Abstract
The central arbiter of cell fate in response to DNA damage is p53, which regulates the expression of genes involved in cell cycle arrest, survival and apoptosis. Although many responses initiated by DNA damage have been characterized, the role of actin cytoskeleton regulators is largely unknown. We now show that RhoC and LIM kinase 2 (LIMK2) are direct p53 target genes induced by genotoxic agents. Although RhoC and LIMK2 have well-established roles in actin cytoskeleton regulation, our results indicate that activation of LIMK2 also has a pro-survival function following DNA damage. LIMK inhibition by siRNA-mediated knockdown or selective pharmacological blockade sensitized cells to radio- or chemotherapy, such that treatments that were sub-lethal when administered singly resulted in cell death when combined with LIMK inhibition. Our findings suggest that combining LIMK inhibitors with genotoxic therapies could be more efficacious than single-agent administration, and highlight a novel connection between actin cytoskeleton regulators and DNA damage-induced cell survival mechanisms.
Keywords: LIMK, RhoC, p53, DNA damage, actin, cofilin, cytoskeleton
Introduction
DNA damage triggers a complex set of cellular responses that have evolved to facilitate rapid recovery or efficient cell death 1. Given that vast numbers of DNA lesions occur in each cell every day, the DNA damage response comprises mechanisms that allow repair to occur. These protective responses include activation of DNA damage checkpoints and the induction of various pro-survival pathways. Ultimately, if the damage exceeds the ability and capacity of repair mechanisms to correct, a typical outcome is apoptosis. Many common cancer therapeutic modalities exploit the DNA damage response by overwhelming repair mechanisms and triggering cell death.
The principal coordinator of the DNA damage response is p53. Following stabilization, p53 accumulates and regulates transcriptional programs that control the expression of genes involved in cell cycle arrest, survival and apoptosis, helping to tailor the response to the magnitude and context of the stress 2, 3. Given the large number of p53-regulated genes, a major challenge is to determine how individual genes contribute to specific cellular outcomes.
The best-studied aspects of the p53-mediated DNA damage response are those that occur as a direct riposte to the initiating event, such as DNA repair. Although there is considerable interest in determining how p53 loss or mutation influences the invasive behavior of tumor cells 4, the consequences of wild-type p53 activation and its effect on cell morphology and the actin cytoskeleton remain unclear. In particular, although relationships between actin cytoskeleton regulators and apoptosis have been described 5, 6, 7, there is little information linking p53 with cell survival and death via cytoskeleton regulators.
The Rho familiy of GTPases regulate a variety of cellular processes, including cell cycle progression and proliferation 8. When in their active GTP-bound form, Rho-GTPases such as RhoA and RhoC recruit effector proteins that are involved in rearranging the actin cytoskeleton. Acting downstream of Rho, ROCK1 and ROCK2 9 phosphorylate and activate the LIM kinases (LIMK1 and LIMK2) 10, 11. Activated LIMK phosphorylates and inactivates the filamentous actin (F-actin)-severing protein cofilin. Spatially and temporally regulated cycles of cofilin inactivation and activation enables dynamic actin rearrangements required for cell motility. Although recent studies have identified the transcription factors Myc 12 and p53 13 as modulators of RhoA and RhoE expression, respectively, little is known about how components of the Rho-ROCK-LIMK pathway are regulated in response to physiological or pathophysiological stimuli.
In examining cytoskeletal responses to genotoxic stresses, we observed significant activation of the Rho-ROCK-LIMK pathway. RHOC expression was induced by direct p53 binding to a regulatory element within the RHOC gene. LIMK2 variant isoforms were also found to be regulated by p53 through direct interaction with regulatory elements within the LIMK2 gene. Repression of LIMK activity by siRNA-mediated knockdown or by selective pharmacological blockade with a first-in-class LIMK inhibitor synergized with genotoxic chemotherapeutic agents or ionizing radiation (IR) to induce cell death. This study reveals novel connections between actin cytoskeleton regulators and p53-mediated cell survival mechanisms. Furthermore, these results suggest that the efficacy of drugs that act by inducing a pro-apoptotic DNA damage response could be increased when combined with LIMK inhibitors.
Results
Genotoxic stress activates the Rho-ROCK-LIMK pathway
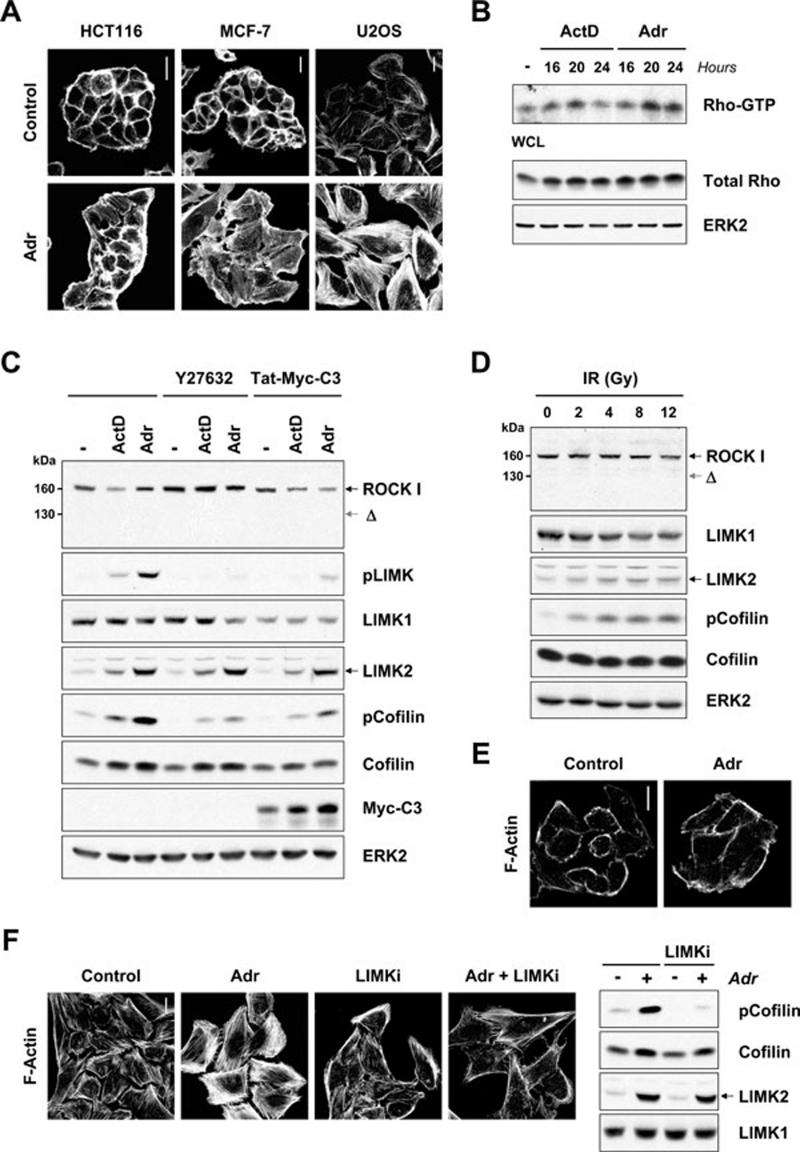
We previously showed that during late stages of apoptosis, robust actin-myosin contractile force resulting from caspase-mediated cleavage and activation of ROCK1 leads to contraction, blebbing and nuclear disintegration 6, 7. These studies also revealed actin rearrangements prior to cell death. To examine the morphological and cytoskeletal responses to activation of intrinsic apoptosis pathways, human tumor cell lines were treated with the clinically used genotoxic agent adriamycin (Adr; also known by its generic name doxorubicin). In contrast to control vehicle-treated HCT116, MCF-7 or U2OS cells, Adr treatment resulted in cell flattening, increased cell size and induction of actin stress fibers (Figure 1A). Analysis of Rho activity by pull-down assay showed that Rho-GTP levels were elevated at 16-24 h following treatment of MCF-7 cells with the genotoxic agents actinomycin D (ActD) or Adr (Figure 1B). Interestingly, genotoxic stress failed to induce actin stress fibers in MDA-MB-231 cells which express mutant p53 (R280K) (Figure 1E). Rho proteins activate numerous effector proteins, including the serine/threonine kinases, ROCK1 and ROCK2, which contribute to stress fiber formation. Treatment with ActD or Adr resulted in increased phosphorylation of the ROCK substrates LIMK1 and LIMK2 on activation loop threonine residues, and of the LIMK substrate cofilin (Figure 1C). Since ROCK1 can be activated via caspase-mediated proteolysis 6, 7, we examined if ROCK1 was cleaved in response to genotoxic stress. Treatment with ActD or Adr did not result in the generation of detectable 130-kDa caspase-cleaved (Δ) ROCK1 (Figure 1C). Concomitant treatment with ROCK inhibitor Y-27632 (10 μM) or Rho inhibitor Tat-Myc-C3 with ActD or Adr reduced both LIMK and cofilin phosphorylation (Figure 1C). Similarly, treatment with increasing doses of IR resulted in a dose-dependent increase in cofilin phosphorylation without evident ROCK1 cleavage (Figure 1D).
Figure 1.
