Abstract
Bax inhibitor-1 (BI-1) is an evolutionarily conserved protein that protects cells against endoplasmic reticulum (ER) stress while also affecting the ER stress response. In this study, we examined BI-1-induced regulation of the ER stress response as well as the control of the protein over cell death under ER stress. In BI-1-overexpressing cells (BI-1 cells), proteasome activity was similar to that of control cells; however, the lysosomal fraction of BI-1 cells showed sensitivity to degradation of BSA. In addition, areas and polygonal lengths of lysosomes were greater in BI-1 cells than in control cells, as assessed by fluorescence and electron microscopy. In BI-1 cells, lysosomal pH was lower than in control cells and lysosomal vacuolar H+-ATPase(V-ATPase), a proton pump, was activated, suggesting high H+ uptake into lysosomes. Even when exposed to ER stress, BI-1 cells maintained high levels of lysosomal activities, including V-ATPase activity. Bafilomycin, a V-ATPase inhibitor, leads to the reversal of BI-1-induced regulation of ER stress response and cell death due to ER stress. In BI-1 knock-out mouse embryo fibroblasts, lysosomal activity and number per cell were relatively lower than in BI-1 wild-type cells. This study suggests that highly maintained lysosomal activity may be one of the mechanisms by which BI-1 exerts its regulatory effects on the ER stress response and cell death.
Keywords: Apoptosis, ATPases, Calcium, Cancer Therapy, Cell pH, ER Stress, Glucose, Glucose Metabolism, Glycolysis, H+-ATPase
Introduction
Eukaryotic cells respond to the threat of protein misfolding in the endoplasmic reticulum (ER)3 by activating either the ER stress response or the unfolded protein response (UPR) (1, 2). The ER stress response consists of pathways that inhibit protein synthesis, up-regulate chaperone proteins, and increase protein degradation activity. Initiation of the ER stress response occurs when the quantity of unfolded proteins exceeds the capacity of chaperone proteins. GRP78 normally binds to the N-terminal ends of the following three transmembrane proteins: RNA-dependent protein kinase-like ER kinase, inositol-requiring enzyme 1α (IRE1α), and activating transcription factor 6 (ATF6) (3, 4). Protein kinase-like ER kinase phosphorylates eukaryotic initiation factor 2α (eIF2α) in the regulation of protein translation (5). IRE1 is related to genes involved in the transport of unfolded proteins out of the ER and in their degradation by ER-associated degradation (ERAD) (6). ATF6α can also induce chaperone proteins and precedes IRE1α-mediated production of the ERAD pathway (6). Lysosomal activity is an ERAD II pathway, whereas ERAD I is a proteasome/ubiquitination pathway (7). The ERAD mechanism increases the protein folding capacity by reducing protein folding loads (7, 8), implying that ERAD is a physiological pathway that can regulate ER stress responses (8, 9).
Bax inhibitor-1 (BI-1, also known as a “testis-enhanced gene transcript”) is an anti-apoptotic protein that inhibits the activation of Bax and its translocation to the mitochondria (10). Functionally, BI-1 affects Ca2+ leakage from the ER, as measured by Ca2+-sensitive ER-targeted fluorescent proteins and Ca2+-sensitive dyes (11). BI-1 also regulates the production of reactive oxygen species by inhibiting Bax (12, 13). BI-1 overexpression induces an increase in heme oxygenase-1 expression, which may regulate reactive oxygen species through activation of the Nrf2 transcription factor (14). In a previous publication, we suggested that BI-1 acts as a pH-dependent Ca2+ channel, which increases Ca2+ leakage via a mechanism that depends on both pH and the C-terminal cytosolic region of the protein (15). Cells isolated from BI-1−/− mice exhibit hypersensitivity to the apoptosis induced by ER stress (11) as well as strong ER stress response after ischemia/reperfusion, leading to increased cell death (16). Reduced ER stress responses have been suggested to allow for protection against ER stress by BI-1 (14, 16, 17). However, the ER stress response occurs as an adaptation of signal transduction rather than a direct mediator for cell damage or death (1, 2, 18). Therefore, the reduced ER stress response in BI-1-overexpressing cells should be carefully interpreted. In this study, we examined the way in which BI-1 reduces ER stress response. More specifically, we examined the involvement of ERAD II, a lysosomal pathway, as a possible mechanism of BI-1-associated regulation of the ER stress response and cell death.
EXPERIMENTAL PROCEDURES
Materials
Antibodies against hemagglutinin antigen (HA) were purchased from Cell Signaling Technologies (Beverly, MA). Antibodies against 20 S core proteasome subunit and carbobenzoxy-Leu-Leu-Glu-7-amino-4-methyl coumarin (Z-LLE-AMC) were purchased from Enzo Life Sciences (Farmingdale, NY). Carbobenzoxy-Gly-Gly-Leu-7-amino-4-methyl coumarin (Z-GGL-AMC) and N-succinyl-Leu-Leu-Val-Tyr-7-amino-4-methyl coumarin (suc-LLVY-AMC) were obtained from Bachem (Bubendorf, Switzerland) and Sigma-Aldrich, respectively. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), trypsin, and other tissue culture reagents were supplied by Invitrogen. Bicinchoninic acid (BCA) protein assay reagents were obtained from Pierce Biotechnology. All other chemicals were at least of analytical grade and were purchased from Sigma-Aldrich.
Cell Culture
Human HT1080 fibrosarcoma cells and BI-1+/+ and BI-1−/− mouse embryo fibroblasts (MEFs) were cultured in DMEM supplemented with 10% FBS, 20 mm glucose, 20 mm HEPES, 100 μg/ml streptomycin, and 100 units/ml penicillin. For construction of the BI-1 stable cell line, HT1080 cells were transfected with pcDNA3 or pcDNA3-BI-1-HA plasmid using the SuperFect transfection reagent (Qiagen). Cells were then cultured for 3 weeks in 1 mg/ml G418 (Invitrogen) for selection.
Immunoblotting
Cell lysates were prepared, and the levels of protein expression were measured as described previously (15). Equal amounts of protein were extracted from cells with radioimmune precipitation assay buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 2 mm sodium fluoride, 2 mm EDTA, 0.1% SDS, and protease inhibitor mixture) and then separated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Proteins were then transferred to nitrocellulose membranes. After probing of each membrane with specific primary antibodies, the blot was stripped and then reprobed with a polyclonal antibody against β-actin to confirm equal protein loading and transfer. An enhanced chemiluminescence (ECL) system (Amersham Biosciences, Buckinghamshire, UK) was used for protein detection.
