Abstract
Previous work from our laboratory showed that the Dictyostelium discoideum SadA protein plays a central role in cell-substrate adhesion. SadA null cells exhibit a loss of adhesion, a disrupted actin cytoskeleton, and a cytokinesis defect. How SadA mediates these phenotypes is unknown. This work addresses the mechanism of SadA function, demonstrating an important role for the C-terminal cytoplasmic tail in SadA function. We found that a SadA tailless mutant was unable to rescue the sadA adhesion deficiency, and overexpression of the SadA tail domain reduced adhesion in wild-type cells. We also show that SadA is closely associated with the actin cytoskeleton. Mutagenesis studies suggested that four serine residues in the tail, S924/S925 and S940/S941, may regulate association of SadA with the actin cytoskeleton. Glutathione S-transferase pull-down assays identified at least one likely interaction partner of the SadA tail, cortexillin I, a known actin bundling protein. Thus, our data demonstrate an important role for the carboxy-terminal cytoplasmic tail in SadA function and strongly suggest that a phosphorylation event in this tail regulates an interaction with cortexillin I. Based on our data, we propose a model for the function of SadA.
INTRODUCTION
Cell-substrate adhesion plays a central role in many biological processes. In nontransformed cells, adhesion is important for cell migration during development and wound healing, for phagocytosis of nutrients or cellular invaders, as well as various other processes. When adhesion is misregulated, under normal circumstances cells may undergo apoptosis (anoikis) or, in the case of tumor cells, this may result in metastasis (18). In metazoans, the protein network involved in adhesion presents an extremely complex process involving adhesion receptors, signaling molecules, and adaptor proteins, to name a few. In excess of 150 individual proteins are known to be present in the adhesive structures and involved in the adhesion process (44). Considering the number of possible posttranslational modifications likely to occur even on just a few of these proteins, the complexity of this system becomes evident.
Perhaps at the center of cell-substrate adhesion in metazoans is a hetero-dimeric set of proteins, singularly known as the α- and β-integrins, which act as receptors linking the extracellular milieu to the intracellular machinery (22). Just as there are numerous types of extracellular matrices to which the cell can bind, so too are there different combinations of α- and β-integrin cognate combinations (22). The intracellular aspect of adhesion links to actin and is mediated by adaptors such as paxillin and talin, as well as many others (43, 44). As alluded to in previous studies, signaling proteins are also important in the adhesion process, and kinases such as integrin-linked kinase (ILK) and focal adhesion kinase (FAK), as well as small G-proteins, such as Rap1, are involved as well (43, 44).
From an evolutionary point of view, it is interesting that although D. discoideum appeared subsequent to the plant/animal split, but before the full metazoan and fungal lineage, its genome contains homologous genes found in less ancient branches of the evolutionary tree (3, 14). Intriguingly, Dictyostelium encodes a number of homologous proteins that are involved in adhesion in higher eukaryotes, such as talin, Rap1, paxillin, and others (7, 21, 24, 32). Recently, a protein with some attributes of metazoan β-integrins was identified in Dictyostelium, but its placement in a cell-substrate adhesion pathway remains to be defined (9, 10). These Dictyostelium proteins likewise have documented roles in adhesion. With a combination of genetic tractability, great cell biology, and excellent biochemistry, D. discoideum provides a unique model system to dissect the process of substrate adhesion in a less complex system.
Interest in identifying proteins involved in cell-substrate adhesion has become more intense in the Dictyostelium community in the last decade. In a screen for such genes, our lab previously reported on a putative adhesion receptor, SadA (16). Cells in which the sadA gene was inactivated demonstrated a complete loss of substrate adhesion, whether it was to the growth surface (a petri dish) or to a phagocytic particle (a latex bead) (16). Furthermore, sadA null cells also exhibited a cytokinesis defect that presumably resulted from the altered F-actin cytoskeleton found in the mutant. SadA is predicted to be a nine-pass transmembrane protein, having no extended sequence similarity to other known proteins, although it does contain 3 epidermal growth factor (EGF)-like repeats in its large amino-terminal extracellular domain. EGF-like domains are found in proteins known to be involved in adhesion, but their function remains unknown (2, 11).
One notable feature of SadA is its 31-residue carboxy-terminal tail, which is similar in length to that of the β-integrins. We hypothesized that this domain of SadA might serve as a link to the actin cytoskeleton. The data we present here demonstrate that SadA is biochemically associated with the actin cytoskeleton. Protein sequence domain analysis suggests that the SadA tail contains sequence motifs that might be targets for posttranslational modifications such as phosphorylation. The results suggest that SadA may be differentially phosphorylated depending on growth conditions. Indeed, mutagenesis of residues S924/S925 and S940/S941 suggests a possible role for phosphorylation as a regulator of SadA-mediated adhesion, as well as for regulated protein-protein interactions. Glutathione S-transferase (GST) pull-down assays using the SadA tail as bait suggest that this domain interacts with cortexillin I, which perhaps serves as a link to the actin cytoskeleton. Based on these results we propose a model outlining a role for SadA as a transmembrane protein-actin cytoskeleton link in Dictyostelium.
MATERIALS AND METHODS
Cell culture.
All experiments were performed using strains that have AX3 as the parental background. Cells were grown axenically in HL5 medium (17) with 300 μg/ml streptomycin sulfate. Drug selection, when required, was with 10 μg/ml blasticidin (Invitrogen) and/or 10 μg/ml G418. For normal growth and passaging, cultures were grown in 10-cm petri dishes and passaged every 2 to 3 days to prevent cell densities reaching 5 × 106 cells/ml. When large quantities of cells were required, cells were grown in 30-cm petri dishes. For growth in suspension, cells were grown in flasks that held five times the volume used for growth at 22°C, 150 to 200 rpm, in a New Brunswick incubator (model G25).
Molecular biology.
A full-length SadA cDNA clone was cloned by reverse transcription-PCR (RT-PCR using mRNA purified from AX3 wild-type adherent cells as input and the Thermoscript RT-PCR system [Invitrogen]). Second-round PCR amplification was performed with Phusion DNA polymerase (New England BioLabs) using sadA gene-specific primers. To simplify downstream cloning, the 5′ primer added an AatII restriction site and the 3′ primer added an NheI restriction site. The gel-purified PCR product was restriction digested with AatII and NheI and cloned into a modified pBluescript KS vector, resulting in the vector pBS-SadA. The modified vector (pBS-MCS) contained an AatII/MluI/NheI polylinker added into the KpnI site of pBluescript KS. Subsequent to verifying the SadA sequence by DNA sequencing, the AatII-SadA-NheI fragment was subcloned into a modified pTXGFP vector (26). The pTXGFP vector was modified by adding a AatII/MluI/NheI polyliner at the KpnI site, resulting in the vector pTX-MCS-GFP. The pTX-SadA-GFP vector was transformed into sadA null cells by using an oscillating electroporation protocol (1) with a Bio-Rad Gene Pulser Xcell electroporator. In our hands, the oscillating electroporation method gave substantially higher transformation efficiencies, especially with sadA null cells.
For GST-tail fusion proteins, the coding sequence for the carboxy-terminal tail of SadA, from nucleotide 2763 (residue 921 [Q]) to nucleotide 2856 (residue 952 [K]), was amplified from SadA cDNA by PCR with the forward primer 5′-ACA ACG ATT TCC CAA ATG AGA TCT TCT TTC ATT-3′, containing an EcoRI site, and the reverse primer 5′-TGT TGT GCG GCC GCT TTA TTT CTT AGA TAA ATC GAT-3′, containing a NotI site. The SadA fragment was cloned into the pGEX 4T3 vector (GE Healthcare) and then verified by DNA sequencing. The pGEX-SadA tail vectors were transformed into BL21 cells (GE Healthcare) for protein expression. Cells were lysed via detergent lysis or in a French press and purified on glutathione-Sepharose 4B resin (GE Healthcare). Purified protein was dialyzed into 1× phosphate (PBS)–10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, flash-frozen in an ethanol-dry ice bath, and stored at −80°C for later use.
