Abstract
Sex and recombination are central processes in life generating genetic diversity. Organisms that rely on asexual propagation risk extinction due to the loss of genetic diversity and the inability to adapt to changing environmental conditions. The fungus-growing ant species Mycocepurus smithii was thought to be obligately asexual because only parthenogenetic populations have been collected from widely separated geographic localities. Nonetheless, M. smithii is ecologically successful, with the most extensive distribution and the highest population densities of any fungus-growing ant. Here we report that M. smithii actually consists of a mosaic of asexual and sexual populations that are nonrandomly distributed geographically. The sexual populations cluster along the Rio Amazonas and the Rio Negro and appear to be the source of independently evolved and widely distributed asexual lineages, or clones. Either apomixis or automixis with central fusion and low recombination rates is inferred to be the cytogenetic mechanism underlying parthenogenesis in M. smithii. Males appear to be entirely absent from asexual populations, but their existence in sexual populations is indicated by the presence of sperm in the reproductive tracts of queens. A phylogenetic analysis of the genus suggests that M. smithii is monophyletic, rendering a hybrid origin of asexuality unlikely. Instead, a mitochondrial phylogeny of sexual and asexual populations suggests multiple independent origins of asexual reproduction, and a divergence-dating analysis indicates that M. smithii evolved 0.5–1.65 million years ago. Understanding the evolutionary origin and maintenance of asexual reproduction in this species contributes to a general understanding of the adaptive significance of sex.
Keywords: Attini, clonality, Formicidae, thelytoky, mutualism
The vast majority of metazoans reproduces sexually, enjoying the benefits of genetic recombination (1–3) such as rapid adaptability to novel ecological conditions (4, 5) and the purging of deleterious mutations from their genomes (6, 7). However, relative to sexually reproducing organisms, an asexual female doubles its fitness by transmitting its entire genetic material to the next generation (8). Despite such obvious short-term fitness advantages, asexual organisms occur only sporadically throughout the tree of life and are predicted to be evolutionarily short-lived and doomed to early extinction (9–11). In contrast to the short-term advantages of asexuality, the adaptive value of sexuality, that is, genetic recombination, is expected to be of long-term benefit (2, 12–14). There remain in evolutionary biology significant unexplored questions about whether sexual reproduction is favored by natural selection over short evolutionary time spans and, if not, why sexual reproduction persists as the prevalent mode of reproduction, given that the selective benefits are deferred. Studying the origin and evolution of parthenogenetic lineages, and understanding how genetic diversity is generated and preserved in such lineages, is essential to answering these questions.
Asexual reproduction by females, or thelytokous parthenogenesis, has recently been reported in queens of the fungus-growing ant Mycocepurus smithii in three geographically distant populations in Latin America: Puerto Rico (15), Panama (16), and Brazil (17). The widespread geographic distribution of asexuality and the complete absence of males from field collections and laboratory colonies suggested that M. smithii might be obligately asexual (16, 17), and one study proposed that asexuality in this species might be ancient (16). Among bees, wasps, and ants, thelytokous parthenogenesis has so far been observed in the Cape honey bee (18, 19) and in 12 distantly related species of ants (17, 20–23). Population-genetic studies of some species revealed a diversity of highly complex genetic systems, including different cytogenetic mechanisms used to produce workers and queens, facultative sexual reproduction, and clonal male lineages (23–27). Asexual eusocial Hymenoptera produce diploid offspring via meiotic parthenogenesis, or automixis, in which a limited amount of genetic variability is generated through fusion of sister nuclei (28–31). In contrast, mitotic parthenogenesis, or apomixis, in which offspring are genetic clones of their mothers, has not been demonstrated unambiguously in social insects.
Although many theoretical studies predict the costs and benefits of sex, little is known about the evolution of asexuality at the organism level (2). To study the origin and maintenance of parthenogenesis and to elucidate the mechanisms generating genetic diversity in parthenogenetic lineages, we investigated the evolutionary history of the asexual fungus-growing ant M. smithii. To test for obligate asexuality in M. smithii, we developed highly variable short tandem repeat (or microsatellite) markers and analyzed colonies from multiple populations across the species's broad range, extending from Mexico to Argentina and including some Caribbean islands (32, 33). To identify the genetic structure within and between populations of M. smithii and to infer the cytogenetic mechanism underlying parthenogenetic reproduction, we genotyped sterile workers and reproductive queens from 234 colonies. Clonality was inferred by genetic identity between nest mates. Controlled laboratory breeding experiments complemented our field observations. To test for a potential hybrid origin of parthenogenesis in M. smithii, we reconstructed a molecular phylogeny of the genus Mycocepurus. An additional fine-scaled mitochondrial phylogeny of asexual and sexual M. smithii populations was used to investigate whether asexuality arose once or multiple times independently from sexually reproducing ancestors. Lastly, we performed a divergence-dating analysis to estimate the time span over which parthenogenesis has persisted in M. smithii, because asexuality was previously proposed to be of ancient origin.
Results
Population-Genetic Analyses.
A total of 1,930 M. smithii individuals from 234 colonies collected at 39 different localities in Latin America (Fig. 1 and Table S1) was genotyped at 12 variable microsatellite loci yielding 106 alleles (range: 2–15 alleles per locus). The number of alleles per locus per individual never exceeded two, indicating diploidy of females. Of the genotyped populations, 89.7% (n = 35) showed population-genetic signatures of clonality, whereas 10.3% (n = 4) showed an increase of unique multilocus genotypes, indicative of genetic recombination caused by sexual reproduction.
