Abstract
Type II DNA topoisomerases (topos) catalyse changes in DNA topology by passing one double-stranded DNA segment through another. This reaction is essential to processes such as replication and transcription, but carries with it the inherent danger of permanent double-strand break (DSB) formation. All type II topos hydrolyse ATP during their reactions; however, only DNA gyrase is able to harness the free energy of hydrolysis to drive DNA supercoiling, an energetically unfavourable process. A long-standing puzzle has been to understand why the majority of type II enzymes consume ATP to support reactions that do not require a net energy input. While certain type II topos are known to ‘simplify’ distributions of DNA topoisomers below thermodynamic equilibrium levels, the energy required for this process is very low, suggesting that this behaviour is not the principal reason for ATP hydrolysis. Instead, we propose that the energy of ATP hydrolysis is needed to control the separation of protein–protein interfaces and prevent the accidental formation of potentially mutagenic or cytotoxic DSBs. This interpretation has parallels with the actions of a variety of molecular machines that catalyse the conformational rearrangement of biological macromolecules.
INTRODUCTION
Type II DNA topoisomerases (topos) perform the remarkable feat of passing one double-stranded segment of DNA through a transient break in another (1). This reaction allows these enzymes to manipulate topological properties of DNA such as supercoiling, unknotting and decatenation (unlinking of DNA circles or loops) (2). DNA topoisomerases are classified into two types, I and II, depending on whether they catalyse reactions involving the breakage of one or both strands of the DNA (3,4). All known cellular organisms have at least one type II topo, whose prototypical reaction is generally considered to be the decatenation of daughter chromosomes after DNA replication. Successful replication and partitioning of chromosomes requires all the double-helical turns linking the parental strands to be removed. This process largely occurs by relaxing positive supercoils ahead of replication forks, which can in principle be carried out by either type I or type II topos (5,6). However, any rotation of the forks or incomplete relaxation also results, after replication is complete, in links and intertwinings between the daughter chromosomes that can only normally be removed by the double-strand passage reaction of type II topos (6). Recent work has also discussed the possible physiological importance of unknotting reactions (7).
Type II topos are divided into two classes on the basis of structural and evolutionary considerations (4). Type IIA enzymes are ubiquitous in eubacteria and eukaryotes and include bacterial DNA gyrase and topo IV, as well as eukaryotic topo IIs. The IIB topos (all designated topo VI) are distant relatives with some domains in common with the IIAs (8–11); they are found in archaea, plants and a few bacteria (12,13).
All type II topos hydrolyse ATP as part of their catalytic reaction cycle. This energetic requirement appeared natural and obvious when the first type II enzyme, DNA gyrase, was discovered in Escherichia coli (14). Gyrase uses nucleotide turnover to introduce negative supercoils into DNA, exploiting the free energy of ATP hydrolysis for the formation of thermodynamically unfavourable reaction products. In contrast, all other type II enzymes (including the type IIBs) catalyse reactions that do not have an obvious energetic cost, such as supercoil relaxation, knotting/unknotting and catenation/decatenation. Indeed, gyrase itself can catalyse the efficient relaxation of positively supercoiled DNA in an ATP-dependent reaction (15). Why type II topos have evolved to rely on a chemical energy source to catalyse otherwise thermodynamically favourable reactions remained a puzzle for many years.
Structure and mechanism of type II topoisomerases
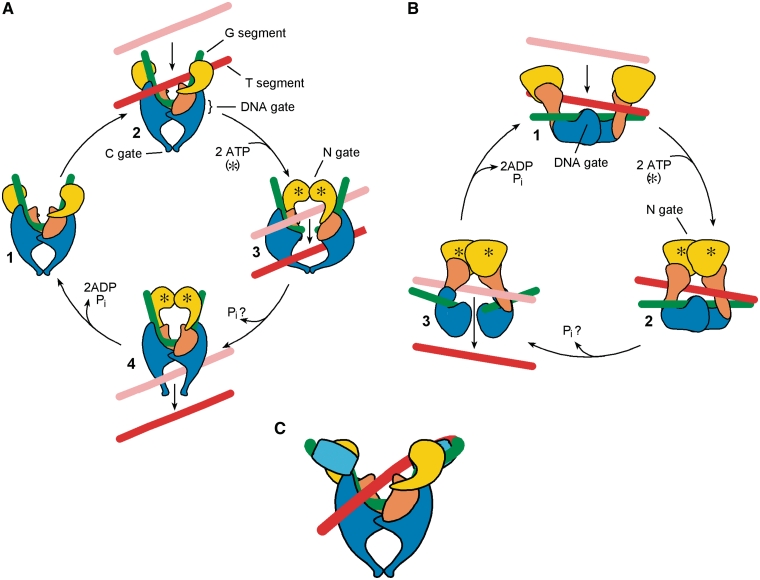
Structural and biochemical studies, particularly on gyrase and yeast topo II, have led to the formulation of a general mechanistic model for type IIA topos (Figure 1A) (3). The enzymes operate as symmetrical dimers; the eukaryotic proteins are homodimers, while the bacterial homologues divide the polypeptide into two distinct gene products and are A2B2 tetramers. Both classes of type IIA topos interact with two DNA segments. The G- (or ‘Gate’-) segment first binds to and is strongly bent by the enzyme [by as much as 150° (16–18)]. Each strand of this DNA is then cleaved by one of a pair of tyrosines, at sites 4 nt apart, forming two covalent, 5′-phosphotyrosine intermediates (19). ATP binding to each monomer results in dimerization of the N-terminal domains to form a new protein–protein interface (termed the N-gate), enclosing a second DNA (the T- or ‘Transported’-segment), which is passed through the G-segment; this process requires not only DNA cleavage, but also the separation of the DNA ends by disruption of an existing protein dimer interface (the DNA gate). The T-segment subsequently leaves the complex through a third protein interface (the C- or exit-gate) (20,21), having passed through the G-segment and across the entire dimer interface of the enzyme (Figure 1A). ATP hydrolysis and product release allows the N-gate to open and resets the enzyme for further rounds of reaction, although hydrolysis of one ATP and release of phosphate also appears to stimulate strand passage (22,23).
Figure 1.
Structure and mechanism of type II topoisomerases. (A) Type IIA topo core mechanism—see text for details. Different domains of the protein are indicated: yellow, ATPase domain (B subunit N-terminus in prokaryotic enzymes); orange, B subunit C-terminus (or homologous region in homodimeric eukaryotic enzymes); blue, A protein N-terminal breakage-reunion domain (or homologue). The A protein C-terminal domain is not shown. The G-segment (green), T-segment (red/pink) and DNA, N- and C-gates are indicated. Movement of the T-segment is shown from the pink to the red position. (B) Type IIB (Topo VI) mechanism. The B subunit N-terminal ATPase domain and C-terminal domain are in yellow and orange, respectively; the A subunit is in blue. The N- and DNA gates are indicated. (C) DNA gyrase structure, showing DNA wrapping around the C-terminal domains of the A subunit (cyan) to deliver a contiguous T-segment (red) to the enzyme, with a right-handed crossing over the G-segment (green). Only the wrapped segment captured by the enzyme is shown for clarity.
The specific reaction carried out by the type II topos depends on the topological relationship between the G- and the T-segment. Intra-molecular strand passage (G- and T- segments on the same circular molecule) leads either to supercoiling or relaxation, with a linking number change of ±2 (24), or to a knotting/unknotting reaction. If the G- and T-segments are on separate molecules, the result is catenation or decatenation.
