Abstract
Cationic antimicrobial peptides (CAMPs) selectively target bacterial membranes by electrostatic interactions with negatively charged lipids. It turned out that for inhibition of microbial growth a high CAMP membrane concentration is required, which can be realized by the incorporation of hydrophobic groups within the peptide. Increasing hydrophobicity, however, reduces the CAMP selectivity for bacterial over eukaryotic host membranes, thereby causing the risk of detrimental side-effects. In this study we addressed how cationic amphipathic peptides—in particular a CAMP with Lysine–Leucine–Lysine repeats (termed KLK)—affect the localization and dynamics of molecules in eukaryotic membranes. We found KLK to selectively inhibit the endocytosis of a subgroup of membrane proteins and lipids by electrostatically interacting with negatively charged sialic acid moieties. Ultrastructural characterization revealed the formation of membrane invaginations representing fission or fusion intermediates, in which the sialylated proteins and lipids were immobilized. Experiments on structurally different cationic amphipathic peptides (KLK, 6-MO-LF11-322 and NK14-2) indicated a cooperation of electrostatic and hydrophobic forces that selectively arrest sialylated membrane constituents.
Keywords: CAMPs, KLK, Sialic acids, Plasma membrane accumulation
Research highlights
► Cationic antimicrobial peptide KLK affects eukaryotic host cells ► KLK induces accumulation of plasma mebrane proteins/lipids ► Dependent on sialic acid residues ► Application of KLK leads to a recycling inhibition of sialyted molecules
1. Introduction
Over the last 30 years antimicrobial peptide research led to the discovery of more than thousand natural and synthetic substances [1], which were described as being suitable to kill or slow down the growth of pathogens [2,3]. Cationic antimicrobial peptides (CAMPs) were found to be key components of the innate immune response, where they play a crucial role in host defense against microbial infection. They are generated by different species including insects, plants, amphibians, prokaryotes and mammals [4–8]. In mammals, four groups of CAMPs have been characterized: α-, ß-, and θ-defensins, and cathelicidins [9–11]. CAMPs kill bacteria very rapidly and thus could play an important role in fighting the uprising microbial resistance to conventional antibiotics [12]. Therefore, great effort has been put into the production of artificial peptides based on key domains of their natural precursors [13] involving also the development of combinatorial libraries and high-throughput screening technologies [14].
Most CAMPs selectively target bacterial membranes due to the presence of negatively-charged lipids, while eukaryotic host cells carrying lipids with no net charge in the outer leaflet remain rather unaffected [15–17]. However, some CAMPs were also shown to act on intracellular targets, such as DNA or ribosomes [18,19] and multiple functions for CAMPs especially expressed in higher organisms like chemotaxis of immune cells, control of the inflammatory response or promoting re-epthelialization and wound closure was reported [19–22]. For our study we selected the CAMP KLK (KLKL5KLK) derived from a core undecapeptide of sapecin B, which is effective against Gram-positive and -negative bacteria as well as fungi with a MIC > 1 μM [23]. Although KLK has been reported to have a profound membrane interacting property [24], it shows further effects: KLK activates human neutrophils [25,26], exhibits chemotherapeutic activity [27], is a potent inducer of adaptive immunity to co-injected antigens [28] and leads to a fast increase of intracellular calcium levels [29]. Interestingly, KLK exerts its effect without being taken up by the cells [30]. The combination of a single stranded oligodeoxynucleotide (ODN1a) and KLK results in a potent immunostimulatory adjuvant termed IC31™ [31].
Generally, high minimum inhibitory concentrations (MIC) in the range of 1 to 10 μΜ [32,33], however, demand for rather high CAMP concentrations in therapeutic applications. Also studies on model phospholipid bilayers have indicated that most CAMPs function only at high membrane-bound concentrations close to full membrane coverage [34]. In this study, we were interested to which extent eukaryotic host cells are affected by such high CAMP concentrations. Indeed, CAMPs are known to interact not only with pathogens, but can also be harmful to mammalian cells. One reason for this may be the hydrophobicity of some peptides, which decreases the selectivity between bacterial and host cell membranes [35–37]. We speculated that the membrane localization of CAMPs containing hydrophobic regions might enable and stabilize their interaction with negatively charged protein modifications present on the eukaryotic cell surface.
Here, we found that application of KLK and other CAMPs led to the accumulation of sialylated proteins and lipids at the plasma membrane of various mammalian cells. The effect was dependent on the presence of hydrophobic peptide moieties, which enables and stabilizes the electrostatic interactions between the CAMP and the negatively charged glycosylations. Ultrastructural investigation revealed the formation of plasma membrane invaginations upon KLK treatment, indicating that KLK immobilizes and arrests specific plasma membrane constituents by blocking the fission or fusion of their carriers.
2. Materials and methods
2.1. Reagents
KLKLLLLLKLK (KLK) and KLKLLPLLKLK (KPK) were provided by Intercell AG (Vienna, Austria). Hank's buffered salt solution (HBSS; 137 mM NaCl, 5.4 mM KCl, 0.25 mM Na2 HPO4, 0.44 mM KH2PO4, 1.3 mM CaCl2, 1 mM MgSO4, 4.2 mM NaHCO3) and phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 1.46 mM KH2PO4) were from PAA Laboratories (Pasching, Austria). Trypan Blue and neuraminidase were purchased from Sigma Aldrich (Schnellendorf, Germany). Alexa647 labeled Cholera toxin B, DiD and Bodipy-GM1 were from Invitrogen (Lofer, Austria). The antimicrobial peptides LF11-322 (PFWRIRIRR-NH2), 6-MO-LF11-322 (6-methyloctanoyl-LF11-322), NK14 (KILRGVSKKIMRTF-NH2), and NK14-2 (KILRGVSKKIMRTFKILRGVSKKIMRTF-NH2) were synthesized in their amidated form by PolyPeptide Laboratories San Diego (CA, USA).
2.2. DNA constructs
The pCR3-GPI-DAF-GFP and pCR3-GPI-TRAIL-GFP plasmids were provided by Daniel Legler (University of Konstanz, Switzerland), the pJB20-GPI-hFR-mGFP, GPI-glycan-YFP (GPI-GL-YFP) and CD59-GFP constructs [38] were a gift by Jennifer Lippincott-Schwartz (NIH, Bethesda, USA). pEYFP-N1-CD43, pEGFP-N1-Lck, pEYFP-N1-Fyn, pEYFP-N1-CD3ζ and pEYFP-N1-CD147 were provided by Hannes Stockinger (Medical University Vienna, Austria), pEGFP-N1-CD71 and pEGFP-C1-CD63 by Katharina Strub (University of Geneva, Switzerland) and Flotillin-1-GFP and Flotillin-2-GFP by Lawrence Rajendran (ETH Zurich, Switzerland). The pEGFP-C1-CLCA1 plasmid encoding Clathrin Light Chain A1 was a gift by Eileen M. Lafer (UTHSC San Antonio, Texas, USA). The pEGFP-N1-Cav1 (encoding Caveolin-1-GFP) and pEGFP-N1-Dyn-2 (encoding Dynamin-2-GFP) constructs were provided by Mark McNiven (Mayo Clinic, Minnesota, USA) and the pcDNA3 4+ and 8+−GFP plasmids were a gift by John Silvius (McGill University, Montreal, Canada). The pEGFP-N1-Orai1 and pEYFP-N1-(PH)δ constructs were from Christoph Romanin. We further obtained pSRα(puro) CD43-45-YFP and pSRα(puro) Thy-1-CD45 plasmids from the lab of Anton van der Merwe (Oxford University, Oxford, UK). The pEGFP-C1-Lact-C2 (GFP fused with C2 domain of bovine Lactadherin) was purchased from Haematologic Technologies Inc., Essex Junction, Vermont, USA). The pEGFP-N1-Orai1 construct has been described previously [39].
