Abstract
Analogous to an assembly line, we employed a modular design for the high-throughput study of 1,536 structurally distinct nanoparticles with cationic cores and variable shells. This enabled elucidation of complexation, internalization, and delivery trends that could only be learned through evaluation of a large library. Using robotic automation, epoxide-functionalized block polymers were combinatorially cross-linked with a diverse library of amines, followed by measurement of molecular weight, diameter, RNA complexation, cellular internalization, and in vitro siRNA and pDNA delivery. Analysis revealed structure-function relationships and beneficial design guidelines, including a higher reactive block weight fraction, stoichiometric equivalence between epoxides and amines, and thin hydrophilic shells. Cross-linkers optimally possessed tertiary dimethylamine or piperazine groups and potential buffering capacity. Covalent cholesterol attachment allowed for transfection in vivo to liver hepatocytes in mice. The ability to tune the chemical nature of the core and shell may afford utility of these materials in additional applications.
Chemically diverse nanoparticles form the basis of a number of emerging applications in materials science (1–3), including drug and gene delivery vehicles (4, 5). However, it is difficult to predict the optimal chemical and physical properties for delivery of a specific drug or biomolecule. A key challenge in nanoparticle development is the need to perform distinct synthesis, characterization, and formulation steps before performance can be evaluated. Although combinatorial methods have driven small molecule drug discovery (6), they have been less explored for the discovery of polymers (7–12) in part due to these inherent challenges. A less studied area in high-throughput (HT) polymeric nanoparticle methodology (13–16) is the facile incorporation of diverse chemical groups into polymers with defined architectures. By applying HT robotic techniques to the controlled cross-linking of block copolymers, we were able to prepare a library of core-shell nanoparticles for nucleic acid complexation and delivery with great variability in the chemical nature of the protonizable amine-based core. The evaluation of a large library revealed that internalization and/or complexation alone is not sufficient for silencing and that certain chemical functionalities including tertiary dimethylamine or piperazine groups with potential buffering capacity may be advantageous for polymer-based delivery.
Advances in the controlled synthesis of polymers with functional group tolerance [particularly controlled/living radical polymerization (CRP) (17–20) methods that enable control over MW and architecture] and in robotic technology (8) motivated us to explore an experimental design based on modules where 96-well plates are shuffled between different HT instruments for synthesis, characterization, and screening. This process allows for examination of key physical and chemical properties in relation to performance. siRNA (21, 22) was selected as an initial agent for delivery; despite its marked potential to treat various diseases, safe and effective delivery remains the most significant barrier to broad clinical use (23). We previously utilized HT methods for the synthesis and screening of materials for delivery including lipids (15, 24) for siRNA delivery and linear poly(β-amino ester)s (PBAEs) (14, 25–27) for pDNA delivery. HT synthesis of star polymers (13) and linear polymers using CRP methods (10, 11, 19) has been reported but not applied for gene therapy where cationic charges are required to bind nucleic acids through electrostatic interactions. Modification of linear polymers for pDNA delivery has been described (12, 28); however, to our knowledge such polymers have not been shown to effectively deliver siRNA. There is a growing interest in sub-100 nm particles for delivery (4) due to the increased ability of smaller particles to access capillaries, penetrate tissues, and cross cell membranes. Because the possible size range for stable liposomes is generally > 50 nm (29), and because star shaped polymers (13, 30, 31) and dendrimers (32) are considerably smaller, we selected a core-shell system that allows for greater size range control and the ability to easily incorporate a multitude of different cationic groups into the core. Distinct polymer-based nanoparticles may have better stability than lipid-based systems and greater potential for attachment of targeting ligands (33).
To address these issues, we prepared a library of nanoparticles composed of cationic cross-linked nanogel cores and variable shells with precise control over particle size, chemical composition, and architecture. siRNA can complex to or inside of the core, while the arms provide a protective shield. To achieve the desired structure, we employed a method of cross-linking block copolymers (34, 35) prepared by reversible addition-fragmentation chain transfer (RAFT) polymerization (18). For cross-linking, we sought an efficient and versatile reaction that would permit library formation. While other click reactions (36) might have been suitable, we chose ring-opening of epoxides with amines (37–39) due to the availability of a large number of amines that can incorporate cationic charge into the core, and the efficiency of this catalyst-free reaction. By changing the physical properties of the core and shell, these particles could also find application in the delivery of proteins and drugs, assembled multilayers (40), electronics (41), as stabilizing particles in polymer blends (42), and catalytic scaffolds and supramolecular hosts (43, 44).
Results and Discussion
Library Design and Synthesis.
