Abstract
Beta-papillomavirus (β-HPV) DNA is present in some cutaneous squamous cell carcinomas (cuSCC), but no mechanism of carcinogenesis has been determined. We used ultra-high throughput sequencing of the cancer transcriptome to assess whether papillomavirus transcripts are present in these cancers. Sixty-seven cuSCC samples were assayed for β-HPV DNA by PCR, and viral loads were measured with type-specific qPCR. Thirty-one SCCs were selected for whole transcriptome sequencing. Transcriptome libraries were prepared in parallel from the HPV18 positive HeLa cervical cancer cell line and HPV16 positive primary cervical and periungual SCC. Thirty percent (20/67) of the tumors were positive for β-HPV DNA, but there was no difference in β-HPV viral load between tumor and normal tissue (p=0.310). Immunosuppression and age were significantly associated with higher viral load (p=0.016 for immunosuppression; p=0.0004 for age). Transcriptome sequencing failed to identify papillomavirus expression in any of the skin tumors. In contrast, HPV 16 and 18 mRNA transcripts were readily identified in primary cervical and periungual cancers and HeLa cells. These data demonstrate that papillomavirus mRNA expression is not a factor in the maintenance of cuSCC.
Introduction
Although 12% of all human cancers are now known to be caused by viruses(Parkin, 2006; Zur Hausen, 2009), the mere presence of viral DNA in a tumor does not necessarily indicate causality. Multiple lines of evidence suggest a viral etiology for cutaneous squamous cell carcinoma (cuSCC). In immunosuppressed solid organ transplant recipients (OTRs), the incidence of cuSCC is 65- to 250-fold higher than in the general population(Hartevelt et al., 1990; Jensen et al., 1999; Lindelof et al., 2000); incidence ratios of this magnitude are commonly seen in other viral cancers, including human herpesvirus-8-mediated Kaposi’s sarcoma and HBV-associated hepatocellular carcinoma(Vajdic et al., 2006). A second line of evidence supporting viral etiology is the behavior of the keratoacanthoma (KA) subtype of cuSCC. KA can spontaneously regress, and has been suggested to lie along a spectrum of carcinogenesis between hyperplastic viral verrucae and neoplastic SCC(LeBoit, 2002).
Previous studies have selectively focused on human papillomavirus (HPV) as a potential etiologic agent in cuSCC. Investigators have hypothesized an analogy between cuSCC and cervical SCC, as the latter has been firmly associated with high-risk α-genus HPV (α-HPV) infection, including HPV 16 and 18(Bouvard et al., 2009; IARC, 2007). However, different HPV types have site-specific tropism for mucosal or cutaneous epithelium; the high-risk mucosal α-HPV are not found in cuSCC, with the exception of genital and periungual tumors(Alam et al., 2003; Dubina and Goldenberg, 2009; Moy et al., 1989). Thus, many studies focus on detection of the cutaneous β-genus HPV types (β-HPV) in cuSCC(Asgari et al., 2008; Berkhout et al., 2000; Forslund et al., 2003b; Harwood et al., 2000; Shamanin et al., 1994; Shamanin et al., 1996; Surentheran et al., 1998).
The association of β-HPV with cuSCC is clearly defined for a specific group of patients with epidermodysplasia verruciformis, an autosomal recessive genodermatosis associated with susceptibility to β-HPV. Patients with epidermodysplasia verruciformis develop widespread viral warts and β-HPV 5- and 8-mediated SCC(Harwood et al., 1999). However, the β-HPV types have not been firmly associated with cuSCC in the general population(IARC, 2007). β-HPV DNA is detected in 27–54% of SCCs from immunocompetent patients and 55–84% of SCCs from immunosuppressed patients(Asgari et al., 2008; Berkhout et al., 2000; Berkhout et al., 1995; Forslund et al., 2007; Forslund et al., 2003a; Harwood et al., 2000; Shamanin et al., 1994; Shamanin et al., 1996). Indeed, in other studies β-HPV has been detected with comparable frequency in normal skin, eyebrow hairs, and premalignant actinic keratoses(Antonsson et al., 2000; Asgari et al., 2008; Boxman et al., 1997; de Koning et al., 2007; de Koning et al., 2009; Forslund et al., 2003b; Hazard et al., 2007). Other studies have reported an association between antibody responses to β-HPV and the development of cuSCC, particularly for patients with antibodies to multiple HPV types(Bouwes Bavinck et al., 2010; Karagas et al., 2006).
High-risk α-HPV, which are present in over 95% of cervical SCC, often integrate into the human genome and express viral proteins that interfere with normal cell cycle control (reviewed in(Bosch et al., 2002; zur Hausen, 1996)). The E6 and E7 proteins of the high-risk α-HPV types interfere with the tumor-suppressor activities of cellular p53 and pRB to drive carcinogenesis(Dyson et al., 1989; Scheffner et al., 1990; Werness et al., 1990). Ongoing expression of E6 and E7 is required for both induction and maintenance of carcinogenesis. By analogy, β-HPV would be expected to utilize the same mechanism of carcinogenesis, but studies using in situ hybridization or RT-PCR to detect HPV mRNA in cuSCC have detected viral transcripts only sporadically and at low levels in occasional tumors, with many other tumors testing negative. (Dang et al., 2006; Purdie et al., 2005). Nevertheless, many authors continue to point to β-HPV as a possible etiologic agent in these tumors.
The goal of our study was to assess whether β-HPV is capable of causing cuSCC through expression of viral oncogenes, using ultra-high throughput sequencing of the SCC transcriptome. This comprehensive, unbiased analysis of total tumor mRNA expression revealed no HPV transcriptional activity, an observation that was further supported by the absence of a substantial viral load in the tumors. These two observations contradict the hypothesis that transcription of viral oncogenes is required for tumor maintenance.
Results
Patient Characteristics
We enrolled 38 patients, including 27 males and 11 females, ranging in age from 41 to 95 years (Table 1). Seventeen patients were immunocompetent and 21 were immunosuppressed due to solid organ transplantation, hematologic malignancy, HIV, or medication for Wegener’s granulomatosis. Eighty-nine tissue samples were collected from these patients, including 71 SCCs (23 KA subtype) and 18 normal skin samples. Four tumor samples did not yield enough tissue for DNA extraction but RNA was obtained for transcriptome analysis. Two α-HPV16-mediated primary tumors were obtained for comparison: a periungual SCC from a 53-year-old immunocompetent man, and a stage I nonkeratinizing cervical SCC from a 35-year-old woman. The α-HPV18-mediated HeLa cervical cancer cell line was used as an additional control.