2.3. Cell culture
Human T24, HeLa, CHO-K1 cells were from American Type Culture Collection Media, and HEK293 cells were provided by Christoph Romanin (JKU Linz, Austria). CHO-K1 cell stably expressing SR-BI-EGFP were a gift by Herbert Stangl (Medical University Vienna, Austria). Fetal bovine serum (FBS), media, antibiotics, trypsin and Geneticin (G418 sulphate) were purchased from PAA Laboratories GmbH, (Pasching, Austria). Culture plates were from Greiner Bio One International (Kremsmuenster, Austria). A Gene Pulser electroporation unit (X-cell) and electroporation cuvettes were from Bio-Rad (CA, USA).
T24 and HeLa cells were cultured in RPMI medium supplemented with 10% FBS and grown at 37 °C in a humidified incubator (≥ 95%) with 5% CO2. For stable expression 70% confluent cells were harvested and transfected with 10 μg plasmid DNA using the X-Cell electroporator with following electroporation conditions: 240 V, 950 μF, unlimited resistance, 4 mm gap cuvettes and RPMI16 without FBS as the electroporation-buffer. Cells were plated into 100 mm culture dishes and grown for 48 h. The medium was removed and replaced with fresh medium supplemented with 400 μg/ml G418. Medium was changed every 3 days, and 15–20 days later individual neomycin-resistant colonies were selected for propagation and analysis. CHO-K1 cells were cultured in DMEM High Glucose medium supplemented with 5% FBS; the same medium supplemented with 10% FBS was used to culture HEK293 cells. Transfection was performed with the X-Cell electroporator following preset protocols according to the manufacturer. Stable clones were established using the same procedure as for T24 cells. For transient expression the same transfection protocols were used. Transfected cells were grown on 3 cm glass coverslips and analyzed 48 h after seeding.
For the preparation of human immature dendritic cells whole human heparinized blood was obtained from the Red Cross and 500 ml of blood was diluted 1:1 in PBS. Diluted blood was placed on LSM 1077 lymphocyte separation medium (PAA, Pasching, Austria) gradient and centrifuged for 20 min at 350 g. Monocytes were harvested and CD14+ cells were selected by MACS purification (Miltenyi Biotec, Bergisch Gladbach, Germany) and incubated in RPMI 1640 supplemented with 10% Fetal bovine serum (PAA, Pasching, Austria), 1% l-glutamine, 1% Sodium-Pyruvate, 0.1% β-mercaptoethanol, 500 U/ml IL-4 and 0.1 μg/ml GM-CSF at 37 °C in 5% CO2 for 7 days. At day 6 of culture, CD83 (below 10%), CD1a (50–70%) and MHCII (70-90%) were assured by FACS analysis. At day 7 non-adherent cells were collected and used in subsequent experiments.
2.4. Fluorescence microscopy and image analysis
The detection system was set up on a home-made ultra-sensitive biochip scanner described in detail elsewhere [40,41]. The device is based on an epi-fluorescence microscope (Axiovert 200/M, Zeiss, Germany). Ar+- and Kr+-ion lasers (Innova, Coherent, USA) were used for selective fluorescence excitation of GFP, YFP and Cy5 at 488 nm, 514 nm and 647 nm, respectively. Samples were illuminated in objective-type total internal reflection (TIR) configuration using a 100× oil immersion objective (NA = 1.45, α-Fluar, Zeiss, Germany). After appropriate filtering using standard filter sets (Chroma Technology Corp., VT, USA), fluorescence was imaged onto a back-illuminated CCD camera (NTE/CCD-1340/100-EMB, Roper Scientific, USA). LSM images were taken by means of a LSM 510 Meta confocal laser scanning microscope using a 40 × 1.2NA water immersion objective (Zeiss, Germany). For microscopy, cells were placed in HBSS.
2.5. Fluorescence recovery after photobleaching (FRAP) experiments
FRAP experiments were performed at 37 °C on a setup described in detail previously [42]. A Zeiss Axiovert 200 microscope was equipped with a 100 × NA = 1.45 Plan-Apo objective (Olympus). Samples were illuminated in objective-type TIR configuration via the epiport using 488 nm (GFP excitation) from an Ar+ Laser (Model 2020, Spectra Physics). A slit diaphragm with a width of 7 μm was used as field stop to confine the illumination area. Images were recorded using a back-illuminated liquid nitrogen cooled CCD camera (Micro Max 1300-PB, Roper Scientific). Images were photobleached with a laser pulse applied for 650 ms; the recovery image was recorded at an illumination time of 1 ms, 30 s after the photobleaching pulse. A pre-bleach image was recorded with an illumination time of 1 ms before the photobleaching pulse in order to calculate the mobile fraction of the fluorescent probe.
2.6. Fluorescent labeling, quenching and neuraminidase treatment
For DiD or Bodipy-GM1 labeling, cells were grown on glass coverslips for 16 h and incubated with loading solution: 1 μM Bodipy-GM1 in HBSS for 30 min at 4 °C or 5 μl of the DiD labeling solution supplied by the manufacturer in 1 ml HBSS for 15 min at 37 °C was added. After loading, cells were rinsed with HBSS buffer for three times and immediately used for further experiments. For fluorescence quenching experiments cells grown on glass coverslips were washed with HBSS buffer and incubated with 0.2% Trypan Blue in HBSS for 2 min. Cells were washed three times with HBSS buffer before performing further experiment. Cleavage of sialic acid residues was achieved by incubation of cells grown on glass cover slips with 1 U/ml neuraminidase in HBSS buffer for 1 h at 37 °C.
2.7. Electron microscopy
T24 cells were grown on Aclar plastics to a cell density of 80% and incubated with 10 μM KLK dissolved in medium for 5 min. Control samples without KLK were treated under the same conditions. For antibody staining samples were incubated with an anti-GFP-antibody (Ab290; 1:1000; in blocking serum) for 1 h followed by washing for 3 times in blocking serum. An anti-rabbit HRP linked antibody in blocking serum was administered for 1 h. Microwave fixation and processing of cell monolayers for transmission electron microscopy was described in detail elsewhere [43]. Cells on Aclar were microwave-fixed in 0.5% glutaraldehyde in 0.15 M Sorensen's buffer, pH 7.4 for 15 s, followed by immersion in 3% glutaraldehyde for 30 min. For subsequent processing, including washing, postfixation in 1% OsO4, dehydration in ethanol, and infiltration with epoxy resin (Agar 100, Agar Scientific; Cambridge, UK), a tissue processor was used (LEICA TP, Microsystems, Vienna, Austria). After polymerization the resin block was separated from the Aclar plastics. Thin sections (60–80 nm) were cut with an ultramicrotome Ultracut S (LEICA Microsystems), mounted on copper grids, stained with uranyl acetate and lead citrate, and examined at 80 kV in a JEM-1210 electron microscope (JEOL; Tokyo, Japan). Images were acquired using a digital camera (Morada; Soft Imaging System GmbH, Münster, Germany) and analySIS FIVE software.
2.8. Cytotoxicity assays
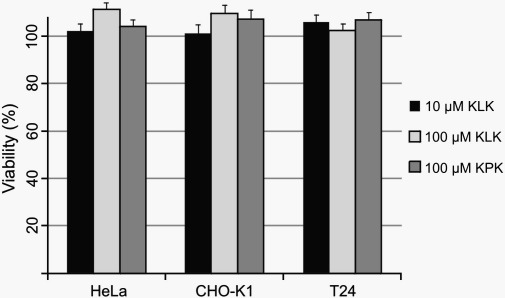
The Resazurin-based cytotoxicity assay (Sigma Aldrich, Schnellendorf, Germany) was performed according to the instructions of the manufacturer. Briefly, cells were seeded in 96-well plates and treated by KLK or KPK diluted in HBSS buffer for 15 min at room temperature. Subsequently, the cells were incubated with medium containing 10% Resazurin for 2 h. The reduced form of Resazurin (Resorufin) was then detected by a fluorescence microplate reader (Polarstar, BMG Labtech). The viability of KLK or KPK treated cells was normalized to the one of non-treated cells grown under the same conditions.
