Abstract
Uridine phosphorylase (UP), a key enzyme in the pyrimidine salvage pathway, catalyzes the reversible phosphorolysis of uridine or 2′-deoxyuridine to uracil and ribose 1-phosphate or 2′-deoxyribose 1-phosphate. This enzyme belongs to the nucleoside phosphorylase I superfamily whose members show diverse specificity for nucleoside substrates. Phylogenetic analysis shows Streptococcus pyogenes uridine phosphorylase (SpUP) is found in a distinct branch of the pyrimidine subfamily of nucleoside phosphorylases. To further characterize SpUP, we determined the crystal structure in complex with the products, ribose 1-phosphate and uracil, at 1.8 Å resolution. Like Escherichia coli UP (EcUP), the biological unit of SpUP is a hexamer with an α/β monomeric fold. A novel feature of the active site is the presence of His169, which structurally aligns with Arg168 of the EcUP structure. A second active site residue, Lys162, is not present in previously determined UP structures and interacts with O2 of uracil. Biochemical studies of wild type SpUP showed that substrate specificity is similar to that of EcUP, while EcUP is about sevenfold more efficient than SpUP. Biochemical studies on active site mutant SpUP showed that mutations of His169 reduced activity, while mutation of Lys162 abolished all activity, suggesting that negative charge in the transition state resides mostly on uracil O2. This is in contrast to EcUP for which transition state stabilization occurs mostly at O4.
Uridine phosphorylase (UP) (EC 2.4.2.3) is a key enzyme in the pyrimidine salvage pathway and is found in most prokaryotes and eukaryotes. The salvage pathway is an alternative to the energetically expensive de novo biosynthetic pathway, which requires six biochemical transformations to make precursors for DNA and RNA biosynthesis (1). Specifically, UP uses free phosphate to catalyze the reversible phosphorolysis of ribonucleosides or 2′-deoxynucleosides of uracil and their analogues to the corresponding nucleobases and (2′-deoxy)ribose 1-phosphate (R1P). In addition, UP is essential for utilizing nucleosides as a carbon source during pyrimidine catabolism (2). Several structures of UPs have been reported and roles for active site residues have been proposed (3–7). All known UPs belong to the nucleoside phosphorylase I (NP-I) superfamily (8); however, mammalian UPs are dimers, while bacterial UPs are hexamers comprised of a trimer of dimers. Other NP-I families include purine nucleoside phosphorylase (PNP), methylthioadenosine phosphorylase, adenosine 5′-monophosphate nucleosidase (AMN) and 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN).
Nucleoside phosphorylases are also known to inactivate certain pyrimidine and purine nucleoside analogues with potential antitumor properties (7, 9). For example, the pyrimidine analogue 5-fluorouracil is used as a chemotherapeutic agent for the treatment of advanced stage colorectal cancer (10). However, the nucleoside analogue 5-fluorouridine is cleaved by UP and consequently, no more effective than 5-fluorouracil itself. Thus, inhibitors for human UP enzymes might enhance the efficacy of certain pyrimidine nucleoside analogues. Mechanisms of inhibition of UPs by analogues have been proposed (7, 11). Despite these studies, the details of the chemical mechanism and the role of active site residues are not as well understood as those of PNP (12–16). For example, in the presence of the sulfate, which has been used previously as an unreactive mimic of phosphate (14), some pyrimidine nucleosides cleave to form a ribosyl intermediate and the free base. Interestingly, the proposed ribosyl intermediate exists as a glycal, a phenomenon that has not been observed previously in nucleoside phosphorylases (6).
The catalytic mechanism of PNPs, which is expected to be similar to that of the Ups, has been extensively explored by enzyme kinetics. The deuterium kinetic isotope effect that occurred during the phosphorolytic reaction supports an SN1 mechanism (17, 18). This mechanism was further validated by studies on the arsenolysis reaction that demonstrated the existence of oxocarbenium ion character during the transition state (15). The resulting high-energy oxocarbenium ion is stabilized by a continuum of electrostatic interactions among the substrate intermediates and conserved residues in the active site (12, 14). The scheme for the proposed transition state stabilization is shown in Figure 1.
Figure 1.
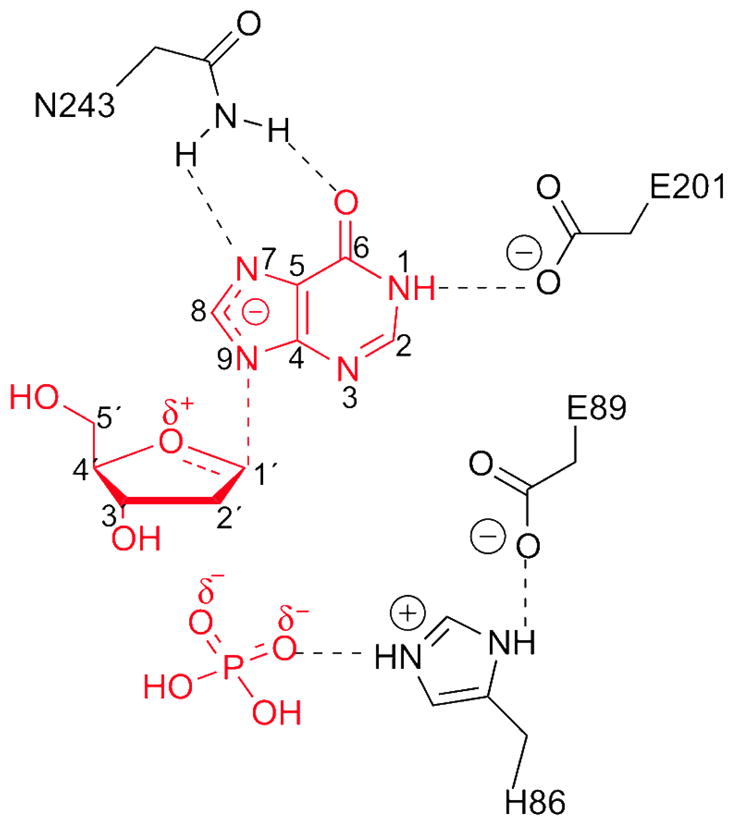
Proposed transition state for PNP from human blood (12).
