by crosses with the polymorphic strain Nd-0, which has a similar arrangement of densely methylated PAI genes as found in WS (Melquist et al. 1999). F2 progeny with weakly fluorescent phenotypes diagnostic of homozygosity for both pai1C251Y and cmt3 were identified by visual inspection under UV light and confirmed by MspI Southern blot for strong PAI cleavage similar to that observed in the parental suppressor isolates. A mapping population of F2 plants that fulfilled these criteria was then used to score for genomic loci linked to the suppressed phenotype. The mapping analysis revealed linkage to a single locus on the lower arm of chromosome 1. Because the CMT3 putative cytosine methyltransferase gene maps to this locus, we focused on this gene as a candidate. Within each mapping population, we found complete linkage to a polymorphic marker that lies within 100 kb of the CMT3 gene. To confirm that the CMT3 gene was in fact the site of the methylation suppressor mutations, we cloned and sequenced the gene from the 11 mutant isolates. Sequencing revealed a single base change in the CMT3 coding sequence in each isolate. Three of the mutant alleles affected absolutely conserved amino acids in the methyltransferase catalytic domain, including the representative cmt3G456D allele used for bisulfite sequencing. Another allele was predicted to prematurely terminate the protein. Two alleles created splice junction mutations. The remaining five alleles affected amino acids between methyltransferase motif IV and the C terminus of the protein that are highly conserved among the CMT genes (Fig. 4).
Figure 4.
Positions of mutations in CMT3. The predicted amino acid sequences of Wassilewskiya (WS) CMT3, WS CMT2, and maize ZMET2 are shown aligned along their conserved C-terminal regions. The N termini, upstream of the backslash at the beginning of each sequence, are unrelated. CMT3 introns are indicated by inverted triangles above the sequence. CMT3 missense mutations are indicated above the affected residues. The stop mutation is indicated by an asterisk, and the splice donor and acceptor site mutations are indicated by an x to the left or right, respectively, above the affected introns. Residues identical between proteins are highlighted in boldface. Conserved sequence motifs are indicated under the alignment. GenBank accession nos. are: AF383170 for WS CMT3 and AF383171 for WS CMT2.
To further confirm that the CMT3 gene was the mutant locus, we transformed the pai1C251Y cmt3 isolates with a wild-type WS genomic clone of the CMT3 gene. Transformant seedlings were strongly fluorescent, similarly to those of the pai1C251Y strain (Fig. 1). Transformant lines assayed by Southern blot analysis in the T2 generation showed remethylation of the PAI2 gene to the levels observed in the original pai1C251Y strain (data not shown). Thus, the cloned CMT3 gene could complement the mutant methylation defects. As a control, the representative pai1C251Y cmt3G456D mutant was also transformed with a wild-type WS genomic clone of the CMT2 gene. CMT2 transformant seedlings were weakly fluorescent, similarly to those of the untransformed parental strain (Fig. 1), and did not display detectable remethylation of PAI2. This analysis shows that CMT2 cannot substitute for CMT3 function. In this regard, it is interesting to note that CMT2 differs from CMT3 primarily in its N-terminal sequence (Fig. 4).
Previously characterized methylation-deficient Arabidopsis strains with defects in either the SWI2/SNF2 chromatin remodeling factor-related gene DDM1 (Jeddeloh et al. 1999) or the Dnmt1-related MET1 cytosine methyltransferase gene display progressive developmental abnormalities (Finnegan et al. 1996; Kakutani et al. 1996; Ronemus et al. 1996). Our preliminary analysis of six-generation-inbred pai1C251Y cmt3 and two-generation-inbred cmt3 strains revealed no obvious segregation of morphological changes. This difference between cmt3 and other methylation-deficient mutants is likely to reflect the fact that CG methylation is retained to a higher degree in cmt3 than in ddm1 or met1 (Fig. 2; Bartee and Bender 2001). Because many of the endogenous methylated sites in the Arabidopsis genome, such as the CEN repeats (Fig. 2; Vongs et al. 1993; Lindroth et al. 2001), and the promoter of the FWA homeo-domain gene (Soppe et al. 2000; Lindroth et al. 2001), carry primarily CG methylation, cmt3 mutations would not be expected to strongly affect these loci. Instead, CMT3 most likely acts as a reinforcing methylase that adds an extra layer of methylation to particular genomic regions such as the PAI genes, in which the increased methylation density leads to increased silencing. A specific model is that the basal layer of CG methylation provided by other functions such as MET1 could serve as a guide for CMT3, which would then decorate the basal layer with extra CG and non-CG methylation. CMT3 recruitment to targeted regions could involve chromatin protein interactions with the chromodomain motif (Henikoff and Comai 1998), along with interactions mediated by the unique N-terminal sequences.
Because fungi such as Neurospora crassa and Ascobolus immersus can maintain non-CG methylation (Selker et al. 1993; Goyon et al. 1994), these organisms might encode CMT genes. Conversely, because animals such as humans and mice lack non-CG methylation, these organisms are predicted to lack CMT genes, as is the case from analyses of current sequence databases. The apparent lack of CMT-like methylases in animal genomes (Finnegan and Kovac 2000) suggests that animals have evolved alternate mechanisms for reinforcing chromatin states.
