Abstract
The transmissible spongiform encephalopathies are characterized by conversion of a host protein, PrPC (cellular prion protein), to a protease-resistant isoform, PrPSc (prion protein scrapie isoform). The importance of the highly flexible, N-terminal region of PrP has recently become more widely appreciated, particularly the biological activities associated with its metal ion-binding domain and its potential to form a poly(l-proline) II (PPII) helix. Circular dichroism spectroscopy of an N-terminal peptide, PrP37–53, showed that the PPII helix is formed in aqueous buffer; as it also contains an Xaa–Pro–Gly consensus sequence, it may act as a substrate for the collagen-modifying enzyme prolyl 4-hydroxylase. Direct evidence for this modification was obtained by mass spectrometry and Edman sequencing in recombinant mouse PrP secreted from stably transfected Chinese hamster ovary cells. Almost complete conversion of proline to 4-hydroxyproline occurs specifically at residue Pro44 of this murine protein; the same hydroxylated residue was detected, at lower levels, in PrPSc from the brains of scrapie-infected mice. Cation binding and/or post-translational hydroxylation of this region of PrP may regulate its role in the physiology and pathobiology of the cell.
Keywords: 4-hydroxyproline/polyproline II helix/post-translational modifications/prion protein/prolyl 4-hydroxylase
Introduction
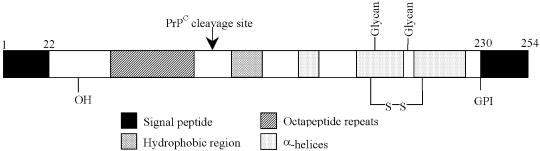
Transmissible spongiform encephalopathies (TSEs) are a family of neurodegenerative disorders that include scrapie of sheep, bovine spongiform encephalopathy (BSE) and Creutzfeldt–Jakob disease (CJD). TSEs are invariably fatal. One of the key processes in their pathogenesis is the conversion of a normal, cell-surface host protein, PrPC (cellular prion protein), to a partially protease-resistant form, PrPSc (prion protein scrapie isoform). PrPC has 208–220 amino acid residues depending on species, two N-glycan linkage sites of variable occupancy and a C-terminal glycosyl-phosphatidylinositol (GPI) membrane anchor (Wopfner et al., 1999) (Figure 1). The C-terminal half of the molecule has a compact three-helix bundle structure while the remaining 80–100 N-terminal amino acids, including a metal ion-binding repeat motif (Viles et al., 1999), adopt a more flexible, largely unstructured shape (Donne et al., 1997). PrPC and PrPSc appear to have the same covalent structure: they are the same in terms of molecular size (Mr 33 000–35 000), N-terminal sequence (Hope et al., 1986), N-glycan structures (Rudd et al., 1999; Stimson et al., 1999), disulfide bridge (Turk et al., 1988) and GPI membrane anchor (Stahl et al., 1992). PrPSc appears to be simply an aggregated, conformational isomer of PrPC. The N-terminal segment of PrP is lost on in vitro proteolytic digestion of PrPSc to the core structure, PrP83–231 (Mr 27 000–30 000). The importance of the N-terminal region has largely been overlooked because it does not appear to be strictly necessary for replication of prions (Fischer et al., 1996). However, most, if not all, PrPSc that accumulates in affected brain includes the mature PrPC N-terminal sequences (Hope et al., 1986; Jeffrey et al., 1996) and the role of this domain of PrP in prion diseases has recently become more widely appreciated (Zulianello et al., 2000).
Fig. 1. Schematic diagram of structural features of PrP (prion protein) including N- and C-terminal signal peptides, metal ion-binding octarepeat region, single disulfide bridge, N-linked glycosylation sites (‘Glycan’) and attachment of the GPI anchor. The site of in vivo PrPC cleavage is indicated. Numbering is according to the murine protein.
Programmed cell death of PrP-deficient (Prnp0/0) cells is suppressed under serum-free conditions by transfection with a heterologous Prnp gene or the Bcl-2 gene (Kuwahara et al., 1999). This has raised the possibility that PrP is a member of the Bcl-2 family of proteins, which modulate one of the molecular cascades leading to apoptosis (Adams and Cory, 1998). PrP-deficient cells show increased sensitivity to reactive oxygen species due to decreased superoxide dismutase (SOD) activity (Brown et al., 1997, 1999). There is also increasing evidence that PrP has SOD activity (Brown et al., 1999) mediated by chelation of copper [Cu(II)] ions to the cation-binding domain of the N-terminal PrP52–89 region, since cells expressing only N-terminally truncated PrP, missing this copper ion-binding motif, are deficient in SOD activity. The extreme N-terminal segment (PrP23–51) has recently been shown to modulate the dominant-negative effects of PrP on the replication of prions (Zulianello et al., 2000), possibly by its interaction with residues 187–193; this part of the C-terminal domain structure has been shown by NMR spectroscopy to be affected by the N-terminal sequence (Donne et al., 1997). The N-terminus may also interact with the co-factor (protein X) implicated by transgenic studies as a determinant of the cross-species transmission of disease (Telling et al., 1995). This X factor is postulated to bind to a discrete epitope in the C-terminus of PrP, which includes residues 187–193 (Zulianello et al., 2000). Structural, biochemical and genetic evidence thus points to the importance of this N-terminal region of PrPC in the physiology of the cell and the pathophysiology of TSEs.