Here we report the X-ray structure of Streptococcus pyogenes UP (SpUP) in complex with the products ribose 1-phosphate and uracil (SpUP/R1P/Ura) at 1.8 Å resolution. The three-dimensional structure shows that the overall monomeric fold is similar to the NP-I superfamily; however, unlike previously reported UP structures, SpUP lacks the two critical active site arginine residues that are proposed to stabilize the negative charge that accumulates on the pyrimidine ring during formation of the oxocarbenium-like transition state (3, 6). Instead SpUP utilizes one histidine residue and one lysine residue for this purpose. These residues are conserved within a group of UPs and thus establish a previously unrecognized subfamily. We also report biochemical studies of wild-type and mutant SpUP to probe substrate specificity, mechanism and catalytic efficiency. Lys162 was found to be essential through electrostatic interactions with uracil O2. As a result of the differences in active site residues and differences in transition state stabilization, Escherichia coli UP (EcUP) is about seven times more efficient than SpUP.
MATERIALS AND METHODS
Cloning of Native SpUP
The up gene was PCR amplified from Streptococcus pyogenes (ATCC 12344) genomic DNA using the following primers: upstream primer 5′-GGG TAG CAT ATG CAA AAT TAT TCA GGT GAA GTC GG-3′ (inserts an NdeI site at the start codon of the up open reading frame); downstream primer 5′-CCC TAC TCG AGT TAT TGT GAT TTA TCA TTT TCA ATA AG-3′ (inserts an XhoI site after the end of the up open reading frame). The purified PCR product was digested with NdeI and XhoI, purified and ligated into similarly digested pTHT (a pET-28 derived vector that incorporates a modified 6×HisTag followed by a TEV protease cleavage site onto the N-terminus of the expressed protein). Kanamycin-resistant colonies were screened for presence of the insert and a representative plasmid was designated pSpUP.THT. The PCR-derived DNA was sequenced and shown to contain no errors.
Mutagenesis of SpUP
Standard methods were used for DNA manipulations (19, 20). Plasmid DNA was purified with the Fermentas GeneJet Miniprep kit. E. coli strain MachI (Invitrogen) was used as a recipient for transformations during plasmid construction and for plasmid propagation and storage.
Site-directed mutagenesis was performed on pSpUP.THT by a standard PCR protocol using Pfu Turbo DNA polymerase per the manufacturer’s instructions (Invitrogen) and DpnI (New England Biolabs) to digest the methylated parental DNA prior to transformation.
In addition to the forward and reverse primers required to introduce the mutation, a third primer was designed to screen for the presence of the mutation by colony PCR (Supporting Table S1).
Expression and Purification of SpUP
The plasmid described above was transformed into expression strain BL21(DE3) E. coli cells. An overnight culture of 10 mL was grown in LB media at 37 °C supplemented with 50 μg/mL kanamycin, and then introduced into 1 L culture containing 50 μg/mL kanamycin. The culture was grown at 37 °C with shaking until an OD600 of 0.6 was reached, at which point the temperature was reduced to 15 °C and isopropyl-1-β-D-galactopyranoside was added to a final concentration of 1 mM. Cells were harvested by centrifugation at 7459 g for 20 min after approximately 16 h growth. The pellet was stored at −80 °C until purification.
The frozen cell pellet was thawed overnight at 4 °C and resuspended in approximately 30 mL of lysis buffer (20 mM Tris (pH 8), 10 mM imidazole, and 300 mM NaCl). The cell suspension was sonicated and then centrifuged at 47, 488 g for 1 h to remove the cellular debris. All the steps after cell lysis were performed at 4 °C. The clarified lysate was loaded onto a pre-equilibrated Ni-NTA gravity column with a volume of 1 mL, after which the column was rinsed with 20 column volumes of lysis buffer. SpUP was then eluted with 10 mL of elution buffer (20 mM Tris (pH 8), 300 mM imidazole, 10% glycerol, and 300 mM NaCl). The eluted protein was loaded directly onto a size exclusion column (Hiload 26/60 Superdex 200 pg, GE Healthcare) for further purification. The protein fractions from the column were pooled together and, using an Amicon Ultra centrifugal filter, were concentrated to 25–30 mg/mL as determined by the method of Bradford (20). The protein was confirmed to be at least 95% pure by SDS-PAGE analysis. The pure protein was buffer exchanged into 20 mM Tris (pH 8), 50 mM NaCl, and 1 mM DTT, flash frozen in liquid nitrogen and stored at −80 °C until use.
Crystallization of SpUP with Products
Frozen SpUP was thawed at 4 °C and incubated for 12 h with 8 mM R1P and 8 mM uracil in Tris buffer pH 8. Cocrystallization trials were initially carried out using the vapor diffusion hanging drop method at 22 °C with sparse matrix screening solutions (Hampton Research, Emerald Biosystems). For each drop, 1 μL of protein solution was combined with an equal volume of well solution. The SpUP/R1P/Ura complex crystallized in 0.1 M sodium citrate (pH 5.2), 18% (w/v) PEG 4000, and 16% isopropanol. The plate-like crystals took 3–7 days to grow and grew to a maximum size of 0.1 mm × 0.2 mm × 0.45 mm. The crystals belonged to space group P1, with a unit cell volume consistent with three complete hexamers per asymmetric unit (Table 1). No additional cryoprotectant was used for crystal freezing.
Table 1.
Data Collection and Refinement Statisticsa
| SpUP/R1P/Ura Complex | |
|---|---|
| beamline | 24-ID-C, NE-CAT, APS |
| wavelength (Å) | 0.97849 |
| space group | P1 |
| unit cell dimensions | a = 90.0 Å, b = 91.7 Å, c = 169.3 Å, α = 78.5°, β = 82.3°, γ = 60.1° |
| chains per asymmetric unit | 18 |
| resolution (Å) | 50 - 1.82 (1.85 - 1.82) |
| total no. of reflections | 793731 (30451) |
| number of unique reflections | 404352 (16027) |
| redundancya | 2.0 (1.9) |
| Rmerge (%)b | 4.9 (20.5) |
| I/σ(I) | 19.3 (4.1) |
| no. of reflections in working set | 383726 |
| completeness (%) | 94.6 (74.7) |
| Rwork/ Rfreeb (%) | 17.7/ 20.3 |
| no. of protein atoms | 33711 |
| no. of ligand atoms | 396 |
| no. of water atoms | 2979 |
| average B-factor protein (Å2) | 13.3 |
| average B-factor water (Å2) | 23.6 |
| average B-factor ligand (Å2) | 17.1 |
| rmsd for bonds (Å) | 0.005 |
| rmsd for angles (°) | 1.283 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ΣΣi | Ii – <I> | / Σ <I>, where <I> is the mean intensity of the N reflections with intensities Ii and common indices h,k,l.