Genotoxic stress activates the Rho-ROCK-LIMK pathway. (A) Genotoxic stress induces actin stress fibers. HCT116, MCF-7 or U2OS cells were treated with adriamycin (Adr) (0.2 μg/ml) for 24 h. Cells were then fixed and F-actin structures visualized with Texas Red-conjugated phalloidin. Scale bars = 20 μm. (B) Genotoxic stress activates Rho. Active Rho-GTP was affinity purified using recombinant Rhotekin Rho-binding domain from MCF-7 cells treated with actinomycin-D (ActD; 2 nM) or Adr (0.2 μg/ml) for 16, 20 or 24 h. ERK2 immunoblotting showed equivalent protein loading. (C) Genotoxic stress leads to LIMK and cofilin phosphorylation. MCF-7 cells were treated with ActD (2 nM) or Adr (0.2 μg/ml) in the presence or absence of Y-27632 (10 μM) or Tat-Myc-C3 (80 μg/ml) for 24 h. Whole cell lysates were immunoblotted with antibodies against ROCK1, phospho-LIMK1 (Thr508)/LIMK2 (Thr505), LIMK1, LIMK2, phosphocofilin (Ser3), cofilin or Myc-epitope. LIMK phosphorylation was blocked by ROCK inhibitor Y-27632 or Rho inhibitor Tat-Myc-C3. Equivalent protein loading was confirmed by ERK2 immunoblotting. (D) Ionizing radiation activates LIMK and leads to cofilin phosphorylation. MCF-7 cells were treated with or without IR (2, 4, 8 and 12 Gy) and whole cell lysates prepared after 24 h. Lysates were immunoblotted with antibodies against ROCK1, LIMK1, LIMK2, phosphocofilin (Ser3) and cofilin. ERK immunoblotting indicated equivalent protein loading. (E) Genotoxic stress does not promote stress fiber formation in cells with mutant p53. MDA-MB-231 cells were treated with Adr (0.2 μg/ml) for 24 h. Cells were then fixed and F-actin structures visualized with Texas Red-conjugated phalloidin. Scale bar = 20 μm. (F) LIMKi inhibits adriamycin-induced actin stress fiber formation and cofilin phosphorylation. U2OS cells were treated with Adr (0.2 μg/ml) in the presence or absence of LIMKi (10 μM) for 24 h. Cells were then fixed and F-actin structures visualized with Texas Red-conjugated phalloidin. Scale bar = 20 μm. MCF-7 cells were treated with Adr (0.2 μg/ml) in the presence or absence of LIMKi (3 μM) for 24 h. Whole cell lysates were immunoblotted with antibodies against phosphocofilin (Ser3), cofilin and LIMK2. Equivalent protein loading was confirmed by LIMK1 immunoblotting.
Small-molecule inhibitors of LIMK with low nanomolar IC50 potencies have recently been described 14. One of these compounds, N-{5-[2-(2,6-dichloro-phenyl)-5-difluoromethyl-2H-pyrazol-3-yl]-thiazol-2-yl}-isobutyramide (compound 3 in reference Ross-Macdonald et al. 14; hereafter termed LIMKi) is a highly selective, non-cytotoxic and potent inhibitor of LIMK1 and LIMK2 (Supplementary information, Figure S1A). Co-administration with LIMKi (3 μM) effectively inhibited Adr-induced actin stress fiber formation and cofilin phosphorylation (Figure 1F); dose-response analysis revealed the LIMKi EC50 for Adr-induced cofilin phosphorylation to be ∼400 nM (Supplementary information, Figure S1B). These results indicate that the DNA damage response is programmed to activate the Rho-ROCK-LIMK pathway independently of caspase-mediated cleavage of ROCK1 and induce actin cytoskeleton reorganization through phosphorylation of key regulatory substrates.
RHOC is a p53 target gene
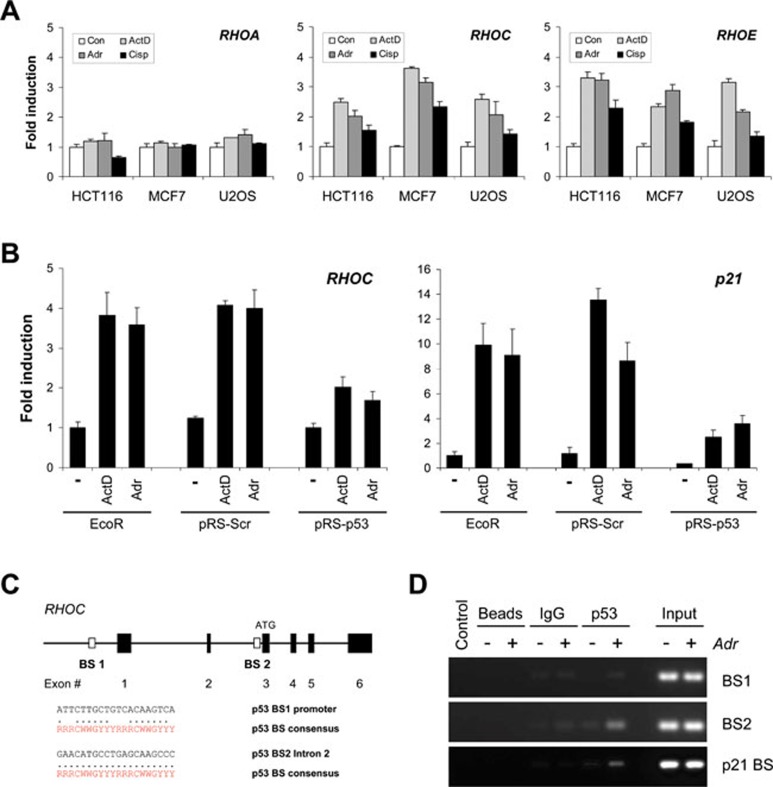
Since genotoxic stress led to the formation of actin stress fibers and C3-sensitive LIMK and cofilin phosphorylation (Figure 1A and 1C), we analyzed what effect these stresses had on the expression of Rho-GTPases. Treatment of HCT116, MCF-7 and U2OS, which express endogenous wild-type p53, with ActD, Adr or cisplatin (Cisp) increased both RHOC and RHOE mRNA expression, as determined by real-time quantitative PCR (qPCR; Figure 2A). In contrast, RHOA mRNA levels were unaffected by treatment with these chemotherapeutic agents (Figure 2A). Given that MDA-MB-231 cells, which express mutant p53, fail to induce stress fibers in response to genotoxic stress (Figure 1E) and that RHOE has previously been identified as a p53 target gene 13, we examined whether increased RHOC expression was similarly mediated by p53. MCF-7 cells (p53 wild-type) that stably express short-hairpin RNA (shRNA) to silence p53 (pRS-p53) or scrambled non-targeting (NT) shRNA (pRS-Scr) were treated with ActD or Adr for 24 h. Similar to the well-validated p53 target gene p21WAF1/CIP1 (p21), the increased expression of RHOC mRNA following genotoxic stress was markedly reduced in cells where p53 levels were knocked down (Figure 2B).
Figure 2.
RHOC is p53 target gene. (A) Genotoxic stress induces RHOC expression. HCT116, MCF-7 or U2OS cells were treated with ActD (2 nM), Adr (0.2 μg/ml) or cisplatin (Cisp; 25 μM) for 24 h. RHOA, RHOC and RHOE mRNA levels were determined by qPCR. Data are presented as mean fold induction ± SEM (n = 3) relative to vehicle control (Con) in each cell line. (B) RHOC induction by genotoxic stress is p53 dependent. Parental MCF-7-Eco, control MCF-7-pRS-Scr and p53-knockdown MCF-7-pRS-p53 cells were treated with ActD (2 nM) or Adr (0.2 μg/ml) for 24 h. RHOC and p21 mRNA levels were determined by qPCR. Data are presented as mean fold induction ± SEM (n = 3), relative to vehicle control. (C) Schematic diagram of the human RHOC gene showing potential p53-binding sites (BS). The p53MH algorithm identified p53-binding sites within the upstream regulatory region (BS1) and the second intron (BS2) of the human RHOC gene. Within each binding site, the individual half-sites are compared with the consensus p53-binding site sequence, where R = purine, Y = pyrimidine and W = adenine or thymine. (D) p53 binds the BS2 element within RHOC intron 2. Chromatin immunoprecipitation was carried out with anti-p53 antibody (DO-7) on chromatin isolated from MCF-7 cells treated with and without Adr (0.2 μg/ml) for 8 h. Ethidium bromide-stained agarose gels show PCR products of RHOC BS2 after immunoprecipitation. Control immunoprecipitations were carried out with beads alone (Beads) or with control mouse IgG (IgG). Input represents 0.5% of the total chromatin used in each condition.
Gene transcription may be regulated by the association of p53 with DNA-binding sites (BS) within target genes 3. Utilizing the p53MH algorithm 15 to search genomic sequences, we identified two elements with significant homology to the consensus p53-binding sequence within the human RHOC gene: one upstream of exon 1 (BS1) and the other within intron 2 (BS2) (Figure 2C). Chromatin immunoprecipitation (ChIP) and RT-PCR analysis of genomic DNA from Adr-treated MCF-7 cells revealed that genomic fragments containing BS2 were specifically immunoprecipitated using a p53 antibody, but not with a non-specific IgG or Protein G-coated magnetic beads (Figure 2D). Binding to BS1 was not detected. Immunoprecipitation of a previously characterized binding site within the p21 promoter 16 was used as a positive control. These data indicate that RHOC is a direct p53 target gene.
