Abstract
Calpain is an intracellular Ca2+-dependent cysteine protease (EC 3.4.22.17; Clan CA, family C02) discovered in 1964. It was also called CANP (Ca2+-activated neutral protease) as well as CASF, CDP, KAF, etc. until 1990. Calpains are found in almost all eukaryotes and a few bacteria, but not in archaebacteria. Calpains have a limited proteolytic activity, and function to transform or modulate their substrates’ structures and activities; they are therefore called, “modulator proteases.” In the human genome, 15 genes—CAPN1, CAPN2, etc.—encode a calpain-like protease domain. Their products are calpain homologs with divergent structures and various combinations of functional domains, including Ca2+-binding and microtubule-interaction domains. Genetic studies have linked calpain deficiencies to a variety of defects in many different organisms, including lethality, muscular dystrophies, gastropathy, and diabetes. This review of the study of calpains focuses especially on recent findings about their structure–function relationships. These discoveries have been greatly aided by the development of 3D structural studies and genetic models.
Keywords: calpain, muscular dystrophy, calpainopathy, gastropathy, intracellular proteolysis, calcium ion
Introduction
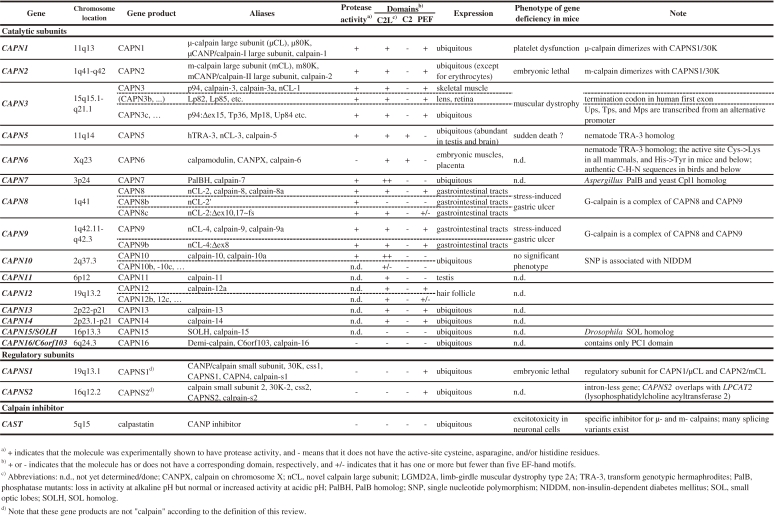
An enzyme corresponding to calpain was first described in 1964 by Gordon Guroff (1933–1999).1,2) Over the next several years, calpain was reported with various names and in various contexts by other researchers, including Edmond H. Fischer (1920–), Edwin G. Krebs (1918–2009),3,4) Darrel E. Goll (1936–2008),5) and Yasutomi Nishizuka (1932–2004).6) In 1978, Kazutomo Imahori (1920–) and his colleagues purified chicken calpain to homogeneity and named it CANP (calcium-activated neutral protease).7) This marked the beginning of serious, focused studies of calpain, and this protease was soon established as one of the most important and intriguing enzymes in the cell.8–10) That same year, an endogenous calpain-specific inhibitor protein was reported by Takashi Murachi (1926–1990) and his colleagues.11) They proposed the names calpain in place of CANP and calpastatin for its inhibitor protein in 1981.12) In 1984, Koichi Suzuki (1939–2010) and his colleagues revealed the complete primary structure of the calpain catalytic subunit,13) and calpain studies turned toward structure–function analyses. For more than ten years, both CANP and calpain were used in the literature. This complication was resolved by Suzuki in accordance with Imahori in 1990. He proposed the usage of calpain as an authorized name at the 8th International Conference on Proteolysis and Protein Turnover.14) Since Suzuki’s 1984 paper,13) cDNA and genomic cloning studies have exploded, and thousands of calpains and related molecules were identified—among them 15 human calpain genes, which are now called CAPNn (n = 1, 2, 3, and 5–16), which is short for calpain.15) There are two genes for small regulatory subunits, CAPNS1 and CAPNS2, short for calpain small subunit, and one for calpastatin, CAST.
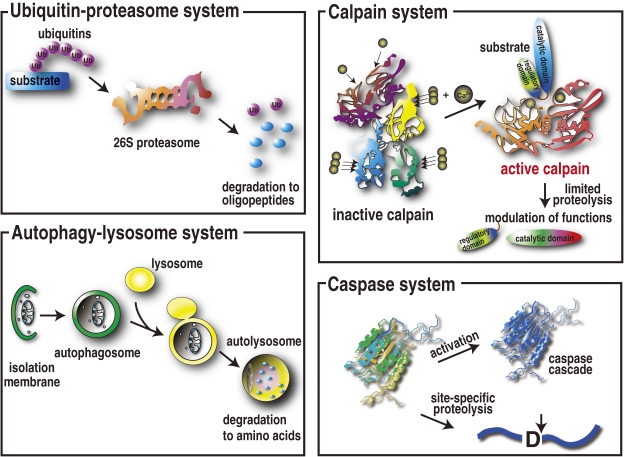
Calpains (EC 3.4.22.17; Clan CA, family C02) constitute a distinct group of intracellular cysteine proteases found in almost all eukaryotes and a few bacteria (Fig. 1 ; for other reviews, see the two landmark reviews in 199116) and 200317) and later reviews18–49)). Calpains are first defined as cytosolic proteases exhibiting Ca2+-dependent proteolytic activity at a neutral pH. Most calpains are intracellular, and their activity is strictly regulated, as is the case with other intracellular proteolytic systems, such as the ubiquitin–proteasome system,50) the autophagy–lysosome system,51) and caspases52) (Fig. 2). Caspases proteolyze substrates sequence-specifically, mostly for apoptosis, and they are activated by signal-dependent oligomerization. The former two systems act as “erasers” that degrade and eliminate substrate proteins. In contrast, calpains act in proteolytic processing, rather than degradation, i.e., calpains proteolyze their substrates at a limited number of sites to transform or modulate their substrates’ structures and activities. Therefore, calpain is referred to as an intracellular “modulator” protease.53,54) Ubiquitin and autophagy systems are regulated in various steps mediated by many proteins, but calpain is distinctive in that a functional entity rests on a single molecule or two.
Figure 1.
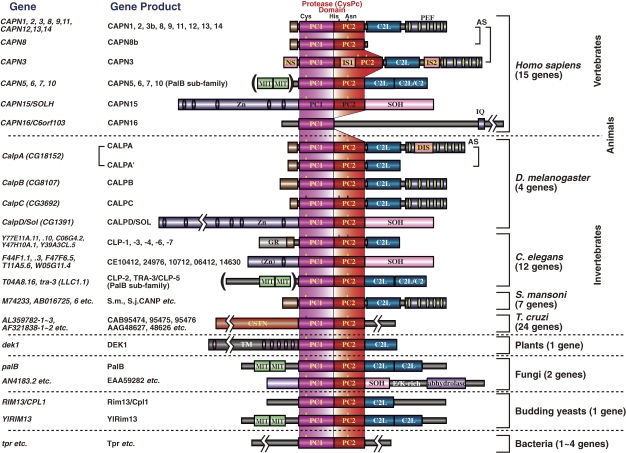
Schematic structures of calpain superfamily members. Calpain homologs have been identified in almost all eukaryotes and in some bacteria. Historically, there are several nomenclatures for domains. To avoid confusion in this review, the domains are called by the abbreviation corresponding to the structure, i.e., PC1 (protease core domain 1), PC2 (protease core domain 2), C2L (C2 domain-like), PEF (penta-EF-hand) etc. Symbols: N, N-terminal region; PEF(L) and PEF(S), PEF domains of large catalytic and small regulatory subunits, respectively; GR, glycine-rich hydrophobic domain; AS, alternative splicing products; NS/IS1/IS2, CAPN3-characteristic sequences; C2, C2 domain; MIT, microtubule-interacting-and-transport motif; Zn, Zn-finger-containing domain; SOH, SOL-homology domain; DIS, CALPA-specific insertion sequence; TM, transmembrane domain; CSTN, calpastatin-like domain; IQ, a motif interactive with calmodulin.
Figure 2.
Major intracellular proteolytic systems. The ubiquitin–proteasome system degrades and eliminates specific substrate proteins in an ubiquitin-tagging system consisting of more than 1,000 ubiquitin ligases. The autophagy–lysosome system primarily degrades non-specific cell components, including proteins and micro-organisms, by compartmentalization by isolation membranes. The caspase system functions mainly for apoptosis. In contrast, calpains primarily use proteolytic processing, rather than degradation, to modulate or modify their substrates’ activity, specificity, longevity, localization, and structure.
The importance of the calpains’ physiological roles is reflected in the variety of defects caused by compromised calpain function in different tissues and organisms, including embryonic lethality (disruption of mouse Capn2),55) muscular dystrophies (mutations in human CAPN3),56) gastropathy (mutated Capn8),57) lissencephaly58) and tumorigenesis59) (indirect defects in calpain functions), impaired neurogenesis in flies,60) deficient sex determination in nematodes,61) defects in aleurone cell development in maize,62) and alkaline stress susceptibility in fungi63) and yeasts.64,65) These effects will be described in more detail in the following sections.
Mammalian gene products can be written as non-italic upper-case letters of their gene name, according to the international nomenclature system for a gene product; for example, mouse Capn1 and Capn2 produce CAPN1 and CAPN2, respectively. Although recently mammalian calpain gene products are often written as calpain-1, calpain-2, etc., this nomenclature is misleading as described below (“calpain” is also used for enzyme names), so we will use the above formal nomenclature (CAPN1, CAPN2, etc.) throughout this review. Some calpains were previously called by independent names, such as μCL and p94. In this review, to clarify old and new mammalian calpain nomenclatures, we will note the old after the current names: e.g., CAPN1/μCL and CAPN3/p94.
1. A research history of calpain and its definition
Because of the scope of this review, it is important to define calpain, and the complexities of defining calpain are best explained in the context of its research history.66–68)
In 1964, Guroff partially purified a Ca2+-dependent neutral protease from rat brain, which afterwards turned out to be calpain.1) In the same year, Meyer, Fischer and Krebs described the activation of rabbit skeletal muscle phosphorylase b kinase by Ca2+ and partially purified the responsible protein, which they called KAF (kinase activating factor).4) Four years later, Huston and Krebs,3) and Drummond and Duncan69) independently reported that this protein was a Ca2+-dependent protease, although they concluded that the proteolytic activation of phosphorylase b kinase was unlikely to be physiological. Krebs and Fishers’ main interest then moved to phosphorylation and its cascade, for which they were awarded a Nobel Prize in 1992.70,71) Since then, Krebs has changed his mind about the physiological importance of proteolytic activation. Indeed, one of calpain’s most important physiological functions is to modulate substrate protein functions by limited proteolysis, and this property must be considered in the definition—or at least, the classification—of calpain.
In 1972, Darrel E. Goll and his colleagues identified a factor that Ca2+-dependently removes the Z-lines of skeletal muscle, and named it CASF (Ca2+-activated sarcoplasmic factor), which also turned out to be calpain.5) This study is the beginning of research on calpain involved in postmortem tenderization of meat.72) In 1977, Yasutomi Nishizuka and his colleagues identified calpain as a protein kinase C (PKC) activating factor,6) and several calpain studies were done in line with PKC.73–76) In 1978, Imahori and his colleagues for the first time in the world purified chicken calpain to homogeneity and called it CANP. Following this remarkable study, Imahori’s group established calpain research field by publishing important studies on enzymology of calpain such as substrate specificity,77) purification from human muscle,78) inhibitors,79,80) autolysis,8,81) and activation.82,83)
Calpain purification studies (ca. 90% purity in 1976,84) and to homogeneity in 19787)) revealed that functional native calpain is composed of large (ca. 80 kDa) and small (ca. 30 kDa) subunits, which is another important property of calpain. The chicken μ/m-calpain catalytic subunit cDNA was cloned in 1984,13) and since then many cDNAs and genomic DNAs corresponding to various calpain species have been cloned and sequenced. Little is known about the nature of calpain proteins as enzymes, however, and most calpains are known only by their sequence.
The most extensively studied calpains are the major ubiquitous mammalian μ- and m-calpains, and the major ubiquitous calpain in chicken, μ/m-calpain.14) These are called “conventional” calpains in the wake of PKC;85) all others are termed “unconventional” calpains. The chicken μ/m-calpain has properties placing itself as an intermediate of μ- and m-calpains.7,77,86) Its catalytic subunit was shown to be an ortholog of mammalian CAPN11 (see also 6.3.3).87)
Among the amino acid (aa) sequences available, there is a group of peptidases that possess a protease domain that is significantly similar to one another but distinct from that of other peptidases.88) A search for “CysPC” in the conserved domain database of NCBI89) extracts the sequences of almost all the calpain homologs from various living organisms, including plants, fungi, yeast, and even bacteria. Calpains belong to the papain superfamily of cysteine proteases and have weak similarity to papains and cysteine cathepsins, although it is clearly less significant than the similarities between calpains. In this superfamily, calpain may evolutionarily be the oldest branch.89)
Considering this situation, it is reasonable to define calpain mainly by the aa sequence in relation to the most-characterized mammalian μ- and m-calpains. Therefore, this review uses the following simple but clear definition: calpains are defined as proteins that have aa sequences significantly similar to the protease domain of the human μ-calpain large subunit (replacing μ-calpain with m-calpain in this definition would give the same result). Additional characteristics reflect calpain classifications: the conventional, classical, non-classical, ubiquitous, and/or tissue-specific calpains should be discussed separately when considering their physiological functions. In other words, sequence similarity does not necessarily reflect functional similarities between calpain species.
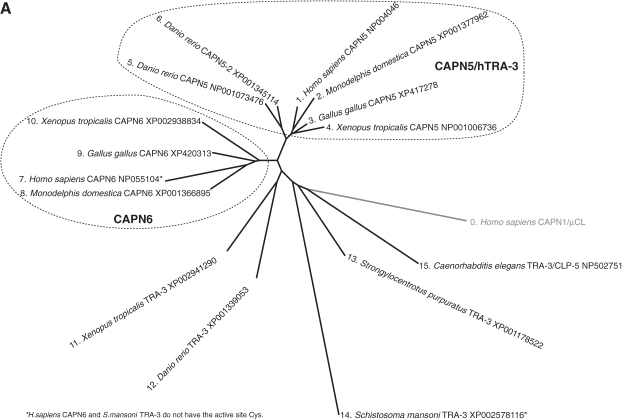
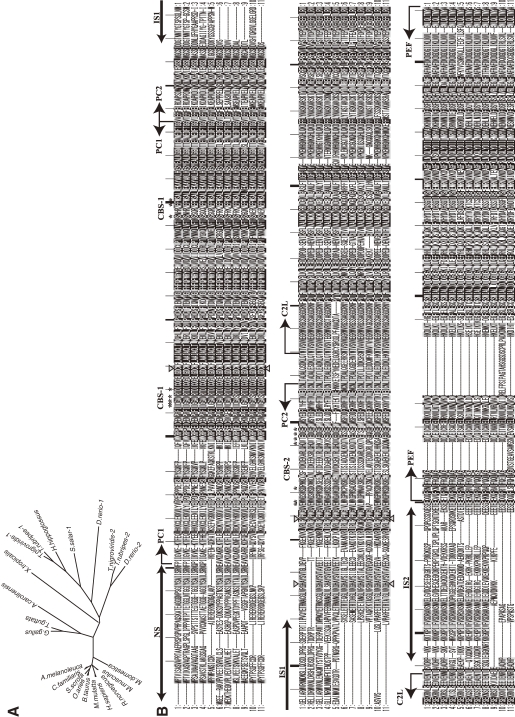
According to this definition, humans have 15 genes that encode calpains (Figs. 3 and 4). In other species, Schistosoma mansoni (Fig. 5), Caenorhabditis elegans (Fig. 6), Anopheles gambiae (Fig. 7), Drosophila melanogaster (Fig. 7), Arabidopsis thaliana (Fig. 1), Emericella (Aspergillus) nidulans (Fig. 1), and Saccharomyces cerevisiae (Fig. 1) have 7, 14, 7, 4, 1, 2, and 1 calpain genes, respectively. There is no calpain gene in Encephalitozoon or Schizosaccharomyces pombe. In prokaryotes, 53 calpains from 42 bacteria were found among the 914 completely sequenced microbial genomes present in the database so far. Each of the bacterial species has 1–4 calpain genes (Fig. 8). However, most genome-sequenced bacteria, including Escherichia coli or any of the archeabacteria, do not have calpain genes.
Figure 3.
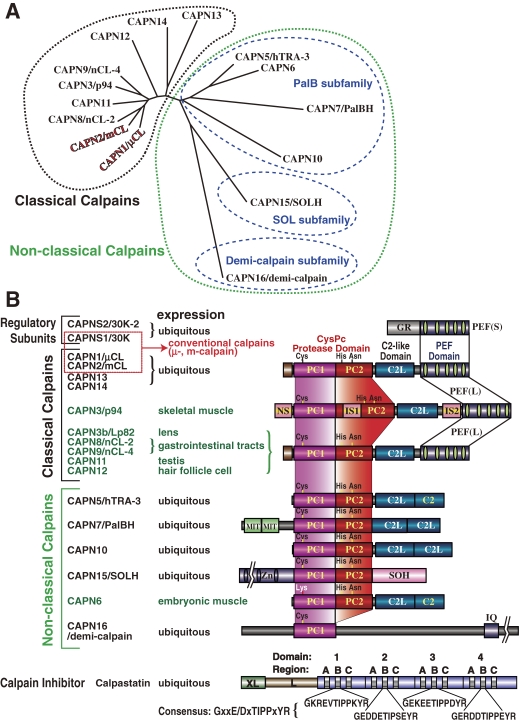
Phylogenetic tree and schematic structures of human calpain-related molecules. A. Phylogenetic tree of human calpains. Method: The tree was drawn by the neighbor-joining/bootstrap method after aligning all the sequences using MAFFT v6.240 (at http://align.genome.jp/mafft/, strategy: E-INS-i). Atypical calpains are more divergent than classical calpains. The PalB subfamily consists of the strict PalB group, the TRA-3 group, and the CAPN10 group. B. Schematic structures: Black and green letters indicate ubiquitous and tissue/organ-specific calpains, respectively. See also Fig. 4. Symbols: L and XL, N-terminal and extended N-terminal regions of calpastatin. See Fig. 1 legend for other symbols.
Figure 4.
Human calpain-related genes. See also Fig. 3.
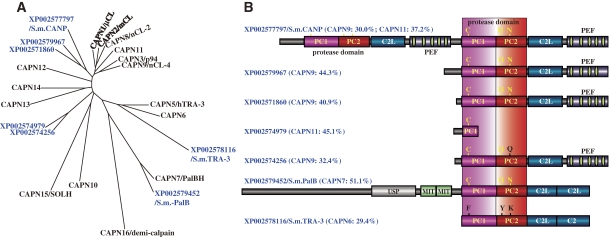
Figure 5.
Phylogenetic tree and schematic structures of S. mansoni calpains. A. Phylogenetic tree of S. mansoni and human calpains. See Fig. 3A for methods. B. Schematic structures. The human calpain most similar to each S. mansoni calpain is indicated in parentheses, with the aa sequence identity. The idnentity is given for each of the tandem classical calpain structures in XP_002577797. USP: ubiquitin-specific peptidase. See Fig. 1 legend for other symbols.
Figure 6.
Phylogenetic tree and schematic structures of C. elegans calpains. A. Phylogenetic tree of C. elegans and human calpains. See Fig. 3A for methods. B. Schematic structures. The human calpain most similar to each C. elegans calpain is indicated in parentheses, with the aa sequence identity. CI and CII stand for conserved regions I and II with unknown function, respectively. See Fig. 1 legend for other symbols.
