Abstract
The complement system is a key component of innate and adaptive immune responses. Complement regulation is critical for prevention and control of disease. We have determined the crystal structure of the complement regulatory enzyme human factor I (fI). FI is in a proteolytically inactive form, demonstrating that it circulates in a zymogen-like state despite being fully processed to the mature sequence. Mapping of functional data from mutants of fI onto the structure suggests that this inactive form is maintained by the noncatalytic heavy-chain allosterically modulating activity of the light chain. Once the ternary complex of fI, a cofactor and a substrate is formed, the allosteric inhibition is released, and fI is oriented for cleavage. In addition to explaining how circulating fI is limited to cleaving only C3b/C4b, our model explains the molecular basis of disease-associated polymorphisms in fI and its cofactors.
Keywords: allostery, innate immunity, atypical hemolytic-uremic syndrome, serine protease
The complement system is a major component of innate and adaptive immunity whose efficient regulation is critical for prevention and control of disease (1). This regulation is effected by host cells expressing and recruiting a series of proteins that protect them from complement-mediated destruction. A key step in this self-protection is the proteolytic cleavage of complement C3b (and its homolog C4b) by the serine protease complement factor I (fI) (2) in the presence of additional regulatory proteins termed “cofactors” (3). In the absence of regulation by fI the alternative pathway of complement leads to continuous generation of fluid-phase and cell-surface–deposited C3b by a self-amplification loop: The more C3 is converted to C3b, the more C3 convertases are formed, resulting in the depletion of C3 (4). With healthy levels of functional fI and its cofactors, the irreversible breakdown of C3b to iC3b has three effects: (i) arrest of the assembly of the C3 convertases on host-cell surfaces, thus avoiding inappropriate complement amplification; (ii) prevention of runaway C3 consumption in the fluid phase; and (iii) generation of the C3b cleavage fragments that go on to bind specific complement receptors and are involved in opsonization and triggering of the adaptive immune response (5). The importance of this regulatory mechanism is highlighted by the fact that fI-deficient individuals suffer from recurring infections and that polymorphisms in or near the genes for fI (and its cofactors) can predispose the carriers to diseases such as systemic lupus erythematosus, atypical hemolytic-uremic syndrome (aHUS), membranoproliferative glomerulonephritis, and age-related macular degeneration (for a recent review, see ref. 6).
During the last few years, biochemical and structural data for complement regulators and C3/C3b in complex with regulators and inhibitors (7–11) have significantly advanced our understanding of the molecular mechanisms underlying complement regulation (12). To date, however, only low-resolution information about fI has been available (13, 14). FI is synthesized as a single 66-kDa chain and is processed posttranslationally by the addition of six Asn-linked glycans (totaling 22 kDa) and the excision of a linker sequence (residues 318RRKR321), splitting the molecule into two chains linked by a single disulfide bond (15–17). The noncatalytic heavy chain comprises an N-terminal region; an fI membrane attack complex (FIMAC) domain; a scavenger receptor cysteine-rich (SRCR) domain; two class A low-density lipoprotein receptor domains (LDLRA1 and LDLRA2); and a C-terminal region of unknown function that is a site of sequence variability across species (SI Appendix, Fig. S1). The catalytic light chain consists of a chymotrypsin-like serine protease (SP) domain.
In the absence of atomic-level structural information, several questions still surround fI-mediated complement regulation. Firstly, fI is unusual among serum proteases in that it neither circulates as an uncleaved proenzyme (16) nor has an endogenous inhibitor. Multiple mechanisms of control of activity have been suggested, such as direct inhibition of the active site (18), conformational changes in both protease and substrate (19), and models based on the transient nature of the substrate/cofactor complex. Second, it is not known how fI acquires its exquisite specificity for the selected sites of cleavage in C3b and C4b (20). Third, a number of fI point mutants have been engineered that show, against synthetic or natural substrates, increased rates of activity with respect to the wild-type enzyme (21, 22), but no explanation for these observations has been proposed. Finally, because homology modeling of the whole fI is rendered unfeasible by its multidomain architecture, the understanding of fI gene polymorphisms associated with disease also is fraught with uncertainty.
Here we describe the crystal structure of intact human fI. Combining this and earlier structures with biochemical assays and with mapping of disease-associated mutations, we propose a model whereby the heavy chain of fI allosterically inhibits the serine protease domain until the C3b/cofactor complex is engaged.
Results and Discussion
Crystal Structure of Human FI.