3. Results
3.1. KLK leads to accumulation of GPI-anchored proteins along the plasma membrane
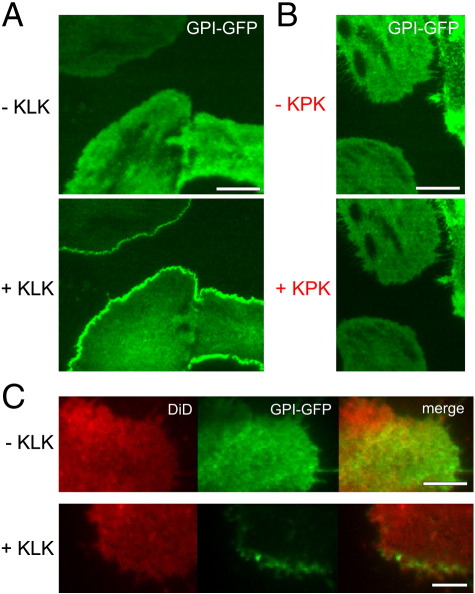
To test for an effect of KLK on glycosylated membrane proteins, we first studied the plasma membrane localization of GPI-anchored proteins in the human bladder carcinoma cell line T24. Fluorescence microscopy was performed under Total Internal Reflection (TIR) excitation to reduce cytosolic background fluorescence. Addition of 100 μM KLK led to a fast redistribution of monomeric GFP fused to the GPI-anchor of the human folate receptor (GPI-hFR-mGFP), resulting in the enrichment of this protein along the plasma membrane (Fig. 1A; Supplementary Movie 1). Similar results were obtained using the GPI-anchor of the decay accelerating factor (DAF or CD55; here termed GPI-DAF-GFP), of the TRAIL-receptor 3 (GPI-TRAIL-GFP), and of the GPI-anchored protein CD59-GFP (data not shown; see Table 1). The effect was also observed at tenfold lower concentrations of KLK (data not shown), but not with an immunologically inert derivative of KLK (KLKLLPLLKLK, KPK) [24] at 100 μM (Fig. 1B). Interestingly, we were not able to compete the effect of 100 μM KLK by simultaneous application of 1 mM KPK (Supplementary Fig. 1). Unless stated 100 μM peptide concentrations were used in all further experiments. Cells remained their viability under these circumstances (Supplementary Fig. 2).
Fig. 1.
KLK induces the accumulation of GPI-anchored proteins along the plasma membrane. T24 cells expressing GPI-hFR-mGFP were seeded on glass slides and analyzed by TIRF microscopy in the absence or presence of 100 μM KLK (A) or a mutant variant of KLK, termed KPK (B). (C) Two-color fluorescence microscopy experiments of the same cells additionally labeled with the fluorescent lipid tracer DiD. The incubation time of KLK or KPK was 5 min. Scale bars 20 μm (A), 25 μm (B), 10 μm (C).
Table 1.
List of all molecules which have been tested for sensitivity to KLK within this study, and their respective membrane localization.
| Molecules affected by KLK | KLK effect | Membrane localization |
|---|---|---|
| GPI-hFR-mGFP | accumulation in plasma membrane | outer leaflet inserted |
| GPI-Trail-GFP | accumulation in plasma membrane | outer leaflet inserted |
| GPI-DAF-GFP | accumulation in plasma membrane | outer leaflet inserted |
| GPI-CD58-GFP | accumulation in plasma membrane | outer leaflet inserted |
| CD59-GFP | accumulation in plasma membrane | outer leaflet inserted |
| Cholera Toxin B | accumulation in plasma membrane | bound to GM1 in outer leaflet |
| Bodipy-GM1 | accumulation in plasma membrane | outer leaflet inserted |
| CD43-YFP | accumulation in plasma membrane | single-pass type I membrane protein |
| CD63-GFP | accumulation in plasma membrane | multi-pass membrane protein |
| 4 + − GFP | cell rupture | inner leaflet associated |
| 8 + − GFP | cell rupture | inner leaflet associated |
| PH-YFP | cell rupture | inner leaflet associated |
| Molecules not affected by KLK | KLK effect | Membrane localization |
| Caveolin-1-GFP | none | inner leaflet inserted |
| Lck-GFP | none | inner leaflet inserted |
| Fyn-YFP | none | inner leaflet inserted |
| Flottilin1-GFP | none | peripheral inner leaflet |
| Flottilin2-GFP | none | peripheral inner leaflet |
| CD71-GFP | none | single-pass type II membrane protein |
| Orai1-GFP | none | multi-pass membrane protein |
| CD147-YFP | none | single-pass type I membrane protein |
| SRBI-GFP | none | multi-pass membrane protein |
| SRBII-GFP | none | multi-pass membrane protein |
| CD3ζ-GFP | none | single-pass type I membrane protein |
| Clathrin light chain-GFP | none | peripheral inner leaflet |
| DiD | none | plasma-membrane integrated |
Supplementary Fig. 1.
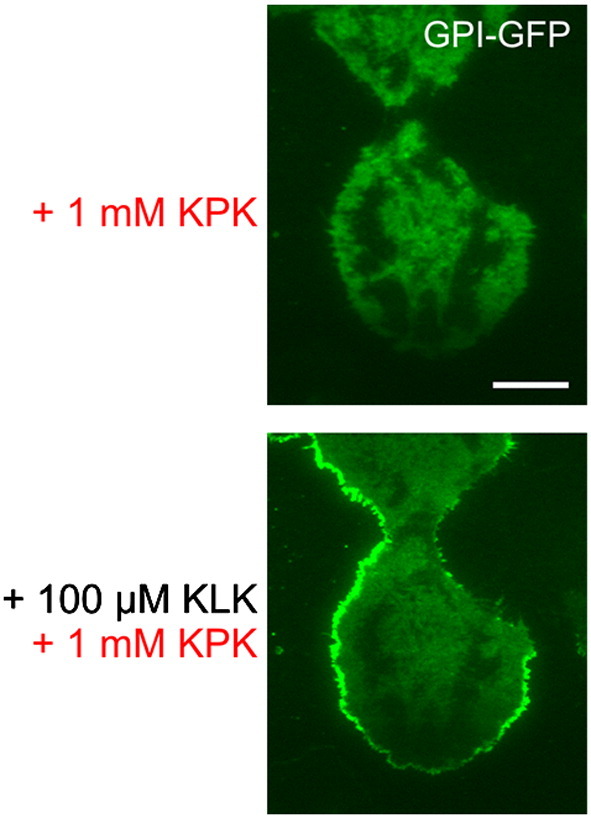
KPK does not compete for the effect of KLK by simultaneous application of KLK and KPK. T24 cells expressing GPI-hFR-mGFP were incubated with 1 mM KPK for 5 min followed by simultaneous application of 1 mM KPK and 100 μM KLK for 10 min. Scale bar 15 μm.
Supplementary Fig. 2.
KLK and KPK do not affect cell-viability. CHO-K1, HeLa and T24 cells were treated with KLK or KPK at the indicated concentrations for 15 min. The ability of all three cell lines to convert the non-fluorescent Resazurin into its fluorescent derivate Resorufin was not reduced for the analyzed cells. The viability of the cells was calculated from 5 individual measurements, and normalized to untreated cells. Error bars are based on the standard error of the mean.
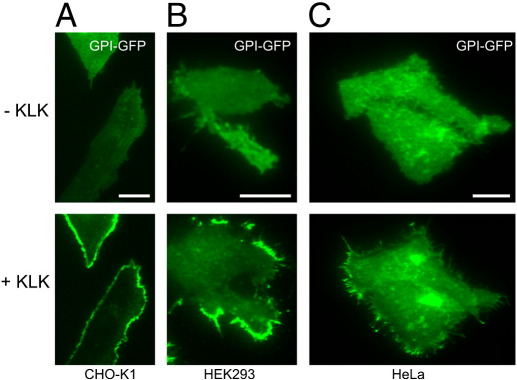
To control for a putative general effect of KLK on the distribution of plasma membrane constituents, we uniformly labeled the plasma membrane of GPI-hFR-mGFP-expressing T24 cells with the fluorescent lipid tracer DiD. Two-color fluorescence microscopy experiments revealed the accumulation of GPI-hFR-GFP upon KLK treatment as described above, while the distribution of DiD remained unaffected (Fig. 1C). In order to analyze the effects of KLK in different cell lines, we expressed GPI-hFR-mGFP in CHO-K1, HEK293 and HeLa cells and treated them with KLK. We found similar accumulation of GPI-hFR-mGFP in all three mammalian cell lines (Fig. 2A–C).