For green fluorescent protein (GFP)-trap validation experiments, two expression vectors were constructed, one for the expression of a GFP-SadA fusion protein and the other for a Flag-GFP fusion. Details of the construction of the SadA signal sequence (SS)-GFP-SadA construct in a modified pTX-MCS-GFP vector as well as for the pFlag-GFP vector will be described elsewhere (unpublished data).
Mutagenesis.
To engineer phosphorylation site mutations in SadA, the QuikChange II mutagenesis kit (Stratagene) was used. PAGE-purified primers from IDT (Coralville, IA) were used to incorporate the desired mutations (S→A, a nonphosphorylated mimetic, or S→E, a phospho-mimetic change) by using pBS-SadA as a template. Mutations were made in pairs (S924/S925 and S940/S941) or in threes (S943/S944/S950). In all cases, 10 colonies were picked from each reaction set, plasmids were purified by minipreparation (Invitrogen), and sequences were verified by DNA sequencing. A validated clone carrying only the desired mutation was subcloned into pTX-MCS-GFP by using the AatII and NheI sites. Plasmid DNA isolated with a maxiprep kit (Invitrogen) was subsequently transformed into sadA− cells by using the oscillating electroporation method.
Triton X-100 solubility assay.
Assays were generally carried out according to (20, 28). Briefly, AX3 wild-type cells were cultured in 10-cm dishes, to yield 2 × 106 cells/ml on the day of the experiment. To initiate the experiment, growth medium was replaced with 10 ml of fresh HL-5 medium containing either (i) no additive, (ii) 0.5% dimethyl sulfoxide (DMSO), or (iii) 5 μM latrunculin A in DMSO. After 45 min of treatment, cells were harvested into 15-ml conical tubes and placed on ice. Cells were harvested at 500 × g at 4°C and washed once with 1 PIPES buffer [20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.8), 1 mM MgCl2, 50 mM KCl, 5 mM EGTA, 5 mM dithiothreitol (DTT)]. The cells were brought up to a density of 2 × 107 cells/ml, and 200 μl of cells was added to 200 μl of 2× PIPES lysis buffer (40 mM PIPES [pH 6.8], 2 mM MgCl2, 100 mM KCl, 10 mM EGTA, 10 mM DTT, 2% Triton X-100). After incubating on ice for 2 min, lysates were centrifuged at 400 × g for 10 min at 4°C. Supernatant and pellet samples were brought up to 1× in SDS-PAGE sample buffer and fractionated on 10% SDS-PAGE gels according to methods described by Laemmli (25). Gels were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) (38), and subsequently probed with a polyclonal rabbit anti-SadA antibody, stripped, and reprobed with an antiactin antibody (C4; catalog number MAB150R; Millipore).
Generation of a SadA-reactive polyclonal antiserum.
The coding sequence for an extracellular region of SadA encompassing amino acids 30 to 273 was amplified by PCR and cloned into the vector pET14b (Novagen). After verification by DNA sequencing, the expression vector was transformed into the E. coli expression strain BL21(DE3). Subsequent to induction the His-tagged SadA protein (His-SadA N1) was purified on nickel beads and used as an immunogen in rabbits (Covance). Resultant antisera were affinity purified on immobilized His-SadA N1 and used for Western blotting applications. The specificity of the SadA N1 antisera was assessed by comparison of reactivity on Western blots containing both wild-type AX3 and sadA− lysates (G. Amargo, P. Fey, and R. L. Chisholm, unpublished data).
Two-dimensional gel electrophoresis.
Wild-type AX3 cells were grown under adherent or nonadherent conditions as described above. Upon reaching the desired cell density, cells were harvested and placed on ice. Subsequent to centrifugation at 500 × g for 5 min at 4°C, medium was removed and two-dimensional (2D) sample buffer was added to the cell pellet (6 M urea, 2 M thiourea, 2% ASBC80, 2% Bio-lyte ampholytes, phosphatase inhibitor cocktail). After solubilizing the pellet, DTT was added to a final concentration of 20 mM and samples were placed at −80°C. First-dimension isoelectric focusing (IEF) was performed using tube gels in a Bio-Rad mini-IEF system. Due to the presence of thiourea in the sample and the gel, an alternative method to polymerize the gel was used, as thiourea interferes with the commonly used APS/Temed method (27, 30). Approximately 4 × 106 cell equivalents were used for focusing, and a subsequent second-dimension SDS-PAGE was performed. After transfer to PVDF, membranes were probed with a rabbit polyclonal anti-SadA antibody.
Adhesion maintenance assay.
Adhesion maintenance assays were performed as described previously (16). Briefly, 5-cm petri dishes were seeded with 5 × 105 cells and allowed to incubate overnight. To initiate the assay, dishes were placed on an orbital rotating shaker (Bellco model 7744-01000 with a 51-cm by 51-cm platform, 2.5-cm orbit), set at a specified speed. After 1 h, nonadherent and adherent cells were harvested into respective 15-ml conical tubes. Cells were counted using a hemocytometer, and the percent cells that were adherent was plotted versus speed of rotation.
GST-tail pull-down assays.
GST or GST-tail proteins were added to glutathione-Sepharose beads (GE Healthcare) in equimolar amounts (∼6.6 nM or 200 μg per 30 μl of packed bead bed) which were subsaturating levels of bead binding capacity. After binding bait proteins, beads were blocked with 1% milk in lysis buffer for 1 to 2 h and subsequently washed before the addition of AX3 wild-type lysates (2.6 × 107 cell equivalents). During the bait binding and blocking steps, cells were harvested, washed twice with ice-cold PBS, and lysed in lysis buffer (50 mM HEPES [pH 7.5], 50 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.5 mM AEBSF [4-(2-aminoethyl)benzenesulfonylfluoride], 5 μg/ml aprotinin, 1 mM benzamidine-HCl, 3 μM E64, 5 μg/ml leupeptin, 8 μM pepstatin A) before being precleared with glutathione-Sepharose beads. Pull-down reactions occurred at 4°C on an end-over-end rotator for no longer than 4 h (longer incubations resulted in higher backgrounds). Beads were washed three times with lysis buffer, and excess liquid was removed using a Hamilton syringe. An equal volume (30 μl) of 2× SDS-PAGE sample buffer was applied to the beads, which were then boiled. Samples were fractionated on either 7.5% or 10% SDS-PAGE gels. For protein identification with mass spectrometry, gels were stained with colloidal Coomassie blue (8), and excised bands were submitted to the CBC-UIC Research Resources Center Proteomics and Informatics Services Facility. A Thermo LTQ liquid chromatography-tandem mass spectrometry (LC/MS/MS) instrument was used for peptide analysis. For Western blot analysis, gels were transferred to PVDF (Millipore or Bio-Rad) and probed with the indicated antibodies.
GST-tail phosphorylation site mutant pull-down assays and immunoprecipitations.