Physiological control of PrP function may be modulated by removal of the N-terminal domain from the membrane-anchored, C-terminal segment of PrPC by proteolysis between Lys111 and His112 (Harris et al., 1993; Chen et al., 1995; Brimacombe et al., 1999) (Figure 1). However, co-factor binding and/or post-translational modification of this N-terminal region offer the cell a fine-tuning mechanism for regulating these processes that is less drastic than proteolysis. Part of the N-terminal sequence has been predicted to have an extended poly(l-proline) II (PPII) helix structure (Smith et al., 1997) of a type involved in regulatory, multiple weak interactions when present in other proteins (Siligardi and Drake, 1995; Wright and Dyson, 1999). This domain has also been implicated in binding to heparan sulfate and other glycosaminoglycans that are important modulators of prion biology (Gabizon et al., 1993; Caughey et al., 1994). In this paper, we show by circular dichroism (CD) spectroscopy, mass spectrometry (MS) and Edman degradation that: (i) a synthetic peptide from this region of PrP can form a PPII structure in aqueous buffers; (ii) this structure (and sequence) is a signal for proline 4-hydroxylation; (iii) almost complete conversion of proline to 4-hydroxyproline occurs specifically at residue Pro44 of recombinant murine PrP secreted from Chinese hamster ovary (CHO) cells; and (iv) the same modification is found, at lower levels, in PrPSc extracted from the brains of scrapie-infected mice. This post-translational modification may provide an epigenetic control mechanism for the cross-species transmission of prions and/or the control of normal PrPC function.
Results
Oxidation of recombinant PrP secreted from CHO cells
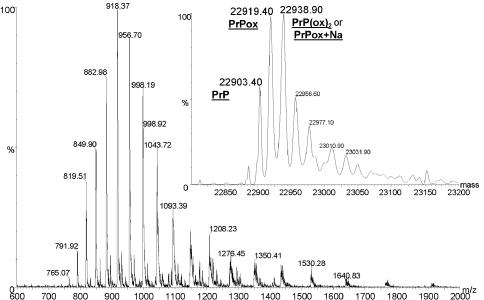
Recombinant prion protein (recPrP) secreted into the culture medium of stably transfected CHO cells (Brimacombe et al., 1999) was purified and analysed by capillary high-performance liquid chromatography (HPLC)–MS. Reversed-phase chromatography of the purified protein separated two UV-absorbing peaks corresponding to mature length PrP and an N-terminal fragment. The mass spectra of the proteins associated with these two peaks are shown in Figures 2 and 3. The peaks in the mass spectrum of mature length recPrP (Figure 2) are broad and include significant tails to high mass. Deconvolution of the mass spectrum (inset) indicates the presence of sodium adducts but there also appears to be a significant level of oxidation of the protein. This is suggested by the presence of a component peak 16 Da higher than the predicted mass of the protein. Only a small portion of the protein appears to have the predicted mass.
Fig. 2. Electrospray mass spectrum and (inset) deconvoluted mass spectrum of mature length, CHO cell-produced, recombinant, murine PrP. The predicted molecular mass (average) of the protein is 22 904.29 Da. Peaks corresponding to unmodified recPrP and oxidized PrP (PrPox) are labelled along with a third peak corresponding to either doubly oxidized recPrP [PrP(ox)2] or singly oxidized PrP with a single sodium adduct (PrPox+Na). Peaks corresponding to higher masses than this peak are assumed to correspond to further sodium adducts.
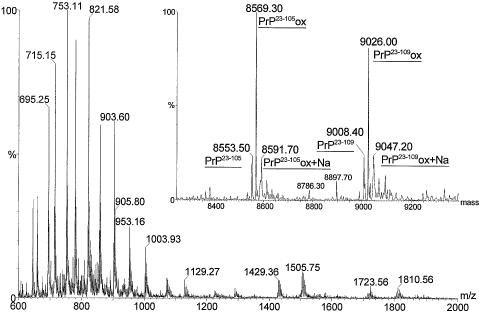
Fig. 3. Electrospray mass spectrum and (inset) deconvoluted mass spectrum of N-terminal recPrP fragments. Peaks corresponding to proteins of residues 23–105 (PrP23–105, theoretical molecular mass 8554.20 Da) and 23–109 (PrP23–109, theoretical molecular mass 9010.74 Da) along with their oxidized equivalents (PrP23–105ox and PrP23–109ox) and single sodium adducts (PrP23–105ox+Na and PrP23–109ox+Na) are labelled.
The mass spectrum of the N-terminal fragment of recPrP (Figure 3) is less dominated by sodium adducts but also indicates that this part of the protein has undergone oxidation. Trimming of the C-terminal residues from this fragment was identified by deconvolution of the mass spectrum (inset). This analysis shows peaks corresponding to PrP residues 23–105 and 23–109. Each peak is also accompanied by a second that is 16 Da higher in mass, indicating that the oxidation observed in full-length PrP is probably localized in this N-terminal fragment. Minor peaks are also evident that correspond to the oxidized forms of residues 23–107 (mass 8786) and 23–108 (mass 8897). The relative heights of the peaks indicate that ≥75% of the protein is oxidized. Non-specific oxidation of methioninyl and cysteinyl residues can occur during purification or storage of proteins, and oxidized methionine residues have previously been detected and reported for recPrP purified from bacterial cells and refolded in the presence of copper ions (Wong et al., 1999). However, both methionine and cysteine residues are lacking in the N-terminal fragment and so the source of the increased mass must lie elsewhere.
Prolyl 4-hydroxylation of recPrP from CHO cells
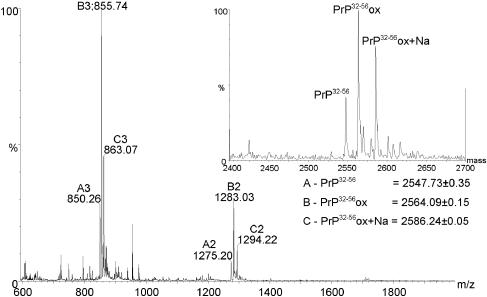
The site of oxidation was localized to a peptide spanning residues 32–56 by capillary HPLC–MS analysis of peptides released by chymotryptic digestion of purified recPrP (Figure 4). This peptide was purified by reversed-phase HPLC and subjected to sequencing by Edman degradation. This technique indicated a modification to Pro44 of recPrP and conclusively identified it as 4-hydroxyproline by comparison with standards. The modification is specific for this proline residue and is the first identification of a 4-hydroxyproline residue in PrP.