Rwork = Σhkl| |Fobs| – k |Fcal| | / Σhkl |Fobs| where Fobs and Fcal are observed and calculated structure factors, respectively, calculated over all reflections used in the refinement. Rfree, is similar to Rwork but calculated over a subset of reflections (5%) excluded from all stages of refinement.
Data Collection and Processing
Data for SpUP/R1P/Ura were collected at 100 K using NE-CAT beamline 24-ID-C at the Advanced Photon Source (APS) at Argonne National Laboratory using a Quantum 315 X-ray detector (Area Detector Systems Corporation). The data were collected at a wavelength of 0.9785 Å over 360° using a 0.5° oscillation range. All data were indexed, integrated, and scaled using the HKL2000 program suite (21). The data collection statistics are shown in Table 1.
Structure Determination
The SpUP structure was determined by molecular replacement with EcUP (PDB ID: 1K3F) (5, 22) as the search model, using MolRep in the CCP4 program suite (23). CHAINSAW in the CCP4 suite was used to prune the side chains of the search model to the last common atom. The relatively large unit cell and predicted solvent content suggested three complete hexamers per asymmetric unit. The three hexamers showed pseudo translational symmetry except that one hexamer was significantly tilted with respect to the other two. After one around of tight rigid body refinement followed by a round of restrained refinement in Refmac5 of the CCP4 program suites, the Rfactor and Rfree dropped to 32.4% and 34.7%, respectively. In subsequent rounds of refinements, the restraints were gradually relaxed. Most of the side chains were built during these rounds of refinement. Further rounds of refinement were performed in CNS (24) starting with rigid body refinement, simulated annealing, and finally B-factor refinement. Difference Fourier maps (Fo-Fc and 2Fo-Fc) and composite omit maps were calculated from models after each round of refinement. Manual model building was done using Coot (25) and the noncrystallographic symmetry (NCS)-averaged composite omit map to minimize model bias. Water molecules were added after convergence of Rfactor and Rfree. The ligands were modeled into all 18 active sites of SpUP. The ligands were generated using PRODRG (26). The geometry of SpUP was validated using PROCHECK (27). The final Rfactor and Rfree converged to 17.7% and 20.3%, respectively. The complete refinement statistics are given in Table 1.
Determination of Substrate Specificity
In 1 mL total reaction volumes containing 100 mM HEPES and 50 mM phosphate buffer (K2HPO4) with a final pH of 7.4, an appropriate amount of SpUP was incubated with 500 μM substrate. Samples of 150 μL were removed from the reaction after incubating for 0, 0.25, 0.5, 1 and 2 hours respectively, and immediately mixed with 150 μL of water, and quenched by boiling. The precipitated protein was removed by filtration using a 0.2 μm syringe filter, and the sample was injected onto a 5 μm BDS Hypersil C-18 column (150 × 4.6 mm) (Keystone Scientific Inc., State College, PA). The mobile phase was a 12.5 mM ammonium dihydrogen phosphate buffer (pH 4.5) containing 1.25% acetonitrile (flow rate of 1 mL/min). Substrates and products were detected by their absorbance at 260 nm as they eluted from the column.
Steady State Kinetic Measurements for SpUP and EcUP
All uridine phosphorolysis reactions were carried out in 500 μL total volume containing 50 mM phosphate buffer (K2HPO4) and 50 mM HEPES with a final pH of 7.5. In the above reaction buffer containing a known amount of uridine substrate, the reaction was initiated by the addition of SpUP or EcUP to final concentrations of 36 nM and 9 nM, respectively. The concentrations are calculated for the monomers. At these concentrations, the enzymes were completely quenched by the addition of 10 μL of 10 M NaOH, which allowed the appearance of the absorbance peak of the uracil product to be monitored at 290 nm (28). The initial rate for each uridine concentration was measured when the reaction had proceded approximately 10% toward completion. All the initial rates were then plotted and fitted to the Michaelis-Menten equation using the Kaleidagraph software package (Synergy Software). In two duplicate sets of experiments, the reaction was quenched with acetic acid and heat (100°C) before NaOH was added to ensure that the measurements were consistent.
Figure Preparation
Figures were prepared using PyMOL (29) and ChemBioDraw (CambridgeSoft).
RESULTS
Quaternary Structure of SpUP
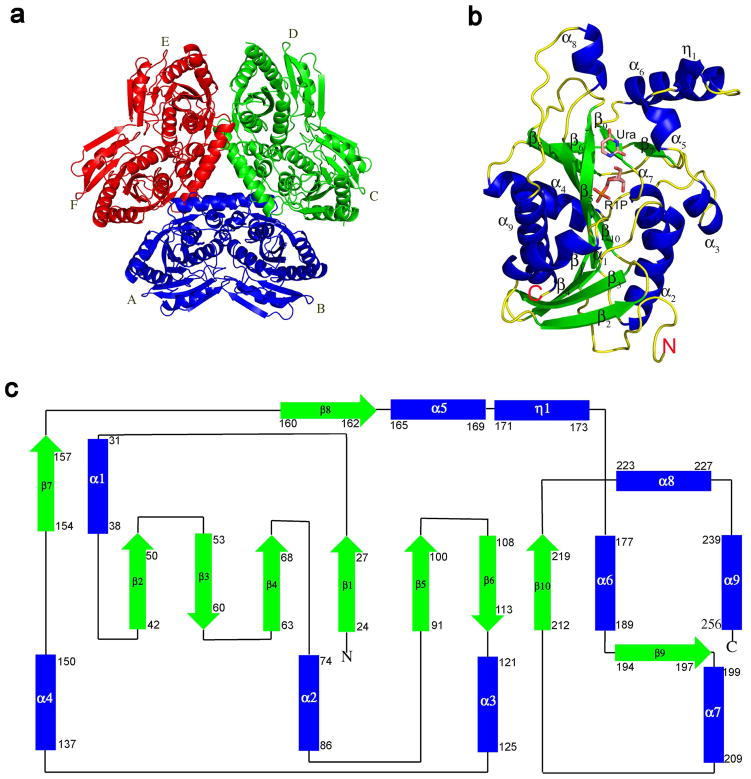
As expected from the relatively high sequence identity with EcUP (40%) and recent classification (8), the biological unit of SpUP is a hexamer having 32 symmetry (Figure 2a). The toroidal hexamer has a diameter of approximately 100 Å and a thickness of 40 Å. The hexamer of SpUP also has a central channel with a diameter of 18 Å. The oligomeric state was also confirmed by the size exclusion chromatography data, which estimated the molecular weight of the SpUP oligomer to be approximately 167 kDa. While each protomer contains an active site, the active site is located at a dimer interface and contains two residues, His13 and Arg51, from the adjacent monomer. Thus, the minimal functional unit is a dimer. The distance between two active sites, which are located on opposite faces of the hexamer, is about 21 Å. In the crystal structure of SpUP/R1P/Ura, three hexamers are found in the asymmetric unit complex.