Table 1.
Demographics
| Total | Male | Female | |
|---|---|---|---|
|
|
|||
| Patients | 38 | 27 | 11 |
| Immunocompetent | 17 | 10 | 7 |
| Immunosuppressed | 21 | 17 | 4 |
| Solid Organ Transplant | 12 | 9 | 3 |
| Hematologic Malignancy | 6 | 5 | 1 |
| HIV | 2 | 2 | 0 |
| Wegener’s Granulomatosis | 1 | 1 | 0 |
| Age Range (Years) | 41–95 | 41–95 | 52–95 |
| Total | Male | Female | |
|---|---|---|---|
|
|
|||
| SCC Samples [KA type] | 71 [23] | 45 [8] | 26 [15] |
| Immunocompetent | 30 [16] | 14 [6] | 16 [10] |
| Immunosuppressed | 41 [7] | 31 [2] | 10 [5] |
| Solid Organ Transplant | 28 | 19 | 9 |
| Hematologic Malignancy | 10 | 9 | 1 |
| HIV | 2 | 2 | 0 |
| Wegener’s Granulomatosis | 1 | 1 | 0 |
| Normal Skin | 18 | 14 | 4 |
| Immunocompetent | 8 | 6 | 2 |
| Immunosuppressed | 10 | 8 | 2 |
| Solid Organ Transplant | 8 | 6 | 2 |
| Hematologic Malignancy | 2 | 2 | 0 |
Low Viral Load of β-HPV in Normal Skin and Cutaneous SCC
Eighty-five DNA samples were assayed for the presence of β-globin DNA by PCR and all demonstrated sufficient quantity and integrity for β-HPV typing. Twenty of 67 cuSCC tumors (30%) were positive for HPV DNA by PCR, 18 of which were confirmed by sequencing. Five of 18 normal skin samples (28%) were also HPV-positive by PCR and sequence confirmation (Table 2). Eleven HPV types and 14 incompletely sequenced fragment types were detected, with no single type predominating. Multivariate regression modeling demonstrated no difference in β-HPV carriage between tumor and normal tissue when controlling for age, sex, and immunosuppression as well as clustering for multiple samples from the same patient (p=0.693). Immunosuppression and older age were significantly associated with β-HPV carriage (p=0.018 for both). PCR and sequencing confirmed the presence of α-HPV16 in the primary cervical and periungual SCC and α-HPV18 in HeLa cells. In addition, the periungual SCC was found to contain β-HPV8 and FA51.2 DNA.
Table 2.
HPV Viral Loads.Patients with HPV PCR positive samples shown.
| Patient | Sex | Age | Immunosuppression | Sample Code |
Histology | Site | DNA HPV Types | Viral Load |
Transcriptome Sequenced |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV Type |
HPV Copies |
Input Cells |
HPV Copies/Cell |
|||||||||
| PC-021 | M | 89 | No | STA01-046 | KA | Scalp | FA7, FA127, FA37 | FA7 | ND | 3921 | ND | |
| FA37 | ND | 3921 | ND | |||||||||
| STA01-051 | KA | Scalp | HPV21, FA127, FA37 | HPV21 | ND | 13952 | ND | |||||
| FA37 | 100 | 13952 | 0.007 | |||||||||
| STA01-053 | Normal skin | Leg | HPV80, FA7 | HPV80 | ND | 6434 | ND | |||||
| FA7 | 28 | 6434 | 0.004 | |||||||||
|
| ||||||||||||
| PC-031 | M | 95 | No | STA01-130 | SCC | Scalp | - | |||||
| STA01-131 | Normal skin | Scalp | HPV5, HPV8 | HPV5 | 220872 | 2313 | 95.501 | |||||
| HPV8 | ND | 2313 | ND | |||||||||
|
| ||||||||||||
| PC-046 | M | 84 | No | STA01-068 | SCC | Hand | HPV75, FA108 | HPV75 | 149 | 14607 | 0.010 | |
| STA01-079 | Normal skin | Leg | HPV75 | HPV75 | 152 | 4651 | 0.033 | |||||
|
| ||||||||||||
| PC-003 | M | 68 | Heart Transplant | STA01-034 | SCC | Forehead | HPV5, HPV49, FAIMVS14 | HPV5 | 696 | 2611 | 0.267 | |
| STA01-122 | Normal skin | Cheek | - | Yes | ||||||||
|
| ||||||||||||
| PC-015 | F | 53 | Heart-Lung Transplant | STA01-018 | SCC | Arm | HPV9 | HPV9 | 984 | 28332 | 0.028 | Yes |
| STA01-076 | Normal skin | Arm | HPV9 | HPV9 | 57 | 33490 | 0.001 | Yes | ||||
| STA01-077 | KA | Arm | HPV9 | HPV9 | ND | 7264 | ND | Yes | ||||
| STA01-094 | Normal skin | Forehead | - | Yes | ||||||||
| STA01-095 | SCC | Forehead | - | Yes | ||||||||
|
| ||||||||||||
| PC-007 | M | 57 | Lung Transplant | STA01-074 | SCC | Scalp | HPV20, HPV21 | HPV20 | 1256 | 51462 | 0.024 | Yes |
| HPV21 | 10407 | 51462 | 0.202 | |||||||||
| STA01-078 | Normal skin | Scalp | HPV21, HPV20 | HPV20 | ND | 47880 | ND | |||||
| HPV21 | ND | 47880 | ND | |||||||||
|
| ||||||||||||
| PC-041 | M | 60 | Lung Transplant | STA01-029 | SCC | Cheek | HPV17 | HPV17 | 26132 | 45136 | 0.579 | |
