Abstract
RNA editing is a collective term referring to enzymatic processes that change RNA sequence apart from splicing, 5′ capping or 3′ extension. In this review, we focus on uridine insertion/deletion mRNA editing found exclusively in mitochondria of kinetoplastid protists. This type of editing corrects frameshifts, introduces start and stops codons, and often adds much of the coding sequence to create an open reading frame. The mitochondrial genome of trypanosomatids, the most extensively studied clade within the order Kinetoplastida, is composed of ~50 maxicircles with limited coding capacity and thousands of minicircles. To produce functional mRNAs, a multitude of nuclear-encoded factors mediate interactions of maxicircle-encoded pre-mRNAs with a vast repertoire of minicircle-encoded guide RNAs. Editing reactions of mRNA cleavage, U-insertions or U-deletions, and ligation are catalyzed by the RNA editing core complex (RECC, the 20S editosome) while each step of this enzymatic cascade is directed by guide RNAs. These 50-60 nucleotide (nt) molecules are 3′ uridylated by RET1 TUTase and stabilized via association with the gRNA binding complex (GRBC). Remarkably, the information transfer between maxicircle and minicircle transcriptomes does not rely on template-dependent polymerization of nucleic acids. Instead, intrinsic substrate specificities of key enzymes are largely responsible for the fidelity of editing. Conversely, the efficiency of editing is enhanced by assembling enzymes and RNA binding proteins into stable multiprotein complexes.
Keywords: RNA editing, Trypanosoma, mitochondria, core editing complex, RNase III, TUTase, exonuclease, RNA ligase, double-stranded RNA, RNA binding proteins
INTRODUCTION
The term “RNA editing” was introduced by Benne and co-workers to describe the insertion of four uridylates correcting the frameshift in the cytochrome c oxidase subunit 2 (CO2) mRNA.1 Subsequent discoveries of massive U-insertion2 and U-deletion3 editing stimulated efforts to find the information source directing these highly-specific changes in RNA sequence. By allowing for non-canonical G-U base-pairing, Simpson and co-workers established that short 3′ uridylated RNAs molecules, astutely termed guide RNAs (gRNAs), are the vehicles of information transfer between maxicircle and minicircle genomes.4, 5 The ensuing “enzymatic cascade”4 and “transesterification” 6, 7 models provided a fruitful foundation for development of in vitro systems. Ultimately, the enzyme cascade model was confirmed by direct visualization of gRNA-directed mRNA cleavage followed by U-insertions or U-deletions, and then by mRNA ligation.8, 9 A biochemical “tour de force” by several laboratories led to purification of the RNA editing core complex (RECC), also referred to as the 20S editosome, and identification of its ~20 stably-associated subunits.10-14 Concurrently, a number of auxiliary factors, such as terminal uridylyl transferase (TUTase) responsible for gRNA 3′ uridylation (RNA editing TUTase 1, RET1 15, 16), RNA chaperones (MRP1/2)17-19, and others have been identified (reviewed in 20, 21).
The editing cascade is initiated upon hybridization of the gRNA's “5′ anchor” region to the pre-edited mRNA just downstream of the first editing site. The editing site selection is accomplished mostly by Watson-Crick base-pairing and may be facilitated by RNA annealing activities. The exact role of the oligo[U] tail remains to be determined. The U-tail is neither required for the in vitro editing8 nor is essential for gRNA stability in vivo;22 however these 15-20 Us may participate in stabilizing gRNA-mRNA binding. Partial complementarity of pre-edited mRNA and gRNA upstream of the “anchor” region results in mismatches that represent two types of substrates. At U-deletion sites unpaired uridines form bulges in the mRNA while insertion sites are characterized by unpaired purines in the gRNA (Figure 1). The editing cascade commences with an endonucleolytic mRNA cleavage by RNase III-like enzymes. The cut is introduced immediately upstream of the “anchor” double helix leaving a free 3′ hydroxyl at the 5′ mRNA fragment and a phosphate at the 5′ end of the 3′ mRNA fragment (Figure 1). During the second step, 3′-5′ exonuclease removes unpaired uridylyl residues from the 5′ fragment23 or RNA editing TUTase 2 (RET2) adds Us to the 5′ cleavage fragment according to the number of guiding purine nucleotides in the gRNA.16, 24 At the final step, the correctly trimmed or extended 5′ fragment and the 3′ fragment are bridged by gRNA thus creating an optimal substrate for the RNA ligase.25-27 Within the editing domain (mRNA fragment covered by overlapping gRNAs), editing events directed by the first gRNA often create a binding site for next guide RNA. This hierarchical mode determines the overall 3′-5′ (mRNA) polarity of editing28 and stipulates that gRNAs must be somehow displaced from the edited mRNA and prevented from re-binding.
Figure 1.
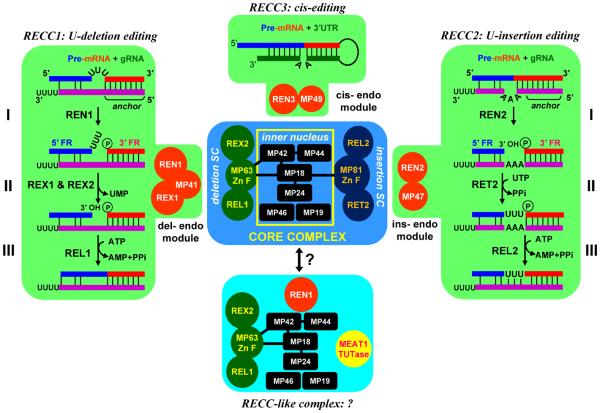
Modular organization of the RNA editing core complex (RECC). Direct protein-protein interactions within core complex are depicted by overlapping squares or black bars. SC, subcomplex; FR: mRNA cleavage fragment. Trans-guided insertion and deletion, and cis-guided insertion pathways are diagramed on the same background as corresponding endonuclease modules.
Although U-insertion/deletion editing is phylogenetically-confined to kinetoplastids extensive studies of this process brought insights into fundamental biological processes, such as small RNA-directed mRNA cleavage, and led to the identification of novel enzymatic activities, such as terminal uridylyl transferases (TUTases). Here we focus on recent advances that deepened our understanding of the core editing complex structure/function. We also review the factors essential for gRNA processing and stability and proteins affecting non-enzymatic editing steps.
COMPOSITION AND ORGANIZATION OF THE RNA EDITING CORE COMPLEX (RECC, ~20S Editosome)
The unambiguous definition of RNA editing complexes remains challenging despite a decade of intense studies. One of the main problems is the technical difficulty of distinguishing protein-protein interactions (PPIs) between complex subunits from transient interactions of individual proteins, or their subsets, with relatively stable core complexes. More trivial issues, such as the definition of “the editosome,” as well as the confusion resulting from assigning different names to the same gene product have made the communication within and outside of editing community difficult. The nomenclature proposed in Ref.29 is used throughout this review.
The available data are consistent with modular organization of editing complexes which follows the general principles derived from large-scale PPI studies in yeast.30, 31 General means of describing a complex assume the existence of core proteins (present in all preparations of the complex), attachment proteins (present in some, but not all preparations), and modules (subsets of co-purifying attachment proteins). These distinctions are based on frequency of protein's occurrence in cross-tagging, affinity purification and mass spectrometric analyses. It is not unusual when different attachments or modules confer a specific biological role to a particular isoform of a core complex. Thus, a multifunctional complex often appears to contain an excess of subunits for any given function, while in fact this reflects the existence of several forms of the core complex that differ by specific function for which not all subunits may be required.
