Abstract
DOT1 (disruptor of telomeric silencing; also called Kmt4) was initially discovered in budding yeast in a genetic screen for genes whose deletion confers defects in telomeric silencing. Since the discovery ∼10 years ago that Dot1 and its mammalian homolog, DOT1L (DOT1-Like), possess histone methyltransferase activity toward histone H3 Lys 79, great progress has been made in characterizing their enzymatic activities and the role of Dot1/DOT1L-mediated H3K79 methylation in transcriptional regulation, cell cycle regulation, and the DNA damage response. In addition, gene disruption in mice has revealed that mouse DOT1L plays an essential role in embryonic development, hematopoiesis, cardiac function, and the development of leukemia. The involvement of DOT1L enzymatic activity in leukemogenesis driven by a subset of MLL (mixed-lineage leukemia) fusion proteins raises the possibility of targeting DOT1L for therapeutic intervention.
Keywords: chromatin, DOT1, epigenetics, histone, leukemia, transcription
Eukaryotic DNA is wrapped around a histone octamer (H3/H4 heterotetramer and two H2A/H2B dimers) to form the nucleosome, the fundamental building block of chromatin (Van Holde et al. 1980; Luger et al. 1997; Kornberg and Lorch 1999; Zhang and Dent 2005). Histone proteins are subject to reversible post-translational modifications, which play an important role in the establishment and maintenance of gene expression patterns within any given cell type of an organism. Studies in the past decade have established that covalent histone modifications influence chromatin structure and function directly or indirectly through the recruitment of effector proteins to specific chromatin regions (Strahl and Allis 2000; Martin and Zhang 2005; Kouzarides 2007).
One of the covalent histone modifications is methylation, which takes place on both lysine (K) and arginine (R) residues (Zhang and Reinberg 2001; Martin and Zhang 2005). Lysine methylation exists in mono, di, and tri states, while arginine methylation only occurs in mono and di states. One way that these methyl marks contribute to transcriptional regulation is to serve as a platform for the recruitment of effector proteins. The well-studied lysine methylation residues include K4, K9, K27, K36, and K79 of histone H3, and K20 of histone H4. In general, methylation at H3K9, H3K27, and H4K20 correlates with transcriptional repression, while methylation at H3K4, H3K36, and H3K79 correlates with gene transcription (Kouzarides 2002; Peterson and Laniel 2004; Martin and Zhang 2005). In addition to its role in transcriptional regulation, methylation has also been linked to X inactivation, cell fate determination, terminal differentiation, and the spatiotemporal patterning of Hox genes (Cavalli 2006; Minard et al. 2009). Moreover, aberrant histone methylation has also been linked to various human cancers (Feinberg et al. 2002; Handel et al. 2010).
Histone methylation is catalyzed by a group of histone methyltransferases. Based on their catalytic domains, the lysine methyltransferases (KMTs) that have been characterized to date can be divided into two classes. The first class contains an evolutionarily conserved SET {Su(var)3-9, Enhancer of Zeste [E(Z)], and Trithorax (trx)} domain (Jenuwein et al. 1998). In contrast, the second class does not possess a SET domain and consists of only an evolutionarily conserved protein named Dot1 (disruptor of telomeric silencing; also called Kmt4) (Singer et al. 1998) and its homologs in other organisms (Feng et al. 2002; Lacoste et al. 2002; van Leeuwen et al. 2002). Dot1 and its homologs contain a catalytic methylase fold resembling that of class I methylases (Ng et al. 2002a; Min et al. 2003; Schubert et al. 2003). Over the years, great progress has been made in elucidating the function of Dot1 and its associated H3K79 methylation mark in various organisms and cellular processes. Here we summarize these studies with an emphasis on Dot1's enzymatic activity and its diverse biological functions.
Dot1/DOT1L (DOT1-Like) enzymatic activity and its regulation
Dot1 and its homologs possess H3K79 methyltransferase activity
Using different approaches, several groups independently discovered that Lys 79 within the globular domain of histone H3 (H3K79) is subject to methylation, and that the yeast protein Dot1 and its human homolog, DOT1L, are responsible for catalyzing the methylation reaction (Feng et al. 2002; Lacoste et al. 2002; Ng et al. 2002a; van Leeuwen et al. 2002). Both enzymes are capable of catalyzing mono-, di-, and trimethylation in a nonprocessive manner (Min et al. 2003; Frederiks et al. 2008). Dot1 and its homologs appear to be solely responsible for H3K79 methylation, since knockout of Dot1 in yeast, flies, and mice results in complete loss of H3K79 methylation (van Leeuwen et al. 2002; Shanower et al. 2005; Jones et al. 2008).
Dot1 homologs share a conserved region with four sequence motifs—I, post I, II, and III—found in SAM methyltransferases (Fig. 1A). The crystal structures of both yeast Dot1and human DOT1L also reveal an open α/β structure composed of a seven-stranded β sheet that is characteristic of the class I SAM-dependent methyltransferases (Fig. 1B; Cheng and Roberts 2001; Min et al. 2003; Sawada et al. 2004). Despite the fact that both human and yeast Dot1 proteins are structurally more similar to arginine methyltransferases (Fig. 1B), members of the Dot1 family proteins catalyze histone lysine methylation rather than arginine methylation (Feng et al. 2002; Lacoste et al. 2002; van Leeuwen et al. 2002; Zhang et al. 2004; Shanower et al. 2005). Due to the structural similarities between Dot1 and arginine methyltransferases (Fig. 1B), there has been some speculation as to whether Dot1 can also catalyze arginine methylation. However, extensive efforts using reverse-phase high-performance liquid chromatography (HPLC) coupled with nano-liquid chromatography electrospray ionization mass spectrometry (LC-ESMS) and tandem mass spectrometry (MS/MS) have failed to demonstrate that Dot1 has arginine methyltransferase activity (van Leeuwen et al. 2002).
Figure 1.
Dot1 is a conserved class I SAM-dependent methylase. (A) Sequence alignment of the conserved catalytic core region of Dot1 across multiple species that include yeast (NP_010728), worm (NM_058569), fly (AE003675), mouse (BB678539), and human (NP_115871). Secondary structure elements are shown above the alignment. The dashed line indicates disordered regions. Invariant amino acids are shown in white letters against a gray background, while conserved residues are in bold. Amino acids conserved across all five species are in bold against a green background. Every 10 amino acids is marked by a plus sign. The conserved methyltransferase fold motifs—I, post I, II, and III—are labeled and boxed in red. (B) The crystal structure of the substrate-binding pocket of human DOT1L (Protein Data Bank [PDB] code 1NW3) demonstrates that it is more similar to that of rat PRMT1 (PDB code 1ORI), a class I arginine methyltransferase, than to the KMT hNSD1 (PDB code 3OOI).
Interplay between H2B ubiquitination and H3K79 methylation
Characterization of DOTL enzymatic activity indicated that it preferentially methylates H3K79 in the context of nucleosomes rather than core histones or recombinant H3 (Feng et al. 2002). This observation suggests that DOT1L may recognize other features of the nucleosome. Based on nucleosome structure, K79 lies within loop 1 of histone H3, which is solvent-accessible and lies adjacent to the interface between the H3/H4 tetramer and H2A/H2B dimer (Luger et al. 1997). This unique position of H3K79 and the preference of DOT1L for nucleosomal substrates suggested that trans-histone cross-talk may play a role in regulating DOT1L enzymatic activity, which indeed turned out to be the case.
