Abstract
Calcium ions (Ca2+) and Ca2+-related proteins mediate a wide array of downstream processes involved in plant responses to abiotic stresses. In Arabidopsis (Arabidopsis thaliana), disruption of the vacuolar Ca2+/H+ transporters CAX1 and CAX3 causes notable alterations in the shoot ionome, including phosphate (Pi) content. In this study, we showed that the cax1/cax3 double mutant displays an elevated Pi level in shoots as a result of increased Pi uptake in a miR399/PHO2-independent signaling pathway. Microarray analysis of the cax1/cax3 mutant suggests the regulatory function of CAX1 and CAX3 in suppressing the expression of a subset of shoot Pi starvation-responsive genes, including genes encoding the PHT1;4 Pi transporter and two SPX domain-containing proteins, SPX1 and SPX3. Moreover, although the expression of several PHT1 genes and PHT1;1/2/3 proteins is not up-regulated in the root of cax1/cax3, results from reciprocal grafting experiments indicate that the cax1/cax3 scion is responsible for high Pi accumulation in grafted plants and that the pht1;1 rootstock is sufficient to moderately repress such Pi accumulation. Based on these findings, we propose that CAX1 and CAX3 mediate a shoot-derived signal that modulates the activity of the root Pi transporter system, likely in part via posttranslational regulation of PHT1;1 Pi transporters.
Transient increases in cytoplasmic calcium concentrations ([Ca2+]cyt) or the spatial and temporal dynamics of stimulus-induced alterations in [Ca2+]cyt constitute a signal that mediates a wide array of downstream processes involved in plant responses to many developmental cues and environmental stresses (Knight, 2000; McAinsh and Pittman, 2009). The generation of such stimulus-specific Ca2+ signatures is associated with various Ca2+ channels, transporters, and pumps throughout the membrane system. In particular, tonoplast-localized Ca2+/H+ exchangers and Ca2+-ATPase pumps play a key role in the sequestration of Ca2+ into the vacuole, the primary pool for Ca2+ buffering and release, and are assumed to participate in resetting the [Ca2+]cyt following stimuli (Hirschi, 2004; McAinsh and Pittman, 2009).
In yeast, the vacuolar Ca2+/H+ exchanger VCX1, as a high-capacity and low-affinity Ca2+ transporter, functions to rapidly sequester cytosolic Ca2+ and supposedly attenuates the activation of Ca2+ signaling pathways, as the vcx1Δ strain displayed a transient and strongly elevated [Ca2+]cyt followed by a slow and weak recovery from a Ca2+ shock (Miseta et al., 1999). In Arabidopsis (Arabidopsis thaliana), the cation/H+ exchangers CAX1 (the ortholog of VCX1), CAX3, and CAX4 are phylogenetically grouped into type IA, whereas CAX2, CAX5, and CAX6 belong to type IB (Shigaki et al., 2006). However, only CAX1 to CAX4 have been functionally characterized to possess a vacuolar Ca2+/H+ exchange activity. Whereas CAX1 and CAX3 mediated specifically Ca2+ transport (Hirschi, 1999; Catala et al., 2003; Cheng et al., 2003; Mei et al., 2007; Zhao et al., 2008), CAX2 and CAX4 were documented to have high transport and selectivity for cadmium ions (Cd2+) over Ca2+ in tonoplast vesicles (Hirschi et al., 2000; Cheng et al., 2002; Pittman et al., 2004; Koren’kov et al., 2007).
Knockout of CAX1 in Arabidopsis increased the tolerance to high concentrations of various ions, Ca2+-depleted conditions, and freezing after cold acclimation (Catala et al., 2003; Cheng et al., 2003). Conversely, transgenic tobacco plants overexpressing CAX1 were hypersensitive to ion imbalance and cold shock and exhibited Ca2+-deficient symptoms in spite of increased accumulation of Ca2+ (Hirschi, 1999). Loss-of-function of CAX3, a close homolog of CAX1, increased the sensitivity to salt stress, lithium, and low pH (Zhao et al., 2008). While cax1 and cax3 single mutants displayed subtle phenotypes, the cax1/cax3 double mutant showed stunted growth with chlorosis on the leaf tips and a drastic reduction in silique size and seed numbers (Cheng et al., 2005). In addition to a decreased activity of vacuolar Ca2+/H+ antiport, the V-ATPase and P-ATPase activities were reduced in cax1/cax3 (Cheng et al., 2005; Zhao et al., 2008). Elemental analysis has also revealed that impairment of CAX1 and CAX3 caused dramatic alterations in the shoot ionome, namely elevated levels of phosphate (PO43–Pi), manganese (Mn2+), and zinc (Zn2+) and decreased Ca2+ and magnesium (Mg2+) ion concentrations (Cheng et al., 2005). A recent study further showed that the leaf apoplastic free Ca2+ concentration ([Ca2+]) of cax1/cax3 was 3-fold greater than that of the wild type, accounting for the phenotypes of reduced cell wall extensibility, stomatal aperture, transpiration, CO2 assimilation, and leaf growth rate (Conn et al., 2011b). Despite no evidence yet of a direct involvement of Ca2+ in Pi signaling, the observation of an increased accumulation of Pi in the cax1/cax3 mutant has provided the first link between Ca2+ and Pi homeostasis and thus drew our attention and interest to investigate the potential role of CAX1 and CAX3 in Pi signaling.
Phosphorus (P), one of the mineral nutrients essential for plant survival and productivity, is a major structural constituent of fundamental macromolecules such as nucleic acids and phospholipids and is involved in energy transfer, metabolic regulation, and protein activation. However, most of the P in the soil is unavailable for plant uptake because of adsorption, precipitation, or conversion to organic forms (Marschner, 1995). As a result, plants constantly encounter Pi limitation and have developed a number of adaptive strategies to maintain Pi homeostasis, including enhancing acquisition of Pi, coordinating allocation of Pi among different organs, and remobilizing Pi from old to young tissues (Poirier and Bucher, 2002; Ticconi and Abel, 2004). Although many plant responses to Pi starvation have been extensively explored, the molecular mechanisms by which plants sense the Pi signal and elicit these responses remain largely unknown.
Identification and characterization of several mutants with aberrant responses to Pi starvation or with altered levels of Pi has advanced our understanding of the molecular components involved in Pi homeostasis (Lin et al., 2009; Chiou and Lin, 2011). For example, the Arabidopsis pho2 mutant accumulated high levels of Pi in the shoot and showed symptoms of Pi toxicity as a result of increased Pi uptake and translocation of Pi from roots to shoots (Delhaize and Randall, 1995; Dong et al., 1998). The PHO2 gene was identified to encode an E2 ubiquitin-conjugating enzyme (UBC24), whose expression during Pi deficiency is posttranscriptionally suppressed by a specific microRNA, miR399 (Fujii et al., 2005; Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). In accordance with the inverse correlation between miR399 and PHO2 mRNA levels, transgenic plants overexpressing miR399 phenocopied the pho2 mutant and PHO2 T-DNA knockout lines (Aung et al., 2006; Chiou et al., 2006). Reciprocal micrografting experiments further demonstrated that a pho2 root genotype is sufficient and necessary for Pi accumulation in the shoot, whereas the shoot-to-root movement of mature miR399 is responsible for the degradation of PHO2 mRNA in roots (Bari et al., 2006; Lin et al., 2008; Pant et al., 2008). Moreover, up-regulation of miR399 by Pi deprivation is mediated by the PHOSPHATE STARVATION RESPONSE1 (PHR1) transcription factor, a key positive regulator of multiple Pi starvation-induced (PSi) genes (Rubio et al., 2001; Bari et al., 2006). These findings suggest that the Pi starvation signaling involving PHR1, miR399, and PHO2 is crucial for the maintenance of Pi homeostasis.
Recently, a unique gene family in Arabidopsis (AtSPX1–AtSPX4) exclusively harboring the SPX (for SYG1/Pho81/XPR1) domain was identified to be regulated by Pi starvation, in part through PHR1 (Hamburger et al., 2002; Wang et al., 2004; Duan et al., 2008). Overexpression of SPX1 (At5g20150) increased the transcript levels of several PSi genes under both Pi-sufficient and Pi-deficient conditions, whereas RNA interference-mediated partial down-regulation of SPX3 (At2g45130) led to aggravated Pi deficiency symptoms, altered Pi allocation, and enhanced expression of a subset of PSi genes (Duan et al., 2008). In rice (Oryza sativa), OsSPX1 (Os06g40120), the ortholog of SPX3, acts via a negative feedback loop to adjust the expression of several PSi genes under Pi-limited conditions (Wang et al., 2009; Liu et al., 2010). These findings revealed that the plant SPX domain-containing proteins are new players in the regulatory network of Pi signaling.