Regulation of LIMK2 expression by DNA damage and p53
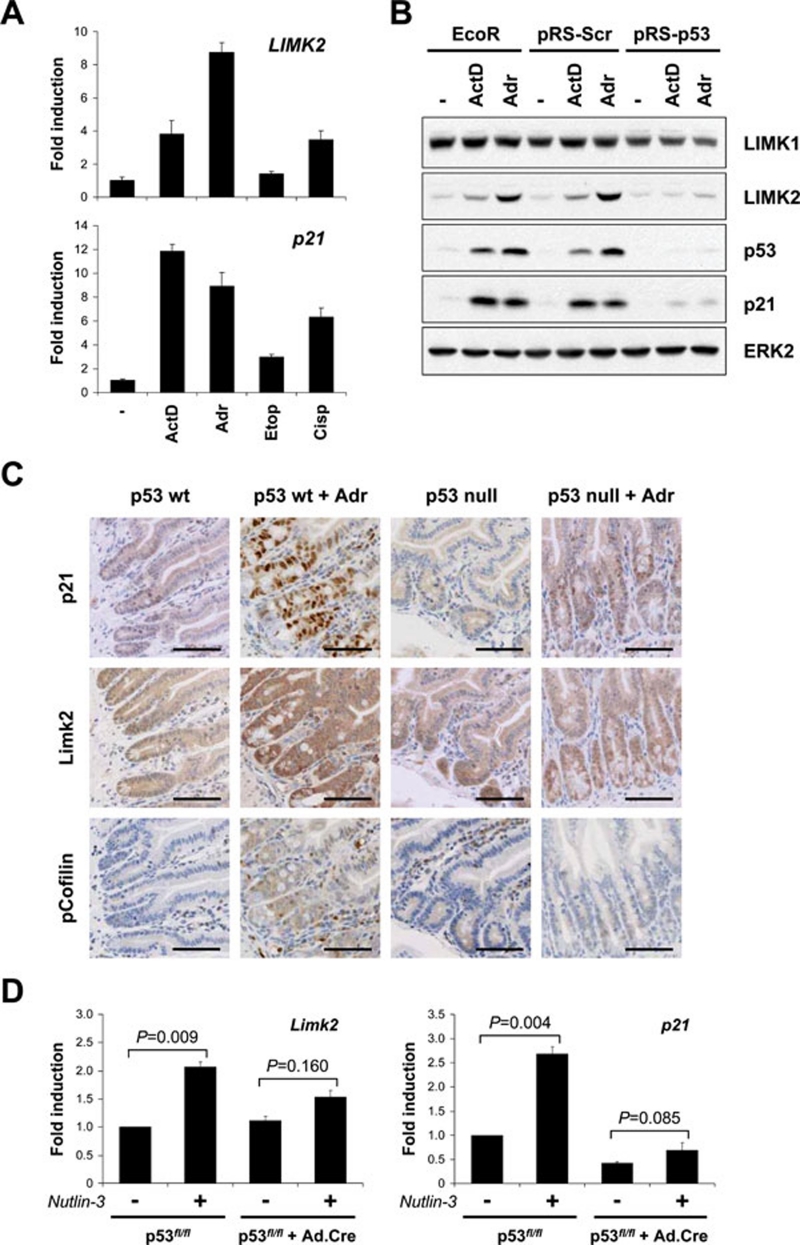
In addition to phosphorylation and activation of LIMK induced by genotoxic stress, we also observed dramatically elevated LIMK2, but not LIMK1, protein levels following treatment with ActD, Adr or IR (Figure 1C and 1D). Elevated LIMK2 protein was associated with increased mRNA expression (Figure 3A), and was unaffected by ROCK or Rho inhibition (Supplementary information, Figure S2). Moreover, treatment with chemotherapeutic agents with differing mechanisms of action, including Adr and Cisp, resulted in increased LIMK2 and p21 mRNA (Figure 3A), but not LIMK1, ROCK1 or ROCK2 mRNA (Supplementary information, Figure S3), suggesting that LIMK2 induction is a general DNA damage response. Neither Y27632 nor Tat-Myc-C3 affected LIMK1, LIMK2, ROCK1, ROCK2 or p21 mRNA, or p21 and p53 protein expression (Supplementary information, Figures S2 and S4).
Figure 3.
LIMK2 induction by DNA damage is p53 dependent. (A) Genotoxic stress induces LIMK2 expression. MCF-7 cells were treated with ActD (2 nM), Adr (0.2 μg/ml), etoposide (Etop; 20 μM) or cisplatin (Cisp; 25 μM) for 24 h. LIMK2 and p21 mRNA levels were determined by qPCR. Data are presented as mean fold induction ± SEM (n = 3) relative to vehicle control. (B) p53 knockdown inhibits genotoxic stress-induced LIMK2 protein expression. Parental MCF-7-Eco, control MCF-7-pRS-Scr and p53-knockdown MCF-7-pRS-p53 cells were treated with ActD (2 nM) or Adr (0.2 μg/ml) for 24 h. Whole cell lysates were immunoblotted with antibodies against LIMK1, LIMK2, p53 or p21. Equivalent protein loading was confirmed by ERK2 immunoblotting. (C) Limk2 induction in mouse small intestinal crypts is p53-dependent. Immunohistochemical analysis of formalin-fixed/paraffin-embedded wild-type or p53-null small intestinal sections taken from mice treated with Adr (single i.p. dose of 10 mg/kg body weight) or saline vehicle control. Scale bars = 100 μm. (D) Nutlin-3 induction of Limk2 is dependent on p53. Following infection of p53fl/fl MEFs with or without Cre-recombinase-expressing adenovirus to delete p53, cells were treated with Nutlin-3 (50 μM) for 24 h. Limk2 and p21 mRNA levels were determined by qPCR. Data are presented as mean fold induction ± SEM in three separate p53fl/fl MEF lines. A two-tailed, paired Student's t-test was used to determine statistical significance.
Microarray analysis that had previously identified DRAM1 also identified LIMK2 as a potential p53-responsive gene 17. Knockdown of p53 by shRNA confirmed that the LIMK2 induction seen following treatment with ActD or Adr was p53 dependent (Figure 3B). To determine whether genotoxic induction of LIMK2 is p53 dependent in normal non-transformed tissues in vivo, we examined Limk2 induction and activation by Adr in AhCre+ p53fl/fl mice, in which Cre-mediated p53 deletion can be induced by β-naphthoflavone treatment 18, 19. In vehicle-treated AhCre+ p53fl/fl mice that express wild-type p53, LIMK2 expression was highest at the base of intestinal crypts (Figure 3C and Supplementary information, Figure S5A). In response to Adr, vehicle-treated AhCre+ p53fl/fl mice showed elevated LIMK2 within the crypts and to a lesser extent throughout the villi (Figure 3C and Supplementary information, Figure S5A), which paralleled increased p21 within the crypts (Figure 3C). Increased phosphocofilin staining in Adr-treated mice indicated that LIMK activation occurs in response to DNA damage in vivo (Figure 3C), consistent with increased cofilin phosphorylation induced by genotoxic agents in vitro (Figure 1C). In contrast, Adr did not increase LIMK2 or p21, or elevate cofilin phosphorylation in β-naphthoflavone-treated AhCre+ p53fl/fl mice that lack intestinal p53 (Figure 3C and Supplementary information, Figure S5A). Similar observations were made in Cisp-treated mice (single i.p. dose of 10 mg/kg; Supplementary information, Figure S5B), indicating that LIMK2 induction and activation in vivo occurs in response to more than one class of DNA-damaging agent, as observed in vitro (Figure 3A). Furthermore, p53 stabilization and activation by the MDM-2 inhibitor Nutlin-3 20 significantly elevated Limk2 and p21 mRNA expression in wild-type MEFs (Figure 3D), but Limk2 and p21 induction was no longer significant following p53 deletion (Figure 3D and Supplementary information, Figure S5C). Interestingly, p53 deletion in untreated MEFs did not affect basal Limk2 expression, indicating that other factors regulate basal Limk2 transcription 21. Taken together, these data show that LIMK2 is induced upon p53 activation in transformed and non-transformed cells.