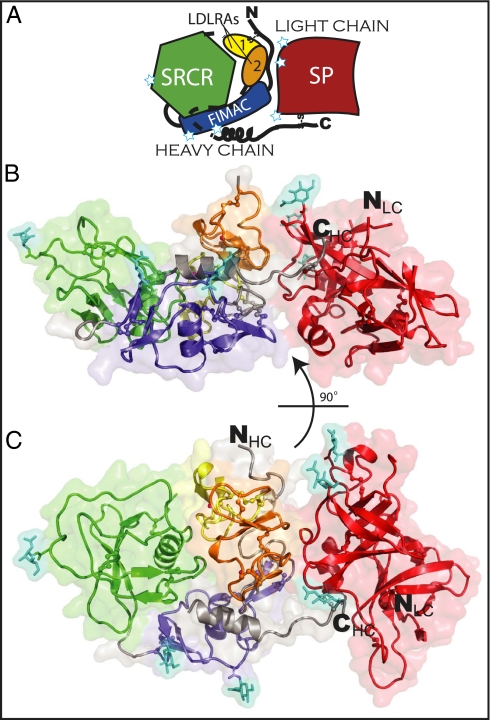
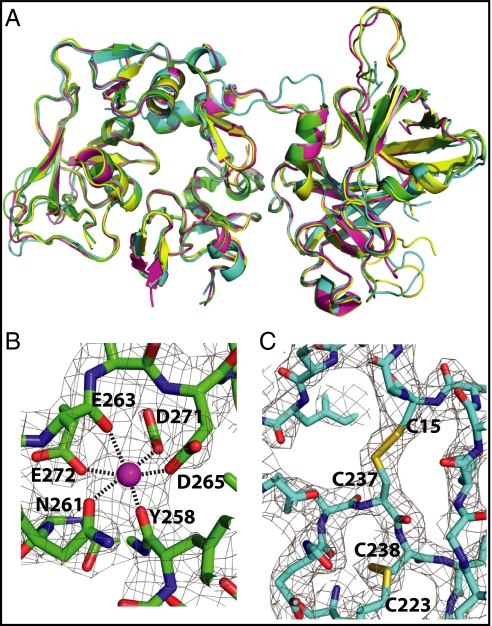
FI contains 20 disulfide bonds and has been expressed in insect and mammalian cells (22), but yields are insufficient for crystallography. We therefore purified fI from pooled human serum, obtained crystals and diffraction data, and determined the structure at 2.7-Å resolution (SI Appendix, Table S1). The final model (Fig. 1) comprises 1,864 (83%) of the 2,244 residues present in four copies of the protein in the asymmetric unit, which overlay well with an rmsd of 0.8 Å over 440 residues (Fig. 2A). Two Ca2+ ions per molecule also are present, each bound to one of the LDLRA domains, and appear to play structural roles, being buried within the molecule (Fig. 2B). The first GlcNAc residue of each of the six N-linked glycosylation sites [Asn residues 52, 85, 159, 446, 476, and 518 (17)] is ordered and visible in the electron density (SI Appendix, Fig. S2). In addition to its four domains, the heavy chain contains two helices that were not predicted from sequence, one in its N-terminal region (residues 13–18) and one toward its C terminus (residues 292–303). Key structural features of the model are summarized in SI Appendix, Figs. S3–S5.
Fig. 1.
The structure of human fI. (A) Cartoon schematic of fI. FIMAC is shown in blue, SRCR in green, LDLRA1 in yellow, LDLRA2 in orange, and SP in red; unmodeled loops are shown by dashed lines; N-linked glycosylation sites are shown by white stars. (B and C) The protein is shown in two views as a cartoon representation with a transparent surface. Disulfide bonds and two bound Ca2+ ions are shown as ball-and-stick representations. The six glycosylated Asn residues and attached GlcNAc residues (cyan) are shown as stick representations. Domains are colored as A.
Fig. 2.
Structural details of human fI. (A) The four crystallographically independent copies of fI are overlaid in cartoon representations revealing no major variation in packing between the heavy and light chains. B and C show the density (weighted 2FO-FC) for the bound Ca2+ in LDLRA2 and novel disulphide between residues 15 and 237 in copy A.
In agreement with the early electron microscopy and solution-scattering data (13, 14), the shape of fI is bilobal, with the heavy and light chains making up the two halves of a “brick” with the approximate dimensions 90 × 45 × 35 Å. The arrangement of the domains in the larger (heavy-chain) lobe forms a ring structure, with the N-terminal FIMAC domain contacting the C-terminal LDLRA domains. This contact is linked covalently by a disulfide bridge between Cys15 and Cys237 (Fig. 2C). The heavy-chain N terminus and LDLRA2 domain also form one of the two interfaces between the heavy and light chains, the second being located around the interchain disulfide bridge Cys309–Cys435 (23). Each chain buries about 720 Å2 of surface in this interface (24). Individually the domains of the heavy chain are mostly canonical (SI Appendix, Figs. S4 and S5). The smaller (light-chain) lobe is the SP domain, with the protease-active site presented on the end of the “brick” opposite the SRCR domain. This positioning, therefore, rules out a mechanism of self-inhibition by direct occlusion of the active site, despite the FIMAC domain’s adopting a fold commonly found in serine protease inhibitors (18).