This basic idea underlying this mechanism, in which the T-segment is captured by the closure of the N-terminal dimer interface, then exits through the cleaved G-segment and its associated protein interface, is known as the ‘two-gate’ model (25,26). However, since subsequent structural studies have shown that the IIA enzymes have three protein dimer interfaces in all [Figure 1A (27,28)], the second gate of the ‘two-gate’ mechanism (the ultimate exit route for the T-segment) is generally taken to be the C-gate. Roca and Wang (29) have described a theoretical alternative ‘one-gate’ scheme (1), in which the enzyme bridges the site of DNA cleavage (30,31) and strand passage of the T-segment through the G-segment into an enclosed protein cavity is followed by partial dissociation of the G-segment to allow the T-segment to leave by the same route without reversing strand passage. In Figure 1A, this mechanism would correspond to a situation where the C-gate remained closed during the reaction. Using a covalently cross-linked dimer of yeast topo II, Lindsley (32) showed that the enzyme could still carry out strand passage. Although this experiment is suggestive of a one-gate mechanism, these experiments do not rule out the two-gate scheme. To date, biochemical and structural evidence largely supports the two-gate mechanism (16,21,29,33,34) and it is generally accepted that an efficient, processive type II reaction requires an enzyme that passes a T-segment between its protein subunits as well as through a DNA gate.
Although type IIB enzymes (topo VI) share the basic structural and mechanistic features of the type IIA enzymes, they differ in some important respects (3). Topo VI is an A2B2 heterotetramer believed to catalyse topo reactions via a two-gate mechanism involving transient double-strand cleavage, albeit with 2 nt rather than 4 nt overhangs (35). However, structural work has shown that the enzyme possesses only two dimer interfaces (36), lacking the separate C-gate found in type IIA enzymes (Figure 1B). This configuration more closely mirrors the original two-gate model than that of the type IIA topos, but raises interesting questions as to how enzyme integrity is maintained during strand passage (see below). In this context, it may be significant that DNA cleavage by topo VI seems to be strictly dependent on ATP binding (35).
DNA gyrase—a special case
With respect to the core type IIA topo mechanism, we can understand in general terms how gyrase operates specifically to reduce linking number, both relaxing positive supercoils and maintaining a steady-state level of negative supercoiling in the presence of ATP. Gyrase is known to wrap ∼130 bp of DNA around itself (Figure 1C) (37–43), presenting a T-segment to the enzyme that is immediately contiguous with the G-segment (44). The selection of a T-segment by this juxtaposition facilitates an intra-molecular reaction with the appropriate orientation for unidirectional strand passage to result in a reduction in linking number (24,44). The ability of gyrase to perform directional strand passage, i.e. to reduce linking number, is crucially dependent on the C-terminal domain of the GyrA protein, the so-called DNA-wrapping domain (Figure 1). Deletion of this domain essentially converts gyrase into a conventional (DNA-relaxing) type II topo, hence implicating wrapping in T-segment selection (45). Topo IV possesses a degenerate version of this domain (the C-terminal domain of ParC) that is unable to stabilize DNA wraps (46) and this enzyme does not supercoil DNA, but is a very efficient decatenase (47).
G-segment cleavage and DNA gate opening
Two essential and separate features of the strand-passage mechanism of type II topos are the transient, double-stranded cleavage of the G-segment and the physical opening of the DNA gate to allow the T-segment through. The cleaved-DNA species is generally believed to represent only a small fraction of the topo complexes, although many agents are known that stimulate cleavage by stabilizing this intermediate [e.g. quinolone antibacterials and antitumour agents such as etoposide (48,49), as well as ATP itself and its non-hydrolysable analogue, 5′-adenylyl-β,γ-imidodiphosphate (ADPNP) (50–53)]. Experiments with several eukaryotic topo IIs have shown that the normal equilibrium level of DNA cleavage is generally less than ∼1% (54,55), although Chlorella virus topo IIs appear to be exceptions (55,56). Work with E. coli gyrase suggests that the level of cleavage is also low, but measurable (57). The fraction of complexes that additionally have the DNA gate interface in a physically open state is likely to be even lower than these values, since, for almost all type IIA enzymes, strand-passage reactions do not occur in the absence of ATP (gyrase being a notable exception) (58).
Recent single-molecule studies have begun to investigate DNA gate opening further. Fluorescence resonance energy transfer experiments using Drosophila topo II and a labelled 28 bp double-stranded DNA oligo have monitored the interconversion of the topo II–DNA complex between open and closed states during ATP hydrolysis and found that the DNA gate may be open ∼50% of the time (59) in the presence of ATP. In contrast, single-molecule experiments using Bacillus subtilis gyrase in which either the enzyme or DNA substrate (60 bp linear or supercoiled plasmid DNA) was labelled, found that the enzyme predominantly resides in the DNA gate-closed conformation (60) and could not detect a sizable gate-open population in the presence of ATP or ADPNP. At present, it is not clear why the Drosophila and B. subtilis studies produced different results, although it is possible that these differences could reflect either a genuine mechanistic difference between enzymes from different sources or distinctions between the experimental set-ups. For example, in the experiments with Drosophila topo II, only short DNA fragments were used, which may represent a non-optimal substrate unlikely to be able to serve simultaneously as a G- and T-segment. The presence of a T-segment is reported to influence both cleavage and DNA gate opening (61–63).
The role of ATP in strand passage
It has been assumed that the central DNA strand-passage mechanism is common to gyrase and non-supercoiling type IIA enzymes (Figure 1). Consequently, attention has focused on the role of ATP binding and hydrolysis– in driving unidirectional transport through the DNA gate and the enzyme subunits. This directionality is a clear requirement of the gyrase supercoiling reaction. It has long been known that substitution of ATP with ADPNP can result in a single-turnover (non-catalytic) strand-passage reaction (64,65). Studies with yeast topo II and ADPNP in particular have suggested that the ATP-dependent closure of the N-terminal domains may physically drive DNA cleavage (61), opening of the DNA gate and transport of the T-segment (58,63). This latter result has been taken to suggest that the cavity between the dimerized N-terminal domains may be too small to stably retain a T-segment (66,67). A recent structure of the dimerized C-terminal domain of the Mycobacterium tuberculosis gyrase B subunit has also been interpreted to suggest a possible role for the T-segment in the opening of the DNA gate, via a steric-based mechanism (62). Interestingly, the situation is somewhat different for E. coli gyrase, where there is good evidence that ATPase activity and strand passage can be uncoupled (15,68). Structural data for gyrase also suggest that a T-segment may be retained between the dimerized N-terminal domains (69), while limited amounts of DNA relaxation can occur when the enzyme is pre-incubated with ADPNP (70), suggesting that strand passage can be reversed even when the N-gate is closed.
Pre-steady-state kinetics and mutagenesis experiments with yeast topo II have indicated that the hydrolysis of one ATP and concomitant release of product (phosphate) may precede and accelerate transport of the T-segment (3,23). Structural studies of gyrase (71) and human topo IIα (67), as well as the type IIB topo, topo VI (36,72), have revealed that nucleotide binding, hydrolysis and phosphate release may transmit conformational changes to the DNA gate region through a link between the ATP binding domain and an adjacent ‘transducer’ domain, thus providing a potential physical context for the kinetic results. However, the precise interaction between these components is unclear, as no crystal structure is available for an intact gyrase B subunit or the homologous region of a non-supercoiling type IIA enzyme.
The role of ATP in topology simplification by type II topoisomerases
In 1997, Rybenkov et al. (73) described a process of topology ‘simplification’ that is carried out by non-supercoiling type IIA topos. In this process, the enzymes use the free energy of ATP hydrolysis to reduce the steady-state levels of supercoiling, knotting or catenation below those seen at thermodynamic equilibrium (73). This finding suggested that standard type IIA topos might employ an energy transduction function analogous to that used in the supercoiling reaction of gyrase. The simplification process has been proposed to be physiologically important, since complete decatenation of replication products is absolutely required for chromosome partitioning during cell division. There is likewise some evidence that the removal of knots may also be an important topo function (7,74,75).