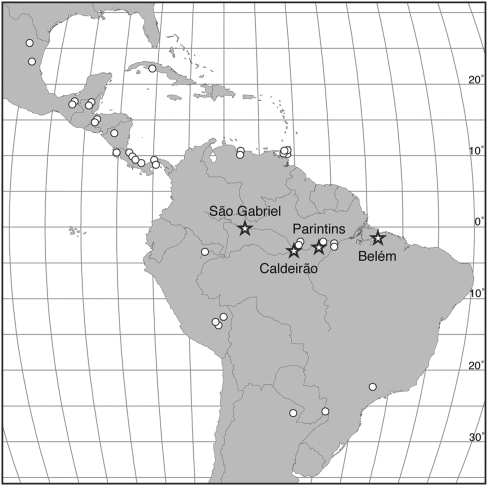
Fig. 1.
Geographic distribution of sexual (stars) and asexual (circles) M. smithii populations. Localities refer to the sexual populations, distributed along the Rio Amazonas and the Rio Negro. Asexual populations are widely distributed in Latin America, ranging from northern Mexico to northern Argentina. Lines of longitude and latitude are separated by units of 5°.
Asexual populations.
A total of 1,647 individuals from 218 colonies in 35 populations exhibited genetic signatures of clonal reproduction. Asexual reproduction was characterized by sharing of repeated multilocus genotypes among individuals (Table S1), maximum deviation from random mating (FIS = −1; Table S2), and a low genotype-to-individual ratio (i.e., G:N approaching 0, whereas a G:N of 1 indicates that each individual is genetically distinct from another) (Table S1). To determine the number of independently evolved asexual lineages that arose at different localities from the sexual population, we estimated the probability that slightly different multilocus genotypes originated from separate sexual events (psex > 0.01) instead of arising from accumulated mutations or scoring errors (psex < 0.01). In addition, clonal diversity (R) was calculated.
Among all M. smithii populations, 66 asexual genotypes were identified, 57 of them representing unique multilocus genotypes (R = 0.86; Tables S2 and S3). Five repeated multilocus genotypes were shared between 10 geographically proximate populations (∼10–40 km distance), and three unique genotypes were identified in seven geographically distant populations (∼700–2,600 km distance; Tables S2 and S3). Calculating the probability that repeated multilocus genotypes from different populations originated from distinct sexual events revealed that identical multilocus genotypes belong to the same clonal lineage (psex < 0.01), indicating long-distance dispersal events of individuals from the same asexual lineage. No genetic variation was present within repeated multilocus genotypes (FIS = −1), but significant genetic variance was structured among them [analysis of molecular variance (AMOVA); FST = 0.624, P = 0.01].
A comparison of the 57 unique multilocus genotypes revealed high frequencies of low genetic distances between genotypes, resulting in a bimodal frequency distribution of genetic distances and indicating the potential existence of mutations or scoring errors in clones (34). Eleven multilocus genotype pairs differed from one other genotype only by a single allele, reducing the number of asexual lineages that potentially originated from distinct sexual events to 46 (psex < 0.01, R = 0.69). Further lowering the threshold and allowing two to six alleles to be shared among multilocus genotypes within an independently evolved clonal lineage, we identified 43 (R = 0.65) to minimally 38 (R = 0.57) independently evolved clonal lineages.
In 20 clonal populations, only a single multilocus genotype was encountered across different colonies. In 15 populations, two to maximally six multilocus genotypes coexisted at a single site (Table S1). In five populations, all or a subset of multilocus genotypes differed by one to six alleles, suggesting a single colonization event followed by diversification within clonal lineages due to the accumulation of mutations or scoring errors (Table S1). In contrast, 12 populations harbored multilocus genotypes differing by 7–15 alleles, indicating independent colonization events of these sites by distantly related clonal foundress queens. The highest diversity of clonal lineages (n = 5) was discovered at a Peruvian lowland rainforest site (Los Amigos).
Genetic uniformity across all loci within colonies suggests either mitotic parthenogenesis (apomixis) as the cytogenetic mechanism underlying thelytokous parthenogenesis in M. smithii or, alternatively, automixis with central fusion and low recombination rates. To trace the genotypes of reproductive individuals over multiple generations, we propagated M. smithii colonies in the laboratory for six consecutive generations and genotyped all 93 queens at the end of the experiment. All queens were genetically identical across generations, and transitions from a heterozygous locus in the mother to a homozygous locus in the offspring was not observed, as would be expected under automixis with central fusion. Interestingly, in field-collected populations in which 7 of the 11 multilocus genotype pairs differ by only a single allele and are identical at all other loci, we observed that one genotype was heterozygous at a given locus whereas the other was homozygous at the same locus. These transitions could indicate a switch from heterozygosity to homozygosity, as expected under automixis. Without knowing which one of these two is the maternal or the offspring genotype, however, it is not possible to distinguish between a transition from a heterozygous to a homozygous state caused by infrequent recombination or an accumulation of “somatic” mutations.
Recombining populations.
Four Amazonian populations, distributed along the Rio Amazonas and the Rio Negro (Fig. 1), exhibited population-genetic signatures of genetic recombination, indicative of sexual reproduction (Tables S1 and S2). Among 283 genotyped individuals, 210 multilocus genotypes were identified, resulting in high genotype-to-individual (G:N) ratios, ranging from 0.71 to 1 (Table S1). Recombinant populations were characterized by inbreeding indices diverging from genetic fixation (FIS = −1), ranging from 0.03 to −0.77, and observed and expected heterozygosities were similar, as expected for populations under Hardy–Weinberg conditions (Table S2).