Mature FI Is Zymogen-Like.
As expected, the SP domain adopts a chymotrypsin-like fold organized around two six-stranded β-barrels with the active site at the barrel interface. Importantly, none of the four independent copies of fI in the crystal contains an SP domain in a proteolytically competent state. Many of the loops crucial to the formation of the serine protease active-site triad and oxyanion hole are disordered, and others are traceable only by reducing the electron-density contour level below 1.0 σ. These loops include residues 322–331, 377–389, 419–425, 454–466, 497–506, and 529–534. Thermal motion, as parameterized by atomic B factors, is also higher in these regions than in the core.
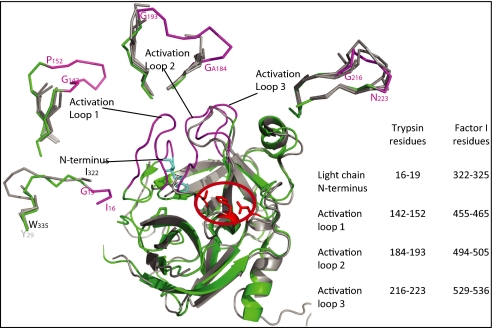
Most serum serine proteases are produced in a zymogen form with low activity and thus circulate without cleaving inappropriate substrates. Activation is triggered by proteolysis of the bond between residues 321 and 322 (residues 15 and 16 in chymotrypsin numbering). The newly generated N terminus of residue 322 then forms a salt bridge with Asp506 (chymotrypsin Asp194) and facilitates the correct positioning of the neighboring catalytic Ser507 (Ser195). Structural studies of the transition from zymogen to active protease have demonstrated that the transition also entails a major reordering of several loops around the active site, the zymogen-activation domain (25). Strikingly, four of the six flexible regions of fI correspond to this zymogen-activation domain (Fig. 3). Our crystals therefore contain fI in a zymogen-like state, despite its having been processed at Ile322. Limited proteolysis of native fI under physiological conditions results in cleavage of the SP domain at residues flexible in our structure (26), including the five N-terminal residues that would be buried in the protease in its fully active form, suggesting that the crystal structure is representative of the structure in solution.
Fig. 3.
Zymogenicity of the fI serine protease domain. Overlay of trypsin (Protein Data Bank ID 3MI4) (green and purple) with the fI serine protease domain (gray). The catalytic triad of trypsin is shown as red sticks, and the region of the active site is circled in red. The activation domain loops and N terminus, which are mobile and disordered in trypsinogen (25), are shown in purple and are labeled. Surrounding the overlay, the Cα trace of each component of the trypsin activation domain (purple: activation domain; green: surrounding region) is overlaid with the coordinates of the equivalent regions from the four independent molecules of fI. The fI N terminus is disordered in all but one copy of the molecule, and even here it is mobile (average B factor, 85 Å2) and is not in an active conformation. Factor I activation loops 1 and 2 are not visible in the electron density in any of the four copies, and activation loop 3 can be built only in two copies (average B factor, 66 Å2).
However, it is very unusual to observe a zymogen-like structure for a processed serine protease in a crystal, and it has been proposed that packing into the crystal environment induces order in loops that are unstructured in solution studies (23).
Although fI is extreme in the zymogen-like nature of the proteolytically processed form of the enzyme, it is related most closely in terms of activity and regulation to allosterically regulated serine proteases such as thrombin, which recently has been demonstrated to have a dynamic zymogen-activation domain in solution that reorders in the presence of allosteric activators or substrate (23, 27). High concentrations of peptide substrates, in the absence of cofactor, can induce low levels of fI activity (28, 29), and substrate analog inhibitors also can bind in the absence of either C3b or cofactor (28, 30). However, the nonsubstrate-like, broad-spectrum protease inhibitor diisopropyl fluorophosphate is able to react with the active site serine only if fI is preincubated with C3b (19). This requirement strongly suggests that substrate-induced remodeling of the active site is key to fI activity, as has been demonstrated recently for another complement system serine protease, factor D (8, 31).