Proteasome Activity
Chymotrypsin-like, trypsin-like, and caspase-like activities of the proteasome were assayed by incubation of cell extracts (30 μg) with 0.1 mm suc-LLVY-AMC, 0.1 mm Z-GGL-AMC, and 0.1 mm Z-LLE-AMC, respectively, in 100 mm Tris-HCl buffer (pH 8.0) containing 5 mm MgCl2, 0.5 mm EDTA, and 2 mm ATP at 37 °C. The fluorescence (excitation = 355 nm, emission = 460 nm) of released AMC was measured continuously with a fluorometer (FLUOStar, BMG Labtech) equipped with a temperature controller. The rate of peptide hydrolysis was then calculated from the slope within a linear range.
Lysosomal Isolation
Lysosomal isolation was performed as follows. Briefly, cells were rinsed in cold STE buffer (0.25 m sucrose, 0.01 m Tris-HCl, 1 mm EDTA, and 0.1% ethanol) and scraped into a 1-ml dish of STE buffer containing protease inhibitors (Sigma-Aldrich). The cell suspension was placed in a cell disruption chamber (Kontes) and disrupted using three passes of 20 min each at 150 p.s.i. The use of this method consistently resulted in disruption of >95% of cells but left lysosomes intact. The suspension was centrifuged at 1,500 rpm to separate postnuclear supernatant from nuclear pellet. The postnuclear supernatant density was raised to 1.15 g/ml through the addition of sucrose and then applied to a sucrose density gradient ranging from 1.28 to 1.00 g/ml. The gradient was centrifuged at 19,400 rpm for 4 h at 4 °C to separate lysosomal fractions based on their buoyant density. Purity of the lysosomal populations was assessed further by Western blotting for markers of cellular organelles.
Lysosomal Protease-induced Protein Lysis Assay
Purified lysosomes were resuspended in buffer A solution (20 mm HEPES, pH 7.5, 100 mm NaCl, 0.1 mm dithiothreitol). Prestained bovine serum albumin (BSA) was encapsulated into artificial membranes consisting of phosphatidylserine and phosphatidic acid (50:50, by a molar ratio) under acidic buffer conditions (50 mm sodium phosphate, pH 4.7, 50 mm NaCl, 0.1 mm dithiothreitol) using a reverse evaporation method as described previously (19). After liposome preparation, membranes were passed through a protein desalting spin column (Pierce) that was equilibrated with buffer A. For analysis of lysosomal protease-induced protein lysis, lysosomal fractions were fused with BSA-encapsulated membranes, and the fusion efficiencies were evaluated using previously described methods (20). The ratio of encapsulated BSA to lysosome (w/w) for fusion was ∼2; in more detail, 60 μg of BSA in liposome was mixed with 30 μg of lysosomal proteins, and fusion was started by adding 1 mm CaCl2. Each indicated time represents the number of minutes for which samples were incubated at 30 °C for vesicle fusion and concomitant substrate (prestained BSA) lysis. Reaction termination and removal of lipid components were undertaken by the addition of a chloroform/methanol (2:1, v/v) solution. Collected protein fractions were analyzed by 12.5% SDS-PAGE for evaluation of BSA degradation. As control experiments, fusion efficiency reached a plateau after a 10-min incubation and was determined to be ∼80%.
Lysosome Activity Assessment with Microscopy
LysoTracker probes are fluorescent acidotropic probes for the labeling and tracking of acidic organelles in live cells. These probes have high selectivity for acidic organelles and are effective for the labeling of live cells (21). Cells were grown in a cell culture dish, rinsed with PBS, and stained with 100 nm LysoTracker Green DND-26 (Molecular Probes, Eugene, OR) in serum-free medium for 30 min at room temperature. Cells were then washed with PBS. Lysosomal intensity and size were analyzed by fluorescence microscopy (Zeiss IM 35, Pleasanton, CA) at 488 nm. Images of red fluorescent cells were acquired using a digital CCD color video camera CCS-212 (Samsung, Seoul, Korea) and then transferred to a compatible computer with a WinFast 3D S680 frame grabber (Leadtek, Taipei, Taiwan).
Lysosomal V-ATPase Activity Assessment with Lysosome Vesicles
For the measurement of lysosomal V-ATPase activity, acridine orange uptake assay was performed with lysosome vesicles (23, 24). The activation buffer contained 6 μm acridine orange, 150 mm KCl, 2 mm MgCl2, and 10 mm Bis-Tris-propane. After achievement of a steady spectrofluorometric baseline, V-ATPase was activated by the addition of ATP (1.4 μm final concentration) and 2.5 μm valinomycin (pH 7.0) (to promote the movement of K+ from the inside to the outside of the lysosome for membrane potential generation). V-ATPase-driven pumping of hydrogen ions into lysosomes (acridine orange dye uptake) was measured by stimulation of intralysosomal fluorescence (excited at 495 nm and recorded at 530 nm), which was determined using a fluorescence system (Photon Technology International).
Separately, isolated lysosomes were placed in a cuvette containing the same activation buffer. Extralysosomal quenching of acridine orange fluorescence was determined using the fluorescence system (Photon Technology International). To clarify V-ATPase activity, stimulation of intralysosomal fluorescence and the quenching of the extralysosomal fluorescence were measured with or without 1 μm bafilomycin.
Measurements of Lysosomal pH and Lysosomal V-ATPase Activity through the Measurement of Lysosomal pH
Intralysosomal pH of Neo and BI-1 cells was measured by FITC-conjugated dextran methods (25). Neo and BI-1 cells grown on coverslips were exposed to 1 mg/ml FITC-conjugated dextran for 24 h at 37 °C in tissue culture medium, washed with DMEM twice, and then incubated at 37 °C for 1 h with 1 μm bafilomycin A in the same medium or for 10 min with ER stress agents. After washing the cells with PBS, the fluorescence emission intensity at 520 nm was measured with excitation at 440 and 490 nm using a spectrofluorometer (PerkinElmer Life Sciences). For the time course experiment, Neo and BI-1 cells were incubated with FITC-dextran, and the change in intralysosomal pH was investigated in serum-free complete medium using a spectrofluorometer at a fluorescence emission of 520 nm with excitation at 450 and 490 nm. The standard curve was generated using 220 μg/ml FITC-dextran in 0.1 m Tris-HCl or sodium acetate buffers at various pH values.
For the measurement of lysosomal V-ATPase activity based on lysosomal pH, after achievement of a steady spectrofluorometric baseline, V-ATPase was activated by the addition of ATP (1.4 μm final concentration) and 2.5 μm valinomycin (pH 7.0). V-ATPase-driven pumping of hydrogen ions into lysosomes (lysosomal pH) was measured in the presence or absence of 1 μm bafilomycin. To clarify V-ATPase activity, bafilomycin was later added to the ATP-(without bafilomycin) treated Neo and BI-1 cells. Relative fluorescence was then compared between Neo and BI-1 cells.