Although Ca2+ and Ca2+-related proteins are indispensable messengers in the signal transduction of many stress responses, the role of Ca2+ in Pi signaling and the cross talk between Ca2+ and Pi homeostasis are barely understood. The association between loss of CAX1 and CAX3 activities and increased Pi accumulation as seen in cax1/cax3 (Cheng et al., 2005) provides an opportunity to tackle this issue. It is also of interest to compare cax1/cax3 and pho2 mutants in terms of regulatory pathways, because both mutants show elevated shoot Pi concentrations. Here, our study of the cax1/cax3 mutant suggests that vacuolar Ca2+/H+ transporters exert a negative regulation of Pi starvation responses, as revealed by suppression of the expression of a subset of shoot Pi starvation-responsive (PSR) genes and inhibition of Pi uptake activity in the root. Our results also suggest that the effects of CAX1/CAX3- and PHO2-mediated signaling pathways on the suppression of Pi uptake are different. Moreover, results from reciprocal grafting experiments demonstrate that CAX1 and CAX3 mediate a shoot-derived signal that modulates the activity of the root Pi transporter system, likely in part via posttranslational regulation of PHT1;1.
RESULTS
cax1/cax3 Mutant Accumulates High Levels of Pi in Shoot and Displays Increased Pi Transport Activity
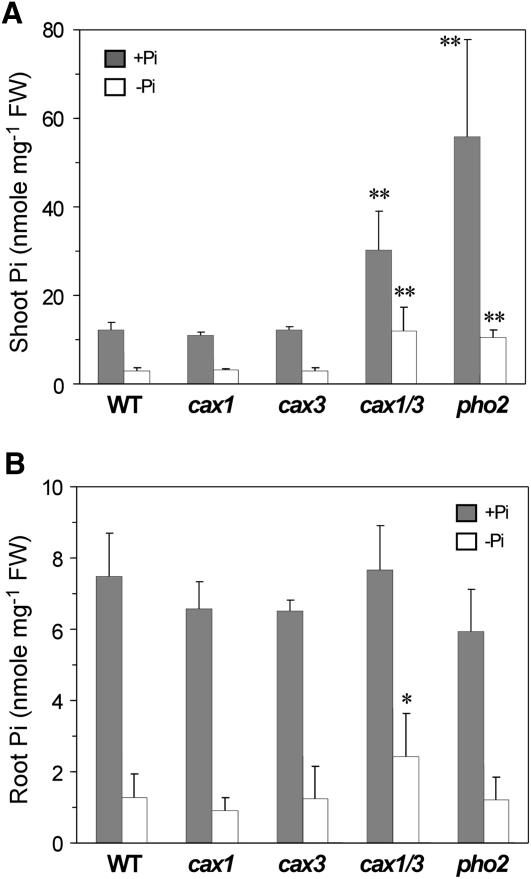
The shoot Pi concentration has been shown to increase by 66% in the cax1/cax3 mutant but to remain unchanged in the single cax1 and cax3 mutants (Cheng et al., 2005). To confirm this finding, we grew cax1, cax3, and cax1/cax3 along with the pho2 mutant and wild-type controls in Pi-sufficient (+Pi) or Pi-deficient (−Pi) half-strength modified Hoagland hydroponic solution for Pi concentration measurement. Under both +Pi and −Pi conditions, the shoot and root Pi concentrations of cax1 and cax3 were similar to those of the wild type; by contrast, the cax1/cax3 double mutant accumulated high levels of Pi in the shoot under both conditions and showed modestly increased Pi concentrations in the root under Pi deficiency (Fig. 1). Of note, cax1/cax3 did not accumulate Pi in the shoot to a level as high as pho2 under +Pi conditions but maintained a comparable level of shoot Pi as pho2 under −Pi conditions (Fig. 1A).
Figure 1.
Analysis of Pi concentrations in the cax1/cax3 mutant. The Pi concentrations are shown in the shoot (A) and root (B) of 23-d-old wild-type (WT), cax1, cax3, cax1/cax3, and pho2 plants grown under +Pi or −Pi conditions. Values represent means + sd of three to five biological replicates. Data significantly different from the corresponding wild-type controls are indicated (* P < 0.05, ** P < 0.01; Student’s t test). FW, Fresh weight.
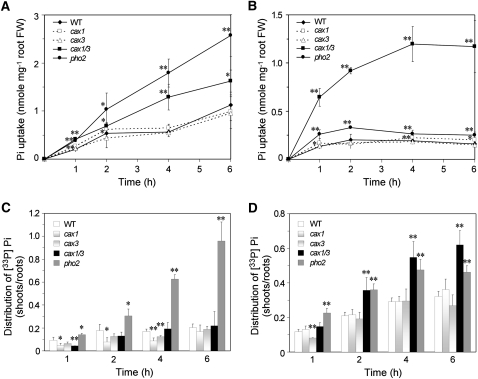
We then performed Pi transport assays to determine whether Pi accumulation in cax1/cax3 can be attributed to an enhanced Pi uptake rate. As expected, cax1/cax3 exhibited a higher Pi uptake activity than wild-type plants, regardless of external Pi concentrations (Fig. 2, A and B). By contrast, pho2 exhibited a higher Pi uptake activity than cax1/cax3 when the Pi supply was adequate (Fig. 2A) but exhibited only a slightly enhanced Pi uptake activity as compared with the wild type when Pi was limited (Fig. 2B). Under +Pi conditions, pho2 but not cax1/cax3 showed an increased shoot-to-root ratio of Pi distribution (Fig. 2C), indicating that the Pi translocation activity of cax1/cax3 from roots to shoots was not changed even though the Pi uptake activity was increased. However, under –Pi conditions, the shoot-to-root ratio of Pi distribution in cax1/cax3 was increased (Fig. 2D). Taken together, cax1/cax3 displayed increased shoot Pi accumulation, increased Pi uptake activity regardless of external Pi concentrations, and greater Pi translocation from roots to shoots when Pi was limited.
Figure 2.
Analysis of Pi uptake rate and Pi translocation in the cax1/cax3 mutant. A and B, The uptake rate of [33P]Pi in wild-type (WT), cax1, cax3, cax1/cax3, and pho2 plants grown under ±Pi (250 μm KH2PO4; A) or −Pi (10 μm KH2PO4; B) conditions. C and D, The shoot-to-root ratio of [33P]Pi accumulation during the transport period under +Pi (C) or −Pi (D) conditions. Values represent means ± sd of three biological replicates. Data significantly different from the corresponding wild-type controls are indicated (* P < 0.05, ** P < 0.01; Student’s t test). FW, Fresh weight.
Pi Accumulation in cax1/cax3 Mutant Increases in an Exogenous [Ca2+]-Dependent Manner
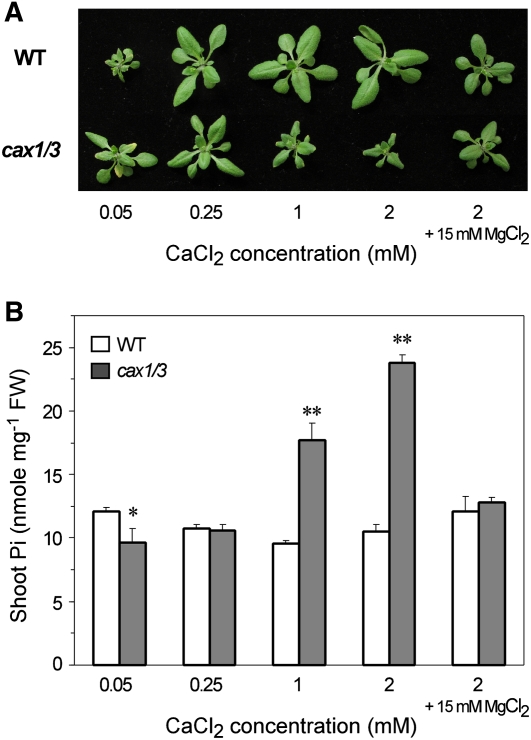
It has been reported that cax1/cax3 showed a higher sensitivity to Ca2+ stress when grown in medium supplemented with high [Ca2+] (Cheng et al., 2005). To address whether the impaired cellular Ca2+ homeostasis due to the loss of CAX1 and CAX3 plays a direct role in enhancing Pi uptake activity, we examined the effect of exogenous Ca2+ on Pi accumulation of cax1/cax3 by growing mutants in +Pi hydroponic medium containing different concentrations of Ca2+ (0.05 mm, 0.25 mm, 1 mm, and 2 mm [Ca2+]), contrasting with 2.5 mm [Ca2+] in half-strength modified Hoagland solution. Interestingly, Pi accumulation in the shoot of cax1/cax3 was increased in a [Ca2+]-dependent manner (Fig. 3B) and displayed a negative correlation with the leaf size of mutants (Fig. 3A). Consistent with previous results showing that the growth defects of cax1/cax3 mutant were suppressed by supplemented exogenous Mg2+ (Cheng et al., 2005), we also found that the exacerbated Pi accumulation in cax1/cax3 mutants resulting from exogenous Ca2+ stress could be attenuated by supplementing high concentrations of Mg2+ (Fig. 3). These results suggest an antagonistic relationship between Ca2+ and Mg2+ regarding their interplay in Pi accumulation of cax1/cax3.