There has been considerable interest in the detail of the mechanism behind topology simplification, which requires both unidirectional strand passage and a selection step amongst potential T-segments to directionally perturb the equilibria (a situation again analogous to that of gyrase) (76). Topology simplification probably arises, at least in part, from bending of the G-segment by the enzyme, which helps to bias the enzyme to select T-segments from within the curved contour of the DNA (18,77–79). However, it has recently been calculated that the energy required for topology simplification is very low. Specifically, the excess free energy of the steady-state distribution of topoisomers (in the presence of a topo IIA and ATP) over that of the equilibrium state is <1 kJ mol−1 (79,80), substantially less than kT. Thus, type II topos are at least a thousand times less efficient at shifting chemical energy into the state of their DNA products than gyrase, which can maintain a steady-state supercoiling free energy of >500 kJ mol−1 (79). Moreover, some data suggest that topology simplification may not be crucial physiologically (79). For example, in chromosome segregation, topo II has been shown to be responsible for decatenation (81), but partitioning forces tend to pull daughter replicons apart, perturbing the equilibrium towards complete decatenation, suggesting that there would not be a need for topology simplification (81,82). Supercoiling is thought to promote decatenation (47,83,84) and unknotting (85) and a recent report has suggested that yeast mitotic chromosomes become positively supercoiled to drive efficient decatenation (86), suggesting again that chromosome separation may be more efficient in vivo than suggested by the equilibration levels determined for dilute solutions of nicked DNAs in vitro. Furthermore, recent experiments (36,79) show that Methanosarcina mazei topo VI does not carry out topology simplification, suggesting that archaeal species can adequately unknot and partition their DNA without active simplification on the part of a topo. In summary, it is clear that while non-supercoiling type IIA topos can simplify DNA topology in an ATP-dependent manner, the energy requirements of the effect are extremely small; thus, topology simplification, if it is physiologically important, is very poorly optimized in energy transduction terms. Given that there is considerable circumstantial evidence to suggest that other factors may promote decatenation and unknotting beyond levels seen in vitro, we have considered an alternative, more fundamental role for ATP.
An ancestral role for ATP hydrolysis?
In common with other enzymes that push a complex mechanical reaction forward (87,88), it has often been stated that in type II topos ATP hydrolysis serves to ‘drive conformational changes’ (23,89,90). However, while one can imagine how nucleotide binding and turnover promote structural rearrangements to facilitate unidirectional strand passage, it is not clear whether or how the free energy of hydrolysis is being channelled to perform some type of otherwise unfavourable molecular ‘work’ to increase the free energy of the DNA product (gyrase being the exception).
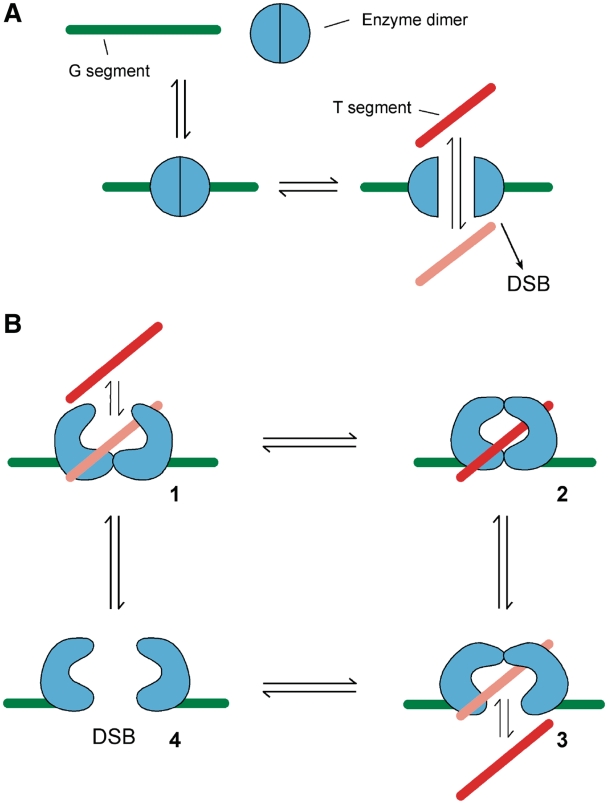
Indeed, a re-consideration of the type II topo reaction cycle suggests that this line of reasoning derived from gyrase, where the enzyme must transduce energy into the DNA substrate, may obscure the real reason why evolution selected for a nucleotide-regulated enzyme. Rather than an energy transduction issue per se, we suggest that the primary ancestral requirement for ATP by the type II enzymes may derive from an evolutionary pressure to avoid aberrant double-strand breaks (DSBs). Type II topos play an essential but potentially dangerous role in the manipulation of DNA (91). The enzymes must produce a transient break in both strands of the DNA, perhaps as many as a million times per human cell cycle (92) and break re-sealing must occur reliably to prevent the accumulation of mutagenic or cytotoxic lesions. Implicit in the notion of a two-gate mechanism is the idea of protection against DSBs: a simple one-gate type II enzyme, such as that shown as a cartoon in Figure 2A, would clearly dissociate to give permanent DSBs at every strand-passage event (this is distinct from the C-shaped, one-gate model discussed above) (29,58). This issue has been alluded to recently (3,63,92), but the role of ATP hydrolysis in ensuring the prevention of DSBs has not been explicitly elaborated. We can explore these issues by imagining the hypothetical evolution of a non-ATP-dependent type II topo.
Figure 2.
Models for an ATP-independent type II topoisomerase. Outline mechanistic schemes for hypothetical type II topos operating independently of ATP hydrolysis. (A) A one-gate enzyme. (B) A two-gate enzyme where the gates equilibrate independently. The numbers are referred to in the text. G-segment, green; T-segment, red/pink (as Figure 1); DSB, double strand break.
Type II topos clearly require more than one protein gate so that a protomer–protomer interaction can be maintained throughout strand passage (Figure 2B) (25,29). In the simplest such case, two gates (one of which acts as a DNA gate) would operate independently, with the passive bidirectional transfer of a T-segment through each gate in turn (Figure 2B). However, independent operation of these two gates implies the possibility of having both gates open at the same time (Figure 2B, 4), a situation that would result in a permanent DSB. To minimize the probability of this outcome, the association of the two dimer interfaces would have to be as tight as possible, yet this would lead to the converse problem: the enzyme would spend most of its time with both gates closed (Figure 2B, 2) and would be a very inefficient topo. Even if each interface possessed a dissociation constant of say 10−3 M, such that the majority of the enzyme would be doubly closed at any given time, this would still give rise to a topo that would produce a DSB with high frequency (at least 1 per 1000 strand-passage events in this example). A refinement of this scheme would be one in which the gates were hypothetically coordinated, so that one must be closed while the other is open (effectively a description of the two-gate model). This is hard to envisage for two separated interfaces of this kind; the doubly closed form would still have to be a very stable intermediate to avoid the possibility of dissociation. Although a slow topo reaction might be acceptable for certain functions, the essential role of type II topos in replication requires that the rates be sufficient to keep up with the overall speed of the replication process. (It is worth noting that although removal of unwanted supercoiling during replication and transcription may seem like an urgent priority, in vivo measurements suggest that transcription-generated supercoils are only removed slowly by topos (93), thus an inefficient topo may not be out of the question in this case.)
For the initial DNA complex of a type II topo to be resistant to DSB formation, either the DNA gate protein interface must remain very tightly associated or the G-segment must remain uncleaved. In type IIA topos, the evidence suggests that DNA cleavage is independent of the opening of the DNA gate (94) and that the distribution between intact and broken DNA is in equilibrium in the absence of ATP, with the fraction of broken DNA being low but measurable (57,95), suggesting that the enzyme dimer is crucial to DSB resistance. On the other hand, in topo VI, there is evidence that the DNA cleavage reaction is tightly coupled to the binding of ATP (35), indicating that the integrity of the G-segment itself may contribute to the DSB resistance of the initial complex.