LIMK2 is a direct p53 target gene
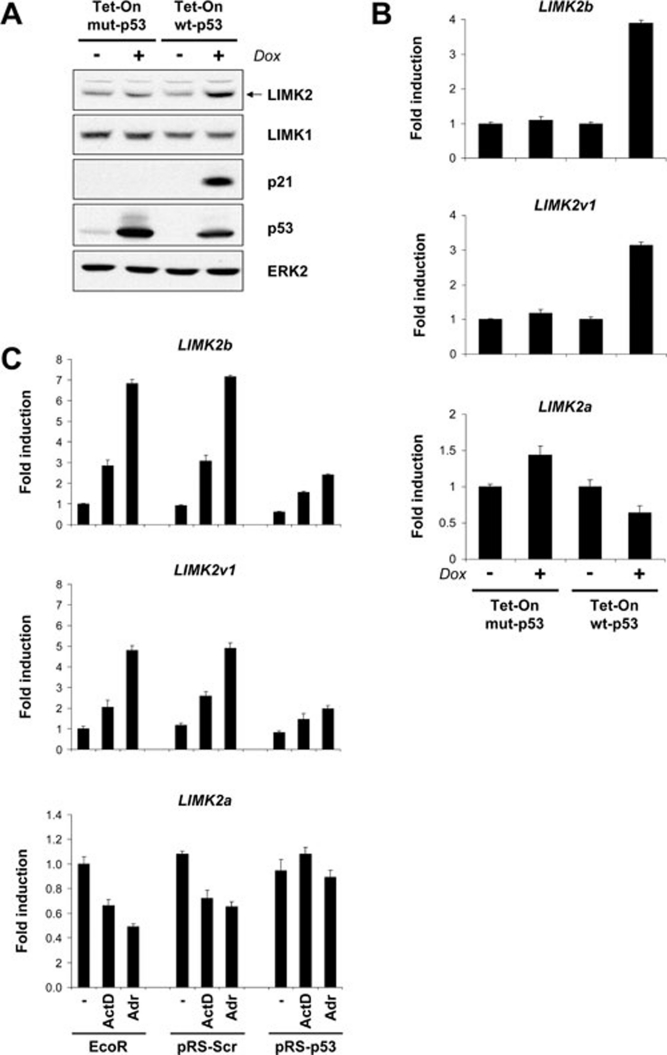
To determine whether elevated p53 is sufficient for LIMK2 induction and whether p53 transcriptional activity is required, Saos-2 cell lines expressing tetracycline-inducible wild-type p53 (Tet-On wt-p53) or a DNA-binding p53 (R175H) mutant (Tet-On mut-p53) 22 were treated with doxycycline (Dox) for 18 h to induce p53. In parallel with p21, LIMK2 protein levels (Figure 4A) were elevated following wt-p53 but not mut-p53 induction, indicating that p53 transcriptional activity is required. Following p53 induction, LIMK2 expression demonstrated similar kinetics of induction to p21 (Supplementary information, Figure S6). The human LIMK2 gene is located on chromosome 22q12.2 and contains 16 coding exons 23. The LIMK2 gene has two distinct promoters that drive expression of the transcript variants 2a and 2b 21 (hereafter termed LIMK2a and LIMK2b, respectively). Alternative splicing of the LIMK2b transcript gives rise to the additional variant 1 (LIMK2v1) 11 (Figure 5A). qPCR analysis of LIMK2 variant expression revealed that LIMK2b and LIMK2v1 mRNA expression was increased following wt-p53 induction, whereas mut-p53 had no apparent effect (Figure 4B). These findings corroborate those of Hsu et al. 24 who recently reported that LIMK2b was a p53 target gene. Surprisingly, expression of LIMK2a was reduced following the induction of wt-53 and modestly elevated by mut-p53 (Figure 4B). qPCR demonstrated that ActD or Adr induced LIMK2b and LIMK2v1 mRNA expression in a p53-dependent manner (Figure 4C). Treatment of MCF-7 cells with these genotoxic agents decreased LIMK2a mRNA expression, while shRNA-mediated knockdown of p53 blocked the decrease in LIMK2a mRNA levels following genotoxic stress, indicating a dependence on p53 (Figure 4C). These data indicate that p53 differentially regulates the expression of variant forms of LIMK2, with the net gain in LIMK2 protein expression due to increased expression of the LIMK2b and LIMK2v1 variants.
Figure 4.
LIMK2 variant isoforms are differentially regulated by p53. (A) Saos-2 Tet-On wt-p53 and Saos-2 Tet-On mut-p53 (R175H) cells were treated with doxycycline (Dox; 1 μg/ml) for 18 h. LIMK2, LIMK1, p21 and p53 protein expression was assessed by western blotting. ERK2 immunoblotting revealed equivalent protein loading. (B) Wild-type p53 induces the expression of LIMK2b and LIMK2v1 mRNA while repressing LIMK2a. mRNA levels were analyzed by qPCR, and data are presented as mean fold induction ± SEM (n = 3) relative to vehicle control. (C) Induction of LIMK2b and LIMK2v1 mRNA and repression of LIMK2a mRNA expression by genotoxic stress is p53 dependent. qPCR data are presented as mean fold induction ± SEM (n = 3) relative to vehicle-treated control.
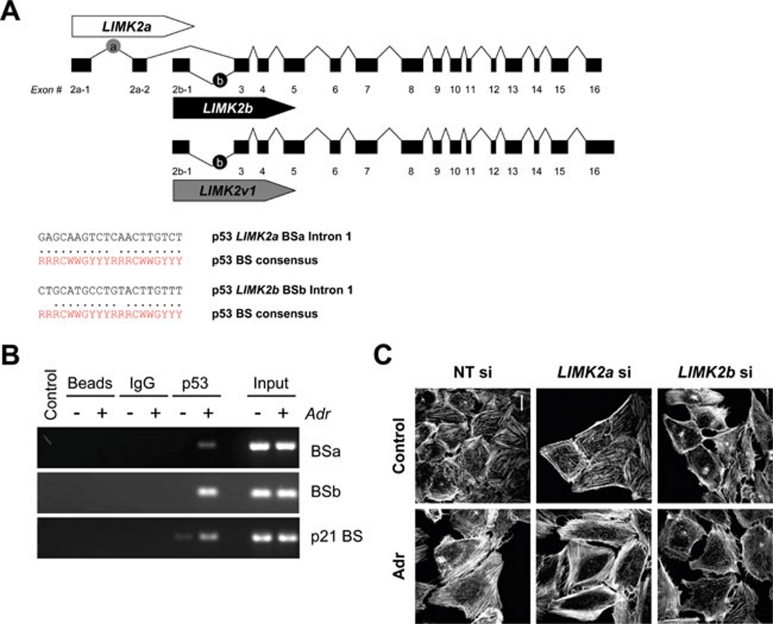
Figure 5.
LIMK2 is a direct p53 target gene. (A) Schematic diagram showing the exon-intron organization of human LIMK2 transcript variants. Potential p53-binding sites (BS) were identified by p53MH within the first intron (BSa) and the alternate first intron (BSb) of human LIMK2a and LIMK2b, respectively. Within each binding site, the individual half-sites are compared with the consensus p53-binding site sequence, where R = purine, Y = pyrimidine and W = adenine or thymine. (B) p53 binds to elements within intron 1 of LIMK2a and LIMK2b. Chromatin immunoprecipitation was performed with anti-p53 antibody (DO-7) on chromatin isolated from MCF-7 cells treated with and without Adr (adriamycin; 0.2 μg/ml) for 8 h. Ethidium bromide-stained agarose gels show PCR products of LIMK2 BSa and BSb after immunoprecipitation. Control immunoprecipitations were carried out with beads alone (Beads) or with control mouse IgG (IgG). Input represents 0.5% of the total chromatin used in each condition. (C) Knockdown of LIMK2b prevents Adr-induced stress fiber formation in U2OS cells. MCF-7 cells transfected with LIMK2b, LIMK2a or non-targeting (NT) control siRNAs were treated with Adr (0.2 μg/ml) for 24 h. Cells were then fixed and F-actin structures visualized with Texas Red-conjugated phalloidin. Scale bars = 20 μm.
Given that there are alternate transcriptional start sties for LIMK2a and LIMK2b, we utilized the p53MH algorithm 15 to identify potential p53-binding sites within the promoter regions and distinct first introns of the LIMK2 variants. ChIP using a p53 antibody and RT-PCR of the immunoprecipitated DNA revealed separate p53-binding sites within intron 1 (BSa) of LIMK2a and the alternate intron 1 (BSb) of LIMK2b (Figure 5A and 5B). These binding sites were not amplified in control immunoprecipitates using Protein G-coated magnetic beads or a nonspecific IgG. Selective siRNA-mediated knockdown of LIMK2 isoforms demonstrated that LIMK2b is responsible for the actin stress fiber formation seen in response to Adr (Figure 5C). Taken together, these data indicate that LIMK2 is a direct p53 target gene and that p53 can either repress or promote the transcription of LIMK2 variants. However, the sum total of these transcriptional events following DNA damage is increased LIMK2 expression and a net gain in signaling output, as indicated by LIMK-dependent increases in cofilin phosphorylation and actin cytoskeleton rearrangements.