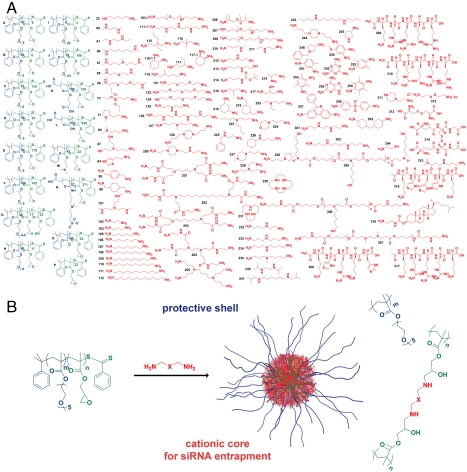
The rationale for employing the HT core-shell design was to create a cationic core to facilitate siRNA complexation, with variation in the nature of the protonizable amine, and a shell with variation in polymer length and chemical properties. Although many techniques exist for the formation of nanoparticles (31, 45–48), we required methods that were amenable to parallel HT synthesis. For this reason, we employed a method of reacting epoxide groups with amines (37–39, 49–51) that avoids the use of salts, surfactants, and mixed solvents. Instead of using homopolymers and random copolymers (38), we chose block copolymers that could form the core-shell morphology. The basic reaction for the synthesis of core-shell nanoparticles involved block copolymers consisting of one nonreactive block and one epoxide-functionalized block, such that the core could be cross-linked with a variety of amines (Fig. 1). These amines varied greatly in structure (linear, ring, branched), reactivity (1–4 reactive amine groups per molecule; 1°, 2°, and 3° amines within the molecule), and MW (small molecule, dendrimer, and polymer). Each amine could form a distinct and structurally unique nanoparticle starting from one block copolymer.
Fig. 1.
Combinatorial synthesis of core-shell nanoparticles. (A) Epoxide-containing block copolymers and amines were used to synthesize a combinatorial library of hairy core-shell nanoparticles. (B) Synthesis occurs via the ring-opening reaction of epoxides by amines forming β-hydroxyl groups and cross-linking points separated by the functionality (X) contained in the amine. Each nanoparticle is named by a letter corresponding to the starting block copolymer and a number corresponding to an amine.
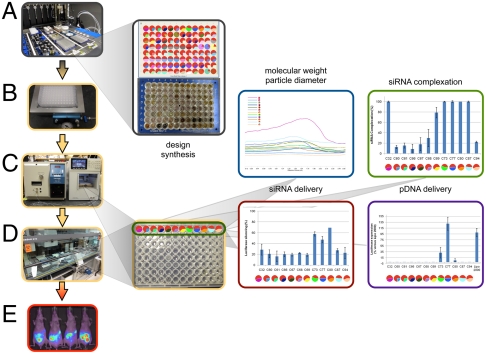
We synthesized a series of different block copolymers to serve as precursors for nanoparticle formation (Table S1). Block copolymers possessing poly(oligo(ethylene oxide) methacrylate) (POEOMA) with different lengths of the PEO side chain, may increase blood circulation time due to the PEO shell of the resulting nanoparticle. Anionic, cationic, zwitterionic, and hydrophobic blocks were also evaluated as shells. Utilizing the 96-well plate footprint, each block copolymer was reacted with 96 amines to form a set of core-shell nanoparticles. The HT approach enabled randomization of the core and shell for elucidation of the key chemical and material properties for intracellular delivery. To facilitate rapid synthesis, characterization, and screening, we developed a method where a microtiter plate was passed through a series of modules (Fig. 2). Periodic analysis by prescreening may lead to an improvement in chemical selection. In this study, we optimized the amine library by replacing the poorer performing amines with new candidate amines during the process. Automation of this process accelerated not only material handling, but also data manipulations and reaction calculations. The MW, density, drawn chemical structure, and dissolved concentration were entered into Library Studio to enable rapid calculation of reaction stoichiometry, taking into account the MW, density, structure (number of amines), and concentration. Through preliminary studies, we found that a mole ratio of 1∶1 of epoxides to 1° or 2° amines and a concentration in dimethyl sulfoxide (DMSO) of 100 mg/mL (approximately 300 mM) was optimal for nanoparticle formation with limited macroscopic gelation.
Fig. 2.
Modular design for the synthesis, characterization, and screening of a library of core-shell nanoparticles. (A) The synthesis of 1,536 core-shell nanoparticles was carried out on a Symyx fluid-handling robot inside of glass vials in a 96-well plate format. (B) The nanoparticles were purified and filtered through a HT filtering apparatus into a standard 96-well tissue culture plate. (C) The MW and particle size of the polymers were measured by a HT GPC system. (D) The same plate format was used for an RNA complexation assay, carried out on a Tecan cell culture robot. HT in vitro screens for siRNA and pDNA delivery were performed. (E) Biodistribution of C227 (left two images) and C80 (right two images) are shown. GPC, siRNA complexation data, and in vitro siRNA and pDNA delivery results with standard deviation (s.d.) for nanoparticles C32-C94 (positions A1-A12 in the plate) are presented.
Nanoparticle-Mediated Delivery in Vitro.