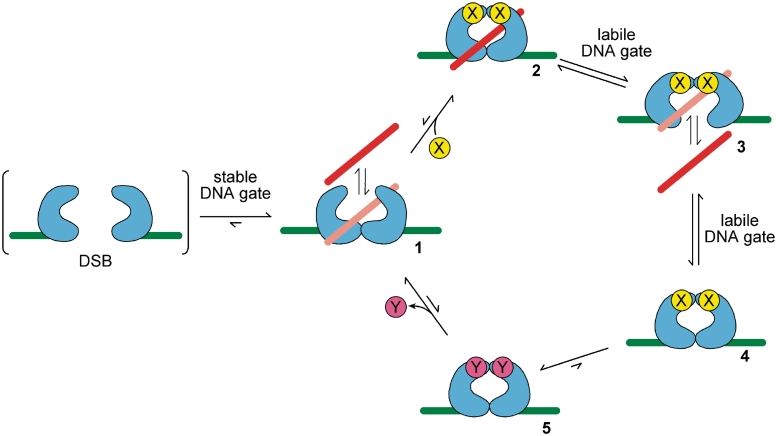
For strand passage to take place, the initially tight DNA gate/G-segment interface must be made sufficiently weak for its dissociation to be frequent, while at the same time a new, strong dimer interface must be formed to maintain the stability of the complex and avoid DSB formation. This situation is essentially equivalent (in Figure 2B) to going from State 1 (where the T- segment is free to enter the enzyme cavity) to State 3 (where strand passage can take place) in a concerted and largely irreversible step, to avoid the ‘both gates open’ (DSB) and ‘both gates closed’ (no reaction) traps. Such a conformational change requires an energy input to render it unidirectional and hence must have an external driver; i.e. the new dimer interface should be essentially as tight as the initial one was. A possible conformational change is shown in Figure 3, with the process driven by the binding of a small molecule X (dyadic symmetry probably requires two such agents). Binding switches the enzyme from a form with a stable DNA gate (Figure 3, 1), to one with a labile DNA gate, with a large negative free energy change (Figure 3, 2–4; the gate does not have to be permanently open).
Figure 3.
Model for a viable type II topoisomerase. Scheme in which the binding of X (1→2) drives a thermodynamically favourable conformational change from a stable (1) to a labile DNA gate (2–3–4), while providing a new strong dimer interface to prevent dissociation to give a DSB. A chemical cycle is required to drive the reverse conformational change (4→5→1); free energy is available from the conversion of X to Y. G-segment, green; T-segment, red/pink (as Figure 1).
If the conformational change on binding molecule X happens with a T-segment in the cavity, then passive strand passage out of the complex can take place safely; in fact in the X-bound form, the T-segment can potentially pass through the G-segment in either direction (Figure 3, 2–4). However, the enzyme is now trapped in the low free energy X-bound state and the conformational change must be reversed to ensure a catalytic reaction. Thus, there must be a second step that drives the re-opening of the top gate. This process could only happen through a thermodynamic cycle (Figure 3, 4→5→1), with free energy input from a chemical change in X (to Y), with the overall free energy of the protein conformational changes being zero over the whole catalytic cycle. Thus, Figure 3 describes a model for the putative simplest viable type II topo.
The binding of X, the (overall) energetically favourable transformation of X to Y and the dissociation of Y now drive the cycle round, dissipating energy. Only in the X-bound form is the DNA gate labile enough to allow strand passage at a significant rate (Figure 3, 2–4) while the new tight dimer interface maintains the complex. Without a change in energy states from a chemical cycle, the alternation of two very strong dimer interfaces could not be accomplished. In principle, the driver for the reaction could be any chemical cycle (e.g. phosphorylation-dephosphorylation, redox change, etc.); however, nucleoside triphosphates are of course a common source of free energy in coupled reactions. We argue that the free energy of ATP hydrolysis in type II topos is driving the sequential breakage and formation of two strong dimer interfaces—the greater the energy available per cycle, the stronger the dimer interfaces can be and the safer the enzyme is from the formation of accidental DSBs. The free energy needed to modulate protein–protein interactions at the DNA gate and ‘stabilization gate’ interfaces should, according to this model, be a significant fraction of the free energy available from the hydrolysis of two ATP molecules. Indeed, a preliminary estimate of the interface free energies (ΔG) for the DNA gates of various type II topos of known structure using the program PISA (96) yields values of around −20 to −70 kJ mol−1 (Lawson, D.M. and A.M., unpublished data), compared with ∼100–150 kJ mol−1 available from the hydrolysis of two ATPs (79), consistent with a potential role for ATP hydrolysis in gate opening. However, it is important to point out that simple dissociation of the DNA gate is unlikely to be the only structural alteration involved. In addition, these calculations are preliminary and based on a number of assumptions and should hence be treated with caution.
It is important to note that in this simplest model case, the T-segment can be transported passively in either direction. For example, in Figure 3, the T-segment could enter at 3 and be carried through 4 and 5 to be released at 1, in a ‘bottom-up’ reaction. This possibility means the enzyme would then behave as a true ‘phantom-chain’ device (97,98), whilst still requiring ATP hydrolysis. In this hypothetical ancestral scheme, the ATP cycle serves (and is required) only to protect against DSBs. However, if we allow the presence of a T-segment to differentially affect the rate of binding of X (ATP) or the rate of dissociation of Y (ADP), then directionality can be introduced into the strand-passage reaction, even though the enzyme would still only equilibrate topoisomers. Further elaborations might include the conformational changes dependent on ATP hydrolysis that have been hypothesized to drive the T-segment unidirectionally through the protein complex (see above) (3). A gyrase or a topology-simplifying enzyme requires some mechanism for T-segment selection in addition to preferential strand passage in one direction.
Overall, the hypothetical scheme in Figure 3 is essentially a representation of the canonical two-gate reaction mechanism proposed for type II topos (Figure 1) and closely matches the proposed mechanism for topo VI (Figure 1B) (36). Type IIA enzymes have an additional protein gate, the C-gate, which may function to increase the security of the DNA gate and further reduce the possibility of DSB formation (63,92). In the presence of the non-hydrolysable analogue ADPNP, only single-turnover strand passage occurs (65); this process is effectively equivalent to the reaction scheme shown in Figure 3, 1–4 only.
As noted above, the addition of ATP or ADPNP can actually promote DNA cleavage, apparently counter to our argument that ATP hydrolysis prevents DSBs. However, this effect is only revealed in experiments involving the destructive denaturation of the whole complex, either in the presence or absence of a drug (53,99,100). This simply means that the addition of nucleotide changes the cleavage–religation equilibrium to produce more cleaved DNA, as would be expected with this model. Our main point is that this inevitable and necessary cleavage of the G-segment and weakening of the DNA gate do not translate into permanent DSBs because the newly closed N-gate maintains the integrity of the overall complex. It is interesting to note that the binding of nucleotide really does ‘form a new tight protein interface’. The ADPNP molecules in structures of the dimerized N-terminal domain actually bridge the gap between the subunits, making substantial contacts with both monomer proteins (101).
Our argument represents a shift of perspective from the idea that the closing of the ATP-operated clamp simply ‘captures the T-segment’, to the concept that it forms a tight new interface, while concomitantly weakening the DNA gate, to allow strand passage while preventing complete dissociation. ‘Capture’ of a T-segment is not specifically required; in the hypothetical ‘bottom-up’ reaction described above (see Figure 3), the binding of X does not capture anything, but sets up the enzyme to allow the T-segment to pass through the G-segment from below and escape from the top, after dissociation of Y. Indeed, it remains a formal possibility that topo VI might be able to work via a ‘bottom-up’ reaction; this idea has not been specifically tested.
The evolution of type II topoisomerases
This rationale also suggests that the simplest successful type II topos in evolutionary terms might have looked something like topo VI, a two-gate type IIB. This enzyme would have been required as soon as genomes evolved to be long enough for tangling or formal linking of the daughter helices to become a problem, but probably before any requirement for active supercoiling or topology simplification. The additional C-gate of the type IIA topos could have evolved later, possibly for additional resistance to DSBs [although a recent discussion of topo phylogenomics has suggested a more complex picture, in which the enzymes may have evolved in an ancient virosphere, with their modern distribution dependent on a number of horizontal gene transfer events (13)]. The ATP-operated clamp domain of the type II topos is not unique, but is a member of the GHKL ATPase family (102) (see below), so its ultimate evolutionary origin may lie in another system. It remains an outstanding question as to whether gyrases or non-supercoiling type IIA enzymes are more primitive. There is evidence that topo IVs are derived from gyrase by loss of the DNA wrap through modification of the C-terminal wrapping domain (13,46,103); however, whether gyrase is also ancestral to the eukaryotic type IIAs, or vice versa, is not clear (103). A circumstantial argument can be made either way. If gyrase is the ancestral form, then the G-segment bend and unidirectional strand passage that leads to topology simplification by the non-supercoiling enzymes could be a relic of the directional gyrase reaction (79). On the other hand, Roca (92) has recently suggested that the G-segment bend might have a role as a sensor of tension in DNA and that an inability to correctly bend DNA could prevent the cleavage reaction under conditions where DNA tension might increase the probability of DSB formation during enzyme action. In the latter instance, the gyrase wrap could have developed from the pre-existing G-segment tension sensor. Interestingly, recent single-molecule experiments support the idea of DNA wrapping by gyrase being sensitive to tension in the DNA (104); however, in some cases, notably at the onset of chromosome partition in the presence of residual catenanes, DNA tension might be thought, a priori, to be a potentially useful trigger of topo action leading to decatenation, rather than an inhibitor of it.