GST-SadA tail (phospho-mutants) pull-down assays were performed as described above with a slight modification. Prior to cell lysis, cells were treated with 5 μm latrunculin A to disrupt the F-actin cytoskeleton. Additionally, lysates were supplemented with 2.5 μm latrunculin A during the GST pull-down assay. Subsequent to SDS-PAGE, gels were transferred to Immobilon P membranes. Western blots were probed with an anti-cortexillin I antibody, stripped, and reprobed with an anti-Abp50 antibody. The GFP-Trap (34) experiment was performed with GFP-TrapA beads (GTA-20) obtained from ChromoTek GmbH. Subsequent to harvesting cells (AX3 plus pFLAG-GFP and sadĀ plus pSS-GFP-SadA), the latter were treated with 5 μM latrunculin A and incubated on ice for 90 min to disrupt the actin cytoskeleton. Cells from each strain were subsequently subjected to Triton X-100 treatment, as described for the solubility assays. Supernatant samples were reserved and diluted 1:2 with 2× immunoprecipitation buffer (1× buffer contains 20 mM HEPES [pH 7.5], 50 mM NaCl, 0.5 mM EGTA, 0.5% Triton X-100, 1% n-dodecyl-β-d-maltoside [catalog number 324355; Calbiochem], 1× protease inhibitor cocktail [catalog number 11836170001; Roche], with or without phosphatase inhibitor cocktail (catalog number 04906845001; Roche]). Diluted lysates (1 × 107 cell equivalents) were incubated with GFP-TrapA beads for 90 min at 4°C, followed by washing. An equal volume of 2× protein sample buffer was added to the beads, and samples were fractionated on 10% SDS-PAGE gels followed by transfer to Immobilon P membranes. The blots were probed with a monoclonal anti-cortexillin I antibody.
Live cell microscopy.
For live cell imaging, cells were harvested from HL5 medium, washed in phosphate buffer, added to glass-bottom petri dishes (WillCo wells; catalog number GWSt-3522; #1.5), and allowed to adhere for at least 30 min prior to imaging. In some instances, the agar overlay technique was employed to aid in imaging (19). Cells were imaged with a Nikon C1Si confocal microscope using either 60× or 100× objectives, and images were collected at a resolution of 512 by 512, 1,024 by 1,024, or 2,048 by 2,048 pixels.
Bioinformatic analysis.
For bioinformatic analysis, a number of individual Web-based programs were employed. For phosphorylation site predictions, two programs were used, NetPhosK and MiniMotif Miner (4, 5, 31). For protein identification, the raw mass spectra data files were converted into an mgf file format and searched against a Dictyostelium database using the program Mascot (29), licensed through the CBC-UIC Research Resources Center Proteomics and Informatics Services Facility at the University of Illinois at Chicago. Search parameters included two missed cleavages with trypsin, carbamidomethyl (C) as fixed and deamidated (NQ) and oxidation (M) as variable modifications, a peptide tolerance of ±2 Da with an MS/MS tolerance of ±0.6 Da, and a peptide charge of +2 and +3, with an ESI-TRAP instrument. To assess the false discovery rate (FDR), the automatic decoy database option was chosen.
RESULTS
SadA associates with the actin cytoskeleton.
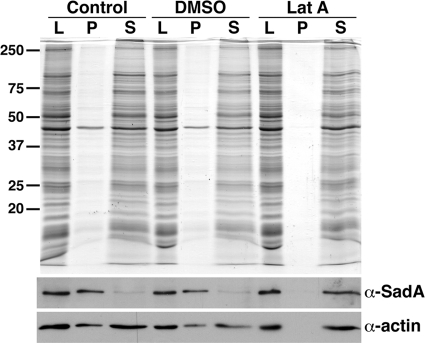
Initial characterization of the sadA mutant phenotype showed that these cells were defective in cytokinesis. Disruptions of the actin cytoskeleton are one common cause of cytokinesis defects. Consistent with this, sadA knockout cells exhibited an altered actin localization pattern (16). To further define the interaction of SadA with the cortical actin cytoskeleton, we employed Triton insolubility assays. In this assay, F-actin, as well as closely associated proteins, partition into the insoluble pellet, while other proteins remain soluble (6, 33, 42). Lysates of wild-type AX3 cells, treated with DMSO or left untreated, were prepared in a buffer containing 1% Triton X-100 and subjected to low-speed centrifugation. Figure 1 shows that the SadA protein was found in the pellet fraction in both untreated and DMSO-treated cells, suggesting a close relationship of SadA with the actin cytoskeleton.
Fig. 1.
SadA associates with the F-actin cytoskeleton. AX3 cells grown under adherent conditions received fresh medium (control) or were treated with DMSO or 5 μM latrunculin A (LatA). Cells lysed with 1% Triton X-100 (L) were subjected to low-speed centrifugation, divided into pellet (P) and supernatant (S) samples, and fractionated by SDS-PAGE followed by Western blotting. Membranes were probed with a polyclonal SadA antibody followed by a monoclonal anti-actin antibody. In control and DMSO-treated cells, SadA partitioned into the Triton-insoluble pellet, and actin was divided between the pellet and supernatant fractions. When cells were treated with latrunculin A, both SadA and actin were observed to fractionate completely into the Triton X-100-soluble fraction.
To verify this possible actin association, we treated cells with the drug latrunculin A, which disrupts the F-actin cytoskeleton (36). When wild-type cells were treated with 5 μM latrunculin A, we observed that the majority of the actin and SadA became soluble. This result is consistent with the hypothesis that SadA is associated with the F-actin cytoskeleton. However, it is not clear whether this association is direct or indirect.
Role for the carboxy-terminal tail in SadA function.
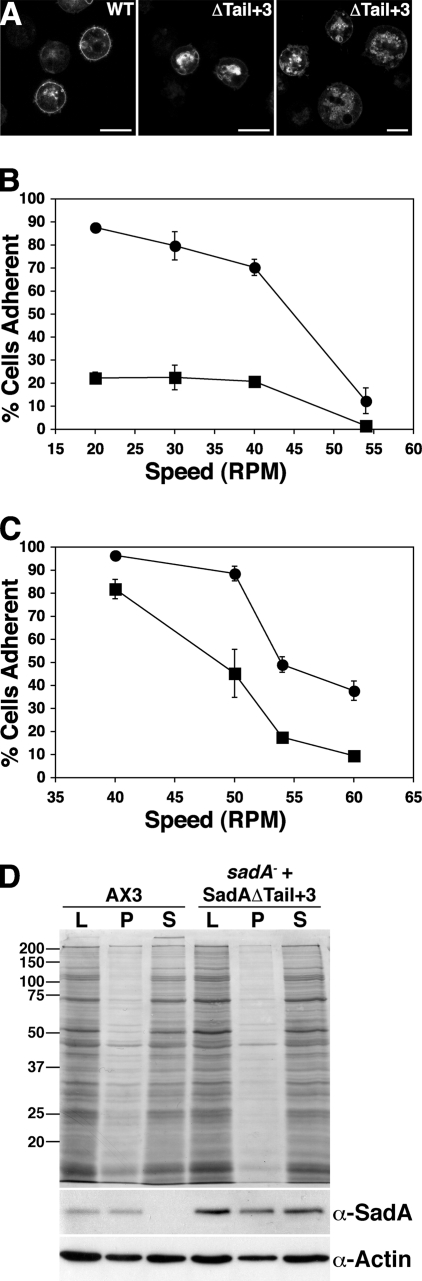
SadA is predicted to be a nine-pass transmembrane protein, with its amino-terminal domain displayed extracellularly and its carboxy-terminal domain in the cytoplasm. Given the interaction with actin, we were interested in identifying the domain that might mediate this interaction. The carboxy-terminal tail was an attractive candidate to play this role. To test this hypothesis, we prepared a SadA expression construct lacking all but the three residues proximal to the COOH-terminal transmembrane domain of the tail and assessed its ability to rescue the adhesion defect of the sadA null mutant in an adhesion maintenance assay as described previously (16). When expressed in sadA mutant cells, we observed that the majority of this truncated SadA did not localize to the plasma membrane but was found predominantly in intracellular vesicles (Fig. 2 A). Not surprisingly, we observed that these cells did not show significant adhesion compared to the sadA− mutant (Fig. 2B). When we assessed Triton solubility of the tail deletion mutant, we observed higher levels of the COOH tail deletion mutant in the Triton X-100-soluble fraction, suggesting a decreased association with actin (Fig. 2D). Therefore, the COOH-terminal tail deletion mutant was unable to fully rescue the sadA null phenotype, likely due in part to disrupted attachments to the actin cytoskeleton.