A conventional column purification of an RNA ligase activity provided initial indications that the editing complex encompasses both U-insertion and U-deletion pathways.14 Rapidly developing affinity purification and mass spectrometric approaches revealed an apparent redundancy of enzymatic components in editing complexes from Trypanosoma brucei10-12 and Leishmania tarentolae.13 Three endonucleases with RNase III and double-stranded RNA binding motifs (RNA editing endonucleases, REN), two 3′-5′ exonucleases bearing exo/endo/phosphatase (EEP) motifs (RNA editing exonucleases, REX), a single TUTase (RNA editing TUTase 2, RET2), and two ligases (RNA editing ligase, REL) more than fulfilled the predictions of the enzymatic cascade model4 (Figure 1). Indeed, these enzymes were rapidly recognized as effectors of gRNA-directed mRNA cleavage, U-deletion from the 5′ mRNA fragment or U-addition to the 5′ fragment, and ligation, respectively.20, 21, 32 Just as perceptible was the potential logistic problem of maintaining competing activities (U-deletion and U-insertion) and divergent endonucleases within a single particle. The fact that a gRNA typically directs several interspersed insertion and deletion events further complicated the matter: an updated editing model would have had to invoke either a pluripotent core complex acting on insertion and deletion sites, or postulate the existence of site-specific isoforms of the core complex. The latter conundrum is also linked to a processivity of editing within a region covered by a single guide RNA (editing block). It seems plausible that a pluripotent complex would catalyze a processive editing, i.e., editing of more than one site upon a single binding event.33 Conversely, site-specific complexes are more likely to act distributively and would be characterized by rapid dissociation kinetics. Recent work suggests that a spatial clustering of deletion/ligation and insertion/ligation enzymes around the ‘inner nucleus’ and deployment of editing type-specific endonuclease modules constitute the basic principles of the editing core complex organization (Figure 1).
Definition of Sub-complexes and the ‘Inner Nucleus’ Within the Core Complex
Converging evidence from affinity purifications,13, 34 pair-wise protein-protein interactions studies34, 35 and RNAi knockdowns of individual components16, 36 firmly established that the core complex contains two tripartite subcomplexes (Figure 1). The U-deletion sub-complex is composed of REX2 exonuclease and the REL1 ligase, each of which is directly bound to the structural protein MP63. Interestingly, in L. tarentolae the homolog of REX2 lacks the EEP domain and is catalytically inactive.13, 37, 38 Indeed, the predominantly structural role for this protein is consistent with a partial loss of REL1 and MP63 upon REX2 RNAi repression.37, 38 Although within the core complex MP63 specifically binds to and stimulates the enzymatic activities of REX2 and REL1,34, 37-39 the full-round U-deletion editing could be reconstituted with unassociated recombinant REN1, REX1 and REL1.40 This indicates that RNA substrate specificities of individual enzymes are chiefly responsible for the fidelity of editing. MP63 is the only component of the deletion sub-complex that is engaged in direct interactions with the ‘inner nucleus’ via contacts with MP42 and MP1835, 41 (Figure 1). Predictably, MP63 is critical for the assembly of the entire core complex.42
Very similar observations have been made for the U-insertion subcomplex. The zinc finger-containing structural protein MP81 directly binds RET2 TUTase and REL2 ligase, thus stimulating both enzymatic activites.24, 34 MP81, similarly to MP63, is essential for the assembly of the entire core complex36, 43, 44 in which it interacts with MP18.34, 35 Remarkably, loss of the U-insertion subcomplex due to repression of RET2 led to a complete elimination of the U-insertion activity while leaving the U-deletion/ligation cascade unaffected.16 Collectively, these observations indicate a spatial clustering of insertion and deletion enzymes around the scaffolding proteins which are directly bound to the ‘inner nucleus’ subunits (Figure 1). The protein-protein interaction landscape within subcomplexes may be even more extensive. For example, the relative abundance of MP63 decreases upon REX2 knockdown38 and the MP81 is largely lost in RET2 RNAi cells,16 indicating a mutual dependence of subcomplex components for assembly into a core complex.
MP63 and MP81 play critical roles in RECC assembly, but the nature of their specific stimulating effects on exonuclease/ligase or TUTase/ligase enzymatic activities remains uncertain. Most enzymes have been produced recombinantly and demonstrated to maintain similar substrate specificities as the core complex-embedded subunits.37, 39, 41, 45-49 Even thought X-ray crystallography revealed apparently stable and ordered structures for REL145 and RET2,46 additional structural studies of reconstituted subcomplexes are necessary to determine whether MP81 and MP63 participate in RNA binding or in stabilizing active enzyme conformations.
Possible Enzymatic and RNA Binding Activities of ‘Inner Nucleus’ Subunits
Five out of the inner nucleus' six components have been assessed for their roles in complex formation; for some RNA and protein binding properties have been also reported. The unifying theme among four of these proteins (MP42, MP24, MP19 and MP18) is the presence of the OB-fold, a conserved domain typically associated with nucleic acid binding (Figure 2).50 The inherent difficulty in distinguishing scaffolding functions within the core complex from binding RNA substrates led to a number of controversies. RNAi knockdowns or gene knockouts of each of these polypeptides tend to disrupt the entire core complex, while placing affinity tags on overexpressed proteins can interfere with their functions51, 52 as is the case for the extensively studied MP42.51-53 In addition to RNA substrate binding it has been proposed that MP42 bridges deletion and insertion subcomplexes via OB-fold mediated contacts with MP44, MP18 and MP63.35, 52 Although the OB-fold and C2H2-zinc fingers motifs are typically associated with protein-nucleic acid or protein-protein interactions, the reported zinc-dependent endo-exo ribonuclease activity of MP42 could not be readily attributed to a discernable catalytic domain.54-56 MP42 preferentially cleaves single-stranded uracil bulges leaving 3′-phosphorylated end, a product incompatible with ligation reaction. It remains to be established whether such nuclease activity and the putative core complex-associated 3′ nucleotidyl phosphatase, which may potentially regenerate the hydroxyl group,56 are involved in the editing process.
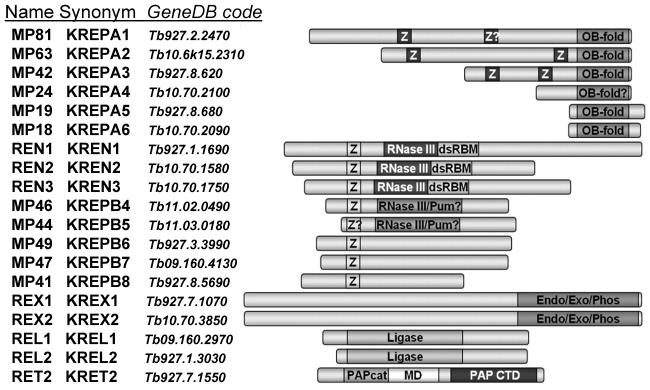
Figure 2.
Components of the RNA editing core complex from T. brucei. Domain organization is reproduced with permission from Ref.35 Pum: Pumilio motif; PAPcat: catalytic domain shared among members of DNA polymerase β superfamily;136 MD: middle domain; PAP CTD: C-terminal base recognition domain.46 White Z on dark background: C2H2-type zinc finger; OB-fold: oligonucleotide binding fold. Black Z on light background: U1-like C2H2 zinc finger; RNase III: RNase III catalytic-like domain; dsRBM: double-stranded RNA binding motif; RNase III-Pum: overlapping RNase III-like and Pumilio motifs; Endo-Exo-Phos: endonuclease-exonuclease-phosphatase family motif. Ligase: polynucleotide ligase-mRNA capping catalytic domain.
Another OB-fold containing protein MP24 may also play a dual role as a scaffolding component of the core complex and an RNA binding subunit. MP24 directly binds to MP1835 and its ablation disrupts the entire core complex.57 In vitro, MP24's OB-fold has been linked to high-affinity binding of uridylated gRNAs. Similar observations regarding the core complex integrity and RNA biding specificity have been made for MP18,58, 59 whereas MP46 evidently acts solely as a structural protein.60
Site-Specific Endonuclease Modules Enable Deletion, Insertion and Cis-Guided Editing
The low efficiency of a guide RNA-directed mRNA cleavage and the subsequent dissociation of the 5′ mRNA fragment have long been recognized as limiting factors of in vitro editing systems.8, 9, 61 Post-cleavage dissociation of the 5′ fragment likely leads to gRNA's U-tail ligation to the 3′ fragment (Figure 1). Formation of such gRNA-mRNA “chimeras” represents a non-productive reaction, which is quite efficient in vitro and was also observed in vivo.62-64 These considerations are further illustrated by the high efficiency of the “pre-cleaved” RNA ligation25 and U-insertion/ligation and U-deletion/ligation26, 27, 65 reactions. Here, the products of endonucleolytic cleavage such as those produced at step I, are synthesized and annealed with a gRNA into a tripartite duplex (Figure 1). Artificially extended double-stranded regions allow the gRNA to maintain the ‘business ends’ of both 5′ and 3′ mRNA fragments in close proximity enabling insertion/ligation and deletion/ ligation steps to proceed with high efficiency. These observations suggest that the initial cleavage is kinetically, and perhaps structurally, detached from downstream reactions and that dedicated mechanisms are engaged to compensate for the entropy cost of tripartite RNA scaffolding in vivo.