Figure 2.
Structures of SpUP. (a) Hexameric quaternary structure of SpUP color-coded to emphasize the 32 point symmetry or a trimer of dimers. Six subunits are labeled A to F. (b) SpUP protomer with the bound products, R1P and Ura, shown in the active site. The helices are colored blue, the strands green, and the loops yellow. The N-terminus and the C-terminus are also indicated with letters N and C. (c) Topology diagram of SpUP protomer. The colors of the helices and strands are the same as Figure 1b, except the loops are depicted as black lines. The first and last residues of each secondary structure are indicated by numbers.
Structure of SpUP Protomer
The SpUP protomer shown in Figure 2b adopts an α/β fold. The core contains a mixed eight-stranded β-sheet with a sharp twist in the following orientation: β2↑β3↓β4↑β1↑β5↑β10↑β8↓β6↓. The topology diagram for SpUP is shown in Figure 2c. The β-sheet is flanked by six α-helices, with α1, α4 and α9 on one side and α2, α3 and α7 on the other. β2, β5 and β10 are longer than the rest of the β-strands, containing nine, ten, and eight residues, respectively. Alpha helices, α5, α6 and α8 as well as a 310 helix (η1) are clustered on one side of the active site. Four α helices, α2, α4, α6, and α9, are much longer than other helices. Helix α9, capping the C-terminus, is the longest with 17 residues, whereas helices α2, α4, and α6 contain 11, 13, and 13 residues, respectively. The loop between α8 and α9, consisting of residues 234–238, generally has weak electron density and high B-factors.
SpUP Active Site
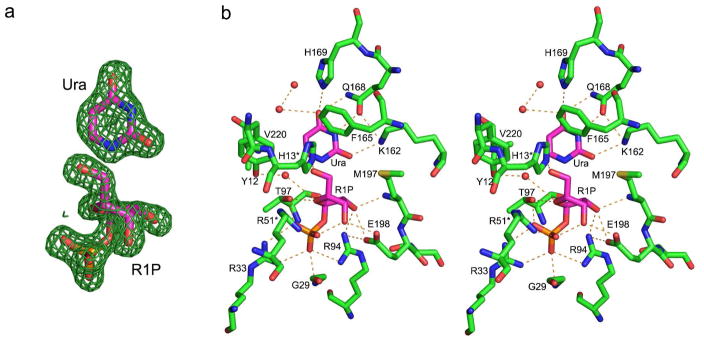
Figure 3 illustrates the active site for the SpUP/R1P/Ura complex. The most significant feature of the phosphate binding site is the presence of three highly conserved arginine residues, Arg33, Arg51*, and Arg94 (* denotes a residue from a neighboring monomer). The three arginine residues position the phosphate moiety of R1P with a strong ionic network. Two other conserved residues also hydrogen bond to the phosphate group: Gly29 via the amide nitrogen atom and Thr97 via its side chain. In addition, the oxygen atom of the phosphate ion closest to the ribose also forms a hydrogen bond to O3′ atom of the sugar moiety.
Figure 3.
Active site of SpUP. (a) Electron density for uracil and R1P. The NCS-averaged Fo-Fc density was calculated with phases from the refined model after the ligands were removed. The map is contoured at 3.0 σ. (b) Stereo diagram of the active site containing R1P and Ura. The red spheres represent water molecules. Hydrogen bonds are indicated by dashed lines.
Figure 3b shows the ribose binding site. The 2′- and 3′-hydroxyl groups form a pair of hydrogen bonds with Glu198. The C2′-hydroxyl forms a second hydrogen bond with Arg94. His13*, which is conserved in all UPs, hydrogen bonds to the hydroxyl group of C5′. Thr97 forms a weak hydrogen bond with O4′ of the ribose ring. The sulfur atom of the highly conserved Met197 interacts with the hydrophobic face of the ribose and could be involved in positioning the substrate within the active site (12). In addition, the main-chain nitrogen atom of Met197 hydrogen bonds to the 2′-hydroxyl group. This methionine residue is conserved throughout the entire NP-I family. A conserved water molecule also hydrogen bonds to the O4′ atom of the sugar group.
While the phosphate and ribose binding sites are both similar to those of EcUP (3), the uracil binding site shows significant differences. Nδ of His169 is located approximately 3 Å away from the O4 atom of the uracil base; however, the N-H…O angle is 120°, suggesting a weak hydrogen bond. Gln168 is strictly conserved among all the known UP structures and makes hydrogen bonds with the N3 and the O4 atoms of the uracil base. Lys162 is coplanar with the uracil and hydrogen bonds to O2 as well as to Gln168. Phe165 forms a herringbone interaction with the aromatic ring of the uracil. Val220 interacts with the hydrophobic portion of the uracil ring.
Substrate Specificity
Because of the variation in the active site residues compared to EcUP, we tested the ability of SpUP to cleave other purine and pyrimidine nucleosides. SpUP cleaves uridine, deoxyuridine and thymidine, but not cytidine, deoxycytidine, adenosine, inosine, or guanosine. Results are shown in Table 2.
Table 2.
SpUP substrate specificity
| substrate | cleavage activity (nmol/mg/h) |
|---|---|
| uridine | 485,000 |
| deoxyuridine | 673,000 |
| thymidine | 352,000 |
| cytidine | < 10,000 |
| deoxycytidine | < 10,000 |
| adenosine | < 10,000 |
| guanosine | < 10,000 |
| inosine | < 10,000 |
Uridine Phosphorolysis Assay
Using the crystal structure, several mutant SpUPs were designed to probe the roles of active site residues in the base binding region. Lys162 was mutated to alanine, and His169 was mutated to alanine, asparagine, and aspartate. In addition, Val220 was mutated to aspartate and glutamate to determine if the enzyme could accommodate a purine base in the presence of these polar, charged groups. Mutation of Lys162 abolished activity, while the other mutations showed various levels of reduction of activity. The results for all mutant SpUPs are shown in Table 3.
Table 3.