Figure 3.
Analysis of cax1/cax3 mutants grown under various exogenous Ca2+ concentrations. The phenotypes (A) and Pi concentrations in the shoot (B) are shown for 21-d-old wild-type (WT) and cax1/cax3 plants grown under +Pi (250 μm KH2PO4) conditions supplemented with 0.05, 0.25, 1, or 2 mm CaCl2. An additional 15 mm MgCl2 was added to the medium containing 2 mm CaCl2. Values represent means + sd of three biological replicates. Data significantly different from the corresponding wild-type controls are indicated (* P < 0.05, ** P < 0.01; Student’s t test). FW, Fresh weight.
Cross-Regulation of PHO2 and CAX1/CAX3 Expression in cax1/cax3 and pho2 Mutants
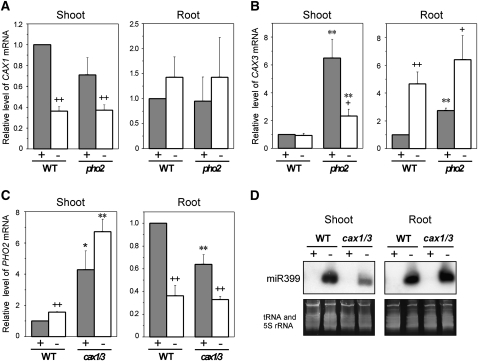
To investigate the role of CAX1 and CAX3 in Pi signaling, we first used quantitative reverse transcription (qRT)-PCR to examine changes in the expression levels of CAX1 and CAX3 in wild-type plants subjected to Pi deficiency. In the shoot, the transcript level of CAX1 was reduced by 60% after 5 d of Pi deprivation (Fig. 4A). However, no difference was observed in the expression level of CAX3 between Pi-sufficient and Pi-deficient shoots (Fig. 4B). In the root, no significant change was found in the transcript level of CAX1 in response to Pi starvation (Fig. 4A), while the CAX3 transcript level was increased (Fig. 4B). Regulation of the expression of CAX1 and CAX3 by Pi deficiency implied the involvement of these genes in Pi starvation responses.
Figure 4.
Pi starvation regulation of CAX1, CAX3, PHO2, and miR399 expression in the cax1/cax3 and pho2 mutants. A and B, qRT-PCR analysis of CAX1 (A) and CAX3 (B) expression in the shoot and root of wild-type (WT) and pho2 plants. C and D, qRT-PCR analysis of PHO2 expression (C) and small RNA gel-blot analysis of mature miR399 (D) in the shoot and root of wild-type and cax1/cax3 plants. Plants were hydroponically grown under +Pi (+) or −Pi (−) conditions. The value for each gene is presented as the fold change relative to the expression in wild-type plants under +Pi conditions. tRNA and 5S rRNA were used as loading controls for small RNA. Error bars represent se of biological replicates from three independent experiments. Data significantly different from the corresponding controls are indicated (mutant versus the wild type, * P < 0.05, ** P < 0.01; Pi sufficient versus Pi deficient, + P < 0.05, ++ P < 0.01; Student’s t test).
Although cax1/cax3 and pho2 mutants showed distinct properties in terms of Pi uptake and translocation (Fig. 2), we wondered whether there is a cross talk between PHO2 and CAX1/CAX3-mediated Pi signaling pathways. Therefore, we examined the expression of CAX1 and CAX3 in pho2 and of PHO2 in cax1/cax3. Under both +Pi and −Pi conditions, the transcript levels of CAX1 in the shoot and root of pho2 were similar to that of wild-type plants (Fig. 4A). Under +Pi conditions, the transcript levels of CAX3 in the shoot and root were higher in pho2 than in wild-type plants; however, under −Pi conditions, the transcript level of CAX3 was higher in the shoot but not in the root of pho2 as compared with wild-type plants (Fig. 4B).
The complementary measurements revealed that the transcript level of PHO2 in the shoot of cax1/cax3 was higher than that of wild-type plants under both +Pi and −Pi conditions (Fig. 4C). As the pho2 rootstock genotype has been shown sufficient for Pi accumulation in the scion (Bari et al., 2006; Lin et al., 2008), the role of PHO2 in the shoot and the implication of increased transcript levels of PHO2 in the shoot of cax1/cax3 remain to be resolved. On the other hand, the level of PHO2 mRNA in the root of cax1/cax3 was reduced by 36% under +Pi conditions as compared with wild-type plants but was not as low as that detected in the root of wild-type plants under Pi deficiency (Fig. 4C). The mature miR399 was not detectable in the shoot or root of cax1/cax3 under +Pi conditions (Fig. 4D), indicating that the moderate reduction of PHO2 mRNA level in the root of cax1/cax3 under +Pi conditions did not result from suppression by miR399. Furthermore, the PHO2 transcripts were reduced to a similar level in the root of cax1/cax3 and wild-type plants under −Pi conditions (Fig. 4C), supporting the idea that the increased Pi uptake activity in the root of cax1/cax3 under Pi deficiency (Fig. 2B) is unlikely mediated by a PHO2-dependent signaling pathway. Of interest, we also found that the induction of mature miR399s under Pi deficiency in the shoot of cax1/cax3 was lower than that of wild-type controls; therefore, it is likely that miR399 induction is inhibited by the elevated concentrations of internal shoot Pi, as shown in Figure 1A. From these results, we conclude that the expression of CAX1/CAX3 and PHO2 appears to be cross-regulated, even though they function to suppress Pi uptake and regulate root-to-shoot Pi translocation in different modes.
Suppression of a Subset of Shoot PSR Genes by CAX1 and CAX3
To gain more insight into the molecular basis for Pi accumulation in cax1/cax3, we analyzed the expression profiles of cax1/cax3 and ecotype Columbia (Col-0) wild-type plants grown in hydroponic solutions under +Pi and −Pi conditions using the Affymetrix ATH1 chip. The numbers of differentially expressed genes (P ≤ 0.01, more than 2-fold change) were identified as follows: 854 and 1,047 under +Pi and −Pi conditions, respectively, between cax1/cax3 and wild-type shoots; and 84 and 655 under +Pi and −Pi conditions, respectively, between cax1/cax3 and wild-type roots (Supplemental Table S1, subgroups 1–4). We also determined the number of PSR genes with more than 2-fold change in expression (P ≤ 0.01) between the +Pi and −Pi conditions (Supplemental Table S1, subgroups 5–8) and compared the expression of the PSR genes between cax1/cax3 and wild-type plants. To evaluate the effects of our Pi starvation regimes on gene expression in wild-type plants, we plotted the fold change ratio of the differentially regulated shoot and root PSR genes from our microarray data against that reported by Morcuende et al. (2007), who subjected liquid culture-grown whole seedlings to Pi deprivation under continuous light. Despite different plant growth stages and growth conditions applied in these two studies, there is a clear positive correlation in the comparison as revealed by the r2 value (Supplemental Fig. S1).
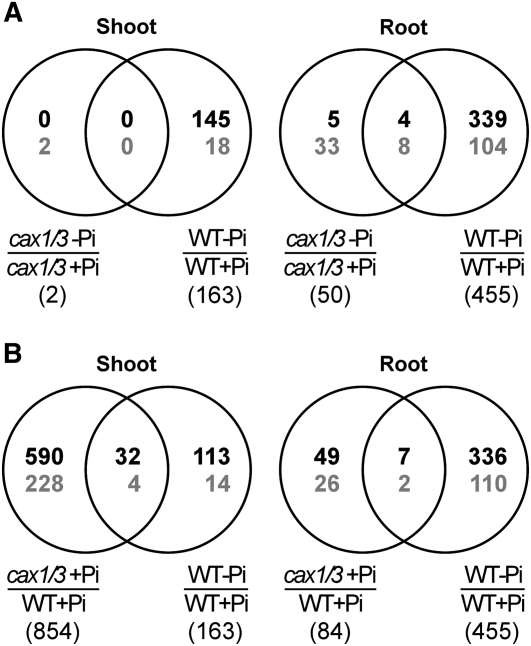
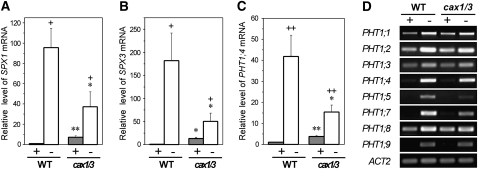
None of the wild-type shoot PSR genes (0 of 163) and only 2.6% (12 of 455) of the wild-type root PSR genes were regulated in the same fashion in cax1/cax3 (Fig. 5A). Strikingly, in the shoot of cax1/cax3 under +Pi conditions, 22.1% (32 of 145) of the wild-type PSi genes were up-regulated and 22.2% (4 of 18) of the wild-type Pi starvation-repressed genes were down-regulated (Fig. 5B), indicating that one-fifth of the PSR genes are constitutively activated in the shoot of cax1/cax3. Among these 36 differentially expressed PSR genes (Table I), we further validated the up-regulation of several PSi genes, including genes implicated in Pi signaling, SPX1 (At5g20150) and SPX3 (At2g45130), and PHT1;4 (At2g38940), a member of the Pi high-affinity transporter (PHT1) gene family, by qRT-PCR (Fig. 6, A–C; Supplemental Fig. S3). Although our microarray data did not reveal gene expression changes of other PHT1 members, we were able to observe by qRT-PCR 4- to 5-fold increased transcript levels for PHT1;1 (At5g43350) and PHT1;3 (At5g43360) in the shoot of cax1/cax3 under +Pi conditions (data not shown). This discrepancy can be explained by the higher sensitivity of qRT-PCR, as the levels of PHT1;1 and PHT1;3 transcripts in the shoot were low. Up-regulation of these PHT1 genes may reflect an increased sink demand for Pi, likely a downstream event of the CAX1/CAX3 Pi signaling cascade. When the expression of the PSR genes was examined in cax1/cax3, most of them, such as SPX1, SPX3, and PHT1;4, were less responsive to Pi starvation compared with wild-type plants (Fig. 6, A–C; Supplemental Table S1, subgroups 5 and 6). This may have resulted from their suppression by the high shoot Pi levels of cax1/cax3 (Fig. 1A). Given that a significant proportion of PSR genes were constitutively activated in the shoot of cax1/cax3, we conclude that the function of CAX1 and CAX3 is required for the suppression of a discrete subset of shoot PSR genes under +Pi conditions.