The molecular details of cleavage reactions emerged from the knockout/knockin studies demonstrating that repression of REN1,66 REN267 or REN368 selectively abolishes U-deletion, U-insertion, or cis-guided editing, respectively. The integrity of the core complex was not compromised in these genetic backgrounds. However, mitochondrial extracts derived from the knockout cell lines lacked endonuclease activities specific for deletion-, insertion-, and cis-editing substrates. In agreement with a modular model (Figure 1), affinity tagging of REN1, REN2, and REN3 produced complexes that contained all core components, yet were distinct due to the exclusive presence of REN1/REX1/MP41, REN2/MP47 or REN3/MP49 modules, respectively. The question remains whether the non-catalytic module components MP41, MP47 and MP49 are responsible for docking the endonucleases into the core complex and whether each module docks into discrete or overlapping sites.
In T. brucei both REX1 (essential for cell viability, part of the deletion endonuclease module) and REX2 (non-essential for cell viability, part of the deletion subcomplex) possess indistinguishable single-stranded U-specific 3′-5′ exonuclease activities, which can be attributed to EEP domains.37, 38 Together with REX1/REN1 comprising the U-deletion module, these results suggest the predominantly structural role for REX2 in deletion sub-complex assembly while REX1 is more likely to carry out U-removal. The tentative “separation of functions” is consistent with the more severe growth phenotype observed in dual REX1/REX2 RNAi dual knockdown cell lines when compared to individual knockdowns.38 However, the possibility that REX1 may compensate for the loss of REX2, but not vice versa, should not be discounted yet. A similar situation has been documented for editing ligases. REL1 is essential, while REL2 is dispensable for RNA editing and cell viability.43, 47, 69-71 The question remains whether REX1 and REL1 are capable of binding to their counterparts' docking sites or, alternatively, whether their position within the complex allows them to act on non-cognate substrates. Considering the highly specific nature of MP63- and MP81-mediated binary interactions and the stimulating effects on bound enzymes, the latter scenario seems more plausible.
Intradomain Interactions and Stoichiometry of the Editing Core Complex
The RNA editing core complex constitutes an interesting ensemble of enzymes, which may give clues to the evolutionary origins of RNA editing. The “construction tool kit” used to build multiprotein complexes is less apparent. It appears that the function-specific domains or extensions have been acquired to integrate the pre-existing catalytic domains into a core complex and to confer editing-specific RNA binding properties. The lack of direct contacts among editing enzymes suggests that scaffolding proteins are responsible for assembly and maintenance of the functional architecture. Interactions between structural components of the deletion subcomplex (MP63), insertion subcomplex (MP81), and the “inner nucleus” are chiefly mediated via OB-folds and/or zinc finger motifs.35, 51, 54, 57, 58 Specific interactions between OB-folds from different proteins are responsible for subcomplexes joining with the ‘inner nucleus’ as well as the assembly of the ‘inner nucleus’ itself.35 Thus, in addition to their demonstrated roles in RNA binding,57, 58, 72 the β-barrel OB-folds50 play a prominent role as PPI interfaces.35 The roles of C2H2 ZnF motifs have been investigated for MP63 in L. tarentolae41 and for MP42 in T. brucei51, 52 by overexpression or inducible knock-ins of metal-binding cysteine mutants. Disruptions of ZnFs adjacent to the OB-fold in MP63 produced a growth inhibition phenotype and breakdown of the entire complex while effects of inactivating the N-terminal ZnF were less severe. In MP42, both ZnF motifs are essential for cell viability, but they may also be involved in RNA binding.
From the enzymatic perspective, several lines of evidence suggest that non-catalytic domains may affect RNA substrate specificities that are critical for fidelity of all three editing steps (Figure 1). Although the 3′-5′ exonuclease activity has been unequivocally assigned to the C-terminal EEP domain,37, 38, 73 the EEP's intrinsic exonuclease activity is non-specific; the N-terminal extension is required to confer a specificity for single-stranded RNAs terminating with Us.74 Next, the recombinant RET2 inserts up to three uracils in a guide RNA-dependent reaction which also requires the presence of a phosphate group at the 5′ end of the 3′ cleavage fragment (Figure 1, step II). A core complex-embedded RET2, however, processively fills gaps approaching the length of maximal U-insertions observed in vivo (11-12 Us).75 This stimulation of RET2 activity is most likely caused by the middle domain (MD)-dependent46 association with MP81.24, 34, 35, 76 The middle domain, which has no similarities to other nucleotidyl transferases (reviewed in77), also constitutes part of the RNA binding interface in RET2.76 Thus, the middle domain in RET2 typifies a dual-function motif that confers both core complex association and affinity for double-stranded RNA to an otherwise highly conserved TUTase catalytic module (Figure 4). The structural basis of MP81-dependent stimulation of RET2 activity remains to be firmly established. At this point its participation in RNA binding seems more plausible than affecting RET2 conformation. Finally, a collective picture from yeast two-hybrid assays34 co-expression in bacteria,35 core complex complementation48, and crystallographic studies45 implicate the C-terminal domain of REL1 ligase as a docking interface with MP63 protein.
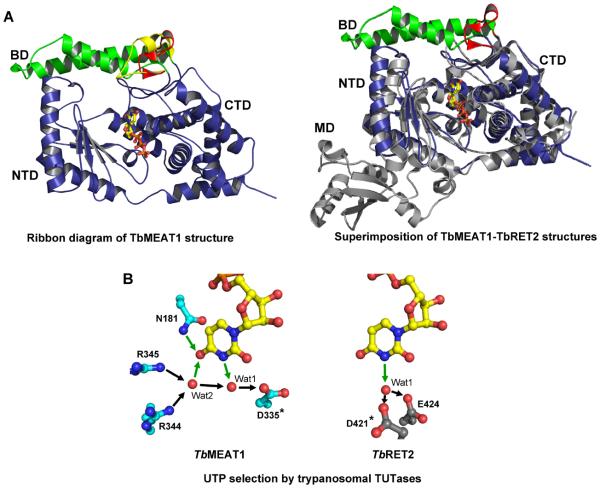
Figure 4.
Domain organization and UTP selectivity of mitochondrial TUTases. A. Ribbon representation illustrating the UTP bound in the deep cleft formed by the NTD and the CTD (left panel). Structural superpositions of TbMEAT1 with TbRET2 (gray) depict the overall differences in these structures, which become increasingly evident away from the active site (right panel). B. Mechanisms of UTP specificity. Hydrogen bonds involved in uracil-base-specific interactions are depicted as green arrows. Invariant aspartic acid residues are indicated by asterisks.
Although the precise stoichiometry of the core complex subunits remains to be determined, the majority of the subunits are present as single copy per core assembly. Depending on purification procedure and analytical methods, the molecular mass of a purified complex varies between 1 and 1.2 MDa78, which is consistent with combined masses of all subunits. Several issues, however, are still unresolved. For example, the oligomerization of the small OB-fold containing MP18 protein has been proposed to explain its high abundance in purified complexes,12 multiple contacts within ‘inner nucleus’35 and cooperative RNA binging activity.58 Indeed, crystallographic and biochemical analyses of the recombinant MP18 demonstrated its tetrameric organization in solution.79 Affinity tagging also suggested that some subcomplex components (REL113 and MP6341) may be present in more then a single copy, but the functional significance of these observations is unclear.