Mapping Mutations onto the FI Structure Implies Allosteric Regulation of Light-Chain Activity by Contact of the Heavy Chain.
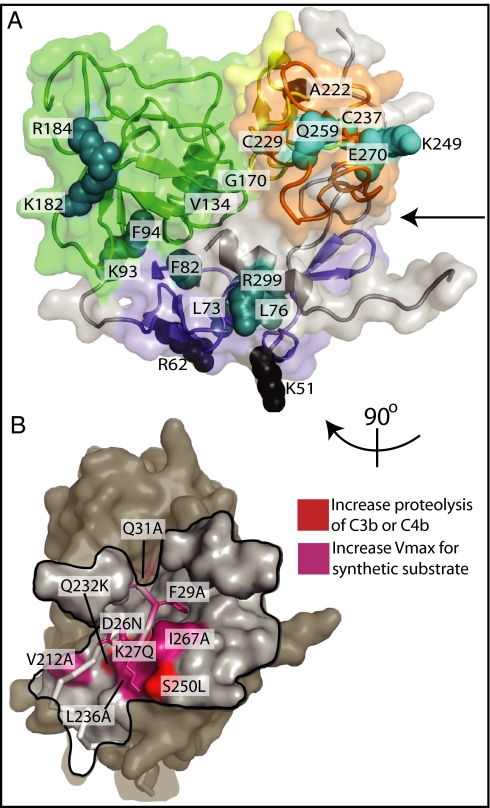
To gain insight into previously unexplained disease-associated gene polymorphisms and mutations known to alter cofactor-assisted C3b/C4b cleavage by fI (21, 22, 32, 33), we mapped them to our crystal structure (Fig. 4 and SI Appendix, Table S2). Many of the mutations are buried in the heavy-chain interdomain interfaces and are expected to affect its structure (Fig. 4A). More importantly, all the mutants that display a significantly increased rate of activity toward synthetic or natural substrates (21, 22) cluster at the interface between the two chains, centered around the N terminus of the heavy chain (Fig. 4B). Alteration of the interchain region by these mutations therefore seems to relieve an inhibitory effect of the heavy chain on the activity of the SP domain. The inhibition suggests an allosteric mechanism whereby contacts between the heavy chain and residues 437–444 and 549–553 of the SP domain maintain the zymogen-activation domain in its zymogenic state. Such a mechanism is consistent with the observation that intact fI will not cleave C3b in the absence of cofactor, whereas purified SP domain does, albeit with altered specificity and low activity (25). The mechanism is also analogous to the regulation of serine proteases such as factor VIIa (34, 35) and thrombin (23), which have allosteric regulator binding sites at locations distant from the zymogen-activation domain or active site (SI Appendix, Fig. S6). Release of the inhibition then would involve alteration of the heavy-chain/light-chain interface on binding to the cofactor:C3b complex.
Fig. 4.
Mapping of heavy-chain mutations that alter fI activity. (A) Residues corresponding to mutations that impair secretion or activity. The fI heavy chain is shown as in Fig. 1C. Mutations that disrupt the fold of the heavy chain [cyan (21, 22)], or impair the fI catalytic activity [teal (21, 33)] are shown in ball representation. The V134M mutant (32), for which no available secretion or activity data are available, also is shown in green. The arrow points to the face of the heavy chain in contact with the light chain. (B) Mutations that increase the rate of activity (21, 22) of the enzyme cluster around the N terminus of the heavy chain and its region of contact with the light chain. The fI heavy chain is shown as a gray surface, except for its N terminus, which is represented by a gray ribbon. The black line marks the footprint of the light chain onto the heavy chain.
Interactions between fI and cofactor/C3b clearly are crucial to the activity of fI (26, 28). It is interesting that several mutations of fI that affect function are surface exposed [Lys51Ala, Arg62Ala, and the aHUS-associated Ala222Gly (21, 36, 37)]. We therefore predict that these residues may map to the cofactor or C3b binding sites. Further understanding of these fI mutations clearly requires more detailed information on the fI–C3b and fI–cofactor interactions.
Modeling the Ternary Complex of FI, Factor H, and C3b.
To understand complement regulation by fI better, we modeled the C3b:factor H1–4 (fH1–4):fI ternary complex by docking our fI crystal structure onto the structure of the C3b:fH1–4 complex. In the presence of one of its cofactors, fI first cleaves C3b between Arg1281–Ser1282 (38), and our modeling focuses on the ternary complex leading to this cleavage, because subsequent proteolysis events are assumed to involve unfolding of the C1r/C1s/Uegf/Bmp1 (CUB) domain of C3b. Existing structures of serine protease/substrate and serine protease/inhibitor complexes were used to define the relative orientation between the C3b peptide targeted for proteolysis and the barrels of the fI serine protease domain (SI Appendix, Fig. S7).