Because multiple colonies were collected from the Caldeirão population in Amazonas, Brazil (Fig. 1), we investigated this population in detail to test for sexual reproduction. Genotyping of 243 individuals (234 workers, 5 queens, 4 spermatheca contents) revealed the existence of 173 unique multilocus genotypes, of which 132 multilocus genotypes were represented by single individuals whereas the remaining 41 multilocus genotypes were shared by 111 individuals. Among the shared multilocus genotypes, two or at most six nestmates carried identical genotypes. After removing identical genotypes from the dataset, we tested whether genotypes that differ by only a single allele are derived from distinct sexual events or from somatic mutations or scoring errors. Among those unique genotypes (n = 173), 55 multilocus genotypes likely belonged to the same clonal lineage (psex < 0.01), whereas 118 multilocus genotypes probably originated from distinct sexual events (psex > 0.01). This result indicates that 48.6% (118 multilocus genotypes out of 243 individuals) of the genotyped individuals result from sexual reproduction. Such a mixture of recombinant and clonal offspring within a single population suggests that sexual M. smithii queens either occasionally reproduce parthenogenetically or, alternatively, that a larger number of clonally reproducing queens coexists with sexual queens in the same colony. Facultative asexual reproduction by otherwise sexual queens seems more likely, however, given the high number of shared genotypes in the Caldeirão population (n = 41), contrasting with the low number of individuals sharing a multilocus genotype (n = 2–6). After excluding repeated genotypes, observed and expected heterozygosities were almost identical (Ho = 0.372, He = 0.369) and the inbreeding index was indicative of random mating (FIS = −0.009) (Table S2).
To directly test whether queens were fertilized, the abdomens of four (out of five) queens were dissected, revealing sperm-filled spermathecae and reproductively active ovaries. The spermatheca contents (n = 4) were identified as sperm under 200× magnification and subsequently genotyped. The sperm from each spermatheca were haploid at all loci, as expected from hymenopteran males developing from unfertilized, haploid eggs. In addition, haploidy at all loci indicates that the queens were singly mated. Furthermore, a subset of paternal alleles matched alleles found in workers which were not present in queens (Table S4). Hence, workers exhibited recombinant genotypes representing both maternal and paternal alleles. The combined evidence demonstrates that the M. smithii population from Caldeirão reproduces sexually and, although males have as far as we know never been collected, sperm content clearly reveals their existence.
The genetically recombinant population from São Gabriel da Cachoeira showed that all nestmates (n = 8) were genetically distinct (G:N = 1, Ho = 0.365, He = 0.315, FIS = −0.172), consistent with strict sexual reproduction (Tables S1 and S2). However, in the Belém colony (G:N = 0.96, Ho = 0.451, He = 0.466, FIS = 0.034) and the Parintins colony (G:N = 0.71, Ho = 0.650, He = 0.398, FIS = −0.773), few individuals shared a multilocus genotype, suggesting mixed sexual and parthenogenetic reproduction in these populations.
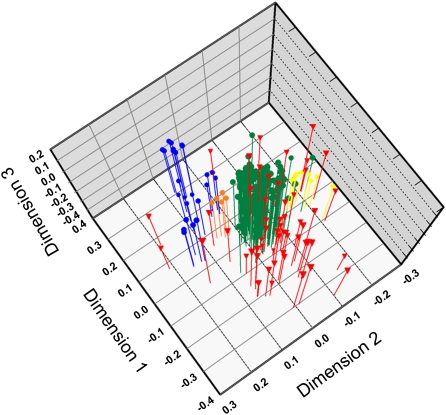
Only a single clonal lineage (from Trinidad) shared a multilocus genotype with the sexual population from São Gabriel da Cachoeira, suggesting that sexual lineages may continuously spawn asexual lineages. To further explore whether sexual populations give rise to asexual lineages, we analyzed the genetic structure of unique multilocus genotypes of asexual and sexual populations. Genotypes of sexual populations group as distinct genetic clusters in the 3D plot generated by a nonmetric multidimensional scaling (NMDS) analysis (Fig. 2). In the discriminate analysis of principal components (DAPC) analysis, the asexual genotypes as a whole and the four clusters of sexual genotypes are significantly different from each other [Wilks's lambda = 0.098, approximate F ratio = 80.247, df (12, 685), P < 0.0001]. Only a few asexual genotypes grouped inside clusters of sexual genotypes, indicating genetic proximity. Greater genetic distances between clones and sexual clusters most likely indicate that the clones originated from sexual source populations other than the four that were sampled, or perhaps that they are of older evolutionary origin and thus highly diverged. Limited overlap between sexual clusters further indicates that the genetic variability of sexual populations was not exhaustively sampled for M. smithii as a species.
Fig. 2.
Plot of 3D object coordinates resulting from an NMDS analysis derived from individual genetic distances. Colored circles represent genotypes of sexual M. smithii populations (blue, Belém; orange, Parintins; green, Caldeirão; yellow, São Gabriel da Cachoeira) and red triangles represent genotypes of asexual lineages.
Phylogenetic Analyses.
To test the monophyly of M. smithii and reconstruct whether asexuality evolved once or multiple times from a sexually reproducing ancestor, we conducted a global phylogenetic analysis of the genus Mycocepurus and a local analysis of only M. smithii taxa representing a sample from each of the genotyped populations (Table S5).
In the global analyses, the monophyly of the genus Mycocepurus was unequivocally supported [Fig. S1; Bayesian posterior probability (BPP) = 1; maximum likelihood bootstrap proportion (MLBP) = 100], which is consistent with a previous analysis (35). Within the genus Mycocepurus, nine reciprocally monophyletic, highly supported groups were recognized [Fig. S1; BPP = 1, MLBP ≥ 92], supporting the existence of five new species (Fig. S1). The monophyly of M. smithii was well-supported [Fig. S1; BPP = 1, MLBP = 92], suggesting that extant M. smithii populations derive from a single most recent common ancestor (MRCA). An undescribed species from the Colombian Amazon was found to be the sister lineage of M. smithii, but with only weak statistical support (Fig. S1; BPP = 0.72, MLBP = 56).
