Abstract
We used an RNAi-mediated loss-of-function screen to study systematically the role of the protein tyrosine phosphatase (PTP) superfamily of enzymes in mammary epithelial cell motility in the absence or presence of the oncoprotein tyrosine kinase ERBB2. We report that although shRNAs directed against most of the PTP family were without effect, suppression of three PTPs—PRPN23, PTPRG, and PTPRR—enhanced cell motility. Furthermore, we found that suppression of PTPN23, but not PTPRG or PTPRR, induced cell invasion. Suppression of PTPN23 increased E-cadherin internalization, impaired early endosome trafficking of E-cadherin, induced the expression of mesenchymal proteins, and caused cell scattering. The activity of SRC and β-catenin was elevated when PTPN23 was suppressed. Moreover, we identified SRC, E-cadherin, and β-catenin as direct substrates of PTPN23. Inhibition of SRC with the small molecular inhibitor SU6656 blocked the effects of PTPN23 depletion. These findings suggest that loss of PTPN23 may increase the activity of SRC and the phosphorylation status of the E-cadherin/β-catenin signaling complex to promote tumor growth and invasive behavior in breast cancer. In addition, our studies highlight functional specificity among PTPs and reveal new roles for PTPs in mammary epithelial cell biology.
Keywords: protein tyrosine phosphatase, RNAi screen, cell motility, invasion, cadherin and catenin, SRC
Disruption of the normal patterns of phosphorylation of tyrosine residues in proteins has been linked with the etiology of several human diseases, including cancer. Protein tyrosine phosphorylation is controlled by the coordinated activity of two families of enzymes: protein tyrosine kinases (PTKs) and the protein tyrosine phosphatases (PTPs). Considerable progress has been made in defining how PTK function is disrupted in disease and in identifying therapeutic targets from within this enzyme family; however, functional analysis of PTPs is not as advanced. Therefore, the development of a “PTP-ome”-directed shRNA library will provide a way to analyze globally the function of PTPs through loss-of-function phenotype studies.
The PTPs, which are encoded by ∼100 genes in humans, are characterized by the presence of the HC(X)5R motif in the protein sequence, in which the Cys residue is essential for catalytic activity (Tonks 2006). Analyses in cell and animal models have implicated some PTPs—such as PTPRE (PTPɛ), PTPRJ (DEP1), and PTPN13 (PTP-BAS)—in the development of breast cancer (Freiss and Vignon 2004). PTPRA is overexpressed in ∼30% of breast tumors (Ardini et al. 2000), and expression of PRL3 (Radke et al. 2006) and LAR (Levea et al. 2000) was found to be up-regulated in metastatic breast cancer. In contrast, the expression of PTPRG is lower in breast tumors (Zheng et al. 2000). Genomic deletions or mutations in PTPs such as PTEN (Liaw et al. 1997), PTPN12 (PTP-PEST) (Streit et al. 2006), and PTPRF (LAR) (Wang et al. 2004) have also been identified in breast tumors. Most recently, PTPN12 was identified as a commonly inactivated tumor suppressor in triple-negative breast cancer (Sun et al. 2011). Breast tumors with loss of PTEN display decreased sensitivity to Herceptin, suggesting that PTEN status may be an indicator of susceptibility to the drug (Berns et al. 2007). Moreover, expression of some PTPs may also be regulated in response to stimuli or oncogenic activation such as estrogen (Liu et al. 2002) or ERBB2 (Zhai et al. 1993), respectively. Therefore, the change in certain PTPs may prove to be useful as a prognostic/diagnostic marker in breast cancer.
Metastasis is the primary cause of mortality in cancer. It is a process that requires a tumor cell to leave its primary site, pass through the blood stream, then invade and break through basement membrane barriers at the secondary site. ERBB2 (HER2 and Neu), which is a member of the ERBB family of receptor tyrosine kinases, is amplified or overexpressed in ∼25% of breast cancer patients, where it correlates with poor prognosis and high invasiveness (Slamon et al. 1989; Makar et al. 1990; Tiwari et al. 1992). To date, there is very limited understanding of the roles of PTPs in the regulation of ERBB2 signaling. It has been shown that PTPN13 (PTP-BAS) (Zhu et al. 2008) and PTPN9 (MEG2) (Yuan et al. 2010) directly dephosphorylate the C-terminal phosphotyrosine of ERBB2, and thereby inhibit ERBB2 signaling in breast cancer cell lines. Overexpression of PTP1B has been reported in breast cancer and correlated with overexpression of ERBB2 (Wiener et al. 1994). The offspring that result from a cross between transgenic mice that express an oncogenically activated form of ERBB2 in mammary glands and mice with targeted deletion of the PTP1B gene display delayed and attenuated tumor development and lung metastasis (Bentires-Alj and Neel 2007; Julien et al. 2007). Moreover, in an in vitro model of breast cancer, the formation of multiacinar structures following activation of ERBB2 requires expression of PTP1B (Arias-Romero et al. 2009). These results implicate PTP1B as a positive regulator of the development and metastasis of ERBB2-positive breast tumors (Tonks and Muthuswamy 2007). However, the possible roles of the other members of the PTP family in regulating the pathophysiology of mammary epithelial cell tumors remains to be investigated.
E-cadherin is a Ca2+-dependent transmembrane protein that functions, together with catenins, in adherens junctions of epithelial cells. It is mapped to human chromosome region 16q22.1, a hot spot for loss of heterozygosity (LOH) in breast cancer (Berx et al. 1995). Loss of E-cadherin expression has been correlated with increased metastasis of breast tumors (Siitonen et al. 1996; Hunt et al. 1997). In addition, down-regulation of E-cadherin expression is a hallmark of the epithelial–mesenchymal transition (EMT), which contributes to cancer metastasis as well as normal embryonic development (Thiery et al. 2009). Interestingly, E-cadherin is not constitutively anchored to the plasma membrane. Instead, it traffics dynamically back and forth from the plasma membrane through endosomal trafficking systems (Le et al. 1999). Phosphorylation of E-cadherin by SRC facilitates internalization and promotes sorting of ubiquitinated E-cadherin to late endosomes and lysosomes for degradation. This leads to the disruption of cell–cell adhesion and promotes cell motility (Fujita et al. 2002; Palacios et al. 2005). To date, however, the role of PTPs in the regulation of E-cadherin internalization and endosomal trafficking remains unknown.
Here, we report an RNAi-mediated loss-of-function screen to study the role of PTPs in the regulation of mammary epithelial cell motility. We used MCF-10A cells expressing an inducible form of ERBB2 to investigate the function of PTPs either in the absence or presence of ERBB2 activation. We identified three PTPs—PTPN23, PTPRG, and PTPRR—which inhibited mammary epithelial cell motility. Interestingly, of these, only PTPN23 was found to affect mammary epithelial cell invasion. In addition, suppression of PTPN23 induced caveolin-1-mediated internalization and blocked early endosome vesicle trafficking, which led to the accumulation of E-cadherin in early endosomes. Loss of E-cadherin from the cell surface may impair cell–cell adherens junctions, induce cell scattering and expression of mesenchymal proteins, and promote cell motility and invasion. Our data reveal that PTPN23 recognized SRC, E-cadherin, and β-catenin directly as substrates, suggesting that PTPN23 may regulate the activity of SRC and the phosphorylation status of E-cadherin/β-catenin signaling complexes to modulate cell motility, invasion, and scattering.