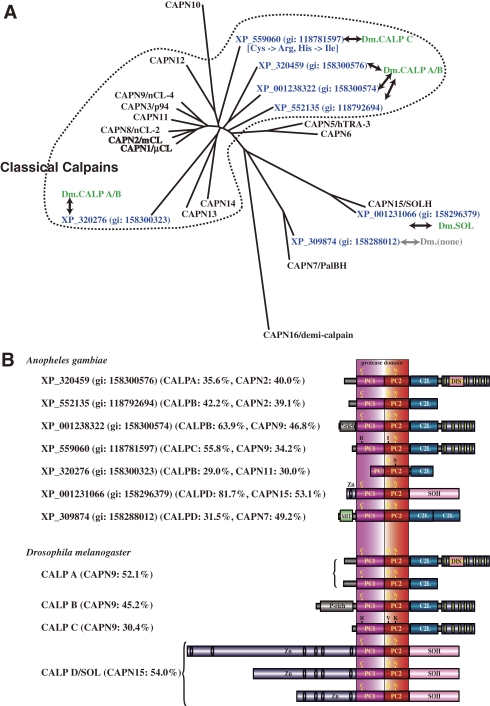
Figure 7.
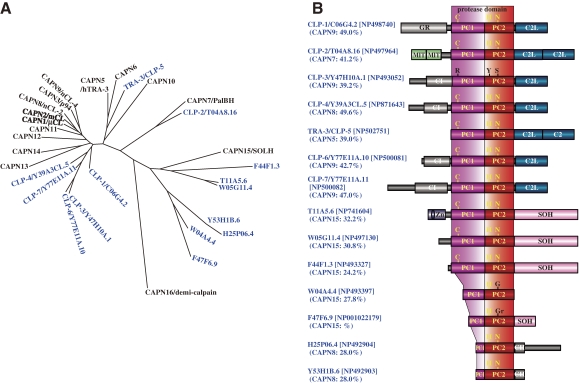
Phylogenetic tree and schematic structures of insect calpains. A. Phylogenetic tree of A. gambiae and human calpains. See Fig. 3A for methods. D. melanogaster (DM) homologs are shown in green letters. B. Schematic structures. The human calpain most similar to each insect calpain is indicated in parentheses, along with the aa sequence identity. The D. melanogaster calpain most similar to A. gambiae calpain is also indicated. P-rich: Pro-rich region. DIS: Drosophila-type insertion sequence. See Fig. 1 legend for other symbols.
Figure 8.
Phylogenetic tree and sequence alignment of bacterial calpains. A. Phylogenetic tree of bacterial calpains and human CAPN1/μCL. See Fig. 3A for methods. B. Sequence alignment of the bacterial calpain protease domains and human CAPN1/μCL. Only the sequences of the protease domains are compared, with the start and end aa residue numbers indicated, as other sequences and lengths are somewhat divergent. Reversed fonts and gray shadow indicate residues conserved in all sequences or more than half, respectively. Triangles: active site residues. Asterisks: residues involved in Ca2+-binding of the protease domain of human CAPN1/μCL. Sequences: 0: H. sapiens NP_001185798; 1: Acaryochloris marina MBIC11017 YP_001520592; 2: Actinomyces odontolyticus ATCC 17982 ZP_02043925; 3: Actinomyces odontolyticus F0309 ZP_06609922; 4: Actinomyces sp. oral taxon 848 str. F0332 ZP_06161744; 5: Actinomyces sp. oral taxon 848 str. F0332 ZP_06161892; 6: Actinomyces sp. oral taxon 848 str. F0332 ZP_06162803; 7: Actinomyces sp. oral taxon 848 str. F0332 ZP_06162809; 8: Anabaena variabilis ATCC 29413 YP_322403; 9: Bacteroides ovatus ATCC 8483 ZP_02064250; 10: Bacteroides sp. 1_1_14 ZP_06994459; 11: Bacteroides sp. 1_1_6 ZP_04847694; 12: Bacteroides sp. D2 ZP_05758284; 13: Bacteroides thetaiotaomicron VPI-5482 NP_812871; 14: Brachybacterium faecium DSM 4810 YP_003153779; 15: Brachybacterium faecium DSM 4810 YP_003155356; 16: Bradyrhizobium sp. BTAi1 YP_001241302; 17: Bradyrhizobium sp. ORS278 YP_001204811; 18: Brevibacterium mcbrellneri ATCC 49030 ZP_06806606; 19: Cyanothece sp. PCC 7822 YP_003887732; 20: Cyanothece sp. PCC 7822 YP_003887741; 21: Frankia alni ACN14a YP_716900; 22: Frankia sp. CcI3 YP_483525; 23: Frankia sp. EAN1pec YP_001504490; 24: Frankia sp. EuI1c YP_004014028; 25: Frankia sp. EUN1f ZP_06411021; 26: Gemmata obscuriglobus UQM 2246 ZP_02733073; 27: Geobacter lovleyi SZ YP_001952615; 28: Gloeobacter violaceus PCC 7421 NP_927086; 29: Granulibacter bethesdensis CGDNIH1 YP_745367; 30: Herpetosiphon aurantiacus ATCC 23779 YP_001544076; 31: Herpetosiphon aurantiacus ATCC 23779 YP_001545619; 32: Herpetosiphon aurantiacus ATCC 23779 YP_001546212; 33: Legionella longbeachae D-4968 ZP_06188344; 34: Legionella longbeachae NSW150 YP_003455673; 35: Microcystis aeruginosa NIES-843 YP_001659697; 36: NC10 bacterium ‘Dutch sediment’ CBE68371; 37: Nostoc sp. PCC 7120 NP_484413; 38: Nostoc sp. PCC 7120 NP_485127; 39: Nostoc sp. PCC 7120 NP_487855; 40: Photorhabdus luminescens subsp. laumondii TTO1 NP_929692; 41: Planctomyces limnophilus DSM 3776 YP_003628665; 42: Porphyromonas gingivalis AAA25652; 43: Porphyromonas gingivalis W83 NP_905271; 44: Rhodopseudomonas palustris BisA53 YP_780017; 45: Salinispora tropica CNB-440 YP_001157346; 46: Segniliparus rotundus DSM 44985 YP_003659885; 47: Streptomyces albus J1074 ZP_06592718; 48: Streptomyces viridochromogenes DSM 40736 ZP_07305717; 49: Synechococcus sp. RS9916 ZP_01472825; 50: Synechococcus sp. RS9916 ZP_01472826; 51: Synechococcus sp. RS9916 ZP_01472827.
2. Conventional calpain—the basics
2.1. Domain structures of conventional calpains.
The conventional calpains, μ- and m-calpains, are heterodimers consisting of a common small regulatory subunit (CAPNS1/30K; ca. 30 kDa) and a distinct, large (ca. 80 kDa) μ- or m-calpain catalytic subunit (CAPN1/μCL or CAPN2/mCL, respectively) (Fig. 3). Calpain-I is an old alternative name for μ-calpain, named as an enzyme, and it is not the same as calpain-1, which is the name often used for a gene product, a subunit of an enzyme. Instead, calpain-I is a heterodimer of CAPN1/μCL and CAPNS1/30K. The distinction is similar for calpain-II and calpain-2.
CAPN1/μCL and CAPN2/mCL have approximately 60% aa identity. Accordingly, they have almost indistinguishable substrate and inhibitor specificities, and both are almost ubiquitously expressed. However, they differ greatly in their in vitro Ca2+-requirement (µM versus mM) for proteolytic activity. Their functions are fundamental and essential, and are implicated in the regulation of signal transduction systems,90,91) cell motility,21,92) membrane repair,93,94) and apoptosis,95,96) although their precise roles in these events remain elusive.
The catalytic and regulatory subunits of conventional calpains can be divided into several domains (Fig. 3). The N-terminal anchor α-helix of the catalytic large subunit (also called anchor region or domain I) is autolyzed when activated by Ca2+, resulting in its ability to function at a lower Ca2+ concentration,81,97–99) with a different substrate specificity,100) and subunit dissociation in some cases100–103) but not others.104–106) Therefore, autolysis of this anchor helix is one of the important regulatory mechanisms for calpain activity.
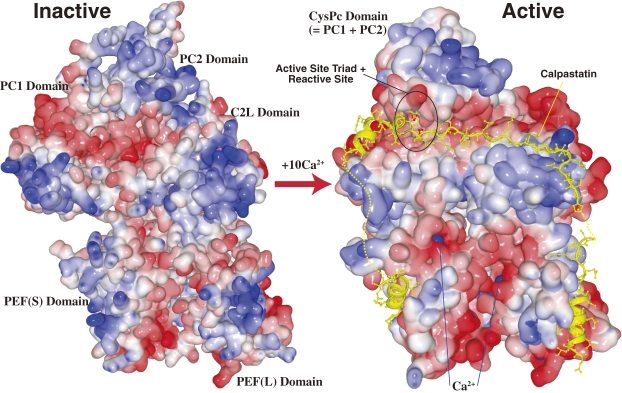
In isolation, m-calpain’s protease (CysPc) domain (also called domain II), has Ca2+-dependent protease activity.107) 3D structural analyses in the absence of Ca2+ showed that the CysPc domain is further divided into two core domains, PC1 (also called D-I or subdomain IIa) and PC2 (D-II or IIb) (Fig. 3). These two core domains fold into a single functional CysPc domain when bound to Ca2+ (Fig. 9).108–113) In other words, calpain remains structurally inactive in the absence of Ca2+. This is reasonable, because calpain resides in the cytosol in direct contact with many other proteins, and its activity must be strictly regulated.
Figure 9.
Schematic of the 3D structure of inactive and active m-calpain. Surface-type schematic 3D structures of inactive (Ca2+ free) and active (Ca2+- and calpastatin-bound) forms of m-calpain using PDB data, 1KFX109) and 3DF0.111) The oligopeptides represented by the yellow ribbon + ball-and-stick indicate portions of calpastatin bound to active m-calpain. The dotted lines indicate portions that were too mobile for the 3D structure to be determined. The active protease domain (CysPc) is formed by the fusion of core domains PC1 and PC2 upon the binding of one Ca2+ to each of the core domains. The active site is circled in black. Blue balls represent Ca2+ (not all are visible).
Ca2+ also binds to C2 domain-like (C2L) domain (also called domain III)114) and penta-EF-hand (PEF) domain (domain IV) of the large catalytic subunit (PEF(L) domain), and that of the small regulatory subunit (PEF(S), also called domain VI).115,116) While C2L and the C2 domains have no sequence similarity, their 3D-structure is very similar (β-sandwich structure composed of eight anti-parallel β-strands). Two PEF domains (PEF(L) and PEF(S)) each contain five EF-hand motifs.48,117–119) The fifth EF-hand motif of each domain contributes to heterodimer formation.120) It should be noted that the N-terminal anchor helix is in contact with the second EF-hand motif (EF-2) of PEF(S) of CAPNS1/30K, and the interaction is broken either by Ca2+-binding to the EF-2 or by autolysis of the N-terminus upon activation.
Gly-rich (GR) domain (also called domain V) in the N-terminus of CAPNS1/30K contains hydrophobic Gly-clusters, most of which are autolyzed during activation. This domain was invisible in the 3D structure, indicating a very “soft” structure. In humans, the CAPNS2 gene encodes a regulatory subunit homolog composed of GR and PEF(S) domains, whose physiological roles remain unclear.121)
2.2. Activation and substrates.
Ca2+ is required for the activation of conventional calpains, raising the question of which part of the calpain molecule contains the Ca2+-binding site that provokes calpain activation. It was previously believed that PEF domains were responsible for activating calpain in response to Ca2+, as they are the only Ca2+-binding sites visible in the primary structure.13) However, the 3D structure of calpain revealed that the PEF domains are somewhat distant from the protease domain (Fig. 9). CD spectrum measurements found no significant changes in calpain’s secondary structure upon activation by Ca2+ binding.122) Biochemical analysis of recombinant m-calpain protease domains revealed that they respond directly to Ca2+.107) Finally, the 3D structures of these domains showed that Ca2+ binds directly to PC1 and PC2, to activate conventional calpain (Fig. 9).112,113)
A number of substrates have been reported for calpains, including protein kinases, phosphatases, phospholipases, cytoskeletal proteins, membrane proteins, cytokines, transcription factors, lens proteins, and calmodulin-binding proteins, but few of them have been shown to be physiological. Due to its complexity and “finesse”, the exact rules governing calpain substrate specificity remain incomplete, although bioinformatics approaches revealed some cleavage-site preferences123,124) (see also http://calpain.org). Substrate 3D structures also seem to be important factors for many cases.
2.3. Novelties deduced from the calpain 3D structure.
The classical calpains CAPN1/μCL, CAPN2/mCL, and CAPN9/nCL-4 have highly conserved protease domains (59–71% aa identity); however, the Ca2+-bound 3D structures of these domains reveal distinguishing features.112,113,125)
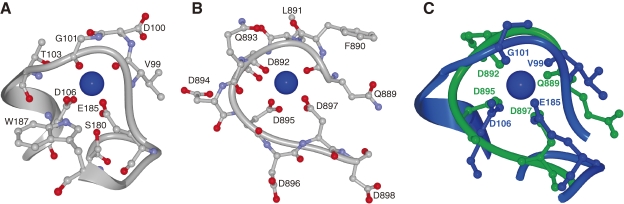
First, comparing the m-calpain protease domains in the presence and absence of Ca2+, all the α-helices and β-strands are conserved, consistent with previous CD spectrum results.122) Both PC1 structures can be aligned very closely (Cα root-mean-squared deviation (r.m.s.d.) = 1.72 Å), whereas those of PC2 are moderately aligned (r.m.s.d. = 3.78 Å). As predicted from PC2’s Ca2+-unbound 3D structure, Ca2+ binding would cause it to be rotated and translated by 50° and 6 Å, if the alignment of PC2 is fixed. Surprisingly, a Ca2+-induced Ca2+-binding site (CBS-1 and CBS-2) is generated in each core domain, and Ca2+ binding to these sites causes the above-described PC2 rotation and translation for activation. These Ca2+-binding sites have a novel and unique structure. Only recently, one structure was shown to have similarity with this site: the 3D structure of the “Ca2+-bowl” of the high-conductance voltage- and Ca2+-activated K+ (BK or SLO1) channel has a similar Ca2+-binding geometry to the μ-calpain CBS-1, although they share no similarity in their primary sequences (Fig. 10).126)
Figure 10.
A comparison of 3D structures of the Ca2+-binding sites of the CAPN1/μCL protease domain IIa and SLO1 (Ca-bowl). Ca2+-binding sites in (A) the CAPN1/μCL protease domain (2R9F) and (B) the high-conductance voltage- and Ca2+-activated K+ channel (BK or SLO1 channel) (3MT5). (C) The structures were superimposed for comparison.126)
Second, the active-site cleft is deeper and narrower than that of papain. Due to this constraint, the substrate must be in a fully extended conformation with its backbone stretched; this was verified by the 3D structures of the Ca2+-bound active μ-calpain protease domain co-crystallized with leupeptin or E64.127) This finding explains calpain’s preference for proteolyzing inter-domain unstructured regions.
Third, although the active protease domains of CAPN1, 2, and 9 are highly similar, a reversible intrinsic inhibitory/safety mechanism exists only for CAPN2/mCL. W106, which is adjacent to the active-site C105 of CAPN2/mCL, positions to interfere with substrates’ access to the active site; in contrast, this mechanism does not occur in rat CAPN1/μCL.113) Moreover, CAPN9/nCL-4 has a novel intrinsic inhibitory mechanism, i.e., the misalignment of the catalytic residues.125) These findings together show that the activation/latency mechanisms of calpains, regardless of their highly conserved sequences, are somewhat specialized for each molecule, and are controlled by inter-domain interactions. In the whole 3D structure of m-calpain,110,111) the above-mentioned W106 mechanism113) was not found, indicating that inter-domain interactions impose extra controls over molecule-specific mechanisms.
Small-angle X-ray scattering analysis revealed direct semi-micro images of both active and inactive whole calpain molecules in the presence of E64,105) showing the following: in the presence of 100 µM Ca2+ and E64, human erythrocyte μ-calpain did not aggregate and had similar dimensions as in the absence of Ca2+ (maximal diameter = 120→130 Å, gyration radius = 36→39 Å). This was consistent with the previous CD spectrum results and with the 3D structure of CAPNS1/30K, whose conformation changes little in the presence or absence of Ca2+.117,119) On the other hand, a small but significant change was observed in the protease domain; it appeared to compact somewhat and partly detach from C2L domain.
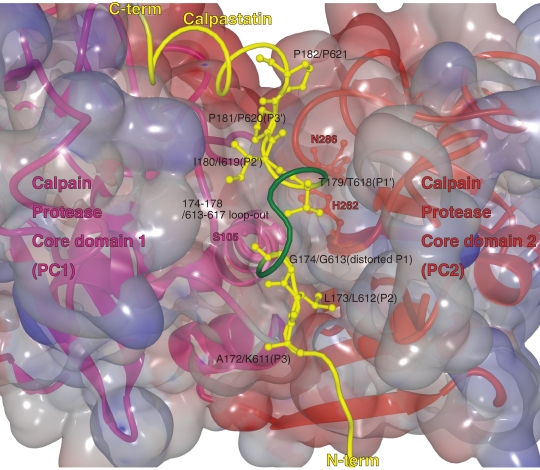
In other words, upon activation, the protease domain dynamically changes its conformation without changing its secondary structure. This feature was finally confirmed when the whole m-calpain 3D structure was determined by co-crystallization with calpastatin repeat 1 or 4 (see 2.6) and Ca2+,110,111) revealing one of the mechanisms used by calpastatin to inhibit conventional calpain activity. G174/G613 of rat calpastatin repeat 1/4, respectively, which is near the conserved TIPPXYR (179–185/618–624) motif in the reactive site of calpastatin, forces a kink (G174-IKE-G178/G613-ERD-D617) between its flanking residues, L173/L612 at the catalytic S2 site and T179/T618 at S1′, to loop-out not to interact with the active site of calpain. A172/K611, L173/L612, T179/T618, I180/I619, and P181/P620 fit into the S3, S2, S1′, S2′, and S3′ pocket of calpain’s active site, respectively (Fig. 11).110,111) Therefore, calpastatin interacts tightly with calpain, but no proteolysis occurs. The importance of this “kink” in resisting proteolysis by calpain was confirmed by calpastatin mutagenesis studies. The deletion of K176, E177, or both, or the insertion of Phe between L612–G613, allows mutant calpastatins to be proteolyzed by calpain.110,111)
Figure 11.
Calpastatin region B binds and inhibits calpain. Enlarged view of the interaction between calpastatin (yellow) and the catalytic cleft in the protease core domain PC1 (pink)–PC2 (red) of m-calpain (the 3D structure is from 3DF0). Calpastatin binds in the substrate orientation indicated by positions P3 to P3′. At P1, calpastatin distorts from the substrate path and projects residues 174–178 or 613–617, which form a kink between the P2 and P1′ anchor sites.
2.4. Genetic studies of conventional calpains.
The first calpain studies using genetically modified mice were reported in 2000, when Arthur et al. and Zimmerman et al. independently demonstrated that disrupting the mouse gene (Capns1) for the conventional calpain regulatory subunit CAPNS1/30K causes embryonic lethality before E11.5.128,129) These reports showed that conventional calpains were necessary for mammalian life, which greatly encouraged researchers struggling to understand this enigmatic enzyme. The disruption of CAPNS1/30K causes the down-regulation of both the CAPN1/μCL and CAPN2/mCL proteins, indicating that CAPNS1/30K is required for the stable presence of both calpain catalytic subunits in vivo, and probably functions as an intramolecular chaperone. In vitro, CAPN2/mCL alone (without CAPNS1/30K) shows full proteolytic activity after being denatured and then renatured by a long incubation with PEG or GroE.130) In cells, unfolded calpain large subunits are probably degraded by other proteases before they form active conformations.