Fig. 2.
The effect of KLK on the localization of GPI-anchored proteins is not restricted to a certain cell line. CHO-K1 (A), HEK293 (B) and HeLa (C) cells expressing GPI-hFR-mGFP were analyzed by TIRF microscopy in the absence or the presence of 100 μM KLK (5 min incubation time). Scale bars 15 μm (A), 20 μm (B) and 10 μm (C).
3.2. KLK modulates the localization of various molecules along the plasma membrane
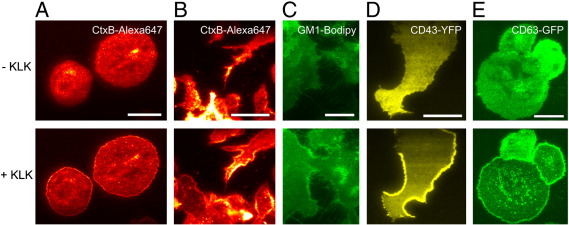
We next tried to corroborate the link between the effects of KLK on membrane targeting and protein or lipid glycosylation. For this, we studied different classes of membrane constituents, among them lipids, transmembrane proteins, inner- or outer leaflet integrated proteins and peripheral membrane proteins, and followed the effects of KLK on the localization of these molecules. The effect was observed for i) the monosialylated lipid GM1, which was either labeled via the fluorescent ligand Cholera Toxin (CtxB) (Fig. 3A and B on T24 and primary dendritic cells, respectively), or applied exogenously as fluorescent analogue (Fig. 3C); ii) the heavily sialylated single pass transmembrane protein CD43-YFP [44] (Fig. 3D); iii) and CD63-GFP, an integrin complexing protein of the tetraspanin family with putative sialylation motifs [45] (Fig. 3E).
Fig. 3.
KLK modulates the localization of various molecules in the plasma membrane. Experiments were performed using TIRF microscopy in the absence or presence of 100 μM KLK (5 min incubation time) on T24 or human immature dendritic cells labeled with CtxB-Alexa647 (A and B, respectively), T24 cells labeled with GM1-Bodipy (C), T24 cells expressing CD43-YFP (D) or CD63-GFP (E). Scale bars 35 (A), 30 (B), 30 (C), 20 (D) and 30 (E) μm, respectively.
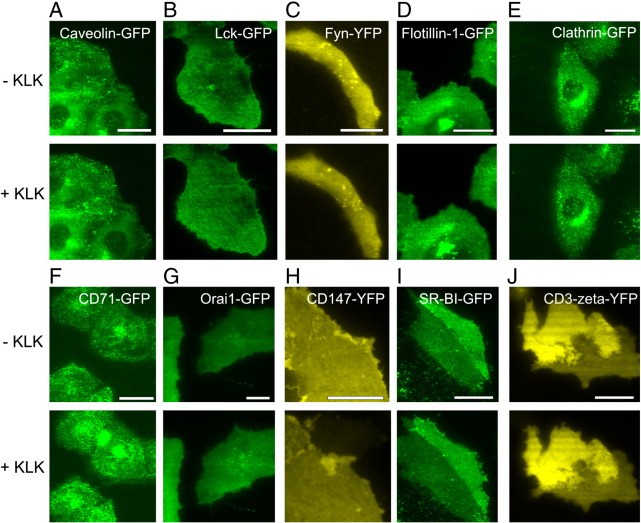
We also tested inner leaflet-associated proteins: i) Caveolin-1-GFP, an integral scaffolding protein (Fig. 4A); ii) Lck-GFP (Fig. 4B) and Fyn-YFP (Fig. 4C), two peripheral kinases involved in early T cell signaling [46,47]; iii) Flotillin-1-GFP (Fig. 4D) and Flotillin-2-GFP (data not shown) [48] and iv) Clathrin light chain-GFP [49] (Fig. 4E). These proteins yielded no effect upon KLK treatment. We also observed no effect for the following transmembrane proteins: i) the transferrin-receptor CD71-GFP [50] (Fig. 4F); ii) the calcium channel-forming polytopic transmembrane protein Orai1-GFP [51] (Fig. 4G); iii) the integral membrane receptor CD147-YFP [52] (Fig. 4H); iv) the polytopic transmembrane and preferentially caveolae-associated receptor SR-BI-GFP [53] (Fig. 4I); v) CD3ζ-YFP [54] (Fig. 4J). Table 1 summarizes all molecules which have been tested in this study.
Fig. 4.
KLK does not affect the localization of specific plasma membrane proteins. Experiments were performed using TIRF microscopy in the absence or presence of 100 μM KLK (5 min incubation time) on T24 cells expressing Caveolin-1-GFP (A), Lck-GFP (B), Fyn-YFP (C), Flotillin-1-GFP (D), Clathrin light chain-GFP (E), CD71-GFP (F), Orai1-GFP (G), CD147-YFP (H), SR-BI-GFP (I) or CD3ζ-YFP (J). Scale bars 30 (A), 35 (B), 10 (C), 30 (D), 20 (E), 30 (F), 20 (G), 10 (H), 20 (I) and 15 (J) μm, respectively.
3.3. KLK interacts with the ectodomain of the affected protein
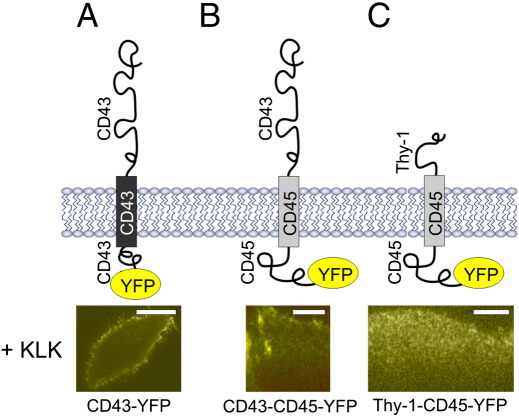
To further confirm the relevance of the exoplasmic protein parts for the observed redistribution, we studied the effect of KLK using chimeric proteins [55]. The transmembrane region of CD45 fused to Thy-1 showed no effect upon KLK treatment. However, when Thy-1 was replaced by the ectodomain of CD43, the KLK effect could be reconstituted (Fig. 5). This result indicates that the extra-cellular domain is responsible for the interaction with the peptide.
Fig. 5.
KLK interacts with the ectodomain of the affected protein. HeLa cells expressing CD43-YFP (A) or the fusion proteins CD43-CD45-YFP (B) or Thy1-CD45-YFP (C) were incubated with 100 μM KLK for 5 min and analyzed using TIRF microscopy. Scales bars 20 (A), 15 (B) and 10 (C) μm, respectively.
3.4. KLK-mediated re-localization requires modification of the respective protein with sialic acids
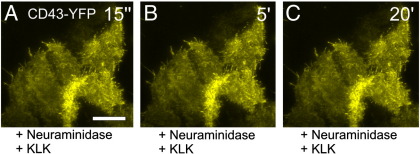
To directly prove the role of sialic acids we pretreated HeLa cells expressing CD43-YFP with neuraminidase, which hydrolyses N-acetyl-neuraminic acid residues from glycoproteins leading to the cleavage of unbranched and branched sialic acids [56]. As already shown in Fig. 3D KLK leads to the accumulation of the highly sialylated protein CD43. We speculated that the cleavage of sialic acids from CD43 would prohibit its accumulation in the plasma membrane upon KLK treatment. Indeed, KLK did not change the localization of CD43-YFP when the cells were pretreated by neuraminidase (Fig. 6). Similar results were found for neuraminidase-treated cells expressing GPI-hFR-mGFP (data not shown). We conclude that KLK interacts with sialic acids resulting in the accumulation of the affected protein in the plasma membrane. Due to the positive charge of KLK a direct binding of the peptide to the negatively charged sialic acids seems likely.
Fig. 6.