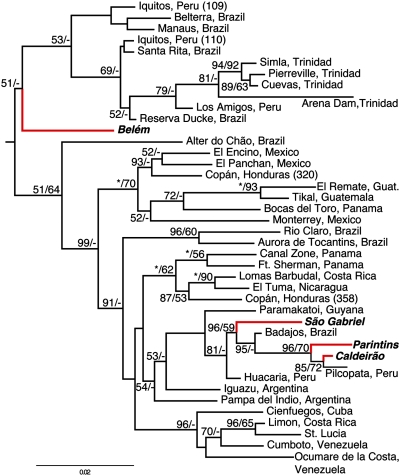
For the mitochondrial gene tree of genotyped M. smithii populations, the statistical support for relationships between sampled individuals is generally low, as expected from the relatively weak phylogenetic informativeness of the mtDNA markers (Table S6; parsimony-informative characters = 169; 11% of mtDNA dataset). Despite this general problem, the monophyly of M. smithii as a species was supported by both the mitochondrial and nuclear data, suggesting that a hybrid origin of asexual reproduction is unlikely in M. smithii. The mitochondrial phylogeny further indicates that the sexual populations are separated into at least two distantly related groups (Fig. 3) and that relationships among asexual populations are in some cases correlated with geography. Three sexual populations form a reasonably well supported clade (BPP = 0.96, MLBP = 59) that also includes two clonal populations (Fig. 3). The sexual population from Belém forms the sister lineage to a clade consisting of asexual populations from the Amazon and Trinidad. This relationship, however, is only weakly supported (BPP = 0.51). Neither the asexual nor the sexual populations are reconstructed as monophyletic under any possible rooting (Fig. 3), consistent with the hypothesis of independent evolutionary origins of asexuality. Based on Bayes factors (BF), the likelihoods of phylogenies resulting from analyses in which the asexual populations are constrained to be monophyletic are significantly worse fitting to the data than those resulting from unconstrained analyses [ML: 2ln(BF) = 137.82; Bayesian: 2ln(BF) = 124.1], further indicating multiple independent origins of asexuality.
Fig. 3.
Midpoint-rooted Bayesian phylogram of M. smithii individuals representing each of the genotyped populations based on analyses of three mitochondrial gene fragments. Bayesian posterior probabilities (×100) and ML bootstrap proportions are indicated as BPP/MLBP. Red branches and bold font indicate taxa from sexually reproducing populations. All other taxa represent asexual populations. (Scale bar, number of substitutions per site.)
Divergence-dating analysis.
The stem-group age (i.e., earliest possible origin) of the fungus-gardening ants was estimated to be 52 million years (Ma) [confidence interval (CI) = 44,60] and the crown-group age was 50 Ma (CI = 43,58), consistent with estimates in Schultz and Brady (35). The estimated crown-group age of the genus Mycocepurus is ∼10 Ma (CI = 6,14), whereas the stem-group age is considerably older with 37 Ma (CI = 27,46), which is also indicated by a long branch leading to the MRCA shared with the sister lineage Myrmicocrypta (Fig. S1). The stem-group age of Mycocepurus smithii is ∼1.65 Ma (CI = 0.57,2.84), whereas the crown-group origin was estimated to be considerably more recent at 0.5 Ma ago (CI = 0.01,1.19). This relatively recent estimate for the evolutionary origin of M. smithii is consistent with the almost complete absence of genetic variability observed in the nuclear DNA sequences.
Discussion
M. smithii consists of a mosaic of sexual and parthenogenetic populations. Although separated by as much as 2,000 km, the sexual populations are located along the Rio Amazonas and the Rio Negro, suggesting the existence of a central widespread sexual (or facultatively sexual/asexual) population that has repeatedly generated asexual, clonally reproducing lineages. These asexual lineages have rapidly dispersed throughout much of Latin America, leading to the current widespread geographic distribution of the species (32, 33). The high clonal diversity in some populations indicates that independently evolved clonal lineages have colonized these habitats separately and repeatedly through time. Once an M. smithii lineage has lost the ability to reproduce sexually, the condition seems irreversible, resulting in our finding of genetically identical individuals in each of the 218 parthenogenetic colonies studied. The mitochondrial phylogeny of M. smithii (Fig. 3) identifies a statistically well-supported group that includes individuals from both asexual and sexual populations, and places the sexual populations in at least two distantly related clades. These patterns, coupled with the results of phylogenetic constraint analyses, are consistent with independent and repeated losses of sexual reproduction. Given the limitations of our sampling, it is nearly certain that additional sexual source populations, from which such closely related groups of asexual clones originated, were not sampled. The divergence-dating analysis provides a recent estimate (crown-group age: 0.5 Ma; CI = 0.01,1.19) for the origin of the presumably sexual most recent common ancestor of extant M. smithii populations, indicating that secondary transitions from sexual to asexual reproduction have occurred recently and possibly continue to occur in the present.
The combined phylogenetic and population-genetic evidence is consistent with the hypothesis that sexual reproduction was lost in ancestors of parthenogenetic M. smithii populations. The spontaneous loss of sexual reproduction has been proposed for the little fire ant Wasmannia auropunctata, in which sexual populations in the native range of this invasive species are likely the source of asexual invasive populations (36). The proximate genetic mechanisms causing the loss of sexuality are not well-understood. However, studies of Cape honey bees (37) and of parthenogenetic lineages of Drosophila melanogaster (38) show that a single recessive allele can cause thelytoky. These examples suggest that the high propensity for switching from sexual to asexual reproduction in M. smithii may be controlled by a small number of genes. Breeding experiments could test whether thelytoky is a qualitative or a quantitative trait in M. smithii by introgressing sexual genes into an asexual genetic background and observing the segregation pattern of the offspring.
Cyclical parthenogenesis, the alteration of asexual and sexual life stages (39, 40), is unlikely to occur in M. smithii. In each of the 218 parthenogenetic colonies collected in different seasons over an 8-y period (2003–2010), nestmates belonged only to one or very few clonal lineages. The nonrandom geographic distribution of sexual and asexual populations likewise suggests that the switch from sexuality to asexuality is unlikely triggered by season.