| STA01-030 | SCC | Cheek | Sequence Not Obtained | Yes | ||||||||
|
| ||||||||||||
| PC-058 | M | 51 | Lung Transplant | STA01-010 | SCC | Lip | HPV5, HPV80, FA14 | HPV5 | ND | 6693 | ND | Yes |
| HPV80 | ND | 6693 | ND | |||||||||
| FA14 | 492 | 6693 | 0.074 | |||||||||
| STA01-045 | SCC | Cheek | FANIMVS11.4, FAIMVS11.3 | NA | ||||||||
| STA01-059 | SCC | Cheek | - | |||||||||
| STA01-132 | SCC | Scalp | - | |||||||||
| STA01-133 | Normal skin | Cheek | - | |||||||||
|
| ||||||||||||
| PC-030 | F | 73 | Renal Transplant | STA01-031 | KA | Leg | FA14, FA16.3 | FA14 | 28006 | 5098 | 5.494 | |
| FA16.3 | 1068 | 5098 | 0.209 | |||||||||
| STA01-032 | KA | Leg | FA16.3, FA75 | FA16.3 | ND | 17901 | ND | |||||
| STA01-090 | KA | Leg | FA14, HPV96, FA140.2, FA16.3 | FA14 | 17216 | 4712 | 3.654 | |||||
| FA16.3 | ND | 4712 | ND | |||||||||
| FA140.2 | ND | 4712 | ND | |||||||||
| STA01-091 | KA | Leg | FA16.3, FA140.2, FA14 | FA14 | 1808 | 3372 | 0.536 | Yes | ||||
| FA16.3 | 44140 | 3372 | 13.089 | |||||||||
| FA140.2 | 110 | 3372 | 0.033 | |||||||||
|
| ||||||||||||
| PC-054 | M | 55 | Renal/Pancreas Transplant | STA01-065 | SCC | Arm | - | |||||
| STA01-066 | SCC | Arm | HPV8 | HPV8 | 4462 | 25593 | 0.174 | Yes | ||||
|
| ||||||||||||
| PC-011 | M | 64 | CLL/SLL | STA01-064 | SCC | Forehead | HPV19, HPV49, FA123 | HPV19 | 1132 | 26360 | 0.043 | Yes |
| FA123 | 4717 | 26360 | 0.179 | |||||||||
| STA01-071 | SCC | Forehead | HPV19, FA33 | HPV19 | 38 | 3883 | 0.010 | |||||
|
| ||||||||||||
| PC-053 | M | 53 | HIV | STA01-099 | SCC | Forehead | HPV5, FA16 | HPV5 | 1126 | 32837 | 0.034 | Yes |
| PC-043 | M | 53 | No | STA01-035 | SCC | Hand | HPV16 | HPV16 | 570172 | 12156 | 46.904 | Yes |
| HPV8, FA51.2 | HPV8 | 3685 | 12157 | 0.303 | Yes | |||||||
| HeLa | F | HeLa | Cell Line | Cervix | HPV18 | HPV18 | 8496079 | 1350389 | 6.291 | Yes | ||
| ILS-19363 | F | 35 | No | ILS-19363-1 | SCC | Cervix | HPV16 | HPV16 | 3403823 | 1403307 | 2.426 | Yes |
ND: Not detected NA: No assay could be designed SCC: Squamous cell carcinoma KA: Keratoacanthoma MDS: Myelodysplastic syndrome ALL: Acute lymphoblastic leukemia CLL/SLL: Chronic lymphocytic leukemia/Small lymphocytic lymphoma SCT: Stem cell transplant GVHD: Graft vs. host disease.
Viral loads were determined for up to 3 HPV types in 24 of the 25 HPV PCR-positive and sequence confirmed samples. Replicate assays were performed for HPV18 in the HeLa cervical cancer cell line, HPV16 in the cervical SCC, and HPV16 and HPV8 in the periungual SCC (Table 2, Figure 1A). With the exception of 4 samples (1 normal skin and 3 tumors), all viral loads were below 1 HPV copy per cell. In contrast, the cervical SCC contained 2.4 HPV16 copies per cell and the HeLa cell line contained 6.3 copies per cell, consistent with viral integration. The periungual SCC contained 46.9 α-HPV16 copies per cell but only 0.3 β-HPV8 copies per cell.
Figure 1. Comparison of HPV DNA viral load and abundance of HPV-derived transcripts for established HPV-driven cancers versus normal skin and cuSCCs.
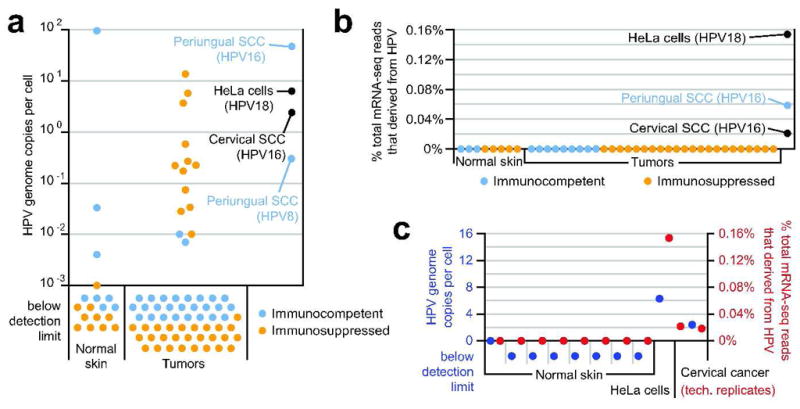
(A) HPV DNA viral loads determined by type-specific qPCR. For each sample with multiple type infection, the sum of the type-specific viral loads is shown (for details, see Table 2.) (B) Abundance of HPV-derived transcripts determined by mRNA-seq. Reads were filtered to remove host-derived and low-complexity sequence prior to viral mapping (see methods), and HPV counts are each presented as a percentage of their total dataset. The most frequently matched HPV type for the HeLa, periungual SCC, and cervical SCC samples is indicated. Cervical cancer is presented as the union of 2 technical replicate datasets. (C) Congruence between HPV genomic load (blue; presented as in panel A) and abundance of HPV-derived transcripts (red; presented as in panel B) among those control samples from which both types of data were collected.