The KLK mediated re-localization of membrane-localized proteins is linked to a post-translational modification with sialic acids. HeLa cells expressing CD43-YFP were incubated with 1 U/ml neuraminidase to cleave sialic acids and treated afterwards with 100 μM KLK for 20 min. Pictures were taken 15 s, 5 min and 20 min after KLK addition. Scale bar 10 μm.
3.5. KLK leads to an immobilization of the accumulated molecules
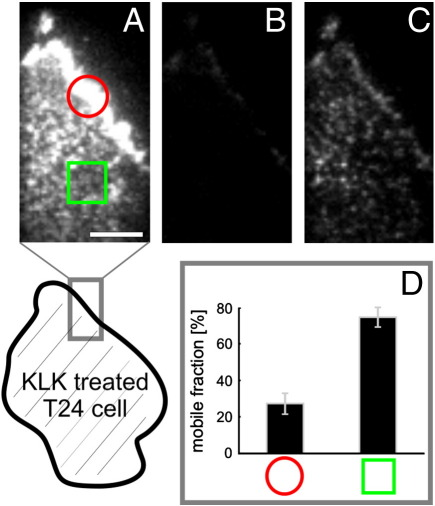
In order to analyze the mobility of proteins that showed altered localization upon KLK treatment, we carried out fluorescence recovery after photobleaching (FRAP) experiments. We treated GPI-hFR-mGFP expressing T24 cells with KLK and determined the mobile fraction after 30 s within defined regions (Fig. 7). When selecting a region of KLK-induced GPI-hFR-mGFP accumulation (marked with a red circle) we determined a mobile fraction of 24% (Fig. 7D), which is substantially lower compared to untreated cells (~ 80%)[42]. Regions in the bottom part of the plasma membrane (marked with a green square) showed a high mobile fraction similar to untreated cells, indicating that the peptide was not able to diffuse into the gap between the cell and the glass slide.
Fig. 7.
KLK leads to an immobilization of the accumulated proteins. T24 cells expressing GPI-hFR-mGFP were treated with 100 μM KLK for 30 s and Fluorescence Recovery After Photobleaching (FRAP) experiments were performed. The mobile fraction within two defined regions (encircled region along the plasma membrane or region marked with a square below the cell) was recorded. Panel A indicates the fluorescence image before, panel B and C the fluorescence image 100 ms and 30 s after the bleach pulse, respectively. Scale bar 3 μm. The mobile fraction within the two regions is plotted in panel D and was calculated from measurements of ~ 15 individual cells. Error bars are based on the standard error of the mean.
3.6. KLK induces invaginations of the plasma membrane
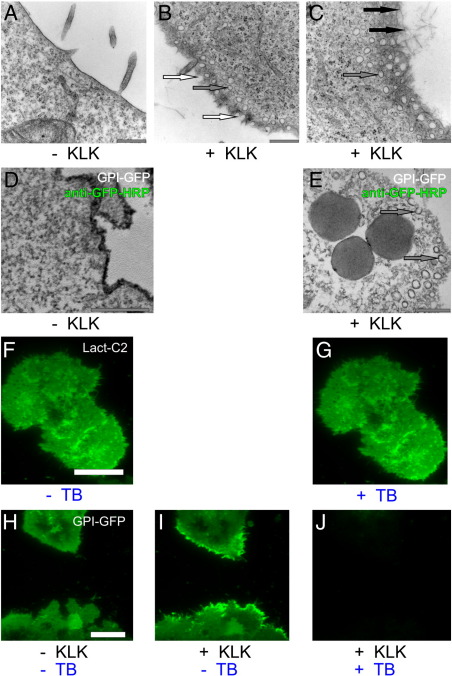
To visualize the effects of KLK on the plasma membrane at high resolution, we used transmission electron microscopy on thin sections (60–80 nm) cut in parallel to the plane of cell growth. When comparing T24 cells incubated with 10 μM KLK to untreated cells, two effects became apparent: Firstly, the plasma membrane showed dramatic morphological changes including numerous membrane invaginations (marked with white arrows), and additional circular structures near the plasma membrane (marked with gray arrows) (Fig. 8A–C). These structures were found to be enriched in GPI-hFR-mGFP as shown by antibody-staining (Fig. 8D–E). Secondly, large filamentous aggregates were formed in the extra-cellular space (marked with black arrows in Fig. 8C).
Fig. 8.
KLK induces the formation of plasma membrane invaginations. Electron microscopic images of cross-sectioned plasma membrane in the periphery of T24 cells were recorded in the absence (A) or after treatment with 10 μM KLK (B–C). Panels D and E show EM images of T24 cells expressing GPI-hFR-mGFP stained with a HRP-conjugated anti-GFP antibody before and after KLK treatment, respectively (incubation time 5 min). White arrows mark membrane invaginations, gray arrows indicate additional circular structures near the plasma membrane and black arrows mark large filamentous aggregates in the extra-cellular space. Additionally, the membrane impermeable quencher Trypan Blue (TB) was used to determine the plasma membrane localization of GPI-anchored proteins after KLK treatment. T24 cells expressing GPI-hFR-mGFP (H) were incubated with 100 μM KLK for 5 min (I) and treated with 0.2% TB (J). As a negative control T24 cells expressing the Phosphatidylserine (PS) binding probe Lact-C2-GFP were imaged by TIRF microscopy without (F) or with 0.2% Trypan Blue (G). Scale bars 20 (A), 10 (C), 1 (F), 0.5 (G and H) and 0.3 (I and J) μm, respectively.
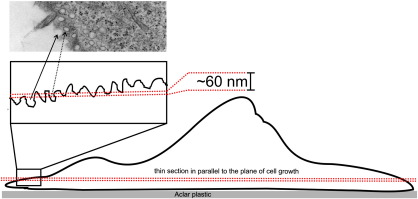
While the invaginations most likely represent fusion or fission intermediates, the interpretation of the circular structures is a priori not clear: they may be interpreted as membrane proximal vesicles, or as the consequence of invaginations cut in the flattened peripheral cell extension at a tangential cutting angle (see Supplementary Fig. 3). To decide which of these interpretations is correct, we performed fluorescence quenching experiments. We used the membrane-impermeable quencher Trypan Blue, which efficiently reduces the fluorescence of various fluorophores. In the negative control, Trypan Blue did not decrease the fluorescence of the inner-leaflet peripheral membrane protein Lact-C2-GFP (Fig. 8F–G). We then used KLK to accumulate GPI-hFR-mGFP in the plasma membrane of T24 cells. Addition of Trypan Blue efficiently decreased the fluorescence, in particular in regions with accumulated GPI-hFR-mGFP (Fig. 8H–J), indicating that the fluorescent protein is accessible to the extracellular milieu. We conclude that the accumulated proteins are located in plasma membrane invaginations, but not in membrane proximal vesicles.
Supplementary Fig. 3.
Sketch displaying invaginations of the plasma membrane cut tangentially to its surface as they were induced by incubation of the T24 cells with 10 μM KLK (5 min incubation time). The resulting EM micrograph of a thin section (60–80 nm), therefore, shows both invaginations at the cell border (solid line arrow) and circular structures surrounded by cytoplasm (dotted line arrow). T24 cells are characterized by flat spreading of the cytoplasm at their periphery [72].
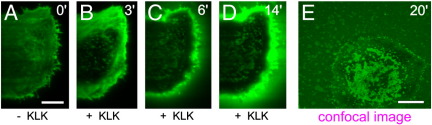
3.7. KLK ruptures the plasma membrane in the presence of positively charged proteins bound to the inner leaflet
When testing the effect of KLK on the various proteins and lipids, we made a surprising observation: KLK treatment of cells overexpressing inner leaflet-associated proteins with positive net charge resulted in a complete rupture of the cells. This result appeared interesting, knowing that there is a manifold of positively charged proteins such as c-Src, Rac1 or K-Ras, which are targeted to the inner leaflet of the plasma membrane via cationic motifs [57], and which can be expressed at high rates—similar to our overexpressed constructs—under specific signaling conditions [58,59]. To explore this phenomenon in more detail we expressed GFP-labeled “charge sensors,” which combine a hydrophobic farnesyl chain with an adjacent sequence of varying net positive charge [60]. These probes bind to negatively charged lipid species at the inner leaflet, particularly phosphatidylserine, phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and phosphatidylinositol (4,5)-bisphosphate (PIP2) [61,62]. The sensor with the highest net charge, 8+-GFP, was localized preferentially to the inner plasma membrane leaflet. Within 3–6 min upon KLK treatment we detected changes in its localization and finally a complete breakup of the cells after 10–15 min (Fig. 9). This phenomenon was also observed for T24 cells expressing a PIP2/PIP3-binding Pleckstrin-homology (PH)-YFP domain, and with a 4+-GFP sensor (data not shown).