Perhaps the most intriguing question is whether REN1, 2 and 3 nucleases follow the RNase III paradigm of forming the active site via inter- or intramolecular catalytic motif dimerization (reviewed in 80). The domain architectures of REN1, 2 and 381 resemble that of the Class 1 RNase III with a single ribonuclease domain adjacent to a dsRNA binding domain (dsRBD). In these typically bacterial or fungal enzymes, the two nuclease domains dimerize to form a long surface cleft serving as a single dsRNA processing center with two active sites at opposite ends of a the ‘catalytic valley.’ Importantly, each domain contributes to the cleavage of one RNA strand, hence two staggered cuts of the dsRNA substrate are produced.82, 83 Available data suggest that only the mRNA is cleaved in mRNA-gRNA imperfect duplexes and the cleavage is introduced immediately upstream of the continuous double-stranded region. Therefore, the stoichiometry of REN1-3 within respective endonucleolytic modules may determine the catalytic mechanism: a monomeric enzyme would be very unusual for the RNase III superfamily whereas one of the active sites must be somehow inactivated in a dimer.
Three-dimensional Models of the RNA Editing Core Complex
Recent efforts combined affinity purification and size fractionation to isolate editing complexes that were sufficiently pure and homogenous for electron microscopy. These major technological advances produced low-resolution structures of the ~20S core complexes from T. brucei84 and L. tarentolae.78 In addition, the less defined 35S-40S particles have been characterized in T. brucei.84 The resultant core complex models are not entirely consistent with each other, most likely because of somewhat disparate sets of orthologous proteins present in final fractions, organism-specific variations, and different imaging techniques (Figure 3).
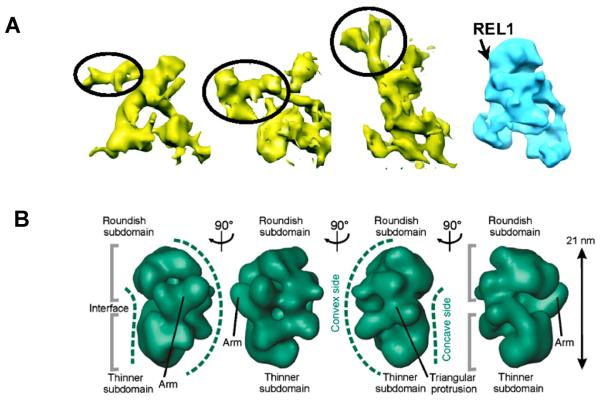
Figure 3.
Low-resolution structures of the editing core complex. A. Reconstitution of the RECC from L. tarentolae. Shaded surface representation of 3 individual anti-REL1 IgG-decorated L-complex particles segmented out from the 3D tomogram (in yellow). The putative IgG densities are indicated by circling. Approximate location of REL1 on unbiased single-particle reconstruction (in blue) is shown by arrow. Reproduced with permission from Ref. 78 B. Consensus model of the T. brucei ~20S complex displaying a bipartite shape. Landmarks are labeled. Reproduced with permission from Ref.84
The 20S particles from T. brucei showed several structural classes with the majority falling into bipartite shapes that may represent deletion and insertion subcomplexes interfaced by the inner nucleus. Partial occupancy of docking sites for endonucleolytic modules and distinct shapes of these modules may have further contributed to topological heterogeneity. The L. tarentolae core complex was more homogenous, thus allowing for single particle reconstitution and averaged molecular tomography models to converge on a single representative architecture. Globular Apex and Base regions have been distinguished from porous Central region within the roughly triangular global structure (~20 × 14 × 8 nm, Figure 3). Simpson and coworkers suggested that channels in the Central region represent possible RNA trafficking paths. Finally, the REL1 ligase was localized to the base of the Apex region by decorating the L. tarentolae core complex with a specific antibody. Such combination of immunochemical and structural approaches is clearly on target to map 3D-positionig of the core complex subunits.
RECC-LIKE COMPLEX
An alternative form of the core complex has been discovered by its association with a third mitochondrial TUTase, MEAT1 (mitochondrial editosome-like complex associated TUTase 1).85 MEAT1 and RET2 are mutually exclusive in their respective complexes, which otherwise share several components. In the MEAT1-associated particle, the entire U-insertion subcomplex is effectively replaced by MEAT1. Accordingly, MEAT1 does not bind to MP81, which forms a direct contact with RET2 in the “canonical” core complex (Figure 1). Similarly to RET2, MEAT1 is exclusively U-specific in vitro and is active on gapped double-stranded RNAs. However, MEAT1 does not require a 5′ phosphate group on the 3′ mRNA cleavage fragment produced by editing endonucleases. RNAi-based knock-in experiments showed that MEAT1 activity is essential for viability of bloodstream and insect parasite forms. However, RNA analyses revealed no gross RNA editing defects in MEAT1-depleted cells,85 suggesting that RET2 is responsible for the majority of U-insertions. It is still conceivable that MEAT1 acts on a small subset of editing sites, but its function in other processes may not be excluded.
The exclusive U-specificity and activity on double-stranded RNAs distinguish RET2 and MEAT1 from RET1,86 TUT387 and TUT488, 89 TUTases which can incorporate limited amounts of CMP and prefer single-stranded RNA substrates. These profound differences bring up interesting questions whether the mechanisms of UTP selection and protein domains responsible for docking into their respective complexes are conserved between RET2 and MEAT1. Notwithstanding a low protein sequence similarity, the high resolution X-ray crystal structures of RET246 and MEAT190 revealed that catalytic bi-domains formed by the N-terminal (NTD) and C-terminal (CTD) domains are similar between these two TUTases (Figure 4). In MEAT1, however, CTD contains an insertion of two α-helices, which form a unique bridge domain (BD) extending across the top of the UTP-binding cleft and interacting with the N-terminal domain. In agreement with the hypothesis that RET2 and MEAT1 interact with different proteins in their respective complexes, MEAT1 lacks the middle domain (MD), which in RET2 is responsible for RET2-MP81 binding.75, 76 Likewise, the mechanism of UTP selection appears to be only partially conserved. In RET2, carboxylate residues coordinate a water molecule to accept a hydrogen bond from position 3 of the uracil base while in MEAT1 an additional hydrogen bonds network involves an oxygen in position 4 (Figure 4). Collectively, the RET2- MEAT1 disparity may point to an evolutionary independent recruitment of distinct enzymes to similar complexes thus enabling divergent functions.
RNA EDITING FACTORS
The defined composition, presence of functional motifs, and stabile protein-protein interactions between subunits were the key to understanding RECC's structure and function. The emerging filed of editing factors, however, is often complicated by the RNA-mediated nature of their interactions with RECC, by the multi-functionality of individual proteins and by the lack of discernable motifs. Furthermore, recent studies have revealed important links between editing, pre-gRNA and pre-mRNA processing, mRNA polyadenylation, and translation.91 Here we survey protein factors implicated in editing and examine experimental findings supporting their possible functions.
RNA Editing TUTase 1 (RET1)
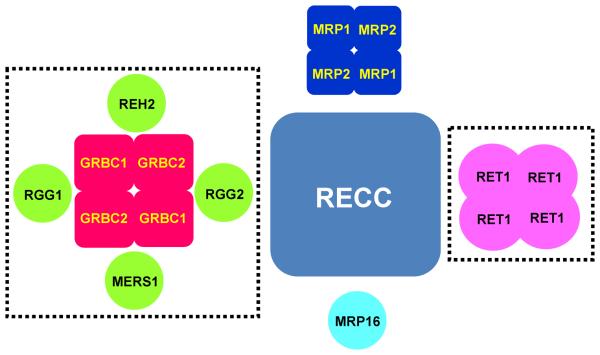
Early discoveries of oligo(U)-tails in gRNAs92 and rRNAs93 led to the detection,94 and purification and gene cloning15 of the first RNA uridylyl transferase. Termed RNA Editing TUTase 1, this enzyme was initially associated with 3′ gRNA uridylation.12, 13 RET1 RNAi knockdown triggered gRNA shortening and concomitant decrease in abundance,16 which is consistent with an inhibitory effect on editing in vivo. Although RET1 repression affects gRNA biogenesis, the mechanism of editing blockade was unclear. The U-tail's participation in the editing process (direct effect) and its requirement for gRNA maintenance in mitochondria (indirect effect) are among the most obvious culprits. Interactions of the U-tail with purine-rich pre-edited mRNA may serve mechanistic purpose,92 which is supported by stabilizing effect of this contact on gRNA-mRNA hybridization in vitro.95-97 In addition, the U-tail may function as a landing pad for protein factors; indeed, a high-affinity for uridylated RNAs has been reported for RET1 itself15, 86 and for several RNA binding proteins.98-101 However, the U-tail is not required for in vitro editing reactions.102 Thus, the participation of the U-tail in the editing process in vivo remains possible, but further work is needed to understand its role, if any, in the editing process.