Fig. 4. Electrospray mass spectrum and (inset) deconvoluted mass spectrum of CHO cell-expressed recombinant PrP residues 32–56, produced by chymotryptic digestion of a mixture of mature length and C-terminally truncated protein. The peptide sequence from bottom to top is NTGGSRYPGQGSPGGNRYPPQGGTW (theoretical monoisotopic mass 2547.16 Da). Peaks corresponding to unmodified peptide (PrP32–56, series A), oxidized peptide (PrP32–56ox, series B) and the sodium adduct of the oxidized peptide (PrP32–56ox+Na, series C) are labelled.
Prolyl 4-hydroxylation of PrPSc
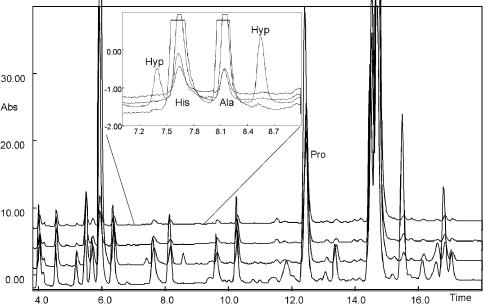
In order to confirm its biological significance, we looked for this hydroxylation in PrPSc purified from the brains of VM mice infected with the 87V strain of scrapie (Hope et al., 1988; Jeffrey et al., 1996). The denatured, alkylated protein was digested with chymotrypsin and the target peptide containing Pro44 was purified by HPLC and subjected to Edman degradation. 4-hydroxyproline was also identified in this protein. Figure 5 shows an overlay of the chromatograms corresponding to proline residues 39, 44, 50 and 51 from the Edman sequencing of PrPSc. Two peaks corresponding to the different isomers of 4-hydroxyproline are labelled, eluting on either side of those resulting from histidine and alanine, and, although small, are clearly and reproducibly detectable at Pro44 and not in the other proline residues within this peptide. The peak corresponding to unmodified proline is also labelled and is of greater intensity, indicating that only a small amount of 4-hydroxyproline is present, estimated to be <1%. The low incidence of 4-hydroxyproline in protein extracted from brain, and its presence in the part of PrPSc routinely cleaved from the protease-resistant core by proteinase K, may explain why previous studies have not identified this modification.
Fig. 5. Overlay of Edman sequencing chromatograms from chymotryptic peptide (residues 32–56) of murine PrPSc corresponding to proline residues 39, 44, 50 and 51. Inset, expansion of the chromatographic region encompassing hydroxyproline peaks.
Synthetic N-terminal PrP37–53 peptide has PPII helix structure
Pro44 is within a short sequence of PrP that has high homology to collagen. Procollagen chains are hydroxylated in vivo within the consensus sequence Xaa–Pro–Gly by the action of prolyl 4-hydroxylase (Kivirikko and Pihlajaniemi, 1998). To date, no other enzyme has been identified that produces 4-hydroxyproline from proline and this modification is difficult to effect non-enzymatically. Pro44 of PrP occurs in the sequence Ser–Pro–Gly and so fits the consensus sequence for hydroxylation by prolyl 4-hydroxylase. In addition to the requirement for a consensus sequence, however, the enzyme has also been reported to require formation of a PPII helix and possibly a β-turn in the substrate (Atreya and Ananthanarayanan, 1991). It has previously been proposed that PPII structure can form in the octarepeat region of PrP (Smith et al., 1997), which starts ∼10 amino acids C-terminal to the hydroxylation site. To investigate whether PPII structure is likely to occur in the residues around Pro44, two short peptides spanning this residue have been synthesized and their conformations probed by CD spectroscopy.
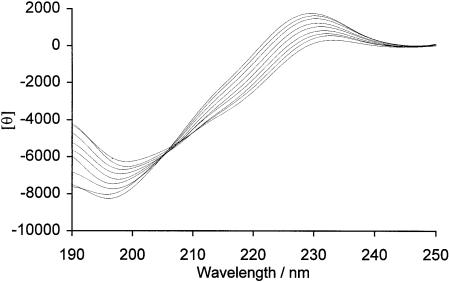
Figure 6 shows the CD spectra of PrP peptide 1 (residues 37–53) at various temperatures. The spectrum at 0°C shows a weak positive maximum at ∼230 nm along with a stronger minimum at ∼195 nm. These two CD spectral features are diagnostic for the presence of PPII helix (Woody, 1992). The spectrum is typical of peptides possessing at least some PPII helical conformation, allowing for a red shift for the maximum at 230 nm due to the tertiary imide of the proline residues. This shift may also be due to a slight negative contribution in the region 215–225 nm, possibly due to a population of peptide molecules or residues in a random coil conformation. A shoulder in the trace at ∼210 nm also indicates a contribution to the spectrum caused by the presence of a second type of structure. As the temperature of the sample is increased, the magnitude of the maximum at 230 nm is reduced, and the peak shifts to longer wavelengths. Likewise, the minimum at 195 nm becomes shallower and moves slightly to longer wavelengths, consistent with an increase of the random coil content of the sample molecules. The temperature gradient produces a set of nested spectra having an isodichroic point at 205 nm. This suggests the presence of two different populations of peptides having two different conformations. The transition with temperature may be rationalized by a shift in the equilibrium from a higher proportion of peptides having PPII structure to a higher proportion having a random coil conformation. A shorter peptide (PrP peptide 2, residues 41–48) also incorporating the hydroxylation site was analysed and was found to have a random coil conformation (data not shown).