In arthropods, the evolution of asexuality is often associated with hybridization (30, 41), a mechanism so far unknown in social Hymenoptera (36). Given the monophyly of M. smithii and the phylogenetic congruence between nuclear and mitochondrial markers, hybridization is also unlikely to explain the origin of asexuality in M. smithii.
Alternatively, microorganisms such as Wolbachia, Cardinium, and Rickettsia have been shown to induce parthenogenesis in parasitoid wasps (42–44). Even though Wolbachia infections have not been detected in social Hymenoptera (45), including M. smithii (16), other parthenogenesis-inducing symbionts cannot be ruled out in M. smithii.
Although we have so far only examined a scenario in which asexual populations of M. smithii have repeatedly arisen from sexual populations, the nonmonophyly of the sexual and asexual populations in the mitochondrial phylogeny equally supports an alternative hypothesis: that sexual populations have repeatedly evolved from widespread asexual populations. Although evolutionary reversals from less complex to more complex ancestral traits have long been deemed unlikely (46, 47), reversals from asexual to sexual reproduction have been suggested for mites and hawkweed (48, 49). The absence of males (17) and the lack of genetic recombination in asexual populations of M. smithii are consistent with the hypothesis that meiosis is dysfunctional in parthenogenetic queens. In species with haplodiploid sex determination, restoring functional meiosis would simultaneously result in recombination and the production of haploid eggs, from which males could develop (41, 50). Therefore, haplodiploid species might theoretically require only a single mutation to reevolve sexuality. However, given (i) that all Mycocepurus species for which we have biological information reproduce sexually, (ii) the high genetic diversity observed in the sexually reproducing M. smithii populations, and (iii) the genetic variability observed between separate clonal lineages, it seems highly unlikely that extant sexual M. smithii individuals descended from asexual ancestors.
Despite the large number of clonal lineages found across the broad geographic distribution of M. smithii, mothers and offspring from field and laboratory colonies were genetically identical across multiple generations and males were completely absent from asexual populations, suggesting apomixis as the cytogenetic mechanism underlying thelytoky. Alternatively, it is possible that M. smithii queens reproduce via meiotic parthenogenesis (automixis) with central fusion, a cytogenetic mechanism characterized by potentially very low recombination rates, depending on the locus's distance to the centromere, as indicated by genotype pairs that differ only at a single locus. Automixis with central fusion has been documented in social Hymenoptera (18, 19, 26, 28, 29, 51, 52), and a recent study of W. auropunctata reported recombination rates as low as 0–2.8% (31). Our current data, however, are insufficient to clearly distinguish between automixis with a low recombination rate and apomixis with rare gene conversion.
Conclusion
M. smithii is a recently evolved, monophyletic species consisting of a mosaic of asexual and sexually reproducing populations. Sex has been lost repeatedly in multiple lineages. Once females have lost the ability to reproduce sexually, the condition seems to be irreversible. The lack of genetic recombination and the complete absence of males in asexual populations and laboratory breeding experiments indicate that meiosis may be dysfunctional in asexual females, and thus that mitotic parthenogenesis (apomixis) is the cytogenetic mechanism underlying parthenogenesis in M. smithii. However, automixis with central fusion and low recombination rates cannot be ruled out as a possible alternative mechanism. Sexually reproducing populations were discovered in the center of M. smithii’s geographic distribution along the Rio Amazonas and the Rio Negro. M. smithii has high local population densities and the most extensive geographic distribution of any fungus-growing ant species, indicating its ecological success. The sympatric existence of sexual and asexual populations in the Amazon suggests that sexual populations continue to enjoy high fitness in the center of the species distribution and are not outcompeted by asexual colonies. The fitness advantage of asexual populations seems to be realized outside the range of sexual populations, where parthenogenetic queens apparently colonize vacant niches and disperse rapidly in the absence of males. Given that kin selection theory predicts that conflict over reproduction should be absent in groups of genetically identical individuals, it would be intriguing to investigate the maintenance of cooperative behavior and social conflict in M. smithii. Finally, given the absence of genetic variation within colonies and the presence of phenotypically distinct queen and worker castes, M. smithii appears to be a study organism that is well-suited for investigating the proximate mechanisms of environmentally based caste determination and for exploring the genetic basis of phenotypic plasticity.
Materials and Methods
Population-Genetic Analyses.
As test statistics for asexuality, we used the existence of repeated multilocus genotypes and maximum deviation from random mating (FIS) (53–55). The genotype-to-individual ratio (G:N ratio) was applied to identify multilocus genotypes (55) (Table S1). Independently evolved asexual lineages (clones) originating from separate sexual events were distinguished from slightly different multilocus genotypes that diversified through accumulation of mutations or scoring errors by calculating the probability, psex, following the methodology outlined in ref. 34 and implemented in GENCLONE 2.0 (56). The observed and expected heterozygosity for each clonal lineage (57), the proportion of clonal genotypes in a population, F statistics, and AMOVA were calculated in GENALEX 6 (58) and Genetic Data Analysis (59). To reveal the underlying population-genetic structure of sexual and asexual populations, we used the multivariate statistical methods (60–62) NMDS, principal component analysis, and DAPC, as implemented in PERMAP (63), GENALEX (58), and SYSTAT (Systat Software).
Phylogenetic Analyses.