Fig. 2.
Evidence for functional significance of the SadA carboxy-terminal tail. (A) Micrographs of sadA null cells expressing either a wild-type SadA or a SadA carboxy-terminal deletion construct (SadA ΔTail+3-GFP construct) imaged with a confocal microscope. Increased levels of GFP signal were generally observed in cytoplasmic intracellular vesicles compared to the plasma membrane compartment. The right panel is a micrograph of cells imaged using the agar overlay technique, and the left panel shows cells without overlay. Bar, 10 μm. (B) Adhesion maintenance assay of AX3 wild-type and sadA null cells expressing the SadA ΔTail+3-GFP construct. Whereas AX3 cells (•) exhibited a sheer force-dependent decrease in substrate adhesion, the SadA ΔTail+3-GFP-expressing cells (▪) had minimal adhesion at all speeds tested. (C) Adhesion maintenance of AX3 cells (•) and AX3 cells overexpressing the SadA carboxy-terminal tail (▪). AX3 cells overexpressing the SadA tail were less resistant to increased sheer stress than AX3 wild-type cells. Error bars represent the standard deviations from triplicate or quadruplicate samples. (D) Triton solubility assay of AX3 and sadA− plus pTXSadAΔtail+3-GFP. The experiment was conducted as described for Fig. 1, but only under control (untreated) conditions. Whereas wild-type SadA partitioned primarily into the Triton X-100-insoluble fraction, SadAΔTail+3 partitioned, almost equally, between the insoluble and soluble fractions.
Next we asked if there might be any consequences of overexpressing this tail domain in a wild-type background. We hypothesized that if the tail was involved in mediating an interaction with actin, an excess of the domain might disrupt endogenous SadA function, perhaps affecting adhesion. To test this hypothesis, we designed a SadA tail expression construct and expressed this domain at high levels in wild-type cells. As can be seen in Fig. 2C, overexpression of the SadA carboxy-terminal tail reduced the adhesive properties of wild-type cells, suggesting that excess SadA tail domain might interfere with the ability of endogenous full-length SadA to interact with the actin cytoskeleton. Collectively, these data suggest that the carboxy-terminal cytoplasmic tail of SadA is functionally significant, justifying further exploration into its role.
SadA is a likely target for posttranslational modification.
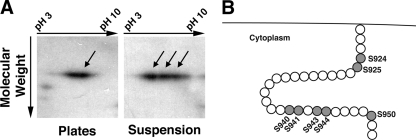
Since the tail domain of SadA seemed to be important for adhesive function, we next tested the hypothesis that SadA might be posttranslationally modified, perhaps depending on the adhesive state of the cells. To assess this question, wild-type cells were grown under adhesive conditions (i.e., in a petri dish), or in shaking suspension, where cells are nonadherent. Lysates were prepared from both growth conditions and subjected to two-dimensional gel electrophoresis followed by Western blotting. As can be observed in Fig. 3 A, when cells were grown in suspension the SadA protein showed up to four isoforms, while in plate-grown cells there were only one or two isoforms visible. This apparent acidic shift is consistent with the possibility of SadA becoming phosphorylated.
Fig. 3.
2D and bioinformatic analysis of SadA. (A) AX3 wild-type cells were grown under adherent conditions (plates) or under nonadherent conditions (suspension). Cell lysates were subjected to 2D gel analysis followed by Western blotting with a SadA antibody. Under adherent conditions, only one or two SadA reactive spots were seen. However, when grown in suspension, SadA reactivity was observed in a trail, consisting of three to four distinct spots. This trail may represent posttranslational modifications in the tail (i.e., phosphorylation) resulting in charge- induced isoforms. (B) Cartoon depiction of the cytoplasmic SadA carboxy-terminal tail. Shaded residues highlight the seven serine residues, which are putative sites of phosphorylation in this region. There are no threonines and only one tyrosine residue at position 933 in the tail region.
SadA S924 S925 and SadA S940 S941 alanine and glutamic acid variants mediate quantitatively different adhesive properties.
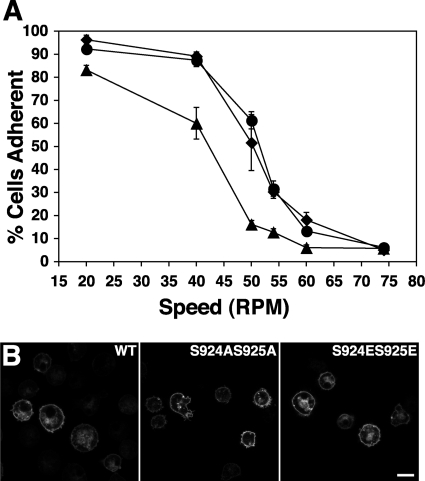
Our data suggest that SadA is closely associated with the F-actin cytoskeleton and appears to be differentially modified depending on the growth state and that the carboxy-terminal cytoplasmic tail is important in adhesion. These findings led us to consider the hypothesis that the tail domain is a target for phosphorylation, perhaps either regulating SadA association with F-actin or in an adhesive function. A scan of the SadA protein sequence using Web-based programs (NetPhos 2.0 server or Mini Motif Miner) (4, 31) identified seven serine residues and one tyrosine residue in the SadA tail region that are potential sites of phosphorylation (Fig. 3B). To test whether any of these sites in the SadA tail might be functionally significant, we systematically produced serine-to-alanine mutations, to mimic a nonphosphorylated state, and serine-to-glutamic acid mutations to mimic a phosphorylated state. These mutations were made in the context of full-length SadA expression constructs that were subsequently expressed in sadA null cells. Table 1 lists the mutants that were tested and summarizes their phenotypic characteristics. Subsequently, these sadA− rescued strains were assayed to determine their adhesive properties. From the seven SadA mutants generated, both the SadA S924/S925 and S940/S941 mutants demonstrated altered adhesion when assayed. As seen in Fig. 4 A, while the wild-type and S924AS925A SadA proteins showed similar levels of adhesive strength, the SadA S924ES925E mutants demonstrated reduced adhesion when subjected to similar shear forces. Similar phenotypes were observed in the S940/S941 mutants, as seen below in Fig. 7A.
Table 1.
SadA mutants generated and their phenotypesa
| Mutant | Mean speed (± SD) that 50% of cells remained adherent (no. of assays) | Actin localization |
|---|---|---|
| S924AS925A | 44.2 ± 6.3 (5) | Cortical |
| S924ES925E | 38.4 ± 10.1 (5) | Cortical |
| S940AS941A | 50.5 ± 4.9 (2) | Cortical |
| S940ES941E | 47 ± 5.7 (2) | Cortical |
| S950A | 63.5 ± 7.8 (2) | ND |
| S943AS944AS950A | ND | ND |
| S943ES944ES950E | ND | ND |
Seven unique SadA tail phosphorylation site mutants were generated during the course of the study. All mutants exhibited primary localization of SadA-GFP at the plasma membrane. Adhesion maintenance assays, as described in Materials and Methods, were conducted with multiple independent clones. The average speeds (in rpm) at which 50% of the cells remained adherent were estimated from graphs and used in the calculations. The number of independent assays performed for each strain is given in parentheses and was used to calculate the standard deviation. Actin localization was visually assessed on fixed cells probed with an antiactin antibody and was deemed to be cortical if strong signals were observed at the cell periphery. ND, not determined.
Fig. 4.