Fig. 6. Temperature variation of the CD spectrum of PrP peptide 1 (sequence RYPGQGSPGGNRYPPQG-NH2). Reading downwards at 220 nm, the temperatures are 0, 10, 20, 30, 40, 50, 60, 70 and 80°C. The units of θ are degrees cm2/dmol residue.
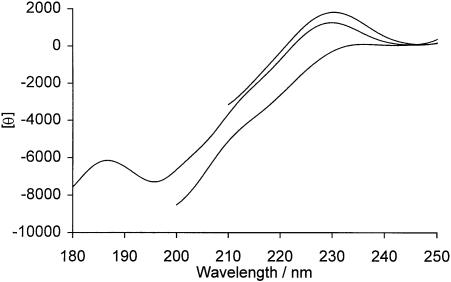
The effects of high concentrations of CaCl2 and the denaturant, guanidine hydrochloride, on the CD spectrum of PrP peptide 1 have also been investigated. These two buffer components have been shown to be useful for discriminating between peptides having PPII structure and those having random coil-like conformations (Woody, 1992; Park et al., 1997). PPII helices are stabilized primarily by hydrogen bonding between side chains and the peptide backbone, and CaCl2 has been shown to disrupt PPII helices by binding preferentially to amino acid side chains, thereby preventing such interactions. Conversely, at high concentrations, guanidine hydrochloride has been shown to stabilize PPII helices by binding to the peptide backbone and inhibiting backbone–backbone interactions. Figure 7 shows the CD spectra of PrP peptide 1 in standard measurement buffer, 5 M guanidine hydrochloride and 4 M CaCl2. The addition of high concentrations of denaturant led to an increase in intensity of the maximum at 230 nm characteristic of an increase in the proportion of PPII structure, whilst high concentrations of CaCl2 reduce the maximum markedly. These findings are consistent with previous work on peptides shown to have PPII structure (Woody, 1992; Park et al., 1997) and provide conclusive evidence for the formation of at least a short section of PPII structure within this region. In preliminary experiments, we have confirmed that Pro44 in this peptide can be specifically hydroxylated by an extract of chicken embryos enriched for proline 4-hydroxylase (Kivirikko and Prockop, 1967) (data not shown).
Fig. 7. Effects of denaturant and high ionic strength on the CD spectrum of PrP peptide 1. Reading downwards at 220 nm, solvents are 5 M guanidine hydrochloride, standard measurement buffer (see Materials and methods) and 4 M calcium chloride. The units of θ are degrees cm2/dmol residue.
Discussion
We have shown that an N-terminal peptide of PrP containing the sequence Ser–Pro44–Gly–Gly can form a PPII structure in aqueous buffers and that this motif can act as a substrate for prolyl 4-hydroxylation in CHO cells and in brain cells of mice infected with prions. These findings raise some interesting questions on the possible biological significance of this structure and the post-translational modification in the control of normal PrPC function as well as its involvement in TSE pathogenesis.
PPII structure and prolyl 4-hydroxylation
Hydroxylation of proline by prolyl 4-hydroxylase takes place during or after translation in the lumen of the endoplasmic reticulum (Kivirikko and Pihlajaniemi, 1998), and may be excessive if the transit of the protein from the endoplasmic reticulum is delayed (see below; Chessler and Byers, 1992). The active form of prolyl 4-hydroxylase is an α2β2 tetramer in which the β chains are identical to protein disulfide isomerase. Its most ubiquitous substrate is collagen where specific prolines occurring in the sequence Xaa–Pro–Gly are hydroxylated in a reaction requiring Fe2+, O2, oxo-glutarate and ascorbic acid. We found site-specific hydroxylation of Pro44 of mammalian-expressed recPrP and murine PrPSc. Two other sites within the N-terminal segment of PrP match the consensus sequence for enzymatic hydroxylation between residues 27–29 (sequence Lys–Pro–Gly) and 38–40 (sequence Tyr–Pro–Gly). Neither tyrosine nor a basic amino acid in the sequence Xaa–Pro–Gly in the C1q complement protein prevents hydroxylation at these sequences (Reid, 1979); therefore, the lack of hydroxyproline at positions 28 or 39 in recPrP or in PrPSc is due to another factor. This other factor is the local conformation of the polypeptide (Myllyharju and Kivirikko, 1999).
Proline-rich peptides with the Xaa–Pro–Gly motif are bound at the N-terminal region of the α-subunit of prolyl 4-hydroxylase and need an extended, principally PPII conformation to bind (Myllyharju and Kivirikko, 1999), and a partial β-turn in order to extend the Xaa–Pro–Gly into the catalytic site for hydroxylation (Atreya and Ananthanarayanan, 1991). The PPII helix is an extended left-handed helix with three residues per turn, the average dihedral angles of residues φ and ψ being approximately –75° and 145° (Macarthur and Thornton, 1991). Our findings that the N-terminal peptide of PrP containing the sequence Ser–Pro44–Gly–Gly can form a PPII structure in aqueous buffers, and that this sequence, and not others, can act as a substrate for prolyl 4-hydroxylation in CHO cells and in brain cells of mice infected with prions, strongly suggest that the polypeptide forms an extended PPII structure in vivo.