Results
Loss-of-function screening of the PTP-ome to identify regulators of mammary epithelial cell motility
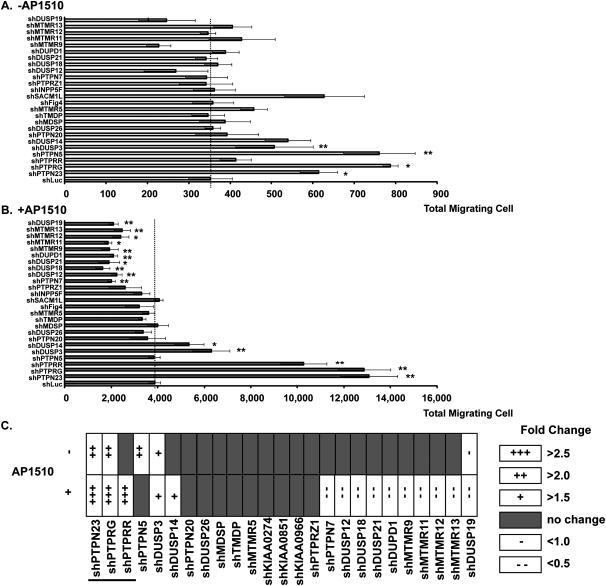
In order to study systematically the function of the PTP family using loss-of-function screens, we constructed an shRNA library specifically designed to target each of the PTPs (Supplemental Material). We used MCF10A/ERBB2 cells, which are MCF10A cells that ectopically express a chimeric form of ERBB2 in which activity can be induced with a small molecule dimerizer, AP1510 (Muthuswamy et al. 2001). The library was expressed in MCF10A/ERBB2 cells and the effect on cell motility was examined. Considering the large number of PTP shRNAs in the library, we scaled down the screening by testing shRNAs in pools. In order to study the collaboration of PTPs with ERBB2, we examined the effects of PTP suppression in conjunction with activation of the chimeric ERBB2. We chose four shRNAs per PTP, grouped the shRNAs into 25 pools according to structural similarity of their target PTPs (Supplemental Table S1), and then examined their effect on MCF10A/ERBB2 cell motility following ERBB2 activation. We identified eight PTP shRNA pools that induced cell motility to ≥2.0-fold (P < 0.01) and one pool of PTP shRNAs that reduced cell motility to ∼50% (P < 0.01) (Supplemental Fig. S1). For further deconvolution, we focused on six pools of PTP shRNAs (five that induced and one that reduced cell motility) that targeted PTPs that had not been characterized extensively to date (Supplemental Fig. S1B).
These six pools targeted 25 PTPs. We tested the effects of suppressing each of these PTPs individually on MCF10A/ERBB2 cell motility either in the absence or presence of ERBB2 activation. We identified four PTPs that induced and one PTP that reduced MCF10A/ERBB2 cell motility (Fig. 1A,C) in the absence of ERBB2 activation. On the other hand, five PTPs were identified as inducing, and 10 as reducing, cell motility in the presence of ERBB2 activation (Fig. 1B,C). It is important to note that suppression of these PTPs did not affect cell proliferation (Supplemental Fig. S2A) or apoptosis (Supplemental Fig. S2B), illustrating that proliferation and death did not contribute to the effects that we observed on motility. In summary, these data reveal that the PTPs may function either positively or negatively to regulate mammary epithelial cell motility, and that they display specificity in their function, rather than exerting pleiotropic effects.
Figure 1.
Loss-of-function screening of the PTP-ome to identify regulators of mammary epithelial cell motility. The effect of suppressing the indicated PTPs on MCF10A/ERBB2 cell motility was assessed in the absence (A) or presence (B) of AP1510 to activate ERBB2. The MCF10A/ERBB2 cells were infected by the indicated PTP shRNAs (pools of four shRNAs to target each PTP) or control (shLuc), which expresses shRNA targeting firefly luciferase. Data are presented as the absolute number of migrating cells. Error bars represent SEM (n = 3); (*) P < 0.05; (**) P < 0.01. (C) Summary of the results presented in A and B, in which a plus sign (+) indicates an increase in cell motility, and a minus sign (−) indicates a decrease in cell motility. The underline marks those PTPs that were selected for further characterization.
Suppression of PTPN23, PTPRG, and PTPRR induced distinct stimulatory effects on mammary epithelial cell motility
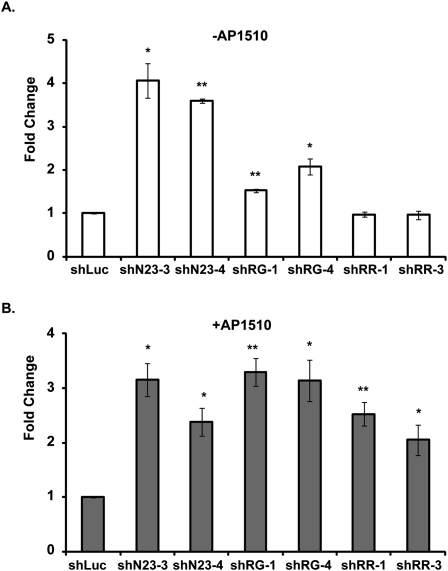
We focused on the three PTPs—PTPN23, PTPRG, and PTPRR—which enhanced MCF10A/ERBB2 cell motility by more than twofold following activation of ERBB2 (Fig. 1C, underlined). We selected two shRNAs for each PTP that efficiently suppressed expression, as analyzed by quantitative PCR (qPCR), and tested their effect individually on MCF10A/ERBB2 cell motility in the absence (Fig. 2A) or presence (Fig. 2B) of ERBB2 activation. PTPRG shRNAs (shRG) and PTPN23 shRNAs (shN23) induced MCF10A/ERBB2 cell motility either in the absence or presence of ERBB2 activation; however, shRG enhanced cell motility preferentially following ERBB2 activation, whereas shN23 had a similar effect in the absence or presence of ERBB2 activation. In contrast, PTPRR shRNAs (shRR) induced cell motility only following the activation of ERBB2. These results suggest that although PTPRG, PTPRR, and PTPN23 are negative regulators of mammary epithelial motility, their effects may be exerted through different mechanisms.
Figure 2.
Suppression of PTPN23, PTPRG, and PTPRR induced distinct stimulatory effects on mammary epithelial cell motility. The motility of MCF10A/ERBB2 cells that expressed the indicated PTP shRNA was measured in the absence (A) or presence (B) of AP1510, the activator of ERBB2. For each condition, the number of migrating cells was counted in 21 random fields. The mean number of migrating cells in the shLuc control was 5.6 cells per field in the absence (A) and 18.2 cells per field in the presence (B) of AP1510. In each case, the number of migrating cells in the shLuc control was normalized to 1. Error bars represent SEM (n = 3); (*) P < 0.05; (**) P < 0.01.
Suppression of PTPN23, but not PTPRG or PTPRR, promoted mammary epithelial cell invasion
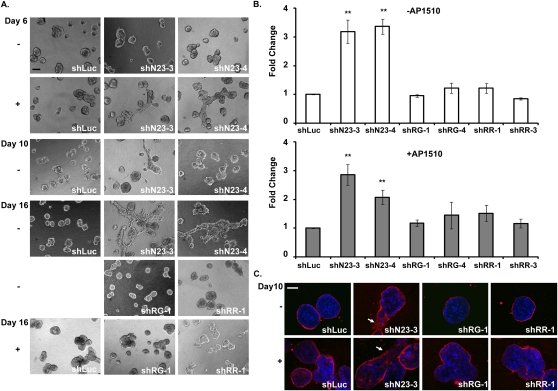
In vitro culture of MCF10A cells on an extracellular matrix (the Matrigel 3D culture system) permits the formation of sphere-like structures, called acini, which recapitulate the architecture of the mammary epithelium in vivo (Debnath et al. 2003). Activation of ERBB2 in these acini through homodimerization induces the formation of multi-acinar structures that resemble an early-stage breast tumor in vivo (Muthuswamy et al. 2001). However, activation of ERBB2 alone is insufficient to induce cell invasion in the acini (Muthuswamy et al. 2001). Therefore, we investigated further the ability of shN23, shRG, and shRR to induce MCF10A/ERBB2 cell invasion in the 3D culture system, in the absence or presence of ERBB2 activation. We observed that PTPN23 shRNA induced MCF10A/ERBB2 cell invasion at day 10 in the absence of dimerizer (Fig. 3A, day 10). Upon activation of ERBB2, the process was accelerated and invasive structures developed by day 6 (Fig. 3A, day 6). In contrast, shRG and shRR did not induce cell invasion, either with or without ERBB2 activation (Fig. 3A, day 16). The effect of these shRNAs on cell invasion was quantitated in the absence (Fig. 3B, top) or presence (Fig. 3B, bottom) of ERBB2 activation. Although the effects of suppressing PTPN23 were accelerated by activation of ERBB2, shN23 was able to induce invasion independently of ERBB2 in parental MCF10A cells (Supplemental Fig. S3), suggesting that ERBB2 activation was not required for shN23-induced cell invasion. In addition, we stained acini that expressed shRG, shN23, and shRR with antibody to laminin V, a marker for the basement membrane that surrounds the acini (Timpl and Brown 1994). As expected, only cells in which PTPN23 was suppressed were able to break through the basement membrane either in the absence or presence of ERBB2 activation (Fig. 3C, arrows), consistent with the observation that only shN23 induced invasion.