Functional and morphological effects of SadA S924 S925 mutations. (A) Adhesion strength of sadA null cells expressing either SadA GFP wild-type protein (•), SadA S924AS925A GFP (♦), or SadA S924ES925E GFP (▴) as measured in an adhesion maintenance assay. Whereas the AX3 and SadA S924AS925A GFP cells exhibited similar sheer force-dependent decreases in adhesion, the SadA S924ES925E mutant was less resistant to the same increases in sheer force. (B) Representative micrographs of confocal sections through sadA null cells expressing the same constructs as used in the adhesion maintenance assay. The sadA-GFP constructs were all capable of localizing to the plasma membrane. Bar, 10 μm.
Fig. 7.
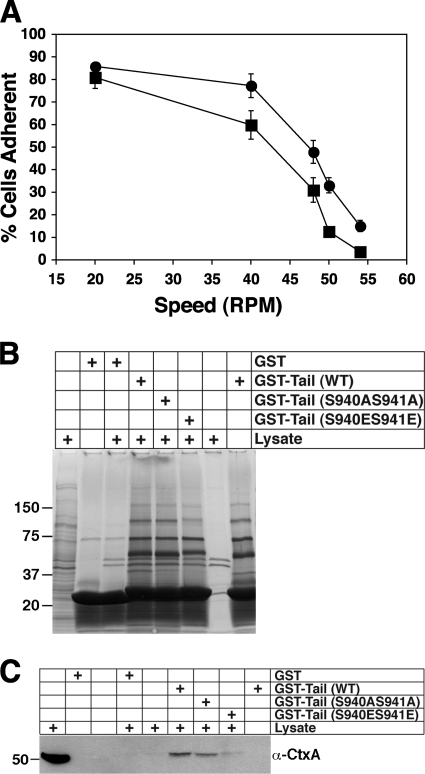
Functional and biochemical effects of SadA S940 S941 mutations. (A) sadA null cells expressing either SadA S940AS941A (•) or SadA S940ES941E (▪) were evaluated in an adhesion maintenance assay, as described for Fig. 4A. The S→E phospho-mimetic mutant exhibited less resistance to sheer force than the S→A non-phospho-mimetic mutant. (B) GST pull-down assays, carried out as described for Fig. 5B. (C) GST pull-down assay, as conducted for Fig. 6A, with anti-cortexillin I antibody as the probe.
To ensure that this reduced adhesion was not due to differences in the ability of the mutant proteins to be properly targeted to the plasma membrane, we observed the localization patterns of SadA-GFP-tagged proteins. As can be seen in Fig. 4B, the wild type and both the S924 S925 alanine and glutamic acid mutants of SadA-GFP were found to be localized to the plasma membrane, suggesting the altered adhesion of the serine mutant is unlikely to be due to differential localization to the plasma membrane. Localization of the SadA S940 S941 mutants also demonstrated plasma membrane localization (data not shown). In toto, these results are consistent with the hypothesis that one or more phosphorylation events in the SadA tail regulate adhesion.
The SadA tail interacts with the actin-associated protein cortexillin I.
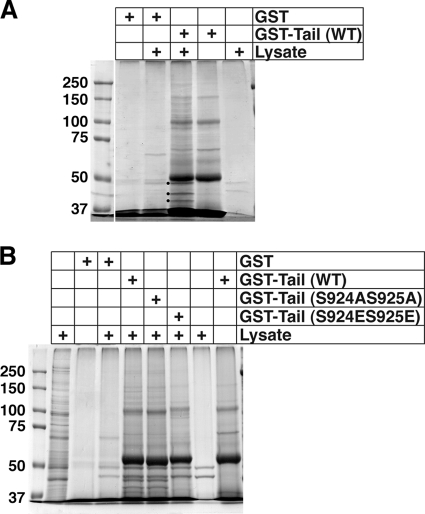
Both the presence of predicted phosphorylation site motifs and the mutational analysis presented above provide evidence that supports a possible role for phosphorylation in the regulation of SadA functions. Next, we wished to determine if the phenotypic differences might result from an altered interaction(s) between the SadA cytoplasmic tail and another possible protein partner(s). To address this question, we performed GST pull-down assays. A GST-SadA tail fusion protein was produced for use as bait in pull-down assays to identify proteins that might interact with the tail. Comparison of the pull-down profiles by using either GST or GST-tail as bait indicated that a number of unique bands were present when the tail was used as bait (Fig. 5 A), but not when GST alone was the bait. Three of these unique bands (indicated with dots) were excised from the gel and subjected to mass spectrometry analysis. A summary of the proteins found in the upper-most band (denoted with a black dot) from the GST-tail pull-down assay is provided in Table 2. Two very interesting candidate interaction partners, EfaAI (Abp50) and CtxA (cortexillin I), were identified. Importantly, both are documented actin binding and/or bundling proteins (12, 15, 41).
Fig. 5.
Identifying SadA tail-interacting proteins. (A) AX3 wild-type lysates were incubated with glutathione-Sepharose beads prebound with either GST (as a negative control) or with a GST-SadA tail fusion protein (wild-type sequence). GST and GST-tail lanes represent beads treated as described for experimental samples but without added lysate, to identify any nonspecific bands from the purified protein sample. The lysate lane represents beads treated as for the experimental samples but without any added bait protein, to observe proteins that nonspecifically bound to the beads. Three of the unique bands were excised from the gel (marked with black dots) and submitted for protein identification via mass spectrometry. (B) GST pull-down assay using GST-tail (WT), GST-tail (S924AS925A), or GST-tail (S924ES925E) fusion proteins as bait. GST, GST-tail, and the far right lysate samples were obtained as described for panel A. The lysate lane next to the molecular weight marker lane represents 1% of the amount of lysate used for pull-down assays.
Table 2.
Top-scoring proteins from the MS analysisa
| Hit | Accession no. | Protein name | Protein mass (Da) | Protein score | Protein coverage (%) |
|---|---|---|---|---|---|
| 1 | DDB_G0269134 | EfaAI | 49,916 | 721 | 33.3 |
| 1a | DDB_G0269136 | EfaAII | 49,916 | 721 | 33.3 |
| 2 | DDB_G0288613 | Erf1 | 49,600 | 121 | 11.1 |
| 3 | DDB_G0267426 | CshA | 55,885 | 117 | 13.4 |
| 4 | DDB_G0290693 | Eif2b3 | 50,656 | 114 | 24.5 |
| 5 | DDB_G0292410 | DDB_G0292410 | 42,966 | 109 | 20.6 |
| 6 | DDB_G0282483 | DDB_G0282483 | 40,363 | 100 | 18.4 |
| 7 | DDB_G0289483 | CtxA | 50,531 | 91 | 18.5 |
| 8 | DDB_G0294012 | Atp1 | 57,543 | 89 | 24.3 |
| 9 | DDB_G0291283 | SptB | 55,149 | 82 | 8.0 |
| 10 | DDB_G0295755 | GlnA2 | 59,506 | 67 | 13.8 |
The data shown are Mascot search results for the unique molecular weight band nearest the 50-kDa marker (denoted with a dot; see Fig. 5A). Search results were output into a csv file, and protein hits were sorted in descending order, based on protein score. The top 10 protein hits, within the 60- to 40-kDa cutoff, are listed. The FDR was computed to be 5.71%.
Due to the possibility of false-positive associations that can result from either the pull-down assay or the MS-based identification, it was critical to confirm any interaction(s) by an independent method. Furthermore, we wished to ascertain biochemically whether the functional differences between the SadA S924S925 nonphosphorylatable (S→A) and the phospho-mimetic (S→E) tail mutants might exhibit different protein-protein interaction profiles.