Disrupting mouse Capn1 or Capn2 leads to contrasting results: Capn2−/− mice die before the blastocyst stage, whereas Capn1−/− mice appear normal and are fertile.55,131) This suggests that μ- and m-calpain differ in their function and/or expression level, at least at specific developmental stage(s). Capns1−/− embryonic stem (ES) cell growth and adhesion were not noticeably different from those of wild-type ES cells, while Capn2−/− ES cells could not be obtained even after extensive screening.55)
Cells from Capns1−/− mice served as ideal tools to unequivocally demonstrate the role of calpain in specific cellular events: Dourdin et al. showed that calpain is required for cell migration;132) Mellgren et al. showed that calpain is required for the rapid, Ca2+-dependent repair of wounded plasma membrane;133) and Demarchi et al. showed that calpain is required for macroautophagy.134)
2.5. Expression of conventional calpains.
Although CAPN1 and CAPN2 shows ubiquitous distribution, their gene expression patterns are slightly different in that CAPN1 is rather constantly expressed whereas expression of CAPN2 varies depending on tissues.135,136) The promoter region of CAPN2 lacks typical promoter elements such as TATA and CAAT boxes and has ca. 70% GC content in the −300 to −20 region with several potential GC boxes.136) The upstream region contains at least four tandemly reiterated regulatory sequences, each of which negatively regulates CAPN2 expression.136) These features are common to several house-keeping genes such as adenosine deaminase137) and hypoxanthine phosphoribosyltransferase,138) and support that CAPN2 gene is a typical house-keeping gene that shows ubiquitous and relatively low-level expression. On the other hand, CAPN2 promoter region also has potential AP-1 binding site, which is responsible for phorbol ester-mediated induction of gene expression.139,140) Indeed, proerythroblastic K562 cell line cells were reported to show upregulation of CAPN2/mCL expression by phorbol 12-myristate 13-acetate.141)
Although CAPN1 gene has not been examined in literature, human genomic sequence showed that CAPN1 has features similar to those of CAPN2, i.e., no significant TATA and CAAT boxes with ca. 70% GC content in the −1000 to −1 region and with potential AP-1 binding site at the −80 position. Analysis of CAPNS1 gene again revealed similar features:142) the promoter region has G/C-rich sequences with several G-C boxes but without TATA or CAT box. Thus, three genes encoding subunits of the conventional calpains are all regarded to be regulated in the same manner as other house-keeping genes.
2.6. Structure and function of calpastatin.
Calpastatin is the only endogenous inhibitor protein for the conventional calpains. It is highly effective and specific, and does not inhibit any other enzymes. Calpastatin has an inhibitor unit that is repeated four times (Fig. 3B), and each unit inhibits one calpain molecule—although their inhibitory efficiencies vary.143–145) Calpastatin inhibits both μ- and m-calpains with similar efficiencies. Among other calpain homologs, CAPN8/nCL-2 and CAPN9/nCL-4, but not CAPN3/p94, are inhibited by calpastatin in vitro.53,146,147)
Disruption of Cast, the mouse gene for calpastatin, does not produce a significant phenotype under normal, unstressed conditions.148) This suggests that conventional calpains are not normally activated dynamically, and that calpastatin is dispensable as a safety for calpain regulation. In accordance with this idea, intra-hippocampal injection of kainic acid (KA), which causes apoptotic neuronal cell death by excitotoxicity, resulted in significantly more DNA fragmentation in Cast−/− mice than in wild-type mice.148) Moreover, the KA effect was reduced in transgenic (Tg) mice overexpressing calpastatin in neuronal cells, whereas Tg mice overexpressing the baculoviral caspase inhibitor p35 showed no change.149) These results indicated that kainic acid-induced apoptotic neuronal cell death is mediated by the conventional calpains, and that caspases are not involved in this process.
Spencer and colleagues developed Tg mice that overexpress calpastatin in muscles.150,151) These mice appear healthy, without observable body or muscle mass changes or gross physiological, morphological, or behavioral defects. These mice were crossed with mdx mice, which have a nonsense Dmd mutation that causes mild muscular dystrophic phenotypes due to a lack of dystrophin; the dystrophic phenotype was significantly ameliorated in the resulting calpastatin-overexpressing mdx mice.150) Even in wild-type mice, calpastatin overexpression slows muscle atrophy during muscle unloading.151)
Studies using Tg mice also impact on food science, e.g., the postmortem tenderization of muscles. Both calpastatin-overexpressing152) and Capn1−/− mice153) showed reduced postmortem proteolysis of muscle proteins. Thus, calpain system has drawn attention to one of the targets for meat quality control.
3. Classification of calpain superfamily members
3.1. Classical and non-classical calpains.
Calpains have a wide variety of domains in addition to their protease domain, such as C2L and PEF domains (Fig. 1). The calpain superfamily is divided into several subfamilies according to the structures of these additional domains. Since the human conventional calpain catalytic subunits (CAPN1/μCL and CAPN2/mCL) are the reference point for calpain structure, calpains having a similar domain structure are called “classical” calpains (Fig. 3) in contrast to “non-classical” ones described below.46,154) Classical calpains are defined as those having C2L and PEF domains in addition to the CysPc domain. Accordingly, non-classical calpains are those having not both of C2L and PEF domains. Among the 15 human calpains, nine are classical and six are non-classical (Fig. 3).
Human classical calpains include CAPN1/μCL, CAPN2/mCL, CAPN3/p94, CAPN8/nCL-2, CAPN9/nCL-4, and CAPN11–14, most of which are conserved among vertebrates; fishes have a duplicate set of most of them.87) In invertebrates, only a few classical calpains have been identified. The blood fluke S. mansoni has four (Fig. 5),155) the fruit fly D. melanogaster has three: CALPA/Dm-calpain, CALPB, and CALPC,34) and the African malaria mosquito A. gambiae has five (Fig. 7). No classical calpain homologs have been found in C. elegans (Fig. 6), trypanosomes,156) plants, fungi, or S. cerevisiae.
Non-classical calpain CysPc domains have aa identities with each other ranging from <30% to >75%, and they may have functional domains on the N- and/or C-terminal sides of CysPc domain (Figs. 1 and 3). These include transmembrane (TM), Zn-finger, and conserved domains with unknown functions such as SOH (SOL homology) domain. This cohort also includes several alternatively spliced products of classical calpain genes, such as Capn8 and CalpA (Fig. 7B). Thus, non-classical calpains probably function differently from classical calpains, and not all are Ca2+-dependent. These features, together with the organization of mammalian calpain genes,157) strongly suggest that calpain molecules were generated evolutionarily by combining an ancestral calpain-type cysteine protease gene with genes encoding other functions.
3.2. Subfamilies.
Non-classical calpains contain several subfamilies grouped according to their additional domain structures. The PalB subfamily is the most evolutionarily conserved, and is found in humans, yeasts, fungi, protists, all insects except Drosophila, and nematodes, but not plants (Fig. 12).63,64,158–161) PalB homologs commonly contain two C2L domains in tandem, each of which diverge greatly or moderately from that of conventional calpains, and have a conserved microtubule-interacting and transport (MIT) motif(s) at the N-terminus (Fig. 3).
Figure 12.
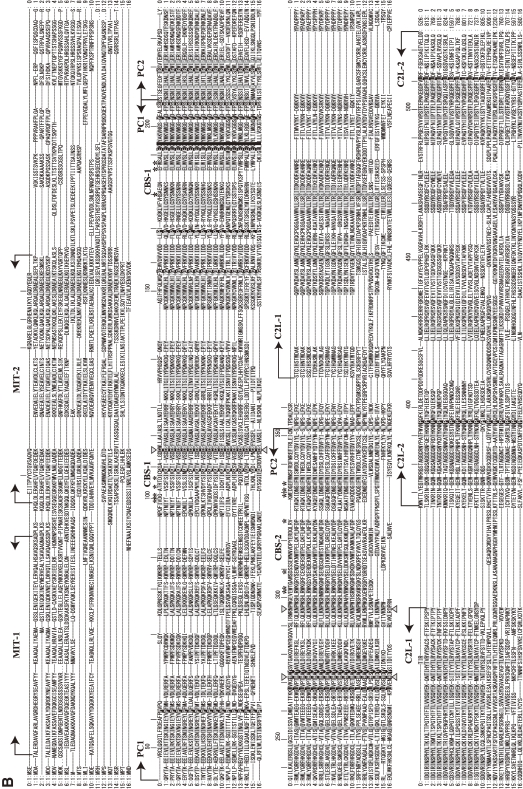
Phylogenetic tree and sequence alignment of PalB calpains. A. Phylogenetic tree of strict PalB calpains and human CAPN1/μCL. See Fig. 3A for methods. B. Sequence alignment of PalBs and human CAPN1/μCL (the start and end aa residue numbers are indicated). Triangles: active site residues. Asterisks: residues involved in Ca2+-binding of the protease domain of CAPN1/μCL. Arrows: domain boundaries. Deletion (−) and multiple (+) residue positions are indicated. Residue numbers of human CAPN1/μCL are given above the sequences. Residues conserved in all the sequences, all except human CAPN1/μCL, and in more than half the sequences are indicated by black reversed fonts, gray reversed fonts, and gray shadow, respectively. Sequences: 0: H. sapiens (human) CAPN1/μCL (NP_005177); 1: H. sapiens CAPN7/PalBH (NP_055111); 2: X. tropicalis (frog) (NP_998853); 3: D. rerio (zebrafish) (NP_001128580); 4: Ciona intestinalis (sea squirt) (XP_002121253); 5: Tribolium castaneum (red flour beetle) (XP_967682); 6: Apis mellifera (honey bee) (XP_001121978); 7: Acyrthosiphon pisum (pea aphid) (XP_001945029); 8: A. gambiae str. PEST (malaria mosquito) (XP_309874); 9: S. mansoni (XP_002579452); 10: C. elegans (NP_497964); 11: T. brucei TREU927 (XP_828540); 12: L. major (CAJ06528); 13: Coprinopsis cinerea okayama7#130 (common ink cap) (EAU91907); 14: E. nidulans FGSC-A4 (XP_657860); 15: C. albicans SC5314 (EAL03290); 16: S. cerevisiae (Q03792).
Human CAPN10, the representative product of CAPN10 from the longest transcript,162) also has both conserved and diverged C2L domains in succession at the C-terminus. The CAPN10 homologs are only found in vertebrates, and have no MIT domain, but they can be included in the PalB subfamily (Fig. 3).46)
TRA-3 is found in nematodes and vertebrates, but not in insects or lower organisms (Fig. 13). Compared with classical calpains, TRA-3 homologs have a C2 domain at the C-terminus, instead of a PEF domain (Figs. 3 and 13). Therefore, TRA-3 homologs can also be included in the PalB subfamily, with its structural consensus of two tandem C2L/C2 domains at the C-terminus. TRA-3 homologs, like CAPN10 homologs, have no MIT domain.
Figure 13.
Phylogenetic tree and sequence alignment of TRA-3 calpains. A. Phylogenetic tree of TRA-3 calpains and human CAPN1/μCL. See Fig. 3A for methods. B. Sequence alignment of TRA-3 and human CAPN1/μCL (the start and end aa residue numbers are indicated). Sequences: 0: H. sapiens CAPN1/μCL NP005177; 1: H. sapiens CAPN5 NP004046; 2: M. domestica CAPN5 XP001377962; 3: G. gallus CAPN5 XP417278; 4: X. tropicalis CAPN5 NP001006736; 5: D. rerio CAPN5 NP001073476; 6: D. rerio CAPN5-2 XP001345114; 7: H. sapiens CAPN6 NP055104; 8: M. domestica CAPN6 XP001366895; 9: G. gallus CAPN6 XP420313; 10: X. tropicalis CAPN6 XP002938834; 11: X. tropicalis TRA-3 XP002941290; 12: D. rerio TRA-3 XP001339053; 13: Strongylocentrotus purpuratus CAPN5 XP001178522; 14: S. mansoni XP002578116; 15: C. elegans TRA-3/CLP-5 NP502751. For other explanation, see Fig. 12B.
The evolutionarily conserved SOL subfamily occurs in almost all animals, including humans, insects, and nematodes, and also in green algae. SOL homolog structures are characterized by having various numbers of Zn-finger motifs in the N-terminal domain. They also share an SOH domain (Figs. 1 and 3).
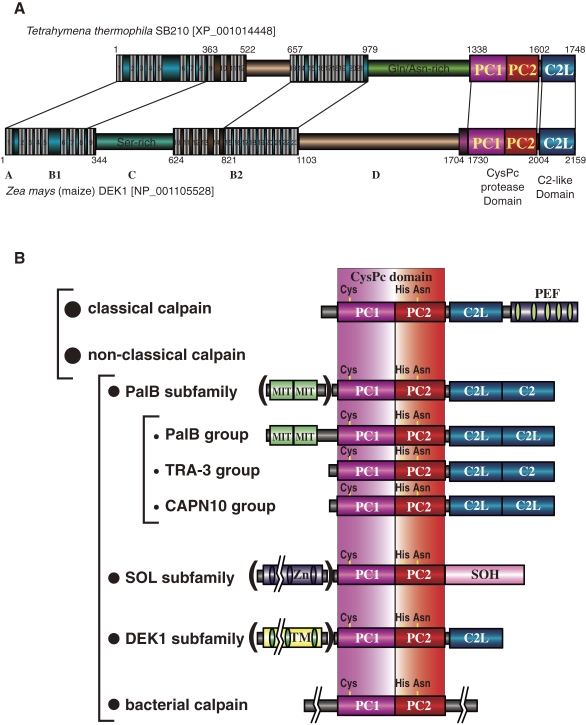
Plant calpains, called phytocalpains,163) were first described in 2002.62) The maize calpain homolog, DEK1 (defective kernel 1), is involved in aleurone cell development. DEK1 homologs are found in various other plants, including rice plants and Arabidopsis.164–166) They have one C2L domain at the C-terminus and a TM domain at the N-terminus. A calpain that is highly similar to DEK1 is also found in Tetrahymena thermophila (Fig. 14A). Some of the nematode calpains (CLP-3, -4, -6, and -7) have a domain structure (CysPc-C2L-COOH) similar to that of DEK1, but they do not have a TM domain (Fig. 6). These calpains are all grouped as the DEK1 subfamily.
Figure 14.
Schematic structures of T. thermophila and Z. mays DEK1s. A. The number of TM spanners is taken from prediction programs and may be corrected in the future. Domain names for Z. mays DEK1 are taken from ref. 62). B. Structural classification and consensus domain structures of calpain super family members. See Fig. 1 legend for symbols.
The first member of the calpain bacterial subfamily was reported in 1992 from Porphyromonas gingivalis; it is called tpr (thiol protease).167) Subsequent bacterial genome projects revealed several bacterial calpain homologs. These are the most divergent calpain species, sharing similarity only in the protease domain (Fig. 8). Although E. coli was once reported to have a calpain-like protease,168) its sequence rather belongs instead to the narrowly defined papain superfamily (clan CA-C1). In summary, all the calpain subfamily consensus structures are shown in Fig. 14B.
3.3. Ubiquitous and tissue-specific calpains.
In addition to their structural features, mammalian calpains are also independently categorized according to their tissue and organ distribution.169) CAPN1/μCL, CAPN2/mCL, CAPN5/hTRA-3, CAPN7/PalBH, CAPN10, CAPN13, and CAPN15/SOLH are ubiquitously expressed, whereas CAPN3/p94, CAPN8/nCL-2, CAPN9/nCL-4, CAPN6, CAPN11, and CAPN12 are expressed in specific tissues or organs (Figs. 3 and 4). Widely accepted assumption is that ubiquitous calpains play fundamental roles in all cells, whereas tissue-specific calpains have tissue-specific roles. In fact, defects in ubiquitous calpains may be lethal, as in Capn2−/− mice,55) whereas defects in tissue-specific calpains may cause tissue-specific diseases, like the muscular dystrophy caused by defective CAPN3.56)
At the same time, under various disease/damaged conditions such as muscular dystrophies, cardiomyopathies, traumatic ischemia, and lissencephaly, conventional calpains tend to be overactivated—probably because the intracellular Ca2+ homeostasis is compromised by the disease—and that overactivity often exacerbates the disease. Conventional calpain inhibitors are currently used to prevent the progression of such diseases, a therapeutic technique first applied by Suzuki and colleagues.58,79,170,171)
3.4. Expression of tissue-specific calpains.
Tissue-specificity of tissue-specific calpains should be regulated by their promoter regions. Human, mouse and rat CAPN3/Capn3 upstream regions have highly conserved structures containing TATA-box and several E-boxes, which probably are responsible for muscle-specific expression of CAPN3/Capn3.172) Intriguingly, CAPN3/Capn3 has an alternative upstream promoter, which drives ubiquitous expression of Up86 etc. starting from an alternative initial exon with slight abundance in the brain and blood cells.173–175) Moreover, rodent Capn3 has additional alternative promoter and initial exon between exons 1 and 2, and these are responsible for lens-specific expression of alternative splicing products of Capn3 such as Lp82 and Rt88.176,177) However, human CAPN3 lost lens-specific products by in-frame termination in exon 1′.178) Capn3 expression is also noted in testis.173)
The promoter region of Capn8 contains two TATA boxes with several GATA boxes, which are probably responsible for stomach expression of Capn8.157) Some cAMP responsive elements is also found in this region. Notably, CAPN8 and CAPN2 are close to each other on human chromosome 1q41 (mouse: chromosome 1), and their transcripts have overlapping complementary sequences, suggesting possible interaction with each other.157) In addition, Capn8 has additional exon in the middle, which generates an alternative splicing product, CAPN8b/nCL-2′. Its physiological relevance, however, is elusive.157,179) As for other tissue-specific calpain genes, their upstream sequences have not been analyzed in the detail.
4. Unconventional calpain functions in health and disease
4.1. Skeletal muscle-specific calpain.
The first tissue-specific calpain, CAPN3/p94, was found in 1989.180) It is expressed predominantly in skeletal muscle. This classical calpain is approximately 50% identical to CAPN1/μCL and CAPN2/mCL (Fig. 3). However, it contains three additional characteristic regions NS, IS1, and IS2, located at the N-terminus, in PC2 domain, and between C2L and PEF domains, respectively. These regions give CAPN3/p94 some unique features.