In yeast, histone H2B can be monoubiquitinated at Lys 123 (H2B-K123ub). This modification is catalyzed by the ubiquitin-conjugating E2 enzyme Rad6 and the ubiquitin E3 ligase Bre1 (Robzyk et al. 2000; Wood et al. 2003a). Bre1 not only serves as an E3 ligase for H2B ubiquitination, but is also required for the recruitment of Rad6 to chromatin (Wood et al. 2003a). Interestingly, both H3K79 and H2B-K123 lie in proximity on the same exposed nucleosome surface (Luger et al. 1997), which provides a structural basis for an interplay between the two modifications (Fig. 2A). Consistently, deletion of Rad6 and members of the Paf1 complex not only blocked H2B ubiquitination, but also prevented H3K4 and H3K79 methylation (Briggs et al. 2002; Ng et al. 2002b; Sun and Allis 2002). Furthermore, mutation of H2B-K123 also prevented H3K79 and H3K4 methylation, indicating that H2B ubiquitination is a prerequisite for H3K79 and H3K4 methylation (Briggs et al. 2002; Ng et al. 2002b, 2003b; Sun and Allis 2002; Krogan et al. 2003; Wood et al. 2003b). This effect appears to be unidirectional, as deletion of Dot1 does not affect H2B ubiquitination (Sun and Allis 2002). Additionally, the relationship between H2B ubiquitination and histone methylation is residue-dependent, as H3K36 methylation is not affected by Rad6 deletion (Briggs et al. 2002; Ng et al. 2002b).
Figure 2.
Ubiquitination of yH2B-K123/hH2B-K120 regulates Dot1-mediated H3K79 methylation. (A) The crystal structure of the human nucleosome (PDB code 3AFA) is shown on the left. Histone H2A is shown in light green, H4 is shown in sand, H2B is shown in hot pink, and H3 is shown in blue. A close-up view of the nucleosome surface is on the right, with the location of H2B-K120 shown as yellow spheres and H3K79 shown as green spheres. Note that H2B-K120 and H3K79 are located in proximity on the same solvent-exposed surface of the nucleosome. (B) Proposed models for trans-histone regulation of Dot1 enzymatic activity by H2B-K123ub. Histone H2B-K123 ubiquitination is mediated by the ubiquitin-conjugating enzyme Rad6 and its E3 ubiquitin ligase, Bre1. Components of the Paf1 complex are required for efficient H2B ubiquitination and di- and trimethylation of H3K4 and H3K79. (Bottom left) The first model suggests that Dot1 can bind directly to H2B-K123ub for intranucleosomal H3K79 methylation. (Top right) The second model suggests that other factors, such as the Set1-containing COMPASS, are required to bridge the interaction between Dot1 and the nucleosome. Rtf1 of the Paf1 complex recruits the COMPASS complex to methylate H3K4. The Cps35 subunit of the COMPASS interacts directly with H2B-K123 and Dot1, thus bringing Dot1 to the nucleosome for H3K79 methylation. (Bottom right) In the third model, ubiquitination of H2B causes a conformational change to the nucleosome, making H3K79 more accessible to DOT1.
The critical role of H2B ubiquitination in mediating H3K79 methylation raised the question of how H2B ubiquitination contributes to H3K79 methylation. Three mutually nonexclusive possibilities have been proposed. They include (1) indirect interaction between Dot1 and H2B-K123ub through a bridging protein, (2) direct interaction between Dot1 and H2B-K123ub, and (3) that ubiquitination of H2B causes chromatin structure changes to facilitate Dot1-mediated H3K79 methylation. Published evidence supports the use of all three mechanisms. For example, H2Bub can recruit Rpt4 and Rpt6 to chromatin to facilitate H3K79 methylation (Ezhkova and Tansey 2004). In addition, Cps35, a component of the Set1 (also called Kmt2)-containing COMPASS complex, interacts directly with both H2B-K123ub and Dot1 and is required for H3K79 trimethylation (Lee et al. 2007). These studies suggest that ubiquitinated H2B indirectly recruits Dot1 to methylate H3K79 through some bridging factors (Fig. 2B, top right). On the other hand, Dot1 can interact directly with ubiquitin through a lysine-rich region (amino acids 101–140) within the N terminus of Dot1, which is also required for nucleosome binding, and deletion of this region resulted in reduced di- and trimethylation (Oh et al. 2010). In addition, chemically engineered ubiquitinated H2B has been shown to stimulate intranucleosomal H3K79 methylation by DOT1L without the presence of other bridging factors (Fig. 2B, bottom left; McGinty et al. 2008; Chatterjee et al. 2010). Finally, in support of the third mechanism, it was shown that ubiquitination of H2B can disrupt chromatin compaction and interchromatin fiber interactions to increase substrate accessibility (Fig. 2B, bottom right; Fierz et al. 2011). This contradicts a previous study demonstrating that H2B ubiquitination regulates nucleosome stability rather than chromatin conformation (Chandrasekharan et al. 2009). The discrepancy between the two studies may be caused by the use of different substrates. Muir and colleagues (Fierz et al. 2011) used chemically engineered and reconstituted nucleosomal substrates, whereas Sun and colleagues (Chandrasekharan et al. 2009) used modified nucleosomes purified from different mutant yeast strains. In summary, current literature supports the proposal that several mutually nonexclusive mechanisms, illustrated in Figure 2B, are used for the regulation of H3K79 methylation by H2B ubiquitination.
Reversibility of H3K79 methylation
Although the establishment of H3K79 methylation is well characterized and is highly regulated by multiple processes, active demethylation of this mark is less well understood. Thus far, with the exception of H3K79, all other known histone lysine residues subject to methylation can be demethylated by at least one histone demethylase (Klose et al. 2006; Klose and Zhang 2007; Cloos et al. 2008; Liu et al. 2010; Qi et al. 2010). However, several lines of evidence suggest that H3K79 methylation might be reversible, as it is subject to dynamic regulation. First, in yeast and human cells, the H3K79me2 methylation level fluctuates with the cell cycle (Feng et al. 2002; Schulze et al. 2009). Second, H3K79me2 is demethylated during early embryonic development in both mice and flies, suggesting that an inactive demethylase might reside in mature oocytes (Shanower et al. 2005; Ooga et al. 2008). Third, a study investigating the antagonistic effects of 2-hydroxyglutarate (2-HG) on α-KG (a cofactor for dioxygenases that include the JmjC domain-containing histone demethylases and the TET family of 5-methylcytosine hydroxylases) demonstrated that global levels of H3K79me2 increased in the presence of synthetic 2-HG and mutant isocitrate dehydrogenase (IDH), a NADP-dependent enzyme that catalyzes the oxidative decarboxylation of isocitrate to α-KG. These observations suggest that a dioxygenase may catalyze the removal of H3K79me2 marks in vivo (Xu et al. 2011). We anticipate that it may not be too long before the identity of this elusive H3K79-specific demethylase is revealed.