On the other hand, the U-tail does not play a significant role in gRNA stabilization. High-resolution in vivo stability assays demonstrated that the loss of gRNAs upon RET1 repression is induced by inhibited gRNA precursor processing rather than accelerated decay of non-uridylated gRNAs.22 Unlike maxicircle-encoded mRNA precursors, which span thousands of bases, gRNAs are transcribed predominantly from minicircles as uniformly-sized (~800 nt) precursors. Mature gRNAs maintain nascent 5′ triphosphates, indicating that processing removes the long 3′ trailer. Although the exact mechanism of RET1-controlled pre-gRNA processing remains to be elucidated, it can be concluded that the TUTase activity is required for both 3′ end precursor processing and uridylation of mature gRNAs. Apparently, the U-tail is rapidly degraded unless it is continuously rebuilt by RET1 while the non-uridylated gRNAs remain relatively stable by virtue of their binding to GRBC1/2 complex22, 103 (see below). A model has been proposed in which anti-sense uridylylated transcripts originating from the opposite minicircle strand direct the pre-gRNA cleavage.22, 91
MRP1/2 Complex
Mitochondrial RNA binding proteins 1 and 2 have been identified independently in T. brucei by UV-induced crosslinking with gRNA (MRP1, originally termed gBP2117), in C. fasciculata as poly(U) binding proteins98 and in L. tarentolae via crosslinking to double-stranded RNA resembling the U-deletion site.19 Extensive biochemical and structural studies concluded that MPR1 and MRP2 assemble into a ~100 kDa α2β2 heterotetramer (dimer of αβ dimers), which binds both single- and double-stranded RNAs with high affinity through nonsequence-specific electrostatic interactions.19, 104 These RNA binding properties are manifested by MRP's RNA annealing activity, making this complex an attractive candidate for promoting gRNA binding to cognate mRNA targets.18, 19, 105 RNA-mediated associations between MRP1/2 , the core editing complex,19, 106 and the gRNA binding complex (GRBC,107 see below) lend additional support to MRP1/2's role in the editing process. However, RNAi knockdowns of MRP1 and MRP2 suggested a more complicated situation: while few edited mRNAs (Cyb and, to lesser extent, RPS12) were downregulated, neither gRNA abundance nor the majority of edited transcripts was affected by MRP1/2 depletion. Remarkably, the never-edited CO1 mRNA was also downregulated,107, 108 which clearly expands RNA targets of this abundant mitochondrial RNA binding complex beyond editing. While much has been learned about the structure and in vitro activities of MRP1/2, the definitive conclusions that stand a critical analysis are few: 1) most, if not all, of MRP1/2 interactions are RNA-mediated; 2) MRP1/2 function(s) in mitochondrial mRNA metabolism somehow affect editing and/or stability of select transcripts.
PBP16
One of the most extensively studied mitochondrial RNA binding proteins, RBP16, belongs to a Y-box protein family and is composed of the N-terminal cold shock and C-terminal RG-rich domains.99 RNAi knockdown studies revealed surprising similarities between mRNA sets negatively affected by MRP1/2 and RBP16 knockdowns. Edited Cyb mRNA, but not other edited transcripts, were severely downregulated while CO1 and ND4 never-edited transcripts also declined.109 Some of RBP16 in vitro properties, such as high-affinity RNA binding, associates with RNA editing substrates in mitochondrial extracts,100, 110, 111 stimulation of RNA annealing112 and in vitro editing113 would be consistent with its participation in the editing process. A possible direct involvement of RBP16 in editing, however, remains to be reconciled with the lack of uniform effects on all edited mRNAs. It seems more plausible that both MRP1/2 and RPB16 exert stabilizing, possibly redundant, effects on the stability of some transcripts, such as never-edited CO1, ND4, and moderately-edited MURF2 mRNAs. The comparative analysis of dual MRP1/2, single RBP16, and triple MRP1/2-RBP16 RNAi knockdowns illustrated that these factors may act as synergists or antagonists in a transcript-specific manner. Additive stabilizing effects of MRP1/2 and RBP16 were observed for edited Cyb mRNA. Conversely, individual removal of either MRP1/2 or RBP16 led to the decline of pan-edited RPS12 mRNAs while a triple knockdown partially restored the mRNA loss.114 With each new study adding another level of complexity, a superficial similarity in phenotypes, in vitro properties, and the lack of stable associations with defined complexes highlight the need for a deeper understanding of MRP1/2 and RBP16 functions. To this end, identification of RNAs bound to MRP1/2 and RBP16 in vivo may be a promising approach to gain mechanistic insights into some of the longest sagas in the editing field.
RGG-box proteins
Two mitochondrial RNA biding proteins with RGG (arginine-glycine-rich) motifs have been identified in T. brucei: TbRGG1 by serendipitous cross-hybridization with nucleolin-specific probe101 and TbRGG2 via co-purification with MRB1 complex (see below).115 Affinity tagging and mass spectrometry analysis of TbRGG1 revealed a significant proteomics overlap with the MRB1 complex.116 Although the interaction networks remain to be established for TbRGG1 and TbRGG2, their contacts with high-molecular mass complexes include both protein-protein and RNA-mediated interactions. RNAi knockdowns of TbRGG1 and TbRGG2 produced similar phenotypes and downregulated edited mRNAs without significantly affecting never-edited mRNAs.116, 117 Because neither gRNA levels nor the integrity of the core editing complex were altered by knockdowns, their functions may belong to poorly understood, non-catalytic steps of the editing process (mRNA binding, gRNA recruitment, stabilization of gRNA-mRNA interaction etc.) or edited mRNA stabilization. Along these lines, TbRGG2 was shown to predominantly affect pan-edited mRNA by stimulating editing progression beyond intrinsic pauses, which typically correspond to sites edited by gRNA's 3′ regions.118
Guide RNA Binding Complex (GRBC)
The early studies of high-molecular mass complexes established that gRNAs are maintained as ribonucleoprotein particles ranging from 10S to 50S.119-121 Several RNA binding proteins have been identified and investigated to various degrees (MRP1/2, RBP16 and RGG box proteins). Although these proteins associate with gRNAs in mitochondrial extracts, their genetic repression did not affect gRNA levels. Of course, functional gRNA-protein interactions during gRNA binding, editing reactions, and gRNA displacement may not necessarily alter the stability of the mature gRNA, but the existence of proteins responsible for stabilizing these short unstructured molecules has been anticipated since gRNA discovery. The first two proteins (associated proteins 1 and 2, AP1 and AP2) that carry out this function have been initially identified in L. tarentolae by co-purification with the MRP1/2 complex and gRNAs.19 In addition to AP1 and AP2, which are homologous to each other but show little similarity to proteins outside of Kinetoplastida, a third associated protein (AP3) with a pronounced NUDIX motif (nucleotide diphosphate linked to an X moiety) was identified in the same complex. Members of the NUDIX superfamily catalyze the hydrolysis of a pyrophosphate attached to a wide variety of nucleosides and polynucleotides.122 Subsequent studies of orthologous proteins in T. brucei demonstrated that AP1 and 2, renamed gRNA binding complex subunits 1 and 2 (GRBC 1 and 2) are required for gRNA stability and, therefore, editing.107 GRBC1 and 2 form a stable α2β2 heterotetramer of ~200 kDa, which was reconstituted from recombinant proteins and demonstrated to bind gRNAs directly, without any preference for the 3′ U-tail. Because the RET1-added U-tail does not significantly affect gRNA stability, it appears that binding to the GRBC1/2 particle is required and sufficient for gRNA stabilization.22 The RNAi of AP3 NUDIX hydrolase, renamed mitochondrial edited RNA stability factor 1 (MERS1), decreased the abundance of edited mRNAs, but the exact function of this factor remains to be determined.