Figure 3.
Suppression of PTPN23, but not PTPRG or PTPRR, promoted mammary epithelial cell invasion. (A) Visualization of cell invasion in 3D Matrigel–collagen (1:1) mixtures. (Top, left panel) Bar, 100 μm. (B) Quantitation of cell invasion. The number of invading cells was counted in 21 random fields. The mean number of invading cells in the shLuc control was 13.9 cells per field in the absence (top) and 16.9 cells per field in the presence (bottom) of AP1510. In each case, the number of invading cells in the shLuc control was normalized to 1. Error bars represent SEM (n = 3); (*) P < 0.05; (**) P < 0.01. (C) Suppression of PTPN23 promoted the ability of the MCF10A/ERBB2 cells to break through the basal membrane. MCF10A/ERBB2 acini that expressed the indicated shRNAs were stained with laminin V antibody in the absence or presence of the ERBB2 activator AP1510. Arrows indicate cell invasion. Bar, 35 μm.
Suppression of PTPN23 induced mammary epithelial cell scattering and impaired cell–cell adhesion
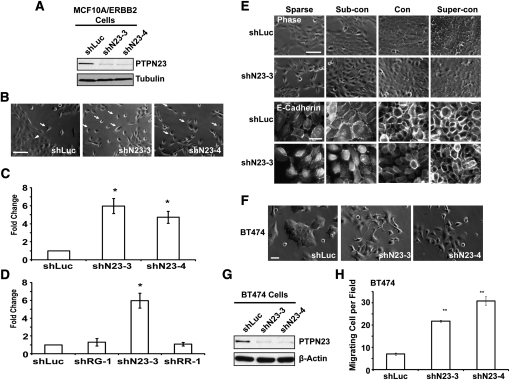
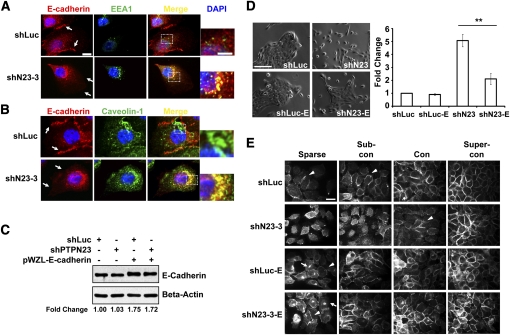
MCF10A/ERBB2 cells normally form cell–cell contacts and cell clusters (Fig. 4B, arrowhead) when grown in subconfluent culture (Debnath et al. 2003). Suppression of PTPN23 protein expression by shRNA (Fig. 4A) induced approximately fivefold more single cells (Fig. 4B, arrows) that do not form cell–cell contacts compared with the control (Fig. 4B,C). Moreover, cell scattering was observed only in MCF10A/ERBB2 cells that expressed shN23, but not shRG or shRR (Fig. 4D). This is consistent with an effect of shN23 on the induction of cell invasion.
Figure 4.
Mammary epithelial cell scattering and impaired cell–cell adhesion induced by suppression of PTPN23. (A) Immunoblot to demonstrate shRNA-induced suppression of the expression of PTPN23 protein. The expression of α-tubulin was used as the loading control. (B) Suppression of PTPN23 induced MCF10A/ERBB2 cell scattering. The photographs illustrate the morphology of the MCF10A/ErbB2 cells that expressed the indicated shRNA in subconfluent culture. Arrows indicate cell scattering. Arrowhead indicates cell cluster. Bar, 100 μm. (C) Cell scattering was quantitated by counting the number of single cells per field. At least five random fields were measured in each condition. Data are presented as change in cell scattering relative to shLuc control. Error bars represent SEM (n = 3); (*) P < 0.05. (D) RNAi-induced suppression of PTPN23, but not PTPRG or PTPRR, induced cell scattering. Cell scattering was quantitated as in C. Error bars represent SEM (n = 3); (*) P < 0.05. (E) Suppression of PTPN23 delayed the formation of adherens junctions. Images are phase (top panels) or immunofluorescence staining of E-cadherin (bottom panels) of MCF10A/ERBB2 cells. Bars, 100 μm or 20 μm, respectively. (F) Morphology of the BT474 cells that expressed the indicated shRNA. Bar, 50 μm. (G) Suppression of PTPN23 protein expression by shRNA in BT474 cells. The expression of β-actin was used as the loading control. (H) Quantitation of cell motility induced by PTPN23-directed shRNA in BT474 cells. Data are presented as the number of migrating cells per field. Error bars represent SEM (n = 3); (**) P < 0.01.
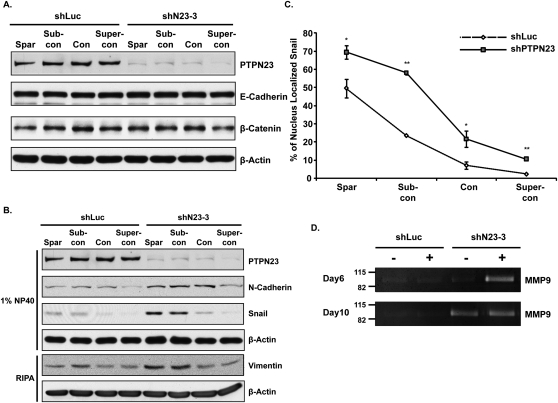
Cadherin-mediated cell–cell adherens junctions are the major mechanical force that tethers cells together. Therefore, we analyzed further the integrity of the adherens junctions in shN23 cells. MCF10A cells undergo spontaneous morphological changes at different cell densities (Sarrio et al. 2008); therefore, we grew the cells in sparse, subconfluent, confluent, or superconfluent cultures, and then examined the effect of shN23 on the formation of cell–cell adherens junctions. As shown in Figure 4E, cell scattering was observed in sparse and subconfluent shN23 cells; however, the cell scattering phenotype was lost when shN23 cells were grown to confluence. Moreover, E-cadherin staining suggested that the formation of cell–cell adherens junctions was impaired in sparse and subconfluent shN23 cells, although junctions were formed when these cells were grown to confluence (Fig. 4E, bottom two panels). We tested further the integrity of the adherens junctions of confluent shN23-expressing cells by challenging them with increasing concentrations of EGTA, which depleted Ca2+ and thus disrupted cadherin-based adhesion. Interestingly, we found that the adherens junctions of the shN23 cells were more sensitive to EGTA than the control cells (Supplemental Fig. S4), indicating impairment of the function of adherens junctions even in confluent shN23 cells.
In order to expand the scope of the analysis to encompass other ERBB2-positive cell lines, we demonstrated that suppression of PTPN23 in BT474 cells induced cell scattering and motility (Fig. 4F–H), whereas there was no change in scattering or motility of PTPN23-depeleted SKBR3 cells (Supplemental Fig. S5). It is interesting to note that BT474 cells are E-cadherin-positive, whereas SKBR3 cells are E-cadherin-negative; consequently, these observations are consistent with the phenotype we observed in MCF10A/ERBB2 cells and highlight the importance of E-cadherin in the mechanism of action of PTPN23.