The most characteristic property of CAPN3/p94 is its extremely rapid autodegradation—its half-life in vitro is less than 10 minutes—starting with autolysis in the NS and IS1 regions.181–183) This autodegradative activity depends on both IS1 and IS2, and conventional calpain inhibitors such as calpastatin and E64 have little effect on it.181) Surprisingly, this autodegradation occurs Na+-dependently in the absence of Ca2+, establishing CAPN3/p94 as the first example of an intracellular Na+-dependent enzyme.54) The physiological relevance of CAPN3/p94’s Na+-dependency is still unclear, but its substrate specificities differ when it is activated by Ca2+ versus Na+. For example, LIM domain-binding protein 3, tropomyosin α-1 chain, and α-actinin-3 are candidate in vivo substrates for Na+-activated CAPN3/p94, whereas troponins T and I and Ras-specific guanine nucleotide-releasing factor 2 are for Ca2+ activation.54) Furthermore, CAPN3/p94 possesses a nuclear localization-signal-like sequence in the IS2 region, and is sometimes found in the nucleus as well as the cytosol.181) CAPN3/p94 binds specifically to the N2A and M-line regions of the gigantic filamentous muscle protein connectin/titin (Mr > 3,000 kDa), and the binding site for N2A connectin/titin is the N-terminal vicinity of the IS2 region.184) Its autodegradative activity is almost completely suppressed in vivo, most probably by binding to N2A connectin/titin. This region also aligns important subcellular structures of muscle, such as the sarcoplasmic reticulum and T-tubules, and therefore represent the potential space for CAPN3/p94 to function.183)
In 1995, CAPN3 mutations were shown to be responsible for limb-girdle muscular dystrophy type 2A (LGMD2A), also called calpainopathy, by an analysis of families of La Réunion Island and by positional cloning.56,185) Capn3−/− mice recapitulate a human calpainopathy-like phenotype,186,187) although milder, indicating that calpainopathy is indeed caused by CAPN3 defects.186) Calpainopathy appears to be primarily caused by compromised CAPN3/p94 protease activity, rather than by damaged structural properties.188) This was confirmed by CAPN3/p94 knock-in (Capn3CS/CS) mice, which express a structurally intact but inactive CAPN3/p94:C129S mutant, and also present a muscular dystrophy phenotype.189) Intriguingly, however, Capn3CS/CS mice have a less severe phenotype than Capn3−/− mice, indicating that the proteolytically inactive CAPN3/p94 retains some function, probably involving its structural properties.189)
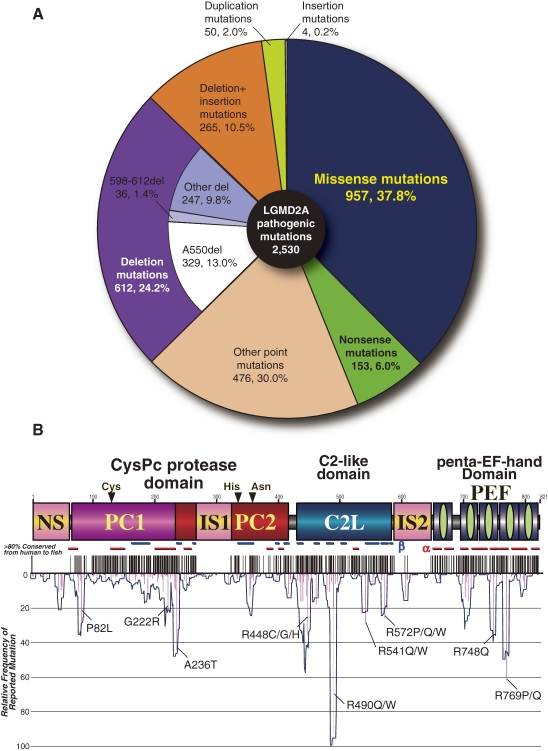
Many subsequent family analyses have been reported, identifying substantial numbers of both pathogenic and polymorphic mutations in CAPN3.190,191) However, no mutational hot spot has been found, so a genetic diagnosis of calpainopathy is not easy. A total of 2,530 mutations, including 456 unique ones, have been reported in the CAPN3 gene so far (available at the Leiden Muscular Dystrophy Pages http://www.dmd.nl/). Among the 80 gene entries with mutations responsible for muscular dystrophy, CAPN3 has the third most, coming only after dystrophin and lamin A/C. One characteristic of CAPN3 gene mutations is that more than half (62.7%) of them are nucleotide substitutions, of which missense mutations are dominant (60.3% among all substitutions, and 37.8% among all pathogenic mutations) (Fig. 15A).
Figure 15.
Pathogenic CAPN3 mutations in calpainopathy patients. A. Summary of calpainopathy mutation types. B. Calpainopathy missense mutation positions and frequencies (relative to the maximum value). Black vertical lines indicate aa residues that are conserved more than 80% of vertebrates as shown in Fig. 16B. α and β indicate α-helix and β-strand secondary structures of the corresponding m-calpain 3D structure. See Fig. 1 legend for other symbols.
Although some missense mutations found in LGMD2A patients are reported relatively frequently, such as R490Q/W, R769Q, and A236T, most of them are distributed all over the protein, between the N-terminus and C-terminus, including 215 unique mutations at 176 independent loci (21.4%) among the 821 aa residues (Fig. 15B). A comparison of vertebrate CAPN3/p94s (including the two gene products from fishes) revealed 241 residues that are conserved in all species (Fig. 16), including 83 of the above 176 missense loci, indicating that conserved aa residues are important for CAPN3/p94 functions. Conservation in the CAPN3/p94-characterizing regions NS, IS1, and IS2 is relatively low; however, it should be noted that some missense mutations, such as R49C/H, P319L, and S606L, which are in the NS, IS1, and IS2 regions respectively, occur relatively frequently. These observations suggest that sequence conservation of the CAPN3/p94-characterizing region is not necessarily required, but its length and a few important aa, usually near the borders of these regions, must be conserved for proper function.
Figure 16.
Phylogenetic tree and sequence alignment of CAPN3 proteins. A. Phylogenetic tree of vertebrate CAPN3. See Fig. 3A for methods. Explicit CAPN3 homologs are found only in vertebrates. B. Sequence alignment of CAPN3s. Fishes have two CAPN3 homologs (type-1 and -2); the latter does not have explicit IS1 or IS2 regions. Sequences: 1: H. sapiens, NP_000061; 2: M. domestic, ENSMODP00000022330; 3: G. gallus, NP_001004405; 4: A. carolinensis, ENSACAP00000015362; 5: X. tropicalis, ENSXETP00000026925; 6: Takifugu rubripes (type-1), ENSTRUP00000046251; 7: Hippoglossus hippoglossus (type-1), ACY78226; 8: Salmo salar (type-1), NP_001158880; 9: D. rerio (type-1), XP_001337065; 10: T. rubripes (type-2), ENSTRUP00000016935; 11: D. rerio (type-2), ENSDARP00000094244. For other explanation, see Fig. 12B.
LGMD2A and CAPN3 mutations are the first—and so far, only—example of a clear cause–effect relationship between human disease and calpain gene mutations.
4.2. Gastrointestinal tract-specific calpains.
CAPN8/nCL-2 and CAPN9/nCL-4 are predominantly expressed in surface mucus cells, called pit cells, in the stomach. Smaller amounts are expressed in the goblet cells in the intestines.179,192,193) Among mammalian calpains, CAPN8/nCL-2 is most similar to CAPN2/mCL (62% identical). Unlike m-calpain, however, E. coli-expressed recombinant CAPN8/nCL-2 exhibits Ca2+-dependent autolytic and caseinolytic activity without CAPNS1/30K, and it forms a homo-di∼oligomer via C2L domain in vitro.146) Xenopus laevis has a CAPN8/nCL-2 ortholog named xCL-2, which causes severe developmental defects when disrupted.194)
CAPN9/nCL-4 is another classical calpain homolog specific for gastrointestinal tracts. It is equally similar to all other classical calpains at the aa level, suggesting that CAPN9/nCL-4 is the molecule closest to the ancestral calpain species.193) CAPN9/nCL-4 is reported to be involved in tumorigenesis195) and in lumen formation in breast epithelial cells induced by an adhesion molecule.196) Recombinant human CAPN9/nCL-4 requires CAPNS1/30K for its activity in vitro, and such a complex has Ca2+-dependent activity that is inhibited by calpastatin and other Cys protease inhibitors, similar to conventional calpains.147)
In contrast, the in vivo characteristics of these calpains are rather different. Experiments using Capn8−/− and Capn9−/− mice showed that neither CAPN8/nCL-2 nor CAPN9/nCL-4 forms a stable complex with CAPNS1/30K; rather, they form a hybrid complex with each other, now designated as gastric calpain (G-calpain).57) As is the case for Capns1−/−, disruption of either Capn8 or Capn9 caused down-regulation of another molecule, suggesting that each species contributes to the stability and the functionality of the other. Both Capn8−/− and Capn9−/− mice seem healthy under normal conditions, but they are significantly more susceptible to ethanol-induced gastric ulcers.57) CAPN8/nCL-2 knock-in (Capn8CS/CS) mice that express the protease-inactive CAPN8/nCL-2:C105S mutant instead of wild-type CAPN8/nCL-2 also show stress-induced gastropathy. This indicates that CAPN8/nCL-2 and CAPN9/nCL-4, in the form of G-calpain, mediate key events for gastric mucosal defense.57)
A single nucleotide polymorphism (SNP) database search revealed that human CAPN8 and CAPN9 have several aa-substituting SNPs.57) An in vitro expression study indicated that some of these SNPs compromised proteolytic activity of G-calpain. Thus, evaluation of the link between these SNPs and susceptibility to gastrointestinal disorders is on list of things to do.
4.3. The PalB subfamily.
PalB was first identified in the ascomycete E. nidulans as a product of the gene responsible for the fungi’s adaptation to alkaline conditions.63) Its orthologs were then widely identified, from S. cerevisiae to humans (Fig. 12).64,158) The structural consensus for PalB subfamily members is (MIT)0–2-CysPc-C2L-C2L/C2 (Fig. 14B). Strict PalB homologs have (MIT)2-CysPc-C2L-C2L (the upstream C2L was once called PBH (PalB-homology) because of its highly divergent sequence48)), whereas CAPN10 and TRA-3 homologs have CysPc-C2L-C2L and CysPc-C2L-C2, respectively.
4.3.1. Strict PalB homologs in fungi.
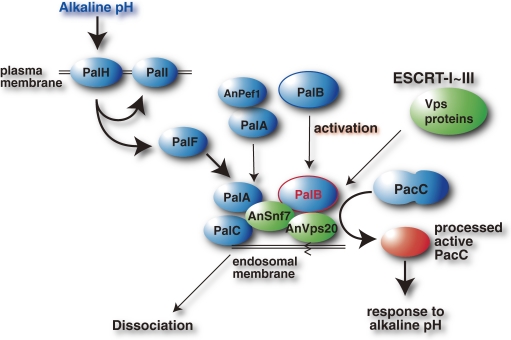
E. nidulans, like many other microorganisms, grows over a wide pH range. The gene locus palB was identified from mutants that mimic the gene expression profiles for acid environments even under normal/neutral conditions, corresponding to pal genes (pal stands for “phosphatase mutants: loss in activity at alkaline pH but normal or increased activity at acidic pH”). PalB is involved in the proteolytic activation of a key transcription factor named PacC, which governs the entire regulation of pH-dependent changes in gene expression, such as the expression of alkaline phosphatase under alkaline conditions. This pH adaptation system is here called the Pal-PacC pathway (Fig. 17).
Figure 17.
Pal-PacC pathway and corresponding molecules in yeast and human. Schematics of signaling pathways involving PalB of fungi. In fungi, a membrane protein (PalH) senses ambient pH and transduces the alkaline signal to PalI. An arrestin homolog, PalF, further transduces the signal to PalA, which forms a complex with AnPef1. At the endosomal membrane, PalA forms the PalB active complex to proteolyze PacC at the C-terminus, thus activating this key transcription factor. Activated PacC regulates gene expression under alkaline conditions. This pathway is highly conserved in yeast and possibly conserved in human; corresponding molecules: PalH (fungi)-Rim21 (yeast)-? (human), PalI-Rim9-?, PalF-Rim8-arrestin, PalA-Rim20-Alix/AIP1, PalC-Ygr122w-?, AnPef1-Pef1/Ygr058w-ALG-2, AnSnf7-Snf7/Vps32-CHMP4s, PalB-Rim13/Cpl1-CAPN7/PalBH, and PacC-Rim101-C2H2 Zn-finger proteins? See text for details.
Mutations in six genes, palA, palB, palC, palF, palH and palI, that mimic the effects of growth at acid pH, and one, pacC, that mimics those at alkaline pH, were identified.197–199) These genes were characterized from 1995, starting with the identification of the palB gene product as a novel and divergent calpain homolog.63,200–203) Primary structures of these pal gene products showed that PalB was a calpain homolog, and PalH and PalI had TM domains. Protein interaction analyses showed that PalF, a distant homolog of mammalian arrestin, binds to the large cytosolic domain of PalH,204) and that PalF’s phosphorylation and ubiquitylation under alkaline conditions depend on PalH and PalI. These findings indicate that PalF–PalH interactions mediate pH signaling, similar to the arrestin–receptor interactions that regulate various processes in mammals, such as photo-signaling.205) These findings strongly imply the existence of an corresponding mammalian signaling pathway(s) involving arrestin and CAPN7/PalBH.
PacC (674 aa) usually forms an inactive, closed conformation, which protects it from activation by intramolecular interactions, and undergoes limited proteolysis to become active.206,207) First, PacC is processed between aa 479–502 by a primary processing protease, PalB, in a pH-regulated manner. This changes PacC to an open form susceptible to proteolysis by the proteasome in a pH-independent constitutive manner, simultaneously creating the processed active form (252–254 aa).207,208) It is thought that ubiquitylation is not involved in this process, which is a rare mode of action for proteasome.209–212)
4.3.2. Strict PalB homologs in yeasts and mammals.
The above-described fungus Pal-PacC pathway is well conserved in budding yeasts. The S. cerevisiae PalB ortholog is Rim13/Cpl1. It was first identified in 1993 as one of the RIM genes, which reduce the expression of sporulation-specific genes when mutated.213,214) Among the RIM gene products, Rim101 corresponds to fungus PacC, and plays a central role in this signaling system, as does PacC. The proteolytic regulation of Rim101 was first discovered in 1997,215) and the responsible protease was identified as Rim13/Cpl1 in 1999.64) Subsequently, RIM8, 9, 20, and 21, which correspond to palF, I, A, and H, were identified.200,201)
Saccharomycetes’ usual environment is rather acidic, and its alkaline adaptations function in near-neutral pH conditions. Therefore, this adaptive system, here called the Rim pathway, has practical importance for human health. Pathogenic and saprophytic yeasts like Candida albicans and Yarrowia lipolytica also have conserved Rim pathways. For example, C. albicans uses this pathway when it infects and invades mammalian skin in neutral pH environments, and disrupting the Rim pathway compromises its pathogenicity.216,217) Y. lipolytica218,219) and Usitlago maydis220) are aslo reported to use an identical system.
The Pal-PacC and Rim pathways relate to membrane trafficking, and involve ESCRT (endosomal sorting complex required for transport) and Vps (vacuolar protein sorting) proteins.65,221–229) Based on a genomic study that showed Rim and Vps interactions, it is demonstrated that these two proteins had a functional relationship in S. cerevisiae and C. albicans.223,228,230) Screening of suppressor mutants for rim21 and rim9 revealed functional relationships between the Rim and Vps proteins.65) These studies clarified components of the Rim signaling cascade and the hierarchy: RIM8, 9, and 21 act upstream of RIM13/CPL1 and RIM20, and Rim13/Cpl1 proteolyzes Rim101 at the endosomal membranes with the aid of the ESCRT-III subcomplex as a scaffold (Fig. 17).
The Pal-PacC and Rim pathways are the first examples that genetics thoroughly uncovered the molecular components and mechanisms of calpain-mediated systems. These pathways also exemplify that calpains function as modulator proteases, affecting their substrates’ functions by limited proteolysis. Theoretical extension of the knowledge of these pathways is anticipated to provide important keys for understanding other calpain systems, including those that involve CAPN7/PalBH.
There are few published reports on CAPN7/PalBH, a mammalian ortholog of PalB.158,231–233) Many studies have examined other homologous components; for instance, arrestin and Alix/AIP1, which correspond to PalA and F, respectively. It should be noted that interactions among Rim20, Fef1/Ygr058w, and Snf7/Vps32 are conserved in mammals as Alix1/AIP1, ALG-2, and CHMP4, respectively.234–237) Although multiple interactions of Alix/AIP1 have been reported,236,238) the physiological roles of these proteins and CAPN7/PalBH have not been clarified. Another message from these studies is that it is essential to take ubiquitous unconventional calpains such as CAPN7/PalBH into account along with the conventional calpains when analyzing various biological phenomena, but very few reports achieve this.239)
4.3.3. TRA-3 homologs in the PalB subfamily.
TRA-3/CLP-5 is classified into the PalB subfamily in the broad sense, but TRA-3/CLP-5 has a distal C2 instead of C2L domain, and no N-terminal MIT domain (Fig. 14B). The distal C2 domain was once called a T domain (T from TRA-3), because its primary sequence rather diverges from that of other C2 domains. TRA-3/CLP-5 was first identified by genetics as the product of the gene, tra-3 (transformer: XX animals transformed into males), which is involved in the sex determination cascade of C. elegans.61) Its action as an enzyme has not been characterized; however, its protease activity is necessary for female development in XX hermaphrodites, in the processing of the TRA-2A membrane protein.240) In contrast, TRA-3/CLP-5 and another C. elegans calpain, CLP-1, are components of a neuronal necrotic death cascade, upstream of the aspartic proteases ASP-3 and ASP-4, which may correspond to mammalian cathepsins D and E.241) An SNP in tra-3 is also reported to be involved in nematode body-size determination.242)
Mammals have two TRA-3 orthologs, CAPN5/hTRA-3 and CAPN6; both share more than 30% aa identity with TRA-3 (Fig. 13).243,244) CAPN5/hTRA-3 has Ca2+-dependent autolytic activity and is sensitive to several calpain inhibitors.245) CAPN5/hTRA-3 is expressed in almost all tissues in varying amounts. Analysis of Capn5−/− mice showed that CAPN5/hTRA-3 is expressed in a subset of T cells, but is dispensable for development.246) SNPs of human CAPN5 were reported to be associated with polycystic ovary syndrome, diastolic blood pressure, and cholesterol levels,247,248) and CAPN5/hTRA-3 and CAPN15/SOLH are upregulated in chronic intermittent, but not constant, heart hypoxia.249)
On the other hand, CAPN6 in eutherians (placental mammals) naturally has aa substitutions in the most important residue in the active site triad (C→K in humans, see Fig. 13B), strongly suggesting that eutherian CAPN6 has no proteolytic activity. Interestingly, CAPN6 in marsupialia (Monodelphis domestica; gray, short-tailed opossum) and in chicken (Gallus gallus) have the active-site residues Cys-His-Asn; moreover, frog (X. tropicalis) and fish (Danio rerio) have three TRA-3 homologs, which also have the active-site residues. This is in contrast to CAPN11, which is a classical calpain expressed specifically in the testis in eutherians, whereas the CAPN11 gene is ubiquitously expressed in chicken but was lost in marsupialia. Mouse CAPN6 is predominantly expressed in embryonic muscles, placenta, and in several cultured cells. It is intriguing to consider the physiological function of eutherian CAPN6 and CAPN11, which are expressed in the placenta and testis, respectively. CAPN6 was shown to be involved in regulating microtubule dynamics,250) and was subsequently shown to regulate cell motility in cultured cells,251) although the in vivo physiological functions of CAPN6 and CAPN11 are still unclear.