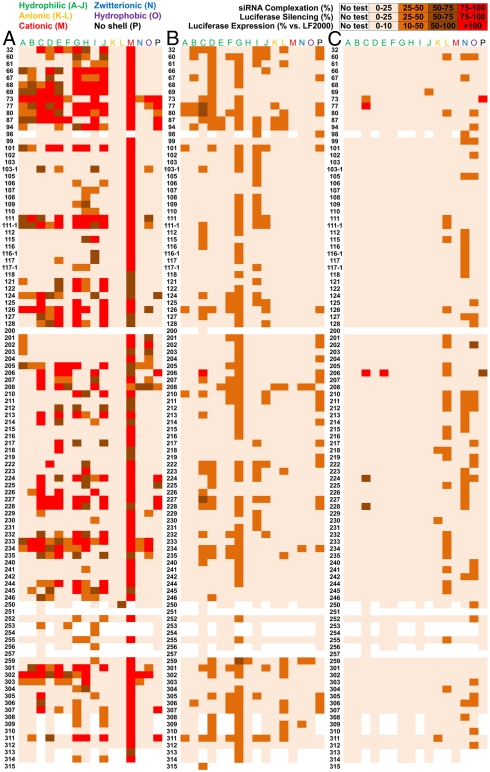
As an initial screen, we evaluated the ability of every nanoparticle to complex RNA (Fig. 3A). Automated methods were developed that allowed for evaluation of 50 microtiter plates per week. The degree of entrapment is expressed as a percentage based on measured fluorescence, where higher values in the figure indicate stronger complexation. For nanoparticles with PEO shells (blocks A–J), a range of complexation was observed that predominantly correlated with the amine. Structurally analogous amines showed that more protonizable nitrogens resulted in better complexation. For example, the linear polyamine 301 complexed well in most cases, whereas the linear aliphatic diamine 112 with similar length did not. Nanoparticles with anionic shells (blocks K and L) repelled the anionic phosphate backbone of siRNA, resulting in almost no measurable complexation. In contrast, nanoparticles with a cationic shell (block M) were able to complex siRNA in every case, implying that the siRNA is located in the shell (and possibly in the core) of these nanoparticles, whereas the siRNA is only complexing with the core of the other nanoparticles.
Fig. 3.
In vitro screening of core-shell nanoparticles. (A) The degree of RNA entrapment was quantified using the RiboGreen assay. The average percent of complexation is shown. (B) HeLa cells stably expressing both firefly and Renilla luciferases were treated with firefly targeting siRNA-nanoparticles complexes. The average percent reduction in firefly luciferase activity after treatment is shown. (C) HeLa cells were treated with pDNA(luc)-nanoparticle complexes. Activity using LF2000 was standardized to 100. The average percent activity versus LF2000 after treatment is shown. To provide a picture of the capability of all nanoparticles, the complexation, siRNA delivery, and pDNA experiments were performed in triplicate at a weight ratio of 50∶1 (nanoparticle:siRNA).
The libraries were then screened for the ability to productively deliver siRNA to a HeLa cell line that stably expressed both firefly and Renilla luciferase (Fig. 3B). Cells were treated with firefly luciferase-targeting siRNA(siLuc)-nanoparticle complexes and the ratio of firefly to Renilla luciferase expression was measured after 24 h. Reduction of only firefly luciferase demonstrates noncytotoxic specific silencing, whereas reduction in expression of both luciferase proteins indicates toxicity or other nonspecific effects. Following the modular HT approach, the complexes were formed in 96-well plates by simple mixing. Because the size, shape, and charge of the nanoparticles vary greatly in this library, we also evaluated them for pDNA delivery. Compared to siRNA, which is rigid and linear, pDNA is a few hundred times longer, circular, and more flexible. HeLa cells were transfected with pDNA coding for luciferase, and luciferase activity was measured after 24 h (Fig. 3C).
When considering the maximum siRNA knockdown for one amine core across all block copolymers, those nanoparticles that were expected to increase in charge as the pH drops with the transition from outside to inside of the endosome were more likely to effectively deliver siRNA. We modeled the microspecies pKas of the GMA-amine products in the core of the nanoparticles, estimating the relative buffering capacity that may be involved in endosomal escape (Table S2). The model system calculations suggest that 62 of the 102 amines will not gain charge during the expected drop in pH within the endosomal compartment. The remaining 40 amines are expected to exhibit buffering. Across all block types, 80 different amines enabled > 30% luciferase silencing in vitro. Considering only the criteria used to segregate buffering from nonbuffering amines, a significantly higher proportion of the modeled buffering amines (77%) are represented amongst the effective group than nonbuffering amines (14%) (Fig. S1).
Analysis of the results also revealed several trends regarding the block copolymer structure in nanoparticles capable of delivering siRNA in vitro. Considering nanoparticles with POEOMA shells (blocks A–J), nanoparticles formed from block C showed the highest knockdown. This was likely due to an optimal block ratio with a short POEOMA block [degree of polymerization (DP) 12] and a longer glycidyl methacrylate (GMA) block (DP 135). Block D, which also had a higher weight fraction of PGMA, demonstrated effective knockdown, although it was less capable of delivering pDNA. In contrast, nanoparticles formed from block H, which possessed a lower weight fraction of POEOMA (DP 48) compared to PGMA (DP 17) were unable to deliver siRNA or pDNA. This trend of weight fraction is likely related to resulting charge density, particle formation, and to surface PEO presentation. J had a long GMA block that resulted in some macroscopic gelation. F, which was a poorly formed block copolymer with high polydispersity, was not effective, suggesting that defined block architecture is important for nanoparticle formation. Many G nanoparticles could deliver siRNA due to their less dense core (via copolymerization of 2-hydroxyethyl methacrylate) but were not superior to the best nanoparticles formed from C. Nanoparticles formed from cationic blocks (H and M) and anionic blocks (K and L) did not effectively deliver siRNA. Some nanoparticles formed from I could deliver siRNA but not pDNA, possibly due to the shorter lengths of the blocks and correspondingly smaller size of the nanoparticles. The opposite trend was also observed, where some nanoparticles with a zwitterionic shell (N) or a hydrophobic shell (O) could deliver pDNA but not siRNA. To a small extent, a few nanoparticles with anionic shells (K and L) mediated transfection of pDNA; however, the anionic nanoparticles were largely ineffectual.