Random-effects interval regression modeling demonstrated no difference in β-HPV viral load between tumor and normal tissue when controlling for age, sex, and immunosuppression as well as clustering for multiple samples from the same patient (p=0.310). Immunosuppression and age were significantly associated with higher viral load (p=0.016 for immunosuppression; p=0.0004 for age).
No HPV Transcripts Observed in Cutaneous SCC
The potential oncogenicity of HPV viruses in our samples was assessed in terms of viral gene expression by mRNA-transcriptome sequencing (mRNA-seq). Thirty-one cuSCC tumors (10 KA type) were assayed by high-throughput mRNA-seq, including 10 β-HPV DNA positive samples with viral loads ranging from undetectable to 13.7 copies per cell. Parallel libraries were prepared from 8 patient-paired normal skin samples as well as the aforementioned periungual SCC tumor, cervical SCC, and HeLa cell samples. Paired-end read counts per library ranged from 1.5 million to 10.6 million reads (corresponding to sequence from 740,000 to 5.3 million cDNA amplicons), with a median count of 3.5 million reads (1.8 million amplicons; Table S2). After removing sequences with low sequence complexity or with high quality matches to the human genome or transcriptome, reads were queried against a database of all fully sequenced viral genomes from RefSeq (see Methods). Abundant HPV-matching reads were detected in the HeLa cell-derived and cervical SCC-derived datasets (0.15% and 0.02% of the total reads, respectively Figure 1B). In both, the HPV subtype identified was an α-HPV subtype known to drive tumorigenesis in that sample type (HPV18 and HPV16, respectively)(Bouvard et al., 2009; IARC, 2007). Read frequencies from 2 technical replicate libraries prepared from cervical SCC were nearly identical (0.022% and 0.018%; Figure 1C, Table S1).
Abundant HPV16 reads were also detected in the transcriptome of the periungual SCC (0.06% of total reads; Figure 1B), with no reads mapping preferentially to HPV8 or any other HPV subtype. No potentially HPV-derived reads were detected in 30 of the cuSCC tumors and 7 of the paired normal skin samples (Figure 1B). One normal skin sample that was HPV-negative by genomic PCR contained 2 HPV-matching reads (0.00008% of the total reads), and 1 skin SCC sample contained 5 HPV-derived reads (0.0001% of the total read set; this sample had no DNA for genomic PCR).
No libraries contained a higher frequency of total viral-matching reads than the lowest HPV-derived read count amongst the HPV-positive control samples (0.018%), and only 5 had viral read frequencies within an order of magnitude of that value (File S2). These 5 samples detected a mixture of phage sequence (likely deriving from bacteria on the skin), human klassevirus 1 (a candidate etiological agent for diarrhea that was isolated from human stool in our lab and is therefore a likely cross-lab contaminant(Greninger et al., 2009); and Moloney murine leukemia virus (MMLV). A reverse transcriptase deriving from the latter was used for library construction (see Methods), making it a likely reagent contaminant. Although MMLV and related viruses do have oncogenic capabilities (Cuypers et al., 1984), the sample in which it was detected at the highest frequency was a normal skin control, not a tumor (sampleID STA01-122, dataset C, barcode CGT, Table S2 and File S2).
In these 39 cases of normal skin and cuSCC, the frequencies of HPV reads were orders of magnitude less than those observed for any of the bona fide HPV-driven tumors. In control samples for which both mRNA-seq and viral genome load data were obtained, the results obtained by these 2 metrics agreed with one another and with prior descriptions of the role of HPV as an oncogenic virus; genomes and transcripts were both abundant in HeLa cells and cervical SCC and both absent in healthy skin (Figure 1C). The similarity of HPV genome and transcriptome quantitation between HeLa cells and cervical SCC versus periungual SCC supported the role for HPV in periungual SCC tumorigenesis, while the quantitative similarity between the cuSCC versus healthy skin samples implied no role for HPV transcription in the maintenance of those tumors.
Discussion
Previous studies have proposed β-HPV as a potential causative agent in cuSCC, citing the presence of viral DNA in tumor tissue, but these have not definitively proved an epidemiologic association or evaluated any particular mechanism of transformation. We used whole transcriptome sequencing to test the hypothesis that HPV is required for the maintenance of cuSCC through expression of viral oncoproteins. Transcriptome sequencing revealed a complete absence of HPV mRNA in these tumors, similar to paired normal skin. This stood in stark contrast to the abundant HPV messages detected in cervical SCC and its derivative HeLa cell line. Our results in fact, contradict the hypothesis that expression of viral oncogenes is required for maintenance of cuSCC.
Periungual SCC represents a special site on the cutaneous epithelium. These tumors are associated with high-risk α-HPV(Alam et al., 2003; Kreuter et al., 2009; Moy et al., 1989), which has been reported as episomal, and in a single case, integrated(Sanchez-Lanier et al., 1994; Theunis et al., 1999). In our control periungual SCC, β-HPV8 and FA51.2 DNA were detected along with α-HPV16. This tumor contained 46.9 α-HPV16 copies per cell but only 0.3 β-HPV8 copies per cell. HPV16 mRNA reads represented 0.06% of the transcriptome, but no β-HPV mRNA reads were detected. Taken together, this control specimen supports our impression of α-HPV as the driver and β-HPV as a mere passenger in periungual SCC.
As in previous studies(Antonsson et al., 2000; Asgari et al., 2008; Berkhout et al., 2000; Berkhout et al., 1995; Boxman et al., 1997; de Koning et al., 2007; de Koning et al., 2009; Forslund et al., 2007; Forslund et al., 2003a; Forslund et al., 2003b; Harwood et al., 2000; Hazard et al., 2007; Shamanin et al., 1994; Shamanin et al., 1996), β-HPV DNA was detectable by nested PCR in 30% of SCC, but was also found in a comparable proportion (28%) of normal skin samples. Moreover, we found extremely low viral loads in tumors that were positive for the viral genome. In all but 3 tumor samples, the viral load was less than 1 copy per cell. Importantly, the 3 contradictory samples all came from a single renal transplant recipient with multiple KAs of the lower leg and may reflect a unique feature of that case. Use of PCR and sequencing allowed identification of a broad range of HPV types although the multiplicity of infection may be limited by the number of clones sequenced. Alternate methods for β-HPV detection such as line blots and microarrays allow simultaneous detection of types but are limited in the types detected. While DNA from other HPV types may be present in these samples, this does not alter the conclusion of this study. The low copy number of β-HPV DNA, combined with the absence of virally-derived oncogenic messages, strongly suggest that β-HPV transcription is not required for tumor maintenance.