Electron Microscopy
Neo cells, BI-1 cells, and BI-1+/+ and BI-1−/− MEFs were prepared for transmission electron microscopy by fixation in a phosphate-buffered solution (Sorensen's phosphate, pH 5.8) of 2.5% glutaraldehyde with 0.15% sucrose and 2% mannitol for maintenance of proper osmosis for 12–24 h at 4 °C. Cells were postfixed in 2% osmium tetroxide in the same buffer for 1 h. After rinsing, cells were exposed to 2% uranyl acetate (aqueous) and subsequently embedded in 2% agar. Samples were cut from agar blocks and then dehydrated in a graded ethanol series, infiltrated with Spurr's resin, embedded, and cured at 70 °C for 24 h. Ultrathin sections were cut and stained with uranyl acetate and Reynold's lead citrate prior to observation on a Zeiss EM10 transmission electron microscope.
Determination of Caspase-3 Activity
Neo and BI-1 cells were washed with PBS and incubated for 30 min on ice with 100 ml of lysis buffer (10 mm Tris-HCl, 10 mm NaH2PO4/NaHPO4, pH 7.5, 130 mm NaCl, 1% Triton X-100, and 10 mm sodium pyrophosphate). Cell lysates were spun down, supernatants were collected, and protein concentrations were determined using the BCA method. For each reaction, 30 μg of protein was added to 1 ml of freshly prepared protease assay buffer (20 mm HEPES, pH 7.5, 10% glycerol, 2 mm dithiothreitol) containing 20 mm Ac-DEVD-AMC (Sigma-Aldrich). Reaction mixtures without cellular extracts were used as negative controls. Reaction mixtures were incubated for 1 h at 37 °C, and aminomethylcoumarin that was liberated from Ac-DEVD-AMC was determined by spectrofluorometry (Hitachi F-2500) at 380 nm for excitation and at 400–550 nm for emission. Readings were corrected for background fluorescence.
Statistical Analysis
Results are presented as means ± S.E. MicroCal Origin software (Northampton, MA) was used for statistical calculations. p values were determined via Student's t tests. Statistical significance was set at p < 0.05.
RESULTS
The ER Stress Response Is Regulated in BI-1 Cells
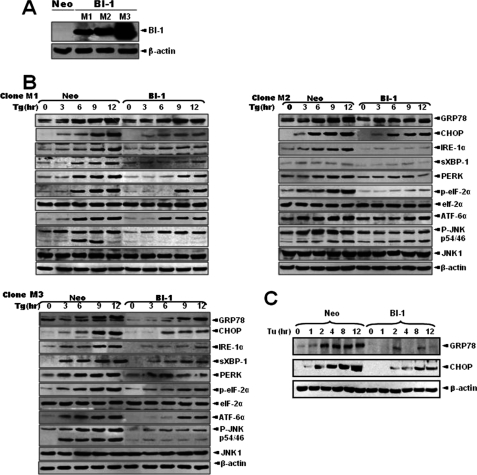
First, the regulatory effect of BI-1 on the ER stress response was confirmed in BI-1-overexpressing HT1080 cells (BI-1 cells). To eliminate the possibility of clonal variation, three independent cell lines (designated as M1, M2, and M3) that overexpress BI-1 were used in this experiment (Fig. 1A). Neo and BI-1 stable transfectant cells were treated with thapsigargin, a Ca2+-ATPase inhibitor, and markers of unfolded protein response were examined. As shown in Fig. 1B, the unfolded protein response was weaker in BI-1 cells than in Neo cells. Induction of ER molecular chaperone glucose-regulated protein (GRP78) and C/EBP homologous protein (CHOP), phosphorylation of eIF-2α, and up-regulation of IRE1 and ATF6 were clearly detected in Neo cells, whereas all of these markers were less detectable in BI-1 cells. ER stress-induced spliced XBP-1 (sXBP-1) protein was also less expressed in BI-1 cells than in Neo cells. In addition, the degree of JNK phosphorylation in Neo cells was more significant than in BI-1 cells, implying reduced unfolded protein response due to BI-1 overexpression.
FIGURE 1.
BI-1 regulates the expression of ER stress proteins. A, HT1080 cells were stably transfected with pcDNA3 or pcDNA3-HA-BI-1 plasmids. Following preparation of cell lysates from monoclonal cells (designated M1, M2, and M3), Western blotting was performed with anti-HA or β-actin antibody. Neo, neomycin-resistant vector-transfected HT1080 cells; BI-1, pcDNA3-HA-BI-1 transfected HT1080 cells. B, Neo and BI-1 monoclonal cells were subjected to 5 μm thapsigargin (Tg) for 0, 3, 6, 9, or 12 h, total proteins were extracted, and the expressions of GRP78, CHOP, IRE1α, spliced XBP-1 (sXBP-1), protein kinase-like ER kinase (PERK), phospho-eIF-2α (p-eIF-2α), eIF-2α, ATF-6α, phospho-JNK (p-JNK), JNK1, and β-actin were analyzed, as described under “Experimental Procedures.” C, representative blots are shown in three panels, respectively. Neo and BI-1 (M1 cells) cells were subjected to 5 μg/ml tunicamycin (Tu) for 0, 1, 2, 4, 8, and 12 h, and then total proteins were extracted and the expressions of GRP78, CHOP, and β-actin were analyzed. Representative blots are shown.
For comparisons with the results of previous studies, Neo and BI-1 (M1) cells were treated with tunicamycin, an N-acetyl glycosylation inhibitor, and inductions of UPR proteins (GRP78 and CHOP) were examined. In accordance with Fig. 1B, GRP78 and CHOP (26) inductions were marginally activated in BI-1 cells when compared with Neo cells (Fig. 1C). The activation of JNK and the phosphorylation of eIF2α were reduced in BI-1 cells, as expected (data not shown, 14).