We conducted analyses of two distinct datasets: first, a global dataset that included 84 M. ingroup taxa, 32 of them M. smithii, and 87 outgroup taxa. The recently described social parasite M. castrator (64) was not included. The alignment consisted of 2,319 bp of protein-coding (exon) sequences of three single-copy nuclear genes and one mitochondrial gene and was divided into 10 partitions. Second, we conducted a local analysis of 41 M. smithii taxa representing one individual from each of the genotyped populations (Table S5). We obtained 1,515 bp of three mitochondrial genes and divided the alignment into two partitions (Table S6). Constrained topologies were estimated using Bayesian and ML analyses, and differences in the likelihoods of constrained versus unconstrained topologies were evaluated using Bayes factors (65–67). All ingroup sequence data were generated for this study (Table S5). Best-fit models of sequence evolution were selected for each partition under the Akaike information criterion (68) and hierarchical likelihood ratio tests as calculated in MODELTEST v3.7 (69) (Table S6). We conducted partitioned Bayesian analyses using MrBayes v3.1.2 (70). Burn-in and convergence were assessed using Tracer v1.5 (71). Partitioned ML analyses were carried out in GARLI 0.97.r737 (72).
Divergence-Dating Analysis.
We used a Bayesian relaxed clock uncorrelated lognormal approach implemented in the program BEAST v1.4.8 with a Yule process as the tree prior (73–75). The root node was given a normal age prior distribution (mean = 73.5, SD = 4.5), following methodology described in ref. 76. Based on fossil data, lognormal age prior distributions were assigned to three internal nodes, as outlined in ref. 35. For more details on analyses and results, see SI Materials and Methods and Tables S1–S8.
Supplementary Material
Acknowledgments
We thank the following scientists for generously contributing specimens and institutions for providing permission and access to study sites: G. Alpert, C. Brandão, S. Cappellari, J. Carpenter, S. Cover, R. Feitosa, F. Fernández, J. Fontenla, A. Harada, A. Henriques, A. Himler, J. Lattke, J. Longino, W. Mackay, J. Maes, J. Martins, B. Merz, N. Pitman, R. Poggi, V. Raineri, C. Samper, S. Sánchez-Peña, J. Santisteban, R. Silva, the late R. Snelling, J. Sosa-Calvo, H. Vasconcelos, P. Ward, and E. Wilson; the Autoridad Nacional del Ambiente and Smithsonian Tropical Research Institute, Panama; Conselho Nacional de Desenvolvimento Científico e Tecnológico and the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis; the Forestry Division and Wildlife Section of Trinidad; the Minesterio del Ambiente y Energia, Costa Rica; and the Office of the President of Guyana. P. Armstrong and E. Okonski kindly provided assistance in the laboratory. L. Hayek and S. Arnaud-Haond provided valuable advice on statistical analyses. D. Bolnik, S. Brady, D. Gotzek, D. Hillis, M. Singer, P. Ward, and two anonymous reviewers improved the manuscript with helpful comments. C.R. gratefully acknowledges financial support from Ernst Mayr grants (Museum of Comparative Zoology) and the Green Fund (Harvard University), a National Science Foundation (NSF) Doctoral Dissertation Improvement Grant (DEB-0808164), the Explorer's Club Exploration Fund, a Lewis and Clark Field Scholarship, and research grants from the Section of Integrative Biology and a Miller Endowed University Continuing Fellowship from the University of Texas at Austin; T.R.S. was supported by the NSF (DEB-0949689 and DEB-0431330), the Smithsonian Scholarly Studies Program, and the Smithsonian Restricted Endowments Fund; M.B. received support from the Fundação de Apoio à Pesquisa do Estado de São Paulo (2008/54386-9) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (476250/2008-0 and 304661/2009-0); M.V.B.G. and M.V. acknowledge the Bundesministerium für Bildung und Forschung (BMBF) and the Conselho National de Pesquisa e Tecnologia (BMBF01LT0014/CNPq 690018/00-2) for kindly providing funding for SHIFT Project ENV52-2; U.G.M. was funded by grants from the NSF (DEB-0639879, DEB-0110073, and DEB-998379) and the Wheeler Lost Pines Endowment. This study is a chapter of C.R.’s doctoral dissertation.
Footnotes
The authors declare no conflict of interest.
Data deposition: DNA sequences reported in this paper have been deposited in the GenBank database (accession nos. JN054745–JN055353).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105467108/-/DCSupplemental.