The ATP-independent relaxation reaction of gyrase
Although almost all of the reactions catalysed by type II topos are dependent upon ATP hydrolysis, one activity of gyrase is a notable exception. In bacterial cells, the primary in vivo roles of this enzyme are thought to be the ATP-dependent relaxation of positive supercoils and introduction of negative supercoils (105). However, it has been established that gyrase can also carry out the ATP-independent relaxation of negative supercoils (99,100). This reaction (for E. coli gyrase) is ∼20-fold slower than ATP-dependent supercoiling and ATP-free gyrase does not relax supercoils processively. Whether ATP-independent relaxation is physiologically important is not known, although this seems unlikely; recent estimates of ATP concentrations in E. coli are sufficiently high to largely saturate the enzyme (106). Nevertheless, the reaction represents a significant distinction between gyrase and non-supercoiling type II enzymes. It has been reported that phage T4 topo II can also carry out non-catalytic strand-passage events in the absence of nucleotide, but this result has not been investigated further (107).
The gyrase relaxation reaction is an obvious counter argument to the idea that ATP hydrolysis is required to avoid DSBs, as it demonstrates that the gyrase DNA gate must be able to open in the absence of a new dimer interface created by ATP binding. However, gyrase is unique amongst type II topos and this exception might be consistent with our hypothesis. In particular, the enzyme can transduce a significant fraction of the free energy of ATP hydrolysis into DNA supercoiling free energy (108–111), suggesting that less free energy may be available to drive the conformational changes necessary for DSB prevention in the enzyme. This, in turn, suggests that the gyrase DNA- and C-gates should be intrinsically more labile, thus allowing slow relaxation, as observed. However, our preliminary consideration of interface binding energies does not so far support this; we did not find any consistent differences in the free energies associated with DNA gyrase and those for other type II topos. These considerations imply that gyrase ought to be more susceptible to forming DSBs than other type II topos, although the unique DNA wrap around the gyrase complex may serve to stabilize the complex and mitigate this effect.
The role of type II topoisomerases in illegitimate recombination
Illegitimate recombination (IR) is a rearrangement of DNA that occurs between nucleic acid segments that bear short regions of homology (<10 bp) or no apparent homology at all (112). It has been appreciated for many years that gyrase can participate in IR reactions (113) and that this reaction is stimulated (2–3 orders of magnitude) by the quinolone drug oxolinic acid (114), which binds at the DNA gate. In contrast, this stimulation is blocked by the coumarin drug coumermycin A1, which prevents ATP binding (115). Gyrase-mediated IR can occur in vitro and requires the presence of an E. coli extract, but is independent of RecA (114). One proposed mechanism for this reaction is that IR can take place when a pair of covalently bound gyrases exchange GyrA subunits (114). Stimulation by oxolinic acid is thought to occur because the drug increases the lifetime of the DNA-cleaved state, enhancing the likelihood that subunit swapping can occur. Support for this model comes from the identification of temperature-sensitive gyrA mutants in E. coli that confer spontaneous illegitimate recombination (116). Of these substitutions, one mutant in particular (Leu492→Pro) displayed normal supercoiling activity, but generated linear DNA during the reaction, suggesting that it has a defect at the DNA rejoining step or in its subunit interactions (116).
IR has also been shown to occur in other type II topos, including T4 and eukaryotic topo II. In the case of the T4 enzyme, it appears that recombination can occur with the purified enzyme alone [i.e. without an added E. coli extract (117–119)]. Purified calf thymus topo II can also mediate IR in a reaction that can be inhibited by the coumarin drug novobiocin (120), whereas in yeast, IR is stimulated by the topo II poison etoposide (VP-16) (121). Subunit exchange is also the suggested mechanism for IR with these type II enzymes, implying that subunit–subunit interactions can be disrupted under certain conditions. Taken together, several lines of data indicate that the aberrant disruption of type II topo subunit interfaces may be a source of illegitimate recombination. If true, these reactions highlight the detrimental consequences to the cell when the security normally afforded by the various topo gates break down.
Spo11 and meiotic recombination
Spo11 is a homologue of the topo VIA subunit (9,122), the dimer of which forms the DNA gate of the type IIB enzymes (11,36) (Figure 1B). Spo11 is present in all eukaryotes, where it is responsible for the initiation of DSBs in meiotic recombination (122,123). Roca (92) has proposed that the relative insecurity of topo VI as a true two-gate (rather than three-gate) enzyme makes the A subunit an appropriate candidate for co-option into a system evolved to produce DSBs. On the other hand, Forterre and Gadelle (13) have suggested that the opposite may be true and Spo11 may have been recruited by the topo VIB subunit to make a functional topo.
To date, there has been no identification of a topo VIB ATPase homologue (or analogue) in the meiotic recombination apparatus. At first glance, our hypothesis suggests that such a component should be necessary to open the protein interface responsible for DSB formation. However, studies of meiotic DSB formation (124) suggest that free 5′-ends are produced after Spo11 binding by nucleolytic cleavage of the covalently bound strands at each side of the Spo11–DNA complex. These data can easily be accommodated by a variation of our model in which separation of Spo11 monomers is not required to generate free DNA ends, but rather binding of Spo11 marks the DNA for homologous recombination repair proteins by generating a covalent protein–DNA link, although it is not yet clear what induces Spo11 to cleave the DNA. Consistent with this line of reasoning, it is known that many DSB repair factors, such as Rad50 and Mre11, are recruited to sites of Spo11 action (125). Alternatively, it may be possible that some as yet undiscovered ‘subunit separation’ component acts on Spo11 in meiotic recombination.
The role of nucleotide hydrolysis in disrupting protein interfaces in other systems
If the primary role of ATP in type II topos is to promote a particular type of mechanical manipulation, DNA gate opening, through the disruption of protein interfaces, then we might expect to see evidence of a similar role for nucleotides in other systems. Although many examples could be chosen, we limit a brief discussion here to other DNA-remodelling systems.
One parallel may be with the Structural Maintenance of Chromosomes (SMC) ATPase superfamily, which are key mediators of chromosome cohesion, condensation and repair in many organisms (126,127). Most bacteria possess a complex involving a dimer of SMC subunits, and a pair of accessory proteins known as ScpA and ScpB (128); γ-proteobacteria possess a diverged form of this assembly comprising the Smc homologue, MukB and two interacting subunits, MukE and MukF (129,130). MukE and MukF form an elongated, dimeric particle, which allows MukF’s C-terminal domains to engage the nucleotide binding ‘head’ regions of MukB and generate either closed ring-like structures or repetitive arrays (131,132); this general organization between SMCs and their accessory subunits is thought to be preserved across different SMC systems (126,127). Nucleotide binding further stabilizes the association of the MukB head domains (a feature shared with other Smc proteins and the ABC ATPase superfamily in general (133–135)) and it has been proposed that disruption of the MukB–MukF interaction depends upon ATP, as well as DNA (131,136). Some studies have even suggested that the ‘hinge’ region of Smc proteins, which lies distal to the ATPase domains, may function as a second regulatable gate to create transient openings within the molecule (137,138).