Dot1-mediated H3K79 methylation in transcriptional regulation
Dot1 and telomeric silencing in Saccharomyces cerevisiae
Four years prior to the characterization of Dot1's enzymatic activity, DOT1 was identified in a genetic screen for genes whose overexpression disrupts telomeric silencing. The establishment of telomere and telomere-proximal DNA silencing is achieved through the recruitment and binding of Sir (silent information regulator) proteins (Norris and Boeke 2010). Interestingly, overexpression and deletion of Dot1, as well as mutation of H3K79, compromise the silencing at telomeres and HML/HMR loci. In all three cases, the level of Sir proteins bound at telomeres is reduced, thus limiting their ability to silence genes. Sir3 interacts with H3K79 through its N-terminal BAH domain, and this interaction is impeded by H3K79 methylation, and, consequently, Sir3 binding and spreading of heterochromatin is affected (Katan-Khaykovich and Struhl 2005; Onishi et al. 2007; van Welsem et al. 2008). Reciprocally, Sir3 inhibits H3K79 methylation by competing with Dot1 for binding to the same basic patch of histone H4 that is required for Dot1 enzymatic activity (Altaf et al. 2007; Fingerman et al. 2007). Thus, balancing the amount of Sir protein binding and the level of H3K79 methylation is important for regulating heterochromatin formation at telomeres.
We note that, in these previous studies, reporter systems were used to draw general conclusions about Dot1-mediated H3K79 methylation and telomere silencing. Interestingly, two recent studies challenge the role of Dot1 in heterochromatin formation. Employing the URA3 telomere reporter system, Rossmann et al. (2011) demonstrated that the variegated silencing observed in Dot1 mutants is due to a disproportion in ribonucleotide reductase and the promoter of URA3 at telomere VII–L rather than a requirement for Dot1 in general telomere silencing. Takahashi et al. (2011) took an alternative approach by analyzing natural telomere position–effect variegation and show that the role of Dot1 in heterochromatin formation is telomere-specific. Nevertheless, interplay between Sir proteins and Dot1 does serve to regulate heterochromatin formation at telomeres, but perhaps to a lesser extent than originally thought.
Genome-wide analysis links H3K79me to active transcription
In addition to participating in telomere silencing, genome-wide analysis of H3K79 methylation has linked this modification to transcription. Approximately 10% of the yeast genome is hypomethylated at H3K79 and bound by Sir proteins (Laurenson and Rine 1992; van Leeuwen et al. 2002), which is consistent with the fact that methylated H3K79 inhibits Sir binding (Onishi et al. 2007; van Welsem et al. 2008). Meanwhile, 90% of the yeast genome bears H3K79 methylation and is actively transcribed (Laurenson and Rine 1992; van Leeuwen et al. 2002), suggesting that H3K79 methylation is a marker of euchromatin. In support of this notion, work by Ng et al. (2003a) showed that H3K79 methylation correlates with euchromatin at sites of active V(D)J recombination, while hypomethylation is found at inactive loci.
The development of high-throughput technology to map specific chromatin modifications on a genome-wide scale has provided an invaluable method to further decipher the role of H3K79 methylation in transcriptional regulation. In Drosophila, chromatin immunoprecipitation (ChIP) coupled with gene expression microarray (ChIP–chip) revealed that histone acetylation, H3K4me2, H3K4me3, and H3K79me2 positively correlated with active gene transcription (Schubeler et al. 2004). In contrast, another study using ChIP–chip demonstrated in S. cerevisiae that, although H3K79me3 is enriched within transcribed gene regions, there was no correlation between the level of H3K79me3 and transcriptional activity (Pokholok et al. 2005). This was further supported by Zhao and colleagues (Barski et al. 2007) using ChIP coupled with deep sequencing (ChIP-seq) to map mono-, di-, and trimethyl H3K79 in human CD4+ T cells. They found a slight correlation between H3K79me1 and gene activation. However, H3K79me3 was more closely associated with gene repression, while H3K79me2 did not correlate with transcriptional status (Barski et al. 2007). These two genome-wide analyses were inconsistent with previous studies linking H3K79 methylation to active gene transcription (Ng et al. 2003a; Schubeler et al. 2004; Okada et al. 2005, 2006; Vakoc et al. 2006).
The discrepancy between different genome-wide profiling studies was later attributed to different methods used for chromatin preparation. The two reports showing no correlation between H3K79 methylation and transcriptional activity performed ChIP in the absence of sodium dodecyl sulfate (SDS), which is required for proper detection of H3K79 methylation in formaldehyde-cross-linked chromatin (Steger et al. 2008). Accordingly, ChIP–chip using mouse 3T3 cells demonstrates that all H3K79 methylation marks are localized within the body of transcribed genes, and that the amount of enrichment correlates with expression level (Steger et al. 2008). Subsequently, following the revised ChIP protocol, Zhao and colleagues (Wang et al. 2008) published a second genome-wide study, again using human CD4+ T cells, which confirmed the findings by Steger et al. (2008). Collectively, analyses of yeast, mouse, fly, and human genomes revealed that H3K79 methylation is indeed a marker of active transcription.
Role of Dot1L in transcriptional elongation
The genome-wide localization studies revealed that H3K79 methylation is enriched in gene coding regions (Kouskouti and Talianidis 2005), implicating a potential role for Dot1 in transcriptional elongation. Consistent with this notion, the Paf1 complex, which is associated with elongating RNA polymerase II (Pol II), regulates Dot1 H3K79 methyltransferase activity (Krogan et al. 2003). In addition, involvement of DOT1L in transcriptional elongation is supported by the purification of various mammalian DOT1L-containing complexes, which are summarized in Table 1.
Table 1.
Comparison of the composition of the DOT1L-relevant transcriptional elongation complexes
The first DOT1L-associated complex, termed ENL-associated proteins (EAPs), was purified by immunoprecipitation using antibodies against ENL. In addition to ENL, the complex contains A4F, AF5q31, LAF4, DOT1L, P-TEFb, Ring1, CBX8, and BCoR (Mueller et al. 2007). A more recent study indicated that the core EAP complex consists of only ENL, AF4, DOT1L, and P-TEFb (Mueller et al. 2009). P-TEFb is a cyclin-dependent kinase complex that phosphorylates Ser2 within the RNA Pol II C-terminal domain (CTD). This phosphorylation event is required for the transition of RNA Pol II from transcription initiation to the transcription elongation phase. The purified EAP complex possesses both H3K79 methyltransferase and RNA Pol II CTD kinase activities. Importantly, knockdown of ENL reduced genome-wide H3K79me2 as well as global transcriptional elongation activity, suggesting a link between H3K79 methylation and transcriptional elongation (Mueller et al. 2007). The second DOT1L-containing complex, which is related to EAP, was purified by using an antibody against AF4 (Bitoun et al. 2007). The complex contains AF4, AF9, AF10, ENL, DOT1L, and P-TEFb. Importantly, overexpression of AF4, AF9, ENL, and AF10 increased H3K79 methylation levels and P-TEFb-dependent transcription elongation (Bitoun et al. 2007). These studies collectively support a role for DOT1L in transcriptional elongation.