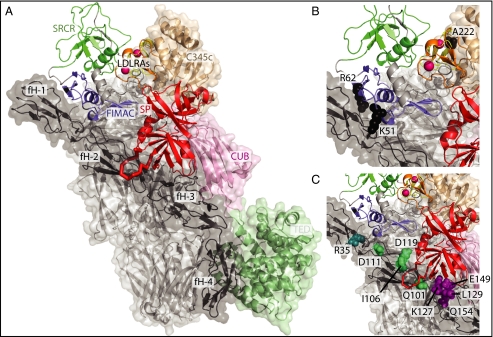
The model shows fI docked in a groove between fH1–3 and C3b (Fig. 5A) and provides a compelling model for the ternary complex as judged by a variety of criteria. First, the SRCR domain is not involved in any major contacts, in agreement with the observation that a monoclonal antibody specific to this domain does not interfere with fI cleavage of C3b (26, 39). However, the rest of the heavy chain is involved intimately in packing against both cofactor and C3b. Second, fI bears six bulky Asn-linked glycans that constitute 25% of the mass of the glycoprotein, and the model places these glycans away from the major interaction surface (SI Appendix, Fig. S8). Third, there is an overall electrostatic complementarity, with the long edges of the fI brick bearing positive charges that pack against negatively charged regions of fH and C3b (SI Appendix, Fig. S9), consistent with the observed salt-dependency of the fI–fH and fI–C3b interactions (14, 40). Most importantly, fI makes significant contacts with both C3b and the cofactor, in agreement with the need for the preformed, binary complex for activity. These interactions include the surface-exposed fI loss-of-function mutants Lys51, Arg62, and Ala222 (21, 36, 37), whose mechanism previously was unexplained (Fig. 5B).
Fig. 5.
Modeling the ternary complex of C3b, fH1–4, and fI. Model for the C3b:fH1–4:fI ternary complex in the arrangement leading to the first C3b cleavage. (A) Model of the C3b:fH1–4:fI ternary complex. FI is shown as a cartoon; C3b and fH1–4 are shown as cartoons and semitransparent surfaces The C3b representation is adapted from ref. 11: C345C domain is shown in light bronze; the CUB domain in pink, the thioester containing domain in light green; and the linker domain, C3 α′ N-terminal domain, and macroglobulin domains of C3b in light gray. (B) Residues corresponding to surface mutations on fI that impair activity and contact C3b (36, 37) or cofactor (21) in the model for the ternary complex. (C) Zoom view of the contact of fI with the cofactor complement control protein domain2–3 junction. The mobile loop in fI (residues 419–425, main chain shown in red) constitutes a point of contact with cofactor surface residues that either impair cofactor activity [vaccine complement control protein (11, 42) shown in green; C4bp (49) shown in purple] or are sites of disease-associated mutations [fH (7, 41) shown in cyan].
The fI footprint on the cofactor also involves residues previously implicated in cofactor activity by either mutational data or disease association in patients but unexplained by the fH1–4/C3b structure (Fig. 5C). These residues include the aHUS-associated Arg35His mutation in fH1 (7) that recently has been demonstrated to have decreased cofactor activity (41). Two other residues associated with altered cofactor activity (fH Gln101 and Ile106) (42) lie at the site of the major interaction region of the cofactor with one of the mobile loops of the SP domain (fI 419–425) (Fig. 5C). Also buried at this contact point are two residues, Ile122 and Glu124, conserved among the cofactors in fH but absent in the noncofactor, complement regulator, CD55.
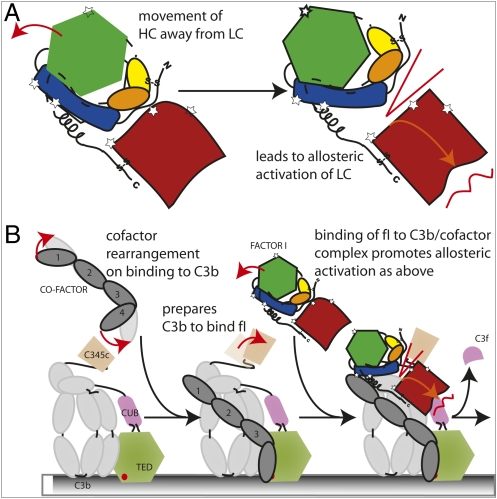
Combining our crystal structures with the existing cofactor/substrate complex and mutagenesis data allows us to propose a general model for fI regulation of complement (Fig. 6 and Movie S1): (i) binding of the cofactor to C3b produces a stable platform onto which the fI can dock (43); (ii) binding of fI releases the allosteric inhibitory effects of the heavy chain and induces remodeling of the zymogen-activation domain; and (iii) the active site forms around the substrate loop for the primary cleavage event. This work thus answers some of the outstanding questions on the mode of fI activity, provides the structural basis for the understanding of the disease-associated fI mutations, and forms the basis for future work on the regulation of immunity by fI proteolysis of C3b/C4b.