References
- 1.Barton N. Evolutionary biology. The geometry of adaptation. Nature. 1998;395:751–752. doi: 10.1038/27338. [DOI] [PubMed] [Google Scholar]
- 2.Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- 3.Avise JC. Clonality: The Genetics, Ecology, and Evolution of Sexual Abstinence in Vertebrate Animals. New York: Oxford Univ Press; 2008. [Google Scholar]
- 4.Hamilton WD. Sex versus non-sex versus parasite. Oikos. 1980;35:282–290. [Google Scholar]
- 5.Lively CM. A review of Red Queen models for the persistence of obligate sexual reproduction. J Hered. 2010;101(Suppl 1):S13–S20. doi: 10.1093/jhered/esq010. [DOI] [PubMed] [Google Scholar]
- 6.Kondrashov AS. Selection against harmful mutations in large sexual and asexual populations. Genet Res. 1982;40:325–332. doi: 10.1017/s0016672300019194. [DOI] [PubMed] [Google Scholar]
- 7.Muller H. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 8.Smith JM. What use is sex? J Theor Biol. 1971;30:319–335. doi: 10.1016/0022-5193(71)90058-0. [DOI] [PubMed] [Google Scholar]
- 9.Bell G. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. Croom Helm, London; 1982. [Google Scholar]
- 10.Maynard Smith J. The Evolution of Sex. Cambridge, UK: Cambridge Univ Press; 1978. [Google Scholar]
- 11.Williams GC. Sex and Evolution. Princeton, NJ: Princeton Univ Press; 1975. [Google Scholar]
- 12.Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281:1986–1990. [PubMed] [Google Scholar]
- 13.Burt A. Perspective: Sex, recombination, and the efficacy of selection—Was Weismann right? Evolution. 2000;54:337–351. doi: 10.1111/j.0014-3820.2000.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 14.Becks L, Agrawal AF. Higher rates of sex evolve in spatially heterogeneous environments. Nature. 2010;468:89–92. doi: 10.1038/nature09449. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Marín H, Zimmerman J, Wcislo W, Rehner S. Colony foundation, nest architecture and demography of a basal fungus-growing ant, Mycocepurus smithii (Hymenoptera, Formicidae) J Nat Hist. 2005;39:1735–1743. [Google Scholar]
- 16.Himler AG, Caldera EJ, Baer BC, Fernández-Marín H, Mueller UG. No sex in fungus-farming ants or their crops. Proc Biol Sci. 2009;276:2611–2616. doi: 10.1098/rspb.2009.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabeling C, et al. Thelytokous parthenogenesis in the fungus-gardening ant Mycocepurus smithii (Hymenoptera: Formicidae) PLoS One. 2009;4:e6781. doi: 10.1371/journal.pone.0006781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma S, Ruttner F. Cytological analysis of the thelytokous parthenogenesis in the Cape honeybee (Apis mellifera capensis Escholtz) Apidologie (Celle) 1983;14:41–57. [Google Scholar]
- 19.Baudry E, et al. Whole-genome scan in thelytokous-laying workers of the Cape honeybee (Apis mellifera capensis): Central fusion, reduced recombination rates and centromere mapping using half-tetrad analysis. Genetics. 2004;167:243–252. doi: 10.1534/genetics.167.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito F, Touyama Y, Gotoh A, Kitahiro S, Billen J. Thelytokous parthenogenesis by queens in the dacetine ant Pyramica membranifera (Hymenoptera: Formicidae) Naturwissenschaften. 2010;97:725–728. doi: 10.1007/s00114-010-0688-5. [DOI] [PubMed] [Google Scholar]
- 21.Gotoh A, Billen J, Tsuji K, Sasaki T, Ito F. Histological study of the spermatheca in three thelytokous parthenogenetic ant species, Pristomyrmex punctatus, Pyramica membranifera and Monomorium triviale (Hymenoptera: Formicidae) Acta Zool. February 18, 2011 10.1111/j.1463-6395.2010.00498.x. [Google Scholar]
- 22.Timmermans I, Hefetz A, Fournier D, Aron S. Population genetic structure, worker reproduction and thelytokous parthenogenesis in the desert ant Cataglyphis sabulosa. Heredity. 2008;101:490–498. doi: 10.1038/hdy.2008.72. [DOI] [PubMed] [Google Scholar]
- 23.Pearcy M, Goodisman MAD, Keller L. Sib mating without inbreeding in the longhorn crazy ant. Proc Biol Sci. February 2, 2011 doi: 10.1098/rspb.2010.2562. 10.1098/rspb.2010.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller L. Uncovering the biodiversity of genetic and reproductive systems: Time for a more open approach. American Society of Naturalists E. O. Wilson Award winner address. Am Nat. 2007;169:1–8. doi: 10.1086/509938. [DOI] [PubMed] [Google Scholar]
- 25.Heinze J. The demise of the standard ant (Hymenoptera: Formicidae) Myrmecol News. 2008;11:9–20. [Google Scholar]
- 26.Fournier D, et al. Clonal reproduction by males and females in the little fire ant. Nature. 2005;435:1230–1234. doi: 10.1038/nature03705. [DOI] [PubMed] [Google Scholar]
- 27.Pearcy M, Aron S, Doums C, Keller L. Conditional use of sex and parthenogenesis for worker and queen production in ants. Science. 2004;306:1780–1783. doi: 10.1126/science.1105453. [DOI] [PubMed] [Google Scholar]
- 28.Pearcy M, Hardy O, Aron S. Thelytokous parthenogenesis and its consequences on inbreeding in an ant. Heredity. 2006;96:377–382. doi: 10.1038/sj.hdy.6800813. [DOI] [PubMed] [Google Scholar]
- 29.Kellner K, Heinze J. Mechanism of facultative parthenogenesis in the ant Platythyrea punctata. Evol Ecol. 2011;25:77–89. [Google Scholar]
- 30.Leach IM, et al. Thelytoky in Hymenoptera with Venturia canescens and Leptopilina clavipes as case studies. In: Schön I, Martens K, van Dijk P, editors. Lost Sex: The Evolutionary Biology of Parthenogenesis. Dordrecht, The Netherlands: Springer; 2009. pp. 347–375. [Google Scholar]
- 31.Rey O, et al. Meiotic recombination dramatically decreased in thelytokous queens of the little fire ant and their sexually produced workers. Mol Biol Evol. March 31, 2011 doi: 10.1093/molbev/msr082. 10.1093/molbev/msr082. [DOI] [PubMed] [Google Scholar]
- 32.Mackay WP, Maes JM, Rojas Fernández P, Luna G. The ants of North and Central America: The genus Mycocepurus (Hymenoptera: Formicidae) J Insect Sci. 2004;4:27. doi: 10.1093/jis/4.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kempf W. A review of the ant genus Mycocepurus Forel, 1893 (Hymenoptera: Formicidae) Stud Entomol. 1963;6:417–432. [Google Scholar]