4.3.4. CAPN10 homologs in the PalB subfamily.
CAPN10 also belongs to the PalB subfamily in the broad sense, but CAPN10 differs from PalB in that it has no N-terminal MIT domain (Fig. 14B). CAPN10 was identified as a risk factor for non-insulin-dependent diabetes mellitus (NIDDM, type 2 diabetes) in a large-scale genetic association study.162) An association was found between G→A SNP (UCSNP-43, the G allele is the at-risk genotype) in intron 3 of CAPN10 and susceptibility to NIDDM in Mexican Americans and in a Northern European population from the Botnia region of Finland. There is no molecular explanation why this SNP causes susceptibility to NIDDM, although G/G homozygotes in UCSNP-43 were reported to have reduced CAPN10 mRNA expression in skeletal muscle.252,253)
The Capn10 gene is also a candidate gene responsible for a phenotype of the Otsuka Long-Evans Tokushima Fatty rat, a NIDDM animal model.254) Quantitative trait locus (QTL) analyses using Capn10−/− mice and two strains, LG/J and SM/J, with low and high obesity phenotypes showed that Capn10 is included in the obesity QTL, Adip1, indicating involvement of CAPN10 in obesity also in mice.255)
Studies using Capn10−/− or CAPN10-overexpressing Tg mice suggest that CAPN10 is involved in type 2 ryanodine receptor-mediated apoptosis.256) Although the phenotypes of these mice have not been described, they do not have embryonic lethality. Ubiquitous distribution and dynamic changes in CAPN10’s cellular localization was shown by a CAPN10-specific antibody.257) CAPN10 was also demonstrated to be involved in GLUT4 vesicle translocation during insulin-stimulated glucose uptake in adipocytes.258) Arrington et al. showed that CAPN10, localized to mitochondria, mediates mitochondrial dysfunction by cleaving Complex I subunits and activating the mitochondrial permeability transition.259)
4.4. The SOL subfamily.
The first genetic calpain study examined the Drosophila gene small optic lobes (sol).60) A mutation in sol, such as sol1078, results in the absence of certain classes of columnar neurons in the optic lobes, leading to specific alterations in flight and walking maneuvers. The longest transcripts from this gene encodes a 1,597-aa protein belonging to the calpain SOL subfamily. Drosophila SOL is composed of the N-terminal six-Zn-finger motif-containing domain and the C-terminal calpain-like protease domain (Fig. 7B). Most of the protease domain is deleted in the sol1078 mutation, demonstrating that SOL as a protease is essential for forming proper optic lobes. Unfortunately, there have been no further reports dissecting the molecular mechanisms of neurodegeneration involving SOL.34) Mammals have one SOL ortholog, CAPN15/SOLH,260,261) but its physiological role is not clear. In parallel with PalB subfamily, this evolutionarily interesting molecules including their mammalian homologs beg further study.
4.5. Phytocalpain.
A plant calpain, now called phytocalpain, was first identified in the sugarcane EST database by Correa et al., and was named SC cal1.262) A sequence similarity search using SC cal1 as bait subsequently identified other phytocalpains: from the dicotyledons, Vitis vinifera (pinot noir grape), A. thaliana, and Solanum melongena (eggplant); from the monocotyledons, Zea mays (maize), Oryza sativa (rice plant), and Hordeum vulgare (barley); and from gymnospermae, Pinus taeda (loblolly pine).163) A genetic study identified phytocalpain as the defective kernel 1 (dek1) gene product, DEK1, required for aleurone cell development in the maize endosperm.62) Since then, DEK1 homologs in A. thaliana and Nicotiana benthamiana (tobacco) have been found to have similar functions.166,263)
A biochemical study using E. coli-expressed recombinant maize DEK1 CysPc-C2L domains showed that its Ca2+-activated caseinolytic activity depends on the predicted active site Cys residue, although this activity was much lower than that of the conventional calpains under the study conditions used.164) DEK1 is the only calpain homolog in the Arabidopsis genome,263) and it is important for regulating growth in this plant. Although the full-length DEK1 protein localizes to membranes, intramolecular autolytic cleavage releases the calpain domain into the cytoplasm, and this domain is sufficient to fully complement dek1 mutants.263,264)
As shown in Fig. 14A, from the N-terminus of maize DEK1, a possible signal peptide is followed by the first TM region (A), eight-TM region (B1), large loop region (C), third TM domain with 14–15 spanners (B2), a domain of unknown function with no homology to other proteins (D), the calpain protease (CysPc) domain, and C2L domain.62,164) The number of TM spanners of B2 region is predicted to be 15 or 14, respectively, by SOSUI or TMHMM Ver.2.0,265,266) which makes the topology of C region and calpain domains on the same or opposite sides of cell membrane. This topology is very important for DEK1 function, and it should be examined biochemically.
Among the 15 human calpains, the protease domain of non-classical CAPN15/SOLH is the most similar to the DEK1 CysPc domain, with which it shows almost 40% aa sequence identity. Surprisingly, DEK1’s C-terminal C2L domain is rather similar to that of classical mammalian calpains, such as CAPN1/μCL, CAPN3/p94, and CAPN8/nCL-2 (25–30% identity). Therefore, DEK1 is considered to consist of three evolutionarily different modules: a TM domain, a CysPc domain, and a C2L domain. In addition, it has been suggested that DEK1 homolog of the protista T. thermophila (Fig. 14A) resulted from a lateral gene-transfer event from a green alga-type endosymbiont of ciliates as an explanation for this inter-kingdom similarity.46)
4.6. Other calpain members.
Leishmania and Trypanosoma have around 20 calpain homologs each, which are likely to contribute to cell morphogenesis, drug resistance, and stress-response mechanisms.267–273) Some trypanosome calpains have N-terminal domains with weak similarity to calpastatin. As described above, some of the calpain homologs have substitutions in one or more of the well-conserved active-site triad residues, Cys-His-Asn. This non-proteolytic family of calpain homologs includes eutherian CAPN6 (Fig. 3), some of the schistosome (Fig. 5) and nematode (Fig. 6) calpains, insect CALPC (Fig. 7B) and all of the Trypanosoma homologs.34,241,267) The generation of these calpain species is interesting from an evolutionary viewpoint, and elucidating their physiological functions will reveal additional roles played by the calpain superfamily, e.g., possible non-proteolytic functions.
Concluding remarks
Gordon Guroff, Darrel E. Goll, Kazutomo Imahori, Koichi Suzuki, and Takashi Murachi established numerous milestones in the study of calpains, including its discovery,1) finding its relationship to skeletal muscle,5) purifying it to homogeneity,7) labeling calpain and calpastatin,12) cloning calpain13) and calpastatin cDNA,143) determining the 3D structure,109) and many more. Their highly comprehensive and educational reviews17,48,49,274,275) have been cited many times. These heroes will always live in the memory of many calpain researchers. They made great strides in the study of calpains, but much work remains to elucidate calpain’s physiological roles. The divergence of its physiological functions and the lack of a good activation marker are major obstacles. Recent progress in genetics and structural biology has led to new and invaluable information about calpains. Considering calpain’s divergent physiological functions, breakthroughs in the field of calpain studies will affect an enormously wide range of life science fields.
Acknowledgments
We dedicate this paper to the memory of Prof. Koichi Suzuki. We thank Dr. Tamio Yamakawa, m.j.a., for giving us the opportunity to write this review, all the laboratory members for their invaluable support, and Drs. Leslie Miglietta and Grace Gray for their excellent English editing. We also thank Drs. Peter L. Davies, Robert L. Campbell, and all calpain researchers who gave opinions at “Calpain Nomenclature Proposal and Comments” web site for valuable discussion about calpain structures and nomenclature. This work was supported in part by MEXT.KAKENHI 18076007 (H.S.), 20780106 (S.H.), 22770139 (Y.O.), JSPS.KAKENHI 20370055, and a Takeda Science Foundation research grant (H.S.).
Abbreviations
- aa
amino acid(s)
- aar
amino acid residue(s)
- C2 domain
PKC conserved domain 2
- C2L domain
C2 domain-like domain
- CANP
calcium-activated neutral protease
- CAPN
calpain gene
- CAPNS
calpain regulatory small subunit gene
- CASF
Ca2+-activated sarcoplasmic factor
- CBS-1 and -2
Ca2+-binding site in PC1 and PC2, respectively
- CL
calpain catalytic large subunit
- CysPc
calpain-like cysteine protease sequence motif defined in the conserved domain database of the National Center for Biotechnology Information (cd00044)
- GR domain
Gly-rich domain
- KAF
kinase activating factor
- MIT motif
microtubule interacting and transport motif
- NIDDM
noninsulin-dependent diabetes mellitus (type 2 diabetes)
- PC1 and PC2 domains
calpain protease core domains 1 and 2, respectively
- PEF
penta-EF-hand
- PEF(L) and PEF(S) domains
PEF domains in calpain catalytic large subunit and regulatory small subunit, respectively
- PKC
protein kinase C
- QTL
quantitative trait locus
Biographies
Profile
Hiroyuki Sorimachi was born in 1963 and started working on calpains in 1988 at The Tokyo Metropolitan Institute of Medical Science (Rinshoken) under the supervision of late Prof. Koichi Suzuki, after receiving his B.Sc. in Biochemistry from The University of Tokyo. In 1992, he moved to The University of Tokyo, and obtained his Ph.D. at The University of Tokyo. He continued to study calpains as a Prof. Suzuki’s assistant professor, and in 1997 moved to the Laboratory of Prof. Keiko Abe at The University of Tokyo as an associate professor. He was awarded Young Investigator Award from Japan Biochemistry Society in 1999. In 2004, he returned to Rinshoken as a project leader of Calpain Project. His research interests include biochemistry and genetics of all kinds of calpains.
Shoji Hata was born in 1973 and started his research career as a post-doctoral researcher of Japan Society for the Promotion of Science (JSPS) in 2001 at The University of Tokyo, after receiving his Ph.D. from The University of Tokyo. In 2004, he moved to Rinshoken joining to Calpain Project. In 2010, he was promoted to a chief researcher at Rinshoken. He was awarded Outstanding Presentation Award and Young Investigator’s Award at Rinshoken Research Workshop (RRW) in 2009 and at the Japanese Society of Proteases in Pathophysiology (JSPP) in 2010. His research interests include biochemistry and genetics of tissue-specific calpains such as stomach-specific, testis-specific, and hair follicle-specific calpains.
Yasuko Ono was born in 1971 and started her research career as a post-doctoral researcher of JSPS in 1999 at The University of Tokyo, after receiving her Ph.D. from The University of Tokyo. From 2000, she studied on muscle regulatory proteins for three years at University of Arizona, USA as a post-doctoral research fellow abroad of JSPS. In 2003, she became a CREST postdoctoral researcher of Japan Science and Technology Agency at The University of Tokyo. In 2004, she moved to Rinshoken joining to Calpain Project. In 2009, she was promoted to a chief researcher at Rinshoken. She was awarded Murachi Award at FASEB Summer Research Conference in 2001, Young Investigator’s Award of the JSPP in 2008, Outstanding Presentation Award at RRW in 2010, and The Young Scientists’ Prize of the Commendation for Science and Technology by the Minister of Education, Culture, Sports, Science and Technology in 2011. She has been trying to enjoy tackling to challenge questions casted by the calpains, especially those by numbers 3 and 7.
References
- 1).Guroff G. (1964) A neutral calcium-activated proteinase from the soluble fraction of rat brain. J. Biol. Chem. 239, 149–155 [PubMed] [Google Scholar]
- 2).Lazarovici P. (2000) Obituary: Dr. Gordon Guroff (1933–1999). J. Mol. Neurosci. 14, 1–2 [DOI] [PubMed] [Google Scholar]
- 3).Huston R.B., Krebs E.G. (1968) Activation of skeletal muscle phosphorylase kinase by Ca2+. II. Identification of the kinase activating factor as a proteolytic enzyme. Biochemistry 7, 2116–2122 [DOI] [PubMed] [Google Scholar]
- 4).Meyer W.L., Fischer E.H., Krebs E.G. (1964) Activation of skeletal muscle phosphorylase kinase by Ca2+. Biochemistry 3, 1033–1039 [DOI] [PubMed] [Google Scholar]
- 5).Busch W.A., Stromer M.H., Goll D.E., Suzuki A. (1972) Ca2+-specific removal of Z lines from rabbit skeletal muscle. J. Cell Biol. 52, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Takai Y., Yamamoto M., Inoue M., Kishimoto A., Nishizuka Y. (1977) A proenzyme of cyclic nucleotide-independent protein kinase and its activation by calcium-dependent neutral protease from rat liver. Biochem. Biophys. Res. Commun. 77, 542–550 [DOI] [PubMed] [Google Scholar]
- 7).Ishiura S., Murofushi H., Suzuki K., Imahori K. (1978) Studies of a calcium-activated neutral protease from chicken skeletal muscle. I. Purification and characterization. J. Biochem. 84, 225–230 [DOI] [PubMed] [Google Scholar]
- 8).Suzuki K., Tsuji S., Ishiura S., Kimura Y., Kubota S., Imahori K. (1981) Autolysis of calcium-activated neutral protease of chicken skeletal muscle. J. Biochem. 90, 1787–1793 [DOI] [PubMed] [Google Scholar]
- 9).Suzuki K. (1983) Reaction of calcium-activated neutral protease (CANP) with an epoxysuccinyl derivative (E64c) and iodoacetic acid. J. Biochem. 93, 1305–1312 [DOI] [PubMed] [Google Scholar]
- 10).Suzuki K., Ishiura S. (1983) Effect of metal ions on the structure and activity of calcium-activated neutral protease (CANP). J. Biochem. 93, 1463–1471 [DOI] [PubMed] [Google Scholar]
- 11).Nishiura I., Tanaka K., Yamato S., Murachi T. (1978) The occurrence of an inhibitor of Ca2+-dependent neutral protease in rat liver. J. Biochem. 84, 1657–1659 [DOI] [PubMed] [Google Scholar]
- 12).Murachi T., Tanaka K., Hatanaka M., Murakami T. (1981) Intracellular Ca2+-dependent protease (calpain) and its high-molecular-weight endogenous inhibitor (calpastatin). Adv. Enzyme Regul. 19, 407–424 [DOI] [PubMed] [Google Scholar]
- 13).Ohno S., Emori Y., Imajoh S., Kawasaki H., Kisaragi M., Suzuki K. (1984) Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature 312, 566–570 [DOI] [PubMed] [Google Scholar]
- 14).Suzuki K. (1991) Nomenclature of calcium dependent proteinase. Biomed. Biochim. Acta 50, 483–484 [PubMed] [Google Scholar]
- 15).Ohno S., Minoshima S., Kudoh J., Fukuyama R., Shimizu Y., Ohmi-Imajoh S., Shimizu N., Suzuki K. (1990) Four genes for the calpain family locate on four distinct human chromosomes. Cytogenet. Cell Genet. 53, 225–229 [DOI] [PubMed] [Google Scholar]
- 16).Croall D.E., DeMartino G.N. (1991) Calcium-activated neutral protease (calpain) system: structure, function, and regulation. Physiol. Rev. 71, 813–847 [DOI] [PubMed] [Google Scholar]
- 17).Goll D.E., Thompson V.F., Li H., Wei W., Cong J. (2003) The calpain system. Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 18).Zatz M., Starling A. (2005) Mechanisms of disease: Calpains and disease. N. Engl. J. Med. 352, 2413–2423 [DOI] [PubMed] [Google Scholar]
- 19).Yamashima T. (2004) Ca2+-dependent proteases in ischemic neuronal death: a conserved ‘calpain-cathepsin cascade’ from nematodes to primates. Cell Calcium 36, 285–293 [DOI] [PubMed] [Google Scholar]
- 20).Wendt A., Thompson V.F., Goll D.E. (2004) Interaction of calpastatin with calpain: a review. Biol. Chem. 385, 465–472 [DOI] [PubMed] [Google Scholar]
- 21).Wells A., Huttenlocher A., Lauffenburger D.A. (2005) Calpain proteases in cell adhesion and motility. Int. Rev. Cytol. 245, 1–16 [DOI] [PubMed] [Google Scholar]
- 22).Turner M.D., Cassell P.G., Hitman G.A. (2005) Calpain-10: from genome search to function. Diabetes Metab. Res. Rev. 21, 505–514 [DOI] [PubMed] [Google Scholar]
- 23).Tian W., Dewitt S., Laffafian I., Hallett M.B. (2004) Ca2+, calpain and 3-phosphorylated phosphatidyl inositides; decision-making signals in neutrophils as potential targets for therapeutics. J. Pharm. Pharmacol. 56, 565–571 [DOI] [PubMed] [Google Scholar]
- 24).Sorimachi H., Kawabata Y. (2003) Calpain and pathology in view of structure–function relationships Nippon Yakurigaku Zasshi 122, 21–29(in Japanese) [DOI] [PubMed] [Google Scholar]
- 25).Ray S.K., Hogan E.L., Banik N.L. (2003) Calpain in the pathophysiology of spinal cord injury: neuroprotection with calpain inhibitors. Brain Res. Rev. 42, 169–185 [DOI] [PubMed] [Google Scholar]
- 26).Ridderstrale M., Parikh H., Groop L. (2005) Calpain 10 and type 2 diabetes: are we getting closer to an explanation? Curr. Opin. Clin. Nutr. Metab. Care 8, 361–366 [DOI] [PubMed] [Google Scholar]
- 27).Ray S.K., Banik N.L. (2003) Calpain and its involvement in the pathophysiology of CNS injuries and diseases: therapeutic potential of calpain inhibitors for prevention of neurodegeneration. Curr. Drug Targets CNS Neurol. Disord. 2, 173–189 [DOI] [PubMed] [Google Scholar]
- 28).Nixon R.A. (2003) The calpains in aging and aging-related diseases. Ageing Res. Rev. 2, 407–418 [DOI] [PubMed] [Google Scholar]
- 29).Mehendale H.M., Limaye P.B. (2005) Calpain: a death protein that mediates progression of liver injury. Trends Pharmacol. Sci. 26, 232–236 [DOI] [PubMed] [Google Scholar]
- 30).Liu X., Van Vleet T., Schnellmann R.G. (2004) The role of calpain in oncotic cell death. Annu. Rev. Pharmacol. Toxicol. 44, 349–370 [DOI] [PubMed] [Google Scholar]
- 31).Laval S.H., Bushby K.M.D. (2004) Limb-girdle muscular dystrophies—From genetics to molecular pathology. Neuropathol. Appl. Neurobiol. 30, 91–105 [DOI] [PubMed] [Google Scholar]
- 32).Horikawa Y. (2005) Type 2 diabetes and genetic variations of calpain 10 gene Nippon Rinsho 63, 155–159(in Japanese) [PubMed] [Google Scholar]
- 33).Harwood S.M., Yaqoob M.M., Allen D.A. (2005) Caspase and calpain function in cell death: bridging the gap between apoptosis and necrosis. Ann. Clin. Biochem. 42, 415–431 [DOI] [PubMed] [Google Scholar]