The set of L. tarentolae GRBC1/2-associated proteins overlaps with the mitochondrial (KPAP1) polyadenylation complex,107, 123 T. brucei mitochondrial RNA binding complex 1 (MRB1)115 and TbRGG1-associated proteins.124 The PPI network involving GRBC1/2 remains to be firmly established, but it seems likely that GRBC1/2-associated proteins and MRB1 represent the same particle. A RECC-like situation in which a “GRBC core complex” interacts with several modules is consistent with limited available data. For example, RNA-mediated contacts are responsible for MERS1 and MRP1/2 co-purification with GRBC1/2.107 In contrast with RECC, most polypeptides detected with high confidence in GRBC lack functional motifs. A detailed analysis is necessary to ascertain whether the GRBC1/2 tetramer represent the sole gRNA binding interface of the GRBC assembly.
RNA Helicases
RNA helicases participate in virtually every RNA processing pathway and U-insertion/deletion editing is no exception. The prominent role of double-stranded RNA formation during initiation and progression of editing, a post-editing unwinding of gRNA-mRNA duplexes and the displacement of RNA-bound protein complexes are only few steps requiring the participation of these ‘professional remodelers.’ To date, two (DExH)-box proteins have been shown to participate in the production of edited mRNAs: mHel61p,125 renamed RNA editing helicase 1 (REH1)29 and RNA editing helicase 2 (REH2).124, 126 REH1 has been detected in some preparations of the core editing complex127 while REH2 co-purifies with GRBC1/2 (MRB1).107, 116, 124, 126 For both helicases RNA-mediated interactions with respective editing complexes are stable enough to withstand affinity purification and their genetic repression causes a specific loss of edited mRNAs. The mechanistic details of REH1's involvement in editing remain to be elucidated while certain observations hint at REH2's involvement in gRNA biogenesis. Two independent studies utilized the labeling of primary transcripts with vaccinia virus guanylyl transferase to detect a REH2 RNAi-triggered decrease of short mitochondrial RNAs.124, 126 These results, however, remain to be substantiated by alternative methods because a short 5′ triphosphorylated mitochondrial RNAs include not only gRNAs, but also molecules on unknown functions.128, 129 In addition, the guanylyl transferase-based detection method reflects a collective 5′ phosphorylation state of short RNAs rather than the abundance of specific gRNA species. In any event, continued investigation of (DExH)-box proteins participation in editing is bound to bring about major advances in our fragmented understanding of editing dynamics.
CONCLUSION
Mitochondrial U-insertion/deletion editing remains one of the most fascinating RNA processing pathways in nature. The complexity of multisubunit assemblies enacting mRNA editing is further exacerbated by their dynamic interactions and functional coupling with upstream and downstream RNA processing events, and translation.130 A conservative estimate of nuclear-encoded proteins required to transcribe, process, and translate eighteen mitochondrial mRNAs exceeds this number by at least ten fold. Despite the recent progress in functional studies, we are only beginning to understand the composition of the RNA editing holoenzyme and how it functions in the context of mitochondrial genome expression. Some important questions remain unresolved.
First, what are those intrinsically stable particles that constitute the RNA editing holoenzyme? In vitro, the ~20S RNA editing core complex catalyzes only a single round of mRNA cleavage, insertion or deletion, and ligation with low efficiency. However, in vivo hundreds of Us are added and deleted in a highly-processive manner. Several groups observed RECC's RNA-dependent participation in particles as large as 50S.84, 120, 131 The missing “half” is unknown, but it appears that the gRNA binding complex (GRBC) may be a promising candidate.
The second problem is generation of RNA substrates for the editing process. The intimate connection between mRNA 3′ processing and editing is manifested by differential effects of mRNA polyadenylation on the stability of pre-edited and edited mRNAs. The short (20-30 nt) A-tails are necessary and sufficient to stabilize transcripts edited beyond few initial sites, but they are dispensable for pre-edited mRNA maintanance.123, 132, 133 Although polycistronic maxicircle transcripts may be edited prior to nucleolytic processing,134 it appears that once the 3′ ends are generated by cleavage such non-adenylated edited molecules likely become very unstable. These observations may reflect the presence of a quality control system that ensures a sequential order of events in which a precursor cleavage is followed by adenylation and only then by editing.
The details of guide RNA biogenesis are even sketchier. The ~800 nt precursor undergoes 3′ processing to remove a long trailer prior to U-tail addition by RET1,22, 135 but the mechanisms defining the mature 3′end and limiting RET1 processivity are unknown. Identification of GRBC1/2 in the polyadenylation (KPAP1) complex123 established physical link between gRNA processing and polyadenylation machineries but the functional significance of these interactions is poorly understood.
To conclude, early studies of mRNA editing introduced the fundamental concepts and discovered the new enzymes which came to prominence as RNA biology progressed in other systems. The editing field has now entered the phase of tedious mechanistic studies but new surprises may still lie ahead.
Figure 5.
Major RNA editing complexes and factors. GRBC- and RET1-centered assemblies are likely to be more complex and participate in biogenesis of editing substrates.
Acknowledgements
We thank Larry Simpson, Ken Stuart, Kestrel Rogers and Jason Carnes for discussions. We thank Dmitri Maslov and Klemens Hertel for reading the manuscript. The work in authors' laboratory is supported by NIH grants AI091914 and AI083863.