Suppression of PTPN23 induced caveolin-1-mediated endocytosis of E-cadherin and caused accumulation of E-cadherin in early endosomes
It has been shown that the BRO1 domain of PTPN23 can associate with proteins that regulate endosome trafficking (Doyotte et al. 2008). Suppression of PTPN23 in HeLa cells inhibits traffic of early endosomal cargo to late endosomes and lysosomes or recycling endosomes and causes the accumulation of internalized cell surface proteins in the early endosomes (Doyotte et al. 2008). In Figure 4E, we observed that E-cadherin failed to localize to the adherens junctions in subconfluent shN23 cells, suggesting that suppression of PTPN23 may cause mislocalization of E-cadherin. To pursue this further, we traced the subcellular localization of E-cadherin in control or shN23 cells. We observed accumulation of EEA1-positive vesicles, an early endosome marker, and colocalization of E-cadherin with EEA1 in the shN23 cells (Fig. 5A, middle and right), suggesting that E-cadherin accumulated in early endosome in the shN23 cells, consistent with the observation in HeLa cells.
Figure 5.
Suppression of PTPN23 induced caveolin-1-mediated endocytosis of E-cadherin and caused accumulation of E-cadherin in early endosomes. (A,B) Colocalization of E-Cadherin with EEA1 (A) and Caveolin-1 (B) in sparse cultured MCF10A/ERBB2 cells that expressed the indicated shRNA. E-cadherin is stained in red, EEA1 and Caveolin-1 in are stained in green, and DAPI is stained in blue. Bar, 10 μm. The selected regions are enlarged in the right column (bar, 5.5 μm). (C) Ectopic expression of E-cadherin in MCF10A/ERBB2 cells in the presence and absence of PTPN23-directed shRNA. The expression of β-actin was used as the loading control. Relative signal intensity (fold change) was quantitated using ImageJ software. (D) Ectopic expression of E-cadherin rescued cell scattering that was induced by suppression of PTPN23. The morphology of the MCF10A/ERBB2 cells that expressed the indicated constructs was imaged (left), and the number of single cells was quantitated (right). Error bars represent SEM (n = 3); (**) P < 0.01. (E) Ectopic expression of E-cadherin rescued the impairment of adherens junctions that was induced by suppression of PTPN23. Images are E-cadherin immunofluorescence staining. Arrowheads indicate adherens junctions. Arrows indicate the accumulation of ectopically expressed E-cadherin. Bar, 20 μm.
Depending on cell type and stimulus, it has been shown that E-cadherin can be internalized via either clathrin-dependent endocytosis or caveolin-1-mediated endocytosis (Zavadil et al. 2001; Lu et al. 2003; Janda et al. 2006). We observed colocalization of E-cadherin with caveolin-1 (Fig. 5B, right), but not with clathrin (Supplemental Fig. S6), suggesting that E-cadherin was internalized by the caveolin-1-mediated endocytosis pathway. There are a similar number of the caveolin-1-positive vesicles in the control and shN23 cells (Fig. 5B, middle), suggesting that PTPN23 does not control the formation of the caveolin-1 vesicles per se. However, the colocalization of E-cadherin with caveolin-1-positive vesicles was increased in shN23 cells (Fig. 5B, right), suggesting that E-cadherin internalization through caveolin-1-mediated endocytosis was promoted in the shN23 cells. Furthermore, the internalization of E-cadherin was decreased when cells became confluent (Supplemental Fig. S7), possibly because homophilic interactions between E-cadherin molecules on apposing cells stabilized localization to the plasma membrane. This was consistent with our observation that the formation of adherens junctions was partially rescued in confluent shN23 cells.
To examine further the importance of E-cadherin (which is the major adhesion molecule of adherens junctions) as a target for these effects of PTPN23 in mammary epithelial cells, we tested the effects of ectopic expression of E-cadherin. We observed that overexpression of E-cadherin in shN23 cells (Fig. 5C) inhibited cell scattering (Fig. 5D) and restored the formation of the adherens junctions (Fig. 5E, arrowheads). Our data suggest that overexpression of E-cadherin may have saturated the internalization system and essentially led to overflow from the endosomes to the plasma membrane (Supplemental Fig. S8), which compensated for the depletion of PTPN23.
Suppression of PTPN23 induced the expression of the mesenchymal proteins
Cell scattering and impairment of cell–cell adhesion is connected with the EMT, which is characterized by the down-regulation of epithelial proteins, such as E-cadherin, and the up-regulation of mesenchymal proteins, such as N-cadherin, vimentin, snail, and matrix metalloproteases (MMPs). In light of the changes we observed in subcellular localization of E-cadherin, we examined whether suppression of PTPN23 altered the expression of epithelial and mesenchymal markers. Neither the expression of E-cadherin nor that of its binding partner, β-catenin, was suppressed in shN23 cells (Fig. 6A). Nevertheless, the expression of mesenchymal proteins—including N-cadherin, vimentin, and snail—was up-regulated in the shN23 cells (Fig. 6B). Moreover, the nuclear localization of snail was also enhanced in the shN23 cells (Fig. 6C; Supplemental Fig. S9). We also analyzed the secretion of MMPs in shN23 cells that were grown in the 3D system. We detected an MMP activity at ∼92k, the expected molecular weight of MMP9, following suppression of PTPN23 (Fig. 6D). Interestingly, MMP9 activity was detected in day 10 acini that were grown both with and without ERBB2 activation (Fig. 6D); however, the day 6 acini secreted MMP9 only when they were grown with ERBB2 activation (Fig. 6D), consistent with a function of ERBB2 to accelerate shN23-induced cell invasion. It is important to note that the expression of these mesenchymal markers was down-regulated when the cells reached confluence. Therefore, these data suggest that although suppression of PTPN23 induced the expression of mesenchymal proteins, this was not sufficient to down-regulate the expression of epithelial proteins or trigger a full-blown EMT.
Figure 6.
Suppression of PTPN23 induced expression of mesenchymal proteins. (A,B) Immunoblot to demonstrate that suppression of PTPN23 did not affect expression of E-cadherin or β-catenin protein (A); nevertheless, it did induce expression of the indicated mesenchymal markers (B). Cells were grown under sparse, subconfluent, confluent, or superconfluent conditions, as described in the text. The expression of β-actin was used as the loading control. (C) Suppression of PTPN23 enhanced nuclear localization of Snail. The percentage of nuclear-localized snail was quantitated as the mean of eight random fields. Error bars represent SEM (n = 3); (*) P < 0.05; (**) P < 0.01. (D) Suppression of PTPN23 induced the expression of MMP9. The MMP activity of the MCF10A/ERBB2 cells was measured by zymogram assay.
Suppression of PTPN23 enhanced downstream signaling through activation of SRC and β-catenin
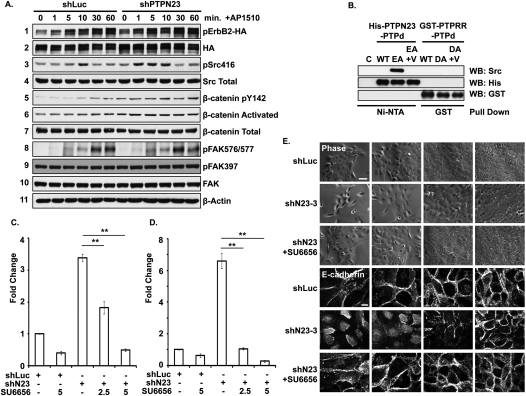
As shown in Figure 7A, the addition of a dimerizer, AP1510, led to activation of chimeric ERBB2 in MCF10A/ERBB2 cells; however, there was no significant difference in ERBB2 phosphorylation between the control and shN23 cells, suggesting that PTPN23 did not regulate the phosphorylation status of ERBB2 directly. In addition, we examined the phosphorylation of 42 different receptor PTKs in control and PTPN23-depleted cells by using a phospho-RPTK antibody microarray kit (R&D Systems). Again, we observed no change in the phosphorylation of these RPTKs (data not shown), suggesting that the effect of PTPN23 lies downstream. However, when we examined the phosphorylation status of SRC in the presence and absence of PTPN23-directed shRNA, we observed that ERBB2-induced autophosphorylation on Tyr 416 occurred both earlier in the time course and to a greater extent in PTPN23-depleted cells (Fig. 7A, panels 3,4), consistent with an inhibitory role for the phosphatase in the regulation of SRC. In contrast, we did not detect a significant change in the phosphorylation of FAK (Fig. 7A, panels 8–10). Having observed mislocalization of E-cadherin in PTPN23-depleted cells (Fig. 4E), we tested whether suppression of PTPN23 induced the activation of β-catenin, which is associated with E-cadherin. It has been shown that β-catenin is activated through phosphorylation of Tyr142 (Piedra et al. 2003; Brembeck et al. 2004) and the dephosphorylation of Ser37/Thr41 (Yost et al. 1996). We observed increased phosphorylation of Tyr142 (Fig. 7A, panel 5) and dephosphorylation of Ser37/Thr41 on β-catenin in shN23 cells (Fig. 7A, panels 6), indicative of activation of β-catenin following suppression of PTPN23.