Our data were consistent with previous evidence that β-HPV merely colonizes the skin. Immunosuppression and older age were associated with a higher prevalence and viral load of β-HPV, consistent with prior studies(Boxman et al., 2001; de Koning et al., 2009; Struijk et al., 2003). These phenomena likely reflected the role of the immune system and age-related immune senescence in controlling epidermal colonization with HPV rather than explaining the increased incidence of SCC in OTRs and older patients. The prevalence of HPV DNA in tape-stripped biopsies is far lower than that on the surface, further supporting a passenger role(Forslund et al., 2004). Support for β-HPV as a passenger also comes from a study of tumors from patients with xeroderma pigmentosum, in which prevalence of viral carriage increases with age and is very low in tumors from children(Luron et al., 2007). A reversed relationship in which SCC somehow results in the presence or increase of β-HPV DNA or antibodies is possible, although further investigation would be required to substantiate this.
Insertional mutagenesis is another mechanism of viral oncogenesis; this mechanism has been described for oncoretroviruses but not for DNA viruses. High-risk α-HPV types can integrate into the host genome but require continual expression of the viral E6 and E7 proteins for their oncogenic activities(Dyson et al., 1989; Scheffner et al., 1990; Werness et al., 1990). The recently described Merkel cell polyomavirus (MCV), another small DNA oncovirus, also integrates into the host genome(Feng et al., 2008), but continued expression of the MCV truncated large T antigen is similarly required for carcinogenesis(Houben et al., 2010). In contrast, there are no reports of β-HPV integration into the genome of cuSCC. The low viral loads of β-HPV in cuSCC reported here further indicate that, even if β-HPV had integrated, only at most only a small proportion of the genomes within any tumor could contain integrated β-HPV, casting doubt upon integration as a carcinogenic mechanism.
It has also been suggested that β-HPV might play a role in induction but not maintenance of cuSCC (based on higher viral load of HPV in precancerous actinic keratoses versus primary SCC, metastatic tumor, or perilesional skin(Weissenborn et al., 2005)). This may occur by interfering with cellular DNA repair or apoptosis following UV-irradiation, creating a pool of genomically unstable cells at risk of oncogenic transformation. Our study was not designed to address this hypothesis. But it should be noted that such a hypothesis would represent a substantial departure from the role played by α-HPV in mucosal SCC, and from the carcinogenic mechanisms known to be employed by other families of DNA tumor viruses in general. Therefore, the most straightforward interpretation of our data is that the sporadic and low-level presence of β-HPV genomic DNA in these tumors, unaccompanied by evidence of active viral gene expression, most likely represents colonization rather than an etiologic association.
Materials and Methods
Sample Collection
All subjects provided informed consent according to procedures approved by the University of California, San Francisco Committee on Human Research and adherent to the Helsinki Guidelines. Tumor tissue was collected from patients during the course of biopsy or excision. All specimens were held for further processing until final pathology confirmed a diagnosis of cuSCC. Normal tissue was collected when surgical discard was available from postoperative reconstruction. All tissue was snap-frozen and stored on liquid nitrogen until nucleic acid extraction.
A primary cervical cancer specimen containing HPV16 was obtained from a commercial tissue bank (ILSBio, LLC, Chestertown, MD).
Nucleic Acid Isolation
Tissue samples were minced, divided, and placed in parallel extraction pathways. DNA was extracted using the QIAamp DNA Mini Kit with RNase A (Qiagen, Valencia, CA) as per manufacturer’s protocol. RNA extractions were carried out using the RNeasy Lipid Tissue Mini Kit with on-column RNase-free DNase I (Qiagen) as per manufacturer’s protocol.
Human Papillomavirus DNA Detection using PCR
A PCR assay for β-globin DNA was performed on each sample to control for DNA integrity and for the presence of adequate quantity of DNA. Five μL of each DNA sample (30–800ng) were tested with primers PCO4 and GH20 as described(Bauer et al., 1991). β-HPV PCR was carried out using the nested primer sets FAP59-FAP64 and FAP6085F-FAP6319R (Forslund et al., 2003a; Forslund et al., 2003b). For the first round of PCR, 5 μL of each DNA sample were amplified using 2μM of the FAP59 and FAP64 primers in a 50μL reaction volume including 1× Taq Buffer, 2mM MgCl2, 0.25mM dNTPs, and 1U Taq polymerase. The reaction was carried out under the following PCR conditions: 94° for 2 minutes followed by 25 cycles of 94° for 30 seconds, 50° for 1 minute, and 72° for 1 minute, with a final extension time of 7 minutes at 72°. A 5μL aliquot of the product was removed for a second round of amplification using the nested FAP 6085F and FAP 6319R primer pair under the same cycling conditions. α-HPV PCR was carried out using the nested primer sets MY09-MY11 and GP5-GP6 as described(Manos et al., 1989; Snijders et al., 2005).
The products were visualized by agarose gel electrophoresis and bands of expected size were isolated using the PureLink Quick Gel Extraction Kit (Invitrogen, Carlsbad, CA) and cloned using the TOPO TA cloning System (Invitrogen). A minimum of 12 colonies were sequenced on the ABI 3130xl Genetic Analyzer (Applied Biosystems, Carlsbad, CA) in order to detect potential multiple infections.
Quantification of HPV Viral Load by Real-time qPCR
Quantitative real-time PCR was performed using the Universal Probe Library (UPL) system (Roche Applied Science, Indianapolis, IN). Primer and probe assay combinations were individually designed for each HPV type and for human β-2-microglobulin (B2M) using the online UPL Design Center software (Roche). For samples with multiple infections, we designed discriminatory assays to measure type-specific viral loads of as many individual types as possible (Table S1). To generate standard curves, assay-specific PCR amplicons were separated on 4% agarose gel and purified using the PureLink Quick Gel Extraction Kit (Invitrogen). Internal standards were generated using 10-fold dilutions of the gel-purified products ranging from 1,000,000 to 10 input copies.