LIMK2 knockdown promotes ionizing radiation-induced G2/M arrest
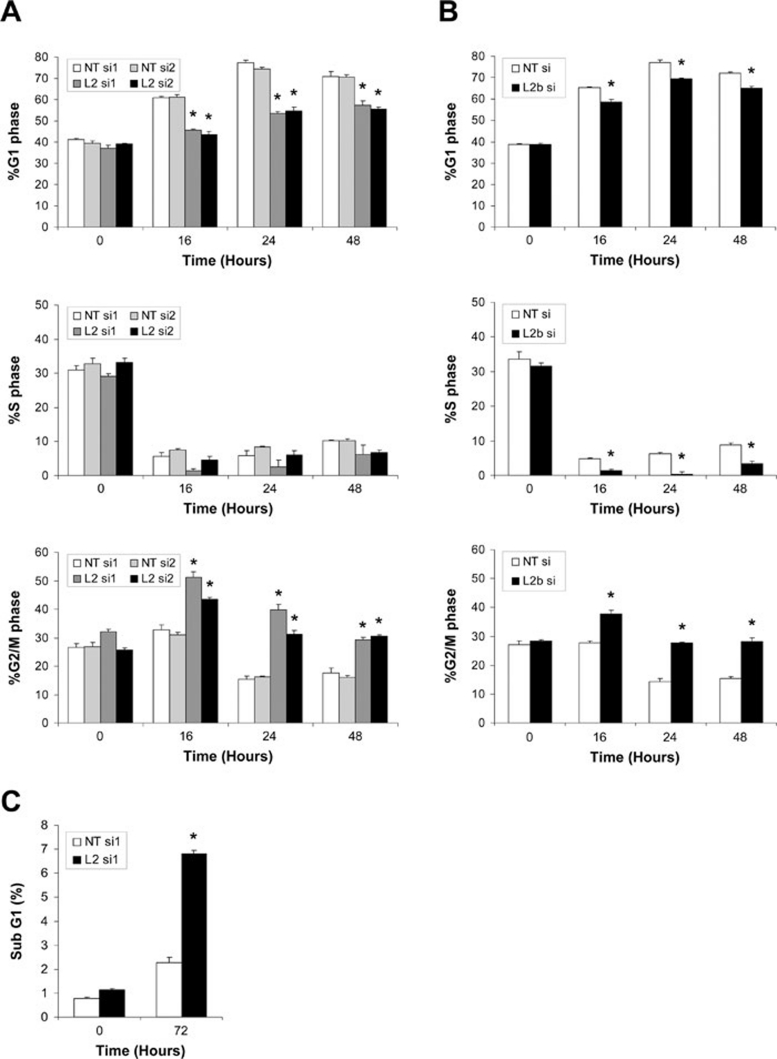
To decipher the role of LIMK2 induction in the p53-mediated DNA damage response, we examined the consequences of siRNA-mediated LIMK2 knockdown on IR-induced cell cycle profiles by staining cells with propidium iodide (PI) followed by flow cytometric analysis. siRNAs against LIMK2 (L2 si1 and si2) effectively knocked down all LIMK2 variants and blocked the induction of LIMK2 protein in response to Adr (Figure 8A and data not shown). Following IR, cells transfected with NT siRNA (NT si1 and si2) exhibited a prominent G1 cell cycle arrest (Figure 6A). In contrast, LIMK2 knockdown significantly reduced arrest in G1 and resulted in a robust G2/M arrest, which was still apparent 48 h after IR treatment (Figure 6A). To determine the role played by the p53-regulated LIMK2b isoform following IR, we utilized a siRNA that specifically reduced LIMK2b expression. Similar to total LIMK2 knockdown, LIMK2b knockdown also resulted in reduced G1 arrest and an arrest in G2/M (Figure 6B). In addition, LIMK2b knockdown prevented entry into S phase following IR (Figure 6B).
Figure 8.
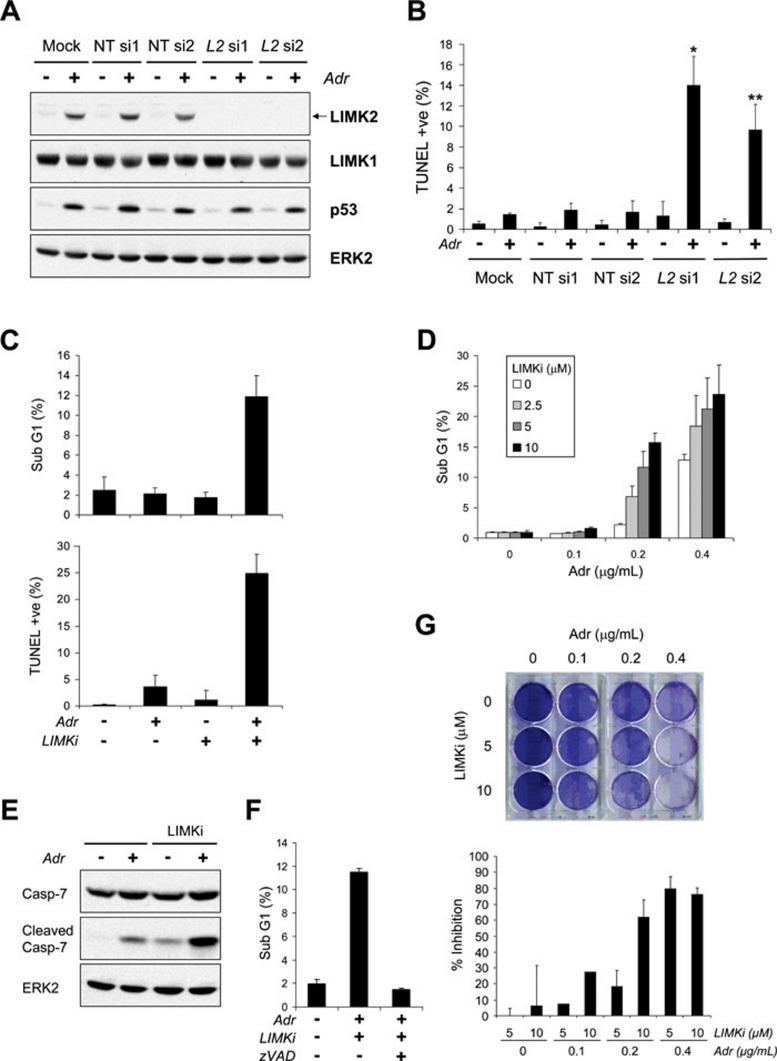
LIMK inhibition synergizes with adriamycin (Adr) to promote apoptosis. (A) siRNA-mediated knockdown of LIMK2. MCF-7 cells were transfected with LIMK2 (L2) or non-targeting control (NT) siRNAs. At 24 h after transfection, cells were treated with Adr (0.2 μg/ml) for a further 72 h. Adherent and non-adherent cells were combined and lysates were subjected to western blotting using antibodies against LIMK2, LIMK1 and p53. Equivalent protein loading was confirmed by ERK2 immunoblotting. (B) LIMK2 knockdown synergizes with adriamycin to promote apoptosis. Apoptotic cell death was measured as mean percentage of TUNEL-positive cells ± SEM (n = 4-5). Statistical significance was determined relative to Mock plus Adr treatment by two-tailed, unpaired Student's t-test (*P = 0.0029 and **P = 0.0016). (C) MCF-7 cells were treated with Adr (0.2 μg/ml) in the presence or absence of LIMKi (10 μM) for 72 h. Adherent and non-adherent cells were collected and processed for PI/TUNEL staining. Apoptotic cell death are shown as the mean percentage of sub-G1 cells ± SEM (n = 4) and mean percentage of TUNEL-positive cells ± SEM (n = 4). (D) LIMKi and adriamycin synergize to affect apoptosis. Apoptotic cell death is shown as the mean percentage of sub-G1 cells ± SEM (n = 5). The combination index (CI) was calculated using the Calcusyn program. (E) Caspase-7 is cleaved and activated following treatment with adriamycin and LIMKi. Adherent and non-adherent cells were collected and immunoblot analysis of whole cell lysates carried out using antibodies against full-length Caspase-7 and cleaved Caspase-7 (Asp198). Equivalent protein loading was confirmed by ERK2 immunoblotting. (F) Combined LIMKi and adriamycin treatment induces caspase-dependent cell death. MCF-7 cells were treated with Adr (0.2 μg/ml) plus or minus LIMKi (10 μM), in the presence or absence of zVAD-fmk (50 μM) for 72 h. Adherent and non-adherent cells were collected and processed for PI staining. Apoptotic cell death is shown as the mean percentage of sub-G1 cells ± SEM (n = 6). (G) Combination treatment of MCF-7 cells affects clonogenic survival. MCF-7 cells were treated with Adr at increasing concentrations in the presence or absence of LIMKi for 48 h. After extensive washing and a 7-day growth period, cells were assessed for clonogenic survival by Giemsa staining and scanning with a Licor Odyssey. Data presented are percentage inhibition relative to the same Adr concentration without LIMKi.
Figure 6.
LIMK2 knockdown promotes IR-induced G2/M arrest. (A) MCF-7 cells transfected with LIMK2 (L2) or non-targeting control (NT) siRNAs were treated with IR (8 Gy). Cells were harvested at 0, 16, 24 and 48 h following irradiation. PI stained cells were analyzed by flow cytometry. Data are shown as the mean percentage of cells in each cell cycle phase ± SEM (n = 3). Statistical significance was determined relative to NT si1 treatment using a one-way ANOVA and Tukey post hoc test (*P < 0.0001). (B) MCF-7 cells transfected with LIMK2b (L2b) or non-targeting control (NT) siRNAs were treated with IR (8 Gy). Cells were harvested at 0, 16, 24 and 48 h following irradiation. Following PI staining, cells were analyzed by flow cytometry. Data are shown as the mean percentage of cells in each cell cycle phase ± SEM (n = 3). Statistical significance was determined relative to control using a one-way ANOVA and Tukey post hoc test (*P < 0.0001). (C) LIMK2 knockdown in combination with IR treatment leads to cell death. MCF-7 cells transfected with LIMK2 (L2) or non-targeting control (NT) siRNAs were treated with IR (8 Gy). Adherent and non-adherent cells were collected after 72 h and stained with PI. Apoptotic cell death is shown as the mean percentage of sub-G1 cells ± SEM (n = 3).