In addition to the two related complexes mentioned above, a recently purified DOT1L-containing compelx, DotCom, contains members of the Wnt pathway in addition to DOT1L, AF10, AF17, and AF9 (Mohan et al. 2010). Despite the absence of P-TEFb in this complex, DOT1L was still required for the expression of Wingless target genes, supporting its role in transcriptional activation (Mohan et al. 2010). Interestingly, another recently purified superelongation complex (SEC) contains multiple transcription elongation factors (ELL1–3, P-TEFb, AFF1/AF4, AFF4, AF9, and ENL) and ELL-associated factors (EAFs), but does not contain DOT1L (Lin et al. 2010). Similarly, another elongation complex, AEP (which is composed of AF4, AF5q31, ENL, and P-TEFb), also lacks DOT1L (Yokoyama et al. 2010). It is likely that the variation between different protein complexes purified may result from the different methods and purification procedures used in these studies. It is possible that multiple transcriptional elongation complexes may exist, and that DOT1L may associate with only a subset of elongation complexes. Although DOT1L may not be required for transcription of all genes, it certainly plays a role for transcribing a subset of genes plausibly through facilitating transcriptional elongation. Based on available information, we propose a model that encompasses sequential events (including H2B-K123ub, H3K4me, H3K79me, and RNA Pol II phosphorylation) that lead to transcriptional activation and elongation (Fig. 3).
Figure 3.
Mechanism for the involvement of DOT1L-mediated H3K79 methylation in transcriptional elongation. For simplicity of the model, only a fraction of the proteins involved in RNA Pol II transcription is shown. In step I, RNA Pol II is phosphorylated on Ser5 of its CTD and has initiated transcription. Negative elongation factor (NELF) and 5,6-dichloro-1b-D-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF) are bound to RNA Pol II for promoter-proximal pausing. In step II, promoter clearance is achieved by the recruitment of P-TEFb, which phosphorylates Ser2 of the RNA Pol II CTD, NELF, and DSIF. Phosphorylated NELF dissociates to release Pol II. The PAF1 complex, along with RAD6 and BRE1, is recruited to ubiquitinate H2B-K120. In step III, which can occur simultaneously with step II, H3K4 di- and trimethylation is achieved by the recruitment of MLL, the human homolog of Set1, by the PAF1 complex and association with H2B-K120ub. During transcription elongation shown in step IV, DOT1L is recruited to elongating RNA Pol II through its association with a network of proteins that include, but are not limited to, the elongation factors ELL, AF4, AF10, AF9, and ENL. DOT1L methylates H3K79 to maintain active transcription and/or serve as a transcription memory.
Role of Dot1 in DNA repair and cell cycle regulation
Role of Dot1 in the DNA damage response
Chromatin structure and nucleosome positioning, which are dynamically regulated by histone modifications, affect transcription by RNA Pol II. In addition, the presence of DNA damage, such as double-strand breaks (DSBs) and lesions, can impede Pol II transcription elongation. Therefore, mechanisms used in the regulation of chromatin conformation and transcription may also be used in repairing DNA damage. Consistent with this notion, studies in yeast and mammalian cells have established a link between Dot1/DOT1L and their associated H3K79 methylation to DNA damage checkpoint control and repair.
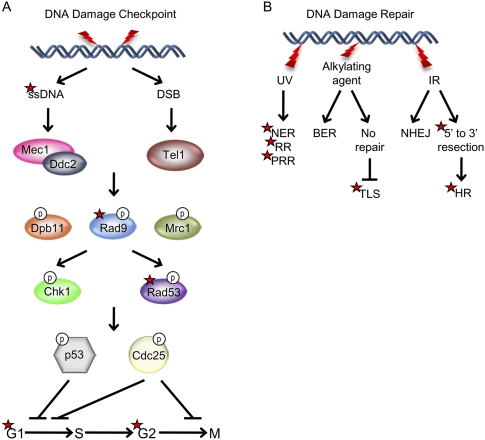
The first evidence that suggested a link between DOT1L-mediated H3K79 methylation and DNA damage repair came from the demonstration that the tandem tudor domain of the human 53BP1 protein binds to methylated H3K79 and is recruited to DNA DSBs (Huyen et al. 2004). Strikingly, both mutation of the tudor domain and blocking H3K79 methylation through either mutating Lys 79 of H3 or knocking down DOT1L inhibited recruitment of 53BP1 to DSBs. The interaction between 53BP1 and methylated H3K79 is evolutionarily conserved, as Rad9, the yeast ortholog of 53BP1, also binds to methylated H3K79 via its tudor domain (Wysocki et al. 2005). Since H3K79 methylation levels do not change upon DNA damage, it is believed that DSBs induce changes in the chromatin structure to expose methylated H3K79, which is then recognized by 53BP1 (Huyen et al. 2004). However, given that Rad9 is still recruited to sites of DSBs in the absence of the chromatin remodeling complexes SWR and INO80 (Javaheri et al. 2006), further studies are needed to prove or disprove this hypothetical model.
Budding yeast cells treated with ionizing radiation (IR) at G1 typically undergo a G1 checkpoint delay. Interestingly, Dot1 mutants are defective in G1- and intra-S-phase checkpoints and progress through the cell cycle at a normal rate even after IR-induced DNA damage (Wysocki et al. 2005). Moreover, mutations that affect Dot1 enzymatic activity, such as disruption of H2B-K123 ubiquitination or mutation of H3K79, display the same checkpoint defects in response to multiple genotoxic stresses (Giannattasio et al. 2005; Game et al. 2006; Chernikova et al. 2010). In response to IR damage, the Dot1/Rad6/Bre1 pathway mediates DNA damage repair through G1 homologous recombination (HR) (Game et al. 2006). Although Dot1 deletion mutants do not display a G2 arrest phenotype (Game et al. 2006), Dot1 is required for Rad9 recruitment to the damage foci and phosphorylation of Rad53 upon IR (Toh et al. 2006). Dot1-mediated H3K79 methylation is believed to play two distinct roles in Rad9-mediated DNA damage response (Grenon et al. 2007): the activation of the G1/S checkpoint, and the repair of DNA damage at late G2 phase.
Repair of DSBs can occur via nonhomologous end-joining (NHEJ) or HR. Resection of DSBs is an important step in DNA damage repair by HR, producing 3′ ssDNA tail intermediates. It is believed that both H3K79 methylation and Rad9 recruitment play an integral role in regulating resection to limit the amount of ssDNA that is produced (Lazzaro et al. 2008). HR with the sister chromatid (SCR) in mitosis ensures faithful transmission of the genetic material. A number of proteins, including cohesin, are required for maintaining chromosome structure and for efficient SCR. Interestingly, not only is Dot1 required for the recruitment of Rad9 to the DSB sites and for DNA resection, it also facilitates the recruitment of cohesin and efficient SCR (Conde et al. 2009).
In addition to DSBs, Dot1-mediated H3K79 methylation plays a role in repairing other types of DNA lesions. Ultraviolet radiation (UV) causes the formation of cis-syn cyclopyrimidine and 6-4 photoproduct dimers, which can be repaired by nucleotide excision repair (NER), recombination repair (RR), or post-replication repair (PRR). Dot1-mediated H3K79 methylation plays a critical role in repairing UV-induced DNA damage (Giannattasio et al. 2005), as loss of H3K79 methylation affects all three repair pathways, resulting in UV hypersensitivity (Bostelman et al. 2007; Lazzaro et al. 2008). When the base excision repair (BER) pathway fails to repair damages caused by alkylating agents, cell cycle progression and cell survival may be achieved by the translesion synthesis (TLS) pathway that uses error-prone polymerases to bypass DNA lesions. Dot1 negatively regulates TLS, thereby maintaining genome integrity upon DNA damage (Conde and San-Segundo 2008; Conde et al. 2010; Levesque et al. 2010).