Figure 7.
PTPN23 functions through SRC family kinases to regulate cell motility, invasion, and scattering. (A) Suppression of PTPN23 activated SRC and β-catenin. A time course of tyrosine phosphorylation of the indicated proteins was measured by immunoblotting with the indicated antibodies following ERBB2 activation in MCF10A/ERBB2 cells that expressed either shLuc or shPTPN23. The expression of β-actin was used as the loading control. (B) Purified recombinant His-tagged forms of the catalytic domain of PTPN23 or GST-tagged forms of the catalytic domain of PTPRR were mixed with pervanadate-treated MCF10A/ERBB2 cell lysates, then precipitated with Ni-NTA agarose or glutathione beads, respectively. Binding of SRC was assessed by blotting the precipitates with anti-SRC antibodies. Anti-His tag and anti-GST antibody blots were used for loading controls.Abbreviations indicate empty bead control (C), wild type (WT), PTPN23-E1357A (EA), or PTPRR-D554A (DA) substrate-trapping mutants in the absence or presence (+V) of vanadate. (C,D) Treatment with the SRC inhibitor SU6656 inhibited MCF10A/ERBB2 cell motility (C) and cell invasion (D) that was induced by suppression of PTPN23. SRC inhibitor SU6656 was added at the indicated micromolar concentrations. Data are presented as change in cell motility and cell invasion relative to shLuc control. Error bars represent SEM (n = 3); (**) P < 0.01. (E) Treatment with the SRC inhibitor SU6656 suppressed cell scattering and disruption of adherens junctions that was induced by depletion of PTPN23. MCF10A/ERBB2 cells, control (shLuc), or PTPN23-depleted (shN23-3) were grown as indicated in the absence or presence of 5 μM SU6656. Images are phase (top) or E-cadherin immunofluorescence staining (bottom). Bars, 100 μm or 20 μm, respectively.
MCF10A/ERBB2 cells are sensitive to serum starvation; therefore, we generated HEK293 cells that ectopically expressed the chimeric ERBB2 receptor (293-B2 cells) as an alternative system in which to analyze the effects of PTPN23. As in MCF10A/ERBB2 cells, suppression of PTPN23 did not affect the phosphorylation of ERBB2 directly, but did lead to enhanced autophosphorylation of SRC and activation of β-catenin (Supplemental Fig. S10A). Furthermore, we demonstrated that suppression of neither PTPRG nor PTPRR led to enhanced activation of SRC or β-catenin, consistent with specificity in the effects of PTPN23 (Supplemental Fig. S10B,C). In fact, suppression of PTPRR both enhanced and sustained the phosphorylation of ERK upon ERBB2 activation (Supplemental Fig. S10D). This is consistent with the role of PTPRR as an inhibitor of MAPK signaling (Chirivi et al. 2007), and with our observation that enhanced cell motility induced by suppression of PTPRR was dependent on ERBB2 activation (Fig. 2).
PTPN23 is an active phosphatase that recognized SRC as a direct substrate
The conserved Asp181 residue of PTP1B plays a crucial role as a general acid in the first step of catalysis (Flint et al. 1997). Our laboratory has shown that mutation of this site to Ala impairs the catalytic activity of PTP1B but maintains its affinity for substrates, thus generating a “substrate-trapping” mutant form of the enzyme (Tonks 2003). Interestingly, the residue in PTPN23 that is equivalent to Asp181 of PTP1B is Glu, rather than Asp. We had shown that mutation of Asp181 to Glu dramatically decreases the activity of PTP1B, but does not inactivate the enzyme (Flint et al. 1997). Therefore, we tested the activity of recombinant PTPN23 toward a small peptide substrate modeled on the pTyr169 phosphorylation site in p130cas. We observed that it dephosphorylated the peptide with a Vmax of ∼245 nmol/min/mg, which is comparable with the specific activity of the Asp181 → Glu mutant form of PTP1B (Flint et al. 1997), illustrating that PTPN23 is an active enzyme (Supplemental Fig. S11).
In order to identify the potential physiological substrates of PTPN23, we generated a recombinant PTPN23 PTP domain as either wild type or the substrate-trapping mutant E1357A (EA). We used the indicated recombinant PTPN23 PTP domain to pull down potential substrates from pervanadate-treated MCF10A/ERBB2 cell lysate and observed coprecipitation of a small number of pTyr proteins with the EA mutant (Supplemental Fig. S12). Addition of the active site-directed inhibitor vanadate reduced the pTyr signal to that associated with the wild type (Supplemental Fig. S12), consistent with these proteins being substrates of PTPN23. Interestingly, three proteins migrated at the expected size for SRC, E-cadherin, and β-catenin. Further characterization by immunoblotting revealed that the PTPN23 EA mutant interacted with these three proteins, whereas the wild type or ED mutant did not (Supplemental Fig. S13A). SRC was not recovered in a stable complex with E-cadherin and/or β-catenin, suggesting that the substrate-trapping mutant form of PTPN23 recognized these proteins independently (Supplemental Fig. S13B). We focused attention on SRC and demonstrated in an in vitro pull-down that the substrate-trapping mutant, but not wild-type, PTPN23 formed a stable complex with SRC that was disrupted by vanadate (Fig. 7B; Supplemental Fig. S13C). In contrast, in a parallel experiment, the Asp554 → Ala substrate-trapping mutant form of PTPRR did not recognize SRC (Fig. 7B), illustrating specificity in the effects of PTPN23. Furthermore, following expression in 293-B2 cells, the full-length substrate-trapping mutant, but not wild-type, form of PTPN23 formed a complex with endogenous SRC (Supplemental Fig. S13D). Overall, the data suggest that SRC together with E-cadherin and β-catenin are potential physiological substrates of PTPN23 in mammary epithelial cells.
Inhibition of SFK activity impaired shN23-induced phenotypes
We showed that SRC autophosphorylation was elevated in MCR10A/ERBB2 and 293-B2 cells in which PTPN23 was suppressed and that SRC was a direct substrate of PTPN23. To understand whether SFK activity is essential for the function of PTPN23, we tested the effect of the inhibitor, SU6656 (Blake et al. 2000). Interestingly, the SRC inhibitor attenuated both the enhanced cell motility (Fig. 7C) and cell invasion (Fig. 7D) that was induced by suppression of PTPN23. Moreover, inhibition of SRC activity promoted cell–cell adhesion (Fig. 7E), inhibited the accumulation of E-cadherin in early endosomes (Supplemental Fig. S14A), and decreased endocytosis of E-cadherin through caveolin-1 vesicles (Supplemental Fig. S14B). Taken together, these results illustrate that SRC plays an important role in mediating the function of PTPN23 in the control of mammary epithelial cell motility, invasion, and the formation of cell–cell adherens junctions.
Discussion
In this study, we examined systematically the role of PTPs in the regulation of mammary epithelial cell motility, in the absence or presence of ERBB2 signaling. We observed that the majority of the PTP shRNAs had little or no effect on mammary epithelial cell motility (Supplemental Fig. S1), consistent with specificity in the effects of PTPs on cell function. In addition, we observed that, in some cases, suppression of particular PTPs enhanced migration, whereas suppression of others was inhibitory, consistent with roles as negative or positive regulators of signaling, respectively. The specificity and subtlety of regulation by PTPs are also seen in the differential effects of suppressing PTPN23, PTPRG, and PTPRR on signaling in the absence or presence of ERBB2 activation, including the fact that suppression of PTPN23, but not PTPRG and PTPRR, induced cell invasion. Taken together, these data highlight that PTPs are not pleiotropic suppressors of tyrosine phosphorylation-dependent signaling that are linked to a housekeeping function; instead the PTPs function as specific regulators of signaling pathways.