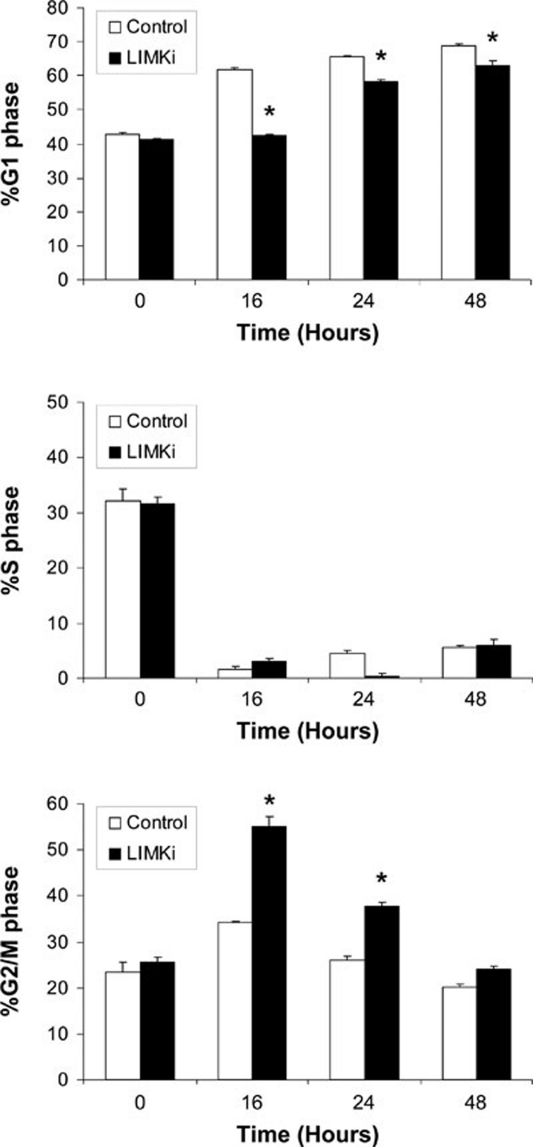
We next used the LIMK inhibitor LIMKi to examine the contribution of LIMK catalytic activity on IR-induced cell cycle arrest. Similar to LIMK2 knockdown, LIMKi (10 μM) significantly reduced G1 arrest together with increased numbers of cells arresting in G2/M (Figure 7). Given that LIMK inhibition and LIMK2b knockdown had similar effects on decreasing the IR-induced G1 arrest with subsequent increased G2/M arrest, the role of increased LIMK2 expression and activity is probably to promote the G1 arrest to allow for DNA repair to occur prior to replication. In the event of reduced G1 arrest, cells proceeded to a G2/M arrest, which when sustained for 72 h promoted apoptosis as measured by increased levels of sub-G1 DNA (Figure 6C).
Figure 7.
LIMK inhibition promotes IR-induced G2/M arrest. MCF-7 cells were treated with or without LIMKi (10 μM) and then exposed to IR (8 Gy). Cells were harvested at 0, 16, 24 and 48 h following irradiation. Following PI staining, cells were analyzed by flow cytometry. Data are shown as the mean percentage of cells in each cell cycle phase ± SEM (n = 3). Statistical significance was determined relative to control using a one-way ANOVA and Tukey post hoc test (*P < 0.0001).
LIMK2 knockdown sensitizes cells to DNA damage-induced apoptosis
Since p53 has a major role in determining whether cells survive or die following DNA damage 25, we explored how LIMK2 knockdown affected DNA damage-induced apoptosis. LIMK2 induction was effectively silenced by specific siRNAs, but not by NT siRNAs, whereas LIMK1 expression and p53 induction by Adr were unaffected (Figure 8A). Apoptotic cell death was measured by flow cytometric analysis of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining, which detects DNA fragmentation following activation of apoptotic DNases. MCF-7 cells transfected with NT or LIMK2 siRNAs demonstrated nonsignficantly low levels of apoptosis (Figure 8B). However, the combination of LIMK2 knockdown with an Adr concentration (0.2 μg/ml) that inefficiently induced apoptosis as a single agent led to significant increases in TUNEL-positive apoptotic cells (Figure 8B). Cells transfected with NT siRNA showed no significant increase in apoptosis relative to mock-transfection controls (Figure 8B). These data indicate that LIMK2 is a pro-survival factor induced as part of the p53-mediated DNA damage response.
LIMK inhibitor synergizes with genotoxic stress to promote apoptosis
We next examined the effect of LIMKi on apoptosis induced by genotoxic stress. Treatment with 10 μM LIMKi, which was sufficient to inhibit cofilin phosphorylation and actin stress fiber formation (Figure 1F and Supplementary information, Figure S1B), had no effect on apoptosis as determined by TUNEL staining and sub-G1 DNA content (Figure 8C and Supplementary information, Figure S7A). Adr induced considerable G2/M phase arrest, but little apoptosis. However, the combination of LIMKi plus Adr resulted in a substantial increase in apoptosis by both measures (Figure 8C and Supplementary information, Figure S7A). Combining LIMKi with Adr also potentiated apoptosis in both A549 lung adenocarcinoma and LNCaP prostate adenocarcinoma cells (Supplementary information, Figure S7B and S7C). Analysis of TUNEL-positive versus PI-positive FACS plots revealed that cell death following combination treatment occurs at all phases of the cell cycle (Supplementary information, Figure S7A). By varying Adr and LIMKi concentrations, the Chou-Talalay median-effect equation 26 revealed combination index (CI) values of < 1 (MCF-7: CI = 0.54, r = 0.96; A549: CI = 0.22, r = 0.99; LNCaP: CI = 0.80, r = 0.95), indicative of a significant synergistic interaction between Adr and LIMKi in the induction of apoptosis (Figure 8D and data not shown). Similar combination effects of LIMKi and the chemotherapeutic drugs camptothecin and Cisp were also observed (Supplementary information, Figure S7D and S7E). Consistent with these measures of apoptosis, Adr plus LIMKi induced greater caspase-7 cleavage than either single treatment (Figure 8E). Moreover, the synergistic increase in cell death was abolished by pre-treatment with zVAD-fmk (Figure 8F), indicating that apoptosis was caspase mediated. Combining Adr with LIMKi did not affect the extrinsic apoptosis pathway, since caspase-8 and BID were not cleaved in response to this drug combination, in contrast to treatment with anti-Fas receptor antibody (Supplementary information, Figure S8). Given that sub-apoptotic doses of Adr induced robust cell cycle arrest, one question is whether there is a long-term benefit in combining Adr with LIMKi. To answer this, we examined the clonogenic survival of cells treated with varying dose combinations of LIMKi and Adr for 48 h, followed by extensive washes and a 7-day growth period. While LIMKi alone had no effect at any concentration, when combined with 0.2 or 0.4 μg/ml Adr, there was a combined effect in decreasing survival (Figure 8G). These data indicate that RHOC and LIMK2 induction, with consequent increased LIMK2 activity, make a major contribution to survival from DNA damage-induced apoptosis. Taken together, these results show that LIMK2 expression and activation are triggered as part of a p53-mediated DNA damage response, which results both in cytoskeletal rearrangements and a previously unknown pro-survival function that protects cells from apoptosis.
Discussion
p53 coordinates a sophisticated program of cellular responses to prevent the deleterious effects of DNA damage 2, 3. In cells with wild-type p53, DNA damage causes activation of repair mechanisms, cell cycle arrest and activation of pro-survival pathways to permit DNA damage repair. If DNA damage is too extensive, p53 activation can also result in cell death. Loss of p53 function contributes to cancer initiation and progression by a reduced ability to initiate DNA damage checkpoints, cell cycle arrest and repair mechanisms, leading to unrestrained proliferation and tolerance for genomic instability. Despite years of research, little is known about how regulators of the actin cytoskeleton contribute to p53-regulated outcomes. We now show that the actin cytoskeleton regulators RhoC and LIMK2 are direct p53 target genes, and that the consequent activation of LIMK2 has a pro-survival function following DNA damage.
Research has shown that actin cytoskeleton disruption with agents such as cytochalasin or latrunculin analogs sensitizes cells to apoptotic stimuli, including staurosporine 27, CD95 28, loss of adhesion 29 and mechanical strain 30. Conversely, actin stress fibers induced by shear stress protect cells from this death inducer 31. Therefore, actin stress fiber formation may be a general stress response that helps protect cells from potential apoptotic stimuli like DNA damage. Interestingly, genetic deletion of the F-actin-stabilizing protein Nd1 increased Adr sensitivity 32, whereas its overexpression protected against Adr -induced apoptosis 33.
As LIMK2 inhibition sensitized cells to DNA damage-induced apoptosis, it follows that its increased expression would be associated with chemoresistance. Chemosensitivity of human cancer cell lines to anti-cancer drugs revealed that elevated LIMK2 expression correlated with resistance to 18 drugs (including topoisomerase I inhibitors and anthracyclines like Adr) 34. Furthermore, neuroblastoma cell lines selected for resistance to vincristine, vinblastine or desoxyepothilone B exhibited increased LIMK2 expression, whereas LIMK2 knockdown increased the sensitivity of NB SH-EP cells to vincristine 35. Increased LIMK2 expression is also associated with progression of myelodysplastic syndrome to chemoresistant leukemia 36. These results suggest that mechanisms that enhance LIMK2 expression may be common contributors to chemotherapeutic drug resistance. As a result, combining LIMK inhibitors with chemotherapeutic agents may enhance their efficacy, and potentially allow chemoresistance to be reversed. Since administration of LIMK inhibitors may be useful as an anti-metastatic strategy 10, their additional possible use in combination therapy as a chemosensitizing agent 37 increases their potential therapeutic value.