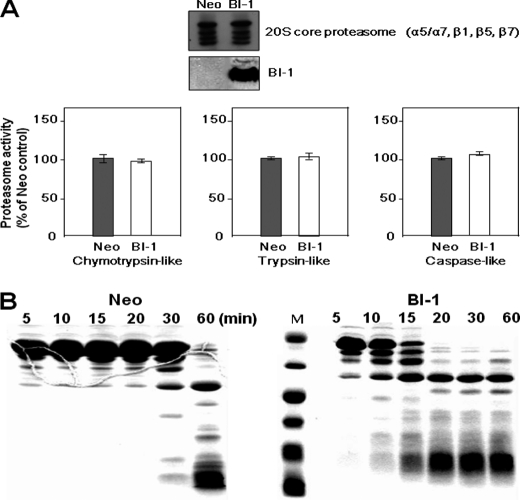
Lysosomal, but Not Proteasomal, Degradation Activity Is Activated in BI-1 Cells
Under ER stress conditions, cells ignite the process of UPR to regulate protein synthesis, enhance the protein degradation capacity and relieve the load of protein in ER (6, 7), and finally adapt to the environmental changes. To examine the role of BI-1 in reduced ER stress response, protein synthesis in Neo and BI-1 cells was compared. However, Neo and BI-1 cells showed similar rates of protein synthesis (supplemental Fig. 1). We next examined the activities of certain protein degradation pathways, namely proteasomal and lysosomal, in Neo and BI-1 cells. The 20 S core proteasome (α5/α7, β1, β5, and β7) complex showed similar activities as chymotrypsin-like, trypsin-like, and caspase-like enzymes in both cell types (Fig. 2A). Next, for analysis of lysosomal protease-induced protein lysis, lysosomal protease activity was determined using BSA-encapsulated membranes. Liposomal BSA was mixed with lysosomal proteins and then incubated at 30 °C for vesicle fusion and concomitant substrate lysis. SDS-PAGE analysis showed that BSA degradation activity was stronger in BI-1 cells than in Neo cells, suggesting that BI-1 cells have stronger lysosomal protease activity than Neo cells (Fig. 2B).
FIGURE 2.
Lysosomal, but not proteasomal, degradation is activated in BI-1-overexpressing cells. A, in both Neo and BI-1 cells, the expressions of proteasome proteins and BI-1 were measured by Western blotting using an antibody against the 20 S core proteasome and HA. Proteasomal activities were calculated by the measurement of peptidolytic activities (chymotrypsin-like, trypsin-like, and caspase-like peptidases). Fluorogenic substrates were incubated with cell extracts for 30 min in the presence of 2 mm ATP, and proteasomal activity was determined by the measurement of fluorescence. Each value is expressed as the mean ± S.E. of three independent experiments. B, after isolation of the lysosomal fraction from Neo and BI-1 cells, 60 μg of BSA in liposome was mixed with 30 μg of lysosomal proteins, and fusion was performed by the addition of 1 mm CaCl2 during the indicated time periods. The collected protein fractions were analyzed by 12.5% SDS-PAGE for evaluation of BSA degradation, as described under “Experimental Procedures.” M, molecular marker.
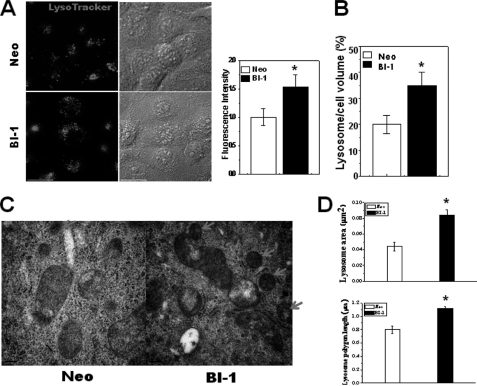
Lysosome Area and Polygon Length Are Increased in BI-1 Cells
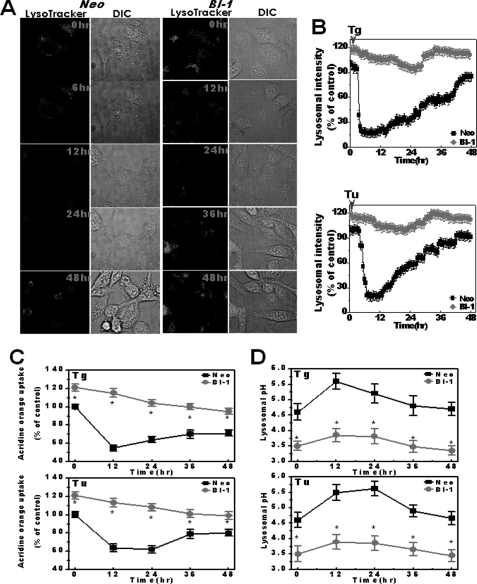
Lysosome morphology is known to be related to lysosomal activity and its functions (29). Therefore, morphological studies are also necessary to confirm the role of lysosome in BI-1 cells. First, we loaded LysoTracker into cells. LysoTracker probes are fluorescent acidotropic probes for the labeling and tracking of acidic organelles in live cells with high selectivity. The fluorescence intensity of LysoTracker is considered to be a marker for lysosomal activity (30, 31). Under baseline conditions, LysoTracker was located at large vesicles in the cytoplasm, showing higher fluorescence intensity in BI-1 cells than in Neo cells (Fig. 3A). The fluorescence intensity quantification results are shown in Fig. 3A (right). BI-1 cells also showed larger lysosome volumes per cell volume than Neo cells (Fig. 3B). Electron microscopy analysis indicated that the lysosome area was larger and the polygon length was longer in BI-1 cells than in Neo cells (Fig. 3C). Quantification data are also shown (Fig. 3D).
FIGURE 3.
Lysosome area and polygon length are increased in BI-1-overexpressing cells. A, Neo and BI-1 cells were exposed to 5 μm LysoTracker and photographed. Fluorescence intensity was quantified (right). B, lysosome volume per cell was also analyzed and quantified. C and D, electron microscopy analysis was performed (C), and the lysosomal area and polygonal length were quantified (D). The arrow in panel C indicates lysosomes. *, p < 0.05; significantly different from Neo cells. In panels A, B, and D, each value is expressed as the mean ± S.E. of three independent experiments.