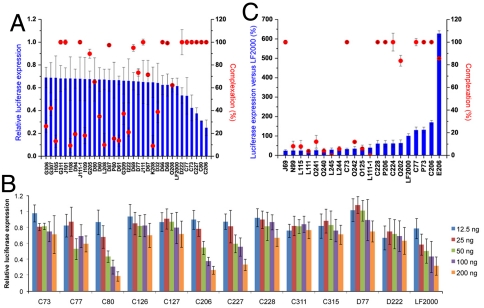
The performance of the top nanoparticles for siRNA delivery is detailed in Fig. 4A. In parallel experiments, these nanoparticles were as effective as Lipofectamine 2000 (LF2000), including C77, C80, C206, and C227, which were more effective at the given dose in the presence of serum. The best performing nanoparticles were able to effectively complex siRNA. Dose response data (Fig. 4B) revealed an increase in gene silencing as the dose increased. In the top 40 nanoparticles that silenced > 30% of firefly expression, five amines appeared in more than one nanoparticle (77, 80, 94, 208, and 222). The top 20 nanoparticles all had amines with 1–2 reactive groups. Having more than two reactive amines possibly led to interparticle cross-linking. E206 was the best nanoparticle for pDNA due to long block lengths and large size, where the cells expressed 625% more luciferase compared to cells transfected with LF2000 (Fig. 4C).
Fig. 4.
In vitro core-shell nanoparticle-mediated delivery trends. (A) RNA complexation and luciferase silencing after delivery of nanoparticle:siRNA complexes (50∶1, wt/wt) is expressed for the top performing nanoparticles. (B) siRNA dose response for the top nanoparticles. (C) Complexation and luciferase activity after delivery of nanoparticle:pDNA complexes (50∶1, wt/wt) is expressed for the top performing nanoparticles. (A–C) n = 4; s.d. is expressed.
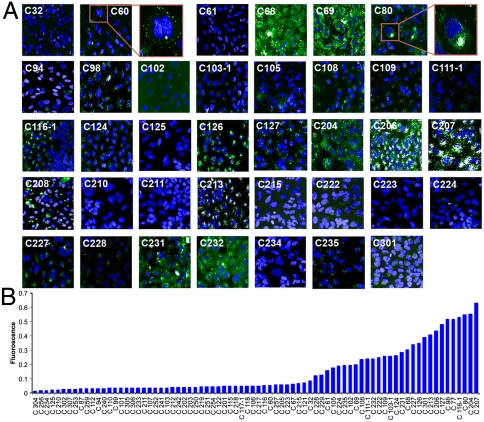
To examine the effect of amine structure on cellular internalization, we quantified the uptake after 1 h of incubation with each of the C-based nanoparticles complexed with fluorescent siRNA (Fig. 5) and used image analysis to identify key structural trends. Nanoparticles containing tertiary amines were more effectively internalized than nanoparticles with secondary amines. Within tertiary alkyl amines, dimethyl (80) > diethyl (222) > dibutyl (223), and within secondary alkyl amines, methyl (211) > ethyl (210). In addition to uptake, siRNA complexation and silencing efficiency was also improved for nanoparticles with pendant dimethyl tertiary amines. Similarly, nanoparticles with piperazine (227) and pyrrolidine (77) structures showed enhanced complexation, delivery, and internalization, as compared to morpholine (224) and imidazole (94) groups. Although complexation remained high for C224, the internalization decreased slightly and the siRNA silencing ability decreased greatly compared to optimal C227. Increased cellular uptake was observed for linear polyamines with internal methyl substitutions (e.g., 235 versus 234; 127 versus 125). PEG-based diamine cross-linkers (103-1, 206, 207, 208) enabled efficient nanoparticle formation, siRNA complexation, and high internalization. However, only C206 exhibited high siRNA silencing ability, possibly due the higher hydrophobicity and methyl group adjacent to the amines. Some nanoparticles (e.g., 207) mediate efficient internalization, but that alone is not sufficient for silencing. In general, incorporation of tertiary amines led to an improvement in complexation, cellular internalization, and siRNA silencing efficiency.
Fig. 5.
In vitro uptake screening. (A) Cellular internalization of selected nanoparticles after 1 hr of incubation is demonstrated by HT automated confocal microscopy. HeLa cells were exposed to all C-based nanoparticles complexed with Alexa-594 siRNA (50∶1, wt/wt). Twenty different fields were imaged and a representative image of nanoparticle/siRNA complex is presented (Alexa 594 is pseudocolored green). Additional images appear in Fig. S4. (B) The total fluorescence was quantified and plotted. False positives were identified by visible aggregation (e.g., C208 included for reference) or significant background and were removed from the graph. Interestingly, C60 nanoparticles interact with the cell membrane but did not enter cells, while C80 nanoparticles were effectively internalized (see zoom insets).
Characterization of the Core-Shell Nanoparticle Library.