DNA samples were assayed in 20μL reactions with a final concentration of 1× LightCycler 480 Probes Master mix, 400nM of each primer, and 200nM of the UPL probe. Using the LightCycler 480 (Roche), samples were heated to 95° for 10 minutes followed by 45 cycles of 95° for 10 seconds, 60° for 30 seconds, and 70° for one second. Data was analyzed with the LightCycler 480 software. DNA samples and standard dilution series were run in duplicate, and total input copy numbers were calculated using the mean crossing point (Cp) values for each sample. Input cellular equivalents were calculated based on 2 copies of β2M copies per cell, and HPV viral loads were calculated as viral copies per cell.
Statistical Analysis
HPV prevalence was analyzed by logistic regression including age, sex, immunosuppression, tissue sample type, and accounting for clustering of multiple samples within patients. HPV viral loads were analyzed with random-effects interval regression where the HPV copy number was left-censored at the lower limit of detection by qPCR. For both prevalence and viral load, univariate regression was performed prior to multivariate modeling. Statistical analysis was performed using Stata 11 (StataCorp LP, College Station, TX).
mRNA-seq Library Preparation and Analysis
Poly(A)+ RNA was isolated from 3 μg of total RNA using the Oligotex Mini kit (Qiagen) according to manufacturer’s instructions. The resulting poly(A) RNA was then amplified using the MessageAmp II aRNA Amplification Kit (Ambion, Austin, TX) using an in vitro transcription time of 14 hours at 37°C to generate aRNA. 200ng of aRNA was used to generate libraries for transcriptome sequencing using an adaptation of the protocol previously described by Yozwiak et al. (Yozwiak et al.). In order to multiplex up to 16 samples within 1 sequencing library, aRNA samples were randomly primed and reverse transcribed using a primer containing a 14-bp sequence common to the 3′ end of both Illumina adapters, a random monomer followed by a unique 3-bp barcode, and a random hexamer (pr1A_barcode). Second-strand synthesis was primed using pr1A followed by PCR amplification using the 18-bp Illumina/barcode sequence without the hexamer (pr1B_barcode) for 25 cycles. PCR products were purified using DNA Clean and Concentrator columns (Zymo Research, Orange, CA). 200ng of each individually barcoded sample were mixed together to generate a library of up to 16 samples, with each sample marked by a unique 3-bp barcode. Library purification, size selection, and amplification proceeded as previously described (Yozwiak et al.). Three multiplexed transcriptome libraries were analyzed on 3 separate paired-end sequencing runs using the Genome Analyzer II (Illumina, San Diego, CA) and designated Datasets A, B, and C. Barcodes and corresponding samples are listed in Table S2. Each run generated pairs of 65nt reads. This data has been submitted to the NCBI Sequence Read Archive under accession number SRA029929.
Read pairs from each library were sorted by 3nt barcode (nucleotides 2–4 of each read), requiring that at least 1 of the 2 reads from each pair contained a perfect match to an input barcode and that the other contained at most 1 mismatch. This yielded the “total” read counts shown in Table S2. For analysis, we removed from each read the nucleotide preceding the barcode, the barcode itself, the 6 nucleotides deriving from the random hexamer used for priming, and the last nucleotide of the read, yielding 54nt reads.
Background model (BGM) DNA sequence datasets included the human genome (UCSC build hg18; BGMhg)(Fujita et al., 2010; Lander et al., 2001), the human mRNA transcriptome (Representative H-Invitational transcripts, 43,159 records; BGMht)(Imanishi et al., 2004), a collection of sequenced human VDJ recombination products (H. sapiens entries from IMGT release 201028-6 67,611 records; BGMvdj)(Lefranc, 2001), the Illumina paired-end adapter sequences ligated to one another (BGMad), and an in vitro-transcribed Xenopus EF1α message that contaminated Dataset A and was reconstructed from that data (File S1; BGMef1a). Matches to BGMhg, BGMht, and BGMef1a of >80% sequence identity across the entire read length were sought using BLAT (-minIdentity=80 -noTrimA)(Kent, 2002), and matches to those datasets plus BGMvdj and BGMad were sought using Blastn (default settings)(Altschul et al., 1990). Matching sequences and their paired ends were filtered from the query pool, leaving the “host-filtered” read counts shown in Table S2. Barcode AGG from Dataset A was excluded from further analysis due to the majority of reads mapping to the BGMef1a contaminating sequence (not shown). Low-complexity sequences were defined as those generating <30 new additions to the string table during an LZW compression (Welch, 1984) and were removed, leaving the “complexity-filtered” read counts shown in Table S2.
Matches to the remaining reads were sought in a database of all complete viral genome sequences in Genbank (3525 records; 72 million nucleotides; downloaded on 1/18/2010; GI’s listed in File S3) (Benson et al., 2009) using tBlastx (-e 1e-3). Read counts were allocated to the viral genome record with the highest alignment bitscore. In the case of a tie, the read count was initially distributed evenly to all records with equal bitscore matches, then re-assigned to whichever record(s) had the greatest total read count for the given dataset. “HPV-matching” read counts for each sample are shown in Table S2.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCRR/OD UCSF-CTSI Grant Number KL2 RR024130, a Canary Foundation/American Cancer Society Postdoctoral Fellowship for the Early Detection of Cancer, a Dermatology Foundation Career Development Award, and an American Society for Dermatologic Surgery Cutting Edge Research Grant to S.T.A. The authors wish to thank Clement Chu for assistance with deep sequencing, Peter Skewes-Cox for assistance with sequence database collection, Amy J. Markowitz for critical reading of this manuscript and Charles McCulloch for advice on statistical analysis.
Abbreviations
- BGM
Background model
- cuSCC
Cutaneous squamous cell carcinoma
- HPV
Human papillomavirus
- KA
Keratoacanthoma
- MCV
Merkel cell polyomavirus
- OTR
Organ transplant recipient
- SCC
Squamous cell carcinoma
- UPL
Universal probe library
Footnotes
Conflict of Interest
The authors state no conflict of interest.
Sequence Read Archive
This mRNA-seq data has been submitted to the NCBI Sequence Read Archive under accession number SRA029929.