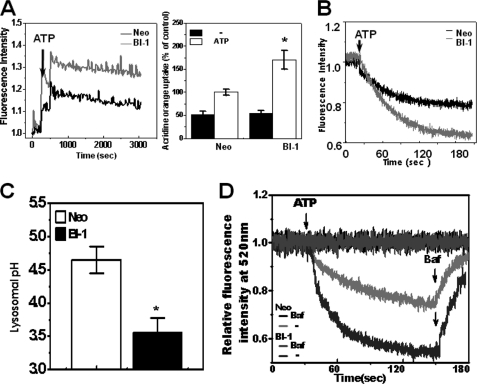
H+ Uptake Is Enhanced, and Lysosomal pH Is Decreased in BI-1 Cells
Acridine orange has been employed to study the acidification of cytoplasmic vesicles. Accumulation of protonated acridine orange in acidic compartments was identified by the resultant orange to red fluorescence emission. Acridine orange was applied to isolated lysosome membranes from Neo and BI-1 cells, and the lysosomal H+ uptake activity was analyzed. In the presence of ATP, H+ uptake was highly increased, especially in the lysosome membranes of BI-1 cells (Fig. 4A). The peak fluorescence of acridine orange dye was normalized, based upon that of Neo cells in the presence of ATP (Fig. 4A, right). Separately, extralysosomal fluorescence quenching was measured in the presence of ATP. In BI-1 cells, the acridine orange fluorescence was more significantly quenched when compared with Neo cells (Fig. 4B). This finding is consistent with the intralysosomal stimulation of acridine orange fluorescence in BI-1 cells (Fig. 4A). In addition, lysosomal pH was also lower in BI-1 cells than in Neo cells (Fig. 4C), in agreement with the lysosomal activity in BI-1 cells (Fig. 4, A and B). With the lysosomal pH analysis, lysosomal H+ uptake was compared between Neo and BI-1 cells. When exposed to ATP, FITC-dextran fluorescence was more sharply decreased in BI-1 cells than in Neo cells, suggesting that the lysosomal V-ATPase activity was higher in BI-1 cells than in Neo cells (Fig. 4D). The inhibition of the lysosomal activity was clearly shown in the presence of bafilomycin, a potent and specific V-ATPase inhibitor (32, 33).
FIGURE 4.
BI-1 enhances lysosomal activity. A, 6 μm acridine orange solution and 2.5 μm valinomycin were added to lysosomal membranes from Neo and BI-1 cells; intravesicular H+ uptake was initiated by the addition of Mg-ATP, and the uptake ratio was quantified. *, p < 0.05; significantly different from ATP-exposed Neo cells. B, separately, extravesicular H+ uptake was also initiated by the addition of Mg-ATP, and the fluorescence quenching was measured as described under “Experimental Procedures.” C, lysosomal pH was measured as described under “Experimental Procedures.” *, p < 0.05; significantly different from Neo cells. D, Neo and BI-1 cells were exposed to ATP with or without 1 μm bafilomycin, and lysosomal pH was measured (as described for panel C). During the measurement of lysosomal pH, bafilomycin (Baf) was added to the ATP treated Neo and BI-1 cells. In panels A and C, each value is expressed as the mean ± S.E. of three independent experiments.
H+ Uptake and Acidic Lysosomal pH Are Stable after ER Stress in BI-1 Cells
ER-associated degradation pathways, along with proteasomal and lysosomal activities, needed to be examined after exposure to ER stress. First, proteasomal activity was compared between Neo and BI-1 cells exposed to ER stress. Proteasomal activities, including chymotrypsin, trypsin, or caspase-like enzymes, were similar between Neo and BI-1 cells (supplemental Fig. 2, A–C). In addition, the 20 S proteasome protein expression was not different with or without ER stress (supplemental Fig. 3, upper and lower). Next, we examined the other ER-associated degradation mechanism, lysosomal activity, with LysoTracker dye. Similar to Fig. 3A, basal fluorescence was higher in BI-1 cells than in Neo cells (Fig. 5A). However, fluorescence was sharply decreased in Neo cells when cells were exposed to thapsigargin. The fluorescence showed recovery to normal levels in Neo cells with the passage of time. However, in BI-1 cells, the high fluorescence of the lysosome was stable even after exposure to ER stress caused by thapsigargin (Fig. 5A). Fig. 5B shows the quantification result of fluorescence in either thapsigargin-treated or tunicamycin-treated Neo and BI-1 cells. As shown in Fig. 5B, lower panel, the fluorescence of LysoTracker in BI-1 cells remained stably high as well. Then, with acridine orange uptake analysis, H+ uptake activity was measured after exposure to ER stress. During the incubation period of 12 h, activity was sharply decreased in Neo cells and slowly recovered to the normal range with the passage of time. In contrast, BI-1 cells stably maintained high H+ uptake throughout the duration of incubation (Fig. 5C). Consistently, BI-1 cells showed more stable acidic lysosomal pH (Fig. 5D) when compared with Neo cells, which showed transiently increased pH followed by recovery of acidic pH. These data suggest that the enhanced lysosomal function of BI-1 cells was little affected, even when exposed to ER stress.
FIGURE 5.
BI-1 maintains high lysosomal activity under ER stress. A, Neo and BI-1 cells were subjected to 5 μm thapsigargin during a period of 48 h followed by exposure to 5 μm LysoTracker and image acquisition. DIC, differential interference contrast microscopy. B, fluorescence intensity in either 5 μm thapsigargin-treated (Tg) or 5 μg/ml tunicamycin-treated (Tu) Neo and BI-1 cells was quantified. C, acridine orange solution and valinomycin were added to cell monolayers; intravesicular H+ uptake was initiated by the addition of Mg-ATP, and the uptake ratio was quantified at the following time points: 12, 24, 36, and 48 h. D, lysosomal pH was measured at the following time points: 12, 24, 36, and 48 h. *, p < 0.05; significantly different from each time point in Neo cells. In panels C and D, each value is expressed as the mean ± S.E. of three independent experiments.
V-ATPase Inhibitor Reverses the Effects of BI-1, Which Are Reduced ER Stress Response and Protection against ER Stress
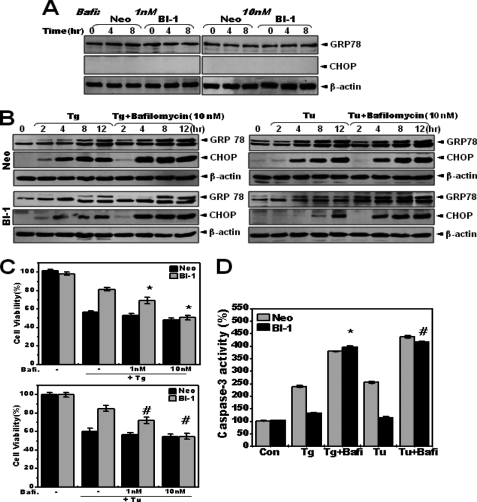
V-ATPase is the primary enzyme that maintains the acidic pH of the lysosome compartment (34). To understand the role of lysosomes in the response of the cell to ER stress and following cell death, bafilomycin, a V-ATPase inhibitor, was utilized. First, for determining the appropriate concentration of bafilomycin for the inhibition of V-ATPase, the response of cells to bafilomycin at concentrations ranging from 10−1 to 10−8 m was examined for inhibitory effect on V-ATPase. Consistent with previous reports (32, 33), the 50% inhibitory concentration (IC50) of bafilomycin for V-ATPase was ∼10–100 nm (supplemental Fig. 4).