References
- 1.Benne R, Van den Burg J, Brakenhoff J, Sloof P, Van Boom J, Tromp M. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 2.Feagin JE, Abraham J, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 3.Shaw J, Feagin JE, Stuart K, Simpson L. Editing of mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988;53:401–411. doi: 10.1016/0092-8674(88)90160-2. [DOI] [PubMed] [Google Scholar]
- 4.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 5.Sturm NR, Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–884. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- 6.Blum B, Sturm NR, Simpson AM, Simpson L. Chimeric gRNA-mRNA molecules with oligo(U) tails covalently linked at sites of RNA editing suggest that U addition occurs by transesterification. Cell. 1991;65:543–550. doi: 10.1016/0092-8674(91)90087-f. [DOI] [PubMed] [Google Scholar]
- 7.Cech TR. RNA editing: World's smallest introns. Cell. 1991;64:667–669. doi: 10.1016/0092-8674(91)90494-j. [DOI] [PubMed] [Google Scholar]
- 8.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 9.Kable ML, Seiwert SD, Heidmann S, Stuart K. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA [see comments] Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- 10.Panigrahi AK, Gygi SP, Ernst NL, Igo RP, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, Stuart KD. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panigrahi AK, Schnaufer A, Carmean N, Igo RP, Jr., Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, Stuart KD. Four Related Proteins of the Trypanosoma brucei RNA Editing Complex. Mol Cell Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 16.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koller J, Muller UF, Schmid B, Missel A, Kruft V, Stuart K, Goringer HU. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 1997;272:3749–3757. doi: 10.1074/jbc.272.6.3749. [DOI] [PubMed] [Google Scholar]
- 18.Muller UF, Lambert L, Goringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Aphasizheva I, Aphasizhev R. RET1-catalyzed Uridylylation Shapes the Mitochondrial Transcriptome in Trypanosoma brucei. Mol. Cell. Biol. 2010;30:1555–1567. doi: 10.1128/MCB.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst NL, Panicucci B, Igo RP, Jr., Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 25.Blanc V, Aphasizhev R, Alfonzo JD, Simpson L. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J. Biol. Chem. 1999;274:24289–24296. doi: 10.1074/jbc.274.34.24289. [DOI] [PubMed] [Google Scholar]
- 26.Igo RP, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igo RP, Jr., Lawson SD, Stuart K. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol Cell Biol. 2002;22:1567–1576. doi: 10.1128/mcb.22.5.1567-1576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maslov DA, Simpson L. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell. 1992;70:459–467. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- 29.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St OP, Ghanny S, Lam MH, Butland G, taf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 31.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 32.Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alatortsev VS, Cruz-Reyes J, Zhelonkina AG, Sollner-Webb B. Trypanosoma brucei RNA editing: coupled cycles of U deletion reveal processive activity of the editing complex. Mol Cell Biol. 2008;28:2437–2445. doi: 10.1128/MCB.01886-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. Separate Insertion and Deletion Subcomplexes of the Trypanosoma brucei RNA Editing Complex. Mol Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 35.Schnaufer A, Wu M, Park YJ, Nakai T, Deng J, Proff R, Hol WG, Stuart KD. A protein-protein interaction map of trypanosome ~20S editosomes. J. Biol. Chem. 2010;285:5282–5295. doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drozdz M, Palazzo SS, Salavati R, O'Rear J, Clayton C, Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers K, Gao G, Simpson L. Uridylate-specific 3′ 5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J. Biol. Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- 38.Ernst NL, Panicucci B, Carnes J, Stuart K. Differential functions of two editosome exoUases in Trypanosoma brucei. RNA. 2009;15:947–957. doi: 10.1261/rna.1373009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao G, Rogers K, Li F, Guo Q, Osato D, Zhou SX, Falick AM, Simpson L. Uridine insertion/deletion RNA editing in Trypanosomatids: specific stimulation in vitro of Leishmania tarentolae REL1 RNA ligase activity by the MP63 zinc finger protein. Protist. 2010;161:489–496. doi: 10.1016/j.protis.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang X, Gao G, Rogers K, Falick AM, Zhou S, Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang X, Falick AM, Nelson RE, Gao G, Rogers K, Aphasizhev R, Simpson L. Disruption of the zinc finger motifs in the Leishmania tarentolae LC-4 (=TbMP63) L-complex editing protein affects the stability of the L-complex 6. J. Biol. Chem. 2004;279:3893–3899. doi: 10.1074/jbc.M310185200. [DOI] [PubMed] [Google Scholar]
- 42.Huang CE, O'Hearn SF, Sollner-Webb B. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Hearn SF, Huang CE, Hemann M, Zhelonkina A, Sollner-Webb B. Trypanosoma brucei RNA editing complex: band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Law JA, Huang CE, O'Hearn SF, Sollner-Webb B. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol Cell Biol. 2005;25:2785–2794. doi: 10.1128/MCB.25.7.2785-2794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 46.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao G, Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- 48.Gao G, Simpson AM, Kang X, Rogers K, Nebohacova M, Li F, Simpson L. Functional complementation of Trypanosoma brucei RNA in vitro editing with recombinant RNA ligase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4712–4717. doi: 10.1073/pnas.0500553102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aphasizhev R, Aphasizheva I. RNA Editing Uridylyltransferases of Trypanosomatids. Methods Enzymol. 2007;424:51–67. doi: 10.1016/S0076-6879(07)24003-0. [DOI] [PubMed] [Google Scholar]
- 50.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo X, Ernst NL, Stuart KD. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei. Mol Cell Biol. 2008;28:6939–6953. doi: 10.1128/MCB.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo X, Ernst NL, Carnes J, Stuart KD. The zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei. PLoS. ONE. 2010;5:e8913. doi: 10.1371/journal.pone.0008913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Law JA, O'Hearn SF, Sollner-Webb B. Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion. RNA. 2008;14:1187–1200. doi: 10.1261/rna.899508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brecht M, Niemann M, Schluter E, Muller UF, Stuart K, Goringer HU. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 55.Niemann M, Brecht M, Schluter E, Weitzel K, Zacharias M, Goringer HU. TbMP42 is a structure-sensitive ribonuclease that likely follows a metal ion catalysis mechanism. Nucleic Acids Res. 2008;36:4465–4473. doi: 10.1093/nar/gkn410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niemann M, Kaibel H, Schluter E, Weitzel K, Brecht M, Goringer HU. Kinetoplastid RNA editing involves a 3′ nucleotidyl phosphatase activity. Nucleic Acids Res. 2009;37:1897–1906. doi: 10.1093/nar/gkp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salavati R, Ernst NL, O'Rear J, Gilliam T, Tarun S, Jr, Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarun SZ, Jr., Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Law JA, O'Hearn S, Sollner-Webb B. In Trypanosoma brucei RNA editing, TbMP18 (band VII) is critical for editosome integrity and for both insertional and deletional cleavages. Mol. Cell Biol. 2007;27:777–787. doi: 10.1128/MCB.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babbarwal VK, Fleck M, Ernst NL, Schnaufer A, Stuart K. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei. RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Byrne EM, Connell GJ, Simpson L. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 1996;15:6758–6765. [PMC free article] [PubMed] [Google Scholar]
- 62.Harris ME, Hajduk SL. Kinetoplastid RNA editing: In vitro formation of cytochrome b gRNA-mRNA chimeras from synthetic substrate RNAs. Cell. 1992;68:1091–1099. doi: 10.1016/0092-8674(92)90080-v. [DOI] [PubMed] [Google Scholar]
- 63.Sabatini R, Hajduk SL. RNA ligase and its involvement in guide RNA/mRNA chimera formation. J. Biol. Chem. 1995;270:7233–7240. doi: 10.1074/jbc.270.13.7233. [DOI] [PubMed] [Google Scholar]
- 64.Rusche LN, Piller KJ, Sollner-Webb B. Guide RNA-mRNA chimeras, which are potential RNA editing intermediates, are formed by endonuclease and RNA ligase in a trypanosome mitochondrial extract. Mol. Cell Biol. 1995;15:2933–2941. doi: 10.1128/mcb.15.6.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Igo, Weston DS, Ernst NL, Panigrahi AK, Salavati R, Stuart K. Role of uridylylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukar. Cell. 2002;1:112–118. doi: 10.1128/EC.1.1.112-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 67.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA Editing in Trypanosoma brucei requires three different editosomes. Mol. Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2161. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 70.Huang CE, Cruz-Reyes J, Zhelonkina AG, O'Hearn S, Wirtz E, Sollner-Webb B. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J. 2001;20:4694–4703. doi: 10.1093/emboj/20.17.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cruz-Reyes J, Zhelonkina AG, Huang CE, Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell. Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kala S, Salavati R. OB-fold domain of KREPA4 mediates high-affinity interaction with guide RNA and possesses annealing activity. RNA. 2010;16:1951–1967. doi: 10.1261/rna.2124610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ringpis GE, Aphasizheva I, Wang X, Huang L, Lathrop RH, Hatfield GW, Aphasizhev R. Mechanism of U Insertion RNA Editing in Trypanosome Mitochondria: The Bimodal TUTase Activity of the Core Complex. J. Mol. Biol. 2010;399:680–695. doi: 10.1016/j.jmb.