Fig. 6.
Schematic of the proposed cofactor model. Red arrows indicate domain rearrangements. (A) Cartoon of the proposed allosteric activation of the SP domain via alteration of the heavy-chain (HC)/light-chain (LC) interface. (B) Schematic of the assembly of the C3b:fH1–4:fI ternary complex, colored as in Fig. 5, extended from cartoon of cofactor–C3b interactions (43).
Materials and Methods
Human FI Protein Purification.
Human fI was purified from citrated, outdated human serum from the blood bank (HD Supplies) with three minor modifications of the protocol described in ref. 44: (i) the serum was clarified by centrifugation; (ii) no protease inhibitors were used; and (iii) size-exclusion chromatography fractions containing fI in Tris-buffered saline (pH 7.2) were pooled and cleaned of IgGs and albumin with a Hi-Trap protein-G column (GE Healthcare) and an anti-human serum albumin column, respectively.
Human FI Crystallization, Data Collection, Data Processing, and Structure Determination.
Crystals of human fI were grown by vapor diffusion in 400-nL sitting drops, using an OryxNano robot (Douglas Instruments), by mixing (3:2) the fI (14 mg/mL) with 20–23.5% (wt/vol) PEG 1,500, 0.1 M Na succinate (pH 4.0), and 1 mM CaCl2. Crystals were harvested, cryoprotected in mother liquor supplemented with 15% ethylene glycol, and flash frozen in liquid nitrogen. Diffraction data were collected at 100 K with X-rays of wavelength λ = 0.9795 Å on beamline I02 at the Diamond Light Source (Harwell). The data are tetartohedrally twinned. All data collection statistics are given in SI Appendix, Table S1.
All datasets were indexed and scaled using XDS (45). CCP4 programs were used throughout for structure determination (46). All model building was done in Coot (47). The structure of fI was determined by molecular replacement using Phaser with models for the individual domains (Protein Data Bank IDs:, in order of placement: SP:1EKB; SRCR:1BY2; LDLRAs:1J8E; FIMAC:3B4V) followed by iterative model building and refinement against intensities modeling the tetartohedral twinning in Refmac5. Fourfold non crystallographic symmetry (NCS) was used during iterative model building. Refinement statistics are reported in SI Appendix, Table S1. The refined model and measured structure factor amplitudes have been deposited in the Protein Data Bank (ID 2XRC).
Although fI proteolysis of C3b with fH as a cofactor is most efficient at pH 5 or lower (48), we used a a C3(NH3) proteolysis assay using fI from washed and redissolved crystals to determine whether the pH 4.5–5.0 range at which the crystals diffracting to high resolution grew had altered activity. SI Appendix, Fig. S10 demonstrates that this material still is fully active in the proteolysis of C3(NH3).
Docking of FI onto the C3b:fH1–4 Crystal Structure.
A model for the C3b:fH1-4:fI ternary complex was generated by first overlaying the 1279PSRSSK1284 region of C3b with the pseudosubstrate peptide 37LCKARF42 from a trypsin/TdPI inhibitor complex (C3b: Protein DataBase ID 2WII; trypsin: Protein Database ID: 2UUY) and then using the orientation of trypsin in this complex to overlay fI onto the binary C3b:fH complex (SI Appendix, Fig. S7).
Supplementary Material
Acknowledgments
C. L. Chow assisted with the fI purification. A. W. Dodds kindly donated C3 for cofactor assays. A. C. Willis helped with mass spectroscopy and sequencing of fI. E. Lowe, M. Noble, E. Garman, E. Rudiño-Piñera, and K. Paithankar helped with X-ray data collection. We are grateful to G. Bricogne and the members of the Global Phasing Consortium for access to a beta-version of the autoPROC software. P.R. and S.J. were funded by Grant 083599 from the Wellcome Trust and S.M.L was supported by Medical Research Council Project Grant G0400775.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.M.H. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB ID 2xrc).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102167108/-/DCSupplemental.