References
- Alam M, Caldwell JB, Eliezri YD. Human papillomavirus-associated digital squamous cell carcinoma: literature review and report of 21 new cases. J Am Acad Dermatol. 2003;48:385–93. doi: 10.1067/mjd.2003.184. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J Virol. 2000;74:11636–41. doi: 10.1128/jvi.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari MM, Kiviat NB, Critchlow CW, Stern JE, Argenyi ZB, Raugi GJ, et al. Detection of human papillomavirus DNA in cutaneous squamous cell carcinoma among immunocompetent individuals. J Invest Dermatol. 2008;128:1409–17. doi: 10.1038/sj.jid.5701227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer HM, Ting Y, Greer CE, Chambers JC, Tashiro CJ, Chimera J, et al. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA. 1991;265:472–7. [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout RJ, Bouwes Bavinck JN, ter Schegget J. Persistence of human papillomavirus DNA in benign and (pre)malignant skin lesions from renal transplant recipients. J Clin Microbiol. 2000;38:2087–96. doi: 10.1128/jcm.38.6.2087-2096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout RJ, Tieben LM, Smits HL, Bavinck JN, Vermeer BJ, ter Schegget J. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J Clin Microbiol. 1995;33:690–5. doi: 10.1128/jcm.33.3.690-695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10:321–2. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- Bouwes Bavinck JN, Neale RE, Abeni D, Euvrard S, Green AC, Harwood CA, et al. Multicenter study of the association between betapapillomavirus infection and cutaneous squamous cell carcinoma. Cancer Res. 2010;70:9777–86. doi: 10.1158/0008-5472.CAN-10-0352. [DOI] [PubMed] [Google Scholar]
- Boxman IL, Berkhout RJ, Mulder LH, Wolkers MC, Bouwes Bavinck JN, Vermeer BJ, et al. Detection of human papillomavirus DNA in plucked hairs from renal transplant recipients and healthy volunteers. J Invest Dermatol. 1997;108:712–5. doi: 10.1111/1523-1747.ep12292090. [DOI] [PubMed] [Google Scholar]
- Boxman IL, Russell A, Mulder LH, Bavinck JN, ter Schegget J, Green A. Association between epidermodysplasia verruciformis-associated human papillomavirus DNA in plucked eyebrow hair and solar keratoses. J Invest Dermatol. 2001;117:1108–12. doi: 10.1046/j.1523-1747.2001.01499.x. [DOI] [PubMed] [Google Scholar]
- Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37:141–50. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Dang C, Koehler A, Forschner T, Sehr P, Michael K, Pawlita M, et al. E6/E7 expression of human papillomavirus types in cutaneous squamous cell dysplasia and carcinoma in immunosuppressed organ transplant recipients. Br J Dermatol. 2006;155:129–36. doi: 10.1111/j.1365-2133.2006.07378.x. [DOI] [PubMed] [Google Scholar]
- de Koning MN, Struijk L, Bavinck JN, Kleter B, ter Schegget J, Quint WG, et al. Betapapillomaviruses frequently persist in the skin of healthy individuals. J Gen Virol. 2007;88:1489–95. doi: 10.1099/vir.0.82732-0. [DOI] [PubMed] [Google Scholar]
- de Koning MN, Weissenborn SJ, Abeni D, Bouwes Bavinck JN, Euvrard S, Green AC, et al. Prevalence and associated factors of betapapillomavirus infections in individuals without cutaneous squamous cell carcinoma. J Gen Virol. 2009;90:1611–21. doi: 10.1099/vir.0.010017-0. [DOI] [PubMed] [Google Scholar]
- Dubina M, Goldenberg G. Viral-associated nonmelanoma skin cancers: a review. Am J Dermatopathol. 2009;31:561–73. doi: 10.1097/DAD.0b013e3181a58234. [DOI] [PubMed] [Google Scholar]
- Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–7. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund O, Iftner T, Andersson K, Lindelof B, Hradil E, Nordin P, et al. Cutaneous human papillomaviruses found in sun-exposed skin: Beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis. 2007;196:876–83. doi: 10.1086/521031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund O, Lindelof B, Hradil E, Nordin P, Stenquist B, Kirnbauer R, et al. High prevalence of cutaneous human papillomavirus DNA on the top of skin tumors but not in “Stripped” biopsies from the same tumors. J Invest Dermatol. 2004;123:388–94. doi: 10.1111/j.0022-202X.2004.23205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund O, Ly H, Higgins G. Improved detection of cutaneous human papillomavirus DNA by single tube nested ‘hanging droplet’ PCR. J Virol Methods. 2003a;110:129–36. doi: 10.1016/s0166-0934(03)00109-5. [DOI] [PubMed] [Google Scholar]
- Forslund O, Ly H, Reid C, Higgins G. A broad spectrum of human papillomavirus types is present in the skin of Australian patients with non-melanoma skin cancers and solar keratosis. Br J Dermatol. 2003b;149:64–73. doi: 10.1046/j.1365-2133.2003.05376.x. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, et al. The complete genome of klassevirus - a novel picornavirus in pediatric stool. Virol J. 2009;6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartevelt MM, Bavinck JN, Kootte AM, Vermeer BJ, Vandenbroucke JP. Incidence of skin cancer after renal transplantation in The Netherlands. Transplantation. 1990;49:506–9. doi: 10.1097/00007890-199003000-00006. [DOI] [PubMed] [Google Scholar]
- Harwood CA, McGregor JM, Proby CM, Breuer J. Human papillomavirus and the development of non-melanoma skin cancer. J Clin Pathol. 1999;52:249–53. doi: 10.1136/jcp.52.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CA, Surentheran T, McGregor JM, Spink PJ, Leigh IM, Breuer J, et al. Human papillomavirus infection and non-melanoma skin cancer in immunosuppressed and immunocompetent individuals. J Med Virol. 2000;61:289–97. doi: 10.1002/1096-9071(200007)61:3<289::aid-jmv2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Hazard K, Karlsson A, Andersson K, Ekberg H, Dillner J, Forslund O. Cutaneous human papillomaviruses persist on healthy skin. J Invest Dermatol. 2007;127:116–9. doi: 10.1038/sj.jid.5700570. [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, et al. Merkel cell polyomavirus infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010 doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. Human Papillomaviruses. IARC Monogr Eval Carcinog Risks Hum. 2007;90:1–636. [PMC free article] [PubMed] [Google Scholar]