Consistent with our results, RHOE was previously reported to be regulated by p53 13. Although RhoE has been shown to inhibit ROCK1 activity through binding to its amino-terminus 38, the increased RhoC expression observed following genotoxic stress has a net greater effect on downstream signaling, since LIMK and cofilin are phosphorylated in a Rho/ROCK-dependent manner. Whether this indicates that ROCK2 is the major mediator of the DNA damage-induced activation of this pathway remains to be determined.
While the results of this study were in preparation, Hsu et al. 24 reported that LIMK2b was a direct p53-target gene. However, there were several major differences with our study. While we concluded that LIMK2 induction and activation contributes to radiation-induced G1 arrest and that knocked down or inhibited cells proceeded to a prolonged G2/M arrest that ultimately led to apoptosis, Hsu et al. reported that depletion of LIMK2 promoted exit from G2/M. One likely explanation for the differences between the two studies stems from the IR dosage used. As we found that cofilin phosphorylation and, therefore, LIMK activity were maximal at 8 Gy (Figure 1D), we used that dose to examine cell cycle dynamics and limit off-target effects. However, Hsu et al. used 20 Gy, which we found resulted in a G2/M arrest (data not shown) that may have masked effects on G1 phase arrest. In addition, while we used siRNA and a selective pharmacological LIMK inhibitor, Hsu et al. often used overexpression of dominant-negative LIMK2 or cofilin mutants to dissect LIMK2 function following DNA damage. Although the studies largely agree, methodological differences likely account for some of the discrepancies. Nonetheless, one of the most exciting conclusions from our study is the potential for LIMK inhibitors as sensitizing agents to increase the efficacy of cancer therapies that act by inducing DNA damage.
Materials and Methods
Antibodies and reagents
Antibodies to BID (2002), caspase-7 (9492), cleaved caspase-7 (Asp198; 9491), caspase-8 (9746), phosphocofilin (Ser3; 3311), LIMK1 (3842), phospho-LIMK1 (Thr508)/LIMK2 (Thr505; 3841) and Myc-Tag (2276) were from Cell Signaling Technologies. Antibodies against Lamin A/C (sc-6215), LIMK2 (sc-5577) and p21 (sc-397G) were from Santa Cruz Biotechnology Inc. Anti-Fas (human, activating; Clone CH11; 05-201) and anti-Rho (-A, -B, -C; Clone 55; 05-778) were obtained from Millipore. Anti-p53 antibodies DO-1 (554293) and DO-7 (554294) were obtained from BD Biosciences. Additional antibodies used in this study were cofilin (ACFL02; Cytoskeleton Inc.) and ERK2 (Ab122; CJ Marshall, Institute of Cancer Research, London). Goat anti-mouse, goat anti-rabbit and rabbit anti-goat horseradish peroxidase (HRP)-conjugated antibodies were from Pierce. Reagent sources: ActD (Sigma); zVAD-fmk (Biomol); Adr, camptothecin, Cisp, etoposide, Nutlin-3 and Y-27632 (Calbiochem). LIMK inhibitor (LIMKi; N-{5-[2-(2,6-dichloro-phenyl)-5-difluoromethyl-2H-pyrazol-3-yl]-thiazol-2-yl}-isobutyramide) was synthesized as previously described 14. Inhibitors: Tat-C3 (used at 80 μg/ml); LIMKi (in DMSO, used at 0.1-10 μM); Nutlin-3 (in DMSO, used at 50 μM); Y-27632 (in water, used at 10 μM); and zVAD-fmk (in DMSO, used at 50 μM).
Generation of cell lines
MCF-7-pRS-Scr and MCF-7-pRS-p53 shRNA-expressing cells were generated by transduction of ecotropic receptor-expressing MCF-7 (MCF-7-Eco) with pRetroSuper retroviruses containing non-silencing shRNA or shRNA targeting p53 17. Cells were selected with 2.5 μg/ml puromycin. Saos-2 Tet-On wt-p53 and Saos-2 Tet-On mut-p53 (R175H) cells have been described previously 17, 22.
Cell culture and siRNA
A549, LNCaP, MBA-MB-231, MCF-7-Eco, MCF-7-pRS-Scr, MCF-7-pRS-p53, Saos-2 Tet-On wt-p53 and Saos-2 Tet-On mut-p53 (R175H) cells were maintained in DMEM supplemented with 10% fetal bovine serum (Autogen Bioclear), 2 mM L-glutamine (Gibco) and 100 U/ml penicillin-streptomycin (Gibco) at 37 °C and 5% CO2. Tet-On lines were induced with 1 μg/ml Dox (Sigma). p53 was deleted in vitro by infection of p53fl/fl MEFs with Ad-CMV-Cre-GFP for 72 h. MCF-7 and U2OS cells were transfected with human LIMK2 (L2 si1 (D-003311-06), L2 si2 (D-003311-07)), human LIMK2a (5′-AGCCAGATATGGTACAGGATT-3′), human LIMK2b (5′-TTCACCTCCAGAGACCTGTAA-3′) and NT (NT si1 (D-001810-02) and NT si2 (D-001810-03)) siRNA (Dharmacon) using Oligofectamine (Invitrogen) according to manufacturer's instructions.
Immunofluorescence and immunohistochemistry
Cells grown on glass coverslips were prepared as described previously 39. F-actin structures were visualized by staining with Texas Red-conjugated phalloidin (Molecular Probes). Confocal laser-scanning microscopy was carried out using a Nikon A1R confocal microscope and NIS Elements software (AR 3.0). Immunohistochemistry was done on paraffin sections as described previously 40 using antibodies against LIMK2 (1:50), phosphocofilin (Ser3; 1:50) and p21 (1:100). Staining was visualized using Envision Dual-link system peroxidase and DAB reagent (Dako). Slides were mounted in DPX (Sigma) and visualized using an Olympus BX51 microscope and CellF imaging software (Olympus).
Cell extraction and immunoblot analysis
RIPA lysates were prepared as described previously 39. Whole cell lysates (60 μg) were separated by SDS-PAGE, transferred to Protran nitrocellulose membranes (Whatman) and probed with primary antibodies and appropriate HRP-conjugated secondary antibodies (Pierce). Blots were visualized using ECL (Pierce) or Supersignal West Femto (Pierce) according to manufacturer's instructions.
Measurement of Rho activity
Cells were lysed in MLB buffer (50 mM Tris (pH 7.2), 150 mM NaCl, 1% (v/v) TX 100, 10 mM MgCl2, 1 mM PMSF and protease inhibitors). Cleared lysates were incubated for 45 min at 4 °C with glutathione-agarose beads coupled to GST-rhotekin RBD. Beads were washed with MLB buffer and bound proteins solubilized by boiling with 3× Laemmli buffer and separated by SDS-PAGE. Rho-GTP and total Rho were detected using anti-Rho (A, B, C) antibody (Upstate).
qPCR analysis
RNA was prepared using an RNeasy Mini kit (Qiagen). cDNA was synthesized and qPCR carried out using the DyNAmo SYBR Green two-step qRT-PCR kit (Finnzymes). Validated QuantiTect human primer sets (Qiagen) for CDKN1A (p21), LIMK1, LIMK2, LIMK2v1, LIMK2a, LIMK2b, RHOA, RHOC, RHOE, ROCK1 and ROCK2 were used. For amplification of MEF cDNA, primer sets for Cdkn1a (Qiagen), Limk2 (Qiagen) and Trp53 (Fwd, 5′-TTATCCGGGTGGAAGGAAAT-3′ Rev, 5′-CACGAACCTCAAAGCTGTCC-3′) were used. Data collection was carried out using a Chromo4 real-time PCR detector (BioRad) and MJ Opticon Monitor software. Expression was normalized relative to 18S rRNA levels (Fwd, 5′-GTAACCCGTTGAACCCCATT-3′ Rev, 5′-CCATCCAATCGGTAGTAGCG-3′).
Mice
Cre-mediated excision of the p53 gene in p53fl/fl;Ah-Cre mice was effected using β-naphthoflavone (80 mg/kg, i.p.). p53fl/fl wild-type and p53-null intestines were isolated, fixed in formalin and embedded in paraffin. Paraffin sections (6 μm) were prepared using citrate-antigen retrieval 40.
ChIP analysis
Chromatin was prepared from MCF-7 cells treated with or without 0.2 μg/ml Adr for 8 h. ChIP assays were performed using the ChIP-IT Express kit (Active Motif) according to manufacturer's instructions. Briefly, sonicated chromatin was immunoprecipitated with anti-p53 monoclonal antibody (3 μg; Clone DO-7, BD Biosciences). Following reversal of DNA-protein crosslinks, DNA was purified using spin columns (Qiagen). Eluted DNA was amplified using primers (Supplementary information, Table S1) spanning the putative p53-binding sites within LIMK2 and RHOC.
Cell cycle analysis
Total populations of cells were harvested and then fixed and permeabilized in 100% ice-cold methanol. PI staining was performed by incubation with PI (50 μg/ml) plus RNAse A (125 μg/ml) for 45 min at room temperature. Flow cytometric analysis was carried out using a FACSCalibur flow cytometer (Becton Dickinson) and CellQuest software. Cell cycle distribution was analyzed using FlowJo (v7.2.2) software.