Another example of ATP regulation of protein–protein interactions occurs in DNA mismatch repair (MMR) (139). In E. coli, three proteins are responsible for the initiation of MMR-directed events: MutS, MutL and MutH. In the case of MutS, an ABC ATPase like the SMCs, it has been shown that DNA-binding domains N- and C-terminal to the nucleotide binding site become predisposed to associate tightly with DNA in the presence of ATP and to loosen their grip following ATP hydrolysis (140,141). For its part, MutL is a GHKL ATPase (102), a protein superfamily that includes the ATPase region of type II topos. The MutL dimer has been proposed to undergo ATP-dependent association of its ATPase domains (142,143), an event that serves to control the endonucleolytic action of MutH. Interestingly, in many bacteria apart from E. coli, as well as in eukaryotes, the domain C-terminal to the MutL ATPase region is in fact an endonuclease whose activity is regulated by nucleotide turnover (144,145). This arrangement and conformational cycle has striking parallels to that seen for the catalytic modules of type II topos, topo VI in particular.
A third example of the role of ATP hydrolysis in DNA remodelling through the formation and resolution of protein–protein interactions occurs with certain enzymes belonging to the AAA+ (ATPases associated with various cellular activities) superfamily, specifically, bacterial enhancer binding proteins (bEBPs) and replication initiation factors such as DnaA and the Origin Recognition Complex (ORC) (146,147)). The bEBPs such as NtrC and PspF initially bind DNA as dimers in an ‘off’ state, but upon phosphorylation via a two-component response regulator system, rearrange into ring-shaped oligomers that loop DNA and engage RNA polymerase/σ54 complexes at adjacent promoters (148,149). Assembly is mediated by the AAA+ ATPase subunits, which further remodel σ54 in a nucleotide-dependent manner to allow the polymerase to clear the promoter. For its part, DnaA binds to replication origins as a monomer, but at elevated concentrations undergoes an ATP-dependent transition to assemble into a helical nucleoprotein complex that melts DNA and assists replisome formation (150–153).
CONCLUDING REMARKS
The type II topos are well-studied molecular machines and many details of their DNA double-strand-passage mechanisms have been elucidated using a combination of structural and biochemical studies. However, their absolute requirement for ATP hydrolysis for their primary reactions has been puzzling. Although it seems that gyrase can transduce much of the free energy of ATP hydrolysis into DNA supercoiling, the non-supercoiling enzymes channel only a tiny fraction of this energy into their substrates. The hypothesis proposed here—that free energy is required to manipulate protein–protein interactions to maintain a high degree of enzyme dimer stability during the strand-passage reaction—explains how these enzymes are able to avoid the formation of permanent DSBs during their reactions. The proposal also provides a framework for explaining a number of diverse features of these enzymes, including the nucleotide-independent relaxation reaction of gyrase, the participation of type II enzymes in illegitimate recombination and the relationship between the meiotic recombination protein Spo11 and the type IIB topos. Further experiments are needed to directly investigate this proposed relationship between ATP hydrolysis and complex stability and the extent to which these ATP-dependent control mechanisms are shared by other DNA-remodelling systems, as well as non-DNA systems such as ATP-dependent transporters and heat-shock proteins.
FUNDING
The Biotechnology and Biological Sciences Research Council (BB/C517376/1 to A.D.B. and A.M.); the National Cancer Institute (CA077373 to J.M.B.). Funding for open access charge: Grant income.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank David Lawson for assistance with protein interface energetics calculations and Stephen Bornemann, Richard Bowater, Kevin Corbett, Patrick Forterre, Steve Halford, John Jenkins, David Lawson and Lynn Zechiedrich for their helpful comments on the manuscript.
REFERENCES
- 1.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 2.Bates AD, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- 3.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 5.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc. Natl Acad. Sci. USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Deibler RW, Chan HS, Zechiedrich L. The why and how of DNA unlinking. Nucleic Acids Res. 2009;37:661–671. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind L, Leipe DD, Koonin EV. Toprim - a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 10.Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. EMBO J. 2003;22:151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo 11. Embo J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadelle D, Filee J, Buhler C, Forterre P. Phylogenomics of type II DNA topoisomerases. Bioessays. 2003;25:232–242. doi: 10.1002/bies.10245. [DOI] [PubMed] [Google Scholar]
- 13.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates AD, O'Dea MH, Gellert M. Energy coupling in Escherichia coli DNA gyrase: the relationship between nucleotide binding, strand passage, and DNA supercoiling. Biochemistry. 1996;35:1408–1416. doi: 10.1021/bi952433y. [DOI] [PubMed] [Google Scholar]
- 16.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 17.Laponogov I, Sohi MK, Veselkov DA, Pan X-S, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 18.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 20.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams NL, Maxwell A. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry. 1999;38:13502–13511. doi: 10.1021/bi9912488. [DOI] [PubMed] [Google Scholar]
- 22.Baird CL, Gordon MS, Andrenyak DM, Marecek JF, Lindsley JE. The ATPase reaction cycle of yeast DNA topoisomerase II. Slow rates of ATP resynthesis and P(i) release. J. Biol. Chem. 2001;276:27893–27898. doi: 10.1074/jbc.M102544200. [DOI] [PubMed] [Google Scholar]
- 23.Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl Acad. Sci. USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 25.Mizuuchi K, Fisher M, O'Dea M, Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl Acad. Sci. USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JC, Gumport RI, Javaherian K, Kirkegaard K, Klevan L, Kotewicz ML, Tse Y-C. In: Mechanistic Studies of DNA Replication and Genetic Recombination. Alberts B, Fox CF, editors. NY: Academic Press; 1980. pp. 769–784. [Google Scholar]
- 27.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–232. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 28.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 29.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 30.Brown PO, Cozzarelli NR. Catenation and knotting of duplex DNA by type I topoisomerases: A mechanistic parallel with type II topoisomerases. Proc. Natl Acad. Sci. USA. 1981;78:843–847. doi: 10.1073/pnas.78.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tse Y-C, Wang JC. E. coli and M. luteus DNA topoisomerase I can catalyse catenation or decatenation of double-stranded DNA rings. Cell. 1980;22:269–276. doi: 10.1016/0092-8674(80)90174-9. [DOI] [PubMed] [Google Scholar]
- 32.Lindsley JE. Intradimerically tethered DNA topoisomerase II is catalytically active in DNA transport. Proc. Natl Acad. Sci. USA. 1996;93:2975–2980. doi: 10.1073/pnas.93.7.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–840. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 35.Buhler C, Lebbink JH, Bocs C, Ladenstein R, Forterre P. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J. Biol. Chem. 2001;276:37215–37222. doi: 10.1074/jbc.M101823200. [DOI] [PubMed] [Google Scholar]
- 36.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 37.Fisher LM, Mizuuchi K, O'Dea MH, Ohmori H, Gellert M. Site-specific interaction of DNA gyrase with DNA. Proc. Natl Acad. Sci. USA. 1981;78:4165–4169. doi: 10.1073/pnas.78.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkegaard K, Wang JC. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981;23:721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Wang J. Micrococcus luteus DNA gyrase: Active components and a model for its supercoiling of DNA. Proc. Natl Acad. Sci. USA. 1978;75:2098–2102. doi: 10.1073/pnas.75.5.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu LF, Wang JC. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978;15:979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- 41.Morrison A, Cozzarelli NR. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc. Natl Acad. Sci. USA. 1981;78:1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orphanides G, Maxwell A. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 1994;22:1567–1575. doi: 10.1093/nar/22.9.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rau DC, Gellert M, Thoma F, Maxwell A. Structure of the DNA gyrase-DNA complex as revealed by transient electric dichroism. J. Mol. Biol. 1987;193:555–569. doi: 10.1016/0022-2836(87)90266-x. [DOI] [PubMed] [Google Scholar]