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ringpis GE, Stagno J, Aphasizhev R. Mechanism of U-insertion RNA Editing in Trypanosome Mitochondria: Characterization of RET2 Functional Domains by Mutational Analysis. J. Mol Biol. 2010;399:696–706. doi: 10.1016/j.jmb.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aphasizhev R, Aphasizheva I. Terminal RNA uridylyltransferases of trypanosomes. Biochim. Biophys. Acta. 2008;1779:270–280. doi: 10.1016/j.bbagrm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li F, Ge P, Hui WH, Atanasov I, Rogers K, Guo Q, Osato D, Falick AM, Zhou ZH, Simpson L. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12306–12310. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu M, Park YJ, Pardon E, Turley S, Hayhurst A, Deng J, Steyaert J, Hol WG. Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies. J. Struct. Biol. 2010 doi: 10.1016/j.jsb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 81.Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blaszczyk J, Tropea JE, Bubunenko M, Routzahn KM, Waugh DS, Court DL, Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 84.Golas MM, Bohm C, Sander B, Effenberger K, Brecht M, Stark H, Goringer HU. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 2009;28:766–778. doi: 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aphasizheva I, Ringpis GE, Weng J, Gershon PD, Lathrop RH, Aphasizhev R. Novel TUTase associates with an editosome-like complex in mitochondria of Trypanosoma brucei. RNA. 2009;15:1322–1337. doi: 10.1261/rna.1538809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J. Biol. Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- 87.Aphasizhev R, Aphasizheva I, Simpson L. Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett. 2004;572:15–18. doi: 10.1016/j.febslet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 88.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual Role of the RNA Substrate in Selectivity and Catalysis by Terminal Uridylyl Transferases. Proc Natl Acad Sci U S A. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stagno J, Aphasizheva I, Bruystens J, Luecke H, Aphasizhev R. Structure of the mitochondrial editosome-like complex associated TUTase 1 reveals divergent mechanisms of UTP selection and domain organization. J. Mol Biol. 2010;399:464–475. doi: 10.1016/j.jmb.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aphasizhev R, Aphasizheva I. Mitochondrial RNA Processing In Trypanosomes. 2011 doi: 10.1016/j.resmic.2011.04.015. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blum B, Simpson L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo-(U) tail involved in recognition of the pre-edited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- 93.Adler BK, Harris ME, Bertrand KI, Hajduk SL. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol. Cell. Biol. 1991;11:5878–5884. doi: 10.1128/mcb.11.12.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bakalara N, Simpson AM, Simpson L. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J. Biol. Chem. 1989;264:18679–18686. [PubMed] [Google Scholar]
- 95.Leung SS, Koslowsky DJ. Mapping contacts between gRNA and mRNA in trypanosome RNA editing. Nucleic Acids Res. 1999;27:778–787. doi: 10.1093/nar/27.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leung SS, Koslowsky DJ. RNA editing in Trypanosoma brucei: characterization of gRNA U-tail interactions with partially edited mRNA substrates. Nucleic Acids Res. 2001;29:703–709. doi: 10.1093/nar/29.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leung SS, Koslowsky DJ. Interactions of mRNAs and gRNAs involved in trypanosome mitochondrial RNA editing: Structure probing of an mRNA bound to its cognate gRNA. RNA. 2001;7:1803–1816. [PMC free article] [PubMed] [Google Scholar]
- 98.Blom D, Burg J, Breek CK, Speijer D, Muijsers AO, Benne R. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 2001;29:2950–2962. doi: 10.1093/nar/29.14.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hayman ML, Read LK. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 1999;274:12067–12074. doi: 10.1074/jbc.274.17.12067. [DOI] [PubMed] [Google Scholar]
- 100.Pelletier M, Miller MM, Read LK. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 2000;28:1266–1275. doi: 10.1093/nar/28.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vanhamme L, Perez-Morga D, Marchal C, Speijer D, Lambert L, Geuskens M, Alexandre S, Ismaïli N, Göringer U, Benne R, Pays E. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 1998;273:21825–21833. doi: 10.1074/jbc.273.34.21825. [DOI] [PubMed] [Google Scholar]
- 102.Cruz-Reyes J, Zhelonkina A, Rusche L, Sollner-Webb B. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell Biol. 2001;21:884–892. doi: 10.1128/MCB.21.3.884-892.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ryan CM, Kao CY, Sleve DA, Read LK. Biphasic decay of guide RNAs in Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;146:68–77. doi: 10.1016/j.molbiopara.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 104.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 105.Muller UF, Goringer HU. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res. 2002;30:447–455. doi: 10.1093/nar/30.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Allen TE, Heidmann S, Reed R, Myler PJ, Goringer HU, Stuart KD. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol Cell Biol. 1998;18:6014–6022. doi: 10.1128/mcb.18.10.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R. Guide RNA-Binding Complex from Mitochondria of Trypanosomatids. Mol. Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vondruskova E, van den BJ, Zikova A, Ernst NL, Stuart K, Benne R, Lukes J. RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J. Biol. Chem. 2005;280:2429–2438. doi: 10.1074/jbc.M405933200. [DOI] [PubMed] [Google Scholar]
- 109.Pelletier M, Read LK. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA. 2003;9:457–468. doi: 10.1261/rna.2160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goulah CC, Read LK. Differential effects of arginine methylation on RBP16 mRNA binding, guide RNA (gRNA) binding, and gRNA-containing ribonucleoprotein complex (gRNP) formation. J. Biol. Chem. 2007;282:7181–7190. doi: 10.1074/jbc.M609485200. [DOI] [PubMed] [Google Scholar]
- 111.Miller MM, Read LK. Trypanosoma brucei: functions of RBP16 cold shock and RGG domains in macromolecular interactions. Exp. Parasitol. 2003;105:140–148. doi: 10.1016/j.exppara.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 112.Ammerman ML, Fisk JC, Read LK. gRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA. 2008 doi: 10.1261/rna.982908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miller MM, Halbig K, Cruz-Reyes J, Read LK. RBP16 stimulates trypanosome RNA editing in vitro at an early step in the editing reaction. RNA. 2006;12:1292–1303. doi: 10.1261/rna.2331506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fisk JC, Presnyak V, Ammerman ML, Read LK. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol. Cell Biol. 2009;29:5214–5225. doi: 10.1128/MCB.00520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. Mitochondrial complexes in trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell Proteomics. 2007;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 116.Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fisk JC, Ammerman ML, Presnyak V, Read LK. TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J. Biol. Chem. 2008;283:23016–23025. doi: 10.1074/jbc.M801021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA. 2010;16:2239–2251. doi: 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goringer HU, Koslowsky DJ, Morales TH, Stuart K. The formation of mitochondrial ribonucleoprotein complexes involving guide RNA molecules in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 1994;91:1776–1780. doi: 10.1073/pnas.91.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pollard VW, Harris ME, Hajduk SL. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992;11:4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peris M, Simpson AM, Grunstein J, Liliental JE, Frech GC, Simpson L. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol. Biochem. Parasitol. 1997;85:9–24. doi: 10.1016/s0166-6851(96)02795-8. [DOI] [PubMed] [Google Scholar]
- 122.McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Missel A, Souza AE, Norskau G, Goringer HU. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J. Biol. Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J. Am. Soc. Mass Spectrom. 2003;14:728–735. doi: 10.1016/S1044-0305(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 128.Madej MJ, Alfonzo JD, Huttenhofer A. Small ncRNA transcriptome analysis from kinetoplast mitochondria of Leishmania tarentolae. Nucleic Acids Res. 2007;35:1544–1554. doi: 10.1093/nar/gkm004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Madej MJ, Niemann M, Huttenhofer A, Goringer HU. Identification of novel guide RNAs from the mitochondria of Trypanosoma brucei. RNA Biol. 2008;5:84–91. doi: 10.4161/rna.5.2.6043. [DOI] [PubMed] [Google Scholar]
- 130.Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide Repeat Proteins Stimulate mRNA Adenylation/Uridylation to Activate Mitochondrial Translation in Trypanosomes. 2011 doi: 10.1016/j.molcel.2011.02.021. Ref Type: In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Osato D, Rogers K, Guo Q, Li F, Richmond G, Klug F, Simpson L. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: In search of the editosome. RNA. 2009;15:1338–1344. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ryan CM, Militello KT, Read LK. Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem. 2003;278:32753–32762. doi: 10.1074/jbc.M303552200. [DOI] [PubMed] [Google Scholar]
- 133.Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNAs in Trypanosoma brucei. Mol. Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koslowsky DJ, Yahampath G. Mitochondrial mRNA 3′ cleavage polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol. Biochem. Parasitol. 1997;90:81–94. doi: 10.1016/s0166-6851(97)00133-3. [DOI] [PubMed] [Google Scholar]
- 135.Grams J, McManus MT, Hajduk SL. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 2000;19:5525–5532. doi: 10.1093/emboj/19.20.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]