The relevance of extended PPII helical structures to biological activity
The biological activity of some proteins depends on a rigid, folded structure while in others their inherent flexibility and ability to change shape on binding with a substrate, ligand or receptor are critical features. From this perspective, the PrP protein is a chimeric molecule with a highly flexible N-terminal segment attached to a more rigid, three-helix bundle, C-terminal domain (Riek et al., 1997; Zahn et al., 2000). The PPII helix N-terminal structure identified in our studies may allow the protein to recognize many different receptors, thereby generating different cellular signals depending on substrate structure and/or on binding specificity. This extended helix structure, and factors influencing its dynamic flexibility, may be critical for a role of PrP in normal cellular function and signalling. Previous suggestions of PPII structure in PrP (Smith et al., 1997) have been largely overlooked mainly because NMR studies on full-length recombinant PrP found no evidence for such a structure (Donne et al., 1997). Unfortunately, the lack of main-chain hydrogen bonding in PPII helices makes NMR determinations of this type of structure difficult, especially if it is in equilibrium with random coil or other types of secondary structure. CD spectroscopy of synthetic peptides comprising residues 48–92 (Smith et al., 1997) and 57–91 (Whittal et al., 2000), both containing the four eight-amino-acid repeats, had previously given spectra with similar features to those shown here for PrP37–53. They also showed that the maximum at 225–230 nm, diagnostic for the presence of the PPII helix, is lost upon addition of copper ions to this metal ion-binding motif (Whittal et al., 2000). Our discovery of 4-hydroxyproline in CHO cell recPrP and PrPSc provides the first proof for the formation of the PPII helix in PrP in vivo. Taken together with the in vitro data, it is clear that this post-translational modification and/or metal ion binding may modulate the conformation of this biologically active sequence of PrP.
The importance of the N-terminus of PrP for its normal function
In vivo, PrPC is attached to the surface of cells by a C-terminal GPI anchor and undergoes surprisingly rapid recycling between the surface and an endosomal compartment by a mechanism involving clathrin-coated pits and vesicles (Shyng et al., 1993). Other GPI-linked proteins, e.g. Thy-1, because they lack a cytoplasmic domain, are unable to interact directly with clathrin and the adaptor proteins of coated pits, and they are generally thought to be internalized via caveolae (Anderson, 1993). Although some workers have suggested that PrPC is also internalized via caveolae (Vey et al., 1996; Kaneko et al., 1997), there is convincing evidence that a second transmembrane signalling molecule mediates the coated-pit trafficking of PrP (Harris, 1999).
Crucially, the N-terminal region of chicken PrPC is essential for its endocytosis via clathrin-coated pits (Shyng et al., 1995) and, most likely, it is this flexible domain of the protein that interacts with a coated-pit signalling molecule controlling its turnover and movement. Chimeric proteins containing the N-terminal region of murine PrP and the C-terminal domain of chicken PrP were internalized at essentially the same rate as chicken PrP in transfected mouse neuroblastoma cells (Shyng et al., 1995), even though the avian and mammalian N-terminal sequences differ considerably. Chicken PrP does, however, retain several consensus sequences for 4-hydroxy-proline formation in the hexarepeat region (Gln–Pro–Gly and Asn–Pro–Gly) of its N-terminal region and may also adopt a PPII helix. Further support for an essential control function for PrP23–80 was seen in a more recent study in which this N-terminal region of PrP has been fused to the C-terminal region of Thy-1. This led to rapid endocytosis and recycling of the fusion protein even though native Thy-1 has a half-life on the cell surface that can be measured in weeks (C.Sunyach and R.G.M.Morris, personal communication).
PrP function may be regulated by proline hydroxylation
Hydroxylated prolines straighten out β-turns in collagen chains and stabilize the collagen triple helix by hydrogen bonding of the hydroxyl oxygen of 4-hydroxyproline to the backbone of a different chain in the triple helix (Brodsky and Ramshaw, 1997). This stabilization of triple helices by 4-hydroxyproline is also found in other, non-collagen proteins (Kivirikko and Pihlajaniemi, 1998). The hydroxylated Pro44 of PrP occurs in a short stretch of sequence that has high homology to collagen; indeed, the PrP sequence Gly42–Ser–Pro–Gly–Gly46 also occurs in many types of collagen. Intriguingly, lyophilization or long-term storage of chicken PrP containing the tandem six-amino-acid repetitive peptides induces the formation of trimeric oligomers and partial resistance to proteolysis has also been found in the repeat region (Marcotte and Eisenberg, 1999). Both findings may be explained by the formation of collagen-like triple helices by the N-terminal regions of different chicken PrP molecules. It remains to be investigated whether such structures have a function, perhaps stabilized by prolyl 4-hydroxylation, and whether they exist in vivo in mammals or birds.
Neurons normally lack prolyl 4-hydroxylase (Blass et al., 1994) and neuronal PrPC is unlikely to be hydroxylated. The low percentage of PrPSc molecules containing 4-hydroxyproline in scrapie-affected mouse brain may derive from astrocytes or microglia; alternatively, and more provocatively, this hydroxylated PrP may be an indicator of prion-infected neurons. In contrast to neurons, normal CHO cells actively produce collagen and express high levels of prolyl 4-hydroxylase. This may be why almost all PrP secreted from these cells is specifically hydroxylated at Pro44. From this perspective, other fibroblastic and non-neuronal cells synthesizing PrP may also do so in a hydroxylated form. However, there may be other explanations. For example, we note that our recombinant PrP lacked sites for its normal post-translational glycosylation and glypiation, and so the prolyl 4-hydroxylation of this molecule, while indicative of PPII structure, could be due to increased cell stress or abnormal protein trafficking (Chessler and Byers, 1992).