Fig. 9.
KLK ruptures the plasma membrane in the presence of positively charged proteins bound to the inner leaflet. T24 cells expressing a GFP-labeled probe that binds to negatively charged lipids in the inner plasma membrane leaflet (8+-GFP) were analyzed by TIRF microscopy after incubation with 100 μM KLK. Images were taken before, and 3, 6 and 14 min upon KLK addition. Additionally, an image was recorded 20 min after KLK treatment using confocal microscopy (E). Scale bars 10 (A) and 15 μm (E).
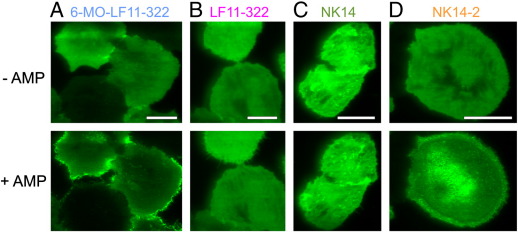
3.8. Hydrophobic regions are required to arrest sialylated proteins at the plasma membrane
To obtain also mechanistic insights into the action of CAMPs on host cell membranes we tested additional substances. LF11-322 is a nonapeptide derived from Lactoferricin [63]; N-terminal acylation with 6-methyloctanoyl to increase its membrane affinity yielded the variant 6-MO-LF11-322. Both CAMPs showed broad spectrum of antimicrobial activity with MICs > 1 μM (Zweytick et al., manuscript submitted). Similar to the treatment with KLK, we found a clear redistribution of CD43 in T24 cells upon incubation with 6-MO-LF11-322 (Fig. 10A). Interestingly, incubation of the cells with the non-acylated version LF11-322 showed no recognizable effect (Fig. 10B). We also tested the CAMP NK14, an amphipathic peptide derived from NK-2 [64,65], and its dimeric version (NK14-2). While NK14 did not show any effect on CD43 localization, we found a clear redistribution upon treatment with NK14-2 (Fig. 10C and D).
Fig. 10.
Effects of further antimicrobial peptides on the localization of sialylated membrane proteins. TIRF microscopy images of T24 cells expressing CD43-GFP were incubated for 10 min with 80 μM 6-MO-LF11-322 (A), 80 μM LF11-322 (B), 40 μM NK14 (C) and 20 μM NK14-2 (D). Scale bars 25 (A), 20 (B), 25 (C) and 15 (D) μm.
4. Discussion
We report here that application of the CAMPs KLK, 6-MO-LF11-322 and NK14-2 affects plasma membrane processes also on mammalian host cells. In particular, we observed two effects:
-
i)
Sialylated membrane proteins or lipids become immobilized and their recycling is arrested. Sialylated proteins are involved in such diverse processes as signaling [66], cellular adhesion and migration [67], receptor-mediated endocytosis and immune response [68,69]. KLK may thus impose a broad spectrum of detrimental nonlethal effects also to host cells. Interestingly, non-sialylated proteins were not affected by these massive rearrangements.
At this stage we cannot rule out indirect effects; however, our observations can be consistently summarized in a mechanistic model of CAMP-induced immobilization. In contrast to the mutant variant KPK, KLK has a strong tendency to multimerize [24]. Moreover, its high number of leucines generates a hydrophobic domain, thereby yielding a great affinity towards membranes; a central proline reduces the membrane affinity for KPK [24]. It thus appears likely that KLK associates with the plasma membrane resulting in a high surface density, so that negatively charged protein modifications such as sialic acids can be efficiently bound by the positive charges on the CAMP. In consequence, sialylated molecules are multimerized and thereby immobilized in the plasma membrane. The free energy of this process includes therefore electrostatic and hydrophobic contributions. Since KPK lacks the membrane affinity, its density at the plasma membrane is substantially lower and not sufficient to mediate the effect; in other words, the hydrophobic term in the free energy is not sufficient for stabilizing the interaction. Consequentially, it was not possible to compete for the KLK-induced crosslink by excess concentrations of KPK.
Similar differential effects were observed for other CAMPs. We found that only the acylated version of LF11-322—termed 6-MO-LF11-322—mediated the arrest of CD43 at the plasma membrane. Also for the amphipathic CAMPs NK14 and NK14-2 we observed strong differences in their ability to arrest CD43: only the dimeric peptide exhibiting a higher hydrophobic moment was able to exert the effect, consistent with the higher membrane affinity of the dimeric construct. In conclusion, cooperation between hydrophobicity and electrostatic interactions may boost adverse effects of CAMPs on eukaryotic host cell membranes. At this stage, we can only speculate about the origin of the observed arrest of sialylated proteins in the plasma membrane: invaginations may represent fission or fusion intermediates, but may also be explained by tension-induced formation of structures which are not related to the cellular uptake machinery.
-
ii)
The plasma membrane gets more susceptible to rupture. We observed KLK to cause the total disintegration of the plasma membrane when positively charged proteins were overexpressed. In our study we used a protein motif derived from K-Ras [60], overexpression of which is involved e.g. in cell proliferation [70]. Potential causes for the observed effect include the reduced elasticity of the membrane due to the crowding of adsorbed proteins on both leaflets, but also the shielding of phosphoinositides, thereby making them unavailable for signaling. Moreover, lactoferricin derived peptides were shown to increase curvature stress in lipid bilayer promoting the formation of highly curved cubic structures [71]. Application of KLK as a CAMP may therefore well disrupt cells containing high levels of cytosolic proteins with polybasic motifs.
Taken together our study demonstrates potential far-reaching effects of the application of CAMPs on the eukaryotic host cell and provides insight into the consequences of peptide binding on the plasma membrane localization and endocytosis of a subgroup of membrane molecules. These findings are of great relevance for the usage of cationic antimicrobial peptides per se: since many of them have to be applied at high concentrations, it is likely that their usage does not only affect the pathogen, but also the host cells. Whether similar effects occur in eukaryotic cells upon exposition to natural CAMPs may be an interesting topic for future studies.
The following are the supplementary materials related to this article.
Temporal resolution of KLK induced accumulation of GPI-anchored proteins. The movie contains 20 TIRF images of T24 cells expressing GPI-hFR-mGFP, which were recorded at 1 min intervals; the overview shows an area of ~ 400 × 400 μm2. 100 μM KLK was added immediately before the first image. The images show two stripes (top and bottom half) that were recorded in succession, yielding a slight incongruity at the assembly line.
Acknowledgments
We thank the following colleagues for providing plasmids and cells: Daniel Legler (University of Konstanz, Switzerland), Jennifer Lippincott-Schwartz (NIH, Bethesda, USA), Hannes Stockinger (Medical University Vienna, Austria), Katharina Strub (University of Geneva, Switzerland), Lawrence Rajendran (ETH Zurich, Switzerland), Eileen M. Lafer (UTHSC San Antonio, Texas, USA), Mark McNiven (Mayo Clinic, Minnesota, USA), John Silvius (McGill University, Montreal, Canada), Christoph Romanin (JKU Linz, Austria), Herbert Stangl (Medical University Vienna, Austria) and Anton van der Merwe (Oxford University, Oxford, UK). We thank Harald Kotisch (MFPL, Vienna) for excellent technical assistance in the processing of samples for electron microscopy and Sergio Grinstein (Hospital for Sick Children Research Institute, Toronto) for fruitful discussions. This work was funded by the GEN-AU project of the Austrian Research Promotion Agency, the Austrian Science Fund (FWF; project Y250-B03) and Intercell AG.
Contributor Information
Julian Weghuber, Email: julian.weghuber@fh-wels.at.
Gerhard J. Schütz, Email: gerhard.schuetz@jku.at.