- 34.Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA. Standardizing methods to address clonality in population studies. Mol Ecol. 2007;16:5115–5139. doi: 10.1111/j.1365-294X.2007.03535.x. [DOI] [PubMed] [Google Scholar]
- 35.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci USA. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foucaud J, et al. Sex and clonality in the little fire ant. Mol Biol Evol. 2007;24:2465–2473. doi: 10.1093/molbev/msm180. [DOI] [PubMed] [Google Scholar]
- 37.Lattorff HMG, Moritz RFA, Fuchs S. A single locus determines thelytokous parthenogenesis of laying honeybee workers (Apis mellifera capensis) Heredity. 2005;94:533–537. doi: 10.1038/sj.hdy.6800654. [DOI] [PubMed] [Google Scholar]
- 38.Fuyama Y. Genetics of parthenogenesis in Drosophila melanogaster. II. Characterization of a gynogenetically reproducing strain. Genetics. 1986;114:495–509. doi: 10.1093/genetics/114.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Normark BB. The evolution of alternative genetic systems in insects. Annu Rev Entomol. 2003;48:397–423. doi: 10.1146/annurev.ento.48.091801.112703. [DOI] [PubMed] [Google Scholar]
- 40.Decaestecker E, De Meester L, Mergeay J. Cyclical parthenogenesis in Daphnia: Sexual versus asexual reproduction. In: Schön I, Martens K, van Dijk P, editors. Lost Sex: The Evolutionary Biology of Parthenogenesis. Dordrecht, The Netherlands: Springer; 2009. [Google Scholar]
- 41.Suomalainen E, Saura A, Lokki J. Cytology and Evolution in Parthenogenesis. Boca Raton, FL: CRC; 1987. [Google Scholar]
- 42.Zchori-Fein E, et al. A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proc Natl Acad Sci USA. 2001;98:12555–12560. doi: 10.1073/pnas.221467498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc Biol Sci. 2006;273:2097–2106. doi: 10.1098/rspb.2006.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 45.Wenseleers T, Billen J. No evidence for Wolbachia-induced parthenogenesis in the social Hymenoptera. J Evol Biol. 2000;13:277–280. [Google Scholar]
- 46.Dollo L. Les lois de l’évolution. Bulletin de la Société Belge de Géologie de Paléontologie et d'Hydrologie. 1893;7:164–166. [Google Scholar]
- 47.Bull J, Charnov E. On irreversible evolution. Evolution. 1985;39:1149–1155. doi: 10.1111/j.1558-5646.1985.tb00455.x. [DOI] [PubMed] [Google Scholar]
- 48.Chapman H, Houliston GJ, Robson B, Iline I. A case of reversal: The evolution and maintenance of sexuals from parthenogenetic clones in Hieracium pilosella. Int J Plant Sci. 2003;164:719–728. [Google Scholar]
- 49.Domes K, Norton RA, Maraun M, Scheu S. Reevolution of sexuality breaks Dollo's law. Proc Natl Acad Sci USA. 2007;104:7139–7144. doi: 10.1073/pnas.0700034104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cook JM. Sex determination in the Hymenoptera: A review of models and evidence. Heredity. 1993;71:421–435. [Google Scholar]
- 51.Foucaud J, et al. Rare sexual reproduction events in the clonal reproduction system of introduced populations of the little fire ant. Evolution. 2006;60:1646–1657. [PubMed] [Google Scholar]
- 52.Foucaud J, Estoup A, Loiseau A, Rey O, Orivel J. Thelytokous parthenogenesis, male clonality and genetic caste determination in the little fire ant: New evidence and insights from the lab. Heredity. 2010;105:205–212. doi: 10.1038/hdy.2009.169. [DOI] [PubMed] [Google Scholar]
- 53.Balloux F, Lehmann L, de Meeûs T. The population genetics of clonal and partially clonal diploids. Genetics. 2003;164:1635–1644. doi: 10.1093/genetics/164.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Meeûs T, Balloux F. F-statistics of clonal diploids structured in numerous demes. Mol Ecol. 2005;14:2695–2702. doi: 10.1111/j.1365-294X.2005.02643.x. [DOI] [PubMed] [Google Scholar]
- 55.Halkett F, Simon JC, Balloux F. Tackling the population genetics of clonal and partially clonal organisms. Trends Ecol Evol. 2005;20:194–201. doi: 10.1016/j.tree.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Arnaud-Haond S, Belkhir K. GENCLONE: A computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes. 2007;7:15–17. [Google Scholar]
- 57.Conner JK, Hartl DL. A Primer of Ecological Genetics. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 58.Peakall R, Smouse PE. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis P, Zaykin D. 2002. GDA (Genetic Data Analysis). http://hydrodictyon.eeb.uconn.edu/people/plewis/software.php.
- 60.Jombart T, Pontier D, Dufour AB. Genetic markers in the playground of multivariate analysis. Heredity. 2009;102:330–341. doi: 10.1038/hdy.2008.130. [DOI] [PubMed] [Google Scholar]
- 61.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lessa EP. Multidimensional analysis of geographic genetic structure. Syst Biol. 1990;39:242–252. [Google Scholar]
- 63.Heady R, Lucas J. 2007. PERMAP: Perceptual MAPping Software (Univ of Louisiana at Lafayette). http://www.ucs.louisiana.edu/∼rbh8900.
- 64.Rabeling C, Bacci M. A new workerless inquiline in the Lower Attini (Hymenoptera: Formicidae), with a discussion of social parasitism in fungus-growing ants. Syst Entomol. 2010;35:379–392. [Google Scholar]
- 65.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 66.Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- 67.Brown JM, Lemmon AR. The importance of data partitioning and the utility of Bayes factors in Bayesian phylogenetics. Syst Biol. 2007;56:643–655. doi: 10.1080/10635150701546249. [DOI] [PubMed] [Google Scholar]
- 68.Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001;50:580–601. [PubMed] [Google Scholar]
- 69.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 70.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 71.Rambaut A, Drummond A. 2007. Tracer v1.5. http://tree.bio.ed.ac.uk/software/tracer.
- 72.Zwickl D. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. 2006. PhD dissertation (Univ of Texas at Austin). http://garli.googlecode.com.
- 73.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brady SG, Schultz TR, Fisher BL, Ward PS. Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc Natl Acad Sci USA. 2006;103:18172–18177. doi: 10.1073/pnas.0605858103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.