Because lysosomal inhibition itself may affect ER function, various concentrations of bafilomycin, from 10−3 to 10 μm, were applied to Neo and BI-1 cells. A single treatment with relatively high concentrations of bafilomycin, 1–10 μm, induced more significant increase in expression of GRP78 and CHOP in BI-1 cells than in Neo cells (supplemental Fig. 5), suggesting that BI-1 cells show greater sensitivity to inhibition of V-ATPase than Neo cells. Relatively low concentrations of bafilomycin (1 and 10 nm) did not induce unfolded protein response (supplemental Fig. 5 and Fig. 6A). Low concentrations of bafilomycin were also effective against V-ATPase activity (supplemental Fig. 4) and were used for inhibition of V-ATPase activity in this study. Neo and BI-1 cells were pretreated with an effective concentration of bafilomycin (10 nm). They were then treated with thapsigargin or tunicamycin. Reduced ER stress response of BI-1 cells was reversed due to inhibition of V-ATPase activity by bafilomycin (Fig. 6B). With a different concentration of bafilomycin, 100 nm, the regulatory effect was clear, especially in BI-1 cells (supplemental Fig. 6).
FIGURE 6.
Bafilomycin reverses the BI-1-regulated ER stress response. A, Neo and BI-1 cells were treated with 1 or 10 nm bafilomycin (Bafi) for the indicated times, and Western blotting was performed with anti-GRP78, CHOP, or β-actin antibody. B, Neo and BI-1 cells were subjected to 5 μm thapsigargin (Tg) or 5 μg/ml tunicamycin (Tu) with or without 10 nm bafilomycin for 0, 2, 4, 8, and 12 h, and Western blotting was performed with anti-GRP78, CHOP, or β-actin antibody. Representative blots for thapsigargin and tunicamycin are shown in the left and right panels, respectively. Neo and BI-1 cells were subjected to 5 μm thapsigargin (left) or 5 μg/ml tunicamycin (right) with or without the indicated concentrations of bafilomycin for 12 h. C and D, cell viability (C) and caspase-3 activity (D) were analyzed. Data represent the mean ± S.E. (n = 4). *, p < 0.05; significantly different from thapsigargin-treated BI-1 cells, #, p < 0.05; significantly different from tunicamycin-treated BI-1 cells. In panels C and D, each value is expressed as the mean ± S.E. of three independent experiments. Con, control.
Next, the protective effect of BI-1 against ER stress was also examined with or without bafilomycin. Bafilomycin reversed the protective effect of BI-1 cells against ER stress (Fig. 6C). Consistently, reduced caspase-3 activity of BI-1 cells was significantly reversed in the presence of bafilomycin (Fig. 6D). These data suggest that enhanced V-ATPase activity is necessary for BI-1-induced regulation of the ER stress response as well as protection against ER stress-induced cell death.
Lysosomal Activity Is Decreased in BI-1 Knock-out Cells
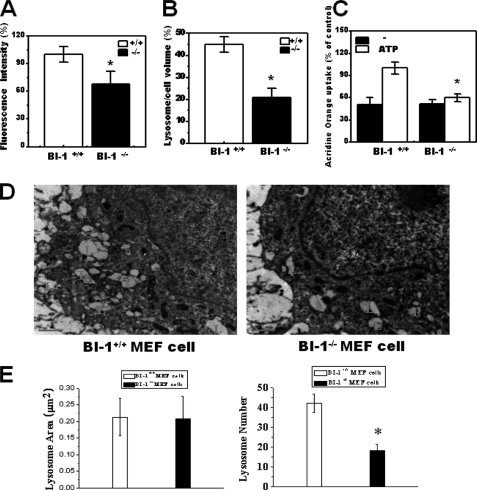
We next examined the lysosomal phenotypes of BI-1 knock-out MEF cells. BI-1 knock-out cells showed lower LysoTracker intensity and lysosome volume per cell (Fig. 7, A and B). Lysosomal H+ uptake was more active in wild-type cells than in BI-1 knock-out cells, indicating an endogenous role of BI-1 in lysosomal H+ uptake (Fig. 7C). EM analysis was performed for determination of the morphological differences between BI-1+/+ and BI-1−/− MEF cells. Although the lysosomal areas were similar, the number of lysosomes was greater in BI-1+/+ cells than in BI-1−/− cells (Fig. 7D). The quantification is also shown in Fig. 7E. These data are consistent with BI-1 overexpression data, suggesting that BI-1 is a key molecule in the determination of lysosomal morphology and activity.
FIGURE 7.
Lysosome activity and lysosomal number are decreased in BI-1 knock-out cells. BI-1 +/+ and BI-1 −/− cells were exposed to 5 μm LysoTracker, and images were captured. A and B, fluorescence intensity (A) and area per cell (B) were quantified. C, acridine orange solution and valinomycin were added to lysosomal membranes; intravesicular H+ uptake was measured in the presence or absence of Mg-ATP, and the uptake ratio was quantified. D and E, electron microscopy was performed (D), and the lysosome area and the number of lysosome per cell were quantified (E). In panel D, L indicates lysosomes. *, p < 0.05; significantly different from BI-1+/+ MEF cells. In panels A–C and E, each value is expressed as the mean ± S.E. of three independent experiments.
DISCUSSION
Here, we identified a regulatory role for BI-1 in the ER stress response and cell death pathways through the lysosome-associated ERAD pathway. The proteasomal activities of BI-1 cells were similar to those of Neo cells; however, lysosomal activities, including V-ATPase activity, were enhanced in BI-1 cells with a stronger H+ uptake than in Neo cells. In this study, increased V-ATPase activity is suggested to regulate ER stress response and cell death in BI-1 cells.