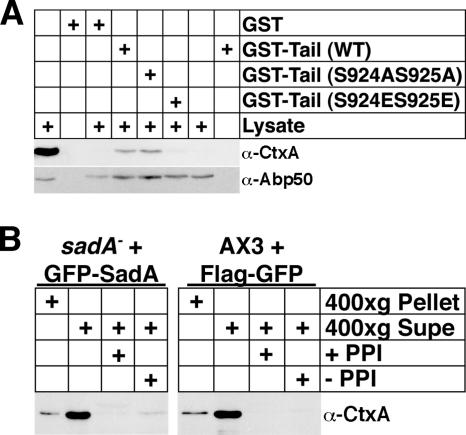
In an effort to validate the SadA tail-cortexillin I interaction, and also to uncover any differences in the interaction profile of wild-type or phospho-mimetic or nonmimetic mutations, we performed the GST-tail pull-down experiments using GST-SadA tail proteins as bait with either a nonmutated sequence or with the phosphorylation site mutations. When resultant samples were run on SDS-PAGE gels and stained with Coomassie blue (Fig. 5B), striking differences were observed in the pull-down profiles. Whereas the GST-tail (wild type [WT]) and GST-tail (S924AS925A) pull-down products exhibited very similar profiles, the GST-tail (S924ES925E) product showed a completely different profile. Samples from GST-tail pull-down assays from replicate experiments were transferred to PVDF following the SDS-PAGE, and Western blots were subsequently probed with antibodies specific to either Abp50 or CtxA (241-438-1). As can be seen in Fig. 6 A, when membranes were probed with either of these antibodies, very clear differences were observed. Whereas cortexillin I reactivity was observed only when GST-tail (WT) or GST-tail (S924AS925A) was used, Abp50 reactivity was seen in both experimental and control (GST-alone) pull-down experiments. The latter result suggests that Abp50 binds nonspecifically with the glutathione-Sepharose beads and is likely a false positive in the MS identification.
Fig. 6.
Testing the interaction between cortexillin I and SadA. (A) GST pull-down assay using GST-SadA tail (WT), GST-SadA tail (S924AS925A), or GST-SadA tail (S924ES925E) as bait. GST pull-down assays were carried out as described for Fig. 5, except that samples were treated with latrunculin A both before and subsequent to lysis to disrupt F-actin. The left-most lysate lane represents 1% of lysate, used as input into the pull-down reaction. The right-most lysate lane represents a mock pull-down sample (input lysate that was reacted with baitless beads). Subsequent to the pull-down assays, samples were fractionated on 10% SDS-PAGE gels, transferred to PVDF membranes, probed with an anti-cortexillin I antibody (241-438-1), stripped, and reprobed with an anti-Abp50 antibody. (B) GFP-trap pull-down assay. Prior to the pull-down assay, cells were subjected to Triton X-100 treatment as described for the solubility assays and divided between soluble and insoluble fractions. The 400 × g supernant (soluble) fraction from either sadA− cells expressing a GFP-SadA fusion protein (treated with latrunculin A) or from AX3 cells expressing a Flag-GFP fusion protein was applied to GFP-Trap beads and allowed to react. Resultant pull-down samples were subjected to SDS-PAGE, followed by transfer to PVDF. The membrane was probed with the anti-cortexillin I antibody.
To independently validate the SadA-cortexillin I interaction, a pull-down assay using GFP-Trap beads was employed (34, 39). When the GFP-SadA protein was immunoprecipitated with GFP-Trap, an interaction with cortexillin I was observed only in the absence of phosphatase inhibitors, but not in their presence or in control pull-down experiments (Fig. 6B). Together these two pieces of data provide evidence supporting cortexillin I as an interaction partner of the SadA tail. However, the experiments do not allow us to determine if the interaction is direct or indirect.
To determine if the SadA S940 S941 mutants also exhibited a similar biochemical phenotype, GST-tail pull-down assays were also conducted with these respective mutants. Interestingly, whereas the protein profiles for the GST-tail (WT) and GST-tail (S924AS925A) were considerably different from that of the GST-tail (S924ES925E) pull-down products, little difference was observed between the GST-tail S940S941 mutants and GST-tail (WT) (Fig. 7 B). However, when Western blots of pull-down products with the GST-tail (S940S941) mutants were probed with an anti-cortexillin I antibody, specific reactivity was observed only in the wild-type and S940AS941A baits, but there was little to no reactivity with the S940ES941E bait (Fig. 7C).
DISCUSSION
In a screen for proteins involved in cell-substrate adhesion in the social amoeba Dictyostelium discoideum, our laboratory previously reported the identification of the SadA protein (16). When the gene encoding SadA was disrupted, cells exhibited a number of intriguing phenotypes. SadA mutants are incapable of cell-substrate adhesion, show strong cytokinesis and phagocytosis defects, and have an abnormal F-actin cytoskeleton. The present work details a preliminary investigation into the mechanisms underlying these phenotypes. Our results demonstrate that SadA is associated with the F-actin cytoskeleton and that the carboxy-terminal cytoplasmic tail is important for SadA adhesive function. This cytoplasmic tail contains several phosphorylation site consensus domains that were assessed for functional roles by using mutagenesis. In addition, we have provided evidence suggesting that this tail associates with the known actin bundling protein cortexillin A. This suggests a model for how SadA might interact with the actin cytoskeleton (Fig. 8).
Fig. 8.
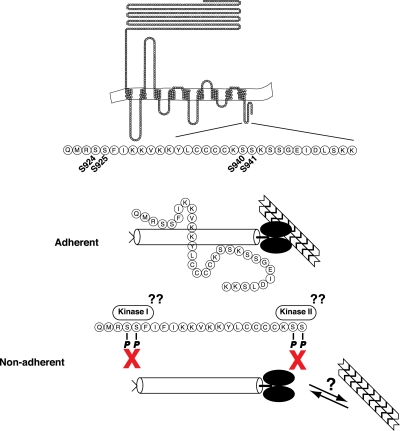
Proposed model for the SadA-cortexillin I interaction. Based on our current data, we propose the following model for the SadA tail-cortexillin I (CtxA) interaction. Under adherent conditions, an interaction occurs between the carboxy-terminal tail of SadA via the S924/5 and S940/1 sites and presumably the coiled-coil region of CtxA. This results in a linkage between SadA and the subcortical actin cytoskeleton. Under nonadherent conditions, or during cellular motility, perhaps, an unknown kinase(s) would phosphorylate sites in the SadA tail, resulting in release of CtxA from the SadA tail. This would temporarily disrupt the link between SadA and the actin cytoskeleton. During the processes of cell motility and phagocytosis, the cell must have a dynamic linkage between the actin cytoskeleton and membrane receptor in order to generate the forces necessary for the cell to move or to engulf a particle, respectively. The interaction between SadA and CtxA in our proposed model might be one mechanism that Dictyostelium has developed to perform these tasks.
Association of SadA with the F-actin cytoskeleton: a role for the tail?
In an effort to understand the association of SadA with the actin cytoskeleton, we employed Triton X-100 solubility studies. It has been well demonstrated that proteins that associate with the actin cytoskeleton pellet with the Triton-insoluble fraction, whereas unassociated proteins will remain in the soluble fraction (6, 33, 42). When AX3 wild-type cells were lysed with Triton X-100, we in fact found that SadA fractionated into the pellet, indicative of an interaction between SadA and F-actin. However, when cells were treated with the F-actin depolymerizing drug latrunculin A prior to lysis with Triton X-100, SadA was found to repartition into the Triton-soluble fraction. This result suggests an association between SadA and F-actin, although it is unknown if this association is direct or indirect.
When the SadA carboxy-terminal tail domain was overexpressed in AX3 wild-type cells, we observed adhesion defects. The most likely explanation for this dominant phenotype is that the overexpressed tail domain competes with the endogenous SadA tail for binding of some partner, effectively reducing its binding to the endogenous tail. Taken together, these observations suggest that the SadA carboxy-terminal tail is important for normal SadA function.
SadA: a putative phosphoprotein.