Materials and methods
Mutant isolation and sequencing
Seeds of WS pai1C251Y were mutagenized with ethylmethane sulfonate (Niyogi et al. 1993) and grown up as 20 pools of ∼500 M1 plants each; M2 progeny seeds were collected from each pool. Approximately 1000 seedlings from each M2 pool were grown on agar medium for 2 wk and then screened with a hand-held short wavelength UV light source for individuals with reduced fluorescence. Putative mutants were transplanted to soil, and genomic DNA prepared from a single leaf was used for Southern blot analysis of methylation patterns. From the screen of 20,000 M2 seedlings, all 11 isolates that displayed cmt3 methylation patterns (Fig. 2) proved to be cmt3 alleles by mapping, sequencing, and complementation analysis. Ten other isolates with distinct methylation patterns as determined by HpaII/MspI Southern blot analysis were also recovered, but these isolates remain to be characterized. Interestingly, none of the other isolates displayed the PAI and CEN Southern blot phenotypes diagnostic of ddm1 mutations in the WS background (Fig. 2). Because we expect to recover ddm1 alleles from the screen (Bartee and Bender 2001), this observation suggests that the screen is not yet saturated.
The cmt3 mutant locus was mapped to the lower arm of chromosome 1 with standard CAPS (Konieczny and Ausubel 1993) and simple sequence length polymorphism (SSLP) (Bell and Ecker 1994) markers that are polymorphic between WS and Nd-0. It was further localized between the markers NF5I14 and NF22K20 (http://www.arabidopsis.org/servlets/mapper). Linkage to CMT3 was determined with the T17F3 marker, which lies within 100 kb of the CMT3 gene: forward primer 5′-gacataataccgagtacccac-3′; reverse primer 5′-ccaccaccttgcactgccgacc-3′; in WS a 354-bp product cleaves once with MspI into 240 bp and 114 bp fragments, whereas the Nd-0 product is uncleaved.
Mutant alleles of CMT3 were amplified by PCR from genomic DNA. Products from two independent PCRs were cloned and sequenced for each allele. Alleles that changed restriction sites (both splice site mutations, cmt3G456D, cmt3G465D, and cmt3R703K) were confirmed by PCR amplification of the mutant region followed by cleavage with the appropriate enzyme.
CMT genomic and cDNA clones
The CMT3 transgene is a WS genomic fragment extending from 2.9 kb upstream of the start codon to 0.8 kb downstream of the stop codon subcloned into the pBIN19 transformation vector (Bevan 1984). The CMT2 transgene is a WS genomic fragment extending from 1.7 kb upstream of the start codon to 0.9 kb downstream of the stop codon subcloned into pBIN19. Both clones were isolated by hybridization from a WS genomic library (Bender and Fink 1995). Both clones were sequenced across the coding region to confirm their structure and determine WS polymorphisms. Transgenes were introduced into the WS pai1C251Y cmt3G456D strain by an in planta transformation method (Clough and Bent 1998).
The CMT3 predicted amino acid sequence was determined by cloning and sequencing a cDNA isolate generated by RT–PCR from WS whole-plant RNA. CMT2 and ZMET2 amino acid sequences are predicted from the WS CMT2 genomic sequence, cDNA sequences available from the database, and alignment with related genes.
Acknowledgments
This work was supported by grants from the Searle Scholars Foundation (97-E-103), March of Dimes Birth Defects Foundation (FY00-418), and NIH (1R01GM61148) to J.B. L.B. was supported by NCI training grant 5-T32-CA09110.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jbender@welchlink.welch.jhu.edu; FAX (410) 955-2926.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.905701.
References
- Bartee L, Bender J. Two Arabidopsis methylation-deficiency mutations confer only partial effects on a methylated endogenous gene family. Nucleic Acids Res. 2001;29:2127–2134. doi: 10.1093/nar/29.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colot V, Rossignol J-L. Eukaryotic DNA methylation as an evolutionary device. BioEssays. 1999;21:402–411. doi: 10.1002/(SICI)1521-1878(199905)21:5<402::AID-BIES7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA. Plant DNA methyltransferases. Plant Mol Biol. 2000;43:189–201. doi: 10.1023/a:1006427226972. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C, Nogueira TI, Faugeron G. Perpetuation of cytosine methylation in Ascobolus immersus implies a novel type of maintenance methylation. J Mol Biol. 1994;240:42–51. doi: 10.1006/jmbi.1994.1416. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Comai L. A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics. 1998;149:307–318. doi: 10.1093/genetics/149.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes & Dev. 1998;12:1714–1725. doi: 10.1101/gad.12.11.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. .. [DOI] [PubMed] [Google Scholar]
- Luff B, Pawlowski L, Bender J. An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol Cell. 1999;3:505–511. doi: 10.1016/s1097-2765(00)80478-5. [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeted screening for induced mutations. Nat Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- Melquist S, Luff B, Bender J. Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics. 1999;153:401–413. doi: 10.1093/genetics/153.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK, Last RL, Fink GR, Keith B. Suppressors of trp1 fluorescence identify a new Arabidopsis gene, TRP4, encoding the anthranilate synthase β subunit. Plant Cell. 1993;5:1011–1027. doi: 10.1105/tpc.5.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa, C.M., Springer, N.M., Muszynski, M.G., Meeley, R., and Kaeppler, S.M. 2001. Maize chromomethylase Zea Methyltransferase2 is required for CpNpG methylation. Plant Cell (in press). [DOI] [PMC free article] [PubMed]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- Rose T M, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU, Fritz DY, Singer MJ. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993;262:1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- Soppe WJJ, Jacobsen SE, Alonso-Blanco C, Jackson JP, Kakutani T, Koornneef M, Peeters AJM. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]