- 34).Friedrich P., Tompa P., Farkas A. (2004) The calpain-system of Drosophila melanogaster: coming of age. Bioessays 26, 1088–1096 [DOI] [PubMed] [Google Scholar]
- 35).Franco S.J., Huttenlocher A. (2005) Regulating cell migration: calpains make the cut. J. Cell Sci. 118, 3829–3838 [DOI] [PubMed] [Google Scholar]
- 36).Costelli P., Reffo P., Penna F., Autelli R., Bonelli G., Baccino F.M. (2005) Ca2+-dependent proteolysis in muscle wasting. Int. J. Biochem. Cell Biol. 37, 2134–2146 [DOI] [PubMed] [Google Scholar]
- 37).Carragher N.O. (2006) Calpain inhibition: a therapeutic strategy targeting multiple disease states. Curr. Pharm. Des. 12, 615–638 [DOI] [PubMed] [Google Scholar]
- 38).Branca D. (2004) Calpain-related diseases. Biochem. Biophys. Res. Commun. 322, 1098–1104 [DOI] [PubMed] [Google Scholar]
- 39).Biswas S., Harris F., Dennison S., Singh J.P., Phoenix D. (2005) Calpains: enzymes of vision? Med. Sci. Monit. 11, RA301–RA310 [PubMed] [Google Scholar]
- 40).Biswas S., Harris F., Dennison S., Singh J., Phoenix D.A. (2004) Calpains: targets of cataract prevention? Trends Mol. Med. 10, 78–84 [DOI] [PubMed] [Google Scholar]
- 41).Bartoli M., Richard I. (2005) Calpains in muscle wasting. Int. J. Biochem. Cell Biol. 37, 2115–2133 [DOI] [PubMed] [Google Scholar]
- 42).Baud L., Fouqueray B., Bellocq A., Peltier J. (2003) Calpains participate in inflammatory reaction development. Med. Sci. (Paris) 19, 71–76(in French) [DOI] [PubMed] [Google Scholar]
- 43).Kuchay S.M., Chishti A.H. (2007) Calpain-mediated regulation of platelet signaling pathways. Curr. Opin. Hematol. 14, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Liu J., Liu M.C., Wang K.K. (2008) Calpain in the CNS: from synaptic function to neurotoxicity. Sci. Signal. 1, re1. [DOI] [PubMed] [Google Scholar]
- 45).Bertipaglia I., Carafoli E. (2007) Calpains and human disease. Subcell. Biochem. 45, 29–53 [DOI] [PubMed] [Google Scholar]
- 46).Croall D.E., Ersfeld K. (2007) The calpains: modular designs and functional diversity. Genome Biol. 8, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Suzuki K., Hata S., Kawabata Y., Sorimachi H. (2004) Structure, activation, and biology of calpain. Diabetes 53, S12–S18 [DOI] [PubMed] [Google Scholar]
- 48).Sorimachi H., Ishiura S., Suzuki K. (1997) Structure and physiological function of calpains. Biochem. J. 328, 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Murachi T. (1983) Calpain and calpastatin. Trends Biochem. Sci. 8, 167–169 [Google Scholar]
- 50).Tanaka K. (2009) The proteasome: overview of structure and functions. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 85, 12–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Mizushima N., Levine B. (2010) Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Tait S.W., Green D.R. (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 [DOI] [PubMed] [Google Scholar]
- 53).Ono Y., Kakinuma K., Torii F., Irie A., Nakagawa K., Labeit S., Abe K., Suzuki K., Sorimachi H. (2004) Possible regulation of the conventional calpain system by skeletal muscle-specific calpain, p94/calpain 3. J. Biol. Chem. 279, 2761–2771 [DOI] [PubMed] [Google Scholar]
- 54).Ono Y., Ojima K., Torii F., Takaya E., Doi N., Nakagawa K., Hata S., Abe K., Sorimachi H. (2010) Skeletal muscle-specific calpain is an intracellular Na+-dependent protease. J. Biol. Chem. 285, 22986–22998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Dutt P., Croall D.E., Arthur S.C., De Veyra T., Williams K., Elce J.S., Greer P.A. (2006) m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 6, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., Hillaire D., Passos-Bueno M.-R., Zats M., Tischfield J.A., Fardeau M., Jackson C.E., Cohen D., Beckmann J.S. (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81, 27–40 [DOI] [PubMed] [Google Scholar]
- 57).Hata S., Abe M., Suzuki H., Kitamura F., Toyama-Sorimachi N., Abe K., Sakimura K., Sorimachi H. (2010) Calpain 8/nCL-2 and Calpain 9/nCL-4 constitute an active protease complex, G-calpain, involved in gastric mucosal defense. PLoS Genet. 6, e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Yamada M., Yoshida Y., Mori D., Takitoh T., Kengaku M., Umeshima H., Takao K., Miyakawa T., Sato M., Sorimachi H., Wynshaw-Boris A., Hirotsune S. (2009) Inhibition of calpain increases LIS1 expression and partially rescues in vivo phenotypes in a mouse model of lissencephaly. Nat. Med. 15, 1202–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Kimura Y., Koga H., Araki N., Mugita N., Fujita N., Takeshima H., Nishi T., Yamashima T., Saido T.C., Yamasaki T., Moritake K., Saya H., Nakao M. (1998) The involvement of calpain-dependent proteolysis of the tumor suppressor NF2 (merlin) in schwannomas and meningiomas. Nat. Med. 4, 915–922 [DOI] [PubMed] [Google Scholar]
- 60).Delaney S.J., Hayward D.C., Barleben F., Fischbach K.F., Miklos G.L. (1991) Molecular cloning and analysis of small optic lobes, a structural brain gene of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 88, 7214–7218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Barnes T.M., Hodgkin J. (1996) The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 15, 4477–4484 [PMC free article] [PubMed] [Google Scholar]
- 62).Lid S.E., Gruis D., Jung R., Lorentzen J.A., Ananiev E., Chamberlin M., Niu X., Meeley R., Nichols S., Olsen O.A. (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 99, 5460–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Denison S.H., Orejas M., Arst H.N., Jr. (1995) Signaling of ambient pH in Aspergillus involves a cysteine protease. J. Biol. Chem. 270, 28519–28522 [DOI] [PubMed] [Google Scholar]
- 64).Futai E., Maeda T., Sorimachi H., Kitamoto K., Ishiura S., Suzuki K. (1999) The protease activity of a calpain-like cysteine protease in Saccharomyces cerevisiae is required for alkaline adaptation and sporulation. Mol. Gen. Genet. 260, 559–568 [DOI] [PubMed] [Google Scholar]
- 65).Hayashi M., Fukuzawa T., Sorimachi H., Maeda T. (2005) Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 25, 9478–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Suzuki K. (1991) Historical background, current status, and future prospect of research on calcium dependent protease. Cell Technol. 10, 545–551(in Japanese) [Google Scholar]
- 67).Suzuki K. (1991) Determination of structure of calpain and East–West problem. Cell Technol. 10, 719–727(in Japanese) [Google Scholar]
- 68).Suzuki K. (1991) In search of physiological function of calpain. Cell Technol. 10, 878–885(in Japanese) [Google Scholar]
- 69).Drummond G.I., Duncan L. (1968) On the mechanism of activation of phosphorylase b kinase by calcium. J. Biol. Chem. 243, 5532–5538 [PubMed] [Google Scholar]
- 70).Fischer E.H., Krebs E.G. (1955) Conversion of phosphorylase b to phosphorylase a in muscle extracts. J. Biol. Chem. 216, 121–132 [PubMed] [Google Scholar]
- 71).Krebs E.G., Fischer E.H. (1955) Phosphorylase activity of skeletal muscle extracts. J. Biol. Chem. 216, 113–120 [PubMed] [Google Scholar]
- 72).Koohmaraie M. (1992) The role of Ca2+-dependent proteases (calpains) in post mortem proteolysis and meat tenderness. Biochimie 74, 239–245 [DOI] [PubMed] [Google Scholar]
- 73).Kishimoto A., Kajikawa N., Tabuchi H., Shiota M., Nishizuka Y. (1981) Calcium-dependent neural proteases, widespread occurrence of a species of protease active at lower concentrations of calcium. J. Biochem. 90, 889–892 [DOI] [PubMed] [Google Scholar]
- 74).Ogawa Y., Takai Y., Kawahara Y., Kimura S., Nishizuka Y. (1981) A new possible regulatory system for protein phosphorylation in human peripheral lymphocytes. I. Characterization of a calcium-activated, phospholipid-dependent protein kinase. J. Immunol. 127, 1369–1374 [PubMed] [Google Scholar]
- 75).Takai Y., Kaibuchi K., Sano K., Nishizuka Y. (1982) Counteraction of calcium-activated, phospholipid-dependent protein kinase activation by adenosine 3′,5′-monophosphate and guanosine 3′,5′-monophosphate in platelets. J. Biochem. 91, 403–406 [DOI] [PubMed] [Google Scholar]
- 76).Kishimoto A., Kajikawa N., Shiota M., Nishizuka Y. (1983) Proteolytic activation of calcium-activated, phospholipid-dependent protein kinase by calcium-dependent neutral protease. J. Biol. Chem. 258, 1156–1164 [PubMed] [Google Scholar]
- 77).Ishiura S., Sugita H., Suzuki K., Imahori K. (1979) Studies of a calcium-activated neutral protease from chicken skeletal muscle. II. Substrate specificity. J. Biochem. 86, 579–581 [DOI] [PubMed] [Google Scholar]
- 78).Suzuki K., Ishiura S., Tsuji S., Katamoto T., Sugita H., Imahori K. (1979) Calcium activated neutral protease from human skeletal muscle. FEBS Lett. 104, 355–358 [DOI] [PubMed] [Google Scholar]
- 79).Sugita H., Ishiura S., Suzuki K., Imahori K. (1980) Ca-activated neutral protease and its inhibitors: in vitro effect on intact myofibrils. Muscle Nerve 3, 335–339 [DOI] [PubMed] [Google Scholar]
- 80).Sugita H., Ishiura S., Suzuki K., Imahori K. (1980) Inhibition of epoxide derivatives on chicken calcium-activated neutral protease (CANP) in vitro and in vivo. J. Biochem. 87, 339–341 [DOI] [PubMed] [Google Scholar]
- 81).Suzuki K., Tsuji S., Kubota S., Kimura Y., Imahori K. (1981) Limited autolysis of Ca2+-activated neutral protease (CANP) changes its sensitivity to Ca2+ ions. J. Biochem. 90, 275–278 [DOI] [PubMed] [Google Scholar]
- 82).Suzuki K. (1982) Activation mechanism of calcium-activated neutral protease. Seikagaku 54, 1264–1270(in Japanese) [PubMed] [Google Scholar]
- 83).Suzuki K., Tsuji S. (1982) Synergistic activation of calcium-activated neutral protease by Mn2+ and Ca2+. FEBS Lett. 140, 16–18 [DOI] [PubMed] [Google Scholar]
- 84).Dayton W.R., Goll D.E., Zeece M.G., Robson R.M., Reville W.J. (1976) A Ca2+-activated protease possibly involved in myofibrillar protein turnover. Purification from porcine muscle. Biochemistry 15, 2150–2158 [DOI] [PubMed] [Google Scholar]
- 85).Ohno S., Akita Y., Hata A., Osada S., Kubo K., Konno Y., Akimoto K., Mizuno K., Saido T., Kuroki T., Suzuki K. (1991) Structural and functional diversities of a family of signal transducing protein kinases, protein kinase C family; two distinct classes of PKC, conventional cPKC and novel nPKC. Adv. Enzyme Regul. 31, 287–303 [DOI] [PubMed] [Google Scholar]
- 86).Sorimachi H., Tsukahara T., Okada-Ban M., Sugita H., Ishiura S., Suzuki K. (1995) Identification of a third ubiquitous calpain species—chicken muscle expresses four distinct calpains. Biochim. Biophys. Acta 1261, 381–393 [DOI] [PubMed] [Google Scholar]
- 87).Macqueen D.J., Delbridge M.L., Manthri S., Johnston I.A. (2010) A newly classified vertebrate calpain protease, directly ancestral to CAPN1 and 2, episodically evolved a restricted physiological function in placental mammals. Mol. Biol. Evol. 27, 1886–1902 [DOI] [PubMed] [Google Scholar]
- 88).Berti P.J., Storer A.C. (1995) Alignment/phylogeny of the papain superfamily of cysteine proteases. J. Mol. Biol. 246, 273–283 [DOI] [PubMed] [Google Scholar]
- 89).Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., Lanczycki C.J., Liebert C.A., Liu C., Lu F., Lu S., Marchler G.H., Mullokandov M., Song J.S., Tasneem A., Thanki N., Yamashita R.A., Zhang D., Zhang N., Bryant S.H. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37, D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Pontremoli S., Melloni E. (1988) The role of calpain and protein kinase C in activation of human neutrophils. Prog. Clin. Biol. Res. 282, 195–208 [PubMed] [Google Scholar]
- 91).Noguchi M., Sarin A., Aman M.J., Nakajima H., Shores E.W., Henkart P.A., Leonard W.J. (1997) Functional cleavage of the common cytokine receptor γ chain (γc) by calpain. Proc. Natl. Acad. Sci. USA 94, 11534–11539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Leloup L., Shao H., Bae Y.H., Deasy B., Stolz D., Roy P., Wells A. (2010) m-Calpain activation is regulated by its membrane localization and by its binding to phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 285, 33549–33566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93).Mellgren R.L., Zhang W., Miyake K., McNeil P.L. (2007) Calpain is required for the rapid, calcium-dependent repair of wounded plasma membrane. J. Biol. Chem. 282, 2567–2575 [DOI] [PubMed] [Google Scholar]
- 94).Mellgren R.L., Huang X. (2007) Fetuin A stabilizes m-calpain and facilitates plasma membrane repair. J. Biol. Chem. 282, 35868–35877 [DOI] [PubMed] [Google Scholar]
- 95).Danial N.N., Korsmeyer S.J. (2004) Cell death: critical control points. Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 96).Nakagawa T., Yuan J. (2000) Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J. Cell Biol. 150, 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97).Hathaway D.R., Werth D.K., Haeberle J.R. (1982) Limited autolysis reduces the Ca2+ requirement of a smooth muscle Ca2+-activated protease. J. Biol. Chem. 257, 9072–9077 [PubMed] [Google Scholar]
- 98).Inomata M., Kasai Y., Nakamura M., Kawashima S. (1988) Activation mechanism of calcium-activated neutral protease. Evidence for the existence of intramolecular and intermolecular autolyses. J. Biol. Chem. 263, 19783–19787 [PubMed] [Google Scholar]
- 99).Cong J., Goll D.E., Peterson A.M., Kapprell H.P. (1989) The role of autolysis in activity of the Ca2+-dependent proteinases (μ-calpain and m-calpain). J. Biol. Chem. 264, 10096–10103 [PubMed] [Google Scholar]
- 100).Nakagawa K., Masumoto H., Sorimachi H., Suzuki K. (2001) Dissociation of m-calpain subunits occurs after autolysis of the N-terminus of the catalytic subunit, and is not required for activation. J. Biochem. 130, 605–611 [DOI] [PubMed] [Google Scholar]
- 101).Pal G.P., Elce J.S., Jia Z. (2001) Dissociation and aggregation of calpain in the presence of calcium. J. Biol. Chem. 276, 47233–47238 [DOI] [PubMed] [Google Scholar]
- 102).Suo S., Koike H., Sorimachi H., Ishiura S., Suzuki K. (1999) Association and dissociation of the calcium-binding domains of calpain by Ca2+. Biochem. Biophys. Res. Commun. 257, 63–66 [DOI] [PubMed] [Google Scholar]
- 103).Li H., Thompson V.F., Goll D.E. (2004) Effects of autolysis on properties of μ- and m-calpain. Biochim. Biophys. Acta 1691, 91–103 [DOI] [PubMed] [Google Scholar]
- 104).Zhang W., Mellgren R.L. (1996) Calpain subunits remain associated during catalysis. Biochem. Biophys. Res. Commun. 227, 890–896 [PubMed] [Google Scholar]
- 105).Dainese E., Minafra R., Sabatucci A., Vachette P., Melloni E., Cozzani I. (2002) Conformational changes of calpain from human erythrocytes in the presence of Ca2+. J. Biol. Chem. 277, 40296–40301 [DOI] [PubMed] [Google Scholar]
- 106).Elce J.S., Davies P.L., Hegadorn C., Maurice D.H., Arthur J.S. (1997) The effects of truncations of the small subunit on m-calpain activity and heterodimer formation. Biochem. J. 326, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107).Hata S., Sorimachi H., Nakagawa K., Maeda T., Abe K., Suzuki K. (2001) Domain II of m-calpain is a Ca2+-dependent cysteine protease. FEBS Lett. 501, 111–114 [DOI] [PubMed] [Google Scholar]
- 108).Hosfield C.M., Elce J.S., Davies P.L., Jia Z. (1999) Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 18, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109).Strobl S., Fernandez-Catalan C., Braun M., Huber R., Masumoto H., Nakagawa K., Irie A., Sorimachi H., Bourenkow G., Bartunik H., Suzuki K., Bode W. (2000) The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc. Natl. Acad. Sci. USA 97, 588–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110).Hanna R.A., Campbell R.L., Davies P.L. (2008) Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature 456, 409–412 [DOI] [PubMed] [Google Scholar]
- 111).Moldoveanu T., Gehring K., Green D.R. (2008) Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature 456, 404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Moldoveanu T., Hosfield C.M., Lim D., Elce J.S., Jia Z., Davies P.L. (2002) A Ca2+ switch aligns the active site of calpain. Cell 108, 649–660 [DOI] [PubMed] [Google Scholar]
- 113).Moldoveanu T., Hosfield C.M., Lim D., Jia Z., Davies P.L. (2003) Calpain silencing by a reversible intrinsic mechanism. Nat. Struct. Biol. 10, 371–378 [DOI] [PubMed] [Google Scholar]
- 114).Tompa P., Emori Y., Sorimachi H., Suzuki K., Friedrich P. (2001) Domain III of calpain is a Ca2+-regulated phospholipid-binding domain. Biochem. Biophys. Res. Commun. 280, 1333–1339 [DOI] [PubMed] [Google Scholar]
- 115).Minami Y., Emori Y., Imajoh-Ohmi S., Kawasaki H., Suzuki K. (1988) Carboxyl-terminal truncation and site-directed mutagenesis of the EF hand structure-domain of the small subunit of rabbit calcium-dependent protease. J. Biochem. 104, 927–933 [DOI] [PubMed] [Google Scholar]
- 116).Minami Y., Emori Y., Kawasaki H., Suzuki K. (1987) E-F hand structure-domain of calcium-activated neutral protease (CANP) can bind Ca2+ ions. J. Biochem. 101, 889–895 [DOI] [PubMed] [Google Scholar]
- 117).Lin G.D., Chattopadhyay D., Maki M., Wang K.K., Carson M., Jin L., Yuen P.W., Takano E., Hatanaka M., DeLucas L.J., Narayana S.V. (1997) Crystal structure of calcium bound domain VI of calpain at 1.9 Å resolution and its role in enzyme assembly, regulation, and inhibitor binding. Nat. Struct. Biol. 4, 539–547 [DOI] [PubMed] [Google Scholar]
- 118).Maki M., Narayana S.V., Hitomi K. (1997) A growing family of the Ca2+-binding proteins with five EF-hand motifs. Biochem. J. 328, 718–720 [PMC free article] [PubMed] [Google Scholar]
- 119).Blanchard H., Grochulski P., Li Y., Arthur J.S., Davies P.L., Elce J.S., Cygler M. (1997) Structure of a calpain Ca2+-binding domain reveals a novel EF-hand and Ca2+-induced conformational changes. Nat. Struct. Biol. 4, 532–538 [DOI] [PubMed] [Google Scholar]
- 120).Imajoh S., Kawasaki H., Suzuki K. (1987) The COOH-terminal E-F hand structure of calcium-activated neutral protease (CANP) is important for the association of subunits and resulting proteolytic activity. J. Biochem. 101, 447–452 [DOI] [PubMed] [Google Scholar]
- 121).Schad E., Farkas A., Jekely G., Tompa P., Friedrich P. (2002) A novel human small subunit of calpains. Biochem. J. 362, 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122).Tsuji S., Ishiura S., Takahashi-Nakamura M., Katamoto T., Suzuki K., Imahori K. (1981) Studies on a Ca2+-activated neutral proteinase of rabbit skeletal muscle. II. Characterization of sulfhydryl groups and a role of Ca2+ ions in this enzyme. J. Biochem. 90, 1405–1411 [DOI] [PubMed] [Google Scholar]