Figure 5.
The number of genes differentially expressed in cax1/cax3 and wild-type (WT) plants under Pi-sufficient and Pi-deficient conditions. A, Overlap of the PSR genes between cax1/cax3 and wild-type plants. B, Overlap of wild-type PSR genes and genes differentially expressed between cax1/cax3 and wild-type plants under +Pi conditions. Numbers designate the genes with significantly differential expression (P ≤ 0.01 and more than 2-fold change) between the indicated data sets derived from microarray analysis. The total number of genes in each data set is shown in parentheses. The numbers of induced and repressed genes are indicated in black and gray, respectively.
Table I. Misregulated PSR genes in cax1/cax3.
| AGIa | Fold Change in Expression |
Gene Description | |
| –Pi:+Pi in the Wild Type | cax1/cax3:Wild Type under +Pi | ||
| Shoots | |||
| At5g20790 | 88.4 | 5.6 | Unknown protein |
| At1g73010 | 62.9 | 17.1 | Pyrophosphate-specific phosphatase 1 (PPsPase1) |
| At2g38940/At3g54700 | 29.1 | 11.4 | Phosphate transporter (PHT1;4/PHT1;7) |
| At5g20150 | 27.6 | 10.9 | SPX domain-containing protein (SPX1) |
| At3g47420 | 20.7 | 9.9 | Glycerol-3-phosphate transporter |
| At3g17790 | 18.5 | 12.1 | Acid phosphatase type 5 (ACP5) |
| At1g23140 | 14.8 | 4.0 | C2 domain-containing protein |
| At3g05630 | 13.6 | 4.9 | Phospholipase Dζ2 (PLDζ2) |
| At1g17710 | 13.1 | 3.2 | Putative phosphatase |
| At2g45130 | 10.1 | 6.9 | SPX domain-containing protein (SPX3) |
| At4g31240 | 8.6 | 7.8 | Protein kinase C-like zinc finger protein |
| At5g20400 | 4.6 | 2.3 | Oxoglutarate/iron-dependent oxygenase |
| At3g11670 | 3.9 | 2.1 | Digalactosyldiacylglycerol synthase (DGD1) |
| At1g61800 | 3.8 | 22.8 | Glc-6-P/phosphate-translocator (GPT2) |
| At4g33550 | 3.4 | 2.7 | Lipid transfer protein |
| At4g19880 | 3.1 | 2.1 | Glutathione S-transferase-related protein |
| At4g35750 | 3.0 | 2.1 | ρ-GTPase-activating protein-related protein |
| At5g44240 | 2.9 | 2.7 | ATPase, Ca2+-transporting (ALA2) |
| At2g46680 | 2.7 | 11.0 | Homeobox-Leu zipper protein 7 (HB-7)/HD-ZIP transcription factor 7 |
| At3g20250 | 2.7 | 3.3 | RNA-binding protein |
| At3g56400 | 2.7 | 2.6 | WRKY family transcription factor (WRKY70) |
| At1g72890 | 2.7 | 5.0 | Similar to disease resistance protein |
| At1g80130 | 2.6 | 2.1 | Unknown protein |
| At2g36790/At2g36800 | 2.5 | 2.3 | Putative glucosyl transferase |
| At1g22070 | 2.4 | 2.0 | bZIP transcription factor (TGA3/AtbZIP22) |
| At5g51070 | 2.3 | 2.0 | ATP-dependent Clp protease ATP-binding subunit (ClpD), (ERD1) |
| Atcg00780 | 2.3 | 3.4 | Ribosomal protein L14 (rpl14) |
| At1g65660 | 2.3 | 2.3 | Zinc knuckle (CCHC-type) family protein; putative step II splicing factor |
| At1g72830 | 2.2 | 3.0 | CCAAT-binding transcription factor B subunit (CBF-B/NF-YA) |
| At1g22930 | 2.2 | 3.0 | Unknown protein |
| At2g30500 | 2.1 | 4.9 | Kinase-interacting family protein |
| At2g17290 | 2.0 | 3.9 | Ca2+-dependent protein kinase isoform 6 (CPK6) |
| At2g19970 | 0.43 | 0.48 | Putative pathogenesis-related protein |
| At5g49560 | 0.42 | 0.44 | Putative methyltransferase family protein |
| At4g16563 | 0.4 | 0.05 | Putative aspartyl protease family protein |
| At1g10550 | 0.3 | 0.11 | Xylogucan:xyloglusyl transferase 33 (XTH33) |
| Roots | |||
| At2g30540 | 11.9 | 2.1 | Putative glutaredoxin |
| At2g21900 | 4.0 | 2.2 | WRKY family transcription factor (WRKY59) |
| At4g01390 | 4.0 | 3.7 | TRAF-like family protein |
| At2g18050 | 3.4 | 3.7 | Histone H1-3 (HIS1-3) |
| At5g13580 | 3.2 | 2.1 | ABC transporter-like protein |
| At1g28480 | 2.6 | 2.4 | GRX480, a member of the glutaredoxin family |
| At4g14060 | 2.2 | 2.6 | Major latex protein (MLP)-related protein |
| At5g59305 | 0.3 | 0.3 | Unknown protein |
| At4g25100 | 0.3 | 0.3 | Fe superoxide dismutase (FSD1) |
Arabidopsis Genome Initiative number. Genes validated by qRT-PCR are underlined.
Figure 6.
Gene expression of SPX1, SPX3, and PHT1;4 Pi transporter in the cax1/cax3 mutant. A to C, qRT-PCR analysis of SPX1 (A), SPX3 (B), and PHT1;4 (C) in the shoot of wild-type (WT) and cax1/cax3 plants under +Pi (+) or −Pi (−) conditions. The value for each gene is presented as the fold change relative to the expression of wild-type plants under +Pi conditions. Error bars represent se of biological replicates from three independent experiments. Data significantly different from the corresponding controls are as indicated (mutant versus the wild type, * P < 0.05, ** P < 0.01; Pi sufficient versus Pi deficient, + P < 0.05, ++ P < 0.01; Student’s t test). D, RT-PCR analysis of members in the PHT1 gene family in the root of wild-type and cax1/cax3 plants under +Pi (+) or −Pi (−) conditions.
In contrast to the gene expression profile in the shoot of cax1/cax3 under +Pi conditions, only 2% (seven of 343) of the wild-type PSi genes and 1.8% (two of 112) of the wild-type Pi starvation-repressed genes were up-regulated and down-regulated, respectively, in the root of cax1/cax3 under +Pi conditions (Fig. 5B; Table I). These results seem contradictory to our speculation that the increased Pi uptake rate in cax1/cax3 may be caused by up-regulation of PHT1 genes in the root, as many members in the PHT1 Pi transporter family are expressed preferentially in root epidermal or cortical cells and function in Pi acquisition (Muchhal and Raghothama, 1996; Mudge et al., 2002). Both results of RT-PCR (Fig. 6D) and qRT-PCR (data not shown) analyses showed that the transcript levels of PHT1 genes were not increased under +Pi or under −Pi conditions in the root of cax1/cax3 as compared with the wild-type controls, indicating that a posttranscriptional regulation of the PHT1 or other unidentified Pi transporters accounts for enhanced Pi transport activity in cax1/cax3.