Beyond its role in the DNA damage response, Dot1 is also important for meiotic checkpoint control. Diploid parental cells undergo meiotic cell division to generate haploid gametes. Proper segregation of chromosomes during meiosis is ensured by the pachytene checkpoint, which monitors completion of chromosome synapsis and recombination to prevent premature nuclear division. In the event of defective meiotic recombination, the pachytene checkpoint induces arrest at mid-meiotic prophase. In yeast, arrest at pachytene can be observed in dmc1 and zip1 mutants, which are required for proper synapsis formation and repair of DSBs. In a genetic screen for pachytene checkpoint components, dmc1 and zip1 mutants failed to arrest in the absence of Dot1. This failure to arrest is partially due to mislocalization of the nucleolar proteins Pch2 and Sir2, both of which are required for pachytene checkpoint function (San-Segundo and Roeder 2000). Collectively, an overwhelming amount of data support a vital role for Dot1 in regulating cell cycle progression in yeast upon DNA damage (Fig. 4). Surprisingly, very little work has been done to determine whether Dot1's role in DNA damage checkpoint control is conserved in mammals.
Figure 4.
Role of Dot1 in DNA damage checkpoint response and repair. The yeast proteins are shown, and the steps in which Dot1 has been implicated are marked by red stars. (A) In yeast, the PI-3 kinases Mec1/Ddc2 (human ATR/ATRIP) sense exposed ssDNA formed during repair of UV-, oxidative-, and alkylating-damaged DNA as well as resected DSBs. The sensor PI-3 kinase Tel (human ATM) recognizes blunt-end DSBs. After recognition and binding of sensor proteins to the site of DNA damage, mediator proteins such as Rad9, Dpb11, and Mrc1 are recruited and activated by phosphorylation. Dot1-mediated H3K79 methylation is required for the recruitment and activation of Rad9 and phosphorylation of the downstream transducer protein Rad53. Dot1 is also required for proper checkpoint function at both G1 and G2. (B) Several different DNA repair mechanisms can be used by the cell in response to different types of DNA damage. Damage by UV radiation can be repaired by NER, RR, and PRR. Loss of H3K79 methylation impairs all three pathways, resulting in hypersensitivity to UV damage. Damage by alkylating agents can be repaired by the BER pathway. In rare instances where the damage cannot be repaired, the error-prone TLS pathway can be activated to bypass the DNA lesion. It was demonstrated that Dot1 maintains DNA integrity by inhibiting TLS. Finally, IR, which generates DSBs, can be repaired by NHEJ. Alternatively, the DSBs can undergo 5′-to-3′ resection and be repaired by HR. Methylation of H3K79 and the subsequent recruitment of Rad9 are required for regulating resection. Additionally, H3K79 methylation is required for the recruitment of cohesin, which is important for maintaining chromosome structure and for efficient SCR to repair DSBs.
Cell cycle-regulated H3K79 methylation
Being able to repair damaged DNA is vital for maintaining genome integrity and stability. DNA repair occurs upon checkpoint activation as a cell progresses through the cell cycle phases: G1, S, G2, and M. Consistent with its role in the DNA damage repair pathways, H3K79 methylation levels fluctuate with the cell cycle. In yeast, H3K79me2 levels are low in G1 and S, begin to increase in late S, and are maintained relatively higher in G2 and M phases (Schulze et al. 2009). Paradoxically, a greater percentage of dot1Δ cells accumulate in G1 (a period with low H3K79me2) compared with wild-type cells. Despite the general correlation between H3K79me2 levels and transcription activity, genes that are expressed specifically in G1 (when H3K79me2 is low) are enriched with H3K79me2 during G2/M (Schulze et al. 2009). These data suggest that, at least in this subset of genes, H3K79me2 is inversely correlated with gene activity and may have a role in epigenetic memory (Schulze et al. 2009). However, the cell cycle-dependent variation of H3K27me2 levels in HeLa cells is different from that observed in yeast. In HeLa cells, the H3K79me2 level is high in G1, decreases in S, reaches the lowest in G2, and increases again in M phase (Feng et al. 2002). Thus, it is apparent that the role of cell cycle-regulated H3K79me2 changes in yeast is not evolutionarily conserved in humans.
Mouse DOT1L has also been implicated in cell cycle progression of differentiating embryonic stem (ES) cells and erythroid progenitor cells. Although DOT1L-depleted ES cells retain their proliferation and pluripotency capacity, they displayed severe proliferation defects upon RA-induced in vitro differentiation, resulting in G2/M accumulation (Barry et al. 2009). Interestingly, both undifferentiated and differentiated DOT1L knockdown ES cells exhibited aneuploidy, with the phenotype being more pronounced upon differentiation (Barry et al. 2009). Consistent with this, DOT1L knockout ES cells also displayed a G2/M cell cycle defect and aneuploidy (Jones et al. 2008). Interestingly, apoptosis was observed in DOT1L knockout ES cells (Jones et al. 2008), but was absent in knockdown cells (Barry et al. 2009). In addition to mouse ES cells, DOT1L plays an important role in cell cycle progression and antagonizing apoptosis in primitive erythroid progenitor cells (Jones et al. 2008). In contrast to that observed in ES cells, DOT1L knockout in the erythroid lineage results in an accumulation at G0/G1 rather than G2/M (Feng et al. 2010), indicating that the cell cycle effect of DOT1L might be cell type-specific. Further studies are needed to understand why cell cycle regulation by Dot1 and DOT1L are organism and cell type-dependent.
Biological functions of DOT1L
Role of Dot1 in embryonic development
In addition to the studies performed in cell lines and lower eukaryotes, the role of Dot1-mediated H3K79 methylation was also analyzed in higher eukaryotes, including animals. For example, the cell cycle regulation of H3K79 methylation levels observed in cell lines is also observed during embryogenesis. During fly embryogenesis, methylation of H3K79 is absent in both transcriptionally active somatic nuclei and quiescent pole cell nuclei at the precellular blastoderm. H3K79me2 begins to emerge after gastrulation at the germband extended embryo stage 8/9 and is restricted to cells undergoing mitosis (Shanower et al. 2005). Increasing levels of H3K79me2 are then observed in epidermal nuclei during dorsal closure and head involution. The re-establishment of H3K79me2 suggests that this modification plays an important role in regulating transcription of developmental genes (Shanower et al. 2005). Interestingly, mutation of Grappa, the fly ortholog of Dot1, causes phenotypes observed in both Polycomb Group (PcG) and trithorax Group (trxG) mutants (Shanower et al. 2005). PcG and trxG genes play antagonistic roles by respectively silencing and activating the expression of Antennapedia and Bithorax homeotic genes, which specify anterior-to-posterior parasegmentation identity (Ringrose and Paro 2004). In addition to a role in parasegmentation, Grappa has been implicated in Wnt/Wingless signaling, which is required for morphogenesis in wing imaginal discs (Mohan et al. 2010). Using knockdown approaches, Grappa was shown to regulate the active expression of Wingless target genes such as senseless, frizzled 3, and homothorax (Mohan et al. 2010). Taken together, the data indicate that Grappa may play a dual role in regulating both the active and silenced states of specific chromatin domains during fly development.