Cell death assays
Total populations of cells were harvested, fixed in 4% (v/v) p-formaldehyde and permeabilized with 80% (v/v) ethanol. PI-TUNEL staining was done using an In Situ Cell Death Detection (fluorescein) kit (Roche) followed by incubation with PI (50 μg/ml) plus RNAse A (125 μg/ml) 22. Flow cytometric analysis was carried out using a FACSCalibur flow cytometer (Becton Dickinson) and CellQuest software. Apoptosis was assessed by sub-G1 DNA content and percentage of TUNEL-positive cells. Clonogenic survival assays were done using MCF-7 cells treated with LIMKi. After 48 h, cells were washed with PBS and the media was replaced. After 7 days, cells were fixed with 4% (v/v) p-formaldehyde and stained with Giemsa (Sigma).
Statistical analysis
Statistical significance was assessed by one-way ANOVA or the two-tailed Student's t-test.
Conflict of interest
Diane Crighton is now employed by Cancer Research Technology, a private company wholly owned by the medical research charity, Cancer Research UK. All other authors declare no conflict of interest.
Acknowledgments
We are grateful to Darren O'Brien and Lindsay Spender for their help with ChIP and FACS analysis. We also thank Arshad Makhdum, Lindsay Spender and Simon Wilkinson for critical reading of the manuscript, Tim Hammonds and Eyal Gottlieb for helpful discussions and Martin Stockley for LIMKi synthesis. We are grateful to the University of Iowa Gene Transfer Vector Core (Iowa City, IA, USA) for Ad-CMV-Cre-GFP. This work was funded by Cancer Research UK.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
LIMK inhibition by a selective small-molecule inhibitor.
Rho or ROCK inhibition does not affect LIMK2 mRNA induction by genotoxic stress.
Genotoxic stress does not induce LIMK1, ROCK1 or ROCK2 mRNA.
Rho or ROCK inhibition do not affect p53 activation. MCF-7 cells were treated with ActD (2 nM) or Adr (0.2 (μg/ml) in the presence or absence of Y-27632 (10 μM) or Tat-C3 (80 μg/ml) for 24 hours.
Limk2 induction in mouse small intestine is p53-dependent.
Time course of LIMK2 induction following p53 activation.
LIMK inhibiton synergizes with DNA-damaging agents to promote apoptosis.
Combination of adriamycin with LIMKi does not activate the extrinsic apoptosis pathway.
Chromatin immunoprecipitation primers
References
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Olson MF. Rho GTPase signalling pathways in the morphological changes associated with apoptosis. Cell Death Differ. 2002;9:493–504. doi: 10.1038/sj.cdd.4400987. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- Croft DR, Coleman ML, Li S, et al. Actin-myosin-based contraction is responsible for apoptotic nuclear disintegration. J Cell Biol. 2005;168:245–255. doi: 10.1083/jcb.200409049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Wickman GR, Samuel MS, Lochhead PA, Olson MF.The Rho-regulated ROCK kinases in cancerIn: van Golen K, ed. The Rho GTPases in CancerNew York: Springer, 2010163–192.
- Bernard O. Lim kinases, regulators of actin dynamics. Int J Biochem Cell Biol. 2007;39:1071–1076. doi: 10.1016/j.biocel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Scott RW, Olson MF. LIM kinases: function, regulation and association with human disease. J Mol Med. 2007;85:555–568. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- Chan CH, Lee SW, Li CF, et al. Deciphering the transcriptional complex critical for RhoA gene expression and cancer metastasis. Nat Cell Biol. 2010;12:457–467. doi: 10.1038/ncb2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongusaha PP, Kim HG, Boswell SA, et al. RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr Biol. 2006;16:2466–2472. doi: 10.1016/j.cub.2006.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ross-Macdonald P, de Silva H, Guo Q, et al. Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol Cancer Ther. 2008;7:3490–3498. doi: 10.1158/1535-7163.MCT-08-0826. [DOI] [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa C, Matsuda K, Fukuda S, Nakamura Y, Arakawa H. p53RDL1 regulates p53-dependent apoptosis. Nat Cell Biol. 2003;5:216–223. doi: 10.1038/ncb943. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Ireland H, Kemp R, Houghton C, et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–1246. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Nomoto S, Tatematsu Y, Takahashi T, Osada H. Cloning and characterization of the alternative promoter regions of the human LIMK2 gene responsible for alternative transcripts with tissue-specific expression. Gene. 1999;236:259–271. doi: 10.1016/s0378-1119(99)00280-2. [DOI] [PubMed] [Google Scholar]
- Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- Osada H, Hasada K, Inazawa J, et al. Subcellular localization and protein interaction of the human LIMK2 gene expressing alternative transcripts with tissue-specific regulation. Biochem Biophys Res Commun. 1996;229:582–589. doi: 10.1006/bbrc.1996.1847. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Lin TY, Chen JY, Shieh SY. p53-Mediated transactivation of LIMK2b links actin dynamics to cell cycle checkpoint control. Oncogene. 2010;29:2864–2876. doi: 10.1038/onc.2010.40. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Chou T-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- Vasilopoulos Y, Gkretsi V, Armaka M, Aidinis V, Kollias G. Actin cytoskeleton dynamics linked to synovial fibroblast activation as a novel pathogenic principle in TNF-driven arthritis. Ann Rheum Dis. 2007;66 Suppl 3:iii23–28. doi: 10.1136/ard.2007.079822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulms D, Dussmann H, Poppelmann B, Stander S, Schwarz A, Schwarz T. Apoptosis induced by disruption of the actin cytoskeleton is mediated via activation of CD95 (Fas/APO-1) Cell Death Differ. 2002;9:598–608. doi: 10.1038/sj.cdd.4401002. [DOI] [PubMed] [Google Scholar]
- Reddig PJ, Juliano RL. Clinging to life: cell to matrix adhesion and cell survival. Cancer Metastasis Rev. 2005;24:425–439. doi: 10.1007/s10555-005-5134-3. [DOI] [PubMed] [Google Scholar]
- Takamatsu H, Takeya R, Naito S, Sumimoto H. On the mechanism of cell lysis by deformation. J Biomech. 2005;38:117–124. doi: 10.1016/j.jbiomech.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Wu CC, Li YS, Haga JH, et al. Directional shear flow and Rho activation prevent the endothelial cell apoptosis induced by micropatterned anisotropic geometry. Proc Natl Acad Sci USA. 2007;104:1254–1259. doi: 10.1073/pnas.0609806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura L, Matsudo Y, Kang M, Takamori Y, Tokuhisa T, Hatano M. Protective role of Nd1 in doxorubicin-induced cardiotoxicity. Cardiovasc Res. 2004;64:315–321. doi: 10.1016/j.cardiores.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Matsudo Y, Takamori Y, Fujimura L, et al. Overexpression of Nd1, a novel Kelch family protein, in the heart of transgenic mice protects against doxorubicin-induced cardiomyopathy. Transgenic Res. 2006;15:573–581. doi: 10.1007/s11248-006-9010-x. [DOI] [PubMed] [Google Scholar]
- Dan S, Tsunoda T, Kitahara O, et al. An integrated database of chemosensitivity to 55 anticancer drugs and gene expression profiles of 39 human cancer cell lines. Cancer Res. 2002;62:1139–1147. [PubMed] [Google Scholar]
- Po'uha ST, Shum MS, Goebel A, Bernard O, Kavallaris M. LIM-kinase 2, a regulator of actin dynamics, is involved in mitotic spindle integrity and sensitivity to microtubule-destabilizing drugs. Oncogene. 2010;29:597–607. doi: 10.1038/onc.2009.367. [DOI] [PubMed] [Google Scholar]
- Miwa H, Teramoto H, Patel V, et al. Genes important for disease progression of MDS-leukemia revealed by 40K cDNA microarray. ASH Annu Meet Abstr. 2005;106:4929. [Google Scholar]
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23:4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft DR, Olson MF. The Rho GTPase effector ROCK regulates cyclin A, cyclin D1, and p27Kip1 levels by distinct mechanisms. Mol Cell Biol. 2006;26:4612–4627. doi: 10.1128/MCB.02061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MS, Suzuki H, Buchert M, et al. Elevated Dnmt3a activity promotes polyposis in Apc(Min) mice by relaxing extracellular restraints on Wnt signaling. Gastroenterology. 2009;137:902–913. doi: 10.1053/j.gastro.2009.05.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LIMK inhibition by a selective small-molecule inhibitor.
Rho or ROCK inhibition does not affect LIMK2 mRNA induction by genotoxic stress.
Genotoxic stress does not induce LIMK1, ROCK1 or ROCK2 mRNA.
Rho or ROCK inhibition do not affect p53 activation. MCF-7 cells were treated with ActD (2 nM) or Adr (0.2 (μg/ml) in the presence or absence of Y-27632 (10 μM) or Tat-C3 (80 μg/ml) for 24 hours.
Limk2 induction in mouse small intestine is p53-dependent.
Time course of LIMK2 induction following p53 activation.
LIMK inhibiton synergizes with DNA-damaging agents to promote apoptosis.
Combination of adriamycin with LIMKi does not activate the extrinsic apoptosis pathway.
Chromatin immunoprecipitation primers