We showed for the first time that suppression of PTPRG and PTPRR enhanced mammary epithelial cell motility (Figs. 1, 2). The PTPRG gene is localized at human chromosome 3p21, a hot spot for deletion in breast cancer, and expression of PTPRG is reduced in lung, ovarian, and breast tumors, suggesting its potential to function as a tumor suppressor (LaForgia et al. 1991; Panagopoulos et al. 1996; van Niekerk and Poels 1999; Vezzalini et al. 2007). In addition, the PTPRG gene has been shown to play a role as an estrogen-regulated tumor suppressor (Liu et al. 2002). These findings are consistent with a role for PTPRG in the development of breast cancer. PTPRR contains a KIM (kinase interaction motif) sequence, which is known to be important in recognition of MAPKs (Szedlacsek et al. 2001). In PTPRR-deficient mice, the phosphorylation of p42/p44 MAPKs is enhanced in the brain (Chirivi et al. 2007). Moreover, a spliced isoform of PTPRR, PTP-SL, has been shown to regulate the phosphorylation and nuclear translocation of ERK5 (Buschbeck et al. 2002). These results suggest that PTPRR is a negative regulator of the MAPK signaling pathways, which is consistent with our finding that suppression of PTPRR increased ERK phosphorylation and only induced cell motility following ERBB2 activation.
We focused our attention on PTPN23 because of the unique links that we observed between this enzyme and cell invasion. The PTPN23 gene is also mapped to human chromosome 3p21 (Toyooka et al. 2000); however, the function of PTPN23 in the regulation of the development of breast cancer has not been characterized. It has been shown that suppression of PTPN23 induces endothelial and bladder carcinoma cell motility (Castiglioni et al. 2007; Mariotti et al. 2009a,b). In those studies, it was suggested that SRC was not a substrate of PTPN23. Instead, it was proposed that PTPN23 could serve as a bridge between the PTKs SRC and FAK, and that SRC regulated this scaffold function by phosphorylating PTPN23. In contrast, it has been shown that the phosphatase activity of the rat ortholog of PTPN23 is important for suppression of RAS-mediated transformation of fibroblasts (Cao et al. 1998). Nevertheless, due to deviation of the active site signature sequences of PTPN23 from the consensus of other active members of the PTP family, there has been a debate about whether it possesses catalytic activity (Gingras et al. 2009). Here, we demonstrated that PTPN23 is an active enzyme (Supplemental Fig. S11), and, furthermore, provide several lines of evidence to support the identification of SRC as an important physiological substrate. When incubated with cell lysates, the substrate-trapping mutant form of the catalytic domain of PTPN23 formed a stable complex with SRC, which was disrupted by the active site-directed inhibitor vanadate (Fig. 7; Supplemental Fig. S13). Upon expression in 293-B2 cells, the full-length, substrate-trapping mutant form of PTPN23 formed a complex with SRC, whereas the full-length wild-type enzyme did not (Supplemental Fig. S13). Suppression of PTPN23 enhanced the autophosphorylation of SRC in MCF10A/ERBB2 cells (Fig. 7) and 293-B2 cells (Supplemental Fig. S10); this was not detected following suppression of either PTPRG or PTPRR in 293-B2 cells (Supplemental Fig. S10), attesting to the specificity of PTPN23. Finally, inclusion of the SRC inhibitor SU6656 blocked the effects of suppression of PTPN23 (Fig. 7; Supplemental Fig. S14). These data suggest that PTPN23 has the potential to be an important regulator of the development of breast cancer that may exert its effects via controlling the activation status of SRC.
SRC is a major regulator of a variety of signaling pathways that control cell proliferation, migration, adhesion, and invasion. Some PTPs—including PTPRA, PTP1B, and PTPRE—have been reported to function as activators of SRC through dephosphorylation of the C-terminal inhibitory tyrosine residue (Try527) (Roskoski 2005). In contrast, PTP-BAS and TCPTP have been shown to be inhibitors of SRC activity through directly dephosphorylating the autophosphorylation site (Tyr416) (Palmer et al. 2002; van Vliet et al. 2005). Here we showed that suppression of PTPN23 increased SRC activity as a consequence of disrupting a direct enzyme–substrate interaction between PTPN23 and SRC (Fig. 7; Supplemental Fig. S13). Therefore, our data suggest that PTPN23 is a direct negative regulator of SFK activity. Overexpression or hyperactivation of SRC is observed in many human cancers, including breast cancer (Summy and Gallick 2003). However, it has been shown in transgenic mouse models that overexpression or hyperactivation of SRC is not sufficient to induce a higher-grade breast tumor and its metastasis (Webster et al. 1995). Interestingly, transgenic overexpression of SRC in a p21−/− background dramatically induces tumor growth and metastasis (Kline et al. 2008), suggesting that the inactivation of tumor suppressors augments SRC transforming activity. Considering the negative effect of PTPN23 on SRC activity, loss of PTPN23 may promote tumor growth and metastasis in breast tumors for which SRC is overexpressed.
Overall, our approach has revealed novel functional insights into members of the PTP family that have remained relatively uncharacterized to date. The data presented here can be integrated into a model of PTPN23 function that highlights its recognition of SRC as a substrate. E-cadherin is a downstream target of SRC (Daugherty and Gottardi 2007). SRC phosphorylates E-cadherin on its cytoplasmic segment, which disrupts its association with β-catenin, leading to the release and activation of β-catenin (Daugherty and Gottardi 2007). In addition, phosphorylation of E-cadherin also promotes its internalization (Fujita et al. 2002; Palacios et al. 2005). Our data illustrate that, in the absence of PTPN23, SRC may become activated, which in turn would increase the phosphorylation of E-cadherin, the release and activation of β-catenin, and the internalization of E-cadherin. In addition, suppression of PTPN23 may also directly regulate the phosphorylation of E-cadherin and β-catenin, disrupt their association and promote β-catenin activation, and contribute to the increase in expression of the mesenchymal proteins that promote mammary epithelial cell motility, scattering, and invasion. The localization of PTPN23 gene at chromosome 3p21, a hot spot for mutation in breast cancer, coupled with our demonstration that PTPN23 may function as a negative regulator of SRC, suggests that inhibition of SRC may be of therapeutic benefit to cancer patients with mutation at the PTPN23 locus. Hopefully, further investigation of this and other PTPs may enhance therapeutic strategies that target tyrosine phosphorylation-dependent signaling pathways that are aberrantly regulated in disease.
Materials and methods
Cell culture
The 293-B2 cell line was generated by infecting HEK293 cells with retrovirus encoding a p75.ERBB2-FKBP chimera (Muthuswamy et al. 1999). The infected cells were sorted by flow cytometry using anti-p75 antibodies to isolate cell populations expressing similar levels of the ERBB2 chimera. A cell population was selected that displayed low basal ERBB2 tyrosine phosphorylation and an increase in tyrosine phosphorylation upon addition of AP1510. To activate ERBB2, 293-B2 cells were serum-starved for 16 h and then stimulated with 1 μM AP1510 for the indicated times. For examining cell morphology and adherens junctions, MCF10A/ERBB2 cells were grown at initial densities of 2 × 104, 4 × 104, 8 × 104, or 12 × 104 cells per well of the eight-well chamber slides (BD Biosciences) for 48 h to generate sparse, subconfluent, confluent, and superconfluent cell monolayers, respectively. For examining mesenchymal protein expression, MCF10A/ERBB2 cells were grown at initial densities of 2 × 105, 4 × 105, 8 × 105, or 12 × 105 cells per 10-cm plate for 48 h to generate sparse, subconfluent, confluent, or superconfluent cell monolayers, respectively.