The finding that the SadA tail is important for adhesive function led us to examine the protein sequence of this region for consensus sequences. Interestingly, 7 out of the 31 (∼23%) amino acids in the tail are serine residues—sites for possible phosphorylation. The observation that SadA in suspension-grown cells showed additional negatively charged isoforms compared to plate-grown cells supports the idea that SadA might be phosphorylated in nonadherent cells. To assess this question more directly, we employed a mutagenesis approach to determine possible functional consequences of changing serines to either alanine (to mimic the nonphosphorylated state) or to glutamic acid (to mimic the phosphorylated state).
Functionally and biochemically (see below), two sites showed very intriguing differences. When resistance to shear stress was assessed we found that the SadA S924AS925A protein was able to rescue adhesion to near-wild-type levels, whereas the phospho-mimetic mutant, SadA S924ES925E, demonstrated a weaker level of adhesion. Although we don't know if modification is required at one or both of these sites, this result suggests that phosphorylation might be important. That both of these mutant proteins were able to localize to the plasma membrane argues against the observed functional difference being due to protein mislocalization. Furthermore, these data coincide with the results from the 2D analysis, in which SadA from adherent wild-type cells showed fewer charge isoforms than did cells grown in suspension. These observations are consistent with the hypothesis that the phosphorylation state correlates with the adhesive state.
SadA interacts with cortexillin.
The dominant negative phenotype of the overexpressed tail domain coupled with the possibility that posttranslational modifications in the tail domain might correlate with adhesion led us to explore potential binding partners for the SadA tail. Using a GST-SadA tail fusion protein as bait in pull-down assays, we showed that the tail is able to pull down multiple partners. GST-SadA tail mutants (S924AS925A or S924ES925E) were also used as bait in pull-down assays, and distinct differences were observed in the protein interaction profiles. Similar to the functional differences observed in the adhesion assays, we observed that the wild-type SadA tail sequence exhibited a similar profile to that of the GST-SadA S924AS925A tail. The GST-SadA S924ES925E tail had a noticeably unique profile: it was clear that the phospho-mimetic mutant was unable to interact with the same proteins as the wild-type or alanine mutant.
Two interesting candidate binding proteins were found in the mass spectrometry-based protein identification experiments: Abp50 (EfaAI) and cortexillin I (CtxA), both of which have known roles in F-actin binding. Abp50, besides being a translational elongation factor, has been found to have F-actin bundling characteristics (12, 13, 41). The exact role for this binding has not been uncovered; however, it has been suggested that F-actin may be used as a sink for Abp50 to regulate protein translation (23).
Cortexillin I was originally identified in a screen for proteins that precipitated with the F-actin cytoskeleton (15). The Dictyostelium genome codes for two cortexillin proteins. Cortexillin I has a carboxy-terminal nonapeptide sequence that interacts with PIP2 (37). Knockout of either proteins results in fairly mild phenotypes; however, ctxA cells show a higher level of cytokinesis defect and are flatter than ctxB cells. Cortexillin I forms homodimers via a coiled-coil domain and bundles F-actin primarily in an antiparallel fashion (15).
Although cortexillin I is found in the cytoplasm of vegetatively growing cells, as its name implies it is enriched at the cortex (15), where SadA is also localized. Subsequently, cortexillin I was found to play an extremely important role during cytokinesis, where it localizes almost exclusively to the cleavage furrow (40). It is interesting that ctxA null cells exhibited increased membrane elasticity (i.e., less energy was required to deform the membrane), similar to that of the membrane-actin linker protein talin A (35). That study also showed that ctxA cells were less adhesive than wild-type cells, although the talin A null cells had the weakest adhesion energy, in agreement with the adhesion phenotype of the talA mutant. We would expect that sadA cells would also exhibit similar defects, due to the mislocalized actin cytoskeleton in this mutant.
To provide independent validation of possible SadA binding of these candidate proteins (Abp50 and CtxA), Western blot analysis was used to identify proteins associated with wild-type and mutant GST-tail protein in pull-down assays and in a GFP-Trap pull-down assay. When blots were probed with an anti-Abp50 antibody (gift of D. Knecht [12]), Abp50 reactivity was observed in all the experimental samples, demonstrating that this was a false positive. In a tour de force experiment attempting to define a “bead proteome,” Trinkle-Mulcahy et al. found that EfaAI was also identified as a false-positive interaction protein, and so our result is perhaps not surprising (39). That anti-cortexillin I antibody reactivity was only observed when GST-tail wild type and S924AS925A were used as bait, but not in any of the other experimental samples, providing additional evidence that the SadA-cortexillin I interaction is significant. It is especially interesting that anti-cortexillin A reactivity was not observed when the GST-SadA S924ES925E tail was used as bait. These data are further supported by the finding in the GFP-Trap pull-down assay, in which cortexillin I reactivity was only observed in the absence of phosphatase inhibitors (Fig. 6B), which would likely increase the amount of a non-phospho-SadA isoform. It must be emphasized, however, that our experimental approach did not allow us to distinguish if the interaction is direct or, alternatively, occurs through an intermediary protein. Therefore, the exact nature of the interaction remains to be demonstrated. Regardless, our results suggest that in vivo phosphorylation could act as a regulator that controls the SadA and cortexillin I interaction.
Conclusions and a proposed model for SadA-CtxA function.
Loss of the sadA gene, besides causing loss of cell-substrate adhesion, also compromises the F-actin cytoskeleton as well as processes that are dependent on an intact cytoskeleton, such as cytokinesis and phagocytosis (16). How SadA mediates these various effects is unknown. The work presented in this report provides some insight into the mechanisms of SadA function. Our data suggest that the carboxy-terminal tail of SadA is functionally important and is a likely site for an interaction with the F-actin bundling protein CtxA. Furthermore, this link between SadA and CtxA is an extremely interesting candidate for providing the connection between SadA and the F-actin cytoskeleton. Evidence suggests that SadA is differentially phosphorylated (Fig. 3A), and that potential phosphorylation events in the carboxy-terminal tail may be used to regulate protein-protein interactions (Fig. 5C and 6C), specifically with CtxA.
We envision that SadA acts as an adhesion receptor, forming a link to the actin cytoskeleton via cortexillin I and perhaps other yet-unidentified proteins. Upon SadA incorporation into a nascent adhesion site, we hypothesize that the SadA tail will be in an unphosphorylated state, competent for interacting with CtxA and thereby mediating SadA's association with the F-actin cytoskeleton. This SadA–CtxA–F-actin linkage would provide mechanical strength to the adhesion site and perhaps be capable of generating tension to aid in migration or strengthened adhesion. Subsequent phosphorylation of the SadA tail would reduce the interaction with CtxA, thereby breaking the link between SadA and the F-actin cytoskeleton and relieving any mechanical tension.
Preliminary evidence from total internal reflection fluorescence microscopy (TIRFM) analysis of sadA null cells rescued with a sadA-GFP expression construct indicates that SadA-GFP is incorporated into transient foci on the surface apposed region of the cell. Characterization of these foci, as well as other potential components of the foci, is of considerable interest and would be useful in testing our hypothesis. It is likely that SadA is not the sole mediator of substrate adhesion during the vegetative stage of Dictyostelium amoebae. The Sib (similar to integrin beta) proteins, of which there are five different genes encoded in the genome, are also receptor candidates (9, 10). While only two Sib proteins (A and C) are expressed during vegetative growth, all five members have been shown to interact with TalA (Dictyostelium talin homologue), which consequently has also been shown to have a role in substrate adhesion. Regardless of the identity or number of receptors involved in cell-substrate adhesion, our model provides an interesting working model for how SadA might be the link between the extracellular substrate and the intracellular actin cytoskeleton.