References
- 1.Wang G., Li X., Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 4.Baba T., Schneewind O. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 1998;6:66–71. doi: 10.1016/S0966-842X(97)01196-7. [DOI] [PubMed] [Google Scholar]
- 5.Bulet P., Stocklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept. Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- 6.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Olmedo F., Molina A., Alamillo J.M., Rodriguez-Palenzuela P. Plant defense peptides. Biopolymers. 1998;47:479–491. doi: 10.1002/(SICI)1097-0282(1998)47:6<479::AID-BIP6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Nascimento A.C., Fontes W., Sebben A., Castro M.S. Antimicrobial peptides from anurans skin secretions. Protein Pept. Lett. 2003;10:227–238. doi: 10.2174/0929866033478933. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsson G.H., Agerberth B. Neutrophil antibacterial peptides, multifunctional effector molecules in the mammalian immune system. J. Immunol. Methods. 1999;232:45–54. doi: 10.1016/s0022-1759(99)00152-0. [DOI] [PubMed] [Google Scholar]
- 10.Lehrer R.I., Ganz T. Defensins of vertebrate animals. Curr. Opin. Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 11.Zaiou M., Gallo R.L. Cathelicidins, essential gene-encoded mammalian antibiotics. J. Mol. Med. 2002;80:549–561. doi: 10.1007/s00109-002-0350-6. [DOI] [PubMed] [Google Scholar]
- 12.Jenssen H., Hamill P., Hancock R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott R.W., DeGrado W.F., Tew G.N. De novo designed synthetic mimics of antimicrobial peptides. Curr. Opin. Biotechnol. 2008;19:620–627. doi: 10.1016/j.copbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blondelle S.E., Lohner K. Optimization and high-throughput screening of antimicrobial peptides. Curr. Pharm. Des. 2010;16:3204–3211. doi: 10.2174/138161210793292438. [DOI] [PubMed] [Google Scholar]
- 15.Lohner K., Latal A., Lehrer R.I., Ganz T. Differential scanning microcalorimetry indicates that human defensin, HNP-2, interacts specifically with biomembrane mimetic systems. Biochemistry. 1997;36:1525–1531. doi: 10.1021/bi961300p. [DOI] [PubMed] [Google Scholar]
- 16.Lohner K., Blondelle S.E. Molecular mechanisms of membrane perturbation by antimicrobial peptides and the use of biophysical studies in the design of novel peptide antibiotics. Comb. Chem. High Throughput Screen. 2005;8:241–256. doi: 10.2174/1386207053764576. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzaki K. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 18.Asaduzzaman S.M., Sonomoto K. Lantibiotics: diverse activities and unique modes of action. J. Biosci. Bioeng. 2009;107:475–487. doi: 10.1016/j.jbiosc.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas P. Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 2009;276:6483–6496. doi: 10.1111/j.1742-4658.2009.07359.x. [DOI] [PubMed] [Google Scholar]
- 20.Dhople V., Krukemeyer A., Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim. Biophys. Acta. 2006;1758:1499–1512. doi: 10.1016/j.bbamem.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Durr U.H., Sudheendra U.S., Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta. 2006;1758:1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 22.Nijnik A., Hancock R.E. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr. Opin. Hematol. 2009;16:41–47. doi: 10.1097/moh.0b013e32831ac517. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Bravo J., Kurata S., Natori S. Novel synthetic antimicrobial peptides effective against methicillin-resistant Staphylococcus aureus. Biochem. J. 1994;302(Pt 2):535–538. doi: 10.1042/bj3020535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aichinger M.C., Ortbauer M., Reipert S., Zauner W., Bogner P., Froschauer E., Nowikovsky K., Lingnau K., von G.A., Schweyen R., Henics T. Unique membrane-interacting properties of the immunostimulatory cationic peptide KLKL(5)KLK (KLK) Cell Biol. Int. 2008;32:1449–1458. doi: 10.1016/j.cellbi.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 25.Cho J.H., Homma K., Kanegasaki S., Natori S. Activation of human neutrophils by a synthetic anti-microbial peptide, KLKLLLLLKLK-NH2, via cell surface calreticulin. Eur. J. Biochem. 1999;266:878–885. doi: 10.1046/j.1432-1327.1999.00920.x. [DOI] [PubMed] [Google Scholar]
- 26.Cho J.H., Homma K.J., Kanegasaki S., Natori S. Activation of human monocyte cell line U937 via cell surface calreticulin. Cell Stress Chaperones. 2001;6:148–152. doi: 10.1379/1466-1268(2001)006<0148:aohmcl>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima Y., Alvarez-Bravo J., Cho J., Homma K., Kanegasaki S., Natori S. Chemotherapeutic activity of synthetic antimicrobial peptides: correlation between chemotherapeutic activity and neutrophil-activating activity. FEBS Lett. 1997;415:64–66. doi: 10.1016/s0014-5793(97)01101-0. [DOI] [PubMed] [Google Scholar]
- 28.Fritz J.H., Brunner S., Birnstiel M.L., Buschle M., Gabain A., Mattner F., Zauner W. The artificial antimicrobial peptide KLKLLLLLKLK induces predominantly a TH2-type immune response to co-injected antigens. Vaccine. 2004;22:3274–3284. doi: 10.1016/j.vaccine.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Weghuber J., Lipp A.M., Stadlbauer J., Aichinger M.C., Ruprecht V., Sonnleitner A., Schutz G.J., Henics T. Antimicrobial and immunostimulatory peptide, KLK, induces an increase in cytosolic Ca2+ concentration by mobilizing Ca2+ from intracellular stores. Cell Biol. Int. 2010;34(11):1109–1112. doi: 10.1042/CBI20100408. [DOI] [PubMed] [Google Scholar]
- 30.Aichinger M.C., Ginzler M., Weghuber J., Zimmermann L., Riedl K., Schutz G., Nagy E., von G.A., Schweyen R., Henics T. Adjuvating the adjuvant: facilitated delivery of an immunomodulatory oligonucleotide to TLR9 by a cationic antimicrobial peptide in dendritic cells. Vaccine. 2011;29:426–436. doi: 10.1016/j.vaccine.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Schellack C., Prinz K., Egyed A., Fritz J.H., Wittmann B., Ginzler M., Swatosch G., Zauner W., Kast C., Akira S., von G.A., Buschle M., Lingnau K. IC31, a novel adjuvant signaling via TLR9, induces potent cellular and humoral immune responses. Vaccine. 2006;24:5461–5472. doi: 10.1016/j.vaccine.2006.03.071. [DOI] [PubMed] [Google Scholar]
- 32.Melo M.N., Ferre R., Castanho M.A. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009;7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- 33.Hancock R.E. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 34.Leontiadou H., Mark A.E., Marrink S.J. Antimicrobial peptides in action. J. Am. Chem. Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- 35.Hwang P.M., Vogel H.J. Structure–function relationships of antimicrobial peptides. Biochem. Cell Biol. 1998;76:235–246. doi: 10.1139/bcb-76-2-3-235. [DOI] [PubMed] [Google Scholar]
- 36.Pistolesi S., Pogni R., Feix J.B. Membrane insertion and bilayer perturbation by antimicrobial peptide CM15. Biophys. J. 2007;93:1651–1660. doi: 10.1529/biophysj.107.104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thennarasu S., Huang R., Lee D.K., Yang P., Maloy L., Chen Z., Ramamoorthy A. Limiting an antimicrobial peptide to the lipid–water interface enhances its bacterial membrane selectivity: a case study of MSI-367. Biochemistry. 2010;49:10595–10605. doi: 10.1021/bi101394r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols B.J., Kenworthy A.K., Polishchuk R.S., Lodge R., Roberts T.H., Hirschberg K., Phair R.D., Lippincott-Schwartz J. Rapid cycling of lipid raft markers between the cell surface and Golgi complex. J. Cell Biol. 2001;153:529–541. doi: 10.1083/jcb.153.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madl J., Weghuber J., Fritsch R., Derler I., Fahrner M., Frischauf I., Lackner B., Romanin C., Schutz G.J. Resting-state Orai1 diffuses as homotetramer in the plasma membrane of live mammalian cells. J. Biol. Chem. 2010;285(52):41135–41142. doi: 10.1074/jbc.M110.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hesse J., Sonnleitner M., Sonnleitner A., Freudenthaler G., Jacak J., Hoglinger O., Schindler H., Schutz G.J. Single-molecule reader for high-throughput bioanalysis. Anal. Chem. 2004;76:5960–5964. doi: 10.1021/ac049300f. [DOI] [PubMed] [Google Scholar]