Similar to that observed in Drosophila, H3K79 methylation is also absent from mouse zygotes and is present at a very low level until the blastocyst stage (Ooga et al. 2008), suggesting that loss of this epigenetic mark is important for early embryogenesis. Although H3K79me2 is detectable from the four-cell to blastocyst stages, H3K79me3 is undetectable even at the blastocyst stage (Ooga et al. 2008). Germline knockout of mDOT1L results in lethality by embryonic day 10.5 (E10.5) during organogenesis of the cardiovascular system (Jones et al. 2008). Consistently, knockout embryos display cardiovascular defects, including heart enlargement, decreased vasculature, and anemia (Jones et al. 2008). These studies collectively demonstrate that DOT1L and Grappa play an important role in embryonic development in mice and Drosophila, respectively.
Role of DOT1L in cardiac function
The cardiovascular defect observed in mDOT1L knockout embryos (Jones et al. 2008) suggested that mDOT1L may play a role in cardiac development, which is consistent with ex vivo studies demonstrating a correlation between increased H3K79 methylation and induced expression of cardiovascular marker genes during myocardiogenesis (Illi et al. 2005). To elucidate the role of mDOT1L in cardiac development and function, we generated a cardiac-specific knockout mouse model by crossing floxed Dot1l mice with α-myosin heavy chain-Cre mice (Nguyen et al. 2011b). Cardiac-specific knockout of mDOT1L caused dilated cardiomyopathy (DCM) (Nguyen et al. 2011b), a disease of the heart muscle manifested by chamber dilation and systolic dysfunction. Mechanistically, mDOT1L-mediated H3K79 methylation directly regulates transcription of dystrophin (Fig. 5). Given that dystrophin is required for forming the dystrophin–glycoprotein complex (DGC) that is involved in signal force transmission and relieving mechanical stress during heart contraction, loss of dystrophin expression causes DGC destabilization, resulting in sarcolemma damage (Kimura 2008). In humans, dystrophin mutations are a known cause of human DCM and muscular dystrophy. Importantly, the DCM phenotype observed in mDOT1L cardiac-specific knockout mice can be rescued by ectopic expression of a functional truncated form of dystrophin (Nguyen et al. 2011b), indicating that dystrophin is a major downstream target that mediates the cardiac function of DOT1L. Interestingly, analysis of DOT1L expression levels revealed that DOT1L is significantly lower in human idiopathic DCM heart tissue compared with normal heart tissue (Nguyen et al. 2011b), raising the possibility that malfunction of DOT1L may be a contributing factor in human DCM pathogenesis.
Figure 5.
DOT1L contributes to cardiac function by activating dystrophin expression. DOT1L-mediated H3K79 methylation is required for dystrophin expression. Dystrophin forms a DGC that plays a pivotal role in lateral force transduction and relieving mechanical stress for optimal cardiac performance. Loss of H3K79 methylation results in silencing of the dystrophin locus, loss of expression of the DGC complex, and sarcolemma damage. The absence of a functional DGC is manifested by systolic dysfunction and DCM.
Role of DOT1L in erythropoiesis
In addition to cardiovascular defects, DOT1L knockout embryos display severe anemia by E10.5, suggesting impaired fetal erythropoiesis (Feng et al. 2010). A possible role for mDOT1L in erythropoiesis would be consistent with a previous report demonstrating that H3K79 methylation is enriched at the erythroid-specific β-globin locus (Im et al. 2003). In support of this, mDOT1L deficiency in yolk sac-derived hematopoietic cells caused a G0/G1 cell cycle arrest and induced apoptosis in erythroid progenitor cells cultured in vitro in the presence of growth stimuli, but had no effect on myeloid lineage growth and differentiation (Feng et al. 2010). RT-qPCR and ChIP analyses indicated that the function of mDOT1L in erythropoiesis was mediated by its role in activation of Gata2. The down-regulation of Gata2 in Dot1l-deficient hematopoietic cells caused derepression of the myeloid transcription factor PU.1 to block erythroid differentiation (Feng et al. 2010).
Role of DOT1L in leukemia
It is apparent that DOT1L is vital for normal cellular and biological processes, including DNA damage repair, cell cycle, development, cardiac muscle function, and hematopoiesis. However, transcriptional activation by DOT1L-mediated H3K79 methylation is also exploited in leukemias that stem from chromosomal rearrangements of MLL (mixed-lineage leukemia). This misregulation of DOT1L is achievable because components of the elongation complexes EAP (Mueller et al. 2007), SEC (Lin et al. 2010), and AEP (Yokoyama et al. 2010) interact with DOT1L and are frequent translocation partners of MLL (Ayton and Cleary 2001). Translocation of the MLL gene results in the expression of oncogenic fusion proteins and is a common cause of acute leukemia (Ayton and Cleary 2001; Hess 2004), accounting for 5%–10% of adult acute leukemias (Daser and Rabbitts 2004) and 60%–80% of infant acute leukemias (Ayton and Cleary 2001; Hess 2004; Krivtsov and Armstrong 2007).
MLL is an H3K4 methyltransferase that maintains the “on” state of Hox genes during embryogenesis and hematopoiesis (Hsieh et al. 2003; Guenther et al. 2005). In leukemia, Hoxa genes remain constitutively active to promote uncontrolled proliferation of leukemic cells. Although MLL fusion proteins lack the catalytic SET domain (Fig. 6A), it is believed that constitutive activation of leukemic oncogenes is mediated by wild-type MLL expressed from the nonmutated allele, as well as the transactivation activity of the MLL fusion partner and other recruited factors such as MENIN (Yokoyama et al. 2005) and LEDGF (Yokoyama and Cleary 2008). Due to the high incidence of MLL-related acute leukemias, a better understanding of the molecular mechanisms used by MLL fusion proteins is vital for the development of effective treatments. However, the heterogeneity of >50 MLL fusion partners makes targeted therapy for MLL-related leukemias difficult.
Figure 6.
MLL fusion proteins recruit DOT1L for transcriptional activation and leukemogenesis. (A, top) MLL is a SET domain-containing H3K4 methyltransferase with multiple functional domains. (AT-hook and CxxC) DNA binding; (SNL) signal nuclear localization; (PHD) protein interactions; (TAD) transactivation domain; (SET) methyltransferase domain. Frequent DSBs occur within the MLL locus at a breakpoint cluster region (BCR). (Bottom) The N terminus of MLL undergoes chromosomal rearrangements and fuses in-frame to >50 different genes, resulting in the expression of leukemogenic MLL fusion proteins. (B) Constitutive activation of a leukemic transcriptional program, including Hoxa and Meis1 genes, is achieved through mistargeting of DOT1L and its associated proteins through the interaction of DOT1L with MLL fusion partners. DOT1L-mediated H3K79 methylation and the subsequent activation of the target genes result in leukemic transformation.