Cell migration and invasion assays
Cell motility was measured using Cell Culture Inserts (8.0-μm pore size) for six-well plates (BD Falcon). To visualize cell invasion, we used eight-well chamber slides (BD Biosciences) precoated with 70 μL of 1:1 mixture of Matrigel and Collagen I (BD Biosciences). On day 1, 4000 cells were grown per well in the presence of 5 ng/mL EGF (Xiang and Muthuswamy 2006). To activate ERBB2, AP1510 (1 μM) was added to the culture medium at day 4. Cell morphology was photographed on days 6, 10, and 16. The phase images were taken by a Zeiss Axiovert 200M using AxioVison 4.4 software. Immunofluorescence staining images were taken by a Perkin Elmer spinning disc confocal microscope using Velocity 5 software (Debnath et al. 2003). To quantitate cell invasion, we used BD BioCoat Matrigel Invasion Chambers, 8.0-μm pore size. MCF10A/ERBB2 cells (1 × 106) were grown in the insert. After 48 h, the cells retained inside the insert were removed, and those that migrated to the other side of the insert were fixed and stained with KARYOMAX Giemsa Stain (GIBCO, Invitrogen) and counted.
Recombinant protein purification and substrate trapping
The wild-type and substrate-trapping mutant E1357A forms of the PTPN23 PTP domain were generated from wild-type full-length PTPN23 (pCDNA3.1-2flag-PTPN23) (Gingras et al. 2009), and the recombinant protein was purified using Ni-NTA column chromatography. Recombinant PTPN23 PTP domain (1 μg) was used to pull down its substrates from 100 μg of pervanadate-treated MCF10A/ERBB2 cell lysate in the presence or absence of 1 mM sodium orthovanadate.
Statistics
All statistics were performed using a standard Student's t-test.
Acknowledgments
We thank Ross Dickins and Scott Lowe (Cold Spring Harbor Laboratory, NY) for use of their shRNA expression construct, pMLP; Jeanette Maier (University of Milan, Italy) for polyclonal PTPN23 antibody; Arnim Pause (McGill University, Montreal, Canada) for the PTPN23 full-length mammalian expression construct; Stefan Knapp and Alastair Barr (Structural Genomics Consortium, University of Oxford, UK) for the GST-PTPRR construct; Todd Miller (Stony Brook University, NY) for phosphotyrosyl peptide; and ARIAD for providing us with AP1510. This study was supported by NIH grant CA53840 and CSHL Cancer Centre Support Grant CA45508.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2018911.
References
- Ardini E, Agresti R, Tagliabue E, Greco M, Aiello P, Yang LT, Menard S, Sap J 2000. Expression of protein tyrosine phosphatase α (RPTPα) in human breast cancer correlates with low tumor grade, and inhibits tumor cell growth in vitro and in vivo. Oncogene 19: 4979–4987 [DOI] [PubMed] [Google Scholar]
- Arias-Romero LE, Saha S, Villamar-Cruz O, Yip SC, Ethier SP, Zhang ZY, Chernoff J 2009. Activation of Src by protein tyrosine phosphatase 1B Is required for ErbB2 transformation of human breast epithelial cells. Cancer Res 69: 4582–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentires-Alj M, Neel BG 2007. Protein-tyrosine phosphatase 1B is required for HER2/Neu-induced breast cancer. Cancer Res 67: 2420–2424 [DOI] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, et al. 2007. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12: 395–402 [DOI] [PubMed] [Google Scholar]
- Berx G, Cleton-Jansen AM, Nollet F, de Leeuw WJ, van de Vijver M, Cornelisse C, van Roy F 1995. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J 14: 6107–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake RA, Broome MA, Liu X, Wu J, Gishizky M, Sun L, Courtneidge SA 2000. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol Cell Biol 20: 9018–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W 2004. Essential role of BCL9-2 in the switch between β-catenin's adhesive and transcriptional functions. Genes Dev 18: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M, Eickhoff J, Sommer MN, Ullrich A 2002. Phosphotyrosine-specific phosphatase PTP-SL regulates the ERK5 signaling pathway. J Biol Chem 277: 29503–29509 [DOI] [PubMed] [Google Scholar]
- Cao L, Zhang L, Ruiz-Lozano P, Yang Q, Chien KR, Graham RM, Zhou M 1998. A novel putative protein-tyrosine phosphatase contains a BRO1-like domain and suppresses Ha-ras-mediated transformation. J Biol Chem 273: 21077–21083 [DOI] [PubMed] [Google Scholar]
- Castiglioni S, Maier JA, Mariotti M 2007. The tyrosine phosphatase HD-PTP: a novel player in endothelial migration. Biochem Biophys Res Commun 364: 534–539 [DOI] [PubMed] [Google Scholar]
- Chirivi RG, Noordman YE, Van der Zee CE, Hendriks WJ 2007. Altered MAP kinase phosphorylation and impaired motor coordination in PTPRR deficient mice. J Neurochem 101: 829–840 [DOI] [PubMed] [Google Scholar]
- Daugherty RL, Gottardi CJ 2007. Phospho-regulation of β-catenin adhesion and signaling functions. Physiology (Bethesda) 22: 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30: 256–268 [DOI] [PubMed] [Google Scholar]
- Doyotte A, Mironov A, McKenzie E, Woodman P 2008. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci 105: 6308–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AJ, Tiganis T, Barford D, Tonks NK 1997. Development of ‘substrate-trapping’ mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci 94: 1680–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiss G, Vignon F 2004. Protein tyrosine phosphatases and breast cancer. Crit Rev Oncol Hematol 52: 9–17 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W 2002. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol 4: 222–231 [DOI] [PubMed] [Google Scholar]
- Gingras MC, Zhang YL, Kharitidi D, Barr AJ, Knapp S, Tremblay ML, Pause A 2009. HD-PTP is a catalytically inactive tyrosine phosphatase due to a conserved divergence in its phosphatase domain. PLoS ONE 4: e5105 doi: 10.1371/journal.pone.0005105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt NC, Douglas-Jones AG, Jasani B, Morgan JM, Pignatelli M 1997. Loss of E-cadherin expression associated with lymph node metastases in small breast carcinomas. Virchows Arch 430: 285–289 [DOI] [PubMed] [Google Scholar]
- Janda E, Nevolo M, Lehmann K, Downward J, Beug H, Grieco M 2006. Raf plus TGFβ-dependent EMT is initiated by endocytosis and lysosomal degradation of E-cadherin. Oncogene 25: 7117–7130 [DOI] [PubMed] [Google Scholar]
- Julien SG, Dube N, Read M, Penney J, Paquet M, Han Y, Kennedy BP, Muller WJ, Tremblay ML 2007. Protein tyrosine phosphatase 1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis and protects from lung metastasis. Nat Genet 39: 338–346 [DOI] [PubMed] [Google Scholar]
- Kline CL, Jackson R, Engelman R, Pledger WJ, Yeatman TJ, Irby RB 2008. Src kinase induces tumor formation in the c-SRC C57BL/6 mouse. Int J Cancer 122: 2665–2673 [DOI] [PubMed] [Google Scholar]
- LaForgia S, Morse B, Levy J, Barnea G, Cannizzaro LA, Li F, Nowell PC, Boghosian-Sell L, Glick J, Weston A, et al. 1991. Receptor protein-tyrosine phosphatase γ is a candidate tumor suppressor gene at human chromosome region 3p21. Proc Natl Acad Sci 88: 5036–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TL, Yap AS, Stow JL 1999. Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J Cell Biol 146: 219–232 [PMC free article] [PubMed] [Google Scholar]
- Levea CM, McGary CT, Symons JR, Mooney RA 2000. PTP LAR expression compared to prognostic indices in metastatic and non-metastatic breast cancer. Breast Cancer Res Treat 64: 221–228 [DOI] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al. 1997. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16: 64–67 [DOI] [PubMed] [Google Scholar]
- Liu S, Sugimoto Y, Kulp SK, Jiang J, Chang HL, Park KY, Kashida Y, Lin YC 2002. Estrogenic down-regulation of protein tyrosine phosphatase γ (PTP γ) in human breast is associated with estrogen receptor α. Anticancer Res 22: 3917–3923 [PubMed] [Google Scholar]
- Lu Z, Ghosh S, Wang Z, Hunter T 2003. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of β-catenin, and enhanced tumor cell invasion. Cancer Cell 4: 499–515 [DOI] [PubMed] [Google Scholar]