- 41.Hesch C., Hesse J., Jacak J., Schutz G.J. Two-stage focus-hold system for rapid ultra-sensitive read-out of large-area biochips. J. Microsc. 2009;234:251–254. doi: 10.1111/j.1365-2818.2009.03165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brameshuber M., Weghuber J., Ruprecht V., Gombos I., Horvath I., Vigh L., Eckerstorfer P., Kiss E., Stockinger H., Schuetz G.J. Imaging of mobile long-lived nanoplatfroms in the live cell plasma membrane. J. Biol. Chem. 2010;285(53):41765–41771. doi: 10.1074/jbc.M110.182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reipert S., Kotisch H., Wysoudil B., Wiche G. Rapid microwave fixation of cell monolayers preserves microtubule-associated cell structures. J. Histochem. Cytochem. 2008;56:697–709. doi: 10.1369/jhc.7A7370.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostberg J.R., Barth R.K., Frelinger J.G. The Roman god Janus: a paradigm for the function of CD43. Immunol. Today. 1998;19:546–550. doi: 10.1016/s0167-5699(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 45.Lewandrowski U., Zahedi R.P., Moebius J., Walter U., Sickmann A. Enhanced N-glycosylation site analysis of sialoglycopeptides by strong cation exchange prefractionation applied to platelet plasma membranes. Mol. Cell. Proteomics. 2007;6:1933–1941. doi: 10.1074/mcp.M600390-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Turner J.M., Brodsky M.H., Irving B.A., Levin S.D., Perlmutter R.M., Littman D.R. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 1990;60:755–765. doi: 10.1016/0092-8674(90)90090-2. [DOI] [PubMed] [Google Scholar]
- 47.Palacios E.H., Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 48.Frick M., Bright N.A., Riento K., Bray A., Merrified C., Nichols B.J. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 2007;17:1151–1156. doi: 10.1016/j.cub.2007.05.078. [DOI] [PubMed] [Google Scholar]
- 49.Brodsky F.M., Hill B.L., Acton S.L., Nathke I., Wong D.H., Ponnambalam S., Parham P. Clathrin light chains: arrays of protein motifs that regulate coated-vesicle dynamics. Trends Biochem. Sci. 1991;16:208–213. doi: 10.1016/0968-0004(91)90087-c. [DOI] [PubMed] [Google Scholar]
- 50.Bleil J.D., Bretscher M.S. Transferrin receptor and its recycling in HeLa cells. EMBO J. 1982;1:351–355. doi: 10.1002/j.1460-2075.1982.tb01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.H., Tanasa B., Hogan P.G., Lewis R.S., Daly M., Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 52.Curtin K.D., Meinertzhagen I.A., Wyman R.J. Basigin (EMMPRIN/CD147) interacts with integrin to affect cellular architecture. J. Cell Sci. 2005;118:2649–2660. doi: 10.1242/jcs.02408. [DOI] [PubMed] [Google Scholar]
- 53.Graf G.A., Matveev S.V., Smart E.J. Class B scavenger receptors, caveolae and cholesterol homeostasis. Trends Cardiovasc. Med. 1999;9:221–225. doi: 10.1016/s1050-1738(00)00031-1. [DOI] [PubMed] [Google Scholar]
- 54.Dave V.P. Hierarchical role of CD3 chains in thymocyte development. Immunol. Rev. 2009;232:22–33. doi: 10.1111/j.1600-065X.2009.00835.x. [DOI] [PubMed] [Google Scholar]
- 55.Irles C., Symons A., Michel F., Bakker T.R., van der Merwe P.A., Acuto O. CD45 ectodomain controls interaction with GEMs and Lck activity for optimal TCR signaling. Nat. Immunol. 2003;4:189–197. doi: 10.1038/ni877. [DOI] [PubMed] [Google Scholar]
- 56.Cassidy J.T., Jourdian G.W., Roseman S. The sialic acids. VI. Purification and properties of sialidase from Clostridium perfringens. J. Biol. Chem. 1965;240:3501–3506. [PubMed] [Google Scholar]
- 57.Yeung T., Gilbert G.E., Shi J., Silvius J., Kapus A., Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 58.Brown M.T., Cooper J.A. Regulation, substrates and functions of src. Biochim. Biophys. Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 59.Ridley A.J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 60.Roy M.O., Leventis R., Silvius J.R. Mutational and biochemical analysis of plasma membrane targeting mediated by the farnesylated, polybasic carboxy terminus of K-ras4B. Biochemistry. 2000;39:8298–8307. doi: 10.1021/bi000512q. [DOI] [PubMed] [Google Scholar]
- 61.Heo W.D., Inoue T., Park W.S., Kim M.L., Park B.O., Wandless T.J., Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vance J.E., Steenbergen R. Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 2005;44:207–234. doi: 10.1016/j.plipres.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Zorko M., Japelj B., Hafner-Bratkovic I., Jerala R. Expression, purification and structural studies of a short antimicrobial peptide. Biochim. Biophys. Acta. 2009;1788:314–323. doi: 10.1016/j.bbamem.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 64.Andra J., Leippe M. Candidacidal activity of shortened synthetic analogs of amoebapores and NK-lysin. Med. Microbiol. Immunol. 1999;188:117–124. doi: 10.1007/s004300050113. [DOI] [PubMed] [Google Scholar]
- 65.Gofman Y., Linser S., Rzeszutek A., Shental-Bechor D., Funari S.S., Ben-Tal N., Willumeit R. Interaction of an antimicrobial peptide with membranes: experiments and simulations with NKCS. J. Phys. Chem. B. 2010;114:4230–4237. doi: 10.1021/jp909154y. [DOI] [PubMed] [Google Scholar]
- 66.Kontou M., Weidemann W., Bork K., Horstkorte R. Beyond glycosylation: sialic acid precursors act as signaling molecules and are involved in cellular control of differentiation of PC12 cells. Biol. Chem. 2009;390:575–579. doi: 10.1515/BC.2009.058. [DOI] [PubMed] [Google Scholar]
- 67.Gascon E., Vutskits L., Kiss J.Z. Polysialic acid-neural cell adhesion molecule in brain plasticity: from synapses to integration of new neurons. Brain Res. Rev. 2007;56:101–118. doi: 10.1016/j.brainresrev.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 68.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 69.Bi S., Baum L.G. Sialic acids in T cell development and function. Biochim. Biophys. Acta. 2009;1790:1599–1610. doi: 10.1016/j.bbagen.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 70.Schubbert S., Bollag G., Shannon K. Deregulated Ras signaling in developmental disorders: new tricks for an old dog. Curr. Opin. Genet. Dev. 2007;17:15–22. doi: 10.1016/j.gde.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 71.Zweytick D., Tumer S., Blondelle S.E., Lohner K. Membrane curvature stress and antibacterial activity of lactoferricin derivatives. Biochem. Biophys. Res. Commun. 2008;369:395–400. doi: 10.1016/j.bbrc.2008.01.176. [DOI] [PubMed] [Google Scholar]
- 72.Weghuber J., Sunzenauer S., Plochberger B., Brameshuber M., Haselgrubler T., Schutz G.J. Temporal resolution of protein–protein interactions in the live-cell plasma membrane. Anal. Bioanal. Chem. 2010;397:3339–3347. doi: 10.1007/s00216-010-3854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Temporal resolution of KLK induced accumulation of GPI-anchored proteins. The movie contains 20 TIRF images of T24 cells expressing GPI-hFR-mGFP, which were recorded at 1 min intervals; the overview shows an area of ~ 400 × 400 μm2. 100 μM KLK was added immediately before the first image. The images show two stripes (top and bottom half) that were recorded in succession, yielding a slight incongruity at the assembly line.