- 123).Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilagyi A., Banoczi Z., Hudecz F., Friedrich P. (2004) On the sequential determinants of calpain cleavage. J. Biol. Chem. 279, 20775–20785 [DOI] [PubMed] [Google Scholar]
- 124).duVerle D., Takigawa I., Ono Y., Sorimachi H., Mamitsuka H. (2010) CaMPDB: a resource for calpain and modulatory proteolysis. Genome Inform. 22, 202–213 [PubMed] [Google Scholar]
- 125).Davis T.L., Walker J.R., Finerty P.J., Jr., Mackenzie F., Newman E.M., Dhe-Paganon S. (2007) The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition. J. Mol. Biol. 366, 216–229 [DOI] [PubMed] [Google Scholar]
- 126).Yuan P., Leonetti M.D., Pico A.R., Hsiung Y., MacKinnon R. (2010) Structure of the human BK channel Ca2+-activation apparatus at 3.0 Å resolution. Science 329, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127).Moldoveanu T., Campbell R.L., Cuerrier D., Davies P.L. (2004) Crystal structures of calpain-E64 and -leupeptin inhibitor complexes reveal mobile loops gating the active site. J. Mol. Biol. 343, 1313–1326 [DOI] [PubMed] [Google Scholar]
- 128).Arthur J.S., Elce J.S., Hegadorn C., Williams K., Greer P.A. (2000) Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol. Cell. Biol. 20, 4474–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129).Zimmerman U.J., Boring L., Pak J.H., Mukerjee N., Wang K.K. (2000) The calpain small subunit gene is essential: its inactivation results in embryonic lethality. IUBMB Life 50, 63–68 [DOI] [PubMed] [Google Scholar]
- 130).Yoshizawa T., Sorimachi H., Tomioka S., Ishiura S., Suzuki K. (1995) A catalytic subunit of calpain possesses full proteolytic activity. FEBS Lett. 358, 101–103 [DOI] [PubMed] [Google Scholar]
- 131).Azam M., Andrabi S.S., Sahr K.E., Kamath L., Kuliopulos A., Chishti A.H. (2001) Disruption of the mouse μ-calpain gene reveals an essential role in platelet function. Mol. Cell. Biol. 21, 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132).Dourdin N., Bhatt A.K., Dutt P., Greer P.A., Arthur J.S., Elce J.S., Huttenlocher A. (2001) Reduced cell migration and disruption of the actin cytoskeleton in calpain-deficient embryonic fibroblasts. J. Biol. Chem. 276, 48382–48388 [DOI] [PubMed] [Google Scholar]
- 133).Mellgren R.L., Zhang W., Miyake K., McNeil P.L. (2006) Calpain is required for the rapid, calcium-dependent repair of wounded plasma membrane. J. Biol. Chem. 282, 2567–2575 [DOI] [PubMed] [Google Scholar]
- 134).Demarchi F., Bertoli C., Copetti T., Tanida I., Brancolini C., Eskelinen E.L., Schneider C. (2006) Calpain is required for macroautophagy in mammalian cells. J. Cell Biol. 175, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135).Farkas A., Tompa P., Friedrich P. (2003) Revisiting ubiquity and tissue specificity of human calpains. Biol. Chem. 384, 945–949 [DOI] [PubMed] [Google Scholar]
- 136).Hata A., Ohno S., Akita Y., Suzuki K. (1989) Tandemly reiterated negative enhancer-like elements regulate transcription of a human gene for the large subunit of calcium-dependent protease. J. Biol. Chem. 264, 6404–6411 [PubMed] [Google Scholar]
- 137).Valerio D., Duyvesteyn M.G., Dekker B.M., Weeda G., Berkvens T.M., van der Voorn L., van Ormondt H., van der Eb A.J. (1985) Adenosine deaminase: characterization and expression of a gene with a remarkable promoter. EMBO J. 4, 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138).Melton D.W., McEwan C., McKie A.B., Reid A.M. (1986) Expression of the mouse HPRT gene: deletional analysis of the promoter region of an X-chromosome linked housekeeping gene. Cell 44, 319–328 [DOI] [PubMed] [Google Scholar]
- 139).Angel P., Imagawa M., Chiu R., Stein B., Imbra R.J., Rahmsdorf H.J., Jonat C., Herrlich P., Karin M. (1987) Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49, 729–739 [DOI] [PubMed] [Google Scholar]
- 140).Lee W., Mitchell P., Tjian R. (1987) Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell 49, 741–752 [DOI] [PubMed] [Google Scholar]
- 141).Nakamura M., Mori M., Morishita Y., Mori S., Kawashima S. (1992) Specific increase in calcium-activated neutral protease with low calcium sensitivity (m-calpain) in proerythroblastic K562 cell line cells induced to differentiation by phorbol 12-myristate 13-acetate. Exp. Cell Res. 200, 513–522 [DOI] [PubMed] [Google Scholar]
- 142).Miyake S., Emori Y., Suzuki K. (1986) Gene organization of the small subunit of human calcium-activated neutral protease. Nucleic Acids Res. 14, 8805–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143).Emori Y., Kawasaki H., Imajoh S., Imahori K., Suzuki K. (1987) Endogenous inhibitor for calcium-dependent cysteine protease contains four internal repeats that could be responsible for its multiple reactive sites. Proc. Natl. Acad. Sci. USA 84, 3590–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144).Maki M., Takano E., Mori H., Kannagi R., Murachi T., Hatanaka M. (1987) Repetitive region of calpastatin is a functional unit of the proteinase inhibitor. Biochem. Biophys. Res. Commun. 143, 300–308 [DOI] [PubMed] [Google Scholar]
- 145).Maki M., Takano E., Mori H., Sato A., Murachi T., Hatanaka M. (1987) All four internally repetitive domains of pig calpastatin possess inhibitory activities against calpains I and II. FEBS Lett. 223, 174–180 [DOI] [PubMed] [Google Scholar]
- 146).Hata S., Doi N., Kitamura F., Sorimachi H. (2007) Stomach-specific calpain, nCL-2/calpain 8, is active without calpain regulatory subunit and oligomerizes through C2-like domains. J. Biol. Chem. 282, 27847–27856 [DOI] [PubMed] [Google Scholar]
- 147).Lee H.J., Tomioka S., Kinbara K., Masumoto H., Jeong S.Y., Sorimachi H., Ishiura S., Suzuki K. (1999) Characterization of a human digestive tract-specific calpain, nCL-4, expressed in the baculovirus system. Arch. Biochem. Biophys. 362, 22–31 [DOI] [PubMed] [Google Scholar]
- 148).Takano J., Tomioka M., Tsubuki S., Higuchi M., Iwata N., Itohara S., Maki M., Saido T.C. (2005) Calpain mediates excitotoxic DNA fragmentation via mitochondrial pathways in adult brains: evidence from calpastatin mutant mice. J. Biol. Chem. 280, 16175–16184 [DOI] [PubMed] [Google Scholar]
- 149).Higuchi M., Tomioka M., Takano J., Shirotani K., Iwata N., Masumoto H., Maki M., Itohara S., Saido T.C. (2005) Distinct mechanistic roles of calpain and caspase activation in neurodegeneration as revealed in mice overexpressing their specific inhibitors. J. Biol. Chem. 280, 15229–15237 [DOI] [PubMed] [Google Scholar]
- 150).Spencer M.J., Mellgren R.L. (2002) Overexpression of a calpastatin transgene in mdx muscle reduces dystrophic pathology. Hum. Mol. Genet. 11, 2645–2655 [DOI] [PubMed] [Google Scholar]
- 151).Tidball J.G., Spencer M.J. (2002) Expression of a calpastatin transgene slows muscle wasting and obviates changes in myosin isoform expression during murine muscle disuse. J. Physiol. 545, 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152).Kent M.P., Spencer M.J., Koohmaraie M. (2004) Postmortem proteolysis is reduced in transgenic mice overexpressing calpastatin. J. Anim. Sci. 82, 794–801 [DOI] [PubMed] [Google Scholar]
- 153).Geesink G.H., Kuchay S., Chishti A.H., Koohmaraie M. (2006) Micro-calpain is essential for postmortem proteolysis of muscle proteins. J. Anim. Sci. 84, 2834–2840 [DOI] [PubMed] [Google Scholar]
- 154).Dear T.N., Boehm T. (2001) Identification and characterization of two novel calpain large subunit genes. Gene 274, 245–252 [DOI] [PubMed] [Google Scholar]
- 155).Andresen K., Tom T.D., Strand M. (1991) Characterization of cDNA clones encoding a novel calcium-activated neutral proteinase from Schistosoma mansoni. J. Biol. Chem. 266, 15085–15090 [PubMed] [Google Scholar]
- 156).Ersfeld K., Barraclough H., Gull K. (2005) Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J. Mol. Evol. 61, 742–757 [DOI] [PubMed] [Google Scholar]
- 157).Hata S., Nishi K., Kawamoto T., Lee H.J., Kawahara H., Maeda T., Shintani Y., Sorimachi H., Suzuki K. (2001) Both the conserved and the unique gene structure of stomach-specific calpains reveal processes of calpain gene evolution. J. Mol. Evol. 53, 191–203 [DOI] [PubMed] [Google Scholar]
- 158).Futai E., Kubo T., Sorimachi H., Suzuki K., Maeda T. (2001) Molecular cloning of PalBH, a mammalian homologue of the Aspergillus atypical calpain PalB. Biochim. Biophys. Acta 1517, 316–319 [DOI] [PubMed] [Google Scholar]
- 159).Li M., Martin S.J., Bruno V.M., Mitchell A.P., Davis D.A. (2004) Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot. Cell 3, 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160).Futai E., Sorimachi H., Jeong S.Y., Kitamoto K., Ishiura S., Suzuki K. (1999) Aspergillus oryzae palBory encodes a calpain-like protease: homology to Emericella nidulans PalB and conservation of functional regions. J. Biosci. Bioeng. 88, 438–440 [DOI] [PubMed] [Google Scholar]
- 161).Villalobo E., Moch C., Fryd-Versavel G., Fleury-Aubusson A., Morin L. (2003) Cysteine proteases and cell differentiation: excystment of the ciliated protist Sterkiella histriomuscorum. Eukaryot. Cell 2, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162).Horikawa Y., Oda N., Cox N.J., Li X., Orho-Melander M., Hara M., Hinokio Y., Lindner T.H., Mashima H., Schwarz P.E., del Bosque-Plata L., Horikawa Y., Oda Y., Yoshiuchi I., Colilla S., Polonsky K.S., Wei S., Concannon P., Iwasaki N., Schulze J., Baier L.J., Bogardus C., Groop L., Boerwinkle E., Hanis C.L., Bell G.I. (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26, 163–175 [DOI] [PubMed] [Google Scholar]
- 163).Margis R., Margis-Pinheiro M. (2003) Phytocalpains: orthologous calcium-dependent cysteine proteinases. Trends Plant Sci. 8, 58–62 [DOI] [PubMed] [Google Scholar]
- 164).Wang C., Barry J.K., Min Z., Tordsen G., Rao A.G., Olsen O.A. (2003) The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J. Biol. Chem. 278, 34467–34474 [DOI] [PubMed] [Google Scholar]
- 165).Lid S.E., Olsen L., Nestestog R., Aukerman M., Brown R.C., Lemmon B., Mucha M., Opsahl-Sorteberg H.G., Olsen O.A. (2005) Mutation in the Arabidopisis thaliana DEK1 calpain gene perturbs endosperm and embryo development while over-expression affects organ development globally. Planta 221, 339–351 [DOI] [PubMed] [Google Scholar]
- 166).Ahn J.W., Kim M., Lim J.H., Kim G.T., Pai H.S. (2004) Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J. 38, 969–981 [DOI] [PubMed] [Google Scholar]
- 167).Bourgeau G., Lapointe H., Peloquin P., Mayrand D. (1992) Cloning, expression, and sequencing of a protease gene (tpr) from Porphyromonas gingivalis W83 in Escherichia coli. Infect. Immun. 60, 3186–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168).Wu K.H., Tai P.C. (2004) Cys32 and His105 are the critical residues for the calcium-dependent cysteine proteolytic activity of CvaB, an ATP-binding cassette transporter. J. Biol. Chem. 279, 901–909 [DOI] [PubMed] [Google Scholar]
- 169).Sorimachi H., Saido T.C., Suzuki K. (1994) New era of calpain research. Discovery of tissue-specific calpains. FEBS Lett. 343, 1–5 [DOI] [PubMed] [Google Scholar]
- 170).Badalamente M.A., Stracher A. (2000) Delay of muscle degeneration and necrosis in mdx mice by calpain inhibition. Muscle Nerve 23, 106–111 [DOI] [PubMed] [Google Scholar]
- 171).Bartus R.T., Hayward N.J., Elliott P.J., Sawyer S.D., Baker K.L., Dean R.L., Akiyama A., Straub J.A., Harbeson S.L., Li Z., Powers J. (1994) Calpain inhibitor AK295 protects neurons from focal brain ischemia. Effects of postocclusion intra-arterial administration. Stroke 25, 2265–2270 [DOI] [PubMed] [Google Scholar]
- 172).Sorimachi H., Forsberg N.E., Lee H.J., Joeng S.Y., Richard I., Beckmann J.S., Ishiura S., Suzuki K. (1996) Highly conserved structure in the promoter region of the gene for muscle-specific calpain, p94. Biol. Chem. 377, 859–864 [PubMed] [Google Scholar]
- 173).Kawabata Y., Hata S., Ono Y., Ito Y., Suzuki K., Abe K., Sorimachi H. (2003) Newly identified exons encoding novel variants of p94/calpain 3 are expressed ubiquitously and overlap the α-glucosidase C gene. FEBS Lett. 555, 623–630 [DOI] [PubMed] [Google Scholar]
- 174).De Tullio R., Stifanese R., Salamino F., Pontremoli S., Melloni E. (2003) Characterization of a new p94-like calpain form in human lymphocytes. Biochem. J. 375, 689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175).König N., Raynaud F., Feane H., Durand M., Mestre-Francès N., Rossel M., Ouali A., Benyamin Y. (2003) Calpain 3 is expressed in astrocytes of rat and Microcebus brain. J. Chem. Neuroanat. 25, 129–136 [DOI] [PubMed] [Google Scholar]
- 176).Azuma M., Fukiage C., Higashine M., Nakajima T., Ma H., Shearer T.R. (2000) Identification and characterization of a retina-specific calpain (Rt88) from rat. Curr. Eye Res. 21, 710–720 [PubMed] [Google Scholar]
- 177).Ma H., Fukiage C., Azuma M., Shearer T.R. (1998) Cloning and expression of mRNA for calpain Lp82 from rat lens: splice variant of p94. Invest. Ophthalmol. Vis. Sci. 39, 454–461 [PubMed] [Google Scholar]
- 178).Fougerousse F., Bullen P., Herasse M., Lindsay S., Richard I., Wilson D., Suel L., Durand M., Robson S., Abitbol M., Beckmann J.S., Strachan T. (2000) Human–mouse differences in the embryonic expression patterns of developmental control genes and disease genes. Hum. Mol. Genet. 9, 165–173 [DOI] [PubMed] [Google Scholar]
- 179).Sorimachi H., Ishiura S., Suzuki K. (1993) A novel tissue-specific calpain species expressed predominantly in the stomach comprises two alternative splicing products with and without Ca2+-binding domain. J. Biol. Chem. 268, 19476–19482 [PubMed] [Google Scholar]
- 180).Sorimachi H., Imajoh-Ohmi S., Emori Y., Kawasaki H., Ohno S., Minami Y., Suzuki K. (1989) Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and μ-types. Specific expression of the mRNA in skeletal muscle. J. Biol. Chem. 264, 20106–20111 [PubMed] [Google Scholar]
- 181).Sorimachi H., Toyama-Sorimachi N., Saido T.C., Kawasaki H., Sugita H., Miyasaka M., Arahata K., Ishiura S., Suzuki K. (1993) Muscle-specific calpain, p94, is degraded by autolysis immediately after translation, resulting in disappearance from muscle. J. Biol. Chem. 268, 10593–10605 [PubMed] [Google Scholar]
- 182).Ono Y., Hayashi C., Doi N., Kitamura F., Shindo M., Kudo K., Tsubata T., Yanagida M., Sorimachi H. (2007) Comprehensive survey of p94/calpain 3 substrates by comparative proteomics—Possible regulation of protein synthesis by p94. Biotechnol. J. 2, 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183).Ono Y., Torii F., Ojima K., Doi N., Yoshioka K., Kawabata Y., Labeit D., Labeit S., Suzuki K., Abe K., Maeda T., Sorimachi H. (2006) Suppressed disassembly of autolyzing p94/CAPN3 by N2A connectin/titin in a genetic reporter system. J. Biol. Chem. 281, 18519–18531 [DOI] [PubMed] [Google Scholar]
- 184).Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., Maruyama K., Suzuki K. (1995) Muscle-specific calpain, p94, responsible for limb girdle muscular dystrophy type 2A, associates with connectin through IS2, a p94-specific sequence. J. Biol. Chem. 270, 31158–31162 [DOI] [PubMed] [Google Scholar]
- 185).Chiannilkulchai N., Pasturaud P., Richard I., Auffray C., Beckmann J.S. (1995) A primary expression map of the chromosome 15q15 region containing the recessive form of limb-girdle muscular dystrophy (LGMD2A) gene. Hum. Mol. Genet. 4, 717–725 [DOI] [PubMed] [Google Scholar]
- 186).Richard I., Roudaut C., Marchand S., Baghdiguian S., Herasse M., Stockholm D., Ono Y., Suel L., Bourg N., Sorimachi H., Lefranc G., Fardeau M., Sebille A., Beckmann J.S. (2000) Loss of calpain 3 proteolytic activity leads to muscular dystrophy and to apoptosis-associated IκBα/nuclear factor κB pathway perturbation in mice. J. Cell Biol. 151, 1583–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187).Kramerova I., Kudryashova E., Tidball J.G., Spencer M.J. (2004) Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 13, 1373–1388 [DOI] [PubMed] [Google Scholar]
- 188).Ono Y., Shimada H., Sorimachi H., Richard I., Saido T.C., Beckmann J.S., Ishiura S., Suzuki K. (1998) Functional defects of a muscle-specific calpain, p94, caused by mutations associated with limb-girdle muscular dystrophy type 2A. J. Biol. Chem. 273, 17073–17078 [DOI] [PubMed] [Google Scholar]
- 189).Ojima K., Kawabata Y., Nakao H., Nakao K., Doi N., Kitamura F., Ono Y., Hata S., Suzuki H., Kawahara H., Bogomolovas J., Witt C., Ottenheijm C., Labeit S., Granzier H., Toyama-Sorimachi N., Sorimachi M., Suzuki K., Maeda T., Abe K., Aiba A., Sorimachi H. (2010) Role of dynamic distribution of muscle-specific calpain in physical-stress adaptation and muscular dystrophy in mice. J. Clin. Invest. 120, 2672–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190).Richard I., Roudaut C., Saenz A., Pogue R., Grimbergen J.E., Anderson L.V., Beley C., Cobo A.M., de Diego C., Eymard B., Gallano P., Ginjaar H.B., Lasa A., Pollitt C., Topaloglu H., Urtizberea J.A., de Visser M., van der Kooi A., Bushby K., Bakker E., Lopez de Munain A., Fardeau M., Beckmann J.S. (1999) Calpainopathy—a survey of mutations and polymorphisms. Am. J. Hum. Genet. 64, 1524–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191).Sáenz A., Leturcq F., Cobo A.M., Poza J.J., Ferrer X., Otaegui D., Camaño P., Urtasun M., Vílchez J., Gutiérrez-Rivas E., Emparanza J., Merlini L., Paisán C., Goicoechea M., Blázquez L., Eymard B., Lochmuller H., Walter M., Bonnemann C., Figarella-Branger D., Kaplan J.C., Urtizberea J.A., Martí-Massó J.F., López de Munain A. (2005) LGMD2A: genotype-phenotype correlations based on a large mutational survey on the calpain 3 gene. Brain 128, 732–742 [DOI] [PubMed] [Google Scholar]
- 192).Hata S., Koyama S., Kawahara H., Doi N., Maeda T., Toyama-Sorimachi N., Abe K., Suzuki K., Sorimachi H. (2006) Stomach-specific calpain, nCL-2, localizes in mucus cells and proteolyzes the β-subunit of coatomer complex, β-COP. J. Biol. Chem. 281, 11214–11224 [DOI] [PubMed] [Google Scholar]