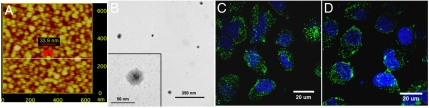
The entire core-shell nanoparticle library was characterized using HT GPC. In order to decrease the total analysis time from months to weeks, we utilized a GPC equipped with a single flow injection polymer analysis column and three detectors capable of measuring both MW and particle size. GPC traces, absolute MW, Rh, and Rg for all nanoparticles appear in Table S3. Preliminary studies were carried out before the full library was synthesized. To achieve the greatest flexibility in choice of monomers, we used cumyl dithiobenzoate as the RAFT chain transfer agent (CTA) in most cases. Conditions were investigated and optimized for the controlled homopolymerization of OEOMA and GMA (Table S4 and Fig. S2). The semilogarithmic plot of ln([M]0/[M]) versus time for the polymerization of GMA was linear, indicating a constant concentration of radicals. Mn also increased linearly with conversion. Chain extension of OEOMA from PGMA was examined, yielding block copolymers with a clean GPC shift in MW, and polydispersity below 1.3. GMA was also polymerized from POEOMA macro-CTAs. Kinetic studies showed trends of a controlled polymerization. In advance of forming nanoparticles, model ring-opening reactions with small molecules were carried out. At 1∶1 molar equivalents of epoxide to amine, reaction with piperidine and sodium azide resulted in complete opening of the epoxides (Fig. S2). To examine the effect of block length on nanoparticle formation, we synthesized a series of block copolymers that systematically varied in weight % of OEOMA and GMA blocks (Table S1). These block copolymers were reacted with different amines, and the particle size of the resulting nanoparticles was measured by DLS. Desired particle size (10–100 nm) was obtained for most block copolymers (15–85 weight % GMA). Atomic force microscopy (AFM) images of nanoparticles prepared by the reaction of FF with the linear aliphatic diamine 110 showed uniform particles with a diameter of 34 nm (Fig. 6A). Polyamines were also able to efficiently cross-link the block copolymers to form the hairy nanoparticle shape. The reaction of R or S with 126 formed particles with a diameter of about 21 nm, as measured by DLS, with the diameter measured by AFM slightly smaller (22.9 nm) for R126 than for S126 (25.4 nm) due to the slightly shorter GMA block (Fig. S3). To verify the core-shell structure, transmission electron microscopy (TEM) was employed. An examination of R126 (Fig. 6B) and C227 nanoparticles (Fig. S3E) revealed a higher electron density in the core, compared with the shell, indicating the core-cross-linked morphology. The measured diameter of C80 and C227 nanoparticles by DLS and TEM was in agreement (Table S5 and Fig. S3). In addition to the HT internalization studies, we examined cellular internalization by synthesizing FITC-labeled nanoparticles via reaction of the isothiocyanate group with dangling amines to covalently label the core. At higher magnification, C80-FITC and C227-FITC can be clearly visualized inside endosomes that are the hallmark for endocytic trafficking (Fig. 6 C and D). Because size and surface charge can also affect cellular uptake, the diameter and zeta potential of top performing nanoparticles were also measured (Table S5).
Fig. 6.
Characterization of core-shell nanoparticles. (A) Dried FF110 nanoparticles on mica showed uniform particle size around 34 nm by AFM. (B) TEM was utilized to visualize the core-shell structure of R126 nanoparticles. The insert clearly showed an electron dense cross-linked core, and a more electron loose shell. These TEM images are unstained. Additional AFM and TEM images appear in Fig. S3. Cell internalization of FITC-C80 (C) and FITC-C227 (D) nanoparticles after 1 hr of incubation was demonstrated by confocal microscopy. Punctate green fluorescence was observed within the cell.
In Vivo Core-Shell Nanoparticle-Mediated Delivery of siRNA to Hepatocytes in Mice.
Based on results from the in vitro screen, we examined the utility of top performing nanoparticles in facilitating siRNA delivery in vivo by employing the mouse Factor VII gene silencing model (15, 24). Factor VII is a useful gene target for the evaluation of hepatocyte-specific delivery because it is only produced in the cells of the liver parenchyma, possesses a relatively short plasma half-life, and is secreted into the blood, enabling facile protein quantification. siRNA-directed against the blood clotting Factor VII was complexed with nanoparticles at a weight ratio of 10∶1 (NP:siRNA) and delivered intravenously. Mouse body weight was monitored over the duration of the experiment because body weight loss can indicate toxicity associated with nanoparticle treatment.
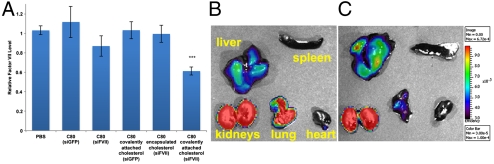
Since in vivo delivery to hepatocytes poses more challenges than the in vitro delivery to HeLa cells, we investigated the inclusion of cholesterol because covalent attachment of cholesterol to siRNA has been shown to improve in vivo delivery to the liver (52, 53). Of all the nanoparticles tested, C80 showed the most improvement after cholesterol attachment and was focused on for this study. 10 Mole % of the epoxide groups were reacted with N′-cholesteryloxycarbonyl-1,2-diaminoethane (318), followed by addition of 80 to stoichiometrically cross-link the remaining epoxide groups. Covalent attachment of cholesterol to C80 nanoparticles enabled 40% silencing of Factor VII (Fig. 7A). Noncovalent encapsulation of unmodified cholesterol had no measured effect. Delivery of control siGFP using these nanoparticles did not result in any measurable silencing, suggesting specific delivery to the liver and silencing of Factor VII. No appreciable weight loss was observed, indicating minimal toxicity. To probe the biodistribution of these nanoparticles with and without cholesterol in vivo, a single bolus dose of 1 mg/kg Cy5-labeled siRNA-nanoparticle complex was administered via tail vein injection (Fig. 7 B and C). C80-Cy5-siRNA nanoparticles were observed in the liver, kidneys, and lungs. The attachment of cholesterol resulted in increased liver accumulation.