Uridine phosphorolysis assay results for SpUP and mutants
| SpUP | uridine cleavage activities (nmol/mg/h) |
|---|---|
| wild-type | 1,500,000 |
| K162A | < 20 |
| H169A | 112,000 |
| H169D | 9,000 |
| H169N | 42,000 |
| V220D | 550 |
| V220E | 3,400 |
Steady State Kinetic Measurements for SpUP and EcUP
The UV spectra of uridine and uracil are very similar. Therefore, to spectrophotometrically monitor the formation of the uracil product, an aliquot of 10 μL of 10 M NaOH (0.2 M) was added to the reaction. This amount of base was sufficient to quench the reaction and to deprotonate the free uracil, and thus to change the aromaticity of the uracil ring. This gave rise to a large absorption peak at 290 nm for the free uracil (28). The same amount of base has no effect on uridine. The robustness of this assay was confirmed by duplicate experiments, in which the reaction was quenched with acetic acid or heat (100 °C) before NaOH was added. Table 4 shows the resulting steady state parameters for SpUP and EcUP.
Table 4.
Steady State Kinetic Parameters for SpUP and EcUPa
| Enzyme | kcat (s−1) | Km (mM) | kcat/Km (mM−1 s−1) |
|---|---|---|---|
| SpUP | 15 ± 3 | 0.158 ± 0.025 | 95 ± 24 |
| EcUP | 23 ± 1 | 0.036 ± 0.004 | 639 ± 76 |
| SpUP-H169A | 1.7 ± 0.2 | 0.171 ± 0.005 | 9.9 ± 1 |
| SpUP-K162Ab | - | - | N/A |
The concentration of uridine was varied and the production of uracil was monitored.
No activity was observed at the level of detection of the assay.
DISCUSSION
Overall Structure of SpUP
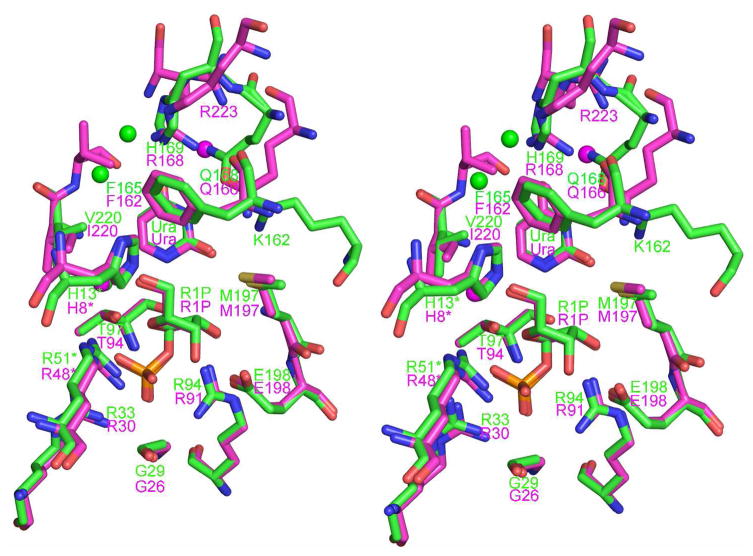
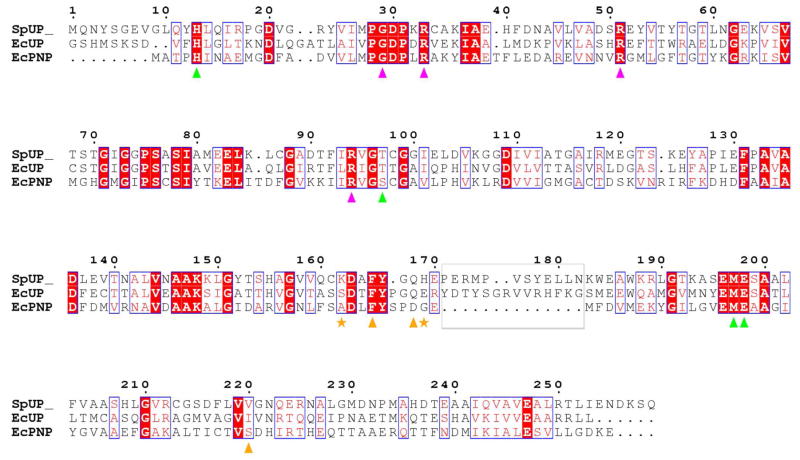
The biological unit of SpUP is a hexamer, as is the case for other prokaryotic UPs (Figure 2a) (3, 30). The asymmetric unit of the SpUP/R1P/Ura complex contains three hexamers. While two of the hexamers show good translational pseudosymmetry, with the center to center separation of about 55 Å, the threefold axis of the third hexamer is tilted 37° with respect to the other two resulting in space group P1 (Table 1). The SpUP protomers superimpose well on the EcUP protomer, especially in the active site, with a root mean square deviation of the Cα carbons ranging from 0.99 to 1.05 Å (Figure 4). SpUP adopts the canonical α/β fold of the NP-I superfamily (8). Residues 170–182, including the 310 helix (η1) and part of α6, comprise an insertion characteristic of UPs (3), which allows pyrimidine nucleosides such as uridine and thymidine, to bind to the active site, while excluding purine nucleosides. The primary sequence alignment demonstrating this insertion is shown in Figure 5. As shown in the alignment, SpUP has two fewer residues in this specificity region compared to EcUP; however, this insertion is absent in E. coli purine nucleoside phosphorylase (EcPNP), and thus allows EcPNP to accommodate purine bases.
Figure 4.
Superposition of the active sites of SpUP (green) and EcUP (magenta) in stereo. The spheres are water molecules found in the active site.
Figure 5.
The insertion region (gray box) of SpUP is revealed by the alignment of SpUP against EcUP and EcPNP. Magenta, green, and orange triangles indicate conserved residues in the phosphate-binding site, ribose-binding site, and uracil-binding site of SpUP, respectively. The residues marked with an asterisk are the two novel residues found in the uracil-binding site of SpUP.
Comparison of SpUP with Other NP-I Enzymes
SpUP was compared with related but divergent NP-I enzymes including EcPNP (PDB ID, 1PR0) (31), E. coli AMN (EcAMN, PDB ID 1T8S) (32), and E. coli MTAN (EcMTAN, PDB ID 1NC1) (33). Examination of the biological unit of SpUP and those of the selected NP-I enzymes reveals that although these enzymes have different substrate specificities and use different nucleophiles for the cleavage reactions, they all share the same monomeric fold. Three of the enzymes are hexamers, while EcMTAN is a dimer.