ACKNOWLEDGMENTS
The cortexillin I antibody (241-438-1) developed in the lab of Gunter Gerisch was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA. Proteomics and informatics services were provided by the CBC-UIC Research Resources Center Proteomics and Informatics Services Facility, which was established by a grant from The Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium.
We also thank David Knecht for providing us with an aliquot of the Dictyostelium Abp50 antibody, as well as Jan Faix for providing an aliquot of the cortexillin I antibody early on in this study. Assistance with imaging was provided by Teng Leong Chew and Satya Kuhon of the Northwestern University Imaging Facility on the Chicago campus. We are grateful to the reviewers of our manuscript for providing constructive suggestions that helped to make this a more robust report.
Footnotes
Published ahead of print on 25 March 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Alibaud L., Cosson P., Benghezal M. 2003. Dictyostelium discoideum transformation by oscillating electric field electroporation. Biotechniques 35:78–80 [DOI] [PubMed] [Google Scholar]
- 2. Appella E., Weber I. T., Blasi F. 1988. Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett. 231:1–4 [DOI] [PubMed] [Google Scholar]
- 3. Baldauf S. L. 2003. The deep roots of eukaryotes. Science 300:1703–1706 [DOI] [PubMed] [Google Scholar]
- 4. Balla S., et al. 2006. Minimotif Miner: a tool for investigating protein function. Nat. Methods 3:175–177 [DOI] [PubMed] [Google Scholar]
- 5. Blom N., Gammeltoft S., Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- 6. Brown S., Levinson W., Spudich J. A. 1976. Cytoskeletal elements of chick embryo fibroblasts revealed by detergent extraction. J. Supramol. Struct. 5:119–130 [DOI] [PubMed] [Google Scholar]
- 7. Bukharova T., et al. 2005. Paxillin is required for cell-substrate adhesion, cell sorting and slug migration during Dictyostelium development. J. Cell Sci. 118:4295–4310 [DOI] [PubMed] [Google Scholar]
- 8. Candiano G., et al. 2004. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25:1327–1333 [DOI] [PubMed] [Google Scholar]
- 9. Cornillon S., Froquet R., Cosson P. 2008. Involvement of Sib proteins in the regulation of cellular adhesion in Dictyostelium discoideum. Eukaryot. Cell 7:1600–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cornillon S., et al. 2006. An adhesion molecule in free-living Dictyostelium amoebae with integrin beta features. EMBO Rep. 7:617–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis C. G. 1990. The many faces of epidermal growth factor repeats. New Biol. 2:410–419 [PubMed] [Google Scholar]
- 12. Demma M., Warren V., Hock R., Dharmawardhane S., Condeelis J. 1990. Isolation of an abundant 50,000-dalton actin filament bundling protein from Dictyostelium amoebae. J. Biol. Chem. 265:2286–2291 [PubMed] [Google Scholar]
- 13. Dharmawardhane S., Demma M., Yang F., Condeelis J. 1991. Compartmentalization and actin binding properties of ABP-50: the elongation factor-1 alpha of Dictyostelium. Cell Motil. Cytoskel. 20:279–288 [DOI] [PubMed] [Google Scholar]
- 14. Eichinger L., et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faix J., et al. 1996. Cortexillins, major determinants of cell shape and size, are actin-bundling proteins with a parallel coiled-coil tail. Cell 86:631–642 [DOI] [PubMed] [Google Scholar]
- 16. Fey P., Stephens S., Titus M. A., Chisholm R. L. 2002. SadA, a novel adhesion receptor in Dictyostelium. J. Cell Biol. 159:1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Franke J., Kessin R. 1977. A defined minimal medium for axenic strains of Dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A. 74:2157–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frisch S. M., Francis H. 1994. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124:619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukui Y., Yumura S., Yumura T. K., Mori H. 1986. Agar overlay method: high-resolution immunofluorescence for the study of the contractile apparatus. Methods Enzymol. 134:573–580 [DOI] [PubMed] [Google Scholar]
- 20. Galvin N. J., Stockhausen D., Meyers-Hutchins B. L., Frazier W. A. 1984. Association of the cyclic AMP chemotaxis receptor with the detergent-insoluble cytoskeleton of Dictyostelium discoideum. J. Cell Biol. 98:584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harwood A., Coates J. C. 2004. A prehistory of cell adhesion. Curr. Opin. Cell Biol. 16:470–476 [DOI] [PubMed] [Google Scholar]
- 22. Hynes R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673–687 [DOI] [PubMed] [Google Scholar]
- 23. Kim S., Coulombe P. A. 2010. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat. Rev. Mol. Cell Biol. 11:75–81 [DOI] [PubMed] [Google Scholar]
- 24. Kreitmeier M., Gerisch G., Heizer C., Muller-Taubenberger A. 1995. A talin homologue of Dictyostelium rapidly assembles at the leading edge of cells in response to chemoattractant. J. Cell Biol. 129:179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 26. Levi S., Polyakov M., Egelhoff T. T. 2000. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid 44:231–238 [DOI] [PubMed] [Google Scholar]
- 27. Lyubimova T., Caglio S., Gelfi C., Righetti P. G., Rabilloud T. 1993. Photopolymerization of polyacrylamide gels with methylene blue. Electrophoresis 14:40–50 [DOI] [PubMed] [Google Scholar]
- 28. McRobbie S. J., Newell P. C. 1983. Changes in actin associated with the cytoskeleton following chemotactic stimulation of dictyostelium discoideum. Biochem. Biophys. Res. Commun. 115:351–359 [DOI] [PubMed] [Google Scholar]
- 29. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- 30. Rabilloud T. 1998. Use of thiourea to increase the solubility of membrane proteins in two-dimensional electrophoresis. Electrophoresis 19:758–760 [DOI] [PubMed] [Google Scholar]
- 31. Rajasekaran S., et al. 2009. Mini Motif Miner 2nd release: a database and web system for motif search. Nucleic Acids Res. 37:D185–D190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robbins S. M., Suttorp V. V., Weeks G., Spiegelman G. B. 1990. A ras-related gene from the lower eukaryote Dictyostelium that is highly conserved relative to the human rap genes. Nucleic Acids Res. 18:5265–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosenberg S., Stracher A., Lucas R. C. 1981. Isolation and characterization of actin and actin-binding protein from human platelets. J. Cell Biol. 91:201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rothbauer U., et al. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell Proteomics 7:282–289 [DOI] [PubMed] [Google Scholar]
- 35. Simson R., et al. 1998. Membrane bending modulus and adhesion energy of wild-type and mutant cells of Dictyostelium lacking talin or cortexillins. Biophys. J. 74:514–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spector I., Shochet N. R., Kashman Y., Groweiss A. 1983. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219:493–495 [DOI] [PubMed] [Google Scholar]
- 37. Stock A., et al. 1999. Domain analysis of cortexillin I: actin-bundling, PIP(2)-binding and the rescue of cytokinesis. EMBO J. 18:5274–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trinkle-Mulcahy L., et al. 2008. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183:223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weber I., et al. 1999. Cytokinesis mediated through the recruitment of cortexillins into the cleavage furrow. EMBO J. 18:586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang F., Demma M., Warren V., Dharmawardhane S., Condeelis J. 1990. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature 347:494–496 [DOI] [PubMed] [Google Scholar]
- 42. Yu J., Fischman D. A., Steck T. L. 1973. Selective solubilization of proteins and phospholipids from red blood cell membranes by nonionic detergents. J. Supramol. Struct. 1:233–248 [DOI] [PubMed] [Google Scholar]
- 43. Zaidel-Bar R., Geiger B. 2010. The switchable integrin adhesome. J. Cell Sci. 123:1385–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zaidel-Bar R., Itzkovitz S., Ma'ayan A., Iyengar R., Geiger B. 2007. Functional atlas of the integrin adhesome. Nat. Cell Biol. 9:858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]