The first evidence that suggested a role for DOT1L in leukemia came from studies looking for functional partners of DOT1L. A yeast two-hybrid screen identified two MLL fusion partners—AF10 and ENL—as DOT1L-interacting proteins (Okada et al. 2005). Further characterization of the AF10 and DOT1L interaction mapped the interaction domain to the OM-LZ region of AF10 required for the transformation activity of MLL-AF10, raising the possibility that AF10 serves as a bridge for the recruitment of DOT1L to MLL-AF10 target genes (Okada et al. 2005). This scenario gained experimental support, as both MLL-AF10 and H3K79me2 were specifically enriched at the Hoxa9 locus in MLL-AF10-transformed cells compared with control cells. Importantly, overexpression of Hoxa9 as well as the transformation capability of MLL-AF10 were dependent on DOT1L enzymatic activity. Moreover, Hoxa9 was shown to be a critical downstream target of MLL-AF10, as deletion of the Hoxa9 gene prevented transformation by MLL-AF10 (Okada et al. 2005). These studies suggest that mistargeting of DOT1L by MLL-AF10 and the subsequent H3K79 methylation and activation of oncogenes such as Hoxa9 are the mechanisms underlying DOT1L's contribution to transformation, and raise the possibility that DOT1L may be a good target for therapeutic intervention. In a follow-up study, the same mechanism was also shown for DOT1L's contribution to CALM-AF10-mediated transformation (Okada et al. 2006). However, Hoxa5, instead of Hoxa9, is the relevant downstream target for CALM-AF10, as bone marrow cells deficient for Hoxa5 cannot be transformed by CALM-AF10, but bone marrow cells deficient for Hoxa9 can (Okada et al. 2006).
In addition to MLL-AF10 and CALM-AF10, DOT1L is involved in transformation by other MLL fusion proteins, such as MLL-ENL (Mueller et al. 2007), MLL-AF4 (Krivtsov et al. 2008), and MLL-AF9 (Chang et al. 2010; Nguyen et al. 2011a). Consistent with the fact that DOT1L is involved in MLL-related transformation, DOT1L associates with a number of MLL fusion proteins and participates in the activation of a leukemic transcriptional program (Bitoun et al. 2007; Mueller et al. 2007, 2009). The exact mechanism regarding how DOT1L contributes to the gene activation process requires further elucidation; however, it is clear that DOT1L is broadly involved in leukemogenesis, particularly those mediated by MLL fusion proteins such as AF4, AF9, AF10, and ENL, which account for two-thirds of all MLL-associated leukemias (Ayton and Cleary 2001).
The above studies collectively establish a broad role for DOT1L in transformation, and suggest a general mechanism by which DOT1L contributes to the leukemogenesis process (Fig. 6B). Interaction between DOT1L and MLL fusion partners leads to mistargeting of DOT1L to the gene targets of MLL fusion proteins, such as the Hoxa cluster and Meis1. Aberrant hypermethylation of H3K79 leads to constitutive transcriptional activation of these genes, which in turn results in leukemic transformation. Inhibition of DOT1L enzymatic activity or disruption of the interaction between DOT1L and the MLL fusion proteins may provide a promising therapeutic strategy for the treatment of MLL fusion-related leukemias. However, given the role of DOT1L in normal hematopoiesis (Feng et al. 2010; Jo et al. 2011), the possible side effects of such drugs have to be carefully evaluated before they can be used in the treatment of leukemia patients.
Concluding remarks
Dot1 is unique not only in its structure, but in its substrate specificity. Although its catalytic domain resembles that of class I arginine methylases, Dot1 methylates histone H3 at Lys 79. H3K79 is also located within the surface-exposed globular domain of H3 rather than its NH3 tail, where all other known methylation residues reside. Paradoxically, Dot1-mediated H3K79 methylation is capable of regulating transcription at both heterochromatin and euchromatin. Globally, H3K79 methylation is enriched in actively transcribing genes. Consistently, H3K79 methylation is required for the expression of Wnt target genes in Drosophila imaginal discs, dystrophin in cardiac and myoblast cells, and the Hoxa cluster in leukemic cells.
Intriguingly, Dot1 is the sole H3K79 methylase capable of catalyzing mono-, di-, and trimethylation in a nonprocessive manner. Thus, in the absence of any other mediator proteins, the likelihood of an H3K79me1 mark becoming H3K79me2 and an H3K79me2 mark becoming H3K79me3 have the same probability. If this were the case, it could be inferred that they all play redundant roles in regulating transcription, and brings into question the need for different H3K79 methylation states (Frederiks et al. 2008). Nevertheless, studies suggest that the various H3K79 methylation states have different functions in transcriptional regulation and/or other processes. For example, in yeast, only H3K79me2 fluctuates with the cell cycle, whereas H3K79me3 levels remain constant. Additionally, in early mouse embryogenesis, H3K79me2 and H3K79me3 emerge at distinct stages of development. How the same enzyme achieves specificity to temporally regulate its catalytic activity at specific chromatin domains remains to be determined. One possibility is that its enzymatic activity might be regulated by other proteins, such as Rad6/Bre1 and the Paf1 complex in yeast and other associated proteins identified in mammals. Another possibility is that the steady-state levels of H3K79 methylation might be regulated by a putative H3K79 demethylase.
In addition to transcription, Dot1 plays an important role in cell cycle regulation and the DNA damage response. Furthermore, mammalian DOT1L is an essential gene that is required for embryogenesis, hematopoiesis, and cardiac function. Finally, DOT1L plays a pivotal role in MLL-related leukemogenesis. Given the importance of its enzymatic activity in these processes, DOT1L emerges as a promising target for therapeutic treatments. It is apparent that DOT1L and its associated H3K79 methylation activity are extremely complex. As such, the regulation of DOT1L enzymatic activity and the functional consequences need to be further elucidated before efficient treatments can be developed. We expect that future studies will reveal details regarding how its enzymatic activity is regulated, and how H3K79 methylation leads to transcriptional activation. We also anticipate a revelation of DOT1L functions outside the cardiovascular and hematopoietic systems. Identification and characterization of DOT1L inhibitors and evaluation of their efficacy in leukemia treatment will provide another example where initial inquiry into the function of a basic histone modification leads to a treatment for a deadly disease.
Acknowledgments
We thank Susan Wu for critical reading of the manuscript. We apologize to colleagues whose work cannot be cited due to space constraints. The DOT1L work in the Zhang laboratory is supported by NIH (CA119133) and HHMI. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2057811.