The secondary structure of these enzymes around the active sites and the geometry of substrate binding are highly conserved among these enzymes. Several critical active site residues are conserved among the four structures: (1) a histidine residue from a neighboring protomer that forms a hydrogen bond with the ribose 5′-hydroxyl group, (2) an aromatic residue that stacks against the nucleobase, (3) a strictly conserved glutamate residue that forms hydrogen bonds with both the 2′ and 3′-hydroxyl groups of the ribose, and (4) a methionine residue that packs against the hydrophobic β-face of the ribose to position it in the active site. Residues in the phosphate binding site of SpUP and EcPNP (as well as EcUP) are also highly conserved but are different from those in human PNP (34) or the nucleosidase (32), which utilize water as the nucleophile.
Substrate Specificity for SpUP
Enzymes in the NP-I family are generally specific for either a purine nucleoside or pyrimidine nucleoside and the nucleophile can be either phosphate or water. An exception is the PNP from Plasmodium falciparum, which can also cleave uridine, but at much lower levels (35). Nucleoside specificity within the NP-I family results from the size of the base binding site and complementarity of hydrogen bonding at the periphery of the purine or pyrimidine base. Two active site residues, Lys162 and His169, that were not previously observed in the active sites of NP-I family members and a slight variation in the size of the loop that normally discriminates between purine and pyrimidine nucleosides led us to determine the SpUP substrate specificity. The results shown in Table 4 indicate that the preferred substrate specificity is deoxyuridine (138%) > uridine (100%) > thymidine (73%). Cleavage of cytidine, deoxcytidine, adenosine, inosine, or guanosine was below detectable levels.
Comparison of the Steady State Kinetic Parameters for SpUP and EcUP
The discovery of SpUP, which catalyzes the same reaction as EcUP but uses different active site residues compared to EcUP, led us to determine if the two UPs had different catalytic efficiencies. Steady state kinetics showed that the turnover number (kcat) of SpUP has a value of 15 s−1 and its apparent substrate binding affinity (Km) of 0.158 mM, which give an overall catalytic efficiency of 95 mM−1 s−1 (Table 4). The corresponding kcat value for EcUP is 23 s−1, which is slightly higher than that of SpUP, whereas its Km value (0.036 mM) is fourfold lower than that of SpUP giving a catalytic efficiency of 639 mM−1 s−1 for EcUP. The previously reported value for the Km of EcUP performed at 25 °C and pH 7.4 is 0.091 mM (36), which is more than twofold higher than the value measured under similar conditions in our base-quench assay. Thus, the catalytic efficiency of EcUP is nearly seven times higher than that of SpUP.
Substrate Specificity and Mechanistic Implications
Mutation of active site residues provided insight into the roles of key residues (Table 3). The most striking result was the complete loss of activity for the K162A mutant. The enzyme still maintains low but detectable levels of activity with the H169A (8%), H169N (3%) and H169D (1%) mutants. In the SpUP complex, Lys162 hydrogen bonds to the O2 of uracil suggesting that this is the major site for accumulation of negative charge. His169 is near O4 but the hydrogen bonding geometry is not optimum, although it is possible that tilting of the base in the substrate complex might provide more favorable geometry. In the H169N mutant, hydrogen bonding to O4 might still be possible and, in the H169A mutant, a water molecule might occupy the vacant side chain space and provide a hydrogen bond. Assuming that the side chain in the H169D mutant is mostly ionized, hydrogen bonding is not favored. These results suggest that a small amount of negative charge accumulates on O4 during catalysis; however, most of the negative charge would be on O2 and stabilized by Lys162. Val220 was mutated to aspartate and glutamate because acidic side chains are sometimes found in this position in the purine nucleoside phosphorylases; however, these mutants had very low activity. In addition, the active site loop insertion is consistent with exclusion of purine nucleosides.
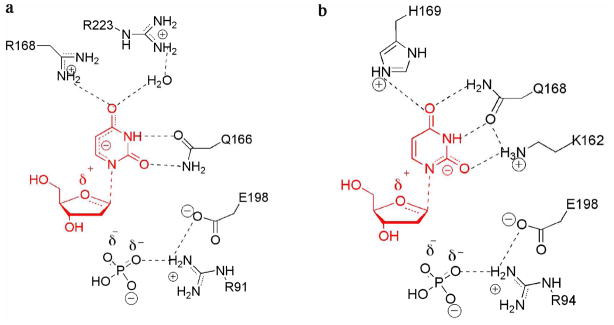
While enzymes in the NP-I family are generally specific for either a purine nucleoside or a pyrimidine nucleoside, and the nucleophile can be either phosphate or water, all family members are believed to function through a high-energy oxycarbenium ion intermediate and require acidic residues or strong hydrogen bond donors to stabilize the negative charge that accumulates on the purine or pyrimidine nucleobase during glycosidic bond cleavage. Various NP-I family members have evolved to promote glycosidic bond cleavage (Figure 6). In the case of purine nucleosides, activation of the leaving group is usually provided by a hydrogen bond to the purine N(7) atom using either an asparagine or protonated aspartic acid residue. SpUP and EcUP both catalyze the phosphorolytic cleavage of the glycosidic bond of uridine and related nucleosides; however, differences in active site residues suggest different mechanisms for activation of the leaving group and stabilization of the negative charge resulting from oxycarbenium ion formation (Figure 7). EcUP and most other UPs utilize two arginine residues (Arg168 and Arg223) to stabilize negative charge and activate the leaving group (3). Arg168 hydrogen bonds directly to O4, while Arg168 hydrogen bonds to O4 through a tightly bound water molecule. Conserved Gln166 donates a hydrogen bond to O2 and accepts a hydrogen bond from N3. In SpUP, Arg168 is replaced by His169 but Arg223 is completely absent. Instead, Gln166 is replaced by Gln168, which shifts to accept a hydrogen bond from N3 and donate a hydrogen bond to O4. The shift of Gln168 makes room for Lys162, which donates a hydrogen bond to O2. Our studies suggest that this hydrogen bond is the primary source of charge stabilization and leaving group activation.
Figure 6.