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lachmann PJ, Müller-Eberhard HJ. The demonstration in human serum of “conglutinogen-activating factor” and its effect on the third component of complement. J Immunol. 1968;100:691–698. [PubMed] [Google Scholar]
- 3.Harris CL, Morgan BP. Complement Regulatory Proteins. London: Academic; 1999. [Google Scholar]
- 4.Nicol PA, Lachmann PJ. The alternate pathway of complement activation. The role of C3 and its inactivator (KAF) Immunology. 1973;24:259–275. [PMC free article] [PubMed] [Google Scholar]
- 5.van Lookeren Campagne M, Wiesmann C, Brown EJ. Macrophage complement receptors and pathogen clearance. Cell Microbiol. 2007;9:2095–2102. doi: 10.1111/j.1462-5822.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 6.Botto M, et al. Complement in human diseases: Lessons from complement deficiencies. Mol Immunol. 2009;46:2774–2783. doi: 10.1016/j.molimm.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 7.Hocking HG, et al. Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations. J Biol Chem. 2008;283:9475–9487. doi: 10.1074/jbc.M709587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jing H, et al. Structural basis of profactor D activation: From a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D. EMBO J. 1999;18:804–814. doi: 10.1093/emboj/18.4.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukacik P, et al. Complement regulation at the molecular level: The structure of decay-accelerating factor. Proc Natl Acad Sci USA. 2004;101:1279–1284. doi: 10.1073/pnas.0307200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, et al. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 13.Chamberlain D, Ullman CG, Perkins SJ. Possible arrangement of the five domains in human complement factor I as determined by a combination of X-ray and neutron scattering and homology modeling. Biochemistry. 1998;37:13918–13929. doi: 10.1021/bi9805184. [DOI] [PubMed] [Google Scholar]
- 14.DiScipio RG. Ultrastructures and interactions of complement factors H and I. J Immunol. 1992;149:2592–2599. [PubMed] [Google Scholar]
- 15.Fearon DT. Purification of C3b inactivator and demonstration of its two polypeptide chain structure. J Immunol. 1977;119:1248–1252. [PubMed] [Google Scholar]
- 16.Goldberger G, et al. Biosynthesis and postsynthetic processing of human C3b/C4b inactivator (factor I) in three hepatoma cell lines. J Biol Chem. 1984;259:6492–6497. [PubMed] [Google Scholar]
- 17.Tsiftsoglou SA, et al. Human complement factor I glycosylation: Structural and functional characterisation of the N-linked oligosaccharides. Biochim Biophys Acta. 2006;1764:1757–1766. doi: 10.1016/j.bbapap.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Ullman CG, Perkins SJ. The Factor I and follistatin domain families: The return of a prodigal son. Biochem J. 1997;326:939–941. doi: 10.1042/bj3260929v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekdahl KN, Nilsson UR, Nilsson B. Inhibition of factor I by diisopropylfluorophosphate. Evidence of conformational changes in factor I induced by C3b and additional studies on the specificity of factor I. J Immunol. 1990;144:4269–4274. [PubMed] [Google Scholar]
- 20.Davis AE, 3rd, Harrison RA. Structural characterization of factor I mediated cleavage of the third component of complement. Biochemistry. 1982;21:5745–5749. doi: 10.1021/bi00266a003. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson SC, et al. Analysis of binding sites on complement factor I that are required for its activity. J Biol Chem. 2010;285:6235–6245. doi: 10.1074/jbc.M109.097212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson SC, et al. Genetic, molecular and functional analyses of complement factor I deficiency. Eur J Immunol. 2009;39:310–323. doi: 10.1002/eji.200838702. [DOI] [PubMed] [Google Scholar]
- 23.Lechtenberg BC, Johnson DJ, Freund SM, Huntington JA. NMR resonance assignments of thrombin reveal the conformational and dynamic effects of ligation. Proc Natl Acad Sci USA. 2010;107:14087–14092. doi: 10.1073/pnas.1005255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Bode W, Fehlhammer H, Huber R. Crystal structure of bovine trypsinogen at 1-8 A resolution. I. Data collection, application of Patterson search techniques and preliminary structural interpretation. J Mol Biol. 1976;106:325–335. doi: 10.1016/0022-2836(76)90089-9. [DOI] [PubMed] [Google Scholar]
- 26.Tsiftsoglou SA, et al. The catalytically active serine protease domain of human complement factor I. Biochemistry. 2005;44:6239–6249. doi: 10.1021/bi047680t. [DOI] [PubMed] [Google Scholar]