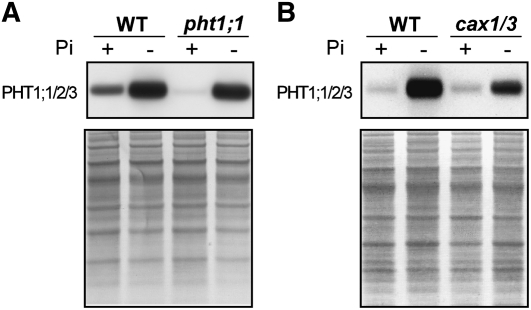
Since the Pi-replete pht1;1 mutant showed reduced Pi uptake activity, PHT1;1 was suggested to play a primary role in Pi acquisition under +Pi conditions (Shin et al., 2004). Thus, we next examined whether PHT1;1 is up-regulated at the protein level in cax1/cax3. Because of the high homology in protein sequence (94%–98% identity) among PHT1;1, PHT1;2, and PHT1;3, we raised an antibody against all three proteins. While the levels of PHT1;1/2/3 were increased in wild-type seedlings in response to low Pi availability, they were greatly decreased in the +Pi but not −Pi root of the pht1;1 knockout mutant (Fig. 7A). This validated the specificity of this antibody against PHT1;1/2/3 and supported the conclusion that PHT1;1 is the major Pi transporter accountable for Pi acquisition under +Pi conditions (Shin et al., 2004). Surprisingly, we did not observe much difference in the protein level of PHT1;1/2/3 in the +Pi root between cax1/cax3 and the wild type (Fig. 7B). Taken together, the enhanced Pi uptake activity in cax1/cax3 does not result from an up-regulation of PHT1;1/2/3 at the protein level.
Figure 7.
Analysis of PHT1;1/2/3 protein in the root of pht1;1 seedlings (A) and cax1/cax3 hydroponically grown plants (B) under +Pi (+) and −Pi (−) conditions. The bottom panels show the protein staining on the membrane. WT, Wild type.
Shoot-Derived Signals Are Responsible for the High Accumulation of Pi in the cax1/cax3 Mutant
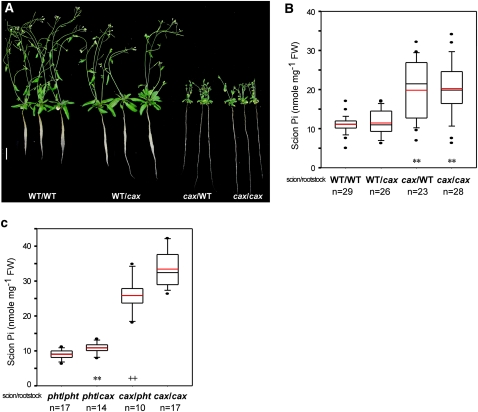
Since our microarray data revealed the activation of about 22% of PSR genes in the shoot of cax1/cax3 under +Pi conditions, we asked next whether a shoot-derived Pi starvation signal mediates the enhanced Pi uptake activity resulting in Pi accumulation of cax1/cax3. To address this issue, we performed reciprocal micrografting between cax1/cax3 and wild-type plants. When grown under +Pi conditions, the grafted plants with cax1/cax3 scions and wild-type rootstocks resembled the phenotype of Pi toxicity and stunted growth seen in the cax1/cax3 mutant, whereas the grafted plants with wild-type scions and cax1/cax3 rootstocks showed a wild-type phenotype (Fig. 8A). In accordance with the phenotype, while cax1/cax3 scions grafted on wild-type rootstocks exhibited high shoot Pi levels (Fig. 8B) and Pi uptake activity (data not shown) as cax1/cax3 self-grafts, wild-type scions grafted on cax1/cax3 rootstocks behaved in a similar way as wild-type self-grafts (Fig. 8B). Although the protein amount of PHT1;1/2/3 was not increased in the +Pi root of cax1/cax3, we checked whether a posttranslational regulation of PHT1;1 is involved in activating Pi uptake of cax1/cax3 by grafting cax1/cax3 scions on pht1;1 rootstocks. The pht1;1 self-grafts showed slightly but significantly reduced shoot Pi levels compared with the grafted plants with pht1;1 scions and cax1/cax3 rootstocks (Fig. 8C). Likewise, the shoot Pi level of the grafted plants with cax1/cax3 scions and pht1;1 rootstocks was also modestly reduced compared with cax1/cax3 self-grafts (Fig. 8C), suggesting that lack of PHT1;1 in the rootstock is sufficient to partially repress Pi accumulation in the cax1/cax3 scion.
Figure 8.
Analysis of reciprocal grafting of wild-type (WT), pht1;1, and cax1/cax3 plants. Shown are the phenotypes (A; bar = 1 cm) and the scion Pi concentrations of 6-week-old grafted plants between wild-type and cax1/cax3 plants (B) and between pht1;1 and cax1/cax3 plants (C). The Pi concentrations are presented as box plots. The boundaries of the boxes indicate the 25th and 75th percentiles. The median and the mean are marked by black and red lines, respectively, within the box. Error bars above and below the box indicate the 90th and 10th percentiles. Each individual plant outside the 10th and 90th percentiles is displayed as a single dot. In B, mean values significantly different from the wild-type self-grafts are indicated by asterisks (** P < 0.01; Student’s t test). In C, mean values showing significant differences between the pht1;1 self-grafts and the grafts with the pht1;1 scion and the cax1/cax3 rootstock (** P < 0.01; Student’s t test) and between the cax1/cax3 self-grafts and the grafts with cax1/cax3 scion and pht1;1 rootstock (++ P < 0.01; Student’s t test) are indicated by asterisks and crosses, respectively. n, The total number of grafted plants.
To further elucidate the systemic role of CAX1 and CAX3 in Pi signaling, split-root experiments were carried out, in which one half of the root system of a single plant was grown in +Pi medium and the other half in –Pi medium (designated as +/–Pi plants). Plants grown in +Pi and –Pi media were included as controls. As expected, in the +Pi and –Pi controls, the expression of SPX1, PHT1;4, and At4 was increased in +Pi cax1/cax3 shoots as compared with the wild type, but their induction by Pi deficiency in cax1/cax3 was not as high as in the wild type in both roots and shoots (Supplemental Fig. S2). The expression level of these PSi genes in the shoot of +/–Pi cax1/cax3 and wild-type plants showed an intermediate level between their own +Pi and –Pi controls (Supplemental Fig. S2, A, C, and E), reflecting the intermediate shoot Pi levels of the +/–Pi plants (data not shown). In agreement with earlier observations of systemic down-regulation of several PSi genes in the −Pi root half of the split-root system (Burleigh and Harrison, 1999; Franco-Zorrilla et al., 2005; Thibaud et al., 2010), we found that the induction of SPX1, PHT1;4, and At4 in the −Pi root half was suppressed in both +/–Pi wild-type and +/–Pi cax1/cax3 plants in a similar fashion (Supplemental Fig. S2, B, D, and F). This suggests that the systemic Pi signal transmitting from the +Pi root half via the shoot to the −Pi root half is not impaired in cax1/cax3.
Taken together, our results here suggest that the cax1/cax3 scion genotype is responsible for high levels of Pi accumulation as a result of increased Pi uptake in the wild-type rootstocks and that a signal originating from cax1/cax3 shoots may move across grafting junctions to activate the Pi transport activity in roots, likely in part through posttranslational up-regulation of PHT1;1. By contrast, CAX1 and CAX3 appear not to be involved in the generation of the putative systemic suppressor in +Pi roots and its long-distance movement.
DISCUSSION
CAX1- and CAX3-Mediated Regulation of Leaf Calcium Ion Homeostasis Is Required for Systemic Pi Homeostasis
To maintain the [Ca2+]cyt levels in the micromolar range (Marty, 1999), plant cells transport Ca2+ out of the cytoplasm across the plasma membrane or into various organelles such as endoplasmic reticulum, chloroplast, and vacuole (Sze et al., 2000). CAX1 and CAX3 are tonoplast-localized Ca2+/H+ antiporters that mediate the sequestration of Ca2+ into the vacuole (Cheng et al., 2003, 2005). Knockout or overexpression of CAX1 or CAX3 alone in planta has been shown to result in perturbations in ion homeostasis and altered responses to salinity and cold stresses, while loss of both CAX1 and CAX3 led to a severe reduction in growth, leaf tip and flower necrosis, and a pronounced sensitivity to exogenous Ca2+ and other ions (Hirschi, 1999; Catala et al., 2003; Cheng et al., 2003, 2005; Mei et al., 2007; Zhao et al., 2009). Intriguingly, alterations in transport properties resulting from overexpression of both CAXs in yeast could not be recapitulated by high-level expression of either transporter individually (Zhao et al., 2009). It was postulated that the differential stress sensitivities of cax mutants are due to specific responses by CAX1 or CAX3 to individual stresses or to distinct transport properties conferred by hetero-CAX complexes formed by CAX1 and CAX3 (Cheng et al., 2005; Zhao et al., 2009). If we suppose that loss of the putative CAX1-CAX3 heteromer is responsible for the enhanced Pi uptake in cax1/cax3, then lack of either CAX1 or CAX3 should have also rendered an increased Pi uptake activity. However, our findings here suggest a functional redundancy of CAX1 and CAX3 regarding their role in regulating Pi uptake, because neither the cax1 nor the cax3 single mutant exhibits an increased uptake rate of Pi. Similarly, no significant change in total leaf [Ca2+] in either the cax1 or the cax3 single mutant was observed (Cheng et al., 2005). Thus, one possible explanation for the impaired Pi homeostasis in cax1/cax3 is that disruption of both vacuolar Ca2+ transporters leads to elevated levels of [Ca2+]cyt and/or aberrant subcellular compartmentation of [Ca2+] due to the inability to move excess cytosolic Ca2+ into the vacuole. Although approximately 50% of wild-type vacuolar Ca2+/H+ transport activity was reported for cax1/cax3 following pretreatment with exogenous Ca2+ (Cheng et al., 2005), whether the [Ca2+]cyt level is indeed increased in cax1/cax3 has not yet been demonstrated.