Theoretically, BI-1 might function by reducing protein synthesis or by increasing chaperone expression or degradation to increase the protein folding capacity and/or decrease the protein folding load as an adaptation mechanism to ER stress, known as the UPR (35, 36). The reduction of protein synthesis would itself lead to reduction of the UPR (36). However, Neo and BI-1 cells showed similar rates of protein synthesis (supplemental Fig. 1). BI-1 may also increase chaperone proteins for the enhancement of protein folding capacity. However, both cell lines showed similar inductions in chaperone genes (data not shown). BI-1 may also reduce UPR by increasing proteolytic degradation and subsequently decreasing the protein folding burden. Slowly folding and folding-incompetent substrates are extracted from chaperone folding machinery and targeted for proteolytic degradation via two routes. The first is retro-translocation of the unfolded polypeptide chain into the cytosol followed by ubiquitination and proteasomal degradation in a process termed ERAD. The second is targeting of parts of the ER to lysosomes or vacuoles in autophagy (37, 38). ER stress also induces autophagy, explaining the protective role against ER stress (39). ER stress induced a significant increase in vesicle formation and conversion of LC3-II in different clones of BI-1 cells (supplemental Fig. 7, A and B), suggesting that ERAD II is highly activated in BI-1 cells. Lysosome ERAD II is an alternative ERAD system for the degradation of excess mutant proteins when the ubiquitin/proteasome ERAD I is unaffected. Proteasome activity was not changed in BI-1 cells (Fig. 2A and supplemental Fig. 2). Although non-lysosomal functions are required for the degradation of short-lived proteins in the cytosol as well as for the stress-induced enhancement of degradation of cellular proteins within lysosomes (40), lysosomal function appears to reflect the reduced ER stress response in BI-1 cells. In BI-1 cells, lysosomal proteolysis, such as degradation of BSA, was greatly enhanced (Fig. 2B) and lysosomal H+ uptake increased (Fig. 4A). Lysosome volume was also markedly larger in BI-1 than in Neo cells (Fig. 3, A–C), showing a correlation between lysosomal structure and activity. The question remains as to how BI-1 induces increases in lysosomal V-ATPase activity. We recently showed that the acidic environment in BI-1 cells is related to mitochondrial dysfunction (41). Increased glucose anaerobic metabolism, leading to H+ production, sodium hydrogen exchanger activity, and lactate production, has also been demonstrated in BI-1 cells. The intracellular accumulation of H+ may induce the activation of V-ATPase, leading to acidic lysosomal pH along with sodium hydrogen exchanger activation for intracellular pH maintenance. Another function, the Ca2+ leak channel-like effect of BI-1 on the ER (11, 15), affects cationic balance through continuous leakage of Ca2+ from the ER. The Ca2+ dynamically active status of BI-1 cells may activate the intracellular Ca2+/H+ antiporter and other H+-associated transporter systems (41, 42). Increasing cytoplasmic H+ may activate lysosomal V-ATPase for intracellular pH homeostasis in BI-1 cells (Fig. 4). BI-1 may also be located on organelles other than the ER, such as the lysosome, where it may directly affect activity. However, further study is required to more clearly identify the role of BI-1 localization.
Throughout this study, observation of high lysosomal activity explained how BI-1 regulates the ER stress response. Basally increased lysosomal function is a key phenomenon of BI-1-overexpressing cells (Figs. 2B, 3, and 4). When exposed to stress, the protein may reduce the folding load of altered/damaged proteins. Our results also demonstrated a clear but transient reduction in lysosomal activity upon exposure to ER stress (Fig. 5, A–C). In BI-1 cells, lysosomal activity changed little despite exposure to ER stress. These data suggest that regulation of the ER stress response is related to the highly activated lysosomal activity of BI-1 cells. Throughout this study, we used high glucose (20 mm)-containing medium rather than 5 mm glucose-containing medium. In high glucose medium, glucose metabolism is more highly activated in BI-1 cells than in Neo cells. Consequently, the lysosomal pH may be easily reduced, and lysosomal degradation activity may also be increased in BI-1 cells. In BI-1-knock-out MEF cells, lysosomal activity is relatively lower than in wild-type MEF cells in high glucose medium. Therefore, the lysosomal degradation pathway (ERAD II) appears to be more increased in medium containing a high concentration of glucose. Although negative and specific interactions of BI-1 with IRE1α in the regulation of the IRE1 pathway have recently been reported (43), BI-1 does not appear to be specifically related to IRE1α, at least in our system where glucose metabolism was highly activated. We should also consider that eIF2α and other signaling pathways are also regulated in BI-1-overexpressing cells (Fig. 1, B and C). Therefore, we were not able to provide a careful and extensive discussion of the different aspects of BI-1 specificity on the IRE1 pathway in this state. A large amount of evidence should be carefully compared and studied with regard to BI-1 characteristics in various conditions and different disease states.
In this study, bafilomycin, a lysosomal V-ATPase inhibitor, was used for the functional study of increased lysosomal V-ATPase activity in BI-1 cells. Bafilomycin-induced cell death and the ER stress response were more enhanced in BI-1 cells than in Neo cells (data not shown, supplemental Fig. 6). In this study, the presence of bafilomycin reversed the lower expressions of ER stress proteins, especially in BI-1 cells (Fig. 6B). In addition, bafilomycin induced reversal of the protective effect of BI-1 cells against ER stress. There were fewer effects on cell death in Neo cells (Fig. 6, C and D). Lysosomal activity-associated protein degradation also functions as a cytoplasmic quality control mechanism for the elimination of protein aggregates and damaged organelles (8, 27). Similar to the role of bafilomycin in this experiment, defects in the ERAD II system can cause the accumulation of cytoplasmic inclusion bodies and protein aggregates in the cytoplasm, leading to toxicity (28). Results of this study suggest that lysosomal activation by BI-1 is a key mechanism in the regulatory function of ER stress and in the protective function of BI-1 against ER stress-induced cell death.
In summary, upon exposure to ER stress, BI-1 reduces UPR through the enhancement of lysosomal activity. BI-1 protects cells via lysosome activation, suggesting a novel mechanism of regulation of the ER stress response and cell death.
Supplementary Material
This work was supported by Grant R01-2007-000-20275-0 from the Korea Science and Engineering Foundation (KOSEF). This work was also supported in part by a National Research Foundation of Korea grant funded by the Korean government (Grant 2010-0029497).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7.