At least two testable ideas spring from our observations and are currently under investigation within our laboratory. The first idea predicts that prolyl 4-hydroxylation of PrP is cell-type specific and that the percentage of hydroxylated PrPSc in infected tissues is an indicator of the cell type in which prion replication takes place. A second scenario to be tested views hydroxylation of PrP as an unnatural event that takes place as a general or specific response to mis-trafficking of this protein in a sick, infected or otherwise stressed cell. Whatever turns out to be true, our finding of site-specific hydroxylation of PrP opens up new avenues for research on the mechanisms regulating its role in the physiology and pathobiology of the cell.
Materials and methods
Expression and purification of CHO cell recPrP
Aglycosyl recPrP lacking its GPI anchor was expressed, produced and purified as described previously (Brimacombe et al., 1999). Cells were grown on Cytodex 1 microcarriers (Pharmacia) in Techne spinner flasks for 3 days in serum-containing medium, followed by a recPrP production phase in serum-free medium. The protein accumulated in the culture medium; after 3 days, soluble protein was purified by cation exchange chromatography, with elution using a sodium chloride gradient. Fractions containing recPrP, as determined by western blotting techniques, were pooled and further purified by immobilized metal affinity chromatography, eluting by means of an imidazole gradient. The final fractions contained mature length recPrP in addition to a 9 kDa N-terminal fragment resulting from a single proteolytic cleavage that has previously been characterized (Brimacombe et al., 1999).
Purification and derivatization of PrPSc
PrPSc was purified from the brains of VM mice infected with the 87V strain of scrapie by detergent lysis, differential centrifugation and size exclusion chromatography according to the procedure described previously (Hope et al., 1988). The protein, which was 95% pure as determined by SDS–PAGE and silver stain analysis, was subsequently reduced with dithiothreitol and alkylated with 4-vinylpyridine in the presence of 6 M guanidine–HCl pH 8.5.
Protein digestion
Reduced and alkylated PrPSc (∼50 µg) was precipitated by the addition of 4 vols of methanol and the pellet was resuspended in digestion buffer (2 M urea, 200 mM ammonium bicarbonate pH 8.5). RecPrP (50 µl, ∼100 µg/ml) was diluted 20-fold in digestion buffer. To each protein solution, ∼5% by weight of chymotrypsin was added and the mixture was incubated overnight at 37°C. Prior to Edman degradation, peptides of interest were purified by reversed-phase HPLC (C18, 1 mm i.d.; Haisil).
On-line capillary HPLC nanospray mass spectrometry
Capillary HPLC columns of 180 µm diameter were constructed using a method similar to that reported previously (Tong et al., 1997; A.C.Gill, M.A.Ritchie and J.Hope, in preparation). Fused silica (180 µm i.d.), used as the column body, was slurry packed with Hichrom C18 reversed-phase packing (3.5 µm beads, 150 Å pore size) and was terminated with a stainless steel screen (1/16 inch diameter, 2 µm pore size; Valco). Samples were injected, via an external port, low dispersion injector (Valco) on to a home-made desalting trap (A.C.Gill, M.A.Ritchie and J.Hope, in preparation) and were washed, eluted on to the capillary column and separated by a linear gradient of increasing solvent B (where solvent A was 95:5 H2O:acetonitrile with 0.05% trifluoroacetic acid and solvent B was 5:95 H2O:acetonitrile with 0.05% trifluoroacetic acid). A dual syringe Microgradient HPLC pump (ABI) provided a flow rate of 50 µl/min, which was split using a ‘T’ connector to ∼750 nl/min. The outlet of the column was connected, via a UV detector (214 nm) equipped with a U-Z View flow cell (LC Packings, Amsterdam, The Netherlands), to the Z-spray source of a Quattro II tandem quadrupole mass spectrometer (Micromass UK Ltd, Altrincham, UK) operated in continuous flow nanospray mode. The mass spectrometer was scanned from m/z 350 to 2200 (4 s/scan) and the cone voltage was ramped from 30 to 75 V over this range to aid detection of ions with high m/z ratios.
Edman degradation
Edman degradation was performed by means of an Applied Biosystems automated protein sequencer. Phenylthio-hydantoin amino acids were separated using the manufacturer’s PTH C18 column and a 140C HPLC system.
Peptide synthesis
Peptides from the N-terminus of PrP were synthesized by standard FMOC chemistry using an automated pioneer synthesizer (Perseptive Biosystems). Peptide sequences of R37YPGQGSPGGNRYPPQG53-NH2, designated PrP peptide 1, and Q41GSPGGNR48, designated PrP peptide 2, were used. After synthesis, peptides were purified on a reversed-phase C18 HPLC column and checked for identity and purity by means of electrospray mass spectrometry.
Circular dichroism
All CD spectra were acquired using a Jasco J-710 spectropolarimeter with a cell of 0.01 cm path length. The PrP peptides were dissolved in analysis buffer (10 mM NaCl, 1 mM phosphate buffer pH 7.0) to a final concentration of 1.66 mg/ml. The accuracy of the concentration was checked by UV absorbance at 280 nm using a Beckman DU 650 spectrophotometer. Measurements were taken between 250 and 180 nm using a temperature-controlled cell. For measurements using different buffers, the peptide was dissolved to the same concentration in either 5 M guanidine hydrochloride or 4 M calcium chloride. Owing to the absorbance and light scattering of the buffer components in these solutions at short wavelengths, measurements were taken to 210 and 200 nm, respectively. Thirty scans were acquired and summed for each sample, and the resulting spectrum was smoothed to remove residual noise.
Acknowledgments
Acknowledgements
We thank Guy Dodson and Peter Bayley (NIMR, Mill Hill, UK) for their support of K.D. and S.B. This work was funded, in part, by grants from the BBSRC under the BSEP IV research programme.
Note added in proof
In collaboration with Blanch and colleagues (J. Mol. Biol., 2000, 301, 553–563) we have now directly confirmed by Raman optical activity spectroscopy that full-length, recombinant sheep PrP contains significant amounts of PPII structure.