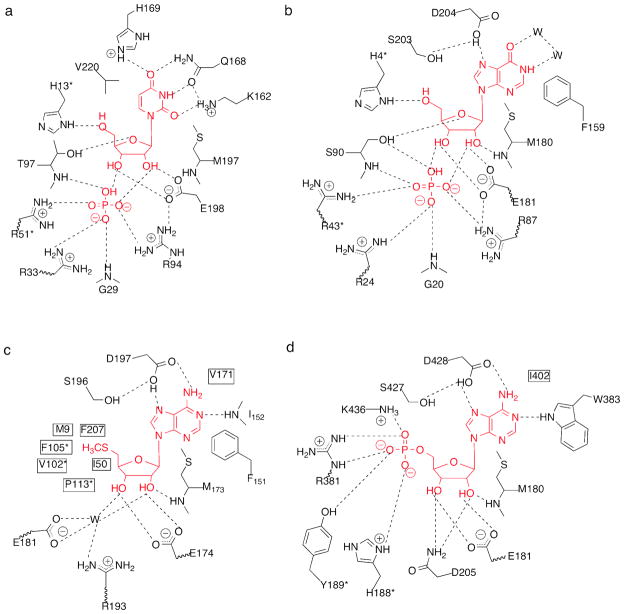
Active sites of SpUP with uridine and phosphate manually positioned in the active site (a), EcPNP/inosine/PO4 complex (b), EcMTAN/5′-methylthiotubercidin (c), and EcAMN/formycin 5′-monophosphate (d), respectively (14, 31–33). ‘W’ stands for water. Residues marked with an asterisk come from an adjacent monomer.
Figure 7.
Transition state stabilization of high energy intermediates of the phosphorolysis reaction in EcUP (a) and SpUP (b) by a continuum of electrostatic and hydrogen bond interactions as illustrated by the dotted line.
Implication for Evolution of the UPs
Comparison of the active sites of SpUP and EcUP suggests that the two enzymes have similar transition states; however, SpUP utilizes different residues to stabilize the negative charge on the base. To determine the distribution of the two subfamilies, a multiple sequence alignment of SpUP and EcUP with sequences found using a BLAST search against the nonredundant database was performed using ClustalW (37). After removal of partial sequences and sequences showing more than 95% sequence identity with an included sequence, 106 UP sequences remained. Among these sequences, 49 UPs were found to contain conserved lysine and histidine residues equivalent to Lys162 and His169 of SpUP. None of these UPs have residues equivalent to Arg168 or Arg223 in EcUP. On the other hand, 57 of the sequences possess the two arginine residues equivalent to Arg168 or Arg223 found in EcUP, but none of these have the residues equivalent to Lys162 and His169 in SpUP. These 106 sequences, including SpUP and EcUP, were placed on a phylogenetic tree using the same ClustalW server. All the SpUP-like species are found in one cluster (Supporting Figure S1), while the EcUP-like species are found in a different cluster (Supporting Figure S2). Further examination of the alignment indicates that most of the SpUP-like species are from Gram-positive bacteria, while most of the EcUP-like species are from Gram-negative bacteria. Two exceptions are the Thermococcus and Fusobacteria species, which fall in the SpUP-like branch.
Interestingly, while UP orthologs are found in both prokaryotes and eukaryotes, no UP othologs were found in archaebacteria. Further examination of available archaebacterial genomes revealed species lacking UP orthologs contained pyrimidine nucleoside phosphorylase (PyNP), which is a member of the nucleoside phosphorylase II superfamily (38). PyNP is structurally homologous to thymidine phosphorylase; however the former accepts both uridine and thymidine as substrates, while the latter is found in most prokaryotes and eukaryotes and is highly specific for thymidine (8).
Our results show clear evidence for evolution of two separate UP subfamilies. Both EcUP and SpUP have similar structures and similar substrate specificities; however, there are critical differences in active site residues and EcUP is nearly seven times as efficient as that of SpUP. The improved catalytic efficiency of EcUP compared to SpUP may provide a competitive advantage for purine metabolism; however, evolution of two distinct UP subfamilies may result in competitive advantages yet to be discovered.
Supplementary Material
Acknowledgments
We would like to thank the National Institutes of Health for the funding support. JP would like to thank Cancerfonden (Sweden) for the funding support. The molecular cloning was provided by Cynthia Kinsland at the Cornell University Protein Production Facility. We are grateful to the staff scientists at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source for their help with the data collection. Finally, we thank Leslie Kinsland for her assistance during manuscript preparation.
ABBREVIATIONS
- UP
uridine phosphorylase
- PNP
purine nucleoside phosphorylase
- SpUP
Streptococcus pyogenes uridine phosphorylase
- EcUP
Escherichia coli uridine phosphorylase
- EcPNP
Escherichia coli purine nucleoside phosphorylase
- EcAMN
Escherichia coli adenosine 5′-monophosphate nucleosidase
- EcMTAN
Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase
- bPNP
bovine purine nucleoside phosphorylase
- APS
Advanced Photon Source
- NP-I
nucleoside phosphorylase I
- Ura
uracil
- R1P
ribose 1-phosphate
- NCS
noncrystallographic symmetry
Footnotes
This work was supported by NIH grant GM73220. This work is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
The coordinates of the SpUP/R1P/Ura complex have been deposited in the Protein Data Bank under accession code 3QPB.