- 44.Heddle JG, Mitelheiser S, Maxwell A, Thomson NH. Nucleotide binding to DNA gyrase causes loss of DNA wrap. J. Mol. Biol. 2004;337:597–610. doi: 10.1016/j.jmb.2004.01.049. [DOI] [PubMed] [Google Scholar]
- 45.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl Acad. Sci. USA. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 47.Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tse-Dinh YC. Exploring DNA topoisomerases as targets of novel therapeutic agents in the treatment of infectious diseases. Infect. Disord. Drug Targets. 2007;7:3–9. doi: 10.2174/187152607780090748. [DOI] [PubMed] [Google Scholar]
- 50.Buhler C, Gadelle D, Forterre P, Wang JC, Bergerat A. Reconstitution of DNA topoisomerase VI of the thermophilic archaeon Sulfolobus shibatae from subunits separately overexpressed in Escherichia coli. Nucleic Acids Res. 1998;26:5157–5162. doi: 10.1093/nar/26.22.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl Acad. Sci. USA. 1999;96:8414–8419. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morrison A, Higgins NP, Cozzarelli NR. Interaction between DNA gyrase and its cleavage site on DNA. J. Biol. Chem. 1980;255:2211–2219. [PubMed] [Google Scholar]
- 53.Sander M, Hsieh T. Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem. 1983;258:8421–8428. [PubMed] [Google Scholar]
- 54.Fortune JM, Dickey JS, Lavrukhin OV, Van Etten JL, Lloyd RS, Osheroff N. Site-specific DNA cleavage by Chlorella virus topoisomerase II. Biochemistry. 2002;41:11761–11769. doi: 10.1021/bi025802g. [DOI] [PubMed] [Google Scholar]
- 55.Fortune JM, Lavrukhin OV, Gurnon JR, Van Etten JL, Lloyd RS, Osheroff N. Topoisomerase II from Chlorella virus PBCV-1 has an exceptionally high DNA cleavage activity. J. Biol. Chem. 2001;276:24401–24408. doi: 10.1074/jbc.M101693200. [DOI] [PubMed] [Google Scholar]
- 56.Dickey JS, Choi TJ, Van Etten JL, Osheroff N. Chlorella virus Marburg topoisomerase II: high DNA cleavage activity as a characteristic of Chlorella virus type II enzymes. Biochemistry. 2005;44:3899–3908. doi: 10.1021/bi047777f. [DOI] [PubMed] [Google Scholar]
- 57.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two-metal-ion mechanism. J. Mol. Biol. 2002;318:361–371. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 58.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Quart. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 59.Smiley RD, Collins TR, Hammes GG, Hsieh TS. Single-molecule measurements of the opening and closing of the DNA gate by eukaryotic topoisomerase II. Proc. Natl Acad. Sci. USA. 2007;104:4840–4845. doi: 10.1073/pnas.0700342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gubaev A, Hilbert M, Klostermeier D. The DNA-gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc. Natl Acad. Sci. USA. 2009;106:13278–13283. doi: 10.1073/pnas.0902493106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corbett AH, Zechiedrich EL, Osheroff N. A role for the passage helix in the DNA cleavage reaction of eukaryotic topoisomerase II. J. Biol. Chem. 1992;267:683–686. [PubMed] [Google Scholar]
- 62.Fu G, Wu J, Liu W, Zhu D, Hu Y, Deng J, Zhang XE, Bi L, Wang DC. Crystal structure of DNA gyrase B' domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009;37:5908–5916. doi: 10.1093/nar/gkp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roca J. The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 2004;279:25783–25788. doi: 10.1074/jbc.M402555200. [DOI] [PubMed] [Google Scholar]
- 64.Osheroff N, Shelton ER, Brutlag DL. DNA topoisomerase II from Drosophila melangaster. Relaxation of supercoiled DNA. J. Biol. Chem. 1983;258:9536–9543. [PubMed] [Google Scholar]
- 65.Sugino A, Higgins NP, Brown PO, Peebles CL, Cozzarelli NR. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc. Natl Acad. Sci. USA. 1978;75:4838–4842. doi: 10.1073/pnas.75.10.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Classen S, Olland S, Berger JM. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc. Natl Acad. Sci. USA. 2003;100:10629–10634. doi: 10.1073/pnas.1832879100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. Nucleotide-dependent domain movement in the ATPase domain of a human type IIA DNA topoisomerase. J. Biol. Chem. 2005;280:37041–37047. doi: 10.1074/jbc.M506520200. [DOI] [PubMed] [Google Scholar]
- 68.Sugino A, Cozzarelli NR. The intrinsic ATPase of DNA gyrase. J. Biol. Chem. 1980;255:6299–6306. [PubMed] [Google Scholar]
- 69.Tingey AP, Maxwell A. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 1996;24:4868–4873. doi: 10.1093/nar/24.24.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams NL, Howells AJ, Maxwell A. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 2001;306:969–984. doi: 10.1006/jmbi.2001.4468. [DOI] [PubMed] [Google Scholar]
- 71.Lamour V, Hoermann L, Jeltsch JM, Oudet P, Moras D. An open conformation of the Thermus thermophilus gyrase B ATP-binding domain. J. Biol. Chem. 2002;277:18947–18953. doi: 10.1074/jbc.M111740200. [DOI] [PubMed] [Google Scholar]
- 72.Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure. 2005;13:873–882. doi: 10.1016/j.str.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 73.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 74.Deibler RW, Mann JK, Sumners de WL, Zechiedrich L. Hin-mediated DNA knotting and recombining promote replicon dysfunction and mutation. BMC Mol. Biol. 2007;8:44. doi: 10.1186/1471-2199-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Portugal J, Rodríguez-Campos A. T7 RNA polymerase cannot transcribe through a highly knotted DNA template. Nucleic Acids Res. 1996;24:4890–4894. doi: 10.1093/nar/24.24.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bates AD, Maxwell A. Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry. 2007;46:7929–7941. doi: 10.1021/bi700789g. [DOI] [PubMed] [Google Scholar]
- 77.Buck GR, Zechiedrich EL. DNA disentangling by type-2 topoisomerases. J. Mol. Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 78.Liu Z, Mann JK, Zechiedrich EL, Chan HS. Topological information embodied in local juxtaposition geometry provides a statistical mechanical basis for unknotting by type-2 DNA topoisomerases. J. Mol. Biol. 2006;361:268–285. doi: 10.1016/j.jmb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 79.Stuchinskaya T, Mitchenall LA, Schoeffler AJ, Corbett KD, Berger JM, Bates AD, Maxwell A. How do type II topoisomerases use ATP hydrolysis to simplify DNA topology beyond equilibrium? Investigating the relaxation reaction of nonsupercoiling type II topoisomerases. J. Mol. Biol. 2009;385:1397–1408. doi: 10.1016/j.jmb.2008.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bates AD, Maxwell A. The role of ATP in the reactions of type II DNA topoisomerases. Biochem. Soc. Trans. 2010;38:438–442. doi: 10.1042/BST0380438. [DOI] [PubMed] [Google Scholar]
- 81.Holm C. Coming undone: how to untangle a chromosome. Cell. 1994;77:955–957. doi: 10.1016/0092-8674(94)90433-2. [DOI] [PubMed] [Google Scholar]
- 82.Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc. Natl Acad. Sci. USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez-Robles ML, Witz G, Hernandez P, Schvartzman JB, Stasiak A, Krimer DB. Interplay of DNA supercoiling and catenation during the segregation of sister duplexes. Nucleic Acids Res. 2009;37:5126–5137. doi: 10.1093/nar/gkp530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vologodskii A. DNA supercoiling helps to unlink sister duplexes after replication. Bioessays. 2010;32:9–12. doi: 10.1002/bies.200900143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burnier Y, Dorier J, Stasiak A. DNA supercoiling inhibits DNA knotting. Nucleic Acids Res. 2008;36:4956–4963. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragon L. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331:1328–1332. doi: 10.1126/science.1201538. [DOI] [PubMed] [Google Scholar]
- 87.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- 88.Muller UF. Re-creating an RNA world. Cell Mol. Life Sci. 2006;63:1278–1293. doi: 10.1007/s00018-006-6047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gellert M. DNA Gyrase and other type II topoismerases. The Enzymes. 1981;XIV:345–366. [Google Scholar]