Recently, Conn et al. (2011b) convincingly demonstrated that the severe phenotypes of cax1/cax3, such as reduced leaf growth rate, mainly result from the increased leaf apoplastic free [Ca2+] rather than from the reduced vacuolar [Ca2+] of mesophyll cells. Their findings prompted us to suspect that the abrogated cellular Ca2+ homeostasis in the leaf of cax1/cax3 brings about misregulation of Pi homeostasis. As qRT-PCR analysis revealed that the transcript level of CAX1 in shoot was decreased upon Pi limitation, it is likely that the leaf vacuolar sequestration of Ca2+ by CAX1 is down-regulated under Pi deficiency (Fig. 4A). We thus surmised that simultaneous loss of CAX1 and CAX3, a gene ectopically expressed in leaves upon knockout of CAX1 (Cheng et al., 2003, 2005), may mimic the demands on reduction of the shoot vacuolar Ca2+/H+ transport activity under Pi deficiency, accompanied by activation of a subset of shoot PSR genes (Fig. 5B). By contrast, since CAX4 is primarily expressed in root tissues and up-regulated in the cax1 mutant (Cheng et al., 2003), the increased expression of CAX4 may compensate for the functional role of CAX1 and CAX3 in the root of cax1/cax3, hinting at why activation of the PSR genes was not observed in the root of cax1/cax3 (Fig. 5B). In fact, overexpression of CAX4 was able to partially suppress the cax1 defect in vacuolar Ca2+/H+ transport (Zhao et al., 2009).
Notably, cax1/cax3 displays 47% and 20% reductions, respectively, in V-ATPase and P-ATPase activities (Cheng et al., 2005; Zhao et al., 2008). As these H+-ATPases generate a pH gradient across membranes that provides the driving force for the H+-coupled transporters and contributes to the maintenance of the cytosolic pH homeostasis, whether the complex interaction of H+-ATPases with other transporters and/or the resulting impaired pH homeostasis is associated with the activation of Pi transporters remains obscure. It is important to note that the vha-a2/vha-a3 double mutant, which lacks the tonoplast V-ATPase, was shown to contain reduced Ca2+ levels in leaves and to display symptoms of Ca2+ deficiency similar to cax1/cax3 (Krebs et al., 2010). Moreover, it has been demonstrated that cytosolic Ca2+ homeostasis is a constitutive function of the yeast V-ATPase. Cellular responses to a brief Ca2+ challenge were affected not only by an acute loss of V-ATPase activity (in temperature-sensitive vma mutants or in wild-type cells treated with a V-ATPase inhibitor) but also by a permanent loss of V-ATPase activity in a vma deletion mutant (Förster and Kane, 2000). In the future, it would be interesting to determine whether V-ATPase contributes to the impaired Pi homeostasis of the cax1/cax3 mutant through the misregulation of Ca2+ homeostasis.
Besides high Pi accumulation, disturbance of other ion homeostases, such as increased levels of Mn2+ and Zn2+ and decreased levels of Mg2+, has also been reported for cax1/cax3 (Cheng et al., 2005). Although an interplay between these various ions within cax1/cax3 cannot be excluded from participating in Pi signaling, our results that Pi accumulation of cax1/cax3 was exacerbated upon exogenous supplement of Ca2+ support the hypothesis that leaf Ca2+ homeostasis is directly involved in Pi signaling. It is unclear why the growth retardation and Pi accumulation of cax1/cax3 can be alleviated when high concentrations of Mg2 are added to growth medium. However, it was argued that with a supplement of Mg2+ to growth medium, more Mg2+ is sequestered to the vacuole from the cytoplasm to compensate for the reduced vacuolar [Ca2+] in cax1/cax3 (Conn et al., 2011a).
Intriguingly, it is known that Ca2+ tends to precipitate with Pi, rendering the soil P unavailable for plant acquisition (Hinsinger, 2001). This aspect is noteworthy, considering that the excess cytosolic Ca2+ in the shoot of cax1/cax3 may potentially decrease the available shoot Pi, which may alternatively but not perfectly explain why only a subset of the shoot PSR genes are activated in cax1/cax3 under +Pi conditions. However, x-ray microanalysis showed that Arabidopsis Col-0 plants preferentially accumulate Ca in the vacuoles of mesophyll cells but P within vacuoles of the epidermis and bundle sheath (Conn et al., 2011b). Cell-specific compartmentation of these two elements makes this possibility unlikely.
Loss of Function of CAX1 and CAX3 Disturbs the Regulation of PSR and Calcium-Related Gene Expression in the Shoot
Under +Pi conditions, one-fifth of the PSR genes are constitutively activated in the shoot of cax1/cax3, including acid phosphatase type 5 (ACP5; At3g17790), digalactosyldiacylglycerol synthase (DGD1; At3g11670), phospholipase Dζ2 (PLDζ2; At3g05630), SPX1, and SPX3 (Table I). ACP5 has been proposed to be involved in Pi mobilization (del Pozo et al., 1999), whereas DGD1 and PLDζ2 participate in the biosynthesis of nonphosphorus lipids during Pi-limited growth (Härtel et al., 2000; Cruz-Ramírez et al., 2006). Loss of PLDζ1 and PLDζ2 reduces primary root elongation under low-Pi conditions (Li et al., 2006), but their roles in the shoot remain unknown. Nucleus-localized SPX1 was suggested to be involved in transcriptional activation of genes related to Pi mobilization and scavenging of reactive oxygen species in response to Pi starvation (Duan et al., 2008). By contrast, SPX3 was shown to localize to intracellular compartments. SPX3 RNA interference lines exhibited higher total P and Pi contents yet stronger induction of several PSi genes, including SPX1, suggesting that SPX3 negatively regulates Pi starvation signaling (Duan et al., 2008). Up-regulation of both genes in the shoot of cax1/cax3 under +Pi conditions may reflect the activation of the SPX1/SPX3-mediated Pi signaling pathway. Moreover, a group of genes encoding Ca2+-ATPases, including ACA2 (At4g37640), ACA10 (At4g29900), ACA11 (At3g57330), ACA12 (At3g63380), and ACA13 (At3g22910), were up-regulated in the shoot but not in the root of cax1/cax3 (Supplemental Table S2), implying that Ca2+-ATPases are activated to balance the decreased Ca2+ efflux in the shoot due to the dysfunction of CAX1 and CAX3. Therefore, it seems that the disturbed cellular Ca2+ homeostasis and misregulated expression of Ca2+-related proteins caused by lack of CAX1 and CAX3, rather than CAX1 and CAX3 per se, are involved in transcriptional regulation of PSR genes.
Common and Distinct Pi Signaling Pathways Mediated by PHO2 and CAX
Because both pho2 and cax1/cax3 mutants displayed an increased level of shoot Pi, it is tempting to know whether PHO2 and CAX1/CAX3 act in the same Pi signaling pathway. Down-regulation of PHO2 in the roots results in increased uptake and root-to-shoot translocation of Pi; however, the role of PHO2 in the shoots is unclear (Lin et al., 2008). Here, we observed that the PHO2 transcript level was greatly increased in the shoot of cax1/cax3 under +Pi and −Pi conditions, whereas the level in the root of cax1/cax3 under +Pi conditions was slightly reduced but still higher than that in the −Pi root of wild-type plants (Fig. 4C). Given that, under Pi deficiency, the level of PHO2 transcript in the root of cax1/cax3 was as low as that in the root of wild-type plants (Fig. 4C), we argue that PHO2 is not involved in enhancing the Pi uptake activity in cax1/cax3 under –Pi conditions. Although both CAX1/CAX3 and PHO2 inhibited the root Pi uptake activity, they seem to mediate different pathways leading to Pi accumulation. First, in contrast to pho2, which exhibits a dramatically increased Pi uptake activity only when Pi remains adequate, cax1/cax3 displayed an enhanced Pi uptake activity under both +Pi and −Pi conditions (Fig. 2, A and B). Notably, although the increase of Pi uptake activity of cax1/cax3 under −Pi conditions was striking, the underlying mechanism is not clear at present. Second, unlike the pho2 mutant, the cax1/cax3 mutant had a similar shoot-to-root ratio of Pi distribution as the wild type under +Pi conditions (Fig. 2C). Third, up-regulation of miR399, which acts upstream of the PHO2-dependent Pi signaling pathway, was not involved in the moderate reduction of PHO2 mRNA level in the root of cax1/cax3 under +Pi conditions (Fig. 4, C and D). These lines of evidence suggest that CAX1/CAX3 and miR399-mediated PHO2 Pi signaling pathways are two distinct pathways attributed to the consequences of Pi uptake and accumulation.