References
- Adams J. and Cory,S. (1998) The Bcl-2 protein family: arbiters of cell survival. Science, 281, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Anderson R.G.W. (1993) Caveolae—where incoming and outgoing messengers meet. Proc. Natl Acad. Sci. USA, 90, 10909–10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atreya P.L. and Ananthanarayanan,V.S. (1991) Interaction of prolyl 4-hydroxylase with synthetic peptide substrates—a conformational model for collagen proline hydroxylation. J. Biol. Chem., 266, 2852–2858. [PubMed] [Google Scholar]
- Blass J.P., Markesbery,W.R., Ko,L.W., Degiorgio,L., Sheu,K.F.R. and Darzynkiewicz,Z. (1994) Presence of neuronal proteins in serially cultured cells from autopsy human brain. J. Neurol. Sci., 121, 132–138. [DOI] [PubMed] [Google Scholar]
- Brimacombe D.B., Bennett,A.D., Wusteman,F.S., Gill,A.C., Dann,J.C. and Bostock,C.J. (1999) Characterization and polyanion-binding properties of purified recombinant prion protein. Biochem. J., 342, 605–613. [PMC free article] [PubMed] [Google Scholar]
- Brodsky B. and Ramshaw,J.A.M. (1997) The collagen triple-helix structure. Matrix Biol., 15, 545–554. [DOI] [PubMed] [Google Scholar]
- Brown D.R., Schultz-Schaeffer,W., Schmidt,B. and Kretschmar,H. (1997) Prion protein-deficient cells show altered response to oxidative stress due to decreased SOD-1 activity. Exp. Neurol., 146, 104–112. [DOI] [PubMed] [Google Scholar]
- Brown D.R., Wong,B.S., Hafiz,F., Clive,C., Haswell,S.J. and Jones,I.M. (1999) Normal prion protein has an activity like that of superoxide dismutase. Biochem. J., 344, 1–5. [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Brown,K., Raymond,G.J., Katzenstein,G.E. and Thresher,W. (1994) Binding of the protease-sensitive form of prion protein PrP to sulfated glycosaminoglycan and Congo red. J. Virol., 68, 2135–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.G., Teplow,D.B., Parchi,P., Teller,J.K., Gambetti,P. and Autilio Gambetti,L. (1995) Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem., 270, 19173–19180. [DOI] [PubMed] [Google Scholar]
- Chessler S. and Byers,P. (1992) Defective folding and stable association with protein disulphide isomerase/prolyl hydroxylase of type I procollagen with a deletion in the pro-α2(1) chain that preserves the Gly-X-Y repeat pattern. J. Biol. Chem., 267, 7751–7757. [PubMed] [Google Scholar]
- Donne D.G., Viles,J.H., Groth,D., Mehlhorn,I., James,T.L., Cohen,F.E., Prusiner,S.B., Wright,P.E. and Dyson,H.J. (1997) Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl Acad. Sci. USA, 94, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Rulicke,T., Raeber,A., Sailer,A., Moser,M., Oesch,B., Brandner,S., Aguzzi,A. and Weissmann,C. (1996) Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J., 15, 1255–1264. [PMC free article] [PubMed] [Google Scholar]
- Gabizon R., Meiner,Z., Halimi,M. and Bensasson,S.A. (1993) Heparin-like molecules bind differentially to prion proteins and change their intracellular metabolic fate. J. Cell. Physiol., 157, 319–325. [DOI] [PubMed] [Google Scholar]
- Harris D.A. (1999) Cellular biology of prion diseases. Clin. Microbiol. Rev., 12, 429–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D.A., Huber,M.T., Vandijken,P., Shyng,S.L., Chait,B.T. and Wang,R. (1993) Processing of a cellular prion protein—identification of N-terminal and C-terminal cleavage sites. Biochemistry, 32, 1009–1016. [DOI] [PubMed] [Google Scholar]
- Hope J., Morton,L.J.D., Farquhar,C.F., Multhaup,G., Beyreuther,K. and Kimberlin,R.H. (1986) The major polypeptide of scrapie-associated fibrils (SAF) has the same size, charge-distribution and N-terminal protein sequence as predicted for the normal brain protein (PrP). EMBO J., 5, 2591–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J., Multhaup,G., Reekie,L.J., Kimberlin,R.H. and Beyreuther,K. (1988) Molecular pathology of scrapie-associated fibril protein (PrP) in mouse brain affected by the ME7 strain of scrapie. Eur. J. Biochem., 172, 271–277. [DOI] [PubMed] [Google Scholar]
- Jeffrey M., Goodsir,C.M., Fowler,N., Hope,J., Bruce,M.E. and McBride,P.A. (1996) Ultrastructural immuno-localization of synthetic prion protein peptide antibodies in 87V murine scrapie. Neurodegeneration, 5, 101–109. [DOI] [PubMed] [Google Scholar]
- Kaneko K., Vey,M., Scott,M., Pilkuhn,S., Cohen,F.E. and Prusiner,S.B. (1997) COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc. Natl Acad. Sci. USA, 94, 2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivirikko K.I. and Pihlajaniemi,T. (1998) Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv. Enzymol. Relat. Areas Mol. Biol., 72, 325–398. [DOI] [PubMed] [Google Scholar]
- Kivirikko K.I. and Prockop,D.J. (1967) Hydroxylation of proline in synthetic polypeptides with purified protocollagen hydroxylase. J. Biol. Chem., 242, 4007–4012. [PubMed] [Google Scholar]
- Kuwahara C. et al. (1999) Prions prevent neuronal cell-line death. Nature, 400, 225–226. [DOI] [PubMed] [Google Scholar]
- Macarthur M.W. and Thornton,J.M. (1991) Influence of proline residues on protein conformation. J. Mol. Biol., 218, 397–412. [DOI] [PubMed] [Google Scholar]