Table S1, which shows the primers used in site-directed mutagenesis, and phylogenetic trees showing the relationship of orthologs of SpUp (Figure S1) and EcUP (Figure S2), are shown in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhang Y, Morar M, Ealick SE. Structural biology of the purine biosynthetic pathway. Cell Mol Life Sci. 2008;65:3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leer JC, Hammer-Jespersen K, Schwartz M. Uridine phosphorylase from Escherichia coli. Physical and chemical characterization. Eur J Biochem. 1977;75:217–224. doi: 10.1111/j.1432-1033.1977.tb11520.x. [DOI] [PubMed] [Google Scholar]
- 3.Caradoc-Davies TT, Cutfield SM, Lamont IL, Cutfield JF. Crystal structures of Escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium. J Mol Biol. 2004;337:337–354. doi: 10.1016/j.jmb.2004.01.039. [DOI] [PubMed] [Google Scholar]
- 4.Lashkov AA, Zhukhlistova NE, Gabdoulkhakov AH, Shtil AA, Efremov RG, Betzel C, Mikhailov AM. The X-ray structure of Salmonella typhimurium uridine nucleoside phosphorylase complexed with 2,2′-anhydrouridine, phosphate and potassium ions at 1.86 Å resolution. Acta Crystallogr D. 2010;66:51–60. doi: 10.1107/S0907444909044175. [DOI] [PubMed] [Google Scholar]
- 5.Morgunova E, Mikhailov AM, Popov AN, Blagova EV, Smirnova EA, Vainshtein BK, Mao C, Armstrong Sh R, Ealick SE, Komissarov AA, et al. Atomic structure at 2.5 Å resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice. FEBS Lett. 1995;367:183–187. doi: 10.1016/0014-5793(95)00489-v. [DOI] [PubMed] [Google Scholar]
- 6.Paul D, O’Leary SE, Rajashankar K, Bu W, Toms A, Settembre EC, Sanders JM, Begley TP, Ealick SE. Glycal formation in crystals of uridine phosphorylase. Biochemistry. 2010;49:3499–3509. doi: 10.1021/bi902073b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roosild TP, Castronovo S, Fabbiani M, Pizzorno G. Implications of the structure of human uridine phosphorylase 1 on the development of novel inhibitors for improving the therapeutic window of fluoropyrimidine chemotherapy. BMC Struct Biol. 2009;9:14. doi: 10.1186/1472-6807-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugmire MJ, Ealick SE. Structural analyses reveal two distinct families of nucleoside phosphorylases. Biochem J. 2002;361:1–25. doi: 10.1042/0264-6021:3610001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renck D, Ducati RG, Palma MS, Santos DS, Basso LA. The kinetic mechanism of human uridine phosphorylase 1: Towards the development of enzyme inhibitors for cancer chemotherapy. Arch Biochem Biophys. 2010;497:35–42. doi: 10.1016/j.abb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 11.Bu W, Settembre EC, el Kouni MH, Ealick SE. Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines. Acta Crystallogr D. 2005;61:863–872. doi: 10.1107/S0907444905007882. [DOI] [PubMed] [Google Scholar]
- 12.Erion MD, Takabayashi K, Smith HB, Kessi J, Wagner S, Honger S, Shames SL, Ealick SE. Purine nucleoside phosphorylase. 1 Structure-function studies. Biochemistry. 1997;36:11725–11734. doi: 10.1021/bi961969w. [DOI] [PubMed] [Google Scholar]
- 13.Erion MD, Stoeckler JD, Guida WC, Walter RL, Ealick SE. Purine nucleoside phosphorylase. 2 Catalytic mechanism. Biochemistry. 1997;36:11735–11748. doi: 10.1021/bi961970v. [DOI] [PubMed] [Google Scholar]
- 14.Federov A, Shi W, Kicska G, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Transition State Structure of Purine Nucleoside Phosphorylase and Principles of Atomic Motion in Enzymatic Catalysis. Biochemistry. 2001;40:853–860. doi: 10.1021/bi002499f. [DOI] [PubMed] [Google Scholar]
- 15.Kline PC, Schramm VL. Purine nucleoside phosphorylase. Catalytic mechanism and transition-state analysis of the arsenolysis reaction. Biochemistry. 1993;32:13212–13219. doi: 10.1021/bi00211a033. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Luo M, Ghanem M, Taylor EA, Schramm VL. Second-sphere amino acids contribute to transition-state structure in bovine purine nucleoside phosphorylase. Biochemistry. 2008;47:2577–2583. doi: 10.1021/bi7021365. [DOI] [PubMed] [Google Scholar]
- 17.Lehikoinen PK, Sinnott ML, Krenitsky TA. Investigation of a-deuterium kinetic isotope effects on the purine nucleoside phosphorylase reaction by the equilibrium-perturbation technique. Biochem J. 1989;257:355–359. doi: 10.1042/bj2570355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein RL, Cordes EH. Kinetic 〈-deuterium isotope effects for Escherichia coli purine nucleoside phosphorylase-catalyzed phosphorolysis of adenosine and inosine. J Biol Chem. 1981;256:767–772. [PubMed] [Google Scholar]
- 19.Ausubel FM, Brent F. Current Protocols in Molecular Biology. John Wiley and Sons; New York: 1987. [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Vol. 3. Cold Spring Harbor Laboratory Press; Plainview, New York: 1989. [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project-Number 4. The CCP-4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Schuettelkopf AW, van Aalten DMF. PRODRG: a Tool for High-Throughput Crystallography of Protein-Ligand Complexes. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 28.Ploeser JM, Loring HS. The ultraviolet absorption spectra of the pyrimidine ribonucleosides and ribonucleotides. J Biol Chem. 1949;178:431–437. [PubMed] [Google Scholar]
- 29.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 30.Dontsova MV, Gabdoulkhakov AG, Molchan OK, Lashkov AA, Garber MB, Mironov AS, Zhukhlistova NE, Morgunova EY, Voelter W, Betzel C, Zhang Y, Ealick SE, Mikhailov AM. Preliminary investigation of the three-dimensional structure of Salmonella typhimurium uridine phosphorylase in the crystalline state. Acta Crystallogr F. 2005;61:337–340. doi: 10.1107/S1744309105007463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett EM, Li C, Allan PW, Parker WB, Ealick SE. Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase. J Biol Chem. 2003;278:47110–47118. doi: 10.1074/jbc.M304622200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Cottet SE, Ealick SE. Structure of Escherichia coli AMP nucleosidase reveals similarity to nucleoside phosphorylases. Structure. 2004;12:1383–1394. doi: 10.1016/j.str.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of Escherichia coli 5′-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis. J Biol Chem. 2003;278:8761–8770. doi: 10.1074/jbc.M210836200. [DOI] [PubMed] [Google Scholar]
- 34.Ealick SE, Rule SA, Carter DC, Greenhough TJ, Babu YS, Cook WJ, Habash J, Helliwell JR, Stoeckler JD, Parks RE, Jr, et al. Three-dimensional structure of human erythrocytic purine nucleoside phosphorylase at 3.2 Å resolution. J Biol Chem. 1990;265:1812–1820. doi: 10.2210/pdb2pnp/pdb. [DOI] [PubMed] [Google Scholar]
- 35.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Kim K, Schramm VL. Transition state analogue inhibitors of purine nucleoside phosphorylase from Plasmodium falciparum. J Biol Chem. 2002;277:3219–3225. doi: 10.1074/jbc.M105905200. [DOI] [PubMed] [Google Scholar]
- 36.Krenitsky TA. Uridine phosphorylase from Escherichia coli. Kinetic properties and mechanism. Biochim Biophys Acta. 1976;429:352–358. doi: 10.1016/0005-2744(76)90283-7. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugmire MJ, Ealick SE. The crystal structure of pyrimidine nucleoside phosphorylase in a closed conformation. Structure. 1998;6:1467–1479. doi: 10.1016/s0969-2126(98)00145-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.