In the pho2 mutant, the expression levels of PHT1;8 and PHT1;9 were shown to be increased under +Pi conditions and assumed to contribute to the establishment of high Pi in the shoot of pho2 (Aung et al., 2006; Bari et al., 2006). However, in the root of cax1/cax3 under both +Pi and −Pi conditions, no up-regulation of PHT1 genes at the transcript level was observed (Fig. 6D). Although the protein level of PHT1;1/2/3 was not increased (Fig. 7B), our results from grafting demonstrate that PHT1;1 is partially responsible for Pi accumulation in the cax1/cax3 scion (Fig. 8C), indicating that a posttranslational regulation of PHT1;1 may be involved. Several consensus sites for N-linked glycosylation and phosphorylation have been predicted in PHT1;1 (Muchhal et al., 1996). Indeed, PHT1;1/2 have been identified in phosphoproteomics of the Arabidopsis plasma membrane, and a phosphorylation site was detected in its C-terminal peptide (Nühse et al., 2004; Hem et al., 2007). Furthermore, as we did not examine the subcellular localization of PHT1;1 in cax1/cax3, changes in the membrane distribution of PHT1;1 cannot be excluded.
CAX1/CAX3-Mediated Shoot-to-Root Pi Signaling
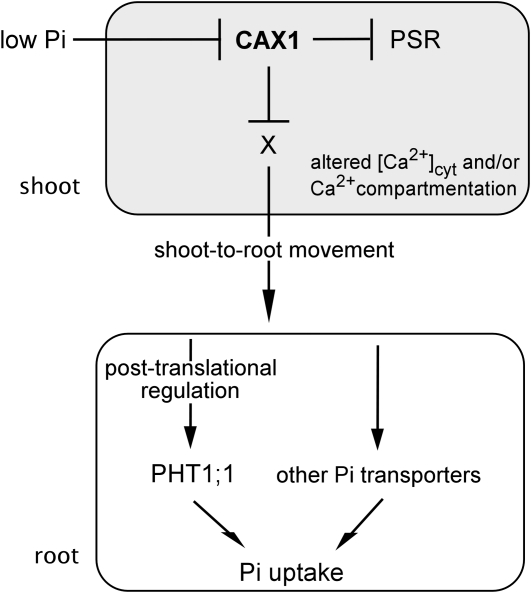
A paradigm for systemic regulation of Pi homeostasis has been recently established. The miR399 generated in shoots after the onset of Pi starvation serves as a long-distance signal to activate Pi transport systems by suppressing PHO2 expression in roots (Lin et al., 2008; Pant et al., 2008). Our results obtained in split-root experiments indicate that CAX1 and CAX3 do not mediate the generation and movement of systemic suppressors from the +Pi root half to the −Pi root half. By contrast, the results of reciprocal grafting experiments clearly suggest the involvement of CAX1 and CAX3 in shoot-to-root Pi signaling. We hypothesize that abrogation of CAX1 and CAX3 in the cax1/cax3 mutant may relieve the repression of shoot PSR genes through alteration of [Ca2+]cyt and/or misregulated compartmentation of Ca2+, thereby triggering a systemic signal that moves from shoots to roots to activate the PHT1 Pi transporters or other unidentified Pi transport systems (Fig. 9). Because mature miR399 was not observed in the shoot and root of cax1/cax3 under +Pi conditions, it is clear that miR399 is not up-regulated to serve as a systemic signal traveling to roots in the CAX1/CAX3-mediated Pi signaling pathway. Several molecules, including hormones, sugars, nutrients themselves or their metabolites, and small RNAs, have been suggested as systemic signals in the long-distance signaling of nutrient status (Liu et al., 2009). In the future, it will be interesting to identify such a shoot-derived signal and the molecular components involved in the up-regulation of the Pi transport system and to establish how those PSR genes misregulated in cax1/cax3 can modulate the Pi transport activity.
Figure 9.
Working hypothesis of the CAX1/CAX3-mediated Pi signaling pathway in Arabidopsis. When Pi is limited, repression of shoot CAX1 leads to abrogated cellular Ca2+ homeostasis involving altered [Ca2+]cyt and/or misregulated compartmentation of Ca2+, which initiates the activation of a subset of shoot PSR genes. A shoot-born signal (X) is thereby generated and moves toward the root to activate the Pi transporter system, including PHT1;1, at the posttranslational level.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) cax1, cax3, and cax1/cax3 were kindly provided as gifts by Drs. Kendal Hirschi and Ning-Hui Cheng (Baylor College of Medicine). Seeds of the wild type (Col-0) and the pht1;1 T-DNA line (SALK_088586) were obtained from the Arabidopsis Biological Resource Center. The +Pi and −Pi media were supplemented with 250 μm KH2PO4 and 0 μm KH2PO4, respectively, unless specified otherwise. For hydroponic growth, 9-d-old seedlings grown on agar plates with half-modified Hoagland nutrient solution containing 250 μm KH2PO4 and 1% Suc solidified with 0.8% agar were transferred to the same nutrient solution containing 250 μm KH2PO4 without Suc for 8 to 10 d. A 5-d treatment of Pi starvation was initiated by replacing 250 μm KH2PO4 with Pi-free medium. For plants grown in the medium supplemented with Ca2+ and Mg2+, 9-d-old seedlings grown on agar plates were transferred to hydroponic medium supplemented with various concentrations of Ca2+ and Mg2+ as indicated and grown for another 12 d. All plants were grown under a 16-h-light/8-h-dark cycle.
Grafting of Arabidopsis Plants
Hypocotyl reciprocal grafting was performed as described previously with minor modifications (Lin et al., 2008). Briefly, micrografting was conducted with 8-d-old seedlings, which were then incubated vertically in the dark for 1 d before being transferred to the culture room under dim light for another 2 d. Two weeks after micrografting, plants were transferred to hydroponic culture and grown for another 2 weeks before sample collection. Lack of contamination of adventitious roots in grafted plants was confirmed by genotyping using PCR.
Affymetrix ATH1 Array Hybridization and Data Analysis
Transcriptomic analyses of plants were conducted using Affymetrix ATH1 arrays. Wild-type and cax1/cax3 plants grown in hydroponic cultures under +Pi and −Pi conditions (see above) were harvested for RNA isolation. Two independent biological replicates were performed. Ten micrograms of total RNA (see below) was used for cDNA synthesis, labeled by in vitro transcription, and followed by fragmentation according to the manufacturer’s recommendations (GeneChip Expression Analysis Technical Manual, Revision 5; Affymetrix). The labeled samples were hybridized to the ATH1 array at 45°C for 16.5 h. Washing and staining were done on a Fluidics Station-450, and the ATH1 array was scanned using the Affymetrix GeneChip Scanner 7G. The results were quantified and analyzed using MicroArray Suite 5.0 software (Affymetrix). The obtained data were normalized using Robust Multichip Average (Irizarry et al., 2003), and the statistical significance of differential expression was determined by Limma analysis (Smyth, 2004).
Pi Concentration and Pi Uptake Analysis
Pi concentration and uptake activity were determined as described (Chiou et al., 2006). To assay the Pi uptake, 4-week-old plants grown under +Pi or –Pi conditions were transferred to medium containing 250 μm KH2PO4 (+Pi) or 10 μm KH2PO4 (−Pi) for the measurement of [33P]Pi uptake.
RNA Isolation, RT-PCR, and qRT-PCR
Total RNA from hydroponic samples was isolated by the use of TRIzol reagent (Invitrogen) and treated with DNase I (Ambion) before qRT-PCR to eliminate genomic DNA contamination. cDNA was synthesized from 0.5 to 1 μg of total RNA by use of Moloney murine leukemia virus reverse transcriptase (Promega) with oligo(dT) primer. RT-PCR conditions and sequences of primers used in our study were identical to those listed in supplemental table S1 of Aung et al. (2006). Sequences of additional primers are listed in Supplemental Table S3. qRT-PCR was performed using the Power SYBR Green PCR Master Mix kit (Applied Biosystems) on a 7300 Real-Time PCR system (Applied Biosystems) according to the manufacturer’s instructions. Relative expression levels were normalized to that of an internal control, UBQ10.