- Imanishi T, Itoh T, Suzuki Y, O’Donovan C, Fukuchi S, Koyanagi KO, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P, Hansen S, Moller B, Leivestad T, Pfeffer P, Geiran O, et al. Skin cancer in kidney and heart transplant recipients and different long-term immunosuppressive therapy regimens. J Am Acad Dermatol. 1999;40:177–86. doi: 10.1016/s0190-9622(99)70185-4. [DOI] [PubMed] [Google Scholar]
- Karagas MR, Nelson HH, Sehr P, Waterboer T, Stukel TA, Andrew A, et al. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J Natl Cancer Inst. 2006;98:389–95. doi: 10.1093/jnci/djj092. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuter A, Gambichler T, Pfister H, Wieland U. Diversity of human papillomavirus types in periungual squamous cell carcinoma. Br J Dermatol. 2009;161:1262–9. doi: 10.1111/j.1365-2133.2009.09343.x. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- LeBoit PE. Can we understand keratoacanthoma? Am J Dermatopathol. 2002;24:166–8. doi: 10.1097/00000372-200204000-00012. [DOI] [PubMed] [Google Scholar]
- Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2001;29:207–9. doi: 10.1093/nar/29.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindelof B, Sigurgeirsson B, Gabel H, Stern RS. Incidence of skin cancer in 5356 patients following organ transplantation. Br J Dermatol. 2000;143:513–9. [PubMed] [Google Scholar]
- Luron L, Avril MF, Sarasin A, Daya-Grosjean L. Prevalence of human papillomavirus in skin tumors from repair deficient xeroderma pigmentosum patients. Cancer Lett. 2007;250:213–9. doi: 10.1016/j.canlet.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Manos M, Ting Y, Wright D, Lewis A, Broker T, Wolinsky S. Cancer Cells 7: Molecular Diagnostics of Human Cancer. Cold Spring Harbor Laboratory; 1989. Use of Polymerase Chain Reaction Amplification for the Detection of Genital Human Papillomaviruses; pp. 209–14. [Google Scholar]
- Moy RL, Eliezri YD, Nuovo GJ, Zitelli JA, Bennett RG, Silverstein S. Human papillomavirus type 16 DNA in periungual squamous cell carcinomas. JAMA. 1989;261:2669–73. [PubMed] [Google Scholar]
- Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–44. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- Purdie KJ, Surentheran T, Sterling JC, Bell L, McGregor JM, Proby CM, et al. Human papillomavirus gene expression in cutaneous squamous cell carcinomas from immunosuppressed and immunocompetent individuals. J Invest Dermatol. 2005;125:98–107. doi: 10.1111/j.0022-202X.2005.23635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Lanier M, Triplett C, Campion M. Possible role for human papillomavirus 16 in squamous cell carcinoma of the finger. J Med Virol. 1994;44:369–78. doi: 10.1002/jmv.1890440410. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–15. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanin V, Glover M, Rausch C, Proby C, Leigh IM, zur Hausen H, et al. Specific types of human papillomavirus found in benign proliferations and carcinomas of the skin in immunosuppressed patients. Cancer Res. 1994;54:4610–3. [PubMed] [Google Scholar]
- Shamanin V, zur Hausen H, Lavergne D, Proby CM, Leigh IM, Neumann C, et al. Human papillomavirus infections in nonmelanoma skin cancers from renal transplant recipients and nonimmunosuppressed patients. J Natl Cancer Inst. 1996;88:802–11. doi: 10.1093/jnci/88.12.802. [DOI] [PubMed] [Google Scholar]
- Snijders PJ, van den Brule AJ, Jacobs MV, Pol RP, Meijer CJ. HPV DNA detection and typing in cervical scrapes. Methods Mol Med. 2005;119:101–14. doi: 10.1385/1-59259-982-6:101. [DOI] [PubMed] [Google Scholar]
- Struijk L, Bouwes Bavinck JN, Wanningen P, van der Meijden E, Westendorp RG, Ter Schegget J, et al. Presence of human papillomavirus DNA in plucked eyebrow hairs is associated with a history of cutaneous squamous cell carcinoma. J Invest Dermatol. 2003;121:1531–5. doi: 10.1046/j.1523-1747.2003.12632.x. [DOI] [PubMed] [Google Scholar]
- Surentheran T, Harwood CA, Spink PJ, Sinclair AL, Leigh IM, Proby CM, et al. Detection and typing of human papillomaviruses in mucosal and cutaneous biopsies from immunosuppressed and immunocompetent patients and patients with epidermodysplasia verruciformis: a unified diagnostic approach. J Clin Pathol. 1998;51:606–10. doi: 10.1136/jcp.51.8.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunis A, Andre J, Noel JC. Evaluation of the role of genital human papillomavirus in the pathogenesis of ungual squamous cell carcinoma. Dermatology. 1999;198:206–8. doi: 10.1159/000018114. [DOI] [PubMed] [Google Scholar]
- Vajdic CM, McDonald SP, McCredie MR, van Leeuwen MT, Stewart JH, Law M, et al. Cancer incidence before and after kidney transplantation. JAMA. 2006;296:2823–31. doi: 10.1001/jama.296.23.2823. [DOI] [PubMed] [Google Scholar]
- Weissenborn SJ, Nindl I, Purdie K, Harwood C, Proby C, Breuer J, et al. Human papillomavirus-DNA loads in actinic keratoses exceed those in non-melanoma skin cancers. J Invest Dermatol. 2005;125:93–7. doi: 10.1111/j.0022-202X.2005.23733.x. [DOI] [PubMed] [Google Scholar]
- Welch TA. A Technique for High-Performance Data-Compression. Computer. 1984;17:8–19. [Google Scholar]
- Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–9. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- Yozwiak NL, Skewes-Cox P, Gordon A, Saborio S, Kuan G, Balmaseda A, et al. Human Enterovirus 109: a novel inter-species recombinant enterovirus discovered in acute pediatric respiratory illness in Nicaragua. J Virol. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomavirus infections--a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