Fig. 7.
In vivo silencing of Factor VII in liver hepatocytes. Nanoparticles were purified by dialysis, complexed with siRNA at a weight ratio of 10∶1 (nanoparticle:siRNA), and delivered intravenously to C57BL/6 mice. Mice received a single bolus administration of 5 mg/kg total siRNA via tail-vein injection and Factor VII levels were quantified 48 hr post injection. (A) Covalent attachment of cholesterol to C80 nanoparticles enabled silencing. Noncovalent encapsulation of cholesterol and delivery of control siGFP resulted in no knockdown. Data points represent group mean ± s.d. Data points marked with asterisks are significant relative to control treated groups (***, P < 0.0001; t-test, double-tailed, n = 14). Representative images of isolated organs for Cy5-siRNA-C80 (b) and Cy5-siRNA-C80-10 mol% cholesterol (C) nanoparticle complexes demonstrated nanoparticle accumulation in the liver, kidneys, and lungs after 120 min. The covalent attachment of cholesterol to C80 resulted in enhanced liver accumulation and reduced lung accumulation.
We believe that the development of this library of core-shell nanoparticles represents an important step forward for the HT synthesis of polymers for intracellular delivery. We aimed to move beyond simple linear polymers by exploring CRP for the formation of complex architectures with great chemical diversity in the core and shell. This synthetic method enables parallel generation of large libraries of chemically distinct core-shell materials and decreases the time from design to analysis. The common structural features of these materials suggest certain design criteria for creating future nanoparticles, including (i) a block weight ratio around 15% nonreactive block and 85% reactive block, (ii) a stoichiometic balance between epoxides and amines, (iii) thin water-soluble hydrophilic shells, (iv) tertiary amines with 1–2 reactive sites, and (v) amines with buffering capacity. Covalent attachment of cholesterol to the nanoparticle was required for in vivo delivery to the liver. Further studies are warranted to extend this technology for the broadest applications of RNAi therapy and drug delivery. We also desire to use what we have learned in this study to aid in the design of fully degradable nanoparticles that satisfy the aforementioned general properties and design criteria. Finally, we believe that the modular design utilized in this report is applicable to many other systems. By changing the composition of the blocks and cross-linkers, a variety of other nanoparticles could be formed with wide-ranging applications.
Methods
HT Nanoparticle Synthesis.
Robotic synthesis was performed on a customized Symyx Core Module equipped with four different liquid dispensing elements (a single tip, a motor driven gripper with a heated piercing tip, a positive displacement tip, and a heated parallel 4-tip), heating and cooling bay elements, a magnetic stirring element, a mass balance, and imaging equipment. Library Studio (Symyx Discovery Tools) was used to design the core-shell nanoparticle libraries. For each nanoparticle, a solution of amine (100 mg/mL in DMSO) was added to a stirring solution of block copolymer (100 mg/mL in DMSO) such that the mole ratio of epoxides to amine was 1∶1 and the total concentration of reactive components was on the order of 300 mM. Reaction blocks with 96 1 mL glass vials equipped with tumbling stir bars and covered with a rubber mat were used. The resulting mixtures were stirred and heated at 50 °C for 24 h on the deck of the robot.
HT RNA Complexation Assay.
The Quant-iT RiboGreen RNA assay (Invitrogen) was performed on a customized Tecan Freedom EVO 200 with 8-Tip, LiHa, MCA-96, and Extended RoMa elements. Fluorescence was measured using a coupled Infinite M200 multimode monochromator-based reader. The assay was programmed in EVOware Plus.
HT Confocal Microscopy.
15,000 HeLa cells were plated in black 96-well plates (Greiner Bio-one). Cells were starved for 30 min in the presence of PBS/BSA and pulsed for 1 h at 37 °C with 50 ng of BLOCK-iT Alexa Fluor Red Fluorescent Oligo (Invitrogen) complexed with the nanoparticle at a nanoparticle/siRNA ratio of 50∶1 (wt/wt). Cells were chased for 30 min in the presence of unlabeled nanoparticles and washed with PBS. Cells were then fixed using 4% paraformaldehyde and counterstained in PBS containing Hoescht (2 μg/mL) for nuclei identification. Cells were then imaged with an automated spinning disk confocal microscope (OPERA, Perkin Elmer). Quantitation of internalized fluorescently labeled siRNA was done using Acapella software. After identification of cell location and perimeter, intracellular siRNA signal intensity over the whole search region (single field) was calculated. Data presented are an average of intracellular intensity from 20 different fields plotted in Excel. The same defined pattern of fields from each well were acquired to eliminate bias during analysis. Quality control was performed on all images and large aggregates or nonspecific background seen in rare fields was excluded from analysis.
Supporting Information.
Additional descriptions of materials, instrumentation, modeling, and experimental details, including the synthesis of all block copolymers, are available in SI Text.
Supplementary Material
Acknowledgments.