Immunoblot Analyses
For extraction of total protein, 10-d-old seedlings of wild-type and pht1;1 plants with or without 5-d treatment of Pi deficiency were ground in liquid nitrogen and dissolved in protein lysis buffer (2% SDS, 60 mm Tris-HCl [pH 8.5], 2.5% glycerol, 0.13 mm EDTA, and 1× complete protease inhibitor [Roche]). Twenty micrograms of total protein was loaded onto the SDS-PAGE apparatus for each sample.
Hydroponically grown cax1/cax3 and wild-type plants were harvested for total membrane protein extraction. One milligram of root tissues was ground with an ice-cold mortar and pestle and dissolved in 3 mL of membrane extraction buffer (330 mm Suc, 50 mm Tris [pH 7.5], 10 mm KCl, 5 mm EDTA, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor [Roche]). The extracts were then collected and centrifuged at 2,000g for 10 min at 4°C. Supernatants were collected and centrifuged at 400,000g for another 40 min at 4°C. Pellets were dissolved in membrane extraction buffer and collected as total membrane proteins. Twenty micrograms of total membrane protein was loaded onto the SDS-PAGE apparatus for each sample. Polyclonal rabbit antibodies were raised and affinity purified against an internal fragment of PHT1;1 corresponding to amino acid residues 266 to 285 (ELEERVEDDVKDPRQNYGLF). The final concentration of 20 to 100 ng mL−1 affinity-purified antibodies was used for immunoblot analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: UBQ10 (At4g05320), PHT1;1 (At5g43350), PHT1;2 (At5g43370), PHT1;3 (At5g43360), PHT1;4 (At2g38940), PHT1;5 (At2g32830), PHT1;7 (At3g54700), PHT1;8 (At1g20860), PHT1;9 (At1g76430), PHO2 (At2g33770), CAX1 (At2g38170), CAX3 (At3g51860), At4 (At5g03545), SPX1 (At5g20150), and SPX3 (At2g45130).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of 163 and 455 differentially expressed genes (more than 2-fold change) in wild-type shoots and roots under –Pi conditions, respectively, with those reported by Morcuende et al. (2007).
Supplemental Figure S2. qRT-PCR analysis of SPX1 (A and B), PHT1;4 (C and D) and At4 (E and F) expression in the shoot (A, C, and E) and root (B, D, and F) of split-root wild-type and cax1/cax3 plants.
Supplemental Figure S3. qRT-PCR analysis of several PSR genes in the shoot of wild-type and cax1/cax3 plants under +Pi or −Pi conditions.
Supplemental Table S1. Genes significantly differentially expressed (P ≤ 0.01, more than 2-fold change) in data sets derived from microarray analysis.
Supplemental Table S2. Ca2+-related genes that are differentially expressed in the shoot and root of cax1/cax3 under Pi-sufficient conditions.
Supplemental Table S3. Sequences of qRT-PCR primers used in this study.
Supplementary Material
Acknowledgments
We thank Drs. Kendal Hirschi and Ning-Hui Cheng for kindly providing the cax mutant seeds. We are grateful to Ya-Shiuan Lai for genotyping the grafted plants, June-Wei Chen for testing the antibody, and Yi-Wei Lee for statistical analysis of microarray data. Affymetrix GeneChip assays were performed by the Affymetrix Gene Expression Service Laboratory (http://ipmb.sinica.edu.tw/affy/) supported by Academia Sinica.
References
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. (1999) The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol 119: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Santos E, Alonso JM, Ecker JR, Martinez-Zapater JM, Salinas J. (2003) Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD. (2003) The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15: 347–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Shigaki T, Hirschi KD. (2002) Characterization of CAX4, an Arabidopsis H+/cation antiporter. Plant Physiol 128: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Shigaki T, Lachmansingh J, LeClere S, Lahner B, Salt DE, Hirschi KD. (2005) Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol 138: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J, Lin S-I. (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Conn V, Tyerman SD, Kaiser BN, Leigh RA, Gilliham M. (2011a) Magnesium transporters, MGT2/MRS2-1 and MGT3/MRS2-5, are important for magnesium partitioning within Arabidopsis thaliana mesophyll vacuoles. New Phytol 190: 583–594 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Gilliham M, Athman A, Schreiber AW, Baumann U, Moller I, Cheng N-H, Stancombe MA, Hirschi KD, Webb AAR, et al. (2011b) Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L. (2006) Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ. (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J. (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Dong B, Rengel Z, Delhaize E. (1998) Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205: 251–256 [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Förster C, Kane PM. (2000) Cytosolic Ca2+ homeostasis is a constitutive function of the V-ATPase in Saccharomyces cerevisiae. J Biol Chem 275: 38245–38253 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Martín AC, Leyva A, Paz-Ares J. (2005) Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol 138: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Dormann P, Benning C. (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hem S, Rofidal V, Sommerer N, Rossignol M. (2007) Novel subsets of the Arabidopsis plasmalemma phosphoproteome identify phosphorylation sites in secondary active transporters. Biochem Biophys Res Commun 363: 375–380 [DOI] [PubMed] [Google Scholar]
- Hinsinger P. (2001) Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237: 173–195 [Google Scholar]
- Hirschi KD. (1999) Expression of Arabidopsis CAX1 in tobacco: altered calcium homeostasis and increased stress sensitivity. Plant Cell 11: 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD. (2004) The calcium conundrum: both versatile nutrient and specific signal. Plant Physiol 136: 2438–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ. (2000) Expression of Arabidopsis CAX2 in tobacco: altered metal accumulation and increased manganese tolerance. Plant Physiol 124: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Knight H. (2000) Calcium signaling during abiotic stress in plants. Int Rev Cytol 195: 269–324 [DOI] [PubMed] [Google Scholar]
- Koren’kov V, Park S, Cheng NH, Sreevidya C, Lachmansingh J, Morris J, Hirschi KD, Wagner GJ. (2007) Enhanced Cd2+ -selective root-tonoplast-transport in tobaccos expressing Arabidopsis cation exchangers. Planta 225: 403–411 [DOI] [PubMed] [Google Scholar]
- Krebs M, Beyhl D, Görlich E, Al-Rasheid KA, Marten I, Stierhof YD, Hedrich R, Schumacher K. (2010) Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA 107: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MY, Qin CB, Welti R, Wang XM. (2006) Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ. (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WY, Lin SI, Chiou TJ. (2009) Molecular regulators of phosphate homeostasis in plants. J Exp Bot 60: 1427–1438 [DOI] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P. (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Liu TY, Chang CY, Chiou TJ. (2009) The long-distance signaling of mineral macronutrients. Curr Opin Plant Biol 12: 312–319 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London [Google Scholar]
- Marty F. (1999) Plant vacuoles. Plant Cell 11: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. (2009) Shaping the calcium signature. New Phytol 181: 275–294 [DOI] [PubMed] [Google Scholar]
- Mei H, Zhao J, Pittman JK, Lachmansingh J, Park S, Hirschi KD. (2007) In planta regulation of the Arabidopsis Ca2+/H+ antiporter CAX1. J Exp Bot 58: 3419–3427 [DOI] [PubMed] [Google Scholar]
- Miseta A, Kellermayer R, Aiello DP, Fu L, Bedwell DM. (1999) The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett 451: 132–136 [DOI] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng WM, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG. (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Raghothama KG. (1996) Cloning and characterization of two high affinity phosphate transporter homologues in Arabidopsis thaliana. Plant Physiol 111: 435 [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nühse TS, Stensballe A, Jensen ON, Peck SC. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK, Shigaki T, Marshall JL, Morris JL, Cheng NH, Hirschi KD. (2004) Functional and regulatory analysis of the Arabidopsis thaliana CAX2 cation transporter. Plant Mol Biol 56: 959–971 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Bucher M. (2002) Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book; 1: e0024, doi/10.1199/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigaki T, Rees I, Nakhleh L, Hirschi KD. (2006) Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J Mol Evol 63: 815–825 [DOI] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Sze H, Liang F, Hwang I, Curran AC, Harper JF. (2000) Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu Rev Plant Physiol Plant Mol Biol 51: 433–462 [DOI] [PubMed] [Google Scholar]
- Thibaud M-C, Arrighi J-F, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. (2010) Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Abel S. (2004) Short on phosphate: plant surveillance and countermeasures. Trends Plant Sci 9: 548–555 [DOI] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang HJ, Li K, Wu P, Shou HX. (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. (2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 135: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Barkla BJ, Marshall J, Pittman JK, Hirschi KD. (2008) The Arabidopsis cax3 mutants display altered salt tolerance, pH sensitivity and reduced plasma membrane H+-ATPase activity. Planta 227: 659–669 [DOI] [PubMed] [Google Scholar]
- Zhao J, Shigaki T, Mei H, Guo YQ, Cheng NH, Hirschi KD. (2009) Interaction between Arabidopsis Ca2+/H+ exchangers CAX1 and CAX3. J Biol Chem 284: 4605–4615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.