- Marcotte E.M. and Eisenberg,D. (1999) Chicken prion tandem repeats form a stable, protease-resistant domain. Biochemistry, 38, 667–676. [DOI] [PubMed] [Google Scholar]
- Myllyharju J. and Kivirikko,K.I. (1999) Identification of a novel proline-rich peptide-binding domain in prolyl 4-hydroxylase. EMBO J., 18, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Shalongo,W. and Stellwagen,E. (1997) The role of PPII conformations in the calculation of peptide fractional helix content. Protein Sci., 6, 1694–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid K.B.M. (1979) Complete amino acid sequences of the three collagen-like regions present in subcomponent C1q of the first component of human complement. J. Biol. Chem., 179, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R., Hornemann,S., Wider,G., Glockshuber,R. and Wuthrich,K. (1997) NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Lett., 413, 282–288. [DOI] [PubMed] [Google Scholar]
- Rudd P.M. et al. (1999) Glycosylation differences between the normal and pathogenic prion protein isoforms. Proc. Natl Acad. Sci. USA, 96, 13044–13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng S.L., Huber,M.T. and Harris,D.A. (1993) A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. J. Biol. Chem., 268, 15922–15928. [PubMed] [Google Scholar]
- Shyng S.L., Moulder,K.L., Lesko,A. and Harris,D.A. (1995) The N-terminal domain of a glycolipid-anchored prion protein is essential for its endocytosis via clathrin-coated pits. J. Biol. Chem., 270, 14793–14800. [DOI] [PubMed] [Google Scholar]
- Siligardi G. and Drake,A.F. (1995) The importance of extended conformations and, in particular, the PPII conformation for the molecular recognition of peptides. Biopolymers, 37, 281–292. [DOI] [PubMed] [Google Scholar]
- Smith C.J., Drake,A.F., Banfield,B.A., Bloomberg,G.B., Palmer,M.S., Clarke,A.R. and Collinge,J. (1997) Conformational properties of the prion octa-repeat and hydrophobic sequences. FEBS Lett., 405, 378–384. [DOI] [PubMed] [Google Scholar]
- Stahl N., Baldwin,M.A., Hecker,R., Pan,K.M., Burlingame,A.L. and Prusiner,S.B. (1992) Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry, 31, 5043–5053. [DOI] [PubMed] [Google Scholar]
- Stimson E., Hope,J., Chong,A. and Burlingame,A.L. (1999) Site-specific characterization of the N-linked glycans of murine prion protein by high-performance liquid chromatography electrospray mass spectrometry and exoglycosidase digestions. Biochemistry, 38, 4885–4895. [DOI] [PubMed] [Google Scholar]
- Telling G.C., Scott,M., Mastrianni,J., Gabizon,R., Torchia,M., Cohen,F.E., Dearmond,S.J. and Prusiner,S.B. (1995) Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell, 83, 79–90. [DOI] [PubMed] [Google Scholar]
- Tong D.X., Moritz,R.L., Eddes,J.S., Reid,G.E., Rasmussen,R.K., Dorow,D.S. and Simpson,R.J. (1997) Fabrication of stable packed capillary reversed-phase columns for protein structural analysis. J. Protein Chem., 16, 425–431. [DOI] [PubMed] [Google Scholar]
- Turk E., Teplow,D.B., Hood,L.E. and Prusiner,S.B. (1988) Purification and properties of the cellular and scrapie hamster prion proteins. Eur. J. Biochem., 176, 21–30. [DOI] [PubMed] [Google Scholar]
- Vey M., Pilkuhn,S., Wille,H., Nixon,R., De Armond,S.J., Smart,E.J., Anderson,R.G.W., Taraboulos,A. and Prusiner,S.B. (1996) Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl Acad. Sci. USA, 93, 14945–14949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viles J.H., Cohen,F.E., Prusiner,S.B., Goodin,D.B., Wright,P.E. and Dyson,H.J. (1999) Copper binding to the prion protein: structural implications of four identical cooperative binding sites. Proc. Natl Acad. Sci. USA, 96, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittal R.M., Ball,H.L., Cohen,F.E., Burlingame,A.L., Prusiner,S.B. and Baldwin,M.A. (2000) Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci., 9, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong B.S., Wang,H., Brown,D.R. and Jones,I.M. (1999) Selective oxidation of methionine residues in prion proteins. Biochem. Biophys. Res. Commun., 259, 352–355. [DOI] [PubMed] [Google Scholar]
- Woody R.W. (1992) Circular dichroism and conformation of unordered polypeptides. Adv. Biophys. Chem., 2, 37–79. [Google Scholar]
- Wopfner F., Weidenhofer,G., Schneider,R., von Brunn,A., Gilch,S., Schwarz,T.F., Werner,T. and Schatzl,M. (1999) Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J. Mol. Biol., 289, 1163–1178. [DOI] [PubMed] [Google Scholar]
- Wright P.E. and Dyson,H.J. (1999) Intrinsically unstructured proteins: re-assessing the protein structure–function paradigm. J. Mol. Biol., 293, 321–331. [DOI] [PubMed] [Google Scholar]
- Zahn R. et al. (2000) NMR solution structure of the human prion protein. Proc. Natl Acad. Sci. USA, 97, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulianello L., Kaneko,K., Scott,M., Erpel,S., Han,D., Cohen,F.E. and Prusiner,S.B. (2000) Dominant-negative inhibition of prion formation diminished by deletion mutagenesis of the prion protein. J. Virol., 74, 4351–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]