- 193).Lee H.J., Sorimachi H., Jeong S.Y., Ishiura S., Suzuki K. (1998) Molecular cloning and characterization of a novel tissue-specific calpain predominantly expressed in the digestive tract. Biol. Chem. 379, 175–183 [DOI] [PubMed] [Google Scholar]
- 194).Cao Y., Zhao H., Grunz H. (2001) xCL-2 is a novel m-type calpain and disrupts morphogenetic movements during embryogenesis in Xenopus laevis. Dev. Growth Differ. 43, 563–571 [DOI] [PubMed] [Google Scholar]
- 195).Liu K., Li L., Cohen S.N. (2000) Antisense RNA-mediated deficiency of the calpain protease, nCL-4, in NIH3T3 cells is associated with neoplastic transformation and tumorigenesis. J. Biol. Chem. 275, 31093–31098 [DOI] [PubMed] [Google Scholar]
- 196).Chen C.J., Nguyen T., Shively J.E. (2010) Role of calpain-9 and PKC-δ in the apoptotic mechanism of lumen formation in CEACAM1 transfected breast epithelial cells. Exp. Cell Res. 316, 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197).Caddick M.X., Brownlee A.G., Arst H.N., Jr. (1986) Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol. Gen. Genet. 203, 346–353 [DOI] [PubMed] [Google Scholar]
- 198).Arst H.N., Jr., Bailey C.R., Penfold H.A. (1980) A possible rôle for acid phosphatase in γ-amino-n-butyrate uptake in Aspergillus nidulans. Arch. Microbiol. 125, 153–158 [DOI] [PubMed] [Google Scholar]
- 199).Arst H.N., Jr., Bignell E., Tilburn J. (1994) Two new genes involved in signalling ambient pH in Aspergillus nidulans. Mol. Gen. Genet. 245, 787–790 [DOI] [PubMed] [Google Scholar]
- 200).Negrete-Urtasun S., Denison S.H., Arst H.N., Jr. (1997) Characterization of the pH signal transduction pathway gene palA of Aspergillus nidulans and identification of possible homologs. J. Bacteriol. 179, 1832–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201).Maccheroni W., Jr., May G.S., Martinez-Rossi N.M., Rossi A. (1997) The sequence of palF, an environmental pH response gene in Aspergillus nidulans. Gene 194, 163–167 [DOI] [PubMed] [Google Scholar]
- 202).Denison S.H., Negrete-Urtasun S., Mingot J.M., Tilburn J., Mayer W.A., Goel A., Espeso E.A., Peñalva M.A., Arst H.N., Jr. (1998) Putative membrane components of signal transduction pathways for ambient pH regulation in Aspergillus and meiosis in Saccharomyces are homologous. Mol. Microbiol. 30, 259–264 [DOI] [PubMed] [Google Scholar]
- 203).Negrete-Urtasun S., Reiter W., Diez E., Denison S.H., Tilburn J., Espeso E.A., Peñalva M.A., Arst H.N., Jr. (1999) Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol. Microbiol. 33, 994–1003 [DOI] [PubMed] [Google Scholar]
- 204).Herranz S., Rodríguez J.M., Bussink H.J., Sanchez-Ferrero J.C., Arst H.N., Jr., Peñalva M.A., Vincent O. (2005) Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. USA 102, 12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205).Ridge K.D., Abdulaev N.G., Sousa M., Palczewski K. (2003) Phototransduction: crystal clear. Trends Biochem. Sci. 28, 479–487 [DOI] [PubMed] [Google Scholar]
- 206).Hervás-Aguilar A., Rodríguez J.M., Tilburn J., Arst H.N., Jr., Peñalva M.A. (2007) Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J. Biol. Chem. 282, 34735–34747 [DOI] [PubMed] [Google Scholar]
- 207).Diez E., Alvaro J., Espeso E.A., Rainbow L., Suarez T., Tilburn J., Arst H.N., Jr., Peñalva M.A. (2002) Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 21, 1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208).Mingot J.M., Tilburn J., Diez E., Bignell E., Orejas M., Widdick D.A., Sarkar S., Brown C.V., Caddick M.X., Espeso E.A., Arst H.N., Jr., Peñalva M.A. (1999) Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol. Cell. Biol. 19, 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209).Attaix D., Ventadour S., Codran A., Bechet D., Taillandier D., Combaret L. (2005) The ubiquitin–proteasome system and skeletal muscle wasting. Essays Biochem. 41, 173–186 [DOI] [PubMed] [Google Scholar]
- 210).Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 211).Orian A., Whiteside S., Israël A., Stancovski I., Schwartz A.L., Ciechanover A. (1995) Ubiquitin-mediated processing of NF-κB transcriptional activator precursor p105. Reconstitution of a cell-free system and identification of the ubiquitin-carrier protein, E2, and a novel ubiquitin-protein ligase, E3, involved in conjugation. J. Biol. Chem. 270, 21707–21714 [DOI] [PubMed] [Google Scholar]
- 212).Palombella V.J., Rando O.J., Goldberg A.L., Maniatis T. (1994) The ubiquitin–proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78, 773–785 [DOI] [PubMed] [Google Scholar]
- 213).Su S.S., Mitchell A.P. (1993) Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 21, 3789–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214).Su S.S., Mitchell A.P. (1993) Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics 133, 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215).Li W., Mitchell A.P. (1997) Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216).Mitchell B.M., Wu T.G., Jackson B.E., Wilhelmus K.R. (2007) Candida albicans strain-dependent virulence and Rim13p-mediated filamentation in experimental keratomycosis. Invest. Ophthalmol. Vis. Sci. 48, 774–780 [DOI] [PubMed] [Google Scholar]
- 217).Davis D., Wilson R.B., Mitchell A.P. (2000) RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20, 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218).Lambert M., Blanchin-Roland S., Le Louedec F., Lépingle A., Gaillardin C. (1997) Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol. Cell. Biol. 17, 3966–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219).González-López C.I., Ortiz-Castellanos L., Ruiz-Herrera J. (2006) The ambient pH response Rim pathway in Yarrowia lipolytica: identification of YlRIM9 and characterization of its role in dimorphism. Curr. Microbiol. 53, 8–12 [DOI] [PubMed] [Google Scholar]
- 220).Aréchiga-Carvajal E.T., Ruiz-Herrera J. (2005) The RIM101/pacC homologue from the basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukaryot. Cell 4, 999–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221).Rodríguez-Galán O., Galindo A., Hervás-Aguilar A., Arst H.N., Jr., Peñalva M.A. (2009) Physiological involvement in pH signaling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J. Biol. Chem. 284, 4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222).Tilburn J., Sánchez-Ferrero J.C., Reoyo E., Arst H.N., Jr., Peñalva M.A. (2005) Mutational analysis of the pH signal transduction component PalC of Aspergillus nidulans supports distant similarity to BRO1 domain family members. Genetics 171, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223).Xu W., Smith F.J., Jr., Subaran R., Mitchell A.P. (2004) Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15, 5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224).Cornet M., Bidard F., Schwarz P., Da Costa G., Blanchin-Roland S., Dromer F., Gaillardin C. (2005) Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect. Immun. 73, 7977–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225).Barwell K.J., Boysen J.H., Xu W., Mitchell A.P. (2005) Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 4, 890–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226).Blanchin-Roland S., Da Costa G., Gaillardin C. (2005) ESCRT-I components of the endocytic machinery are required for Rim101-dependent ambient pH regulation in the yeast Yarrowia lipolytica. Microbiology 151, 3627–3637 [DOI] [PubMed] [Google Scholar]
- 227).Boysen J.H., Mitchell A.P. (2006) Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17, 1344–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228).Kullas A.L., Li M., Davis D.A. (2004) Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229).Rothfels K., Tanny J.C., Molnar E., Friesen H., Commisso C., Segall J. (2005) Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 6772–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230).Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231).Franz T., Vingron M., Boehm T., Dear T.N. (1999) Capn7: a highly divergent vertebrate calpain with a novel C-terminal domain. Mamm. Genome 10, 318–321 [DOI] [PubMed] [Google Scholar]
- 232).Yorikawa C., Takaya E., Osako Y., Tanaka R., Terasawa Y., Hamakubo T., Mochizuki Y., Iwanari H., Kodama T., Maeda T., Hitomi K., Shibata H., Maki M. (2008) Human calpain 7/PalBH associates with a subset of ESCRT-III-related proteins in its N-terminal region and partly localizes to endocytic membrane compartments. J. Biochem. 143, 731–745 [DOI] [PubMed] [Google Scholar]
- 233).Osako Y., Maemoto Y., Tanaka R., Suzuki H., Shibata H., Maki M. (2010) Autolytic activity of human calpain 7 is enhanced by ESCRT-III-related protein IST1 through MIT-MIM interaction. FEBS J. 277, 4412–4426 [DOI] [PubMed] [Google Scholar]
- 234).Katoh K., Shibata H., Suzuki H., Nara A., Ishidoh K., Kominami E., Yoshimori T., Maki M. (2003) The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J. Biol. Chem. 278, 39104–39113 [DOI] [PubMed] [Google Scholar]
- 235).Missotten M., Nichols A., Rieger K., Sadoul R. (1999) Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 6, 124–129 [DOI] [PubMed] [Google Scholar]
- 236).Sadoul R. (2006) Do Alix and ALG-2 really control endosomes for better or for worse? Biol. Cell 98, 69–77 [DOI] [PubMed] [Google Scholar]
- 237).Vito P., Pellegrini L., Guiet C., D’Adamio L. (1999) Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 274, 1533–1540 [DOI] [PubMed] [Google Scholar]
- 238).Odorizzi G. (2006) The multiple personalities of Alix. J. Cell Sci. 119, 3025–3032 [DOI] [PubMed] [Google Scholar]
- 239).Gafni J., Hermel E., Young J.E., Wellington C.L., Hayden M.R., Ellerby L.M. (2004) Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J. Biol. Chem. 279, 20211–20220 [DOI] [PubMed] [Google Scholar]
- 240).Sokol S.B., Kuwabara P.E. (2000) Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev. 14, 901–906 [PMC free article] [PubMed] [Google Scholar]
- 241).Syntichaki P., Xu K., Driscoll M., Tavernarakis N. (2002) Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature 419, 939–944 [DOI] [PubMed] [Google Scholar]
- 242).Kammenga J.E., Doroszuk A., Riksen J.A., Hazendonk E., Spiridon L., Petrescu A.J., Tijsterman M., Plasterk R.H., Bakker J. (2007) A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 3, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243).Dear N., Matena K., Vingron M., Boehm T. (1997) A new subfamily of vertebrate calpains lacking a calmodulin-like domain: implications for calpain regulation and evolution. Genomics 45, 175–184 [DOI] [PubMed] [Google Scholar]
- 244).Mugita N., Kimura Y., Ogawa M., Saya H., Nakao M. (1997) Identification of a novel, tissue-specific calpain htra-3; a human homologue of the Caenorhabditis elegans sex determination gene. Biochem. Biophys. Res. Commun. 239, 845–850 [DOI] [PubMed] [Google Scholar]
- 245).Waghray A., Wang D.S., McKinsey D., Hayes R.L., Wang K.K. (2004) Molecular cloning and characterization of rat and human calpain-5. Biochem. Biophys. Res. Commun. 324, 46–51 [DOI] [PubMed] [Google Scholar]
- 246).Franz T., Winckler L., Boehm T., Dear T.N. (2004) Capn5 is expressed in a subset of T cells and is dispensable for development. Mol. Cell. Biol. 24, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247).Gonzalez A., Saez M.E., Aragon M.J., Galan J.J., Vettori P., Molina L., Rubio C., Real L.M., Ruiz A., Ramirez-Lorca R. (2006) Specific haplotypes of the CALPAIN-5 gene are associated with polycystic ovary syndrome. Hum. Reprod. 21, 943–951 [DOI] [PubMed] [Google Scholar]
- 248).Sáez M.E., Martinez-Larrad M.T., Ramirez-Lorca R., Gonzalez-Sánchez J.L., Zabena C., Martinez-Calatrava M.J., González A., Morón F.J., Ruiz A., Serrano-Ríos M. (2007) Calpain-5 gene variants are associated with diastolic blood pressure and cholesterol levels. BMC Med. Genet. 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249).Fan C., Iacobas D.A., Zhou D., Chen Q., Lai J.K., Gavrialov O., Haddad G.G. (2005) Gene expression and phenotypic characterization of mouse heart after chronic constant or intermittent hypoxia. Physiol. Genomics 22, 292–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250).Tonami K., Kurihara Y., Aburatani H., Uchijima Y., Asano T., Kurihara H. (2007) Calpain 6 is involved in microtubule stabilization and cytoskeletal organization. Mol. Cell. Biol. 27, 2548–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251).Tonami K., Kurihara Y., Arima S., Nishiyama K., Uchijima Y., Asano T., Sorimachi H., Kurihara H. (2011) Calpain 6, a microtubule-stabilizing protein, regulates Rac1 activity and cell motility through interaction with GEF-H1. J. Cell Sci. 124, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 252).Baier L.J., Permana P.A., Yang X., Pratley R.E., Hanson R.L., Shen G.Q., Mott D., Knowler W.C., Cox N.J., Horikawa Y., Oda N., Bell G.I., Bogardus C. (2000) A calpain-10 gene polymorphism is associated with reduced muscle mRNA levels and insulin resistance. J. Clin. Invest. 106, R69–R73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253).Stumvoll M., Fritsche A., Madaus A., Stefan N., Weisser M., Machicao F., Häring H. (2001) Functional significance of the UCSNP-43 polymorphism in the CAPN10 gene for proinsulin processing and insulin secretion in nondiabetic Germans. Diabetes 50, 2161–2163 [DOI] [PubMed] [Google Scholar]
- 254).Muramatsu Y., Taniguchi Y., Kose H., Matsumoto K., Yamada T., Sasaki Y. (2003) Capn10, a candidate gene responsible for type 2 diabetes mellitus in the OLETF rat. IUBMB Life 55, 533–537 [DOI] [PubMed] [Google Scholar]
- 255).Cheverud J.M., Fawcett G.L., Jarvis J.P., Norgard E.A., Pavlicev M., Pletscher L.S., Polonsky K.S., Ye H., Bell G.I., Semenkovich C.F. (2010) Calpain-10 is a component of the obesity-related quantitative trait locus Adip1. J. Lipid Res. 51, 907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256).Johnson J.D., Han Z., Otani K., Ye H., Zhang Y., Wu H., Horikawa Y., Misler S., Bell G.I., Polonsky K.S. (2004) RyR2 and calpain-10 delineate a novel apoptosis pathway in pancreatic islets. J. Biol. Chem. 279, 24794–24802 [DOI] [PubMed] [Google Scholar]
- 257).Ma H., Fukiage C., Kim Y.H., Duncan M.K., Reed N.A., Shih M., Azuma M., Shearer T.R. (2001) Characterization and expression of calpain 10. A novel ubiquitous calpain with nuclear localization. J. Biol. Chem. 276, 28525–28531 [DOI] [PubMed] [Google Scholar]
- 258).Paul D.S., Harmon A.W., Winston C.P., Patel Y.M. (2003) Calpain facilitates GLUT4 vesicle translocation during insulin-stimulated glucose uptake in adipocytes. Biochem. J. 376, 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259).Arrington D.D., Van Vleet T.R., Schnellmann R.G. (2006) Calpain 10: a mitochondrial calpain and its role in calcium-induced mitochondrial dysfunction. Am. J. Physiol. Cell Physiol. 291, C1159–C1171 [DOI] [PubMed] [Google Scholar]
- 260).Kamei M., Webb G.C., Young I.G., Campbell H.D. (1998) SOLH, a human homologue of the Drosophila melanogaster small optic lobes gene is a member of the calpain and zinc-finger gene families and maps to human chromosome 16p13.3 near CATM (cataract with microphthalmia). Genomics 51, 197–206 [DOI] [PubMed] [Google Scholar]
- 261).Kamei M., Webb G.C., Heydon K., Hendry I.A., Young I.G., Campbell H.D. (2000) Solh, the mouse homologue of the Drosophila melanogaster small optic lobes gene: organization, chromosomal mapping, and localization of gene product to the olfactory bulb. Genomics 64, 82–89 [DOI] [PubMed] [Google Scholar]
- 262).Correa G.C., Margis-Pinheiro M., Margis R. (2001) Identification, classification and expression pattern analysis of sugarcane cysteine proteinases. Genet. Mol. Biol. 24, 275–283 [Google Scholar]
- 263).Johnson K.L., Degnan K.A., Ross Walker J., Ingram G.C. (2005) AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 44, 114–127 [DOI] [PubMed] [Google Scholar]
- 264).Tian Q., Olsen L., Sun B., Lid S.E., Brown R.C., Lemmon B.E., Fosnes K., Gruis D.F., Opsahl-Sorteberg H.G., Otegui M.S., Olsen O.A. (2007) Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. Plant Cell 19, 3127–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265).Hirokawa T., Boon-Chieng S., Mitaku S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 266).Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 267).Ersfeld K., Barraclough H., Gull K. (2005) Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J. Mol. Evol. 61, 742–757 [DOI] [PubMed] [Google Scholar]
- 268).Andrade H.M., Murta S.M., Chapeaurouge A., Perales J., Nirdé P., Romanha A.J. (2008) Proteomic analysis of Trypanosoma cruzi resistance to Benznidazole. J. Proteome Res. 7, 2357–2367 [DOI] [PubMed] [Google Scholar]
- 269).Giese V., Dallagiovanna B., Marchini F.K., Pavoni D.P., Krieger M.A., Goldenberg S. (2008) Trypanosoma cruzi: a stage-specific calpain-like protein is induced after various kinds of stress. Mem. Inst. Oswaldo Cruz 103, 598–601 [DOI] [PubMed] [Google Scholar]
- 270).Hertz-Fowler C., Ersfeld K., Gull K. (2001) CAP5.5, a life-cycle-regulated, cytoskeleton-associated protein is a member of a novel family of calpain-related proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 116, 25–34 [DOI] [PubMed] [Google Scholar]
- 271).Olego-Fernandez S., Vaughan S., Shaw M.K., Gull K., Ginger M.L. (2009) Cell morphogenesis of Trypanosoma brucei requires the paralogous, differentially expressed calpain-related proteins CAP5.5 and CAP5.5V. Protist 160, 576–590 [DOI] [PubMed] [Google Scholar]
- 272).Pereira F.M., Elias C.G., d’Avila-Levy C.M., Branquinha M.H., Santos A.L. (2009) Cysteine peptidases in Herpetomonas samuelpessoai are modulated by temperature and dimethylsulfoxide-triggered differentiation. Parasitology 136, 45–54 [DOI] [PubMed] [Google Scholar]
- 273).Sangenito L.S., Ennes-Vidal V., Marinho F.A., Da Mota F.F., Santos A.L., D’Avila-Levy C.M., Branquinha M.H. (2009) Arrested growth of Trypanosoma cruzi by the calpain inhibitor MDL28170 and detection of calpain homologues in epimastigote forms. Parasitology 136, 433–441 [DOI] [PubMed] [Google Scholar]
- 274).Saido T.C., Sorimachi H., Suzuki K. (1994) Calpain: new perspectives in molecular diversity and physiological-pathological involvement. FASEB J. 8, 814–822 [PubMed] [Google Scholar]
- 275).Imahori K. (1987) Calcium-dependent proteases. Tanpakushitsu Kakusan Koso 32, 93–95(in Japanese) [PubMed] [Google Scholar]