The authors thank Boris Klebanov, Manos Karagiannis, Christian J. Kastrup, Avi Schroeder, Carmen Barnes, and Robert S. Siegwart for their help. This work was supported by Alnylam Pharmaceuticals and National Institutes of Health (NIH) Grants R01-EB000244-27 and 5-R01-CA132091-04. D.J.S. acknowledges postdoctoral support from NIH NRSA award F32-EB011867.
Footnotes
Conflict of interest statement: R.L. is a shareholder and member of the Scientific Advisory Board of Alnylam. D.G.A. is a consultant with Alnylam. R.L and D.G.A have sponsored research grants from Alnylam. Alnylam also has a license to certain intellectual property invented at MIT. W.Q., C.Z., and T.N. are employed by Alnylam.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106379108/-/DCSupplemental.
References
- 1.Balazs A, Emrick T, Russell T. Nanoparticle polymer composites: Where two small worlds meet. Science. 2006;314:1107–1110. doi: 10.1126/science.1130557. [DOI] [PubMed] [Google Scholar]
- 2.Taton T, Mirkin C, Letsinger R. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 3.Shipway A, Katz E, Willner I. Nanoparticle arrays on surfaces for electronic, optical, and sensor applications. ChemPhysChem. 2000;1:18–52. doi: 10.1002/1439-7641(20000804)1:1<18::AID-CPHC18>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.LaVan D, Lynn D, Langer R. Moving smaller in drug discovery and delivery. Nat Rev Drug Discov. 2002;1:77–84. doi: 10.1038/nrd707. [DOI] [PubMed] [Google Scholar]
- 5.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7(9):771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 6.Geysen H, Schoenen F, Wagner D, Wagner R. Combinatorial compound libraries for drug discovery: An ongoing challenge. Nat Rev Drug Discov. 2003;2:222–230. doi: 10.1038/nrd1035. [DOI] [PubMed] [Google Scholar]
- 7.Brocchini S, James K, Tangpasuthadol V, Kohn J. A combinatorial approach for polymer design. J Am Chem Soc. 1997;119:4553–4554. [Google Scholar]
- 8.Boussie T, et al. A fully integrated high-throughput screening methodology for the discovery of new polyolefin catalysts: Discovery of a new class of high temperature single-site group (IV) copolymerization catalysts. J Am Chem Soc. 2003;125:4306–4317. doi: 10.1021/ja020868k. [DOI] [PubMed] [Google Scholar]
- 9.Xiang XD, et al. A combinatorial approach to materials discovery. Science. 1995;268:1738–1740. doi: 10.1126/science.268.5218.1738. [DOI] [PubMed] [Google Scholar]
- 10.Hoogenboom R, Meier M, Schubert U. Combinatorial methods, automated synthesis and high-throughput screening in polymer research: Past and present. Macromol Rapid Commun. 2003;24:16–32. [Google Scholar]
- 11.Webster D. Combinatorial and high-throughput methods in macromolecular materials research and development. Macromol Chem Phys. 2008;209:237–246. [Google Scholar]
- 12.Wong S, Sood N, Putnam D. Combinatorial evaluation of cations, pH-sensitive and hydrophobic moieties for polymeric vector design. Mol Ther. 2009;17:480–490. doi: 10.1038/mt.2008.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosman AW, et al. High-throughput synthesis of nanoscale materials: Structural optimization of functionalized one-step star polymers. J Am Chem Soc. 2001;123:6461–6462. doi: 10.1021/ja010405z. [DOI] [PubMed] [Google Scholar]
- 14.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed Engl. 2003;42:3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 15.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hook A, et al. High throughput methods applied in biomaterial development and discovery. Biomaterials. 2010;31:187–198. doi: 10.1016/j.biomaterials.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 17.Braunecker WA, Matyjaszewski K. Controlled/living radical polymerization: Features, developments, and perspectives. Prog Polym Sci. 2007;32:93–146. [Google Scholar]
- 18.Moad G, Rizzardo E, Thang S. Living radical polymerization by the RAFT process. Aust J Chem. 2005;58:379–410. [Google Scholar]
- 19.Zhang H, Marin V, Fijten M, Schubert U. High-throughput experimentation in atom transfer radical polymerization: A general approach toward a directed design and understanding of optimal catalytic systems. J Polym Sci A Polym Chem. 2004;42:1876–1885. [Google Scholar]
- 20.Matyjaszewski K, Tsarevsky N. Nanostructured functional materials prepared by atom transfer radical polymerization. Nat Chem. 2009;1:276–288. doi: 10.1038/nchem.257. [DOI] [PubMed] [Google Scholar]