- Makar AP, Desmedt EJ, De Potter CR, Vanderheyden JS, Schatteman EA 1990. Neu (C-erbB-2) oncogene in breast cancer and its possible association with the risk of distant metastases. A retrospective study and review of literature. Acta Oncol 29: 931–934 [DOI] [PubMed] [Google Scholar]
- Mariotti M, Castiglioni S, Garcia-Manteiga JM, Beguinot L, Maier JA 2009a. HD-PTP inhibits endothelial migration through its interaction with Src. Int J Biochem Cell Biol 41: 687–693 [DOI] [PubMed] [Google Scholar]
- Mariotti M, Castiglioni S, Maier JA 2009b. Inhibition of T24 human bladder carcinoma cell migration by RNA interference suppressing the expression of HD-PTP. Cancer Lett 273: 155–163 [DOI] [PubMed] [Google Scholar]
- Muthuswamy SK, Gilman M, Brugge JS 1999. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol 19: 6845–6857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS 2001. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol 3: 785–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios F, Tushir JS, Fujita Y, D'Souza-Schorey C 2005. Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell–cell adhesion during epithelial to mesenchymal transitions. Mol Cell Biol 25: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A, Zimmer M, Erdmann KS, Eulenburg V, Porthin A, Heumann R, Deutsch U, Klein R 2002. EphrinB phosphorylation and reverse signaling: regulation by Src kinases and PTP-BL phosphatase. Mol Cell 9: 725–737 [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Pandis N, Thelin S, Petersson C, Mertens F, Borg A, Kristoffersson U, Mitelman F, Aman P 1996. The FHIT and PTPRG genes are deleted in benign proliferative breast disease associated with familial breast cancer and cytogenetic rearrangements of chromosome band 3p14. Cancer Res 56: 4871–4875 [PubMed] [Google Scholar]
- Piedra J, Miravet S, Castano J, Palmer HG, Heisterkamp N, Garcia de Herreros A, Dunach M 2003. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate β-catenin Tyr-142 phosphorylation and β-catenin-α-catenin Interaction. Mol Cell Biol 23: 2287–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke I, Gotte M, Kersting C, Mattsson B, Kiesel L, Wulfing P 2006. Expression and prognostic impact of the protein tyrosine phosphatases PRL-1, PRL-2, and PRL-3 in breast cancer. Br J Cancer 95: 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R Jr 2005. Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun 331: 1–14 [DOI] [PubMed] [Google Scholar]
- Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J 2008. Epithelial–mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 68: 989–997 [DOI] [PubMed] [Google Scholar]
- Siitonen SM, Kononen JT, Helin HJ, Rantala IS, Holli KA, Isola JJ 1996. Reduced E-cadherin expression is associated with invasiveness and unfavorable prognosis in breast cancer. Am J Clin Pathol 105: 394–402 [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. 1989. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 244: 707–712 [DOI] [PubMed] [Google Scholar]
- Streit S, Ruhe JE, Knyazev P, Knyazeva T, Iacobelli S, Peter S, Hoefler H, Ullrich A 2006. PTP-PEST phosphatase variations in human cancer. Cancer Genet Cytogenet 170: 48–53 [DOI] [PubMed] [Google Scholar]
- Summy JM, Gallick GE 2003. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev 22: 337–358 [DOI] [PubMed] [Google Scholar]
- Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, Nguyen DX, Pavlova NN, Botero M, Huang J, et al. 2011. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell 144: 703–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szedlacsek SE, Aricescu AR, Fulga TA, Renault L, Scheidig AJ 2001. Crystal structure of PTP-SL/PTPBR7 catalytic domain: implications for MAP kinase regulation. J Mol Biol 311: 557–568 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA 2009. Epithelial–mesenchymal transitions in development and disease. Cell 139: 871–890 [DOI] [PubMed] [Google Scholar]
- Timpl R, Brown JC 1994. The laminins. Matrix Biol 14: 275–281 [DOI] [PubMed] [Google Scholar]
- Tiwari RK, Borgen PI, Wong GY, Cordon-Cardo C, Osborne MP 1992. HER-2/neu amplification and overexpression in primary human breast cancer is associated with early metastasis. Anticancer Res 12: 419–425 [PubMed] [Google Scholar]
- Tonks NK 2003. PTP1B: from the sidelines to the front lines! FEBS Lett 546: 140–148 [DOI] [PubMed] [Google Scholar]
- Tonks NK 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol 7: 833–846 [DOI] [PubMed] [Google Scholar]
- Tonks NK, Muthuswamy SK 2007. A brake becomes an accelerator: PTP1B—a new therapeutic target for breast cancer. Cancer Cell 11: 214–216 [DOI] [PubMed] [Google Scholar]
- Toyooka S, Ouchida M, Jitsumori Y, Tsukuda K, Sakai A, Nakamura A, Shimizu N, Shimizu K 2000. HD-PTP: a novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem Biophys Res Commun 278: 671–678 [DOI] [PubMed] [Google Scholar]
- van Niekerk CC, Poels LG 1999. Reduced expression of protein tyrosine phosphatase γ in lung and ovarian tumors. Cancer Lett 137: 61–73 [DOI] [PubMed] [Google Scholar]
- van Vliet C, Bukczynska PE, Puryer MA, Sadek CM, Shields BJ, Tremblay ML, Tiganis T 2005. Selective regulation of tumor necrosis factor-induced Erk signaling by Src family kinases and the T cell protein tyrosine phosphatase. Nat Immunol 6: 253–260 [DOI] [PubMed] [Google Scholar]
- Vezzalini M, Mombello A, Menestrina F, Mafficini A, Della Peruta M, van Niekerk C, Barbareschi M, Scarpa A, Sorio C 2007. Expression of transmembrane protein tyrosine phosphatase γ (PTPγ) in normal and neoplastic human tissues. Histopathology 50: 615–628 [DOI] [PubMed] [Google Scholar]
- Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS, et al. 2004. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304: 1164–1166 [DOI] [PubMed] [Google Scholar]
- Webster MA, Cardiff RD, Muller WJ 1995. Induction of mammary epithelial hyperplasias and mammary tumors in transgenic mice expressing a murine mammary tumor virus/activated c-src fusion gene. Proc Natl Acad Sci 92: 7849–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener JR, Kerns BJ, Harvey EL, Conaway MR, Iglehart JD, Berchuck A, Bast RC Jr 1994. Overexpression of the protein tyrosine phosphatase PTP1B in human breast cancer: association with p185c-erbB-2 protein expression. J Natl Cancer Inst 86: 372–378 [DOI] [PubMed] [Google Scholar]
- Xiang B, Muthuswamy SK 2006. Using three-dimensional acinar structures for molecular and cell biological assays. Methods Enzymol 406: 692–701 [DOI] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT 1996. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev 10: 1443–1454 [DOI] [PubMed] [Google Scholar]
- Yuan T, Wang Y, Zhao ZJ, Gu H 2010. Protein-tyrosine phosphatase PTPN9 negatively regulates ErbB2 and epidermal growth factor receptor signaling in breast cancer cells. J Biol Chem 285: 14861–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, Piek E, Bottinger EP 2001. Genetic programs of epithelial cell plasticity directed by transforming growth factor-β. Proc Natl Acad Sci 98: 6686–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai YF, Beittenmiller H, Wang B, Gould MN, Oakley C, Esselman WJ, Welsch CW 1993. Increased expression of specific protein tyrosine phosphatases in human breast epithelial cells neoplastically transformed by the neu oncogene. Cancer Res 53: 2272–2278 [PubMed] [Google Scholar]
- Zheng J, Kulp SK, Zhang Y, Sugimoto Y, Dayton MA, Govindan MV, Brueggemeier RW, Lin YC 2000. 17 β-estradiol-regulated expression of protein tyrosine phosphatase γ gene in cultured human normal breast and breast cancer cells. Anticancer Res 20: 11–19 [PubMed] [Google Scholar]
- Zhu JH, Chen R, Yi W, Cantin GT, Fearns C, Yang Y, Yates JR III, Lee JD 2008. Protein tyrosine phosphatase PTPN13 negatively regulates Her2/ErbB2 malignant signaling. Oncogene 27: 2525–2531 [DOI] [PubMed] [Google Scholar]