- 90.Wang JC. DNA topoisomerases as targets of therapeutics: an overview. Adv. Pharmacol. 1994;29A:1–19. doi: 10.1016/s1054-3589(08)60537-2. [DOI] [PubMed] [Google Scholar]
- 91.Wang JC, Caron PR, Kim RA. The role of DNA topoisomerases in recombination and genome stability: a double-edged sword? Cell. 1990;62:403–406. doi: 10.1016/0092-8674(90)90002-v. [DOI] [PubMed] [Google Scholar]
- 92.Roca J. Topoisomerase II: a fitted mechanism for the chromatin landscape. Nucleic Acids Res. 2009;37:721–730. doi: 10.1093/nar/gkn994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 94.Williams NL, Maxwell A. Locking the DNA gate of DNA gyrase: investigating the effects on DNA cleavage and ATP hydrolysis. Biochemistry. 1999;38:14157–14164. doi: 10.1021/bi991478m. [DOI] [PubMed] [Google Scholar]
- 95.Wilstermann AM, Osheroff N. Stabilization of eukaryotic topoisomerase II-DNA cleavage complexes. Curr. Top. Med. Chem. 2003;3:321–338. doi: 10.2174/1568026033452519. [DOI] [PubMed] [Google Scholar]
- 96.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 97.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sikorav JL, Jannink G. Kinetics of chromosome condensation in the presence of topoisomerases: a phantom chain model. Biophys. J. 1994;66:827–837. doi: 10.1016/s0006-3495(94)80859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gellert M, Mizuuchi K, O'Dea MH, Itoh T, Tomizawa J. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl Acad. Sci. USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sugino A, Peebles CL, Kruezer KN, Cozzarelli NR. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl Acad. Sci. USA. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 102.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 103.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 104.Nollmann M, Stone MD, Bryant Z, Gore J, Crisona NJ, Hong SC, Mitelheiser S, Maxwell A, Bustamante C, Cozzarelli NR. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 2007;14:264–271. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- 105.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 106.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu LF, Liu C-C, Alberts BM. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 108.Bates AD, Maxwell A. DNA gyrase can supercoil DNA circles as small as 174 base pairs. EMBO J. 1989;8:1861–1866. doi: 10.1002/j.1460-2075.1989.tb03582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cullis PM, Maxwell A, Weiner DP. Energy coupling in DNA gyrase: a thermodynamic limit to the extent of DNA supercoiling. Biochemistry. 1992;31:9642–9646. doi: 10.1021/bi00155a017. [DOI] [PubMed] [Google Scholar]
- 110.Maxwell A, Gellert M. Mechanistic aspects of DNA topoisomerases. Adv. Prot. Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- 111.Westerhoff HV, O'Dea MH, Maxwell A, Gellert M. DNA supercoiling by DNA gyrase. A static head analysis. Cell Biophys. 1988;12:157–181. doi: 10.1007/BF02918357. [DOI] [PubMed] [Google Scholar]
- 112.Ikeda H, Shiraishi K, Ogata Y. Illegitimate recombination mediated by double-strand break and end-joining in Escherichia coli. Adv. Biophys. 2004;38:3–20. [PubMed] [Google Scholar]
- 113.Ikeda H, Moriya K, Matsumoto T. In vitro study of illegitimate recombination: Involvement of DNA gyrase. Cold Spring Harbor Symp. Quant. Biol. 1981;45:399–408. doi: 10.1101/sqb.1981.045.01.054. [DOI] [PubMed] [Google Scholar]
- 114.Ikeda H, Aoki K, Naito A. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc. Natl Acad. Sci. USA. 1982;7:3724–3728. doi: 10.1073/pnas.79.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ikeda H. DNA topoisomerase-mediated illegitimate recombination. Adv. Pharmacol. 1994;29A:147–165. doi: 10.1016/s1054-3589(08)60544-x. [DOI] [PubMed] [Google Scholar]
- 116.Ashizawa Y, Yokochi T, Ogata Y, Shobuike Y, Kato J, Ikeda H. Mechanism of DNA gyrase-mediated illegitimate recombination: characterization of Escherichia coli gyrA mutations that confer hyper-recombination phenotype. J. Mol. Biol. 1999;289:447–458. doi: 10.1006/jmbi.1999.2758. [DOI] [PubMed] [Google Scholar]
- 117.Chiba M, Shimizu H, Fujimoto A, Nashimoto H, Ikeda H. Common sites for recombination and cleavage mediated by bacteriophage T4 DNA topoisomerase in vitro. J. Biol. Chem. 1989;264:12785–12790. [PubMed] [Google Scholar]
- 118.Ikeda H. Illegitimate recombination mediated by T4 DNA topoisomerase in vitro. Mol. Gen. Genet. 1986;1986:518–520. doi: 10.1007/BF00333287. [DOI] [PubMed] [Google Scholar]
- 119.Ikeda H. Bacteriophage T4 DNA topoisomerase mediates illegitimate recombination in vitro. Proc. Natl Acad. Sci. USA. 1986;83:922–926. doi: 10.1073/pnas.83.4.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bae Y-S, Kawsaki I, Ikeda H, Liu LF. Illegitimate recombination mediated by calf thymus DNA topoisomerase II in vitro. Proc. Natl Acad. Sci. USA. 1988;85:2076–2080. doi: 10.1073/pnas.85.7.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Asami Y, Jia DW, Tatebayashi K, Yamagata K, Tanokura M, Ikeda H. Effect of the DNA topoisomerase II inhibitor VP-16 on illegitimate recombination in yeast chromosomes. Gene. 2002;291:251–257. doi: 10.1016/s0378-1119(02)00622-4. [DOI] [PubMed] [Google Scholar]
- 122.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 123.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 124.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 2006;34:523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 126.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 127.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 128.Lim JH, Oh BH. Structural and functional similarities between two bacterial chromosome compacting machineries. Biochem. Biophys. Res. Commun. 2009;386:415–419. doi: 10.1016/j.bbrc.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 129.Yamazoe M, Onogi T, Sunako Y, Niki H, Yamanaka K, Ichimura T, Hiraga S. Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 1999;18:5873–5884. doi: 10.1093/emboj/18.21.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cobbe N, Heck MM. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol. Biol. Evol. 2004;21:332–347. doi: 10.1093/molbev/msh023. [DOI] [PubMed] [Google Scholar]
- 131.Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 132.Fennell-Fezzie R, Gradia SD, Akey D, Berger JM. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. Embo J. 2005;24:1921–1930. doi: 10.1038/sj.emboj.7600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 134.Thomsen ND, Berger JM. Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases. Mol. Microbiol. 2008;69:1071–1090. doi: 10.1111/j.1365-2958.2008.06364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 136.She W, Wang Q, Mordukhova EA, Rybenkov VV. MukEF Is required for stable association of MukB with the chromosome. J. Bacteriol. 2007;189:7062–7068. doi: 10.1128/JB.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 138.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 139.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 140.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 141.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 142.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 143.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 144.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O'Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 146.Bose D, Joly N, Pape T, Rappas M, Schumacher J, Buck M, Zhang X. Dissecting the ATP hydrolysis pathway of bacterial enhancer-binding proteins. Biochem. Soc. Trans. 2008;36:83–88. doi: 10.1042/BST0360083. [DOI] [PubMed] [Google Scholar]
- 147.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 148.Schumacher J, Joly N, Rappas M, Zhang X, Buck M. Structures and organisation of AAA+ enhancer binding proteins in transcriptional activation. J. Struct. Biol. 2006;156:190–199. doi: 10.1016/j.jsb.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 149.Chen B, Sysoeva TA, Chowdhury S, Nixon BT. Regulation and action of the bacterial enhancer-binding protein AAA+ domains. Biochem. Soc. Trans. 2008;36:89–93. doi: 10.1042/BST0360089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bramhill D, Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988;54:915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- 151.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 152.Margulies C, Kaguni JM. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J. Biol. Chem. 1996;271:17035–17040. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 153.Leonard AC, Grimwade JE. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol. Microbiol. 2005;55:978–985. doi: 10.1111/j.1365-2958.2004.04467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]