- 21.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 22.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 23.Whitehead K, Langer R, Anderson D. Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Love K, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynn DM, Anderson DG, Putnam D, Langer R. Accelerated discovery of synthetic transfection vectors: Parallel synthesis and screening of degradable polymer library. J Am Chem Soc. 2001;123:8155–8156. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 26.Anderson D, et al. A polymer library approach to suicide gene therapy for cancer. Proc Natl Acad Sci USA. 2004;101:16028–16033. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green J, Langer R, Anderson D. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun B, Liu X, Buck M, Lynn D. Azlactone-functionalized polymers as reactive templates for parallel polymer synthesis: Synthesis and screening of a small library of cationic polymers in the context of DNA delivery. Chem Commun. 2010;46:2016–2018. doi: 10.1039/b921664b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagayasu A, Uchiyama K, Kiwada H. The size of liposomes: A factor which affects their targeting efficiency to tumors and therapeutic activity of liposomal antitumor drugs. Adv Drug Delivery Rev. 1999;40:75–87. doi: 10.1016/s0169-409x(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 30.Zheng G, Pan C. Reversible addition-fragmentation transfer polymerization in nanosized micelles formed in situ. Macromolecules. 2006;39:95–102. [Google Scholar]
- 31.Gao H, Matyjaszewski K. Synthesis of functional polymers with controlled architecture by CRP of monomers in the presence of cross-linkers: From stars to gels. Prog Polym Sci. 2009;34:317–350. [Google Scholar]
- 32.Lee C, MacKay J, Frechet J, Szoka F. Designing dendrimers for biological applications. Nat Biotechnol. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 33.Davis M, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–U1140. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, Peng H, Jiang M. A novel one-step approach to core-stabilized nanoparticles at high solid contents. Macromolecules. 2003;36:2576–2578. [Google Scholar]
- 35.Peng H, Chen D, Jiang M. A one-pot approach to the preparation of organic core-shell nanoobjects with different morphologies. Macromolecules. 2005;38:3550–3553. [Google Scholar]
- 36.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 37.Tsarevsky NV, Bencherif SA, Matyjaszewski K. Graft copolymers by a combination of ATRP and two different consecutive click reactions. Macromolecules. 2007;40:4439–4445. [Google Scholar]
- 38.van der Ende A, Kravitz E, Harth E. Approach to formation of multifunctional polyester particles in controlled nanoscopic dimensions. J Am Chem Soc. 2008;130:8706–8713. doi: 10.1021/ja711417h. [DOI] [PubMed] [Google Scholar]
- 39.Kurkuri MD, et al. Multifunctional polymer coatings for cell microarray applications. Biomacromolecules. 2009;10:1163–1172. doi: 10.1021/bm801417s. [DOI] [PubMed] [Google Scholar]
- 40.Kim BS, Gao HF, Argun AA, Matyjaszewski K, Hammond PT. All-star polymer multilayers as pH-responsive nanofilms. Macromolecules. 2009;42:368–375. [Google Scholar]
- 41.Gao H, et al. Site isolation of emitters within cross-linked polymer nanoparticles for white electroluminescence. Nano Lett. 2010;10(4):1440–1444. doi: 10.1021/nl100347p. [DOI] [PubMed] [Google Scholar]
- 42.Stenzel M. Hairy Core-Shell Nanoparticles via RAFT: Where are the opportunities and where are the problems and challenges? Macromol Rapid Commun. 2009;30:1603–1624. doi: 10.1002/marc.200900180. [DOI] [PubMed] [Google Scholar]
- 43.Hecht S, Frechet J. Dendritic encapsulation of function: Applying nature’s site isolation principle from biomimetics to materials science. Angew Chem Int Ed Engl. 2001;40:74–91. doi: 10.1002/1521-3773(20010105)40:1<74::aid-anie74>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 44.Rodionov V, et al. Easy access to a family of polymer catalysts from modular star polymers. J Am Chem Soc. 2010;132:2570–2572. doi: 10.1021/ja9104842. [DOI] [PubMed] [Google Scholar]
- 45.Oh J, Drumright R, Siegwart D, Matyjaszewski K. The development of microgels/nanogels for drug delivery applications. Prog Polym Sci. 2008;33:448–477. [Google Scholar]
- 46.O’Reilly R, Hawker C, Wooley K. Cross-linked block copolymer micelles: Functional nanostructures of great potential and versatility. Chem Soc Rev. 2006;35:1068–1083. doi: 10.1039/b514858h. [DOI] [PubMed] [Google Scholar]
- 47.Zhang K, Fang H, Wang Z, Taylor J, Wooley K. Cationic shell-crosslinked knedel-like nanoparticles for highly efficient gene and oligonucleotide transfection of mammalian cells. Biomaterials. 2009;30:968–977. doi: 10.1016/j.biomaterials.2008.10.057. [DOI] [PubMed] [Google Scholar]
- 48.Matsumoto S, et al. Environment-responsive block copolymer micelles with a disulfide cross-linked core for enhanced siRNA delivery. Biomacromolecules. 2009;10:119–127. doi: 10.1021/bm800985e. [DOI] [PubMed] [Google Scholar]
- 49.Yin X, Stover H. Hydrogel microspheres by thermally induced coacervation of poly(N,N-dimethylacrylamide-co-glycidyl methacrylate) aqueous solutions. Macromolecules. 2003;36:9817–9822. [Google Scholar]
- 50.Suzuki D, Kawaguchi H. Modification of gold nanoparticle composite nanostructures using thermosensitive core-shell particles as a template. Langmuir. 2005;21:8175–8179. doi: 10.1021/la0504356. [DOI] [PubMed] [Google Scholar]
- 51.Hantzschel N, Zhang F, Eckert F, Pich A, Winnik M. Poly(N-vinylcaprolactam-co-glycidyI methacrylate) aqueous microgels labeled with fluorescent LaF3 : Eu nanoparticles. Langmuir. 2007;23:10793–10800. doi: 10.1021/la701691g. [DOI] [PubMed] [Google Scholar]
- 52.Lorenz C, Hadwiger P, John M, Vornlocher H, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg Med Chem Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 53.Wolfrum C, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.