- 27.Kamath P, Huntington JA, Krishnaswamy S. Ligand binding shuttles thrombin along a continuum of zymogen- and proteinase-like states. J Biol Chem. 2010;285:28651–28658. doi: 10.1074/jbc.M110.154914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsiftsoglou SA, Sim RB. Human complement factor I does not require cofactors for cleavage of synthetic substrates. J Immunol. 2004;173:367–375. doi: 10.4049/jimmunol.173.1.367. [DOI] [PubMed] [Google Scholar]
- 29.Pangburn MK, Müller-Eberhard HJ. Kinetic and thermodynamic analysis of the control of C3b by the complement regulatory proteins factors H and I. Biochemistry. 1983;22:178–185. doi: 10.1021/bi00270a026. [DOI] [PubMed] [Google Scholar]
- 30.Fevig JM, et al. Rational design of boropeptide thrombin inhibitors: Beta, beta-dialkyl-phenethylglycine P2 analogs of DuP 714 with greater selectivity over complement factor I and an improved safety profile. Bioorg Med Chem Lett. 1998;8:301–306. doi: 10.1016/s0960-894x(98)00013-4. [DOI] [PubMed] [Google Scholar]
- 31.Forneris F, et al. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westra D, et al. Genetic disorders in complement (regulating) genes in patients with atypical haemolytic uraemic syndrome (aHUS) Nephrol Dial Transplant. 2010;25:2195–2202. doi: 10.1093/ndt/gfq010. [DOI] [PubMed] [Google Scholar]
- 33.Kavanagh D, et al. Characterization of mutations in complement factor I (CFI) associated with hemolytic uremic syndrome. Mol Immunol. 2008;45:95–105. doi: 10.1016/j.molimm.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Eigenbrot C. Structure, function, and activation of coagulation factor VII. Curr Protein Pept Sci. 2002;3:287–299. doi: 10.2174/1389203023380675. [DOI] [PubMed] [Google Scholar]
- 35.Higashi S, Matsumoto N, Iwanaga S. Molecular mechanism of tissue factor-mediated acceleration of factor VIIa activity. J Biol Chem. 1996;271:26569–26574. doi: 10.1074/jbc.271.43.26569. [DOI] [PubMed] [Google Scholar]
- 36.Caprioli J, et al. International Registry of Recurrent and Familial HUS/TTP. Genetics of HUS: The impact of MCP, CFH, and IF mutations on clinical presentation, response to treatment, and outcome. Blood. 2006;108:1267–1279. doi: 10.1182/blood-2005-10-007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Servais A, et al. Primary glomerulonephritis with isolated C3 deposits: A new entity which shares common genetic risk factors with haemolytic uraemic syndrome. J Med Genet. 2007;44:193–199. doi: 10.1136/jmg.2006.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis AE, 3rd, Harrison RA, Lachmann PJ. Physiologic inactivation of fluid phase C3b: Isolation and structural analysis of C3c, C3d,g (alpha 2D), and C3g. J Immunol. 1984;132:1960–1966. [PubMed] [Google Scholar]
- 39.Vĕtvicka V, Reed W, Hoover ML, Ross GD. Complement factors H and I synthesized by B cell lines function to generate a growth factor activity from C3. J Immunol. 1993;150:4052–4060. [PubMed] [Google Scholar]
- 40.Soames CJ, Sim RB. Interactions between human complement components factor H, factor I and C3b. Biochem J. 1997;326:553–561. doi: 10.1042/bj3260553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pechtl IC, Kavanagh D, McIntosh N, Harris CL, Barlow PN. Disease-associated N-terminal complement factor H mutations perturb cofactor and decay-accelerating activities. J Biol Chem. 2011;286:11082–11090. doi: 10.1074/jbc.M110.211839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yadav VN, Pyaram K, Mullick J, Sahu A. Identification of hot spots in the variola virus complement inhibitor (SPICE) for human complement regulation. J Virol. 2008;82:3283–3294. doi: 10.1128/JVI.01935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roversi P, et al. Structures of the rat complement regulator CrrY. Acta Crystallographica Section. 2011;F67:739–743. doi: 10.1107/S1744309111016551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sim RB, Day AJ, Moffatt BE, Fontaine M. Complement factor I and cofactors in control of complement system convertase enzymes. Methods Enzymol. 1993;223:13–35. doi: 10.1016/0076-6879(93)23035-l. [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sim E, Sim RB. Enzymic assay of C3b receptor on intact cells and solubilized cells. Biochem J. 1983;210:567–576. doi: 10.1042/bj2100567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blom AM, Villoutreix BO, Dahlbäck B. Mutations in alpha-chain of C4BP that selectively affect its factor I cofactor function. J Biol Chem. 2003;278:43437–43442. doi: 10.1074/jbc.M306620200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.