References
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell 28: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20: 5695–5707 [DOI] [PubMed] [Google Scholar]
- Barry ER, Krueger W, Jakuba CM, Veilleux E, Ambrosi DJ, Nelson CE, Rasmussen TP 2009. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells 27: 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE 2007. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet 16: 92–106 [DOI] [PubMed] [Google Scholar]
- Bostelman LJ, Keller AM, Albrecht AM, Arat A, Thompson JS 2007. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst) 6: 383–395 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Cavalli G 2006. Chromatin and epigenetics in development: blending cellular memory with cell fate plasticity. Development 133: 2089–2094 [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW 2009. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci 106: 16686–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ, Hemenway CS, Zhang W 2010. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res 70: 10234–10242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, Muir TW 2010. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol 6: 267–269 [DOI] [PubMed] [Google Scholar]
- Cheng X, Roberts RJ 2001. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res 29: 3784–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova SB, Dorth JA, Razorenova OV, Game JC, Brown JM 2010. Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat Res 174: 558–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K 2008. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22: 1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F, San-Segundo PA 2008. Role of Dot1 in the response to alkylating DNA damage in Saccharomyces cerevisiae: regulation of DNA damage tolerance by the error-prone polymerases Polzeta/Rev1. Genetics 179: 1197–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F, Refolio E, Cordon-Preciado V, Cortes-Ledesma F, Aragon L, Aguilera A, San-Segundo PA 2009. The Dot1 histone methyltransferase and the Rad9 checkpoint adaptor contribute to cohesin-dependent double-strand break repair by sister chromatid recombination in Saccharomyces cerevisiae. Genetics 182: 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F, Ontoso D, Acosta I, Gallego-Sanchez A, Bueno A, San-Segundo PA 2010. Regulation of tolerance to DNA alkylating damage by Dot1 and Rad53 in Saccharomyces cerevisiae. DNA Repair (Amst) 9: 1038–1049 [DOI] [PubMed] [Google Scholar]
- Daser A, Rabbitts TH 2004. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev 18: 965–974 [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP 2004. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell 13: 435–442 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Oshimura M, Barrett JC 2002. Epigenetic mechanisms in human disease. Cancer Res 62: 6784–6787 [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12: 1052–1058 [DOI] [PubMed] [Google Scholar]
- Feng Y, Yang Y, Ortega MM, Copeland JN, Zhang M, Jacob JB, Fields TA, Vivian JL, Fields PE 2010. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood 116: 4483–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW 2011. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerman IM, Li HC, Briggs SD 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev 21: 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks F, Tzouros M, Oudgenoeg G, van Welsem T, Fornerod M, Krijgsveld J, van Leeuwen F 2008. Nonprocessive methylation by Dot1 leads to functional redundancy of histone H3K79 methylation states. Nat Struct Mol Biol 15: 550–557 [DOI] [PubMed] [Google Scholar]
- Game JC, Williamson MS, Spicakova T, Brown JM 2006. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics 173: 1951–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6–Bre1 and H3 methylation by Dot1. J Biol Chem 280: 9879–9886 [DOI] [PubMed] [Google Scholar]
- Grenon M, Costelloe T, Jimeno S, O'Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF 2007. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24: 105–119 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA 2005. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci 102: 8603–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel AE, Ebers GC, Ramagopalan SV 2010. Epigenetics: molecular mechanisms and implications for disease. Trends Mol Med 16: 7–16 [DOI] [PubMed] [Google Scholar]
- Hess JL 2004. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med 10: 500–507 [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Cheng EH, Korsmeyer SJ 2003. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 115: 293–303 [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Illi B, Scopece A, Nanni S, Farsetti A, Morgante L, Biglioli P, Capogrossi MC, Gaetano C 2005. Epigenetic histone modification and cardiovascular lineage programming in mouse embryonic stem cells exposed to laminar shear stress. Circ Res 96: 501–508 [DOI] [PubMed] [Google Scholar]
- Im H, Park C, Feng Q, Johnson KD, Kiekhaefer CM, Choi K, Zhang Y, Bresnick EH 2003. Dynamic regulation of histone H3 methylated at lysine 79 within a tissue-specific chromatin domain. J Biol Chem 278: 18346–18352 [DOI] [PubMed] [Google Scholar]
- Javaheri A, Wysocki R, Jobin-Robitaille O, Altaf M, Cote J, Kron SJ 2006. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc Natl Acad Sci 103: 13771–13776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G 1998. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci 54: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL 2011. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 117: 4759–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, Baltus GA, Kadam S, Zhai H, Valdez R, et al. 2008. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 4: e1000190 doi: 10.1371/journal.pgen.1000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J 24: 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A 2008. Molecular etiology and pathogenesis of hereditary cardiomyopathy. Circ J 72: A38–A48 doi: 10.1253/circj.CJ-08-0050 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y 2007. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8: 307–318 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y 2006. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285–294 [DOI] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J 24: 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2002. Histone methylation in transcriptional control. Curr Opin Genet Dev 12: 198–209 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA 2007. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7: 823–833 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. 2008. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14: 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem 277: 30421–30424 [DOI] [PubMed] [Google Scholar]
- Laurenson P, Rine J 1992. Silencers, silencing, and heritable transcriptional states. Microbiol Rev 56: 543–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M 2008. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 27: 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Levesque N, Leung GP, Fok AK, Schmidt TI, Kobor MS 2010. Loss of H3 K79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway. J Biol Chem 285: 35113–35122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37: 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y 2005. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849 [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW 2008. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Z, Zhang Y, Xu RM 2003. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112: 711–723 [DOI] [PubMed] [Google Scholar]
- Minard ME, Jain AK, Barton MC 2009. Analysis of epigenetic alterations to chromatin during development. Genesis 47: 559–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A 2010. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev 24: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al. 2007. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110: 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK 2009. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol 7: e1000249 doi: 10.1371/jounral.pbio.1000249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K 2002a. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K 2002b. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277: 34655–34657 [DOI] [PubMed] [Google Scholar]
- Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K 2003a. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci 100: 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K 2003b. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem 278: 33625–33628 [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Taranova O, He J, Zhang Y 2011a. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9 leukemogenesis. Blood. doi: 10.1182/blood-2011-02-334369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Xiao B, Neppl RL, Kallin EM, Li J, Chen T, Wang DZ, Xiao X, Zhang Y 2011b. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev 25: 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A, Boeke JD 2010. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev 24: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Jeong K, Kim H, Kwon CS, Lee D 2010. A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem Biophys Res Commun 399: 512–517 [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121: 167–178 [DOI] [PubMed] [Google Scholar]
- Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y 2006. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol 8: 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell 28: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Ooga M, Inoue A, Kageyama S, Akiyama T, Nagata M, Aoki F 2008. Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod 78: 413–424 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA 2004. Histones and histone modifications. Curr Biol 14: R546–R551 doi: 10.1016/j.cub.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. 2010. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443 [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504 [DOI] [PubMed] [Google Scholar]
- Rossmann MP, Luo W, Tsaponina O, Chabes A, Stillman B 2011. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol Cell 42: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS 2000. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol Biol Cell 11: 3601–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X 2004. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem 279: 43296–43306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X 2003. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci 28: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A 2009. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell 35: 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28: 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Schulze JM, Jackson J, Hentrich T, Seidel C, Jaspersen SL, Kobor MS, Shilatifard A 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol Cell 42: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh GW, O'Shaughnessy AM, Jimeno S, Dobbie IM, Grenon M, Maffini S, O'Rorke A, Lowndes NF 2006. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 5: 693–703 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA 2006. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26: 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde KE, Allen JR, Tatchell K, Weischet WO, Lohr D 1980. DNA–histone interactions in nucleosomes. Biophys J 32: 271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- van Welsem T, Frederiks F, Verzijlbergen KF, Faber AW, Nelson ZW, Egan DA, Gottschling DE, van Leeuwen F 2008. Synthetic lethal screens identify gene silencing processes in yeast and implicate the acetylated amino terminus of Sir3 in recognition of the nucleosome core. Mol Cell Biol 28: 3861–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. 2003a. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11: 267–274 [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A 2003b. The Paf1 complex is essential for histone monoubiquitination by the Rad6–Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ 2005. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25: 8430–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML 2008. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML 2005. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123: 207–218 [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML 2010. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17: 198–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Dent SY 2005. Histone modifying enzymes and cancer: going beyond histones. J Cell Biochem 96: 1137–1148 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360 [DOI] [PubMed] [Google Scholar]
- Zhang W, Hayashizaki Y, Kone BC 2004. Structure and regulation of the mDot1 gene, a mouse histone H3 methyltransferase. Biochem J 377: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]