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- BI-1
- BAX inhibitor-1
- UPR
- unfolded protein response
- GRP78
- glucose response protein 78
- IRE1α
- inositol-requiring enzyme 1α
- ATF6
- activating transcription factor 6
- CHOP
- C/EBP homologous protein
- C/EBP
- CCAAAT enhancer-binding protein
- V-ATPase
- vacuolar H+-ATPase
- MEF
- mouse embryonic fibroblast
- suc
- succinyl
- Z
- carbobenzoxy
- AMC
- 7-amino-4-methyl coumarin
- Ac
- aminomethylcoumarin
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hersey P., Zhang X. D. (2008) Pigment Cell Melanoma Res. 21, 358–367 [DOI] [PubMed] [Google Scholar]
- 3. Kim R., Emi M., Tanabe K., Murakami S. (2006) Apoptosis 11, 5–13 [DOI] [PubMed] [Google Scholar]
- 4. Szegezdi E., Logue S. E., Gorman A. M., Samali A. (2006) EMBO Rep. 7, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carnevalli L. S., Pereira C. M., Jaqueta C. B., Alves V. S., Paiva V. N., Vattem K. M., Wek R. C., Mello L. E., Castilho B. A. (2006) Biochem. J. 397, 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Foufelle F., Ferré P. (2007) Med. Sci. (Paris) 23, 291–296 [DOI] [PubMed] [Google Scholar]
- 7. Ishida Y., Yamamoto A., Kitamura A., Lamandé S. R., Yoshimori T., Bateman J. F., Kubota H., Nagata K. (2009) Mol. Biol. Cell 20, 2744–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujita E., Kouroku Y., Isoai A., Kumagai H., Misutani A., Matsuda C., Hayashi Y. K., Momoi T. (2007) Hum. Mol. Genet. 16, 618–629 [DOI] [PubMed] [Google Scholar]
- 9. Friedlander R., Jarosch E., Urban J., Volkwein C., Sommer T. (2000) Nat. Cell. Biol. 2, 379–384 [DOI] [PubMed] [Google Scholar]
- 10. Xu Q., Reed J. C. (1998) Mol. Cell 1, 337–346 [DOI] [PubMed] [Google Scholar]
- 11. Chae H. J., Kim H. R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., Banares S., Cui J., Digicaylioglu M., Ke N., Kitada S., Monosov E., Thomas M., Kress C. L., Babendure J. R., Tsien R. Y., Lipton S. A., Reed J. C. (2004) Mol. Cell 15, 355–366 [DOI] [PubMed] [Google Scholar]
- 12. Kawai-Yamada M., Ohori Y., Uchimiya H. (2004) Plant Cell 16, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baek D., Nam J., Koo Y. D., Kim D. H., Lee J., Jeong J. C., Kwak S. S., Chung W. S., Lim C. O., Bahk J. D., Hong J. C., Lee S. Y., Kawai-Yamada M., Uchimiya H., Yun D. J. (2004) Plant Mol. Biol. 56, 15–27 [DOI] [PubMed] [Google Scholar]
- 14. Lee G. H., Kim H. K., Chae S. W., Kim D. S., Ha K. C., Cuddy M., Kress C., Reed J. C., Kim H. R., Chae H. J. (2007) J. Biol. Chem. 282, 21618–21628 [DOI] [PubMed] [Google Scholar]
- 15. Kim H. R., Lee G. H., Ha K. C., Ahn T., Moon J. Y., Lee B. J., Cho S. G., Kim S., Seo Y. R., Shin Y. J., Chae S. W., Reed J. C., Chae H. J. (2008) J. Biol. Chem. 283, 15946–15955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bailly-Maitre B., Fondevila C., Kaldas F., Droin N., Luciano F., Ricci J. E., Croxton R., Krajewska M., Zapata J. M., Kupiec-Weglinski J. W., Farmer D., Reed J. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2809–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krajewska M., Xu L., Xu W., Krajewski S., Kress C. L., Cui J., Yang L., Irie F., Yamaguchi Y., Lipton S. A., Reed J. C. (2011) Brain Res. 1370, 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hetz C. A. (2007) Antioxid. Redox Signal. 9, 2345–2355 [DOI] [PubMed] [Google Scholar]
- 19. Szoka F., Jr., Papahadjopoulos D. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 4194–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corazzi L., Pistolesi R., Arienti G. (1991) J. Neurochem. 56, 207–212 [DOI] [PubMed] [Google Scholar]
- 21. Trivedi N. S., Wang H. W., Nieminen A. L., Oleinick N. L., Izatt J. A. (2000) Photochem. Photobiol. 71, 634–639 [DOI] [PubMed] [Google Scholar]
- 22. Deleted in proof.
- 23. Cox B. E., Griffin E. E., Ullery J. C., Jerome W. G. (2007) J. Lipid Res. 48, 1012–1021 [DOI] [PubMed] [Google Scholar]
- 24. Crider B. P., Xie X. S. (2003) J. Biol. Chem. 278, 44281–44288 [DOI] [PubMed] [Google Scholar]
- 25. Ohkuma S., Poole B. (1978) Proc. Natl. Acad. Sci. U.S.A. 75, 3327–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohoka N., Yoshii S., Hattori T., Onozaki K., Hayashi H. (2005) EMBO J. 24, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klionsky D. J., Emr S. D. (2000) Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 29. Hamanaka R., Shinohara T., Yano S., Nakamura M., Yasuda A., Yokoyama S., Fan J. Q., Kawasaki K., Watanabe M., Ishii S. (2008) Biochim. Biophys. Acta 1782, 408–413 [DOI] [PubMed] [Google Scholar]
- 30. Yoon J., Kim K. J., Choi Y. W., Shin H. S., Kim Y. H., Min J. (2010) Mol. Cell. Biochem. 340, 175–178 [DOI] [PubMed] [Google Scholar]
- 31. Arsham A. M., Neufeld T. P. (2009) PLoS One 4, e6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Youmans S. J., Barry. C. R. (2001) J. Exp. Biol. 204, 2911–2919 [DOI] [PubMed] [Google Scholar]
- 33. Farina C., Gagliardi S. (2002) Curr. Pharm. Des. 8, 2033–2048 [DOI] [PubMed] [Google Scholar]
- 34. Kawai A., Uchiyama H., Takano S., Nakamura N., Ohkuma S. (2007) Autophagy 3, 154–157 [DOI] [PubMed] [Google Scholar]
- 35. Welihinda A. A., Tirasophon W., Kaufman R. J. (1999) Gene Expr. 7, 293–300 [PMC free article] [PubMed] [Google Scholar]
- 36. Yan W., Frank C. L., Korth M. J., Sopher B. L., Novoa I., Ron D., Katze M. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15920–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshida Y., Tanaka K. (2010) Biochim. Biophys. Acta 1800, 172–180 [DOI] [PubMed] [Google Scholar]
- 38. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 39. Kim K. W., Moretti L., Mitchell L. R., Jung D. K., Lu B. (2010) Oncogene 29, 3241–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kurz T., Terman A., Gustafsson B., Brunk U. T. (2008) Histochem. Cell Biol. 129, 389–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee G. H., Yan C., Shin S. J., Hong S. C., Ahn T., Moon A., Park S. J., Lee Y. C., Yoo W. H., Kim H. T., Kim D. S., Chae S. W., Kim H. R., Chae H. J. (2010) Oncogene 29, 2130–2141 [DOI] [PubMed] [Google Scholar]
- 42. Ahn T., Yun C. H., Kim H. R., Chae H. J. (2010) Cell Calcium 47, 387–396 [DOI] [PubMed] [Google Scholar]
- 43. Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J. H., Walter P., Reed J. C., Glimcher L. H., Hetz C. (2009) Mol. Cell 33, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.